- College of Agriculture and Biology, Liaocheng University, Liaocheng, China
This review examines the genetic basis of meat production phenotypic traits in sheep, addressing the challenge of enhancing carcass and meat quality to meet global demand. The article identifies key potential genes associated with vertebral traits, body size, muscle development, and fat deposition across diverse sheep breeds worldwide. Through comprehensive analysis of recent literature (2018–2025), the study synthesizes findings from genome-wide association studies, candidate gene approaches, and transcriptomic analyses. Specific potential genes like VRTN, NR6A1, MSTN, ADIPOQ, LCORL, MEF2B, FASN, FABP4, SCD, DGAT1, BMP and HOX family genes demonstrate significant associations with economically valuable traits. The potential genes influencing meat production phenotypic traits (intramuscular fat contents, growth, vertebral traits and body size traits) have been highlighted in this review. This comprehensive genetic marker catalog serves as a critical resource repository for implementing marker-assisted selection programs, providing breeders and researchers with validated genetic targets to accelerate breeding efficiency and enhance meat production in sheep worldwide.
1 Introduction
Sheep farming plays a critical role in global agricultural production, serving as a significant source of meat, wool, and other essential products (1). Global food consumption is projected to witness a substantial rise by 2050, particularly in the demand for animal protein products. This demand, however, will not only be driven by quantity but also by the quality of animal protein products desired by consumers. Notably, tenderness stands out as a paramount sensory attribute for consumers when it comes to meat consumption (2, 3). As global food demand continues to rise, there is an increasing imperative to enhance livestock productivity through advanced genetic approaches. The genetic improvement of meat production traits in sheep represents a crucial strategy for addressing these challenges, offering the potential to develop more efficient, high-quality meat-producing breeds that can contribute to global food security.
The complex nature of meat production traits in sheep involves multiple genetic and environmental factors that influence characteristics such as muscle growth, carcass quality, fat deposition, and overall meat quantity and quality (4, 5). Recent advances in molecular genetics and genomic technologies have opened unprecedented opportunities for understanding the genetic mechanisms underlying these important phenotypic traits (6–10). Genome-wide association studies (GWAS), transcriptome and candidate gene approaches have increasingly revealed the intricate genetic architecture that controls meat production characteristics, providing researchers and animal breeders with valuable insights into potential genetic markers and selection strategies (11).
This review article aims to comprehensively explore the current landscape of genetic research related to meat production traits in sheep. By systematically examining recent scientific literature, we will synthesize the most significant candidate genes associated with critical meat production phenotypes across various sheep breeds worldwide. Our analysis will not only highlight the genetic diversity and potential for genetic improvement but also provide a roadmap for future marker-assisted selection (MAS) programs. Through this comprehensive review, we seek to contribute to the ongoing efforts to optimize sheep breeding strategies, ultimately supporting more sustainable and productive livestock farming practices.
2 Literature search and selection criteria
This review article was designed to overview the potential candidate genes linked to various meat production traits in sheep. For this purpose, we selected articles published within the last 5 years (2018–2025), reflecting the contemporary landscape of research in the field. However, for the introductory section of this review, we extended our purview to include articles dating back to the year 2015. This comprehensive approach allowed us to establish a robust historical context for the subject matter.
The keywords employed in our search strategy were thoughtfully chosen to capture the multifaceted dimensions of the topic. These keywords included “carcass weight,” “muscle pH,” “muscle tenderness,” “meat quality and quantity,” “vertebrae,” “body size,” “body weight,” “Sheep “molecular breeding,” and “genetic markers, potential genes.” The selection of genes reported by any article for inclusion in this review was underpinned by their recognition as significant (p < 0.05) potential candidate genes associated with meat quality and quality-related traits. This recognition was based on the declarations made by authors in their respective published articles, signifying the genes’ significance in the field. To perform functional enrichment analysis and identify biological pathways associated with the genes examined in this review, we used ShinyGO online software (12).
In order to maintain a rigorous standard, we excluded articles published in non-science citation index–(SCI) journals and those not published in the English language. This deliberate choice was made to ensure that the articles included in our review were subjected to peer-review processes and accessible to a wider academic audience. Furthermore, it is important to note that book chapters and unpublished data were excluded from our discussion. However, we did consider the foundational insights from previously published review articles pertaining to specific genes associated with meat production traits in small ruminants. The summary of articles used in the current review is provided in Figure 1.
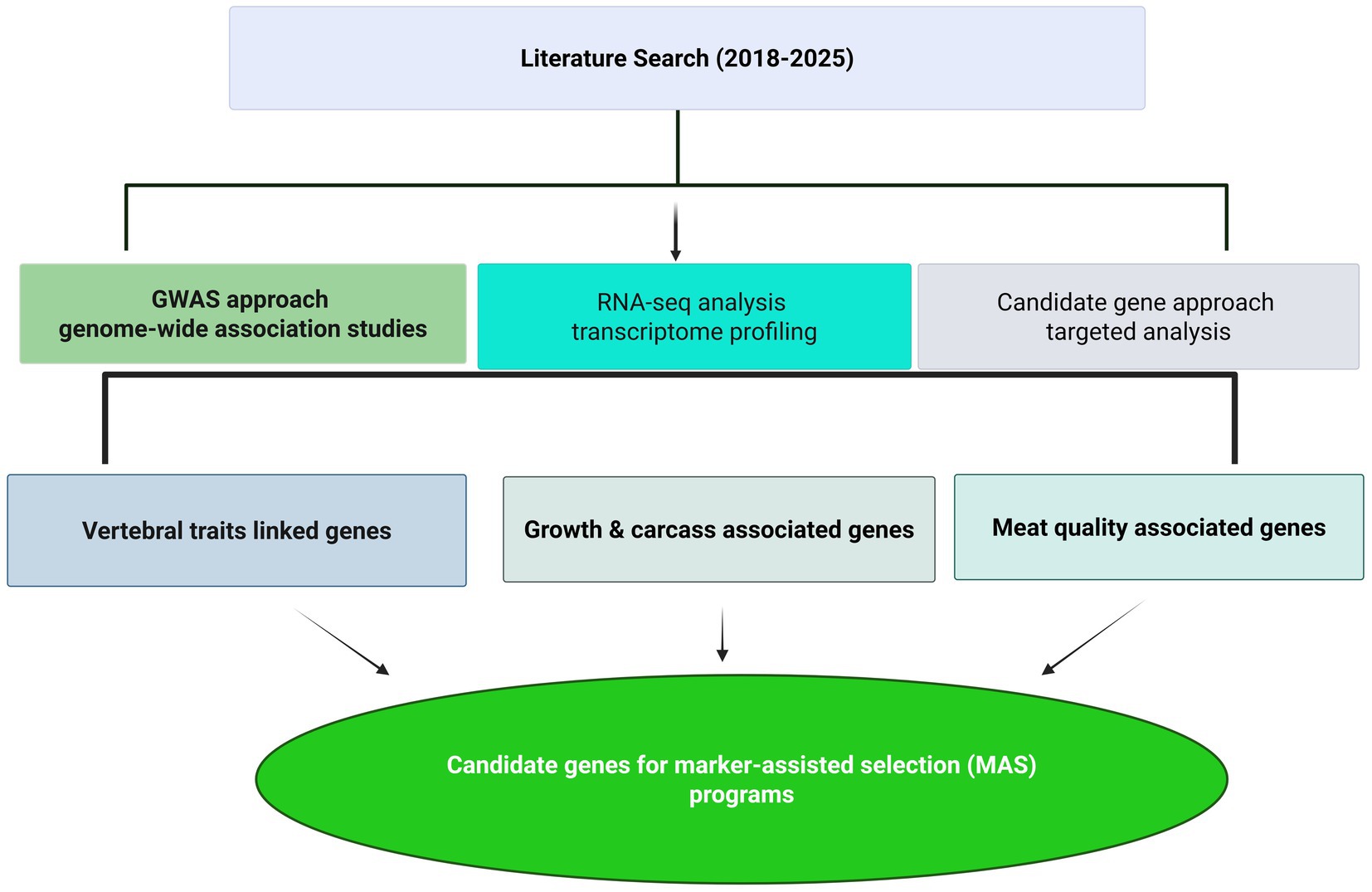
Figure 1. Schematic methodological framework showing the literature strategy and the three main approaches (GWAS, RNA-seq analysis, and candidate gene approaches) used to identify candidate genes for meat production traits in sheep.
3 Overview of potential genes associated with meat production phenotypic traits in sheep
The study of genes associated with meat production phenotypic traits in sheep has significant agricultural and economic importance, as identifying these genetic markers enables more efficient selective breeding programs that can improve meat quality, yield, and production efficiency. Consistently, the association of genes associated with meat production traits have already been documented in previous studies (13, 14). By understanding the genetic basis of traits like muscle growth, fat deposition, tenderness, and flavor profile, researchers can develop genomic-based selection tools that allow producers to make breeding decisions earlier in an animal’s life, reducing costs while increasing genetic gains. Additionally, this genetic knowledge helps address consumer demands for consistent, high-quality meat products while potentially improving animal welfare through selecting for traits that enhance health and reduce stress susceptibility. Such research also contributes to broader food security goals by helping develop more efficient and sustainable sheep production systems that can adapt to changing environmental conditions and market demands.
3.1 Potential genes associated with number of vertebral traits in sheep
During the course of livestock evolution, there has been significant variation in the body size of domestic animals, both between and within species or breeds. Among the traits of economic importance, the number of vertebrae is noteworthy due to its association with body length and carcass characteristics. Notably, the association of variations in the number of thoracic and lumbar vertebrae thoracolumbar vertebrae with carcass length have been observed across different breeds of pigs (15), donkey (16–20), sheep (21) and cattle (22). It is worth mentioning that variations in the number of thoracolumbar vertebrae have been considered a selection trait in commercial animal breeding due to its correlations with growth and meat production.
In a general context, the arrangement of vertebrae in sheep typically includes 7 cervical vertebrae (C), 13 thoracic vertebrae (T), 6 lumbar vertebrae (L), and 4 sacral vertebrae (S), resulting in a total of 30 vertebrae. Among these, mutations in the thoracolumbar region, such as T14L6 or T13L7, have been reported as the most common (23). Multi-vertebrae sheep, exhibiting such mutations, demonstrate advantages in terms of adaptability and meat production performance (23). In the case of Kazakh sheep, which are indigenous to west Xinjiang of China, it is observed that there is variation in the number of lumbar vertebrae. Typically, for most sheep, the count includes 13 thoracic vertebrae and 6 lumbar vertebrae, often labeled as T13L6. However, in the case of Kazakh sheep, variations have been found, specifically T13L7 and T14L6, which, respectively, result in increased carcass length by 2.22 cm and 2.93 cm compared to normal T13L6 Kazakh sheep. Additionally, carcass weight is raised by 1.68 kg and 1.90 kg, respectively (23–26). Given the significant economic and productive advantages associated with vertebral variations in sheep, particularly the increased carcass length and weight observed in T13L7 and T14L6 configurations, understanding the underlying genetic mechanisms controlling these traits has become a priority in livestock genomics research. Recent advances in genomic technologies, have enabled researchers to identify candidate genes associated (SYNDIG1L, VRTN, NR6A1, LTBP2, BMP4) with vertebral development and segmentation (23, 26–30). Table 1 presents a comprehensive overview of genes associated with vertebral development and bone formation in various sheep breeds. This research area is particularly significant for the sheep industry as the number and structure of vertebrae directly influence carcass length, meat yield, and overall productivity.
3.2 Screening potential genes associated with growth, carcass and body size traits using RNA sequencing (RNA-seq) and GWAS in sheep
The integration of RNA-seq and GWAS represents a powerful approach for identifying genes and genetic variants associated with economically important meat production traits in sheep. This comprehensive strategy combines transcriptomic profiling to reveal differentially expressed genes in relevant tissues with population-based association analyses to pinpoint significant genetic variants. By correlating expression pat-terns with phenotypic data and genetic polymorphisms, researchers can identify candidate genes influencing key traits such as muscle growth, fat deposition, meat quality, and carcass composition. Understanding the genetic basis of growth and carcass-related traits in sheep plays a pivotal role in enhancing muscle growth, hypertrophy, and, ultimately, meat production (31, 32). Recently, several meat production associated genetic markers have been identified in various meat sheep breeds (Uruguayan Merino sheep, Romney, Karachaevsky Sheep, Hu, Dorper, Awassi, Afghani, Bandur, Baluch etc.) (Table 2). Consistently, a study has highlighted several genes (LHX3, LHX4, CAPN, MEF2B, TRHDE, MEF2A, MEF2C, MEF2D, FTO, APOBR, TP53, DRB1 2001, MSTN, GH, GRM1, MBD5, UBR2, RPL7, SMC2, and SHISA9) associated with various meat quality traits, including body weight, growth, and chest girth in sheep (33). Additionally, this study identified genes (CAST, LEP, MSTN, RFXANK, RIPK2, DGAT1, UCP1, and MCPs) linked to carcass and fat traits in sheep. The genetic analysis of sheep from Table 2 reveals a comprehensive landscape of genes controlling economically valuable production traits. The myostatin gene (MSTN) emerges as a critical regulator of muscle development, while LCORL and NCAPG appear repeatedly as major determinants of growth and body size traits. Fat metabolism and deposition are primarily influenced by DGAT2, FABP4, and SCD, which regulate lipid biosynthesis and transport. The bone morphogenetic protein (BMP) family, particularly BMP2, plays a significant role in both skeletal development and fat deposition in tail regions. Growth hormone pathways involving GHR and IGF1 control overall growth performance, while muscle-specific genes like MYL2 and TNNC2 influence meat quality characteristics. Notably, these candidate genes have been validated across multiple sheep populations worldwide using both GWAS and RNA-seq approaches, providing robust genetic markers that could be incorporated into breeding programs aimed at enhancing meat production efficiency and quality in commercial sheep operations. The summary of potential genes affecting meat production phenotypic traits in sheep is provided in Table 2.
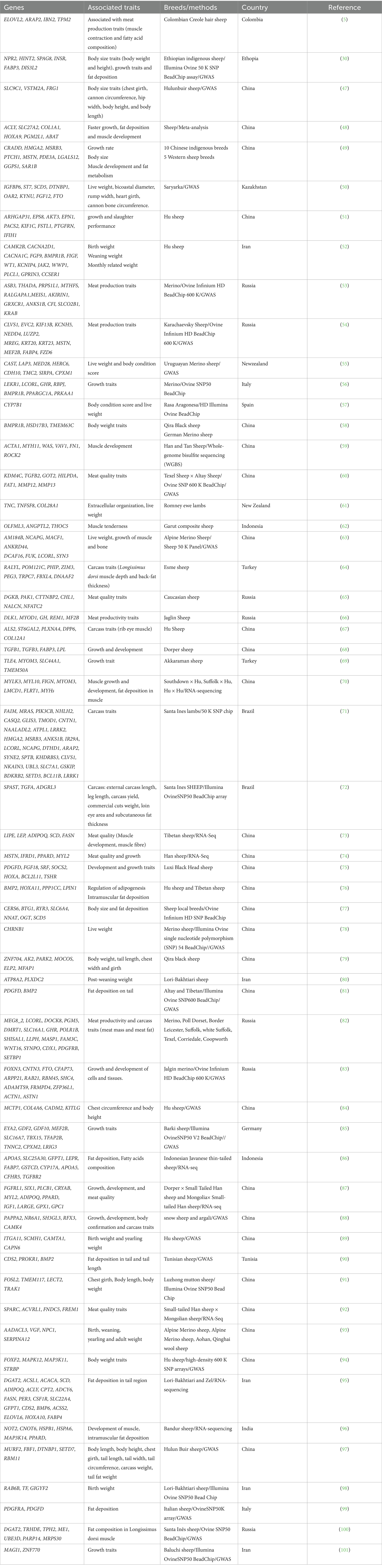
Table 2. Potential genes associated with growth, carcass and body confirmation phenotypic traits using GWAS and RNA-seq in sheep.
3.3 Candidate gene approach to screen potential genetic markers associated with meat production phenotypic traits in sheep
The candidate gene approach represents a targeted strategy in sheep genetics research that focuses on identifying and analyzing specific genes with potential influence on economically important meat production traits. This method selectively examines genes with known biological functions related to muscle development, growth, fat deposition, and meat quality characteristics based on prior physiological knowledge or findings from other livestock species. For example, researchers typically analyze polymorphisms within these candidate genes—such as myostatin (MSTN), calpain (CAPN), calpastatin (CAST), leptin (LEP), DGAT1 and growth hormone (GH)—to establish associations with phenotypic traits including carcass weight, muscle mass, intramuscular fat content, tenderness, and meat flavor profile (Table 3). Consistently, our previously published research extensively examined the role of DGAT1 K232A polymorphism in enhancing sheep meat quality traits (34). Fatty acid-binding protein 4 (FABP4) is involved in fatty acid transportation, and variations in this gene have been reported to influence fat deposition in mammals. Several studies have consistently demonstrated the involvement of FABP4 in regulating meat quality traits in sheep (35). Additionally, Alwan et al. (35) observed a detrimental effect of p.61Thr > Asp on FABP4, resulting in reduced fatty acid binding efficiency and increased carcass traits in Karakul and Awassi Sheep. Furthermore, other studies have documented associations between FABP4 variations and various economic traits in sheep, such as carcass and growth traits in New Zealand Romney lambs (36), morphometric traits in Albanian sheep (37), body weight, final weight, and average daily gain in three Egyptian sheep breeds (38), as well as intramuscular and internal fat weight in two Russian sheep breeds (39). The approach has proven valuable for marker-assisted selection programs in sheep breeding, allowing producers to make informed breeding decisions that enhance meat production efficiency and quality while reducing the time and resources required compared to genome-wide studies. Despite limitations in detecting novel genes, the candidate gene approach continues to provide practical applications in sheep breeding programs focused on improving commercially relevant meat production traits. The summary of determinant genes associated with meat production phenotypic traits in sheep is provided in Table 3.
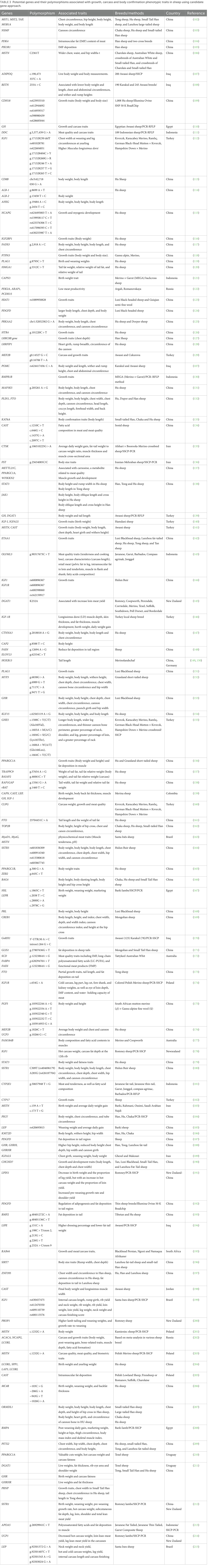
Table 3. Potential genes and their polymorphisms associated with growth, carcass and body confirmation phenotypic traits in sheep using candidate gene approach.
4 Discussion
The genetic architecture underlying meat production traits in sheep represents a sophisticated biological system wherein multiple interconnected pathways coordinate growth, muscle development, fat deposition, and skeletal formation. Brief information about the genes documented in this review and their related pathways is provided in Supplementary Files 1, 2. This complex network involves numerous candidate genes that have been consistently reported across diverse sheep populations (Figure 2; Tables 1–3) and breeding programs worldwide, each contributing specific functional roles while participating in broader regulatory circuits that determine economically valuable traits.
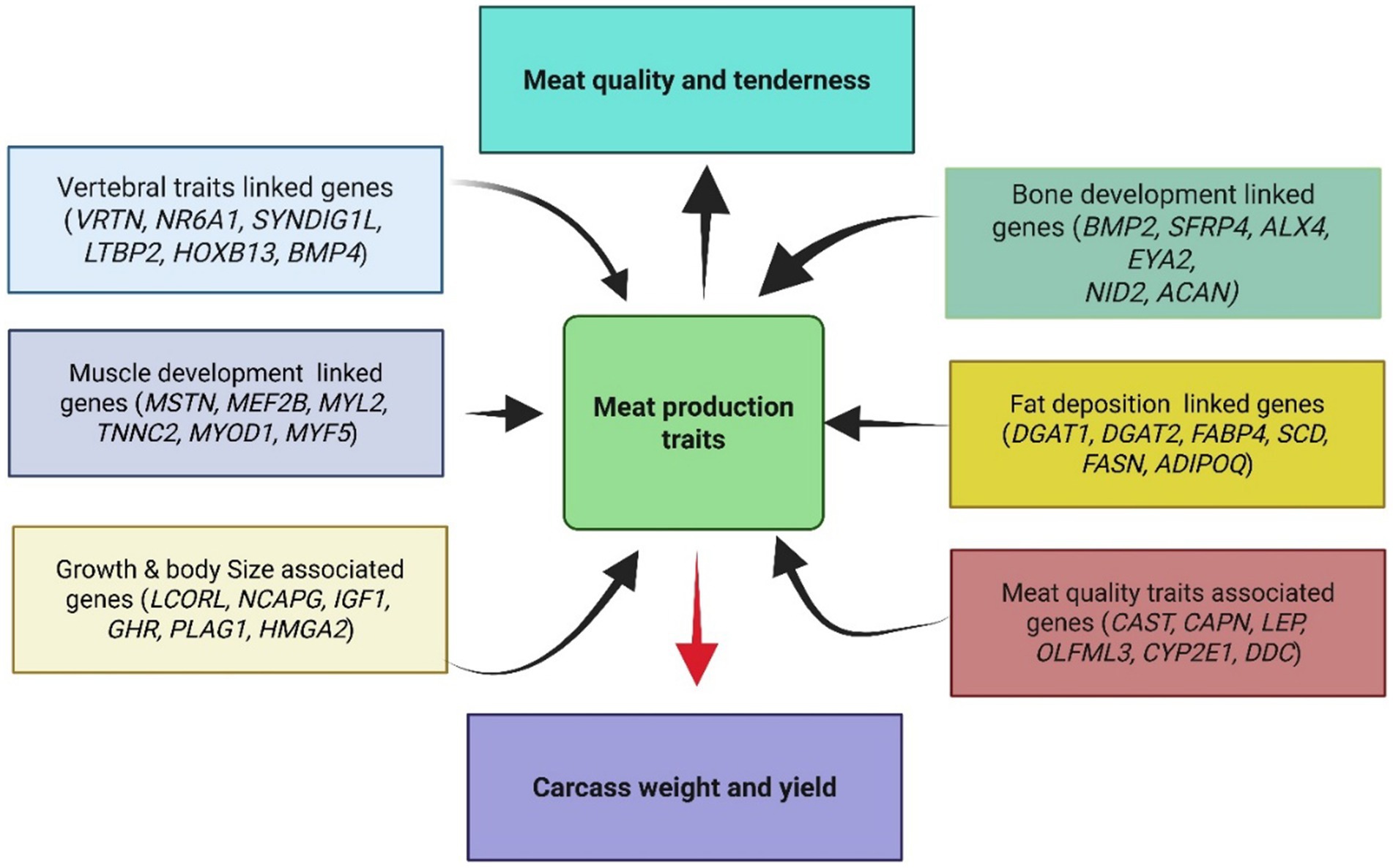
Figure 2. Conceptual framework showing the major gene categories affecting meat production traits in sheep. Genes are grouped by their primary biological functions, with arrows indicating their influence on final phenotypic outcomes.
Central to this genetic framework, myogenesis pathways control the fundamental processes of muscle development and ultimately determine muscle mass and composition that defines meat yield. The MSTN gene operates as a negative regulator within the transforming growth factor-beta signaling network, where its expression limits muscle growth through inhibition of satellite cell activation and myoblast proliferation. Consequently, when MSTN signaling is reduced through genetic variants, normal growth constraints are released, resulting in increased muscle fiber number and size, which translates directly to enhanced muscle mass and improved carcass composition. Furthermore, muscle-specific transcription factors MEF2B, MYOD1, and MYF5 coordinate myogenic differentiation programs, controlling the expression of muscle-specific genes that determine fiber type characteristics and contractile properties. These regulatory networks interact synergistically with calcium-dependent signaling pathways involving troponin components such as TNNC2 and myosin light chains including MYL2, which collectively determine muscle fiber contractility and ultimately influence meat texture and quality attributes. Complementing the myostatin pathway, the growth hormone regulatory network represents another critical system controlling overall growth performance and carcass development. This integrated pathway encompasses insulin-like growth factor 1 and its receptor, along with growth hormone and its corresponding receptor, functioning as a master regulator of somatic growth and metabolic processes through a sophisticated feedback system that regulates traits ranging from birth weight to final carcass characteristics. The signaling mechanism initiates with growth hormone binding to its receptor, triggering downstream activation of IGF1 synthesis in the liver and peripheral tissues. Subsequently, IGF1 binds to its receptor, initiating intracellular signaling cascades that promote protein synthesis, muscle fiber development, and overall growth performance. This pathway directly influences carcass weight and yield by regulating cell proliferation, differentiation, and metabolism throughout the animal’s development, demonstrating dual influence on both muscle development and fat metabolism through intricate feedback mechanisms that ensure balanced growth processes responsive to physiological demands. In parallel, lipid metabolism pathways represent equally critical regulatory systems determining fat deposition patterns and meat quality characteristics. The triglyceride synthesis pathway, culminating in DGAT1 and DGAT2 enzymatic activity, controls the final steps of fat formation and storage. Notably, the diacylglycerol O-acyltransferase 1 gene catalyzes the final enzymatic step in triglyceride synthesis, demonstrating remarkable consistency in its associations with meat quality traits across sheep populations. Specific polymorphisms, particularly the K232A variant, have been extensively validated for their positive effects on loin meat yield and intramuscular fat content, directly influencing consumer-perceived meat quality. Concurrently, fatty acid-binding protein 4 plays a crucial role in fatty acid transport and cellular uptake, with genetic variations affecting both fat deposition patterns and meat quality characteristics. The fatty acid synthesis pathway, regulated by FASN, controls the production of fatty acids from acetyl-CoA precursors, while stearoyl-CoA desaturase introduces unsaturation into fatty acid chains, influencing membrane fluidity and meat quality attributes. Additionally, the bone morphogenetic protein family introduces an intriguing dimension to meat production genetics through its dual functionality in both skeletal development and adipogenesis. Specifically, BMP2 and BMP4 operate through specialized signaling pathways that simultaneously regulate fat tail development in certain sheep breeds while affecting bone formation and overall body size determination. This dual role becomes particularly relevant for breeds adapted to harsh environmental conditions, where fat reserves serve as critical survival mechanisms during periods of feed scarcity, thus representing an evolutionary adaptation that balances immediate production goals with long-term survival capacity. Moreover, skeletal development pathways contribute significantly to carcass characteristics through their control of bone formation and vertebral segmentation. The vertebral development genes VRTN and NR6A1 regulate axial skeleton segmentation during embryogenesis, with genetic variants affecting the number of thoracic and lumbar vertebrae. Increased vertebral number directly correlates with longer carcass length and greater total carcass weight, providing measurable economic benefits. The HOX gene family provides positional information during development, ensuring proper spatial organization of skeletal structures that determine final body conformation and carcass geometry. Transcending individual pathway effects, master regulatory genes emerge as overarching controllers of multiple production traits through their influence on chromatin remodeling and transcriptional regulation. The LCORL and NCAPG genes appear consistently across genome-wide association studies investigating body size and growth traits, suggesting fundamental roles in determining mature body size and growth rate. These genes operate through epigenetic modifications and transcriptional control mechanisms, influencing the expression of numerous downstream targets involved in muscle development, bone growth, and overall body size determination. Similarly, HMGA2 and PLAG1 contribute additional layers of transcriptional control, particularly influencing growth-related gene expression patterns that determine mature body size and growth trajectory. The calpain-calpastatin proteolytic system represents a specialized post-mortem pathway that significantly influences meat quality and consumer acceptance. The calpain proteases, regulated by the calpastatin inhibitor encoded by the CAST gene, control protein degradation processes that occur after slaughter, determining the extent of myofibrillar protein breakdown that directly affects meat tenderness development during aging. Genetic variants affecting calpastatin expression influence the balance between protease activity and inhibition, ultimately determining the rate and extent of tenderization during post-mortem storage.
Furthermore, metabolic regulation pathways connect nutritional status with growth performance and carcass composition through genes such as adiponectin and leptin. These genes regulate energy homeostasis, fat distribution, appetite control, and energy expenditure, creating essential links between metabolic efficiency and production outcomes. The adiponectin pathway influences energy balance and fat distribution patterns, while leptin regulates appetite and energy expenditure, ensuring that growth processes remain aligned with nutritional resources and metabolic capacity.
Environmental interactions add considerable complexity to these genetic systems, wherein genes like the fat mass and obesity-associated gene respond to nutritional status and environmental stressors, modulating their effects on growth and fat deposition based on external conditions. This environmental responsiveness indicates that gene expression can be influenced by factors including nutrition quality, health issues, temperature stress, and management practices, suggesting that optimal genetic selection programs must account for genotype-by-environment interactions to achieve consistent performance across diverse production systems (Figure 3).
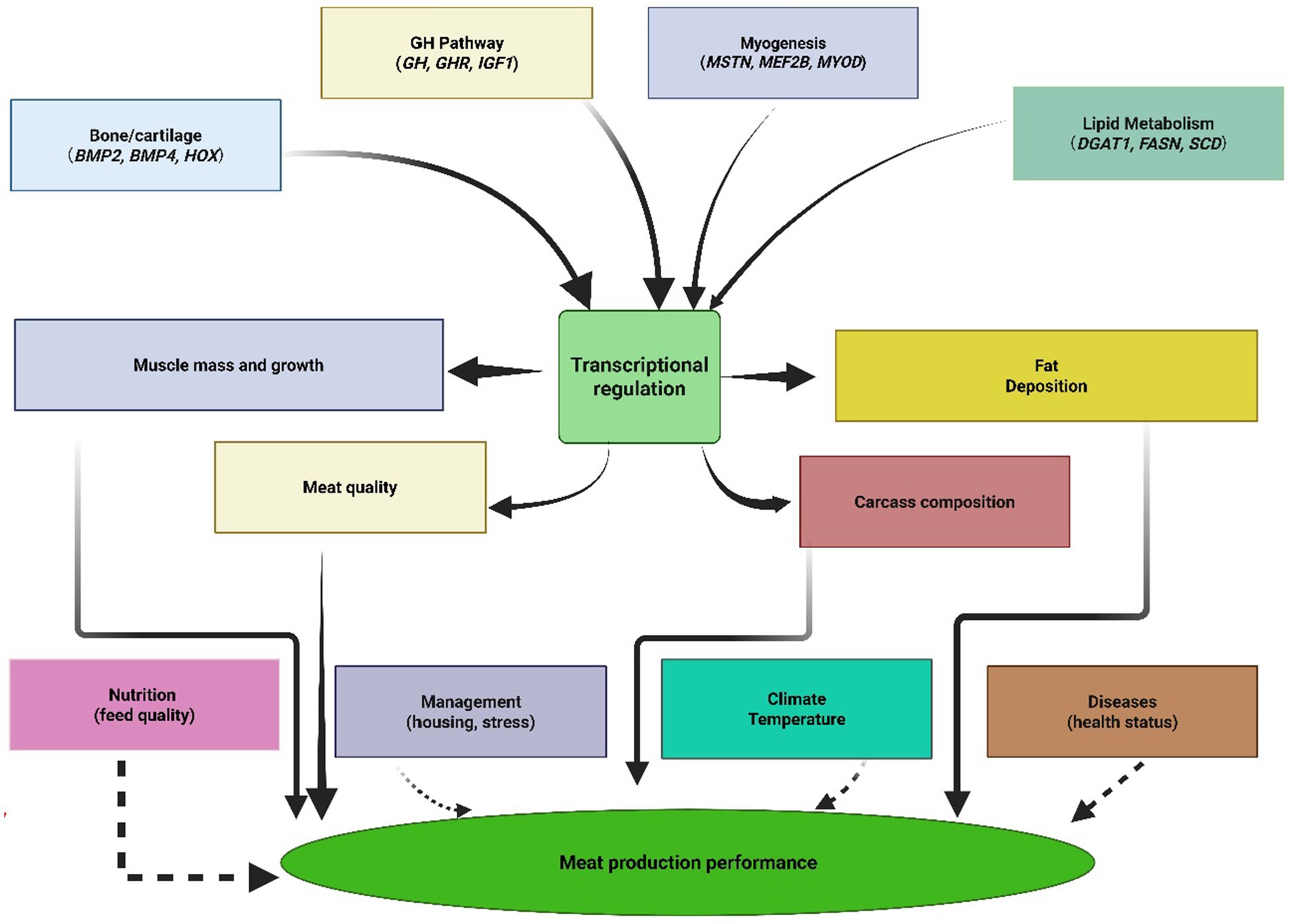
Figure 3. Biological pathway network showing how genetic factors interact with environmental influences to determine meat production traits. Solid arrows indicate direct genetic effects, while dashed lines show environmental modulation.
The integration of these multiple pathways reveals that successful meat production genetics requires a systems-level approach rather than optimization of individual genes. Growth hormone signaling pathways interact extensively with muscle development regulators, while fat metabolism genes simultaneously influence meat quality characteristics and adaptive capacity. Bone morphogenetic proteins affect both skeletal development and fat deposition patterns, demonstrating the interconnected nature of physiological systems underlying meat production traits. Consequently, modern genomic selection approaches increasingly recognize these pathway interactions, moving beyond single-gene effects toward polygenic selection strategies that capture cumulative effects across multiple biological systems. This systems-level understanding provides the foundation for developing comprehensive genetic evaluation programs that can enhance meat production efficiency while maintaining genetic diversity and adaptive capacity essential for sustainable sheep production worldwide.
Based on published data, we concluded that current genetic research faces several significant limitations. Primarily, most genetic associations are discovered within specific breeds but lack validation across diverse populations. This creates limited applicability due to varying genetic backgrounds, distinct linkage disequilibrium patterns, and divergent population histories that cause population stratification effects. Furthermore, the field suffers from insufficient attention to epigenetic factors. DNA methylation patterns are largely ignored despite their significant influence on gene expression. Similarly, environmental interactions remain poorly understood, particularly how nutrition, climate, and management practices interact with genetic variants through complex epigenetic mechanisms. Moreover, the inheritance and influence of epigenetic marks across generations through transgenerational effects remains inadequately investigated. Consequently, the lack of comprehensive epigenome mapping across relevant tissues such as muscle, fat, and liver creates substantial knowledge gaps. This subsequently limits our understanding of tissue-specific regulatory mechanisms. Another critical limitation involves functional validation, where many identified single nucleotide polymorphisms may merely be in linkage disequilibrium with actual causal variants rather than being functionally relevant themselves. Additionally, insufficient experimental validation of how genetic variants actually affect gene function and protein activity perpetuates an oversimplified understanding of gene interactions within complex biological pathways.
To address these multifaceted challenges, future research must embrace multi-omics integration approaches. This includes combining epigenomics data such as DNA methylation, histone modifications, and chromatin accessibility with transcriptomics through expression quantitative trait loci mapping. Furthermore, incorporating proteomics and metabolomics will effectively link genetic variants to protein abundance and metabolite levels. Finally, investigating host-microbiome interactions that significantly affect production traits represents a critical research priority for advancing the field.
5 Conclusion and future research directions
This review has cataloged an extensive array of potential genes associated with meat production traits in sheep breeds globally. The identified genes—particularly those affecting vertebral development, muscle growth, and fat deposition—provide valuable targets for marker-assisted selection strategies to enhance sheep meat production efficiency. Future research should focus on validating these genetic associations across diverse populations and production environments to ensure broader applicability. Integration of advanced genomic technologies, including whole-genome sequencing and multi-omics approaches, will be crucial to understand the functional mechanisms underlying these genetic markers. Additionally, research examining gene–environment interactions and the role of epigenetic modifications on meat production traits deserves attention. Development of cost-effective genotyping platforms suitable for implementing these findings in resource-limited settings would further extend their practical value. Finally, the integration of consumer preferences with genetic selection represents a critical pathway for sustainable sheep breeding programs, where market demands increasingly favor specific meat quality attributes that should directly inform trait selection priorities. Consumer preference for leaner cuts drives selection for enhanced muscle development while reducing excessive fat deposition, while market premiums for higher carcass yield support prioritizing traits that increase carcass length and overall meat yield through improved skeletal development. Premium markets increasingly value optimal marbling for tenderness and flavor, requiring breeding programs to focus not just on fat deposition, but on achieving consumer-preferred intramuscular fat distribution that enhances both meat yield and quality characteristics. Growing consumer awareness of health benefits drives demand for favorable omega-3 to omega-6 fatty acid ratios, necessitating selection for optimized fatty acid synthesis and desaturation pathways to improve nutritional profiles, while market differentiation through functional meat products requires targeted selection of lipid metabolism traits. Post-mortem tenderization processes directly affect meat tenderness, a primary consumer concern, requiring selection programs to balance rapid growth with meat quality attributes that determine consumer satisfaction and repeat purchases. Future strategies should develop market-responsive breeding indices that weight genetic markers based on current consumer preferences and price premiums, establish feedback loops between consumer testing, market analysis, and breeding decisions, and consider regional market variations in trait preferences when implementing marker-assisted selection programs, ensuring that genetic improvements translate into economic value throughout the supply chain while meeting evolving consumer expectations for meat quality, nutrition, and eating experience.
Author contributions
YH: Writing – review & editing, Writing – original draft, Data curation, Investigation, Conceptualization, Methodology, Validation. MA: Visualization, Writing – review & editing, Investigation, Validation, Conceptualization. WC: Methodology, Writing – review & editing, Validation, Investigation, Data curation. XL: Data curation, Conceptualization, Investigation, Writing – review & editing, Visualization. MZ: Data curation, Investigation, Visualization, Methodology, Writing – review & editing. LS: Methodology, Investigation, Data curation, Writing – review & editing. MK: Writing – review & editing, Funding acquisition, Writing – original draft, Conceptualization, Software, Investigation, Resources, Formal analysis, Project administration, Supervision, Data curation, Visualization, Validation, Methodology. CW: Funding acquisition, Software, Visualization, Conceptualization, Resources, Writing – review & editing, Investigation, Writing – original draft, Formal analysis, Project administration, Validation, Supervision, Data curation, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Key R&D Program of China (grant numbers 2022YFD1600103; 2023YFD1302004), the Shandong Province Modern Agricultural Technology System Donkey Industrial Innovation Team (grant no. SDAIT-27), Livestock and Poultry Breeding Industry Project of the Ministry of Agriculture and Rural Affairs (grant number 19211162), Shandong Province Agricultural Major Technology Collaborative Promotion Plan (SDNYXTTG-2024-13), and Liaocheng Municipal Bureau of Science and Technology, High-talented Foreign Expert Introduction Program (GDWZ202401).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1616533/full#supplementary-material
References
1. Ma, K, Song, J, Li, D, Liu, Z, Wang, C, Li, T, et al. Insights into the differences in meat quality among different sheep breeds in the Qilian Mountains from the perspective of metabolomics and transcriptomics. Food Biosci. (2025) 63:105693. doi: 10.1016/j.fbio.2024.105693
2. Naserkheil, M, Lee, DH, Kong, HS, Seong, J, and Mehrban, H. Estimation of genetic parameters and correlation between yearling ultrasound measurements and carcass traits in Hanwoo cattle. Animals. (2021) 11:1425. doi: 10.3390/ani11051425
3. Liu, J, Ellies-Oury, MP, Stoyanchev, T, and Hocquette, JF. Consumer perception of beef quality and how to control, improve and predict it? Focus on eating quality. Food Secur. (2022) 11:1732. doi: 10.3390/foods11121732
4. Kaseja, K, Lambe, N, Yates, J, Smith, E, and Conington, J. Genome wide association studies for carcass traits measured by video image analysis in crossbred lambs. Meat Sci. (2024) 214:109518. doi: 10.1016/j.meatsci.2024.109518
5. Revelo, HA, López-Alvarez, D, Palacios, YA, Vergara, OD, Yánez, MB, Ariza, MF, et al. Genome-wide association study reveals candidate genes for traits related to meat quality in Colombian creole hair sheep. Trop Anim Health Prod. (2023) 55:357. doi: 10.1007/s11250-023-03688-z
6. Zhu, Q, Peng, Y, Liu, X, Chen, W, Geng, M, Na, J, et al. Application of omics in donkey meat research: a review. Animals. (2025) 15:991. doi: 10.3390/ani15070991
7. Khan, MZ, Chen, W, Wang, X, Liang, H, Wei, L, Huang, B, et al. A review of genetic resources and trends of omics applications in donkey research: focus on China. Front Vet Sci. (2024) 11:1366128. doi: 10.3389/fvets.2024.1366128
8. Lou, M, Zhang, S, Yang, W, Li, S, Cao, H, Zhang, Z, et al. Transcriptome analysis revealed the mechanism of skeletal muscle growth and development in different hybrid sheep. Anim Biosci. (2024) 38:408. doi: 10.5713/ab.24.0269
9. Hosseini, SF, Bakhtiarizadeh, MR, and Salehi, A. Meta-analysis of RNA-Seq datasets highlights novel genes/pathways involved in fat deposition in fat-tail of sheep. Front Vet Sci. (2023) 10:1159921. doi: 10.3389/fvets.2023.1159921
10. Zhang, W, Xu, M, Wang, J, Wang, S, Wang, X, Yang, J, et al. Comparative transcriptome analysis of key genes and pathways activated in response to fat deposition in two sheep breeds with distinct tail phenotype. Front Genet. (2021) 12:639030. doi: 10.3389/fgene.2021.639030
11. Liu, D, Li, X, Wang, L, Pei, Q, Zhao, J, Sun, D, et al. Genome-wide association studies of body size traits in Tibetan sheep. BMC Genomics. (2024) 25:739. doi: 10.1186/s12864-024-10633-3
12. Ge, SX, Jung, D, and Yao, R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. (2020) 36:2628–9. doi: 10.1093/bioinformatics/btz931
13. Khan, MZ, Chen, W, Huang, B, Liu, X, Wang, X, Liu, Y, et al. Advancements in genetic marker exploration for livestock vertebral traits with a focus on China. Animals. (2024) 14:594. doi: 10.3390/ani14040594
14. Yıldırır, M, Nurlan, M, Seilkanovna, MA, Seisenovna, OZ, Sholpan, B, Yasemin, Ö, et al. A review of thoracolumbar vertebrae number variation in sheep breeding. Small Rumin Res. (2024):107405. doi: 10.1016/j.smallrumres.2024.107405
15. Burgos, C, Latorre, P, Altarriba, J, Carrodeguas, A, Varona, L, and Buesa, L. Allelic frequencies of NR6A1 and VRTN, two genes that affect vertebrae number in diverse pig breeds: a study of the effects of the VRTN insertion on phenotypic traits of a Duroc×landrace-large White cross. Meat Sci. (2015) 100:150–5. doi: 10.1016/j.meatsci.2014.09.143
16. Wang, T, Wang, X, Liu, Z, Shi, X, Ren, W, Huang, B, et al. Genotypes and haplotype combination of DCAF7 gene sequence variants are associated with number of thoracolumbar vertebrae and carcass traits in Dezhou donkey. J Appl Anim Res. (2023) 51:31–9. doi: 10.1080/09712119.2022.2149538
17. Wang, T, Liu, Z, Wang, X, Li, Y, Akhtar, F, Li, M, et al. Polymorphism detection of PRKG2 gene and its association with the number of thoracolumbar vertebrae and carcass traits in Dezhou donkey. BMC Genomic Data. (2023) 24:2. doi: 10.1186/s12863-022-01101-6
18. Liu, Z, Wang, T, Shi, X, Wang, X, Ren, W, Huang, B, et al. Identification of LTBP2 gene polymorphisms and their association with thoracolumbar vertebrae number, body size, and carcass traits in Dezhou donkeys. Front Genet. (2022) 13:969959. doi: 10.3389/fgene.2022.969959
19. Liu, Z, Gao, Q, Wang, T, Chai, W, Zhan, Y, Akhtar, F, et al. Multi-thoracolumbar variations and NR6A1 gene polymorphisms potentially associated with body size and carcass traits of Dezhou donkey. Animals. (2022) 12:1349. doi: 10.3390/ani12111349
20. Shi, X, Li, Y, Wang, T, Ren, W, Huang, B, Wang, X, et al. Association of HOXC8 genetic polymorphisms with multi-vertebral number and carcass weight in Dezhou donkey. Genes. (2022) 13:2175. doi: 10.3390/genes13112175
21. Donaldson, L, Lambe, R, Maltin, A, Knott, S, and Bunger, L. Between-and within-breed variations of spine characteristics in sheep. J Anim Sci. (2013) 91:995–1004. doi: 10.2527/jas.2012-5456
22. Yang, J, Wen, Y, Yong, L, Feng, Z, Ke, M, Gao, X, et al. Correlation analysis for beef performance and multi-vertebra properties of Jinchuan yak. J Domestic Animal Ecol. (2015) 36:26–30.
23. Zhong, YJ, Yang, Y, Wang, XY, Di, R, Chu, MX, and Liu, QY. Expression analysis and single-nucleotide polymorphisms of SYNDIG1L and UNC13C genes associated with thoracic vertebral numbers in sheep (Ovis aries). Arch Animal Breed. (2021) 64:131–8. doi: 10.5194/aab-64-131-2021
24. Li, S, Luo, R, Lai, D, Ma, M, Hao, F, Qi, X, et al. Whole-genome resequencing of Ujumqin sheep to investigate the determinants of the multi-vertebral trait. Genome. (2018) 61:653–61. doi: 10.1139/gen-2017-0267
25. Li, C, Zhang, X, Cao, Y, Wei, J, You, S, Jiang, Y, et al. Multivertebrae variation potentially contribute to carcass length and weight of Kazakh sheep. Small Rumin Res. (2017) 150:8–10. doi: 10.1016/j.smallrumres.2017.02.021
26. Zhang, Z, Sun, Y, Du, W, He, S, Liu, M, and Tian, C. Effects of vertebral number variations on carcass traits and genotyping of Vertnin candidate gene in Kazakh sheep. Asian Australas J Anim Sci. (2017) 30:1234–8. doi: 10.5713/ajas.16.0959
27. Zhou, C, Zhang, Y, Ma, T, Wu, D, Yang, Y, Wang, D, et al. Whole-genome resequencing of Ujimqin Sheep identifies genes associated with vertebral number. Animals. (2024) 14:677. doi: 10.3390/ani14050677
28. Purev, C, Wu, H, Lkhagva, K, and Tumendemberel, O. Understanding molecular mechanisms of vertebral number of variations on Mongolian sheep using candidate genes analysis. Anim Biosci. (2024) 38:247. doi: 10.5713/ab.24.0212
29. Mi, T, Liu, K, Guo, T, Li, L, Wang, Y, Li, C, et al. Analysis of the eighth intron polymorphism of NR6A1 gene in sheep and its correlation with lumbar spine number. Anim Biotechnol. (2023) 34:218–24. doi: 10.1080/10495398.2021.1954529
30. Ahbara, A, Bahbahani, H, Almathen, F, Al Abri, M, Agoub, MO, Abeba, A, et al. Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous Sheep. Front Genet. (2019) 9:699. doi: 10.3389/fgene.2018.00699
31. Talebi, R, Ghaffari, MR, Zeinalabedini, M, Abdoli, R, and Mardi, M. Genetic basis of muscle-related traits in sheep: a review. Anim Genet. (2022) 53:723–39. doi: 10.1111/age.13266
32. Knapik, J, Ropka-Molik, K, and Pieszka, M. Genetic and nutritional factors determining the production and quality of sheep meat-a review. Ann Anim Sci. (2017) 17:23. doi: 10.1515/aoas-2016-0036
33. Gebreselassie, G, Berihulay, H, Jiang, L, and Ma, Y. Review on genomic regions and candidate genes associated with economically important production and reproduction traits in sheep (Ovies aries). Animals. (2019) 10:33. doi: 10.3390/ani10010033
34. Khan, MZ, Ma, Y, Ma, J, Xiao, J, Liu, Y, Liu, S, et al. Association of DGAT1 with cattle, buffalo, goat, and sheep milk and meat production traits. Front Vet Sci. (2021) 8:712470. doi: 10.3389/fvets.2021.712470
35. Alwan, IH, Aljubouri, TR, and Al-Shuhaib, MB. A novel missense SNP in the fatty acid-binding protein 4 (FABP4) gene is associated with growth traits in karakul and Awassi sheep. Biochem Genet. (2023) 62:1. doi: 10.1007/s10528-023-10504-8
36. Yan, W, Zhou, H, Hu, J, Luo, Y, and Hickford, JG. Variation in the FABP4 gene affects carcass and growth traits in sheep. Meat Sci. (2018) 145:334–9. doi: 10.1016/j.meatsci.2018.07.007
37. Anila, H, Hajno, L, Lorena, H, and Bixheku, X. Variation in FABP4 gene associated with the morphometric traits in Albanian sheep. Acta Biol Turc. (2022) 35:25–8.
38. Shafey, HI, Mahrous, KF, Hassan, AA, Rushdi, HE, and Ibrahim, MA. Single-nucleotide polymorphisms in FABP4 gene associated with growth traits in Egyptian sheep. Vet World. (2020) 13:1126. doi: 10.14202/vetworld.2020.1126-1132
39. Gorlov, IF, Shirokova, NV, Anisimova, EY, Slozhenkina, MI, Kolosov, YA, Natyrov, AK, et al. MC4R gene polymorphism and its association with meat traits of Karachai sheep grown in Russian Federation. J Appl Anim Res. (2021) 49:68–74. doi: 10.1080/09712119.2021.1883624
40. Li, C, Liu, K, Dai, J, Li, X, Liu, X, Ni, W, et al. Whole-genome resequencing to investigate the determinants of the multi-lumbar vertebrae trait in sheep. Gene. (2022) 809:146020. doi: 10.1016/j.gene.2021.146020
41. Moradi, MH, Mahmodi, R, Farahani, AH, and Karimi, MO. Genome-wide evaluation of copy gain and loss variations in three afghan sheep breeds. Sci Rep. (2022) 12:14286. doi: 10.1038/s41598-022-18571-4
42. Kalds, P, Luo, Q, Sun, K, Zhou, S, Chen, Y, and Wang, X. Trends towards revealing the genetic architecture of sheep tail patterning: promising genes and investigatory pathways. Anim Genet. (2021) 52:799–812. doi: 10.1111/age.13133
43. Zhang, D, Zhang, X, Li, F, Liu, T, Hu, Z, Gao, N, et al. Whole-genome resequencing identified candidate genes associated with the number of ribs in Hu sheep. Genomics. (2021) 113:2077–84. doi: 10.1016/j.ygeno.2021.05.004
44. Zhao, F, Deng, T, Shi, L, Wang, W, Zhang, Q, Du, L, et al. Genomic scan for selection signature reveals fat deposition in Chinese indigenous sheep with extreme tail types. Animals. (2020) 10:773. doi: 10.3390/ani10050773
45. Li, C, Li, M, Li, X, Ni, W, Xu, Y, Yao, R, et al. Whole-genome resequencing reveals loci associated with thoracic vertebrae number in sheep. Front Genet. (2019) 10:674. doi: 10.3389/fgene.2019.00674
46. Zhang, X, Li, C, Li, X, Liu, Z, Ni, W, Hazi, W, et al. Expression profiles of MicroRNAs from multiple lumbar spine in sheep. Gene. (2018) 678:105–14. doi: 10.1016/j.gene.2018.08.020
47. Yang, H, Li, T, Zhang, N, Chen, J, Zhang, Y, Peng, S, et al. Identification of candidate genes and functional pathways associated with body size traits in Hulunbuir sheep through GWAS analysis. Genes. (2025) 16:410. doi: 10.3390/genes16040410
48. Rehman, SU, Zhen, Y, Ding, L, Saleh, AA, Zhang, Y, Zhang, J, et al. Integrative Meta-analysis: unveiling genetic factors in meat Sheep growth and muscular development through QTL and transcriptome studies. Animals. (2024) 14:1679. doi: 10.3390/ani14111679
49. Li, T, Jin, M, Wang, H, Zhang, W, Yuan, Z, and Wei, C. Whole-genome scanning for selection signatures reveals candidate genes associated with growth and tail length in sheep. Animals. (2024) 14:687. doi: 10.3390/ani14050687
50. Dossybayev, K, Amandykova, M, Orakbayeva, A, Adylkanova, S, Kozhakhmet, A, Yergali, K, et al. Genome-wide association studies revealed several candidate genes of meat productivity in Saryarka fat-tailed coarse-wool Sheep breed. Genes. (2024) 15:1549. doi: 10.3390/genes15121549
51. Wang, Q, Xu, J, Bao, M, Wang, H, Sun, X, Ji, D, et al. Weighted gene co-expression network analysis reveals genes related to growth performance in Hu sheep. Sci Rep. (2024) 14:13043. doi: 10.1038/s41598-024-63850-x
52. Khazaei-Koohpar, H, Gholizadeh, M, Hafezian, SH, and Esmaeili-Fard, SM. Weighted single-step genome-wide association study for direct and maternal genetic effects associated with birth and weaning weights in sheep. Sci Rep. (2024) 14:13120. doi: 10.1038/s41598-024-63974-0
53. Krivoruchko, A, Surov, A, Kanibolotskaya, A, Sheludko, P, Likhovid, N, Yatsyk, O, et al. A genome-wide search of meat productivity candidate genes in Russian meat merino breed. Anim Gene. (2023) 27:200146. doi: 10.1016/j.angen.2023.200146
54. Krivoruchko, A, Likhovid, A, Kanibolotskaya, A, Saprikina, T, Safaryan, E, and Yatsyk, O. Genome-wide search for associations with meat production parameters in Karachaevsky Sheep breed using the Illumina BeadChip 600 K. Genes. (2023) 14:1288. doi: 10.3390/genes14061288
55. Ramos, Z, Garrick, DJ, Blair, HT, Vera, B, Ciappesoni, G, and Kenyon, PR. Genomic regions associated with wool, growth and reproduction traits in Uruguayan merino Sheep. Genes. (2023) 14:167. doi: 10.3390/genes14010167
56. Ceccobelli, S, Landi, V, Senczuk, G, Mastrangelo, S, Sardina, MT, Ben-Jemaa, S, et al. A comprehensive analysis of the genetic diversity and environmental adaptability in worldwide merino and merino-derived sheep breeds. Genet Sel Evol. (2023) 55:1–8. doi: 10.1186/s12711-023-00797-z
57. Lakhssassi, K, Meneses, C, Sarto, MP, Serrano, M, and Calvo, JH. Genome-wide analysis reveals that the cytochrome P450 family 7 subfamily B member 1 gene is implicated in growth traits in rasa Aragonesa ewes. Animal. (2023) 17:100975. doi: 10.1016/j.animal.2023.100975
58. Tuersuntuoheti, M, Zhang, J, Zhou, W, Zhang, CL, Liu, C, Chang, Q, et al. Exploring the growth trait molecular markers in two sheep breeds based on genome-wide association analysis. PLoS One. (2023) 18:e0283383. doi: 10.1371/journal.pone.0283383
59. Yue, C, Wang, J, Shen, Y, Zhang, J, Liu, J, Xiao, A, et al. Whole-genome DNA methylation profiling reveals epigenetic signatures in developing muscle in Tan and Hu sheep and their offspring. Front Vet Sci. (2023) 10:1186040. doi: 10.3389/fvets.2023.1186040
60. Zhao, Y, He, S, Huang, J, and Liu, M. Genome-wide association analysis of muscle pH in Texel Sheep× Altay Sheep F2 resource population. Animals. (2023) 13:2162. doi: 10.3390/ani13132162
61. Haslin, E, Corner-Thomas, RA, Kenyon, PR, Pettigrew, EJ, Hickson, RE, Morris, ST, et al. Effect of breeding heavier Romney ewe lambs at seven months of age on lamb production and efficiency over their first three breeding seasons. Animals. (2021) 11:3486. doi: 10.3390/ani11123486
62. Listyarini, K, Sumantri, C, Rahayu, S, Islam, MA, Akter, SH, Uddin, MJ, et al. Hepatic transcriptome analysis reveals genes, polymorphisms, and molecules related to lamb tenderness. Animals. (2023) 13:674. doi: 10.3390/ani13040674
63. Li, C, Li, J, Wang, H, Zhang, R, An, X, Yuan, C, et al. Genomic selection for live weight in the 14th month in alpine merino Sheep combining GWAS information. Animals. (2023) 13:3516. doi: 10.3390/ani13223516
64. Yilmaz, O, Kizilaslan, M, Arzik, Y, Behrem, S, Ata, N, Karaca, O, et al. Genome-wide association studies of preweaning growth and in vivo carcass composition traits in Esme sheep. J Anim Breed Genet. (2022) 139:26–39. doi: 10.1111/jbg.12640
65. Krivoruchko, A, Yatsyk, O, and Kanibolockaya, A. New candidate genes of high productivity in north-Caucasian sheep using genome-wide association study (GWAS). Anim Gene. (2022) 23:200119. doi: 10.1016/j.angen.2021.200119
66. Krivoruchko, A, Surov, A, Skokova, A, Kanibolotskaya, A, Saprikina, T, Kukharuk, M, et al. A genome-wide search for candidate genes of meat production in Jalgin merino considering known productivity genes. Genes. (2022) 13:1337. doi: 10.3390/genes13081337
67. Zhao, Y, Zhang, X, Li, F, Zhang, D, Zhang, Y, Li, X, et al. Whole genome sequencing analysis to identify candidate genes associated with the rib eye muscle area in Hu sheep. Front Genet. (2022) 13:824742. doi: 10.3389/fgene.2022.824742
68. Peng, H, Hu, M, Liu, Z, Lai, W, Shi, L, Zhao, Z, et al. Transcriptome analysis of the liver and muscle tissues of Dorper and small-tailed Han Sheep. Front Genet. (2022) 13:868717. doi: 10.3389/fgene.2022.868717
69. Kizilaslan, M, Arzik, Y, White, SN, Piel, LM, and Cinar, MU. Genetic parameters and genomic regions underlying growth and linear type traits in Akkaraman sheep. Genes. (2022) 13:1414. doi: 10.3390/genes13081414
70. Chen, B, Yue, Y, Li, J, Liu, J, Yuan, C, Guo, T, et al. Transcriptome-metabolome analysis reveals how sires affect meat quality in hybrid sheep populations. Front Nutr. (2022) 9:967985. doi: 10.3389/fnut.2022.967985
71. de Souza, TC, de Souza, TC, da Cruz, VA, Mourão, GB, Pedrosa, VB, Rovadoscki, GA, et al. Estimates of heritability and candidate genes for primal cuts and dressing percentage in Santa Ines sheep. Livest Sci. (2022) 264:105048. doi: 10.1016/j.livsci.2022.105048
72. Ladeira, GC, Pilonetto, F, Fernandes, AC, Bóscollo, PP, Dauria, BD, Titto, CG, et al. CNV detection and their association with growth, efficiency and carcass traits in Santa Inês sheep. J Anim Breed Genet. (2022) 139:476–87. doi: 10.1111/jbg.12671
73. Wen, Y, Li, S, Bao, G, Wang, J, Liu, X, Hu, J, et al. Comparative transcriptome analysis reveals the mechanism associated with dynamic changes in meat quality of the longissimus Thoracis muscle in Tibetan Sheep at different growth stages. Front Vet Sci. (2022) 9:926725. doi: 10.3389/fvets.2022.926725
74. Song, Y, Zhang, Q, Shi, J, Fu, L, and Cheng, S. Screening of genes related to growth, development and meat quality of Sahan crossbred F1 Sheep based on RNA-Seq technology. Front Vet Sci. (2022) 9:831519. doi: 10.3389/fvets.2022.831519
75. Liu, Z, Tan, X, Wang, J, Jin, Q, Meng, X, Cai, Z, et al. Whole genome sequencing of Luxi black head sheep for screening selection signatures associated with important traits. Animal Biosci. (2022) 35:1340–50. doi: 10.5713/ab.21.0533
76. Jin, M, Fei, X, Li, T, Lu, Z, Chu, M, Di, R, et al. Transcriptome study digs out BMP2 involved in adipogenesis in sheep tails. BMC Genomics. (2022) 23:1. doi: 10.1186/s12864-022-08657-8
77. Xu, SS, Gao, L, Shen, M, and Lyu, F. Whole-genome selective scans detect genes associated with important phenotypic traits in sheep (Ovis aries). Front Genet. (2021) 12:738879. doi: 10.3389/fgene.2021.738879
78. Zhao, B, Luo, H, Huang, X, Wei, C, Di, J, Tian, Y, et al. Integration of a single-step genome-wide association study with a multi-tissue transcriptome analysis provides novel insights into the genetic basis of wool and weight traits in sheep. Genet Sel Evol. (2021) 53:1–4. doi: 10.1186/s12711-021-00649-8
79. Tao, L, Liu, YF, Zhang, H, Li, HZ, Zhao, FP, Wang, FY, et al. Genome-wide association study and inbreeding depression on body size traits in Qira black sheep (Ovis aries). Anim Genet. (2021) 52:560–4. doi: 10.1111/age.13099
80. Almasi, M, Zamani, P, Mirhoseini, SZ, and Moradi, MH. Genome-wide association study for postweaning weight traits in Lori-Bakhtiari sheep. Trop Anim Health Prod. (2021) 53:1–8. doi: 10.1007/s11250-021-02595-5
81. Zhu, C, Li, N, Cheng, H, and Ma, Y. Genome wide association study for the identification of genes associated with tail fat deposition in Chinese sheep breeds. Biology Open. (2021) 10:bio054932. doi: 10.1242/bio.054932
82. Zlobin, AS, Nikulin, PS, Volkova, NA, Zinovieva, NA, Iolchiev, BS, Bagirov, VA, et al. Multivariate analysis identifies eight novel loci associated with meat productivity traits in sheep. Genes. (2021) 12:367. doi: 10.3390/genes12030367
83. Krivoruchko, A, Sermyagin, A, Saprikina, T, Golovanova, N, Kvochko, A, and Yatsyk, O. Genome wide associations study of single nucleotide polymorphisms with productivity parameters in Jalgin merino for identification of new candidate genes. Gene Rep. (2021) 23:101065. doi: 10.1016/j.genrep.2021.101065
84. Jiang, J, Cao, Y, Shan, H, Wu, J, Song, X, and Jiang, Y. The GWAS analysis of body size and population verification of related SNPs in Hu sheep. Front Genet. (2021) 12:642552. doi: 10.3389/fgene.2021.642552
85. Abousoliman, I, Reyer, H, Oster, M, Murani, E, Mohamed, I, and Wimmers, K. Genome-wide analysis for early growth-related traits of the locally adapted Egyptian Barki Sheep. Genes. (2021) 12:1243. doi: 10.3390/genes12081243
86. Gunawan, A, Listyarini, K, Harahap, RS, Jakaria,, Roosita, K, Sumantri, C, et al. Hepatic transcriptome analysis identifies genes, polymorphisms and pathways involved in the fatty acids metabolism in sheep. PLoS One. (2021) 16:e0260514. doi: 10.1371/journal.pone.0260514
87. Shi, J, Wang, X, Song, Y, Liu, T, Cheng, S, and Zhang, Q. Excavation of genes related to the mining of growth, development, and meat quality of two crossbred sheep populations based on comparative transcriptomes. Animals. (2021) 11:1492. doi: 10.3390/ani11061492
88. Chen, ZH, Xu, YX, Xie, XL, Wang, DF, Aguilar-Gómez, D, Liu, GJ, et al. Whole-genome sequence analysis unveils different origins of European and Asiatic mouflon and domestication-related genes in sheep. Commun Biol. (2021) 4:1307. doi: 10.1038/s42003-021-02817-4
89. Cao, Y, Song, X, Shan, H, Jiang, J, Xiong, P, Wu, J, et al. Genome-wide association study of body weights in Hu Sheep and population verification of related single-nucleotide polymorphisms. Front Genet. (2020) 11:588. doi: 10.3389/fgene.2020.00588
90. Baazaoui, I, Bedhiaf-Romdhani, S, Mastrangelo, S, and Ciani, E. Genome-wide analyses reveal population structure and identify candidate genes associated with tail fatness in local sheep from a semi-arid area. Animal. (2021) 15:100193. doi: 10.1016/j.animal.2021.100193
91. Tao, L, He, XY, Pan, LX, Wang, JW, Gan, SQ, and Chu, MX. Genome-wide association study of body weight and conformation traits in neonatal sheep. Anim Genet. (2020) 51:336–40. doi: 10.1111/age.12904
92. Cheng, S, Wang, X, Zhang, Q, He, Y, Zhang, X, Yang, L, et al. Comparative transcriptome analysis identifying the different molecular genetic markers related to production performance and meat quality in longissimus dorsi tissues of MG× STH and STH sheep. Genes. (2020) 11:183. doi: 10.3390/genes11020183
93. Lu, Z, Yue, Y, Yuan, C, Liu, J, Chen, Z, Niu, C, et al. Genome-wide association study of body weight traits in chinese fine-wool sheep. Animals. (2020) 10:170. doi: 10.3390/ani10010170
94. Wang, Z, Guo, J, Guo, Y, Yang, Y, Teng, T, Yu, Q, et al. Genome-wide detection of CNVs and association with body weight in Sheep based on 600K SNP arrays. Front Genet. (2020) 11:558. doi: 10.3389/fgene.2020.00558
95. Bakhtiarizadeh, MR, and Alamouti, AA. RNA-Seq based genetic variant discovery provides new insights into controlling fat deposition in the tail of sheep. Sci Rep. (2020) 10:13525. doi: 10.1038/s41598-020-70527-8
96. Arora, R, Fairoze, MN, Kaur, M, Sharma, A, Girdhar, Y, Devatkal, SK, et al. Transcriptome profiling of longissimus thoracis muscles identifies highly connected differentially expressed genes in meat type sheep of India. PLoS One. (2019) 14:e0217461. doi: 10.1371/journal.pone.0217461
97. Zhang, T, Gao, H, Sahana, G, Zan, Y, Fan, H, Liu, J, et al. Genome-wide association studies revealed candidate genes for tail fat deposition and body size in the Hulun Buir sheep. J Anim Breed Genet. (2019) 136:362–70. doi: 10.1111/jbg.12402
98. Ghasemi, M, Zamani, P, Vatankhah, M, and Abdoli, R. Genome-wide association study of birth weight in sheep. Animal. (2019) 13:1797–803. doi: 10.1017/S1751731118003610
99. Mastrangelo, S, Moioli, B, Ahbara, A, Latairish, S, Portolano, B, Pilla, F, et al. Genome-wide scan of fat-tail sheep identifies signals of selection for fat deposition and adaptation. Anim Prod Sci. (2018) 59:835–48. doi: 10.1071/AN17753
100. Rovadoscki, GA, Pertile, SF, Alvarenga, AB, Cesar, AS, Pértille, F, Petrini, J, et al. Estimates of genomic heritability and genome-wide association study for fatty acids profile in Santa Inês sheep. BMC Genomics. (2018) 19:1–4. doi: 10.1186/s12864-018-4777-8
101. Pasandideh, M, Rahimi-Mianji, G, and Gholizadeh, M. A genome scan for quantitative trait loci affecting average daily gain and Kleiber ratio in Baluchi Sheep. J Genet. (2018) 97:493–503. doi: 10.1007/s12041-018-0941-9
102. Zhang, B, Zhao, W, Tang, X, Zhou, M, Qiu, Y, Wang, S, et al. Identification and analysis of InDel variants in key hippo pathway genes and their association with growth traits in four Chinese Sheep breeds. Vet Sci. (2025) 12:283. doi: 10.3390/vetsci12030283
103. Cao, X, Liu, Y, Cheng, J, Ling, C, Huang, J, and Sun, W. Copy number variations of the NSMF gene and Their associations with growth traits in three Chinese Sheep breeds. Genes. (2025) 16:218. doi: 10.3390/genes16020218
104. Xiao, C, Liu, Y, Zhao, W, Liang, Y, Cui, C, Yang, S, et al. The comparison of meat yield, quality, and flavor between small-tailed Han sheep and two crossbred sheep and the verification of related candidate genes. Front Nutr. (2024) 11:1399390. doi: 10.3389/fnut.2024.1399390
105. Fu, L, Shi, J, Meng, Q, Tang, Z, Liu, T, Zhang, Q, et al. Verification of key target molecules for intramuscular fat deposition and screening of SNP sites in Sheep from small-tail Han Sheep breed and its cross with Suffolk. Int J Mol Sci. (2024) 25:2951. doi: 10.3390/ijms25052951
106. Pan, Y, Li, S, Zhang, Q, Li, J, Song, C, Kong, L, et al. Production performance analysis of sheep MSTN gene C2361T locus. J Genet Eng Biotechnol. (2024) 22:100372. doi: 10.1016/j.jgeb.2024.100372
107. Aljubouri, TR, and Al-Shuhaib, MB. A missense SNP in the proopiomelanocortin (POMC) gene is associated with growth traits in Awassi and karakul sheep. Anim Biotechnol. (2023) 34:4837–4850. doi: 10.1080/10495398.2023.2197469
108. Al-Jumaili, WS, Kadhim, AH, and Al-Thuwaini, TM. Polymorphism of the ADIPOQ gene and its association with productive traits in Awassi ewes. Mol Biol Rep. (2023) 50:913–7. doi: 10.1007/s11033-022-07975-0
109. Liu, T, Bi, Y, Bao, J, Shang, M, Hu, W, and Zhang, L. Single nucleotide polymorphisms in the CDH18 gene affect growth traits in Hu sheep. Animal Res One Health. (2023). doi: 10.1002/aro2.22
110. El-Mansy, SA, Naiel, MA, El-Naser, IA, De Waard, M, Babalghith, AO, Ogaly, HA, et al. The growth hormone gene polymorphism and its relationship to performance and carcass features in Egyptian Awassi lambs. Heliyon. (2023) 9:e14194. doi: 10.1016/j.heliyon.2023.e14194
111. Harahap, RS, Noor, RR, and Gunawan, A. The polymorphism and expression of CYP2E1 gene and its relation to carcass and meat quality of Indonesian lamb. Trop Anim Sci J. (2021) 44:377–85. doi: 10.5398/tasj.2021.44.4.377
112. Kader Esen, V, and Esen, S. Association of the IGF1 5′ UTR polymorphism in meat-type Sheep breeds considering growth, body size, slaughter, and meat quality traits in Turkey. Vet Sci. (2023) 10:270. doi: 10.3390/vetsci10040270
113. Lin, C, Li, F, Zhang, X, Zhang, D, Li, X, Zhang, Y, et al. Expression and polymorphisms of CD8B gene and its associations with body weight and size traits in sheep. Anim Biotechnol. (2023) 34:1214–22. doi: 10.1080/10495398.2021.2016432
114. Zhao, L, Wang, W, Wang, X, Zhang, D, Li, X, Zhao, Y, et al. Identification of SNPs and expression patterns of ALB, AHSG and GC genes and their association with growth traits in Hu sheep. Gene. (2023) 853:147100. doi: 10.1016/j.gene.2022.147100
115. Yuan, Z, Ge, L, Su, P, Gu, Y, Chen, W, Cao, X, et al. NCAPG regulates myogenesis in sheep, and SNPs located in its putative promoter region are associated with growth and development traits. Animals. (2023) 13:3173. doi: 10.3390/ani13203173
116. Liu, H, Xu, H, Lan, X, Cao, X, and Pan, C. The indel variants of sheep IGF2BP1 gene are associated with growth traits. Anim Biotechnol. (2023) 34:134–42. doi: 10.1080/10495398.2021.1942029
117. Zeng, X, Wang, W, Zhang, D, Li, X, Zhang, Y, Zhao, Y, et al. Polymorphism and expression level of the FADS3 gene and associated with the growth traits in Hu sheep. Anim Biotechnol. (2023):1.
118. Wang, C, Yuan, Z, Hu, R, Li, F, and Yue, X. Association of SNPs within PTPN3 gene with wool production and growth traits in a dual-purpose sheep population. Anim Biotechnol. (2023) 34:1429–35. doi: 10.1080/10495398.2022.2029465
119. Wang, Y, Li, YX, Zhang, J, Qian, Y, Meng, CH, Zhong, JF, et al. PLAG1 g. 8795C> T mutation regulates early body weight in Hu Sheep by weakening miR-139 binding. Genes. (2023) 14:467. doi: 10.3390/genes14020467
120. Wang, J, Zhang, X, Wang, X, Li, F, Zhang, D, Li, X, et al. Polymorphism and expression of the HMGA1 gene and association with tail fat deposition in Hu sheep. Anim Biotechnol. (2023) 34:1626–34. doi: 10.1080/10495398.2021.1998093
121. Puruhita,, Noor, RR, Margawati, ET, and Raadsma, HW. Association of the single nucleotide polymorphism in CAPN3 gene with growth performance in merino and Garut (MEGA) backcross sheep. J Genet Eng Biotechnol. (2023) 21:77. doi: 10.1186/s43141-023-00524-7
122. Zlobin, AS, Volkova, NA, Zinovieva, NA, Iolchiev, BS, Bagirov, VA, Borodin, PM, et al. Loci associated with negative Heterosis for viability and meat productivity in interspecific Sheep hybrids. Animals. (2023) 13:184. doi: 10.3390/ani13010184
123. Luo, Y, Akhatayeva, Z, Mao, C, Jiang, F, Guo, Z, Xu, H, et al. The ovine HIAT1 gene: mRNA expression, InDel mutations, and growth trait associations. Front Vet Sci. (2023) 10:1134903. doi: 10.3389/fvets.2023.1134903
124. Luo, Y, Zhang, M, Guo, Z, Wijayanti, D, Xu, H, Jiang, F, et al. Insertion/deletion (InDel) variants within the Sheep fat-deposition-related PDGFD gene strongly affect morphological traits. Animals. (2023) 13:1485. doi: 10.3390/ani13091485
125. Li, W, Wang, X, Zhang, X, Li, F, Zhang, D, Li, X, et al. Polymorphism of sheep PRKAA2 gene and its association with growth traits. Anim Biotechnol. (2023) 34:1324–30. doi: 10.1080/10495398.2021.2021215
126. Xu, D, Wang, X, Wang, W, Zhang, D, Li, X, Zhang, Y, et al. Detection of single nucleotide polymorphism in HTR4 and its relationship with growth traits in sheep. Anim Biotechnol. (2023) 34:1–8. doi: 10.1080/10495398.2023.2174877
127. Zhang, J, Toremurat, Z, Liang, Y, Cheng, J, Sun, Z, Huang, Y, et al. Study on the association between LRRC8B gene InDel and sheep body conformation traits. Genes. (2023) 14:356. doi: 10.3390/genes14020356
128. Wen, Y, Wang, E, Wang, X, Qing, S, Chaogetu, B, Wang, C, et al. Copy number variations of LRRFIP1 gene and the relationship with growth traits in four Chinese sheep. Anim Biotechnol. (2023) 34:3008–15. doi: 10.1080/10495398.2022.2126981
129. Bayraktar, M, Durmuş, M, and Al-Shuhaib, MB. Identification of two novel SNPs in the myocyte enhancer factor 2B (MEF2B) gene and its association with growth traits in two breeds of Turkish sheep. Small Rumin Res. (2023) 218:106867. doi: 10.1016/j.smallrumres.2022.106867
130. Margawati, ET, Putra, WP, Rizki, M, Soetrisno, E, and Raadsma, HW. Detection of carrier Booroola (FecB) allele in BMPR1B gene of MEGA (merino× Garut) sheep and its association with growth traits. J Genet Eng Biotechnol. (2023) 21:1–7. doi: 10.1186/s43141-023-00475-z
131. Yang, X, Wang, W, Zhang, D, Li, X, Zhang, Y, Zhao, Y, et al. Genetic polymorphism of the ovine MAP3K5 gene and its association with body size traits in Hu sheep of China. Arch Anim Breed. (2023) 66:71–9. doi: 10.5194/aab-66-71-2023
132. Wang, X, Li, J, Bai, J, Chen, M, Wang, L, Fan, H, et al. Exploring the impact of insertion/deletion in FTO and PLIN1 genes on morphometric traits in Sheep. Animals. (2023) 13:3032. doi: 10.3390/ani13193032
133. Wang, X, Wang, Y, Cao, X, Huang, Y, Li, P, Lan, X, et al. Copy number variations of the KAT6A gene are associated with body measurements of Chinese sheep breeds. Anim Biotechnol. (2023) 34:947–54. doi: 10.1080/10495398.2021.2005616
134. Guo, X, Li, T, Lu, D, Yamada, T, Li, X, Bao, S, et al. Effects of the expressions and variants of the CAST gene on the fatty acid composition of the longissimus Thoracis muscle of grazing Sonid Sheep. Animals. (2023) 13:195. doi: 10.3390/ani13020195
135. Pasandideh, M, Harkinezhad, T, and Mohammadi, L. A SNP in the ovine cathepsin K (CTSK) gene is associated with yearling growth performance in a crossbred sheep population. Anim Biotechnol. (2023) 34:5155–9. doi: 10.1080/10495398.2023.2174873
136. Talebi, R, Ahmadi, A, Hajiloei, Z, Ghaffari, MR, Zeinalabedini, M, Saki, AA, et al. Association of ovine follistatin gene polymorphisms with body measurements, fat-tail traits and morphometric of head in Iranian Mehraban sheep. Small Rumin Res. (2023) 225:107020. doi: 10.1016/j.smallrumres.2023.107020
137. Kong, L, Yue, Y, Li, J, Yang, B, Chen, B, Liu, J, et al. Transcriptomics and metabolomics reveal improved performance of Hu sheep on hybridization with Southdown sheep. Food Res Int. (2023) 173:113240. doi: 10.1016/j.foodres.2023.113240
138. Yang, M, Zhao, W, Wang, Z, Liu, J, Sun, X, and Wang, S. Detection of key gene InDels in JAK/STAT pathway and their associations with growth traits in four Chinese sheep breeds. Gene. (2023) 888:147750. doi: 10.1016/j.gene.2023.147750
139. Bayraktar, M, and Shoshin, O. Estimation of the associations between GH and DGAT1 genes and growth traits by using decision tree in Awassi sheep. Anim Biotechnol. (2022) 33:167–73. doi: 10.1080/10495398.2021.1975727
140. Bayraktar, M, and Shoshin, O. Estimate of the association of IGF-I and IGFALS genes with growth traits in Hamdani sheep. Braz Arch Biol Technol. (2022) 64:e21210262. doi: 10.1590/1678-4324-2021210262
141. Bayraktar, M, and Shoshin, O. Association between CAST and MSTN gene polymorphisms with growth traits in Awassi sheep. Kuwait J Sci. (2022) 49:1–15. doi: 10.48129/kjs.10955
142. Wijayanti, D, Erdenee, S, Akhatayeva, Z, Li, H, Li, J, Cai, Y, et al. Genetic polymorphisms within the ETAA1 gene associated with growth traits in Chinese sheep breeds. Anim Genet. (2022) 53:460–5. doi: 10.1111/age.13197
143. Listyarini, K, Sumantri, C, Rahayu, S, Uddin, MJ, and Gunawan, A. Association study and expression analysis of olfactomedin like 3 gene related to meat quality, carcass characteristics, retail meat cut, and fatty acid composition in sheep. Animal Biosci. (2022) 35:1489–98. doi: 10.5713/ab.21.0406
144. Ding, N, Tian, D, Li, X, Zhang, Z, Tian, F, Liu, S, et al. Genetic polymorphisms of IGF1 and IGF1R genes and their effects on growth traits in Hulun Buir sheep. Genes. (2022) 13:666. doi: 10.3390/genes13040666
145. Dai, R, Zhou, H, Fang, Q, Zhou, P, Yang, Y, Jiang, S, et al. Variation in ovine DGAT1 and its association with carcass muscle traits in Southdown sheep. Genes. (2022) 13:1670. doi: 10.3390/genes13091670
146. Karadag, O. The polymorphism of insulin-like growth factor-1 receptor (IGF-1R) gene in meat-type lambs in Turkey: I. Effect on growth traits and body measurements. Small Rumin Res. (2022) 215:106765. doi: 10.1016/j.smallrumres.2022.106765
147. Zhao, L, Li, F, Liu, T, Yuan, L, Zhang, X, Zhang, D, et al. Ovine ELOVL5 and FASN genes polymorphisms and their correlations with sheep tail fat deposition. Gene. (2022) 807:145954. doi: 10.1016/j.gene.2021.145954
148. Zhao, L, Li, F, Yuan, L, Zhang, X, Zhang, D, Li, X, et al. Expression of ovine CTNNA3 and CAP2 genes and their association with growth traits. Gene. (2022) 807:145949. doi: 10.1016/j.gene.2021.145949
149. Kalds, P, Huang, S, Chen, Y, and Wang, X. Ovine HOXB13: expanding the gene repertoire of sheep tail patterning and implications in genetic improvement. Commun Biol. (2022) 5:1196. doi: 10.1038/s42003-022-04199-7
150. Lagler, DK, Hannemann, E, Eck, K, Klawatsch, J, Seichter, D, Russ, I, et al. Fine-mapping and identification of candidate causal genes for tail length in the Merinolandschaf breed. Commun Biol. (2022) 5:918. doi: 10.1038/s42003-022-03854-3
151. Pan, Y, Wang, M, Wu, H, Akhatayeva, Z, Lan, X, Fei, P, et al. Indel mutations of sheep PLAG1 gene and their associations with growth traits. Anim Biotechnol. (2022) 33:1459–65. doi: 10.1080/10495398.2021.1906265
152. Zhang, Z, Liu, C, Hao, W, Yin, W, Ai, S, Zhao, Y, et al. Novel single nucleotide polymorphisms and haplotype of MYF5 gene are associated with body measurements and ultrasound traits in grassland short-tailed sheep. Genes. (2022) 13:483. doi: 10.3390/genes13030483
153. Akhatayeva, Z, Li, H, Mao, C, Cheng, H, Zhang, G, Jiang, F, et al. Detecting novel Indel variants within the GHR gene and their associations with growth traits in Luxi blackhead sheep. Anim Biotechnol. (2022) 33:214–22. doi: 10.1080/10495398.2020.1784184
154. Zhai, R, Wang, W, Zhang, D, Li, X, Zhang, Y, Zhao, Y, et al. Novel polymorphism at KLF15 gene and its association with growth traits in Hu sheep. Anim Biotechnol. (2022) 34:1–7. doi: 10.1080/10495398.2022.2138413
155. Esen, VK, and Elmacı, C. Effect of growth hormone exon-5 polymorphism on growth traits, body measurements, slaughter and carcass characteristics, and meat quality in meat-type lambs in Turkey. Ruminants. (2022) 2:420–34. doi: 10.3390/ruminants2040029
156. Chen, P, Zhao, H, Wu, M, He, S, Yuan, T, Yi, X, et al. A novel 17 bp InDel polymorphism within the PPARGC1A gene is significantly associated with growth traits in sheep. Anim Biotechnol. (2022) 33:312–20. doi: 10.1080/10495398.2020.1796697
157. Cui, P, Wang, W, Zhang, D, Li, C, Huang, Y, Ma, Z, et al. Identification of TRAPPC9 and BAIAP2 gene polymorphisms and their association with fat deposition-related traits in Hu sheep. Front Vet Sci. (2022) 9:928375. doi: 10.3389/fvets.2022.928375
158. Ma, Z, Wang, W, Zhang, D, Zhang, Y, Zhao, Y, Li, X, et al. Ovine RAP1GAP and rBAT gene polymorphisms and their association with tail fat deposition in Hu sheep. Front Vet Sci. (2022) 9:974513. doi: 10.3389/fvets.2022.974513
159. Valencia, CP, Franco, LÁ, and Herrera, DH. Association of single nucleotide polymorphisms in the CAPN, CAST, LEP, GH, and IGF-1 genes with growth parameters and ultrasound characteristics of the longissimus dorsi muscle in Colombian hair sheep. Trop Anim Health Prod. (2022) 54:82. doi: 10.1007/s11250-022-03086-x
160. Esen, VK, Esen, S, Karadağ, O, Önenç, A, and Elmaci, C. Genotypic characterization of meat-type lambs expressing the callipyge gene in Turkey: I. Carcass characteristics and retail yield. Turk J Vet Anim Sci. (2022) 46:157–64. doi: 10.3906/vet-2112-7
161. Zhao, Y, Zhang, D, Zhang, X, Li, F, Xu, D, Zhao, L, et al. Expression features of the ovine FTO gene and association between FTO polymorphism and tail fat deposition related-traits in Hu sheep. Gene. (2022) 826:146451. doi: 10.1016/j.gene.2022.146451
162. Toremurat, Z, Ibrahim, EE, Huang, YZ, Lan, X, Pi, L, Chaogetu, B, et al. Copy number variations of TOP2B gene are associated with growth traits in Chinese sheep breeds. Anim Biotechnol. (2022) 33:85–9. doi: 10.1080/10495398.2020.1773490
163. Sousa-Junior, LP, Meira, AN, Azevedo, HC, Muniz, EN, Coutinho, LL, Mourão, GB, et al. Variants in myostatin and MyoD family genes are associated with meat quality traits in Santa Inês sheep. Anim Biotechnol. (2022) 33:201–13. doi: 10.1080/10495398.2020.1781651
164. Li, X, Ding, N, Zhang, Z, Tian, D, Han, B, Liu, D, et al. Identification of SSTR5 gene polymorphisms and their association with growth traits in Hulun Buir Sheep. Front Genet. (2022) 13:831599. doi: 10.3389/fgene.2022.831599
165. Zhang, D, Zhang, X, Li, F, La, Y, Li, G, Zhang, Y, et al. The association of polymorphisms in the ovine PPARGC1B and ZEB2 genes with body weight in Hu sheep. Anim Biotechnol. (2022) 33:90–7. doi: 10.1080/10495398.2020.1775626
166. Yang, Z, Cao, X, Ma, Y, Cheng, J, Song, C, Jiang, R, et al. Novel copy number variation of the BAG4 gene is associated with growth traits in three Chinese sheep populations. Anim Biotechnol. (2021) 32:461–9. doi: 10.1080/10495398.2020.1719124
167. Ibrahim, AH. Polymorphisms in hormone-sensitive lipase and leptin receptor genes and their association with growth traits in Barki lambs. Vet World. (2021) 14:515. doi: 10.14202/vetworld.2021.515-522
168. Mao, C, Akhatayeva, Z, Cheng, H, Zhang, G, Jiang, F, Meng, X, et al. A novel 23 bp indel mutation in PRL gene is associated with growth traits in Luxi blackhead sheep. Anim Biotechnol. (2021) 32:740–7. doi: 10.1080/10495398.2020.1753757
169. Erdenee, S, Akhatayeva, Z, Pan, C, Cai, Y, Xu, H, Chen, H, et al. An insertion/deletion within the CREB1 gene identified using the RNA-sequencing is associated with sheep body morphometric traits. Gene. (2021) 775:145444. doi: 10.1016/j.gene.2021.145444
170. Aljubouri, TR, Hassan, AF, Al-Shuhaib, MB, and Mahyari, SA. Association of GnRH1 gene with growth traits in two breeds of sheep. Agric Res. (2021) 10:285–93. doi: 10.1007/s40003-020-00501-3
171. Luo, R, Zhang, X, Wang, L, Zhang, L, Li, G, and Zheng, Z. GLIS1, a potential candidate gene affect fat deposition in sheep tail. Mol Biol Rep. (2021) 48:4925–31. doi: 10.1007/s11033-021-06468-w
172. Pewan, SB, Otto, JR, Huerlimann, R, Budd, AM, Mwangi, FW, Edmunds, RC, et al. Next generation sequencing of single nucleotide polymorphic DNA-markers in selecting for intramuscular fat, fat melting point, omega-3 long-chain polyunsaturated fatty acids and meat eating quality in Tattykeel Australian White MARGRA lamb. Food Secur. (2021) 10:2288. doi: 10.3390/foods10102288
173. Wang, S, Liu, S, Yuan, T, and Sun, X. Genetic effects of FTO gene insertion/deletion (InDel) on fat-tail measurements and growth traits in Tong sheep. Anim Biotechnol. (2021) 32:229–39. doi: 10.1080/10495398.2019.1680379
174. Grochowska, E, Lisiak, D, Akram, MZ, Adeniyi, OO, Lühken, G, and Borys, B. Association of a polymorphism in exon 3 of the IGF1R gene with growth, body size, slaughter and meat quality traits in colored polish merino sheep. Meat Sci. (2021) 172:108314. doi: 10.1016/j.meatsci.2020.108314
175. Zhao, H, Hu, R, Li, F, and Yue, X. Five SNPs within the FGF5 gene significantly affect both wool traits and growth performance in fine-wool sheep (Ovis aries). Front Genet. (2021) 12:732097. doi: 10.3389/fgene.2021.732097
176. Cheng, J, Zhang, X, Li, F, Yuan, L, Zhang, D, Zhang, Y, et al. Detecting single nucleotide polymorphisms in MEF2B and UCP3 and elucidating their association with sheep growth traits. DNA Cell Biol. (2021) 40:1554–62. doi: 10.1089/dna.2021.0782
177. Yuan, Z, Sunduimijid, B, Xiang, R, Behrendt, R, Knight, MI, Mason, BA, et al. Expression quantitative trait loci in sheep liver and muscle contribute to variations in meat traits. Genet Sel Evol. (2021) 53:1–4. doi: 10.1186/s12711-021-00602-9
178. Li, S, Zhou, H, Zhao, F, Fang, Q, Wang, J, Liu, X, et al. Nucleotide sequence variation in the insulin-like growth factor 1 gene affects growth and carcass traits in New Zealand Romney sheep. DNA Cell Biol. (2021) 40:265–71. doi: 10.1089/dna.2020.6166
179. Chong, Y, Liu, G, Girmay, S, and Jiang, X. Novel mutations in the signal transducer and activator of transcription 3 gene are associated with sheep body weight and fatness traits. Mamm Genome. (2021) 32:38–49. doi: 10.1007/s00335-020-09850-4
180. Li, X, Ding, N, Zhang, Z, Tian, D, Han, B, Liu, S, et al. Identification of somatostatin receptor subtype 1 (SSTR1) gene polymorphism and their association with growth traits in Hulun Buir sheep. Genes. (2021) 13:77. doi: 10.3390/genes13010077
181. Harahap, RS, Noor, RR, and Gunawan, A. Effect of CYP2E1 gene polymorphisms on lamb odor and flavor in Indonesian sheep. IOP Conf Series Earth Environ Sci. (2021) 788:012022. doi: 10.1088/1755-1315/788/1/012022
182. Karaca, S, Ser, G, Ülker, H, Yılmaz, O, Çakmakçı, C, Ata, N, et al. Associations between CYP17 gene polymorphisms, temperament and maternal behavior in ewes, and growth in their lambs. J Vet Behav. (2021) 45:1–9. doi: 10.1016/j.jveb.2021.05.004
183. Osman, NM, Shafey, HI, Abdelhafez, MA, Sallam, AM, and Mahrous, KF. Genetic variations in the Myostatin gene affecting growth traits in sheep. Vet World. (2021) 14:475–82. doi: 10.14202/vetworld.2021.475-482
184. Feng, Z, Li, X, Cheng, J, Jiang, R, Huang, R, Wang, D, et al. Copy number variation of the PIGY gene in sheep and its association analysis with growth traits. Animals. (2020) 10:688. doi: 10.3390/ani10040688
185. Abousoliman, I, Reyer, H, Oster, M, Muráni, E, Mourad, M, Abdel-Salam Rashed, M, et al. Analysis of candidate genes for growth and milk performance traits in the Egyptian Barki sheep. Animals. (2020) 10:197. doi: 10.3390/ani10020197
186. Cheng, J, Jiang, R, Yang, Y, Cao, X, Huang, Y, Lan, X, et al. Association analysis of KMT2D copy number variation as a positional candidate for growth traits. Gene. (2020) 753:144799. doi: 10.1016/j.gene.2020.144799
187. Li, X, Yang, JI, Shen, M, Xie, XL, Liu, GJ, Xu, YX, et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat Commun. (2020) 11:2815. doi: 10.1038/s41467-020-16485-1
188. Wu, M, Zhao, H, Tang, X, Li, Q, Yi, X, Liu, S, et al. Novel InDels of GHR, GHRH, GHRHR and their association with growth traits in seven Chinese sheep breeds. Animals. (2020) 10:1883. doi: 10.3390/ani10101883
189. Alizadeh, F, Moradian, F, and Farhadi, A. Association of allelic polymorphisms of IGFALS gene with growth traits in Makouei and Ghezel sheep breeds. Trop Anim Health Prod. (2020) 52:3027–34. doi: 10.1007/s11250-020-02321-7
190. Xu, H, Li, H, Wang, Z, Abudureyimu, A, Yang, J, Cao, X, et al. A deletion downstream of the CHCHD7 gene is associated with growth traits in sheep. Animals. (2020) 10:1472. doi: 10.3390/ani10091472
191. Wang, G, Zhou, H, Gong, H, He, J, Luo, Y, Hickford, JG, et al. Variation in the Lipin 1 gene is associated with birth weight and selected carcass traits in New Zealand Romney sheep. Animals. (2020) 10:237. doi: 10.3390/ani10020237
192. Dong, K, Yang, M, Han, J, Ma, Q, Han, J, Song, Z, et al. Genomic analysis of worldwide sheep breeds reveals PDGFD as a major target of fat-tail selection in sheep. BMC Genomics. (2020) 21:1–2. doi: 10.1186/s12864-020-07210-9
193. Lu, Z, Liu, J, Han, J, and Yang, B. Association between BMP2 functional polymorphisms and sheep tail type. Animals. (2020) 10:739. doi: 10.3390/ani10040739
194. Al-Thuwaini, TM, Al-Shuhaib, MB, Lepretre, F, and Mahdi, ZA. Co-inherited novel SNPs of the LIPE gene associated with increased carcass dressing and decreased fat-tail weight in Awassi breed. Trop Anim Health Prod. (2020) 52:3631–8. doi: 10.1007/s11250-020-02400-9
195. Dzomba, EF, Chimonyo, M, Snyman, MA, and Muchadeyi, FC. The genomic architecture of south African mutton, pelt, dual-purpose and nondescript sheep breeds relative to global sheep populations. Anim Genet. (2020) 51:910–23. doi: 10.1111/age.12991
196. Xu, H, Zhang, X, Zang, R, Cai, Y, Cao, X, Yang, J, et al. Genetic variations in the sheep SIRT7 gene and their correlation with body size traits. Arch Animal Breed. (2019) 62:189–97. doi: 10.5194/aab-62-189-2019
197. Erdenee, S, Li, J, Kang, Z, Xu, H, Zang, R, Cao, X, et al. Sheep zinc finger proteins 395 (ZNF395): insertion/deletion variations, associations with growth traits, and mRNA expression. Anim Biotechnol. (2019) 31:1–8. doi: 10.1080/10495398.2019.1585865
198. Jawasreh, KI, Al-Amareen, AH, and Aad, PY. Relationships between Hha 1 Calpastatin gene polymorphism, growth performance, and meat characteristics of Awassi Sheep. Animals. (2019) 9:667. doi: 10.3390/ani9090667
199. Meira, AN, Montenegro, H, Coutinho, LL, Mourão, GB, Azevedo, HC, Muniz, EN, et al. Single nucleotide polymorphisms in the growth hormone and IGF type-1 (IGF1) genes associated with carcass traits in Santa Ines sheep. Animal. (2019) 13:460–8. doi: 10.1017/S1751731118001362
200. Ekegbu, UJ, Burrows, L, Amirpour-Najafabadi, H, Zhou, H, and Hickford, JG. Gene polymorphisms in PROP1 associated with growth traits in sheep. Gene. (2019) 683:41–6. doi: 10.1016/j.gene.2018.10.024
201. Kolenda, M, Grochowska, E, Milewski, S, and Mroczkowski, S. The association between the polymorphism in the myostatin gene and growth traits in Kamieniec and Pomeranian sheep breeds. Small Rumin Res. (2019) 177:29–35. doi: 10.1016/j.smallrumres.2019.06.007
202. Zlobin, AS, Volkova, NA, Borodin, PM, Aksenovich, TI, and Tsepilov, YA. Recent advances in understanding genetic variants associated with growth, carcass and meat productivity traits in sheep (Ovis aries): an update. Arch Animal Breed. (2019) 62:579–83. doi: 10.5194/aab-62-579-2019
203. Grochowska, E, Borys, B, Lisiak, D, and Mroczkowski, S. Genotypic and allelic effects of the myostatin gene (MSTN) on carcass, meat quality, and biometric traits in colored polish merino sheep. Meat Sci. (2019) 151:4–17. doi: 10.1016/j.meatsci.2018.12.010
204. La, Y, Zhang, X, Li, F, Zhang, D, Li, C, Mo, F, et al. Molecular characterization and expression of SPP1, LAP3 and LCORL and their association with growth traits in sheep. Genes. (2019) 10:616. doi: 10.3390/genes10080616
205. Greguła-Kania, M, Gruszecki, TM, Junkuszew, A, Juszczuk-Kubiak, E, and Florek, M. Association of CAST gene polymorphism with carcass value and meat quality in two synthetic lines of sheep. Meat Sci. (2019) 154:69–74. doi: 10.1016/j.meatsci.2019.04.007
206. Shishay, G, Liu, G, Jiang, X, Yu, Y, Teketay, W, Du, D, et al. Variation in the promoter region of the MC4R gene elucidates the association of body measurement traits in hu sheep. Int J Mol Sci. (2019) 20:240. doi: 10.3390/ijms20020240
207. Wang, X, Cao, X, Wen, Y, Ma, Y, Elnour, IE, Huang, Y, et al. Associations of ORMDL1 gene copy number variations with growth traits in four Chinese sheep breeds. Arch Animal Breed. (2019) 62:571–8. doi: 10.5194/aab-62-571-2019
208. Ibrahim, AH. Association of growth performance and body conformational traits with BMP4 gene variation in Barki lambs. Growth Factors. (2019) 37:153–63. doi: 10.1080/08977194.2019.1662417
209. Zhao, H, He, S, Wang, S, Zhu, Y, Xu, H, Luo, R, et al. Two new insertion/deletion variants of the PITX2 gene and their effects on growth traits in sheep. Anim Biotechnol. (2018) 29:276–82. doi: 10.1080/10495398.2017.1379415
210. Armstrong, E, Ciappesoni, G, Iriarte, W, Da Silva, C, Macedo, F, Navajas, EA, et al. Novel genetic polymorphisms associated with carcass traits in grazing Texel sheep. Meat Sci. (2018) 145:202–8. doi: 10.1016/j.meatsci.2018.06.014
211. Li, J, Erdenee, S, Zhang, S, Wei, Z, Zhang, M, Jin, Y, et al. Genetic effects of PRNP gene insertion/deletion (indel) on phenotypic traits in sheep. Prion. (2018) 12:42–53. doi: 10.1080/19336896.2017.1405886
212. Zhao, F, Zhou, H, Li, S, Fang, Q, Luo, Y, and Hickford, JG. Growth and carcass trait association with variation in the somatostatin receptor 1 (SSTR1) gene in New Zealand Romney sheep. N Z J Agric Res. (2018) 61:477–86. doi: 10.1080/00288233.2017.1415942
213. Gunawan, A, Anggrela, D, Listyarini, K, Abuzahra, MA, Jakaria, J, Yamin, M, et al. Identification of single nucleotide polymorphism and pathway analysis of apolipoprotein A5 (APOA5) related to fatty acid traits in Indonesian sheep. Trop Anim Sci J. (2018) 41:165–73. doi: 10.5398/tasj.2018.41.3.165
214. An, Q, Zhou, H, Hu, J, Luo, Y, and Hickford, JG. Sequence and haplotypes variation of the ovine uncoupling protein-1 gene (UCP1) and their association with growth and carcass traits in New Zealand Romney lambs. Genes. (2018) 9:189. doi: 10.3390/genes9040189
Keywords: meat production, carcass weight, vertebral traits, small ruminants, genetic markers
Citation: Han Y, Akhtar MF, Chen W, Liu X, Zhao M, Shi L, Khan MZ and Wang C (2025) Potential candidate genes influencing meat production phenotypic traits in sheep: a review. Front. Vet. Sci. 12:1616533. doi: 10.3389/fvets.2025.1616533
Edited by:
Zhihong Liu, Inner Mongolia Agricultural University, ChinaReviewed by:
Sena Ardicli, Bursa Uludağ University, TürkiyeRongsong Luo, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Han, Akhtar, Chen, Liu, Zhao, Shi, Khan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changfa Wang, d2FuZ2NoYW5nZmFAbGN1LmVkdS5jbg==; Muhammad Zahoor Khan, emFob29ya2hhdHRhazlAeWFob28uY29t
 Ying Han
Ying Han Muhammad Faheem Akhtar
Muhammad Faheem Akhtar Wenting Chen
Wenting Chen Muhammad Zahoor Khan
Muhammad Zahoor Khan