- 1Veterinary Public Health Research Laboratory, Department of Veterinary Medicine, College of Agriculture and Veterinary Medicine, United Arab Emirates University, Al Ain, United Arab Emirates
- 2Food Research Section, Applied Research and Capacity Building Division, Abu Dhabi Agriculture and Food Safety Authority (ADAFSA), Abu Dhabi, United Arab Emirates
- 3Department of Food Technology, Arid Lands Cultivation Research Institute (ALCRI), City of Scientific Research and Technological Applications (SRTACITY), Alexandria, Egypt
- 4Department of Evolutionary Biology, Institute for Biology, Freie Universität Berlin, Berlin, Germany
- 5Animal Health Research Institute, Agriculture Research Centre, Cairo, Egypt
This study reports the first detection of mcr-1.1-mediated colistin resistance in Salmonella enterica serovar Infantis from a commercial broiler farm in the United Arab Emirates (UAE). Two S. infantis isolates (SAL_93 and SAL_94) were recovered from caecal droppings and characterized using whole-genome sequencing (WGS). Genomic analysis revealed a single-nucleotide polymorphism (SNP) difference between them, confirming their close epidemiological relationship. Both isolates belonged to multilocus sequence type 32 and exhibited multidrug resistance (MDR), including resistance to colistin (MIC = 4 mg/L) and ciprofloxacin (MIC = 0.5 mg/L). Notably, the mcr-1.1 gene was detected on a conjugative IncX4 plasmid. Additionally, the isolates harbored a large (275,043 bp) conjugative IncFIB plasmid carrying multiple AMR genes, including aadA1, sul1, tet(A), qacEdelta1. Bioinformatic analysis showed a high identity for globally reported mcr-1.1-carrying IncX4 plasmids. The investigation of virulence-associated factors in the studied isolates identified 162 potential virulence-related genes. These included genes linked to the type 3 secretion system, specifically those encoded by pathogenicity island-1 (SPI-1). However, multiple genes linked to the second type 3 secretion system, encoded by SPI-2, were absent in all isolates. These findings suggest a potential risk of horizontal gene transfer in poultry production. Given these risks, the UAE’s recent ban on colistin in veterinary medicine marks a crucial step in mitigating AMR transmission within a One Health framework.
1 Introduction
The emergence of colistin resistance has become a global concern, affecting both human health and food-producing animals. In human health, colistin has been recommended as a last-line defense against infections by multidrug-resistant (MDR) pathogens (1). Within animal health, colistin has long served as a therapeutic and prophylactic antimicrobial against infections caused by Gram-negative bacteria, such as Salmonella spp., which are prevalent in both human and animal health sectors (2). Among the various Salmonella serovars, Salmonella enterica serovar Infantis ranks among the leading causes of human infections worldwide (3). The increasing frequency of S. infantis illnesses in some regions is further complicated by the proliferation of MDR strains, including those capable of producing β-lactamases, as seen in the recent dissemination of S. infantis serovar in broiler farms, poultry meat, and human gastroenteritis cases (4).
In addition to its impact on animal health, S. infantis poses a significant zoonotic risk, with multiple outbreaks linked to contaminated poultry products and subsequent human infections reported globally (e.g., Europe, North America, and Asia) (3–5). The serovar’s ability to harbor resistance and virulence determinants makes it a priority pathogen in integrated food safety monitoring programs. Despite the growing poultry industry in the United Arab Emirates (UAE), data on the epidemiology of S. infantis in broiler production and its potential role in zoonotic transmission remain scarce, underscoring a critical research gap.
Since the identification of the gene mcr-1 in China in 2015, this plasmid-mediated colistin resistance determinant has been linked to the rising prevalence of colistin resistance in food, livestock, and people internationally (1). In the UAE, previous studies have documented the presence of the gene mcr-1 in colistin-resistant E. coli and S. Minnesota strains isolated from supermarket poultry (6, 7). This study presents the first documented occurrence of mcr-1-mediated colistin non-susceptibility in an MDR S. infantis isolate, which also exhibits resistance to broad-spectrum cephalosporins. To our knowledge, this is the first report of mcr-1-positive MDR S. infantis in UAE broiler farms, highlighting the urgent need to monitor emergent resistance in local production systems and its potential implications for food safety and public health. The resistant strains were recovered from one broiler farm as part of a baseline microbiological assessment of the broiler supply chain in Abu Dhabi.
2 Materials and methods
2.1 Study setting
The study site was a standard medium-sized broiler farm in the UAE, housing approximately 20,000 Lohmann broilers in a closed, climate-controlled system. The farm followed standard all-in/all-out flock management, automated feeding and watering systems, and strict biosecurity measures with a contracted (not resident) routine veterinary oversight. The birds were reared on deep litter flooring composed of wood shavings, with a stocking density of approximately 33 kg/m2. The farm operated on a 30 to 35-day production cycle; such a short production cycle is a typical practice in the UAE broiler farming sector in response to consumer preference of small-sized (net weight of ≤1,000 gm). For sample collection, caecal droppings were systematically collected from various locations within the broiler house to ensure a representative sampling of the flock’s gut microbiota. Sampling sites were selected to cover different zones, including areas near feeders and water lines, corners of the house, and the center where birds tend to aggregate. Fresh caecal droppings, distinguishable by their typical dark brown to mahogany color, pasty, and more homogenous texture than the firmer, segmented regular feces, were identified and collected using sterile scoops. A pooled sample of a total of 50 caecal droppings was collected to reach a target quantity of approximately 150 g (or about the size of a tennis ball, ~8 cm diameter), as typically applied in national and regional monitoring programs for Salmonella detection in poultry (8). The samples were immediately placed into a sterile capped container, transported in a cold chain (4°C), and processed within four to six hours of collection for microbiological and genomic analysis.
2.2 Microbiological isolation and characterization
The container with collected caecal droppings was weighed, and an equal portion of buffered peptone water (BPW) (Oxoid, Basingstoke, Hampshire, UK) was added to attain a 1:1 diluent-to-sample ratio. The mixture was homogenized for one minute using a BagMixer (Interscience, St Nom, France). Then, 50 mL of this homogenized sample was combined with 200 mL of BPW and homogenized again, resulting in a final dilution factor of 1:10 (w/v). From this suspension, 25 mL was directly incubated at 37°C for 18 h for pre-enrichment. No plating of serial dilutions was performed at this stage, as the method relies on selective enrichment and subsequent isolation on selective media following pre-enrichment, in accordance with Salmonella detection protocol of the ISO 6579-1 (2017) (9). For pre-enrichment, 100 μL was sub-cultured in modified semi-solid Rappaport Vassiliadis (MSRV) medium (Oxoid, UK) and incubated at 41.5°C for 48 h. MSRV plates were checked at 24 h for a migration zone (turbid, white halo >10 mm) and rechecked at 48 h if absent. Following pre-enrichment, streaking was performed on xylose lysine deoxycholate agar (Oxoid, UK) with incubation at 37°C for 24 h (10). Up to five suspected colonies from selective media were transferred to nutrient agar (Oxoid, UK) and incubated at 37°C for 24 h. Purified colonies were identified at the species level using MALDI-TOF MS with the Autobio-MS-1000 (Autobio Diagnostics, China).
The characterization of antimicrobial resistance using minimum inhibitory concentrations (MICs) technique was carried out utilizing Sensititre™ EUVSEC3 plates (Thermofisher Scientific, UK), following the protocol provided by the manufacturer. The assessment of resistance was based on epidemiological cut-off values (ECOFFs) stipulated by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (11).
2.3 Whole-genome sequencing (WGS)
Genomic DNA was prepared using the Wizard® DNA Kit (Promega, USA), followed by a quality assessment (12). Short-read sequencing was outsourced to Novogene (UK) and conducted on the NovaSeq platform using 2 × 150 bp paired-end reads. Library preparation followed standard Illumina protocols. Sequencing generated assemblies with an N50 of 203,983 bp, genome sizes of ~5.00 Mbp, and 52–56 contigs per genome (Table 1), consistent with high-quality Salmonella WGS data reported in previous studies (3, 12). Genome analysis was performed with the cloud-based Solu platform v1.0.229 (accessed 15 February 2025) (13), which integrates a suite of validated pipelines for bacterial genomics. Species confirmation was achieved with Kraken2 v2.1.2 followed by Bracken re-estimation, serovar prediction employed the SISTR v1.1.1 workflow, and multilocus sequence types were assigned with mlst v2.23.0 using the Salmonella PubMLST scheme (13). Antimicrobial-resistance genes and point mutations were detected in parallel with AMRFinderPlus v3.11, plasmid replicons were identified with PlasmidFinder v2.1, and virulence loci were screened using VirulenceFinder. Default parameters were applied throughout, ensuring reproducible and transparent outputs for each analytical step (13). All raw sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) under BioProject number PRJNA1231376.
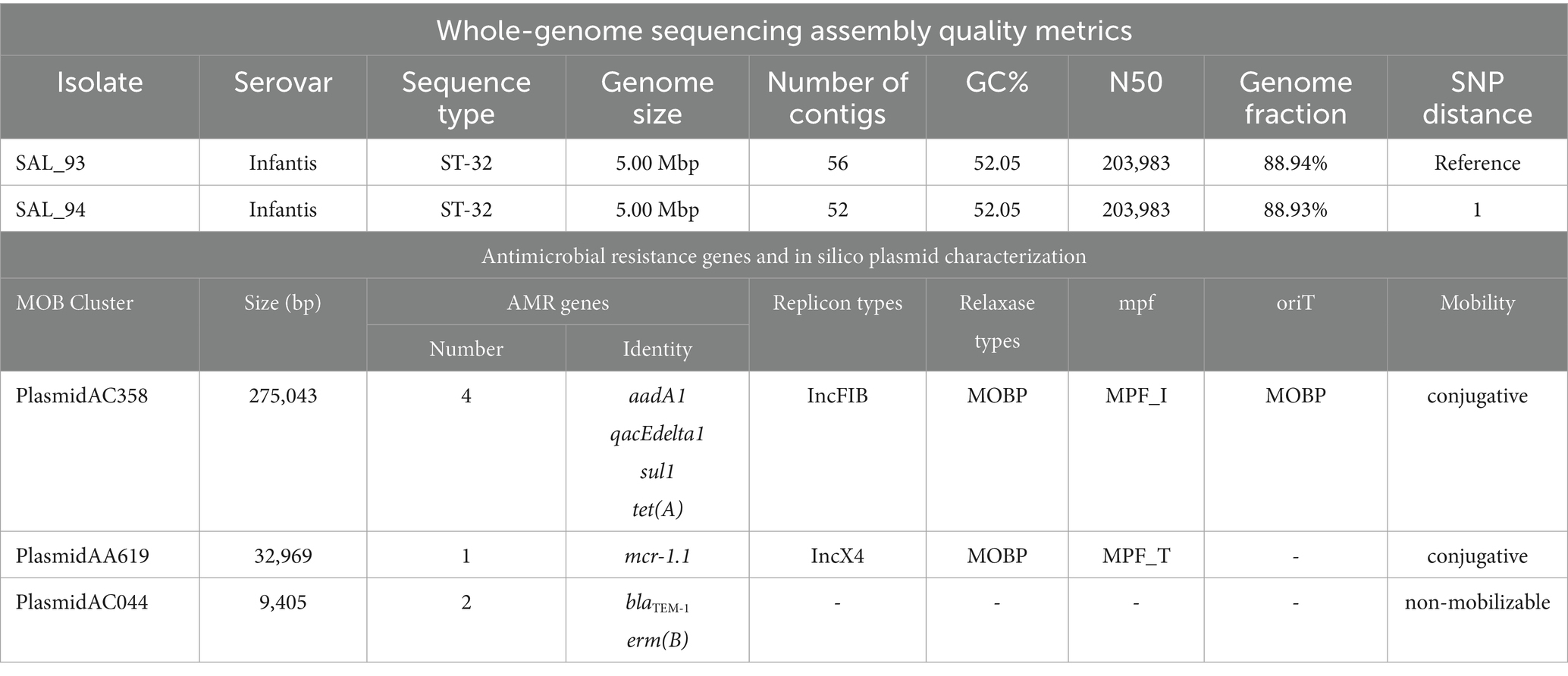
Table 1. Whole-genome sequencing assembly details and antimicrobial resistance genes and MOB-suite based predicted plasmid characters.
2.4 In silico plasmid characterization and mobility prediction
Plasmids carrying mcr-1 were analyzed by extracting plasmid-associated contigs from genome sequence data using Plasmid SPAdes with default settings (14). The reconstructed contigs were then compared against the non-redundant NCBI database to identify the closest matching plasmid, ensuring 99–100% coverage and 100% sequence identity (12).
Plasmid mobility was analyzed using the MOB-suite platform (v3.1.9). Identified plasmid scaffolds from WGS data were matched against the MOB-clusters database to determine their closest counterparts (15). Plasmids were classified into MOB-clusters and categorized as “Conjugative,” “Mobilizable,” or “Non-mobilizable” based on their mobility potential. Additionally, relaxase gene clusters were identified, as they play a key role in plasmid transfer by recognizing and cleaving the origin of transfer (oriT) sites, enabling the horizontal spread of antimicrobial resistance genes (15).
3 Results
3.1 Characteristics of mcr-1- harboring Salmonella Infantis isolates
Two Salmonella enterica serovar infantis strains (SAL_93 and SAL_94) were identified from a pooled sample of fresh caecal droppings collected from the environment of a commercial broiler farm in the UAE (Table 1). WGS confirmed their classification using a computational analysis performed using Solu platform v1.0.229 with standard settings (13). The genome-level relatedness of these strains was evident, with only a Single Nucleotide Polymorphism (SNP) difference between them (Table 1). As shown in Table 1, both strains belonged to multilocus sequence type 32.
The minimum inhibitory concentration (MIC, Sensititre™ EUVSEC3 panel) of colistin for these isolates was determined to be 4 mg/L. Additionally, both isolates exhibited MDR profile to a wide range of antimicrobials, including resistance to ampicillin (MIC = 32 mg/L), azithromycin (MIC = 32 mg/L), chloramphenicol (MIC = 32 mg/L), ciprofloxacin (MIC = 0.5 mg/L), nalidixic acid (MIC = 64 mg/L), sulfamethoxazole (MIC = 512 mg/L), and tetracycline (MIC = 32 mg/L).
3.2 In silico plasmids reconstruction and mcr-1.1 gene context
As indicated in Table 1, the mcr-1.1 gene identified in these isolates was found on a predicted conjugative plasmid classified as MOB-cluster AA619 (replicon type IncX4). These S. infantis isolates also carried a conjugative plasmid classified as MOB-cluster AC358 (replicon type IncFIB), which harbored multiple antimicrobial resistance (AMR) genes, including aadA1 (conferring streptomycin resistance), qacEdelta1 (associated with quaternary ammonium compound resistance), sul1 (resistance to sulfonamides), and tet(A) (tetracycline resistance). The former predicted plasmid is notably large, measuring 275,043 base pairs. Additionally, a chromosomal mutation (gyrA_S83Y) was identified in the gyrase gene of the two strains. These strains also carried mdsA and mdsB, which encode the MdsABC efflux system. Moreover, the β-lactamase gene blaTEM-1 and the erm(B) gene were identified and predicted to be on a non-mobilizable plasmid (Table 1).
3.3 Comparison of mcr-1.1 gene–environment with internationally related plasmids
Plasmid contigs from these isolates exhibited a 100% identity match with previously documented IncX4 plasmids carrying mcr-1.1, including pCFSAN061769_01 (100% coverage, detected in E. coli from beef sausage in Egypt) and pMFDS1318.1 and pMFDS2258.1 (99% coverage, identified in E. coli from pig farms in Korea) (Figure 1). The genetic localization of the mcr-1.1 gene showed that a PAP-2 family protein-coding gene was positioned downstream of mcr-1.1 in S. infantis, while the nearby transposable element was identified as IS-26 (Figure 1).
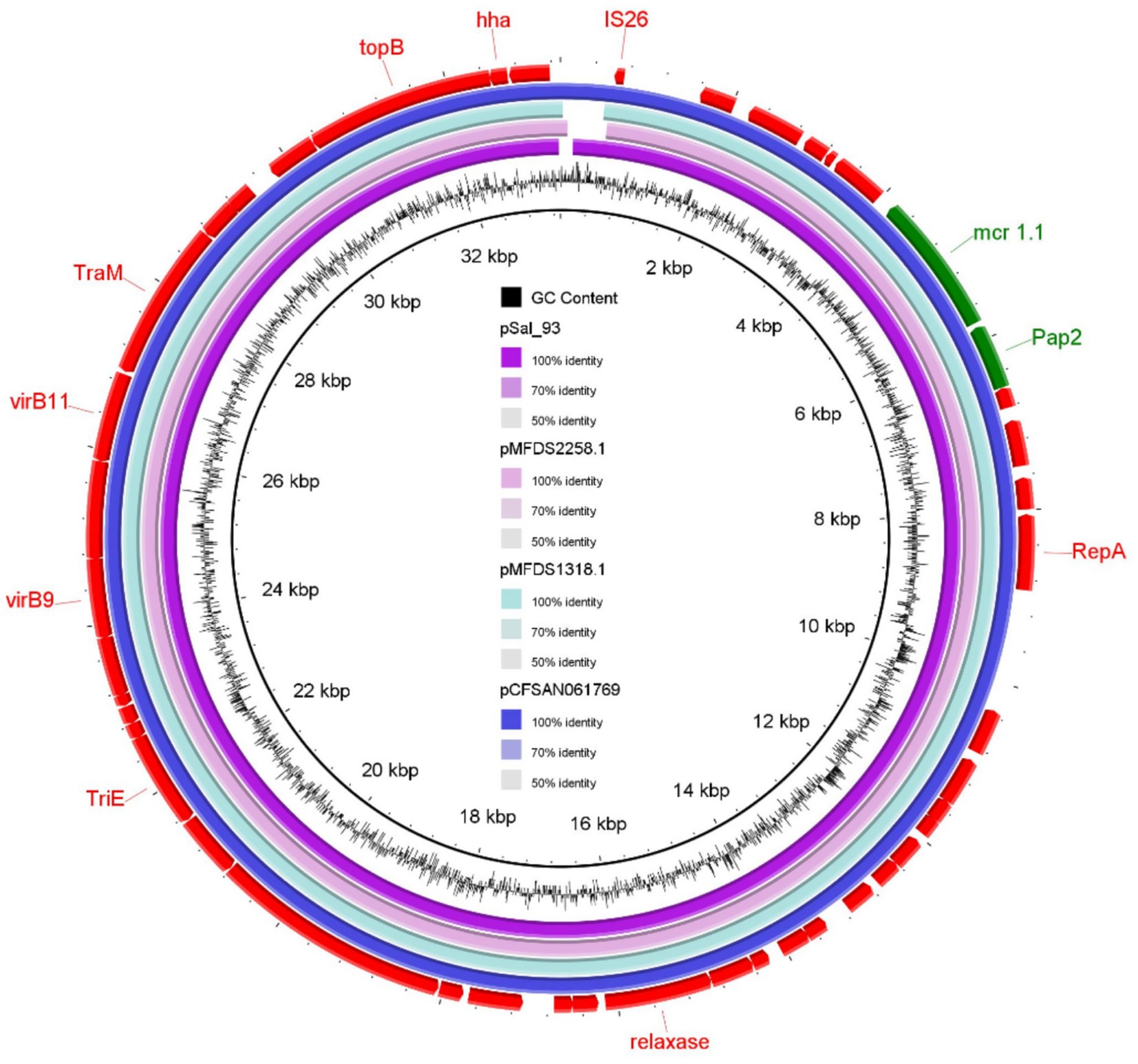
Figure 1. Genomic comparison of mcr-1.1 gene–environment between Salmonella Infantis strain SAL_93 isolated from a broiler farm in the United Arab Emirates and particular internationally concorded plasmids.
3.4 Putative virulence determinants
WGS uncovered 162 potential virulence-related genes in the mcr-1.1-bearing S. infantis strains (Sal_93 and Sal_94) (Table 2). These isolates lacked genes typically linked to capsule formation, invasion, immune evasion, antiphagocytic activity, autotransport, serum resistance, stress adaptation, or toxin production. However, they retained virulence factors associated with adhesion (fimbrial and nonfimbrial) and biofilm formation. The csg (curli-specific gene) operons involved in surface adherence, cell aggregation, environmental persistence, and biofilm development were present. Both isolates possessed virulence elements characteristic of Salmonella pathogenicity islands SPI-1 and SPI-2 but lacked the SPI-2-associated genes ssaS and sseA (Table 2).
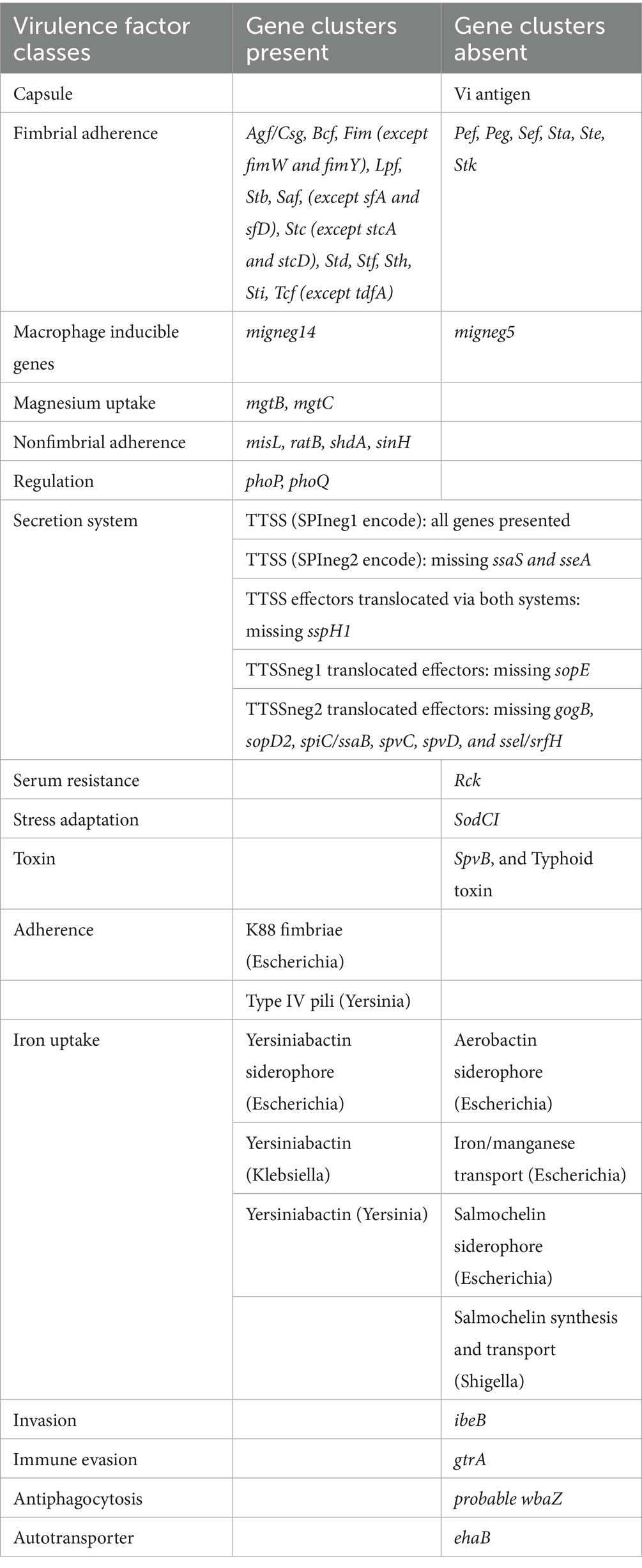
Table 2. Putative virulence factors and their associated gene clusters presented in the mcr-1.1 positive Salmonella Infantis strain SAL_93 isolated from a broiler farm in the United Arab Emirates and particular internationally concorded plasmids.
4 Discussion
This study reports for the first time in the UAE the detection of colistin-resistant S. infantis strains harboring the mcr-1.1 gene recovered from broiler farms. Previously, colistin resistance linked to mcr genes in UAE poultry had only been associated with Salmonella Minnesota isolates from farms and retail meat products (6). The mcr-1.1 gene context here was located on an IncX4 conjugative plasmid, well-documented for its capability to facilitate horizontal transfer of mcr-mediated colistin resistance across Enterobacteriaceae and Salmonella serovars (6, 16). Additionally, the isolates contained an exceptionally large IncFIB plasmid, aligning closely with pESI-like plasmids, known for enhancing the adaptability and survival of S. infantis in various environments (17).
S. infantis identified in this study belonged to the dominant sequence type commonly reported before as the most prevalent infantis subtype in UAE retail chicken products (3). In Europe, S. infantis has been recognized as an emerging serovar., frequently found in poultry production systems, ranking just behind S. enteritidis and S. typhimurium in prevalence (5, 17). The antimicrobial resistance profile of these isolates mirrors earlier observations from the UAE poultry sector, where over 95% of S. infantis isolates exhibited multidrug resistance (MDR) (3). While colistin resistance had not previously been identified in S. infantis in the UAE, its detection in this study raises significant public health concerns, as it potentially restricts available therapeutic options for treating severe human infections, especially those involving MDR strains transmitted through the food chain (18).
The genetic context of the mcr-1.1 gene was notably close to the transposable element IS-26, a mobile genetic element well-known for facilitating genetic transposition through replicative or conservative mechanisms, playing a crucial role in the spread of antimicrobial resistance (AMR) genes (19). This genetic context, coupled with the presence of conjugative plasmids harboring multiple AMR genes, highlights the potential for further dissemination and challenges efforts aimed at controlling MDR bacterial pathogens, including S. infantis.
The isolates resisted critically important antibiotics, such as colistin, ciprofloxacin, and azithromycin, underlining the potential implications for human health. Furthermore, the detection of genes promoting the MdsABC efflux system (mdsA and mdsB), members of the resistance-nodulation-cell division (RND) superfamily, further emphasizes the robustness of resistance mechanisms in these strains (20). Identifying conjugative plasmids carrying such AMR genes in mcr-positive S. infantis from UAE broiler farm environment raises concerns regarding potential cross-transmission between poultry and humans and underscores the role poultry-associated strains may play in facilitating horizontal gene transfer (21).
Despite the absence of traditional virulence determinants associated with immune evasion, toxin production, and serum resistance, the isolates retained virulence factors related to adhesion and biofilm formation, notably the csg operons (22). The presence of virulence genes characteristic of Salmonella pathogenicity islands SPI-1 and SPI-2 (excluding specific SPI-2-associated genes ssaS and sseA) suggests these strains maintain pathogenic potential primarily through biofilm-mediated environmental persistence and intracellular survival (23, 24). These findings indicate that pathogenicity in these S. infantis isolates may rely more on colonization efficiency and persistence rather than conventional virulence pathways.
5 Conclusion
These findings highlight the importance of strengthening antimicrobial resistance surveillance through a One Health approach, integrating human, food, and environmental monitoring systems. In a proactive step toward addressing the growing threat of colistin resistance, the UAE Ministry of Climate Change and Environment implemented a complete ban on its use in veterinary medicine as of February 2024 (https://www.moccae.gov.ae/en/our-services/product-and-materials-page.aspx). By adopting this measure, the UAE contributes proactively to international efforts to combat antimicrobial resistance, potentially reducing the spread of resistant bacteria and preserving colistin as a vital last-resort antibiotic for human medicine.
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) under BioProject number PRJNA1231376 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1231376).
Author contributions
IH: Formal analysis, Writing – original draft, Funding acquisition, Writing – review & editing, Conceptualization, Methodology. M-YM: Project administration, Investigation, Writing – review & editing, Formal analysis. GL: Writing – review & editing, Formal analysis, Project administration, Investigation. HMA: Writing – review & editing, Supervision, Project administration, Funding acquisition. HSA: Project administration, Funding acquisition, Writing – review & editing, Supervision. MS: Writing – review & editing, Project administration, Supervision, Data curation. ME: Methodology, Resources, Writing – review & editing, Visualization, Supervision.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research was funded by the Abu Dhabi Agriculture and Food Safety Authority (ADAFSA) (grant code: 21F059). Publication fees and genome sequencing costs were covered by project One Health Poultry (grant code: 21F064) supported by an anonymous veterinary pharmaceutical company and managed by the United Arab Emirates University Research Office.
Acknowledgments
We thank the poultry farm team for their support in granting access and assisting with sample collection. We thank Ms. Afra Abdalla and Ms. Febin Anes for their valuable assistance in the laboratory work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. During the preparation of this work, the authors used OpenAI. (2025). ChatGPT [Large language model]. https://chatgpt.com, in order to correct sentence structure and grammatical errors. After using this tool, the authors reviewed and edited proposed changes and take full responsibility for the content of the publication.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Barlaam, A, Parisi, A, Spinelli, E, Caruso, M, Taranto, P Di, and Normanno, G. Global emergence of colistin-resistant Escherichia coli in food chains and associated food safety implications: a review. J Food Prot. (2019) 82:1440–8. doi: 10.4315/0362-028X.JFP-19-116
2. Jansen, W, van Hout, J, Wiegel, J, Iatridou, D, Chantziaras, I, and De Briyne, N. Colistin use in European livestock: veterinary field data on trends and perspectives for further reduction. Vet Sci. (2022) 9:650. doi: 10.3390/vetsci9110650
3. Habib, I, Mohamed, M-YI, Lakshmi, GB, Ghazawi, A, Khan, M, Abdalla, A, et al. High prevalence and genomic features of multidrug-resistant Salmonella enterica isolated from chilled broiler chicken on retail sale in the United Arab Emirates. Int J Food Microbiol. (2024) 423:110828. doi: 10.1016/j.ijfoodmicro.2024.110828
4. García-Soto, S, Abdel-Glil, MY, Tomaso, H, Linde, J, and Methner, U. Emergence of multidrug-resistant Salmonella enterica subspecies enterica serovar Infantis of multilocus sequence type 2283 in German broiler farms. Front Microbiol. (2020) 11:11. doi: 10.3389/fmicb.2020.01741
5. Alvarez, DM, Barrón-Montenegro, R, Conejeros, J, Rivera, D, Undurraga, EA, and Moreno-Switt, AI. A review of the global emergence of multidrug-resistant Salmonella enterica subsp. enterica Serovar Infantis. Int J Food Microbiol. (2023) 403:110297. doi: 10.1016/j.ijfoodmicro.2023.110297
6. Habib, I, Elbediwi, M, Ghazawi, A, Mohamed, MYI, Lakshmi, GB, and Khan, M. First report from supermarket chicken meat and genomic characterization of colistin resistance mediated by mcr-1.1 in ESBL-producing, multidrug-resistant Salmonella Minnesota. Int J Food Microbiol. (2022) 379:109835. doi: 10.1016/j.ijfoodmicro.2022.109835
7. Habib, I, Mohamed, M-YI, Elbediwi, M, Ghazawi, A, Khan, M, Abdalla, A, et al. Genomics characterization of colistin resistant Escherichia coli from chicken meat—the first report in the United Arab Emirates. Foodborne Pathog Dis. (2024) 21:521–4. doi: 10.1089/fpd.2024.0021
8. Carrique-Mas, JJ, Breslin, M, Sayers, AR, McLaren, I, Arnold, M, and Davies, R. Comparison of environmental sampling methods for detecting Salmonella in commercial laying flocks in the UK. Lett Appl Microbiol. (2008) 47:514–9. doi: 10.1111/j.1472-765X.2008.02450.x
9. Mooijman, KA, Pielaat, A, and Kuijpers, AFA. Validation of EN ISO 6579-1 - microbiology of the food chain - horizontal method for the detection, enumeration and serotyping of Salmonella - part 1 detection of Salmonella spp. Int J Food Microbiol. (2019) 288:3–12. doi: 10.1016/j.ijfoodmicro.2018.03.022
10. Worcman-Barninka, D, Destro, MT, Fernandes, SA, and Landgraf, M. Evaluation of motility enrichment on modified semi-solid Rappaport–Vassiladis medium (MSRV) for the detection of Salmonella in foods. Int J Food Microbiol. (2001) 64:387–93. doi: 10.1016/S0168-1605(00)00484-0
11. EUCAST. (2025). Data from the EUCAST MIC distribution website. Available online at: http://www.eucast.org (Accessed February 25, 2025).
12. Elbediwi, M, Tang, Y, and Yue, M. Genomic characterization of ESBL-producing Salmonella Thompson isolates harboring mcr-9 from dead chick embryos in China. Vet Microbiol. (2023) 278:109634. doi: 10.1016/j.vetmic.2022.109634
13. Saratto, T, Visuri, K, Lehtinen, J, Ortega-Sanz, I, Steenwyk, JL, and Sihvonen, S. Solu: a cloud platform for real-time genomic pathogen surveillance. BMC Bioinformatics. (2025) 26:12. doi: 10.1186/s12859-024-06005-z
14. Antipov, D, Hartwick, N, Shen, M, Raiko, M, Lapidus, A, and Pevzner, PA. PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. (2016) 32:3380–7. doi: 10.1093/bioinformatics/btw493
15. Robertson, J, and Nash, JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. (2018) 4:4. doi: 10.1099/mgen.0.000206
16. Ghazawi, A, Strepis, N, Anes, F, Yaaqeib, D, Ahmed, A, AlHosani, A, et al. First report of colistin-resistant Escherichia coli carrying mcr-1 IncI2(delta) and IncX4 plasmids from camels (camelus dromedarius) in the Gulf region. Antibiotics. (2024) 13:227. doi: 10.3390/antibiotics13030227
17. Hindermann, D, Gopinath, G, Chase, H, Negrete, F, Althaus, D, Zurfluh, K, et al. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front Microbiol. (2017) 8. doi: 10.3389/fmicb.2017.01322
18. Alba, P, Leekitcharoenphon, P, Carfora, V, Amoruso, R, Cordaro, G, Di Matteo, P, et al. Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb Genom. (2020) 6. doi: 10.1099/mgen.0.000365
19. Song, S, Lee, B, Yeom, J-H, Hwang, S, Kang, I, Cho, J-C, et al. MdsABC-mediated pathway for pathogenicity in Salmonella enterica serovar Typhimurium. Infect Immun. (2015) 83:4266–76. doi: 10.1128/IAI.00653-15
20. Wong, MH, Chan, EW, and Chen, S. IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J Antimicrob Chemother. (2017) 72:2750–4. doi: 10.1093/jac/dkx238
21. Carfora, V, Alba, P, Leekitcharoenphon, P, Ballarò, D, Cordaro, G, Di Matteo, P, et al. Colistin resistance mediated by mcr-1 in ESBL-producing, multidrug resistant Salmonella Infantis in broiler chicken industry. Front Microbiol Italy, (2018) 9. doi: 10.3389/fmicb.2018.01880
22. González, JF, Laipply, B, Sadowski, VA, Price, M, and Gunn, JS. Functional role of the biofilm regulator CsgD in Salmonella enterica sv. Typhi. Front Cell Infect Microbiol. (2024) 14. doi: 10.3389/fcimb.2024.1478488
23. Amavisit, P, Lightfoot, D, Browning, GF, and Markham, PF. Variation between pathogenic Serovars within Salmonella Pathogenicity Islands. J Bacteriol. (2003) 185:3624–35. doi: 10.1128/JB.185.12.3624-3635.2003
Keywords: United Arab Emirates, antibiotic resistance, Salmonella, IncX4, Colistin
Citation: Habib I, Mohamed M-YI, Lakshmi GB, Al Marzooqi HM, Afifi HS, Shehata MG and Elbediwi M (2025) First detection and genomic analysis of mcr-1-positive Salmonella Infantis isolated from a broiler production system in the United Arab Emirates. Front. Vet. Sci. 12:1592955. doi: 10.3389/fvets.2025.1592955
Edited by:
Benti Deresa Gelalcha, The University of Tennessee, United StatesReviewed by:
Muhammad Asif Zahoor, Government College University, PakistanRachel Amanda Hickman, Independent researcher, Uppsala, Sweden
Copyright © 2025 Habib, Mohamed, Lakshmi, Al Marzooqi, Afifi, Shehata and Elbediwi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ihab Habib, aS5oYWJpYkB1YWV1LmFjLmFl
 Ihab Habib
Ihab Habib Mohamed-Yousif Ibrahim Mohamed
Mohamed-Yousif Ibrahim Mohamed Glindya Bhagya Lakshmi1
Glindya Bhagya Lakshmi1 Mohammed Elbediwi
Mohammed Elbediwi