- 1College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea
- 2Division of Bacterial Diseases, Bureau of Infectious Disease Diagnosis Control, Korea Disease Control and Prevention Agency, Cheongju, Republic of Korea
- 3School of Veterinary Medicine, Mongolian University of Life Sciences, Ulaanbaatar, Mongolia
Salmonella enterica serotype Enteritidis (S. Enteritidis) is a leading foodborne pathogen associated with poultry products, and the emergence of antimicrobial resistance (AMR) in this serotype poses a growing public health concern, particularly in regions with increasing poultry trade. Between April and June 2024, we collected 114 poultry meat samples (Mongolian domestic and Chinese imported) from retail markets in Mongolia and isolated 45 S. Enteritidis strains. Antimicrobial susceptibility testing revealed high resistance rates to nalidixic acid (100%), ampicillin (93.3%), and streptomycin (88.9%). Whole-genome sequencing (WGS) identified major resistance genes, including aac(6′)-Iaa, aph(3″)-Ib, blaTEM-1B, and sul2. Mongolian domestic isolates additionally harbored extended-spectrum β-lactamase (ESBL) blaCTX-M-14 and plasmid-mediated quinolone resistance (PMQR) qnrS1, both of which are clinically significant. Plasmid replicon typing revealed IncF as the most prevalent type across isolates, while IncI1- α was predominantly found in multidrug-resistant (MDR) domestic strains. Phylogenetic analysis using whole-genome SNPs (wgSNPs) demonstrated that domestic and imported isolates clustered separately, indicating that Chinese S. Enteritidis strains have not yet been introduced into Mongolia’s domestic poultry industry. This study represents the first comprehensive analysis of prevalence and resistance mechanisms of S. Enteritidis in Mongolia poultry production. The findings underscore the necessity of continuous surveillance and the implementation of effective antibiotic stewardship in the poultry production sector.
1 Introduction
Non-typhoidal Salmonella enterica (S. enterica) is a major foodborne pathogen that causes salmonellosis worldwide (1). Among its serotypes, Salmonella Enteritidis (S. Enteritidis) is a leading serovar and is a major cause of foodborne illness, particularly primarily transmitted through poultry products such as chicken meat and eggs (2, 3). According to the Centers for Disease Control and Prevention (CDC), S. Enteritidis infections have increased globally, particularly in regions where poultry consumption is widespread (4).
Mongolia has traditionally relied on locally sourced meats such as lamb, goat, and beef, supported by its robust pastoral economy. According to the Mongolian Meat Association, the country produced 420,000 tons of meat in 2017, with per capita meat consumption reaching 16.4 kg, reflecting increased demand driven by economic growth and rising living standards (5). Recently, changing dietary habits and urbanization have led to growing poultry consumption in Mongolia, despite limited local poultry production capacity. Poultry meat, however, constitutes only a minor fraction of domestic production, highlighting Mongolia’s heavy reliance on imports. FAOSTAT data indicate that Mongolia’s chicken meat imports from China have steadily increased, rising more than five-fold from 2015 to 2022. This dependence on Chinese poultry raises food safety concerns, as studies have identified a growing prevalence of multidrug-resistant (MDR) Salmonella in Chinese poultry production (6–8). Such reliance increases the risk of introducing antimicrobial-resistant S. Enteritidis into Mongolia’s food supply, posing substantial threats to public health.
Recent advances in whole-genome sequencing (WGS) technology and bioinformatics have provided powerful tools for genetic analysis of Salmonella, studying public health surveillance, antimicrobial resistance detection, and epidemiological tracking (9–11). However, no WGS-based studies on S. Enteritidis have been conducted in Mongolia, leaving its prevalence and antimicrobial resistance profiles in the local food supply chain unexplored. A previous study in Mongolia employed direct polymerase chain reaction (PCR) to assess microbial contamination in poultry meat, reporting a Salmonella spp. prevalence of 7.6% (12). Given Mongolia’s heavy reliance on poultry imports from China, there is a critical need to investigate whether MDR S. Enteritidis strains are being introduced and established in the national poultry industry. Therefore, this study aims to employ WGS to investigate the prevalence and resistance profiles of Salmonella spp., representing the first genomic assessment of MDR S. Enteritidis in poultry meat sold in Mongolian retail markets.
In this study, we collected poultry meat samples from Mongolian retail markets, including both domestic products and imports from China and Russia, to determine the prevalence of Salmonella spp. in the national poultry supply. Additionally, we employed WGS to characterize antimicrobial resistance profiles, plasmid types, and phylogenetic relationships among S. Enteritidis isolates. This research represents the first WGS-based prevalence study of S. Enteritidis in Mongolia’s poultry supply chain.
2 Materials and methods
2.1 Salmonella isolation from poultry sources
From April 15th to June 17th, 2024, a total of 114 samples of retail chicken meats were collected and tested, all purchased from retail markets in Ulaanbaatar, Mongolia. These included domestic Mongolian whole chickens and imported Chinese and Russian sliced chicken meats. Retail chicken meats were aseptically placed in a sterile plastic bag containing 400 mL of buffered peptone water broth (BPW, Difco, Detroit, MI, USA) and shaken for 2 mins. From this initial rinse, a 20 mL aliquot of the BPW-sample mixture was transferred, vortex-mixed with fresh 20 mL of BPW for 15 s, then incubated at 36 ± 1°C for 18–24 h. Subsequently, 100 μL of the incubated culture was vortex-mixed for 15 s in 10 mL of Rappaport-Vassiliadis (RV) broth, then incubated at 41.5 ± 1°C for 20–24 h. Cultures exhibiting a color change were streaked onto Salmonella ChromoSelect agar (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 37°C for 24 h. Pink colonies presumptively identified as Salmonella spp. on the agar were confirmed using the VITEK-MS system (bioMérieux, Marcy-L’Etoile, France). Confirmed isolates were stored at −80°C in glycerol. All Salmonella spp. isolates obtained during the study period were included for the further studies, following their transport to Konkuk University, Seoul, Korea.
2.2 Whole-genome sequencing and genome assembly
Genomic DNA was extracted from overnight cultures of Salmonella spp. using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The purity and concentration of the extracted DNA were measured with the NanoDrop™ spectrophotometer (Thermo Fisher Scientific) and the Quantus™ fluorometer (Promega, Madison, WI, USA), respectively. Library preparation was performed with the TruSeq Nano DNA Library Prep kit (Illumina, San Diego, CA, USA) following the manufacturer’s instructions. The library was then sequenced on a NovaSeq 6,000 platform (Illumina, San Diego, CA, USA), generating 150 bp paired-end reads. Read quality was assessed using fastp v.0.20.1 (13). Low-quality bases and adapter sequences were trimmed using Trimmomatic v0.39 (14). Trimmed reads were assembled using SPAdes v4.1.0 (15) with the “—careful” option. Serotype prediction was performed using SeqSero v1.3.1 (16) on the assembled contigs, and only isolates identified as serovar Enteritidis were included in further analyses. The whole-genome sequencing data generated in this study has been deposited in the NCBI BioProject database under accession number PRJNA1236736.
2.3 Antimicrobial susceptibility test
Antimicrobial susceptibility testing was determined using the Sensititre™ ARIS HiQ AST system (Thermo Fisher Scientific, Waltham, MA, USA) with the Sensititre™ panel (KRCDC2F; Thermo Fisher Scientific, Waltham, MA, USA). The following antimicrobials were tested: ciprofloxacin (CIP, 0.03–0.5 μg), nalidixic acid (NAL, 2–128 μg), imipenem (IMI, 1–8 μg), colistin (COL, 2–16 μg), ampicillin (AMP, 2–64 μg), tetracycline (TET, 2–128 μg), chloramphenicol (CHL, 2–32 μg), azithromycin (AZI, 2–32 μg), gentamicin (GEN, 1–64 μg), streptomycin (STR, 2–128 μg; Ref. National Antimicrobial Resistance Monitoring System, 2014) (17), amikacin (AMI, 4–64 μg), trimethoprim/sulfamethoxazole (SXT, 1/19–16/304), cefotaxime (FOT, 1–32 μg), ceftriaxone (AXO, 1–32 μg), cefoxitin (FOX, 4–32 μg), and ceftazidime (TAZ, 1–16 μg).
Purified bacterial isolates were suspended in distilled pure water and adjusted to a 0.5 McFarland standard using a nephelometer. A 10 μL aliquot of the adjusted suspension was inoculated into 11 mL of cation-adjusted Mueller-Hinton (MH) medium and dispensed into the KRCDC2F Sensititre™ panel using the Sensititre AIM automated dispensing system. The inoculated microplates were sealed with a microplate-sealing film and incubated at 37°C for 18 h in the Sensititre ARIS HiQ automated analysis system.
After incubation, a fluorescence auto-read system determined minimum inhibitory concentration (MIC) values. These MIC values were determined according to Clinical and Laboratory Standards Institute (CLSI) M100-2018 28th edition guidelines (Wayne, PA, USA) (18), except for Streptomycin, which followed the National Antimicrobial Resistance Monitoring System (NARMS) criteria (17). Escherichia coli ATCC25922 was used as a quality control standard. Salmonella isolates resistant to more than three classes and more than one antimicrobial in a single class were designated MDR strains, following standard criteria (19).
2.4 Phylogenetic analysis
Whole-genome single nucleotide polymorphisms (wgSNPs) were identified among S. Enteritidis (n = 45) in this study using Snippy v4.6.01 (20), with S. Enteritidis strain P125109 as the reference for SNP calling (GenBank accession no. AM933172.1, NCBI RefSeq accession no.NC_011294.1). Prophage regions, repetitive genomic regions, and recombination regions in the reference genome were identified and excluded from the analysis. Specifically, prophage regions were identified using PHASTEST (21), repetitive genomic regions were determined by self-alignment of the reference genome using the nucmer tool from the Mummer package v.4.0.1 (22) using the --maxmatch and --nosimplify options, and recombination regions were filtered out using Gubbins v.3.4 (23). A maximum likelihood phylogenetic tree was constructed from the cleaned alignment using a TVMe model selected by ModelFinder (24) in IQ-TREE (25) under 1,000 bootstrap replicates. The phylogenetic tree was visualized using the interactive Tree of Life version 5 (iTOLv7)2. Genotype clustering was performed using Bayesian Analysis of Population Structure (BAPS) through the “FastBAPS” R package.
For phylogenetic analysis with Chinese isolates, genomic sequences of S. Enteritidis isolates (n = 816) from China were downloaded from EnteroBase3 (26). A phylogenetic tree was constructed using Mashtree v.1.4.6 (27) with bootstrapping using Mash distances. To reduce redundancy and computational complexity, Treemmer v0.3 (28) was applied with the -lmc option, pruning the tree to maintain its overall topology while preserving all Mongolian S. Enteritidis isolates and selectively reducing the number of Chinese isolates by removing leaves that contribute minimally to the tree’s diversity.
The final dataset included a total of 106 isolates comprising 45 Mongolian isolates from this study and 61 Chinese isolates (detailed in Supplementary Table S3). These isolates were analyzed using wgSNP, followed the construction of a maximum-likelihood phylogenetic tree using a TVM + F + ASC model, selected by ModelFinder (24) in IQ-TREE (25) under 1,000 bootstrap replicates and determination of population structure with FastBAPS as detailed above (29).
2.5 Antimicrobial resistance genes and comparative genome analysis
All isolates were screened for antimicrobial resistance genes (ARGs) using a pipeline available at github.com/maxlcummins/pipelord. Gene screening was performed using abricate v1.0.14 in conjunction with the following databases: ResFinder (30) and PlasmidFinder (31). Genes were marked as present when detected by abricate and filtered using a custom R script5 to have 90% identity and 90% coverage.
3 Results
3.1 Prevalence of Salmonella spp.
A total of 56 Salmonella spp. isolates were recovered from 114 retail chicken meat samples: 48 isolates (63.2%, 48/76) from Mongolian domestic whole chickens and eight isolates (23.5%, 8/34) from Chinese imported sliced chicken meats. No Salmonella spp. was detected in Russian imported sliced chicken meat (n = 4). Sampling metadata for all Salmonella spp. are provided in Supplementary Table S1.
3.2 Serovars prediction
Whole-genome sequencing (WGS) analysis identified four distinct serovars among the 56 studied Salmonella isolates (Table 1). The predominant serovar was S. Enteritidis (80.4% 45/56), with 41 isolates recovered from Mongolian domestic chicken and four from Chinese imported chicken. S. Typhimurium (12.5% 7/56) was detected exclusively in Mongolian domestic chicken, while S. Agona (3.6% 2/56) and S. Indiana (3.6% 2/56) were only identified in imported samples from China.

Table 1. Prevalence and predicted serovar distribution of 56 Salmonella spp. isolates from Mongolian domestic and Chinese imported retail chicken meats collected in Ulaanbaatar, Mongolia, from April 15th to June 17th, 2024.
3.3 Antimicrobial resistance profiles of S. Enteritidis
The antimicrobial resistance profiles of 45 S. Enteritidis isolates were determined using the broth dilution method against 16 antimicrobial agents, and the results are presented in Table 2 (Supplementary Table S2). The highest resistance was observed against nalidixic acid (100% 45/45), followed by ampicillin (93.3% 42/45), and streptomycin (88.9% 40/45). Resistance to third-generation cephalosporins (cefotaxime and ceftriaxone) was observed in 4 isolates (8.9%), all of which concurrently resistant to ampicillin, nalidixic acid, streptomycin, tetracycline, and trimethoprim/sulfamethoxazole (Table 2). Colistin resistance was also detected in four isolates (8.9%), two originating from Mongolia domestic chicken and the other two from imported Chinese chicken. The remaining 41 isolates (91.1%) were classified as intermediate to colistin.
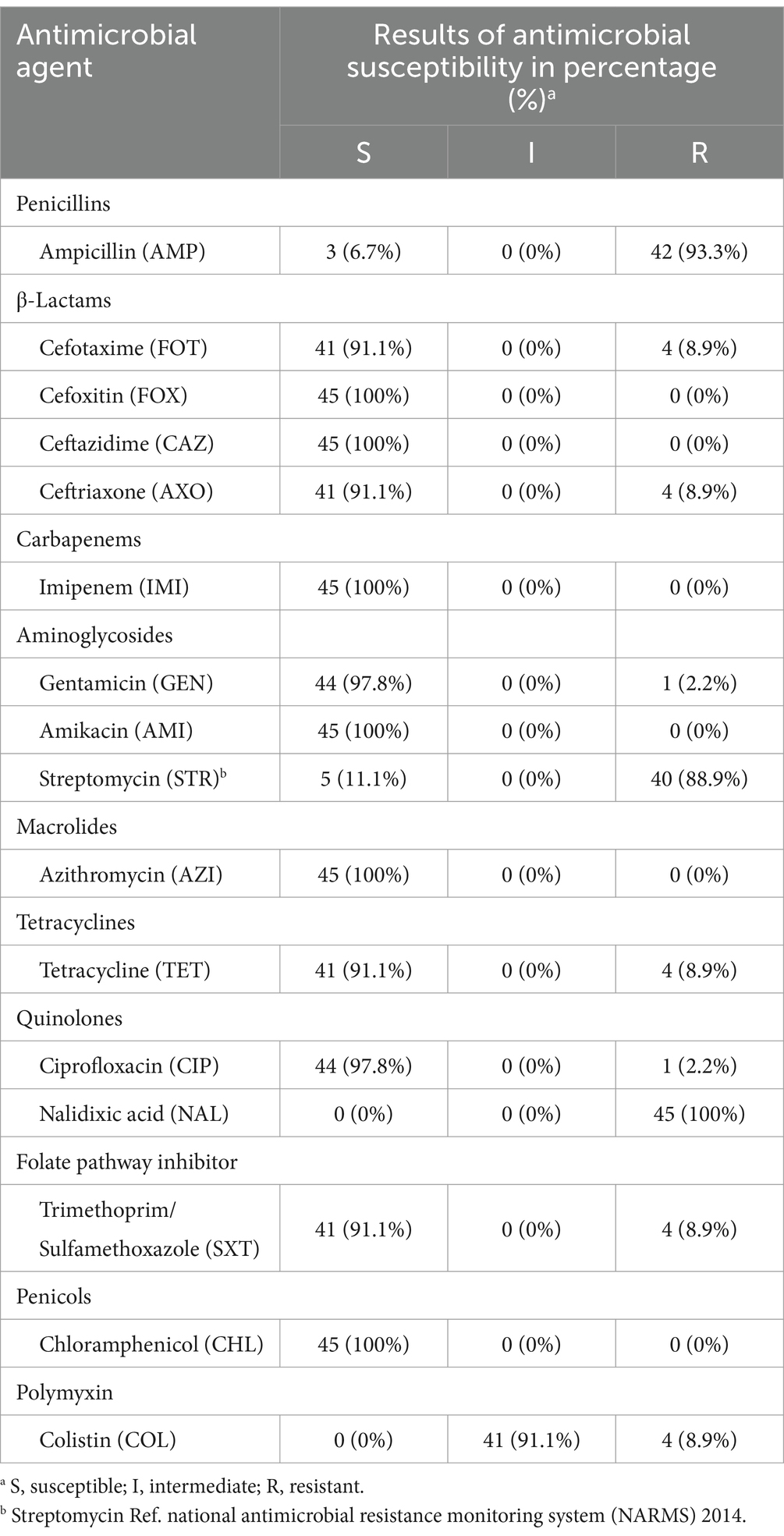
Table 2. Antimicrobial resistance profiles of 45 S. Enteritidis isolates in this study tested against 16 antimicrobial agents using the Sensititre™ ARIS HiQ AST system.
3.4 Antimicrobial resistance gene profile in S. Enteritidis
The WGS analysis results for the 45 S. Enteritidis isolates in this study are summarized in Table 3 (Supplementary Table S4). The most frequently detected resistance genes were aac(6′)-Iaa (100% 45/45), blaTEM-1B (93.3% 42/45), aph(3″)-Ib (88.9% 40/45), and sul2 (88.9% 40/45), which confer resistance to aminoglycosides, penicillins, and sulfonamides, respectively. The blaCTX-M-14 gene, which confers resistance to third-generation cephalosporins, was detected in 4 isolates (8.9%). These isolates are harbored aadA5, dfrA17, fosA3, qnrS1, and tet(A).
In addition to antimicrobial resistance gene detection, chromosomal mutations associated with quinolone resistance were also investigated in Table 3. All isolates carried at least one non-synonymous point mutation in the gyrA gene including gyrA D87Y (Aspartic acid to Tyrosine) in 93.3% (42/45) of isolates, gyrA D87G (Aspartic acid to Glycine) in one isolate (2.2%), gyrA D87N (Aspartic acid to Asparagine) and gyrA S83Y (Serine to Tyrosine) in two isolates (4.4%), respectively. None of the isolates had a point mutation in the gyrB, parC, and parE genes (data not shown). Although four isolates exhibited phenotypic resistance to colistin in antimicrobial susceptibility testing, the mobile colistin resistance (mcr) gene was not detected in any of them. A single chromosomal mutation, pmrB L14F, which is associated with colistin resistance, was identified in one isolate; However, this mutation did not correspond with the observed phenotypic resistance.
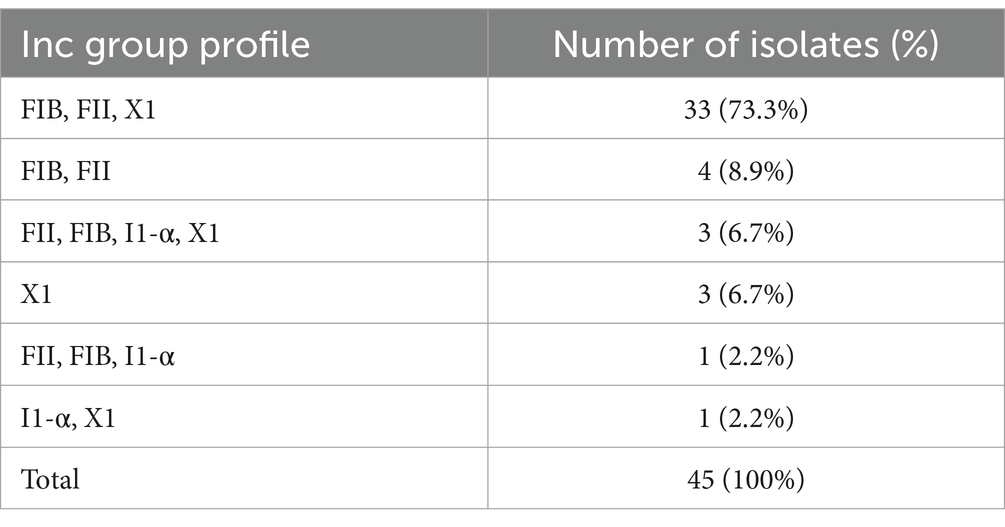
Two dominant AMR profiles were identified among the S. Enteritidis isolates. The AMP-NAL-STR profile, observed in the majority of isolates (77.8%, 35/45),was associated with the presence of blaTEM-1B, aac(6′)-Iaa, aph(3″)-Ib, and sul2, as well as the gyrA D87Y chromosomal mutation. The AMP-FOX-AXO-NAL-STR-TET-STX profile, which conferred resistance to the highest number of antimicrobial classes, was found in 8.9% (4/45) of isolates, and was characterized by blaTEM-1B, blaCTX-M-14, fosA3, qnrS1, tet(A), sul2, and dfrA17.
3.5 Plasmid replicon profile in S. Enteritidis
WGS analysis of 45 S. Enteritidis isolates in this study revealed four plasmid incompatibility (Inc) types: IncFIB(S) and IncFII were the most common, detected in 91.1% (41/45), followed by IncX1_4 in 88.9% (32/45), and IncI1-α in 11.1% (5/45). Seven distinct Inc. group profiles were identified, as summarized in Table 4 (Supplementary Table S1). The predominant profile -IncFIB(S), IncFII(S), and IncX1- was observed in 71.1% (32/45), followed by IncFIB(S) and IncFII(S) in 8.9% (4/45), and three profiles each accounting for 6.7% (3/45): IncFIB(s), IncFII(S), and IncI1-α; IncFII(S), IncX1; and IncX1 alone. The remaining profiles were each identified in a single isolate, reflecting a diverse plasmid landscape among the isolates.
3.6 wgSNP phylogenetic analysis of S. Enteritidis isolates
A midpoint-rooted maximum-likelihood (ML) phylogenetic tree was constructed based on whole-genome single nucleotide polymorphism (wgSNP) analysis of 396 SNP sites to assess the genetic diversity among 45 S. Enteritidis isolates obtained in this study (Figure 1). Based on the SNP alignment and phylogenetic structure, FastBAPS analysis grouped the 45 S. Enteritidis isolates into two BAPS clusters: BAPS 1, consisting of Chinese imported isolates, and BAPS 2, comprising Mongolian domestic isolates.

Figure 1. Phylogenetic analysis of 45 S. Enteritidis strains isolated in this study. A midpoint-rooted maximum-likelihood (ML) phylogenetic tree was constructed based on whole-genome SNP analysis of 396 SNP sites. The analysis used 1,000 bootstrap replicates and support values greater than 0.9 were indicated by black dots. To determine clustering between S. Enteritidis according to genomic similarity, we used FastBAPS. Mongolian domestic chicken isolates were visualized in blue and Chinese imported chicken isolates in red. AMR gene presence was indicated in red, and absence was uncolored. The presence of Plasmid Inc type was indicated in green and absence was uncolored.
Mongolian domestic S. Enteritidis isolates exhibited distinct clustering patterns in the phylogenetic analysis. To further investigate the genetic basis of this separation, AMR gene profiles and plasmid incompatibility (Inc) groups were analyzed, revealing three genetically distinct clusters. Notably, isolates within BAPS 2 (Mongolian domestic group), were further subdivided into a major and minor subgroup based on difference in AMR gene content and Inc. group composition.
The analysis of AMR genes showed that isolates in BAPS 1 harbored fewer AMR genes than those in BAPS 2, with the aac(6′)-Iaa gene consistently present in both clusters. Most isolates (35/45) in BAPS 2 group carried the key antimicrobial resistance genes, including aac(6′)-Iaa, aph(3″)-IIa, aph(6)-Id, blaTEM-1B, and sul2. In contrast, the minor domestic subgroup harbored additional AMR genes, including clinically significant antimicrobial resistance genes like the extended-spectrum beta-lactamase (ESBL) genes blaCTX-M-14, as well as plasmid-mediated quinolone resistance (PMQR) genes qnrS1.
Plasmid analysis revealed a strong association between Inc. group composition and the distribution of AMR genes. In the BAPS 1, most isolates harbored only IncF plasmid. In contrast, the major Mongolian domestic subgroup primarily exhibited a combination of IncF and IncX1 plasmids, except for three isolates that contained solely IncX1. The minor domestic subgroup also showed a distinct profile characterized by the presence of IncF, IncI1-α, and IncX1 plasmids. This variation suggests a potential role of IncI1-α in facilitating the acquisition and dissemination of AMR genes, warranting further investigation.
The distinct clustering profile also highlighted a notable genetic divergence between Mongolian domestic S. Enteritidis isolates and those sourced from China, as further supported by a comparative analysis with 61 publicly available Chinese S. Enteritidis genomes (Figure 2). A midpoint-rooted ML phylogenetic tree was constructed based on wgSNP analysis of 1,502 SNP sites. FastBAPS grouped the combined dataset into four clusters (BAPS A to D), clearly separating Mongolian isolates (BAPS 2) from Chinese ones (BAPS 1), consistent with their AMR gene profiles and Inc. group compositions.
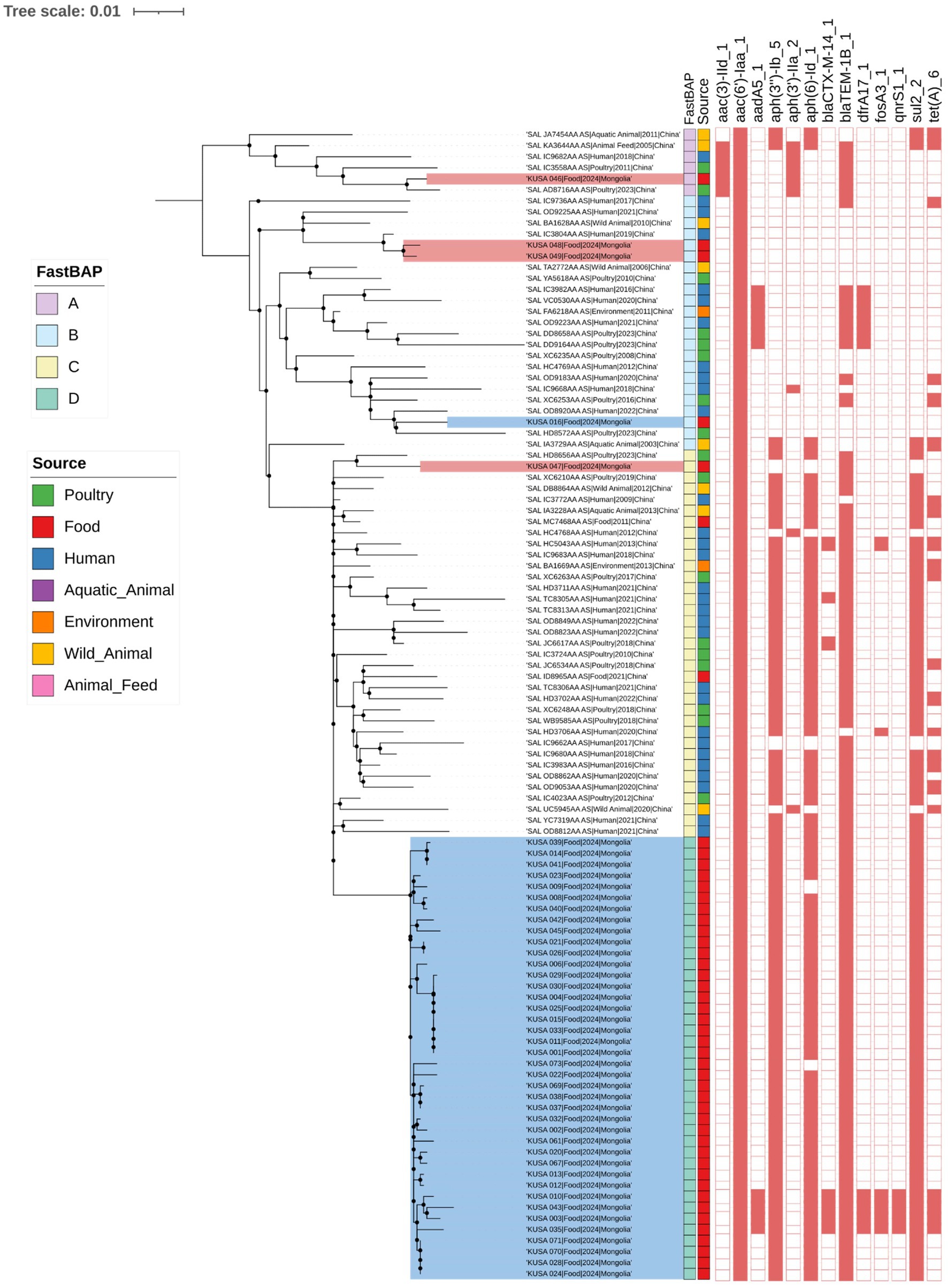
Figure 2. Phylogenetic analysis of 106 S. Enteritidis isolate from this study with Chinese S. Enteritidis isolates from public database. A midpoint-rooted ML phylogenetic tree was constructed based on whole-genome SNP analysis of 1,502 SNP sites. The analysis used 1,000 bootstrap replicates and support values greater than 0.9 were indicated by black dots. To determine clustering between S. Enteritidis according to genomic similarity, we used FastBAPS. Information of source of S. Enteritidis was indicated by different colors. The presence of antimicrobial resistance genes is marked with the red square. Antimicrobial resistance gene presence was indicated in red and absence was uncolored.
Notably, all five Chinese imported S. Enteritidis isolates clustered closely with isolates from China, indicating high genetic similarity. In BAPS A, one Mongolian domestic isolate (KUSA_046) grouped with four Chinese isolates obtained from animal feed (n = 1), aquatic animal (n = 1), human (n = 1), and poultry (n = 2). BAPS B comprised two Chinese imported isolates (KUSA_048 and KUSA_049), which clustered with three Chinese isolates from humans (n = 2) and wild animals (n = 1), as well as one Mongolian domestic isolate associated with samples from the environment (n = 1), human (n = 7), wild animal (n = 1), and poultry (n = 6).
BAPS C included one Chinese imported isolate that clustered with 34 Chinese isolates from various sources. In contrast, BAPS D consisted exclusively of Mongolian domestic isolates and formed a distinct phylogenetic cluster. Despite this separation, BAPS C and BAPS D shared similar AMR gene profiles, including aac(6′)-Iaa, aph(3″)-Ib, aph(6)-Id, blaTEM-1B, and sul2, which are associated with resistance to aminoglycosides, β-lactams, and sulfonamides.
4 Discussion
For the purpose of this study, we collected 114 retail chicken meats -including Mongolian, Chinese and Russian products- from market in Mongolia, with the primary aim of investigating the prevalence of Salmonella spp. in poultry meat sold in local retail settings. In addition, further characterization of the isolates was performed through antimicrobial susceptibility testing and WGS.
A previous study in Mongolia employed direct multiplex PCR with specific primers to assess the prevalence of Salmonella spp. in poultry meat, reporting a low prevalence of 7.6% (12). In contrast, studies conducted in China have reported a wide range of contamination rates in chicken meat, varying from 10.4 to 57% depending on region and sampling conditions (32–35). The lower prevalence in the prior Mongolian study may be attributed to the use of direct PCR without culture enrichment, which likely reduced the sensitivity of Salmonella detection. Pre-enrichment steps using buffered peptone water (BPW) or selenite broth have been shown to significantly improve the sensitivity and specificity of PCR-based detection methods, especially in samples with low bacterial loads, such as food and stool (36–39). Accordingly, standardized culture-based enrichment protocols, such as those recommended by the United States Department of Agriculture Food Safety and Inspection Service (USDA FSIS) Microbiology Laboratory Guidebook (MLG 4.13) (40), are considered essential for accurate Salmonella spp. detection. By incorporating standardized enrichment methods, our study achieved a more reliable detection rate, revealing a Salmonella spp. prevalence of 49.1%. This isolation rate is notably higher than that reported in previous studies conducted in Mongolia and falls on the higher end of the prevalence range reported in various regions of China. Moreover, S. Enteritidis was identified as the predominant serovar, with the majority of isolates classified as MDR. These findings indicate that retail chicken meat sold in Mongolia is frequently contaminated with MDR Salmonella, representing a potential threat to food safety and public health.
The wgSNP phylogenetic analysis indicated that 39 S. Enteritidis isolates from Mongolian domestic chickens exhibited a monophyletic relationship, characterized by relatively extensive root-to-tip distance and supported by 1,000 bootstrap replicates, in contrast to isolates from imported Chinese poultry (Figure 2). In comparison, a study conducted in the United Kingdom found that Salmonella Heidelberg and S. Minnesota strains isolated from Brazilian and UK-imported poultry formed a single monophyletic group, distinct from domestic UK strains (41). Unlike the UK, where Salmonella from imported poultry was introduced into the local poultry industry, there is no evidence of S. Enteritidis transmission from Chinese poultry to Mongolian domestic chickens. This may be explained by Mongolia’s importation of frozen poultry meat rather than live birds (12), which likely reduces the risks of introducing foreign Salmonella strains into domestic production. These findings underscore the importance of maintaining and strengthening quarantine and biosecurity measures to prevent the introduction of Salmonella and other foodborne pathogens through imported poultry products.
Notably, four isolates within the Mongolian domestic cluster carried IncI1-type plasmids with distinct MDR profiles, including genes conferring resistance to critically important antibiotics such as the ESBL gene blaCTX-M-14, and PMQR gene qnrS1. The IncI1 plasmid is widely distributed among Enterobacteriaceae and has been recognized as a major driver for transferring AMR genes in Salmonella spp. (42–44). Globally, IncI1 plasmids carrying blaCTX-M-14 have been reported in the UK and Japan (45) and co-carriage of fosA3 alongside blaCTX-M-14 has been documented in China (46), reflecting their widespread role of IncI1 plasmids in AMR dissemination. Although our findings do not confirm that IncI1 plasmids directly carry antimicrobial resistance genes, four S. Enteritidis isolates harboring a high number of resistance genes were also found to possess IncI1-type plasmids. This co-occurrence suggests a potential association between IncI1 plasmids and multidrug resistance, which may pose a public health risk if such strains become more prevalent. Therefore, reducing the use of third-generation cephalosporins and quinolones in Mongolia’s poultry industry is crucial to mitigate the potential spread of these MDR strains.
This study has several limitations that should be considered when interpreting the findings. First, the sampling period was relatively short and may not fully capture temporal variation in Salmonella prevalence across seasons. Second, the recovery rate of S. Enteritidis from Chinese-imported poultry was low, which limited the ability to perform direct genomic comparisons with domestic strains. To partially address this, we supplemented our analysis with publicly available S. Enteritidis genomes from EnteroBase, enabling broader phylogenetic contextualization and comparison.
Given the high prevalence of MDR S. Enteritidis among Mongolian isolates, including resistance to clinically important antibiotics such as ESBLs and PMQR, and the increasing consumption of poultry meat in Mongolia, the implementation of effective antibiotic stewardship in domestic poultry production is urgently needed to mitigate potential public health risks. To support these efforts, future studies should aim to conduct longitudinal and geographically broader surveillance to better understand temporal and spatial trends in Salmonella contamination.
In conclusion, this study represents the first prevalence-based investigation of S. Enteritidis isolates from poultry in Mongolia, providing valuable insights into their genetic diversity, antimicrobial resistance profiles, and phylogenetic relationships. These finding establish critical baseline data to inform future monitoring and control strategies to combat the spread of MDR S. Enteritidis in the Mongolian poultry industry.
Data availability statement
The whole-genome sequencing data generated in this study has been deposited in the NCBI BioProject database under accession number PRJNA1236736.
Author contributions
S-uS: Investigation, Writing – original draft, Formal analysis, Software. T-ML: Formal analysis, Visualization, Writing – review & editing. TK: Data curation, Writing – original draft. JK: Resources, Validation, Writing – review & editing. ES: Resources, Validation, Writing – review & editing. UT: Data curation, Investigation, Writing – review & editing. J-YH: Formal analysis, Methodology, Writing – review & editing. D-HL: Methodology, Supervision, Writing – review & editing. S-WL: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Korea Disease Control and Prevention Agency. This paper was also supported by Konkuk University in 2022.
Acknowledgments
We would like to thank Uyangaa Temuujin from the Mongolian University of Life Sciences, for their help with collecting Salmonella spp. from Mongolian chickens.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1595674/full#supplementary-material
Footnotes
1. ^http://github.com/tseemann/snippy
2. ^https://itol.embl.de; accessed on 21, February 2025
3. ^https://enterobase.warwick.ac.uk
References
1. Teklemariam, AD, Al-Hindi, RR, Albiheyri, RS, Alharbi, MG, Alghamdi, MA, Filimban, AAR, et al. Human salmonellosis: a continuous global threat in the farm-to-fork food safety continuum. Food Secur. (2023) 12:1756. doi: 10.3390/foods12091756
2. Sher, AA, Mustafa, BE, Grady, SC, Gardiner, JC, and Saeed, AM. Outbreaks of foodborne Salmonella enteritidis in the United States between 1990 and 2015: an analysis of epidemiological and spatial-temporal trends. Int J Infect Dis. (2021) 105:54–61. doi: 10.1016/j.ijid.2021.02.022
3. Peng, S, Xiong, H, Lu, J, Luo, F, Liu, C, Zhou, H, et al. Epidemiological and whole genome sequencing analysis of restaurant Salmonella Enteritidis outbreak associated with an infected food handler in Jiangxi Province, China, 2023. Foodborne Pathog Dis. (2024) 21:316–22. doi: 10.1089/fpd.2023.0123
4. About National Enteric Disease Surveillance | National Surveillance of Bacterial Foodborne Illnesses (Enteric Diseases) | CDC. Available online at: https://www.cdc.gov/national-enteric-surveillance/about/index.html?CDC_AAref_Val=https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf (Accessed March 10, 2025).
5. Munkhdelger, B. The meat processing industry in Mongolia. Int J Scientific Res Public. (2020) 10:563–568. doi: 10.29322/ijsrp.10.03.2020.p9963
6. Wang, W, Zhao, L, Hu, Y, Dottorini, T, Fanning, S, Fanning, S, et al. Epidemiological study on prevalence, serovar diversity, multidrug resistance, and CTX-M-type extended-spectrum β-lactamases of salmonella spp. from patients with diarrhea, food of animal origin, and pets in several provinces of China. Antimicrob Agents Chemother. (2020) 64:e00092-20. doi: 10.1128/AAC.00092-20
7. Wang, W, Cui, J, Liu, F, Hu, Y, Li, F, Zhou, Z, et al. Genomic characterization of Salmonella isolated from retail chicken and humans with diarrhea in Qingdao, China. Front Microbiol. (2023) 14:1295769. doi: 10.3389/fmicb.2023.1295769
8. Wang, Z, Jiang, Y, Xu, H, Jiao, X, Wang, J, and Li, Q. Poultry production as the main reservoir of ciprofloxacin- and tigecycline-resistant extended-spectrum β-lactamase (ESBL)producing Salmonella enterica serovar Kentucky ST198.2-2 causing human infections in China. Appl Environ Microbiol. (2023) 89:e0094423. doi: 10.1128/aem.00944-23
9. Cooper, AL, Low, AJ, Koziol, AG, Thomas, MC, Leclair, D, Tamber, S, et al. Systematic evaluation of whole genome sequence-based predictions of Salmonella serotype and antimicrobial resistance. Front Microbiol. (2020) 11:549. doi: 10.3389/fmicb.2020.00549
10. Deng, Y, Jiang, M, Kwan, PSL, Yang, C, Chen, Q, Lin, Y, et al. Integrated whole-genome sequencing infrastructure for outbreak detection and source tracing of Salmonella enterica serotype Enteritidis. Foodborne Pathog Dis. (2021) 18:582–9. doi: 10.1089/fpd.2020.2856
11. Hu, L, Cao, G, Brown, EW, Allard, MW, Ma, LM, and Zhang, G. Whole genome sequencing and protein structure analyses of target genes for the detection of Salmonella. Sci Rep. (2021) 11:20887. doi: 10.1038/s41598-021-00224-7
12. Zhanabayeva, DK, Paritova, AY, Murzakaeva, GK, Zhanabayev, AA, Kereev, A, Asauova, ZS, et al. Pcr diagnosis for the identification of the virulent gene of salmonella in poultry meat. Online J Biol Sci. (2021) 21:235–44. doi: 10.3844/ojbsci.2021.235.244
13. Chen, S, Zhou, Y, Chen, Y, and Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. (2018) 34:i884–90. doi: 10.1093/bioinformatics/bty560
14. Bolger, AM, Lohse, M, and Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
15. Bankevich, A, Nurk, S, Antipov, D, Gurevich, AA, Dvorkin, M, Kulikov, AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:455–77. doi: 10.1089/cmb.2012.0021
16. Zhang, S, den Bakker, HC, Li, S, Chen, J, Dinsmore, BA, Lane, C, et al. SeqSero2: rapid and improved salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol. (2019) 85:e01746-19. doi: 10.1128/AEM.01746-19
17. NARMS. National Antimicrobial Resistance Monitoring System (NARMS): 2014 human isolates final report. (2016) Available online at: https://stacks.cdc.gov/view/cdc/41823 (Accessed March 10, 2025)
18. Limbago, B. CLSI. Performance standards for antimicrobial susceptibility testing, vol. 23. 29th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2019).
19. Magiorakos, AP, Srinivasan, A, Carey, RB, Carmeli, Y, Falagas, ME, Giske, CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. European Society of Clinical Microbiology and Infectious Diseases (ESCMID), published by Elsevier on behalf of ESCMID. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
20. Seemann, T. Snippy, Rapid haploid variant calling and core genome alignment. GitHub repository (2015)
21. Wishart, DS, Han, S, Saha, S, Oler, E, Peters, H, Grant, JR, et al. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res. (2023) 51:W443–50. doi: 10.1093/nar/gkad382
22. Marçais, G, Delcher, AL, Phillippy, AM, Coston, R, Salzberg, SL, and Zimin, A. MUMmer4: A fast and versatile genome alignment system. PLoS Comput Biol. (2018) 14:e1005944. doi: 10.1371/journal.pcbi.1005944
23. Croucher, NJ, Page, AJ, Connor, TR, Delaney, AJ, Keane, JA, Bentley, SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. (2015) 43:e15. doi: 10.1093/nar/gku1196
24. Kalyaanamoorthy, S, Minh, BQ, Wong, TKF, Von Haeseler, A, and Jermiin, LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. (2017) 14:587–9. doi: 10.1038/nmeth.4285
25. Nguyen, LT, Schmidt, HA, Von Haeseler, A, and Minh, BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
26. Achtman, M, Zhou, Z, Alikhan, NF, Tyne, W, Parkhill, J, Cormican, M, et al. Genomic diversity of Salmonella enterica -the UoWUCC 10K genomes project. Wellcome Open Res. (2021) 5:223. doi: 10.12688/wellcomeopenres.16291.2
27. Katz, L, Griswold, T, Morrison, S, Caravas, J, Zhang, S, Bakker, H, et al. Mashtree: a rapid comparison of whole genome sequence files. J Open Source Softw. (2019) 4:1762. doi: 10.21105/joss.01762
28. Menardo, F, Loiseau, C, Brites, D, Coscolla, M, Gygli, SM, Rutaihwa, LK, et al. Treemmer: a tool to reduce large phylogenetic datasets with minimal loss of diversity. BMC Bioinformatics. (2018) 19:164. doi: 10.1186/s12859-018-2164-8
29. Tonkin-Hill, G, Lees, JA, Bentley, SD, Frost, SDW, and Corander, J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. (2019) 47:5539–49. doi: 10.1093/nar/gkz361
30. Bortolaia, V, Kaas, RS, Ruppe, E, Roberts, MC, Schwarz, S, Cattoir, V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
31. Carattoli, A, Zankari, E, Garciá-Fernández, A, Larsen, MV, Lund, O, Villa, L, et al. PlasmidFinder and pMLST: in silico detection and typing of plasmid. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
32. Sun, T, Liu, Y, Qin, X, Aspridou, Z, Zheng, J, Wang, X, et al. The prevalence and epidemiology of salmonella in retail raw poultry meat in China: a systematic review and meta-analysis. Food Secur. (2021) 10:2757. doi: 10.3390/foods10112757
33. Liu, C, Yao, K, Ren, D, and Xiao, Y. Prevalence and characterization of Salmonella from meat in slaughterhouses in Hangzhou, China. Int J Food Microbiol. (2022) 371:109649. doi: 10.1016/j.ijfoodmicro.2022.109649
34. Yang, X, Huang, J, Zhang, Y, Liu, S, Chen, L, Xiao, C, et al. Prevalence, abundance, serovars and antimicrobial resistance of Salmonella isolated from retail raw poultry meat in China. Sci Total Environ. (2020) 713:136385. doi: 10.1016/j.scitotenv.2019.136385
35. Gu, D, Wang, Z, Tian, Y, Kang, X, Meng, C, Chen, X, et al. Prevalence of Salmonella isolates and their distribution based on whole-genome sequence in a chicken slaughterhouse in Jiangsu, China. Front Vet Sci. (2020) 7:29. doi: 10.3389/fvets.2020.00029
36. Vinayaka, AC, Ngo, TA, Kant, K, Engelsmann, P, Dave, VP, Shahbazi, MA, et al. Rapid detection of Salmonella enterica in food samples by a novel approach with combination of sample concentration and direct PCR. Biosens Bioelectron. (2019) 129:224–30. doi: 10.1016/j.bios.2018.09.078
37. Chirambo, AC, Nyirenda, TS, Jambo, N, Msefula, C, Kamng’ona, A, Molina, S, et al. Performance of molecular methods for the detection of Salmonella in human stool specimens. Wellcome Open Res. (2021) 5:237. doi: 10.12688/wellcomeopenres.16305.2
38. Parker, AM, Mohler, VL, Gunn, AA, and House, JK. Development of a qPCR for the detection and quantification of Salmonella spp. in sheep feces and tissues. J Vet Diagn Invest. (2020) 32:835–43. doi: 10.1177/1040638720952359
39. Ríos-Castillo, AG, Ripolles-Avila, C, and Rodríguez-Jerez, JJ. Detection of Salmonella typhimurium and Listeria monocytogenes biofilm cells exposed to different drying and pre-enrichment times using conventional and rapid methods. Int J Food Microbiol. (2020) 324:108611. doi: 10.1016/j.ijfoodmicro.2020.108611
40. Rose, BE. Isolation and identification of Salmonella from meat, poultry, pasteurized egg, and catfish products and carcass and environmental sponges. Washington, DC, USA: USDA Microbiology Laboratory Guidebook (2014).
41. Alikhan, NF, Moreno, LZ, Castellanos, LR, Chattaway, MA, McLauchlin, J, Lodge, M, et al. Dynamics of Salmonella enterica and antimicrobial resistance in the Brazilian poultry industry and global impacts on public health. PLoS Genet. (2022) 18:e1010174. doi: 10.1371/journal.pgen.1010174
42. Oladeinde, A, Abdo, Z, Press, MO, Cook, K, Cox, NA, Zwirzitz, B, et al. Horizontal gene transfer is the Main driver of antimicrobial resistance in broiler chicks infected with Salmonella enterica Serovar Heidelberg. mSystems. (2021) 6:e00729-21. doi: 10.1128/msystems.00729-21
43. Carattoli, A, Villa, L, Fortini, D, and García-Fernández, A. Contemporary IncI1 plasmids involved in the transmission and spread of antimicrobial resistance in Enterobacteriaceae. Plasmid. (2021) 118:102392. doi: 10.1016/j.plasmid.2018.12.001
44. Foley, SL, Kaldhone, PR, Ricke, SC, and Han, J. Incompatibility group I1 (IncI1) plasmids: their genetics, biology, and public health relevance. Microbiol Mol Biol Rev. (2021) 85:e00031-20. doi: 10.1128/mmbr.00031-20
45. Kameyama, M, Chuma, T, Yokoi, T, Yabata, J, Tominaga, K, Miyasako, D, et al. Emergence of Salmonella enterica serovar infantis harboring IncI1 plasmid with blaCTX-M-14 in a broiler farm in Japan. J Vet Med Sci. (2012) 74:1213–6. doi: 10.1292/jvms.11-0488
46. Wang, XM, Dong, Z, Schwarz, S, Zhu, Y, Hua, X, Zhang, Y, et al. Plasmids of diverse Inc groups disseminate the fosfomycin resistance gene fosA3 among Escherichia coli isolates from pigs, chickens, and dairy cows in Northeast China. Antimicrob Agents Chemother. (2017) 61:e00859-17. doi: 10.1128/AAC.00859-17
Keywords: Salmonella Enteritidis, antimicrobial susceptibility test, whole-genome sequencing, antimicrobial resistance gene, whole-genome SNP analysis, poultry
Citation: Song S-u, La T-M, Kim T, Kim J, Shin E, Temuujin U, Hyeon J-Y, Lee D-H and Lee S-W (2025) Whole-genome sequencing analysis of Salmonella enterica serotype Enteritidis isolated from poultry sources in Mongolia. Front. Vet. Sci. 12:1595674. doi: 10.3389/fvets.2025.1595674
Edited by:
Ulises Garza-Ramos, National Institute of Public Health, MexicoReviewed by:
Rachel Amanda Hickman, Independent Researcher, Uppsala, SwedenNaim Deniz Ayaz, Kırıkkale University, Türkiye
Copyright © 2025 Song, La, Kim, Kim, Shin, Temuujin, Hyeon, Lee and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Won Lee, b2Rzc2V5QGtvbmt1ay5hYy5rcg==
 Seung-un Song
Seung-un Song Tae-Min La
Tae-Min La Taesoo Kim1
Taesoo Kim1 Dong-Hun Lee
Dong-Hun Lee