- 1Center for Food Animal Health, Food Safety and Defense, College of Veterinary Medicine, Department of Pathobiology, Tuskegee University, Tuskegee, AL, United States
- 2Department of Large Animal Clinical Science, College of Veterinary Medicine, Tuskegee University, Tuskegee, AL, United States
- 3Poultry Microbiological Safety and Processing Research Unit, United States Department of Agriculture-Agriculture Research Service (USDA-ARS), U.S. National Poultry Research Center, Athens, GA, United States
- 4Faculty of Medical Sciences, School of Veterinary Medicine, University of the West Indies, St. Augustine, Trinidad and Tobago
- 5Department of Anatomy and Physiology, School of Veterinary Medicine, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
Introduction: Cattle are well-recognized reservoirs of Salmonella; however, reports of atypical hydrogen sulfide (H2S)-negative strains from bovine sources remain scarce. This cross-sectional study aimed to investigate the antimicrobial resistance profiles and epidemiology of Salmonella among rural cow-calf herds in Alabama, United States, with a particular focus on isolating emerging H2S-negative variants.
Method: Between April and May 2024, a total of 311 fecal samples were collected from cattle across 18 farm operations in six counties. Samples were cultured for Salmonella, and recovered isolates were identified using whole genome sequencing and the Salmonella In Silico Typing Resource. H2S production was assessed using Xylose Lysine Deoxycholate agar, Lysine Iron Agar, and Triple Sugar Iron agar. Antimicrobial susceptibility testing was performed against 24 antimicrobial agents. A mixed-effects logistic regression model was used for statistical analysis.
Results: Overall, 3.5% (11 out of 311) of animals from 27.8% (5 out of 18) of the farms tested positive for Salmonella. Fifteen isolates representing six serovars were identified: Salmonella Thompson (5 out of 15), Salmonella Hadar (4 out of 15), Salmonella Braenderup (3 out of 15), Salmonella Enteritidis (1 out of 15), Salmonella Bareilly (1 out of 15), and Salmonella Typhimurium (1 out of 15). Notably, the tet(A) gene conferring tetracycline resistance was detected exclusively in the Salmonella Hadar isolates. Diarrheic animals were significantly more likely to shed Salmonella in their feces (p value = 0.0192). Importantly, the Salmonella Typhimurium isolate was identified as an H2S-negative strain, carrying an A > C missense mutation in the phsC gene and a C > T synonymous mutation in the cysI gene.
Conclusion: To our knowledge, this is the first report of an H2S-negative Salmonella Typhimurium isolate from cattle feces. These findings also reveal a notable prevalence of Salmonella shedding among an underexplored population of rural cow-calf herds in the southeastern United States. The potential public health implications of these findings merit further investigation.
1 Introduction
Pathogen surveillance and cross-sectional studies are critical epidemiological tools for investigating infectious agents and are essential for informing public health interventions (1–3). In the United States (U.S.), for instance, the National Animal Health Monitoring System (NAHMS) routinely conducts nationwide assessments to evaluate the health and management practices of various domestic animal populations (4). In fulfilling this role, NAHMS frequently employs surveillance methodologies, as demonstrated by its most recent 2021–2022 nationwide study of the U.S. swine industry (5). Within the cattle sector, NAHMS has successfully conducted four such national studies to date (6). Similarly, in China, surveillance efforts have led to the identification of 46 hydrogen sulfide (H2S)-negative Salmonella serovars from a range of human, animal, and environmental sources (7).
H2S-negative Salmonella serovars are considered “atypical” because “typical” Salmonella isolates produce H2S (8). In recent years, the global incidence of H2S-negative Salmonella isolates has increased (7), prompting increased calls for enhanced surveillance to support their detection (9, 10). Unfortunately, these atypical variants often go undetected in traditional Salmonella isolation procedures due to their deviation from the conventional phenotypic characteristics of the bacteria (11, 12). Specifically, H2S production on various culture media, manifested as “black colonies,” has long served as a key phenotypic marker for identifying Salmonella and distinguishing it from other members of the Enterobacteriaceae family (7, 8, 13, 14). However, the absence of this trait in H2S-negative strains increases the likelihood of false negatives during routine culture-based identification, unless detection methods that do not rely on H2S production, such as those reviewed by Yang et al. (15), are employed.
Furthermore, these atypical strains have been associated with clinical diseases and outbreaks (16, 17). In Salmonella, the phs, cys, and asr operons are key genomic regions involved in H2S production (17). The H2S-negative phenotype is primarily attributed to missense mutations in the phs operon, which plays a central role in the reduction of thiosulfate to H2S (12). Additional mutations, including those in the cysJ gene, have also been documented (17). In some isolates, mutations have been identified in other genes such as tetR (a putative transcriptional regulator of the TetR family), moaC (involved in molybdenum cofactor biosynthesis), and sph (streptomycin phosphotransferase) (12). Multiple studies across the globe have reported the isolation of these atypical Salmonella serovars from live animals and animal-derived products (8, 9, 14, 18, 19). However, aside from the identification of an H2S-negative Salmonella Cerro isolate from cattle and cattle farm environments in the U.S. (20), no other such isolates have been reported from bovine sources or their environments. This is particularly notable given that cattle are well-established reservoirs of Salmonella (21).
In the U. S., cattle production accounts for the largest share of total cash receipts from agricultural commodities, making it the most significant sector in U.S. agriculture (22). The two primary production systems in the U.S. beef industry are cow–calf operations and cattle-feeding operations (23). Compared to cow–calf operations, cattle-feeding operations are higher-risk ventures that economically favor large-scale establishments, whereas cow–calf systems are more adaptable for small-scale, part-time farmers (24). These cow–calf operations are widely distributed across the country (25), with the southeastern U.S. serving as home to approximately one-third of all cow–calf producers (26). This makes the region an ideal geographic area for conducting research on cow–calf populations.
Cow–calf operations focus on producing annual calf crops, which are typically weaned or sold at ~6 months of age, either as stockers (weaned calves up to 1 year of age) or as yearlings destined for cattle-feeding operations (27). These systems serve as an important source of income for many part-time farmers, who often rely on local extension services to manage their animals.
Despite the critical role cow–calf operations play in U.S. agriculture, research specifically focused on rural cow–calf systems and their contribution to the prevalence of key pathogens, such as Salmonella, remains limited. This gap is partly due to their underrepresentation in national surveillance studies, which often overlook rural operations because of geographic and demographic constraints.
To address this gap, we conducted a study during a scheduled animal extension activity in rural Alabama, as part of the Cooperative Extension Program (CEP) (28), that involved deworming and vaccinating local cow–calf herds. The objective of this study was to investigate the presence of H2S-negative Salmonella in cattle feces, as well as serovar diversity, antimicrobial resistance (AMR) profiles, and overall epidemiology of Salmonella among cow–calf farms in the southeastern U.S.
2 Materials and methods
2.1 Study area and sampling strategy
This cross-sectional study was conducted between April and May 2024 among small-scale cow–calf farms located across six counties in Alabama, in the southeastern U.S. A collaborative team of researchers and personnel from the Center for Food Animal Health, Food Safety, and Defense Laboratory at Tuskegee University, along with the Tuskegee University Large Animal Clinic, worked with county coordinators from the CEP to identify and select farms for participation.
The selected farms had herd sizes ranging from 1 to 50 cattle, with 70% of the herds consisting of at least 10 animals. County CEP coordinators assisted in enrolling eligible farms based on the following criteria: owner willingness to participate, the geographic location, ease of access, and the presence of at least two cattle in the herd. To minimize selection bias across farms, more than 50% of the animals in each selected herd were sampled.
Given an estimated total herd population of 1,500 cattle across the selected counties and the absence of prior prevalence studies in this population, the sample size was calculated using the formula provided by Thrusfield (29), and the following parameters were applied: a 95% confidence interval (z-score = 1.96); an expected animal-level prevalence of 28%, and a precision level of 0.05. The formula used was as follows:
where n represents the sample size, Z represents the Z statistic for a confidence level of 95%, P represents the expected prevalence, and d represents the estimated precision.
In summary, 311 fecal samples were collected from apparently healthy cattle across 18 different cow–calf operations during single, one-time farm visits, with no repeat sampling. The fecal grab technique was used for sample collection (30), with all samples obtained using sterile disposable gloves and deposited into sterile 50-ml container tubes. Sampling was conducted while the animals were restrained in a squeeze chute for routine deworming and vaccination. The collected samples were immediately placed in a cooler box on ice and transported to the laboratory within 3–4 h of collection for immediate bacterial culture and isolation. Sampled animals were classified by sex (male or female) and age group: calves (≤12 months), including both pre-weaned and weaned individuals, and adults (>12 months). Additionally, based on fecal consistency evaluated by the same personnel at each farm visit, animals were categorized as either non-diarrheic (producing firm or fairly consistent feces) or diarrheic (producing loose or watery feces). Of the 311 samples collected, 76 were from calves and 235 from adults; 273 were from females and 38 from males; and 76 were from diarrheic animals, while 235 were from non-diarrheic animals.
2.2 Salmonella culture and isolation
Screening of fecal samples for Salmonella was performed following the guidelines outlined in the U.S. Department of Agriculture, Food Safety and Inspection Service Microbiological Laboratory Guide (version 4.13) (31), with slight modifications. Briefly, to achieve a 1:9 sample-to-buffer ratio, 1 g of fecal matter was inoculated into 9 ml of Buffered Peptone Water (BPW) (Millipore Sigma, Danvers, MA, U.S.) in sterile 15-ml tubes and homogenized by vortexing for 2 min. The resulting fecal suspension was incubated in a shaker incubator at 37°C for 24 h.
Following this pre-enrichment step in BPW, selective enrichment for Salmonella was carried out. Specifically, 1.0 and 0.1 ml aliquots of the pre-enriched culture were transferred to 9 ml of Tetrathionate (TT) broth (Neogen, Lansing, MI, U.S.) and 10 ml of Rappaport-Vassiliadis (RV) broth (Millipore Sigma, Danvers, MA, U.S.), respectively. Both enrichment broths were incubated at 42°C ± 0.5°C for 24 h.
After incubation, ~20 μl from each TT and RV broth culture was streaked separately onto Xylose-Lysine-Desoxycholate (XLD) agar (Millipore Sigma, Danvers, MA, U.S.) and Hektoen Enteric (HE) agar (Neogen, Lansing, MI, U.S.). The plates were incubated at 37°C ± 2°C for 24–48 h. A minimum of three presumptive Salmonella colonies per plate, selected based on their characteristic colony morphology (32), were picked and subjected to biochemical confirmation.
Biochemical screening was performed by inoculating the colonies onto Triple Sugar Iron (TSI) agar slants (Neogen, Lansing, MI, U.S.) and Lysine Iron Agar (LIA) plates (Neogen, Lansing, MI, U.S.), followed by incubation at 37°C ± 2°C for 24 h. Isolates exhibiting typical biochemical profiles consistent with those of Salmonella (33) were further confirmed using Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF-MS; bioMérieux, Marcy-l'Étoile, France).
To confirm the H2S-negative phenotype, suspect isolates were plated on TSI, LIA, and XLD, using the Salmonella Typhimurium ATCC 13311 strain as a positive control. Finally, all confirmed isolates were preserved in Luria broth (Millipore Sigma, Danvers, MA, U.S.) supplemented with 30% glycerol and stored at −80°C until further analyses were performed.
2.3 DNA extraction from isolates, whole genome sequencing, and assembly
Genomic DNA was extracted from pure Salmonella isolates using the DNeasy UltraClean Microbial Kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. The quality and quantity of the extracted DNA were assessed using a Bioanalyzer (Agilent Technologies, Santa Clara, CA, U.S.) and a Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham, MA, U.S.). Whole-genome sequencing was performed on the Illumina platform. Sample libraries were prepared using the Illumina DNA Preparation Kit and Nextera DNA CD Index Kits (Illumina, San Diego, CA, U.S.), followed by sequencing on the MiSeq platform using the 250 bp paired-end protocol.
Sequence quality was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/), and high-quality reads were assembled using the A5 pipeline (34). This pipeline includes five steps: read cleanup, read assembly, crude scaffolding, misassembly correction, and final scaffolding. The resulting contigs were used for downstream analyses.
2.4 Serovar identification, multi-locus sequence typing (MLST) of isolates and phylogeny
Serovar identification, including the determination of the somatic “O” and flagellar “H” antigens, was performed using the Salmonella In silico Typing Resource (SISTR) tool (version 1.1.2) (35). Sequence types (STs) were determined using the Achtman MLST scheme (36) via the PubMLST platform (37).
A phylogenetic tree based on single-nucleotide polymorphisms (SNPs) was constructed using the SNP Phylogeny tool of the Center for Food Safety and Applied Nutrition (CFSAN) pipeline via the Galaxy Platform (https://galaxy.sciensano.be/root). Salmonella enterica subsp. enterica serovar Typhimurium strain LT2 (NCBI accession number: AE006468.2) was used as the reference genome. The resulting phylogenetic tree, along with associated metadata, was visualized using the Interactive Tree of Life (iTOL) tool (38).
2.5 Antimicrobial susceptibility testing and resistance gene analysis
Antimicrobial susceptibility testing of the recovered isolates was conducted using the Sensititre™ National Antimicrobial Resistance Monitoring System (NARMS) Gram-negative GN4F plate (Thermo Scientific), which assesses susceptibility to 24 antibiotics. These antibiotics included amikacin, ampicillin, ampicillin/sulbactam, aztreonam, cefazolin, cefepime, ceftazidime, ceftriaxone, ciprofloxacin, doripenem, ertapenem, gentamicin, imipenem, levofloxacin, meropenem, minocycline, nitrofurantoin, piperacillin, tetracycline, ticarcillin/clavulanic acid, tigecycline, tobramycin, and trimethoprim/sulfamethoxazole.
Susceptibility interpretations followed the breakpoint criteria for Enterobacteriaceae, as outlined in the Clinical and Laboratory Standards Institute (CLSI) guidelines (39), except for nitrofurantoin and tigecycline, for which interpretation was based on previous studies (40–42).
To identify antimicrobial resistance genes, assembled genome sequences were screened against the Comprehensive Antimicrobial Resistance Database (CARD, version 4.0.0) (43) and ResFinder (version 4.3.3) (44), both accessed via ABRicate (version 1.0.1) (Seemann, Abricate, GitHub: https://github.com/tseemann/abricate) on 14 January 2025. The detection threshold for resistance genes was set at a minimum of 80% nucleotide identity and 80% sequence coverage.
2.6 Variation analysis on the hydrogen sulfide negative isolate
SNPs in the H2S-negative Salmonella Typhimurium isolate were analyzed using the variation analysis pipeline available on the Bacterial and Viral Bioinformatics Resource Center (BV-BRC) web-based platform (45), accessed on 19 February 2025. The sequence reads from the isolate were aligned to the reference genome of Salmonella enterica subsp. enterica serovar Typhimurium strain LT2 (NCBI Accession: AE006468.2) using the BWA-MEM algorithm (46). SNPs were then identified using FreeBayes (47), and the resulting variants, along with their predicted genetic effects, were evaluated using SNP-effect (48). Special focus was given to SNPs located in the phs, cysJIH, and asr operons, which are key genomic regions associated with H2S production in Salmonella.
2.7 Statistical analysis
A mixed-effects logistic regression analysis was performed using the generalized linear mixed model (GLMM) framework via the (lme4) package in R software (version 4.5.0) (49, 50). Farm groups of the animals were treated as a random effect, while age, sex, and diarrhea status were included as fixed effects. A p-value of < 0.05 was deemed statistically significant.
3 Results
3.1 Shedding of Salmonella among cattle from various farms
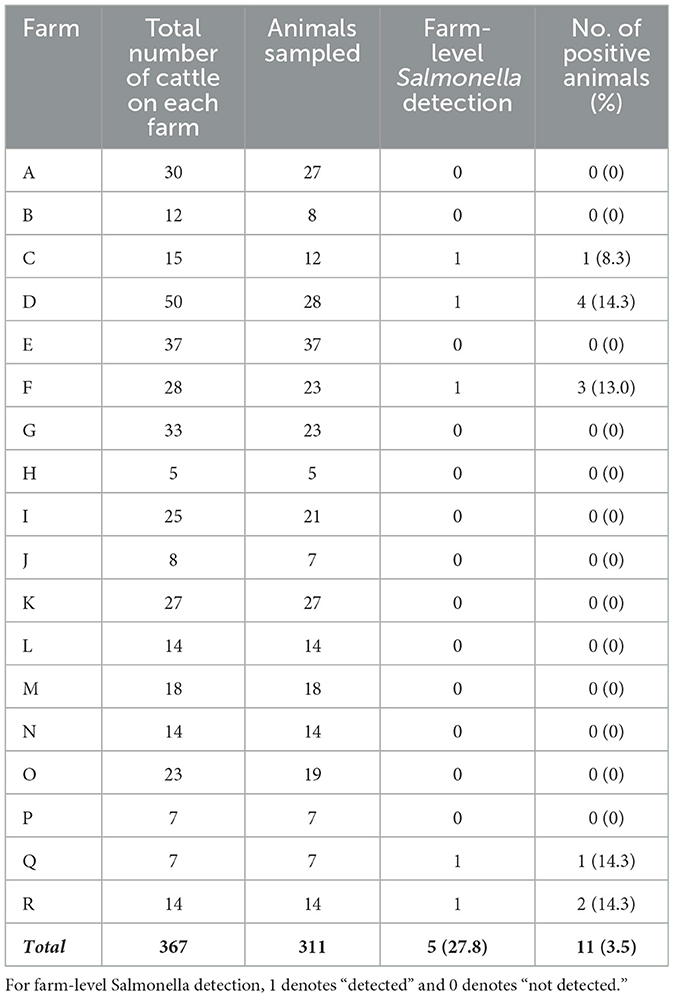
To maintain anonymity, the 18 farm-herds included in this study were labeled alphabetically from A to R (Table 1). At the herd level, Salmonella was isolated from 27.8% (five out of 18) of the farms visited. At the individual animal level, 3.5% (11 out of 311) of all cattle sampled tested positive for Salmonella (Table 1).
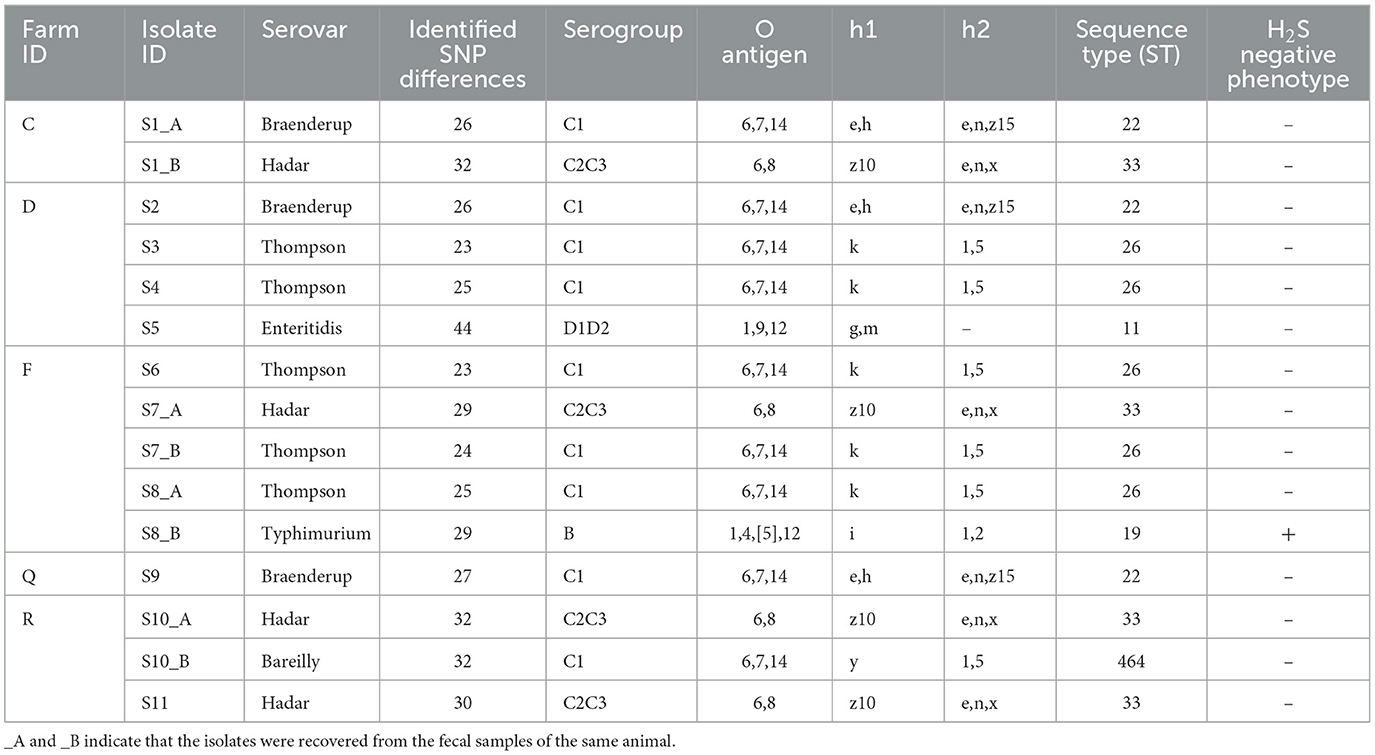
For isolate identification, each isolate was labeled with the prefix “S” (for sample), followed by the isolate number and the suffix “_A” or “_B” to distinguish multiple isolates recovered from the same sample (Table 2). From the 11 Salmonella-positive animals, a total of 15 isolates were recovered, representing six distinct Salmonella serovars with their corresponding sequence types (STs). These isolates included the following: five Salmonella Thompson isolates (ST26), four Salmonella Hadar isolates (ST33), three Salmonella Braenderup isolates (ST22), and one isolate each of Salmonella Bareilly (ST464), Salmonella Enteritidis (ST11), and Salmonella Typhimurium (ST19; Table 2).
Notably, four animals from three different farms were found to be co-shedding two distinct Salmonella serovars. The co-shedding combinations observed were as follows: Braenderup/Hadar, Thompson/Hadar, Bareilly/Hadar, and Thompson/Typhimurium (Table 2). The Salmonella Typhimurium isolate was identified as H2S-negative, based on its atypical appearance on XLD agar, LIA, and TSI agar (Figure 1).
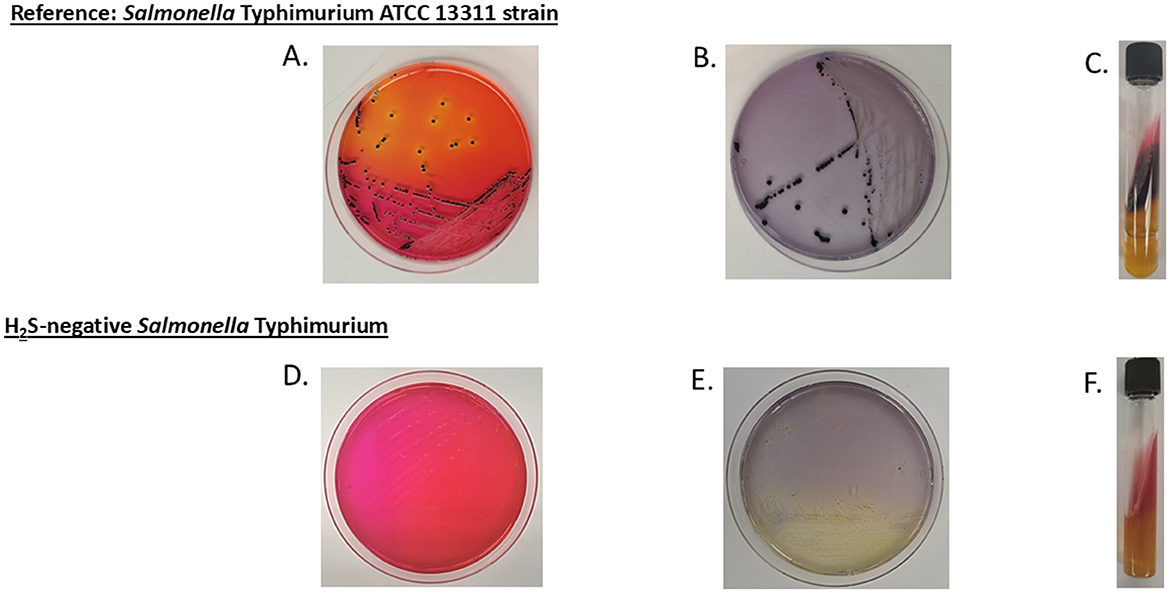
Figure 1. Colony morphology of reference and H2S-negative Salmonella on selective media. (A–C) display Salmonella Typhimurium ATCC 13311 (reference strain) cultured on xylose lysine deoxycholate (XLD) agar, lysine iron agar (LIA), and triple sugar iron (TSI) agar slant, respectively, showing the typical dark coloration morphology indicative of H2S production. (D–F) show the H2S-negative Salmonella Typhimurium isolate from this study cultured on the same respective media, exhibiting the absence of dark coloration due to the lack of H2S production.
3.2 Epidemiology of Salmonella on the sampled farms
Among the adult cattle sampled, 3.8% (nine out of 235) tested positive for Salmonella, compared to 2.6% (two out of 76) among calves (Table 3). All positive isolates were recovered from female cattle, representing 4% (11 out of 273) of the females tested. Based on fecal consistency, 6.6% (five out of 76) of diarrheic animals and 2.6% (six out of 235) of non-diarrheic animals tested positive for Salmonella (Table 3).
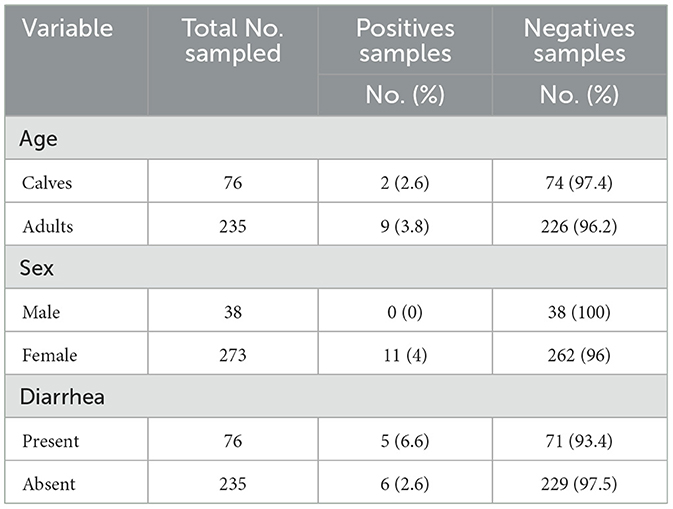
Table 3. Frequency of Salmonella detection among sampled cattle categorized by age, sex, and fecal consistency.
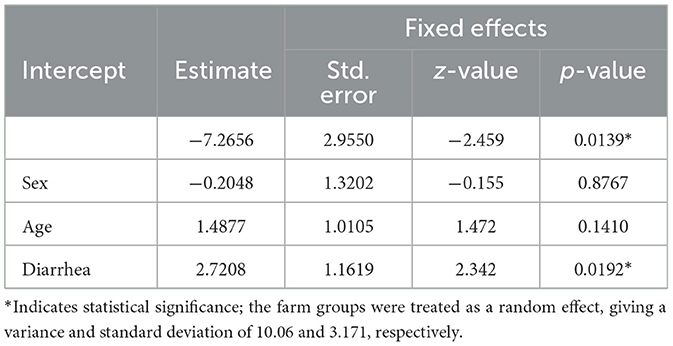
The results from the mixed-effects logistic regression analysis revealed considerable variation in the number of Salmonella-positive animals across farms. Diarrheic status was significantly associated with Salmonella shedding (p-value = 0.0192). However, neither the sex nor the age of the animals showed a statistically significant association with fecal shedding of Salmonella (Table 4).
3.3 Antibiotic-resistance pattern of Salmonella isolates
The major AMR genes and phenotypic resistance profiles identified among the Salmonella isolates are summarized in relation to the SNP-based phylogenetic tree (Figure 2). The number of SNP differences identified for each isolate, along with the isolate IDs used in the phylogeny, is presented in Table 2.
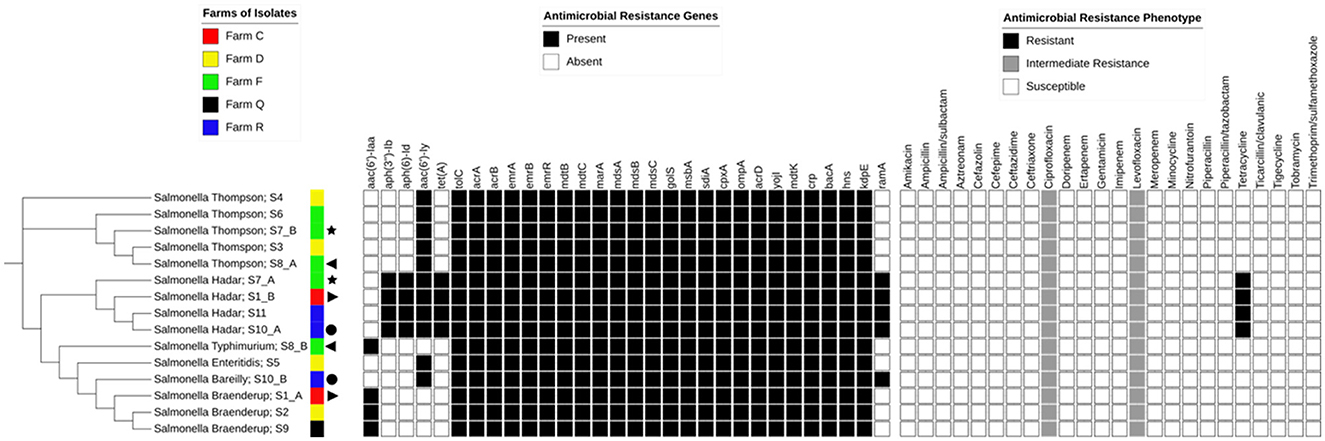
Figure 2. Antimicrobial resistance profiles and resistance genes among Salmonella isolates. A single-nucleotide polymorphism (SNP)-based phylogenetic tree is shown alongside the corresponding farm of origin, the antimicrobial resistance (AMR) genes, and phenotypic susceptibility profiles of the Salmonella isolates. Each isolate is labeled with its serovar name and isolate ID (as shown in Table 2), separated by a semicolon. The legends above each matrix provide an interpretation of the color-coded tiles, and the same symbols depict isolates from the same animal.
Notably, resistance was detected exclusively among the Salmonella Hadar isolates, all of which were resistant to tetracycline and harbored the tet(A) gene. Additionally, all isolates showed intermediate resistance to ciprofloxacin and levofloxacin. Detailed antimicrobial susceptibility profiles for each isolate are provided in the Supplementary material (Antimicrobial sensitivity).
The aminoglycoside resistance gene aac(6′)-Iy was detected in 11 out of 15 isolates. However, only four of the 15 isolates carried the aph(3″)-Ib, aph(6)-Id, and aac(6′)-Iaa genes (Figure 2).
3.4 Variation analysis of the H2S-negative isolate
Within the phs operon, a missense mutation (A > C) was identified in the phsC gene, resulting in an amino acid substitution from valine (V) to glycine (G) (Figure 3A). In the cys operon, a synonymous mutation (C > T) was detected in the cysI gene; however, this mutation did not lead to an amino acid change (Figure 3B). No genetic variations were observed in the asr operon. A complete list of the identified genetic variations and their predicted functional effects is provided in the Supplementary material (Variation analysis).
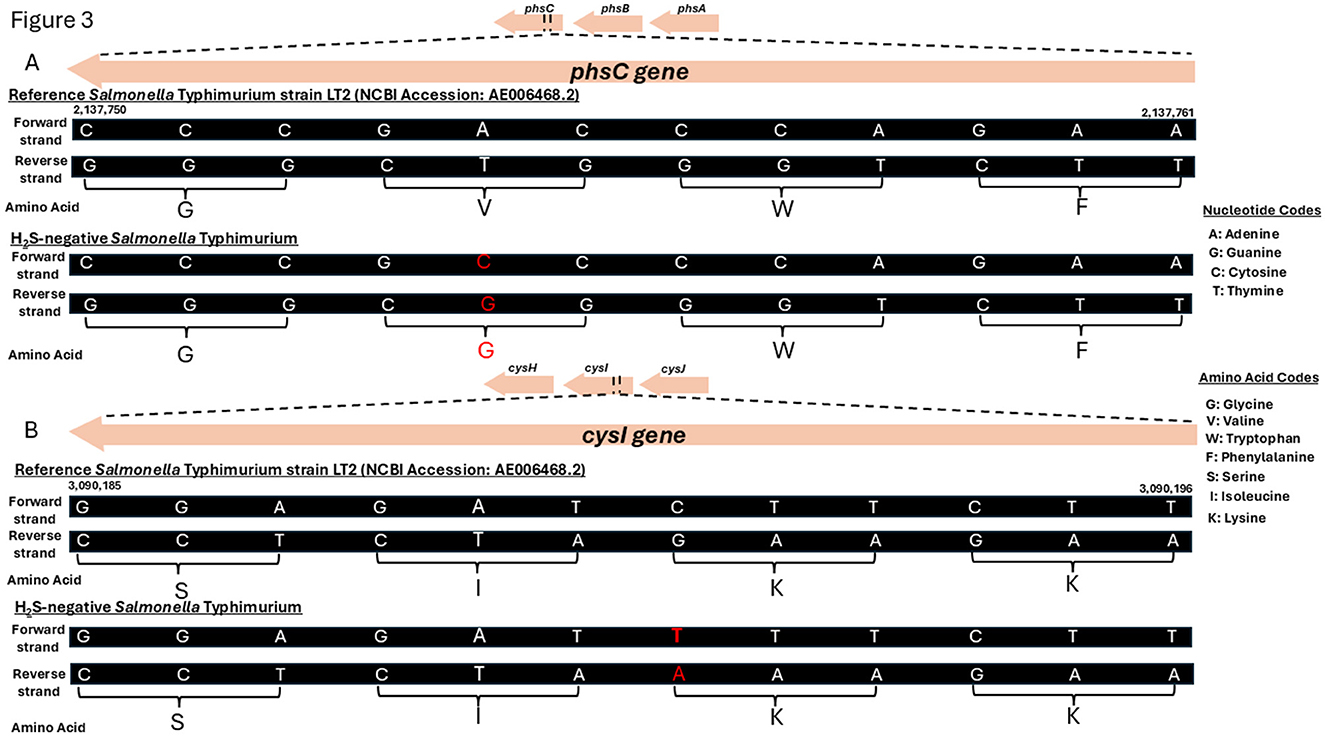
Figure 3. Genetic mutations identified in the phsC and cysI genes of the H2S-negative Salmonella Typhimurium isolate. (A) A schematic genome segment showing the phsC gene region (nucleotides 2,137,750 to 2,137,761) for both reference strain Salmonella enterica subsp. enterica serovar Typhimurium LT2 (NCBI Accession: AE006468.2) and the H2S-negative Salmonella Typhimurium isolate from this study. (B) A schematic genome segment showing the cysI gene region (nucleotides 3,090,185 to 3,090,196) in both the reference and the H2S-negative Salmonella Typhimurium isolate. Nucleotides and amino acids are shown in their single-letter codes. Mutated nucleotides and the corresponding amino acids (where a change in amino acids occurred) are highlighted in red.
4 Discussion
This cross-sectional study targeted rural, small-scale cow–calf operations, a population often underrepresented in national-level surveillance due to geographic and demographic constraints. The most recent cow–calf study, organized by the National Animal Health Monitoring System (NAHMS) in the U.S., took place in 2017 (51). That survey reported a Salmonella prevalence of 1.0% at the herd level and 4.4% at the animal level among cow–calf populations (51). Alabama, one of the major cow–calf states included in the NAHMS survey, recorded 20,004 beef operations as of December 2017 (51).
Although our study does not match the scale of national surveillance efforts, the Salmonella prevalence we observed, 27.8% at the herd level and 3.5% at the animal level, is notably high, given our sample size of 311 animals. This discrepancy may arise from methodological differences, some of which are discussed below.
National studies typically rely on stratified sampling strategies designed to generate broad, generalizable inferences across states (52). In contrast, our investigation focused on multiple herds within a defined subregion of a single state, specifically targeting rural, small-scale cow–calf operations. Additionally, inherent sampling biases arising from farm selection criteria and reliance on owner consent were not controlled in this study. National-level surveillance efforts often offer incentives and resources that help reduce the impact of such limitations.
Moreover, findings from localized studies may reflect regional and time-dependent factors, as noted in earlier research (53). For example, cattle in the southern U.S. have been shown to shed Salmonella in feces at higher rates than those in the northern regions (54). Seasonal variation may also influence shedding; previous studies have reported increased Salmonella prevalence in dairy herds during the summer months (55). Since our study was conducted as a one-time, cross-sectional survey in the spring, future studies conducted in other seasons could provide valuable insights into seasonal trends in Salmonella shedding among these populations.
Additional confounding factors likely contributed to the observed prevalence. Notably, poor biosecurity practices were evident across several farms. For instance, none of the operations required visitors to wear boot covers, a basic yet critical preventive measure. Based on these observations, we recommend increased outreach through veterinary extension services and biosecurity training programs targeted at rural cattle producers. Such efforts could substantially reduce Salmonella transmission risk.
Finally, public education regarding the importance of research participation is essential in these communities. Engaging rural farmers in ongoing disease surveillance through community-centered outreach can enhance participation and enable more comprehensive studies that better capture the burden of disease among underrepresented cattle populations.
The higher frequency of Salmonella shedding observed among adult cattle (3.8%) compared to calves (2.6%) in our study was not statistically significant. Although sampling bias related to animal age may have influenced this outcome, a similar study investigating clinical salmonellosis in dairy cattle reported a markedly higher prevalence rate of 49.3% in adults and 23.8% in calves (56). In the same study, the prevalence of fecal Salmonella shedding in apparently healthy cattle was 10.1% for adults and 5.4% for calves (56). This age-related dynamic in Salmonella shedding may be partly attributed to the passive immunity that calves acquire through colostrum, which offers temporary protection against infection (57, 58). In our study, calves were not classified as either pre-weaned or weaned, limiting our ability to assess this effect more precisely. Future research that specifically distinguishes between pre-weaned and weaned calves would provide a more comprehensive understanding of Salmonella shedding dynamics in this age group.
Nevertheless, the high frequency of Salmonella shedding among adult cattle may be associated with waning immunity, underlying health conditions, or physiological stressors such as pregnancy. Additionally, adult cattle outnumbered calves in all the herds visited, resulting in a smaller calf sample size. This sampling imbalance may also have contributed to the apparent disparity in Salmonella shedding between age groups.
Our findings contrast with those reported by Cummings et al. (59), who observed a higher prevalence of Salmonella in calves compared to adult cattle. A potential explanation for this discrepancy is that all of the animals in their study were clinically ill and admitted to a veterinary hospital, a setting that may have influenced infection rates. The role of age as a risk factor for Salmonella infection in cattle remains inconclusive. In a previous study, a recent investigation also found no significant association between age and Salmonella shedding (60).
Interestingly, with regard to antimicrobial resistance, two of the four tetracycline-resistant isolates in our study were recovered from calves. Previous studies have shown that calves are more likely to harbor resistant Escherichia coli strains than adults (61, 62). This pattern has been consistently reported over decades across diverse geographic regions and farming practices (61). One explanation offered is the unique composition of the intestinal microbiome in young calves, which may favor the persistence of multidrug-resistant strains (61). While the presence of tetracycline-resistant Salmonella in calves, as observed in our study supports this hypothesis, further research is needed to substantiate this relationship, especially regarding Salmonella isolates from cattle.
Similar to age, the sex of cattle was not significantly associated with Salmonella shedding. However, just as there were more adults than calves in our sampled herds, there were also more females than males, which likely introduced sampling bias. These limitations undoubtedly influenced our findings. Therefore, the lack of significant associations between age or sex and Salmonella shedding in cow–calf operations in this study should be interpreted cautiously, taking these methodological constraints into account.
The association between diarrhea and fecal shedding of Salmonella was also investigated in this study. It is important to emphasize that the diarrheic state of animals cannot be conclusively attributed to either nutritional or infectious causes, as both factors can influence fecal consistency in cattle. However, all animals sampled appeared clinically healthy, and most farms maintained their herds on grass pastures, supplementing with Bermuda grass hay and soy hull pellets twice daily, along with ad libitum access to water. Sampling occurred at various times of the day, including before, during, and after feeding.
Despite these variables, fecal shedding of Salmonella among diarrheic animals was the only statistically significant parameter in our study. This finding aligns with earlier studies that have linked bovine enteritis to Salmonella shedding, a common clinical manifestation of salmonellosis in adult cattle (63). Similar associations have been documented in other research (64, 65). Notably, some non-diarrheic animals in our study were also found to be shedding Salmonella, supporting the notion that asymptomatic carriers can serve as a persistent source of infection (54). These subclinical shedders pose a risk of undetected transmission, emphasizing the importance of maintaining strict hygiene protocols for farmworkers handling both symptomatic and asymptomatic animals.
Our study also demonstrated that individual animals can shed multiple Salmonella serovars simultaneously, a phenomenon supported by previous research (66). This underscores the importance of selecting more than one presumptive colony during Salmonella isolation, when resources allow. However, this recommendation must be balanced with practical limitations, as dominant serovars may be repeatedly isolated due to the often indistinguishable colony morphology on selective agar media. The number of colonies selected for further analysis should therefore be guided by the researcher's discretion, resource availability, and specific study objectives.
A noteworthy observation from our study was the prevalence of Salmonella Thompson, Salmonella Hadar, and Salmonella Braenderup—serovars not commonly associated with cattle globally (67). Nonetheless, Salmonella Thompson and Salmonella Braenderup have been isolated from bovine lymph nodes (68), which are significant reservoirs of Salmonella and may contribute to human infection (69), particularly when these infected lymph nodes are inadvertently incorporated into ground beef (68). Recent studies have shown that a considerable proportion of human salmonellosis cases linked to beef are associated with ground beef products (70).
In addition to direct transmission through meat, these serovars have also been implicated in environmental contamination, leading to outbreaks. For example, a recent multistate outbreak of Salmonella Braenderup in the U.S. affected 551 individuals across 34 states (71–73). Traceback investigations and comparative genomic analyses linked the outbreak strain to a water source used on a vegetable farm (71). These findings highlight the potential for Salmonella-infected cattle feces to contaminate nearby water bodies through runoff, posing broader public health risks. Research suggests that reintroduction events by host animals may contribute to the persistent contamination of farm water systems (74).
As expected, the presence of the tet(A) gene in Salmonella Hadar isolates conferred phenotypic resistance to tetracycline. This finding is consistent with previous studies that identified tet(A) as the most prevalent tetracycline resistance gene in Salmonella (75). Interestingly, although aminoglycoside resistance genes such as aph(3″)-Ib, aph(6)-Id, aac(6′)-Iy, and aac(6′)-Iaa were detected in several isolates, none exhibited phenotypic resistance to aminoglycosides. This discrepancy aligns with earlier research in which resistance genes were identified in 37 Salmonella isolates without corresponding phenotypic resistance (76). These findings suggest that resistance genes may be functionally inactive or “silenced” due to regulatory mutations, disrupted expression pathways, or the presence of cryptic or non-functional elements (77, 78). In particular, the aac (6′)-Iaa gene has been previously reported as a cryptic gene (79).
A contrasting phenomenon was observed with ciprofloxacin and levofloxacin. Despite the absence of known quinolone resistance genes, such as qnr, qep, or aac(6′)-Ib-cr (80), and no impactful point mutations detected, all isolates exhibited intermediate resistance to these fluoroquinolones. Resistance to quinolones is multifaceted and is commonly associated with mutations in genes encoding DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) (80). In our study, however, the intermediate resistance observed may instead be attributed to the activity of multiple efflux pumps, particularly those belonging to the SOS-box family, including mdtABC-tolC, emrAB-tolC, and acrAB-tolC (81). The emrA and emrB genes specifically encode efflux systems capable of expelling fluoroquinolones, thereby contributing to reduced susceptibility (81). Although some resistance patterns were shared among serovars isolated from different farms, further studies are needed to understand the dynamics of antimicrobial resistance and selection in small-scale farming systems.
To the best of our knowledge, this study is the first to report the isolation of an H2S-negative Salmonella Typhimurium strain from cattle feces. Globally, H2S-negative Salmonella Typhimurium isolates have been reported from various animal and human sources. For instance, these isolates have been recovered from retail chicken and pork products in Japan and China (8, 18), while a national surveillance effort in China identified H2S-negative Salmonella Typhimurium in humans and an unspecified livestock source (7). More recently, the monophasic variant of this serovar exhibiting the H2S-negative phenotype was isolated from raw chicken samples in Portugal (9), and in Sweden, the monophasic variant was implicated in a human salmonellosis outbreak (16). However, none of these reports documented the isolation of this atypical serovar from cattle feces. In the U.S., H2S-negative Salmonella has previously been detected in cattle, but the isolate belonged to the Salmonella Cerro serovar (20). The rarity of these atypical isolates likely reflects their ability to evade detection by conventional culture methods, although emerging evidence suggests that they are more widespread than previously assumed.
In terms of genetic mechanisms, the synonymous mutation observed in the cysI gene of our isolate is unlikely to have functional consequences, as such mutations are generally considered neutral (82). However, the missense mutation in the phsC gene may have functional implications, as such changes can affect protein stability and protein-protein interactions at the cellular level (83). Of the operons involved in H2S production in Salmonella, the phs operon, which includes the phsA, phsB, and phsC genes, is considered the most critical (14). Mutations in this operon, particularly in the phsA, have been reported to result in premature stop codons (9, 14), which truncates the thiosulfate reductase enzyme required for H2S production (12). Similar missense mutations in the phs and cys operons have also been described in previous studies (7, 14, 17). Nonetheless, it remains uncertain whether the phsC mutation identified in our isolate alone is sufficient to account for the H2S-negative phenotype observed. Further functional studies are needed to clarify the impact of these mutations on Salmonella metabolism and detectability.
We acknowledge that biases associated with our sampling strategy, particularly in relation to age group and sex, represent limitations of this study. These biases were largely due to the voluntary participation of farmers, which constrained our ability to randomize the study population. This challenge has also been noted in similar surveillance efforts (56). Despite these constraints, our study provides important insights into a largely underrepresented population of rural cattle farms and reports, for the first time, the isolation of an H2S-negative Salmonella Typhimurium serovar from cattle feces. Given the observed prevalence of Salmonella shedding, we recommend that extension services actively promote improved hygiene practices and biosecurity measures within cow–calf operations.
Furthermore, expanded studies involving larger sample sizes and broader geographic coverage are needed to provide a comprehensive overview of Salmonella prevalence and diversity in U.S. cattle and to better assess their public health implications. In conclusion, through an integrative approach combining classical microbiology, antimicrobial resistance profiling, and whole-genome sequencing, this study underscores the significant role that rural cow–calf herds may play as reservoirs for Salmonella.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material. Additional information can be found in the Supplementary materials. All raw sequence data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) under the BioProject accession number PRJNA1253333 and are accessible at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1253333/.
Ethics statement
The animal studies were approved by Office of the Associate Dean for Research and Advanced Studies. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
KB: Methodology, Writing – original draft, Formal analysis, Data curation, Visualization, Conceptualization, Investigation, Writing – review & editing. EK: Writing – review & editing, Visualization, Formal analysis. RN: Writing – review & editing, Visualization, Formal analysis. CW: Resources, Writing – review & editing, Data curation. KA: Writing – review & editing, Data curation, Resources. LM: Resources, Data curation, Writing – review & editing. DM: Writing – review & editing, Data curation, Resources. CJ: Writing – review & editing, Resources, Data curation. AA: Writing – review & editing, Data curation, Resources. TO-A: Resources, Writing – review & editing, Data curation. TS: Supervision, Funding acquisition, Conceptualization, Writing – review & editing. GR: Funding acquisition, Writing – review & editing, Supervision, Conceptualization. WA: Conceptualization, Writing – review & editing, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by grants from USDA/NIFA CBG 2021-38821-34710 and USDA/NIFA/AFRI 2022-67017-36982.
Acknowledgments
The authors express their sincere appreciation to Dr. Ruby Perry, Dean of the College of Veterinary Medicine at Tuskegee University, for her support of graduate research. Gratitude is also extended to the Center for Food Animal Health, Food Safety and Defense Laboratory, for its institutional support, and to Viona Osei and Asmaa Elrefaey for their support provided in diverse ways to this project. The authors especially thank Sandra House and Woodley Tiffanie, technicians at the USDA-ARS laboratory in Athens, Georgia, for their invaluable technical assistance throughout this project. Finally, the authors extend appreciation to Mr. George Hunter, the Agriculture and Natural Resources Coordinator for the Cooperative Extension Program at Tuskegee University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1619880/full#supplementary-material
References
1. Crone MA, Freemont PS. Thinking beyond pathogen surveillance: building resilient biotech ecosystems to combat the next pandemic. Front Sci (2024) 2:1412291. doi: 10.3389/fsci.2024.1412291
2. Dohoo, IR, Martin SW, Stryhn H. Veterinary Epidemiologic Research. Vol. 2. Charlottetown: VER, Incorporated (2009).
3. Zhang M, Kozlowski H, Chew R, Htun NSN, Morris SK, Akladious C, et al. The spectrum of health conditions in community-based cross-sectional surveys in Southeast Asia 2010-21: a scoping review. BMC Public Health. (2024) 24:1853. doi: 10.1186/s12889-024-19347-3
4. United States Department of Agriculture - Animal and Plant Health Inspection Service (USDA-APHIS). National Animal Health Monitoring System (NAHMS). (2025). Available online at: https://www.aphis.usda.gov/livestock-poultry-disease/nahms (Accessed March 19, 2025).
5. Hempstead SC, Gensler CA, Haley CA, Wiedenheft AM, Robertson JB, Fedorka-Cray PJ, et al. Prevalence and characterization of Salmonella species on U.S. swine sites as part of the NAHMS 2021 swine enteric study. J Food Prot. (2025) 88:100435. doi: 10.1016/j.jfp.2024.100435
6. United States Department of Agriculture - Animal and Plant Health Inspection Service (USDA-APHIS). NAHMS Beef Cow-Calf Studies. (2024). Available online at: https://www.aphis.usda.gov/livestock-poultry-disease/nahms/beef-cow-calf (Accessed June 4, 2024).
7. Xie J, Wu F, Xu X, Yang X, Zhao R, Ma Q, et al. Antibiotic resistance and molecular characterization of the hydrogen sulfide-negative phenotype among diverse Salmonella serovars in China. BMC Infect Dis. (2018) 18:1–9. doi: 10.1186/s12879-018-3209-3
8. Lin D, Yan M, Lin S, Chen S. Increasing prevalence of hydrogen sulfide negative Salmonella in retail meats. Food Microbiol. (2014) 43:1–4. doi: 10.1016/j.fm.2014.04.010
9. Mourão J, Rebelo A, Ribeiro S, Peixe L, Novais C, Antunes P. Atypical non-H2S-producing monophasic Salmonella Typhimurium ST3478 strains from chicken meat at processing stage are adapted to diverse stresses. Pathogens. (2020) 9:1–18. doi: 10.3390/pathogens9090701
10. Yi S, Xie J, Liu N, Li P, Xu X, Li H, et al. Emergence and prevalence of non-H2S-producing Salmonella enterica serovar Senftenberg isolates belonging to novel sequence type 1751 in China. J Clin Microbiol. (2014) 52:2557–66. doi: 10.1128/JCM.00377-14
11. Wu F, Xu X, Xie J, Yi S, Wang J, Yang X, et al. Molecular characterization of Salmonella enterica serovar aberdeen negative for H2S production in China. PLoS ONE. (2016) 11:e0161352. doi: 10.1371/journal.pone.0161352
12. Drauch V, Palmieri N, Spergser J, Hummel K, Brandstetter M, Kornschober C, et al. Comprehensive phenotyping combined with multi-omics of Salmonella Infantis and its H2S negative variant - resolving adaption mechanisms to environmental changes. Food Microbiol. (2025) 129:104744. doi: 10.1016/j.fm.2025.104744
13. Shelef LA, Tan W. Automated detection of hydrogen sulfide release from thiosulfate by Salmonella spp. J Food Prot. (1998) 61:620–2. doi: 10.4315/0362-028X-61.5.620
14. Müştak IB, Müştak HK, Sariçam S. Molecular characterisation of hydrogen sulfide negative Salmonella enterica serovar Havana. Int J Gen Mol Microbiol. (2020) 113:1241–6. doi: 10.1007/s10482-020-01432-3
15. Yang Q, Zu J, Zhang S, Liu C, Qin X, Xu W. An overview of rapid detection methods for Salmonella. Food Control. (2025) 167:110771. doi: 10.1016/j.foodcont.2024.110771
16. Colombe S, Jernberg C, Löf E, Angervall AL, Mellström-Dahlgren H, Dotevall L, et al. Outbreak of unusual H2S-negative monophasic Salmonella Typhimurium strain likely associated with small tomatoes, Sweden, August to October 2019. Eurosurveillance. (2019) 24:1900643. doi: 10.2807/1560-7917.ES.2019.24.47.1900643
17. Lee KS, Kim D, Lee H, Lee K, Yong D. Isolation of non-hydrogen sulfide-producing Salmonella enterica serovar Infantis from a clinical sample: the first case in Korea. Ann Lab Med. (2020) 40:334–6. doi: 10.3343/alm.2020.40.4.334
18. Sakano C, Kuroda M, Sekizuka T, Ishioka T, Morita Y, Ryo A, et al. Genetic analysis of non-hydrogen sulfide-producing Salmonella enterica serovar Typhimurium and S. enterica serovar Infantis isolates in Japan. J Clin Microbiol. (2013) 51:328–30. doi: 10.1128/JCM.02225-12
19. Albert MJ, Obaid K. Al, Alfouzan W, Sheikh AR, Udo E, Izumiya H, Bulach DM, Seemann T. Isolation of Salmonella enterica serovar Kentucky strain ST 198 and its H2S-negative variant from a patient: implications for diagnosis. J Clin Microbiol. (2014) 52:4090–3. doi: 10.1128/JCM.01775-14
20. Kovac J, Cummings KJ, Rodriguez-Rivera LD, Carroll LM, Thachil A, Wiedmann M. Temporal genomic phylogeny reconstruction indicates a geospatial transmission path of Salmonella Cerro in the United States and a clade-specific loss of hydrogen sulfide production. Front Microbiol. (2017) 8:261935. doi: 10.3389/fmicb.2017.00737
21. Gutema FD, Agga GE, Abdi RD, De Zutter L, Duchateau L, Gabriël S. Prevalence and serotype diversity of Salmonella in apparently healthy cattle: systematic review and meta-analysis of published studies, 2000–2017. Front Vet Sci. (2019) 6:102. doi: 10.3389/fvets.2019.00102
22. United States Department of Agriculture -Economic Research Service (USDA-ERS). Cattle & beef - Sector at a glance | Economic Research Service. Available online at: https://www.ers.usda.gov/topics/animal-products/cattle-beef/sector-at-a-glance (Accessed June 7, 2025).
23. United States Department of Agriculture -Economic Research Service (USDA-ERS) - Cattle & Beef. (2024). Available online at: https://www.ers.usda.gov/topics/animal-products/cattle-beef/ (Accessed April 15, 2024).
24. PennState Extension - Feeding Beef Cattle. (2025). Available online at: https://extension.psu.edu/feeding-beef-cattle (Accessed June 7, 2025).
25. PennState Extension - Beef Cow-calf Production. (2024). Available online at: https://extension.psu.edu/beef-cow-calf-production (Accessed April 17, 2024).
26. Rourke C, Waggie R, Hill N, Ellis JD, Starzec K. Risk information sufficiency & seeking of southeastern United States beef producers. Adv Agric Dev. (2023) 4:10–23. doi: 10.37433/aad.v4i4.309
27. Endres MI, Schwartzkopf-Genswein K. Overview of cattle production systems. Adv Cattle Welf . (2018) 1−26. doi: 10.1016/B978-0-08-100938-3.00001-2
28. United States Department of Agriculture - National Institute of Food and Agriculture (USDA-NIFA). Cooperative Extension System. (2025). Available online at: https://www.nifa.usda.gov/about-nifa/what-we-do/extension/cooperative-extension-system (Accessed June 2, 2025).
29. Thrusfield M. Veterinary Epidemiology. Hoboken, NJ: John Wiley & Sons (2013). Available online at: https://books.google.com/books?hl=en&lr=&id=JPRqOU_fgWUC&oi=fnd&pg=PP17&ots=c8PTNaEz4Y&sig=hR2wNj6mbA8jZM-0ptXuEZR3VaQ#v=onepage&q&f=false (Accessed June 9, 2025).
30. Agga GE, Arthur TM, Schmidt JW, Wang R, Brichta-Harhay DM. Diagnostic accuracy of rectoanal mucosal swab of feedlot cattle for detection and enumeration of Salmonella enterica. J Food Prot. (2016) 79:531–7. doi: 10.4315/0362-028X.JFP-15-409
31. United States Department of Agriculture - Food Safety and Inspection Service (USDA-FSIS). MLG 4.13 Isolation and Identification of Salmonella from Meat, Poultry, Pasteurized Egg, Carcass, and Environmental Sponges | Enhanced Reader. (2024). Available online at: https://www.fsis.usda.gov/sites/default/files/media_file/documents/MLG-4.13.pdf (Accessed November 19, 2024).
32. Oludairo OO, Kwaga JKP, Junaid K, Abdu PA, Gitanjali A, Perets A, et al. A review of the International Organization for Standardization (ISO) guidelines for the detection of Salmonella from faeces. J Appl Vet Sci. (2022) 7:14–22. doi: 10.21608/javs.2022.146858.1158
33. Cox NA, Williams JE. A simplified biochemical system to screen Salmonella isolates from poultry for serotyping. Poult Sci. (1976) 55:1968–71. doi: 10.3382/ps.0551968
34. Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. (2014) 31:587–9. doi: 10.1093/bioinformatics/btu661
35. Yoshida C, Kruczkiewicz P, Laing C, Lingohr E, Gannon V, Nash J. The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS ONE. (2016) 11:e0147101. doi: 10.1371/journal.pone.0147101
36. Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. (2012) 8:1002776. doi: 10.1371/journal.ppat.1002776
37. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. (2018) 3:124. doi: 10.12688/wellcomeopenres.14826.1
38. Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. (2021) 49:W293–6. doi: 10.1093/nar/gkab301
39. Clinical Laboratory Standards Institute (CLSI). CLSI M100 30th Edition Performance Standards for Antimicrobial Susceptibility Testing. (2020). p. 282. Available online at: https://www.nih.org.pk/wp-content/uploads/2021/02/CLSI-2020.pdf (Accessed January 15, 2025).
40. Liao CH, Kung HC, Hsu GJ, Lu PL, Liu YC, Chen CM, et al. In-vitro activity of tigecycline against clinical isolates of Acinetobacter baumannii in Taiwan determined by the broth microdilution and disk diffusion methods. Int J Antimicrob Agents. (2008) 32:192–6. doi: 10.1016/j.ijid.2008.05.1068
41. Hirsch EB, Zucchi PC, Chen A, Raux BR, Kirby JE, McCoy C, et al. Susceptibility of multidrug-resistant gram-negative urine isolates to oral antibiotics. Antimicrob Agents Chemother. (2016) 60:3138–40. doi: 10.1128/AAC.02961-15
42. Li H, Zhou M, Chen X, Zhang Y, Jian Z, Yan Q, et al. Comparative evaluation of seven tigecycline susceptibility testing methods for carbapenem-resistant Enterobacteriaceae. Infect Drug Resist. (2021) 14:1511–6. doi: 10.2147/IDR.S289499
43. Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. (2017) 45:D566–73. doi: 10.1093/nar/gkw1004
44. Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, et al. ResFinder 40 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
45. Olson RD, Assaf R, Brettin T, Conrad N, Cucinell C, Davis JJ, et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. (2023) 51:D678–89. doi: 10.1093/nar/gkac1003
46. Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. (2013). Available online at: https://arxiv.org/abs/1303.3997v2 (Accessed March 1, 2025).
47. Garrison E, Marth G. Haplotype-based Variant Detection from Short-read Sequencing. (2012). Available online at: http://arxiv.org/abs/1207.3907 (Accessed September 2, 2024).
48. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (2012) 6:80–92. doi: 10.4161/fly.19695
49. Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. (2015) 67:1–48. doi: 10.18637/jss.v067.i01
50. R Core Team. A Language and Environment for Statistical Computing. (2013). Available online at: https://www.r-project.org/ (Accessed June 5, 2025).
51. United States Department of Agriculture - Animal and Plant Health Inspection Service (USDA-APHIS). Salmonella on U.S. Beef Cow-calf Operations NAHMS Beef 2017 Study Information Brief . (2023). Available online at: https://www.aphis.usda.gov/sites/default/files/beef2017-Salmonella-infobrief.pdf (Accessed September 2, 2024).
52. United States Department of Agriculture. Beef 2017: Beef Cow-calf Management Practices in the United States, 2017. (2020). Available online at: https://www.aphis.usda.gov/sites/default/files/beef2017_dr_parti.pdf (Accessed March 1, 2025).
53. Alam MJ, Renter DG, Ives SE, Thomson DU, Sanderson MW, Hollis LC, et al. Potential associations between fecal shedding of Salmonella in feedlot cattle treated for apparent respiratory disease and subsequent adverse health outcomes. Vet Res. (2009) 40:2. doi: 10.1051/vetres:2008040
54. Hanson DL, Loneragan GH, Brown TR, Edrington TS. Salmonella prevalence varies over time and space in three large, adjacent cattle operations in the southwestern United States. Front Anim Sci. (2022) 3:878408. doi: 10.3389/fanim.2022.878408
55. Fossler CP, Wells SJ, Kaneene JB, Ruegg PL, Warnick LD, Eberly LE, et al. Cattle and environmental sample-level factors associated with the presence of Salmonella in a multi-state study of conventional and organic dairy farms. Prev Vet Med. (2005) 67:39–53. doi: 10.1016/j.prevetmed.2004.10.005
56. Cummings KJ, Warnick LD, Elton M, Gröhn YT, McDonough PL, Siler JD. The effect of clinical outbreaks of salmonellosis on the prevalence of fecal Salmonella shedding among dairy cattle in New York. Foodborne Pathog Dis. (2010) 7:815–23. doi: 10.1089/fpd.2009.0481
57. Huston CL, Wittum TE, Love BC, Keen JE. Prevalence of fecal shedding of Salmonella spp in dairy herds. J Am Vet Med Assoc. (2002) 220:645–9. doi: 10.2460/javma.2002.220.645
58. Mohler VL, Izzo MM, House JK. Salmonella in calves. Vet Clin North Am - Food Anim Pract. (2009) 25:37–54. doi: 10.1016/j.cvfa.2008.10.009
59. Cummings KJ, Divers TJ, McDonough PL, Warnick LD. Fecal shedding of Salmonella spp among cattle admitted to a veterinary medical teaching hospital. J Am Vet Med Assoc. (2009) 234:1578–85. doi: 10.2460/javma.234.12.1578
60. Bonifait L, Thépault A, Baugé L, Rouxel S, Le Gall F, Chemaly M. Occurrence of Salmonella in the cattle production in France. Microorganisms. (2021) 9:872. doi: 10.3390/microorganisms9040872
61. Gaire TN, Scott HM, Sellers L, Nagaraja TG, Volkova VV. Age dependence of antimicrobial resistance among fecal bacteria in animals: a scoping review. Front Vet Sci. (2021) 7:622495. doi: 10.3389/fvets.2020.622495
62. Khachatryan AR, Hancock DD, Besser TE, Call DR. Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl Environ Microbiol. (2004) 70:752–7. doi: 10.1128/AEM.70.2.752-757.2004
63. Cummings KJ, Warnick LD, Alexander KA, Cripps CJ, Gröhn YT, James KL, et al. The duration of fecal Salmonella shedding following clinical disease among dairy cattle in the northeastern USA. Prev Vet Med. (2009) 92:134–9. doi: 10.1016/j.prevetmed.2009.07.002
64. Eguale T, Engidawork E, Gebreyes WA, Asrat D, Alemayehu H, Medhin G, et al. Fecal prevalence, serotype distribution and antimicrobial resistance of Salmonellae in dairy cattle in central Ethiopia. BMC Microbiol (2016) 16:20. doi: 10.1186/s12866-016-0638-2
65. Abd El-Rahman AM, Mahmoud AE-KA, Khadr AM, El-Shemy TM. Some studies on Salmonella enterica associated with diarrhea in cattle. Alexandria J Vet Sci. (2016) 48:54–60. doi: 10.5455/ajvs.197182
66. Gorski L. Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLoS ONE. (2012) 7:e0034722. doi: 10.1371/journal.pone.0034722
67. Nickodem C, Arnold AN, Gehring KB, Gill JJ, Richeson JT, Samuelson KL, et al. A longitudinal study on the dynamics of Salmonella enterica prevalence and serovar composition in beef cattle feces and lymph nodes and potential contributing sources from the feedlot environment. Appl Environ Microbiol. (2023) 89:e00033–23. doi: 10.1128/aem.00033-23
68. Gragg SE, Loneragan GH, Brashears MM, Arthur TM, Bosilevac JM, Kalchayanand N, et al. Cross-sectional study examining Salmonella enterica carriage in subiliac lymph nodes of cull and feedlot cattle at Harvest. Foodborne Pathog Dis. (2013) 10:368–74. doi: 10.1089/fpd.2012.1275
69. Locke SR, Pempek JA, Meyer R, Portillo-Gonzalez R, Sockett D, Aulik N, et al. Prevalence and sources of Salmonella lymph node infection in special-fed veal calves. J Food Prot. (2022) 85:906–17. doi: 10.4315/JFP-21-410
70. Canning M, Birhane MG, Dewey-Mattia D, Lawinger H, Cote A, Gieraltowski L, et al. Salmonella outbreaks linked to beef, United States, 2012–2019. J Food Prot. (2023) 86:100071. doi: 10.1016/j.jfp.2023.100071
71. United States-Food and Drug Administration (US-FDA). Outbreak Investigation of Salmonella: Cucumbers (June 2024) | FDA. (2024). Available online aat: https://www.fda.gov/food/outbreaks-foodborne-illness/outbreak-investigation-Salmonella-cucumbers-june-2024 (Accessed November 20, 2024).
72. Brandenburg JM, Stapleton GS, Kline KE, Khoury J, Mallory K, Machesky KD, et al. Salmonella Hadar linked to two distinct transmission vehicles highlights challenges to enteric disease outbreak investigations. Epidemiol Infect. (2024) 152:e86. doi: 10.1017/S0950268824000682
73. Shen AQ, Dalen A, Bankers L, Matzinger SR, Schwensohn C, Patel K, et al. Multistate outbreak of Salmonella Thompson infections linked to seafood exposure — United States, 2021. MMWR Morb Mortal Wkly Rep. (2023) 72:513–6. doi: 10.15585/mmwr.mm7219a2
74. Liu H, Whitehouse CA Li B. Presence and persistence of Salmonella in water: the impact on microbial quality of water and food safety. Front Public Heal. (2018) 6:159. doi: 10.3389/fpubh.2018.00159
75. Pavelquesi SLS, Ferreira ACAO, Rodrigues ARM, Silva CMS, Orsi DC, da Silva ICR. Presence of tetracycline and sulfonamide resistance genes in Salmonella spp: literature review. Antibiotics. (2021) 10:1314. doi: 10.3390/antibiotics10111314
76. Schwan CL, Lomonaco S, Bastos LM, Cook PW, Maher J, Trinetta V, et al. Genotypic and phenotypic characterization of antimicrobial resistance profiles in non-typhoidal Salmonella enterica strains isolated from Cambodian informal markets. Front Microbiol. (2021) 12:711472. doi: 10.3389/fmicb.2021.711472
77. Deekshit VK, Srikumar S. ‘To be, or not to be'—the dilemma of ‘silent' antimicrobial resistance genes in bacteria. J Appl Microbiol. (2022) 133:2902–14. doi: 10.1111/jam.15738
78. Stasiak M, Maćkiw E, Kowalska J, Kucharek K, Postupolski J. Silent genes: antimicrobial resistance and antibiotic production. Polish J Microbiol. (2021) 70:421. doi: 10.33073/pjm-2021-040
79. Kagambèga A, McMillan EA, Bouda SC, Hiott LM, Ramadan H, Soro DK, et al. Resistance genes, plasmids, multilocus sequence typing (MLST), and phenotypic resistance of Non-Typhoidal Salmonella (NTS) isolated from slaughtered chickens in Burkina Faso. Antibiotics. (2022) 11:782. doi: 10.3390/antibiotics11060782
80. Brandis G, Gockel J, Garoff L, Guy L, Hughes D. Expression of the qepA1 gene is induced under antibiotic exposure. J Antimicrob Chemother. (2021) 76:1433–40. doi: 10.1093/jac/dkab045
81. Kerek Á, Török B, Laczkó L, Somogyi Z, Kardos G, Bányai K, et al. In vitro microevolution and co-selection assessment of amoxicillin and cefotaxime impact on Escherichia coli resistance development. Antibiotics. (2024) 13:1728. doi: 10.3390/antibiotics13030247
82. Bailey SF, Morales LAA, Kassen R. Effects of synonymous mutations beyond codon bias: the evidence for adaptive synonymous substitutions from microbial evolution experiments. Genome Biol Evol. (2021) 13:evab141. doi: 10.1093/gbe/evab141
Keywords: Salmonella, antimicrobial resistance, fecal shedding, H2S-negative, mutation
Citation: Bentum KE, Kuufire E, Nyarku R, Woods C, Ale K, McKie L, McKenzie D, Jackson CR, Adesiyun A, Opoku-Agyemang T, Samuel T, Reddy G and Abebe W (2025) Detection of a hydrogen sulfide-negative Salmonella Typhimurium from cattle feces in a cross-sectional study of cow–calf herds in the Southeastern United States. Front. Vet. Sci. 12:1619880. doi: 10.3389/fvets.2025.1619880
Received: 28 April 2025; Accepted: 23 June 2025;
Published: 29 July 2025.
Edited by:
Filippo Biscarini, National Research Council (CNR), ItalyReviewed by:
Samantha Locke, The Ohio State University, United StatesDelower Hossain, Sher-e-Bangla Agricultural University, Bangladesh
Mahsa Dehnavi, Universidad de Leon Instituto de Ganaderia de Montana, Spain
Copyright © 2025 Bentum, Kuufire, Nyarku, Woods, Ale, McKie, McKenzie, Jackson, Adesiyun, Opoku-Agyemang, Samuel, Reddy and Abebe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woubit Abebe, d2FiZWJlQHR1c2tlZ2VlLmVkdQ==
 Kingsley E. Bentum
Kingsley E. Bentum Emmanuel Kuufire
Emmanuel Kuufire Rejoice Nyarku1
Rejoice Nyarku1 Khim Ale
Khim Ale Charlene R. Jackson
Charlene R. Jackson Temesgen Samuel
Temesgen Samuel