- 1Laboratory of Translational Immunology, University Medical Center Utrecht, Utrecht, Netherlands
- 2Department of Cardiology, University Medical Center Utrecht, Utrecht, Netherlands
- 3Department of Respiratory Medicine, University Medical Center Utrecht, Utrecht, Netherlands
- 4Center of Interstitial Lung Diseases, St. Antonius Hospital, Nieuwegein, Netherlands
- 5Department of Rheumatology, University Medical Center Utrecht, Utrecht, Netherlands
- 6Department of Dermatology, University Medical Center Utrecht, Utrecht, Netherlands
Lung transplantation (LTx) outcome is hampered by development of chronic rejection, often manifested as the bronchiolitis obliterans syndrome (BOS). Low serum levels of thymus and activation-regulated chemokine (TARC/CCL17), a chemoattractant, measured during the first month post-LTx are predictive for BOS development. Since TARC/CCL17 promotor polymorphisms correlate with serum TARC/CCL17 levels, we investigated seven single-nucleotide polymorphisms (SNPs) within this region and their potential association with LTx outcome. We analyzed donor and patient SNP configurations and haplotypes and observed a trend between a donor SNP (rs223899) configuration and patient TARC/CCL17 serum levels post-LTx (p = 0.066). Interestingly, this SNP configuration in patients did not show any correlation with pre-LTx TARC/CCL17 serum levels (p = 0.776). Survival analysis showed that receiving a graft from a donor heterozygous for rs223899 has a disadvantageous impact on transplantation outcome. When stratified per donor SNP genotype, patients receiving a transplant from a heterozygous donor showed a lower BOS-free survival (p = 0.023) and survival rate (p = 0.0079). Since rs223899 is located within a NFκB binding site, heterozygosity at this position could result in a reduced TARC/CCL17 expression. Our data indicate that a single TARC/CCL17 promotor SNP in the donor correlates with lower serum TARC/CCL17 levels measured 1 month after LTx and affects clinical outcome after LTx.
Introduction
For patients suffering from end-stage lung disease, lung transplantation (LTx) can be the final treatment modality. Currently, 5-year survival after LTx is 50%, predominantly due to the development of chronic lung allograft dysfunction (CLAD) (1). CLAD can present an obstructive (bronchiolitis obliterans syndrome, BOS) and a restrictive form (restrictive allograft syndrome, RAS) (2). CLAD pathogenesis is poorly understood; however, various donor and patient risk factors associated with disease development have been identified, particularly regarding development of BOS (3, 4). A clinical diagnosis of BOS is often made using a 20% decline of the forced expiratory volume in 1 s compared to baseline in the absence of any other disease etiology (5). Thus, a clinical diagnosis is made at the time that obliterative bronchiolitis has fully developed. To prevent BOS, novel biomarkers reflecting preclinical development identifying patients at risk early after transplantation are urgently needed (6).
Thymus and activation-regulated chemokine (TARC/CCL17) is a chemoattractant, which is secreted by various cell types, including endothelial cells, dendritic cells, keratinocytes, bronchial epithelial cells, and fibroblasts (7–10). It mainly functions as a chemoattractant for Th2 cells via the interaction with its receptor CCR4 (11, 12). TARC/CCL17 serum levels are associated with various types of lung diseases including idiopathic pulmonary fibrosis (13) and eosinophilic pneumonia (14), and as risk marker for lung cancer (15). Interestingly, previous results from our group have shown that serum levels of TARC/CCL17 in the first month post-transplantation are predictive for BOS development after LTx (16).
The TARC/CCL17 gene is located on chromosome 16q13, in near proximity of the CCR4 interacting chemokine CCL22 and CX3CL1 (17). TARC/CCL17 expression is controlled by multiple pro-inflammatory cytokines, including tumor necrosis factor-α, interferon (IFN)-γ, interleukin (IL)-1, and IL-4 (18). The transcriptional regulation of the TARC/CCL17 gene has partly been elucidated. Both the transcription factors STAT6 and NFκB have binding sites in the promotor region of TARC/CCL17 (18, 19). Several single-nucleotide polymorphisms (SNPs) in the TARC/CCL17 promotor region correlate with serum levels of TARC/CCL17 and are associated with a risk for Kawasaki disease and different allergic diseases (20–22).
As low early post-transplantation serum levels of TARC/CCL17 predict a risk for post-LTx BOS, we hypothesized that TARC/CCL17 polymorphisms may be correlated to outcome after LTx. In this study, we genotyped and analyzed several SNPs in the TARC promotor region of patients undergoing LTx as well as in that of the donor. We show that a single donor SNP configuration in the promotor region of TARC/CC17 of the donor correlates with recipient TARC/CCL17 serum levels and relates to BOS development and overall survival after LTx.
Patients and Methods
Patients
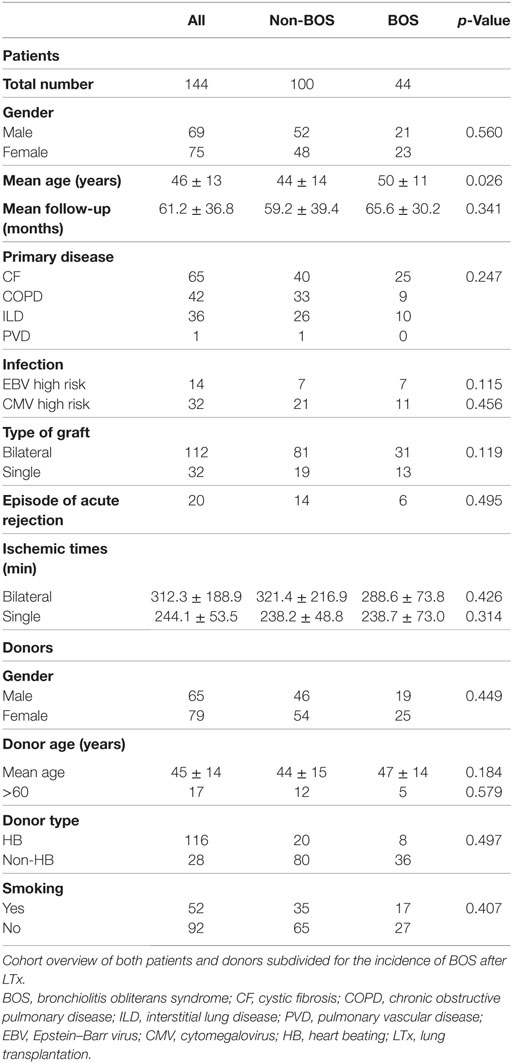
A total of 144 patients undergoing LTx between January 2004 and March 2013 in the Heart Lung Center of the University Medical Center Utrecht, The Netherlands, were included in this retrospective study. Written informed consent was obtained from all study participants, and the study was approved by the medical ethical committee of the University Medical Center Utrecht (METC 06-144), and all methods were carried out in accordance with the approved guidelines. Post-transplantation follow-up therapy was standardized and consisted of tacrolimus, prednisolone, and mofetil mycophenolate. In the first year after transplantation, spirometry was performed every week during the first 3 months reducing it to every 4 weeks after a year. Lung volumes were routinely assessed every 6 months and on indication when spirometry or X-ray changed. Patients at high risk for cytomegalovirus (CMV) or Epstein–Barr virus (EBV) activation, i.e., CMV- or EBV-negative patients transplanted with a EBV- or CMV-positive donor, were treated with valganciclovir for 6 months after transplantation. A clinical diagnosis of BOS was made when FEV1 had declined by 20% of more compared to baseline (5). Since surveillance biopsies were not performed, acute rejection (AR) was defined as a spontaneous decline of lung function that was reversed after steroid pulse treatment and for which other causes of lung function decline were excluded.
Prior to transplantation, blood was obtained from donor and patient, as well as a spleen samples from the donor. Mononuclear cells from patient and donor samples were isolated using Ficoll-paque (GE Healthcare, Little Chalfont, UK), which were then aliquoted and stored in liquid nitrogen until further use. In addition, serum from the patient was collected and stored at −80°C.
DNA Extraction
Frozen mononuclear cells were used for DNA isolation via the MagnaPure Compact System (Roche Diagnostics, Switzerland) according to protocol. Cell samples were thawed at 37°C, dissolved in 9 ml RPMI-1640 (Lonza, Basel, Switzerland) supplemented with 20%, v/v, fetal bovine serum (Bodinco, Alkmaar, The Netherlands), and centrifuged for 10 min at 1,800 RPM. Prior to DNA extraction, cells ware dissolved in phosphate-buffered saline (Sigma-Aldrich, USA) at a concentration of 5 × 106 cells/ml. After DNA extraction, both concentration and purity were analyzed using the NanoDrop™ system (Thermo Fischer Scientific, Waltham, MA, USA).
SNP Selection and Genotyping
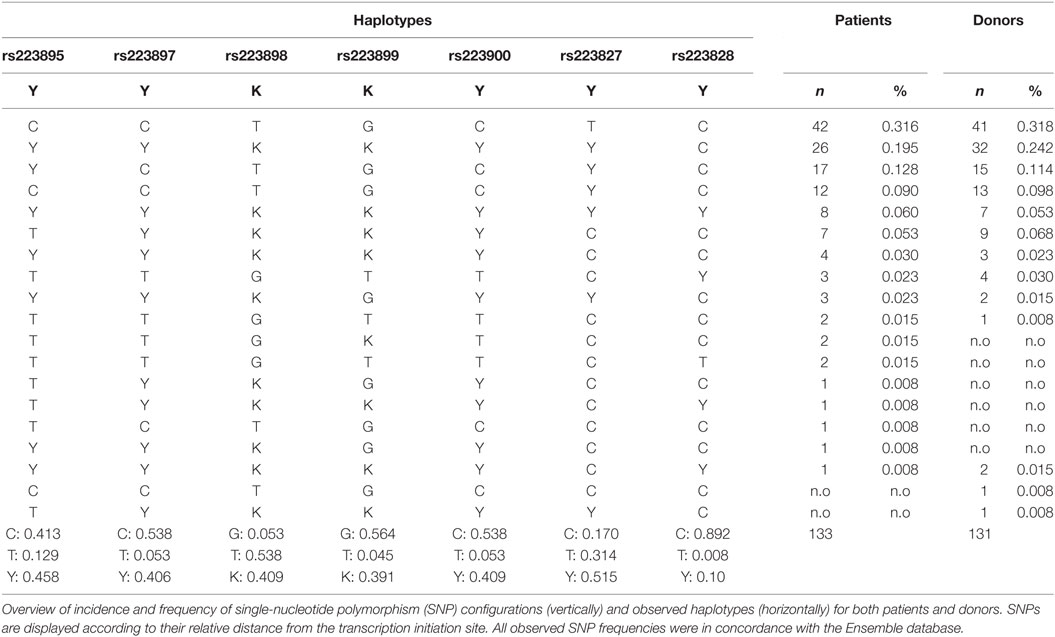
Six SNPs (rs223895, rs223897, rs223898, rs223899, rs223900, and rs229827) in the promotor region of TARC/CCL17 that are frequent in the western European population were selected from the HapMap (http://hapmap.ncbi.nlm.nih.gov/) and the Ensemble databases (23). We also analyzed the configuration of rs229828, which configuration has previously been associated with CCCL17/TARC serum levels (20). Samples were genotyped using the Affymetrix “TxArray” (24, 25) containing 767,203 variants, and stringent quality control (QC) was conducted to remove low-quality SNPs and samples. Samples with a missing rate >3% were removed. We created a subset of high-quality, independent SNPs with missing rate <1%, Hardy–Weinberg p > 0.001, minor allele frequency >0.1, and LD pruning leaving no SNP pairs with r2 > 0.2. Using this subset, we removed samples with heterozygosity >2 SD from the mean of all samples, related samples (keeping only one samples of each pair with proportion of IBD > 0.2), and samples of non-European ancestry [based on principle component analysis using the 1000 Genomes Project (Phase 1) populations as reference (26)]. SNPs were removed if they had a missing rate >5%, Hardy–Weinberg p < 0.01, or if they were monomorphic. After QC, 543,637 SNPs and 133 patients and 131 donor samples remained. Untyped SNPs were imputed using a combined reference panel of the 1000 Genomes Project (Phase 3) (27) and the Genomes of the Netherlands (v5) (28). Samples were first phased with SHAPEIT (29) and then imputed with IMPUTE v2 (30).
Measurement of Serum TARC/CCL17 Concentrations
Serum concentrations of TARC/CCL17 were determined via a solid-phase ELISA kit (R&D systems, Minneapolis, MN, USA) according to protocol. Briefly, wells were first incubated with serum samples for 2 h, then with conjugate for 1 h, and finally with substrate. From OD450 values, levels were calculated by reference to a standard curve. Serum samples were briefly centrifuged prior to analysis. All samples where measured in duplicate. Inter- and intra-assay variability’s of the assay were 8.3 and 4.4%, respectively.
Statistics
All statistical analyses were performed using GraphPad Prism version 6.02 (GraphPad Software Inc., San Diego, CA, USA) and SPSS version 21 (IBM Corp., Armonk, NY, USA). Data were tested for Gaussian distribution via the D’Agostino and Pearson omnibus normality test. Normally distributed data are represented as mean value ± SEM whereas data not following a Gaussian distribution are represented as median ± interquartile range. Depending on the distribution of the data, differences between groups were analyzed with the unpaired t test or the Mann–Whitney test, indicated in the respective figure legend. Differences in categorical data were analyzed using the Fischer’s exact test and in continuous variables via ANOVA. Survival analyses were conducted using Kaplan–Meier analysis with both BOS incidence and overall survival as endpoint parameters. A Cox-regression model was used for multivariate analysis including known risk factors in patients and donors for BOS development. A p-value <0.05 was considered to be statistical significant.
Results
Patient Demographics
From the total cohort of 144 patients transplanted in our center, 65 were treated with LTx because of chronic obstructive pulmonary disease, 42 because of cystic fibrosis, 36 because of interstitial lung disease, and 1 patient was diagnosed with pulmonary vascular disease prior to transplantation. Besides the fact that BOS + patients were slightly older at the time of transplantation, no significant demographic and clinical differences were observed between BOS+ and BOS− groups (Table 1). During transplantation follow-up, 44 patients developed BOS. No RAS was observed. In total, 44 patients deceased during the study period, whereas 20 patients presented with one or more AR episodes.
TARC/CCL17 Promotor Polymorphisms
All extracted DNA samples from patient/donor couples were analyzed on the Affymetrix-based TxArray and selected SNPs were imputed as described in Section “Patients and Methods.” After stringent pre- and post-imputation QC, including deviation from Hardy–Weinberg equilibrium, sample and SNP missingness, heterozygosity checks, and principle component analyses (data not shown) (31), 133 patients and 131 donor could be genotyped for the selected TARC/CCL17 promotor SNPs (92.4 and 91.7% of the total cohort, respectively). From all samples identified, SNP genotypes were stratified per haplotype. Table 2 describes these results as well as haplotype and genotype frequencies of the individual SNPs. We observed no significant differences in either SNP or haplotype distribution between patients and donors. Also, the genotype frequencies of the selected TARC/CCL17 SNPs were in concordance with frequencies found in the HapMap and the Ensemble databases.
rs223899 Influences Serum TARC/CCL17 Concentrations Post-Transplantation
From a subset of 67 representative patients (no differences on clinical and demographic parameters compared to the total cohort), serum samples obtained during the first month after transplantation were analyzed for TARC/CCL17 levels, based on serum sample availability. The relation between both donor and patient haplotypes and SNP genotypes, and serum TARC/CCL17 levels were then analyzed. The strongest association was observed between donor SNP rs223899 and serum levels. Serum TARC/CCL17 levels in patients with lungs from donors with the homozygous (G/G) SNP configuration of rs223899, tended to be higher than those in patients with a lung from a donor with heterozygous (G/T) configuration (p = 0.066, Figure 1A). Notably, serum TARC/CCL17 levels before transplantation were not different in these patient groups (n = 38, p = 0.776, Figure 1B).
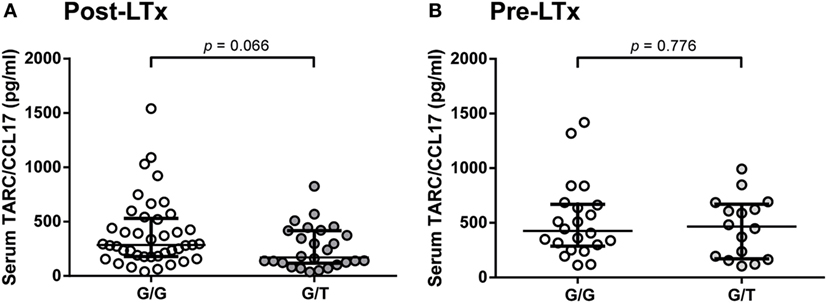
Figure 1. Donor rs223899 correlates with serum thymus and activation-regulated chemokine (TARC/CCL17) concentrations post-lung transplantation (LTx) LTx but not pre-LTx. (A) Serum TARC/CCL17 levels were measured in LTx patients 1 month after LTx stratified for the configuration of donor single-nucleotide polymorphism (SNP) rs223899 (homozygous, G/G, 286.0 pg/ml vs. heterozygous, G/T 170.5 pg/ml). A trend toward significance (p = 0.066) was observed with increased TARC/CCL17 serum concentrations in patients who received a graft genotyped homozygous for SNP position rs223899. Non-Gaussian distribution, median ± interquartile range, Mann–Whitney test, homozygous n = 41, heterozygous n = 26. (B) Serum TARC/CCL17 levels were also assessed pre-LTx in patients based on serum availability. No differences in serum TARC/CCL17 could be observed when patients were stratified for the respective rs223899 SNP genotype. Non-Gaussian distribution, median ± interquartile range, Mann–Whitney test, homozygous n = 22, heterozygous n = 16.
Donor SNP rs223899 Influences the Clinical Outcome after LTx
Since we observed that patient serum TARC/CCL17 levels correlate with the configuration of donor SNP rs223899, and decreased serum levels of TARC/CCL17 predict a higher risk for BOS development after transplantation (16), we analyzed the genotyped donor haplotypes and individual donor SNP configurations in a Kaplan–Meier survival analysis. For overall survival, all 131 patients for which the imputed donor SNP passed QC were included. For the analyses of BOS development, we excluded patients who had deceased within the first 4 months after transplantation or from whom SNP analysis did not pass QC, resulting in the inclusion of 122 patients.
In total, six different donor haplotypes had a frequency above 5% and were analyzed for correlation to outcome after LTx. None of the donor or patient haplotypes of the seven selected TARC/CCL17 SNPs showed a correlation with either AR episodes, BOS incidence, or survival after LTx (data not shown). In contrast, we observed a significant difference in the development of chronic rejection when patients were stratified by donor SNP rs223899 genotype. Of the patients who received a transplant from a heterozygous (G/T) donor at position rs223899, 50% remained free from BOS within the first 100 months after transplantation. This percentage was significantly higher, 75%, in the patients who had received a graft from a homozygous (G/G) donor (p = 0.023, Figure 2A). This was confirmed in a multivariate analysis using a Cox proportional-hazards model for BOS development, which included both donor and patient risk factors for BOS development, and designated the risk variant of rs223899 as a significant predictor for the development of BOS after LTx (p = 0.018, Table 3). Also, this nucleotide substitution in the promotor region of TARC/CCL17 in the donor correlated with a lower survival rate of recipients’ post-transplantation (50 vs. 80%, respectively, p = 0.0079, Figure 2B). None of the other individual donor SNPs correlated with BOS development or survival after LTx.
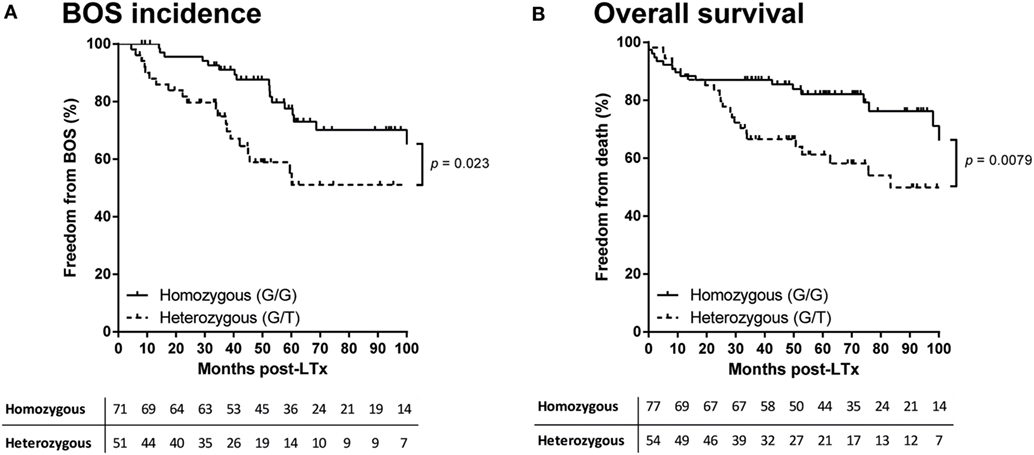
Figure 2. Donor rs223899 affects clinical outcome after lung transplantation (LTx). (A) Kaplan–Meier analysis on bronchiolitis obliterans syndrome (BOS) incidence after LTx. Patients were stratified according to the single-nucleotide polymorphism (SNP) configuration of rs223899 in the received allograft. Patients who received a graft genotyped as heterozygous (G/T) for this specific SNP have a lower BOS-free survival rate measured over the first 100 months after transplantation (p = 0.023). Lower table represents numbers at risk. All 131 patients for which the imputed donor SNP passed quality control (QC) were included. (B) Kaplan–Meier analysis on survival after LTx. Patients were stratified as mentioned earlier. Additional to an increase of chronic rejection after LTx, stratification of LTx patients for receiving a grafted organ genotyped heterozygous (G/T) at SNP position rs223899 resulted in a lower survival rate post-LTx (p = 0.0079). Lower table represents numbers at risk. Patients who had deceased within the first 4 months after transplantation or from whom SNP analysis did not pass QC were excluded, resulting in the inclusion of 122 patients. Log-rank test used in both analyses.
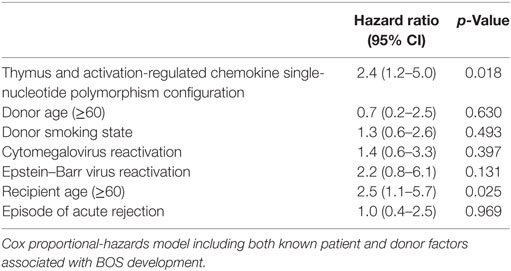
Table 3. Multivariate analysis on bronchiolitis obliterans syndrome (BOS) incidence in patients treated with lung transplantation.
Discussion
In this study, we analyzed the relation of both donor and patient TARC/CCL17 genotypes to clinical parameters, but only found one single donor SNP to be correlated with BOS development. Interestingly, this specific promotor SNP configuration, heterozygosity for rs223899, also correlated with lower serum TARC/CCL17 levels, which is in concordance with previous observations that low serum TARC/CCL17 levels in the first month after transplantation are predictive for BOS development (16). We have also assessed patient–donor combinations stratified by rs223899 genotype. We observed higher concentrations of serum TARC/CCL17 in the first month after LTx when patients heterozygous for SNP rs223899 were transplanted with a homozygous donor. This was not observed in the other three combinations (patient/donor homozygous, patient/donor heterozygous, and patient homozygous/donor heterozygous).
We only analyzed SNPs that are frequent in the European population. Therefore, validation in an external cohort, and re-analysis of our findings in populations with other genetic backgrounds is expedient. In this cohort, BOS was diagnosed according to international guidelines. However, surveillance bronchoscopy and transbronchial lung biopsies are not performed, which is a limitation of this cohort. Unresolved infections or undiagnosed episodes of AR could be a possibility of persistent lung function decline. Also, due to cohort constraints, we were not able to discriminate between early and late BOS onset and potential TARC/CCL17 SNP configurations.
Since TARC/CCL17 serum levels measured at month 1 post-LTx are increased and a predictor for BOS development (16), we investigated these serum levels in relation to the donor SNP configuration of rs223899. Unfortunately, bronchoalveolar lavage samples were not available. TARC/CCL17 expression is controlled by various pro-inflammatory cytokines. The insignificant trend of lower serum TARC/CCL17 levels observed could be because of the immunosuppressive treatment which could potentially influence cytokine production and regulation of TARC/CCL17 production. Also, infections with community-acquired respiratory viruses could impact expression. However, we observed no differences in treatment regimen or the incidence of community-acquired respiratory viral infections in patients stratified for donor SNP genotype (Table S1 in Supplementary Material).
Most studies on the role of genetics in lung transplant complications have focused on the obstructive form of CLAD, BOS (32). These results are mainly obtained using patient DNA, illustrated by studies of Awad et al. concerning SNPs in IFN-γ and TGF-β1, in which the authors correlate gene polymorphisms with increased allograft fibrosis (33, 34). Also, an association between an IL-6 polymorphism and BOS development was observed (35). However, these findings could not be validated in independent cohorts (36). Recently, our group has shown that a SNP in the promotor region of complement regulatory protein CD59 in the donor correlates with a higher risk for chronic rejection after LTx (37). Furthermore, a specific donor MBL promotor haplotype has been associated with graft survival and BOS development after transplantation (38). Taken together, these data stress the potential importance of both patient and donor SNPs on the clinical outcome after LTx.
The correlation between rs223828, another TARC/CCL17 promotor polymorphism, and protein serum levels has been described previously in a cohort of Japanese patients (10). This polymorphism was also found to be associated with atopy and asthma in children, as well as with higher circulating levels of TARC/CCL17 (21). We could not confirm the correlation of this SNP with serum TARC/CCL17 levels in our cohort of western European LTx patients, presumably due to low minor allele frequency in our patient cohort. Observations in patients suffering from Kawasaki disease have shown that rs223899 is associated with disease progression. However, individual SNPs, including rs223899, did not correlate with serum levels when stratified by genotype (22).
The genetic regulation of the TARC/CCL17 gene has partly been elucidated. Two STAT6 binding sites have been identified at position -213/-223 and -177/-187 relatively to ATG upstream of Exon 1. Furthermore, a binding-motif for NFκB is present upstream of the two STAT6 binding sites (18). Interestingly, using a RSV-inducible mice epithelial cell model, Monick et al. have shown that optimal TARC expression is achieved via the combined activation of both transcription factors, which would involve the recruitment of CREB-binding protein/p300 via NFκB and is essential for STAT-mediated transcription (19). The identified SNP rs223899 lies within consensus binding sequence for NFκB (39). Thus, the heterozygous configuration of rs223899 could result in a less optimal NFκB binding, which would lead to a reduced expression of TARC/CCL17.
The role of serum levels of TARC/CCL17 in LTx outcome remains speculative. Bronchial epithelial cells have the potency to secrete large amounts of TARC/CCL17 when activated (7) and considering the small size of TARC/CCL17 (10.5 kDa), it seems logical to assume leakage from the allograft into the circulation that can subsequently be quantified in serum. Immunoregulatory functions have been attributed to TARC/CCL17 due to the presence of its receptor, CCR4, on a specific subset of regulatory T cells (40). A reduced secretion of TARC/CCL17 could lead to a diminished influx of regulatory T cells, which would result in less regulation of the overall immune response associated with transplant rejection (41, 42). Additional experiments are expedient to support this hypothesis.
In summary, our data indicate that heterozygosity for a single SNP in the promotor region of TARC/CCL17 located within the consensus sequence of the binding site of transcription factor NFκB correlates with serum levels of the TARC/CCL17 protein. Low serum TARC/CCL17 levels are predictive for BOS development following LTx. In line with these observations, we show that patients who receive a heterozygous allograft for SNP rs223899 present with a higher BOS incidence and impaired survival after LTx.
Ethics Statement
Informed consent was obtained from all study participants, and the study was approved by the medical ethical committee of the University Medical Center Utrecht (METC 06-144), and all methods were carried out in accordance with the approved guidelines.
Author Contributions
KB, JS, TK-H, and OR performed the research; KB, JS, EG, OR, CH, and HO participated in data analysis; EG and E-JO contributed patient material; EG and HO participated in research design; KB, JS, EG, CH, and HO wrote the paper. All the authors provided final approval of the version to be published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the technicians of the diagnostic department of the Laboratory for Translational Immunology, UMC Utrecht, The Netherlands, for conducting the TARC/CCL17 serum quantification experiments.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.01109/full#supplementary-material.
Abbreviations
AR, acute rejection; BOS, bronchiolitis obliterans syndrome; CF, cystic fibrosis; CLAD, chronic lung allograft dysfunction; COPD, chronic obstructive pulmonary disease; ILD, interstitial lung disease; LTx, lung transplantation; PVD, pulmonary vascular disease; RAS, restrictive allograft syndrome; SNP, single-nucleotide polymorphism; TARC, thymus and activation-regulated chemokine.
References
1. Yusen RD, Edwards LB, Kucheryavaya AY, Benden C, Dipchand AI, Dobbels F, et al. The registry of the International Society for Heart and Lung Transplantation: thirty-first adult lung and heart-lung transplant report – 2014; focus theme: retransplantation. J Heart Lung Transplant (2014) 33(10):1009–24. doi:10.1016/j.healun.2014.08.004
2. Verleden GM, Raghu G, Meyer KC, Glanville AR, Corris P. A new classification system for chronic lung allograft dysfunction. J Heart Lung Transplant (2014) 33(2):127–33. doi:10.1016/j.healun.2014.01.316
3. Hennessy SA, Hranjec T, Swenson BR, Kozower BD, Jones DR, Ailawadi G, et al. Donor factors are associated with bronchiolitis obliterans syndrome after lung transplantation. Ann Thorac Surg (2010) 89(5):1555–62. doi:10.1016/j.athoracsur.2010.01.060
4. Todd JL, Palmer SM. Bronchiolitis obliterans syndrome: the final frontier for lung transplantation. Chest (2011) 140(2):502–8. doi:10.1378/chest.10-2838
5. Estenne M, Maurer JR, Boehler A, Egan JJ, Frost A, Hertz M, et al. Bronchiolitis obliterans syndrome 2001: an update of the diagnostic criteria. J Heart Lung Transplant (2002) 21(3):297–310. doi:10.1016/S1053-2498(02)00398-4
6. Berastegui C, Román J, Monforte V, Bravo C, López-Meseguer M, Montero MÁ, et al. Biomarkers of pulmonary rejection. Transplant Proc (2013) 45(9):3163–9. doi:10.1016/j.transproceed.2013.06.013
7. Berin MC, Eckmann L, Broide DH, Kagnoff MF. Regulated production of the T helper 2-type T-cell chemoattractant TARC by human bronchial epithelial cells in vitro and in human lung xenografts. Am J Respir Cell Mol Biol (2001) 24(4):382–9. doi:10.1165/ajrcmb.24.4.4360
8. Fujisawa T, Fujisawa R, Kato Y, Nakayama T, Morita A, Katsumata H, et al. Presence of high contents of thymus and activation-regulated chemokine in platelets and elevated plasma levels of thymus and activation-regulated chemokine and macrophage-derived chemokine in patients with atopic dermatitis. J Allergy Clin Immunol (2002) 110(1):139–46. doi:10.1067/mai.2002.126079
9. Panina-Bordignon P, Papi A, Mariani M, Di Lucia P, Casoni G, Bellettato C, et al. The C-C chemokine receptors CCR4 and CCR8 identify airway T cells of allergen-challenged atopic asthmatics. J Clin Invest (2001) 107(11):1357–64. doi:10.1172/JCI12655
10. Sekiya T, Miyamasu M, Imanishi M, Yamada H, Nakajima T, Yamaguchi M, et al. Inducible expression of a Th2-type CC chemokine thymus- and activation-regulated chemokine by human bronchial epithelial cells. J Immunol (2000) 165(4):2205–13. doi:10.4049/jimmunol.165.4.2205
11. Bonecchi R, Bianchi G, Bordignon PP, D’Ambrosio D, Lang R, Borsatti A, et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med (1998) 187(1):129–34. doi:10.1084/jem.187.1.129
12. Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem (1997) 272(23):15036–42. doi:10.1074/jbc.272.23.15036
13. Kawashima T, Tada Y, Asano Y, Yazawa N, Tomita M, Tamaki Z, et al. Serum TARC/CCL17 levels are increased in dermatomyositis associated with interstitial lung disease. J Dermatol Sci (2010) 60(1):52–4. doi:10.1016/j.jdermsci.2010.07.012
14. Miyazaki E, Nureki S, Fukami T, Shigenaga T, Ando M, Ito K, et al. Elevated levels of thymus- and activation-regulated chemokine in bronchoalveolar lavage fluid from patients with eosinophilic pneumonia. Am J Respir Crit Care Med (2002) 165(8):1125–31. doi:10.1164/ajrccm.165.8.2106110
15. Shiels MS, Pfeiffer RM, Hildesheim A, Engels EA, Kemp TJ, Park JH, et al. Circulating inflammation markers and prospective risk for lung cancer. J Natl Cancer Inst (2013) 105(24):1871–80. doi:10.1093/jnci/djt309
16. Paantjens AW, Kwakkel-van Erp JM, van Ginkel WG, van Kessel DA, van den Bosch JM, van de Graaf EA, et al. Serum thymus and activation regulated chemokine levels post-lung transplantation as a predictor for the bronchiolitis obliterans syndrome. Clin Exp Immunol (2008) 154(2):202–8. doi:10.1111/j.1365-2249.2008.03764.x
17. Nomiyama H, Imai T, Kusuda J, Miura R, Callen DF, Yoshie O. Assignment of the human CC chemokine gene TARC (SCYA17) to chromosome 16q13. Genomics (1997) 40(1):211–3. doi:10.1006/geno.1996.4552
18. Wirnsberger G, Hebenstreit D, Posselt G, Horejs-Hoeck J, Duschl A. IL-4 induces expression of TARC/CCL17 via two STAT6 binding sites. Eur J Immunol (2006) 36(7):1882–91. doi:10.1002/eji.200635972
19. Monick MM, Powers LS, Hassan I, Groskreutz D, Yarovinsky TO, Barrett CW, et al. Respiratory syncytial virus synergizes with Th2 cytokines to induce optimal levels of TARC/CCL17. J Immunol (2007) 179(3):1648–58. doi:10.4049/jimmunol.179.3.1648
20. Sekiya T, Tsunemi Y, Miyamasu M, Ohta K, Morita A, Saeki H, et al. Variations in the human Th2-specific chemokine TARC gene. Immunogenetics (2003) 54(10):742–5. doi:10.1007/s00251-002-0520-2
21. Leung TF, Tang NL, Li CY, Lam CW, Wong GW, Fok TF. Association between TARC C-431T and atopy and asthma in children. J Allergy Clin Immunol (2004) 114(1):199–202. doi:10.1016/j.jaci.2004.03.048
22. Lee CP, Huang YH, Hsu YW, Yang KD, Chien HC, Yu HR, et al. TARC/CCL17 gene polymorphisms and expression associated with susceptibility and coronary artery aneurysm formation in Kawasaki disease. Pediatr Res (2013) 74(5):545–51. doi:10.1038/pr.2013.134
23. Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2014. Nucleic Acids Res (2014) 42(Database issue):D749–55. doi:10.1093/nar/gkt1196
24. International Genetics & Translational Research in Transplantation Network (iGeneTRAiN). Design and implementation of the international genetics and translational research in transplantation network. Transplantation (2015) 99:2401–12. doi:10.1097/TP.0000000000000913
25. Li YR, van Setten J, Verma SS, Lu Y, Holmes MV, Gao H, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Med (2015) 7(1):90. doi:10.1186/s13073-015-0211-x
26. Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature (2012) 491(7422):56–65. doi:10.1038/nature11632
27. 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature (2015) 526(7571):68–74. doi:10.1038/nature15393
28. Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet (2014) 46(8):818–25. doi:10.1038/ng.3021
29. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods (2011) 9(2):179–81. doi:10.1038/nmeth.1785
30. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet (2007) 39(7):906–13. doi:10.1038/ng2088
31. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc (2010) 5(9):1564–73. doi:10.1038/nprot.2010.116
32. Ruttens D, Vandermeulen E, Verleden SE, Bellon H, Vos R, Van Raemdonck DE, et al. Role of genetics in lung transplant complications. Ann Med (2015) 47(2):106–15. doi:10.3109/07853890.2015.1004359
33. Awad M, Pravica V, Perrey C, El-Gamel A, Yonan N, Sinnott PJ, et al. CA repeat allele polymorphism in the first intron of the human interferon-gamma gene is associated with lung allograft fibrosis. Hum Immunol (1999) 60(4):343–6. doi:10.1016/S0198-8859(98)00133-5
34. El-Gamel A, Awad MR, Hasleton PS, Yonan NA, Hutchinson JA, Campbell CS, et al. Transforming growth factor-beta (TGF-beta1) genotype and lung allograft fibrosis. J Heart Lung Transplant (1999) 18(6):517–23. doi:10.1016/S1053-2498(98)00024-2
35. Lu KC, Jaramillo A, Lecha RL, Schuessler RB, Aloush A, Trulock EP, et al. Interleukin-6 and interferon-gamma gene polymorphisms in the development of bronchiolitis obliterans syndrome after lung transplantation. Transplantation (2002) 74(9):1297–302. doi:10.1097/00007890-200211150-00017
36. Snyder LD, Hartwig MG, Ganous T, Davis RD, Herczyk WF, Reinsmoen NL, et al. Cytokine gene polymorphisms are not associated with bronchiolitis obliterans syndrome or survival after lung transplant. J Heart Lung Transplant (2006) 25(11):1330–5. doi:10.1016/j.healun.2006.07.001
37. Budding K, van de Graaf EA, Kardol-Hoefnagel T, Broen JC, Kwakkel-van Erp JM, Oudijk EJ, et al. A promoter polymorphism in the CD59 complement regulatory protein gene in donor lungs correlates with a higher risk for chronic rejection after lung transplantation. Am J Transplant (2016) 16:987–98. doi:10.1111/ajt.13497
38. Munster JM, van der Bij W, Breukink MB, van der Steege G, Zuurman MW, Hepkema BG, et al. Association between donor MBL promoter haplotype and graft survival and the development of BOS after lung transplantation. Transplantation (2008) 86(12):1857–63. doi:10.1097/TP.0b013e31819064b8
39. Huang SP, Lin VC, Lee YC, Yu CC, Huang CY, Chang TY, et al. Genetic variants in nuclear factor-kappa B binding sites are associated with clinical outcomes in prostate cancer patients. Eur J Cancer (2013) 49(17):3729–37. doi:10.1016/j.ejca.2013.07.012
40. Iellem A, Mariani M, Lang R, Recalde H, Panina-Bordignon P, Sinigaglia F, et al. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J Exp Med (2001) 194(6):847–53. doi:10.1084/jem.194.6.847
41. Karczewski J, Karczewski M, Glyda M, Wiktorowicz K. Role of TH1/TH2 cytokines in kidney allograft rejection. Transplant Proc (2008) 40(10):3390–2. doi:10.1016/j.transproceed.2008.07.125
Keywords: lung transplantation, thymus and activation-regulated chemokine, chronic lung allograft dysfunction, bronchiolitis obliterans syndrome, chronic rejection
Citation: Budding K, van Setten J, van de Graaf EA, van Rossum OA, Kardol-Hoefnagel T, Oudijk E-JD, Hack CE and Otten HG (2017) Association between a Single Donor TARC/CCL17 Promotor Polymorphism and Obstructive Chronic Lung Allograft Dysfunction after Lung Transplantation. Front. Immunol. 8:1109. doi: 10.3389/fimmu.2017.01109
Received: 15 February 2017; Accepted: 23 August 2017;
Published: 06 September 2017
Edited by:
Gianfranco Pittari, Hamad Medical Corporation, QatarReviewed by:
Ralf Dressel, Universitätsmedizin Göttingen, GermanyJackson Y. W. Wong, Peak Pulmonary Specialty Clinic, Canada
Copyright: © 2017 Budding, van Setten, van de Graaf, van Rossum, Kardol-Hoefnagel, Oudijk, Hack and Otten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin Budding, ay5idWRkaW5nQHVtY3V0cmVjaHQubmw=
 Kevin Budding
Kevin Budding Jessica van Setten2
Jessica van Setten2 Tineke Kardol-Hoefnagel
Tineke Kardol-Hoefnagel Henderikus G. Otten
Henderikus G. Otten