- 1Department of Internal Medicine, Reproduction and Population Medicine, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium
- 2Department of Pathobiology, Pharmacology and Zoological Medicine, Faculty of Veterinary Medicine, Ghent University, Merelbeke, Belgium
- 3Transversal Activities in Applied Genomics, Sciensano, Brussels, Belgium
Introduction: Sustained resistance against fluoroquinolones (FQ), without the use of FQ in broilers, raises important questions about other possible factors contributing to the persistence in farms. Therefore, the current study investigates the prevalence of FQ non-susceptibility in Escherichia coli on broiler farms and examines the roles of day-old chicks, the farm environment, and the antimicrobial use (AMU) in the dynamics of the within-flock spread.
Methods: On 29 Belgian broiler farms, AMU was monitored, while environmental and day-old chick intestinal samples (day 0) were collected before their arrival. On days 3 and 36 of the production round, 30 cloacal swabs were taken per farm. In all samples, total E. coli and FQ non susceptible E. coli isolates were quantified by non-selective and FQ selective isolation. A selection of the isolates was analyzed using whole-genome sequencing to characterize their resistance and virulence-associated determinants and to investigate their phylogenetic relatedness using core-genome multi-locus sequence typing (cgMLST) and whole-genome single-nucleotide polymorphism (SNP) analysis.
Results: Before entering the stable, day-old chicks carried FQ non-susceptible E. coli on 79.3% of the farms, while FQ non-susceptible E. coli were found in 48.3% of the sampled environments. According to cgMLST, identical FQ non-susceptible isolates were found on day 0 and 36, suggesting that FQ non-susceptible isolates present in the environment at the start of a production round or in day-old chicks, can remain present until slaughter, even when no FQs were used. Total AMU was positively correlated with the proportion of FQ non-susceptible E. coli [r = 0.42, 95% CI (0.06, 0.68), p = 0.03], often also multidrug-resistant, at the end of the production cycle.
Conclusion: The continued presence of FQ non-susceptible E. coli in broiler farms is likely the result of both a historical contamination at the farm level and a continuous influx along the production chain. AMU contributes to the continued presence of FQ non-susceptible E. coli in broiler farms, but accounted for only a small proportion of the variability in FQ non-susceptibility in currently investigated farms. The role of certain virulence-associated genes in the persistence of FQ non-susceptible E. coli in broiler farms deserves more in-depth research.
1 Introduction
Antimicrobial resistance (AMR) is of critical concern for both human and animal health. The emergence of AMR is a complex issue with antimicrobial usage (AMU) often being the main driver (1). Several studies have demonstrated a link between lowering AMU and a reduction in AMR in food producing animals (2–4). However, in countries or farms not using any (fluoro)quinolones anymore, quinolone resistance in Escherichia coli has been shown to persist (5–8).
The sustained resistance against fluoroquinolones (FQ), without the use of FQ in broilers, raises important questions about possible other factors contributing to the persistence in these farms. Several studies showed that day-old chicks and the broiler house environment are involved in the introduction and transmission of extended-spectrum β-lactamase (ESBL) producing E. coli in broiler flocks (9–13). For instance, Huijbers et al. (3) demonstrated that both introduction routes contributed to flock colonization, where nearly all broilers tested positive for ESBL-producing E. coli 3 days after the start of the production cycle. Also Dierickx et al. (10) confirmed this rapid spread within a broiler flock, and was able to link isolates from day-old chicks to isolates in parent stock, suggesting a transmission route from breeding flocks to hatcheries and subsequent uptake by chicks from the hatchery environment. This form of pseudo-vertical transmission in the hatchery was also described by Projahn et al. (12). On top of that, environmental recirculation of resistant strains from previous production cycles was suggested, as resistant strains were detected even after cleaning and disinfection, a finding also supported by Daehre et al. (13). Furthermore, insects have been identified as potential vectors contributing to the introduction and dissemination of ESBL-producing E. coli within broiler houses (11).
As far as FQ resistance is concerned, several studies suggest vertical transmission of FQ resistant E. coli within the broiler production pyramid, highlighting the potential role of breeding animals (8, 14–17). This has been demonstrated by linking isolates from (grand)parent stock to broilers samples retrieved at the end of the production cycle (8, 14–17). Notably, these studies often focussed on sampling at the end of the broiler production cycle and often did not investigate transmission within a flock during the production cycle and the contribution of other potential risk factors such as AMU at the farm level. Petersen et al. (16) was able to link FQ resistant E. coli in parent stock to hatchery fluff and hatching eggs causing an outbreak of colibacillosis in broiler flocks, indicating vertical transmission. Also, several studies highlighted the role of the environment in the circulation of FQ resistant E. coli among broilers (14, 15, 18, 19). Börjesson et al. (15) could trace strains from grandparent bird to broilers and found resistant E. coli with identical patterns in hatcheries and poultry houses, suggesting environmental involvement. However, no direct link was visible between broiler environment and the housed broiler batch. This persistence in the broiler house environment was also suggested by Kaspersen et al. (14). In an experimental setting, Leclerq et al. (20) observed that multi-drug-resistant E. coli could survive in the environment and potentially recirculate. However, they found no evidence of introduction of resistant strains through day-old chicks. The study did emphasize the need for research under commercial farm conditions, where biosecurity measures would presumably be less controlled than in experimental environments. While several introduction and persistence routes for FQ-resistant E. coli in broiler production have been proposed, including vertical transmission and environmental contamination, most existing studies have examined these factors in isolation. Large, comprehensive studies that simultaneously take into account the role of the environment, day-old chicks and AMU, are lacking. To effectively combat FQ resistance in the broiler industry, a more holistic understanding of its presence and dissemination within broiler flocks is essential, allowing for targeted interventions to reduce its prevalence.
The purpose of this study was to investigate the role of AMU, day-old chicks and the environment in the spread of FQ non-susceptible E. coli within broiler flocks. Furthermore, the goal was to see how this FQ resistance persists throughout the production cycle and what the (genetic) AMR traits are of the persisting non-susceptible E. coli strains.
2 Materials and methods
Approval for this study was granted by the ethical committee of the Faculties of Veterinary Medicine and Bioscience engineering of Ghent University (EC2020/040).
2.1 Farm characteristics
This study included 29 commercial broiler farms from the Flanders region of Belgium. Farms were included based on convenience sampling, as they were recruited through outreach efforts facilitated by local veterinary poultry practices. Participation was on a voluntary basis and only conventional broiler (Ross 308) farms in Belgium were included. Per farm, one house was randomly selected for follow-up during one production cycle and samples were collected to determine prevalence of FQ non-susceptible E. coli on day 0, 3 ± 2 days, and 36 ± 3 days of that production round. Also, information was collected on farm size, hatchery, age of the parent stock, and AMU during that specific production round. AMU of the specific production cycle was collected by a form which was filled in by the farmer during the sampling period. Quantification of AMU was done through the calculation of the Treatment Incidence (TI) per 100 days, as described by Persoons et al. (20, 21). The TI is equal to the number of days animals are treated with antimicrobials in a theoretical period of 100 days. Sampling was conducted between October 2020 and December 2021.
2.2 Sample collection environment
Environmental swabs from the stable were collected on day 0 before the day-old chicks arrived, when bedding material was already placed and the feeding pans were filled. For the environmental swabs premoistened sponge swabs with 10 mL Buffered Pepton Water (BPW) were used (3 M, SLL10BPW, St-Paul, United States). Five locations were sampled in duplicate: the floor and boots in the anteroom, and the floor, drinking cups, and feeding pans in the house. An A4-sized floor area was sampled per swab, while three drinking cups, one feeding pan per swab, or both soles of a pair of boots were sampled per swab. These locations were chosen based on previous research, which identified these locations as high risk areas for contamination with bacteria, even after cleaning and disinfection (22). Also, drinking water samples were collected at the end of the drinking line in each house after cleaning the line with a disinfectant and running the water for around 3 min. The sample was sent to an external laboratory for bacteriological analysis.
2.3 Sample collection broilers
On day 0, 3, and 36, chicks were sampled to screen for gastrointestinal carriage of FQ non-susceptible E. coli. On day 0, 30 day-old chicks were randomly collected before they were unloaded in the house. The chicks were transported to the laboratory in a cardboard transport box, followed by euthanasia through cervical dislocation and collection of the entire gastrointestinal tract and contents. On day 3 and 36, cloacal samples were collected from 30 randomly selected chicks using sterile rayon-tipped swabs (Vacutest Kima, Italy). Also, on day 36, three broilers were euthanized and feather samples were collected to screen for FQ residues. All collected samples were cooled directly, transported at a temperature of 2–7°C and processed within 4 h after collection.
2.4 FQ residues in feathers
To detect residues of FQ in the feathers of broilers, a validated UPHLC-MS/MS method was used (23). The feathers were screened for the presence of ciprofloxacin, enrofloxacin and flumequine residues. The residue concentration of the complete feather sample was determined by weighing around 500 mg of feathers, followed by cutting the feathers into smaller pieces and grinding. From every grinded feather sample, around 100 ± 1 mg was weighed and further processing took place as described in Ringenier et al. (23). The limit of quantification (LOQ) of the used method was 5 ng/g feather for enrofloxacin and ciprofloxacin, and 1 ng/g feather for flumequine.
2.5 Bacteriological analysis
In the day-old chicks, the entire gastrointestinal tracts were removed with sterile material, cut into smaller pieces, weighed and transferred into a stomacher bag. Subsequently, sterile BPW (Oxoid, Thermo Fisher Scientific, Merelbeke, Belgium) was added (in ratio of 9 mL of BPW per gram of intestines) and thoroughly mixed using a stomacher. Next, 100 μL of the mixture was inoculated on FQ selective (MacConkey agar Nr 3 (Oxoid, Thermo Fisher Scientific, Merelbeke, Belgium) supplemented with 0.25 mg/L enrofloxacin) and FQ non-selective (MacConkey agar Nr 3) media. These plates were aerobically incubated overnight at 37°C, as well as the intestine suspensions in BPW. If no growth was visible on the (supplemented) MacConkey agar plates, 100 μL of the enriched broth was inoculated on a MacConkey plate and/or a MacConkey plate supplemented with 0.25 mg/L enrofloxacin. The concentration of enrofloxacin in the supplemented plates was used to select for non-wild type isolates (0.25 mg/L is the epidemiological cut-off value (ECOFF) for enrofloxacin in E. coli). “FQ non-susceptible E. coli isolates” will be used in this article for FQ non-wild type isolates.
To the environmental swabs, 10 mL of sterile BPW was added immediately after transportation to the laboratory. After homogenization, using a stomacher, the swab suspensions were aerobically incubated for 24 h at 37°C. After incubation, 10 μL of this suspension was plated on FQ selective and FQ-non-selective media as described above and aerobically incubated overnight at 37°C.
One hundred milliliter of the collected water samples was filtered through a 0.45 μm filter. The filter was placed on non-supplemented Rapid E. coli (Bio-Rad) agar plates and Rapid E. coli agar plates supplemented with 0.25 mg/L enrofloxacin to screen for FQ non-susceptible E. coli. Plates were aerobically incubated at 37°C for 24 h. The colonies were counted and the number of E. coli isolates was determined per 100 mL.
Cloacal swabs were weighed before and after collecting feces to determine the amount of feces collected. Three milliliter of BPW was added and the swab was thoroughly vortexed. Next, 100 μL of the feces suspension was used to make 10 fold serial dilutions (from 10−1 to 10−4) in Phosphate Buffered Salt solution (PBS). Of every dilution, 100 μL was inoculated on FQ selective and FQ non-selective media. These plates were aerobically incubated overnight at 37°C, as were the swabs containing BPW broth for microbial enrichment. The concentration of FQ non-susceptible E. coli and total E. coli in the cloacal samples were determined (CFU/gram feces) based on the counts on plates with 20–200 CFU and taking into account the weight of the feces on the swab (ranging between 0.01 and 0.34 gram). The proportion of CFU grown on FQ selective over FQ non-selective MacConkey plates allowed to determine the prevalence of FQ non-susceptible E. coli strains in a specific cloacal sample. Identification of the isolates at the species level were done by MALDI-TOF mass spectrometry, as previously described in Vereecke et al. (24). When no growth was visible on the FQ selective or the non-selective MacConkey plates, 100 μL of the overnight enriched broth was inoculated on a second MacConkey plate (supplemented with enrofloxacin) and aerobically incubated overnight at 37°C. When colonies were detected after enrichment only, it was assumed that the concentration was below the detection limit of direct plating. For analysis, the value of such samples was set at the detection limit (for day-old chicks this was 10 CFU/mL BPW and for cloacal swabs 100 CFU/mL BPW) and corrected for the amount of feces on the swabs, as described previously (25).
Of every farm from each sampling point, around 10 randomly selected FQ non-susceptible isolates originating from 10 different animal samples were stored at −20°C. From each positive environmental sample, one FQ non-susceptible E. coli isolate was stored. Due to practical and financial constraints, only a selection of isolates was further analyzed.
2.6 MIC
For the determination of the minimum inhibitory concentration (MIC), five farms were included based on the results of the prevalence of FQ non-susceptible E. coli in the environment and in day-old chicks. Only isolates of day 0 and 36 were included for MIC determination, which resulted in a selection of 92 E. coli isolates obtained from FQ supplemented plates. Antimicrobial susceptibility testing was performed using a Sensititre EU Surveillance E. coli EUVSEC Plate (Trek Diagnostic Systems, Thermofisher Scientific, Merelbeke, Belgium) according to the manufacturer’s guidelines. The protocol was followed as described in Ibrahim et al. (26). In brief, the Sensititre plate was aerobically incubated at 35°C for 18–24 h. The MIC was determined as the lowest concentration at which no visible growth was observed. The quality control strain used was E. coli ATCC 25922. The ECOFFs from the European Committee on Antimicrobial Susceptibility Testing were used for interpretation (27).
2.7 WGS
Based on MIC results, a further selection of E. coli isolates was made for whole genome sequencing (WGS). Isolates with different resistance profiles from the same sample time point within a farm were included to undergo WGS, resulting in a selection of 66 FQ non-susceptible E. coli isolates, originating from four farms. DNA extraction was done using the GenElute™ Bacterial Genomic DNA Kit from Sigma-Aldrich (Merck, Overijse, Belgium) following the manufacturer’s protocol. The quantity and purity of the DNA was determined spectrophotometrically using a NanoDrop (Thermo Scientific, Waltham, MA, United States) and the quantity using fluorometry (Qubit 4, Invitrogen, Carlsbad, CA). A DNA-library was made using the Nextera XT library kit (Illumina). Library fragmentation length and library DNA concentration were evaluated using the TapeStation 4,200 with the HS D5000 ScreenTape and Reagent kits (Agilent Technologies, Santa Clara, CA) and a dsDNA HS assay kit for the Qubit 4 fluorometer, respectively. All libraries were sequenced on a MiSeq instrument (Illumina, San Diego, CA) using the MiSeq V3 chemistry, as described by the manufacturer’s protocol, for the production of 2 × 250 bp paired-end reads. Pre-processing and quality control of the sequenced data were performed using the Shiga-toxin producing E. coli pipeline described by Bogaerts et al. (28). NCBI AMRFinder+ (v3.10.18) was used to screen the assembled contigs for genes and mutations associated with AMR using the 2021-12-21.1 database. This database includes genomic markers commonly associated with FQ resistance, such as the often plasmid-encoded qnr and qepA genes, as well as chromosomal mutations in for example gyrA and parC. Additionally, the assemblies were screened for the presence of virulence-associated genes using blastn v2.6.0 and the VirulenceFinder E. coli database (accessed on July 23, 2023). Sequence typing was performed using the MLST and cgMLST schemes obtained from EnteroBase (29) (accessed on July 7th 2024) as described previously (28). Additional SNP-based clustering was performed for sequence types (STs) with multiple isolates using SnapperDB v1.0.6 (30), of which the selected reference genomes are listed in Supplementary Table S1. Isolates were considered to be clusters if their pairwise SNP distances were 5 or less (31).
2.8 Validation of WGS-based taxonomic identification for Escherichia fergusonii strains
The WGS data analysis workflow includes taxonomic classification of the trimmed reads using Kraken 2 v2.0.7 and a custom database containing all NCBI RefSeq genome entries (database accessed 2nd November 2021) annotated as “complete genome” with accession prefixes NC, NW, AC, NG, NT, NS, and NZ of the following taxonomic groups: archaea, bacteria, fungi, human, protozoa, and viruses (32). For nine strains, the proportion of reads classified to an unexpected species (i.e., not E. coli) was greater than 5%, which is the failure threshold enforced in the workflow (28). For these datasets, the majority of reads were classified as E. fergusonii. Additional verification was performed by constructing a core genome MLST phylogeny containing all in-house strains, complemented with 50 randomly selected complete E. coli and randomly selected complete E. fergusonii genomes from NCBI. In the resulting topology, the nine strains classified as E. fergusonii by Kraken 2 clustered with the E. fergusonii genomes obtained from NCBI, indicating that this taxonomic identification is likely to be correct.
2.9 Statistical analysis
For descriptive statistics, IBM SPSS Statistics 29.0 (IBM, New York, United States) was used. To investigate the relationship between the proportion of FQ non-susceptible E. coli at the end of the production cycle and total TI of the production cycle, age of the parent stock, capacity of the stable, proportion of positive day-old chicks, and sum of FQ positive environmental locations; a backwards stepwise regression model was conducted, using p < 0.1 for inclusion in the model, resulting in a univariable linear regression model. An independent t-test was conducted to determine if there was a significant difference in proportion of FQ non-susceptible E. coli on day 3 between the flocks that were treated with lincomycin and spectinomycin and the flocks that did not receive such treatment. An independent samples Kruskal-Wallis test was performed to compare the average number of day-old chicks carrying FQ non-susceptible E. coli per farm across different hatcheries.
3 Results
This study included 29 commercial broiler farms with an average house capacity of 35,453 birds [min 14,400 - max 80,000]. The batches of day-old chicks from all farms originated from a total of seven different hatcheries. The average age of the parent stock was 48.82 weeks (SD ± 12.80). Farm characteristics are provided in Supplementary Table S2.
3.1 AMU
On 72.4% (21/29) of the farms, the production cycle started with a course of antimicrobials (Figure 1), of which 95.5% used lincomycin in combination with spectinomycin. Five farms started using antibiotics some days after the start of the production cycle. Only three farms did not administer antimicrobials during the entire production period (Supplementary Table S3). The average TIDDDvet was 4.18 [min 0 - max 33.72], meaning that broilers in this study received antimicrobials during on average 4.18% of their lifespan, which equals to 1.8 treatment days within a production cycle of 42 days. Only one farm reported using a FQ (flumequine) during the observed production cycle. The concentration of flumequine in the feathers from this farm ranged from 50 to 600 ng/g feather (Supplementary Table S4). In the feather samples of the other farms, no FQ concentrations above LOQ could be detected, except for one farm where very low levels of flumequine above LOQ were found, ranging from 1.2 to 3.7 ng/g feather. According to the AMU records, only lincomycin in combination with spectinomycin was administered on this farm in the first 3 days of the production cycle.
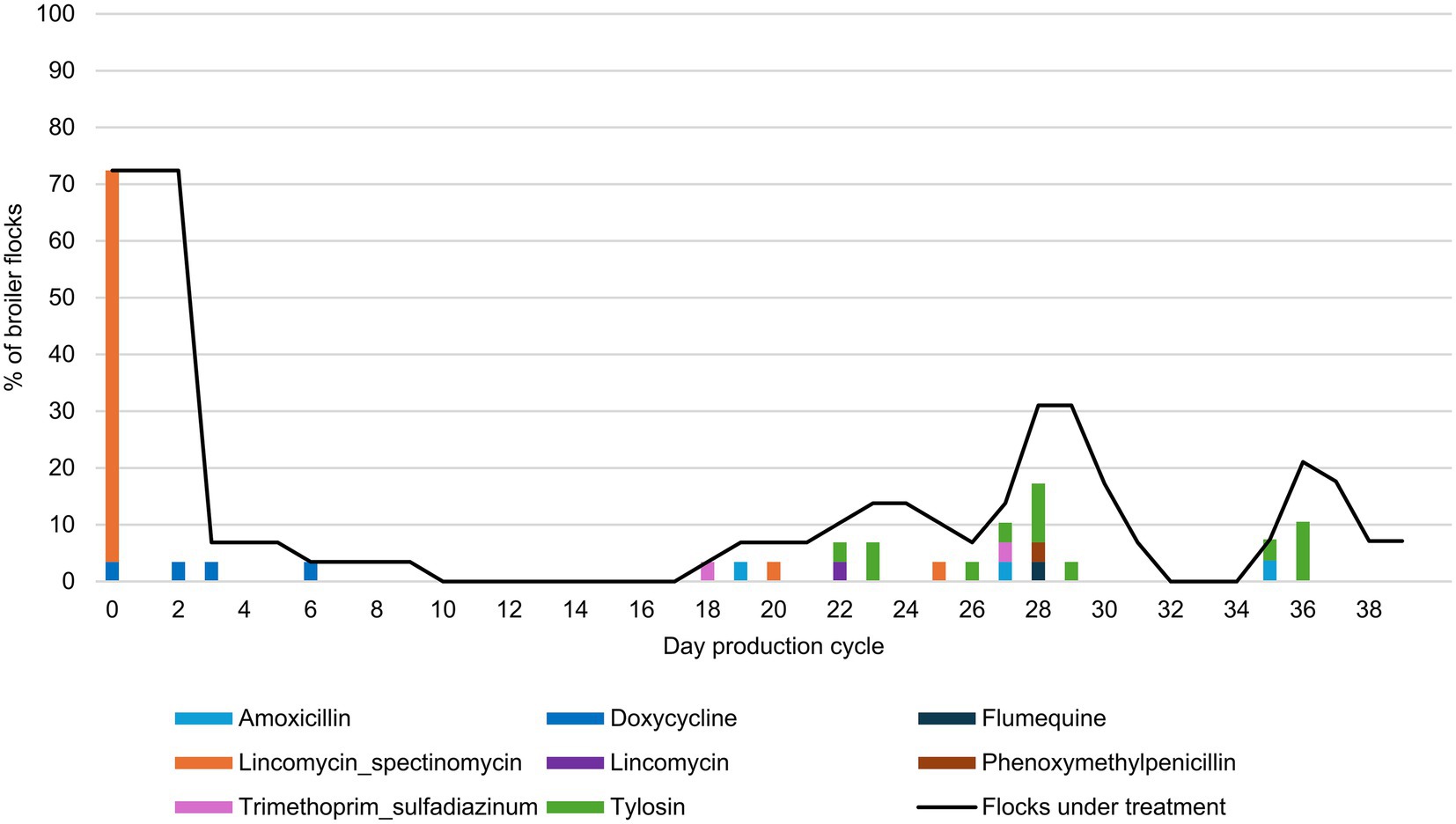
Figure 1. Antimicrobial treatments in the broiler flocks during the observed production cycle (n = 29). The results show the percentage of flocks starting treatment per antimicrobial group, together with percentage of flocks under treatment during each day of the production cycle (continuous black line).
3.2 FQ non-susceptibility day 0
On 14 farms, at least one FQ non-susceptible E. coli isolate could be obtained from the environment before the day-old chicks entered the stable (Table 1; Supplementary Table S5). On 79.3% (23/29) of the farms, FQ non-susceptible E. coli were detected in the gastro-intestinal tracts from day-old chicks before entering the stable (Table 2). Of these positive flocks, the percentage of day-old chicks carrying FQ non-susceptible E. coli was on average 55.3% [min 6.7 - max 100] (Figure 2). There was no statistically significant difference in the percentage of positive day-old chicks between the different hatcheries (p = 0.38), indicating that there was no specific hatchery providing a significant higher or lower number of day-old chicks carrying FQ non-susceptible E. coli.
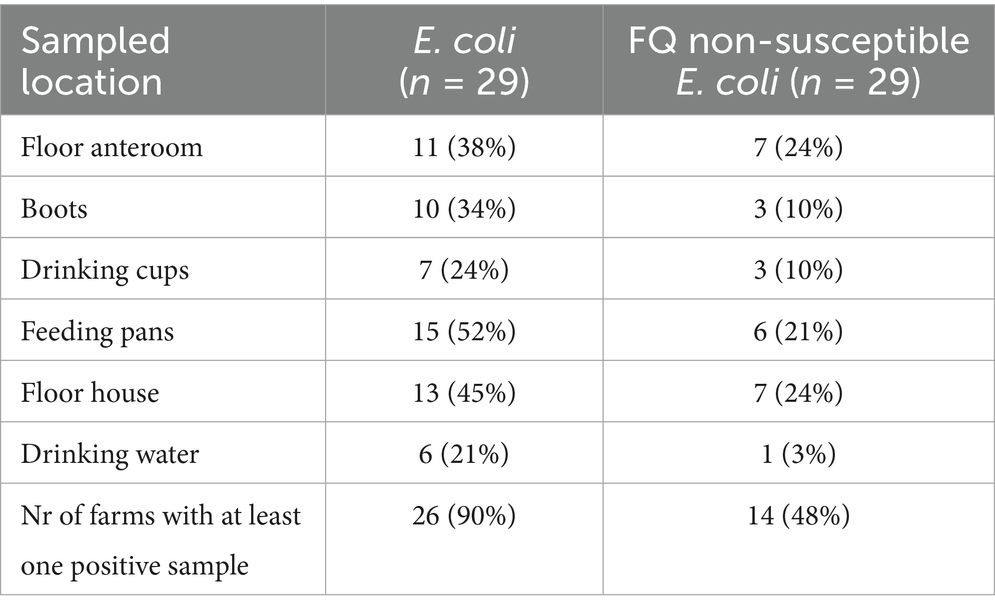
Table 1. Number of farms with E. coli and FQ non-susceptible E. coli in the environment (broiler house) before arrival of day-old chicks, specified for the different sampled locations within the house.
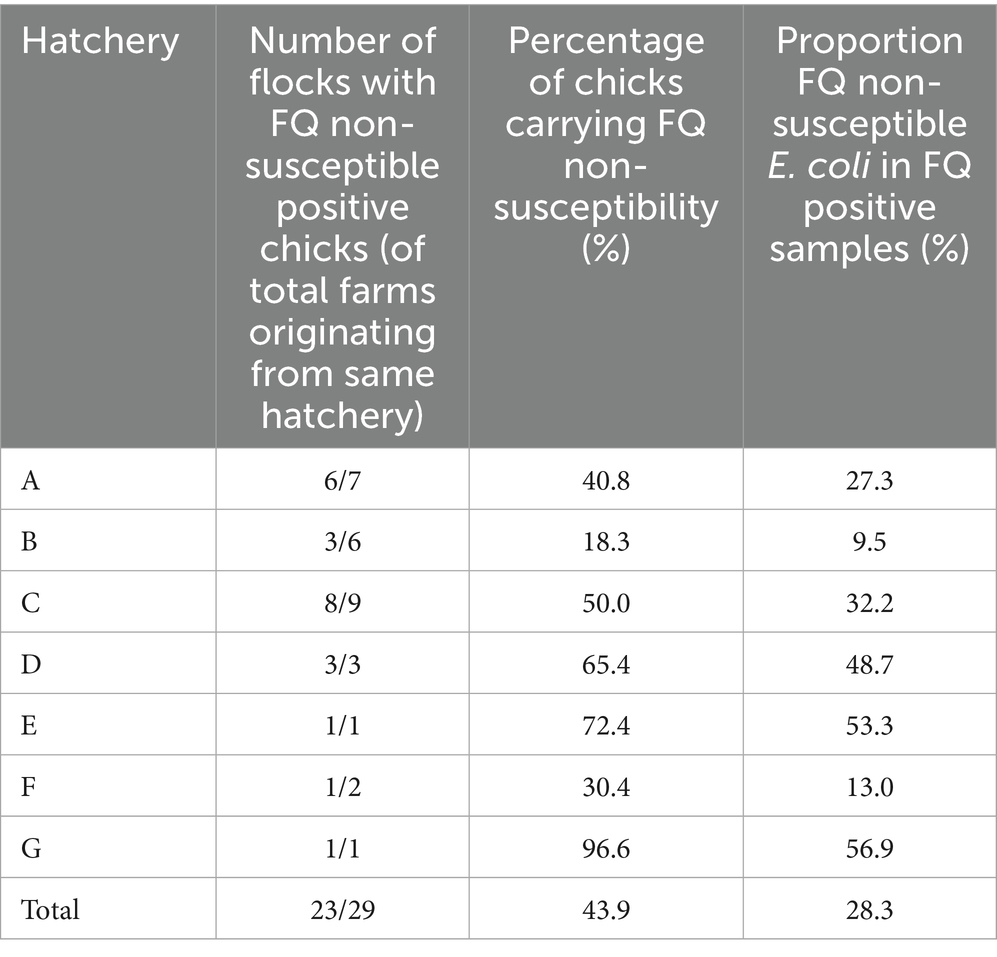
Table 2. The number of farms that received day-old chicks with FQ non-susceptible E. coli in the gastrointestinal tract, the average percentage of chicks carrying FQ non-susceptibility, and the proportion of FQ non-susceptible E. coli (of total E. coli) at farm level, for each hatchery from which the flocks originated. The flocks of the 29 participating farms originated from 7 different hatcheries (A to G).
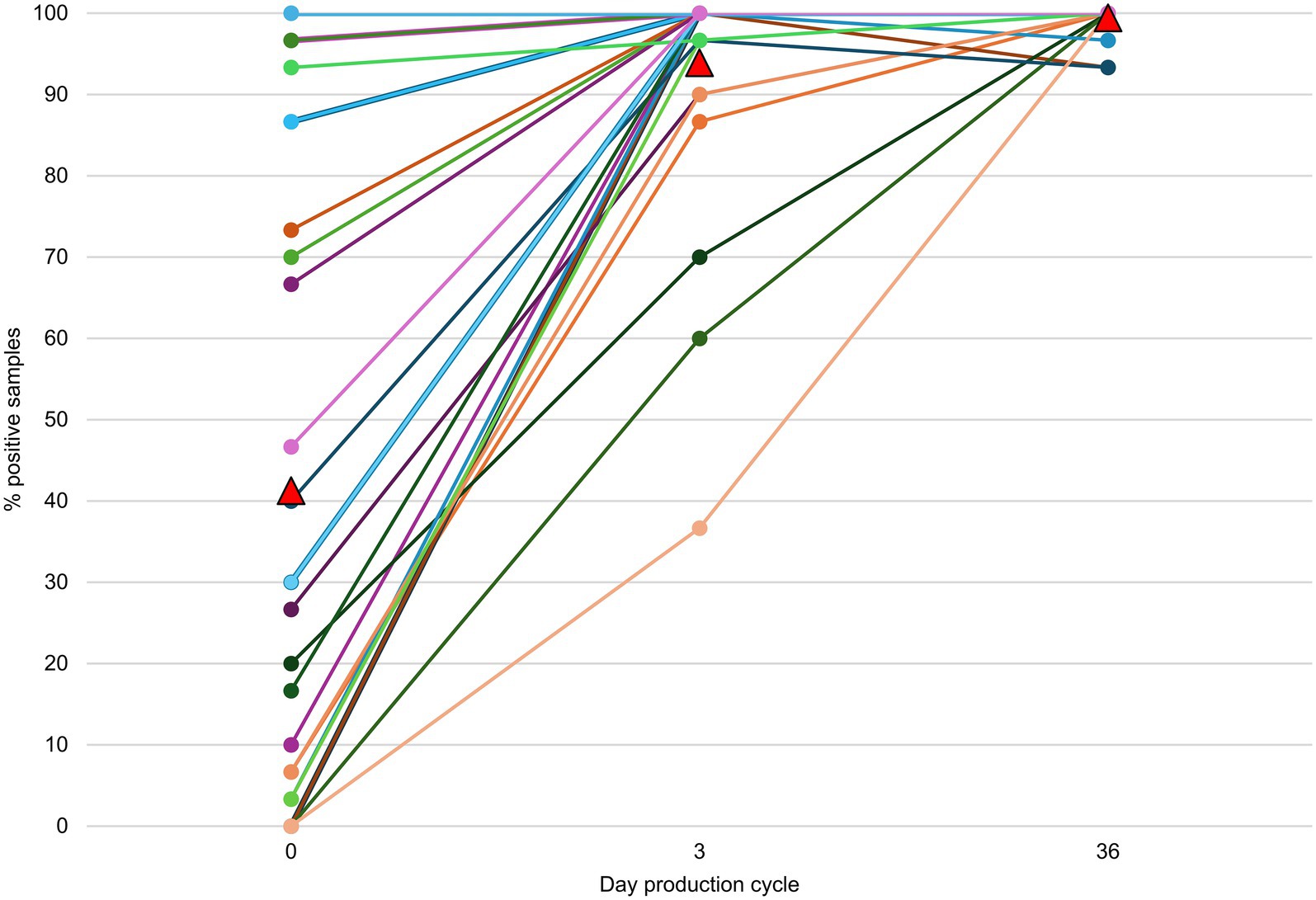
Figure 2. Farm level percentage of samples harboring fluoroquinolone (FQ) non-susceptible E. coli per sampling point during one production cycle. On day 0, gastrointestinal contents were sampled and on day 3 (±2) and 36 (±3), cloacal swabs were taken. In six batches of day-old chicks no FQ non-susceptible E. coli could be detected. The mean percentage for each time point is depicted as a red triangle.
3.3 FQ non-susceptibility during production cycle
On days 3 and 36, FQ non-susceptible E. coli were present at 100% of the farms (and in 94.0 and 99.4% of the broiler samples, respectively) (Figure 2). On days 0, 3 and 36, the proportion of FQ non-susceptible E. coli in the total E. coli population (only taking into account samples that contained E. coli) was 28.3%, 26.7, and 30.6%, respectively (Figure 3; Supplementary Table S6). The total TI of the production cycle was significantly associated with the proportion of FQ non-susceptible E. coli at the end of the production cycle (r = 0.42, 95% CI [0.06, 0.68], p = 0.03) (Figure 4). There was no significant difference in mean proportion of FQ non-susceptible E. coli at day 3 between the flocks treated with lincomycin and spectinomycin and the flocks that did not receive such treatment, with a mean difference of 4.87% (95% CI [−10.38, 20.10%], p = 0.52).
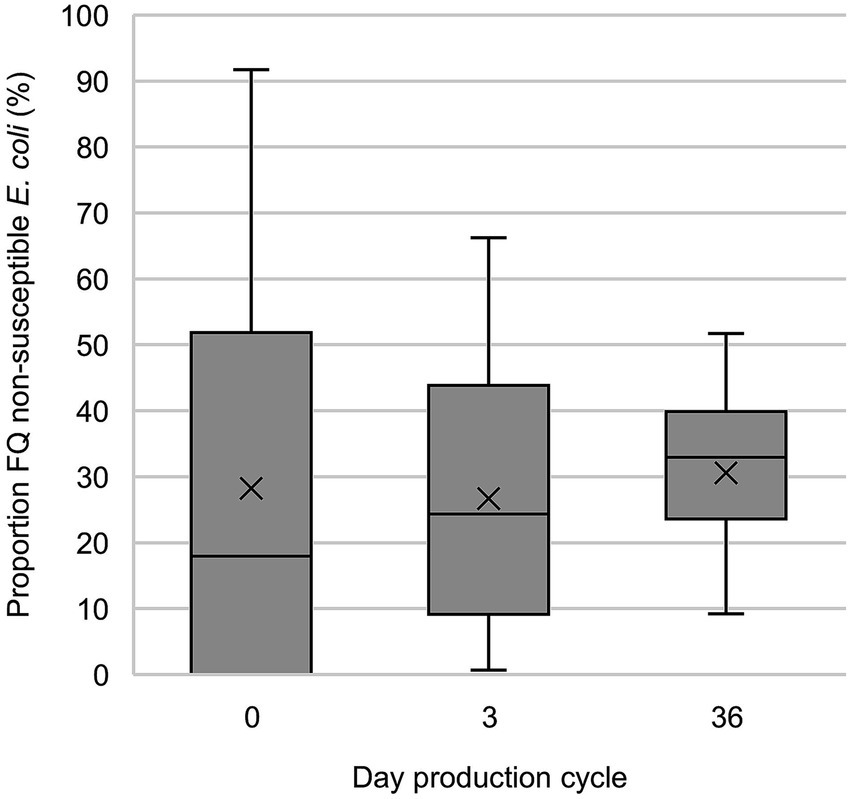
Figure 3. Proportion of fluoroquinolone (FQ) non-susceptible E. coli in the total E. coli population in fecal samples of broilers containing FQ non-susceptible E. coli (only taking into account samples that contained E. coli). On day 0, intestines and contents were sampled and on day 3 (±2) and 36 (± 3), cloacal swabs were investigated.

Figure 4. Scatter plot and corresponding regression line for the relationship between the dependant variable proportion of FQ non-susceptible E. coli at the end of the production cycle (day 36) and the independent variable total antimicrobial usage expressed in treatment incidence (TI) at the farm level.
3.4 Multidrug-resistance
Of the 92 selected isolates (obtained from FQ supplemented plates) which underwent MIC determination, 63.6% was multidrug-resistant when applying ECOFF values (resistant against 3 or more antimicrobial classes) (Supplementary Table S7). Of the 66 isolates selected for WGS, nine isolates which were previously identified as E. coli by MALDI-TOF MS, were identified as Escherichia fergusonii based on WGS data (Supplementary Table S8). Of the remaining 57 E. coli isolates, 14 isolates originated from environmental samples, 22 isolates from day-old chicks and 21 isolates from broilers at day 36. The concordance between the detected genetic traits and the observed phenotypic resistance, known as the phenotypic-genotypic relationship, ranged from 93.0% for beta-lactam antibiotics to 100% for sulfonamides, trimethoprim and quinolones. About 80.7% of the isolates carried resistance genes against beta-lactam antimicrobials and 71.9% against aminoglycosides. Only one isolate, from a broiler at day 36, carried an ESBL gene (blaCTX-M-55). WGS detected genes and/or mutations associated with three or more classes of antibiotics in over 86% of the isolates (Supplementary Table S8).
3.5 Genetic traits FQ non-susceptibility
Of the FQ non-susceptible isolates, 61.4% carried point mutations in the Quinolone Resistance-Determining Region (QRDR) and 38.6% carried qnr resistance genes (Figure 5). No other FQ resistance traits were detected. The proportions of qnr genes and QRDR mutations per sampling time point are listed in Table 3. All isolates with triple or quadruple QRDR mutations (n = 11) had ciprofloxacin MIC values > 4 mg/L (Supplementary Table S8). Of the 14 strains carrying qnr genes, two carried an additional AMR gene on the corresponding contig, being blaCTX-M-55 and tet(A) (Supplementary Table S8). All E. fergusonii isolates (n = 9), consisting of two clusters of identical isolates carried the gyrA S83L mutation. One cluster of E. fergusonii isolates (n = 7) additionally carried the parE I355T mutation.
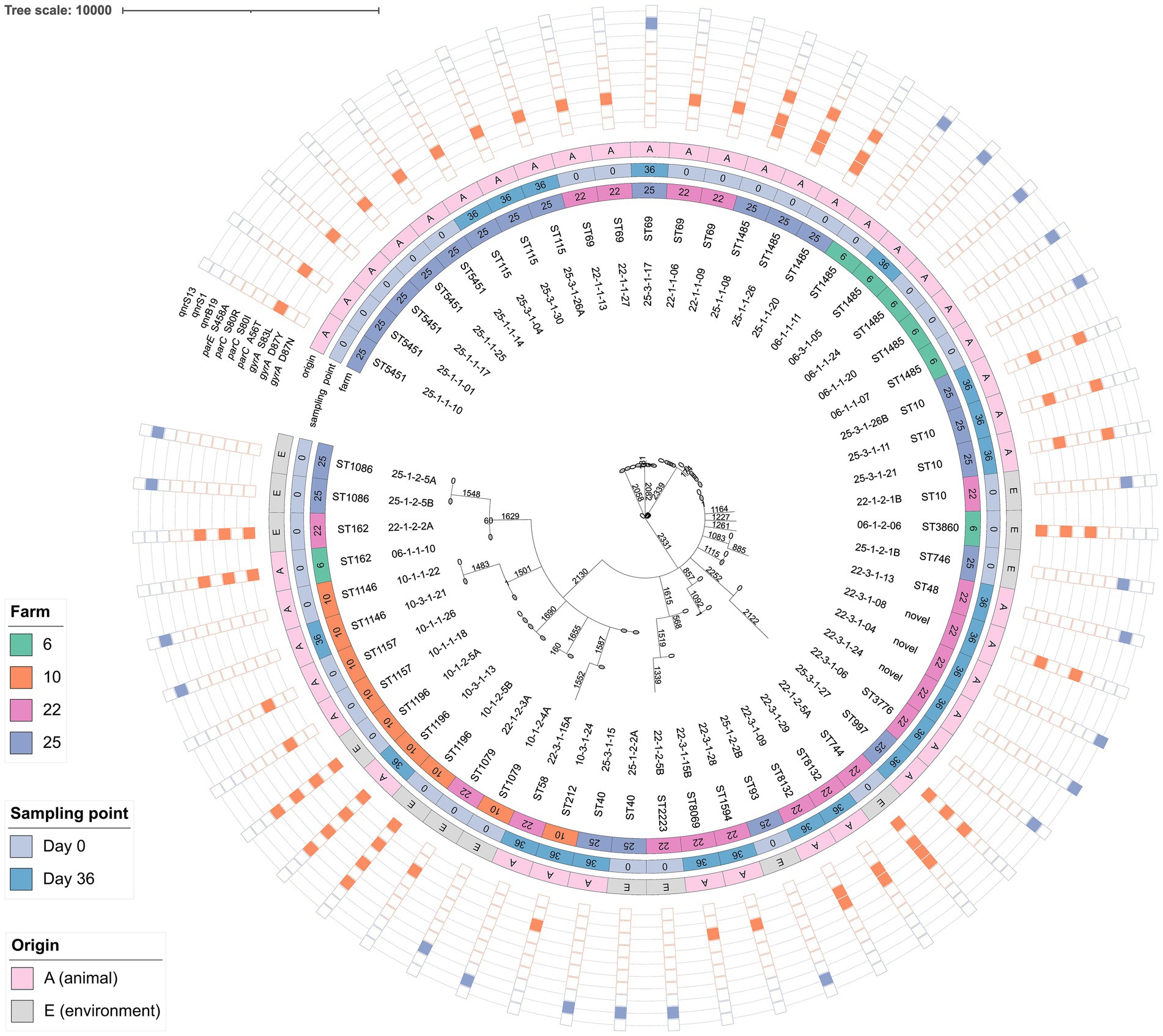
Figure 5. Phylogenetic tree of fluoroquinolone (FQ) non-susceptible E. coli based on core genome MLST. The FQ resistance genes and/or mutations are shown in the outer band: orange is the presence of QRDR mutations, purple is presence of qnr genes. Isolates from four farms were included, originating from sampling point day 0 and day 36. For every isolate the sampling name and sequence type are given. The letter A or E indicates whether the sample is taken from an animal (A) or the environment (E). The tree was constructed using the “MSTreeV2” method of GrapeTree. Branch lengths and the scale bar are expressed as the number of allelic differences.
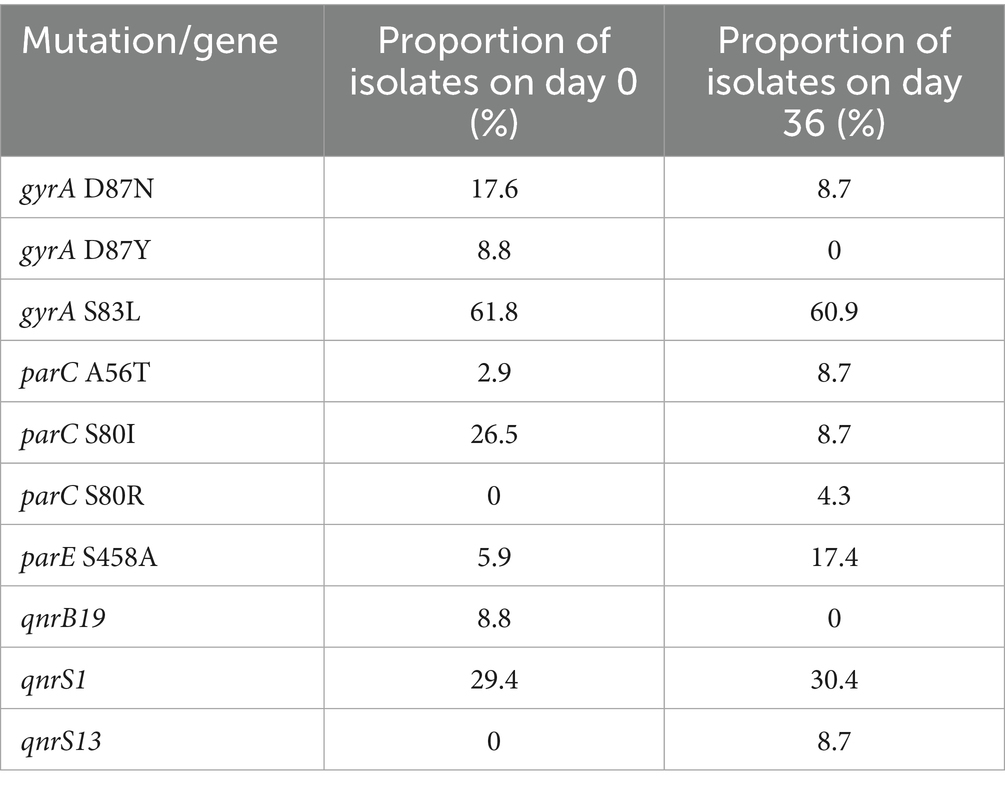
Table 3. Proportion of E. coli isolates with QRDR mutations and qnr genes on day 0 (n = 34) and day 36 (n = 23).
3.6 Virulence-associated genes
The 57 analyzed E. coli isolates carried on average 13 virulence-associated genes (VAGs) (SD ± 5.13). Of the 57 E. coli isolates, most common VAGs were terC (93.0%), nlpI (87.7%), iss (86.0%), traT (77.2%), sitA (75.4%), hlyF (64.9%), csgA (56.1%), iutA (56.1%), iucC (54.4%), and lpfA (52.6%). Other genes were present in less than 50% of the isolates (Supplementary Table S8).
3.7 Clustering
There was a large variation in the STs of FQ non-susceptible E. coli, both within and between farms (Figure 5). Three isolates were assigned to different novel STs (i.e., STs not yet introduced in the EnteroBase database). On farm 10 (ST1196) and 25 (ST40), FQ non-susceptible E. coli isolates collected on day 0 from the environment, showed identical cgMLST profiles and <5 pairwise SNP differences to isolates obtained from cloacal swabs of broilers on the same farm (Figure 5; Supplementary Table S6). In addition, on farms 6 (ST1485) and 10 (ST1146), isolates from day-old chicks clustered together with those from the broiler at the end of the production cycle. On farm 6, five ST1485 isolates exhibited ≤3 pairwise SNP differences, while on farm 10, two ST1146 isolates, for which SNP analysis was not possible due to the lack of a suitable reference genome, had identical cgMLST profiles. Moreover, isolates from different day-old chicks within the same farm clustered together, which was the case on farm 25 (three ST1485 isolates with 0 pairwise SNP differences), farm 22 (three ST69 isolates with 0–4 pairwise SNP differences), and farm 6 (four ST1485 isolates with 1–4 pairwise SNP differences) (Supplementary Table S9). For ST5451, five isolates collected from chicks on day 0 shared identical cgMLST profiles. However, no suitable reference genome was available on EnteroBase, and SNP analysis was not performed.
4 Discussion
FQ are classified as critically important antimicrobials by both the World Health Organization and the World Organization of Animal Health (33, 34). To reduce FQ resistance prevalence through targeted interventions, more insight into the role of the environment, day-old chicks and AMU in the spread of FQ resistance in the broiler industry is required. Our study revealed that E. coli isolates with non-wild type FQ phenotype were present on all sampled broiler farms at day 3 until the end of the production cycle. The proportion of FQ non-susceptible E. coli stayed relatively stable at around 30% from day of hatch till day 36. Notably, these non-susceptible strains persisted even on 29 farms where no FQ were used, suggesting the influence of other contributing factors. Our study showed that FQ non-susceptible E. coli are often multidrug-resistant strains, as also previously published (7, 35). This could explain the persistence of FQ non-susceptibility through co-selection by the use of other antimicrobials (36–39). This hypothesis is supported by our WGS results, showing a high number of isolates carrying resistance genes against aminoglycosides and beta-lactam antibiotics. These antimicrobial classes are frequently used in the broiler industry, including in the farms in our study. In particular, aminoglycoside usage was common, as 20 farms in our study started their production cycle with a treatment of lincomycin combined with spectinomycin. Additionally, one farm showed substantially higher antibiotic usage compared to the others. After careful review, this observation was retained, as there was no indication of an error and it accurately reflected the variability in antibiotic use across farms. All in all, co-selection could account for the significant, though weak, positive correlation we found between the total TI during the production cycle and the proportion of FQ non-susceptible E. coli at the end of the cycle, indicating that reducing only FQ use is insufficient to limit resistance selection pressure and a broader reduction in total AMU is probably warranted. In addition, two strains harboring a qnr gene carried an additional AMR gene on the corresponding contig, being blaCTX-M-55 and a tet(A) gene. Both genes are commonly plasmid-associated, suggesting possible co-selection of AMR genes (40, 41). However, as only short-read sequencing data is available, plasmid sequences are likely fragmented and additional co-localizations of qnrS with other AMR genes may have been missed. Further studies are needed to determine the effect of co-selection and antibiotic treatment strategies on the emergence of multidrug resistance in E. coli.
However, AMU accounted for only a small portion of the variability in the proportion of FQ non-susceptible E. coli, indicating that other factors also have an impact on the occurrence of FQ resistance. On the three farms that did not use any antimicrobials, FQ non-susceptible E. coli were found in (almost) all sampled broilers at day 3 and 36. The fact that even without any antimicrobial selection pressure, the FQ non-susceptible E. coli strains can persist in the broiler population, suggests that the fitness costs of the present FQ resistance mechanisms are low or even non-existent (42, 43). The study by Marcusson et al. (42) showed that an initial mutation (mostly gyrA) had a fitness cost, but later compensatory parC mutations could confer an increase in both resistance and fitness. This was later confirmed by the study of Machuca et al. (43), where an enhanced bacterial fitness was observed when there was a combination of mutations in the QRDR region or in the presence of only the qnrS1 gene. In our study, three of the four clusters of persistent isolates carried only a qnrS1 gene, and the other cluster carried a combination of QRDR mutations, including mutations in gyrA and parC. Also, the FQ non-susceptible E. coli isolates carried on average 13 virulence-associated genes. VAGs are commonly used to differentiate pathogenic E. coli strains from commensal ones. However, making a clear distinction remains challenging due to the extensive genetic diversity within the E. coli species (44–46). Ovi et al. (46) identified 10 key VAGs in avian pathogenic E. coli isolates compared to non-pathogenic strains, serving three main functional roles, being secretion-related functions (cvaC, episomal ompT, hlyF), iron acquisition (iroN, iutA, iucD), and host attachment, invasion, and survival (tsh, papG, hlyF, iss). However, other studies reported different genes being common in APEC strains, like traT, sitA, csgA, lpfA, iucC (44, 45, 47, 48). Notably, many of these possible pathogenic related VAGs were also present in the FQ non-susceptible isolates from our study, being iss, traT, sitA, hlyF, csgA, iutA, iucC, lpfA, and cvaC.
However, commensal E. coli strains can also harbor VAGs which can offer a competitive advantage in the gut microbiota (44). These genes may not necessarily confer pathogenicity but can improve survival and competitiveness in various ecological niches (49). Pakbin et al. (48) reported the presence of several VAGs from human enteric E. coli pathotypes which play a role in survival influencing colonization (e.g., csgA), fitness (e.g., iutA, kpsMTII, fyuA), toxin production (e.g., astA, hlyE), and effector functions promoting adherence and biofilm formation (e.g., espB, espA, espF, espJ). The presence of these VAGs could enhance survival in both gut and environmental settings (44, 48). For example, sitA is linked to increased resistance to oxidative stress, and iron acquisition systems is considered being beneficial for environmental persistence (44). Moreover, commensal isolates may act as reservoirs for transferable VAGs (50), facilitating the spread of virulence traits within microbial communities. The FQ non-susceptible E. coli isolates in this study carried on average 13 VAGs, which does not necessarily indicate pathogenic potential. However, the presence of these genes could contribute to enhanced fitness, colonization, and survival in both host-associated and external environments (44, 45, 47, 48, 51), which could be an explanation for their survival within the broiler population. Further research is needed to investigate the fitness of these FQ non-susceptible strains and to link specific resistance mechanisms and virulence-associated genes to their potential fitness effects.
Furthermore, according to our cgMLST clustering, FQ non-susceptible E. coli isolates obtained from both day-old chicks and the environment present at day 0 can persist in chicks throughout the whole production cycle, indicating that these strains play a role in the persistence of FQ resistance in broilers. Regarding environmental dissemination, previous studies have already described the persistence of quinolone resistant E. coli in the broiler production environment and hypothesized that the occurrence in broilers may be influenced by on-farm recirculation (15, 18, 52, 53). Our findings support this hypothesis, as in 45% of the farms, FQ non-susceptible E. coli were found in the house environment before the animals even entered the stable and were able to persist until the end of the production cycle. In Belgium it is common practice to clean and disinfect the house between production cycles, although the used methods can vary in effectiveness (22, 54). The high prevalence of FQ non-susceptible E. coli in the environment highlights the critical need for performing the cleaning and disinfection more thoroughly to prevent cross-contamination between production rounds. However, contamination could also occur through the introduction of materials into the house after cleaning and disinfection, such as through feed (55), or due to inadequate biosecurity measures during preparation of the house (5). A multifactorial approach is therefore needed, improving biosecurity measures and management (56).
Yet, our study also clearly indicated that biosecurity measures on the farm level alone will not be sufficient to stop the introduction of FQ non-susceptible E. coli. In 79.3% of the farms, day-old chicks arrived with FQ non-susceptible E. coli in their gastrointestinal tracts, confirming previous reports of vertical transmission of these isolates throughout the broiler production pyramid (14–17). In our study, the isolates from day-old chicks within the farm clustered together for three out of four farms, from which the isolates were analyzed through WGS. This suggests that a batch of day-old chicks may acquire FQ non-susceptible E. coli from a common source or there is clonal spread within a batch of chicks, which could be facilitated through the high density in which the chicks are being held in hatcheries and during transport. A previous study showed the potential entry of Enterobacterales from the parent flock into the hatchery, where ESBL-producing E. coli were detected on 0.9% of egg surfaces and in the hatchery environment, however, no positive isolates were found in hatchlings (12). The presence of resistant isolates on eggshells of broiler hatching eggs was also detected by Mezhoud et al. (57). A different study focused specifically on quinolone-resistant E. coli and conducted longitudinal sampling across all levels of the production pyramid and highlighted the role of environmental recirculation (15). To better understand how and when these chicks are colonized with FQ non-susceptible E. coli isolates, further research is needed, particularly investigating the role of the hatchery and (grand)parent stock.
Regarding underlying FQ resistance mechanisms, our study found a substantial number of isolates carrying PMQR genes. Previous studies on FQ dissemination in the broiler production chain did not identify PMQR genes in broiler E. coli isolates (14–16), likely due to differences in screening methods. For example, these studies selected isolates based on nalidixic acid resistance (>16 mg/L), but E. coli isolates carrying PMQR genes often exhibit only a minor reduction in susceptibility to nalidixic acid (58). Our findings corroborate this, as the isolates carrying PMQR genes exhibited lower MIC values to FQ compared to those with QRDR mutations. Although Myrenås et al. (17) specifically selected FQ isolates with MIC profiles indicative of the presence of PMQR genes (ciprofloxacin MIC ≥ 0.06 mg/L combined with nalidixic acid MICs 4–32 mg/L), they could identify only one isolate carrying a qnrS gene. De Koster et al. (59) found PMQR genes (qnrS1) in 8.9% of the ciprofloxacin resistant E. coli broiler isolates, using selective plates supplemented with ciprofloxacin (2 mg/L) according to the clinical breakpoint for the detection of FQ isolates. These isolates carried PMQR genes in combination with QRDR mutations. In the study of Temmerman et al. (35), which only included avian pathogenic E. coli isolates collected from colibacillosis outbreaks, PMQR associated genes were found in 18% of non-wild type FQ resistant isolates, of which qnrS was detected in 14% and qnrB in 4% of the isolates. These different study outcomes suggest that the screening method might play a role in the detection of PMQR genes. Due to the specific selection of isolates in our study, our prevalence rates are not directly comparable to those of other studies.
Additionally, other Enterobacterales may contribute to the circulation of AMR in the environment and animals. After a pre-selection for E. coli and using WGS, we identified nine FQ non-susceptible E. fergusonii isolates, which can be explained by the fact that the MALDI-TOF database only contained one E. fergusonii entry and the related software confirmed it as non-distinguishable from E. coli due to the close relatedness. Previous studies have confirmed the potential of E. fergusonii as a reservoir for AMR (60–63). Ferreira et al. (64) described E. fergusonii isolates as being multidrug-resistant and also carrying the qnrB19 gene. In our study, only QRDR mutations were found in E. fergusonii isolates, but further research is needed to determine the role of E. fergusonii and other Enterobacterales in the dissemination of FQ non-susceptibility.
Regarding the screening of FQ residues, the FQ usage on one farm was confirmed through residue detection in collected feather samples well above the LOQ (50–600 ng/g feather). On another farm, very low (1.2–3.7 ng/g feather) flumequine residue concentrations were detected, despite no registered usage. A recent study showed that broilers treated with flumequine in the first 3 days of life had residue levels in feathers ranging from 3.4 to 15 ng/g feather on day 38 of the production cycle (23). The low residue levels in the current study could suggest unreported use at the beginning of the production cycle or uptake of residues from the surroundings which could originate from usage in a previous cycle, as FQ have been described to be notoriously stable in the environment (65, 66).
5 Conclusion
The continued presence of FQ non-susceptible E. coli on broiler farms is likely due to both historical contamination at the farm level and a continuous influx along the production chain. AMU contributes to the continued presence of FQ non-susceptible E. coli in broiler farms, but accounted for only a small proportion of the variability in FQ non-susceptibility in the currently investigated farms. The role of certain virulence-associated genes in the persistence of FQ non-susceptible E. coli in broiler farms deserves more in-depth research. To effectively combat this issue, efforts must focus on preventing the transmission of resistant bacteria, both at the farm level and throughout the broiler production chain, together with further reductions in AMU.
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) under BioProject number PRJNA1192038. The accession numbers can be found in Supplementary Table S8.
Ethics statement
The animal studies were approved by Ethical committee of the Faculties of Veterinary Medicine and Bioscience engineering of Ghent University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MR: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. FB: Conceptualization, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing. BB: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – review & editing. MG: Investigation, Writing – review & editing. KV: Formal analysis, Methodology, Writing – review & editing. SK: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. IC: Formal analysis, Writing – review & editing. NI: Investigation, Writing – review & editing. MD: Funding acquisition, Supervision, Writing – review & editing. JD: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Belgian Federal Public Service Health, Food Chain Safety and Environment (grant no. RF 19/6334).
Acknowledgments
We want to thank the veterinarians and farmers for their help and participation in this study. Also, we are grateful to the microbiology technicians in the participating laboratories for their contribution (to the collection of the microbiological data) and to the technicians of the service Transversal activities in Applied Genomics at Sciensano, Belgium to conduct the NGS. The MALDI-TOF mass spectrometer was financed by the Research Foundation Flanders (FWO-Vlaanderen, Brussels, Belgium) as Hercules project G0H2516N (AUGE/15/05). Fluoroquinolones in feather samples were determined using an UHPLC–MS/MS instrument as part of the Ghent University MSsmall Expertise Center for mass spectrometry analysis of small organic molecules.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1610997/full#supplementary-material
References
1. Holmes, AH, Moore, LSP, Sundsfjord, A, Steinbakk, M, Regmi, S, Karkey, A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. (2016) 387:176–87. doi: 10.1016/S0140-6736(15)00473-0
2. Roth, N, Käsbohrer, A, Mayrhofer, S, Zitz, U, Hofacre, C, and Domig, KJ. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: a global overview. Poult Sci. (2019) 98:1791–804. doi: 10.3382/ps/pey539
3. Callens, B, Cargnel, M, Sarrazin, S, Dewulf, J, Hoet, B, Vermeersch, K, et al. Associations between a decreased veterinary antimicrobial use and resistance in commensal Escherichia coli from Belgian livestock species (2011–2015). Prev Vet Med. (2018) 157:50–8. doi: 10.1016/j.prevetmed.2017.10.013
4. Huber, L, Agunos, A, Gow, SP, Carson, CC, and Van Boeckel, TP. Reduction in antimicrobial use and resistance to Salmonella, Campylobacter, and Escherichia coli in broiler chickens, Canada, 2013–2019. Emerg Infect Dis. (2021) 27:2434. doi: 10.3201/eid2709.204395
5. Taylor, NM, Wales, AD, Ridley, AM, and Davies, RH. Farm level risk factors for fluoroquinolone resistance in E. coli and thermophilic Campylobacter spp. on poultry farms. Avian Pathol. (2016) 45:559–68. doi: 10.1080/03079457.2016.1185510
6. Perrin-Guyomard, A, Jouy, E, Urban, D, Chauvin, C, Granier, SA, Mourand, G, et al. Decrease in fluoroquinolone use in French poultry and pig production and changes in resistance among E. Coli and Campylobacter. Vet Microbiol. (2020) 243:108637. doi: 10.1016/j.vetmic.2020.108637
7. De Koster, S, Ringenier, M, Lammens, C, Stegeman, A, Tobias, T, Velkers, F, et al. ESBL-producing, carbapenem-and ciprofloxacin-resistant Escherichia coli in Belgian and Dutch broiler and pig farms: a cross-sectional and cross-border study. Antibiotics. (2021) 10:945. doi: 10.3390/antibiotics10080945
8. Bortolaia, V, Bisgaard, M, and Bojesen, AM. Distribution and possible transmission of ampicillin-and nalidixic acid-resistant Escherichia coli within the broiler industry. Vet Microbiol. (2010) 142:379–86. doi: 10.1016/j.vetmic.2009.10.024
9. Huijbers, PMC, Graat, EAM, van Hoek, AHAM, Veenman, C, De Jong, MCM, and Van Duijkeren, E. Transmission dynamics of extended-spectrum β-lactamase and AmpC β-lactamase-producing Escherichia coli in a broiler flock without antibiotic use. Prev Vet Med. (2016) 131:12–9. doi: 10.1016/j.prevetmed.2016.07.001
10. Dierikx, C, Van der Goot, J, Fabri, T, van Essen-Zandbergen, A, Smith, H, and Mevius, D. Extended-spectrum-β-lactamase-and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother. (2013) 68:60–7. doi: 10.1093/jac/dks349
11. Gazal, LES, Medeiros, LP, Dibo, M, Nishio, EK, Koga, VL, Gonçalves, BC, et al. Detection of ESBL/AmpC-producing and fosfomycin-resistant Escherichia coli from different sources in poultry production in southern Brazil. Front Microbiol. (2021) 11:604544. doi: 10.3389/fmicb.2020.604544
12. Projahn, M, Daehre, K, Roesler, U, and Friese, A. Extended-spectrum-beta-lactamase-and plasmid-encoded cephamycinase-producing enterobacteria in the broiler hatchery as a potential mode of pseudo-vertical transmission. Appl Environ Microbiol. (2017) 83:e02364-16. doi: 10.1128/AEM.02364-16
13. Daehre, K, Projahn, M, Semmler, T, Roesler, U, and Friese, A. Extended-spectrum beta-lactamase-/AmpC beta-lactamase-producing Enterobacteriaceae in broiler farms: transmission dynamics at farm level. Microb Drug Resist. (2018) 24:511–8. doi: 10.1089/mdr.2017.0150
14. Kaspersen, H, Fiskebeck, EZ, Sekse, C, Slettemeås, JS, Urdahl, AM, Norström, M, et al. Comparative genome analyses of wild type-and quinolone resistant Escherichia coli indicate dissemination of QREC in the Norwegian broiler breeding pyramid. Front Microbiol. (2020) 11:938. doi: 10.3389/fmicb.2020.00938
15. Börjesson, S, Guillard, T, Landén, A, Bengtsson, B, and Nilsson, O. Introduction of quinolone resistant Escherichia coli to Swedish broiler population by imported breeding animals. Vet Microbiol. (2016) 194:74–8. doi: 10.1016/j.vetmic.2015.11.004
16. Petersen, A, Christensen, JP, Kuhnert, P, Bisgaard, M, and Olsen, JE. Vertical transmission of a fluoroquinolone-resistant Escherichia coli within an integrated broiler operation. Vet Microbiol. (2006) 116:120–8. doi: 10.1016/j.vetmic.2006.03.015
17. Myrenås, M, Slettemeås, JS, Thorsteinsdottir, TR, Bengtsson, B, Börjesson, S, Nilsson, O, et al. Clonal spread of Escherichia coli resistant to cephalosporins and quinolones in the Nordic broiler production. Vet Microbiol. (2018) 213:123–8. doi: 10.1016/j.vetmic.2017.11.015
18. Leclercq, SO, Bochereau, P, Foubert, I, Baumard, Y, Travel, A, Doublet, B, et al. Persistence of commensal multidrug-resistant Escherichia coli in the broiler production pyramid is best explained by strain recirculation from the rearing environment. Front Microbiol. (2024) 15:1406854. doi: 10.3389/fmicb.2024.1406854
19. Poirel, L, Cattoir, V, and Nordmann, P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front Microbiol. (2012) 3:24. doi: 10.3389/fmicb.2012.00024
20. Persoons, D, Dewulf, J, Smet, A, Herman, L, Heyndricx, M, Martel, A, et al. Antimicrobial use in Belgian broiler production. Prev Vet Med. (2012) 105:320–5. doi: 10.1016/j.prevetmed.2012.02.020
21. Joosten, P, Sarrazin, S, Van Gompel, L, Luiken, REC, Mevius, DJ, Wagenaar, JA, et al. Quantitative and qualitative analysis of antimicrobial usage at farm and flock level on 181 broiler farms in nine European countries. J Antimicrob Chemother. (2019) 74:798–806. doi: 10.1093/jac/dky498
22. Luyckx, K, Van Weyenberg, S, Dewulf, J, Herman, L, Zoons, J, Vervaet, E, et al. On-farm comparisons of different cleaning protocols in broiler houses. Poult Sci. (2015) 94:1986–93. doi: 10.3382/ps/pev143
23. Ringenier, M, Cherlet, M, Dewulf, J, and Devreese, M. Residue depletion of enrofloxacin and flumequine in feathers of broilers based on quantitative UHPLC-MS/MS detection. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. (2024) 41:1065–76. doi: 10.1080/19440049.2024.2370367
24. Vereecke, N, Vandekerckhove, A, Theuns, S, Haesebrouck, F, and Boyen, F. Whole genome sequencing to study antimicrobial resistance and RTX virulence genes in equine Actinobacillus isolates. Vet Res. (2023) 54:33. doi: 10.1186/s13567-023-01160-2
25. Dame-Korevaar, A, Fischer, EA, Van der Goot, J, Velkers, F, Van den Broek, J, Veldman, K, et al. Effect of challenge dose of plasmid-mediated extended-spectrum β-lactamase and AmpC β-lactamase producing Escherichia coli on time-until-colonization and level of excretion in young broilers. Vet Microbiol. (2019) 239:108446. doi: 10.1016/j.vetmic.2019.108446
26. Ibrahim, N, Boyen, F, Mohsin, MAS, Ringenier, M, Berge, AC, Chantziaras, I, et al. Antimicrobial resistance in Escherichia coli and its correlation with antimicrobial use on commercial poultry farms in Bangladesh. Antibiotics. (2023) 12:1361. doi: 10.3390/antibiotics12091361
27. EUCAST. Antimicrobial wild type distributions of microorganisms. Available online at: https://mic.eucast.org.
28. Bogaerts, B, Nouws, S, Verhaegen, B, Denayer, S, Van Braekel, J, Winand, R, et al. Validation strategy of a bioinformatics whole genome sequencing workflow for Shiga toxin-producing Escherichia coli using a reference collection extensively characterized with conventional methods. Microb Genom. (2021) 7:000531. doi: 10.1099/mgen.0.000531
29. Zhou, Z, Alikhan, NF, Mohamed, K, Fan, Y, and Achtman, M. The entero base user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. (2020) 30:138–52. doi: 10.1101/gr.251678.119
30. Dallman, T, Ashton, P, Schafer, U, Jironkin, A, Painset, A, Shaaban, S, et al. A database solution for routine sequencing analysis of bacterial isolates. Bioinformatics. (2018) 34:3028–9. doi: 10.1093/bioinformatics/bty212
31. Dallman, TJ, Greig, DR, Gharbia, SE, and Jenkins, C. Phylogenetic structure of Shiga toxin-producing Escherichia coli O157: H7 from sub-lineage to SNPs. Microb Genom. (2021) 7:000544. doi: 10.1099/mgen.0.000544
32. Wood, DE, Lu, J, and Langmead, B. Improved metagenomic analysis with kraken 2. Genome Biol. (2019) 20:257. doi: 10.1186/s13059-019-1891-0
33. World Health Organization (WHO). Critically important antimicrobials for human medicine. (2017). Available online at: https://iris.who.int/bitstream/handle/10665/255027/9789241512220-eng.pdf
34. World Organisation for Animal Health (WOAH). WOAH list of antimicrobial agents of veterinary importance (2024). Available online at: http://www.woah.org/app/uploads/2021/06/amended-91gs-tech-03-amr-working-group-report-en.pdf
35. Temmerman, R, Garmyn, A, Antonissen, G, Vanantwerpen, G, Vanrobaeys, M, Haesebrouck, F, et al. Evaluation of fluoroquinolone resistance in clinical avian pathogenic Escherichia coli isolates from Flanders (Belgium). Antibiotics. (2020) 9:800. doi: 10.3390/antibiotics9110800
36. Ingram, PR, Rogers, BA, Sidjabat, HE, Gibson, JS, and Inglis, TJJ. Co-selection may explain high rates of ciprofloxacin non-susceptible Escherichia coli from retail poultry reared without prior fluoroquinolone exposure. J Med Microbiol. (2013) 62:1743–6. doi: 10.1099/jmm.0.062729-0
37. Jiang, HX, Song, L, Liu, J, Zhang, XH, Ren, YN, Zhang, WH, et al. Multiple transmissible genes encoding fluoroquinolone and third-generation cephalosporin resistance co-located in non-typhoidal Salmonella isolated from food-producing animals in China. Int J Antimicrob Agents. (2014) 43:242–7. doi: 10.1016/j.ijantimicag.2013.12.005
38. Apostolakos, I, Feudi, C, Eichhorn, I, Palmieri, N, Fasolato, L, Schwarz, S, et al. High-resolution characterisation of ESBL/pAmpC-producing Escherichia coli isolated from the broiler production pyramid. Sci Rep. (2020) 10:11123. doi: 10.1038/s41598-020-68036-9
39. Taylor, E, Ossa-Trujillo, C, Vinasco, J, Jordan, ER, García Buitrago, JA, Hagevoort, R, et al. Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extended-spectrum beta-lactam use in dairy cattle. Lett Appl Microbiol. (2021) 72:220–4. doi: 10.1111/lam.13419
40. Hellweger, FL. Escherichia coli adapts to tetracycline resistance plasmid (pBR322) by mutating endogenous potassium transport: in silico hypothesis testing. FEMS Microbiol Ecol. (2013) 83:622–31. doi: 10.1111/1574-6941.12019
41. Nakayama, T, Yamamoto, S, Ohata, N, Yamaguchi, T, Jinnai, M, Minh, DT, et al. Common presence of plasmid encoding blaCTX-M-55 in extended-spectrum β-lactamase-producing Salmonella enterica and Escherichia coli isolates from the same edible river fish. J Microorg Control. (2023) 28:49–56. doi: 10.4265/jmc.28.2_49
42. Marcusson, LL, Frimodt-Møller, N, and Hughes, D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. (2009) 5:e1000541. doi: 10.1371/journal.ppat.1000541
43. Machuca, J, Briales, A, Labrador, G, Díaz-de-Alba, P, López-Rojas, R, Docobo-Pérez, F, et al. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother. (2014) 69:3203–15. doi: 10.1093/jac/dku308
44. Kemmett, K, Humphrey, T, Rushton, S, Close, A, Wigley, P, and Williams, NJ. A longitudinal study simultaneously exploring the carriage of APEC virulence associated genes and the molecular epidemiology of faecal and systemic E. coli in commercial broiler chickens. PLoS One. (2013) 8:e67749. doi: 10.1371/journal.pone.0067749
45. Rehman, MA, Rempel, H, Carrillo, CD, Ziebell, K, Allen, K, Manges, AR, et al. Virulence genotype and phenotype of multiple antimicrobial-resistant Escherichia coli isolates from broilers assessed from a “one-health” perspective. J Food Prot. (2022) 85:336–54. doi: 10.4315/JFP-21-273
46. Ovi, F, Zhang, L, Nabors, H, Jia, L, and Adhikari, P. A compilation of virulence-associated genes that are frequently reported in avian pathogenic Escherichia coli (APEC) compared to other E. coli. J Appl Microbiol. (2023) 134:lxad 014. doi: 10.1093/jambio/lxad014
47. Projahn, M, Daehre, K, Semmler, T, Guenther, S, Roesler, U, and Friese, A. Environmental adaptation and vertical dissemination of ESBL−/pA mp C-producing Escherichia coli in an integrated broiler production chain in the absence of an antibiotic treatment. Microb Biotechnol. (2018) 11:1017–26. doi: 10.1111/1751-7915.13040
48. Pakbin, B, Brück, WM, and Rossen, JWA. Virulence factors of enteric pathogenic Escherichia coli: a review. Int J Mol Sci. (2021) 22:9922. doi: 10.3390/ijms22189922
49. Van Elsas, JD, Semenov, AV, Costa, R, and Trevors, JT. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J. (2011) 5:173–83. doi: 10.1038/ismej.2010.80
50. de Oliveira, AL, Rocha, DA, Finkler, F, de Moraes, LB, Barbieri, NL, Pavanelo, DB, et al. Prevalence of col V plasmid-linked genes and in vivo pathogenicity of avian strains of Escherichia coli. Foodborne Pathog Dis. (2015) 12:679–85. doi: 10.1089/fpd.2014.1934
51. Vila, J, Sáez-López, E, Johnson, JR, Römling, U, Dobrindt, U, Cantón, R, et al. Escherichia coli: an old friend with new tidings. FEMS Microbiol Rev. (2016) 40:437–63. doi: 10.1093/femsre/fuw005
52. Kaspersen, H, Sekse, C, Fiskebeck, EZ, Slettemeås, JS, Simm, R, Norström, M, et al. Dissemination of quinolone-resistant Escherichia coli in the Norwegian broiler and pig production chains and possible persistence in the broiler production environment. Appl Environ Microbiol. (2020) 86:e02769-02719. doi: 10.1128/AEM.02769-19
53. Luyckx, K, Van Coillie, E, Dewulf, J, Van Weyenberg, S, Herman, L, Zoons, J, et al. Identification and biocide susceptibility of dominant bacteria after cleaning and disinfection of broiler houses. Poult Sci. (2017) 96:938–49. doi: 10.3382/ps/pew355
54. Maertens, H, De Reu, K, Van Weyenberg, S, Van Coillie, E, Meyer, E, Van Meirhaeghe, H, et al. Evaluation of the hygienogram scores and related data obtained after cleaning and disinfection of poultry houses in Flanders during the period 2007 to 2014. Poult Sci. (2018) 97:620–7. doi: 10.3382/ps/pex327
55. Da Costa, PM, Oliveira, M, Bica, A, Vaz-Pires, P, and Bernardo, F. Antimicrobial resistance in Enterococcus spp. and Escherichia coli isolated from poultry feed and feed ingredients. Vet Microbiol. (2007) 120:122–31. doi: 10.1016/j.vetmic.2006.10.005
56. Robé, C, Projahn, M, Boll, K, Blasse, A, Merle, R, Roesler, U, et al. Survival of highly related ESBL-and pAmpC-producing Escherichia coli in broiler farms identified before and after cleaning and disinfection using cg MLST. BMC Microbiol. (2024) 24:143. doi: 10.1186/s12866-024-03292-7
57. Mezhoud, H, Chantziaras, I, Iguer-Ouada, M, Moula, N, Garmyn, A, Martel, A, et al. Presence of antimicrobial resistance in coliform bacteria from hatching broiler eggs with emphasis on ESBL/AmpC-producing bacteria. Avian Pathol. (2016) 45:493–500. doi: 10.1080/03079457.2016.1167837
58. Veldman, K, Cavaco, LM, Mevius, D, Battisti, A, Franco, A, Botteldoorn, N, et al. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother. (2011) 66:1278–86. doi: 10.1093/jac/dkr084
59. De Koster, S, Ringenier, M, Xavier, BB, Lammens, C, De Coninck, D, De Bruyne, K, et al. Genetic characterization of ESBL-producing and ciprofloxacin-resistant Escherichia coli from Belgian broilers and pigs. Front Microbiol. (2023) 14:1150470. doi: 10.3389/fmicb.2023.1150470
60. Srinivas, K, Ghatak, S, Pyngrope, DA, Angappan, M, Milton, AAP, Das, S, et al. Avian strains of emerging pathogen Escherichia fergusonii are phylogenetically diverse and harbor the greatest AMR dissemination potential among different sources: comparative genomic evidence. Front Microbiol. (2023) 13:1080677. doi: 10.3389/fmicb.2022.1080677
61. Galetti, R, Penha Filho, RAC, Ferreira, JC, Am, V, and Darini, ALC. Antibiotic resistance and heavy metal tolerance plasmids: the antimicrobial bulletproof properties of Escherichia fergusonii isolated from poultry. Infect Drug Resist. (2019) 12:1029–33. doi: 10.2147/IDR.S196411
62. Tang, B, Chang, J, Chen, Y, Lin, J, Xiao, X, Xia, X, et al. Escherichia fergusonii, an underrated repository for antimicrobial resistance in food animals. Microbiol Spectr. (2022) 10:e01617-21. doi: 10.1128/spectrum.01617-21
63. Forgetta, V, Rempel, H, Malouin, F, Vaillancourt, R Jr, Topp, E, and Dewar, K. Pathogenic and multidrug-resistant Escherichia fergusonii from broiler chicken. Poult Sci. (2012) 91:512–25. doi: 10.3382/ps.2011-01738
64. Ferreira, JC, Penha Filho, RAC, Kuaye, APY, Andrade, LN, Berchieri Junior, A, and Da Costa Darini, AL. Identification and characterization of plasmid-mediated quinolone resistance determinants in Enterobacteriaceae isolated from healthy poultry in Brazil. Infect Genet Evol. (2018) 60:66–70. doi: 10.1016/j.meegid.2018.02.003
65. Bhatt, S, and Chatterjee, S. Fluoroquinolone antibiotics: occurrence, mode of action, resistance, environmental detection, and remediation–a comprehensive review. Environ Pollut. (2022) 315:120440. doi: 10.1016/j.envpol.2022.120440
Keywords: fluoroquinolone, Escherichia coli , poultry, antimicrobial resistance, whole genome sequencing, antimicrobial usage
Citation: Ringenier M, Boyen F, Bogaerts B, Gand M, Vanneste K, De Keersmaecker SCJ, Chantziaras I, Ibrahim N, Devreese M and Dewulf J (2025) Mapping the spread of fluoroquinolone resistance: continued presence of non-susceptible Escherichia coli in broilers. Front. Vet. Sci. 12:1610997. doi: 10.3389/fvets.2025.1610997
Edited by:
Frederique Pasquali, University of Bologna, ItalyReviewed by:
Rogers Ruyu Azabo, Sokoine University of Agriculture, TanzaniaAlois Cizek, University of Veterinary and Pharmaceutical Sciences Brno, Czechia
Copyright © 2025 Ringenier, Boyen, Bogaerts, Gand, Vanneste, De Keersmaecker, Chantziaras, Ibrahim, Devreese and Dewulf. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moniek Ringenier, TW9uaWVrLnJpbmdlbmllckB1Z2VudC5iZQ==
 Moniek Ringenier
Moniek Ringenier Filip Boyen
Filip Boyen Bert Bogaerts
Bert Bogaerts Mathieu Gand
Mathieu Gand Kevin Vanneste
Kevin Vanneste Sigrid C. J. De Keersmaecker
Sigrid C. J. De Keersmaecker Ilias Chantziaras
Ilias Chantziaras Nelima Ibrahim
Nelima Ibrahim Mathias Devreese
Mathias Devreese Jeroen Dewulf
Jeroen Dewulf