- 1School of Mathematics, Sciences and Health Professions, Harrogate, TN, United States
- 2Cumberland Mountain Research Center, Harrogate, TN, United States
- 3College of Veterinary Medicine, Lincoln Memorial University, Harrogate, TN, United States
- 4Tifton Veterinary Diagnostic and Investigational Lab, College of Veterinary Medicine, University of Georgia, Tifton, GA, United States
- 5J. Frank White Academy, Harrogate, TN, United States
- 6Center for Infectious, Zoonotic and Vector-Borne Diseases, Lincoln Memorial University, Harrogate, TN, United States
Leptospirosis is an important zoonotic disease that is maintained in populations due to chronic kidney infection of reservoir mammals. Previous work from our lab has identified rodents, voles, shrews, chipmunks and several species of amphibians and reptiles as hosts of Leptospira spp. in the Cumberland Gap Region of Kentucky, Tennessee, and Virginia. The aim of this study was to determine if fish contribute to the maintenance of the pathogen in the aquatic environment. Fish (n = 238), belonging to 19 genera, were collected from seven different locations in the Powell River in East Tennessee. Fish kidneys were harvested and screened for leptospiral DNA using a TaqMan quantitative polymerase chain reaction (qPCR) assay that targets pathogenic Leptospira spp. Blood samples were collected for measuring leptospiral antibodies using microscopic agglutination test (MAT). Of the 238 fish screened, 11 were positive by either qPCR or MAT (4.62%; 95% CI: 2.33–8.12). Of these 3 (3/238; 1.26%; 95% CI: 0.26–3.64) were positive by qPCR and 8 (8/237; 3.38%; 95% CI: 1.47–6.54) were found to have antibodies to at least one leptospiral serovar by MAT. This is the first report of leptospiral DNA detection in fish kidneys, providing insights on the potential role of fish in the epidemiology of leptospirosis in the region.
1 Introduction
Leptospirosis is a water-borne zoonotic disease primarily caused by the pathogenic species of Leptospira. The disease in humans is characterized by protean manifestations with clinical signs ranging from flu-like symptoms in most cases to life-threatening multisystemic organ failure in a smaller subset of patients. In animals, leptospirosis leads to significant production losses through decreased milk production, infertility, spontaneous abortion, hepatorenal failure, and death (1, 2). The etiological agent lives in the proximal renal tubules of chronically infected animals and is shed in their urine, thus contaminating surface water, soil, streams, ponds, and rivers. The infection is acquired by other animals, and humans when they come in direct contact with urine from infected animals or water or soil contaminated by such urine (3–5). Pathogenic Leptospira enter hosts through small cuts on the skin, or intact mucus membranes.
Although rodents are the primary reservoir host of Leptospira, many species of small mammals have been known to carry the pathogen (6). Historically, mammals have been the primary host species maintaining leptospires in the environment; however, several studies have shown that various species of reptiles and amphibians can also carry this pathogen in their kidneys (7–14). These host species hold an ecological niche both on land and in water, thus potentially expanding the reach of this pathogen to aquatic life.
Previous studies from our lab suggest that Leptospira is enzootic in the Cumberland Gap Region (CGR) of South-Central Appalachia, continuously circulating among small wild mammals, herpetofauna, livestock and shelter dogs (14–16). One of those studies also provided evidence of leptospiral contamination in environmental water in the region (16). Since no information from the United States is available on Leptospira infection in fish, we conducted this study to investigate the association by testing freshwater fish for the presence of pathogenic Leptospira and leptospiral antibodies. Kidneys of fish were screened for the presence of leptospiral DNA using quantitative polymerase chain reaction (qPCR), and leptospiral antibodies were measured using microscopic agglutination test, the gold standard in leptospiral serology.
2 Materials and methods
2.1 Ethics statement
All fish capture protocols were based on the Guidelines for the Use of Fishes in Research established by the American Fisheries Society (17) and were reviewed and approved by the Animal Care and Use Committee at the Lincoln Memorial University (IACUC # 2102-RES). Fish in this study were collected under an aquatic collection permit issued through Tennessee Wildlife Resources Agency.
2.2 Study area and sample collection
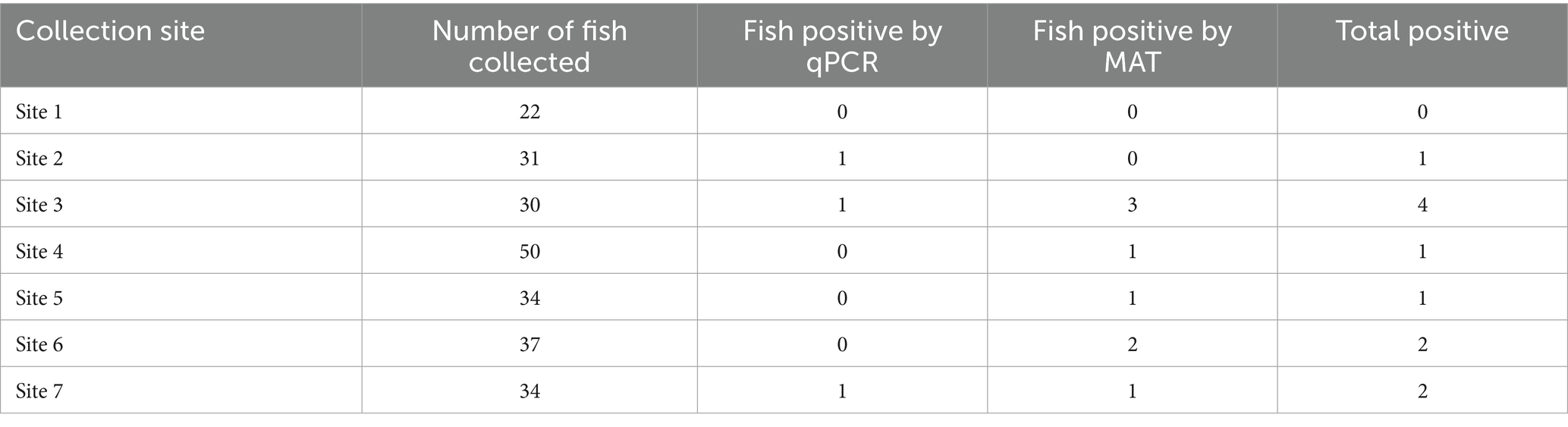
The sampling area included seven different sites (Sites 1–7) within a 47-mile segment of the Powell River in Claiborne County, Tennessee, extending from River Mile 112 to 65 (Figure 1). From each of the seven sites, no more than five fish per species were collected and the total number of fish collected across all species did not exceed 50 (Table 1). Only common species of fish within the Powell River were collected.

Figure 1. Map showing the seven fish collection sites along the Powell River in Tennessee, including the number of fish collected at each site, the number of positive fish, and the diagnostic methods used. Map created with Google My Maps.
A total of 238 fish of 19 genera were collected from seven different locations along the Powell River in Tennessee. Fish were collected in-stream with the use of a Smith-Root electrofishing backpack unit, and as described elsewhere (18–23). Electrofishing introduces an electric current into the water to momentarily stun fish to allow for collection by hand-net. Once collected by hand-net, fish were identified to species using dichotomous keys provided in (24). Fish collected for research were euthanized by exposure to lethal levels of the sedative MS-222, tricaine methane sulfonate. Once the animals were anesthetized and humanely euthanized, blood samples were collected from the tail vein, transported to lab on ice, centrifuged and stored at –20 °C until tested for MAT. The fish were also transported to the laboratory on ice, where the renal tissue was harvested and stored at –20 °C until further processing.
2.3 DNA extraction and quantitative polymerase chain reaction (qPCR)
Renal tissue from one or both kidneys was pooled and processed for DNA extraction using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s instructions. Leptospiral DNA in fish renal tissue was detected using a TaqMan based qPCR that targets a 242 bp region of leptospiral lipl32 gene as described by Stoddard et al. (25). The assay was performed in a MicroAmp Fast Optical 96-well reaction plate (Applied Biosystems, Foster City, CA, USA) using QuantStudio 3 and Platinum Quantiative PCR SuperMix-UDG (Invitrogen, Carlsbad, CA, USA). Each reaction was performed in a 25 μL final volume, using 500 nM of LipL32-45F (forward primer; 5’-AAGCATTACCGCTTGTGGTG-3′), 500 nM of LipL32-286R (reverse primer; 5’-GAACTCCCATTTCAGCGATT-3′) and 100 nM of LipL32-189P (probe; FAM-5’-AAAGCCAGGACAAGCGCCG-3’-BHQ1) (25). The standard curve was generated with leptospiral DNA equivalent to 107–100 genome units. Each column on the assay plate had a no-template control. Thermal conditions used were as follows: a holding stage of 95 °C for 20 s, and 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Each test DNA sample was tested in duplicate and repeated twice or more.
2.4 Microscopic agglutination test (MAT)
A total of 237 fish sera were screened for the presence of leptospiral antibodies by Microscopic Agglutination Test (MAT). The MAT was performed following the standardized protocol recommended by the World Organization for Animal Health (26). Briefly, two-fold serum dilutions from 1:100 to 1:6,400 were tested against leptospiral serovars Pomona strain Pomona (serogroup Pomona), Hardjo type Prajitno strain Hardjoprajitno (serogroup Sejroe), Grippotyphosa strain Andaman (serogroup Grippotyphosa), Icterohaemorrhagiae strain M20 (serogroup Icterohaemorrhagiae), Canicola strain Hond Utrecht IV (serogroup Canicola), Bratislava strain Jez Bratislava (serogroup Australis), and Autumnalis strain Akiyami A (serogroup Autumnalis) (National Veterinary Services Laboratories, Ames, Iowa, IA, USA). MAT titer was defined as the reciprocal of the highest dilution of a serum sample that agglutinated more than half of leptospires. Titers of more than or equal to 1:100 were considered positive for the presence of leptospiral antibodies.
2.5 Statistical analysis
The ‘cii” command in STATA version 17.0 (College Station, TX) was used to calculate 95% confidence intervals for the prevalence estimates.
3 Results
The collected fish included 40 Sunfish (Lepomis), 34 Rockbass (Ambloplites), 34 Chubs (Erimystax, Nacomis, Semotilus, Hybopsis), 30 Stonerollers (Campostoma), 24 Shiners (Cyprinella, Notemigonus, Luxilus), 22 Hogsuckers (Hypentelium), 16 Bass (Micropterus), 13 Shortnosed Redhorse (Moxostoma), 12 Common Log Perch (Percina caprodes), 6 Darters (Percina aurantiaca, Etheostoma), 5 Longnose Gar (Lepisosteus), 1 Catfish (Pylodictus), and 1 Bluntnose Minnow (Pimephales).
Of the 238 fish kidneys screened, leptospiral DNA was detected in three fish (3/238; 1.26%; 95% CI: 0.26–3.64) (Table 2). The qPCR-positive fish included two Green Sunfish (Lepomis cynellis) and one Rockbass (Ambloplites rupestris). Leptospiral concentration in fish kidneys ranged from 42–1.5×103 genomic equivalents (GE)/gram of renal tissue, with an average concentration of 626 GE/gram of renal tissue. Although two of the three positive fish were Green Sunfish, the data are insufficient to determine species-specific predisposition to leptospiral carriage. Additionally, due to the small kidney size in some fish species, renal tissues from both kidneys were pooled for DNA extraction. As a result, the current data cannot differentiate between unilateral and bilateral kidney infections.
Of the 237 fish sera tested, 8 (3.38%; 95% CI: 1.47–6.54) had antibodies to at least one leptospiral serovar (Table 2). The majority of positive fish (n = 7) reacted with one serovar., with 50 % of the MAT-positive fish (3 Rockbass and 1 Sunfish) reactive to serovar Icterohaemorrhagiae (Table 3). While two serum samples belonging to a Smallmouth Bass (Micropterus dolomieu) and a River Chub (Nacomis micropogon) reacted with serovar Autumnalis, one fish (Sunfish) contained antibodies to serovar Grippotyphosa. A Shortnosed Redhorse (Moxostoma macrolepidotum) reacted with serovars Pomona, Hardjo, and Grippotyphosa. All positive sera had titers greater than or equal to 1:100, except for one sera that had a titer greater than or equal to 1:200 for serovar Autumnalis (Table 3). None of the screened fish sera contained antibodies to serovars Canicola or Bratislava. No tested fish had both leptospiral antibodies and leptospiral DNA.
In total, 11 fish (11/238; 4.62%; 95% CI: 2.33–8.12) had either leptospiral DNA present in their kidneys or serum antibodies reactive to at least one leptospiral serovar tested. Six of the seven collection sites (Figure 1) had at least one fish positive for the presence of leptospiral antibodies or leptospiral DNA. The highest number of fish testing positive for leptospiral DNA or antibodies was observed at Site 3 (n = 4), followed by Sites 6 and 7 (n = 2) and Sites 2, 4, 5 (n = 1). No fish collected from Site 1 tested positive for either test (Table 1).
4 Discussion
Previous studies from our laboratory have shown in the Cumberland Gap Region of South-Central Appalachia, a variety of small wild mammals (rodents, shrews, voles, cottontails and chipmunks) as well as several species of amphibians and reptiles (Green frog, American bullfrog, American toad, Northern slimy salamander, Eastern newt, Common snapping turtle, Garter snake, Northern water snake, and Ringneck snake) can carry Leptospira spp. in their kidneys. Since many species of amphibians that tested positive in our previous study live in or around water bodies, we investigated if fish too contribute to maintenance of leptospires in the aquatic environment.
Our study provides evidence of leptospiral presence in common freshwater fish species found in the Powell River. Leptospiral DNA was detected in the kidneys of Green Sunfish and Rockbass. Additionally, leptospiral antibodies were found in the sera of Rockbass, Sunfish, Smallmouth Bass, River Chub, and Shortnosed Redhorse. In our knowledge, there are only two published reports that directly investigated Leptospira in fish (27, 28), although a few studies have examined the infection as an occupational hazard associated with fish farming. (29–31). In a 2014 study from Tanzania, 26 of 48 (54.2%) tested Catfish, Tilapia and Eel fish were positive when tested serologically against locally prevalent serovars Sokoine, Kenya, Pomona, and Hebdomadis (28). The high prevalence in fish in that study (28) could be due to small sample size, overall high prevalence of the disease across multiple reservoir species in that region, or low MAT cut-off values. Although the seroprevalence observed in our study was lower than that reported in the study by Mgode et al., our study provides the first documented evidence of leptospiral renal carriage in fish. These findings suggest that fish may play a role in the maintenance of pathogenic Leptospira spp. within aquatic ecosystems. Moreover, the detection of leptospiral DNA in the kidneys of freshwater fish underscores the importance of proper food safety measures. Thorough cooking and, in case of raw fish dishes, freezing at sufficiently low temperatures are critical to reduce exposure to this zoonotic pathogen.
The Powell River, a 195-mile-long body of water spanning from the US states of Virginia to Tennessee, is named as the second most diverse aquatic system in the nation by the Environmental Protection Agency and serves as a highly populated crossroad for wildlife and humans (32). The 47-mile-long stretch of the river sampled in this study runs through Claibourne County, Tennessee. It is notable for its high foot-traffic as community members kayak, fish, and swim in the river throughout late summer and early fall. In addition to aquatic activities, many bring household pets to walk or swim in the area. The Powell River is also home to highly diverse fauna, and acts as an overlapping point of community, domesticated animals, and wildlife. The wildlife in this area, which includes deer, foxes, and small mammals, rely upon this body of water for survival. The overlapping habitats of various potential hosts, and use of this environment by humans for recreational activities, may facilitate the continuous circulation of this pathogen among terrestrial, amphibian, and aquatic species.
The seven collection sites across the Powell River in Tennessee maintained varying degrees of flowing water, with portions being still and the river itself being home to a variety of fish species. The collected nineteen genera of fish included both bottom-dwelling fish as well as species that spend a majority of their time at the surface of the water. Although this study provided evidence for renal carriage of Leptospira spp. in fish, there are still many unknown aspects of this host-pathogen relationship. For example, how do structural and physiological differences in fish kidneys impact their role in leptospiral maintenance and shedding? Do other organs in fish harbor leptospires? Additionally, there is a lack of data on the prevalent serovars in fish. We screened fish sera against the most prevalent leptospiral serovars in the US, but there is a possibility that other serovars are more prevalent in aquatic species and there may also be regional variations. All or some of these factors may have a role in relatively low leptospiral prevalence in fish in this study.
We did not attempt leptospiral culture, nor did we include intermediate leptospiral strains in the MAT panel or used a molecular assay that can detect intermediate strains. These limitations, along with the absence of genotyping of Leptospira in qPCR-positive kidney samples, should be addressed in future studies to further characterize the diversity and potential pathogenicity of leptospiral strain in fish.
In summary, this work confirms the presence of leptospiral antibodies and DNA in freshwater fish, adding to the growing body of evidence that non-mammalian species may play a role in the ecology of leptospirosis. Whether fish contribute to the transmission cycle or act as a mere bystander of infection remains to be investigated.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Animal Care and Use Committee at the Lincoln Memorial University (IACUC # 2102-RES). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LB: Data curation, Investigation, Methodology, Supervision, Writing – review & editing. EW: Investigation, Writing – original draft. JM: Data curation, Investigation, Writing – review & editing. LW: Formal analysis, Writing – review & editing. SBo: Writing – review & editing. AG: Investigation, Writing – review & editing. CD: Investigation, Writing – review & editing. SBr: Investigation, Writing – review & editing. BN: Investigation, Writing – review & editing. MC: Investigation, Writing – review & editing. HN: Investigation, Writing – review & editing. SV: Investigation, Writing – review & editing. AMV: Investigation, Methodology, Writing – review & editing. AV: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by the LMU-CVM intramural research grant.
Acknowledgments
We dedicate this work to the memory of Noah Canady.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Orjuela, AG, Parra-Arango, JL, and Sarmiento-Rubiano, LA. Bovine leptospirosis: effects on reproduction and an approach to research in Colombia. Trop Anim Health Prod. (2022) 54:251. doi: 10.1007/s11250-022-03235-2
2. Sykes, JE, Hartmann, K, Lunn, KF, Moore, GE, Stoddard, RA, and Goldstein, RE. 2010 ACVIM small animal consensus statement on leptospirosis: diagnosis, epidemiology, treatment, and prevention. J Vet Intern Med. (2011) 25:1–13. doi: 10.1111/j.1939-1676.2010.0654.x
3. Adler, B, and de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet Microbiol. (2010) 140:287–96. doi: 10.1016/j.vetmic.2009.03.012
4. Ko, AI, Goarant, C, and Picardeau, M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. (2009) 7:736–47. doi: 10.1038/nrmicro2208
5. Yanagihara, Y, Villanueva, SYAM, Nomura, N, and Masuzawa, T. Leptospira is an environmental bacterium that grows in waterlogged soil. Microbiol Spectr. (2022) 10:e02157–21. doi: 10.1128/spectrum.02157-21
6. Haake, DA, and Levett, PN. Leptospirosis in humans. Curr Top Microbiol Immunol. (2015) 387:65–97. doi: 10.1007/978-3-662-45059-8_5
7. Biscola, NP, Fornazari, F, Saad, E, Richini-Pereira, VB, Campagner, MV, Langoni, H, et al. Serological investigation and PCR in detection of pathogenic leptospires in snakes. Pesqui Vet Bras. (2011) 31:806–11. doi: 10.1590/S0100-736X2011000900013
8. Dezzutto, D, Barbero, R, Canale, G, Acutis, P, Biolatti, C, Dogliero, A, et al. Detection of Leptospira spp. in water turtle (Trachemys scripta) living in ponds of urban parks. Vet Sci. (2017) 4:51. doi: 10.3390/vetsci4040051
9. Lindtner-Knific, R, Vergles-Rataj, A, Vlahović, K, Zrimšek, P, and Dovč, A. Prevalence of antibodies against Leptospira sp. in snakes, lizards and turtles in Slovenia. Acta Vet Scand. (2013) 55:65. doi: 10.1186/1751-0147-55-65
10. Miranda, JMS, Rocha, KS, Monteiro, LH, Baia, IWM, Monteiro, TRM, Brito, JS, et al. Presence of anti-Leptospira spp. antibodies in captive yellow-spotted river turtles (Podocnemis unifilis) in the eastern Amazon. Cienc Rural. (2020) 50:e20190088. doi: 10.1590/0103-8478cr20190088
11. Oliveira, JP, Kawanami, AE, Silva, ASL, Chung, DG, and Werther, K. Detection of Leptospira spp. in wild Phrynops geoffroanus in urban environment. Acta Trop. (2016) 164:165–8. doi: 10.1016/j.actatropica.2016.08.019
12. Rocha, KS, Baia, IWM, Monteiro, LH, Miranda, JMS, Monteiro, TRM, Silva, AF, et al. Identification of antibodies to Leptospira spp. in the spot-legged turtle (Rhinoclemmys punctularia) maintained in captivity. Semina. (2019) 40:3763. doi: 10.5433/1679-0359.2019v40n6Supl3p3763
13. Silva, ÉF, Seyffert, N, Cerqueira, GM, Leihs, KP, Athanazio, DA, Valente, ALS, et al. Serum antileptospiral agglutinins in freshwater turtles from southern Brazil. Braz J Microbiol. (2009) 40:227–30. doi: 10.1590/S1517-83822009000200003
14. Verma, A, Brandt, L, Runser, S, Gruszynski, K, Gallatin, K, Morgan, J, et al. Detection of pathogenic Leptospira spp. in herpetofauna in central Appalachia. Zoonoses Public Health. (2022) 69:325–32. doi: 10.1111/zph.12921
15. Spangler, D, Kish, D, Beigel, B, Morgan, J, Gruszynski, K, Naikare, H, et al. Leptospiral shedding and seropositivity in shelter dogs in the Cumberland Gap region of southeastern Appalachia. PLoS One. (2020) 15:e0228038. doi: 10.1371/journal.pone.0228038
16. Verma, A, Beigel, B, Smola, CC, Kitts-Morgan, S, Kish, D, Nader, P, et al. Evidence of leptospiral presence in the Cumberland Gap region. PLoS Negl Trop Dis. (2019) 13:e0007990. doi: 10.1371/journal.pntd.0007990
17. Jenkins, JA, Bart, HL Jr, Bowker, JD, Bowser, PR, MacMillan, JR, Nickum, JG, et al. Guidelines for use of fishes in research—revised and expanded, 2014. Fisheries. (2014) 39:415–6. doi: 10.1080/03632415.2014.924408
18. Freund, JG, and Petty, JT. Response of fish and macroinvertebrate bioassessment indices to water chemistry in a mined Appalachian watershed. Environ Manag. (2007) 39:707–20. doi: 10.1007/s00267-005-0116-3
19. Hense, Z, Martin, RW, and Petty, JT. Electrofishing capture efficiencies for common stream fish species to support watershed-scale studies in the Central Appalachians. N Am J Fish Manage. (2010) 30:1041–50. doi: 10.1577/M09-029.1
20. McCormick, FH, Hughes, RM, Kaufmann, PR, Peck, DV, Stoddard, JL, and Herlihy, AT. Development of an index of biotic integrity for the mid-Atlantic highlands region. Trans Am Fish Soc. (2001) 130:857–77. doi: 10.1577/1548-8659(2001)130<>2.0.CO;2
21. Petty, JT, Lamothe, PJ, and Mazik, PM. Spatial and seasonal dynamics of brook trout populations inhabiting a central Appalachian watershed. Trans Am Fish Soc. (2005) 134:572–87. doi: 10.1577/T03-229.1
22. Reynolds, JB. Electrofishing In: BR Murphy and DW Willis, editors. Fisheries techniques. 2nd ed. Bethesda, MD, USA: American Fisheries Society (1996). 221–53.
23. White, SM, and Rahel, FJ. Complementation of habitats for Bonneville cutthroat trout in watersheds influenced by beavers, livestock, and drought. Trans Am Fish Soc. (2008) 137:881–94. doi: 10.1577/T06-207.1
24. Etnier, DA, and Starnes, WC. The fishes of Tennessee. Knoxville: University of Tennessee Press (1993).
25. Stoddard, RA, Gee, JE, Wilkins, PP, McCaustland, K, and Hoffmaster, AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn Microbiol Infect Dis. (2009) 64:247–55. doi: 10.1016/j.diagmicrobio.2009.03.014. Epub 2009 Apr 22.19395218
26. World Organisation for Animal Health (WOAH). Leptospirosis. In: Manual of diagnostic tests and vaccines for terrestrial animals, chapter 3.1.12. Paris: WOAH (2021).
27. Maestrone, G, and Benjaminson, M. Leptospira infection in the gold fish (Carassius auratus). Nature. (1962) 195:719–20. doi: 10.1038/195719a0
28. Mgode, GF, Mhamphi, GG, Katkweba, AS, and Thomas, M. Leptospira infections in freshwater fish in Morogoro Tanzania: a hidden public health threat. Tanzan J Health Res. (2014) 16:112–7. doi: 10.4314/thrb.v16i2.7
29. Gill, N, Waitkins, SA, and Calder, IM. Further update on leptospirosis: continuing risk in fish farmers. Br Med J (Clin Res Ed). (1985) 290:1988. doi: 10.1136/bmj.290.6486.1988
30. Loiseau, PM, and German, M. La leptospirose: Une menace sanitaire croissante pour les activités piscicoles et récréatives [leptospirosis: a growing health threat to fish farming and recreational activities]. Ann Pharm Fr. (2022) 80:778–81. doi: 10.1016/j.pharma.2022.04.003
31. Robertson, MH, Clarke, IR, Coghlan, JD, and Gill, ON. Leptospirosis in trout farmers. Lancet. (1981) 2:626–7. doi: 10.1016/s0140-6736(81)92757-4
32. United States Forest Service (2004) Jefferson National Forest (N.F.), revised land and resource management plan: environmental impact statement. Available online at: https://books.google.com/books?id=K7c2AQAAMAAJ (Accessed on June 25, 2025).
Keywords: Leptospira spp., fish, Powell River, Appalachia, Cumberland Gap Region
Citation: Brandt L, Willems E, Morgan J, Wisnieski L, Boukobza S, Geer A, Duke C, Brovarney S, Noah B, Coarsey MD, Naikare H, Verma SS, Vanderpool AM and Verma A (2025) Detection of leptospiral antibodies and DNA in freshwater fish. Front. Vet. Sci. 12:1663896. doi: 10.3389/fvets.2025.1663896
Edited by:
Fabrizio Bertelloni, University of Pisa, ItalyReviewed by:
Francesca Grippi, Experimental Zooprophylactic Institute of Sicily (IZSSi), ItalyAlda Natale, Experimental Zooprophylactic Institute of the Venezie (IZSVe), Italy
Israel Barbosa Guedes, University of São Paulo, Brazil
Martin Wainaina, Federal Institute for Risk Assessment (BfR), Germany
Copyright © 2025 Brandt, Willems, Morgan, Wisnieski, Boukobza, Geer, Duke, Brovarney, Noah, Coarsey, Naikare, Verma, Vanderpool and Verma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashutosh Verma, YXNodXRvc2gudmVybWFAbG11bmV0LmVkdQ==
 LaRoy Brandt
LaRoy Brandt Emily Willems3
Emily Willems3 Joey Morgan
Joey Morgan Lauren Wisnieski
Lauren Wisnieski Sloane Boukobza
Sloane Boukobza Michele D. Coarsey
Michele D. Coarsey Ashutosh Verma
Ashutosh Verma