- 1State Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, China
- 2University of Chinese Academy of Sciences, Beijing, China
As a crucial reciprocal partner, arbuscular mycorrhizal fungi (AMF) can alleviate the negative effects of a variety of pollutants on their hosts and soil microbes. In our previous studies, such characteristics of AMF on plant growth were documented in response to metal engineered nanoparticle (ENP) treatments. However, the role of AMF in influencing ENP effects on soil microbes remains a matter of debate. To address this issue, we investigated the responses of soil microorganisms to iron oxide nanoparticles (Fe3O4NPs) along a concentration gradient (0.1, 1.0, and 10.0 mg kg−1) in maize plants inoculated with and without AMF. The results showed that a high concentration of Fe3O4NPs significantly decreased the soil bacterial abundance and shifted the community composition, and these negative responses were associated with decreased soil dissolved organic C (DOC) contents in non-AMF-inoculated treatments. However, in AMF-inoculated treatments, no significant changes in soil biota or DOC contents were observed under Fe3O4NPs treatment. These results indicate that AMF alter the effects of Fe3O4NPs on soil microorganisms, possibly by influencing plant growth and organic matter released from plant roots, as DOC contents were impacted by AMF. Our findings suggest that AMF can influence Fe3O4NP-plant-microbe interactions; therefore, more attention should be focused on plant-associated microbes when evaluating the biological effects of nanoparticles.
Introduction
Engineered nanoparticles (ENPs) are man-made particles that measure from 1 to 100 nm in diameter and possess novel properties that are not shared by corresponding bulk materials (Auffan et al., 2009). Because of their unique physicochemical properties, ENPs are increasingly used in a wide range of technical applications and consumer products. According to “The project of emerging nanotechnologies,” the number of nanoproducts worldwide is expected to increase three-fold by 2020 (Roco, 2011). With this increase in production and uses, the release of ENPs into the environment is inevitable (Nowack, 2009; Lee et al., 2010). ENPs can enter the soil through applications of nano-containing pesticides and fertilizers and land applications of sewage sludge (Nowack et al., 2012); thus, they may pose risks to agricultural ecosystems (Klaine et al., 2008; Qiu, 2012).
Soil microorganisms are an indispensable component of agricultural ecosystems and are drivers of global biogeochemical cycles, including the cycling of carbon, nitrogen, sulfur, and phosphorus. Therefore, microbial responses to ENPs are of great interest and provide valuable information that may aid in our understanding of the influence of ENPs on soil health. Many ENPs, such as carbon nanotubes, graphene-based nanomaterials, iron-based nanoparticles, silver nanoparticles and copper, zinc and titanium oxide nanoparticles, have been used to assess the ecotoxicity of ENPs. In general, most of these studies have found limited (Tong et al., 2007; Johansen et al., 2008) or negative effects (Ge et al., 2011) on soil microbes. However, these studies are mainly in the absence of plants and ignore the potential impacts of plants on microbes. Because of the interactions between soil microbes and plants, investigations of the effects of ENPs on soil microbes in situ are more meaningful than those in the absence of plants. Recently, the effects of ENPs on soil microbial communities have been found to be modified by plants via effects on belowground biogeochemistry (Ge et al., 2014). Because plant roots can release many types of organic compounds (Philippot et al., 2013), they may alter soil processes in the following ways: (i) by changing the carbon sources' bioavailability to bacteria, because plants provide the main input of carbon and energy to the plant-soil-microbe web, (ii) by modifying ENP toxicity, because organic compounds can adsorb and immobilize some ENPs (Li et al., 2008). These findings suggest that a change in plant growth and subsequently, a change in carbon sources and/or ENP toxicity would directly or indirectly alter both the structural and functional aspects of soil microbes.
Arbuscular mycorrhizal fungi (AMF) are important and ubiquitous soil microorganisms that can form symbiotic associations with 90% of land plants (Smith and Read, 2008). These fungi are beneficial for plant growth, because they can improve plant nutrition acquisition by supplying mineral nutrients. Indeed, the roles of AMF in stress attenuation in plants that grow in soils contaminated with a variety of pollutants, such as heavy metals (Andrade et al., 2010), salt or alkali (Labidi et al., 2012), and organic pollutants (Wu et al., 2011), have been widely reported. Because plant physiology is affected by AMF symbiosis, root exudates are also influenced by the production of a new biochemical, resulting in the alteration of microbial activities (Zarea et al., 2009). For example, Aghababaei et al. (2014) revealed that the presence of AMF increased soil microbial activity that might be partially attributable to the greater plant biomass under heavy metal stress. Moreover, AMF play many other roles, such as improving soil structure and producing C sources for soil microbes. Despite recognition of AMF-based amelioration of the negative effects of many soil stresses on plant growth and soil microbes, the impacts of such plant-associated microorganisms on ENP-plant-soil microbe systems have not been well studied. Our previous study indicates that AMF alter the biological effects of Fe3O4NPs on clover (Trifolium repens) (Feng et al., 2013). However, the roles of AMF in influencing the effects of Fe3O4NPs on soil microbes in plant-soil systems remain subject to debate.
To evaluate how AMF alter the effects of ENPs on soil microbial communities, we studied how Fe3O4NPs affect soil microbial community structure and activity and how those impacts are altered by AMF in plant-soil systems. We chose to study Fe3O4NPs because these particles are commonly used as additives in medical diagnostics, the targeted delivery of drugs, controlled drug release, and pollution remediation (Sjögren et al., 1997; Prashant et al., 2010; Wang and Irudayaraj, 2010) due to their novel properties, such as enhanced surface-to-volume ratios, superparamagnetism, and inherent biocompatibility (Perez et al., 2002; Auffan et al., 2009). Accordingly, Fe3O4NPs are increasingly being introduced into the environment. We investigated the influences of AMF on the responses of soil biota to a concentration gradient of Fe3O4NPs (0.1, 1.0, and 10.0 mg kg−1) in a maize-soil system. The microcosm soils were harvested when the effects of Fe3O4NPs on plants were manifest and soil microbial activity, bacterial abundance, community composition and diversity were analyzed in AMF and non-AMF-inoculated treatments. We hypothesized that (1) plant growth, AMF status and soil bacterial community would differ between control and Fe3O4NPs treated soils; and (2) the inoculation of AMF would influence the toxicity of Fe3O4NPs to soil microbes due to the interactions of AMF-plant-soil microbe.
Materials and Methods
Fe3O4NPs Preparation and Characterization
Fe3O4NPs were synthesized by chemical co-precipitation of Molday (1984). Typically, a solution of FeCl3 and FeSO4 (molar ratio 2:1) was prepared under an N2 atmosphere followed by the slow addition of ammonia aqueous solution with vigorous stirring for 30 min. The black Fe3O4 precipitate was obtained and washed immediately with distilled water five times using a magnetic separation technique. The final precipitate was dispersed in distilled water at a concentration of 0.128 M and pH 3.0. These Fe3O4NPs were dispersed in deionized water, and the pH was adjusted to 2.7. To remove residual iron, the nanoparticles were washed five times with distilled water by magnetic separation. Finally, the Fe3O4NPs were re-suspended in distilled water (obtaining ~2 mg ml−1), sterilized through a 0.22 μm sterilized filter and stored at 4°C for use. Bulk Fe3O4 (particle size 5.0 μm, purity 99%, AR) were purchased from Aladdin Co., P. R. China.
The morphology and size of the Fe3O4NPs were investigated using a JEOL/JEM-2000E transmission electronic microscope (TEM) operated at 120 kV. The nanoparticles diameter distribution was determined with SAED (selected area electron diffraction) (ED, JEOL, JEM-200EX) by analyzing greater than 400 particles. The zeta potential (ζ) of the Fe3O4NPs was determined using a zeta potential analyzer (Delsa 440SX, Beckman Coulter, USA). Prior to their addition to the growth matrix, the Fe3O4NPs or bulk Fe3O4 were dispersed in double distilled water using a sonicator (600 W, 40 kHz) for 20 min.
Soil Description and Sampling
The soils used for this study were collected from Fengqiu County, Henan Province, China (35°00′N, 114°24′E). The soil is derived from the alluvial sediments of the Yellow River and is classified as Aquic Inceptisol (a calcareous fluvo-aquic soil). The relevant surface soil properties were analyzed as previously described by Lin et al. (2012) and are as follows: soil organic matter 5.83 g kg−1, total nitrogen 0.45 g kg−1, available phosphorus 1.93 mg kg−1, available potassium 78.5 mg kg−1 dry weight soil (d.w.s.) and pH (H2O) 8.6. The crop succession was winter wheat (Triticum aestivum L.) and summer maize (Zea mays L.).
After the wheat harvest, soil samples were collected from a depth of 0-15 cm, packed on-site into sealed polythene bags and then transported to the laboratory. Large roots, macro-fauna and stones were removed from the soil samples, which were then air-dried and sieved (2 mm) prior to application in the experiments.
AMF Inocula
The AMF inocula, Glomus caledonium 90036, were chosen in this experiment because they were isolated from a fluvo-aquic soil in Fengqiu County, Henan Province, China (35°00′N, 114°24′E) where the soil matrix was collected. The AMF specie was identified morphologically using current taxonomic criteria (Schenck and Perez, 1990) and Internet information from INVAM (http://invam.caf.wvu.edu); the inocula were then deposited in the greenhouse. The inocula were propagated on cover (Trifolium repens) grown in an autoclaved (121°C for 1 h on three successive days) sandy soil for three successive propagation cycles, each 4 months long. At the same time, sterilized inocula (121°C, 2 h) plus mycorrhizal fungal-free filtrate (20 ml) from the inocula suspension (to provide a similar microflora except for the AMF) were added to the same autoclaved sandy soil on which cover was cultivated for three successive cycles under the same conditions. Then, both the AMF and non-AMF inoculum were collected, air-dried and sieved (2 mm), prior to their addition to the soil matrix. The AMF inocula consisted of a mixture of rhizospheric soil that contained spores, hyphae, and infected root fragments.
Experimental Design and Setup
There were three Fe3O4NP levels (L: 0.1 mg kg−1; M: 1.0 mg kg−1; H: 10.0 mg kg−1 d.w.s.), three corresponding bulk Fe3O4 levels (to demonstrate the effects of nanoparticles) and a common control (without particles amendment); all these levels were either with (M) or without (NM) AMF inoculation. 10 g of AMF/non-AMF inocula were thoroughly mixed with 180 g d.w.s. by hand for even distribution. Thereafter, either Fe3O4NPs or bulk Fe3O4 was added to the inocula amendment mixture. To ensure that the particles were thoroughly mixed into the soil, distilled water containing the appropriate amount of Fe3O4NPs or bulk Fe3O4 was added dropwise to the soil surface, following the protocol of Jia and Conrad (2009). Each treatment had eight replicates.
Seeds of maize, Zea mays L. cv. ChengHai-1, were surface-sterilized with 0.5% NaClO, rinsed five times in autoclaved distilled water and germinated at 28°C (48 h) before sowing. Two healthy seeds of maize were transplanted into 250-ml plastic pots filled with growth medium and the amendment mixture at a depth of ~0.5 cm. All plants were grown in a sunlight greenhouse for a 16 h photoperiod, at a 25/18°C day/night temperature, with 60% relative humidity. Each pot was irrigated each week with 20 ml of 50% Hoagland's nutrient solution to maintain normal growth. After 20 days, a time at which the effects of Fe3O4NPs on plant growth are apparent, three pots of plant performance in unity per treatments were selected to harvest. Shoots and roots were sampled separately and weighed after oven drying at 70°C for 48 h. All roots were thoroughly rinsed with tap water before drying, and weighed subsamples of fresh roots were used for mycorrhizal colonization assessment by the grid-line intersect method (Giovannetti and Mosse, 1980) after clearing with 10% (m/m) KOH and staining with acid fuchsin (Phillips and Hayman, 1970). After the final plant harvest, the soils were collected for analysis. Because the taproot and fibrous roots of maize extended throughout the soil, the entire pot of soil was thoroughly mixed after the roots were removed, and the result was considered the rhizospheric soil. Samples for molecular studies were maintained at −40°C until used. Sub-samples for chemical and microcalorimetric determinations were maintained at 4°C.
Soil Characteristic Determination
Soil dissolved organic C (DOC) was extracted by adding 50 ml of 0.5 M K2SO4 to 10 g of fresh soil and shaking for 1 h. The extracts were centrifuged at 4000 rpm for 10 min, and the supernatants were filtered through glass fiber filters (Fisher G4, 1.2 μm pore space). The filtrate was stored at −20°C until further analyzed. DOC was determined using a TOC-TN analyzer (Skalar, The Netherlands).
Microcalorimetric Measurements
Soil microorganism metabolic activities were determined using a third-generation thermal activity monitor (Thermometric AB, Newcastle, DE, USA). All 4-ml steel ampoules were cleaned and sterilized in an oven before use. A soil sample (1.2 g) was added to 0.2 ml of a solution containing 5 mg of glucose and 5 mg of ammonium sulfate to stimulate soil microbial activity and to provide nitrogen and sulfur for amino acids synthesis. Before the addition of the amendments, the soil samples were incubated at 28°C for 1 day to activate the microorganisms. The ampoules were then hermetically sealed using Teflon sealing discs to control evaporation and energy loss, they were subjected to microcalorimetric measurements. The heat flow rate of microbial growth was monitored and recorded in a real-time mode, eventually obtaining a power-time curve.
The thermodynamic parameters, that is, the maximum thermal power (Pmax), time to reach the maximum of the peak (tmax), total heat dissipation (QT) and growth rate constant (k), were obtained by integrating the power-time curves (Zheng et al., 2009). Pmax and tmax were obtained directly from the power-time curve. Qt is the sum of metabolic processes that occur during substrate consumption. The k-value was calculated by fitting a logarithmic growth model based on data from the power-time curve in the logarithmic growth stage, which obeys the following thermokinetic equation: lnPt = lnP0 + kt, where t is the time, Pt is the power output at time t, and P0 is the power at the beginning of the exponential growth phase (Critter et al., 2004).
Soil DNA Extraction
For each soil sample, total DNA was extracted from 0.5 g soil subsampled from each of the three end point replicates for each treatment using the FastDNA® SPIN Kit for soil (MP Biomedicals, Santa Ana, CA), according to the manufacturer's instructions. The extracted soil DNA was dissolved in 100 μl of TE buffer, and then quantified by spectrophotometer (NanoDrop ND-1000, Thermo Scientific, USA) and stored at −20°C until further use.
Real-Time Quantitative PCR of the Bacterial 16S rRNA Gene
For each soil sample, real-time quantitative polymerase chain reaction (qPCR) was used to determine the abundance of the bacterial 16S rRNA gene. The primers were 519F (5′-CAGCMGCCGCGGTAATWC-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) (Feng et al., 2011). Briefly, the target gene copy number was quantified by qPCR analysis using a C1000TM Thermal Cycler equipped with a CFX96TM Real-Time system (Bio-Rad, USA). To generate the qPCR standard curve, a single clone containing the correct insert was grown in Luria-Bertani medium; plasmid DNA was then extracted, purified, and quantified. A 10-fold dilution series of the linear plasmid DNA was prepared to generate a standard curve covering seven orders of magnitude, from 102 to 108 copies of template. Assays were set up using SYBR Premix Ex Taq™ Kit (TaKaRa Biotechnology, Dalian, Co., Ltd) with a 20-μl reaction mixture containing 10 μl of SYBR® Premix Ex Taq™, 0.8 μl of the primer set (initial concentration of 10 μM each), 7.2 μl of sterile deionized water, and 2.0 μl of template, which contained ~2–9 ng of DNA. Blanks were run with water as the template instead of the soil DNA extract. Specific amplification of the target gene was confirmed by agarose gel electrophoresis of qPCR amplicons showing the band of the expected size and melting curve analysis resulting in a single peak. The qPCR was performed in triplicate, and amplification efficiencies of 97.4–104% were obtained, with R2-values of 0.966–0.977. The final bacterial 16S rRNA gene quantities were obtained by calibrating against the total DNA concentration extracted and the soil water content. For each treatment of the fourteen treatments, three biological replicates were quantitated.
PCR and Preparation of Amplification Libraries
The bacterial 16S rRNA gene and AMF 18S rRNA gene were amplified with primers set 519F/907R) and AMV4.5NF (5′-AAGCTCGTAGTTGAATTTCG-3′)/AMDGR (5′-CCCAACTATCCCTATTAATCAT-3′) for sequencing on the Illumina Miseq pyrosequencing platform. PCR was performed in 50-μl reaction mixtures containing 4 μl (initial 2.5 mM each) of deoxynucleoside triphosphates, 2 μl (initial 10 μM each) of forward and reverse primers, and 0.4 μl (2 U) of Taq DNA polymerase (TaKaRa, Japan). Each reaction mixture included a 1 μl template that contained ~50 ng of genomic community DNA. Thirty-five cycles were performed at 95°C for 45 s, 56°C for 45 s and 72°C for 1 min, with a final extension at 72°C for 7 min. The oligo nucleotides sequences of included 5-bp barcode fused to the primer set: barcode + forward primer to separate the corresponding reads from the data pool generated in a single pyrosequencing run. Triplicate reaction mixtures per sample were pooled, purified using the QIAquick PCR Purification kit (QIAGEN), and quantified using a NanoDrop ND-1000 (Thermo Scientific, USA). The PCR products were normalized in equimolar amounts, prepared using the TruSeq™ DNA Sample Prep LT Kit and pyrosequenced using the MiSeq Reagent Kit (500-cycles-PE) following the manufacturer's protocols. For each of the fourteen treatments, three biological replicates were sequenced.
Processing of Pyrosequencing Data
After pyrosequencing, the 16S rRNA and 18S rRNA data were processed using Quantitative Insights into Microbial Ecology (QIIME) (Caporaso et al., 2010) before further statistical analysis. Briefly, according to the nucleotide barcode, the reads were assigned to the designated samples, and any low-quality (average score < 25, or reads length shorter than 200 bp) or ambiguous sequences (containing two or more nucleotides) were trimmed and removed using the default parameters set by QIIME. The sequences were binned into operational taxonomic units (OTUs) using a 97% identity threshold and the most abundant sequence from each OTU was selected as a representative sequence for that OTU. The taxonomic classification was assigned against a subset of the Silva 104 database (http://www.arb-silva.de/download/archive/qiime/). OTU representative sequences were aligned using PyNAST, and chimeric sequences were removed using QIIME.
The composition and richness of the microbial community were investigated at a single level of taxonomic resolution. In this diversity analysis, 485,390 bacterial sequences that passed QIIME's quality filtering were included. We obtained between 4422 and 20,621 sequences per sample for all the soil samples. Because an even depth of sampling is required for beta diversity calculations, we reduced the datasets to the lowest number available to correct for different survey efforts among the samples. Namely, we calculated both diversity metrics using a randomly selected subset of 4000 sequences per soil sample; this approach allowed us to compare general diversity patterns among sites, although it is highly unlikely that we surveyed the full extent of diversity in each community (Shaw et al., 2008). Based on the subsets of 4000 sequences per soil sample, the response ratios of soil bacterial guilds under a high concentration of Fe3O4NPs were analyzed following the statistical method of Luo et al. (2006). The pyrosequencing reads have been deposited at National Center for Biotechnology Information Search database (NCBI) with the accession number PRJNA305184.
Statistical Data Analysis
The statistical procedures were carried out using an analysis of variance (both one-way and two-way ANOVA) with the software package SPSS 18.0 (SPSS Inc., Chicago, U.S.A.) for Windows. Data are expressed as the means with a standard deviation (SD). Mean separation was performed based on Tukey's multiple range tests; Student's-t test was performed for changes in the relative abundance of the dominant bacterial phyla. Two diversity indices, Faith's index of phylogenetic diversity (PD) (Faith, 1992) and Chao 1 index (Chao, 1984), were used to compare soil bacterial alpha diversity. The response ratios of bacterial classes between the high Fe3O4NPs and control were analyzed following the statistical method of Luo et al. (2006). Differences at p < 0.05 were considered to be statistically significant.
Results
TEM Analysis of Fe3O4NPs
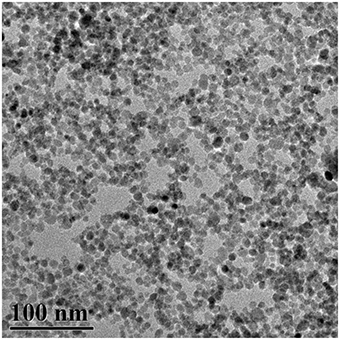
The typical TEM visualization of the Fe3O4NPs (Figure 1) shows the aggregations of Fe3O4NPs due to drying on the TEM grid or aggregation in the suspension. Fe3O4NPs were quasi-spherical in shape, and their diameters were ~10.2 ± 2.6 nm. The zeta potential (ζ) values of Fe3O4NPs were evaluated to be −52.0 ± 1.1 mV, indicating that these particles repelled each other and that the particles did not have a tendency to aggregate.
Changes in Soil Dissolved Organic C Content, Plant Biomass, AMF Colonization and Assemblages
The effects of Fe3O4NPs on DOC contents are shown in Figure 2. To demonstrate the toxicity that resulted from nanoparticles, the effects of the corresponding bulk Fe3O4 on DOC contents were compared, and it was found that bulk Fe3O4 at three concentrations did not influence soil DOC contents. In contrast, compared to the control, Fe3O4NPs had a negative influence on soil DOC contents (Figure 2). With an increase in content, the adverse effect of Fe3O4NPs on DOC contents worsened, and significant decreases (by 13.7%) were finally observed under a high concentration of Fe3O4NPs in non-AMF-inoculated treatments. Correspondingly, significant decreases (p < 0.05) in plant shoot and root biomass (by 17.0 and 19.6%, respectively) (Supplementary Figure 3) and mycorrhizal colonization (by 34.1%) (Supplementary Figure 1) were eventually observed in high concentration of Fe3O4NPs. Moreover, the AMF assemblages of Fe3O4NPs-treated soils were shifted (Supplementary Figure 2). Compared to the control, Fe3O4NPs at the high concentration exerted higher relative abundance in the genus of Glomus, and lower relative abundance in Acaulosporaceae and Gigasporaceae. In AMF-inoculated treatments, however, no significant effects of a high Fe3O4NPs concentration on soil DOC content, plant biomass, and mycorrhizal colonization were detected in comparison with the corresponding control. Correspondingly, the degree of the changes in AMF community was decreased. The effects on DOC content of AMF inoculation and the interaction between AMF inoculation and particle amendment were all significant (Figure 2).
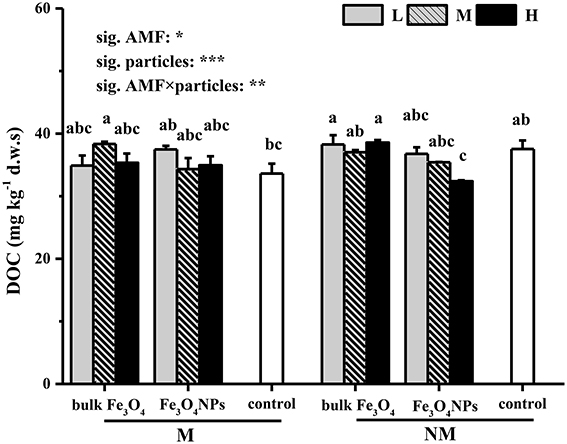
Figure 2. Soil DOC (mg kg−1 d.w.s.) contents in response to Fe3O4NPs in soils cultivated with maize with or without AMF inoculation. M, AMF inoculation; NM, non-AMF-inoculation; L, M, and H indicate 0.1, 1.0, and 10 mg kg−1 soil Fe3O4NPs and bulk Fe3O4 amendment respectively; control indicates the treatment without nanoparticle amendment. Vertical T bars indicate standard deviations (SD). Bars not topped by the same letter indicate a significant difference in values (p < 0.05). ANOVA results are shown in the figure with ***, **, and * representing significant at the 1o, 1%, and 5% level, respectively.
Soil Microbial Activity Revealed by Microcalorimetry
The activity of soil microorganisms was evaluated by measuring microbial metabolic activity using a microcalorimetric analyzer. The power-time curves obtained from soils amended with Fe3O4NPs in AMF or non-AMF-inoculated treatments are illustrated in Figure 3, which shows the typical processes of various soil microbial metabolic activities. All curves presented typical microbial growth, in four phases: a lag phase, an exponential growth phase, a short stationary phase, and a decline phase. From these power-time curves, the total heat released per gram of soil by microbial growth reactions, Q (J g−1); the power at the maximum of the peak, Pmax (μW); the microbial growth rate constant, k (h−1); and the time to reach the maximum peak, tmax(h); were attained in three replications through the integration of each curve, shown in Supplementary Table 1. Overall, the effect of AMF inoculation on Pmax was significant (p < 0.05); the effect of particle amendment on Q was also significant (p < 0.01). The tmax were systematically affected by their interaction (p < 0.05) (Supplementary Table 1). Comparison with control, no significant differences in Pmax (μW), tmax(h), Q (J g−1), and k (h−1) were observed among the treatments (Supplementary Table 1), suggesting no significant influence of Fe3O4NPs and bulk Fe3O4 on soil microbial metabolic activity.
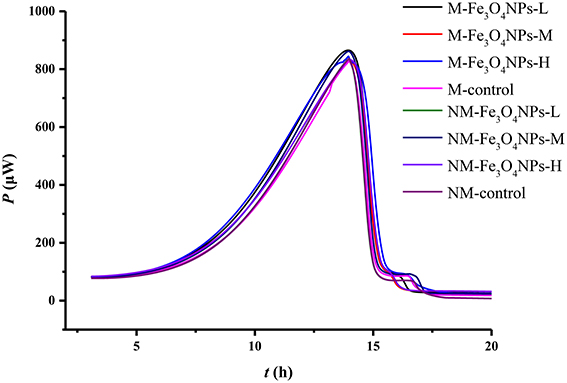
Figure 3. Power-time curves recorded microcalorimetrically from soil samples amended with glucose and ammonium sulfate. M, AMF inoculation; NM, non-inoculation; Fe3O4NPs -L, M, and H indicate 0.1, 1.0, and 10 mg kg−1 soil Fe3O4NPs amendment respectively; control indicates the treatment without nanoparticle amendment.
Quantitative Analysis of Soil Bacterial Abundance by qPCR
The abundance (or 16S copy number) of the soil bacterial community and changes under nanoparticle treatments were evaluated using qPCR (Figure 4). First, compared to the corresponding control, the bulk Fe3O4 at three concentrations did not have any influence on soil bacterial abundance. Second, Fe3O4NPs had a significant negative influence on bacterial abundance (Figure 4). Under a high concentration of Fe3O4NPs amendment, soil bacterial abundance was significantly decreased (p < 0.05) in non-AMF-inoculated treatments. Similar phenomena were observed in the treatments with low and medium concentrations of Fe3O4NPs, although the differences in soil bacterial abundance were not significant. Furthermore, effects on the bacterial abundance content of AMF inoculation were significant (Figure 4). In AMF-inoculated treatments, no significant effects of three Fe3O4NPs concentrations on soil bacterial abundance were detected in comparison with the corresponding control. The interaction between AMF inoculation and particle amendment on bacterial abundance was significant (Figure 4).
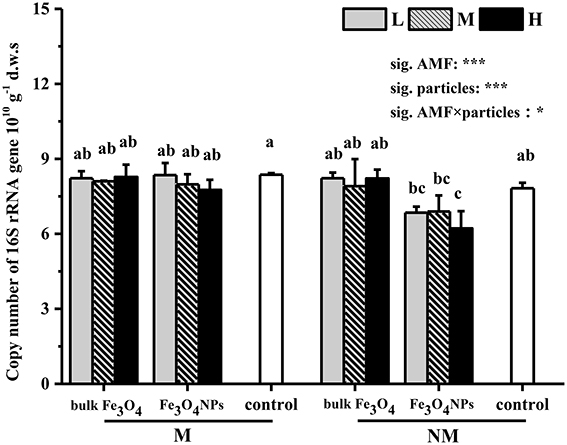
Figure 4. Shifts in copy numbers of bacterial 16S rRNA genes in response to Fe3O4NPs as well as respective bulk Fe3O4 materials, in comparison to the control without nanoparticle amendment. M, AMF inoculation; NM, non- inoculation; L, M, and H indicate 0.1, 1.0, and 10 mg kg−1 soil Fe3O4NPs and bulk Fe3O4 amendment respectively; control indicates the treatment without nanoparticle amendment. Vertical T bars indicate standard deviations (SD). Bars not topped by the same letter indicate a significant difference in values (p < 0.05). ANOVA results are shown in the figure with *** and * representing significant at the 1o and 5% level, respectively.
Taxonomic Distribution of Bacterial Community in Soils
Overall, 485,390 bacterial sequences were obtained. Pyrosequencing revealed that the soil bacterial community was dominated by several phyla, including Proteobacteria (32.7%), Actinobacteria (16.1%), Acidobacteria (15.3%), Bacteroidetes (6.9%), Chloroflexi (7.0%), Planctomycetes (5.0%), Gemmatimonadetes (3.3%), Firmicutes (1.6%), and Nitrospirae (0.81%) (Figure 5).
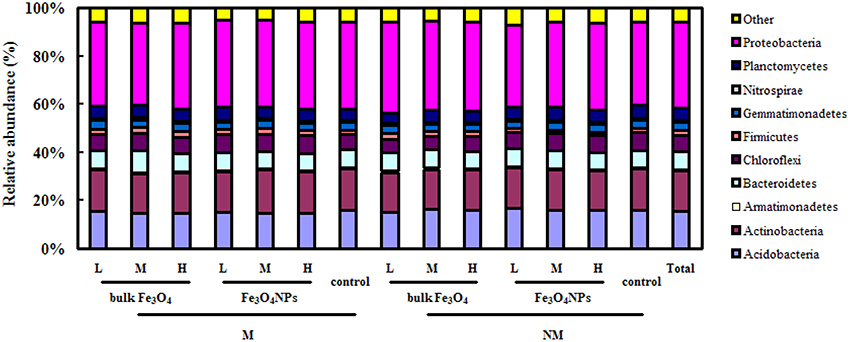
Figure 5. The 100% stacked column chart of relative abundances of the dominant bacteria phyla derived from 16S rRNA genes in soils combined and in each soil sample. The value of each phylum percentage is the mean of triplicates. M, AMF inoculation; NM, non-inoculation; L, M, and H indicate 0.1, 1, and 10 mg kg−1 soil Fe3O4NPs and bulk Fe3O4 amendment respectively; control indicates the treatment without nanoparticle amendment.
Pyrosequencing also enabled us to track shifts in the structure of the soil bacterial community in response to Fe3O4NPs in AMF-inoculated or non-AMF-inoculated treatments. For example, according to a pairwise comparison, compared to the corresponding control, only a high concentration of Fe3O4NPs in the non-AMF-inoculated treatments significantly decreased (p < 0.05) the relative abundance of Chloroflexi (from 7.0 ± 0.3% to 6.2 ± 0.6%, p = 0.0274), Gemmatimonadetes (from 3.3 ± 0.1% to 2.9 ± 0.2%, p = 0.0189) and Nitrospirae (from 0.81 ± 0.1% to 0.74 ± 0.1%, p = 0.0443). Interestingly, in the AMF-inoculated treatments, no significant negative effects of a high concentration of Fe3O4NPs on the relative abundance of these bacteria were found. Additionally, a high concentration of Fe3O4NPs increased the percentage of Chloroflexi from 5.6 ± 0.3% to 6.8 ± 0.3% (p = 0.00428) (Figure 5).
Changes in the Diversity Indices of Soil Bacteria
The bacterial diversity among different samples was evaluated based on the phylogenetic diversity (PD) and Chao1 values (Table 1). In general, none of the particle amendments had an influence on PD and Chao1 values. Neither AMF inoculation nor particle amendment had an influence on PD and Chao1. However, their interaction on PD and Chao1 was statistically significant (p < 0.05) (Table 1).
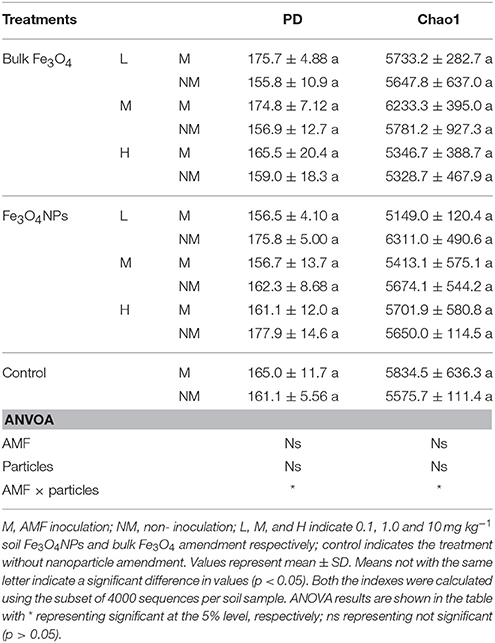
Table 1. The bacterial phylogenetic diversity indices under different concentration of Fe3O4NPs with or without AMF inoculation.
Shifts in Bacterial Assemblages
Variations in the composition of the bacterial community under the various treatments were statistically evaluated using principal component analysis (PCA) (Figure 6). To demonstrate the effects of nanoparticles, the influence of bulk Fe3O4 treatments on the composition of soil bacterial community was statistically evaluated first (Supplementary Figure 4). As expected, no significant shifts in bacterial assemblages were observed among all the treatments with bulk Fe3O4 amendment. In contrast, significant shifts in soil bacterial assemblages were observed between the high concentration of Fe3O4NPs and the corresponding control in non-AMF-inoculated treatments (Figure 6). The distances between the different concentrations of Fe3O4NPs-treated soils and the corresponding control soil along axis 1 indicate that a high Fe3O4NPs concentration had the greatest influence on the composition of the soil bacterial community. Notably, the degree of variation in the composition of the soil bacterial community between the high concentration of Fe3O4NPs and the corresponding control was reduced by AMF-inoculation (Figure 6).
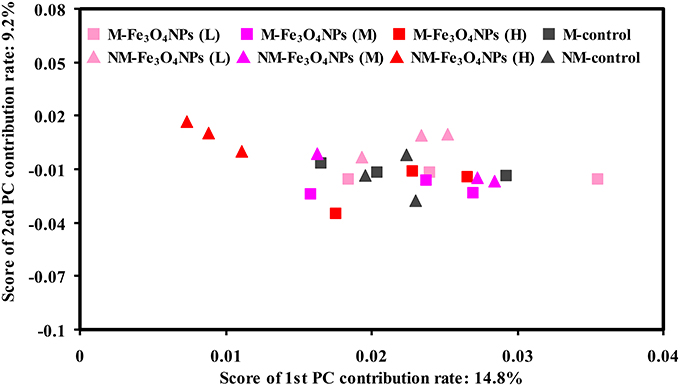
Figure 6. Bacterial community compositional structure in soils under different concentrations of Fe3O4NPs amendments as indicated by a principal component analyses (PCA). M, AMF inoculation; NM, non-inoculation; L, M, and H indicate 0.1, 1.0, and 10 mg kg−1 soil Fe3O4NPs amendment respectively; control indicates the treatment without nanoparticle amendment.
Changed Bacterial OTUs
To verify the effects of AMF inoculation on the response of Fe3O4NPs in the composition of the soil bacterial community, response ratios were generated to identify changes in taxa-specific OTUs. Because of the consistent significant differences in soil bacterial abundance (Figure 4) and soil bacterial community composition (Figure 6) between the high concentration of Fe3O4NPs and the corresponding control, the response ratio was calculated for only this concentration based on the sequence size of each class (Figure 7). The 95% confidence interval (CI) under the high concentration of Fe3O4NPs in non-AMF-inoculated treatments ranged from −0.10 to −0.07 and did not overlap 0, which indicates that Fe3O4NPs has a significant negative influence on the composition of the soil bacterial community (p < 0.05). Compared to the control, 12 classes were significantly decreased at a high concentration of Fe3O4NPs. The dominant class, namely Alphaproteobacteria, Actinobacteria, Acidobacteria, Chloracidobacteria, Planctomycea, Gemmatimonadetes, Anaerolineae, and Nitrospira were significantly decreased (p < 0.05) by a high concentration of Fe3O4NPs. In AMF-inoculated treatments, the 95% CI, which ranged from −0.01 to 0.01, revealed insignificant effects of Fe3O4NPs on the composition of the soil bacterial community (Figure 6). For specific guilds, it was found that the diversities of Alphaproteobacteria and Chloracidobacteria were significantly decreased, whereas the diversities of Anaerolineae, and Nitrospira were significantly increased. The results for the response ratios were consistent with the general trends of the taxonomic distribution of the soil bacterial community (Figure 5) and PCA plots (Figure 6). In summary, AMF alleviated the negative effects of Fe3O4NPs on the composition of the soil bacterial community.
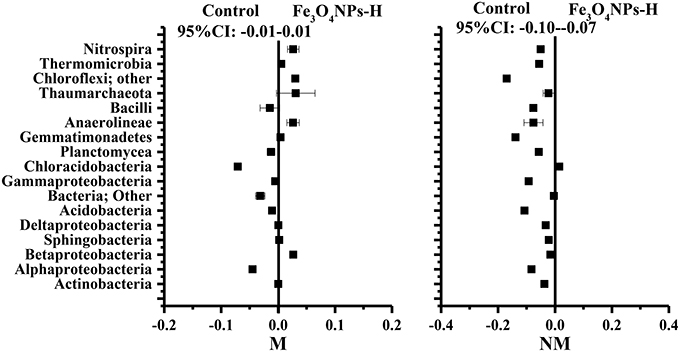
Figure 7. Significant changes in bacterial classes under the high concentration of Fe3O4NPs according to the response ratio method at a 95% confidence interval (CI). M, AMF inoculation; NM, non-inoculation; control indicates the treatment without nanoparticle amendment.
Discussion
The community structure of soil bacterial assemblages has been previously studied (He et al., 2011; Kumar et al., 2012). To display a high resolution of the composition of the soil bacterial community and to follow their shifts in response to Fe3O4NPs, Miseq pyrosequencing technology was utilized in the present investigation. Overall, 485,390 bacterial sequences were obtained that provided detailed information on the composition of the bacterial community, which was dominated by Proteobacteria, Actinobacteria, Acidobacteria, Bacteroidetes, Chloroflexi, Planctomycetes, etc. (Figure 5). This information about the dominant phyla supports the finding from denaturing gradient gel electrophoresis (DGGE) fingerprinting and 454 pyrosequencing. For example, Khodakovskaya et al. (2013) have found that the soil bacterial community under carbon nanotubes is dominated by the phylum of Bacteroidetes, Proteobacteria, Firmicutes, Actinobacteria and Acidobacteria. However, the proportion of each phylum in this investigation is different from those reports. These differences could be attributed to the characteristics of nanoparticles and different soil physicochemical properties.
Fe3O4NPs at 10.0 mg kg−1 d.w.s. Significantly Influence the Soil Bacterial Community
In general, the soil rhizospheric biota was negative influenced by Fe3O4NPs in a dose-dependent manner. Compared to the corresponding control, the high concentration of Fe3O4NPs (10 mg kg−1 d.w.s.) significantly decreased (p < 0.05) the bacterial abundances (Figure 4). Correspondingly, the community composition was shifted (Figure 6), and the response ratios to the high concentration of Fe3O4NPs were less than zero (Figure 7), although the microbial activity and bacterial community diversity were not influenced (Figure 3, Table 1). This impact of Fe3O4NPs on soil biota might result from the chemical characteristics of nanoparticles and their influences on microorganisms' metabolism. It is well known that Fe3O4NPs have enhanced surface-to-volume ratios and high mobility of electrons within the structure. Therefore, Fe3O4NPs have the ability to release iron ions (Fe2+, Fe3+) and sustainably generate reactive oxygen species (ROS) because iron ions (Fe2+) are efficient ROS producers (Auffan et al., 2008). The ROS production and resulting oxidant stress for microbes may cause cell apoptosis by damaging cellular lipids, carbohydrates, proteins and DNA (Kelly et al., 1998; Kahru et al., 2008). For example, Auffan et al. (2008) studied the cytotoxicity of Fe3O4NPs on Escherichia coli and analyzed the underlying mechanisms, and found that Fe3O4NPs decreased the survival of Escherichia coli mainly due to oxidative stress. In this investigation, we found that the high concentration of Fe3O4NPs decreased the proportion of Acaulosporaceae and Gigasporaceae and increased the proportion of Glomus, and subsequently shifted the composition of AMF community in Fe3O4NPs-treated soils (Supplementary Figure 2). It is possible that Glomus could colonize through pieces of mycelium or mycorrhizal root fragments and as a result survive and propagate more easily than other members under changed soil conditions (Hassan et al., 2011). However, Cornejo et al. (2013) found that heavy metal can enter Glomus spores and decrease their vitality. Therefore, although the relative abundance of Glomus in Fe3O4NPs-treated soils was increased, the colonization of AMF (Supplementary Figure 1) was decreased, indicating that the symbiosis between AMF and plant was disturbed. Consequently, a significant decrease (p < 0.05) in the maize shoot and root biomass (by 17.0 and 19.6%, respectively) (Supplementary Figure 3) was observed under a high concentration of Fe3O4NPs. The changing pattern of plant growth (Supplementary Figure 3) in response to Fe3O4NPs was similar to that observed for soil biota (Figures 4–7). Furthermore, lower concentrations of Fe3O4NPs had little effect on plant growth (Supplementary Figure 3) and AMF colonization (Supplementary Figure 1), which also accompanies the minimal effects of the medium and low concentrations of Fe3O4NPs on the soil bacterial abundance (Figure 4) and community composition (Figure 6). Although the root exudates were not determined directly, they are likely reduced as the decreased plant biomass (Supplementary Figure 3) under the high Fe3O4NPs concentration because it has been reported that root exudates positively correlated with plant biomass (Lynch and Whipps, 1990). As partially evidence, we found a significant decrease in DOC contents (Figure 2) under the high concentration of Fe3O4NPs. Because carbon sources are the substrate for microorganisms' metabolisms, a decrease in soil DOC contents could result in changes in both the structural and functional aspects of soil biodiversity. Similarly, Ge et al. (2014) have found that a decrease in the input of organic materials introduced by plant roots exert shifts in the bacterial community composition under CeO2NPs. Hence, it can be concluded that the decreased bioavailability of carbon sources is related to the negative response of the soil biota to a high concentration of Fe3O4NPs. Together, it is possible that soil biota might be impacted directly and through plant-microbe interactions in the presence of Fe3O4NPs.
The negative response of the soil bacterial community resulted from the responses of the dominant groups, and this observation was more obvious for several keystone guilds. For example, the diversity of Anaerolineae, Planctomycea, Sphingobacteria, Chloracidobacteria, Acidobacteria, and Nitrospira were all decreased under a high concentration of Fe3O4NPs (Figure 7). Hackl et al. (2004) have reported that the composition of the Acidobacteria community in soil is influenced by the soil organic carbon concentration. Therefore, it is possible that the decrease in the relative abundance of Acidobacteria might have resulted from the reduction in the soil DOC content under the high concentration of Fe3O4NPs (Figure 2). Moreover, Planctomycetes are a remarkable guild in soil biota assemblages, and the diversity of this phylum is positively associated with the global nitrogen cycle (Kalyuzhnyi et al., 2006). Therefore, the decrease in Planctomycetes suggests that the soil's carbon and nitrogen cycles in soil were affected by a high concentration of Fe3O4NPs. Collectively, decreases in keystone microbial guilds in response to a high concentration of Fe3O4NPs could deteriorate the sustainability of soil fertility, creating feedback that alters the nanoparticle-plant-microbe interactions.
AMF Alleviate the Negative Influences of Fe3O4NPs on the Soil Bacterial Community
As initially hypothesized, the adverse effects of Fe3O4NPs on the soil bacterial community were altered by AMF. Fe3O4NPs at three concentrations have no influence on the soil biota in AMF-inoculated treatments. These insignificant influences were characterized by levels of metabolic activity (Figure 3, Supplementary Table 1), bacterial abundance (Figure 3) and bacterial community diversity indices (Table 1) in the Fe3O4NPs treatments that were similar to those in the corresponding control. Correspondingly, the degree of variation in the community composition was reduced (Figure 6), and the response ratios to the high Fe3O4NPs concentration overlapped zero (Figure 7), which indicates that Fe3O4NPs did not influence on soil biota in the AMF-inoculated treatments. A similar phenomenon under heavy metal stress is reported by Aghababaei et al. (2014), who find that AMF alleviated the negative effects of cadmium (Cd) on soil microbial biomass and enzyme activities. They further speculated that the beneficial effects of AMF on soil microbes might be partially attributed to the stimulation of plant growth. Indeed, it has been widely documented that AMF can confer plant some degree of tolerance to exotic stresses by improving plant nutrient acquisition and altering metal uptake. For example, it has been reported that some Glomus spores can accumulate the metal in their cytoplasm (Cornejo et al., 2013) and thus decrease the toxicity of heavy metals to plants and other microorganisms. Therefore, the amelioration of phytotoxicity by AMF has long been recognized as one of the benefits of these fungi for host plants growing in a variety of pollutant-contaminated soils, such as those containing AgNPs and heavy metals (Andrade et al., 2010; Miransari, 2010; Feng et al., 2013; Wang et al., 2014). In this investigation, the inoculation of AMF inocula, Glomus caledonium, might result in an increase in the relative abundance of Glomus in AMF assemblages. As a result, insignificant changes in AMF colonization and plant biomass under Fe3O4NPs treatments in the inoculation of AMF were observed in this study (Supplementary Figures 2, 3). In this sense, carbon input from plant to soil, including root-derived compounds, could also be different in AMF-inoculated treatments compared to that in non-AMF-inoculated treatments. Also, it should be noted that AMF hyphae can produce carbon compounds as a source of energy for soil microbes in the mycorrhizosphere (Andrade et al., 1997). Possible related, less (or no) reduction in the soil DOC content, substrate for microbial metabolism, was found in AMF-inoculated treatments than in non-AMF-inoculated treatments (Figure 2). In addition to providing fuel for soil microbial metabolism, soil organic matter can adsorb and immobilize some ENPs or the ions released from ENPs (Tong et al., 2007). For example, Philippot et al. (2013) reported that chelators exuded by plant roots can sequester metal ions. The toxicity of ENPs to microorganisms is found to be mitigated by algal-produced organic compounds (Stevenson et al., 2013). Similarly, Martineau et al. (2014) find that CuO or ZnO was non-toxic to microorganisms in root washes of exudates extracted from plants. Fe3O4NPs have the most active surface sites (mainly the Fe-OH site), with high affinity for organic compounds (Liu et al., 2008). Because of this property, some Fe3O4NPs or their released irons might be immobilized and resulting decreased direct toxicity to microbes caused by the relatively greater amount of organic material available in the soil in AMF-inoculated treatments than that in non-AMF-inoculated treatments (Figure 2). Therefore, we speculate that AMF might alleviate the negative influence of Fe3O4NPs on soil biota by stimulating the plant growth. Collectively, the high concentration of Fe3O4NPs ultimately resulted in insignificant changes in soil biota in AMF-inoculated treatments, which contrasts with the negative responses in non-AMF-inoculated treatments.
The alleviation of the negative responses of soil biota to Fe3O4NPs by AMF was also characterized by several dominant groups. For example, compared to the corresponding control, no significant changes in the relative abundance of Planctomycea, Sphingobacteria, Chloracidobacteria, Acidobacteria and Actinobacteria were found under the high concentration of Fe3O4NPs in AMF-inoculated treatments. Moreover, the relative abundance of Nitrospira and Anaerolineae increased significantly (Figure 7). Nitrospira are nitrite-oxidizing bacteria that are ubiquitous in terrestrial environments and play a major role in the biological nitrogen cycling (Daims et al., 2001). Because Nitrospira's diversity is positively associated with nutrient turnover (Li et al., 2014), increased in Nitrospira indicates enhanced nutrient turnover and a sufficient nutrient supply to sustain plant growth. Moreover, Yamada and Sekiguchi (2009) report that Anaerolineae can decompose carbohydrates, especially relatively recalcitrant carbon compounds; therefore, an increase in Anaerolineae would positively influence carbon cycling and promote nutrient release from organic compounds in soils. Similarly, Cao et al. (2015) reported that AMF increase the abundance of Flavobacteriaceae, subsequently enhancing the availability and utilization of phosphorus in soils (Martinez et al., 2014). Therefore, it should be emphasized that AMF have positive effects not only on plants but also on soil microorganisms and soil function under ENP treatments.
Conclusion
In conclusion, we found that a high concentration of Fe3O4NPs (10.0 mg kg−1) did not influence the soil microbial activity or bacterial community richness, but instead significantly decreased the soil bacterial abundance and shifted their community composition. The observed negative responses were associated with decreased DOC contents. Such negative responses to Fe3O4NPs by several keystone guilds suggest that Fe3O4NPs might inhibit C and N cycles in soils. These negative effects of Fe3O4NPs on soil biota were alleviated by AMF. It is possible that AMF altered the adverse effects of Fe3O4NPs on soil biota by influencing plant growth and the release of organic matter from plant roots because the DOC content was impacted. As ENPs increasingly enter the soil environment, they will alter how soils function and may therefore affect plant growth. Furthermore, the ecotoxicity of ENPs will be directly altered by microbes or through plant-microbe interactions, creating feedback that alters interactions in plant-soil-ENP systems. Therefore, as an important constituent of plant systems, more attention should be focused on the responses and feedback of soil microorganisms when evaluating the effects of nanoparticles on agricultural ecosystems.
Author Contributions
JC and XL designed the study. JC and YF performed the experiments. JC, YF, and JW analyzed the data. JC and XL wrote the paper. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Fengqiu Agro-Ecological Experimental Station, Institute of Soil Science, Chinese Academy of Sciences, for the kind support on soil sample collection. This work was supported by the National Natural Science Foundation of China (No. 41371255, 41301267, 41271256).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fenvs.2016.00010
References
Aghababaei, F., Raiesi, F., and Hosseinpur, A. (2014). The combined effects of earthworms and arbuscular mycorrhizal fungi on microbial biomass and enzyme activities in a calcareous soil spiked with cadmium. Appl. Soil Ecol. 75, 33–42. doi: 10.1016/j.apsoil.2013.10.006
Andrade, G., Mihara, K. L., Linderman, R. G., and Bethlenfalvay, G. J. (1997). Bacteria from rhizosphere and hyphosphere soils of different arbuscular mycorrhizal fungi. Plant Soil. 192, 71–79. doi: 10.1023/A:1004249629643
Andrade, S. A. L., Gratao, P. L., Azevedo, R. A., Silveira, A. P. D., Schiavinato, M. A., and Mazzafera, P. (2010). Biochemical and physiological changes in jack bean under mycorrhizal symbiosis growing in soil with increasing Cu concentrations. Environ. Exp. Bot. 68, 198–207. doi: 10.1016/j.envexpbot.2009.11.009
Auffan, M., Achouak, W., Rose, J., Roncato, M. A., Chanéac, C., Waite, D. T., et al. (2008). Relation between the redox state of iron-based nanoparticles and their cytotoxicity toward Escherichia coli. Environ. Sci. Technol. 42, 6730–6735. doi: 10.1021/es800086f
Auffan, M., Rose, J., Wiesner, M. R., and Bottero, J. Y. (2009). Chemical stability of metallic nanoparticles: a parameter controlling their potential cellular toxicity in vitro. Environ. Pollut. 157, 1127–1133. doi: 10.1016/j.envpol.2008.10.002
Cao, J., Huang, Y., and Wang, C. (2015). Rhizosphere interactions between earthworms (Eisenia fetida) and arbuscular mycorrhizal fungus (Funneliformis mosseae) promote utilization efficiency of phytate phosphorus in maize. Appl. Soil Ecol. 94, 30–39. doi: 10.1016/j.apsoil.2015.05.001
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chao, A. (1984). Nonparametric-estimation of the number of classes in a Population. Scand. J. Stat. 11, 265–270.
Cornejo, P., Perez-Tienda, J., Meier, S., Valderas, A., Borie, F., Azcon-Aguilar, C., et al. (2013). Copper compartmentalization in spores as a survival strategy of arbuscular mycorrhizal fungi in Cu-polluted environments. Soil. Biol. Biochem. 57, 925–928. doi: 10.1016/j.soilbio.2012.10.031
Critter, S. A. M., Freitas, S. S., and Airoldi, C. (2004). Comparison of microbial activity in some Brazilian soils by microcalorimetric and respirometric methods. Thermochim. Acta 410, 35–46. doi: 10.1016/S0040-6031(03)00371-X
Daims, H., Nielsen, J. L., Nielsen, P. H., Schleifer, K. H., and Wagner, M. (2001). In situ characterization of Nitrospira-like nitrite oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microb. 67, 5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001
Faith, D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Feng, Y. Z., Cui, X. C., He, S. Y., Dong, G., Chen, M., Wang, J. H., et al. (2013). The role of metal nanoparticles in influencing arbuscular mycorrhizal fungi effects on plant growth. Environ. Sci. Technol. 47, 9496–9504. doi: 10.1021/es402109n
Feng, Y. Z., Lin, X. G., Zhu, J. G., and Jia, Z. J. (2011). A phototrophy-driven microbial food web in a rice soil. J. Soil. Sediment. 11, 301–311. doi: 10.1007/s11368-010-0303-6
Ge, Y., Priester, J. H., Van De Werfhorst, L. C., Walker, S. L., Nisbet, R. M., An, Y.-J., et al. (2014). Soybean plants modify metal oxide nanoparticle effects on soil bacterial communities. Environ. Sci. Technol. 48, 13489–13496. doi: 10.1021/es5031646
Ge, Y., Schimel, J. P., and Holden, P. A. (2011). Evidence for negative effects of TiO2 and ZnO nanoparticles on soil bacterial communities. Environ. Sci. Technol. 45, 1659–1664. doi: 10.1021/es103040t
Giovannetti, M., and Mosse, B. (1980). An evaluation of techniques for measuring vesicular-arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Hackl, E., Zechmeister-Boltenstern, S., Bodrossy, L., and Sessitsch, A. (2004). Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl. Environ. Microb. 70, 5057–5065. doi: 10.1128/AEM.70.9.5057-5065.2004
Hassan, S. E. D., Boon, E., St-Arnaud, M., and Hijri, M. (2011). Molecular biodiversity of arbuscular mycorrhizal fungi in trace metal-polluted soils. Mol. Ecol. 20, 3469–3483. doi: 10.1111/j.1365-294X.2011.05142.x
He, S. Y., Feng, Y. Z., Ren, H. X., Zhang, Y., Gu, N., and Lin, X. G. (2011). The impact of iron oxide magnetic nanoparticles on the soil bacterial community. J. Soil. Sediment. 11, 1408–1417. doi: 10.1007/s11368-011-0415-7
Jia, Z. J., and Conrad, R. (2009). Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11, 1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x
Johansen, A., Pedersen, A. L., Jensen, K. A., Karlson, U., Hansen, B. M., Scott-Fordsmand, J. J., et al. (2008). Effects of C(60) fullerene nanoparticles on soil bacteria and protozoans. Environ. Toxicol. Chem. 27, 1895–1903. doi: 10.1897/07-375.1
Kahru, A., Dubourguier, H.-C., Blinova, I., Ivask, A., and Kasemets, K. (2008). Biotests and biosensors for ecotoxicology of metal oxide nanoparticles: a minireview. Sensors 8, 5153–5170. doi: 10.3390/s8085153
Kalyuzhnyi, S., Gladchenko, M., Mulder, A., and Versprille, B. (2006). New anaerobic process of nitrogen removal. Water Sci. Technol. 54, 163–170. doi: 10.2166/wst.2006.729
Kelly, S. A., Havrilla, C. M., Brady, T. C., Abramo, K. H., and Levin, E. D. (1998). Oxidative stress in toxicology: established mammalian and emerging piscine model systems. Environ. Health Persp. 106, 375–384. doi: 10.1289/ehp.98106375
Khodakovskaya, M. V., Kim, B. S., Kim, J. N., Alimohammadi, M., Dervishi, E., Mustafa, T., et al. (2013). Carbon nanotubes as plant growth regulators: effects on tomato growth, reproductive system, and soil microbial community. Small 9, 115–123. doi: 10.1002/smll.201201225
Klaine, S. J., Alvarez, P. J. J., Batley, G. E., Fernandes, T. F., Handy, R. D., Lyon, D. Y., et al. (2008). Nanomaterials in the environment: behavior, fate, bioavailability, and effects. Environ. Toxicol. Chem. 27, 1825–1851. doi: 10.1897/08-090.1
Kumar, N., Shah, V., and Walker, V. K. (2012). Influence of a nanoparticle mixture on an arctic soil community. Environ. Toxicol. Chem. 31, 131–135. doi: 10.1002/etc.721
Labidi, S., Ben Jeddi, F., Tisserant, B., Debiane, D., Rezgui, S., Grandmougin-Ferjani, A., et al. (2012). Role of arbuscular mycorrhizal symbiosis in root mineral uptake under CaCO3 stress. Mycorrhiza 22, 337–345. doi: 10.1007/s00572-011-0405-z
Lee, J., Mahendra, S., and Alvarez, P. J. J. (2010). Nanomaterials in the construction industry: a review of their applications and environmental health and safety considerations. Acs Nano 4, 3580–3590. doi: 10.1021/nn100866w
Li, D., Lyon, D. Y., Li, Q., and Alvarez, P. J. J. (2008). Effect of soil sorption and aquatic natural organic matter on the antibacterial activity of a fullerene water suspension. Environ. Toxicol. Chem. 27, 1888–1894. doi: 10.1897/07-548.1
Li, Y. Y., Chen, L. Q., Wen, H. Y., Zhou, T. Y., Zhang, T., and Gao, X. L. (2014). 454 Pyrosequencing analysis of bacterial diversity revealed by a comparative study of soils from mining subsidence and reclamation areas. J. Microbiol. Biotechn. 24, 313–323. doi: 10.4014/jmb.1309.09001
Lin, X. G., Feng, Y. Z., Zhang, H. Y., Chen, R. R., Wang, J. H., Zhang, J. B., et al. (2012). Long-term balanced fertilization decreases arbuscular mycorrhizal fungal diversity in an arable soil in north china revealed by 454 pyrosequencing. Environ. Sci. Technol. 46, 5764–5771. doi: 10.1021/es3001695
Liu, J. F., Zhao, Z. S., and Jiang, G. B. (2008). Coating Fe3O4 magnetic nanoparticles with humic acid for high efficient removal of heavy metals in water. Environ. Sci. Technol. 42, 6949–6954. doi: 10.1021/es800924c
Luo, Y. Q., Hui, D. F., and Zhang, D. Q. (2006). Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: a meta-analysis. Ecology 87, 53–63. doi: 10.1890/04-1724
Lynch, J. M., and Whipps, J. M. (1990). Substrate flow in the rhizosphere. Plant Soil. 129, 1–10. doi: 10.1007/BF00011685
Martineau, N., McLean, J. E., Dimkpa, C. O., Britt, D. W., and Anderson, A. J. (2014). Components from wheat rootsmodify the bioactivity of ZnO and CuO NPs in a soilbacterium. Environ. Pollut. 187, 65–72. doi: 10.1016/j.envpol.2013.12.022
Martinez, R. J., Wu, C. H., Beazley, M. J., Andersen, G. L., Conrad, M. E., Hazen, T. C., et al. (2014). Microbial community responses to organophosphate substrate additions in contaminated subsurface sediments. PLoS ONE 9:e100383. doi: 10.1371/journal.pone.0100383
Miransari, M. (2010). Contribution of arbuscular mycorrhizal symbiosis to plant growth under different types of soil stress. Plant Biol. 12, 563–569. doi: 10.1111/j.1438-8677.2009.00308.x
Molday, R. S. (1984). Magnetic Iron-Dextran Microspheres. U.S. Patent No. 4,452,773. Canadian Patents and Development Limited.
Nowack, B. (2009). The behavior and effects of nanoparticles in the environment. Environ. Pollut. 157, 1063–1064. doi: 10.1016/j.envpol.2008.12.019
Nowack, B., Ranville, J. F., Diamond, S., Gallego-Urrea, J. A., Metcalfe, C., Rose, J., et al. (2012). Potential scenarios for nanomaterial release and subsequent alteration in the environment. Environ. Toxicol. Chem. 31, 50–59. doi: 10.1002/etc.726
Perez, J. M., O'Loughin, T., Simeone, F. J., Weissleder, R., and Josephson, L. (2002). DNA-based magnetic nanoparticle assembly acts as a magnetic relaxation nanoswitch allowing screening of DNA cleaving agents. J. Am. Chem. Soc. 124, 2856–2857. doi: 10.1021/ja017773n
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Phillips, J. M., and Hayman, D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular arbuscular mycorrhizal fungi for rapid assessment of infection. Transac. British Mycol. Soci. 55, 158–161. doi: 10.1016/S0007-1536(70)80110-3
Prashant, C., Dipak, M., Yang, C. T., Chuang, K. H., Jun, D., and Feng, S. S. (2010). Superparamagnetic iron oxide-Loaded poly (lactic acid)-D-alpha-tocopherol polyethylene glycol 1000 succinate copolymer nanoparticles as MRI contrast agent. Biomaterials 31, 5588–5597. doi: 10.1016/j.biomaterials.2010.03.070
Roco, M. C. (2011). The long view of nanotechnology development: the National Nanotechnology Initiative at 10 years. J. Nanopart. Res. 13, 427–445. doi: 10.1007/s11051-010-0192-z
Schenck, N. C., and Perez, Y. (1990). Manual for Identification of Vesicular Arbuscular Mycorrhizal Fung, 2rd Edn. Gainesville, FL: Synergistic Publication, University of Florida, 286.
Shaw, A. K., Halpern, A. L., Beeson, K., Tran, B., Venter, J. C., and Martiny, J. B. H. (2008). It's all relative: ranking the diversity of aquatic bacterial communities. Environ. Microbiol. 10, 2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x
Sjögren, C. E., Johansson, C., Naevestad, A., Sontum, P. C., BrileySaebo, K., and Fahlvik, A. K. (1997). Crystal size and properties of superparamagnetic iron oxide (SPIO) particles. Magn. Reson. Imaging 15, 55–67. doi: 10.1016/S0730-725X(96)00335-9
Stevenson, L. M., Dickson, H., Klanjscek, T., Keller, A. A., McCauley, E., and Nisbet, R. M. (2013). Environmental feedbacks and engineered nanoparticles: mitigation of silver nanoparticle toxicity to Chlamydo-monas reinhardtii by algal-produced organic compounds. PLoS ONE 8:e74456. doi: 10.1371/journal.pone.0074456
Tong, Z. H., Bischoff, M., Nies, L., Applegate, B., and Turco, R. F. (2007). Impact of fullerene (C60) on a soil microbial community. Environ. Sci. Technol. 41, 2985–2991. doi: 10.1021/es061953l
Wang, C. G., and Irudayaraj, J. (2010). Multifunctional magnetic-optical nanoparticle probes for simultaneous detection, separation, and thermal ablation of multiple pathogens. Small 6, 283–289. doi: 10.1002/smll.200901596
Wang, W. Z., Wang, F. Y., Li, S., and Liu, X. Q. (2014). Arbuscular mycorrhizal symbiosis influences the biological effects of nano-ZnO on Maize. Environ. Sci. 35, 3135–3141.
Wu, F. Y., Yu, X. Z., Wu, S. C., Lin, X. G., and Wong, M. H. (2011). Phenanthrene and pyrene uptake by arbuscular mycorrhizal maize and their dissipation in soil. J. Hazard. Mater. 187, 341–347. doi: 10.1016/j.jhazmat.2011.01.024
Yamada, T., and Sekiguchi, Y. (2009). Cultivation of uncultured chloroflexi subphyla: significance and ecophysiology of formerly uncultured chloroflexi ‘Subphylum I’ with natural and biotechnological relevance. Microbes. Environ. 24, 205–216. doi: 10.1264/jsme2.ME09151S
Zarea, M. J., Ghalavand, A., Mohammadi Goltapeh, E., Rejali, F., and Zamaniyan, M. (2009). Effects of mixed cropping, earthworms (Pheretima sp.), and arbuscular mycorrhizal fungi (Glomus mosseae) on plant yield, mycorrhizal colonization rate, soil microbial biomass, and nitrogenase activity of free-living rhizosphere bacteria. Pedobiologia 52, 223–235. doi: 10.1016/j.pedobi.2008.10.004
Keywords: iron oxide nanoparticles, arbuscular mycorrhizal fungi, soil biota, microcalorimetry, pyrosequencing
Citation: Cao J, Feng Y, Lin X and Wang J (2016) Arbuscular Mycorrhizal Fungi Alleviate the Negative Effects of Iron Oxide Nanoparticles on Bacterial Community in Rhizospheric Soils. Front. Environ. Sci. 4:10. doi: 10.3389/fenvs.2016.00010
Received: 09 September 2015; Accepted: 01 February 2016;
Published: 18 February 2016.
Edited by:
Rene Kizek, Central European Institute of Technology in Brno, Czech RepublicReviewed by:
Yutao Wang, South China Normal University, ChinaMiguel Angel Merlos, Brno University of Technology, Czech Republic
Copyright © 2016 Cao, Feng, Lin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangui Lin, xglin@issas.ac.cn
 Jiling Cao
Jiling Cao Youzhi Feng
Youzhi Feng Xiangui Lin1*
Xiangui Lin1*