- 1Parasitology Laboratory, Veterinary College, Xinjiang Agricultural University, Ürümqi, China
- 2Muwang Veterinary Service Co., Ltd, Jimusaer, China
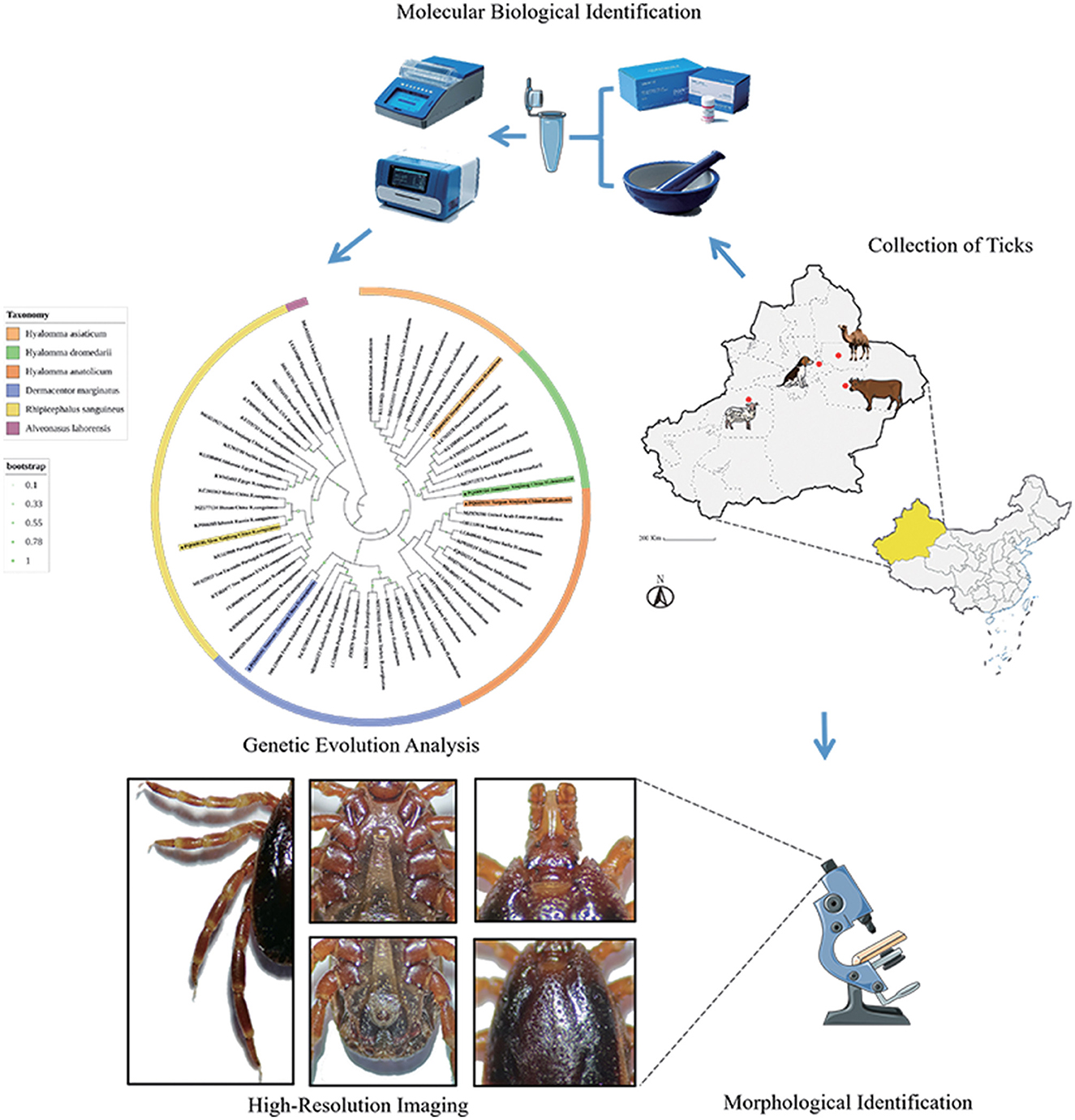
Ticks serve as major vectors of zoonotic pathogens, posing significant threats to public health and livestock. However, morphological similarity among closely related species complicates accurate identification. This study employed an integrative taxonomic approach combining morphological and molecular methods to delineate five medically important tick species in Xinjiang, China. From April to July 2024, a total of 1,128 ticks were collected from livestock across four ecological regions (Ürümqi, Turpan, Jimsar, and Aksu). Morphological features—including basis capituli, scutum, and genital aperture—were analyzed via stereomicroscopy, while molecular characterization targeted mitochondrial (16S rDNA, COI) and nuclear (ITS2) markers. Phylogenetic trees were reconstructed using the maximum likelihood method. Morphological identification confirmed five species: Hyalomma anatolicum, Hyalomma asiaticum, Hyalomma dromedarii, Rhipicephalus turanicus, and Dermacentor marginatus, supported by high-resolution imaging. Molecular data revealed notable interregional genetic affinities: H. anatolicum from Turpan shared COI similarity with strains from Kazakhstan; H. asiaticum from Turpan clustered with Iranian populations; H. dromedarii from Jimsar showed 16S rDNA similarity to Saudi Arabian lineages; R. turanicus from Aksu grouped with Egyptian COI sequences; and D. marginatus from Jimsar aligned with populations from Alashankou, China. Notably, single-gene phylogenies posed taxonomic limitations—for example, ITS2 misclassified R. turanicus as Rhipicephalus sanguineus sensu stricto. These issues were mitigated through morphological diagnostics such as scutal enamel spots in D. marginatus and the distinctive genital plates in H. dromedarii. The integrative approach improved taxonomic resolution, with 16S rDNA providing genus-level insight, COI enabling species-level discrimination, and ITS2 offering strain-level resolution. Additionally, high-resolution morphological imaging addressed gaps in existing reference databases. Overall, multi-locus strategies combined with morphological validation are essential for accurate tick identification, improving our capacity to monitor vector-borne pathogen transmission and contributing to One Health surveillance frameworks.
1 Introduction
Ticks are globally distributed, with ~896 species identified to date. In China alone, over 110 species of hard ticks (Ixodidae) have been recorded (1–4). Xinjiang, the largest province in China by land area, encompasses diverse geographical and environmental conditions that favor the proliferation of multiple tick species (5, 6). As obligate hematophagous ectoparasites, ticks represent the second most significant group of biological vectors after mosquitoes, requiring blood meals at every developmental stage (1, 2, 7). Their feeding behavior causes tissue damage through skin penetration and facilitates the transmission of a wide range of tick-borne pathogens (8–10). Notably, some pathogens can be vertically transmitted via tick eggs (7), thereby expanding their spatial and temporal transmission potential. These attributes collectively pose substantial threats to public health and the livestock industry (8, 11, 12). Different tick species are associated with distinct pathogen profiles—for instance, Hyalomma anatolicum primarily transmits Theileria annulata, whereas its congener Hyalomma asiaticum is a major vector of Crimean-Congo Hemorrhagic Fever in Central Asia and China. Accurate tick species identification is therefore crucial from both medical and veterinary perspectives, enabling early intervention and containment of emerging tick-borne diseases.
Morphological identification of tick species relies on key structural features, including the shape of the basis capituli, the sulci and ornamentation of the dorsal scutum, leg spur configurations, and ventral plate morphology. However, this process requires significant taxonomic expertise due to the subtle and often overlapping morphological traits among species (13–15). Molecular identification methods, primarily based on polymerase chain reaction (PCR) amplification of targeted genetic regions, have become increasingly prevalent. Commonly employed genetic markers for tick identification and phylogenetic analysis include the 16S ribosomal RNA gene (16S rDNA), cytochrome c oxidase subunit I (COI), and the internal transcribed spacer 2 (ITS2). The 16S rDNA comprises 10 conserved and nine variable regions; the former display high sequence homology across taxa, reflecting deep phylogenetic relationships, while the latter exhibit species-specific variations that align with evolutionary divergence patterns (16, 17). COI, a key component of the mitochondrial respiratory chain, is widely used as a molecular barcode for accurate tick species classification and is often referred to as the “DNA barcode” for ticks (18, 19). ITS2, a non-coding nuclear region within the eukaryotic ribosomal RNA gene cluster, incorporates biparental genetic contributions and offers robust phylogenetic resolution due to its high interspecific variability (20, 21).
Northern Xinjiang's Urumqi region represents a hotspot for tick-borne disease diversity, predominantly involving Hyalomma and Rhipicephalus species, and is associated with Lyme disease, tick-borne encephalitis, and emerging bunyaviral infections (22). In contrast, Turpan, dominated by Hyalomma ticks, is a key region for investigating livestock-associated pathogens such as bovine theileriosis, Crimean-Congo hemorrhagic fever, and Q fever (6, 23). Jimsar, influenced by climate change-driven northward tick expansion, has become a pivotal transitional zone for monitoring the genotypic evolution of Borrelia burgdorferi and the cross-border transmission of tick-borne relapsing fever (24). Aksu, situated near Central Asian epidemic zones, serves as a strategic frontier for tracking transboundary pathogen incursions and studying tick–symbiont coevolution dynamics (25). Together, these regions span Xinjiang's major ecological gradients—forest-steppe, extreme arid zones, transitional belts, and cross-border interfaces—providing an ideal framework for systematically investigating tick-borne pathogen diversity, transmission dynamics, and control strategies. Enhancing the precision of tick identification in these regions is thus of paramount importance.
A major challenge in current tick taxonomy stems from limited access to high-resolution morphological imaging and the presence of intraspecific morphological variability, which complicates identification and has driven a growing dependence on molecular methods. However, this reliance has also led to taxonomic ambiguities, particularly among closely related congeneric species. To address these issues, this study integrates traditional morphological identification with molecular phylogenetic analysis of ticks collected from the four aforementioned regions. High-resolution morphological imaging is combined with mitochondrial (e.g., cox1) and nuclear (e.g., ITS2) genetic markers to resolve evolutionary relationships and minimize misclassification. Moreover, this integrative approach facilitates elucidation of the genetic relatedness of ticks across the Central Asia–Middle East region and addresses existing gaps in diagnostic imaging datasets.
2 Materials and methods
2.1 Sample collection
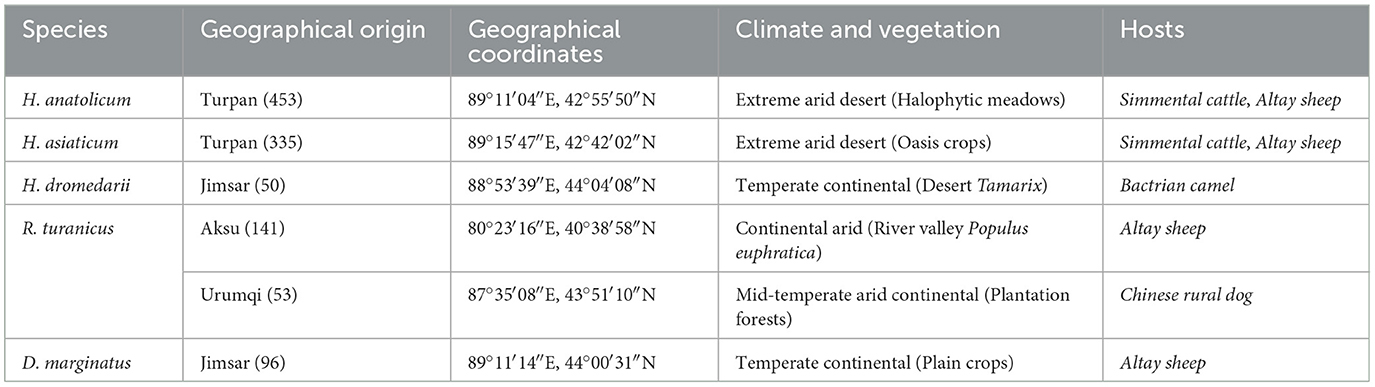
Between April and July 2024, a total of 1,128 ticks were collected across Xinjiang (see Table 1; distribution shown in Figure 1) from hosts including Simmental cattle, Altay sheep, Bactrian camels, and Chinese rural dogs. Ticks were housed in perforated containers sealed with cotton gauze and transported to the Parasitology Laboratory, College of Veterinary Medicine, Xinjiang Agricultural University, for preservation and analysis.
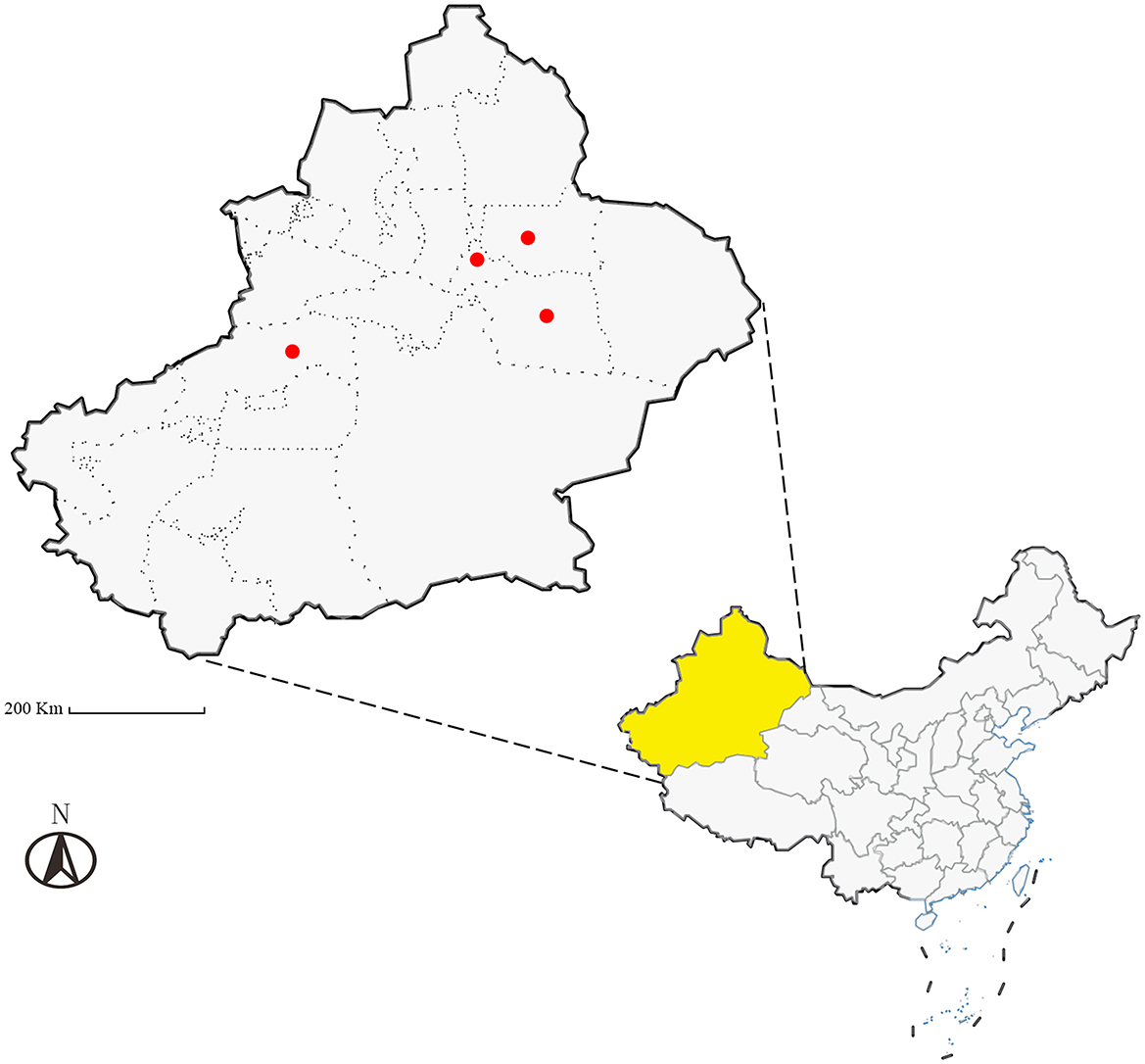
Figure 1. Schematic map of the study sampling sites (Ürümqi City, Turpan City, Jimsar County, and Aksu Prefecture in Xinjiang, China), Scale bar = 200 Km.
2.2 Morphological identification and high-resolution imaging
Specimens were alternately rinsed with 75% ethanol and physiological saline to eliminate surface contaminants. Morphological examination was conducted using an SMZ161 stereomicroscope, following standardized protocols from relevant literature [references (26–29) and Books (13–15)]. Key morphological features—including the capitulum, scutum, spiracular plates, anal plates, legs, and pulvilli—were assessed for preliminary taxonomic classification. Specimens were preserved in 75% ethanol and subsequently subjected to molecular validation. High-resolution imaging was performed post-genetic confirmation.
2.3 Genomic DNA extraction
In each sampling area, three live ticks per species exhibiting consistent morphological characteristics were selected. Specimens were flash-frozen in liquid nitrogen and homogenized. Genomic DNA was extracted under low-temperature conditions using a commercial kit (Tiangen Biochemical Technology Co., Ltd.), according to the manufacturer's protocol. Extracted DNA was stored at −20°C for downstream applications.
2.4 PCR amplification of the target gene
PCR amplification targeted three genetic loci using the following primer sets: 16S rDNA: Forward (F): 5′-TTAAATTGCTGTRGTATT-3′; Reverse (R): 5′-CCGGTCTGAACTCASAWC-3′. Tick COI gene: Forward (F): 5′-GGTCAACAAATCATAAAGATATTGG-3′; Reverse (R): 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′. Tick ITS2 gene: Forward (F): 5′-TCGTCTGTCTGAGGGTCGGA-3′; Reverse (R): 5′-ATCGTCTCGTGTAGCGTCG-3′.
Each 25-μL PCR reaction system included 12.5 μl of 2 × Taq PCR Master Mix, 1.0 μl each of forward and reverse primers (10 μmol/L), 1.0 μl DNA template, and 9.5 μl ddH2O. Thermal cycling conditions for 16S rDNA gene amplification were as follows: 94°C initial denaturation for 3 min; 35 cycles of 94°C denaturation for 30 s, 43.6°C annealing for 30 s, and 72°C extension for 1 min; followed by a final extension step at 72°C for 10 min. For the COI gene, the conditions were as follows: 94°C initial denaturation for 3 min; 35 cycles of 94°C denaturation for 30 s, 51.5°C annealing for 30 s, and 72°C extension for 1 min; with a final extension step at 72°C for 10 min. The ITS2 gene amplification protocol was as follows: 94°C initial denaturation for 3 min; 35 cycles of 94°C denaturation for 30 s, 57.2°C annealing for 30 s, and 72°C extension for 1 min; concluding with a final extension at 72°C for 10 min.
Negative controls using genomic DNA from Mus musculus (Kunming mouse strain) blood samples were included in all PCR assays. Amplicons were separated by 1.5% agarose gel electrophoresis. Target DNA fragments were extracted, purified using a standard agarose gel DNA extraction kit (Tiangen Biochemical Technology Co., Ltd.) following the manufacturer's protocol and subsequently subjected to Sanger sequencing (Sangon Biotech Co., Ltd.).
2.5 Sequence alignment and phylogenetic analysis
Raw sequences were aligned against the National Center for Biotechnology Information (NCBI) database to identify homologous tick sequences. Primer sequences were trimmed, and curated sequences were uploaded to the NCBI repository. Reference sequences corresponding to conspecific ticks were retrieved, while those with consecutive accession numbers, incomplete metadata (e.g., uncertain collection localities), or ambiguous nucleotide content or length were excluded.
Multiple sequence alignments were performed using the ClustalW algorithm in MEGA 11. Optimal nucleotide substitution models were determined as follows: Kimura 2-parameter with gamma distribution and invariant sites (G+I) for 16S rDNA; Tamura-Nei with G+I for COI; and Tamura 3-parameter with gamma distribution (G) for ITS2. Phylogenetic trees were constructed using the Maximum Likelihood method. Alveonasus lahorensis (for 16S rDNA and COI) and Argas persicus (for ITS2) were designated as outgroups for phylogenetic inference.
3 Results
3.1 Morphological identification results
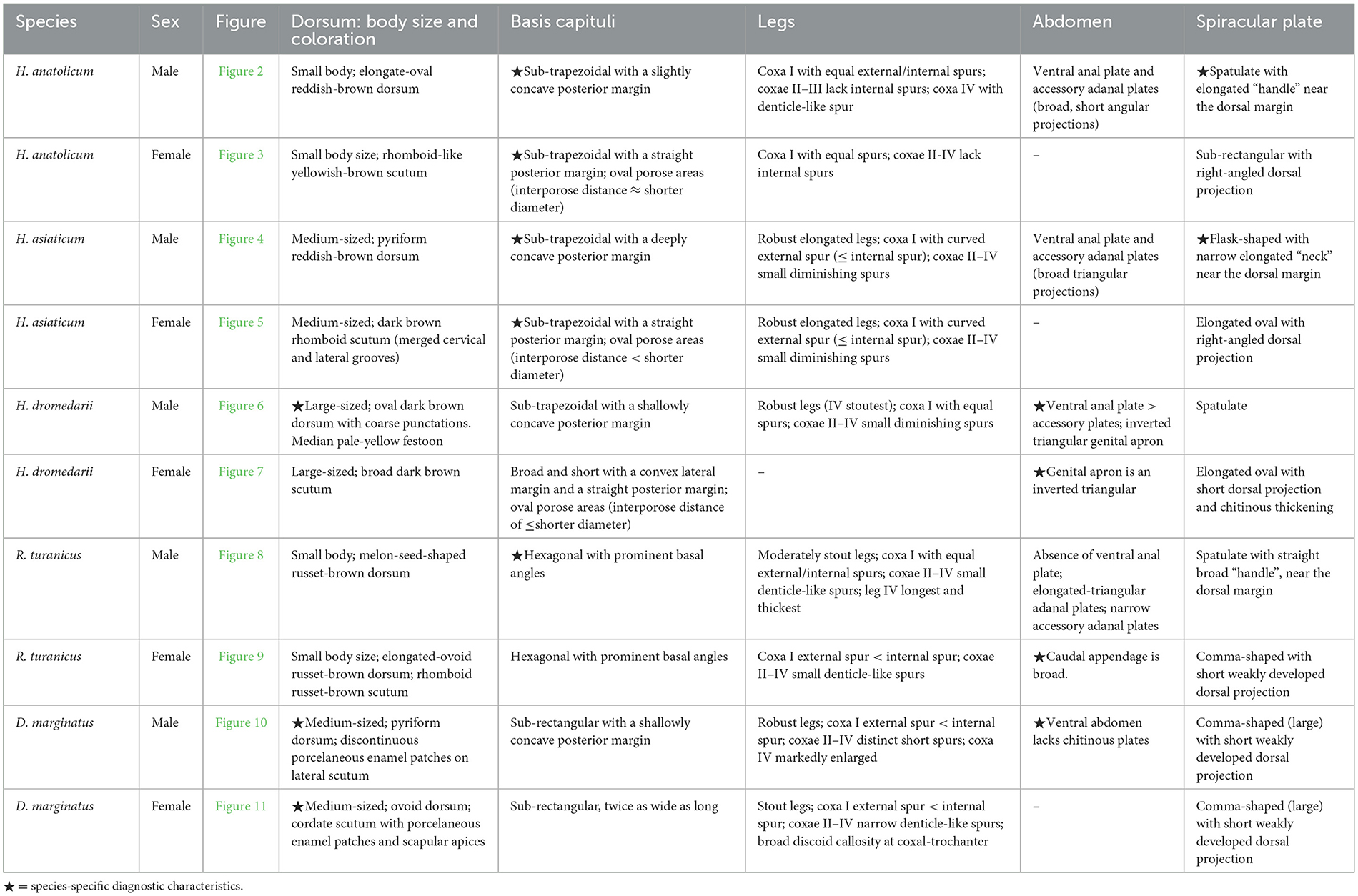
The capitulum, scutum, and spiracular plates of the collected tick specimens—Hyalomma anatolicum (H. anatolicum), Hyalomma asiaticum (H. asiaticum), Hyalomma dromedarii (H. dromedarii), Rhipicephalus turanicus (R. turanicus), and Dermacentor marginatus (D. marginatus)—were examined to determine morphological characteristics and capture high-resolution images, as detailed in Figures 2–11, Table 2 and Supplementary Table S1.
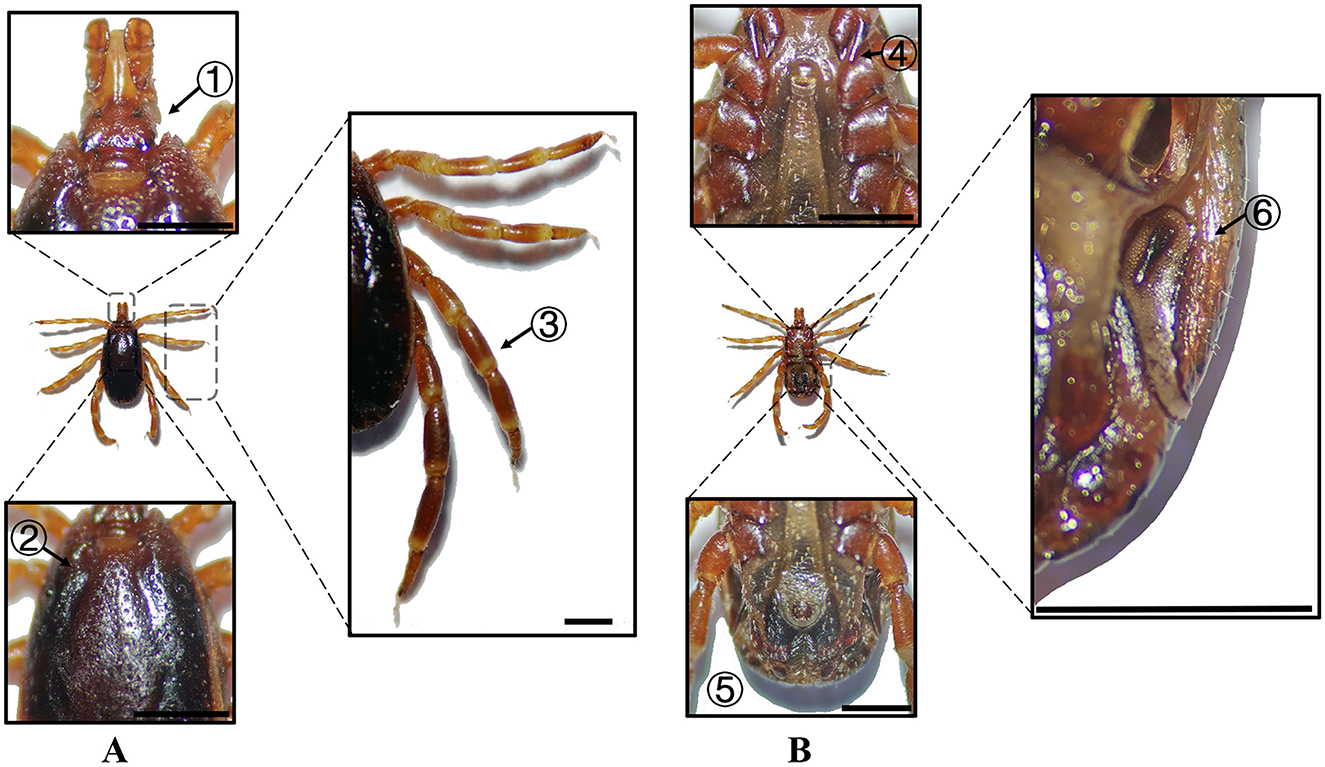
Figure 2. (A) Dorsal and (B) ventral views of a male H. anatolicum. Labels: (1) Basis capituli; (2) Cervical and lateral grooves; (3) Leg articulations; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
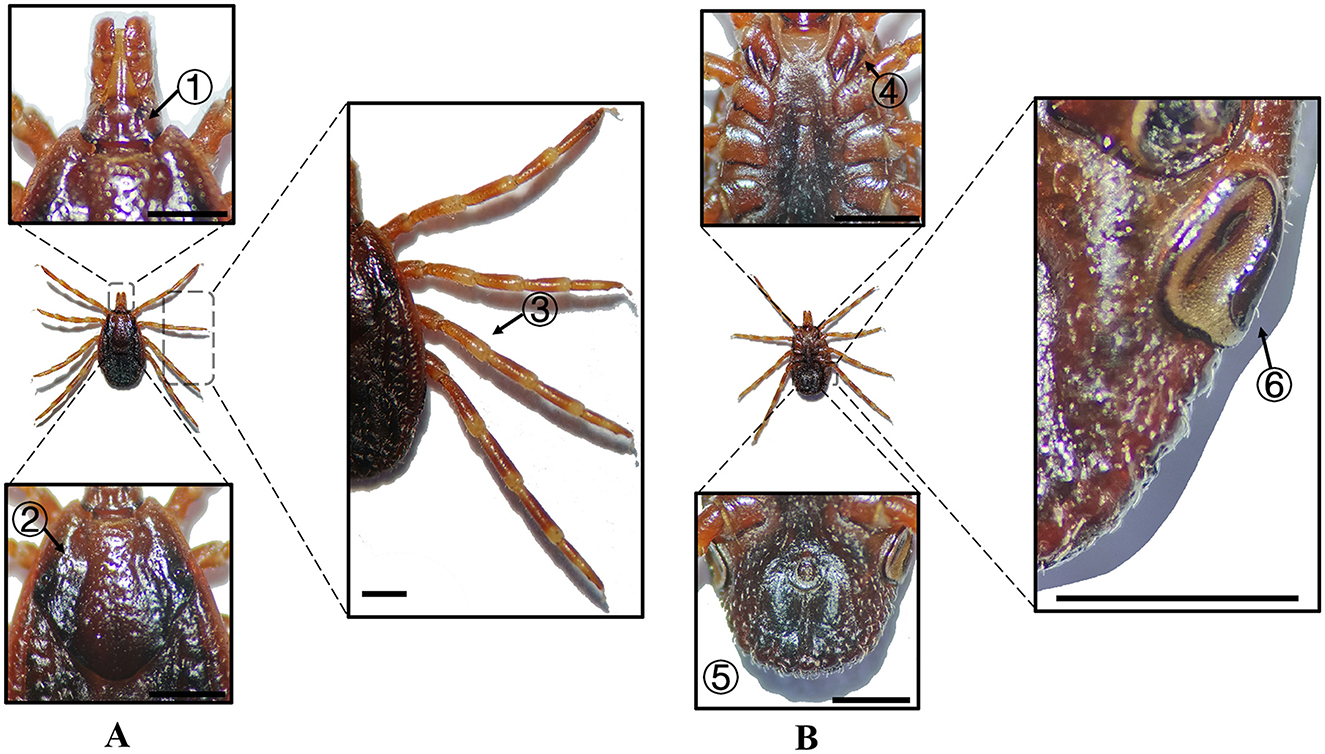
Figure 3. (A) Dorsal and (B) ventral views of a female H. anatolicum. Labels: (1) Basis capituli; (2) Cervical and lateral grooves; (3) Leg articulations; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
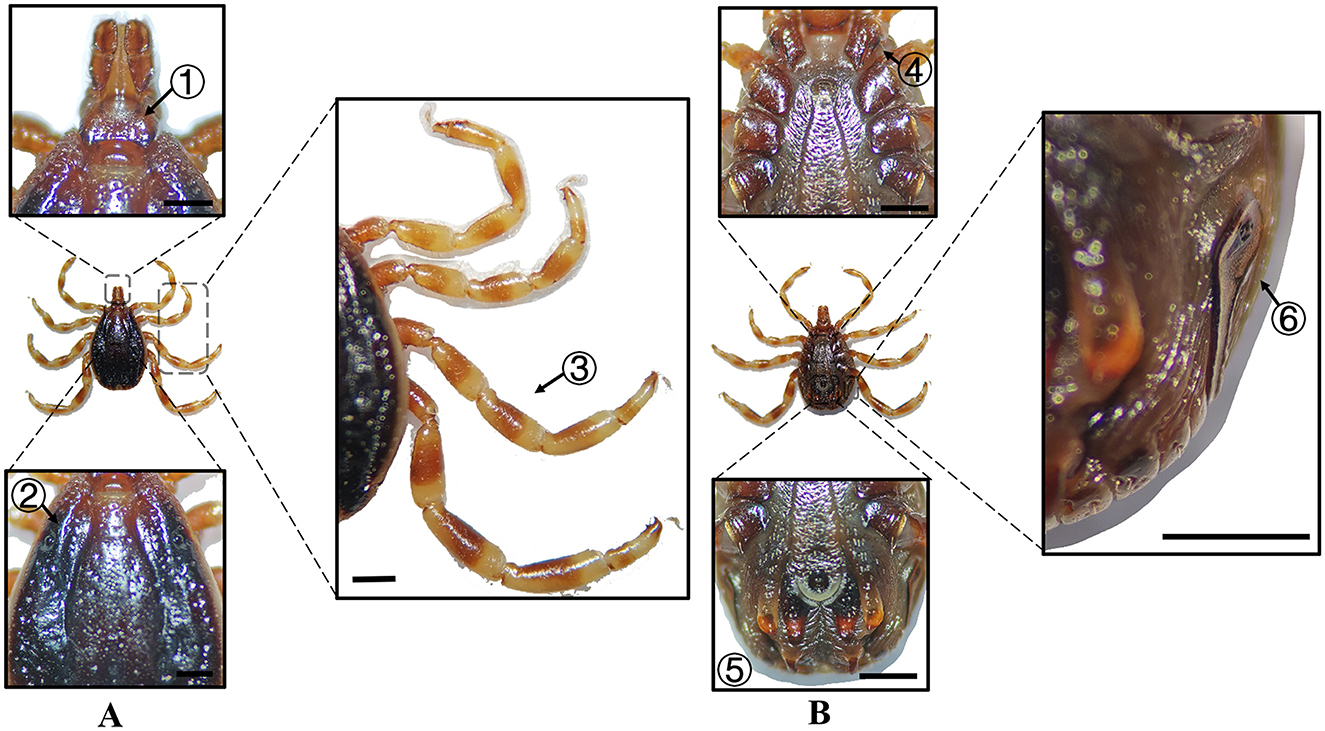
Figure 4. (A) Dorsal and (B) ventral views of a male H. asiaticum. Labels: (1) Basis capituli; (2) Cervical and lateral grooves; (3) Leg articulations; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
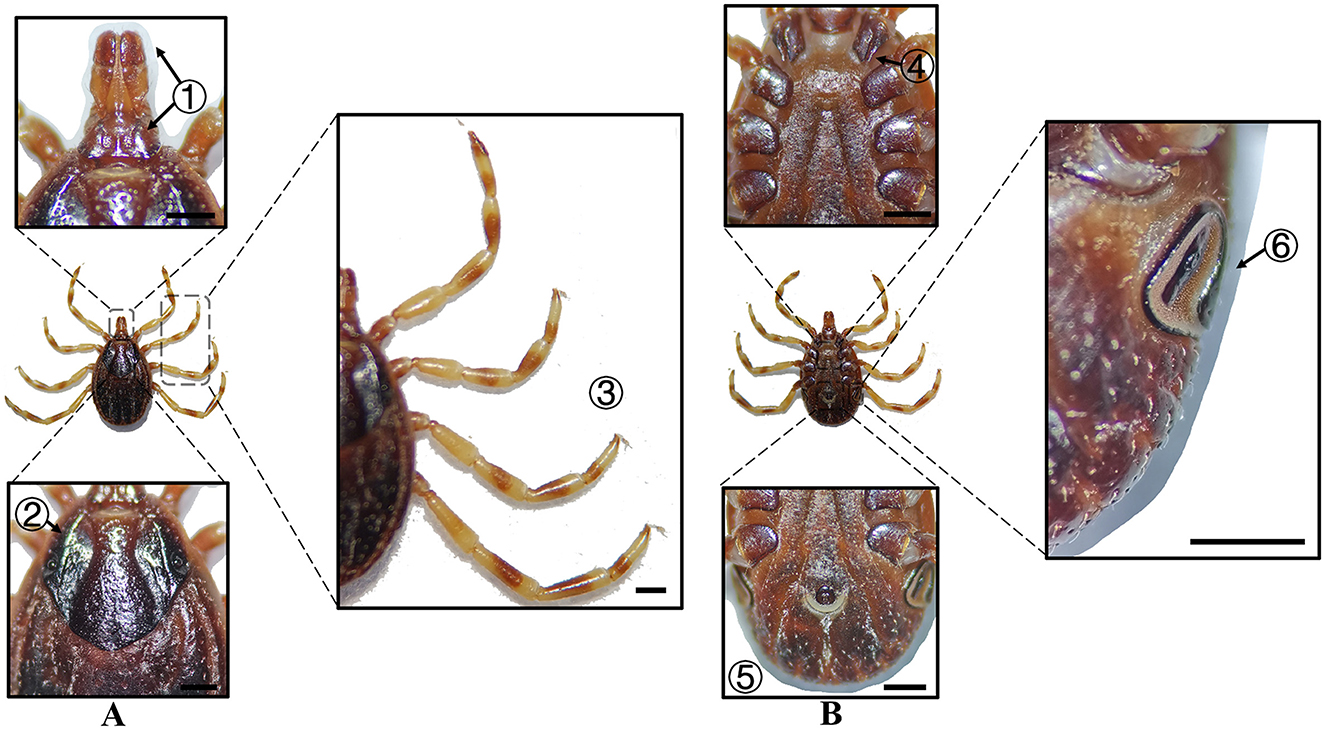
Figure 5. (A) Dorsal and (B) ventral views of a female H. asiaticum. Labels: (1) Basis capituli; (2) Cervical and lateral grooves; (3) Legs; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
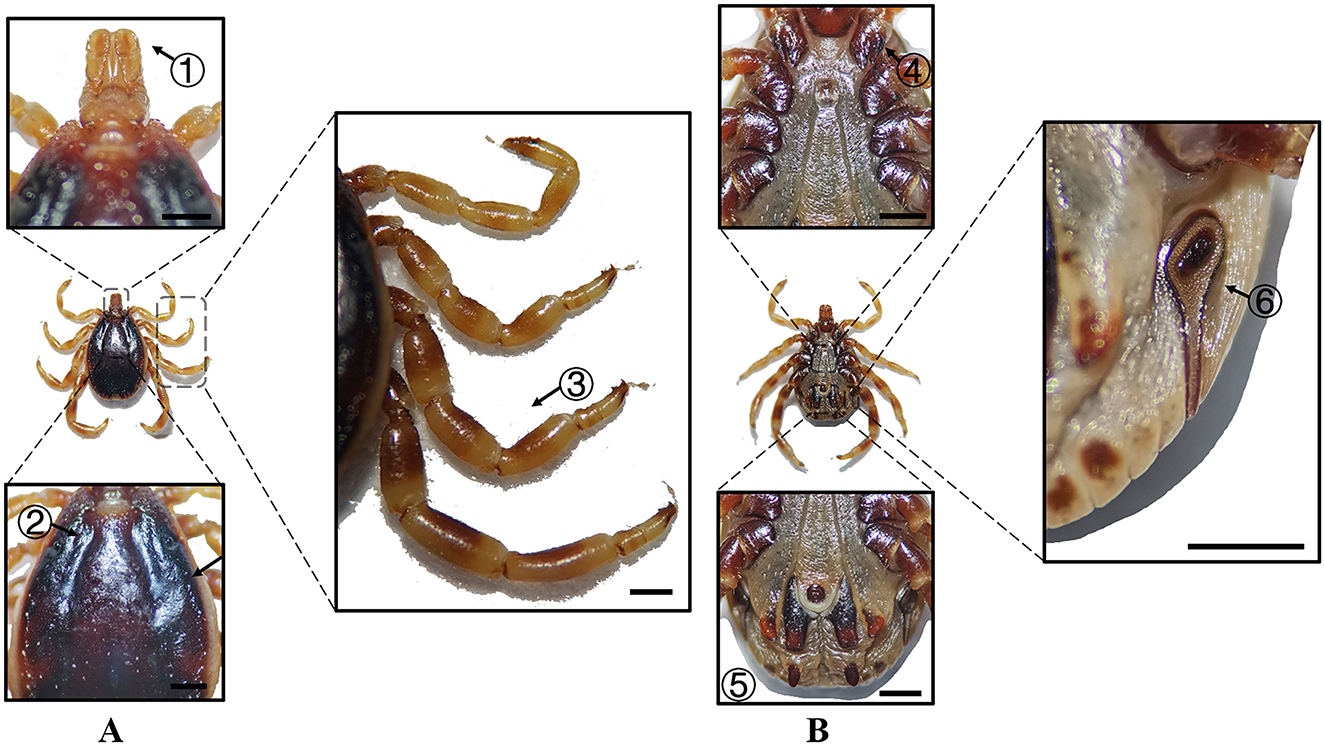
Figure 6. (A) Dorsal and (B) ventral views of a male H. dromedarii. Labels: (1) Palpi; (2) Cervical and lateral grooves; (3) Legs; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
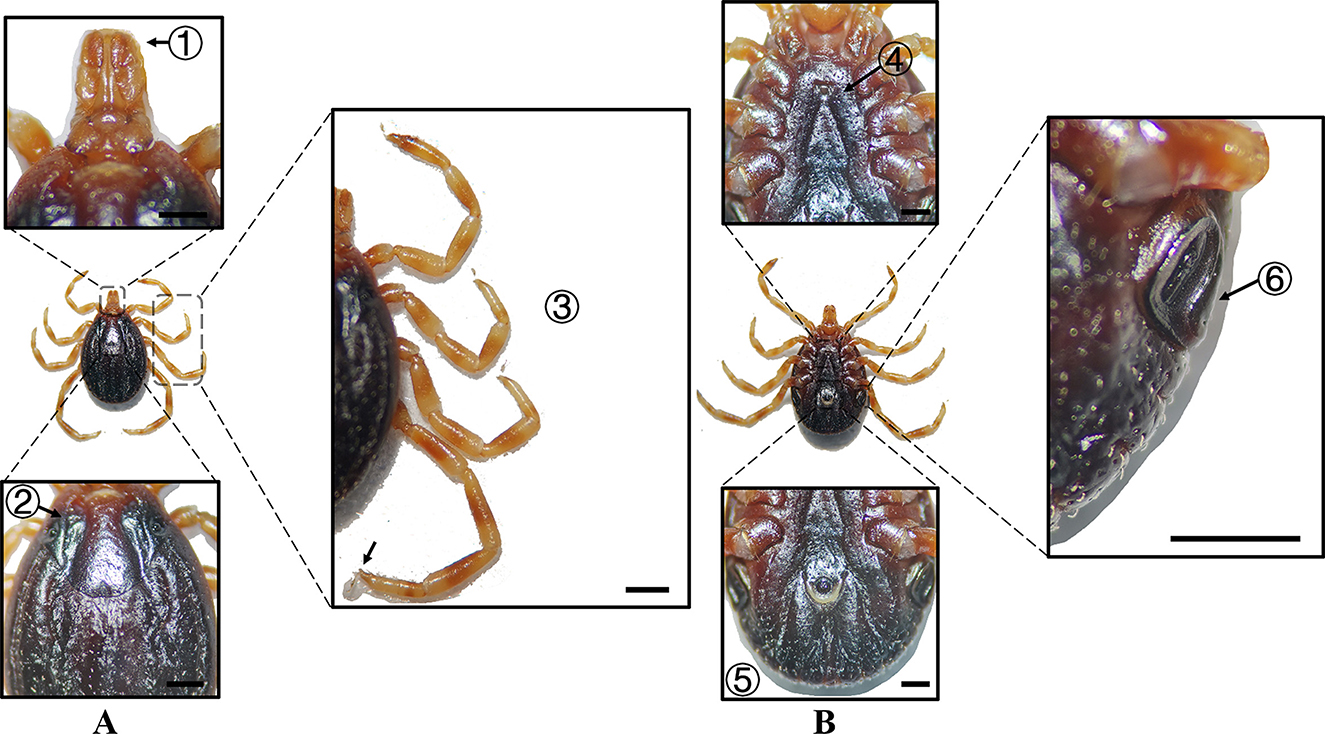
Figure 7. (A) Dorsal and (B) ventral views of a female H. dromedarii. Labels: (1) Palpi; (2) Cervical and lateral grooves; (3) Legs; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
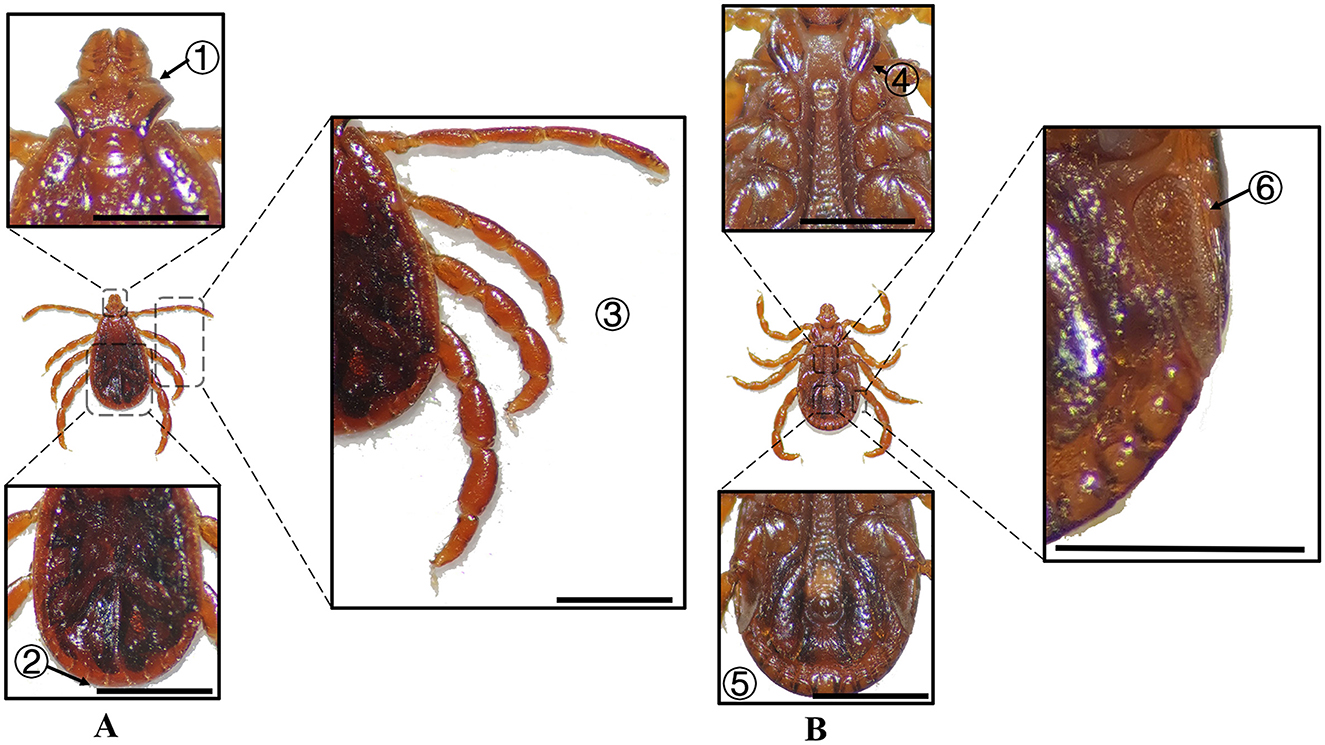
Figure 8. (A) Dorsal and (B) ventral views of a male R. turanicus. Labels: (1) Basis capituli; (2) Festoons; (3) Legs; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
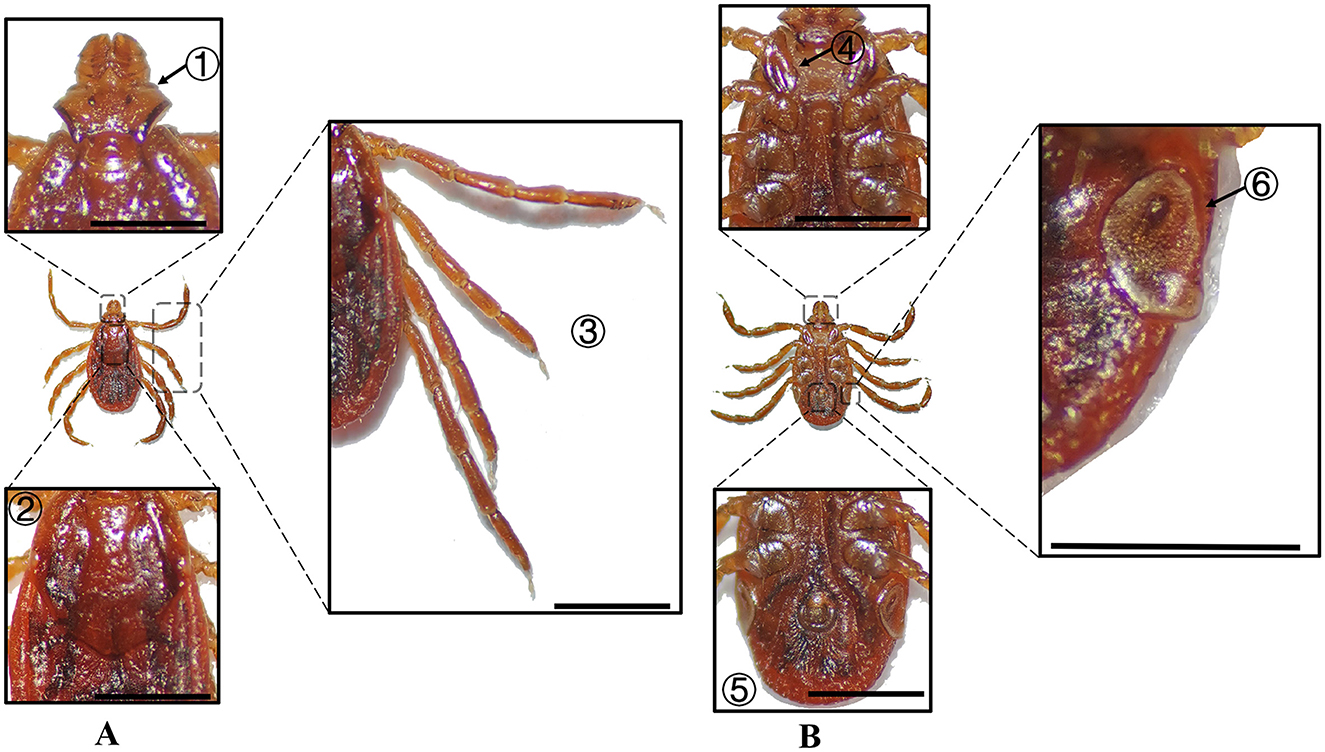
Figure 9. (A) Dorsal and (B) ventral views of a female R. turanicus. Labels: (1) Basis capituli; (2) Scutum; (3) Legs; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
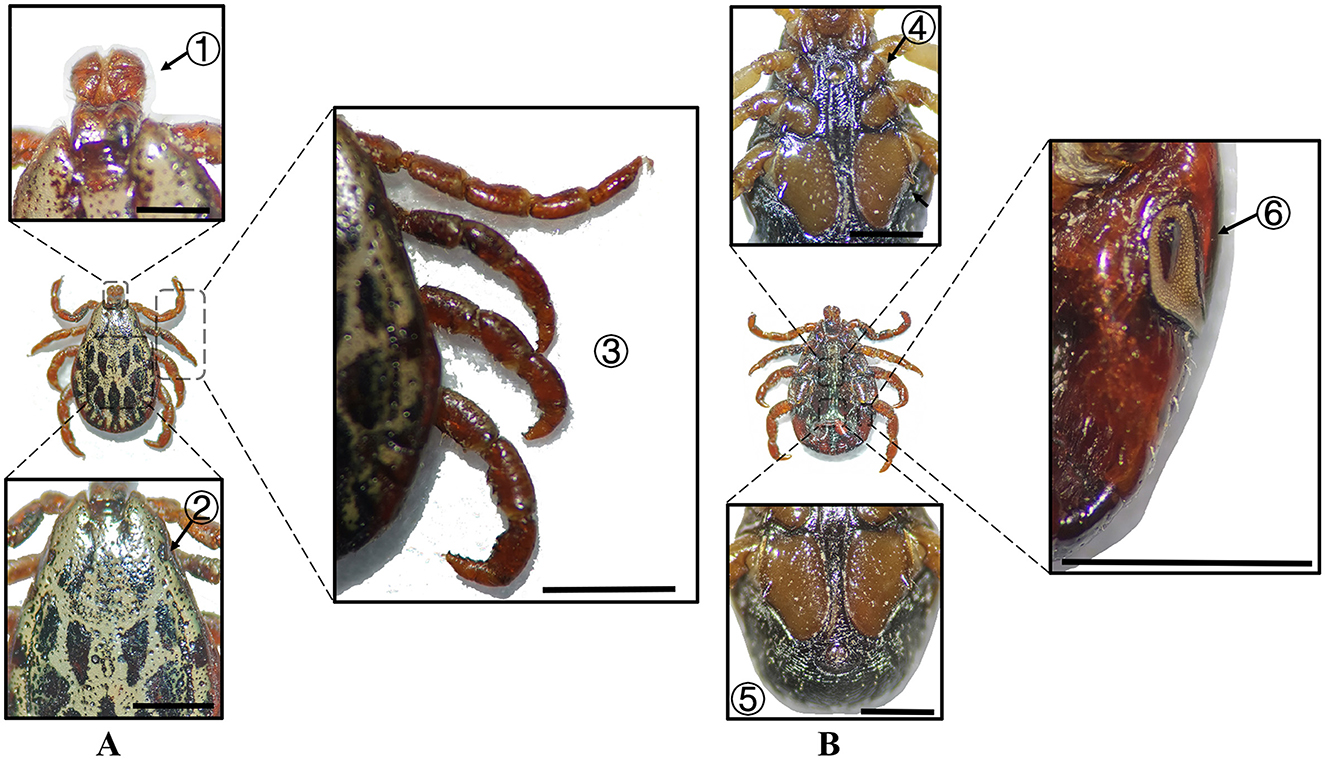
Figure 10. (A) Dorsal and (B) ventral views of a male D. marginatus. Labels: (1) Palps; (2) Eyes; (3) Legs; (4) External and internal spurs of coxae I; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
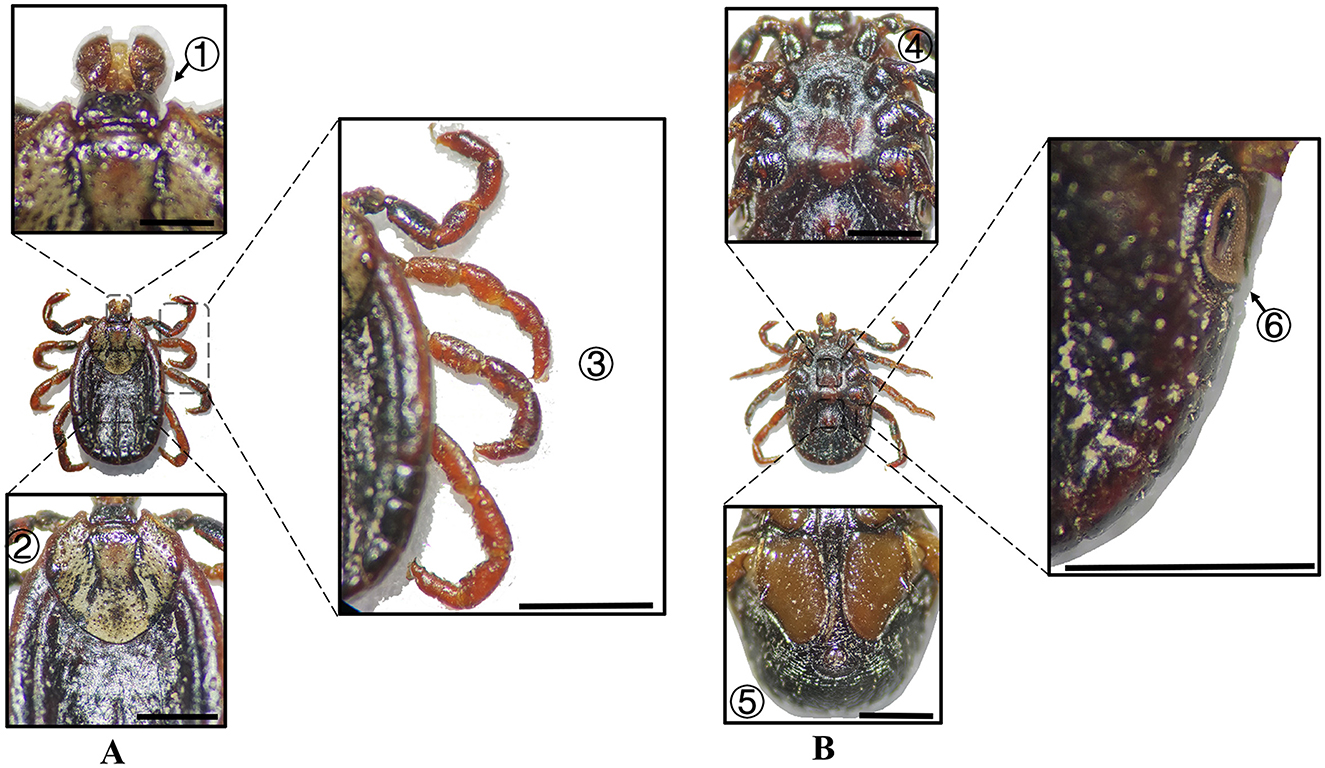
Figure 11. (A) Dorsal and (B) ventral views of a female D. marginatus. Labels: (1) Basis capituli; (2) Scutum; (3) Legs; (4) Genital aperture; (5) Anal sulcus; (6) Spiracular plate. Scale bar = 1 mm.
3.2 Phylogenetic analysis results
PCR amplification targeting the mitochondrial 16S rDNA and COI genes, along with the nuclear ITS2 region, was conducted using whole-genome DNA extracted from the ticks. Agarose gel electrophoresis revealed distinct bands of approximately 461 bp (16S rDNA), 709 bp (COI), and 821 bp (ITS2), corresponding to the expected fragment sizes (Supplementary Figures S1–S3 and Supplementary Table S2).
Phylogenetic analysis based on the 16S rDNA gene showed that H. anatolicum (PQ669102) from Turpan clustered with sequences from the United Arab Emirates; H. asiaticum (PQ669103) from Turpan was closely related to populations in Wensu, Xinjiang; H. dromedarii (PQ669104) from Jimsar grouped with sequences from Saudi Arabia; R. turanicus (PQ669105) from Aksu aligned with Kuqa, Xinjiang; and D. marginatus (PQ669101) from Jimsar clustered with sequences from Alashankou, Xinjiang (Figure 12).
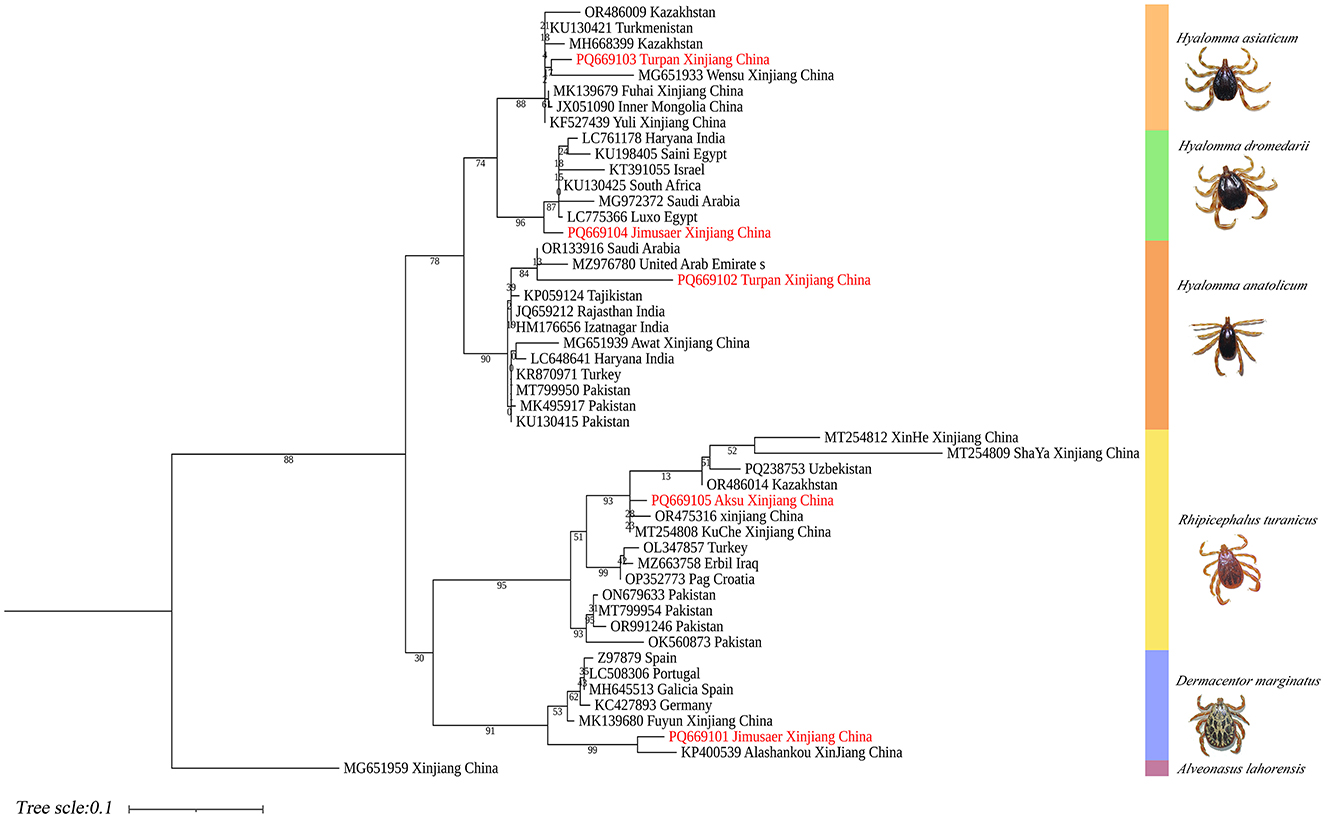
Figure 12. Phylogenetic tree of 16S rDNA genes from five tick species: H. anatolicum (PQ669102), H. asiaticum (PQ669103), H. dromedarii (PQ669104), R. turanicus (PQ669105), and D. marginatus (PQ669101). Alveonasus lahorensis (MG651959) was used as the outgroup. The tree was constructed using the maximum likelihood method under the Kimura 2-parameter + G + I model with 1,000 bootstrap replicates.
COI-based phylogenetic analysis revealed that H. anatolicum (PP886500) from Turpan clustered with specimens from the Zhanbyl Region, Kazakhstan; H. asiaticum (PQ669080) aligned with sequences from Kostanay City, Kazakhstan; H. dromedarii (PQ669079) grouped with sequences from the United Arab Emirates; R. turanicus (PP971648) aligned with samples from Luxor, Egypt; and D. marginatus (PP962438) clustered closely with specimens from Almaty, Kazakhstan (Figure 13).
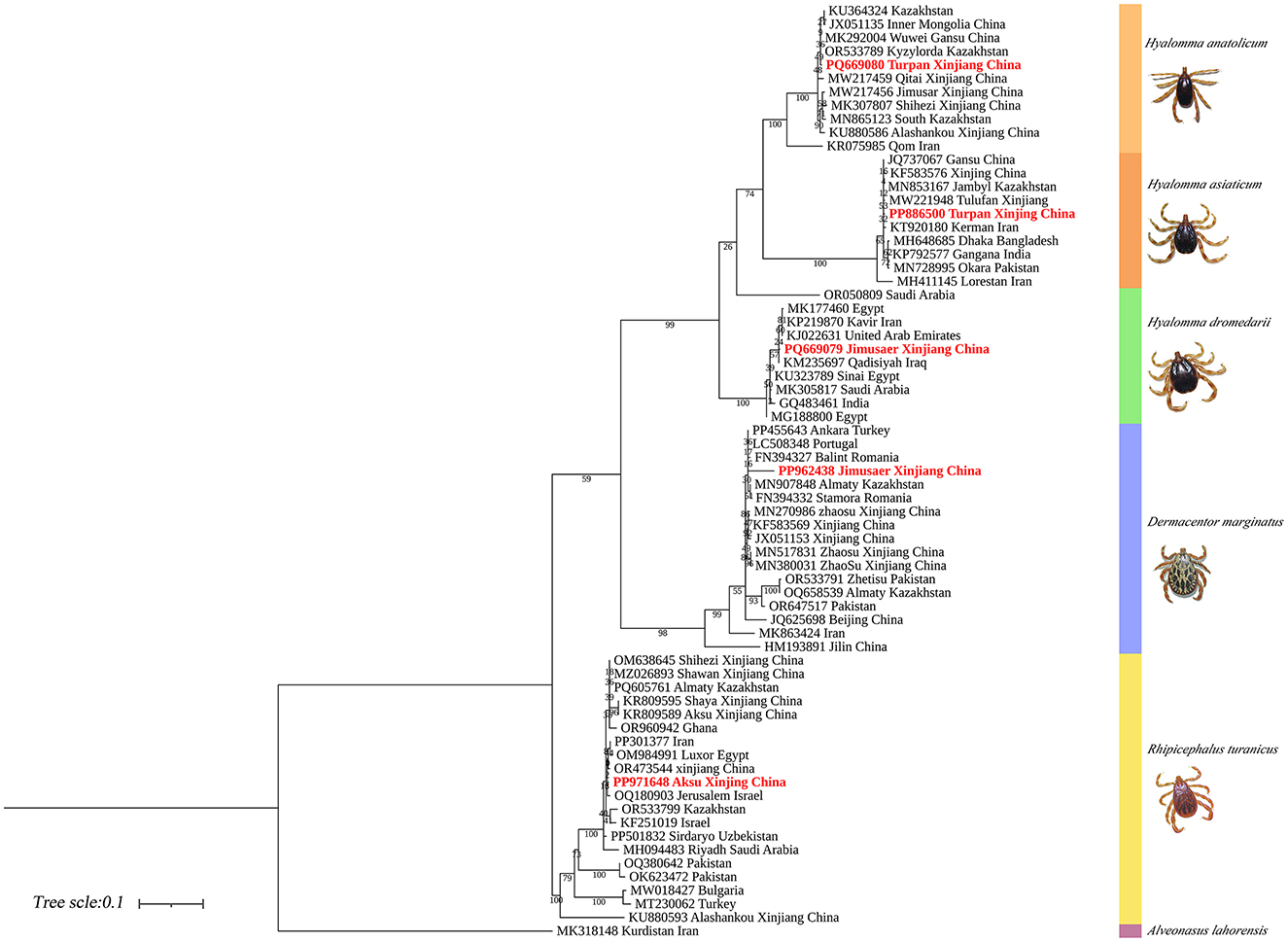
Figure 13. Phylogenetic tree of the COI gene for five tick species: H. anatolicum (PP886500), H. asiaticum (PQ669080), H. dromedarii (PQ669079), R. turanicus (PP971648), and D. marginatus (PP962438). Alveonasus lahorensis (MK318148) was used as the outgroup. The tree was constructed using the maximum likelihood method based on the Tamura-Nei + G + I model with 1,000 bootstrap replicates.
ITS2-based phylogenetic reconstruction demonstrated that H. anatolicum (PQ671043) from Turpan clustered with sequences from Iran; H. asiaticum (PQ671044) showed high genetic similarity to populations in Kerman Province, Iran; H. dromedarii (PQ671045) grouped with Iranian sequences; R. turanicus (PQ671046) aligned with other Iranian samples; and D. marginatus (PQ671042) from Jimsar exhibited strong homology with specimens from Alashankou, Xinjiang (Figure 14).
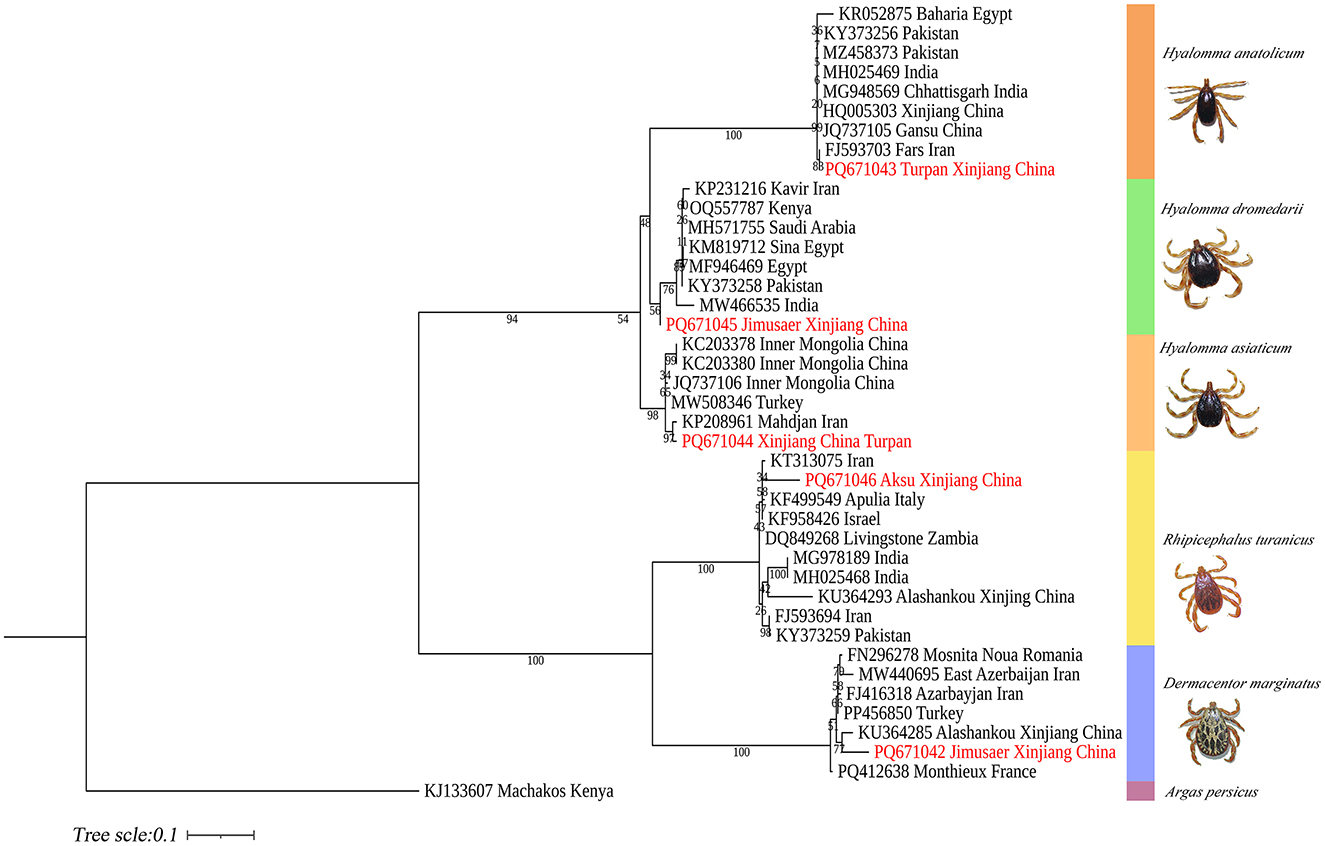
Figure 14. Phylogenetic tree of the ITS2 gene from five tick species: H. anatolicum (PQ671043), H. asiaticum (PQ671044), H. dromedarii (PQ671045), R. turanicus (PQ671046), and D. marginatus (PQ671042). Argas persicus (KJ133607) was used as the outgroup. The tree was constructed using the maximum likelihood method under the Tamura 3-parameter + G model with 1,000 bootstrap replicates.
4 Discussion
The Xinjiang Uygur Autonomous Region is located in northwest China, bordering the Eurasian continental core with over 5,600 kilometers of international boundaries shared with Russia, Kazakhstan, Kyrgyzstan, Tajikistan, Pakistan, Mongolia, India, and Afghanistan. Xinjiang encompasses extensive natural grasslands and supports a large livestock population. In recent years, its strategic geographic location and the rapid advancement of the Belt and Road Initiative have accelerated cross-border livestock movement, promoting the dissemination of ticks and increasing the risk of tick-borne pathogen transmission (9). Rapid shifts in tick population distributions, coupled with subtle interspecific morphological differences and a lack of high-resolution reference images, have substantially hindered morphological identification—particularly for congeneric species and for researchers without specialized taxonomic training.
In Xinjiang, H. anatolicum primarily transmits T. annulata, Babesia spp., and Anaplasma bovis. Meanwhile (30, 31), H. asiaticum serves as a vector for Rickettsia spp., Severe Fever with Thrombocytopenia Syndrome Virus, Babesia caballi, among others (22). These two species frequently co-occur due to overlapping host preferences and shared ecological niches (32). Morphological differences between H. anatolicum and H. asiaticum are subtle, particularly in immature or undersized H. asiaticum specimens, often leading to misidentification. Key distinguishing features include the posterior margin of the basis capituli in males, which is relatively straight in H. anatolicum but deeply concave in H. asiaticum. Additionally, the cervical groove in H. anatolicum males is shallow and tapers posteriorly, whereas in H. asiaticum, it is elongated and prominent, extending to the posterior third of the scutum (13–15). Contrary to previous reports (15), microscopic examination of H. anatolicum reveals distinct pale bands near the leg joints and a longitudinal pale stripe along the dorsal margin—features not readily visible under macroscopic observation. In contrast, H. asiaticum exhibits clearly visible pale bands at the leg joints under both microscopic and macroscopic conditions. Hyalomma dromedarii transmits several pathogens, including Coxiella burnetii, Borrelia spp., and Crimean-Congo hemorrhagic fever virus (33–35). It is the largest among the collected Hyalomma specimens, a trait likely reflecting its adaptation to parasitize camels (26), which possess larger body size, denser fur, and inhabit more extreme environments. Its robust legs, specialized mouthparts, and enlarged digestive system likely enhance its parasitic efficiency and survival. Misidentification with H. asiaticum is common; diagnostic features for H. dromedarii include: (1) in males, a dark brown ovoid scutum with coarse punctations that are sparsely distributed medially and concentrated laterally; (2) in females, an inverted triangular genital aperture—an important taxonomic marker. Rhipicephalus turanicus is a known vector for Babesia ovis, Babesia caballi, and other pathogens (6, 36). It exhibits a compact body and is often confused with R. sanguineus sensu stricto (s.s.), particularly in females. Both sexes of R. turanicus have short, stout palps, and males present distinct lamellar projections on the ventral side of the basis capituli, with dorsal extensions visible (26, 27, 37). Although broad festoons and caudal processes are typical diagnostic traits in males, the R. turanicus specimens collected in this study lacked these features. Nevertheless, they clustered clearly in molecular phylogenetic analyses (28). Dermacentor marginatus primarily transmits Rickettsia and Anaplasma spp., among other pathogens (38, 39). It displays enamel markings characteristic of the Dermacentor genus, facilitating rapid differentiation from other genera (29). However, D. marginatus exhibits minimal morphological divergence from its congeners D. nuttalli and D. silvarum. Therefore, molecular approaches are recommended to improve taxonomic resolution.
In this study, 788 ticks (~70% of the total) were collected from cattle in Turpan City. Among them, H. anatolicum exhibited close phylogenetic (9, 24, 32, 40) relationships with populations from Kazakhstan and Iran, likely due to the shared arid, high-temperature temperate continental climate that favors its survival. H. asiaticum specimens also showed close genetic affinities with Iranian populations. Despite sharing ecological niches with H. anatolicum, its three-host life cycle and longer developmental duration may constrain dispersal, limiting its genomic representation. H. dromedarii from Jimsar County clustered phylogenetically with Iranian populations. This may be attributed to historical camel husbandry and trade, facilitating gene flow. R. turanicus specimens from Aksu—geographically near Central Asia—exhibited close affinities with Iranian and Egyptian populations. Ancient host migration routes, such as the Silk Road, and convergent ecological adaptations may have maintained genetic connectivity (41). Dermacentor marginatus from Jimsar County showed strong phylogenetic similarity with specimens from Alashankou City, Xinjiang. This may result from environmental similarities (e.g., arid semi-desert steppe), seasonal livestock migration, or tick dispersal via migratory birds along the Alashankou corridor (38). Most ticks collected in this study were from bovines and ovines in the Turpan region (70% of total), while samples from camelids (Junggar Bactrian camels) and canines (Chinese rural dogs) were limited. This sampling imbalance may restrict interpretations of host-specific transmission dynamics of tick-borne pathogens.
Molecular identification is unaffected by developmental stage (larva, nymph, adult) or morphological integrity, thus overcoming the limitations of morphology-based taxonomy. By targeting signature genes, interspecies differentiation can be achieved, even among morphologically similar taxa. Three molecular markers offer complementary advantages: The 16S rDNA gene effectively reveals genetic differentiation driven by geographic isolation in population studies, but offers limited resolution for closely related species (17, 20, 42, 43). The COI gene (cytochrome oxidase I): a mitochondrial gene with high copy numbers, enabling efficient PCR amplification even from degraded or low-quantity samples (20). ITS2: a nuclear ribosomal DNA marker subject to lower selective pressure and higher evolutionary rates, resulting in greater interspecific variability than mitochondrial genes, making it ideal for resolving closely related taxa (20, 44). However, overreliance on ITS2 alignments in open-access databases such as NCBI can introduce misidentification due to annotation errors (45). For instance, in this study, R. turanicus ITS2 sequences showed 99% similarity with R. sanguineus in GenBank, likely stemming from early taxonomic ambiguities. The R. sanguineus group has long been taxonomically unstable due to overlapping morphological features with R. turanicus, lack of designated type specimens, potential hybridization, and limited diagnostic tools. Consequently, R. sanguineus s.s. and R. turanicus were frequently misidentified under the umbrella term “R. sanguineus” (27, 37). Recent taxonomic revisions now recognize R. sanguineus s.s. and R. turanicus as distinct species. The term “R. sanguineus sensu lato (s.l.)” is recommended for ticks morphologically resembling R. sanguineus but lacking genetic confirmation or showing divergent genotypes (37). To resolve these challenges, integrative analyses incorporating both mitochondrial and nuclear markers are strongly recommended to improve species delimitation and mitigate database inconsistencies among cryptic taxa (20, 45, 46).
Accurate identification of tick species requires the integration of morphological characterization and molecular diagnostics. By combining analyses of morphological traits with genetic sequence alignment, this dual approach enables precise taxonomic classification and robust phylogenetic inference. This multidimensional strategy addresses the limitations inherent in single-method identifications and generates critical data for elucidating ecological transmission pathways of tick-borne diseases and pathogen evolution.
5 Conclusions
The integrative morphological-molecular framework effectively resolves taxonomic ambiguities among five tick species, wherein diagnostic traits—such as capitulum morphology and spiracular plate structure—complement multi-locus phylogenetic analyses based on 16S rDNA, COI, and ITS2 sequences. Genetic affinities observed between tick populations in Xinjiang and lineages from Central Asia and the Middle East suggest that transboundary dispersal is likely facilitated by livestock trade and climate-driven habitat shifts. The limitations of single-gene identification methods and the presence of cryptic species (e.g., Rhipicephalus turanicus vs. R. sanguineus sensu stricto) underscore the necessity for high-resolution imaging and multi-gene validation to ensure accurate species delimitation. This framework enhances global tick surveillance capacity and informs targeted control strategies in increasingly dynamic ecological landscapes.
Data availability statement
The sequences obtained in this study have been deposited in the NCBI GenBank database under the following accession numbers: 16S rDNA gene (PQ669101-PQ669105), COI gene (PP886500, PQ669080, PQ669079, PP971648, PP962438), and ITS2 gene (PQ671042-PQ671046). All other data generated or analyzed during this study are available in the published article and its Supplementary Information files.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
GS: Methodology, Writing – original draft, Investigation, Formal analysis, Data curation. LX: Resources, Writing – review & editing, Investigation. ZL: Writing – review & editing, Resources, Investigation. WM: Writing – review & editing, Investigation, Resources. DY: Investigation, Resources, Writing – review & editing. CL: Software, Writing – review & editing, Data curation, Methodology, Validation. BC: Funding acquisition, Methodology, Supervision, Writing – review & editing, Conceptualization. QG: Supervision, Data curation, Methodology, Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the following projects National Key Research and Development Program of China (No. 2021YFD1600702-3), and Innovative Research Team on Biological Vectors and Transmission of Vector-borne Zoonotic Diseases (No. 2023TSYCTD0008).
Acknowledgments
The authors sincerely thank all participants involved in this study, particularly the farmers and veterinarians for their contributions. The authors declare that they have no competing interests.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2025.1623318/full#supplementary-material
References
1. de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. (2008) 13:6938–46. doi: 10.2741/3200
2. Eisen L. Tick species infesting humans in the United States. Ticks Tick Borne Dis. (2022) 13:102025. doi: 10.1016/j.ttbdis.2022.102025
3. Wu X-B, Na R-H, Wei S-S, Zhu J-S, Peng H-J. Distribution of tick-borne diseases in China. Parasit Vectors. (2013) 6:119. doi: 10.1186/1756-3305-6-119
4. Zhuang L, Du J, Cui XM, Li H, Tang F, Zhang PH, et al. Identification of tick-borne pathogen diversity by metagenomic analysis in Haemaphysalis Longicornis from Xinyang, China. Infect Dis Poverty. (2018) 7:45. doi: 10.1186/s40249-018-0417-4
5. Sheng J, Jiang M, Yang M, Bo X, Zhao S, Zhang Y, et al. Tick distribution in border regions of Northwestern China. Ticks Tick Borne Dis. (2019) 10:665–9. doi: 10.1016/j.ttbdis.2019.02.011
6. Li Y, Wen X, Li M, Moumouni PFA, Galon EM, Guo Q, et al. Molecular detection of tick-borne pathogens harbored by ticks collected from livestock in the Xinjiang Uygur Autonomous Region, China. Ticks Tick Borne Dis. (2020) 11:101478. doi: 10.1016/j.ttbdis.2020.101478
7. Schön MP. The tick and I: parasite-host interactions between ticks and humans. J Dtsch Dermatol Ges. (2022) 20:818–53. doi: 10.1111/ddg.14821
8. Madison-Antenucci S, Kramer LD, Gebhardt LL, Kauffman E. Emerging tick-borne diseases. Clin Microbiol Rev. (2020) 33:e00083–18. doi: 10.1128/CMR.00083-18
9. Zhao GP, Wang YX, Fan ZW, Ji Y, Liu MJ, Zhang WH, et al. Mapping ticks and tick-borne pathogens in China. Nat Commun. (2021) 12:1075. doi: 10.1038/s41467-021-21375-1
10. Zhong Z, Wang K, Wang J. Tick symbiosis. Curr Opin Insect Sci. (2024) 62:101163. doi: 10.1016/j.cois.2024.101163
11. Gilbert L. The impacts of climate change on ticks and tick-borne disease risk. Annu Rev Entomol. (2021) 66:373–88. doi: 10.1146/annurev-ento-052720-094533
12. Sakamoto JM. Progress, challenges, and the role of public engagement to improve tick-borne disease literacy. Curr Opin Insect Sci. (2018) 28:81–9. doi: 10.1016/j.cois.2018.05.011
15. Yu X, Ye R-Y, Zheng-da G. Acarina of Xinjiang. Ürümqi: Xinjiang Science, Technology and Health Publishing House (1997).
16. Wang T, Zhang S, Pei T, Yu Z, Liu J. Tick mitochondrial genomes: structural characteristics and phylogenetic implications. Parasit Vectors. (2019) 12:451. doi: 10.1186/s13071-019-3705-3
17. Black WC, Piesman J. Phylogeny of hard- and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc Nat Acad Sci U S A. (1994) 91:10034–8. doi: 10.1073/pnas.91.21.10034
18. Zhang RL, Zhang B. Prospects of using DNA barcoding for species identification and evaluation of the accuracy of sequence databases for ticks (Acari: Ixodida). Ticks Tick Borne Dis. (2014) 5:352–8. doi: 10.1016/j.ttbdis.2014.01.001
19. Young MR, deWaard JR, Hebert PDN. DNA barcodes enable higher taxonomic assignments in the Acari. Sci Rep. (2021) 11:15922. doi: 10.1038/s41598-021-95147-8
20. Lv J, Wu S, Zhang Y, Chen Y, Feng C, Yuan X, et al. Assessment of four DNA fragments (Coi, 16s Rdna, Its2, 12s Rdna) for species identification of the Ixodida (Acari: Ixodida). Parasit Vectors. (2014) 7:93. doi: 10.1186/1756-3305-7-93
21. Song S, Shao R, Atwell R, Barker S, Vankan D. Phylogenetic and phylogeographic relationships in ixodes holocyclus and ixodes cornuatus (Acari: Ixodidae) inferred from Cox1 and ITS2 Sequences. Int J Parasitol. (2011) 41:871–80. doi: 10.1016/j.ijpara.2011.03.008
22. Wang B, Liu Z, Zhu S, Zhang J, Qi W, Wang J, et al. Molecular characterization of tick-borne bacterial and protozoan pathogens in parasitic ticks from Xinjiang, China. Parasit Vectors. (2025) 18:207. doi: 10.1186/s13071-025-06857-1
23. Zhang B, Zhang N, Zheng T, Lu M, Baoli B, Jie R, et al. Tick-borne bacterial agents in Hyalomma asiaticum ticks from Xinjiang Uygur Autonomous Region, Northwest China. Parasit Vectors. (2024) 17:167. doi: 10.1186/s13071-024-06256-y
24. Ma R, Li C, Gao A, Jiang N, Li J, Hu W, et al. Tick species diversity and potential distribution alternation of dominant ticks under different climate scenarios in Xinjiang, China. PLoS Negl Trop Dis. (2024) 18:e0012108. doi: 10.1371/journal.pntd.0012108
25. Zhao L, Lv J, Li F, Li K, He B, Zhang L, et al. Identification and molecular analysis of ixodid ticks (Acari: Ixodidae) infesting domestic animals and tick-borne pathogens at the Tarim Basin of Southern Xinjiang, China. Korean J Parasitol. (2020) 58:37–46. doi: 10.3347/kjp.2020.58.1.37
26. Senbill H, Tanaka T, Karawia D, Rahman S, Zeb J, Sparagano O, et al. Morphological identification and molecular characterization of economically important ticks (Acari: Ixodidae) from North and North-Western Egypt. Acta Trop. (2022) 231:106438. doi: 10.1016/j.actatropica.2022.106438
27. Nava S, Beati L, Venzal JM, Labruna MB, Szabó MPJ, Petney T, et al. Rhipicephalus sanguineus (Latreille, 1806): neotype designation, morphological re-description of all parasitic stages and molecular characterization. Ticks Tick Borne Dis. (2018) 9:1573–85. doi: 10.1016/j.ttbdis.2018.08.001
28. Millán J, Rodríguez-Pastor R, Estrada-Peña A. Description of Rhipicephalus hibericus sp. nov. (Ixodoidea: Ixodidae), a species of the Rhipicephalus sanguineus group in southwestern Europe. Ticks Tick Borne Dis. (2024) 15:102340. doi: 10.1016/j.ttbdis.2024.102340
29. Johnson N. Chapter 2 - tick classification and diversity. In:Johnson N, , editor. Ticks. Cambridge, MA: Academic Press (2023). p. 9–23. doi: 10.1016/B978-0-323-91148-1.00013-7
30. Yu P, Liu Z, Niu Q, Yang J, Abdallah MO, Chen Z, et al. Molecular evidence of tick-borne pathogens in Hyalomma anatolicum ticks infesting cattle in Xinjiang Uygur Autonomous Region, Northwestern China. Exp Appl Acarol. (2017) 73:269–81. doi: 10.1007/s10493-017-0162-6
31. Li Y, Li J, Xieripu G, Rizk MA, Macalanda AMC, Gan L, et al. Molecular detection of Theileria ovis, Anaplasma ovis, and Rickettsia Spp. in Rhipicephalus turanicus and Hyalomma anatolicum collected from sheep in Southern Xinjiang, China. Pathogens. (2024) 13:680. doi: 10.3390/pathogens13080680
32. Sands AF, Apanaskevich DA, Matthee S, Horak IG, Harrison A, Karim S, et al. Effects of tectonics and large scale climatic changes on the evolutionary history of Hyalomma ticks. Mol Phylogenet Evol. (2017) 114:153–65. doi: 10.1016/j.ympev.2017.06.002
33. El Tigani-Asil ETA, Blanda V, Abdelwahab GE, Hammadi ZMA, Habeeba S, Khalafalla AI, et al. Molecular investigation on tick-borne hemoparasites and Coxiella burnetii in dromedary camels (Camelusdromedarius) in Al Dhafra Region of Abu Dhabi, UAE. Animals. (2021) 11:666. doi: 10.3390/ani11030666
34. Elhelw R, Elhariri M, Hamza D, Abuowarda M, Ismael E, Farag H. Evidence of the presence of Borrelia burgdorferi in dogs and associated ticks in Egypt. BMC Vet Res. (2021) 17:49. doi: 10.1186/s12917-020-02733-5
35. Schulz A, Karger A, Bettin B, Eisenbarth A, Sas MA, Silaghi C, et al. Molecular discrimination of Hyalomma tick species serving as reservoirs and vectors for crimean-congo hemorrhagic fever virus in sub-Saharan Africa. Ticks Tick Borne Dis. (2020) 11:101382. doi: 10.1016/j.ttbdis.2020.101382
36. Wang N, Lu S, Ye RZ, Li C, Huang JH, Ye G, et al. Virome of Hyalomma and Rhipicephalus ticks from Desert of Northwestern China. Virus Evol. (2025) 11:veaf022. doi: 10.1093/ve/veaf022
37. Dantas-Torres F, de Sousa-Paula LC, Otranto D. The Rhipicephalus sanguineus group: updated list of species, geographical distribution, and vector competence. Parasit Vectors. (2024) 17:540. doi: 10.1186/s13071-024-06572-3
38. Song R, Ma Y, Hu Z, Li Y, Li M, Wu L, et al. Maxent modeling of Dermacentor marginatus (Acari: Ixodidae) distribution in Xinjiang, China. J Med Entomol. (2020) 57:1659–67. doi: 10.1093/jme/tjaa063
39. Huercha, Song R, Li M, Fan X, Hu Z, Wu L, et al. Caracterization of glutathione S-transferase of Dermacantor marginatus and effect of the recombinant antigen as a potential anti-tick vaccine. Vet Parasitol. (2020) 279:109043. doi: 10.1016/j.vetpar.2020.109043
40. Hu E, Hu Z, Mi X, Li C, He W, Gan L, et al. Distribution prediction of Hyalomma asiaticum (Acari: Ixodidae) in a localized region in Northwestern China. J Parasitol. (2022) 108:330–6. doi: 10.1645/21-96
41. Petrović A, Stanić K, Popović A, Ivanović I, Supić D, Marinković D, et al. Seasonal dynamics and physiological age of ixodid ticks collected from dogs. Animals. (2023) 13:3026. doi: 10.3390/ani13193026
42. Li LH, Zhang Y, Wang JZ Li XS, Yin SQ, Zhu D, et al. High genetic diversity in hard ticks from a China-Myanmar Border County. Parasit Vectors. (2018) 11:469. doi: 10.1186/s13071-018-3048-5
43. Filipe D, Parreira R, Pereira A, Galvão N, Cristóvão JM, Nunes M, et al. Preliminary comparative analysis of the resolving power of Cox1 and 16s-Rdna as molecular markers for the identification of ticks from Portugal. Vet Parasitol Reg Stud Reports. (2021) 24:100551. doi: 10.1016/j.vprsr.2021.100551
44. Etiang P, Atim SA, Nkamwesiga J, Nalumenya D, Byaruhanga C, Odongo S, et al. Identification and distribution of Rhipicephalus microplus in selected high-cattle density districts in Uganda: signaling future demand for novel tick control approaches. BMC Vet Res. (2024) 20:119. doi: 10.1186/s12917-024-03979-z
45. Khumalo N, Chaisi M, Magoro R, Mwale M. An analysis of the gaps in the South African DNA barcoding library of ticks of veterinary and public health importance. Genome. (2024) 67:392–402. doi: 10.1139/gen-2024-0052
Keywords: tick, Hyalomma anatolicum, Hyalomma asiaticum, Hyalomma dromedarii, Rhipicephalus turanicus, Dermacentor marginatus, Identification
Citation: Shi G, Xin L, Li Z, Ma W, Yang D, Li C, Chahan B and Guo Q (2025) Integrative morpho-molecular delineation of five medically significant tick species: facilitating precision-based vector surveillance. Front. Vet. Sci. 12:1623318. doi: 10.3389/fvets.2025.1623318
Received: 05 May 2025; Accepted: 01 July 2025;
Published: 08 August 2025.
Edited by:
Nicola Pugliese, University of Bari Aldo Moro, ItalyReviewed by:
Kokouvi Kassegne, Shanghai Jiao Tong University, ChinaAshley Kennedy, Delaware Division of Fish and Wildlife, United States
Copyright © 2025 Shi, Xin, Li, Ma, Yang, Li, Chahan and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bayin Chahan, MjUxNDA2Mjg4MUBxcS5jb20=; Qingyong Guo, ZHlncXlAZWR1LnhqYXUuY24=
 Guangxin Shi
Guangxin Shi Lianxi Xin1
Lianxi Xin1 Wanpeng Ma
Wanpeng Ma Caishan Li
Caishan Li