- 1College of Food Science and Engineering, Hainan University, Haikou, Hainan, China
- 2College of Materials Science and Engineering, Hainan University, Haikou, Hainan, China
To improve the silage quality of wilted king grass, the study investigated the effects of Limosilactobacillus fermentum HHL-5 alone and L. fermentum complex citric acid fermentation on wilted king grass silage. Four experimental groups were designed as follows: no additive (CK), citric acid addition (CA), L. fermentum addition (L), and combined L. fermentum and citric acid addition (LCA). The fermentation quality, microbial composition, and aerobic stability of the silage in each group were analyzed. After 30 days of ensiling, LCA had the highest protein content, and L had the lowest ADF content (p < 0.05). Lactic acid and acetic acid contents were significantly increased in the LCA group (p < 0.05), whereas lactic acid content was increased and acetic acid content was significantly decreased in the L group (p < 0.05). Ammonia nitrogen content was significantly decreased in the CA and LCA groups (p < 0.05). The L group was not significantly different from the CK group in terms of bacterial diversity and relative abundance, whereas the LCA group showed markedly higher bacterial diversity and was considerably different from the CK group. The relative abundance of Lactobacillus in the LCA group was higher than that in the L group, while that of Enterobacterales was lower. Compared to the addition of L. fermentum alone, complex citric acid silage significantly enhanced the aerobic stability of this feed. In summary, the application of L. fermentum combined with citric acid can more effectively improve the quality of wilted king grass silage.
1 Introduction
In recent years, the rapid expansion of ruminant farming has significantly increased the demand for roughage. The supply of high-quality roughage is essential for advancing grass-fed animal husbandry, with silage serving as a practical method to ensure year-round feed availability (1). King grass (Pennisetum purpureum × P. glaucum, KG) is a hybrid species of the Poaceae family, derived from an intergeneric cross between elephant grass (P. purpureum, 2n = 28) and pearl millet (P. glaucum, 2n = 14), king grass inherits the beneficial traits of both parent species and is widely recognized as one of the “top high-yield forage grasses”. Its exceptional biomass productivity, wide adaptability, and diverse applications make it an essential economic and ecological resources in tropical and subtropical regions (2). King grass exhibits relatively poor silage quality when directly ensiled. Therefore, the addition of appropriate additives is required for improved preservation. Silage additives are typically classified into two main types: fermentation enhancers and inhibitors. Fermentation enhancers are additives that enhance the fermentation process by lactic acid bacteria. These primarily include fermentable substrates (e.g., sugars and molasses), enzymatic preparations, and specific bacterial inoculants. While fermentation inhibitors increase acidity, lower the pH of silage, and suppress the proliferation of harmful microorganisms. Common inhibitors include inorganic compounds such as aldehydes, salts, and acid–base substances, as well as organic acids such as formic, acetic, citric, and malic acids (3).
In livestock and poultry production, the use of low-dose antibiotics is often unavoidable. Nevertheless, their extensive application in animal feed has led to a range of detrimental consequences, such as antibiotic resistance (4), drug residues in food (5), and environmental pollution (6). Therefore, selecting the appropriate silage additives is crucial for improving feed quality and mitigating these negative effects. In this context, lactic acid bacteria (LAB) are essential for silage fermentation, as they produce organic acids that rapidly lower pH (7), shorten fermentation time, improve feed palatability, and reduce nutrient losses in forage (8). Among the various LAB species, Limosilactobacillus fermentum is particularly notable. Difference ordinary lactic acid bacteria, L. fermentum strains are highly resistant to acidic and bile conditions (9), making them well-suited for silage fermentation. Research indicates that combinations of propionic acid + Limosilactobacillus plantarum + L. fermentum, and propionic acid + L. fermentum are particularly effective in improving silage fermentation quality. Additionally, the combination of propionic acid + L. fermentum has shown superior potential in reducing yeast and mold growth after aerobic exposure, making it an effective approach for improving silage preservation (9). However, research on using L. fermentum as an additive is limited, with most studies focusing on L. plantarum. One such promising additive is citric acid, which is considered safer and milder than other commonly used acids such as formic or acetic acid (10). Citric acid not only improves feed efficiency, animal health, and productivity (11), but is also cost-effective and can be easily produced through microbial fermentation (12). Studies have demonstrated that adding malic or citric acid, in combination with L. plantarum, can improve fermentation quality, limit protein hydrolysis, and enhance the fatty acid composition of alfalfa silage (13). Citric acid also inhibits undesirable microorganisms and works synergistically with LAB to enhance silage quality (13).
Building on this, the present study aimed to assess the physicochemical properties of king grass silage from four treatment groups: without additives (CK), with citric acid (CA), with L. fermentum HHL-5 (L), and with the combination of L. fermentum and citric acid fermentation (LCA). High-throughput sequencing was employed to analyze the microbial communities in these silage groups, allowing for a deeper understanding of how different conditions affect bacterial populations. Additionally, the aerobic stability and shelf life of the silages were measured, and findings providing a solid theoretical foundation for the research and production of high-quality king grass silage.
2 Materials and methods
2.1 Limosilactobacillus fermentum HHL-5 and citric acid
L. fermentum HHL-5: Isolated from the Food Biotechnology Laboratory, School of Food Science and Engineering, Hainan University.
Citric acid: Purchased from China National Pharmaceutical Group Corporation (Sinopharm Group Chemical Reagent Co., Ltd.).
2.2 Collection of king grass samples
King grass (Thermal Research No. 4) was collected from the Montenegrin sheep breeding farm at the Institute of Tropical Crop Variety Resources, Chinese Academy of Tropical Agricultural Sciences (CATAS). The collected king grass was chopped into 2–5 cm pieces. After chopping, the grass was transferred to a sterile bags and transported to the laboratory, where it was air-dried within 48 h to achieve a moisture content of less than 65%. Select the period time without precipitation for 7 consecutive days for the sample, and the sampling time was November 2022.
2.3 King grass silage preparation and grouping
The experimental design included four groups, as shown in Table 1. The groups were as follows: CK (no additives), CA (citric acid added), L (L. fermentum HHL-5 added), and LCA (L. fermentum HHL-5 added 0.1% and citric acid added 0.15%). Each group included three replicates, and the sealed silage was stored at room temperature, protected from light. Silage temperature: 30 °C.
2.4 Analysis of the composition of king grass silage
2.4.1 Nutrient analysis of king grass
Dry matter (DM) content was determined according to the method outlined in GB/T6435-2014 for Determination of Moisture in Feed.
Crude protein content was determined using the Kjeldahl method for king grass silage.
Acid detergent fiber (ADF) content was determined following the method outlined in NY/T 1459-2022 for Determination of Acid Detergent Fiber in Feed.
Neutral detergent fiber (NDF) content was determined according to the method specified in GB/T 20806–2022 for Determination of Neutral Detergent Fiber (NDF) in Feed.
Soluble carbohydrate (WSC) content was determined using the anthrone colorimetric method (14).
2.4.2 Measurement of silage fermentation quality
Determination of silage pH: At each designated sampling time, silage bags from each group were opened, and the upper layer of the sample was mixed. A 10 g portion of the mixed silage was transferred to a 150 mL sterile conical flask, followed by the addition of 90 mL of sterile water. The mixture was stirred until the sample was fully submerged, then sealed. After shaking for 24 h, the mixture was filtered to separate the solid residue, obtaining the leachate. A portion of the leachate was used to measure the pH with a pH meter, while the remainder was stored in a sterile centrifuge tube.
Determination of organic acid content: Centrifuge the prepared silage sample supernatant at 10,000 rpm for 10 min using a high-speed centrifuge to obtain the supernatant. Withdraw the extract using a disposable syringe and filter it through a 0.22 μm cellulose membrane (organic type) to remove impurities. Carefully withdraw the filtered sample using a 10 μL injector, ensuring that no air bubbles are present in the injector. Analyze the sample using high-performance liquid chromatography (HPLC) (15).
Ammonia nitrogen content was determined using the phenol-hypochlorite method (14).
2.5 Sequencing the bacterial composition of king grass silage
2.5.1 Extraction of microbial genomic DNA of king grass
For each group of king grass silage, a 25 g of samples was weighed, mixed with 225 mL of sterile water, and incubated in a shaker for 2 h. The silage residue was filtered, and the filtrate was centrifuged for 15 min to collect the precipitate. The microbial genomic DNA was then extracted from the king grass silage samples using the CTBA method.
2.5.2 PCR amplification and sequencing
The bacterial DNA extracted from king grass silage was amplified by PCR. The sequence information of the PCR primers is provided in Table 2, and the PCR reaction system is detailed in Table 3. The amplified products were then sent to Guangdong Mega Gene Technology Co., Ltd. for sequencing.
2.5.3 Bioinformatics and data analysis
Based on the effective data and OTU annotation table provided by the sequencing company, R software was used to analyze the community composition at the kingdom, phylum, class, order, family, genus, and species levels. Alpha diversity analysis was performed using the OTU relative abundance table and the alpha_div command in usearch (V10). A custom Python script (Python v2.7) was used to plot the dilution curve and Rank Abundance curve. PCA analysis was performed using the prcomp function in R, and PCoA plots were generated using the vegan package in R. Species composition and relative abundance information of the research samples were extracted using GraPhlAn software. The sequencing data from this study have been submitted to the National Center for Biotechnology Information Sequence Read Archive database under Bioproject accession number of PRJNA1333438.
2.6 Determination of the aerobic stability of king grass silage
After 30 days of silage fermentation, the sample bags were opened, mixed, and placed at room temperature. The pH was measured at 72, 144, and 216 h post-opening to assess aerobic stability based on pH changes.
2.7 Data processing and analysis
Experimental data were processed in Excel 2020. Statistical analysis of variance was performed using SPSS 26. Graphs were poltted using Origin 2017. The bacterial and fungal communities sequencing data were generated by the Illumina HiSeq sequencing platform.
3 Results
3.1 Nutrient analysis of king grass silage
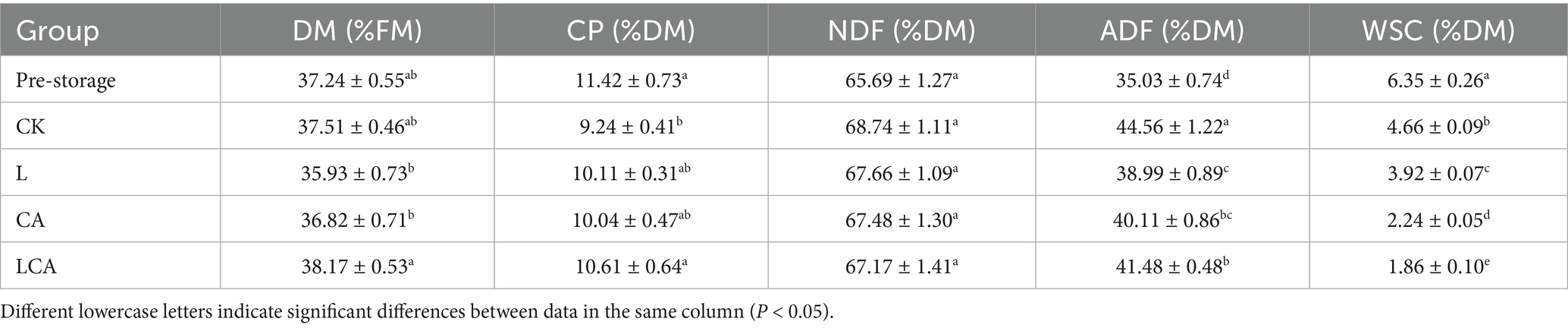
Table 4 presented the nutrient composition of king grass silage for different treatment groups. No significant difference in dry matter (DM) content was observed among the four treatment groups after 30 days of silage compared to pre-silage (p > 0.05) (Table 4). The DM content in the LCA group was significantly higher than in the L and CA groups (p < 0.05). After 30 days of ensiling, the crude protein content decreased significantly in the CK group compared to pre-silage levels (p < 0.05), whereas no significant change in crude protein content was observed in the L, CA, and LCA groups (p > 0.05). The crude protein content in the LCA group was significantly higher than that in the CK group (p < 0.05). However, no significant difference was observed between the LCA, L, and CA groups (p > 0.05). Additionally, there was no significant difference between the crude protein contents of the L and CA groups (p > 0.05). This result suggested that the addition of citric acid or L. fermentum alone did not reduce crude protein loss during silage. However, combining both additives effectively reduced crude protein loss. After 30 days of ensiling, the contents of NDF and ADF were significantly higher in all four treatment groups compared to pre-silage levels (p < 0.05). No significant difference in NDF content was observed among the four treatment groups (p > 0.05). However, ADF content was significantly lower in the L, CA, and LCA groups, with the L group showing the lowest ADF content (p < 0.05). After 30 days of ensiling, the WSC content was significantly lower in all four treatment groups compared to pre-silage levels (p < 0.05). The WSC content ranked from highest to lowest as follows: CK > L > CA > LCA.
3.2 Quality and quality analysis of king grass silage
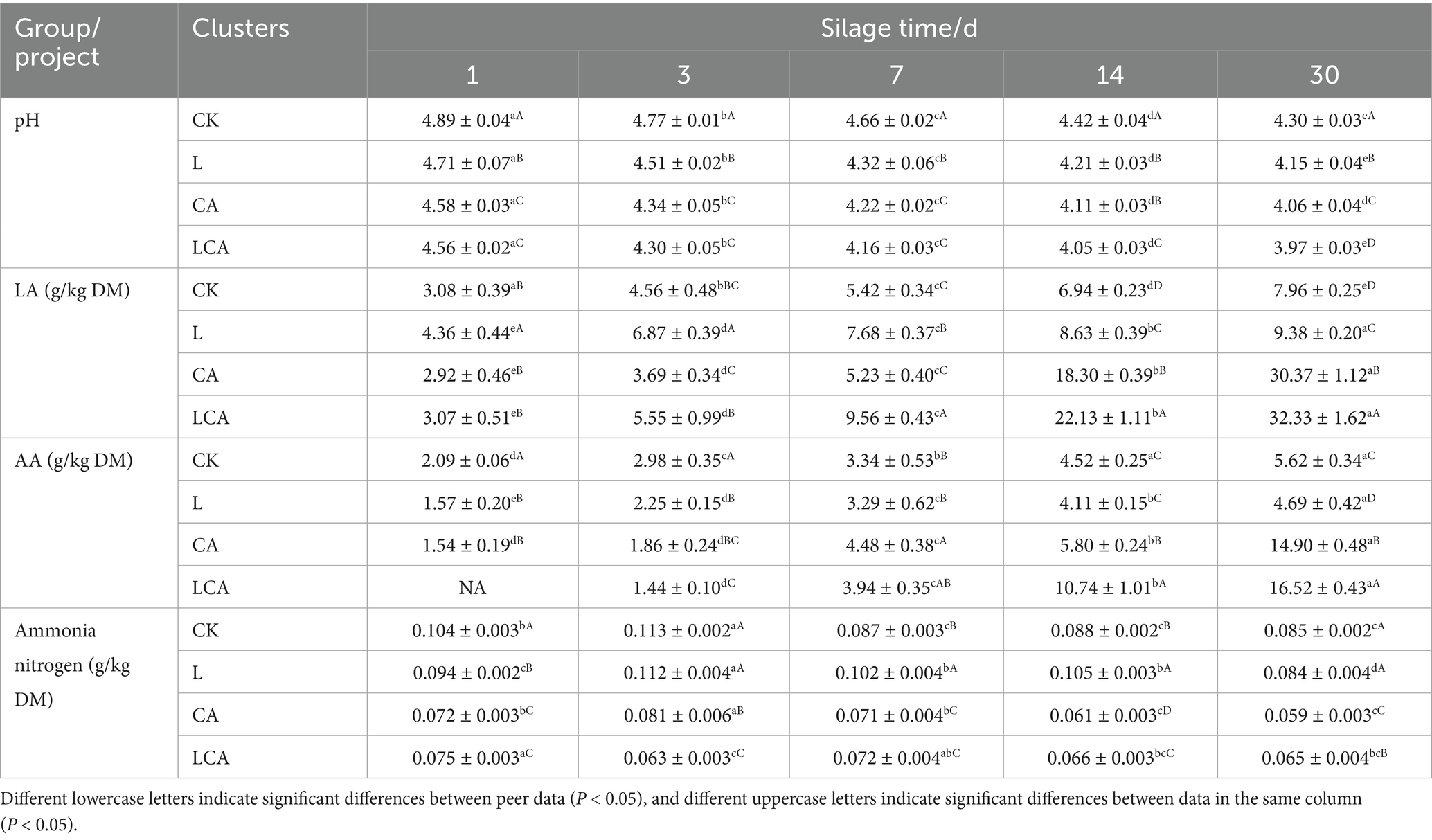
The dynamics of changes in fermentation quality of king grass silage in different treatment groups was shown in Table 5. With the prolongation of silage time, the pH in the four treatment groups of CK, L, CA and LCA showed a decreasing trend. On the 1st day of silage, the pH in the three treatment groups L, CA and LCA was significantly lower than that in the CK group (p < 0.05). After 30 days of ensiling, the pH in the CK group was significantly different from that in the three groups of L, CA, and LCA (p < 0.05), and the final pH of the four silages of CK, L, CA, and LCA was 4.30, 4.15, 4.06, and 3.97, respectively. Suggesting that the composite additives were better able to reduce the pH of the silage.
During the 30 days of fermentation, the contents of lactic acid and acetic acid both showed an increasing trend (p < 0.05) and reached the maximum value on the 30th day, propionic acid and butyric acid were not detected in the silage. At the end of ensiling, the contents of the lactic and acetic acid differed significantly (p < 0.05) among the four treatment groups (CK, L, CA, LCA). The LCA group exhibited the highest concentrations, with 32.33 g/kg of lactic acid and 16.52 g/kg of acetic acid. The analysis of individual effects revealed that both L. fermentum and citric acid alone significantly increased lactic acid content compared to CK (p < 0.05). Notably, the LCA group exhibited a synergistic effect, yielding significantly higher lactic acid and acetic acid contents than either additive alone (p < 0.05), which conclusively demonstrates the superior efficacy of the composite additive in enhancing fermentation quality. Additionally, the ammonia nitrogen content in king grass silage showed irregular changes during fermentation. Overall, the ammonia nitrogen content in groups CA and LCA was significantly lower than that in groups CK and L (p < 0.05), while no significant difference was observed between groups CK and L (p > 0.05).
3.3 High-throughput sequencing of king grass silage
3.3.1 Sequencing data analysis
Processing of the raw sequencing data yielded 1,231,788 valid sequences (an average of 102,649 per sample), representing a validity rate of over 95%. According to Figure 1A, the sequencing depth of all samples covered the majority of microorganisms, fulfilling the sequencing requirements. The rank abundance curve (Figure 1B) displayed the sample diversity, which meets the requirements for subsequent analysis.

Figure 1. Dilution curves (A) and rank relative abundance curves (B) for different groups of king grass silage. Panel (A) shows dilution curves illustrating the relationship between sample dilution and observed diversity across the silage groups. Panel (B) presents rank relative abundance curves, displaying the distribution of species relative abundance, with the x-axis representing species rank and the y-axis their relative abundance. These curves compare microbial diversity and species richness among the silage groups.
The petal plots for different treatment groups of king grass silage were shown in Figure 2. The observed OTU counts were as follows: CK group, 620; L group, 549; CA group, 561; LCA group, 596. Among them, the CK and LCA groups had a higher number of OTUs. Additionally, Figure 2 showed that the number of core OTUs was 405.

Figure 2. Venn diagram of OTUs (operational taxonomic units) shared between different treatment groups of king grass silage. The diagram highlights the overlap and unique OTUs among the treatment groups, showing the distribution of microbial taxa. The circles represent the distinct treatment groups, with the intersecting areas indicating common OTUs. This diagram provides a visual comparison of microbial community composition across the silage treatments.
3.3.2 Alpha diversity analysis
The Alpha diversity indices for each sample were shown in Table 6. A high Shannon index indicated high species richness, while a low Simpson index suggested the presence of dominant species within a diverse microbial community. The Shannon index of the LCA group was significantly higher than that of the CK group (p < 0.05), while the Simpson index was significantly lower (p < 0.05). This indicated that the combined addition of L. fermentum and citric acid enhanced microbial richness and promoted dominant species in king grass silage. Furthermore, the Chao 1 and Ace indices in the CA group were significantly lower than those in the CK group (p < 0.05). However, there were no significant differences in the Chao 1 and Ace indices between the L, LCA, and CK groups (p > 0.05).
3.3.3 Beta diversity analysis
Beta diversity is a method used to assess microbial community differences between samples based on the distance between species communities. Analyzing beta diversity revealed differences in the bacterial community composition of king grass silage. The principal coordinate analysis (PCoA) of differences of bacterial composition differences in king grass silage was shown in Figure 3. The first principal component (PCoA1) and the second principal component (PCoA2) accounted for 71.8 and 11% of the total variance, respectively. The bacterial compositions of the CK and L groups were located closely on the PCoA plot, indicating similarity in their bacterial compositions. The bacterial communities of the CK group were distinctly separated from those of the CA and LCA groups, indicating significant compositional differences, while CA and LCA exhibited highly similar bacterial profiles. Overall, the addition of L. fermentum alone did not significantly alter the bacterial composition of king grass silage, while citric acid or the combination of L. fermentum and citric acid significantly changed the composition.

Figure 3. PCoA (Principal Coordinate Analysis) of king grass silage with different treatments. The plot visualizes the similarity and dissimilarity of microbial communities across treatment groups based on multivariate analysis of OTU composition. Each point represents a treatment group, with distances between points reflecting the degree of similarity in microbial profiles. The axes represent the principal coordinates that capture the most variation in the data, providing insights into the effects of different treatments on the microbial community structure.
3.3.4 Analysis of bacterial community composition at the phylum level
The relative abundance of bacterial species at the phylum level in king grass silage under different treatments was shown in Figure 4. As shown in Figure 4, Proteobacteria, Bacteroidete, and Firmicutes were the dominant phyla in king grass silage. The relative abundance of Proteobacteria in the CK, L, CA, and LCA groups were 68.25, 71.74, 46.11, and 44.27%, respectively. The relative abundance of Bacteroidete were 25.79, 21.8, 34.01, and 31.06%, and those of Firmicutes were 5.59, 5.99, 19.25, and 23.1%. The addition of L. fermentum alone did not significantly alter the bacterial composition at the phylum level in wilted king grass silage. In contrast, the inclusion of citric acid, either alone or in combination with L. fermentum, notably modified the bacterial community structure, with the combined treatment exhibiting the most pronounced effect.
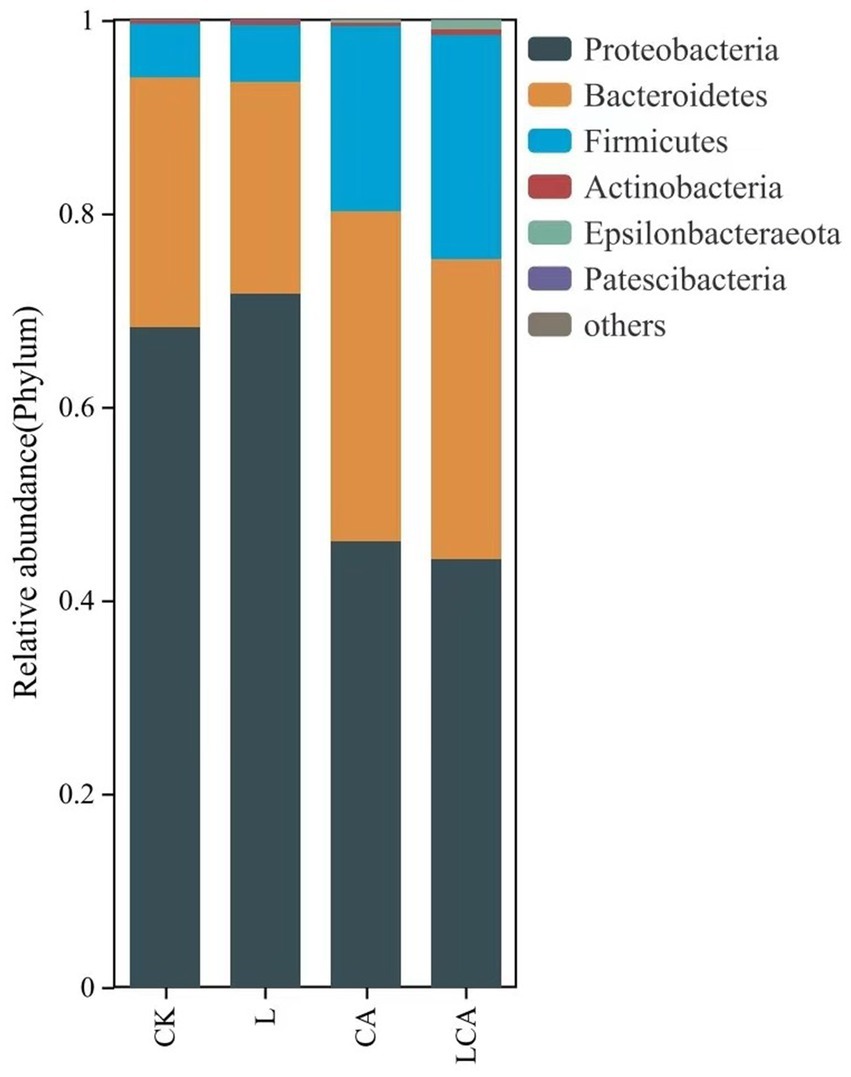
Figure 4. Relative abundance of species at the phylum level in different treatment groups of king grass silage. This bar chart illustrates the distribution of microbial phyla across the treatment groups, with the height of each bar representing the relative abundance of each phylum. The colors correspond to different phyla, providing a clear comparison of microbial composition between the groups. This analysis helps to reveal shifts in microbial community structure in response to different treatments.
3.3.5 Analysis of bacterial community composition at the genus level
The relative abundance of bacterial species at the genus level in king grass silage under different treatments was shown in Figure 5. Enterobacter, Sphingobacterium, Acinetobacter, and Lactobacillus were the dominant genera in king grass silage across the different treatments. The relative abundance of Enterobacter in the CK, L, CA, and LCA groups were 36.92, 40.18, 18.13, and 17.44%, respectively. The relative abundance of Sphingobacterium in the CK, L, CA, and LCA groups were 19.85, 16.60, 29.09, and 23.51%, respectively. The relative abundance of Acinetobacter in the CK, L, CA, and LCA groups were 11.14, 11.96, 11.13, and 11.51%, respectively. The relative abundance of Lactobacillus in the CK, L, CA, and LCA groups were 2.66, 2.65, 15.53, and 18.65%, respectively. Compared to the CK group, the relative abundance of Enterobacter decreased in the CA and LCA groups, while that of Lactobacillus increased. In the L group, the relative abundance of Enterobacter increased, while Lactobacillus showed no significant change. This suggested that the addition of L. fermentum alone did not significantly increase the relative abundance of Lactobacillus in king grass silage, whereas the addition of citric acid alone or the combination of L. fermentum significantly increased its relative abundance. The main bacteria involved in silage fermentation were lactic acid bacteria. Comparative analysis revealed that the addition of lactic acid bacteria decreased the relative abundance of Sphingobacterium, while the addition of citric acid increased its relative abundance. Data comparison showed that the addition of L. fermentum, citric acid, or their combination in wilted king grass silage did not significantly alter the relative abundance of Acinetobacter. Additionally, Lactococcus was also detected in king grass silage.
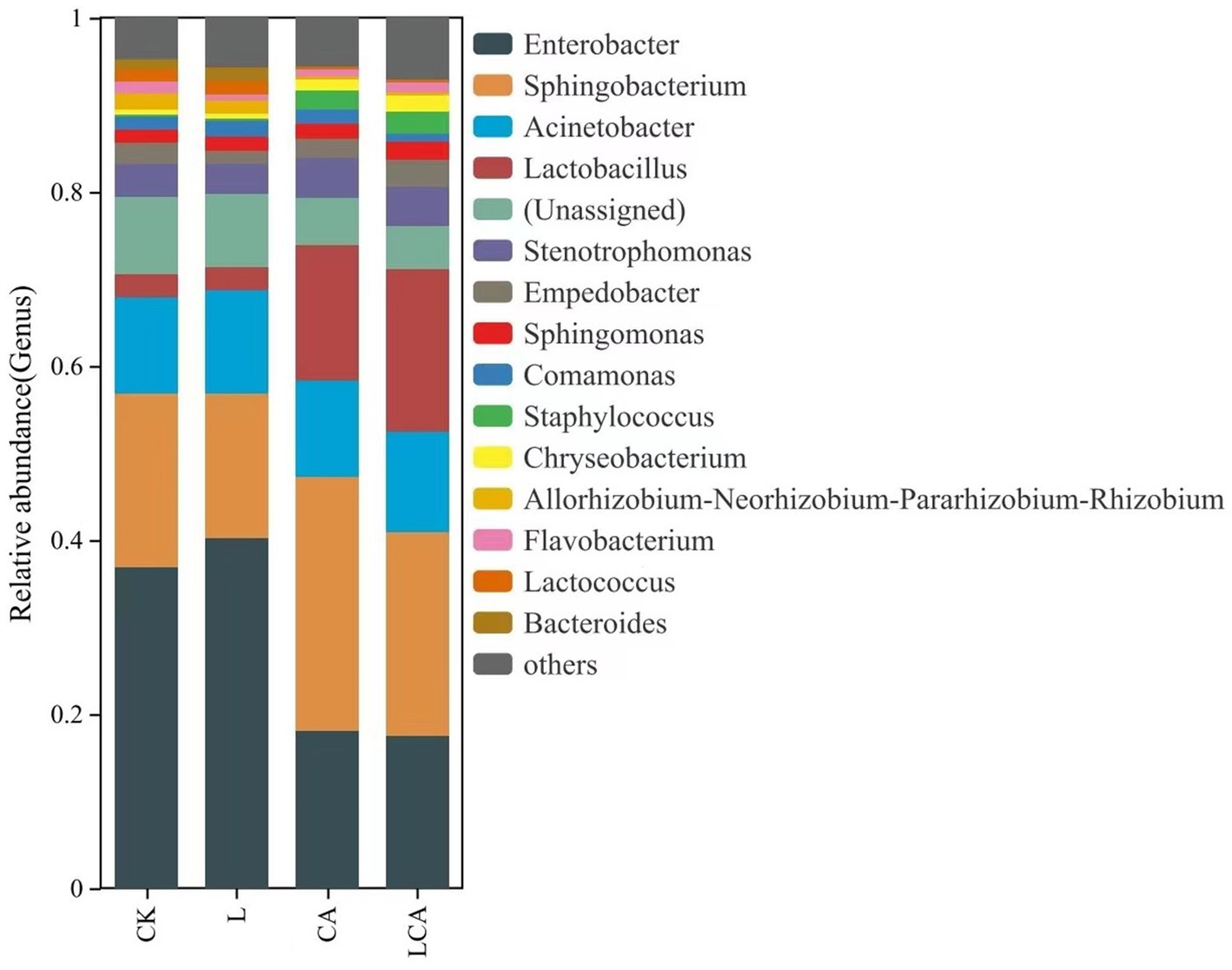
Figure 5. Relative abundance of species at the genus level in different treatment groups of king grass silage. The bar chart shows the distribution of microbial genera within each treatment group, with the length of each bar indicating the relative abundance of the corresponding genus. Different colors represent distinct genera, allowing for a clear comparison of microbial composition across the treatment groups. This analysis provides insights into the genus-level shifts in microbial communities due to the different treatments.
3.4 Aerobic stability of king grass silage
Figure 6 illustrated the pH changes of king grass silage under different treatments during aerobic exposure. After the initiation of aerobic exposure, the pH values of all groups began to rise, starting at 72 h. The rate of pH increase was higher in the CK and L groups compared to the CA and LCA groups. After 216 h of aerobic exposure, the pH values for the CK, L, CA, and LCA groups were 6.68, 6.92, 4.51, and 4.35, respectively. Furthermore, the figure showed that the aerobic stability of the L group was significantly lower than that of the CK group (p < 0.05), while the aerobic stability of the LCA group was significantly higher than that of the CA group (p < 0.05). The analysis indicated that the addition of L. fermentum alone did not improve the aerobic stability of king grass silage, while the addition of citric acid or the combination of citric acid and L. fermentum significantly enhanced its aerobic stability.

Figure 6. pH changes during aerobic exposure of king grass silage. This graph illustrates the variation in pH levels over time as the silage undergoes aerobic exposure. The x-axis represents the duration of exposure, while the y-axis shows the corresponding pH values.
4 Discussion
4.1 Effects of Limosilactobacillus fermentum and citric acid on the chemical composition of king grass silage
This study demonstrated that after wilting treatment, king grass silage exhibited a relatively high dry matter content. The moisture content of silage material was a critical factor affecting silage quality. Excessive moisture promoted the growth and proliferation of undesirable microorganisms (16). Research indicated that reducing the moisture content of silage material helped improve silage quality (17). In this study, the crude protein content in the CA and LCA groups was significantly higher than in the CK group, possibly because citric acid inhibited the hydrolysis of crude protein in king grass. Studies had shown that citric acid could inhibit the hydrolysis of crude protein in silage (13). This study further confirmed this hypothesis. The crude protein content in the L group did not differ significantly from the CK group, indicating that the addition of L. fermentum alone did not inhibit protein hydrolysis in king grass silage. These findings suggested that the combination of L. fermentum and citric acid could effectively improve protein preservation in silage. Excessive NDF content reduced feed intake in ruminants, while high ADF content affected digestibility. In this study, there was no significant difference in NDF content among the L, CA, LCA, and CK groups. However, the ADF content differed significantly, suggesting that L. fermentum and citric acid improved the digestibility of king grass silage for ruminants. WSC was the primary energy source for lactic acid bacteria fermentation during silage process. In this study, significant differences in carbohydrate content were observed among the L, CA, LCA, and CK groups. These findings suggest that L. fermentum and citric acid not only enhance the nutritional quality of the silage but also create a more favorable fermentation environment.
4.2 Effects of Limosilactobacillus fermentum and citric acid on the fermentation quality of king grass silage
The pH is an important indicator for assessing the fermentation quality of silage. The pH level reflected the quality of the silage. According to China’s silage standards, a pH of 3.4–4.4 indicated good silage quality (18). In this study, the pH of king grass silage was within this range, with the LCA group having a pH of 3.97. This suggested that the combined treatment of L. fermentum and citric acid contributed to a favorable fermentation environment. The lower pH of king grass silage could be attributed to three factors: (1) wilting reduced moisture content, inhibiting the growth of undesirable microorganisms and preventing nutritional competition with lactic acid bacteria; (2) the proliferation of lactic acid bacteria accelerated fermentation and increased lactic acid production, thereby inhibiting competing microorganisms; (3) citric acid had antimicrobial properties, and its lower pH further suppressed the growth of undesirable microorganisms.
Organic acids are another important indicator of silage quality. Lactic acid was the primary product of the silage fermentation process, and a higher lactic acid content generally indicated better quality silage (19). In this study, the lactic acid content in the CK, L, CA, and LCA groups differed significantly, with the LCA group showing the highest lactic acid content, suggesting that adding L. fermentum could increase lactic acid content, thereby improving silage quality. Additionally, studies have shown that appropriately adding citric acid could promote an increase in lactic acid content (20). This study further verified this viewpoint. The enhanced lactic acid production not only contributed to the overall quality of silage but also directly affected its preservation and digestibility. Lactic acid bacteria ferment water-soluble carbohydrates (WSC) to produce lactic acid, and the increase in lactic acid content resulted in a decrease in pH, which was also the main reason for the lower WSC content in the L, CA, and LCA groups. Acetic acid is the primary organic acid in tropical grass silage, and a higher concentration of acetic acid leads to an increase in pH (21). Moreover, research indicated that acetic acid content was positively correlated with the aerobic stability of silage (22). The significantly lower acetic acid content in the L group compared to the CK group is primarily attributed to the efficient fermentation driven by the added L. fermentum. This process rapidly consumed available WSC to produce lactic acid, lowering the pH and suppressing the activity of acetic acid-producing bacteria. Propionic acid and butyric acid are undesirable organic acids in silage, they damaged silage fermentation by stimulating Clostridium growth, resulting in nutrient loss and reduced palatability (23). Neither was detected in this study. Ammonia nitrogen is an important indicator for evaluating silage quality (24). The proteins in silage were hydrolyzed into ammonia nitrogen and other substances, thereby lowering the nutritional value and quality of the feed. In this study, the ammonia nitrogen content in the LCA group was significantly lower than that in the CK group, which was consistent with the crude protein content in the silage.
4.3 Effects of Limosilactobacillus fermentum and citric acid on bacterial composition in king grass silage
The silage process is a microbial fermentation process. High-throughput sequencing can reveal the microbial composition in silage and provide an in-depth analysis of the interactions between microbes during the silage process (25). In this study, the LCA group showed a lower Simpson index, indicating that the combined treatment with L. fermentum and citric acid increased bacterial diversity in king grass silage and promoted the dominance of specific beneficial taxa. L. fermentum and citric acid might have synergistically inhibited the growth of many undesirable microorganisms, with lactic acid bacteria becoming the dominant microbial group in the later stages of fermentation. Under anaerobic conditions, bacteria that could not adapt disappeared, resulting in increased bacterial diversity. Principal coordinate analysis (PCoA) results showed that the addition of L. fermentum alone did not significantly alter the bacterial composition of king grass silage, while the citric acid or L. fermentum + citric acid treatments significantly changed its bacterial composition. The PCoA results suggested that L. fermentum had a limited effect on microbial community structure. The similarity between CA and LCA groups implied that citric acid was the main factor influencing microbial composition.
In this study, the major bacterial phyla in king grass silage were Proteobacteria, Bacteroidete, and Firmicutes. After ensiling, the increase in Firmicutes and decrease in Proteobacteria could be ascribed to the anaerobic and acidic microenvironment in silage, which limited aerobic microorganisms and promoted LAB strains (26) The addition of L. fermentum alone did not significantly affect the major bacterial phyla in king grass silage, possibly due to the low moisture content after the wilting treatment, which inhibited the growth of lactic acid bacteria. This highlights the necessity of citric acid to drive significant changed in microbial composition. In the groups with citric acid or L. fermentum + citric acid treatment, the relative abundance of Firmicutes and Bacteroidete increased significantly. This result was consistent with previous studies (27). Enterobacter was considered a harmful bacterium in silage, capable of converting lactic acid into acetic acid and other substances (28). In this study, the relative abundance of Enterobacter was higher in the CK and L groups, but the acetic acid content was lower, possibly due to the reduced release of WSC from the king grass material with low moisture content, which inhibited its growth and metabolism. This study showed that the relative abundance of Enterobacter decreased after adding citric acid, which was consistent with other studies (12). Furthermore, after adding citric acid, the relative abundance of Lactobacillus increased, with higher relative abundance in the L. fermentum and citric acid combined group. The addition of citric acid seemed to have enhanced the growth of Lactobacillus while suppressing Enterobacter, a harmful bacterium. This phenomenon might be due to lactic acid bacteria using citric acid as a fermentation substrate. The acidic environment formed which was consistent with the growth of acid-sensitive microorganisms, thereby creating favorable conditions for lactic acid bacteria growth and forming a synergistic effect (29). Sphingomonas was a common genus in tropical grass silage (30), and researchers speculated it might be a characteristic genus of tropical silage (31). Sphingomonas was considered a beneficial bacterium in tropical grass silage due to its ability to degrade biogenic amines. Acinetobacter was an aerobic bacterium, but studies have shown that it can survive under anaerobic conditions containing acetic acid (32). Our previous studies also detected the presence of Acinetobacter. This study showed that the addition of L. fermentum and citric acid did not inhibit the growth of Acinetobacter.
4.4 Effects of Limosilactobacillus fermentum and citric acid on the aerobic stability of king grass silage
Aerobic stability is the ability of silage to resist spoilage from air exposure after fermentation is completed. Aerobic deterioration of silage feed can affect feeding effectiveness and, in severe cases, may damage the health of livestock and poultry. Aerobic deterioration is primarily caused by aerobic microorganisms, yeast, molds, and other microbes. Acetic acid can inhibit yeast growth, thereby alleviating aerobic spoilage of silage feed (33). In this study, the CA and LCA groups exhibited better aerobic stability, primarily due to their higher acetic acid content. After exposure to air, acetic acid inhibited the growth of some harmful microorganisms. The lactic acid bacteria added in this study were L. fermentum, which belongs to heterofermentative lactic acid bacteria. However, studies have shown that the addition of L. fermentum alone failed to improve the aerobic stability of silage. The possible reason for this phenomenon was that excessive wilting treatment resulted in excessively low moisture content, which not only inhibited the growth of undesirable microorganisms but also suppressed the growth and metabolism of L. fermentum.
5 Conclusion
In this study, L. fermentum HHL-5 was combined with citric acid as a silage additive and added to king grass silage. However, adding L. fermentum alone has a limited effect on improving the quality of wilted king grass silage, as low moisture content (<65%) inhibits the growth and metabolism of L. fermentum. Additionally, the results show that ensiling with L. fermentum combined with citric acid can significantly improve the quality of wilted king grass silage. Adding L. fermentum alone to wilted king grass has no significant effect on the microbial composition of king grass silage. However, ensiling with citric acid significantly alters the microbial composition, increased bacterial diversity, increasing the relative abundance of Lactobacillus, and decreasing the relative abundance of Enterobacteriaceae. In wilted king grass silage, adding L. fermentum alone does not improve aerobic stability, while ensiling with citric acid significantly enhances its aerobic stability.
Data availability statement
The sequencing data from this study have been submitted to the National Center for Biotechnology Information Sequence Read Archive database under Bioproject accession number of PRJNA1333438.
Author contributions
YD: Validation, Data curation, Supervision, Formal analysis, Methodology, Writing – review & editing, Conceptualization, Investigation, Writing – original draft. XX: Validation, Data curation, Supervision, Formal analysis, Writing – original draft, Conceptualization, Investigation, Writing – review & editing. RC: Conceptualization, Methodology, Writing – review & editing, Data curation, Investigation. JY: Project administration, Formal analysis, Funding acquisition, Supervision, Resources, Writing – review & editing, Conceptualization. WL: Conceptualization, Visualization, Supervision, Data curation, Writing – original draft. HT: Formal analysis, Resources, Writing – review & editing, Supervision, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 31960678) and the Hainan Province Science and Technology Special Fund (ZDYF2022XDNY186).
Acknowledgments
The authors gratefully acknowledge the technical support provided by the laboratory and farm staff and also thank funding support from the National Natural Science Foundation of China and the Key Research and Development Program of Hainan Province.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Lai, X, Wang, H, Peng, R, Chen, Z, Xiang, Y, and Yan, L. Different commercial microbial additives influence fermentation quality and microbial community of king grass silage. Fermentation. (2025) 11:264. doi: 10.3390/fermentation11050264
2. Zhao, J, Xia, B, Meng, Y, Yang, Z, Pan, L, Zhou, M, et al. Transcriptome analysis to shed light on the molecular mechanisms of early responses to cadmium in roots and leaves of king grass (Pennisetum americanum × P. purpureum). IJMS. (2019) 20:2532. doi: 10.3390/ijms20102532
3. Qiu, C, Yang, K, Diao, X, Zhang, W, Lv, R, and He, L. Effects of kinds of additives on fermentation quality, nutrient content, aerobic stability, and microbial community of the mixed silage of king grass and rice straw. Front Microbiol. (2024) 15:1420022. doi: 10.3389/fmicb.2024.1420022
4. Tian, M, He, X, Feng, Y, Wang, W, Chen, H, Gong, M, et al. Pollution by antibiotics and antimicrobial resistance in LiveStock and poultry manure in China, and countermeasures. Antibiotics. (2021) 10:539. doi: 10.3390/antibiotics10050539
5. Khalifa, HO, Shikoray, L, Mohamed, M-YI, Habib, I, and Matsumoto, T. Veterinary drug residues in the food chain as an emerging public health threat: sources, analytical methods, health impacts, and preventive measures. Foods. (2024) 13:1629. doi: 10.3390/foods13111629
6. Khmaissa, M, Zouari-Mechichi, H, Sciara, G, Record, E, and Mechichi, T. Pollution from livestock farming antibiotics an emerging environmental and human health concern: a review. J Hazardous Mater Adv. (2024) 13:100410. doi: 10.1016/j.hazadv.2024.100410
7. Gulahmadov, SG, Abdullaeva, NF, Guseinova, NF, Kuliev, AA, Ivanova, IV, Dalgalarondo, M, et al. Isolation and characterization of bacteriocin-like inhibitory substances from lactic acid bacteria isolated from Azerbaijan cheeses. Appl Biochem Microbiol. (2009) 45:266–71. doi: 10.1134/S0003683809030053
8. Borreani, G, Tabacco, E, Schmidt, RJ, Holmes, BJ, and Muck, RE. Silage review: factors affecting dry matter and quality losses in silages. J Dairy Sci. (2018) 101:3952–79. doi: 10.3168/jds.2017-13837
9. Chauhan, N, Kumari, N, Mani, V, Pradhan, D, Gowane, GR, Kumar, S, et al. Effects of Lactiplantibacillus plantarum, Limosilactobacillus fermentum, and propionic acid on the fermentation process of sugarcane tops silages along with variations in pH, yeast and mould count after aerobic exposure. Waste Biomass Valorizat. (2024) 15:2215–30. doi: 10.1007/s12649-023-02280-8
10. Ma, J, Zi, X, Wu, S, Ma, Y, Liang, R, Yang, J, et al. Multi-omics insights into the regulatory mechanism of citric acid in silage fermentation. Bioresour Technol. (2025) 436:133025. doi: 10.1016/j.biortech.2025.133025
11. Mahdizadeh, S, Sawford, K, van Andel, M, and Browning, GF. Efficacy of citric acid and sodium hypochlorite as disinfectants against Mycoplasma bovis. Vet Microbiol. (2020) 243:108630. doi: 10.1016/j.vetmic.2020.108630
12. Lv, H, Pian, R, Xing, Y, Zhou, W, Yang, F, Chen, X, et al. Effects of citric acid on fermentation characteristics and bacterial diversity of Amomum villosum silage. Bioresour Technol. (2020) 307:123290. doi: 10.1016/j.biortech.2020.123290
13. Ke, W, Ding, W, Xu, D, Shah, MN, Zhang, P, and Guo, X. Influences of addition of malic acid or citric acid, Lactobacillus plantarum and their mixtures on fermentation quality, proteolysis and fatty acid composition of ensiled alfalfa. Arch Anim Nutr. (2018) 72:492–502. doi: 10.1080/1745039X.2018.1510156
14. Broderick, GA, and Kang, JH. Automated simultaneous determination of Ammonia and Total amino acids in ruminal fluid and in vitro media. J Dairy Sci. (1980) 63:64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
15. Li, Y. Effects of mixed silage of corn and alfalfa on nutritional value and quality. Xianyang: Northwest A&F University (2019).
16. Zhang, YC, Li, DX, Wang, XK, Lin, YL, Zhang, Q, Chen, XY, et al. Fermentation dynamics and diversity of bacterial community in four typical woody forages. Ann Microbiol. (2019) 69:233–40. doi: 10.1007/s13213-018-1398-z
17. Nicholson, JK, Holmes, E, Kinross, J, Burcelin, R, Gibson, G, Jia, W, et al. Host-gut microbiota metabolic interactions. Science. (2012) 336:1262–7. doi: 10.1126/science.1223813
18. Wang, S, Ding, C, Tian, J, Cheng, Y, Xu, N, Zhang, W, et al. Evaluation of growth stage and storage time on fermentation characteristics, microbial community structure, co-occurrence networks, and their functional shifts and pathogenic risk of fermented Italian ryegrass. LWT. (2025) 215:117272. doi: 10.1016/j.lwt.2024.117272
19. De O, AIP, De Oliveira, MF, MAPO, J, Retore, M, Fernandes, T, Da Silva, YA, et al. Modulating fermentation in Total mixed ration silages using Lasalocid sodium and essential oils. Fermentation. (2025) 11:468. doi: 10.3390/fermentation11080468
20. He, L, Wang, C, Zhou, W, Zhang, Q, and Chen, X. Effect of additives on fermentation characteristics, nutrient values and in vitro fermentation profile of Neolamarckia cadamba leaf silage. South Afr J Anim Sci. (2019) 49:644–53. doi: 10.4314/sajas.v49i4.6
21. Li, D, Ni, K, Zhang, Y, Lin, Y, and Yang, F. Fermentation characteristics, chemical composition and microbial community of tropical forage silage under different temperatures. Asian Australas J Anim Sci. (2018) 32:665–74. doi: 10.5713/ajas.18.0085
22. Filya, I. The effect of Lactobacillus buchneri and Lactobacillus plantarum on the fermentation, aerobic stability, and ruminal degradability of low dry matter corn and Sorghum silages. J Dairy Sci. (2003) 86:3575–81. doi: 10.3168/jds.S0022-0302(03)73963-0
23. Xu, S, Yin, G, and Yu, X. Dynamics of microorganisms and metabolites in the mixed silage of oats and vetch in alpine pastures, and their regulatory mechanisms under low temperatures. Microorganisms. (2025) 13:1535. doi: 10.3390/microorganisms13071535
24. Zhang, Y, Liao, J, Pan, Y, Zhang, Q, Lu, Q, Chen, X, et al. Recycling of Lycium barbarum by-products and bioactive substance application in silage—insight into antioxidant activity and the regulation mechanism of anaerobic fermentation. BMC Microbiol. (2025) 25:431–19. doi: 10.1186/s12866-025-04033-0
25. Adams, IP, Glover, RH, Monger, WA, Mumford, R, Jackeviciene, E, Navalinskiene, M, et al. Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol Plant Pathol. (2009) 10:537–45. doi: 10.1111/j.1364-3703.2009.00545.x
26. Wang, S, Ding, C, Tian, J, Cheng, Y, Xu, N, Zhang, W, et al. An evaluation of storage length on ensiling characteristics, bacterial community compositions, co-occurrence networks, and their functional shifts and pathogenic risk in high-moisture oat silage. Chem Biol Technol Agric. (2024) 11:1–13. doi: 10.1186/s40538-024-00702-w
27. Chen, R, Li, M, Yang, J, Chen, L, Zi, X, Zhou, H, et al. Exploring the effect of wilting on fermentation profiles and microbial community structure during ensiling and air exposure of king grass silage. Front Microbiol. (2022) 13:971426. doi: 10.3389/fmicb.2022.971426
28. Santos, AO, Ávila, CLS, Pinto, JC, Carvalho, BF, Dias, DR, and Schwan, RF. Fermentative profile and bacterial diversity of corn silages inoculated with new tropical lactic acid bacteria. J Appl Microbiol. (2016) 120:266–79. doi: 10.1111/jam.12980
29. Li, M, Zhang, L, Zhang, Q, Zi, X, Lv, R, Tang, J, et al. Impacts of citric acid and malic acid on fermentation quality and bacterial Community of Cassava Foliage Silage. Front Microbiol. (2020) 11:595622. doi: 10.3389/fmicb.2020.595622
30. Wang, C, Sun, L, Xu, H, Na, N, Yin, G, Liu, S, et al. Microbial communities, metabolites, fermentation quality and aerobic stability of whole-plant corn silage collected from family farms in desert steppe of North China. PRO. (2021) 9:784. doi: 10.3390/pr9050784
31. Ali, N, Wang, S, Zhao, J, Dong, Z, Li, J, Nazar, M, et al. Microbial diversity and fermentation profile of red clover silage inoculated with reconstituted indigenous and exogenous epiphytic microbiota. Bioresour Technol. (2020) 314:123606. doi: 10.1016/j.biortech.2020.123606
32. Ogunade, IM, Jiang, Y, Kim, DH, Cervantes, AAP, Arriola, KG, Vyas, D, et al. Fate of Escherichia coli O157:H7 and bacterial diversity in corn silage contaminated with the pathogen and treated with chemical or microbial additives. J Dairy Sci. (2017) 100:1780–94. doi: 10.3168/jds.2016-11745
Keywords: Limosilactobacillus fermentum, wilted king grass, silage, microbial communities, aerobic exposure
Citation: Dou Y, Xu X, Chen R, Yang J, Liu W and Tan H (2025) Effects of compound fermentation with Limosilactobacillus fermentum HHL-5 and citric acid on wilted king grass silage. Front. Vet. Sci. 12:1660833. doi: 10.3389/fvets.2025.1660833
Edited by:
Izhar Hyder Qazi, South China Agricultural University, ChinaReviewed by:
Siran Wang, Jiangsu Academy of Agricultural Sciences, ChinaYong Liu, Chinese Academy of Sciences (CAS), China
Shuo Wu, South China Agricultural University, China
Copyright © 2025 Dou, Xu, Chen, Yang, Liu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinsong Yang, aG55YW5namluc29uZ0BoYWluYW51LmVkdS5jbg==; Haisheng Tan, dGhzNjg4QDE2My5jb20=
†These authors have contributed equally to this work
‡ORCID: Ying Dou, https://orcid.org/0009-0003-2703-4331
Xindan Xu, https://orcid.org/0009-0004-9882-3757
Rong Chen, https://orcid.org/0000-0001-9438-0769
Jinsong Yang, https://orcid.org/0000-0002-8789-5008
Liu Wei, https://orcid.org/0009-0000-8283-3994
Haisheng Tan, https://orcid.org/0000-0002-4088-3915
 Ying Dou
Ying Dou Xindan Xu1†‡
Xindan Xu1†‡ Rong Chen
Rong Chen