- Marine Biotechnology and Ecology Division, CSIR-Central Salt and Marine Chemicals Research Institute, Bhavnagar, India
Multidrug-resistance bacteria commonly use cell-to-cell communication that leads to biofilm formation as one of the mechanisms for developing resistance. Quorum sensing inhibition (QSI) is an effective approach for the prevention of biofilm formation. A Gram-negative bacterium, Delftia tsuruhatensis SJ01, was isolated from the rhizosphere of a species of sedge (Cyperus laevigatus) grown along the coastal-saline area. The isolate SJ01 culture and bacterial crude extract showed QSI activity in the biosensor plate containing the reference strain Chromobacterium violaceum CV026. A decrease in the violacein production of approximately 98% was detected with the reference strain C. violaceum CV026. The bacterial extract (strain SJ01) exhibited anti-quorum sensing activity and inhibited the biofilm formation of clinical isolates wild-type Pseudomonas aeruginosa PAO1 and P. aeruginosa PAH. A non-toxic effect of the bacterial extract (SJ01) was detected on the cell growth of the reference strains as P. aeruginosa viable cells were present within the biofilm. It is hypothesized that the extract (SJ01) may change the topography of the biofilm and thus prevent bacterial adherence on the biofilm surface. The extract also inhibits the motility, virulence factors (pyocyanin and rhamnolipid) and activity (elastase and protease) in P. aeruginosa treated with SJ01 extract. The potential active compound present was identified as 1,2-benzenedicarboxylic acid, diisooctyl ester. Microarray and transcript expression analysis unveiled differential expression of quorum sensing regulatory genes. The key regulatory genes, LasI, LasR, RhlI, and RhlR were down-regulated in the P. aeruginosa analyzed by quantitative RT-PCR. A hypothetical model was generated of the transcriptional regulatory mechanism inferred in P. aeruginosa for quorum sensing, which will provide useful insight to develop preventive strategies against the biofilm formation. The potential active compound identified, 1,2-benzenedicarboxylic acid, diisooctyl ester, has the potential to be used as an anti-pathogenic drug for the treatment of biofilm-forming pathogenic bacteria. For that, a detailed study is needed to investigate the possible applications.
Introduction
The biggest challenge for the healthcare sector is drug resistance in pathogenic bacteria. The efficiency of antibiotics against pathogenic bacteria is currently decreasing because of the emergence of multidrug-resistance (Adonizio et al., 2008). Biofilm formation is one of the mechanisms, used by bacteria for developing such resistance (Fuqua and Greenberg, 1998). It is well-established that curing of diseases caused by biofilm-forming bacteria requires prolonged treatment, which may also lead to antibiotic resistance due to high evolutionary pressure. The biofilm formation is controlled by cell-to-cell communication, which is widely known as quorum sensing. The inhibition of quorum sensing is one of the methods among the different strategies deployed to control biofilm forming microorganisms without causing drug resistance (Singh et al., 2013, 2016b). In recent years, several anti-quorum sensing compounds were reported in plants and microbes (Choo et al., 2006; Adonizio et al., 2008; Ni et al., 2009; Kalia and Purohit, 2011; Kalia, 2012).
The ubiquitous gram-negative bacterium Pseudomonas aeruginosa is an opportunistic pathogen, having a wide range of hosts such as insects, plants, animals, and humans (Rahme et al., 2000; Vandeputte et al., 2010). The bacterium P. aeruginosa causes very severe infection in immunocompromised patients (Driscoll et al., 2007; Vandeputte et al., 2010; Sarabhai et al., 2013) and is responsible for about 57% of all nosocomial infections (Oncul et al., 2009; Sarabhai et al., 2013).
It was observed that P. aeruginosa uses a range of virulence factors and multiple mechanisms, including biofilm formation, to successfully infect a diverse range of hosts and to protect itself from environmental stress and antibiotics (Driscoll et al., 2007; Vandeputte et al., 2010; Lee and Zhang, 2015). Quorum sensing controls the virulence factors and biofilm formation of P. aeruginosa. Therefore, anti-quorum sensing strategies could be a potential target to prevent P. aeruginosa infection.
The rhizosphere, a region of soil that surrounds the plant roots, possess a diverse bacterial community that containing molecules with both quorum sensing and quorum quenching activities (Christiaen et al., 2011), including anti-biofilm activity against P. aeruginosa (Christiaen et al., 2014). The anti-quorum sensing activity of Acinetobacter sp. strain C1010 (isolated from cucumber rhizosphere) was evaluated and found to degrade the acyl-homoserine lactones (AHLs) produced by P. chlororaphis O6 (Kang et al., 2004). A large number of AHL-degrading bacteria, including Sphingomonas sp. and Bosea sp., were isolated from the tobacco rhizosphere (D'Angelo-Picard et al., 2005). The bacteria Acinetobacter (GG2), Burkholderia (GG4), and Klebsiella (Se14) isolated from the ginger rhizosphere also showed AHL-degrading activity (Chan et al., 2011). Bacterial consortia isolated from the rhizosphere of potato contained anti-quorum sensing and plant growth promoting potential (Cirou et al., 2007). To date, however, there is no report on rhizospheric bacteria with anti-quorum sensing and anti-biofilm activity from the saline ecosystem.
In the present study, the rhizosphere of a monocot Cyperus laevigatus, a species of sedge from the coastal saline area, was explored and the bacterium Delftia tsuruhatensis SJ01 was isolated. Members of the Delftia genus are Gram-negative, aerobic, rod-shaped and motile bacteria comprised of five species: Delftia acidovorans (Wen et al., 1999), D. tsuruhatensis (Shigematsu et al., 2003), Delftia lacustris (Jørgensen et al., 2009), Delftia litopenaei (Chen et al., 2012), and Delftia deserti (Li et al., 2015). The coral-associated bacterial strain D. tsuruhatensis from the Gulf of Mannar was reported for its anti-quorum sensing activity. However, a detailed study and the identification of compounds has still not been performed (Bakkiyaraj et al., 2012, 2013). The isolated bacterium was explored for anti-quorum sensing and anti-biofilm potential. The active fraction was identified, regulatory key genes were studied, and a possible mechanism was inferred.
Materials and Methods
Isolation and Screening of Bacteria
A monocot, C. laevigatus, growing luxuriantly in the wet coastal areas of New-port, Bhavnagar, India (Latitude N 21° 45.124″, Longitude E 72° 13.579″), was collected. Bacteria were isolated from rhizosphere using a standard method, and axenic cultures were made for each isolate. Isolated axenic cultures were subjected to the screening of anti-quorum sensing activity using the reference strain Chromobacterium violaceum (CV026), cinnamaldehyde (Sigma-Aldrich, USA) as a positive control and methanol as a negative control in a plate-based bioassay (Singh et al., 2013). Bacterial isolates showing quorum sensing inhibition (QSI) activity were selected and checked further for antibacterial activity on Mueller-Hinton agar (MHA), along with tobramycin, which used as a positive control (Choo et al., 2006). A bacterial isolate showing promising positive QSI and negative anti-bacterial activities was selected further. The QSI and anti-bacterial activities of the selected isolate were repeated five times independently.
Identification of Bacteria and Fatty Acid Methyl Ester Profiling
Genomic DNA of selected bacteria was isolated, and the 16S rRNA gene amplified with universal primers fD1-5′-AGA GTT TGA TCC TGG CTC AG-3′ and rP2-5′-ACG GCT ACC TTG TTA CGA CTT-3′ (Weisburg et al., 1991) and optimized PCR conditions (Keshri et al., 2013, 2015). The PCR product was purified, sequenced (M/s Macrogen Inc., South Korea) and subjected to BLAST analysis. Phylogenetic analysis was performed using MEGA (Molecular Evolutionary Genetics Analysis) version 6.0 software (Tamura et al., 2013). The phylogenetic tree was reconstructed using neighbor-joining methods (Saitou and Nei, 1987), bootstrap analysis was performed (Felsenstein, 1985), and evolutionary distances were determined using maximum composite likelihood algorithms (Tamura et al., 2004). The bacterial isolate was identified as D. tsuruhatensis strain SJ01, and the 16S rRNA gene sequence was deposited in the NCBI GenBank (KX130769).
Fatty acid methyl ester (FAME) profiling of identified bacteria was performed using Microbial Identification System (MIDI; Microbial ID) coupled with gas chromatography (GC system-6850, Agilent Technologies, USA). For whole cell fatty acid methyl ester profiling, the bacteria were grown on tryptic soy yeast agar for 24 h at 30°C, and fatty acid methyl esters were prepared according to the instruction manual of the Microbial Identification System (MIDI; Microbial ID). Peaks were identified and matched with RTSBA6 6.10 database (Jha et al., 2015).
Preparation of Bacterial Extract
Bacterial culture (D. tsuruhatensis strain SJ01, 500 ml in nutrient broth, NB), grown for 48 h, 180 rpm at 30°C was centrifuged for 15 min. at 10,000 × g, 4°C, and the supernatant was collected in a flask. The supernatant was filtered through 0.45 and 0.22 μm vacuum filters for the complete removal of bacterial cells. The filtrate was extracted twice with an equal volume of ethyl acetate. Ethyl acetate extract was evaporated to dryness under vacuum in a rotary evaporator (Büchi, Switzerland) and dissolved in methanol for further studies (Nithya et al., 2010).
Anti-quorum Sensing Activity
The anti-quorum sensing activity of a methanolic extract of bacteria was tested by quantifying violacein (Choo et al., 2006). In brief, 1 ml of the freshly grown (OD600nm 0.7) reference strain C. violaceum (CV026) was added to 20 ml NB Hi-veg media (Hi-media, India) containing hexonyl homoserine lactone (0.0625 μg/ml) and different concentrations of bacterial extract (0.01, 0.02, 0.03, 0.04, 0.05, 0.075, or 0.1 mg/ml). Cultures without extract and with methanol were considered the control and negative control, respectively. All cultures (controls and experimental) were incubated for 24 h at 30°C and 180 rpm (Choo et al., 2006). One milliliter of overnight grown culture from each flask was centrifuged 16,000 × g for 10 min, and the pellet containing violacein (produced by CV026) was suspended in 1 ml of dimethylsulfoxide (DMSO). The solution was centrifuged at 16,000 × g for 10 min to remove cell debris and absorbance was read at 585 nm in a microplate reader (Spectra Max Plus, USA).
Biofilm Formation Assay
A measure of 200 μl of overnight grown cultures (OD600nm 0.1) of clinical isolates P. aeruginosa PAO1 (ATCC 15692) or P. aeruginosa PAH (by courtesy from Govt. Medical College, Bhavnagar; Goswami et al., 2011) was added to a 96-well microtiter plate with different concentrations of bacterial (strain SJ01) extracts (0.01, 0.02, 0.03, 0.04, 0.05, 0.075, and 0.1 mg/ml). The plate was incubated at 37°C, 100 rpm for 24 h, after which the growth of bacteria was measured at 600 nm and colony forming units (CFU) were also determined. Wells were washed after removing planktonic bacterial cells, dried and stained with 1% crystal violet. Excess dye was taken out after 20 min, wells were washed (with sterile distilled water), 200 μl ethanol (aqueous 96%) was added, and absorbance was measured at 590 nm (Andersson et al., 2009; Singh et al., 2013; Kavita et al., 2014). The experiments were performed thrice with five replicates each.
Fluorescence Microscopy
Cell viability within the biofilm was examined at different time points (24, 48, and 72 h) and compared with the control (Singh et al., 2013). Cells inhabiting the biofilm were stained with a fluorescent dye using the FilmTracer™Live/Dead® Biofilm Viability Kit (Invitrogen, USA) following manufacturer's instructions and visualized under an epi-fluorescence microscope (Axio Imager, Carl Zeiss AG, Germany).
Scanning Electron Microscopy
The effect of bacterial extract (SJ01) on biofilm formation was visualized by scanning electron microscopy (SEM; Andersson et al., 2009; Singh et al., 2013). Biofilms of P. aeruginosa PAO1 and P. aeruginosa PAH, grown on glass coverslips (11 mm) submerged in nutrient broth with (0.1 mg/ml) or without bacterial extract were gently washed with 0.9% NaCl to remove planktonic cells. Samples were kept in 2.5% glutaraldehyde for 20 min followed by 4% OsO4 in 0.1 M phosphate buffer for 30 min. Samples were dehydrated with a gradient ethanol series (10–95%) for 10 min. The dried biofilms were coated with gold and visualized under a scanning electron microscope (SEM, LEO series VP1430, Germany).
Atomic Force Microscopy
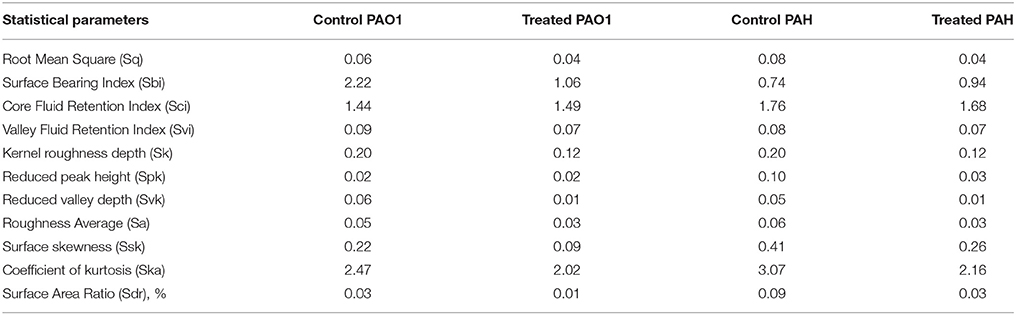
For atomic force microscopy (AFM), biofilms developed on glass coverslips were rinsed gently with phosphate buffer saline (pH 7.4) and kept in a desiccator for drying completely. The biofilm was scanned under AFM (NT-MDT, Russia) in a semi-contact mode at the speed of 1 Hz (Oh et al., 2009; Nithya et al., 2010). The surface bearing index (Sbi), core fluid retention index (Sci), valley fluid retention index (Svi), kernel roughness depth (Sk), reduced peak height (Spk), reduced valley depth (Svk), average roughness (Sa), root mean square (Sq), surface skewness (Ssk), coefficient of kurtosis (Ska), and surface area ratio (Sdr) were calculated.
Bacterial Motility Assay
Bacterial extract (SJ01) was tested on the swarming and swimming motility of P. aeruginosa. For the swarming motility assay, P. aeruginosa strains were spotted on a plate containing BM2 swarming medium (62 mM PBS at pH 7, 2 mM MgSO4, 10 μM FeSO4, 0.4% glucose, 0.1% casamino acids, and 0.5% agar) supplemented with (0.1 mg/ml) or without extract (Overhage et al., 2007). For the swimming motility assay, P. aeruginosa strains were spotted on a plate containing tryptone broth (10 g/l tryptone, 5 g/l NaCl, and 0.3% agar) supplemented with (0.1 mg/ml) or without extract (Rashid and Kornberg, 2000). Plates were analyzed after incubation of 24 h at 37°C.
Virulence Factor Analysis
The effect of bacterial extracts (SJ01; 0.1 mg/ml) was studied on the production of virulence factors of reference P. aeruginosa strains by quantifying pyocyanin and rhamnolipid, and analyzing elastase and protease activities. Briefly, P. aeruginosa PAO1 and P. aeruginosa PAH were grown overnight in 5 ml of PB medium (20 g/l peptone, 1.4 g/l MgCl2 and 10 g/l K2SO4) supplemented with extract of strain SJ01 (0.1 mg/ml) and without extract (control) at 37°C (180 rpm). The culture was centrifuged at 10,000 × g for 10 min, and pyocyanin was extracted first from the supernatant in 3 ml of chloroform, followed by 1 ml of 0.2 N HCl. The absorbance was measured spectrophotometrically at 520 nm (Essar et al., 1990).
For rhamnolipid, reference strains (P. aeruginosa) were grown in nutrient broth supplemented with bacterial extract (SJ01; 0.1 mg/ml) or without extract (control). The culture was centrifuged at 10,000 × g for 10 min, supernatants were collected, acidified with HCl (to pH 2) and absorbance was measured at 570 nm (McClure and Schiller, 1992). Supernatants (750 μl) of overnight grown (with 0.1 mg/ml or without extract of strain SJ01) P. aeruginosa were incubated with 250 μl elastin Congo-red solution (5 mg/ml in 0.1 M tris-HCl pH 8; 1 mM CaCl2) at 37°C, 180 rpm for 16 h. After incubation, the mixture was centrifuged at 3,000 × g for 10 min, and absorbance was measured at 490 nm for elastase activity (Zhu et al., 2002). For protease activity, supernatant (400 μl) was incubated with an equal volume of 2% azocasein solution (prepared in 50 mM phosphate buffer saline, pH 7) at 37°C for 1 h. The reaction was stopped by adding 500 μl of 10% trichloroacetic acid (TCA), and reaction mix was centrifuged at 8,000 g for 5 min to remove residual azocasein. The absorbance of the supernatant was read at 400 nm (Adonizio et al., 2008).
Fractionation and Identification of Active Compound
Bacterial extract (D. tsuruhatensis SJ01) was fractionated by the solid phase extraction (SPE) method using different cartridges (non-polar C18, polar SI, anion exchanger DAE and cation mixed Plexa PCX) and each fraction was screened for anti-quorum sensing activity. The positive fraction was further analyzed, and an active compound was identified by GC-MS. Briefly, crude bacterial extract (1 ml) was loaded to the preconditioned (by 5 ml methanol, 10 ml water and 5 ml acidified water pH 2.0) SPE cartridges (Agilent, USA). The elution was performed with a different concentration of 1 ml methanol (20, 40, 60, 80, and 100% v/v in water) and different fractions were collected (Singh et al., 2013). Each fraction was screened for plate based anti-quorum sensing activity (as described above) using the reference strain C. violaceum (CV026). The positive fraction was subjected to GC-MS (GC-2010, Shimadzu, Japan) and the identification of compounds was done by comparing the mass spectra with the reference mass spectra library. The mass of the fractionated compound identified was further confirmed by electrospray ionization mass spectrometry (ESI-MS; Q-Tof micro TM, Micromass, UK), performed in a positive mode.
Microarray and Expression Analysis
Differential expression of regulatory genes of reference strain P. aeruginosa PAO1, involved in the quorum sensing was analyzed using microarray. Total RNA was isolated from reference strain P. aeruginosa PAO1, grown with or without bacterial extracts (0.1 mg/ml) using TRI reagent (Sigma, USA). Total RNA was quantified, and 10 μg RNA was converted to cDNA, befor being fragmented and labeled by following the GeneChip® P. aeruginosa PAO1 genome array user manual (Affymetrix, USA). Labeled cDNAs were hybridized with the P. aeruginosa genome array gene chip (containing total 5,886 gene probes), and then washed and stained (Singh et al., 2016a). Hybridized chips were scanned (Scanner 3000 7G, Affymetrix, USA), processed and analyzed using the expression console and the transcriptome analysis console (Affymetrix, USA). Microarray analysis was performed in duplicate (n = 2) and genes exhibiting significant fold expression (ANOVA p < 0.05) were considered for the study. All microarray data are available with Array-Express accession number E-MTAB-5693. For expression profiling, key regulatory genes (LasI, LasR, RhlI, and RhlR) were selected. Total RNA was extracted from control and treated P. aeruginosa (PAO1 and PAH strains) converted to cDNA and then quantitative real-time PCR was performed (Wang et al., 2005). A melt curve analysis was also done for the validation of specificity of the qRT-PCR reaction, and the relative fold expression change was calculated using the CT method (Livak and Schmittgen, 2001). The 16S rRNA gene was used as a reference gene (Wang et al., 2005).
Results
Isolation and Screening of Bacteria for Anti-quorum Sensing Activity
A total of 56 bacterial axenic cultures were obtained from the rhizosphere of C. laevigatus L., of which two axenic cultures showed anti-quorum sensing activity in a plate-based bioassay. The isolate SJ01 showed promising anti-quorum sensing activity and a clear white opaque zone of inhibition was observed in the biosensor plate containing reference strain C. violaceum CV026 (Figure 1). Furthermore, the bacterial crude extract also showed QSI, whereas the zone of inhibition was not detected with the negative control (methanol). The disc diffusion antibacterial assay confirmed that selected bacterial isolates did not show antibacterial activity against the reference strain C. violaceum CV026 (Figure S1).
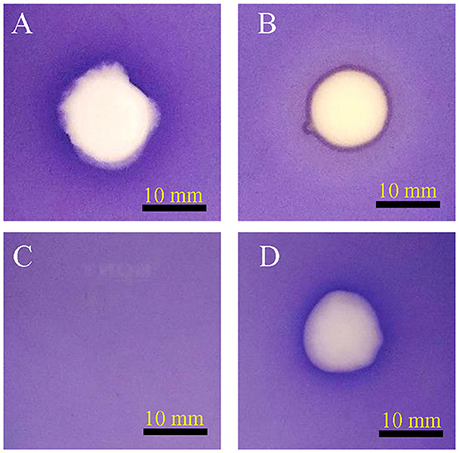
Figure 1. Anti-quorum sensing activity of isolate SJ01. The biosensor plates containing reference strain C. violaceum CV026 were spotted with (A) cinnamaldehyde, (B) SJ01 axenic culture, (C) methanol, and (D) crude bacterial (SJ01) extract. Cinnamaldehyde was used as a positive control. The isolate SJ01 and its extract showed the anti-quorum sensing activity and a clear white opaque zone of inhibition.
Identification of Bacteria, Fatty Acid Methyl Ester Profiling, and Phylogenetic Analysis
The 16S rRNA gene sequence (accession no. KX130769) of the selected bacterial isolate showed 99% similarity to D. tsuruhatensis, with 100% query coverage; therefore, this was designated D. tsuruhatensis SJ01. The phylogenetic tree reconstructed using the neighbor-joining algorithm shows the taxonomic position of identified bacterium with other species (Figure S2). The whole cell fatty acid profiling of the bacterium D. tsuruhatensis SJ01 revealed the abundance of C16:0 fatty acids (Figure S3).
Delftia tsuruhatensis SJ01 Extract Shows Anti-quorum Sensing Activity by Inhibiting Violacein Production
The bacterium D. tsuruhatensis SJ01 and its methanolic extract showed anti-quorum sensing activity with the reference strain on a biosensor plate. Different concentrations of bacterial extract were used to quantify the inhibition of violacein, an indicator of quorum sensing activity (Figure 2). The violacein production decreased concomitantly with the increasing concentration of the extract, and about 98% inhibition was observed with 0.1 mg/ml extract.
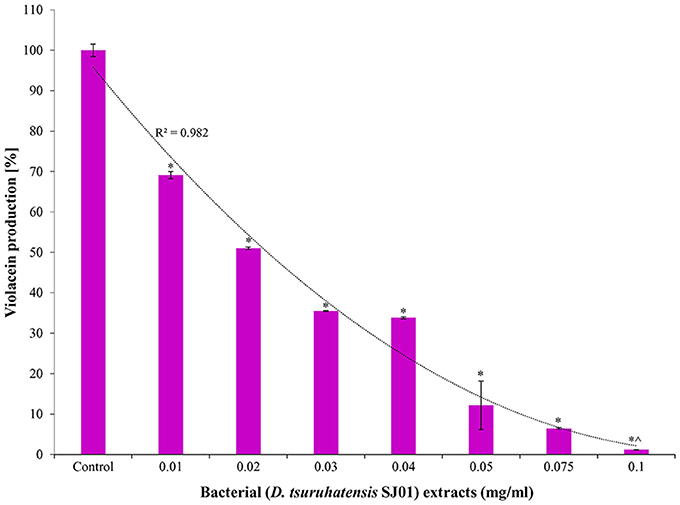
Figure 2. Effect of different concentration of D. tsuruhatensis SJ01 extract on violacein production. Different concentration of bacterial extract (0.01−0.1 mg/ml) was used to quantify the inhibition of violacein, an indicator of quorum sensing activity. Cultures without extract were considered as a control. *Indicates significant differences from the control at P < 0.05 and ∧ indicates maximum significant differences from the control at P < 0.05.
Delftia tsuruhatensis SJ01 Extract Inhibits Biofilm Formation
The anti-biofilm activity of the extract (D. tsuruhatensis SJ01) was tested against the wild-type, widely used biofilm forming clinical isolate P. aeruginosa PAO1 and a local clinical isolate P. aeruginosa PAH. The biofilm formation decreased concurrently in both reference strains with increasing concentration of bacterial extracts (Figure 3). About 60–64% inhibition of the biofilm formation was observed with 0.1 mg/ml extract. The possibility of an inhibitory effect of D. tsuruhatensis SJ01 extract on the growth of reference strains (P. aeruginosa) was also analyzed (Figures S4, S5). No significant effect was observed on the planktonic growth of P. aeruginosa in the presence of different concentration of bacterial extracts (0.01–0.1 mg/ml). Further, the disc diffusion antibacterial assay performed with SJ01 extract confirmed that bacterial extract did not show antibacterial activity against the clinical isolates P. aeruginosa (Figure S6).
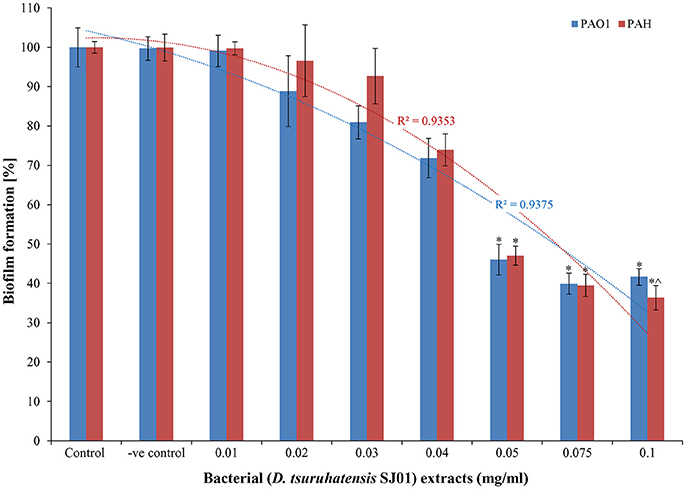
Figure 3. The antibiofilm activity of D. tsuruhatensis SJ01 extract. Different concentration of bacterial extracts (0.01−0.1 mg/ml) was tested against wild-type, widely used biofilm forming reference strain P. aeruginosa strains. Tests without extract and with methanol were considered as control and negative control, respectively. *Indicates significant differences from the control at P < 0.05 and ∧ indicates maximum significant differences from the control at P < 0.05.
Fluorescence Microscopy Analysis Confirms That Biofilm Inhabiting Viable Cells
The effect of the bacterial extract on the viability of the reference strain in the biofilm (24–72 h) was studied with an epi-fluorescence microscope (Figure 4). The dead P. aeruginosa cells were labeled with propidium iodide whereas live cells stained with SYTO 9, which produced red and green fluorescence, respectively. Less attachment of P. aeruginosa cells to the surface was observed even up to 72 h in the treated biofilm compared to control, and an insignificant number of dead cells was detected in the biofilms.
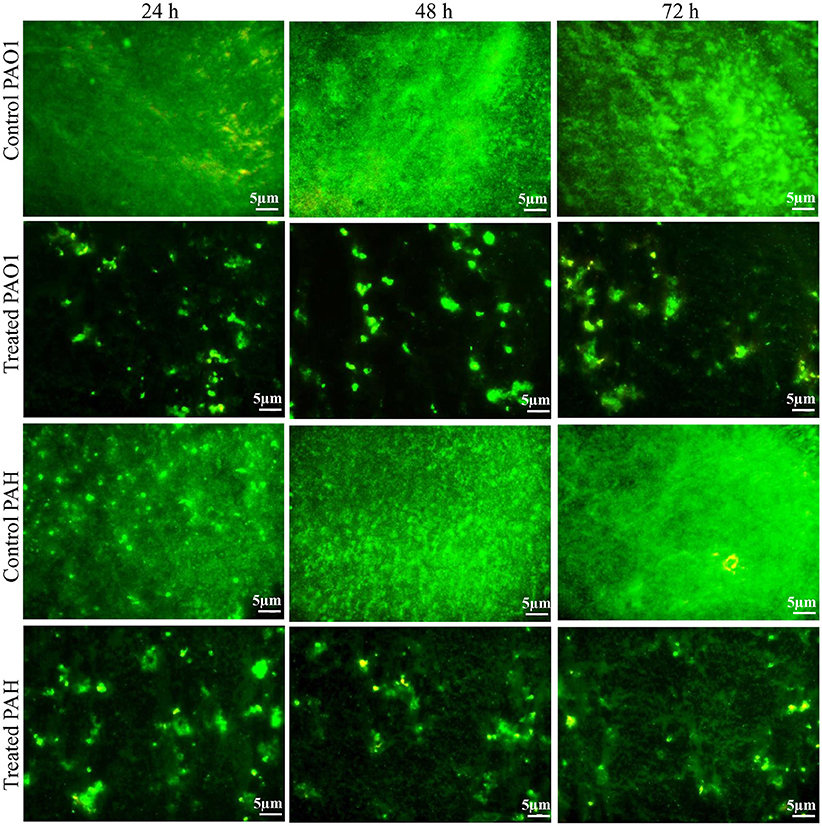
Figure 4. Epi-fluorescence micrographs of biofilms developed by P. aeruginosa. The effect of the bacterial extract (0.1 mg/ml) on the viability of reference P. aeruginosa strains in the biofilm was examined at different time points (24, 48, and 72 h) and compared with control. The dead bacterial cells were labeled with propidium iodide whereas live cells stained with SYTO 9, which produced red and green fluorescence, respectively.
Delftia tsuruhatensis SJ01 Extract Disrupts the Architecture of the Biofilm
The topology of the biofilm developed by P. aeruginosa and the effect of D. tsuruhatensis SJ01 extract on it was analyzed by SEM and AFM. A well-grown biofilm along with adhering bacterial cells was observed in controls (normal biofilm developed by P. aeruginosa) in the SEM analysis, whereas dispersed bacterial cells were observed in treated samples (Figure 5). Similarly, AFM clearly showed the disrupted surface topology and height distribution profile of the biofilm developed in the presence of D. tsuruhatensis SJ01 extract compared to the control biofilm (Figure 6). The surface bearing indices, roughness analysis, and functional parameters based on the linear material ratio curve showed alterations of the biofilm developed in treated samples (Table 1).
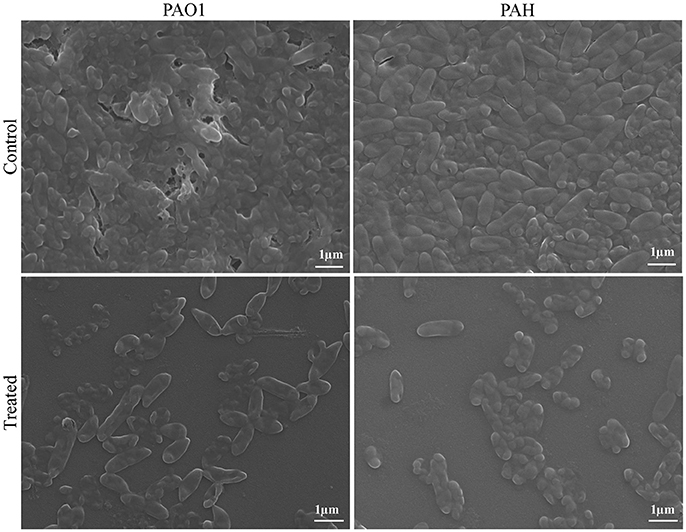
Figure 5. SEM images of biofilms developed by P. aeruginosa. SEM images illustrating the effect of bacterial extract (0.1 mg/ml) on biofilm formation. A well-grown biofilm along with adhering bacterial cells was observed in control (normal biofilm developed by P. aeruginosa), whereas dispersed bacterial cells were observed in treated samples.
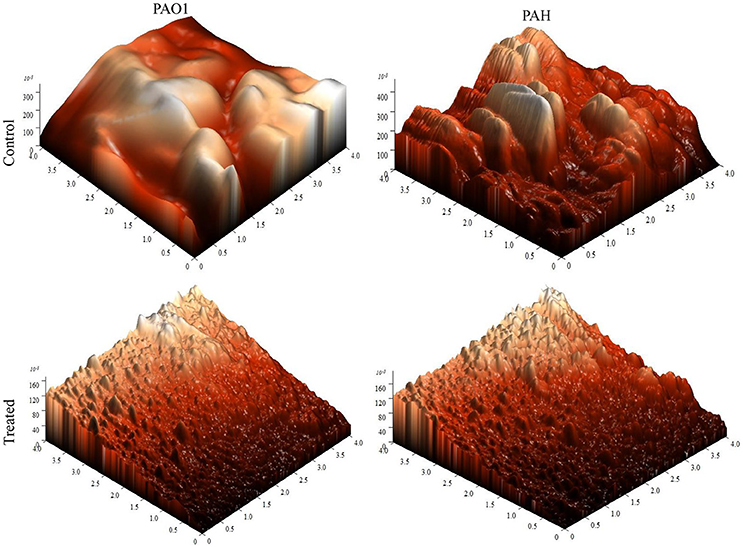
Figure 6. AFM images illustrating the effect of D. tsuruhatensis SJ01 extract on P. aeruginosa biofilms. AFM showed a disrupt surface topology and height distribution profile of the biofilm developed by reference P. aeruginosa strains in the presence of bacterial extract (0.1 mg/ml) compared to the control biofilm.
Delftia tsuruhatensis SJ01 Extract Shows Inhibitory Effect on the Motility of P. aeruginosa
Bacterial invasion is a prerequisite for biofilm formation. Therefore, the effect of bacterial extract (D. tsuruhatensis SJ01) was studied on the motility of biofilm forming P. aeruginosa bacterial cells. It was observed that bacterial extract (0.1 mg/ml) inhibits the swarming and swimming motility of P. aeruginosa strains in the plate assay (Figure 7). The extract reduced flagellum driven motility of P. aeruginosa in the treated sample compared to the control.
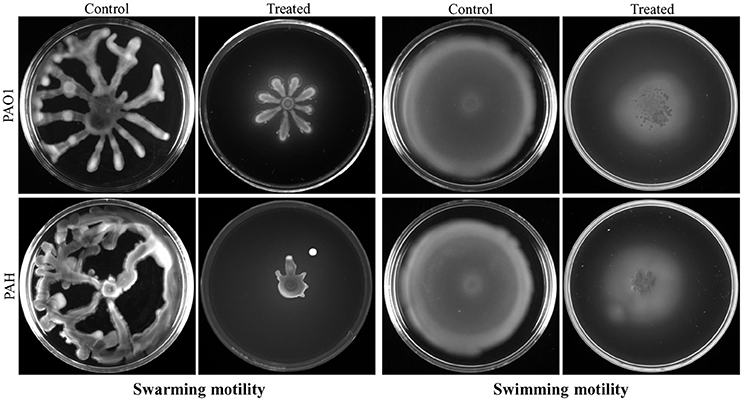
Figure 7. Study of cell motility of P. aeruginosa. The effect of bacterial extract (D. tsuruhatensis SJ01) on the swarming and swimming motility of reference P. aeruginosa strains was studied. P. aeruginosa was spotted on a plate supplemented with (0.1 mg/ml) or without extract. Plates were analyzed after incubation of 24 h at 37 °C.
Delftia tsuruhatensis SJ01 Extract Relegates the Virulence Activities
It was observed that bacterial extract (D. tsuruhatensis SJ01) reduced the production of virulence factors; pyocyanin and rhamnolipid (Figure 8). Pyocyanin production decreased about 70 and 55% in PAO1 and PAH strains, respectively with the treatment of 0.1 mg/ml bacterial extract. Similarly, rhamnolipid production was also decreased by 85 and 67% in PAO1 and PAH strains, respectively, in the presence of bacterial extract (0.1 mg/ml). The effect D. tsuruhatensis SJ01 extract on the elastase and protease activities of cell-free P. aeruginosa bacterial culture supernatant were also assessed (Figure 8). About 32–35% decrease in elastase activities was detected for both strains compared to the control. However, about 23–24% inhibition in the protease activity was found in both strains with 0.1 mg/ml bacterial extract compared to untreated samples.
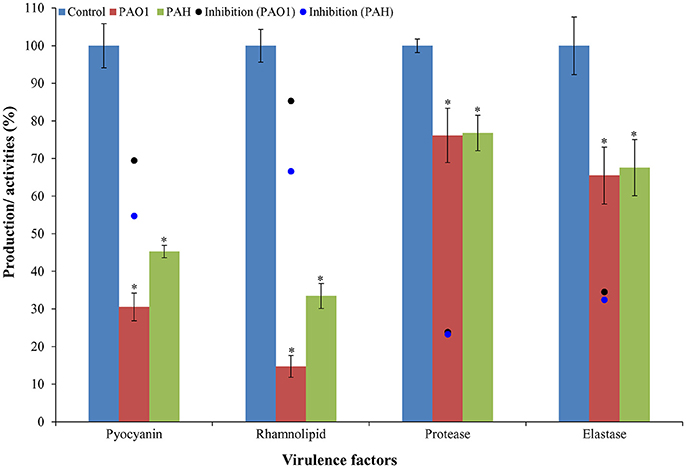
Figure 8. Effect of D. tsuruhatensis SJ01 extract on the virulence factors of P. aeruginosa. The effect of bacterial extracts (SJ01; 0.1 mg/ml) was studied on the production of virulence factors of reference P. aeruginosa strains by quantifying pyocyanin and rhamnolipid, and analyzing elastase and protease activities. *Indicates significant differences from the control at P< 0.05.
Identification of Quorum Sensing Inhibitor Compound
In total, five fractions (in 20, 40, 60, 80, and 100% methanol) were collected through each SPE cartridge (non-polar C18, polar SI, anion exchanger DAE, and cation mixed Plexa PCX); all were screened individually for QSI using a biosensor plate containing C. violaceum CV026. Fraction (C18-100), collected through the C18 cartridge with 100% methanol, showed a maximum zone of QSI; therefore, this was selected for further characterization. Fraction C18-100 was subjected to GC-MS analysis, and the chromatogram showed a single peak at the retention time 16.518 min (Figure 9). The detected mass spectra showed some resemblance to 1,2-benzenedicarboxylic acid, diisooctyl ester, in the GC-MS library (NIST 27. LB). The calculated (theoretical) or expected molecular mass of compound 1,2-benzenedicarboxylic acid, diisooctyl ester (C24H38O4) is 390.55. The molecular mass of the active fraction (C18-100) was further confirmed by ESI-MS. A mass spectral peak, detected at m/z 397.1852, was considered the corresponding experimental mass of the active fraction (Figure 9).
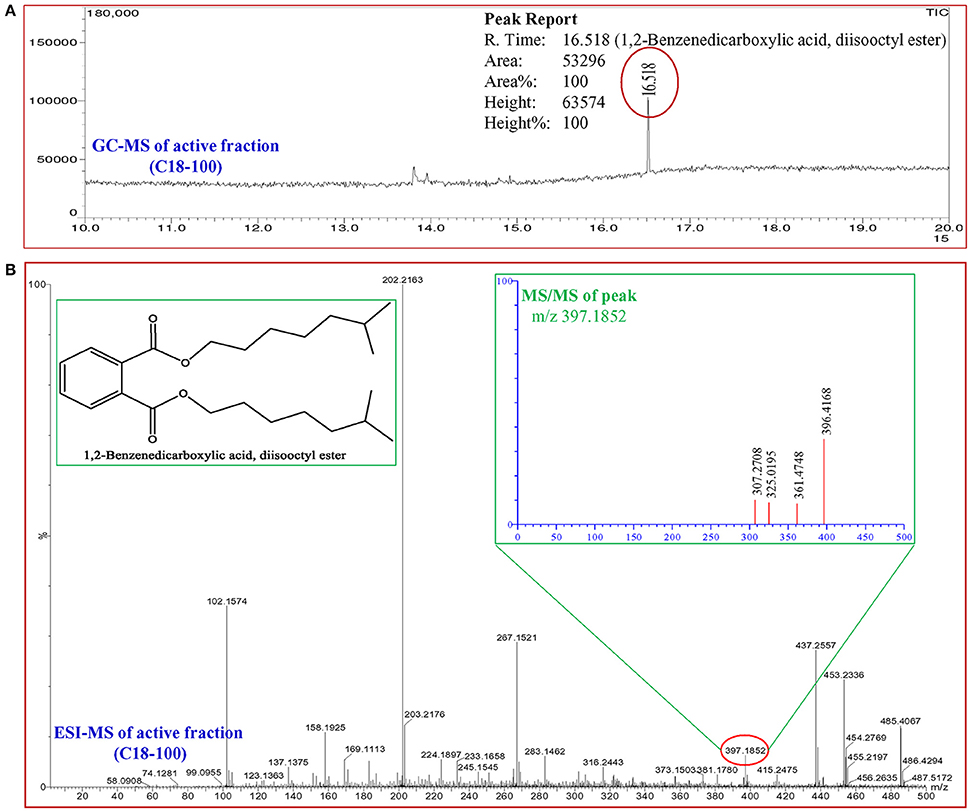
Figure 9. Analysis of active fraction showing quorum sensing inhibition. (A) GC chromatograms and (B) ESI-MS/MS of the C18-100 active fraction of D. tsuruhatensis SJ01 extract and structure of 1,2-benzenedicarboxylic acid, diisooctyl ester (redrawn by ChemBioDraw Ultra 12.0).
Microarray and Transcript Expression Analyses Exhibit Differential Expression of QS Regulatory Genes
Differential expression of quorum sensing regulatory genes of reference strain P. aeruginosa PAO1 treated with a bacterial fraction (C18-100) containing 1,2-benzenedicarboxylic acid, diisooctyl ester as a probable bioactive compound was analyzed using P. aeruginosa PAO1 genome array gene chip. Out of the 5,886 gene probe sets, 1,434 genes were differentially expressed (Table S1; Array-Express accession E-MTAB-5693) and showed at least 2-fold up- (>2) or down-(< −2) expression at p < 0.05 (Figure 10). Of these, 734 genes were up-regulated, whereas 700 genes were down-regulated. Some differentially expressed important genes (as observed in microarray analysis) involved in the quorum sensing and general metabolic pathways are listed in Table 2. The microarray scattered plot showed the differential expression of genes; up-regulation of genes was indicated by blue marks whereas green-colored dots represented down-regulation (Figure S7). The quantitative RT-PCR revealed that the genes LasI, LasR, RhlI, and RhlR were down-regulated in the treated P. aeruginosa compared to the control (Figure 10). About, 9.7-, 3.9-, 3-, and 5.9-fold down-regulation of the genes LasI, LasR, RhlI and RhlR, respectively, was observed in P. aeruginosa PAO1 strain. Similarly, 5.7-, 3.1-, 5.2-, and 4-fold decrease in gene expression was found in P. aeruginosa PAH strain.
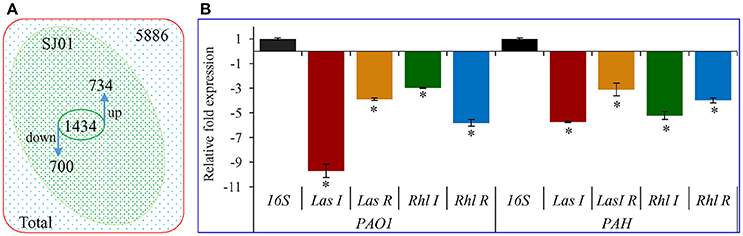
Figure 10. Transcript expression analysis of P. aeruginosa treated with D. tsuruhatensis SJ01 extract. (A) Venn diagram showing genes differentially expressed in P. aeruginosa PAO1 as studied by microarray and (B) expression profiling of some QSI regulatory genes from P. aeruginosa (strains PAO1 and PAH). *Indicates significant differences from the control at P < 0.05.
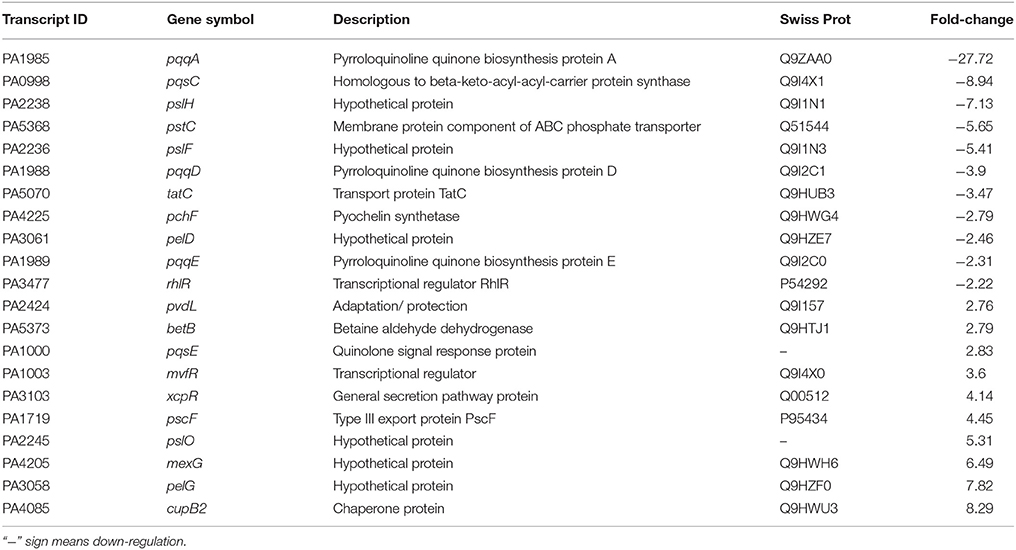
Table 2. Selected transcripts that differentially expressed (up- or down- regulated) in P. aeruginosa PAO1, treated with bacterial (D. tsuruhatensis SJ01) active fraction (C18-100; containing 1,2-benzenedicarboxylic acid, diisooctyl ester) compared with control (untreated PAO1 strain).
Discussion
Natural products are an imperative source for the discovery of novel therapeutics, and microbes are therefore considered a primary source for drug discovery (Gillespie et al., 2002; Courtois et al., 2003). Biofilm forming bacteria are shown to be resistant toward a broad spectrum of antibiotics and make it difficult to cure biofilm-related infections (Høiby et al., 2010). It has been demonstrated that the social behavior of bacterial life depends on two interrelated phenomena: quorum sensing and biofilm formation (Nadell et al., 2008). Biofilm formation of pathogenic P. aeruginosa is controlled by the quorum sensing (QS) regulatory genes, and anti-quorum sensing compounds are explored to inhibit the biofilm formation. These compounds intervene in the QS mechanism and inhibit the expression of virulence factors. Recently, it has been shown that commercially available anti-QS compounds could increase the susceptibility of bacterial biofilm to antibiotics, both in vitro and in vivo (Brackman et al., 2008). Anti-QS properties have been reported from several rhizospheric bacteria, and Stenotrophomona rhizosphila reduced the AHL level (Christiaen et al., 2011). The rhizosphere of different plants (cucumber, tobacco, and ginger) was also exploited to isolate bacteria with anti-quorum sensing activity (Kang et al., 2004; D'Angelo-Picard et al., 2005; Chan et al., 2011). In this study, D. tsuruhatensis SJ01 was isolated from the rhizosphere of C. laevigatus L. collected from the coastal saline area. Previously, we have demonstrated that Stenotrophomonas maltophilia, isolated from C. laevigatus rhizosphere, showed quorum quenching and anti-biofilm forming activity (Singh et al., 2013).
Violacein production is a prerequisite for quorum sensing that leads to biofilm formation. A reference strain C. violaceum CV026 is well known for the production of violacein in the presence of external AHL and is widely used for quorum sensing studies. Extracts of D. tsuruhatensis SJ01 showed anti-QS activity against C. violaceum CV026 on biosensor plates (Figure 1) and inhibited violacein production in a concentration-dependent manner (Figure 2). About 98% inhibition of violacein production was detected with 0.1 mg/ml D. tsuruhatensis extract. However, it is difficult to compare the results with previous reports because of variation in the extraction methods and other parameters. About 90–94% reduction in the violacein production was reported with 3–4 mg/ml extract of S. maltophilia and Melicope lunu-ankenda extracts (Tan et al., 2012; Singh et al., 2013). Furthermore, the zone of inhibition was not observed when D. tsuruhatensis was spotted onto a plate containing C. violaceum culture (Figure S1). This rules out the possibility of antibacterial (C. violaceum) activity of D. tsuruhatensis. Inhibition of the AHL-dependent quorum sensing mechanism of CV026 (Figure 2) revealed the anti-quorum sensing potential of the extract at very low concentration (0.1 mg/ml).
The extract of D. tsuruhatensis SJ01 inhibits the biofilm formation of clinical isolates P. aeruginosa PAO1 as well as P. aeruginosa PAH (Figure 3) without affecting planktonic growth (Figures S4, S5). Strain PAO1 showed about 15% increase in planktonic cell growth (with a higher concentration of extracts), possibly because of the inability of strains to attach to the surface and subsequently to form a biofilm. This may lead to an increase of planktonic cell growth. However, a detailed study is required to ascertain the exact reason behind it. The viable P. aeruginosa cells were observed under epi-fluorescence microscopy (Figure 4) which confirmed that extract (SJ01) does not have a toxic effect on cells within the biofilm (Figure 4). The functional indices of biofilm exhibited physical characteristics (Ţǎlu, 2013). The AFM-based statistical analysis indicated a decrease in the bearing property, fluid retention and roughness of the biofilm (Table 1). The AFM topographs suggest full grown biofilm in control compared to treated conditions (Figure 6). Alterations in the physical property under treated conditions led to loosely packed polymers which are not supportive for bacterial adherence; as a result, delicate biofilms are formed. Similarly, a discreet biofilm was visualized under a scanning electron microscopy (Figure 5). The steady decrease of biofilm formation was associated with an increase of extract concentration and about 60% biofilm inhibition was observed with 0.1 mg/ml SJ01 extract. The motility of bacteria plays a vital role in biofilm formation, for which bacteria need to attach to the surface or substratum. They utilize their flagellum driven motility to reach substratum; once attached to the surface, they were spared all around via swimming and swarming, which led to the biofilm formation (O'May and Tufenkji, 2011). The extract of SJ01 inhibits the motility of the P. aeruginosa (Figure 7) and thus decreases the possibility of biofilm formation.
A compound 1,2-benzenedicarboxylic acid, diisooctyl ester was identified in the active fraction of the SJ01 extract by GC-MS and ESI (Figure 9). A similar compound, 1,2-benzenedicarboxylic acid bis (2α-methylheptyl) ester, was isolated from Alcaligenes faecalis YMF 3.175 and reported to have antibacterial activity against Escherichia coli and Staphylococcus aureus (Zhu et al., 2011). The antibacterial activity was also reported for 1,2-benzenedicarboxylic acid, mono (2-ethylhexyl) ester isolated from the endophytic fungus Muscodor tigerii (Saxena et al., 2015). However, in this study, antibacterial activity was not detected for 1,2-benzenedicarboxylic acid, diisooctyl ester (Figure 4 and Figure S4). Secondary infections caused by P. aeruginosa are difficult to eradicate due to their high levels of resistance to most conventional antibiotics. The challenge of combatting the infection becomes more complex due to the ability of the pathogen to form a biofilm matrix which protects bacterial cells from environmental stress as well as antibiotics (Driscoll et al., 2007; Lee and Zhang, 2015). It is the first report of anti-quorum sensing and anti-biofilm activity of 1,2-benzenedicarboxylic acid, diisooctyl ester on P. aeruginosa however, a detailed study is required to develop this compound as an anti-pathogenic drug for the treatment of the biofilm forming pathogenic bacteria.
Early colonization on host tissues is initiated by elastase and protease, whereas pyocyanin interferes with multiple cellular functions, chelates iron uptake, and promotes virulence expression (Lau et al., 2004; Stehling et al., 2008). The rhamnolipids facilitate surface motility of P. aeruginosa for biofilm formation and are also involved in the dispersal of mature biofilm (O'May and Tufenkji, 2011). Thus, the pathogenicity of P. aeruginosa depends on the virulence factor, and pyocyanin plays a key role in this infection (Lau et al., 2004). It was observed that pyocyanin production decreased by about 70 and 55% in strain PAO1 and PAH, respectively, by SJ01 extract (Figure 8). Furthermore, rhamnolipid, protease, and elastase are also regarded as important indicators for quorum sensing (Sarabhai et al., 2013). About 85 and 67% reduction of rhamnolipid production was noticed for P. aeruginosa PAO1 and PAH, respectively; however, a significant decrease (24–35%) was observed for protease and elastase activity by SJ01 extract (Figure 8). The production and activity of virulence factors is controlled by the las and rhl regulatory system in P. aeruginosa (De Kievit and Iglewski, 2000; Kohler et al., 2000).
The GeneChip probe array is a powerful tool for monitoring transcriptional regulation of any organism. The array used in this study represents the annotated genome of P. aeruginosa strain PAO1 and includes 5,549 protein-coding sequences, 18 tRNA genes, a representative of the ribosomal RNA cluster and 117 genes present in strains other than PAO1. The microarray analysis showed the differential expression of 1,434 genes and revealed that a large number of genes are directly or indirectly involved in biofilm formation (Figure 10, Table 2, and Table S1). Most of these genes are involved in quorum sensing, virulence, motility, and transport. Transcriptional regulators and hypothetical proteins were also differentially expressed and thus may play an important role in biofilm formation. The key genes, LasI, LasR, RhlI, and RhlR, were down-regulated in P. aeruginosa compared to the control (Figure 10).
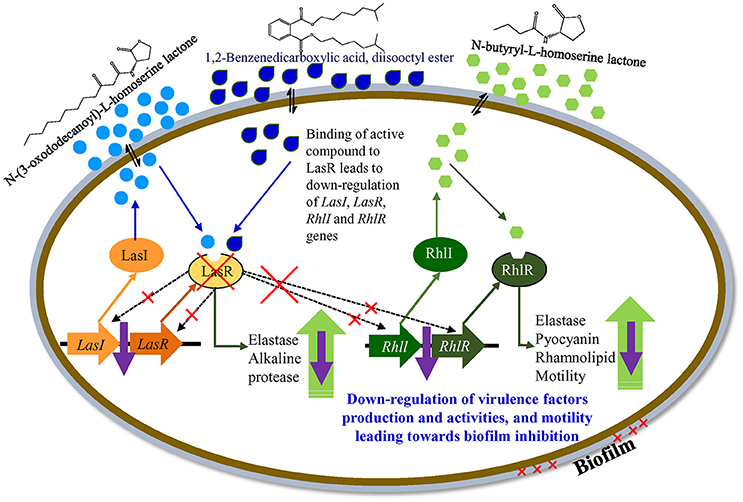
The las regulatory system of P. aeruginosa consists of the LasI synthase protein and LasR transcriptional regulator. LasI is essential for the production of the AHL signal molecule N-(3-oxododecanoyl)-l-homoserine lactone (3O-C12-HSL), and LasR requires 3O-C12-HSL to become an active transcription factor (Gambello and Iglewski, 1991; Pearson et al., 1994; Kiratisin et al., 2002). A second QS system (of P. aeruginosa), rhl, is also comprised of the RhlI and RhlR proteins. RhlI synthase produces the AHL N-butyryl-L-homoserine lactone (C4-HSL) and the transcriptional regulator RhlR becomes activated when complexed with C4-HSL (Ochsner et al., 1994; Pearson et al., 1995). Both lasR and rhlR regulate the expression of several genes and activity including, pyocyanin, rhamnolipid, elastase, protease, and motility.
Based on the differential gene expression (microarray and qRT-PCR) of quorum sensing key regulatory gene(s) a theoretical model for the transcriptional regulatory mechanism in P. aeruginosa was inferred (Figure 11). The proposed model is just a schematic representation (based on available literature) in the form of a hypothetical model explaining transcriptional regulation of QSI in P. aeruginosa. However, a detailed study is needed to confirm the exact role of the identified compound in the QSI regulation mechanism. It was hypothesized that the identified compound 1,2-benzenedicarboxylic acid, diisooctyl ester (showing structural similarity with AHL) may compete with AHL and bind to LasR. Binding with LasR down-regulates the protease and elastase activity, along with expression of the rhl regulatory system. Down-regulation of the rhl QS system leads to the lower activity of pyocyanin and rhamnolipid production along with elastase, protease, and motility. Results indicate that the active compound may decrease the production of virulence factors through transcriptional regulation of the expression of las and rhl QS systems.
Conclusion
A bacterium, D. tsuruhatensis SJ01, isolated from the rhizosphere of C. laevigatus showed anti-quorum sensing and anti-biofilm activities. Furthermore, SJ01 extract does not possess anti-bacterial properties. A compound 1,2-benzenedicarboxylic acid, diisooctyl ester was identified as a probable active compound in the bacterial fraction. The compound inhibits the biofilm formation of clinical isolate P. aeruginosa PAO1 and human pathogenic strain P. aeruginosa PAH by decreasing the swimming and swarming motility and regulating virulence factors such as pyocyanin, rhamnolipid, elastase, and protease. The compound may intervene in the QS system of P. aeruginosa and down-regulate the gene(s) responsible for the quorum sensing mechanism. Our results demonstrate that the active compound may target the QS systems. Targeting a QS system is important for therapeutics, and this may be used for the effective treatment of biofilm-related infection. The inhibitor may be a potent drug for the eradication of P. aeruginosa infections, and the active compound has the potential to be developed as an anti-pathogenic drug; however, a detailed study is still needed to investigate potential pharmaceutical applications.
Author Contributions
Conceived and designed the experiments: AM and BJ; Performed the experiments: VS; Analyzed the data: VS and AM; Wrote the manuscript: AM and VS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CSIR-CSMCRI Communication No.: PRIS-15/2017. This study was supported by the Ministry of Earth Sciences (MoES), Government of India, New Delhi (Sanction No. MoES/16/06/2013-RDEAS). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. Authors are duly acknowledged Prof. Anton Hartmann (Helmholtz Zentrum, München, Germany) for providing reference strain Chromobacterium violaceum CV026. Authors are also thankful to Govt. Medical College, Bhavnagar (India) for giving clinical isolate P. aeruginosa PAH. Analytical Discipline and Centralized Instrument Facility of the Institute is duly acknowledged for running the samples.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00337/full#supplementary-material
Figure S1. Antibacterial disc diffusion assay of D. tsuruhatensis SJ01 against C. violaceum CV026. The Mueller-Hinton agar (MHA) plate containing reference strain C. violaceum CV026 were tested for antibacterial activity of D. tsuruhatensis SJ01. Strain SJ01 represents the culture (5 μl) and the antibiotic tobramycin (5 μl) was used as a positive control.
Figure S2. Phylogenetic position of D. tsuruhatensis SJ01 (KX130769) with taxonomic neighbors. Numbers at nodes are percentage bootstrap values. The phylogenetic tress was computed using the maximum composite likelihood method and are in the units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated by the complete deletion option. Phylogenetic analysis was conducted in MEGA (ver 6). Bar indicates 0.005 substitutions per nucleotide position.
Figure S3. Whole cell fatty acid profiling of the bacterium D. tsuruhatensis SJ01. The whole cell fatty acid profile of strain SJ01 was performed by GC coupled with MIDI. The name of the fatty acids was assigned on the basis of corresponding fatty acids of RTSBA6 6.10 library match.
Figure S4. Effect of extract of D. tsuruhatensis SJ01 extract on planktonic cell growth of P. aeruginosa. Different concentration of bacterial extracts (SJ01; 0.01−0.1 mg/ml) was tested against biofilm forming reference strain P. aeruginosa PAO1 and pathogenic strain P. aeruginosa PAH. Tests without extract and with methanol were considered as control and negative control, respectively.
Figure S5. Effect of extract (0.1 mg/ml) of D. tsuruhatensis SJ01 extract on the growth curve of P. aeruginosa. Bacterial extracts (SJ01; 0.1 mg/ml) was tested for effect on growth of biofilm forming reference strain P. aeruginosa PAO1 and pathogenic strain P. aeruginosa PAH. The OD was taken up to 24 h at 600 nm using spectrophotometer. Growth of bacteria without treatment of extract (SJ01) was considered control.
Figure S6. Antibacterial disc diffusion assay of D. tsuruhatensis SJ01 against P. aeruginosa. Clinical isolates of P. aeruginosa PAO1 and PAH were tested for antibacterial activity of D. tsuruhatensis SJ01 extract. The bacterial extract did not show any antibacterial activity against the clinical isolates P. aeruginosa.
Figure S7. A microarray scattered plot showing differential expression of genes. Up- and down- regulation of genes are indicated by blue and green colored marks, respectively. The analysis was performed in expression console and transcriptome analysis console, and genes exhibiting significant fold expression (ANOVA p < 0.05) were considered for the study.
Table S1. Total transcripts that differentially expressed (up- or down- regulated) in P. aeruginosa PAO1, treated with bacterial (D. tsuruhatensis SJ01) active fraction (C18-100; containing 1,2-benzenedicarboxylic acid, diisooctyl ester) compared with control (untreated PAO1 strain). Array-Express accession E-MTAB-5693.
References
Adonizio, A., Kong, K. F., and Mathee, K. (2008). Inhibition of quorum sensing controlled virulence factor production in Pseudomonas aeruginosa by south florida plant extracts. Antimicrob. Agents Chemother. 52, 198–203. doi: 10.1128/AAC.00612-07
Andersson, S., Dalhammar, G., Land, C., and Rajarao, G. (2009). Characterization of extracellular polymeric substances from denitrifying organism Comamonas denitrificans. Appl. Microbiol. Biotechnol. 82, 535–543. doi: 10.1007/s00253-008-1817-3
Bakkiyaraj, D., Sivasankar, C., and Pandian, S. K. (2012). Inhibition of quorum sensing regulated biofilm formation in Serratia marcescens causing nosocomial infections. Bioorg. Med. Chem. Lett. 22, 3089–3094. doi: 10.1016/j.bmcl.2012.03.063
Bakkiyaraj, D., Sivasankar, C., and Pandian, S. K. (2013). Anti-pathogenic potential of coral associated bacteria isolated from Gulf of Mannar against Pseudomonas aeruginosa. Indian J. Microbiol. 53, 111–113. doi: 10.1007/s12088-012-0342-3
Brackman, G., Defoirdt, T., Miyamoto, C., Bossier, P., Calenbergh, S. V., Nelis, H., et al. (2008). Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 8:149. doi: 10.1186/1471-2180-8-149
Chan, K., Atkinson, S., Mathee, K., Sam, C., Chhabra, S., Camara, M., et al. (2011). Characterization of N-acylho- moserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: co-existence of quorum quenching and quorum sensing Acinetobacter and Burkholderia. BMC Microbiol. 11:51. doi: 10.1186/1471-2180-11-51
Chen, W.-M., Lin, Y.-S., Sheu, D.-S., and Sheu, S.-Y. (2012). Delftia litopenaei sp. nov., a poly-β-hydroxybutyrate-accumulating bacterium isolated from a freshwater shrimp culture pond. Int. J. Syst. Evol. Microbiol. 62, 2315–2321. doi: 10.1099/ijs.0.037507-0
Choo, J., Rukayadi, Y., and Hwang, J. (2006). Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 42, 637–641. doi: 10.1111/j.1472-765x.2006.01928.x
Christiaen, S. E., Brackman, G., Nelis, H. J., and Coenye, T. (2011). Isolation and identification of quorum quenching bacteria from environmental samples. J. Microbiol. Meth. 87, 213–219. doi: 10.1016/j.mimet.2011.08.002
Christiaen, S. E., Matthijs, N., Zhang, X. H., Nelis, H. J., Bossier, P., and Coenye, T. (2014). Bacteria that inhibit quorum sensing decrease biofilm formation and virulence in Pseudomonas aeruginosa PAO1. Pathog. Dis. 70, 271–279. doi: 10.1111/2049-632X.12124
Cirou, A., Diallo, S., Kurt, C., Latour, X., and Faure, D. (2007). Growth promotion of quorum-quenching bacteria in the rhizosphere of Solanum tuberosum. Environ. Microbiol. 9, 1511–1522. doi: 10.1111/j.1462-2920.2007.01270.x
Courtois, S., Cappellano, C. M., Ball, M., et al. (2003). Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69, 49–55. doi: 10.1128/AEM.69.1.49-55.2003
D'Angelo-Picard, C., Faure, D., Penot, I., and Dessaux, Y. (2005). Diversity of N-acyl homoserine lactone-producing and degrading bacteria in soil and tobacco rhizosphere. Environ. Microbiol. 7, 1796–1808. doi: 10.1111/j.1462-2920.2005.00886.x
De Kievit, T. R., and Iglewski, B. H. (2000). Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68, 4839–4849. doi: 10.1128/IAI.68.9.4839-4849.2000
Driscoll, J. A., Brody, S. L., and Kollef, M. H. (2007). The epidemiology, pathogenesis, and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368. doi: 10.2165/00003495-200767030-00003
Essar, D. W., Eberly, L., Hadero, A., and Crawford, I. P. (1990). Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172, 884–900. doi: 10.1128/jb.172.2.884-900.1990
Felsenstein, J. (1985). Confidence limits on phylogenesis: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Fuqua, C., and Greenberg, E. P. (1998). Self perception in bacteria: quorum sensing with acylated homoserine lactones. Curr. Opin. Microbiol. 2, 183–189. doi: 10.1016/S1369-5274(98)80009-X
Gambello, M. J., and Iglewski, B. H. (1991). Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173, 3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991
Gillespie, D. E., Brady, S. F., Bettermann, A. D., et al. (2002). Isolation of antibiotics turbomycin A and B fromametagenomic library of soil microbial DNA. Appl. Environ. Microb. 68, 4301–4306. doi: 10.1128/AEM.68.9.4301-4306.2002
Goswami, N. N., Trivedi, H. R., Goswami, A. P. P., Patel, T. K., and Tripathi, C. B. (2011). Antibiotic sensitivity profile of bacterial pathogens in postoperative wound infections at a tertiary care hospital in Gujarat, India. J. Pharmacol. Pharmacother. 2, 158. doi: 10.4103/0976-500X.83279
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Jha, B., Singh, V. K., Weiss, A., Hartmann, A., and Schmid, M. (2015). Zhihengliuella somnathii sp. nov., a halotolerant actinobacterium from the rhizosphere of a halophyte Salicornia brachiata. Int. J. Syst. Evol. Microbiol. 65, 3137–3142. doi: 10.1099/ijsem.0.000391
Jørgensen, N. O., Brandt, K. K., Nybroe, O., and Hansen, M. (2009). Delftia lacustris sp. nov., a peptidoglycan-degrading bacterium from fresh water, and emended description of Delftia tsuruhatensis as a peptidoglycan-degrading bacterium. Int. J. Syst. Evol. Microbiol. 59, 2195–2199. doi: 10.1099/ijs.0.008375-0
Kalia, V. C. (2012). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. doi: 10.1016/j.biotechadv.2012.10.004
Kalia, V. C., and Purohit, H. J. (2011). Quenching the quorum sensing system: potential antibacterial drug targets. Crit. Rev. Microbiol. 37, 121–140. doi: 10.3109/1040841X.2010.532479
Kang, B. R., Lee, J. H., Ko, S. J., Lee, Y. H., Cha, J. S., Cho, B. H., et al. (2004). Degradation of acyl-homoserine lactone molecules by Acinetobacter sp. strain C1010. Can. J. Microbiol. 50, 935–941. doi: 10.1139/w04-083
Kavita, K., Singh, V. K., Mishra, A., and Jha, B. (2014). Characterisation and anti-biofilm activity of extracellular polymeric substances from Oceanobacillus iheyensis. Carbohydr. Polym. 101, 39–35. doi: 10.1016/j.carbpol.2013.08.099
Keshri, J., Mishra, A., and Jha, B. (2013). Microbial population index and community structure in saline-alkaline soil using gene targeted metagenomics. Microbiol. Res. 168, 165–173. doi: 10.1016/j.micres.2012.09.005
Keshri, J., Yousuf, B., Mishra, A., and Jha, B. (2015). The abundance of functional genes, cbbL, nifH, amoA and apsA, and bacterial community structure of intertidal soil from Arabian Sea. Microbiol. Res. 175, 57–66. doi: 10.1016/j.micres.2015.02.007
Kiratisin, P., Tucker, K. D., and Passador, L. (2002). LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 184, 4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002
Kohler, T., Curty, L. K., Barja, F., Delden, C. V., and Pechere, J. C. (2000). Swarming of Pseudomonas aeruginosa is dependent on cell to cell signalling and requires flagella and pili. J. Bacteriol. 182, 5990–5996. doi: 10.1128/JB.182.21.5990-5996.2000
Lau, G. W., Hassett, D. J., Ran, H., and Kong, F. (2004). The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol. Med. 10, 599–606. doi: 10.1016/j.molmed.2004.10.002
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Li, C. T., Yan, Z. F., Chu, X., Hussain, F., Xian, W. D., Yunus, Z., et al. (2015). Delftia deserti sp. nov., isolated from a desert soil sample. Antonie Van Leeuwenhoek 107, 1445–1450. doi: 10.1007/s10482-015-0440-4
Livak, K. J., and Schmittgen, T. D. (2001). Analyses of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C (T) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McClure, C. D., and Schiller, N. L. (1992). Effects of Pseudomonas aeruginosa rhamnolipids on human monocyte-derived macrophages. J. Leukocyte Biol. 51, 97–102.
Nadell, C., Xavier, J., Levin, S., and Foster, K. (2008). The evolution of quorum sensing in bacterial biofilms. PLoS Biol. 6:e14. doi: 10.1371/journal.pbio.0060014
Ni, N., Li, M., Wang, J., and Wang, B. (2009). Inhibitors and antagonists of bacterial quorum sensing. Med. Res. Rev. 29, 65–124. doi: 10.1002/med.20145
Nithya, C., Begum, M. F., and Pandian, S. K. (2010). Marine bacterial isolates inhibit biofilm formation and disrupt mature biofilms of Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 88, 341–358. doi: 10.1007/s00253-010-2777-y
Ochsner, U. A., Koch, A. K., Fiechter, A., and Reiser, J. (1994). Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176, 2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994
Oh, Y., Lee, N., Jo, W., Jung, W., and Lim, J. (2009). Effects of substrates on biofilm formation observed by atomic force microscopy. Ultramicroscopy 109, 874–880. doi: 10.1016/j.ultramic.2009.03.042
O'May, C., and Tufenkji, N. (2011). The swarming motility of Pseudomonas aeruginosa is blocked by cranberry proanthocyanidins and other tannin-containing materials. Appl. Environ. Microbiol. 77, 3061–3067. doi: 10.1128/AEM.02677-10
Oncul, O., Ulkur, E., Acar, A., Turhan, V., Yeniz, E., Karacaer, Z., et al. (2009). Prospective analysis of nosocomial infections in a burn care unit, Turkey. Indian J. Med. Res. 130, 758–764. Available online at: http://icmr.nic.in/ijmr/2009/december/1214.pdf
Overhage, J., Lewenza, S., Marr, A. K., and Hancock, R. E. (2007). Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 Mini-Tn5-lux mutant library. J. Bacteriol. 189, 2164–2169. doi: 10.1128/JB.01623-06
Pearson, J. P., Gray, K. M., Passador, L., Tucker, K. D., Eberhard, A., Iglewski, B. H., et al. (1994). Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U.S.A. 91, 197–201. doi: 10.1073/pnas.91.1.197
Pearson, J. P., Passador, L., Iglewski, B. H., and Greenberg, E. P. (1995). A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 92, 1490–1494. doi: 10.1073/pnas.92.5.1490
Rahme, L. G., Ausubel, F. M., Cao, H., Drenkard, E., Goumnerov, B. C., Lau, G. W., et al. (2000). Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. U.S.A. 97, 8815–8821. doi: 10.1073/pnas.97.16.8815
Rashid, M. H., and Kornberg, A. (2000). Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 97, 4885–4890. doi: 10.1073/pnas.060030097
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sarabhai, S., Sharma, P., and Capalash, N. (2013). Ellagic acid derivatives from terminalia chebula retz. Downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS ONE 8:e53441. doi: 10.1371/journal.pone.0053441
Saxena, S., Meshram, V., and Kapoor, N. (2015). Muscodor tigerii sp. nov.-Volatile antibiotic producing endophytic fungus from the Northeastern Himalayas. Ann. Microbiol. 65, 47–57. doi: 10.1007/s13213-014-0834-y
Shigematsu, T., Yumihara, K., Ueda, Y., Numaguchi, M., Morimura, S., and Kida, K. (2003). Delftia tsuruhatensis sp. nov., a terephthalate-assimilating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 53, 1479–1483. doi: 10.1099/ijs.0.02285-0
Singh, V. K., Kavita, K., Prabhakaran, R., and Jha, B. (2013). Cis-9-octadecenoic acid from the rhizospheric bacterium Stenotrophomonas maltophilia BJ01 shows quorum quenching and anti-biofilm activities. Biofouling 29, 855–867. doi: 10.1080/08927014.2013.807914
Singh, V. K., Mishra, A., Haque, I., and Jha, B. (2016a). A novel transcription factor-like gene SbSDR1 acts as a molecular switch and confers salt and osmotic endurance to transgenic tobacco. Sci. Rep. 6:31686. doi: 10.1038/srep31686
Singh, V. K., Mishra, A., and Jha, B. (2016b). “Marine bacterial extracellular polymeric substances: characteristics and applications,” in Marine Glycobiology: Principles and Applications, ed S. Kim (Boca Raton, FL: Taylor and Francis Group; CRC Press), 369–377. doi: 10.1201/9781315371399-27
Stehling, E. G., da Silveira, W. D., and da Silva Leite, D. (2008). Study of biological characteristics of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis and from patients with extra-pulmonary infections. Braz. J. Infect. Dis. 12, 86–88. doi: 10.1590/S1413-86702008000100018
Ţǎlu, Ş. (2013). Characterization of surface roughness of unworn hydrogel contact lenses at a nanometric scale using methods of modern metrology. Polym. Eng. Sci. 53, 2141–2150. doi: 10.1002/pen.23481
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, L. Y., Yin, W. F., and Chan, K. G. (2012). Silencing quorum sensing through extracts of Melicope lunu-ankenda. Sensors 12, 4339–4351. doi: 10.3390/s120404339
Vandeputte, O. M., Kiendrebeogo, M., Rajaonson, S., Diallo, B., Mol, A., El Jaziri, M., et al. (2010). Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 76, 243–253. doi: 10.1128/AEM.01059-09
Wang, E. W., Jung, J. Y., Pashia, M. E., Nason, R., Scholnick, S., and Chole, R. A. (2005). Otopathogenic Pseudomonas aeruginosa strains as competent biofilm formers. Arch. Otolaryngol. Head Neck Surg. 131, 983–989. doi: 10.1001/archotol.131.11.983
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Wen, A., Fegan, M., Hayward, C., Chakraborty, S., and Sly, L. I. (1999). Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926 and Tamaoka et al. 1987) gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 49, 567–576. doi: 10.1099/00207713-49-2-567
Zhu, H., He, C.-C., and Chu, Q.-H. (2011). Inhibition of quorum sensing in Chromobacterium violaceum by pigments extracted from Auricularia auricular. Lett. Appl. Microbiol. 52, 269–274. doi: 10.1111/j.1472-765X.2010.02993.x
Keywords: anti-biofilm, anti-quorum, microarray, quorum network, quorum quenching, quorum sensing, virulence factors
Citation: Singh VK, Mishra A and Jha B (2017) Anti-quorum Sensing and Anti-biofilm Activity of Delftia tsuruhatensis Extract by Attenuating the Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 7:337. doi: 10.3389/fcimb.2017.00337
Received: 11 May 2017; Accepted: 10 July 2017;
Published: 26 July 2017.
Edited by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoReviewed by:
Anil Kumar Singh, Indian Institute of Agricultural Biotechnology (ICAR), IndiaAnanda Mustafiz, South Asian University, India
Thibault Géry Sana, Stanford University, United States
Israeñ Castillo Juárez, College of Postgraduates Montecillo, Mexico
Copyright © 2017 Singh, Mishra and Jha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Avinash Mishra, YXZpbmFzaEBjc21jcmkub3Jn; YXZpbmFzaEBjc21jcmkucmVzLmlu; YXZpbmFzaG1pc2hyYTExQHJlZGlmZm1haWwuY29t
Bhavanath Jha, YmpoYUBjc21jcmkucmVzLmlu
 Vijay K. Singh
Vijay K. Singh Avinash Mishra
Avinash Mishra Bhavanath Jha
Bhavanath Jha