- 1Department of Microbiology and Immunology, University of Miami Miller School of Medicine, Miami, FL, United States
- 2Division of Pediatric Infectious Diseases, Department of Pediatrics, University of Miami Miller School of Medicine, Miami, FL, United States
- 3Department of Mathematics, Charles E. Schmidt College of Science, Florida Atlantic University, Boca Raton, FL, United States
- 4Department of Biomedical Science, Charles E. Schmidt College of Medicine, Florida Atlantic University, Boca Raton, FL, United States
Yersinia pestis, the causative agent of plague, possesses a number of virulence mechanisms that allows it to survive and proliferate during its interaction with the host. To discover additional infection-specific Y. pestis factors, a transposon site hybridization (TraSH)-based genome-wide screen was employed to identify genomic regions required for its survival during cellular infection. In addition to several well-characterized infection-specific genes, this screen identified three chromosomal genes (y3397, y3399, and y3400), located in an apparent operon, that promoted successful infection. Each of these genes is predicted to encode a leucine-rich repeat family protein with or without an associated ubiquitin E3 ligase domain. These genes were designated Yersinia leucine-rich repeat gene A (ylrA), B (ylrB), and C (ylrC). Engineered strains with deletions of y3397 (ylrC), y3399 (ylrB), or y3400 (ylrA), exhibited infection defects both in cultured cells and in the mouse. C-terminal FLAG-tagged YlrA, YlrB, and YlrC were secreted by Y. pestis in the absence but not the presence of extracellular calcium and deletions of the DNA sequences encoding the predicted N-terminal type III secretion signals of YlrA, YlrB, and YlrC prevented their secretion, indicating that these proteins are substrates of the type III secretion system (T3SS). Further strengthening the connection with the T3SS, YlrB was readily translocated into HeLa cells and expression of the YlrA and YlrC proteins in yeast inhibited yeast growth, indicating that these proteins may function as anti-host T3S effector proteins.
Introduction
Yersinia pestis, the etiologic agent of plague, causes a variety of serious diseases in humans and animals. The clinical syndromes in humans include bubonic, pneumonic, and septicemic plague (Perry and Fetherston, 1997). Fleas transmit Y. pestis from infected domestic rats to humans causing bubonic plague. Y. pestis also can be transmitted via respiratory secretions following contact with another infected human, leading to pneumonic plague. Currently, plague is still a public health threat in certain regions of Asia, Africa, North and South America (Centers for Disease Control Prevention., 2013). In recent years in the U.S., most cases of plague have occurred in children in whom diagnosis has been delayed. Among 183 U.S. pediatric cases from 1947 to 2001, 91% presented primarily as bubonic and one third of these cases developed secondary complications, such as sepsis, meningitis, and pneumonia. Children were more likely than adults to manifest with bubonic plague (91 vs. 79%), develop complications (32 vs. 27%), and to die (17 vs. 14%) (Dennis and Chow, 2004). Because plague is highly contagious, Y. pestis can be used in biological warfare and is considered a Category A agent of bioterrorism (Inglesby et al., 2000).
Among the virulence factors identified in Y. pestis, the plasmid pCD1 encoded type III secretion system (T3SS) is one of the best-studied. The T3SS is a complex protein machinery shared by numerous Gram-negative pathogens that injects effector proteins directly into the cytosol of an infected eukaryotic cell (Cornelis, 2002a). The translocation of effector proteins requires direct contact between the bacterium and the host cell. In Y. pestis, the T3S apparatus is called the Ysc injectosome and the effector proteins are termed Yops (Yersinia outer proteins). The Yops disrupt host signaling pathways that normally lead to bacterial phagocytosis and the production of pro-inflammatory cytokines (Cornelis and Van Gijsegem, 2000). To date, six Yop effectors have been identified—YopH, YopE, YopT, YpkA/YopO, YopP/YopJ, and YopM (Cornelis, 2002b). Following its identification as a virulence factor in the mouse model, YopM was initially described as an inhibitor of platelet aggregation (Leung and Straley, 1989). Later YopM was linked to a variety of activities associated with the host innate immune response including depletion of inflammatory monocytes and natural killer (NK) cells and induction of specific cytokines (Kerschen et al., 2004; Ye et al., 2009; McPhee et al., 2010). Once translocated into eukaryotic cells, YopM directly interacts with a number of host proteins including kinases and capase-1 to inhibit signaling pathways and inflammasome activation (McDonald et al., 2003; Hentschke et al., 2010; LaRock and Cookson, 2012; Chung et al., 2014).
Structurally YopM is primarily composed of a number of leucine-rich repeats (LRR) (Evdokimov et al., 2001). Recently it has been shown that Y. pestis, as well as a number of other Yersinia species (but not Y. enterocolitica), possesses a chromosomal locus that potentially encodes three YopM-like proteins (Hu et al., 2016). Sequence analysis showed that the plasmid-encoded YopM was evolutionarily distinct from these three chromosomal-encoded YopM-like proteins and that two of these YopM-like proteins possessed E3 ligase domains in their C-terminal regions (Hu et al., 2016). Here we describe the independent identification and characterization of the genes encoding these three chromosomally-encoded YopM-like proteins in an unbiased screen for Y. pestis factors that promote its survival following its infection of macrophages.
Materials and Methods
Bacterial Strains
The Escherichia coli DH5α and Y. pestis strains were routinely grown in heart infusion broth (HIB) or on tryptose blood agar (TBA) base plates (Difco, Detroit, MI) at 27°C (Y. pestis) or 37°C (E. coli). Appropriate antibiotics were added to culture media when needed with ampicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (20 μg/ml), and streptomycin (50 μg/ml). Non-polar deletion of Y. pestis KIM5 chromosomal DNA sequences ylrC (y3397; codons 29-515), ylrB (y3399; codons 29-261), and ylrA (y3400; codons 23-529) was accomplished using lambda Red recombination as described by Datsenko and Wanner (2000). PCR products used to construct the gene replacement were amplified using the template plasmid pKD4 (Kmr). The resulting PCR products were gel purified, ethanol precipitated, and resuspended in 10 μl of distilled water. Y. pestis KIM5 strain carrying plasmid pKD46, which encodes the Red recombinase, was induced with 0.2% L-arabinose for 2 h prior to harvest. Electrocompetent cells were electroporated with the purified PCR products. The transformations were plated onto TBA plates containing kanamycin (50 μg/ml). Plasmid pCP20, which encodes the FLP recombinase, was electroporated into the Kmr resistant strains to facilitate the removal of the FLP recognition target-flanked kan cassette and plasmid pKD46 simultaneously. Plasmid pCP20 was cured from the Y. pestis deletion strains by overnight growth at 39°C. Additionally, a deletion strain of the three gene sequences ylrC-codon 29 to ylrA-codon 529 was constructed using lambda Red-recombination as described above.
Transposon Site Hybridization (TraSH)-Based Screening
A TraSH-based approach was used to map and quantify the relative abundance of the different transposon mutants in order to identify transposon mutant insertion sites that are underrepresented in the population of mutants exposed to RAW264.7 murine macrophage-like cells. The details of the mutagenesis, infection, and TraSH-screen and data analysis are described in Bartra et al. (2012).
Infection Assays
Cell infection assays were performed essentially as described by Rosenzweig et al. (2005). RAW 264.7 murine macrophage-like cells and HeLa cells were cultured in Dubelcco's modified Eagle's medium (DMEM, Gibco) containing 10% heat-inactivated fetal bovine serum (Cellgro) at 37°C in the presence of 5% CO2. Cells were seeded in 24-well plates at densities of 2.5–4.0 × 105 per well. Bacterial cell cultures were grown in tissue culture (TC) medium overnight at 27°C, diluted into fresh TC media and incubated with shaking at 27°C for 2 h and then shifted to 37°C for 1 h prior to being added to the cultured macrophages at a multiplicity of infection (MOI) of 30. After a 30 min attachment period, fresh DMEM medium was added to each well. At the 0 h and 8 h time points, the infected cells were lysed with 500 μl of distilled H2O and a portion of which was plated on TBA plates. Colony forming units (CFU) were counted after 2–3 days. For mouse infections, all procedures were in strict accordance with federal and state government guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health and their use was approved for this entire study by the University of Miami institutional animal care and use committee (protocol number 15-081). Mice were infected intravenously with a total of 2000 CFU (1000 CFU each of the parental Y. pestis KIM5 and the isogenic ΔyopB or ΔylrABC strains). Mice were humanely sacrificed at 48 h post infection, spleens were removed and homogenized in sterile water containing 0.05% triton X-100 by grinding through a fine wire mesh. The resulting homogenates were diluted and plated on media containing either chloramphenicol (CM) to select for the CM-resistant parental strain, as well as antibiotic-free media that allowed growth of both the parental strain and the CM-sensitive mutant strains. Two to three days later colonies were enumerated and the competition index (CI) for the parental/ ΔyopB and parental/ΔylrABC co-infected animals was computed by dividing the CFU of the mutant by the CFU of the parental strain.
Construction of YlrA, YlrB, and YlrC Expression Plasmids
DNA fragments used encoding YlrA, YlrB, and YlrC were PCR amplified from chromosomal DNA of Y. pestis KIM5. The resultant DNA fragments were digested with HindIII and BglII and inserted into HindIII- and BglII-digested pFLAG-CTC vector (Sigma-Aldrich). These vectors express full-length C-terminal FLAG-tagged YlrA-FLAG, YlrB-FLAG, and YlrC-FLAG. In addition, DNA sequences predicted to encode the YlrA, YlrB, and YlrC N-terminal T3S signal (SS) (amino acid residues 2 to 10) were deleted from each expression vector using whole plasmid PCR (Imai et al., 1991), generating plasmids pYlrA-FLAG-ΔSS, pYlrB-FLAG-ΔSS, and pYlrC-FLAG-ΔSS. Oligonucleotide pairs used were YlrASS-F and YlrASS-R, YlrBSS-F and YlrBSS-R, and YlrCSS-F and YlrCSS-R The resultant Ylr expression plasmids were transformed into Y. pestis KIM8Δ4 (Bartra et al., 2006).
Construction of Vectors for β-Lactamase Translocation Studies
Expression plasmids encoding full length YlrA, YlrB, and YlrC carrying a C-terminal β-lactamase gene were constructed by the PCR-ligation-PCR technique (Ali and Steinkasserer, 1995). Individual Y. pestis KIM genes and upstream sequences that include each gene's ribosomal binding site were amplified by PCR from vectors encoding full-length FLAG-YlrA, FLAG-YlrB, and FLAG-YlrC, respectively, using oligonucleotides primer pairs YlrA-KpnI-F and YlrA-FL-R, YlrB-KpnI-F and YlrB-FL-R, and YlrC-KpnI-F and YlrC-FL-R. The DNA fragments encoding the Bla gene were amplified from plasmid pBSKII- using primers Bla-25-F and Bla-STOP-HindIII-R. Obtained DNA fragments were gel purified, kinased and ligated. The reaction was used for a second PCR using primers YlrA-KpnI-F and Bla-STOP-HindIII-R, YlrB-KpnI-F and Bla-STOP-HindIII-R, and YlrC-KpnI-F and Bla-STOP-HindIII-R. The resulting DNA fragments were ethanol precipitated, digested with KpnI and HindIII, and inserted into KpnI and HindIII-digested pBad18-Cmr. The resultant plasmids were transformed into a Y. pestis strain lacking YopE, YopJ, YopM, YopT, YopH, and YpkA.
Yeast Studies
DNA fragments encoding YlrA, YlrB and YlrC were amplified by PCR from pFLAG-YlrA, pFLAG-YlrB, and pFLAG-YlrC, respectively, using oligonucleotides primer pairs YlrA-XhoI and YlrA-BamHI, YlrB-XhoI and YlrB-BamHI, and YlrC-XhoI and YlrC-BamHI listed in Table 2. The resultant DNA fragments were digested with BamHI and XhoI and inserted into BamHI- and XhoI-digested pREP3X. The pREP-3X vector contains the inducible nmt1 promoter (Maundrell, 1993) and the presence of thiamine (15 μM) in the medium represses the promoter. If thiamine is not present in the media, the promoter becomes activated resulting in the transcription of the ylr gene cloned downstream. Plasmids generated were pREP-3X-YlrA, pREP-3X-YlrB, and pREP-3X-YlrC. In addition, site-specific mutation of three catalytic residues of the putative NEL domain of YlrA was done using the whole plasmid PCR method (Imai et al., 1991) generating plasmids pREP-3X-YlrA-C386S, pREP-3X-YlrA-R389A, and pREP-3X-YlrA-S449F. Schizosaccharomyces pombe h− ade6-704 leu1-32 ura4-D18 was grown in Pombe Glutamate medium (PMG) at 32°C. The plasmid vector pREP3x (nmt13x Thiamine repressible promoter) was used to express the Ylr genes in fission yeast. The plasmid vectors were transformed using the lithium acetate protocol (Morita and Takegawa, 2004). The yeast strains were grown in PMG supplemented with the appropriate amino acids as well as thiamine to maintain selection for 2 days in logarithmic growth to an optical density (OD) of 0.1–0.4 at 595 nm. After growth, the cells were centrifuged at 4,000 × g, washed 3 times with distilled sterile water, and diluted for an overnight growth in media lacking thiamine. The following day, the OD of the cultures at 595 nm was measured and the cultures were diluted to an OD of 0.1. Three 10-fold serial dilutions of the cultures were made, and 5 μL of each dilution was spotted onto an agar plate with and without thiamine and grown at 32°C for 3 days.
Secretion Assay
For standard secretion assays, Y. pestis strains were grown in thoroughly modified Higuchi's (TMH) medium in the presence or absence of 2.5 mM CaCl2 for 1 h at 27°C and then shifted to 37°C for the next 5 h as previously described (Jackson et al., 1998). FLAG-tagged Ylr proteins (pFLAG-CTC vector) were induced with 0.05 mM IPTG (isopropyl-β-D-thiogalactopyranoside) at the temperature shift.
SDS-PAGE and Immunoblotting
Cultures of bacteria were harvested by centrifugation at 14,000 × g for 10 min at room temperature. Pellets of whole-cell bacteria and trichloroacetic acid (TCA)-precipitated supernatant proteins were resuspended according to the harvest OD620 and subjected to SDS-PAGE and immunoblotting as previously described (Jackson et al., 1998). LcrV was detected with rabbit polyclonal antisera (1:20,000) raised against the full-length LcrV proteins. FLAG-tagged proteins were detected with anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) (1:1,000).
β-Lactamase Translocation Assay
HeLa cells were seeded to a six-well plate to achieve a 40–50% confluence 1 day prior to infection. Y. pestis strains were grown at 27°C for 2 h in HIB with antibiotics as appropriate and then shifted to 37°C for 1 h to induce expression of T3SS proteins. At temperature shift, 0.2% L-arabinose was added to the cultures. Bacteria were added at MOI = 50 to HeLa cells. The infected cells were incubated at 37°C with 5% CO2. Two negative controls (without Y. pestis cells and Y. pestis with pBAD18-pYscF1-Bla) and a positive control were used to determine the background blue and green fluorescence. After infection for 3 h, the growth medium from the cells was removed and the cells were washed with 500 μL of 1x of GIBCO Hank's Balanced Salt Solution (HBSS). 6X CCF-AM (Invitrogen) was added to each sample to obtain a final concentration of 1X per manufacturer's instructions. The plate was covered and incubated in room temperature for 1 h. Cells were then visualized by confocal-fluorescent microscopy (Leica TCS SP5 Inverted Confocal Microscope).
Results
Pro-survival Activity of Y. pestis Leucine-Rich Proteins During Cellular Infection
Previously we described a transposon site hybridization (TraSH)-based screen to identify Y. pestis loci that are required during infection of cultured mouse macrophage-like RAW 264.7 cells (Bartra et al., 2012). From a pool of approximately 90,000 unique variants, 44 Y. pestis ORFs were identified that appeared to be required for pathogen survival during its interaction with the macrophage. A number of these ORFs (17) are encoded on the extrachromosomal plasmids pCD1 and pPCP1 and express components of the T3SS and Pla protease that both play critical roles in Y. pestis virulence (Viboud and Bliska, 2005). One such pCD1-encoded locus among this set of 17 identified ORFs, YopM, possesses Leucine-Rich Repeats (LRRs), and is associated with a variety of infection phenotypes (see Introduction). Among the 27 chromosomally-encoded pro-survival ORFs, there were three, y3397, y3399, and y3400, that were previously predicted to possess LRRs (Hu et al., 2016) and here will be designated as Yersinia leucine-rich repeat A (ylrA), B (ylrB), and C (ylrC), respectively (Figure 1). In addition to LRR-encoding sequences, these three ORFs also possess predicted secretion signals (SS) for the T3SS at their N-termini and YlrA and YlrC are additionally predicted to encode novel ubiquitin E3 ligase (NEL) domains (Wang et al., 2013; Marchler-Bauer et al., 2017).
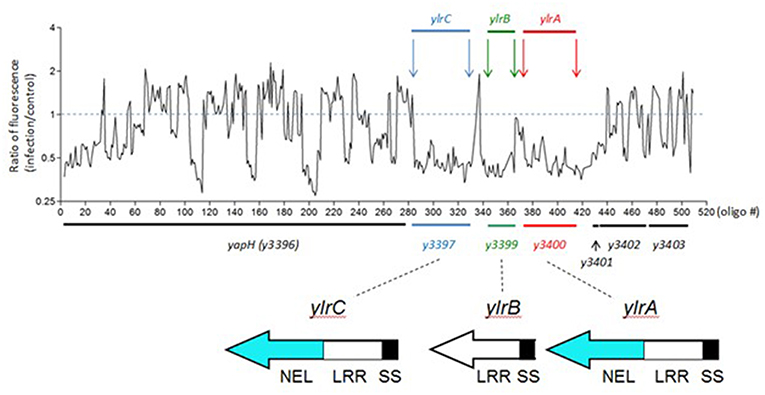
Figure 1. A TraSH screen indicates that the ylrABC locus promotes the survival of Y. pestis during infection. A pool of 90,000 unique Y. pestis transposon variants were used to infect cultured RAW 264.7 macrophages. Following infection bacterial DNA was isolated and the relative abundance of specific regions of the Y. pestis genome was determined. Shown in the abundance at the ylr locus indicating that Y. pestis variants possessing transposon insertion at this locus were under-represented at the termination of the infection period. Protein domains are labeled as Novel E3 Ubiquitin Ligase (NEL), Leucine-Rich Repeat (LRR), and T3SS Secretion and Translocation Signal (SS) domains.
Defective Infection Phenotypes of Engineered Δylr Strains
The aforementioned TraSH screen involves infecting macrophages with a mixture of Y. pestis variants followed by a quantification of under-represented genetic loci. To test whether the Ylr-encoding loci play a pro-survival function for Y. pestis during a single-strain infection, individual ΔylrA, ΔylrB, and ΔylrC mutant strains were constructed and tested in a colony-forming unit (CFU)-based infection assay. Similar to what has been shown previously, the fold-increases of the parental Y. pestis KIM5 strain and the T3SS mutant stain (ΔyopB) during a 8 h infection, were 170 and 3, respectively (Figure 2A; Rosenzweig et al., 2005). The infectivity of the single gene deletion ΔylrA, ΔylrB, and ΔylrC mutant strains, as well as the triple gene deletion ΔylrABC mutant strain were intermediate between the parental and T3SS mutant strains (Figure 2A). This intermediate in vitro infection phenotype is similar to that observed in Yersinia mutants individually deleted for the YopE and YopH T3SS effectors (Bartra et al., 2001). These data validated the findings of the TraSH screen indicating that the Ylr-encoding genes promote the survival of Y. pestis during their interaction with macrophages.
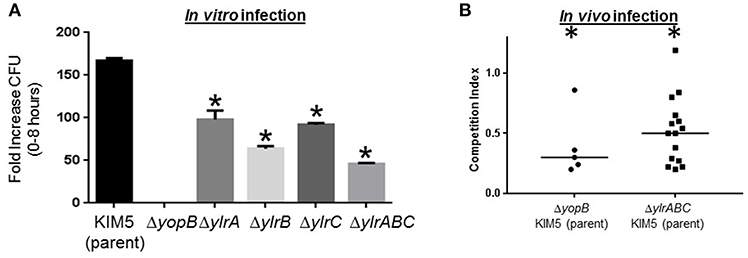
Figure 2. Infection defects of engineered ylr mutants. (A) Cultured RAW264.7 macrophages were infected with Y. pestis KIM5, ΔyopB, ΔylrA, ΔylrB, ΔylrC, or ΔylrABC strains at an MOI of 1. After a 30 min attachment period, unattached bacteria were removed and cell-associated bacteria (cfu) were determined by plating at the 0 and 8 h time points. Three independent wells per strain were analyzed and the average fold increase in cfu recovered for each strain over the 8 h infection period is shown (*P < 0.05 by Student t-test of a single representative experiment performed multiple times). (B) Mice were infected intravenously with an equal mixture of wild-type Y. pestis KIM5 and either the ΔyopB or ΔylrABC strains and 2 days later mice were humanly euthanized and the relative abundance of each strain in the spleen was determined. Each data point represents an individual mouse and the medium competition index is indicated. Statistically significant reductions in splenic colonization by the mutant strain is labeled by an asterisk (P < 0.05 by Student t-test of multiple cohorts of mice in separate infections).
The Y. pestis ΔylrABC strain was tested in a mouse-based infection model to determine whether the Ylr-encoding loci promote the pathogen survival in vivo. A competition infection assay was used to compare the proliferation of the parental KIM5 strain to that of the mutant strain within individual mice. Accordingly, in the control experiment mice were infected with an equal mixture of the parental KIM5 and the isogenic T3SS mutant ΔyopB strains and after 2 days the mice were humanely sacrificed and the presence of each strain in the spleen was determined by CFU assay. In contrast to the equal ratio of the parental and ΔyopB stains in the “input” (i.e., the dosed inoculum), the parental strain was greatly over-represented in the “output” (i.e., the splenic homogenates derived from mice infected for 2 days) (Figure 2B). Mice similarly infected with an equal mixture of the parental KIM5 and the isogenic ΔylrABC strains also yielded a statistically significant over-representation of the parental strain following 2 days of infection (Figure 2B). There was no detectable differences in growth rates between the parental KIM5 and the isogenic ΔylrABC strains (Supplementary Figure 1). Collectively these data indicate that the ylr locus promotes Y. pestis infection both in cells and in mice.
YlrA, Ylrb, and Ylrc Are Secreted by the Y. pestis Ysc Type III Secretion System
Sequence analysis of the ylrA, ylrB, and ylrC genes indicated that they each potentially encode at their amino-terminus a recognition sequence for the type III secretion system (T3SS) (Hu et al., 2016). To determine if the YlrA, YlrB, and YlrC proteins are secreted by the T3SS, expression vectors containing full-length C-terminal FLAG-tagged proteins were constructed. In addition, DNA sequences predicted to encode YlrA, YlrB, and YlrC N-terminal T3SS signal residues 2 to 10 were deleted from each of the expression vectors. Generated plasmids were placed in Y. pestis KIM8Δ4, a strain containing a modified pCD1 virulence plasmid that contains all of the genes necessary to assemble a functional T3SS but lacks all other effector proteins (Bartra et al., 2006). The strains were tested for expression and secretion in the presence and absence of 2.5 mM of calcium and bacterial pellet and supernatant were analyzed. FLAG-tagged YlrA, YlrB, and YlrC were expressed in the presence and absence of calcium, but were secreted only in the absence of calcium (Figure 3; red boxes; and Data Sheet 1). There was no secretion of the YlrA, YlrB, and YlrC proteins when the putative T3SS signal of each protein was deleted. Finally, there was no secretion of YlrA-FLAG, YlrB-FLAG, or YlrC-FLAG detected by Y. pestis strain KIM8 (pCD1-) or Y. pestis ΔyscF, both of which are unable to assemble a functional Ysc injectisome (Supplementary Figures 2–4). Together, these results demonstrate that YlrA, YlrB and YlrC are T3S substrates and that they are recognized and secreted by the Y. pestis T3SS.
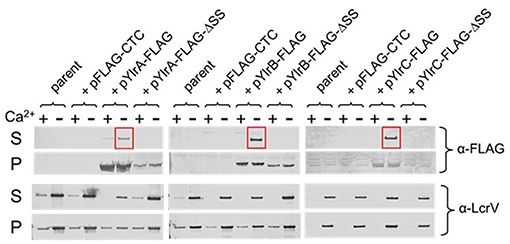
Figure 3. YlrA, YlrB, and YlrC are secreted by the Y. pestis Ysc type III secretion system. Y. pestis KIM8Δ4 (parent) carrying either plasmid pYlrA-FLAG, pYlrB-FLAG, or pYlrC-FLAG as well as plasmids deleted for sequences encoding the predicted Ylr secretion signals (ΔSS) were grown for 5 h in the presence (+) and absence (–) of 2.5 mM Ca2+. Cell pellet (P) and culture supernatant (S) fractions were evaluated by SDS-PAGE and immunoblot analysis with anti-FLAG and anti-LcrV (control) antibodies.
YlrB Is Translocated Into Mammalian Cells
A β-lactamase reporter system was used to determine whether Ylr proteins are translocated into cultured cells during infection (Marketon et al., 2005). Upon diffusion into cultured cells, CCF2-AM is cleaved into the membrane-impermeable CCF2 by endogenous cytoplasmic esterases, emitting a green fluorescence. When β-lactamase fusion proteins are injected into the cells, CCF2 can be cleaved further, changing the fluorescent emission to blue (Charpentier and Oswald, 2004). Cells that were either uninfected or infected with a Y. pestis strain expressing YscF1-Bla, which is expressed but not secreted by the T3SS, retained their initial green fluorescence (Figure 4). In striking contrast, essentially all CCF2-labeled cells infected with a Y. pestis strain expressing YopM-Bla, which, as discussed in the Introduction, is a plasmid-encoded virulence factor, fluoresce blue indicating that YopM-Bla is readily translocated into the infected cell cytosol. Cells infected with Y. pestis strains expressing either YlrA-Bla or YlrC-Bla retained their green fluorescence indicating that these proteins were not detectably translocated as assayed by this method despite being readily expressed as assayed by western blotting (Figure 4; Supplementary Figure 5). In contrast, blue fluorescing cells were readily detected following infection with a Y. pestis strain expressing YlrB-Bla. These show that the YlrB is translocated by Y. pestis during infection.
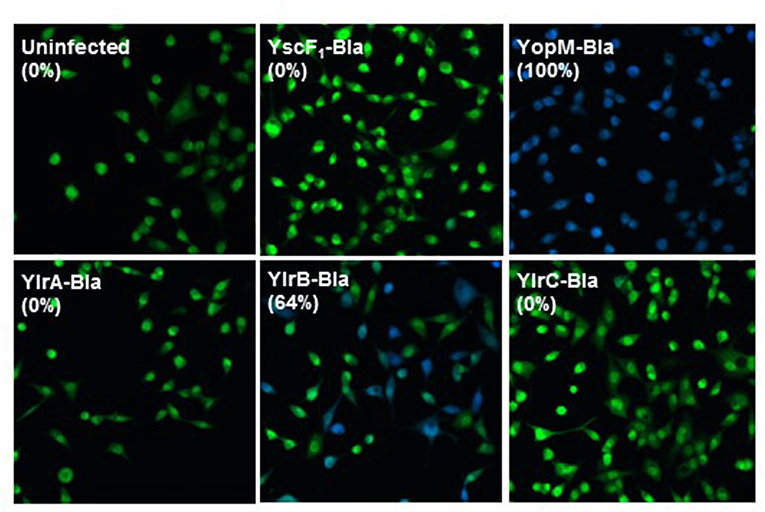
Figure 4. YlrB is translocated into eukaryotic cells. HeLa cells were infected with Yp589 (Y. pestis strain with deletion of YopE, YopJ, YopM, SycT, and YpKA) carrying YscF1-Bla (possessing the yscF 5′ UTR and the initiation codon), pMM85 YopM-Bla, YlrA-Bla, YlrB-Bla, or YlrC-Bla and were then incubated at 37°C for 3 h. CCF2-AM was added to HeLa cells and visualized by fluorescence microscopy. HeLa cells infected with the strain carrying the YlrB-Bla were positive for cytoplasmic β-lactamase activity (blue fluorescence), indicating that these cells had been injected with YlrB-Bla. Shown is a single representative experiment performed multiple times.
Intracellular Activity of Ylr
Previous studies of the Yersinia Yop virulence factors have shown a robust correlation between their growth inhibition in yeast and their activity in mammalian cells (Lesser and Miller, 2001; Wiley et al., 2009). Yeast provide a relatively simple genetic model system for the study of the eukaryotic genes and is based on the premise that yeast share many molecular, genetic, and biochemical features with mammalian cells (Zhao and Lieberman, 1995). Accordingly, the fission yeast Schizosaccharomyces pombe was transformed with either an empty vector control plasmid or inducible expression plasmids encoding either YlrA, YlrB, or YlrC and plated either on media in which the Ylr proteins were not expressed (“uninduced”) or on media in which Ylr expression occurs (“induced”). All S. pombe transformant strains grew equally well on uninduced media (Figure 5). However, on induced media, there was a marked differences between the relatively rapid growth of the empty vector and YlrB transformants (rows 1 and 6) and that of the relatively poorer growth of the YlrA and YlrC transformants (rows 2 and 7). Previous work has shown that IpaH effectors and SspH2 have a conserved catalytic cysteine residue that is absolutely required for ubiquitin ligase activity (Rohde et al., 2007; Quezada et al., 2009). To determine if this residue and other conserved residues of the NEL domain of YlrA will affect its growth-inhibiting activity in yeast, these residues were substituted generating plasmids expressing YlrA-C386S, YlrA-R389A, and YlrA-S449F. There was a clear reduction in growth inhibition for the S. pombe strain expressing the YlrA-C386S variant compared to the strain expressing wild-type YlrA (Figure 5, rows 2 and 3). In contrast, there was no detectable loss of growth inhibitory activity in the S. pombe strains expressing either the YlrA-R389A or YlrA-S449F (rows 4 and 5). These data suggest that the ubiquitin ligase activity of YlrA is required for its growth inhibitory activity in eukaryotic cells.
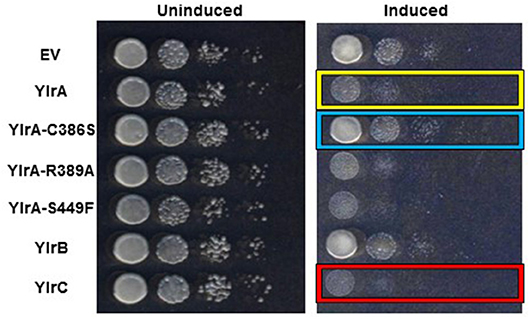
Figure 5. YlrA and YlrC, but not YlrB, disrupt yeast growth. Yeast transformed with either an empty vector (EV) control plasmid or the indicated Ylr proteins under the control of an inducible promoter. Comparable growth was observed when strains were plated on non-inducing media (left panel). However, when strains were plated on inducing media (right panel), the YlrA- and YlrC-expressing strains (yellow and red boxes, respectively) displayed a growth defect compared to the EV and YlrB-expressing strains. Also, mutation of the putative catalytic cysteine residue of the NEL domain of YlrA reversed the disruptive effect of YlrA on yeast growth (blue box); whereas mutation of the other residues did not have any effect. Shown is the result of a single experiment performed three times with similar outcomes.
Discussion
In this study, we demonstrate that the Y. pestis chromosomally-encoded proteins YlrA, YlrB, and YlrC are secreted and recognized by the T3SS and are required for the optimal survival of this pathogen both in the presence of cells as well as in mice. Although many chromosomally-encoded Y. pestis virulence factors have been described, to the best of our knowledge the Ylr are the first chromosomally-encoded virulence factors secreted by the pCD1-encoded T3SS.
YlrA, YlrB, and YlrC are leucine rich repeat proteins (LRR) with (YlrA and YlrC) or without (YlrB) an E3 ligase domain (NEL). The Ylr proteins belong to a diverse group of molecules distinguished by a consensus sequence consisting predominantly of leucines, hence the term leucine-rich repeat. Similar proteins are found in other Gram-negative bacteria that possess T3SSs, including Yersinia (YopM) (Kobe and Deisenhofer, 1994), Shigella (IpaH family proteins) (Ashida et al., 2007), Salmonella (SspH1, SspH2 and SlrP) (Miao et al., 1999; Tsolis et al., 1999; Bernal-Bayard et al., 2010) and other bacterial pathogens. In addition to possessing detectable membrane-penetrating activity, LRR domains offer adaptable structural frameworks that mediate protein-protein interactions, coupling enzymatic domains to substrate-binding domains, function to determine substrate specificity, and to suppress the enzymatic activity of the NEL domain prior to substrate binding (Kobe and Kajava, 2001; Quezada et al., 2009; Rüter et al., 2010). The NEL domains function as ubiquitin E3 ligases which mimic the activities of host E3 ubiquitin ligases and ubiquitinate specific target proteins (Rohde et al., 2007; Quezada et al., 2009). YlrA and YlrC contain an N-terminal LRR domain and a predicted C-terminal novel E3 ligase domain (NEL) (Hu et al., 2016). E3 ligase activity has been demonstrated for the Shigella effectors IpaH4.5 and IpaH9.8 and Salmonella effectors SspH1, SspH2, and SlrP (Rohde et al., 2007; Singer et al., 2008; Zhu et al., 2008; Bernal-Bayard and Ramos-Morales, 2009; Quezada et al., 2009). Bacterial E3 ubiquitin ligases transported into host cells mimic the activities of host E3 ubiquitin ligases and ubiquitinate specific target proteins. For example, IpaH 4.5 appears to dampen the innate immune system of human cells by inhibiting nuclear factor KB (NF-KB) signaling in response to an intracellular infection with Shigella (Ashida et al., 2010). The Salmonella virulence effector SlrP targets the mammalian thioredoxin-1 (Trx) leading to ubiquitination of Trx and causes a decrease in its redox activity (Bernal-Bayard and Ramos-Morales, 2009).
We have demonstrated that YlrB is translocated into the eukaryotic cell, whereas there was no evidence of translocation of the YlrA and YlrC using a β-lactamase reporter system. These results are difficult to interpret. Both full-length YlrA and YlrC are expressed and secreted, albeit at lower levels than YlrB (see Figure 3 and Supplementary Figure 5); therefore we expect them to be translocated as well. In addition, the fact that they contain an E3 ligase domain suggests that these proteins work inside the eukaryotic cell. Lack of detectable translocation in our experiments could be due to the large size of the proteins (~600 amino acids). Further experiments are needed using smaller-sized constructs expressing the translocation domains of both proteins. The most compelling data we present that YlrA and YlrC actually function within eukaryotic cells is the fact that they both disrupt the growth of the fission yeast S. pombe. Furthermore, to test if the conserved catalytic cysteine residue in YlrA is important to the activity of the protein, it was replaced with a serine residue. This resulted to the loss of the growth-inhibiting activity of YlrA. We speculate that the deleterious effect of YlrA and YlrC is due to the activity of their NEL domain. In a study using a yeast model, Rohde et al. (2007) have shown that S. flexneri IpaH9.8 inhibit yeast pheromone response signaling by ubiquitination and by promoting proteasome-dependent destruction of the mitogen-activated protein kinase kinase (MAPKK) Ste7. It was also demonstrated in the same study that the cysteine residue conserved in the NEL domain of all IpaH family members is required for the IpaH9.8 activities in yeast. The Salmonella effector SspH2 also has a conserved cysteine residue (Quezada et al., 2009). This catalytic cysteine residue and all other NEL active site residues are absolutely conserved between YlrA, YlrC and SspH2. We conclude that both YlrA and YlrC are strong candidates to join the list of bacterial virulence proteins that target the mammalian ubiquitination system.
To date, little is known about the leucine-reach repeat proteins in Yersinia with the exception of YopM. Miao et al. (1999) described three leucine-rich repeat open reading frames (ORFs) in Y. pestis that share significant homology with SspH2. These ORFs seem to be ylrA, ylrB and ylrC, since the ORFs were described to be present in an operon-like structure in the chromosome. Later, Evdokimov et al. (2001) described that these three ORFs share similar structure with Salmonella effectors SlrP, SspH1, SspH2; Shigella effector IpaH; and Yersinia effector YopM. Chou et al. (2012) PCR amplified Y. pestis y3400 (ylrA), expressed and purified the previously uncharacterized protein in E. coli and demonstrated that the purified protein possessed constitutive, not autoinhibited, ubiquitin E3 ligase activity. Finally, Soundararajan et al. (2011) were the first to identify and analyze the structure-function relation of a protein which appears to be the protein product of y3397 (YlrC) (McPhee and Bliska, 2011). More systematic studies are needed to characterize the expression, secretion, translocation, molecular functions and host target of these novel effector proteins. Research on these ylr gene products will lead to a better understanding of Y. pestis pathogenesis and possibly to the identification of the proteins as novel vaccine candidates or potential targets for therapeutics.
In conclusion, the ylrA, ylrB, and ylrC gene products represent the first chromosome-encoded T3S effector proteins of Y. pestis and are required for the optimal survival of this pathogen in the presence of macrophages. The gene products share significant similarity with T3S effector proteins from other bacterial pathogens and hence they are strong candidates to join the list of bacterial virulence proteins.
Author Contributions
SS, CL, XG, WB, ZL, GP, and KS planned and performed the majority of the experiments. HB and RH provided technical assistance. LQ analyzed the results of the TraSH screen. SS, CL, and KS wrote the manuscript.
Funding
This study was funded in part by NIH AI119450 (GP/KS) and U.S. Army Research, Development and Engineering Command (RDECOM), Contract W911SR-07-C-0084 (ZL).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00023/full#supplementary-material
References
Ali, S. A., and Steinkasserer, A. (1995). PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. BioTechniques 18, 746–750.
Ashida, H., Kim, M., Schmidt-Supprian, M., Ma, A., Ogawa, M., and Sasakawa, C. (2010). A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NF-κB-mediated inflammatory response. Nat. Cell Biol. 12, 66-73. doi: 10.1038/ncb2006
Ashida, H., Toyotome, T., Nagai, T., and Sasakawa, C. (2007). Shigella chromosomal IpaH proteins are secreted via the type III secretion system and act as effectors. Mol. Microbiol. 63, 680–693. doi: 10.1111/j.1365-2958.2006.05547.x
Bartra, S., Cherepanov, P., Forsberg, A., and Schesser, K. (2001). The Yersinia YopE and YopH type III effector proteins enhance bacterial proliferation following contact with eukaryotic cells. BMC Microbiol. 1:22. doi: 10.1186/1471-2180-1-22
Bartra, S. S., Gong, X., Lorica, C. D., Jain, C., Nair, M. K., Schifferli, D., et al. (2012). The outer membrane protein A (OmpA) of yersinia pestis promotes intracellular survival and virulence in mice. Microb. Pathog. 52, 41–46. doi: 10.1016/j.micpath.2011.09.009
Bartra, S. S., Jackson, M. W., Ross, J. A., and Plano, G. V. (2006). Calcium-regulated type III secretion of yop proteins by an Escherichia coli hha mutant carrying a yersinia pestis pCD1 virulence plasmid. Infect. Immun. 74, 1381–1386. doi: 10.1128/IAI.74.2.1381-1386.2006
Bernal-Bayard, J., Cardenal-Munoz, E., and Ramos-Morales, F. (2010). The salmonella type III secretion effector, salmonella leucine-rich repeat protein (SlrP), targets the human chaperone ERdj3. J. Biol. Chem. 285, 16360–16368. doi: 10.1074/jbc.M110.100669
Bernal-Bayard, J., and Ramos-Morales, F. (2009). Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J. Biol. Chem. 284, 27587–27595. doi: 10.1074/jbc.M109.010363
Centers for Disease Control Prevention. (2013). Plague: Map and Statistics. Available online at: http://www.cdc.gov/plague/maps/index.html
Charpentier, X., and Oswald, E. (2004). Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186, 5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004
Chou, Y. C., Keszei, A. F., Rohde, J. R., Tyers, M., and Sicheri, F. (2012). Conserved structural mechanisms for autoinhibition in IpaH ubiquitin ligases. J. Biol. Chem. 287, 268–275. doi: 10.1074/jbc.M111.316265
Chung, L. K., Philip, N. H., Schmidt, V. A., Koller, A., Strowig, T., Flavell, R. A., et al. (2014). IQGAP1 is important for activation of caspase-1 in macrophages and is targeted by Yersinia pestis type III effector YopM. MBio 5:e01402-14. doi: 10.1128/mBio.01402-14
Cornelis, G. R. (2002a). The yersinia ysc-yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3, 742–752. doi: 10.1038/nrm932
Cornelis, G. R. (2002b). Yersinia type III secretion: send in the effectors. J. Cell Biol. 158, 401–408. doi: 10.1083/jcb.200205077
Cornelis, G. R., and Van Gijsegem, F. (2000). Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774. doi: 10.1146/annurev.micro.54.1.735
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U S A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dennis, D. T., and Chow, C. C. (2004). Plague. Pediatr. Infect. Dis. J. 23, 69–71. doi: 10.1097/01.inf.0000106918.18570.dd
Evdokimov, A. G., Anderson, D. E., Routzahn, K. M., and Waugh, D. S. (2001). Unusual molecular architecture of the yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J. Mol. Biol. 312, 807–821. doi: 10.1006/jmbi.2001.4973
Hentschke, M., Berneking, L., Belmar Campos, C., Buck, F., Ruckdeschel, K., and Aepfelbacher, M. (2010). Yersinia virulence factor YopM induces sustained RSK activation by interfering with dephosphorylation. PLoS ONE 5:e13165. doi: 10.1371/journal.pone.0013165
Hu, Y., Huang, H., Hui, X., Cheng, X., White, A. P., Zhao, Z., et al. (2016). Distribution and evolution of yersinia leucine-rich repeat proteins. Infect Immun. 84, 2243–2254. doi: 10.1128/IAI.00324-16
Imai, Y., Matsushima, Y., Sugimura, T., and Terada, M. (1991). A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res. 19:2785. doi: 10.1093/nar/19.10.2785
Inglesby, T. V., Dennis, D. T., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., et al. (2000). Plague as a biological weapon: medical and public health management. Working group on civilian biodefense. JAMA 283, 2281–2290. doi: 10.1001/jama.283.17.2281
Jackson, M. W., Day, J. B., and Plano, G. V. (1998). YscB of yersinia pestis functions as a specific chaperone for YopN. J. Bacteriol. 180, 4912–4921.
Kerschen, E. J., Cohen, D. A., Kaplan, A. M., and Straley, S. C. (2004). The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells. Infect. Immun. 72, 4589–4602. doi: 10.1128/IAI.72.8.4589-4602.2004
Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19, 415–421. doi: 10.1016/0968-0004(94)90090-6
Kobe, B., and Kajava, A. V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. doi: 10.1016/S0959-440X(01)00266-4
LaRock, C. N., and Cookson, B. T. (2012). The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe 12, 799–805. doi: 10.1016/j.chom.2012.10.020
Lesser, C. F., and Miller, S. I. (2001). Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20, 1840-1849. doi: 10.1093/emboj/20.8.1840
Leung, K. Y., and Straley, S. C. (1989). The yopM gene of Yersinia pestis encodes a released protein having homology with the human platelet surface protein GPIb alpha. J. Bacteriol. 171, 4623–4632. doi: 10.1128/jb.171.9.4623-4632.1989
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Marketon, M. M., DePaolo, R. W., DeBord, K. L., Jabri, B., and Schneewind, O. (2005). Plague bacteria target immune cells during infection. Science 309, 1739–1741. doi: 10.1126/science.1114580
Maundrell, K. (1993). Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123, 127–130. doi: 10.1016/0378-1119(93)90551-D
McDonald, C., Vacratsis, P. O., Bliska, J. B., and Dixon, J. E. (2003). The yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J. Biol. Chem. 278, 18514–18523. doi: 10.1074/jbc.M301226200
McPhee, J. B., and Bliska, J. B. (2011). Letter to the editor and response. Innate Immun. 17, 558–559. doi: 10.1177/1753425911426251
McPhee, J. B., Mena, P., and Bliska, J. B. (2010). Delineation of regions of the Yersinia YopM protein required for interaction with the RSK1 and PRK2 host kinases and their requirement for interleukin-10 production and virulence. Infect. Immun. 78, 3529–3539. doi: 10.1128/IAI.00269-10
Miao, E. A., Scherer, C. A., Tsolis, R. M., Kingsley, R. A., Adams, L. G., Bäumler, A. J., et al. (1999). Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34, 850–864. doi: 10.1046/j.1365-2958.1999.01651.x
Morita, T., and Takegawa, K. (2004). A simple and efficient procedure for transformation of schizosaccharomyces pombe. Yeast 21, 613–617. doi: 10.1002/yea.1104
Perry, R. D., and Fetherston, J. D. (1997). Yersinia pestis–etiologic agent of plague. Clin. Microbiol. Rev. 10, 35–66. doi: 10.1128/CMR.10.1.35
Quezada, C. M., Hicks, S. W., Galan, J. E., and Stebbins, C. E. (2009). A family of salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc. Natl. Acad. Sci. USA. 106, 4864–4869. doi: 10.1073/pnas.0811058106
Rohde, J. R., Breitkreutz, A., Chenal, A., Sansonetti, P. J., and Parsot, C. (2007). Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 1, 77–83. doi: 10.1016/j.chom.2007.02.002
Rosenzweig, J. A., Weltman, G., Plano, G. V., and Schesser, K. (2005). Modulation of yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280, 156–163. doi: 10.1074/jbc.M405662200
Rüter, C., Buss, C., Scharnert, J., Heusipp, G., and Schmidt, M. A. (2010). A newly identified bacterial cell-penetrating peptide that reduces the transcription of pro-inflammatory cytokines. J. Cell. Sci. 123(Pt 13):2190–2198. doi: 10.1242/jcs.063016
Singer, A. U., Rohde, J. R., Lam, R., Skarina, T., Kagan, O., Dileo, R., et al. (2008). Structure of the shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat. Struct. Mol. Biol. 15, 1293–1301. doi: 10.1038/nsmb.1511
Soundararajan, V., Patel, N., Subramanian, V., Sasisekharan, V., and Sasisekharan, R. (2011). The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immun. 17, 548–557. doi: 10.1177/1753425910377099
Tsolis, R. M., Townsend, S. M., Miao, E. A., Miller, S. I., Ficht, T. A., Adams, L. G., et al. (1999). Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67, 6385–6393.
Viboud, G. I., and Bliska, J. B. (2005). Yersinia outer proteins: role in modulation of host cell signaling responses and pathogenesis. Annu. Rev. Microbiol. 59, 69–89. doi: 10.1146/annurev.micro.59.030804.121320
Wang, Y., Sun, M., Bao, H., Zhang, Q., and Guo, D. (2013). Effective identification of bacterial type III secretion signals using joint element features. PLoS ONE 8:e59754 doi: 10.1371/journal.pone.0059754
Wiley, D. J., Shrestha, N., Yang, J., Atis, N., Dayton, K., and Schesser, K (2009). The activities of the Yersinia protein kinase A (YpkA) and outer protein J (YopJ) virulence factors converge on an eIF2α kinase. J. Biol. Chem. 284, 24744–24753. doi: 10.1074/jbc.M109.010140
Ye, Z., Kerschen, E. J., Cohen, D. A., Kaplan, A. M., van Rooijen, N., and Straley, S. C. (2009). Gr1+ cells control growth of YopM-negative yersinia pestis during systemic plague. Infect. Immun. 77, 3791–3806. doi: 10.1128/IAI.00284-09
Zhao, Y., and Lieberman, H. B. (1995). Schizosaccharomyces pombe: a model for molecular studies of eukaryotic genes. DNA Cell Biol. 14, 359–371. doi: 10.1089/dna.1995.14.359
Keywords: Yersinia, type III secretion, pathogenesis, leucine rich repeats, plague, mice
Citation: Schesser Bartra S, Lorica C, Qian L, Gong X, Bahnan W, Barreras H Jr, Hernandez R, Li Z, Plano GV and Schesser K (2019) Chromosomally-Encoded Yersinia pestis Type III Secretion Effector Proteins Promote Infection in Cells and in Mice. Front. Cell. Infect. Microbiol. 9:23. doi: 10.3389/fcimb.2019.00023
Received: 09 August 2018; Accepted: 22 January 2019;
Published: 22 February 2019.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Nikhil A. Thomas, Dalhousie University, CanadaAndreas Diepold, Max-Planck-Institut für terrestrische Mikrobiologie, Germany
Luís Jaime Mota, Universidade Nova de Lisboa, Portugal
Copyright © 2019 Schesser Bartra, Lorica, Qian, Gong, Bahnan, Barreras, Hernandez, Li, Plano and Schesser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kurt Schesser, a3NjaGVzc2VyQG1lZC5taWFtaS5lZHU=
 Sara Schesser Bartra
Sara Schesser Bartra Cherish Lorica1,2
Cherish Lorica1,2 Henry Barreras Jr.
Henry Barreras Jr. Gregory V. Plano
Gregory V. Plano Kurt Schesser
Kurt Schesser