- Anhui Province Key Laboratory of Embryo Development and Reproductive Regulation, College of Biological and Food Engineering, Fuyang Normal University, Fuyang, China
Introduction: Enterocytozoon bieneusi (E. bieneusi) and Giardia duodenalis (G. duodenalis) are common intestinal pathogens in humans and farmed animals. There is limited data available on the positivity rates and genetic identity of E. bieneusi and G. duodenalis in dairy cattle from Anhui, China.
Methods: To understand the transmission of E. bieneusi and G. duodenalis in these animals, a total of 1,043 fecal samples were collected from cattle on five farms (Fuyang, Huainan, Huaibei, Bengbu, and Jieshou) in Anhui province of China between May 2023 and August 2024. The G. duodenalis in fecal samples was detected by nested PCR targeting a 511-bp fragment of the β-giardin (bg) gene, a 599-bp fragment of the glutamate dehydrogenase (gdh) gene, and a 530-bp fragment of the triosephosphate isomerase (tpi) gene. The E. bieneusi was detected by nested PCR targeting a 392-bp fragment of the internal transcribed spacer (ITS) of the rRNA gene.
Results: The PCR analysis revealed positivity rates of 29.7% (310/1043) for E. bieneusi and 2.8% (29/1043) for G. duodenalis. The cattle from the Bengbu farm had significantly higher positivity rates of E. bieneusi than Fuyang, Huainan, and Huaibei farms (χ2 = 61.6, df = 1, p < 0.0001; χ2 = 76.4, df = 1, p < 0.0001; χ2 = 20.6, df = 1, p < 0.0001). A total of 11 known genotypes of E. bieneusi have been identified: J (n = 154), BEB4 (n = 76), I (n = 63), CGC1 (n = 8), N (n = 2), BEB8 (n = 2), ALP1 (n = 1), BLC13 (n = 1), CHC13 (n = 1), CHN6 (n = 1), and D (n = 1). Additionally, two genotypes of G. duodenalis have been identified, including assemblage A (n = 6) and assemblage E (n = 23).
Discussion: The results indicate that known zoonotic E. bieneusi and G. duodenalis are prevalent in dairy cattle, thereby enhancing our understanding of the genetic diversity and transmission of these pathogens in these animals.
1 Introduction
Enterocytozoon bieneusi (E. bieneusi) and Giardia duodenalis (G. duodenalis) are common intestinal pathogens in humans and other farmed animals, especially cattle (1, 2). The life cycle of E. bieneusi and G. duodenalis consists of two main stages. Spores of the former and cysts of the latter are ingested by susceptible hosts, invade the epithelial cells that line the intestinal lumen, and replicate intracellularly (3, 4). Diarrhea, malnutrition, and weight loss are the primary symptoms of giardiasis and microsporidiosis (5, 6). These two zoonotic pathogens cause outbreaks of diarrhea diseases in both humans and animals worldwide (7, 8).
At least 500 genotypes of E. bieneusi and 11 major genetic groups have been identified by conducting phylogenetic analysis of nucleotide sequences of the internal transcribed spacer (ITS), with the genotypes of Group 1 being the major zoonotic pathogens (1, 4, 5). A large number of studies indicate that cattle serve as a prevalent reservoir host for E. bieneusi (4, 5). Genotypes BEB4, BEB6, I, and J in Group 2 are frequently reported in cattle (5, 8–12). The genotypes BEB6 and BEB4 are more common in cattle. Genotypes I and J have been found in a wide range of ruminants (5). While they are predominantly reported in ruminants, genotypes BEB4, BEB6, I, and J have been shown to have the capacity to infect humans (13–15). Therefore, E. bieneusi genotypes in cattle possess zoonotic potential.
To date, a total of eight assemblages (A-H) of G. duodenalis have been identified through sequence analysis of β-giardin (bg), triosephosphate isomerase (tpi), and glutamate dehydrogenase (gdh) genes (7, 16). Of the eight assemblages of G. duodenalis, assemblages A and B have the widest host ranges, including humans and cattle (17). However, assemblages C-H of G. duodenalis are host-specific (18). The most common genotype of G. duodenalis in cattle is the assemblage E, whereas assemblages A and B occur sporadically (7). Assemblage A of G. duodenalis is responsible for the majority of cases of giardiasis in humans (7). Recent studies indicate that more than 50 cases of human giardiasis attributed to assemblage E have been reported in Egypt, New Zealand, Brazil, Vietnam, and Australia (6, 19–22). Therefore, there is a zoonotic potential for assemblages of G. duodenalis in bovine animals.
Bovine animals are widely farmed in Anhui, China, which is a large agricultural province. To date, only a limited number of studies have been carried out to determine the identity of G. duodenalis and E. bieneusi in cattle. To understand the transmission of E. bieneusi and G. duodenalis in these animals, we examined the prevalence and genetic identity of G. duodenalis and E. bieneusi in dairy cattle in Anhui, China. The data show a prevalent occurrence of zoonotic genotypes in these animals. The aim of the present study was to investigate the distribution and genetic identity of two pathogens in cattle in Anhui, China. The data obtained suggest that the zoonotic genotypes of both pathogens are commonly found in these animals.
2 Materials and methods
2.1 Specimens
A total of 1,043 fecal samples were collected from cattle on five farms (Fuyang, Huainan, Huaibei, Bengbu, and Jieshou) in Anhui province of China between May 2023 and August 2024. Farms in Bengbu, Huaibei, Huainan, and Fuyang were established in 2011, 2018, 2021, and 2023, respectively; however, the year of establishment for Jieshou Farm was unclear. Depending on the number of animals on the farm, 234, 167, 238, 309, and 95 were randomly collected from Fuyang, Huainan, Huaibei, Bengbu, and Jieshou, respectively. These cattle did not originate from the tested areas and were imported from the Netherlands. All fecal samples were obtained from intensive dairy farming systems, and all cattle exhibited no obvious clinical signs during the study period. All cattle were housed separately according to their age, with calves under 2 months housed separately. The cattle were divided into four age groups, based on their age: < 2 months (n = 323), 2–6 months (n = 237), 6–12 months (n = 249), and >12 months (n = 234). All fecal samples were collected from the rectum of each animal, and the age of cattle, the time, and place of the sample collection was recorded. All samples were stored in 2.5% potassium dichromate at 4°C prior to use in the DNA extraction.
2.2 DNA extraction
Approximately 200 mg fecal samples from cattle were washed three times with distilled water by centrifugation. Genomic DNA was extracted using the E. Z. N. A.®Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA). The extracted genomic DNA was stored at −20°C before being used in PCR analysis.
2.3 Detection of G. duodenalis and E. bieneusi
Assemblages of G. duodenalis in fecal samples were detected by nested PCR targeting a 511-bp fragment of the β-giardin (bg) gene, a 599-bp fragment of the glutamate dehydrogenase (gdh) gene, and a 530-bp fragment of the triosephosphate isomerase (tpi) gene (23–25). In contrast to G. duodenalis, E. bieneusi was detected by nested PCR targeting a 392-bp fragment of the ITS of the rRNA gene (26). Each PCR assay included both positive and negative samples. Two replicates were used for PCR analysis of each sample at each locus.
2.4 Sequence analysis
All positive products of the second PCR were sequenced bi-directionally on an ABI 3730 Auto sequencer (Applied Biosystems, Foster City, CA, USA) to identify the presence of G. duodenalis and E. bieneusi. The nucleotide sequences were assembled using ChromasPro 2.1.5.0,1 edited using BioEdit 7.1.3.0,2 and aligned using ClustalX 2.0.11.3 The phylogenetic relationship of the E. bieneusi was analyzed using maximum likelihood analysis implemented in Mega 7.0.4
2.5 Statistical analysis
The chi-squared test, implemented in SPSS 20.0 (IBM Inc., Chicago, IL, USA), was performed to compare positivity rates of G. duodenalis and E. bieneusi between geographical areas and age groups. Comparisons of positivity rates were made using the Mann–Whitney U test, and the p-values were calculated. Differences with p ≤ 0.05 were considered significant.
3 Results
3.1 Distribution of genotypes of E. bieneusi and G. duodenalis in cattle
The PCR analysis revealed that the positivity rates of E. bieneusi from five farms was 29.7% (310/1043; Table 1). The positivity rates of E. bieneusi were 15.8% (37/234), 8.4% (14/167), 29.4% (70/238), 47.9% (148/309), and 43.2% (41/95) in Fuyang, Huainan, Huaibei, Bengbu, and Jieshou, respectively. Thus, cattle from Bengbu had significantly higher positivity rates of E. bieneusi than Fuyang, Huainan, and Huaibei cities (χ2 = 61.6, df = 1, p < 0.0001; χ2 = 76.4, df = 1, p < 0.0001; χ2 = 20.6, df = 1, p < 0.0001). Conversely, no significant difference was found in the positivity rates of E. bieneusi between Bengbu and Jieshou (χ2 = 0.66, df = 1, p = 0.4182; Table 1). By age, the positivity rates of E. bieneusi were 14.8% (48/323), 49.8% (118/237), 30.5% (76/249), and 29.1% (68/234) in < 2 months animals, 2–6 months animals, 6–12 months animals, and > 12 months animals, respectively. Furthermore, 2–6 months cattle had significantly higher positivity rate of E. bieneusi than < 2 months animals, 6–12 months animals, and > 12 months animals (χ2 = 80.2, df = 1, p < 0.0001; χ2 = 18.8, df = 1, p < 0.0001; χ2 = 21.2, df = 1, p < 0.0001; Table 2).
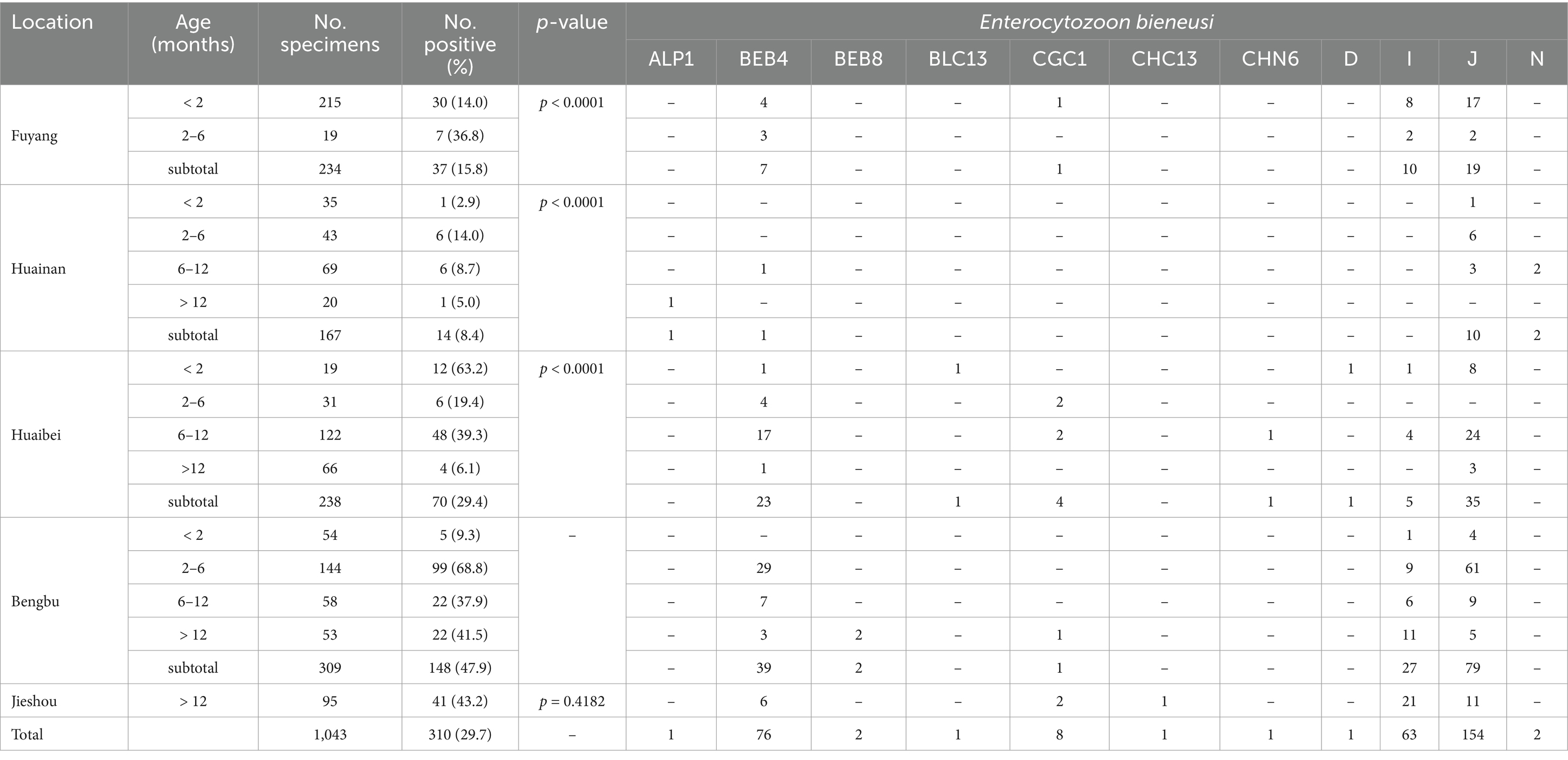
Table 1. Distribution of Enterocytozoon bieneusi genotypes in cattle on farms from Anhui province, China.
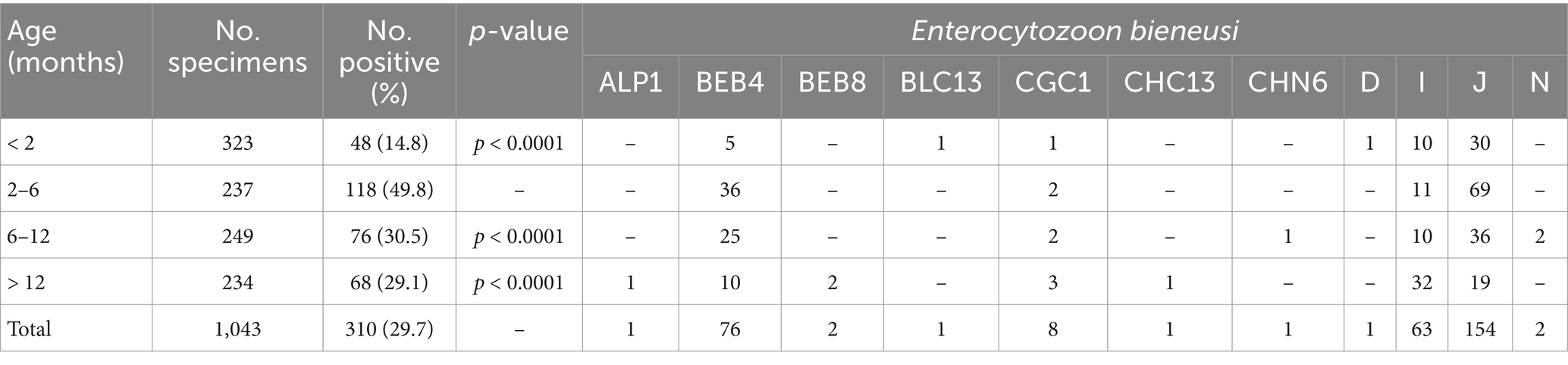
Table 2. Occurrence of Enterocytozoon bieneusi genotypes in cattle on farms from Anhui province, China, broken down by age.
The PCR analysis revealed a positivity rate of G. duodenalis from five farms was 2.8% (29/1043; Table 3). The positivity rates of G. duodenalis were 6.5% (14/234), 2.4% (4/167), 1.3% (3/238), 2.3% (7/309), and 1.1% (1/95) in Fuyang, Huainan, Huaibei, Bengbu, and Jieshou, respectively. Thus, cattle from Fuyang had significantly higher positivity rates for G. duodenalis than Huaibei and Bengbu cities (χ2 = 7.6, df = 1, p = 0.0059; χ2 = 4.9, df = 1, p = 0.0261). In contrast, no significant difference was found in the positivity rates of G. duodenalis in Huainan and Jieshou (Table 3).
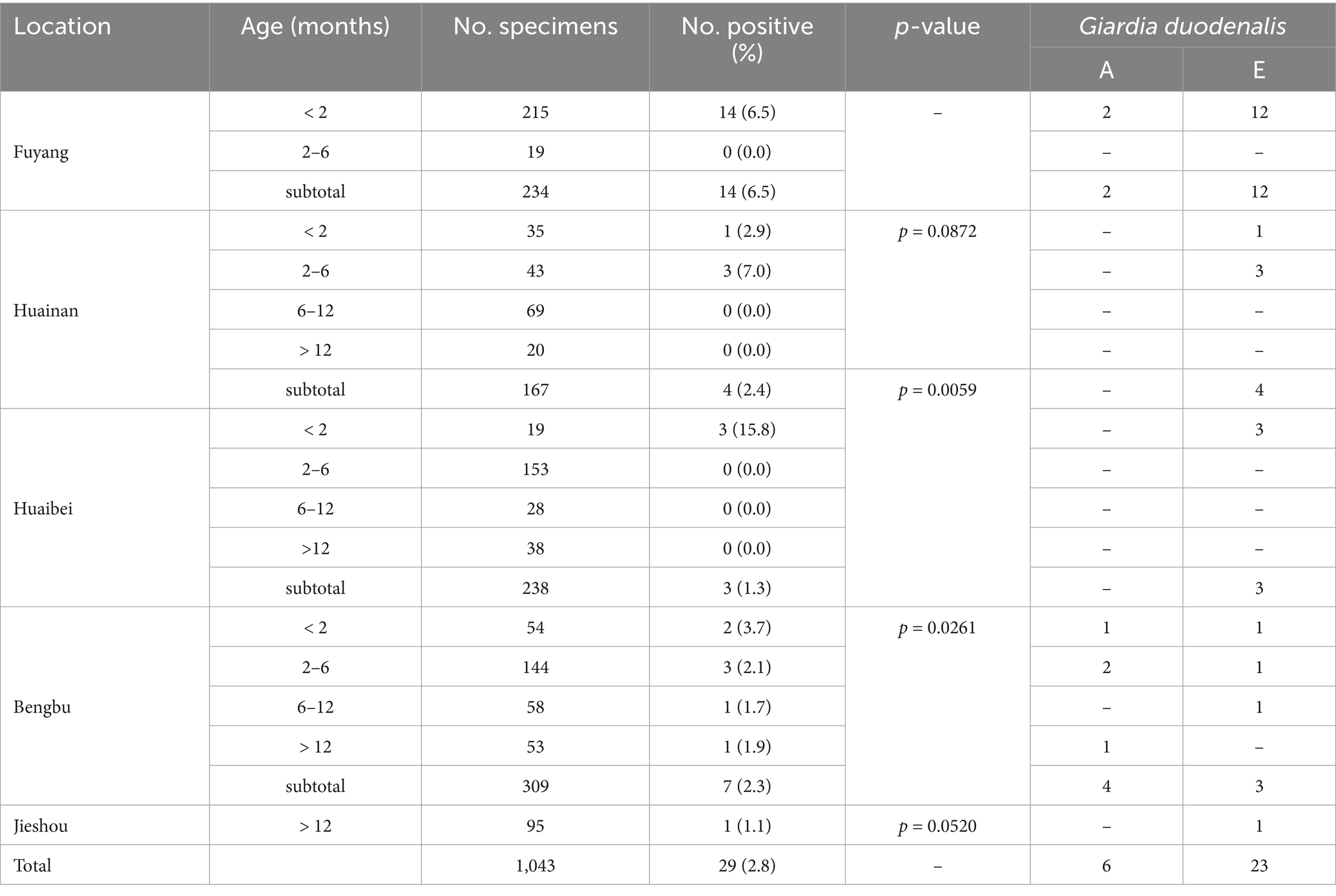
Table 3. Distribution of Giardia duodenalis genotypes in cattle on farms from Anhui province, China.
3.2 Distribution of E. bieneusi and G. duodenalis genotypes in cattle
The ITS secondary PCR products from all 310 E. bieneusi-positive samples were successfully sequenced. A total of 11 known genotypes were identified, including J (n = 154), BEB4 (n = 76), I (n = 63), CGC1 (n = 8), N (n = 2), BEB8 (n = 2), ALP1 (n = 1), BLC13 (n = 1), CHC13 (n = 1), CHN6 (n = 1), and D (n = 1). Among these genotypes, J (154/310) was dominant over E. bieneusi in animals. Furthermore, genotype N, BEB8, ALP1, BLC13, CHC13, CHN6, and D were only detected in a small number of animals. The ITS sequences from 154 J samples, 76 BEB4 samples, 63 I samples, 8 CGC1 samples, 2 N samples, 2 BEB8 samples, 1 ALP1 sample, 1 BLC13 sample, 1 CHC13 sample, 1 CHN6 sample, and 1 D sample were identical to the GenBank reference sequence KU55767, MH732750, MT231513, OK416087, MN178159, OK416086, KC860908, MN758760, OR491688, MN136733, and OK117963, respectively (Table 1).
The secondary PCR products from 29 G. duodenalis-positive samples have been successfully sequenced. A total of two known genotypes have been identified, including assemblage A (n = 6) and assemblage E (n = 23). Among them, assemblage E was detected in cattle in all five cities; however, assemblage A was only detected in cattle from Fuyang and Bengbu cities. In this study, assemblage E was the dominant genotype of G. duodenalis in animals. The obtained sequences from 23 assemblage E samples and 6 assemblage A samples were identical to the GenBank reference sequences GQ337965 and MN996372, respectively (Table 3).
3.3 Phylogenetic relationship of E. bieneusi genotypes
In the maximum likelihood analysis of the ITS sequences obtained, genotypes D and BLC13 were clustered with Group 1. The other genotypes (I, J, CGC1, ALP1, CHC13, BEB4, N, CHN6, and BEB8) were clustered together with Group 2 (Figure 1).
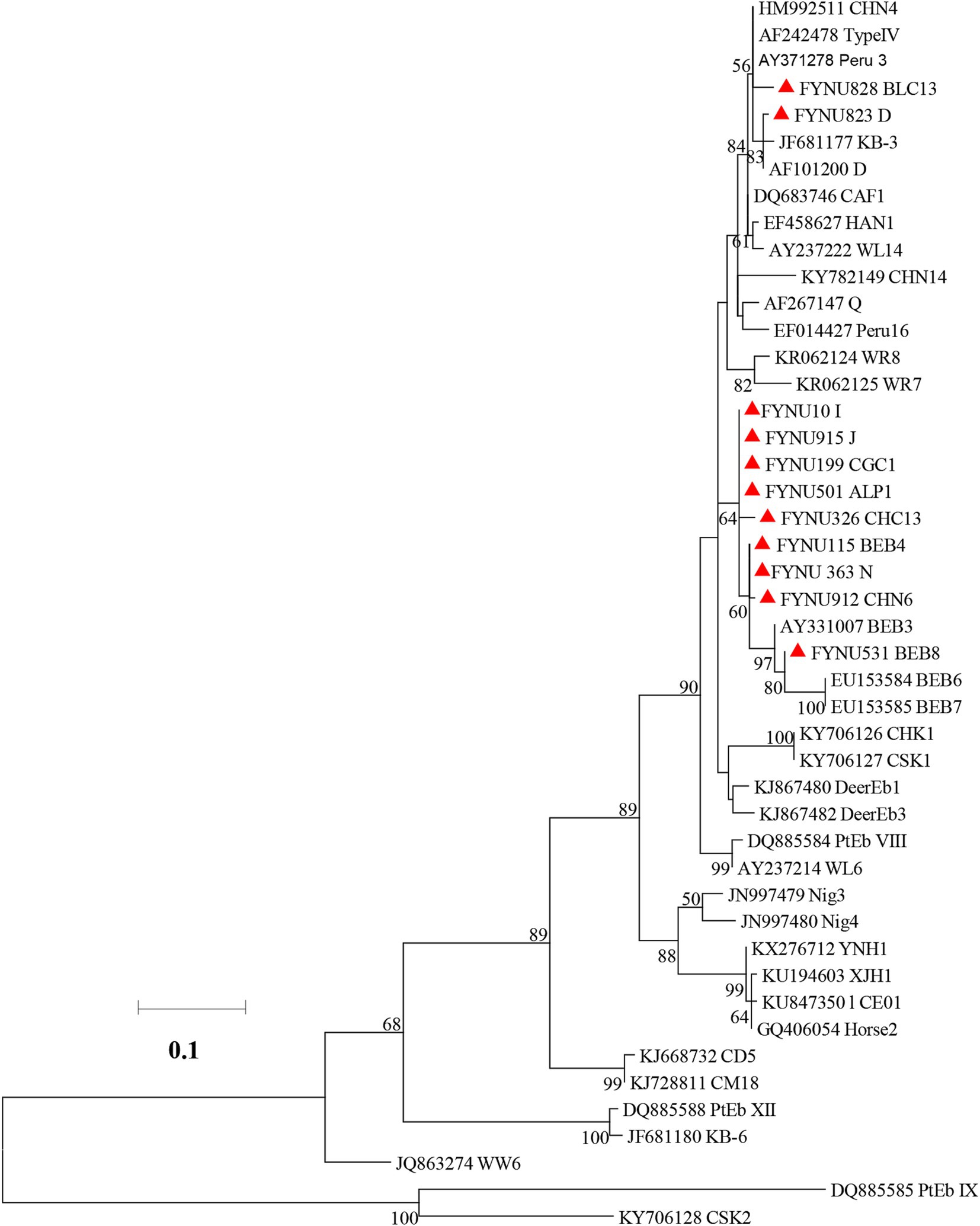
Figure 1. Phylogenetic relationships of Enterocytozoon bieneusi genotypes based on maximum likelihood analysis. The genotypes of E. bieneusi that have been identified in this study are indicated by red triangles. Bootstrap values below 50% are not shown. Bar = 0.02 substitutions per site.
4 Discussion
The results of the present study indicate that E. bieneusi and G. duodenalis are prevalent in cattle in Anhui. By sampling location, the E. bieneusi detection rate was found to be higher in Bengbu (47.9%, 148/309) compared to Fuyang (15.8%, 37/234), Huainan (8.4%, 14/167), and Huaibei (29.4%, 238). The long history of animal farming could have contributed to the high prevalence of E. bieneusi in animals in the present study. Farm patterns of long-term intensive farming lead to high animal densities and increased fecal and environmental contamination, which increase the spread of E. bieneusi. The E. bieneusi may be transmitted by other animals such as birds or rodents. By age, the detection rate (49.8%, 118/237) was significantly higher in cattle aged 2–6 months compared to other age groups, probably due to the group feeding practices of this age group of cattle. The positivity rate of G. duodenalis, when analyzed by sampling location and animal age, showed no significant difference between cities and animal age.
The overall detection rate of 29.7% for E. bieneusi is much higher than that reported in a previous study in Anhui (3.0%, 16/526), Xingjiang (16.5%, 85/514), Guangdong (15.7%, 61/388), Jiangxi (7.9%, 13/165), Yunan (0.6%, 3/490), Heilongjiang (6.5%, 75/1155), and Jiangsu (13.0%, 177/1366) (27–31). The higher detection rate of E. bieneusi in Anhui could also be attributed to the long history of animal farming and the presence of underage animals. The overall detection rate of 2.8% for G. duodenalis is much lower than that reported in a previous study from Xingjiang (24.0%, 180/749), Jiangxi (9.4%, 52/556), Yunnan (27.5%, 144/524), Taiwan (19.8%, 31/156), and Inner Mongolia (29.5%, 149/505) (32, 33–36). The lower detection rate of G. duodenalis in this study could also be attributed to the number of samples analyzed and the time of sampling.
The significance of E. bieneusi in cattle for public health is unclear. This study identified 11 distinct genotypes from 310 E. bieneusi-positive samples based on the ITS gene locus in cattle, which include BLC13 (n = 1) and D (n = 1), from group 1, and J (n = 154), as well as BEB4 (n = 76), I (n = 63), CGC1 (n = 8), N (n = 2), BEB8 (n = 2), ALP1 (n = 1), CHC13 (n = 1), and CHN6 (n = 1), from group 2. The dominant genotype of these was J (49.7%, 154/310). Genotype D of group 1 is the most common in both humans and cattle globally (4, 5, 37, 38). Genotype BLC13 was only detected in edible bullfrogs (Lithobates catesbeiana) in China (39). Genotypes J, I, N, BEB4, and BEB8 have been subsequently found in cattle and humans worldwide (5, 29, 40, 41). However, the genotype CHC13, which is less common, is also found in dairy cattle. In contrast, the ALP1 genotype has only been found in newborn alpacas in Peru (Unpublished data). These results suggest that cattle may be a potential source of human infection with this pathogen.
The genotypes of G. duodenalis detected in cattle in this study also appear to be primary zoonotic genotypes. In this study, two genotypes were identified in 29 G. duodenali-positive samples in cattle, including A (n = 6) and E (n = 23), with the dominant genotype being E (79.3%, 23/29). Similar to observations in other studies in recent publications, assemblages A and E were found to be the dominant genotypes in cattle (7, 32). Previous studies have shown that assemblage A is the dominant genotype in humans (2, 7). Although assemblage E is generally considered to be a host-specific genotype in bovine animals, and this genotype has been reported to occur in humans in many countries, including New Zealand, Egypt, Australia, Brazil, and Vietnam (6, 19–22). This study suggests that the presence of assemblage A and assemblage E of G. duodenalis in animals may have implications for public health.
5 Conclusion
The results of the study indicate a common occurrence of E. bieneusi and G. duodenalis in dairy cattle from Anhui, China. The results suggest that known zoonotic E. bieneusi and G. duodenalis are prevalent in dairy cattle, thereby enhancing our understanding of the genetic diversity and transmission of these pathogens in these animals. Attention should be paid to the monitoring of the spread of these zoonotic E. bieneusi and G. duodenalis in animals. To better understand the transmission of these two pathogens, future molecular epidemiological studies should involve sampling from a larger number of farms.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PQ738189–PQ738196.
Ethics statement
The animal studies were approved by Research Ethics Committee of the Fuyang Normal University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
FL: Writing – original draft, Writing – review & editing. SC: Writing – review & editing, Investigation. CH: Writing – review & editing, Investigation. LM: Software, Writing – review & editing. AW: Software, Writing – review & editing. MS: Writing – review & editing, Software. GX: Writing – review & editing, Funding acquisition, Project administration. HZ: Project administration, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Key Projects of Scientific Research Plan of Colleges and Universities of Anhui Province (2023AH050427), the National Undergraduate Training Program for Innovation and Entrepreneurship (202410371016), the Scientific research project of Fuyang Normal University (2023KYQD0003), and the Biological and Medical Sciences of Applied Summit Nurturing Disciplines in Anhui Province (Anhui Education Secretary Department [2023]13).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Li, W, Feng, Y, and Xiao, L. Enterocytozoon bieneusi. Trends Parasitol. (2022) 38:95–6. doi: 10.1016/j.pt.2021.08.003
2. Ryan, UM, Feng, Y, Fayer, R, and Xiao, L. Taxonomy and molecular epidemiology of Cryptosporidium and Giardia - a 50 year perspective (1971-2021). Int J Parasitol. (2021) 51:1099–119. doi: 10.1016/j.ijpara.2021.08.007
3. Dixon, BR. Giardia duodenalis in humans and animals - transmission and disease. Res Vet Sci. (2021) 135:283–9. doi: 10.1016/j.rvsc.2020.09.034
4. Li, W, and Xiao, L. Ecological and public health significance of Enterocytozoon bieneusi. One Health. (2021) 12:100209. doi: 10.1016/j.onehlt.2020.100209
5. Li, W, Feng, Y, and Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. (2019) 35:436–51. doi: 10.1016/j.pt.2019.04.004
6. Zahedi, A, Field, D, and Ryan, U. Molecular typing of Giardia duodenalis in humans in Queensland - first report of assemblage E. Parasitology. (2017) 144:1154–61. doi: 10.1017/S0031182017000439
7. Cai, W, Ryan, U, Xiao, L, and Feng, Y. Zoonotic giardiasis: an update. Parasitol Res. (2021) 120:4199–218. doi: 10.1007/s00436-021-07325-2
8. Decraene, V, Lebbad, M, Botero-Kleiven, S, Gustavsson, AM, and Löfdahl, M. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol Infect. (2012) 140:519–27. doi: 10.1017/S095026881100077X
9. Duan, J, Qin, H, Sun, M, Fu, Y, Lang, J, Zhang, A, et al. Occurrence and genotypic identification of Blastocystis sp., Enterocytozoon bieneusi, and Giardia duodenalis in dairy cattle in Heilongjiang Province, China. Parasitol Int. (2024) 100:102871. doi: 10.1016/j.parint.2024.102871
10. Fayer, R, Santín, M, and Trout, JM. Enterocytozoon bieneusi in mature dairy cattle on farms in the eastern United States. Parasitol Res. (2007) 102:15–20. doi: 10.1007/s00436-007-0746-x
11. Karim, MR, Rume, FI, Li, D, Li, J, and Zhang, L. First molecular characterization of Enterocytozoon bieneusi in children and calves in Bangladesh. Transbound Emerg Dis. (2022) 69:1999–2007. doi: 10.1111/tbed.14187
12. Yang, X, Fan, YY, Yang, DJ, Huang, S, Wang, JW, Chen, X, et al. High genotype diversity and zoonotic potential of Enterocytozoon bieneusi in yaks (Bos grunniens) from Ganzi tibetan autonomous prefecture, Sichuan province. Parasite. (2023) 30:39. doi: 10.1051/parasite/2023044
13. Sak, B, Brady, D, Pelikánová, M, Květoňová, D, Rost, M, Kostka, M, et al. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J Clin Microbiol. (2011) 49:1064–70. doi: 10.1128/JCM.01147-10
14. Wang, L, Xiao, L, Duan, L, Ye, J, Guo, Y, Guo, M, et al. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl Trop Dis. (2013) 7:e2437. doi: 10.1371/journal.pntd.0002437
15. Zhang, X, Wang, Z, Su, Y, Liang, X, Sun, X, Peng, S, et al. Identification and genotyping of Enterocytozoon bieneusi in China. J Clin Microbiol. (2011) 49:2006–8. doi: 10.1128/JCM.00372-11
16. Ryan, U, and Zahedi, A. Molecular epidemiology of giardiasis from a veterinary perspective. Adv Parasitol. (2019) 106:209–54. doi: 10.1016/bs.apar.2019.07.002
17. Feng, Y, and Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. (2011) 24:110–40. doi: 10.1128/CMR.00033-10
18. Cacciò, SM, Lalle, M, and Svärd, SG. Host specificity in the Giardia duodenalis species complex. Infect Genet Evol. (2018) 66:335–45. doi: 10.1016/j.meegid.2017.12.001
19. Abdel-Moein, KA, and Saeed, H. The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol Res. (2016) 115:3197–202. doi: 10.1007/s00436-016-5081-7
20. Fantinatti, M, Bello, AR, Fernandes, O, and Da-Cruz, AM. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J Infect Dis. (2016) 214:1256–9. doi: 10.1093/infdis/jiw361
21. Garcia, RJ, Ogbuigwe, P, Pita, AB, Velathanthiri, N, Knox, MA, Biggs, PJ, et al. First report of novel assemblages and mixed infections of Giardia duodenalis in human isolates from New Zealand. Acta Trop. (2021) 220:105969. doi: 10.1016/j.actatropica.2021.105969
22. Iwashita, H, Sugamoto, T, Takemura, T, Tokizawa, A, Vu, TD, Nguyen, TH, et al. Molecular epidemiology of Giardia spp. in northern Vietnam: potential transmission between animals and humans. Parasite Epidemiol Control. (2021) 12:e00193. doi: 10.1016/j.parepi.2020.e00193
23. Caccio, SM, and Ryan, U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. (2008) 160:75–80. doi: 10.1016/j.molbiopara.2008.04.006
24. Sulaiman, IM, Fayer, R, Bern, C, Gilman, RH, Trout, JM, Schantz, PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. (2003) 9:1444–52. doi: 10.3201/eid0911.030084
25. Ye, J, Xiao, L, Li, J, Huang, W, Amer, SE, Guo, Y, et al. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. (2014) 63:132–7. doi: 10.1016/j.parint.2013.10.007
26. Sulaiman, IM, Fayer, R, Lal, AA, Trout, JM, Schaefer, FW 3rd, and Xiao, L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl Environ Microbiol. (2003) 69:4495–501. doi: 10.1128/AEM.69.8.4495-4501.2003
27. Feng, Y, Gong, X, Zhu, K, Li, N, Yu, Z, Guo, Y, et al. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasit Vectors. (2019) 12:41. doi: 10.1186/s13071-019-3310-5
28. Gao, JF, Zhou, L, Zhang, AH, Hou, MR, Liu, XW, Zhang, XH, et al. Prevalence and molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in cattle in Heilongjiang Province, Northeast China. Animals (Basel). (2024) 14:1635. doi: 10.3390/ani14111635
29. Liu, X, Tang, L, Li, W, Li, C, and Gu, Y. Prevalence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J Vet Med Sci. (2022) 84:40–7. doi: 10.1292/jvms.21-0425
30. Qi, M, Jing, B, Jian, F, Wang, R, Zhang, S, Wang, H, et al. Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, Northwest China. Parasitol Int. (2017) 66:960–3. doi: 10.1016/j.parint.2016.10.019
31. Song, HY, Wang, KS, Yang, JF, Mao, HM, Pu, LH, Zou, Y, et al. Prevalence and novel genotypes identification of Enterocytozoon bieneusi in dairy cattle in Yunnan Province, China. Animals (Basel). (2021) 11:3014. doi: 10.3390/ani11113014
32. Zhao, Q, Yang, B, Huang, M, Qi, M, Xu, C, Jing, B, et al. Molecular detection and genetic characteristics of Giardia duodenalis in dairy cattle from large-scale breeding farms in Xinjiang, China. Parasitol Res. (2024) 123:106. doi: 10.1007/s00436-024-08123-2
33. Heng, ZJ, Yang, JF, Xie, XY, Xu, CR, Chen, JR, Ma, J, et al. Prevalence and multilocus genotyping of Giardia duodenalis in Holstein cattle in Yunnan, China. Front Vet Sci. (2022) 9:949462. doi: 10.3389/fvets.2022.949462
34. Lam, HYP, Chen, TT, Tseng, YC, Chang, KC, Yang, TH, and Peng, SY. Detection and genotyping of Giardia duodenalis from cattle and pigs in Hualien country, eastern Taiwan. J Microbiol Immunol Infect. (2021) 54:718–27. doi: 10.1016/j.jmii.2020.05.009
35. Qi, M, Wang, H, Jing, B, Wang, R, Jian, F, Ning, C, et al. Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, northwestern China. Parasit Vectors. (2016) 9:546. doi: 10.1186/s13071-016-1828-3
36. Zhao, L, Zhang, ZS, Han, WX, Yang, B, Chai, HL, Wang, MY, et al. Prevalence and molecular characterization of Giardia duodenalis in dairy cattle in Central Inner Mongolia, northern China. Sci Rep. (2023) 13:13960. doi: 10.1038/s41598-023-40987-9
37. Jiang, Y, Tao, W, Wan, Q, Li, Q, Yang, Y, Lin, Y, et al. Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in Northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl Environ Microbiol. (2015) 81:5278. doi: 10.1128/AEM.00328-15
38. Ma, J, Li, P, Zhao, X, Xu, H, Wu, W, Wang, Y, et al. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet Parasitol. (2015) 207:220–7. doi: 10.1016/j.vetpar.2014.10.011
39. Ding, H, Zhao, A, Wang, L, Gao, N, Sun, Y, Li, J, et al. Genotypes and zoonotic potential of Enterocytozoon bieneusi in edible bullfrogs (Lithobates catesbeiana) in China. Int J Parasitol Parasites Wildl. (2020) 11:103–7. doi: 10.1016/j.ijppaw.2020.01.004
40. Li, S, Wang, P, Zhu, XQ, Zou, Y, and Chen, XQ. Prevalence and genotypes/subtypes of Enterocytozoon bieneusi and Blastocystis sp. in different breeds of cattle in Jiangxi Province, southeastern China. Infect Genet Evol. (2022) 98:105216. doi: 10.1016/j.meegid.2022.105216
41. Wang, MY, Zhang, S, Zhang, ZS, Qian, XY, Chai, HL, Wang, Y, et al. Prevalence and molecular characterization of Cryptosporidium spp., Enterocytozoon bieneusi, and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. Vet Res Commun. (2024) 48:2629–43. doi: 10.1007/s11259-024-10364-6
Keywords: Enterocytozoon bieneusi, Giardia duodenalis, dairy cattle, zoonosis, China
Citation: Li F, Cheng S, He C, Meng L, Wang A, Shao M, Xu G and Zhang H (2025) Prevalence and zoonotic potential of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle from Anhui Province, China. Front. Vet. Sci. 12:1613342. doi: 10.3389/fvets.2025.1613342
Edited by:
Vikrant Sudan, Guru Angad Dev Veterinary and Animal Sciences University, IndiaReviewed by:
Qingxia Wu, Tibet Agricultural and Animal Husbandry University, ChinaZorica D. Dakić, University of Belgrade, Serbia
Copyright © 2025 Li, Cheng, He, Meng, Wang, Shao, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gaoxiao Xu, eGd4MTM4QDEyNi5jb20=; Huilin Zhang, MjAyMjEyQGZ5bnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Falei Li
Falei Li Shimei Cheng†
Shimei Cheng† Libing Meng
Libing Meng Huilin Zhang
Huilin Zhang