- 1Department of Experimental Therapeutics, BC Cancer Research Centre, Vancouver, BC, Canada
- 2Vancouver Prostate Centre, Vancouver, BC, Canada
- 3Department of Urologic Sciences, The University of British Columbia, Vancouver, BC, Canada
- 4Department of Microbiology, School of Basic Medicine, Fourth Military Medical University, Xi'an, China
Many global infectious diseases are not well-controlled, underlining a critical need for new, more effective therapies. Pathogens and pathogen-infected host cells, like cancer cells, evade immune surveillance via immune evasion mechanisms. The present study indicates that pathogenic bacteria, endoparasites, and virus-infected host cells can have immune evasion mechanisms in common with cancers. These include entry into dormancy and metabolic reprogramming to aerobic glycolysis leading to excessive secretion of lactic acid and immobilization of local host immunity. The latter evasion tactic provides a therapeutic target for cancer, as shown by our recent finding that patient-derived cancer xenografts can be growth-arrested, without major host toxicity, by inhibiting their lactic acid secretion (as mediated by the MCT4 transporter)-with evidence of host immunity restoration. Accordingly, the multiplication of bacteria, endoparasites, and viruses that primarily depend on metabolic reprogramming to aerobic glycolysis for survival may be arrested using cancer treatment strategies that inhibit their lactic acid secretion. Immune evasion mechanisms shared by pathogens and cancer cells likely represent fundamental, evolutionarily-conserved mechanisms that may be particularly critical to their welfare. As such, their targeting may lead to novel therapies for infectious diseases.
Introduction
Infectious diseases constitute a major health problem worldwide and impose a substantial burden on societies in human suffering and economic loss. While diseases like smallpox have essentially been eliminated, many other pathogen-induced diseases are not well-controlled and new infectious diseases are continually emerging, thus underlining a critical need for novel therapeutic targets leading to more effective therapies (Fonkwo, 2008).
The human body is constantly under attack by foreign microorganisms such as pathogenic bacteria, parasites, and viruses, i.e., pathogens whose invasion of the body and subsequent multiplication can lead to a wide variety of global infectious diseases. In defense of the body, invading pathogens are identified by the innate immune system as “non-self” via cell surface pattern recognition receptors (PRRs) on innate immune cells which recognize pathogen-associated molecular patterns (PAMPs) on the foreign microbes; the latter are then eliminated by, for example, macrophage-mediated phagocytosis or secretion of cytotoxins by NK cells (Chaplin, 2010; Vaure and Liu, 2014).
Recognition of foreign microbes by adaptive immunity is based on the ability of T- and B-lymphocytes to distinguish self from non-self-antigens via cell surface antigen-specific receptors, i.e., T-cell receptors (TCRs) and B-cell receptors (BCRs). Identification of pathogen-infected host cells is achieved by T cells via recognition of antigen fragments presented by major histocompatibility complex (MHC) molecules. Elimination of the intruders is accomplished via cytotoxic responses by CD8+ T cells and CD45+ lymphocytes, recruitment of neutrophils, monocytes, and mature B lymphocytes (mediated by CD4+ helper T cells), as well as antibody production by plasma cells.
In addition to foreign microorganisms, host cancer cells can be recognized by the immune system as changes in their cell surface structure render them foreign (Chaplin, 2010).
Despite vigorous immune surveillance by healthy, immunocompetent hosts, pathogens often manage to survive in the body. They accomplish this by employing a large variety of immune evasion mechanisms based on, for example, immobilizing immune responses or avoiding recognition by host immune cells (Finlay and McFadden, 2006; Reddick and Alto, 2014). Such survival tactics can also be used by cancer cells (Chaplin, 2010; Vinay et al., 2015). In the last decade it has become evident that metabolic reprogramming to aerobic glycolysis (the Warburg effect) is a key mechanism that most cancer types use to evade host immune surveillance, as the resulting increases in lactic acid levels and acidity of the tumor micro-environment immobilize host immune cell activity (Fischer et al., 2007; Choi et al., 2013; Brand et al., 2016). As recently shown, this immune evasion tactic provides a target for cancer therapy (Choi et al., 2016, 2018).
In the present study, we have compared immune evasion mechanisms of pathogens (i.e., bacteria, endoparasites, and virus-infected host cells) and human cancer cells in search of mechanisms common to both groups. Such shared tactics may represent fundamental, evolutionarily-conserved mechanisms of immune evasion, which could in turn be useful as targets for novel therapies of both cancer and infectious diseases.
Immune Evasion Mechanisms Employed By Pathogens and Cancer Cells
A literature survey led to the following information.
Pathogenic Bacteria
A small percentage of bacterial species can cause globally significant diseases such as tuberculosis and pneumonia. A typical immune response to pathogenic bacteria consists of opsonisation of the microbes followed by phagocytosis by host macrophages and fusion of the phagosomes with lysosomes. Intra-lysosomal acid hydrolases, reactive oxygen species (ROS) and nitric oxide ultimately kill the pathogens (van Kessel et al., 2014).
Bacteria have developed a number of immune evasion mechanisms and methods to manipulate host machinery to promote their survival and proliferation. They can avoid recognition by TLRs by masking their surface antigens with a carbohydrate capsule (Finlay and McFadden, 2006) or through alterations of cell surface lipids (Cambier et al., 2014). Some bacterial species are able to subvert the phagocytic process by secreting proteinaceous effectors into the host cell via their “type three secretion systems” (T3SS) (Quitard et al., 2006) to target host actin remodeling required for phagocytosis (Finlay and McFadden, 2006) and to promote entry into host cells (Betts et al., 2009; da Cunha et al., 2014). Other species manage to use the host intracellular environment for proliferation, with Mycobacterium tuberculosis, Coxiella burnetii, and Listeria monocytogenes residing in macrophages, Salmonella enterica in the intestinal epithelium, Mycobacterium leprae in connective and nerve tissues, and Chlamydia trachomatis in ocular and urogenital tissues (Betts et al., 2009; da Cunha et al., 2014). The MAPK/NFκB pro-inflammatory pathway can also be targeted by bacterial proteases to inhibit an inflammatory response, and TLR signaling can be modulated to secrete the immunosuppressive cytokine IL-10 (Finlay and McFadden, 2006).
When phagocytized, bacteria employ various strategies to survive. Commonly, they escape the phagosomal environment prior to its lysosome-mediated acidification: Shigella and L. monocytogenes escape by remodeling host actin and secreting phospholipases to cleave the phagosomal membrane (Small et al., 1994; Baxt et al., 2013). Some bacteria, such as M. tuberculosis, prevent the acidification of the phagosome by disrupting phagosome-lysosome fusion using the PtpA tyrosine phosphatase (Bach et al., 2008). Others exploit the phagosome to create a suitable microenvironment in which to proliferate: Legionella pneumophila creates a vacuolar environment specifically lacking MHC class II molecules, thereby protecting itself from innate immunity (Clemens and Horwitz, 1992), whereas C. burnetii requires an acidic environment for growth and virulence (Maurin et al., 1992).
In response to attack by the immune system, bacteria can enter a hardy, non-replicating state, termed dormancy, for protection (Rittershaus et al., 2013). Dormancy comes as two different types: (i) cellular quiescence, a period of reduced cellular growth that maintains a basal metabolism as observed for persistent M. tuberculosis (Betts et al., 2002) and (ii) true dormancy, a metabolically-arrested spore state promoting survival under adverse conditions, as exhibited by the difficult-to-treat Clostridium genus (Rittershaus et al., 2013). Cellular quiescence is achieved through stress-triggered expression of dormancy-related genes, e.g., activation of signaling system-related genes such as the type II toxin-antitoxin (TA) module and tnaA, a gene responsible for the conversion of tryptophan to indole, an intracellular signaling molecule responsible for mediating persistence (Wood et al., 2013; Pu et al., 2017).
Bacteria can also evade innate immunity by reprogramming their energy metabolisms. Staphylococcus aureus, in its hypoxic phagosomal environment, upregulates glycolysis and lactic acid fermentation in order to maintain a redox balance in response to immune-induced nitric oxide stress (Vitko et al., 2015). When glucose is abundant, Escherichia coli reprograms glycolysis toward the production of acetate in its exponential growth (Dittrich et al., 2005). Intra-macrophagic Brucella abortus induces a shift toward aerobic glycolysis in its host—a mechanism shown to be required for its survival (Czyz et al., 2017). The lactic acid and derivative short-chain fatty acids (SCFAs) produced by the bacteria are secreted in order to maintain an alkaline intracellular pH (Konings et al., 1997). The lactic acid and SCFAs may also act as immune-modulatory signaling molecules, as their production by the gut microbiota generates an immune-tolerogenic environment by promoting Treg cell differentiation and inhibiting epithelial cell proliferation (Thangaraju et al., 2009; Iraporda et al., 2015; Asarat et al., 2016; Corrêa-Oliveira et al., 2016). Other evidence shows that volatile bacterial SCFAs can markedly inhibit T- and B-cell proliferation (Kurita-Ochiai et al., 1995).
Pathogenic Endoparasites
Endoparasites, i.e., parasites living inside the human body, can be grouped into two main categories: single-celled protozoa and multicellular worms, helminths. Both require successful evasion of the host immune system for survival and reproduction (Cox, 2002).
Protozoa employ several adaptive mechanisms to avoid contact with immune cells, often seeking residence in immune privileged sites. Plasmodium falciparum, which causes malaria in humans, produces SPECT-1 and -2 proteins allowing it to evade humoral immunity while traveling to the liver where it can mature in a relative immune privileged environment (Patarroyo et al., 2011). It then travels in the peripheral blood and infects erythrocytes, which lack MHC I receptors, and hence cannot be targeted by cytotoxic immune cells (Gomes et al., 2016). Trypanosoma Brucei, causing African sleeping disease, proliferates in blood and lymph and invades the central nervous system—another immune privileged site (Masocha et al., 2007). Migrating helminths also evade immune attack by remaining in the central nervous system and liver (Pockros and Capozza, 2005). Polymorphism also plays a role in immune evasion by protozoa. P. falciparum, for example, go through multiple stages during their lifetime and alter their surface antigens after every stage, whereas T. Brucei survives through subsurface protein remodeling which is involved in signaling transitions during developmental stages of dormancy and disease progression (Batram et al., 2014). These polymorphic modifications downgrade the ability of B cells to make highly specific antibodies (Zambrano-Villa et al., 2002).
Another mechanism via which endoparasites survive the host defense is through immunomodulation; some protozoa are able to induce host T cell anergy through suppression of co-stimulators and cytokines (Zambrano-Villa et al., 2002; Rodrigues et al., 2014). T. brucei uses its “vector host” to its advantage, in which the saliva of the Tsetse fly transmitted along with the parasite contains a peptide which suppresses human host release of cytokines TNF-α, IFN-γ, IL-6, and IL-10 (Bai et al., 2015; Stijlemans et al., 2016). Helminths are able to survive in humans for many years due to their ability to secrete immunomodulatory products (Hewitson et al., 2009).
Entry into a dormant state is also a survival strategy of some endoparasites, with stress-induced dormancy allowing their persistence under adverse conditions. In particular, induction of cell cycle arrest by ring-stage P. falciparum has been observed following use of traditional anti-malarial artemisinin-based combination treatments (Witkowski et al., 2010) and amino acid starvation (McLean and Jacobs-Lorena, 2017). The dormant subpopulations upregulate key survival genes (e.g., genes encoding heat shock proteins), downregulate cell cycle regulators and DNA biosynthesis proteins, and sustain their energy needs through fatty acid synthesis and pyruvate metabolism. Importantly, drug-selected parasites were able to re-enter growth and development upon stress removal (Witkowski et al., 2010), accentuating an ever-pressing issue of parasitic persistence in disease treatment.
The metabolic profiles of endoparasites can also be modulated in response to variations in their environment and developmental stage. In proliferative stages, protozoa and helminths can display a preferred dependence on anaerobic glycolysis and lactic acid production for ATP generation. The intra-erythrocytic Plasmodium parasite displays deregulated glycolytic activity coupled to impaired mitochondrial metabolism, hypothesized to provide a distinct growth advantage during its proliferative stage (Salcedo-Sora et al., 2014). Parasitic helminths can proliferate in high oxygen environments with or without aerobic respiration, indicating a preference for aerobic glycolysis as well (Tielens, 1994).
Pathogenic Viruses
Viruses can cause a wide variety of diseases such as influenza, HIV/AIDS and Ebola. They employ various strategies to evade and suppress the human host immune response. Primarily, viruses contain high sequence variability, which contributes to immune escape via interruption and alteration of antigen presentations from virus-infected cells (Balamurugan et al., 2017; Karlsson Hedestam et al., 2017). Influenza viruses, for example, evolve rapidly in a process called “antigenic drift,” gaining mutations in genes involved in antigen binding (Peacock et al., 2017; Wu and Wilson, 2017). Other mechanisms include T cell exhaustion leading to loss of cytotoxic function (Wieland et al., 2017) and reduced presentation of MHC class I and NKG2D molecules resulting in inadequate immune activation (Pymm et al., 2017; Schmiedel and Mandelboim, 2017).
Immune surveillance of viral pathogens is largely accomplished via intracellular sensors that detect viral RNA and DNA (Ma and Damania, 2016; Roy et al., 2016; Nerbøvik et al., 2017). Viruses can suppress the immune response by interfering with antiviral signaling pathways, particularly by modifying their nucleic acids, inhibiting proper expression of PRRs and their adaptors (Chan and Gack, 2016), and suppressing the production of antiviral cytokines (Zou et al., 2016). For example, the Seneca Valley virus can inhibit interferon responses by targeting host adaptors (Qian et al., 2017), and Dengue and West Nile viruses can block IFN-α/β receptors (Gack and Diamond, 2016). Furthermore, the role of gene-silencing microRNAs produced by human polyomaviruses has been found to downregulate expression of large T antigen, a target of antiviral immunity, thus mediating the immune escape and survival of the viruses (Martelli and Giannecchini, 2017).
Another immune evasion tactic of viruses consists of entering a dormant state within a host cell, with subsequent viral reactivation once cell immunity wanes. Cells infected with viruses that have become dormant upregulate expression of latency-associated genes, but not lytic viral genes, making immune detection of the viruses difficult (Phelan et al., 2017). Notably, HPV can, upon infection, either actively replicate or assume a state of dormancy (Hoppe-Seyler et al., 2017a,b). It has also been shown that migrating HIV-infected cells can reactivate following dormancy with differential response to drugs (Bohn-Wippert et al., 2017). Mechanisms underlying viral dormancy indicate epigenetic regulation in response to host-induced stressors; for example, binding of the innate immune sensor, IFI16, to the lytic gene promoter in KSHV-infected cells results in transcriptional repression of latent KSHV (Roy et al., 2016). Furthermore, post-translational histone modifications have also been associated with decreased antigen expression (Bloom et al., 2010; Arbuckle et al., 2017).
To optimize their environment for infection, viruses can alter many host cellular metabolic pathways; in particular, cells infected by actively replicating viruses exhibit cancer phenotypes such as aerobic glycolysis, upregulated glutaminolysis and fatty acid synthesis (Sanchez and Lagunoff, 2015). Viruses such as HBV, HIV, and Zika have been shown to dysregulate glycolysis by increasing GLUT1 expression, glucose influx and lactic acid production in their host cells (Masson et al., 2017). Moreover, an acidified microenvironment resulting from increased lactic acid secretion may contribute to virus-induced pathogenesis, as enveloped viruses' fusion to host cell membrane was observed to be more effective at a low pH (Desai et al., 2017). The fusion rate of the Ebola virus can also be increased by a brief exposure to an acidic environment (Markosyan et al., 2016).
Cancer Cells
Although cancer cells originate from the human host, they can be recognized by the immune system as non-self due to changes in their cell surface structures, and must evade immune recognition to survive. They can accomplish this by masking their cancer-specific surface neoantigens to prevent recognition by NK and effector T cells, and by suppressing anticancer immune responses (Mohme et al., 2017). MHC class I molecules on tumor cell surfaces are often weakly expressed or lacking, thus precluding cytotoxic T-cell responses (Garrido et al., 1993). In another strategy, cancer cells incorporate host platelet-derived vesicles exhibiting MHC I molecules on their plasma membrane to disguise themselves with regular host antigens (Placke et al., 2012). Moreover, immune recognition was found to be reduced via downregulation of NKG2D, a transmembrane NK cell receptor complex in pancreatic, gastric, colorectal, and breast cancers (Wang et al., 2016; Bi and Tian, 2017), while the inhibitory PD-1/PD-L1 signaling pathway was upregulated (Munn and Bronte, 2016). Suppression of the immune system can also be achieved through activation of indoleamine 2,3-dioxygenase (IDO), an inducible enzyme that catalyzes the rate-limiting step in tryptophan catabolism. Elevated levels of IDO are produced by a variety of malignancies (Munn and Mellor, 2016) and result in IDO-induced degradation of tryptophan to kynurenine. This metabolite promotes Treg differentiation (Chen et al., 2008), while tryptophan at diminished levels cannot activate mTORC1, an enzyme required for T-cell proliferation (Moon et al., 2015).
Cancer cells are also able to avoid immune surveillance by entering a latent state, termed tumor dormancy (Aguirre-Ghiso, 2007; Crea et al., 2015; Dong et al., 2017). Dormancy-capable cells, which express growth-inhibitory signals, self-impose a slow-cycling state and upregulate expression of pluripotency and self-renewal genes (Sosa et al., 2015; Malladi et al., 2016). Circulating tumor cells also adopt this dormant pro-survival phenotype (Mohme et al., 2017). Changes in the surrounding environment may awaken dormant cells into an immune-tolerogenic proliferative state.
A major immune evasion mechanism of cancers consists of reprogramming glucose metabolism to aerobic glycolysis (the Warburg effect) as distinct from the mitochondrial oxidative phosphorylation pathway used by normal cells (Warburg, 1956; Gatenby and Gillies, 2004; Choi et al., 2013; Agrawal and Rangarajan, 2015; Wong et al., 2015). The resulting excessive secretion of lactic acid by cancer cells, mediated by monocarboxylate transporters, MCT1 and MCT4, leads to abnormally high extracellular lactic acid levels and acidification of the tumor microenvironment; elevated MCT1 and MCT4 levels have been associated with poor cancer prognosis (Polanski et al., 2014; Marchiq and Pouysségur, 2016; Noble et al., 2017), with MCT4 being a main exporter of lactate from glycolytic tissues (Ullah et al., 2006). The highly elevated levels of extracellular lactic acid and decreased pH in the microenvironment act in combination to inhibit lactic acid secretion by T lymphocytes of both innate and adaptive immunity (Fischer et al., 2007; Calcinotto et al., 2012), thus reducing their glycolytic flux-dependent immune functions (Fischer et al., 2007; Calcinotto et al., 2012; Loftus and Finlay, 2016). The resultant intracellularly acidified immune cells undergo decreased metabolism and acidosis while the low-pH environment indirectly impairs cytokine production and secretion, both effects resulting in local immunosuppression (Fischer et al., 2007; Calcinotto et al., 2012; Chang et al., 2013; Tan et al., 2015; Paul et al., 2016; Bi and Tian, 2017). Aerobic glycolysis also has several proliferative advantages for cancers as the glycolytic intermediates can be used for biosynthesis of nucleotides and lipids essential for rapid cell proliferation (Lunt and Vander Heiden, 2011). The conversion of pyruvate to lactate is coupled to the reconversion of cofactor NADH to NAD+, which is essential for maintaining the glycolytic flux and intracellular redox status (Lunt and Vander Heiden, 2011). The acidic tumor microenvironment may also promote cancer growth by inducing epigenetic changes toward a proliferative stem cell-like profile in solid tumors (Som et al., 2016). Moreover, lactate itself can act as a signaling molecule responsible for immune modulation, mediating M2-polarization of tumor-associated macrophages, resulting in secretion of angiogenic factors and inducing tissue-remodeling inflammation (Shime et al., 2008; Colegio et al., 2014; Roszer, 2015).
Discussion
As highlighted by this study, pathogenic bacteria, endoparasites and virus-infected host cells employ, like cancer cells, a large variety of mechanisms to evade immune surveillance. These mechanisms are diverse and multifaceted and, even within the four classes, various species may have their own unique methods of evading immune surveillance. As such, it is beyond the scope of this perspective article to elaborate on them. Further information can be obtained from the following literature references for pathogenic bacteria (Finlay and McFadden, 2006; Flannagan et al., 2015; Thammavongsa et al., 2015; Fisher et al., 2017; Karkhah et al., 2019), endoparasites (Cooper and Eleftherianos, 2016; Gomes et al., 2016; Nakada-Tsukui and Nozaki, 2016; Stijlemans et al., 2016; Wahlgren et al., 2017; Martínez-López et al., 2018), viruses (Finlay and McFadden, 2006; Chan and Gack, 2016; Agrawal et al., 2017; Felix and Savvides, 2017; Hsu, 2018; Soto et al., 2018), and cancer cells (Bhatia and Kumar, 2014; Vinay et al., 2015; Goodman et al., 2017; Bates et al., 2018).
Of special interest to us are the immune evasion mechanisms that the various pathogens share with cancer cells, as they likely represent fundamental, evolutionarily-conserved mechanisms that may be particularly critical to their welfare. Thus, such mechanisms could be useful as targets for novel therapies of both cancer and infectious diseases. In searching for such shared tactics two were found to be of paramount importance: (i) Metabolic reprogramming to aerobic glycolysis leading to excessive lactic acid secretion and consequent local host immunity suppression, a well-defined mechanism used by most human cancers (Gatenby and Gillies, 2004; Choi et al., 2013; Agrawal and Rangarajan, 2015) (Figure 1A). This tactic can also be used by pathogenic bacteria (Konings et al., 1997; Thangaraju et al., 2009; Iraporda et al., 2015; Vitko et al., 2015; Asarat et al., 2016; Corrêa-Oliveira et al., 2016; Czyz et al., 2017), endoparasites (Tielens, 1994; Salcedo-Sora et al., 2014), and virus-infected cells (Sanchez and Lagunoff, 2015; Markosyan et al., 2016; Desai et al., 2017; Masson et al., 2017). (ii) Entry into dormancy of pathogens or pathogen-infected host cells (Figure 1B), a less defined mechanism to avoid recognition by the immune system (Aguirre-Ghiso, 2007; Crea et al., 2015; Dong et al., 2017). It can be used by pathogenic bacteria (Betts et al., 2002; Rittershaus et al., 2013; Wood et al., 2013; Pu et al., 2017), endoparasites (Witkowski et al., 2010; McLean and Jacobs-Lorena, 2017), and virus-infected cells (Bohn-Wippert et al., 2017; Hoppe-Seyler et al., 2017a,b; Phelan et al., 2017).
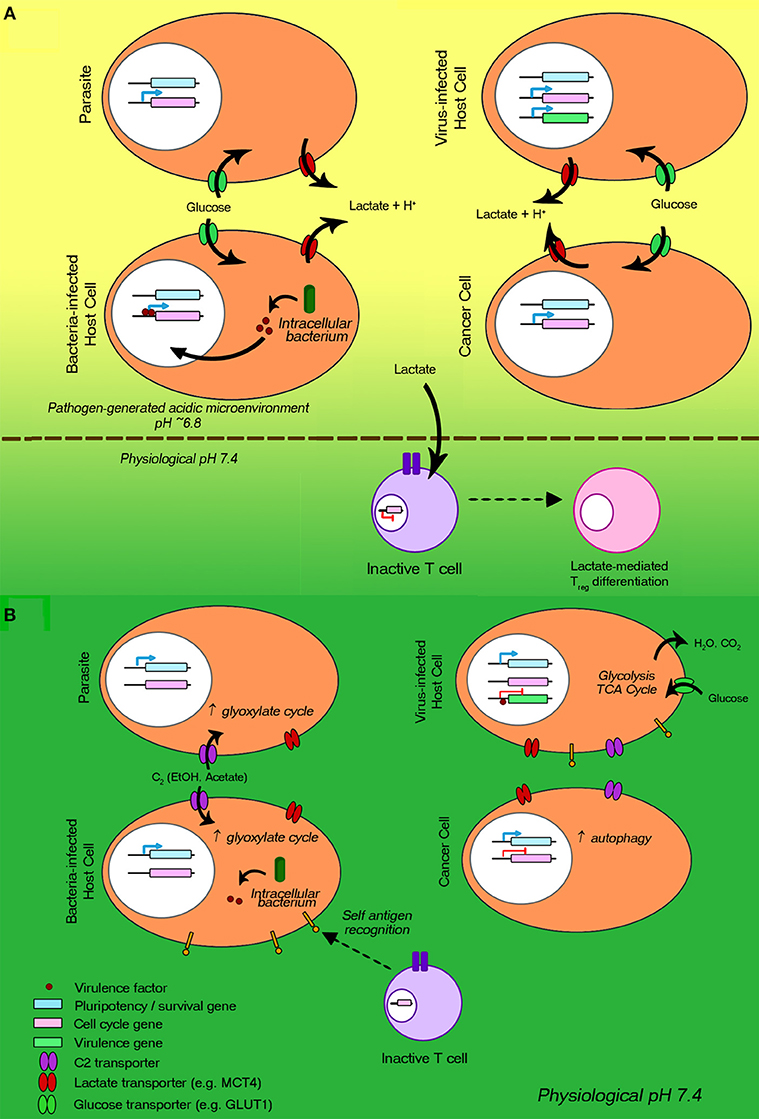
Figure 1. Metabolic reprogramming to aerobic glycolysis and entry into dormancy: two common immune evasion mechanisms employed by proliferating bacteria, endoparasites, viruses, and cancer cells. The host immune evasion/suppression processes are regulated via gene mutation and epigenetic reprogramming. (A) Processes involved in aerobic glycolysis are represented. Cell cycle genes are greatly upregulated. The reprogrammed metabolism leads to increases in (i) glucose uptake via glucose transporters (green) and (ii) lactic acid and short chain fatty acid secretion via monocarboxylate transporters (red), generating an acidic microenvironment. The lowered pH and increased lactate levels are immunosuppressive, e.g., inhibiting cytotoxic T cell activation. (B) Processes underlying entry into and maintenance of dormancy are illustrated. Pluripotency genes (blue) are expressed, while the expression of cell-cycle progression genes (pink) is arrested. Viral genes (green) of virus-infected cells are not expressed. Limited-nutrient metabolism states are induced (e.g., autophagy, glyoxylate cycle). T cells, recognizing self-antigens, are not activated.
Excessive secretion of lactic acid by cancer cells due to metabolic reprogramming to aerobic glycolysis provides a new potential target for cancer therapy. This has been demonstrated by our recent discovery that treatment of advanced prostate cancer cells with antisense oligonucleotides (ASOs) targeting MCT4-mediated lactic acid secretion led to a marked reduction in MCT4 expression and lactic acid secretion, increased intracellular lactic acid levels, and marked reductions in aerobic glycolysis, cell proliferation, cell migration, and tissue invasion (Choi et al., 2016, 2018). Treatment of PC-3 tumor-bearing nude mice with the anti-MCT4 ASOs markedly inhibited tumor growth without inducing major host toxicity (Choi et al., 2016; Choi, 2017). In addition, a limited study of the effect of MCT4-targeting on restoration of host immunity was carried out using clinically relevant, first-generation subrenal capsule xenograft models of patient-derived advanced prostate cancer. Such first-generation models lack systemic functional host immunity, but exhibit viable patient tumor-associated immune cells (Dong et al., 2017). Importantly, treatment of these models with anti-MCT4 ASOs led to an increase in the number of human CD45+ lymphocytes and CD8+ cytotoxic T cells, indicative of restoration of the host anticancer immunity (Choi, 2017). These findings raise the possibility that pathogens, which use metabolic reprogramming to aerobic glycolysis as a main immune evasion tactic, can also benefit from therapy based on inhibition of lactic acid secretion, particularly as the mechanism underlying transporter-mediated lactic acid secretion is relatively simple.
Although we here have outlined our perspective regarding potential therapeutic intervention resulting from inhibiting lactic acid secretion, we realize that possible approaches to reversing immune evasion are as vast and varied as the mechanisms themselves (Beatty and Gladney, 2015; Spranger and Gajewski, 2018). Alternative methods of reactivating the anti-cancer immune response are being actively investigated, the most successful of which involve neutralizing antibodies targeting immune checkpoint inhibitors such as PD-1 and CTLA-4 (Alsaab et al., 2017; Sharpe and Pauken, 2018; Tang et al., 2018). Furthermore, multiple components within a particular key pathway can also be considered therapeutic targets. Beyond MCT4, additional transporters as well as enzymes associated with aerobic glycolysis are also considered candidates for therapeutic inhibition of cancer (Tennant et al., 2010; Cairns et al., 2011; Zhao et al., 2013). Taken together, strategies of reactivating anti-cancer immunity have recently been shown to be successful across a number of cancer types (Goodman et al., 2017; Cascone et al., 2018; Seidel et al., 2018), offering evidence both scientifically and clinically that immunomodulation can translate into effective therapies. Similar considerations may apply to pathogen-induced diseases.
Finally, an additional aspect worth exploring as a potential therapeutic approach would be the use of combination therapies targeting multiple biological phenomena as surveyed here. In particular, targeting the dormant cell/organism population in conjunction with the actively proliferating population could offer enhanced efficacy. Given the importance of aerobic glycolysis to rapid cellular proliferation and the significance of cells entering dormancy to avoid immune detection, a combined effort to inhibit both lactic acid secretion (e.g., by MCT4 inhibition as mentioned above) and the unique metabolic properties of dormant cells could eliminate a greater proportion of cancers and pathogenic microorganisms. While mechanisms underlying cellular entry into dormancy remain to be more fully elucidated, preliminary investigations in our laboratory have uncovered changes in amino acid metabolism as a potentially unique metabolic profile in dormant cancer cells post-therapy (Nabavi et al., 2017). As such, targeting the metabolic rewiring that occurs within the dormant state could help eliminate even difficult-to-target residual cells.
Conclusion
This study indicates that pathogenic bacteria, endoparasites, virus-infected host cells, and human cancer cells can share immune evasion mechanisms, i.e., (i) metabolic reprogramming to aerobic glycolysis leading to excessive lactic acid secretion and host immunity suppression and (ii) entry into dormancy. Targeting fundamental, evolutionarily-conserved mechanisms of immune evasion could be particularly effective for treatments of both cancer and multiple infectious diseases.
Author Contributions
YW initiated the topic in July 2016. JW, SC, and PG organized the manuscript. RL and EX provided published evidence. JW, SC, JK, PG, and YW were involved in the further development of the ideas and the writing of the paper. PG finalized the write-up.
Funding
This study was supported in part by grants awarded to YW by the Canadian Institutes of Health Research (Grant No. 141635, 144159, 153081), the Urology Foundation and the Terry Fox Research Institute (Grant No. 1062).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agrawal, A., and Rangarajan, V. (2015). Appropriateness criteria of FDG PET/CT in oncology. Indian J. Radiol. Imag. 25, 88–101. doi: 10.4103/0971-3026.155823
Agrawal, P., Nawadkar, R., Ojha, H., Kumar, J., and Sahu, A. (2017). Complement evasion strategies of viruses: an overview. Front. Microbiol. 8:1117. doi: 10.3389/fmicb.2017.01117
Aguirre-Ghiso, J. A. (2007). Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer 7, 834–846. doi: 10.1038/nrc2256
Alsaab, H. O., Sau, S., Alzhrani, R., Tatiparti, K., Bhise, K., Kashaw, S. K., et al. (2017). PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front. Pharmacol. 8:561. doi: 10.3389/fphar.2017.00561
Arbuckle, J. H., Gardina, P. J., Gordon, D. N., Hickman, H. D., Yewdell, J. W., Pierson, T. C., et al. (2017). Inhibitors of the histone methyltransferases EZH2/1 induce a potent antiviral state and suppress infection by diverse viral pathogens. MBio 8:e01141–17. doi: 10.1128/mBio.01141-17
Asarat, M., Apostolopoulos, V., Vasiljevic, T., and Donkor, O. (2016). Short-chain fatty acids regulate cytokines and Th17/treg cells in human peripheral blood mononuclear cells in vitro. Immunol. Invest. 45, 205–222. doi: 10.3109/08820139.2015.1122613
Bach, H., Papavinasasundaram, K. G., Wong, D., Hmama, Z., and Av-Gay, Y. (2008). Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe 3, 316–322. doi: 10.1016/j.chom.2008.03.008
Bai, X., Yao, H., Du, C., Chen, Y., Lai, R., and Rong, M. (2015). An immunoregulatory peptide from tsetse fly salivary glands of glossina morsitans morsitans. Biochimie 118, 123–128. doi: 10.1016/j.biochi.2015.09.001
Balamurugan, A., Claiborne, D., Ng, H. L., and Yang, O. O. (2017). HIV-1 epitope variability is associated with t cell receptor repertoire instability and breadth. J. Virol. 91:e00771–17. doi: 10.1128/JVI.00771-17
Bates, J. P., Derakhshandeh, R., Jones, L., and Webb, T. J. (2018). Mechanisms of immune evasion in breast cancer. BMC Cancer 18:556. doi: 10.1186/s12885-018-4441-3
Batram, C., Jones, N. G., Janzen, C. J., Markert, S. M., and Engstler, M. (2014). Expression site attenuation mechanistically links antigenic variation and development in Trypanosoma brucei. Elife 3:e02324. doi: 10.7554/eLife.02324
Baxt, L. A., Garza-Mayers, A. C., and Goldberg, M. B. (2013). Bacterial subversion of host innate immune pathways. Science 340, 697–701. doi: 10.1126/science.1235771
Beatty, G. L., and Gladney, W. L. (2015). Immune escape mechanisms as a guide for cancer immunotherapy. Clin. Cancer Res. 21, 687–692. doi: 10.1158/1078-0432.CCR-14-1860
Betts, H. J., Wolf, K., and Fields, K. A. (2009). Effector protein modulation of host cells: examples in the Chlamydia spp. Arsenal. Curr. Opin. Microbiol. 12, 81–87. doi: 10.1016/j.mib.2008.11.009
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Bhatia, A., and Kumar, Y. (2014). Cellular and molecular mechanisms in cancer immune escape: a comprehensive review. Exp. Rev. Clan. Immunol. 10, 41–62. doi: 10.1586/1744666X.2014.865519
Bi, J., and Tian, Z. (2017). NK cell exhaustion. Front. Immunol. 8:760. doi: 10.3389/fimmu.2017.00760
Bloom, D. C., Giordani, N. V., and Kwiatkowski, D. L. (2010). Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 1799, 246–256. doi: 10.1016/j.bbagrm.2009.12.001
Bohn-Wippert, K., Tevonian, E. N., Megaridis, M. R., and Dar, R. D. (2017). Similarity in viral and host promoters couples viral reactivation with host cell migration. Nat. Commun. 8:15006. doi: 10.1038/ncomms15006
Brand, A., Singer, K., Koehl, G. E., Kolitzus, M., Schoenhammer, G., Thiel, A., et al. (2016). LDHA-associated lactic acid production blunts tumor immunosurveillance by T and NK Cells. Cell Metab. 24, 657–671. doi: 10.1016/j.cmet.2016.08.011
Cairns, R. A., Harris, I. S., and Mak, T. W. (2011). Regulation of cancer cell metabolism. Nat. Rev. Cancer 11, 85–95. doi: 10.1038/nrc2981
Calcinotto, A., Filipazzi, P., Grioni, M., Iero, M., De Milito, A., Ricupito, A., et al. (2012). Modulation of microenvironment acidity reverses anergy in human and murine tumor-infiltrating T lymphocytes. Cancer Res. 72, 2746–2756. doi: 10.1158/0008-5472.CAN-11-1272
Cambier, C. J., Falkow, S., and Ramakrishnan, L. (2014). Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159, 1497–1509. doi: 10.1016/j.cell.2014.11.024
Cascone, T., McKenzie, J. A., Mbofung, R. M., Punt, S., Wang, Z., Xu, C., et al. (2018). Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27:e974. doi: 10.1016/j.cmet.2018.02.024
Chan, Y. K., and Gack, M. U. (2016). Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 14, 360–373. doi: 10.1038/nrmicro.2016.45
Chang, C. H., Curtis, J. D., Maggi, L. B., Faubert, B., Villarino, A. V., O'Sullivan, D., et al. (2013). Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251. doi: 10.1016/j.cell.2013.05.016
Chaplin, D. D. (2010). Overview of the immune response. J Allergy Clan. Immunol. 125(Suppl. 2), S3–23. doi: 10.1016/j.jaci.2009.12.980
Chen, W., Liang, X., Peterson, A. J., Munn, D. H., and Blazar, B. R. (2008). The indoleamine 2,3-dioxygenase pathway is essential for human plasmacytoid dendritic cell-induced adaptive T regulatory cell generation. J. Immunol. 181, 5396–5404. doi: 10.4049/jimmunol.181.8.5396
Choi, S. Y., Collins, C. C., Gout, P. W., and Wang, Y. (2013). Cancer-generated lactic acid: a regulatory, immunosuppressive metabolite? J. Pathol. 230, 350–355. doi: 10.1002/path.4218
Choi, S. Y., Xue, H., Wu, R., Fazli, L., Lin, D., Collins, C. C., et al. (2016). The MCT4 gene: a novel, potential target for therapy of advanced prostate cancer. Clin. Cancer Res. 22, 2721–2733. doi: 10.1158/1078-0432.CCR-15-1624
Choi, S. Y. C. (2017). Targeting MCT4 for Treatment of Advanced Prostate Cancers: Inhibiting Cell Proliferation and Enhancing Anticancer Immunity Through Suppressing Lactic Acid Secretion and Elevated Glycolysis. Ph.D. dissertation, The University of British Columbia. doi: 10.14288/1.0361768
Choi, S. Y. C., Ettinger, S. L., Lin, D., Xue, H., Ci, X., Nabavi, N., et al. (2018). Targeting MCT4 to reduce lactic acid secretion and glycolysis for treatment of neuroendocrine prostate cancer. Cancer Med. 7, 3385–3392. doi: 10.1002/cam4.1587
Clemens, D. L., and Horwitz, M. A. (1992). Membrane sorting during phagocytosis: selective exclusion of major histocompatibility complex molecules but not complement receptor CR3 during conventional and coiling phagocytosis. J. Exp. Med. 175, 1317–1326. doi: 10.1084/jem.175.5.1317
Colegio, O. R., Chu, N. Q., Szabo, A. L., Chu, T., Rhebergen, A. M., Jairam, V., et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 513, 559–563. doi: 10.1038/nature13490
Cooper, D., and Eleftherianos, I. (2016). Parasitic nematode immunomodulatory strategies: recent advances and perspectives. Pathogens 5:E58. doi: 10.3390/pathogens5030058
Corrêa-Oliveira, R., Fachi, J. L., Vieira, A., Sato, F. T., and Vinolo, M. A. (2016). Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 5:e73. doi: 10.1038/cti.2016.17
Cox, F. E. (2002). History of human parasitology. Clin. Microbiol. Rev. 15, 595–612. doi: 10.1128/CMR.15.4.595-612.2002
Crea, F., Nur Saidy, N. R., Collins, C. C., and Wang, Y. (2015). The epigenetic/noncoding origin of tumor dormancy. Trends Mol. Med. 21, 206–211. doi: 10.1016/j.molmed.2015.02.005
Czyz, D. M., Willett, J. W., and Crosson, S. (2017). Brucella abortus induces a warburg shift in host metabolism that is linked to enhanced intracellular survival of the pathogen. J. Bacteriol. 199:e00227–17. doi: 10.1128/JB.00227-17
da Cunha, M., Milho, C., Almeida, F., Pais, S. V., Borges, V., Maurício, R., et al. (2014). Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol. 14:40. doi: 10.1186/1471-2180-14-40
Desai, T. M., Marin, M., Mason, C., and Melikyan, G. B. (2017). pH regulation in early endosomes and interferon-inducible transmembrane proteins control avian retrovirus fusion. J. Biol. Chem. 292, 7817–7827. doi: 10.1074/jbc.M117.783878
Dittrich, C. R., Bennett, G. N., and San, K. Y. (2005). Characterization of the acetate-producing pathways in Escherichia coli. Biotechnol. Prog. 21, 1062–1067. doi: 10.1021/bp050073s
Dong, X., Gout, P. W., Yi, L., Wang, Y., Xu, Y., and Yang, K. (2017). “First-Generation Tumor Xenografts: A Link Between Patient-Derived Xenograft Models and Clinical Disease,” in Patient-Derived Xenograft Models of Human Cancer, eds Y. Wang, D. Lin and P. W. Gout (Cham: Springer International Publishing), 155–176.
Felix, J., and Savvides, S. N. (2017). Mechanisms of immunomodulation by mammalian and viral decoy receptors: insights from structures. Nat Rev Immunol 17, 112–129. doi: 10.1038/nri.2016.134
Finlay, B. B., and McFadden, G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782. doi: 10.1016/j.cell.2006.01.034
Fischer, K., Hoffmann, P., Voelkl, S., Meidenbauer, N., Ammer, J., Edinger, M., et al. (2007). Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109, 3812–3819. doi: 10.1182/blood-2006-07-035972
Fisher, R. A., Gollan, B., and Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464. doi: 10.1038/nrmicro.2017.42
Flannagan, R. S., Heit, B., and Heinrichs, D. E. (2015). Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens 4, 826–868. doi: 10.3390/pathogens4040826
Fonkwo, P. N. (2008). Pricing infectious disease. The economic and health implications of infectious diseases. EMBO Rep. 9(Suppl. 1), S13–S17. doi: 10.1038/embor.2008.110
Gack, M. U., and Diamond, M. S. (2016). Innate immune escape by dengue and west nile viruses. Curr. Opin. Virol. 20, 119–128. doi: 10.1016/j.coviro.2016.09.013
Garrido, F., Cabrera, T., Concha, A., Glew, S., Ruiz-Cabello, F., and Stern, P. L. (1993). Natural history of HLA expression during tumour development. Immunol. Today 14, 491–499. doi: 10.1016/0167-5699(93)90264-L
Gatenby, R. A., and Gillies, R. J. (2004). Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899. doi: 10.1038/nrc1478
Gomes, P. S., Bhardwaj, J., Rivera-Correa, J., Freire-De-Lima, C. G., and Morrot, A. (2016). Immune escape strategies of malaria parasites. Front. Microbiol. 7:1617. doi: 10.3389/fmicb.2016.01617
Goodman, A., Patel, S. P., and Kurzrock, R. (2017). PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat. Rev. Clin. Oncol. 14, 203–220. doi: 10.1038/nrclinonc.2016.168
Hewitson, J. P., Grainger, J. R., and Maizels, R. M. (2009). Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol. Biochem. Parasitol. 167, 1–11. doi: 10.1016/j.molbiopara.2009.04.008
Hoppe-Seyler, K., Bossler, F., Lohrey, C., Bulkescher, J., Rösl, F., Jansen, L., et al. (2017a). Induction of dormancy in hypoxic human papillomavirus-positive cancer cells. Proc. Natl. Acad. Sci. U.S.A. 114, E990–E998. doi: 10.1073/pnas.1615758114
Hoppe-Seyler, K., Mändl, J., Adrian, S., Kuhn, B. J., and Hoppe-Seyler, F. (2017b). Virus/host cell crosstalk in hypoxic hpv-positive cancer cells. Viruses 9:174. doi: 10.3390/v9070174
Hsu, A. C. (2018). Influenza virus: a master tactician in innate immune evasion and novel therapeutic interventions. Front. Immunol. 9:743. doi: 10.3389/fimmu.2018.00743
Iraporda, C., Errea, A., Romanin, D. E., Cayet, D., Pereyra, E., Pignataro, O., et al. (2015). Lactate and short chain fatty acids produced by microbial fermentation downregulate proinflammatory responses in intestinal epithelial cells and myeloid cells. Immunobiology 220, 1161–1169. doi: 10.1016/j.imbio.2015.06.004
Karkhah, A., Ebrahimpour, S., Rostamtabar, M., Koppolu, V., Darvish, S., Vasigala, V. K. R., et al. (2019). Helicobacter pylori evasion strategies of the host innate and adaptive immune responses to survive and develop gastrointestinal diseases. Microbiol. Res. 218, 49–57. doi: 10.1016/j.micres.2018.09.011
Karlsson Hedestam, G. B., Guenaga, J., Corcoran, M., and Wyatt, R. T. (2017). Evolution of B cell analysis and Env trimer redesign. Immunol. Rev. 275, 183–202. doi: 10.1111/imr.12515
Konings, W. N., Lolkema, J. S., Bolhuis, H., van Veen, H. W., Poolman, B., and Driessen, A. J. (1997). The role of transport processes in survival of lactic acid bacteria. Energy transduction and multidrug resistance. Antonie Van Leeuwenhoek 71, 117–128. doi: 10.1023/A:1000143525601
Kurita-Ochiai, T., Fukushima, K., and Ochiai, K. (1995). Volatile fatty acids, metabolic by-products of periodontopathic bacteria, inhibit lymphocyte proliferation and cytokine production. J. Dent. Res. 74, 1367–1373. doi: 10.1177/00220345950740070801
Loftus, R. M., and Finlay, D. K. (2016). Immunometabolism: cellular metabolism turns immune regulator. J. Biol. Chem. 291, 1–10. doi: 10.1074/jbc.R115.693903
Lunt, S. Y., and Vander Heiden, M. G. (2011). Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464. doi: 10.1146/annurev-cellbio-092910-154237
Ma, Z., and Damania, B. (2016). The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe 19, 150–158. doi: 10.1016/j.chom.2016.01.010
Malladi, S., Macalinao, D. G., Jin, X., He, L., Basnet, H., Zou, Y., et al. (2016). Metastatic latency and immune evasion through autocrine inhibition of WNT. Cell 165, 45–60. doi: 10.1016/j.cell.2016.02.025
Marchiq, I., and Pouysségur, J. (2016). Hypoxia, cancer metabolism and the therapeutic benefit of targeting lactate/H(+) symporters. J. Mol. Med. 94, 155–171. doi: 10.1007/s00109-015-1307-x
Markosyan, R. M., Miao, C., Zheng, Y. M., Melikyan, G. B., Liu, S. L., and Cohen, F. S. (2016). Induction of cell-cell fusion by ebola virus glycoprotein: low pH is not a trigger. PLoS Pathog. 12:e1005373. doi: 10.1371/journal.ppat.1005373
Martelli, F., and Giannecchini, S. (2017). Polyomavirus microRNAs circulating in biological fluids during viral persistence. Rev. Med. Virol. 27:e1927. doi: 10.1002/rmv.1927
Martínez-López, M., Soto, M., Iborra, S., and Sancho, D. (2018). Hijacks myeloid cells for immune escape. Front. Microbiol. 9:883. doi: 10.3389/fmicb.2018.00883
Masocha, W., Rottenberg, M. E., and Kristensson, K. (2007). Migration of African trypanosomes across the blood-brain barrier. Physiol. Behav. 92, 110–114. doi: 10.1016/j.physbeh.2007.05.045
Masson, J. J., Billings, H. W., and Palmer, C. S. (2017). Metabolic reprogramming during hepatitis B disease progression offers novel diagnostic and therapeutic opportunities. Antivir. Chem. Chemother. 25, 53–57. doi: 10.1177/2040206617701372
Maurin, M., Benoliel, A. M., Bongrand, P., and Raoult, D. (1992). Phagolysosomes of Coxiella burnetii-infected cell lines maintain an acidic pH during persistent infection. Infect. Immun. 60, 5013–5016.
McLean, K. J., and Jacobs-Lorena, M. (2017). Confers survival upon amino acid starvation. MBio 8:e02317–16. doi: 10.1128/mBio.02317-16
Mohme, M., Riethdorf, S., and Pantel, K. (2017). Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 14, 155–167. doi: 10.1038/nrclinonc.2016.144
Moon, Y. W., Hajjar, J., Hwu, P., and Naing, A. (2015). Targeting the indoleamine 2,3-dioxygenase pathway in cancer. J. Immunother. Cancer 3:51. doi: 10.1186/s40425-015-0094-9
Munn, D. H., and Bronte, V. (2016). Immune suppressive mechanisms in the tumor microenvironment. Curr. Opin. Immunol. 39, 1–6. doi: 10.1016/j.coi.2015.10.009
Munn, D. H., and Mellor, A. L. (2016). IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 37, 193–207. doi: 10.1016/j.it.2016.01.002
Nabavi, N., Ettinger, S. L., Crea, F., Wang, Y., and Collins, C. C. (2017). “Biological and Clinical Evidence for Metabolic Dormancy in Solid Tumors Post Therapy,” in Tumor Dormancy and Recurrence, eds Y. Wang and F. Crea (Cham: Springer International Publishing), 17–29.
Nakada-Tsukui, K., and Nozaki, T. (2016). Immune response of amebiasis and immune evasion by entamoeba histolytica. Front. Immunol. 7:175. doi: 10.3389/fimmu.2016.00175
Nerbøvik, I. G., Solheim, M. A., Eggestøl, H., Rønneseth, A., Jakobsen, R. A., Wergeland, H. I., et al. (2017). Molecular cloning of MDA5, phylogenetic analysis of RIG-I-like receptors (RLRs) and differential gene expression of RLRs, interferons and proinflammatory cytokines after in vitro challenge with IPNV, ISAV and SAV in the salmonid cell line TO. J. Fish Dis. 40, 1529–1544. doi: 10.1111/jfd.12622
Noble, R. A., Bell, N., Blair, H., Sikka, A., Thomas, H., Phillips, N., et al. (2017). Inhibition of monocarboxyate transporter 1 by AZD3965 as a novel therapeutic approach for diffuse large B-cell lymphoma and Burkitt lymphoma. Haematologica 102, 1247–1257. doi: 10.3324/haematol.2016.163030
Patarroyo, M. E., Alba, M. P., and Curtidor, H. (2011). Biological and structural characteristics of the binding peptides from the sporozoite proteins essential for cell traversal (SPECT)-1 and -2. Peptides 32, 154–160. doi: 10.1016/j.peptides.2010.09.026
Paul, S., Kulkarni, N., and Shilpi Lal, G. (2016). Intratumoral natural killer cells show reduced effector and cytolytic properties and control the differentiation of effector Th1 cells. Oncoimmunology 5:e1235106. doi: 10.1080/2162402X.2016.1235106
Peacock, T. P., Benton, D. J., James, J., Sadeyen, J. R., Chang, P., Sealy, J. E., et al. (2017). Immune escape variants of H9N2 influenza viruses containing deletions at the hemagglutinin receptor binding site retain fitness. J. Virol. 91:e00218–17. doi: 10.1128/JVI.00218-17
Phelan, D., Barrozo, E. R., and Bloom, D. C. (2017). HSV1 latent transcription and non-coding RNA: a critical retrospective. J. Neuroimmunol. 308, 65–101. doi: 10.1016/j.jneuroim.2017.03.002
Placke, T., Örgel, M., Schaller, M., Jung, G., Rammensee, H. G., Kopp, H. G., et al. (2012). Platelet-derived MHC class I confers a pseudonormal phenotype to cancer cells that subverts the antitumor reactivity of natural killer immune cells. Cancer Res. 72, 440–448. doi: 10.1158/0008-5472.CAN-11-1872
Pockros, P. J., and Capozza, T. A. (2005). Helminthic infections of the liver. Curr. Infect. Dis. Rep. 7, 61–70. doi: 10.1007/s11908-005-0025-x
Polanski, R., Hodgkinson, C. L., Fusi, A., Nonaka, D., Priest, L., Kelly, P., et al. (2014). Activity of the monocarboxylate transporter 1 inhibitor AZD3965 in small cell lung cancer. Clin. Cancer Res. 20, 926–937. doi: 10.1158/1078-0432.CCR-13-2270
Pu, Y., Ke, Y., and Bai, F. (2017). Active efflux in dormant bacterial cells - New insights into antibiotic persistence. Drug. Resist. Updat. 30, 7–14. doi: 10.1016/j.drup.2016.11.002
Pymm, P., Illing, P. T., Ramarathinam, S. H., O'Connor, G. M., Hughes, V. A., Hitchen, C., et al. (2017). MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat. Struct. Mol. Biol. 24, 387–394. doi: 10.1038/nsmb.3381
Qian, S., Fan, W., Liu, T., Wu, M., Zhang, H., Cui, X., et al. (2017). Seneca valley virus suppresses host type i interferon production by targeting adaptor proteins MAVS, TRIF, and TANK for cleavage. J. Virol. 91:e00823–17. doi: 10.1128/JVI.00823-17
Quitard, S., Dean, P., Maresca, M., and Kenny, B. (2006). The enteropathogenic Escherichia coli EspF effector molecule inhibits PI-3 kinase-mediated uptake independently of mitochondrial targeting. Cell Microbiol. 8, 972–981. doi: 10.1111/j.1462-5822.2005.00680.x
Reddick, L. E., and Alto, N. M. (2014). Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol. Cell 54, 321–328. doi: 10.1016/j.molcel.2014.03.010
Rittershaus, E. S., Baek, S. H., and Sassetti, C. M. (2013). The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe 13, 643–651. doi: 10.1016/j.chom.2013.05.012
Rodrigues, V., Cordeiro-da-Silva, A., Laforge, M., Ouaissi, A., Akharid, K., Silvestre, R., et al. (2014). Impairment of T cell function in parasitic infections. PLoS Negl. Trop. Dis. 8:e2567. doi: 10.1371/journal.pntd.0002567
Roszer, T. (2015). Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015:816460. doi: 10.1155/2015/816460
Roy, A., Dutta, D., Iqbal, J., Pisano, G., Gjyshi, O., Ansari, M. A., et al. (2016). Nuclear innate immune DNA sensor IFI16 is degraded during lytic reactivation of kaposi's sarcoma-associated herpesvirus (KSHV): role of IFI16 in maintenance of KSHV latency. J. Virol. 90, 8822–8841. doi: 10.1128/JVI.01003-16
Salcedo-Sora, J. E., Caamano-Gutierrez, E., Ward, S. A., and Biagini, G. A. (2014). The proliferating cell hypothesis: a metabolic framework for Plasmodium growth and development. Trends Parasitol. 30, 170–175. doi: 10.1016/j.pt.2014.02.001
Sanchez, E. L., and Lagunoff, M. (2015). Viral activation of cellular metabolism. Virology 479, 609–618. doi: 10.1016/j.virol.2015.02.038
Schmiedel, D., and Mandelboim, O. (2017). Disarming cellular alarm systems-manipulation of stress-induced NKG2D ligands by human herpesviruses. Front. Immunol. 8:390. doi: 10.3389/fimmu.2017.00390
Seidel, J. A., Otsuka, A., and Kabashima, K. (2018). Anti-PD-1 and Anti-CTLA-4 therapies in cancer: mechanisms of action, efficacy, and Limitations. Front. Oncol. 8:86. doi: 10.3389/fonc.2018.00086
Sharpe, A. H., and Pauken, K. E. (2018). The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 18, 153–167. doi: 10.1038/nri.2017.108
Shime, H., Yabu, M., Akazawa, T., Kodama, K., Matsumoto, M., Seya, T., et al. (2008). Tumor-secreted lactic acid promotes IL-23/IL-17 proinflammatory pathway. J. Immunol. 180, 7175–7183. doi: 10.4049/jimmunol.180.11.7175
Small, P. L., Ramakrishnan, L., and Falkow, S. (1994). Remodeling schemes of intracellular pathogens. Science 263, 637–639. doi: 10.1126/science.8303269
Som, A., Bloch, S., Ippolito, J. E., and Achilefu, S. (2016). Acidic extracellular pH of tumors induces octamer-binding transcription factor 4 expression in murine fibroblasts in vitro and in vivo. Sci. Rep. 6:27803. doi: 10.1038/srep27803
Sosa, M. S., Parikh, F., Maia, A. G., Estrada, Y., Bosch, A., Bragado, P., et al. (2015). NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes. Nat. Commun. 6:6170. doi: 10.1038/ncomms7170
Soto, J. A., Gálvez, N. M. S., Benavente, F. M., Pizarro-Ortega, M. S., Lay, M. K., Riedel, C., et al. (2018). Human metapneumovirus: mechanisms and molecular targets used by the virus to avoid the immune system. Front. Immunol. 9:2466. doi: 10.3389/fimmu.2018.02466
Spranger, S., and Gajewski, T. F. (2018). Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 18, 139–147. doi: 10.1038/nrc.2017.117
Stijlemans, B., Caljon, G., Van Den Abbeele, J., Van Ginderachter, J. A., Magez, S., and De Trez, C. (2016). Immune evasion strategies of trypanosoma brucei within the mammalian host: progression to pathogenicity. Front. Immunol. 7:233. doi: 10.3389/fimmu.2016.00233
Tan, Z., Xie, N., Banerjee, S., Cui, H., Fu, M., Thannickal, V. J., et al. (2015). The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J. Biol. Chem. 290, 46–55. doi: 10.1074/jbc.M114.603589
Tang, J., Yu, J. X., Hubbard-Lucey, V. M., Neftelinov, S. T., Hodge, J. P., and Lin, Y. (2018). Trial watch: The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug. Discov. 17, 854–855. doi: 10.1038/nrd.2018.210
Tennant, D. A., Durán, R. V., and Gottlieb, E. (2010). Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer 10, 267–277. doi: 10.1038/nrc2817
Thammavongsa, V., Kim, H. K., Missiakas, D., and Schneewind, O. (2015). Staphylococcal manipulation of host immune responses. Nat. Rev. Microbiol. 13, 529–543. doi: 10.1038/nrmicro3521
Thangaraju, M., Cresci, G. A., Liu, K., Ananth, S., Gnanaprakasam, J. P., Browning, D. D., et al. (2009). GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 69, 2826–2832. doi: 10.1158/0008-5472.CAN-08-4466
Tielens, A. G. (1994). Energy generation in parasitic helminths. Parasitol. Today 10, 346–352. doi: 10.1016/0169-4758(94)90245-3
Ullah, M. S., Davies, A. J., and Halestrap, A. P. (2006). The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J. Biol. Chem. 281, 9030–9037. doi: 10.1074/jbc.M511397200
van Kessel, K. P., Bestebroer, J., and van Strijp, J. A. (2014). Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front. Immunol. 5:467. doi: 10.3389/fimmu.2014.00467
Vaure, C., and Liu, Y. (2014). A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 5:316. doi: 10.3389/fimmu.2014.00316
Vinay, D. S., Ryan, E. P., Pawelec, G., Talib, W. H., Stagg, J., Elkord, E., et al. (2015). Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin. Cancer Biol. 35, S185–S198. doi: 10.1016/j.semcancer.2015.03.004
Vitko, N. P., Spahich, N. A., and Richardson, A. R. (2015). Glycolytic dependency of high-level nitric oxide resistance and virulence in Staphylococcus aureus. MBio 6:e00045–15. doi: 10.1128/mBio.00045-15
Wahlgren, M., Goel, S., and Akhouri, R. R. (2017). Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat. Rev. Microbiol. 15, 479–491. doi: 10.1038/nrmicro.2017.47
Wang, S., Zhang, Y., Cong, W., Liu, J., Fan, H., Xu, Y., et al. (2016). Breast cancer stem-like cells can promote metastasis by activating platelets and down-regulating antitumor activity of natural killer cells. J. Tradit. Chin. Med. 36, 530–537. doi: 10.1016/S0254-6272(16)30071-1
Wieland, D., Hofmann, M., and Thimme, R. (2017). Overcoming CD8+ T-cell exhaustion in viral hepatitis: lessons from the mouse model and clinical perspectives. Dig. Dis. 35, 334–338. doi: 10.1159/000456584
Witkowski, B., Lelièvre, J., Barragán, M. J., Laurent, V., Su, X. Z., Berry, A., et al. (2010). Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 54, 1872–1877. doi: 10.1128/AAC.01636-09
Wong, N., Ojo, D., Yan, J., and Tang, D. (2015). PKM2 contributes to cancer metabolism. Cancer Lett. 356(2 Pt A), 184–191. doi: 10.1016/j.canlet.2014.01.031
Wood, T. K., Knabel, S. J., and Kwan, B. W. (2013). Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121. doi: 10.1128/AEM.02636-13
Wu, N. C., and Wilson, I. A. (2017). A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J. Mol. Biol. 429, 2694–2709. doi: 10.1016/j.jmb.2017.06.015
Zambrano-Villa, S., Rosales-Borjas, D., Carrero, J. C., and Ortiz-Ortiz, L. (2002). How protozoan parasites evade the immune response. Trends Parasitol. 18, 272–278. doi: 10.1016/S1471-4922(02)02289-4
Zhao, Y., Butler, E. B., and Tan, M. (2013). Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 4:e532. doi: 10.1038/cddis.2013.60
Keywords: immune evasion, pathogen, bacteria, endoparasite, virus, cancer, metabolic reprogramming, aerobic glycolysis
Citation: Wong J, Choi SYC, Liu R, Xu E, Killam J, Gout PW and Wang Y (2019) Potential Therapies for Infectious Diseases Based on Targeting Immune Evasion Mechanisms That Pathogens Have in Common With Cancer Cells. Front. Cell. Infect. Microbiol. 9:25. doi: 10.3389/fcimb.2019.00025
Received: 22 October 2018; Accepted: 24 January 2019;
Published: 12 February 2019.
Edited by:
Stephanie M. Seveau, The Ohio State University, United StatesReviewed by:
Yang Zhang, University of Pennsylvania, United StatesTonyia Eaves-Pyles, The University of Texas Medical Branch at Galveston, United States
Copyright © 2019 Wong, Choi, Liu, Xu, Killam, Gout and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter W. Gout, cGdvdXRAYmNjcmMuY2E=
Yuzhuo Wang, eXdhbmdAYmNjcmMuY2E=
†These authors have contributed equally to this work
 Jodi Wong1†
Jodi Wong1† Rongrong Liu
Rongrong Liu Peter W. Gout
Peter W. Gout Yuzhuo Wang
Yuzhuo Wang