- 1UOC Immunologia, IRCCS Ospedale Policlinico San Martino, Genova, Italy
- 2Laboratorio di Biologia Molecolare, IRCCS Istituto Giannina Gaslini, Genova, Italy
- 3Laboratorio di Immunologia Clinica e Sperimentale, IRCCS Istituto Giannina Gaslini, Genova, Italy
- 4Dipartimento di Medicina Sperimentale, Università di Genova, Genova, Italy
- 5Immunology Area, Ospedale Pediatrico Bambin Gesù, Rome, Italy
- 6Center of Excellence for Biomedical Research, University of Genoa, Genova, Italy
Hypoxia, which characterizes most tumor tissues, can alter the function of different immune cell types, favoring tumor escape mechanisms. In this study, we show that hypoxia profoundly acts on NK cells by influencing their transcriptome, affecting their immunoregulatory functions, and changing the chemotactic responses of different NK cell subsets. Exposure of human peripheral blood NK cells to hypoxia for 16 or 96 h caused significant changes in the expression of 729 or 1,100 genes, respectively. Gene Set Enrichment Analysis demonstrated that these changes followed a consensus hypoxia transcriptional profile. As assessed by Gene Ontology annotation, hypoxia-targeted genes were implicated in several biological processes: metabolism, cell cycle, differentiation, apoptosis, cell stress, and cytoskeleton organization. The hypoxic transcriptome also showed changes in genes with immunological relevance including those coding for proinflammatory cytokines, chemokines, and chemokine-receptors. Quantitative RT-PCR analysis confirmed the modulation of several immune-related genes, prompting further immunophenotypic and functional studies. Multiplex ELISA demonstrated that hypoxia could variably reduce NK cell ability to release IFNγ, TNFα, GM-CSF, CCL3, and CCL5 following PMA+Ionomycin or IL15+IL18 stimulation, while it poorly affected the response to IL12+IL18. Cytofluorimetric analysis showed that hypoxia could influence NK chemokine receptor pattern by sustaining the expression of CCR7 and CXCR4. Remarkably, this effect occurred selectively (CCR7) or preferentially (CXCR4) on CD56bright NK cells, which indeed showed higher chemotaxis to CCL19, CCL21, or CXCL12. Collectively, our data suggest that the hypoxic environment may profoundly influence the nature of the NK cell infiltrate and its effects on immune-mediated responses within tumor tissues.
Introduction
NK cells are powerful effectors of the innate immunity with anti-tumor activity (1–5). They are endowed with a unique pattern of receptors sensing changes in MHC-I expression levels (which are often decreased in tumor cells) or recognizing ligands induced by tumor transformation, cell stress, and DNA damage (1, 5–8). By these receptors NK cells can direct their potent lytic machinery to target and eliminate many tumor cell types (6). In addition, NK cells can release pro-inflammatory cytokines and various chemotactic factors (IFNγ, TNFα, GM-CSF, CCL3/CCL4) potentially amplifying immune responses to the tumor (1, 6, 9–11). The cytolytic function and the capability of releasing cytokines and chemokines appear to be differently represented in the two major subsets of peripheral blood (PB)-NK cells, characterized by the CD56dim/CD16bright (CD56dim) or the CD56bright/CD16dim/neg (CD56bright) phenotype (1, 12–14). The CD56dim cells are strongly cytotoxic and can also produce cytokines in response to specific stimuli. These cells represent the large majority of the PB-NK cell population and express chemokine receptors, mainly CXCR1 and CX3CR1, that enable their recruitment to inflamed tissues (1, 9, 11, 15, 16). Conversely, CD56bright cells are poorly cytotoxic and release high amount of cytokines, especially in response to monokines. According to their expression of CCR7 and CD62L, these cells are mainly located within secondary lymphoid compartments while they account for only 10% of PB-NK cells (1, 9, 11, 15).
The increasing interest on NK cells as potential tools for immunotherapy has recently inspired many studies aimed at defining how their anti-tumor activity can be influenced by the tumor microenvironment. Along this line, different suppressive interactions mediated by tumor cells, tumor-associated fibroblasts, or regulatory immune cells have been described and characterized (17–22). In addition, it has been shown that tumor cells can escape NK cell attack by modulating the surface expression of various NK-receptor ligands (2, 23–27). In spite of these important advances in the field, a crucial issue that still remains to be investigated for an effective exploitation of NK cells in the therapy of solid tumors is the recruitment of NK cells to tumor tissues. Few recent studies have shown that higher NK cell infiltration correlates with a better prognosis of the disease (23, 28, 29), but have also indicated that the NK cell infiltrate in tumor tissues is often poor and, in some cases, mostly represented by poorly cytotoxic CD56bright cells (5, 30, 31). Specific chemokine milieus, or alteration of chemokine receptor patterns, may account for these findings. However, an exhaustive explanation on how the tumor microenvironment can influence NK cell infiltration has not yet been achieved.
Reduced partial O2 tension (pO2, 0–20 mm Hg, hypoxia), which often affects tumor tissues, may play role in this context. Hypoxia is an important driver of malignant progression and resistance to therapy (23, 32, 33). It can influence the function of different cell types within the tumor lesion and affect the recruitment of immune cells, favoring tumor escape mechanisms (33). Indeed, exposure to hypoxia can induce different immune and non-immune cells to change the expression of pro-angiogenetic factors, cytokines, and chemokines (including VEGF, SPP1, IL-1β, MIF, CXCL12, and CXCL8), or chemokine receptors (including CXCR4, CCR2, and CCR5) (34–38). In spite of many studies on this issue, limited information is currently available on the impact of hypoxia on NK cells and their subsets, notably on their ability to respond to specific chemotactic stimuli or to release immune-active soluble factors (39–41). We have previously shown that, in NK cells exposed to IL-2, hypoxia can down-regulate expression and function of most NK cell receptors that activate cytolytic activity against tumor or virally infected cells, but preserves NK cell ability to kill targets via ADCC (39), suggesting that NK cells may be effective even in hypoxic niches in the context of combined immunotherapeutic approaches. In this study, we integrate previous data and provide an overview of the effect of hypoxia on NK cells stimulated with IL-2. Moreover, we indicate some clues on how the composition and the function of NK cell infiltrate may be influenced by the hypoxic environment in tumor tissues.
Materials and Methods
NK Cells Isolation and Culture
NK cells were obtained from PB of healthy donors provided by the transfusion center of the Ospedale Policlinico San Martino following approved internal operational procedures (IOH78). Written informed consent from the donors was provided according to the Declaration of Helsinki. Briefly: NK cells from healthy donors were isolated from PB mononuclear cells using RosetteSep NK Cell Enrichment Cocktail (StemCell Technologies, 15025 Vancouver, Canada). Only preparations displaying more than 95% of CD56+ CD3– CD14– NK cells were selected for the experiments. After isolation, NK cells were cultured for the indicated time points in RPMI 1640 (Lonza Verviers, Belgium) supplemented with 10% Fetal Bovine Serum (FBS, Voden Medical S.p.a. Meda MB, Italy), antibiotic mixture (0.05 mg/mL penicillin, 0.05 mg/mL streptomycin Lonza, Verviers, Belgium), and 100 U/mL recombinant human IL-2 (Proleukin, Novartis Basilea, Switzerland) at 2X 106cells/mL in round bottom 96-well microtiter plates. The cultures were performed either under normoxic conditions in a humidified incubator containing 20% O2, 5% CO2, and 75% N2 or under hypoxic conditions. Hypoxic conditions were obtained by culturing cells in a sealed anaerobic workstation incubator (Ruskinn, INVIVO2 400, CARLI Biotec, Roma, Italy), incorporating a gas mixing system (Ruskinn Gas Mixer Q) and flushed with a mixture of 1% O2, 5% CO2, and 94% N2.
RNA Isolation and cRNA Synthesis
Total RNA was purified from NK cells derived from three healthy donors using the RNeasy MiniKit from Qiagen (Milano, Italy). RNA was controlled for integrity by nanoelectrophoresis with an Agilent 2100 Bioanalyzer (Agilent Technologies Europe, Waldbroon, Germany), quantified by spectrophotometry using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, USA), and reverse-transcribed into double-stranded cDNA on a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Milano) using the one-cycle cDNA synthesis kit (Affymetrix, Milano). cDNA derived from three donors/time point was purified and biotin labeled using the GeneChip IVT kit (Affymetrix). Labeled cRNA was fragmented according to Affymetrix's instructions.
GeneChip Hybridization and Array Data Analysis
Gene expression profiling was performed as described previously (42). Briefly, Fragmented cRNA was hybridized on the Affymetrix HG-U133 plus 2.0 GeneChips (Genopolis Corporation, Milano) containing 54,000 probe sets (coding for 47,000 transcripts and variants, including 38,500 unique human genes) on a single array. Chips were stained with streptavidin-phycoerythrin (Invitrogen Life Technologies, Milano) and scanned using an Affymetrix GeneChip Scanner 3000, as described. Data were processed by RMA normalization utilizing the “Affy” R package. Statistical analysis using paired t-test was performed to identify differentially expressed genes. We corrected the p-value for multiple hypothesis testing by Benjamini–Hochberg method to false discovery rate control. Only gene differences that passed the test at a confidence level of 95% (P < 0.05) and a false discovery rate of 0.05% were considered significant. Fold-change (FC) was calculated as the ratio between the average expression level under hypoxia and normoxia. Genes were defined as being differentially regulated by hypoxia if they exhibited more than 2-fold increase in gene expression or down-regulated if they showed <0.5-fold change compared with normoxic cultures. We converted the Affymetrix probe sets into the corresponding gene symbol by Netaffx tool. When multiple probe sets were associated with the same gene symbol, the probe set with the highest expression signal was considered. The full set of data from each microarray experiment has been deposited in the Gene Expression Omnibus public repository at NCBI (www.ncbi.nlm.nih.gov) and is accessible through GEO (Accession number GSE116660). Biological processes were assessed by DAVID Gene Ontology (GO) enrichment analysis (http://david.niaid.nih.gov). The significant GO terms were defined as p < 0.05 and FDR < 0.05.
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) was performed on all probe sets of the Affymetrix HG-U133 Plus 2.0 GeneChip, as described previously (43). An enrichment score (ES) and a normalized enrichment score (NES) were calculated for every gene set. The statistical significance of NES was estimated by an empirical test using 1,000 gene set permutations to obtain the nominal p-value. A false discovery rate (FDR) q value was estimated to control the probability that a NES could represent a false positive finding. The gene sets used in the GSEA belong to the C2.CGP collection of the Broad Institute Molecular Signature v5 Database (MSigDB) (44). The analysis used Signal-to-Noise metric and considered gene sets containing at least 15 and up to 250 probe sets. An enrichment with FDR q-values lower than 0.05 and nominal p < 0.05 was considered significant. Leading Edge Analysis (LEA) of enriched gene sets was used to identify key genes related to NK response to hypoxia.
Real-Time RT-PCR
cDNA was prepared from purified total RNA using SuperScript Double-Stranded cDNA synthesis kit (Invitrogen). Real time PCR (qRT-PCR) was performed on a 7500 Real Time PCR System (Applied) in triplicate for each target transcript using SYBR Green PCR Master Mix and sense/antisense oligonucleotide primers synthesized by TIBMolbiol (Genova) or purchased from Quiagen, as detailed before (45). Expression data were normalized on the values obtained in parallel for three reference genes (actin related protein 2/3 complex subunit 1B, ARCP1B; ribosomal proteins S18, RSP18; and RSP19), using the Bestkeeper software, and relative expression values were calculated using Q-gene software, as detailed (45).
mAbs and Flow Cytofluorimetric Analysis
The following mAbs were used in this study: anti-CCR1 (R&D System, MAB 145-100, Minneapolis U.S.A.), anti-CCR5 (R&D System, MAB 182-100 Minneapolis U.S.A.), anti-CCR7 (R&D System, MAB 197-100 Minneapolis U.S.A.), anti-CXCR1/IL-8 RA (R&D System, MAB 173-100 Minneapolis U.S.A.), anti-CXCR3 (R&D System, MAB 160-100), anti-CXCR4 (R&D System, MAB 173-100), PE-conjugated anti-CX3CR1 (Medical & Biological Laboratories Co., LTD, D070-5), FITC-conjugated anti-CD3 (eBioscience, 11-0038-42 Thermofisher scientific, Waltham, Massachusetts, Stati Uniti), PE-cyanine 7-conjugated anti-CD56 (Beckman Coulter, A21692, Brea, California U.S.A.), PE-conjugated anti-CD16 (130-106-704, Miltenyi Biotec Bergisch Gladbach, Germany). The staining with the appropriate unlabeled mAbs are followed by PE-conjugated isotype-specific goat anti-mouse second reagent (Southern Biotechnology Associated, Birmingham, AL, U.S.A.), and fluorescence was quantified on a Gallios™ Flow Cytometer (Beckman Coulter, Brea, California U.S.A.).
Multiplex ELISA Analyses
Freshly isolated NK cells were cultured for 20 h at 5X 105/mL in flat bottom 96-well microtiter plates in the presence of the following recombinant human cytokines: IL-2, IL-12+IL-18, or IL-15+IL-18. The cytokine concentrations were: 100 U/mL IL-2 (Proleukin, Novartis Basilea, Switzerland); 2.5 ng/mL IL-12 (Peprotech, 200-12 London, UK); 20 ng/mL IL-15 (Peprotech, 200-15 London, UK); 200 ng/mL IL-18 (Medical & Biological Laboratories Co. LTD, B001-5, Japan). In the “PMA+IONO IL-2” condition, NK cells were cultured in the presence of IL-2 for 26 h, and 100 ng/mL PMA (Phorbol 12-myristate 13 acetate, SIGMA-Aldrich Saint Louis, Missouri, U.S.A.) and 500 ng/mL IONO (Ionomycin, SIGMA-Aldrich, Missouri, U.S.A.) were added to the cultures for the last 6 h. The cultures were performed in parallel under normoxic and hypoxic conditions (see above). Culture supernatants were then collected and analyzed for their cytokine content by MAGPIX® System (Luminex® xMAP® Technology, Merck Millipore, Germany).
Chemotaxis Assay
NK cells freshly isolated from peripheral blood and then cultured for different time points (24, 48, and 96 h) with IL-2 under hypoxic or normoxic conditions were seeded at 2.5 × 106/mL in the upper chamber of a Transwell system (3 mm pore size; Corning Costar, 3415). 10% FBS RPMI 1640 medium alone or supplemented with recombinant human CXCL12 [100 ng/mL] (Peprotech, 300-28A), or CCL19 [0.3 μg/mL], or CCL21 [0.6 μg/mL] was added to the lower compartment. Cells were allowed to migrate for 2 h at 37°C under normoxic condition. Cells migrated in the lower chamber were collected and counted using the MACSQuant Analyzer (Miltenyi Biotec Bergisch Gladbach, Germany) or analyzed with the Gallios™ Flow Cytometer after surface double staining of CD56/CD16 markers. Cells migrated in the lower chamber containing medium alone (w/o chemokines) represented spontaneous migration due to unspecific cell motility. Chemotactic response was assessed as percentage of spontaneous migration and was calculated as follows: (number of migrated cells in the presence of chemotactic stimulus/number of migrated cells in the absence of stimulus) × 100. The chemotactic response of CD56brightCD16dim/neg NK cells to CCL19, CCL21, and CXCL12 was assessed as enrichment of this specific cell subset within migrated cells and was calculated as follows: CD56brightCD16dim/neg cell percentage within cells migrated in response to chemokines/CD56brightCD16dim/neg cell percentage within spontaneously migrated cells.
Statistical Analysis
Statistical analyses were performed using the Prism software package (GraphPad Software). Data are expressed as the mean ± SEM of at least three independent experiments, unless differently specified. Statistical significance was evaluated by two-tailed paired Student's t-test. A p < 0.05 (*), < 0.01(**), or < 0.001(***) was considered statistically significant.
Results
Gene Expression Profile of Hypoxic NK Cells
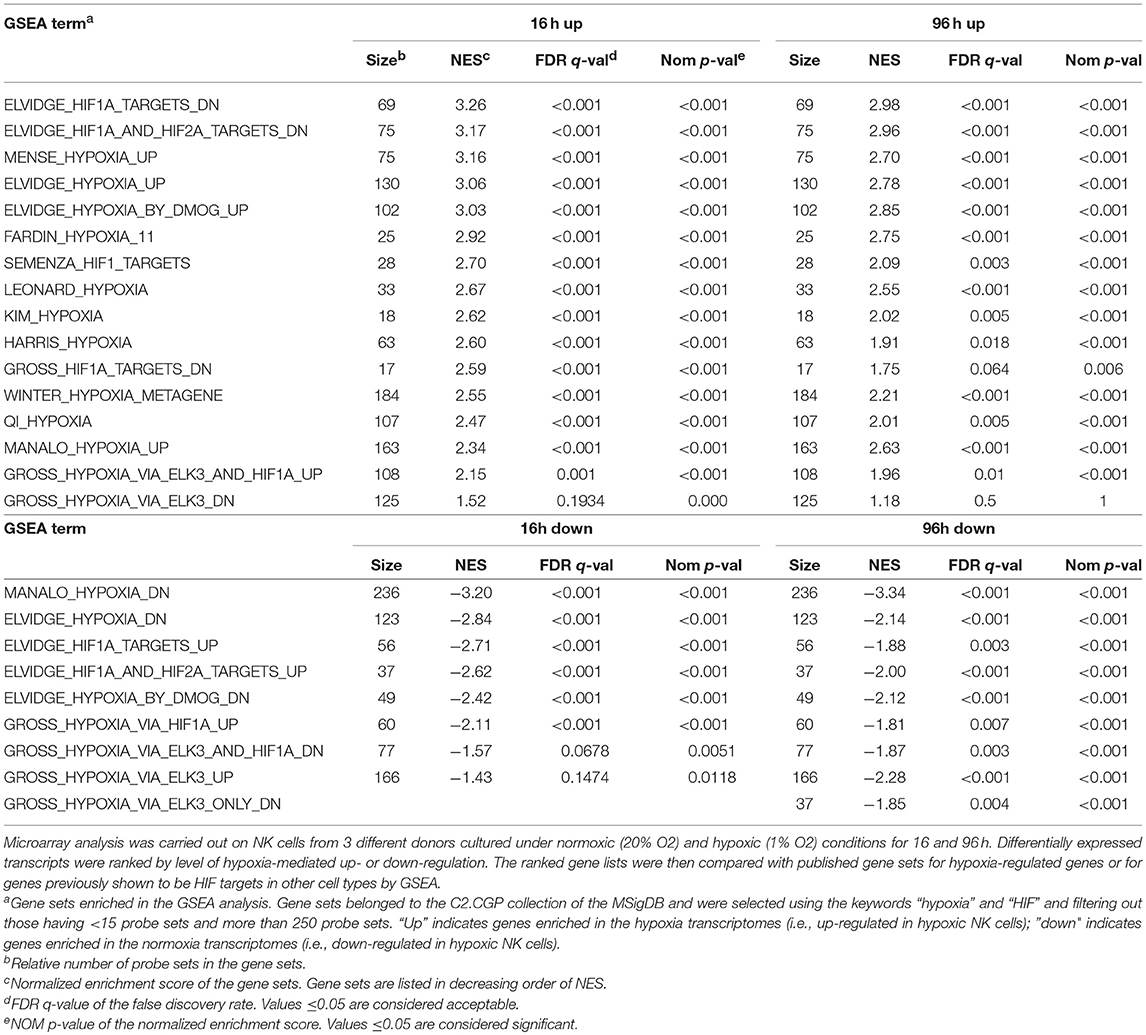
To obtain an overview of NK cell response to hypoxia, we assessed the gene expression profile of NK cells isolated from the PB of three independent healthy donors and cultured with IL-2 for 16 or 96 h under hypoxic (1% O2) or normoxic (20% O2) conditions. mRNA was individually hybridized to human Affymetrix HG-U133 plus 2.0 GeneChips, obtaining three biological replicates for each experimental condition. Raw data were processed as described in the section Materials and Methods. We used GSEA to determine the enrichment of the published C2.CGP gene set collection (44) in the 16 and 96 h transcriptomes of hypoxic NK cells (Hy-NK) as compared to their normoxic counterparts. We selected 25 gene sets using “hypoxia” and “hypoxia-inducible factor (HIF)” as keywords (see section Materials and Methods for details). The list of gene sets, their normalized enrichment score (NES), false discovery rate q-values (FDR q-val), and nominal p-values (NOM p-val) are reported in Table 1. Among selected gene sets, 20 were significantly enriched in both Hy-NK cell transcriptomes (p < 0.05; FDR q < 0.05), 1 and 3 additional gene sets were specifically enriched in the 16 h (upregulated) and the 96 h (downregulated) hypoxic transcriptomes, respectively, and only 1 gene set (upregulated) was not significantly enriched at either time points. Representative 16 and 96 h plots showing clear enrichment of the gene sets at the top or the bottom of the ranked list are presented in Figure 1A for a visual inspection of the GSEA results (46). These data demonstrate that gene expression changes in Hy-NK cells follow a consensus hypoxia transcriptional profile.
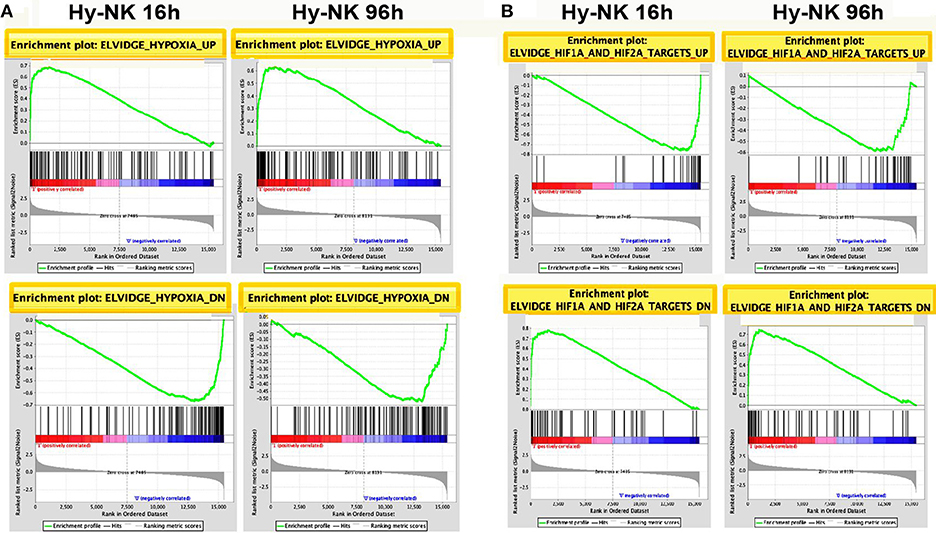
Figure 1. Gene Set Enrichment Analysis (GSEA) Plots for representative hypoxia- or HIF1α/2α-related gene sets in Hy-NK cell transcriptomes. The transcripts identified by microarray analysis in NK cells were ranked by level of hypoxia-mediated up- or down-regulation. The ranked gene lists were then compared by GSEA with previously published gene sets for hypoxia-regulated genes or for genes previously shown to be HIF targets in other cell types. (A) GSEA plots of representative sets of up- or down-regulated genes from cells exposed to hypoxia (ELVIDGE_HYPOXIA_UP or _DN, respectively). (B) GSEA plots of representative sets of up- or down-regulated genes from cells undergoing HIF-1α and HIF-2α silencing (Elvidge_HIF1A_and_HIF2A_TARGETS_UP or _DN, respectively). Note that in the case HIF1α/2α-silencing the sets of up- or down-regulated genes resulted inversely enriched in the Hy-NK cell transcriptomes. The enrichment score is calculated by walking down a list of genes ranked by their correlation with the phenotype, increasing a running-sum statistic when a gene in that gene set is encountered (each black vertical line underneath the enrichment plot) and decreasing it when a gene that isn't in the gene set is encountered. The enrichment score is the maximum deviation from zero encountered in the walk.
Gene transcriptional activation by hypoxia is mediated primarily by HIF, a heterodimer of a constitutive HIF-1β subunit and an O2-sensitive α-subunit (HIF-1α or HIF-2α) (32, 34). Interestingly, some of the selected gene sets were from cells undergoing HIF-1α or HIF-2α silencing (46). The reported sets of down- or up-regulated genes were found inversely enriched in the Hy-NK cell transcriptomes (Table 1—gene sets 1, 2, 19, 20—and Figure 1B), suggesting that HIF-1α and HIF-2α and their target genes could play an important role in NK cell response to hypoxia.
Leading Edge Analysis (LEA) applied to the significantly enriched gene sets allowed to define the subsets of hypoxia-related genes with the highest impact on the enrichment score (referred to as the leading edge subset) at 16 or 96 h (Figure S1). These subsets include genes involved in glycolysis, gluconeogenesis, and glucose transport (ALDOA, ALDOC, ENO1, ENO2, GAPDH, GPI, HK1, HK2, LDHA, PDK1, PGK1, SLC2A1, TPI1), non-glycolytic metabolism and ion transport (P4HA1, P4HA2, PAM, VLDLR), apoptosis, stress response, and proliferation (BNIP3, BNIP3L, CCNG2, DDIT3, EGLN1, EGLN3, NDRG1), transcription and signaling activity (FOSL2, JAK2, JUN, MXI1, SOCS2). These results extend to Hy-NK cells the expression of a large cluster of hypoxia-related genes previously identified in other cell types, including tumor and immune cells (32, 34, 47–49).
Functional Assessment of Genes Modulated by Hypoxia in NK Cells
To identify novel genes affected by hypoxia in NK cells, we performed differential expression analysis of microarray data. We filtered transcripts for a differential expression of at least 2-fold changes and a p ≤ 0.05. Using these selection criteria, we identified a total of 1,474 transcripts that were significantly modulated under hypoxic vs. normoxic conditions, with expression changes ranging from 106-fold upregulation to 25-fold downregulation (Table S1). The majority of differentially-expressed transcripts were identified as unique genes named in the GenBank™, whereas the remaining transcripts were either unnamed expressed sequence tags or hypothetical. As shown by the Venn diagram in Figure 2A, 355 transcripts were up- [179] or down-[176] regulated at both time points, whereas 374 (139 induced and 235 repressed) and 745 (334 induced and 411 repressed) transcripts were specifically modulated at 16 or 96 h, respectively. These results provide the first indication that Hy-NK cell signature varies with the duration of exposure to hypoxia.
Figure 2. Identification of genes significantly modulated by hypoxia in NK cells by differential expression analysis. (A) Graphical representation of transcripts differentially expressed in hypoxic vs. normoxic NK cells. The gene expression profile of NK cells isolated from 3 different donors and exposed to hypoxia for 16 h (top) or 96 h (bottom) was analyzed by microarray analysis, as described in the section Materials and Methods. The Venn diagram depicts the number of transcripts exhibiting ≥2 fold up- or down-regulation in hypoxic vs. normoxic cells at the two time points. About 24% of differentially expressed transcripts are common to the 16 and 96 h transcriptomes. (B) Functional assessment of hypoxia-responsive genes by GO enrichment analysis. Unique genes showing at least 2-fold change in expression levels between Hy-NK and NK cells were clustered into different biological processes using the DAVID GO enrichment analysis. Based on this classification scheme, genes were placed in more than one biological process if more than one function of the encoded protein was established. The y-axis shows the GO terms. The x-axis shows the percentage of genes within each process relative to the total amount of genes belonging to that process: bars on the right of the y axis represent upregulated genes; bars on the left of the y axis represent downregulated genes. The blue columns represent genes modulated at 16 h whereas the red column represent genes modulated at 96 h.
To gain insights into the biological processes modulated by hypoxia, we carried out a Gene Ontology (GO) enrichment analysis on the lists of up- and down-regulated transcripts. We identified 28 biological processes containing a statistically significant enrichment of hypoxia-modulated genes (HMGs) (Figure 2B). Most processes were represented at both time points, although with variable HMG enrichment. Metabolism and biosynthesis resulted as the most enriched processes (in both up- and down-regulated genes), followed by response to stimulus. Additionally, Hy-NK cell transcriptional profile was related to regulation of apoptosis, and response to stress, but also to cell proliferation, signaling, and chromatine/chromosome organization. Certain processes, including regulation of gene transcription and expression, and cell differentiation, were selectively enriched in upregulated genes, whereas processes related to cell cycle, cellular component organization, and DNA replication and repair were exclusively enriched in downregulated genes at both time-points.
Importantly, different immune-related processes, including immune system development/response, hemopoiesis, leukocyte activation and differentiation, angiogenesis, regulation of cell motility and communication, and immune effector processes, were enriched in a statistically significant percentage of HMGs, suggesting regulatory effects of hypoxia on NK cell-mediated immune responses.
Characterization of Cytokine/Chemokine and Receptor Gene Modulation in Hy-NK Cells
As the effect of hypoxia on the NK cell capability of killing target cells has been previously assessed (39), we focused our analysis on genes involved in immunoregulation and migration. The evaluation of immune-related gene clusters highlighted by GO analysis led to the identification of 43 HMGs coding for cytokines and chemokines, their receptors, and/or associated signaling molecules (Table 2). Some of these genes (26) were rapidly modulated by hypoxia (at 16 h time-point), while the remaining genes (17) were modulated after longer exposure (96 h).
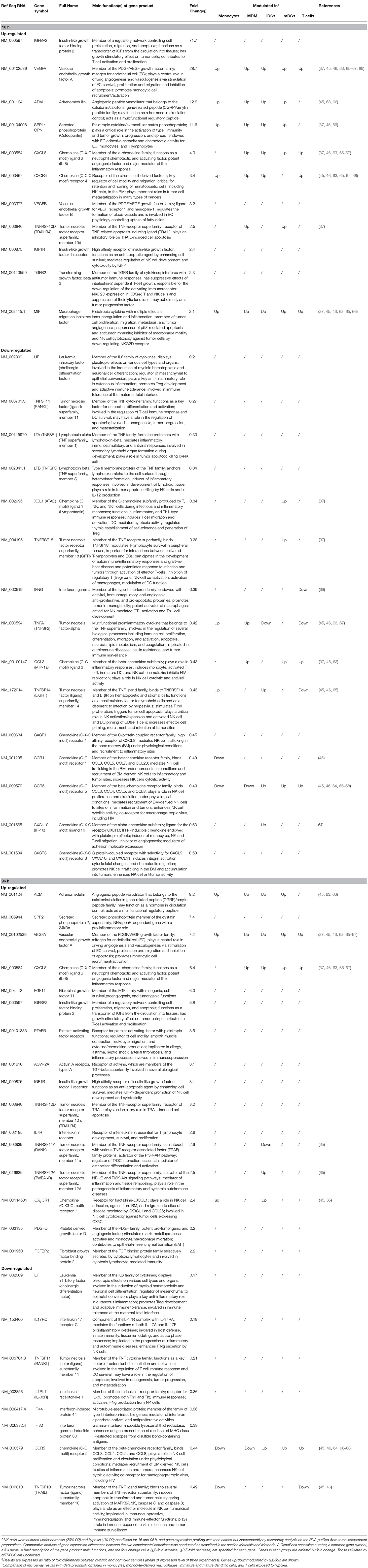
Table 2. Relative expression of genes encoding cytokines/chemokines and their receptors in H-NK vs. NK cells∧.
Compared to their normoxic counterparts, Hy-NK cells showed increased expression of genes coding for molecules with a primary role in angiogenesis (VEGFA, VEGFB, ADM, SPP1, FGF11) and promotion of inflammation or lymphocye cytotoxic responses (SPP1, SPP2, VEGF, TNFRSF11A and 12A, IGFBP2, FGFBP2, PTAFR), but also in apoptosis inhibition (IGF1R, TNFRSF10D, 11A, and 12A), tumor progression (IGFBP2, SPP1, MIF, PDGFD, TGFβ2, FGF11), and immunosuppression (MIF and TGFβ2) (34, 37, 48, 50–55). On the other hand, hypoxia inhibited mRNAs coding for cytokines and/or receptors mainly involved in anti-tumor and anti-viral immune responses including IFNγ, IFI30, IFI44, IL-17RC, IL1RL1, and various components of the tumor necrosis factor (TNF) superfamily, such as TNFα, LTA, LTB, TNFSF10,11,14, and TNFRSF18 (55–63). The only exception was represented by the inhibition of mRNA coding for LIF, a pleiotropic factor involved in the regulation of inflammation.
The Hy-NK transcriptome was also characterized by the differential modulation of genes coding for chemokines and chemokine receptors (11, 15). Specifically, we observed hypoxia-dependent upregulation of the mRNA for CXCL8 (which also has proangiogenic properties) (11, 37, 52), and downregulation of mRNAs for CXCL10, CCL3, and XCL1 (9, 11, 52, 64). Regarding the chemokine receptors known to be important for NK cell migratory activity mRNAs coding for CXCR4 and CX3CR1 were selectively up-regulated, whereas those coding for CXCR1, CCR1, CCR5, and CXCR3 were downregulated.
Microarray results were validated by qRT-PCR analysis of a subset of HMGs (17 belonging to the 16 h transcriptome and 9 to the 96 h transcriptome). As shown in Figure 3 and Table 2 there was almost full concordance between qRT-PCR and Affymetrix data with respect to the direction of the expression changes, with the only exception of CX3CR1 whose upregulation by hypoxia was not confirmed by qRT-PCR. For about half of validated genes, the extent of modulation was also comparable to that shown by microarray data, whereas it was higher for nine genes and lower for three genes. Such discrepancies, however, are consistent with previous findings showing that these techniques can often differently estimate the extent of gene regulation (42, 48).
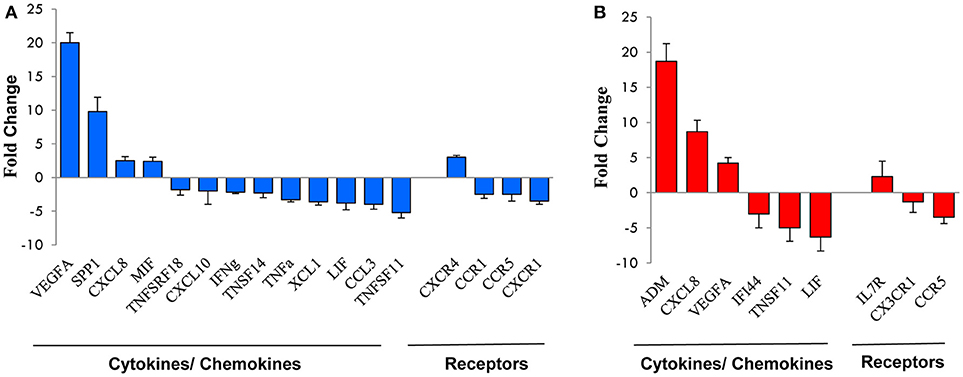
Figure 3. qRT-PCR validation of genes selected from the microarray profile. Total RNA from the NK cell preparations analyzed by microarray was subjected to qRT-PCR for the expression of a subset of genes randomly selected from those up- or down-modulated at 16 h (A) or 96 h (B). Expression changes were evaluated in relation to the values obtained for three reference genes, as detailed in the section Materials and Methods. Results are expressed as fold-changes (Hy-NK relative to NK cells) and represent the mean of three determinations for each transcript. Positive values indicate that the mRNA levels of a specific gene was up-regulated, whereas negative values indicate that the transcript was down-regulated. Genes are ordered by fold-change within each group.
A literature survey indicated that some of the HMGs in NK cells were targeted by hypoxia in different cell types including T lymphocytes (49), primary monocytes (48), monocyte-derived macrophages (MDMs) (38, 65, 66), immature (i)DCs (67–70), and mature (m)DCs (37, 68) (Table 2). In particular, MIF- and VEGFA-coding genes were upregulated by hypoxia in all the immune cell types analyzed, ADM and SPP1 were increased in the innate immune cells, while CXCL8 and CXCR4 upregulation was reported in T cells and in some mononuclear phagocyte populations. Other genes were variably modulated depending on the analyzed cell type. To our knowledge, a consistent part of the cytokine/chemokine- and receptor-coding genes identified in Hy-NK cells have not been reported to be modulated by hypoxia in other immune cell populations analyzed.
Taken together, these data indicate that hypoxia can induce a specific cytokine/chemokine and receptor gene signature in NK cells with possible functional consequences. To assess this possibility, we proceeded with the overall evaluation of the effects of hypoxia on NK cell immune-regulatory and migratory functions.
Effects of Hypoxia on NK Cell-Mediated Release of Chemokines and Cytokines
To assess the effect of hypoxia on cytokine/chemokine release, PB-NK cells were freshly isolated from additional donors and cultured in the presence of IL-2 under hypoxic or normoxic conditions. After 20 h, supernatants were collected and analyzed by multiplex immunoassay for the content of IFNγ, TNFα, CCL3, GM-CSF, CCL5, CXCL8, VEGF, and MIF (Figure 4), namely those factors that are typically released by NK cells (6, 12) and/or that were shown to be transcriptionally affected in microarray analysis (Table 2). As shown in Figure 4A, upon exposure to IL-2 NK cells released low levels of different factors, including IFNγ, CCL3, GM-CSF, CCL5, and MIF. Under hypoxic conditions, release of IFNγ, CCL3, GM-CSF, and CCL5 was decreased (although differences reached statistical significance only for CCL3 and GM-CSF). Since IL-2 has typically limited direct effects on cytokine release while it primes NK cells to respond to other stimuli, we assessed the effect of hypoxia on NK cells cultured for 20 h with IL-2 and stimulated for further 6 h with PMA + Ionomycin (PMA+IONO). As shown in Figure 4B, an inhibitory effect on CCL3 and (slightly) on GM-CSF release was induced by hypoxia also on PMA+IONO-stimulated NK cells.
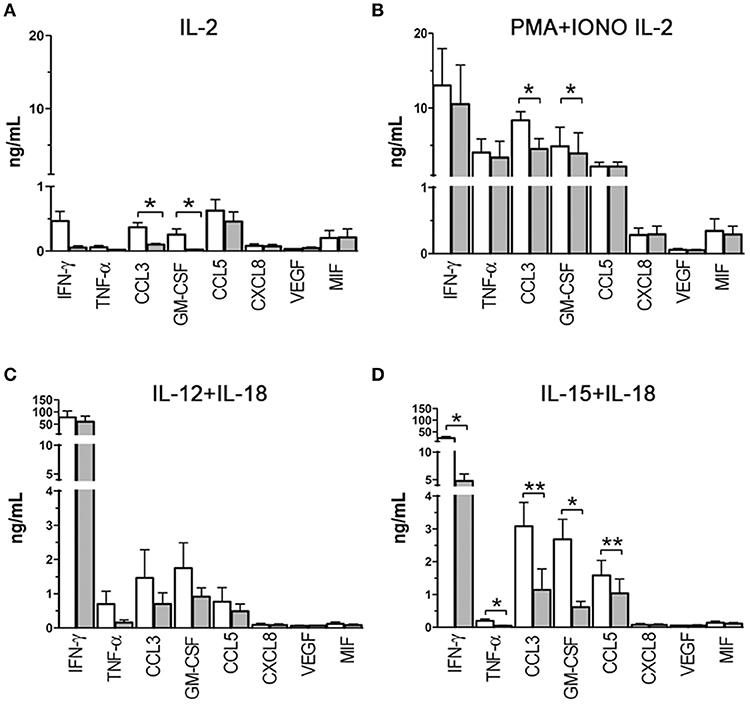
Figure 4. Cytokine/chemokine release capability of NK cells exposed to hypoxia or normoxia. Cell-free supernatants from NK cells cultured in the presence of IL-2 (A), IL-2 and PMA+ionomycin (B), IL-12+IL-18 (C), IL-15+IL-18 (D) under normoxic (white columns) or hypoxic (gray columns) conditions were analyzed for 8 cytokine/chemokine content using the MAGPIX® System. Results are expressed as ng/ml and are the mean + SEM of 5 independent experiments. In (A,C,D), the cells were cultured for 20 h in the presence of the indicated stimulating cytokines, in (B), the cells were cultured for 26 h in the presence of IL-2 with the addition of PMA+ionomycin in the latter 6 h. * p < 0.05, ** p < 0.01.
We next assessed the effect of hypoxia on NK cells cultured with other classical NK-activating stimuli, such as the monokines, IL-12, IL-15, and IL-18. In particular, we set monokine combinations known to potently stimulate NK cell cytokine secretion (i.e., IL-12+IL-18, and IL-15+IL-18). As shown in Figure 4C, NK cells cultured in the presence of IL-12 + IL-18 for 20 h released into the culture supernatant very high amounts of IFNγ and moderate to low amounts of CCL3, GM-CSF, TNFα, and CCL5. Hypoxia did not modify significantly NK cell ability to release cytokines in response to IL-12+IL-18, although a trend toward inhibition was observed for CCL3, GM-CSF, and TNFα release. Compared to IL-12+IL-18, IL-15+IL-18 stimulation induced lower release of IFNγ and TNFα and higher secreted levels of CCL3, GM-CSF, and CCL5 under normoxic conditions (Figure 4D). Exposure to hypoxic conditions resulted in the significant inhibition of IFNγ, TNFα, CCL3, GM-CSF, and CCL5 release.
We conclude from these data that hypoxia can differently affect cytokine/chemokine release depending on the type of stimulus. Specifically, it can modulate the release of only a few cytokines/chemokines in NK cells cultured in the presence of IL-2 or IL-2+PMA+IONO, exert a more general inhibition of cytokine/chemokine secretion on NK cells exposed to IL-15+IL-18, while it has no significant effects on NK cells exposed to IL-12+IL-18.
Effect of Hypoxia on Chemokine Receptor Expression
We next assessed whether hypoxia could modulate chemokine receptor expression on NK cell surface. To this end, freshly isolated PB-NK cells were cultured in the presence of IL-2 under hypoxic or normoxic conditions and analyzed by flow cytometry for the expression of CCR5, CCR7, CCR1, CX3CR1, CXCR1, CXCR4, and CXCR3 immediately after isolation or after 24, 48, or 96 h of culture. Upon exposure to IL-2 (under normoxic conditions), NK cells progressively down-regulated expression of CXCR1, up-regulated that of CXCR3 and (transiently) that of CXCR4, while they minimally modified the expression of CCR5, CCR1, and CX3CR1. Hypoxia further significantly increased the up-regulation of CXCR4 expression, slightly decreased CCR5 expression, while it did not substantially modify the expression trend of CCR1, CX3CR1, CXCR1, and CXCR3 (Figures 5A,C).
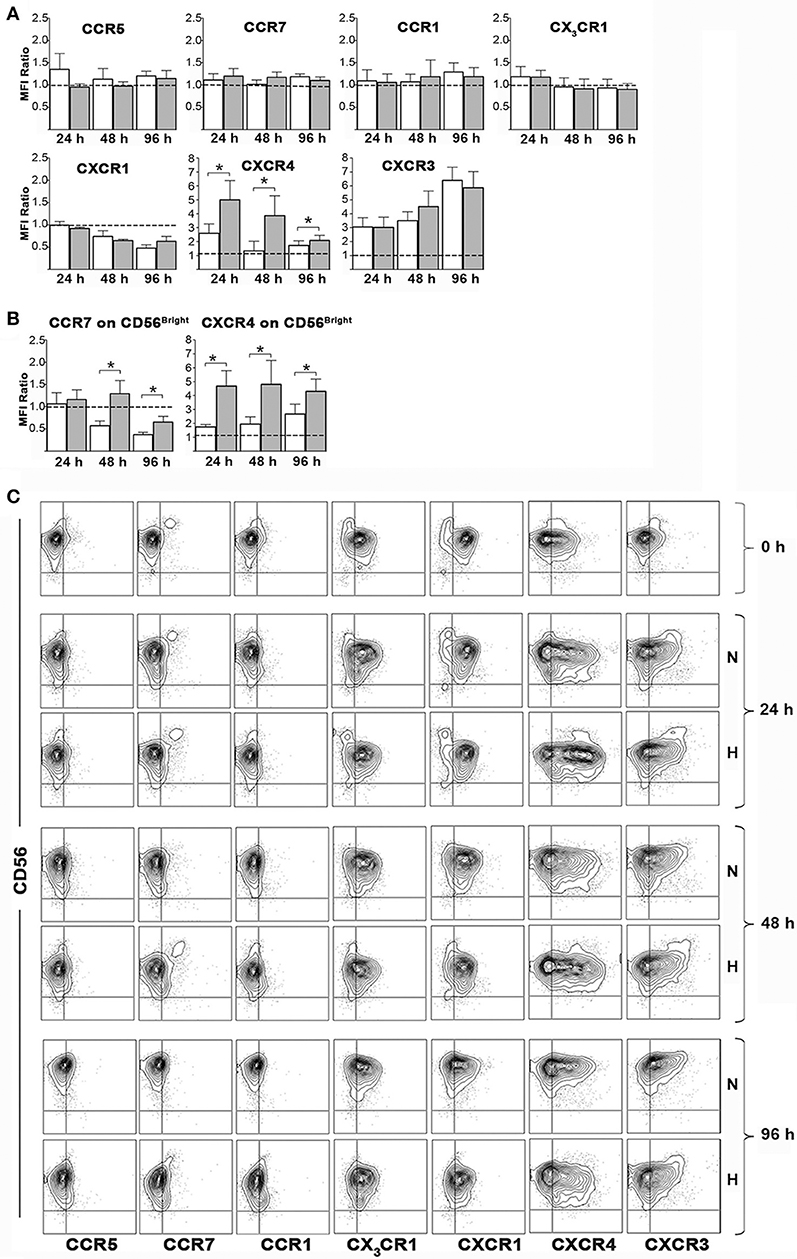
Figure 5. Effects of hypoxia on NK cell chemokine receptor expression. Freshly isolated PB-NK cells were cultured in the presence of IL-2 for 24, 48, or 96 h under normoxic or hypoxic conditions and then analyzed by flow cytometry for surface expression of the indicated receptors. (A,B) The ratio between the MFI observed at the indicated culture time points and that at t0 (i.e., on freshly isolated cells) is reported for each receptor. Horizontal dotted lines indicate no changes. White and gray bars are referred to NK cells cultured under normoxic or hypoxic conditions, respectively. Data are the mean + SEM of 5 independent experiments. In (B) data on CD56bright gated cells are reported for CCR7 and CXCR4 expression. (C) FACS profiles of a representative donor are shown. * p < 0.05.
The analysis of CCR7 on the whole PB-NK cell population didn't give meaningful data, as CCR7 expression is generally confined to the small fraction of CD56bright cells (12, 17) (Figures 5A,C). On the other hand, CD56bright NK cells showed progressive decrease of CCR7 expression during culture with IL-2. Hypoxia significantly reversed such effect sustaining CCR7 expression on CD56bright cells (Figures 5B,C). Remarkably, a careful analysis of such NK cell subset revealed that also CXCR4 expression could be sustained by hypoxia in CD56bright cells (Figures 5B,C).
Effects of Hypoxia on NK Cell Chemotaxis
Experiments were then carried out to assess whether hypoxia-induced changes of CCR7 and CXCR4 expression could affect specific chemotactic activity of NK cells. To this end, NK cells cultured under normoxic or hypoxic conditions for 24, 48, and 96 h were analyzed in classical migration assays using CCL19, CCL21 (CCR7 ligands) and CXCL12 (CXCR4 ligand) as chemoattractants.
Given the peculiar distribution of CCR7 within the PB NK cells, we analyzed whether CCL19 or CCL21 could induce the preferential migration of the CD56brightCD16dim/neg cell subset, resulting in the enrichment of such population within migrated cells. Before performing this analysis, we evaluated by FACS whether the percentage of CD56brightCD16dim/neg cells could be modified over time under hypoxic or normoxic culture conditions. As shown in Figure 6A the percentage of CD56bright cells slightly decreased during culture under normoxic conditions, while hypoxia preserved such a population at the 24 and 48 h time points. This observation suggests that hypoxia could contribute to increase the absolute number of migrated CD56bright cells by preserving them over time. In order to selectively evaluate the effect of hypoxia on specific chemotactic properties of the cells, the chemotactic response to chemokines was calculated as ratio of the CD56brightCD16dim/neg cell percentages within cells migrated to specific chemokines or spontaneously. Among NK cells that have been cultured under normoxic conditions for 24 h, CD56bright cells were able to migrate in response to both chemokines and enrich the population of migrated cells. However, this ability progressively disappeared at later culture time points. By contrast, Hy-NK cells gave rise to higher enrichment of CD56bright cells within cells that migrated in response to CCL19/21 and maintained this capability over time (Figures 6B,C).
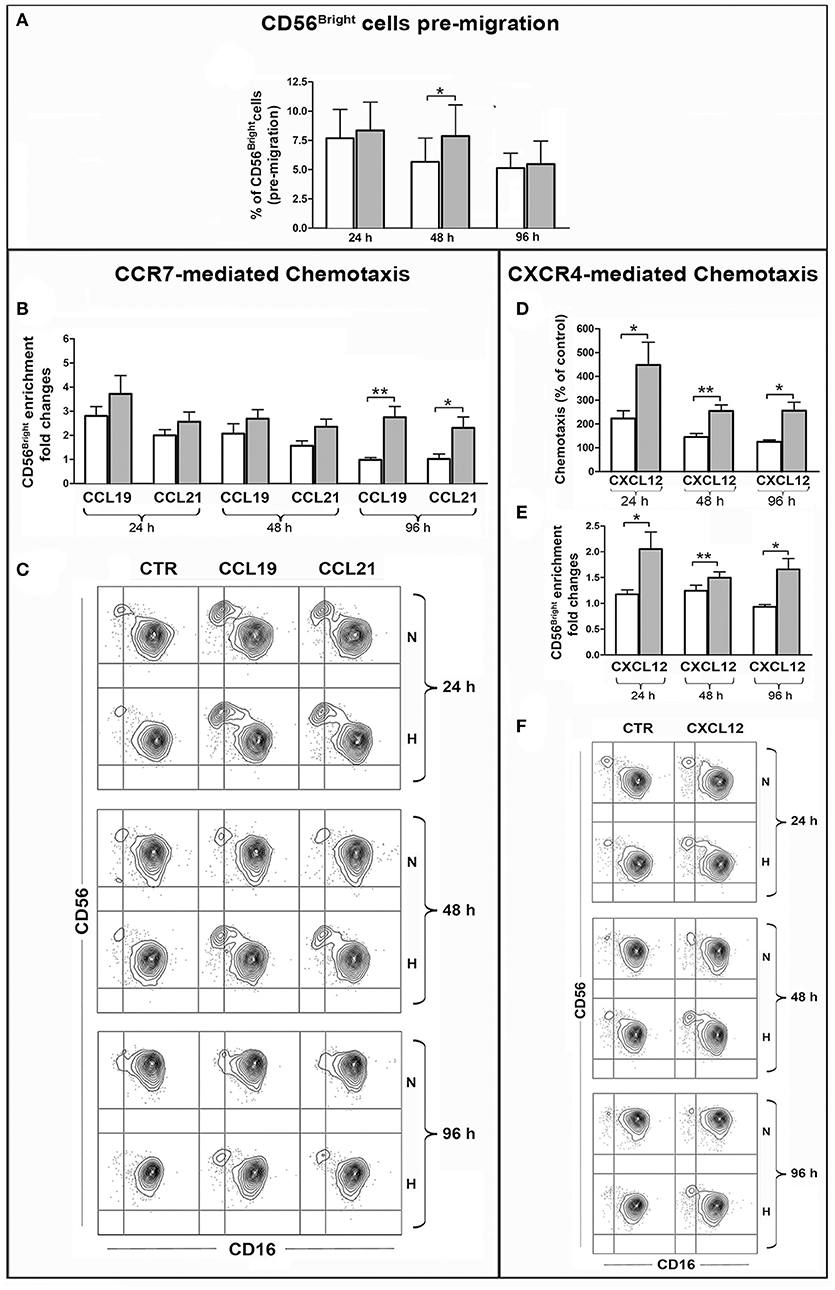
Figure 6. Effect of hypoxia on chemotaxis of PB-NK cells and their CD56bright/CD56dim subset to CXCL12, CCL19, CCL21. PB-NK cells were cultured in the presence of IL-2 for 24, 48, or 96 h under normoxic or hypoxic conditions and analyzed by FACS for the combined expression of CD56 and CD16 markers in order to assess the percentage of the CD56brightCD16dim/neg cells before migration (A). Cells were then assessed for chemotaxis to the indicated chemokines under normoxic conditions for 2 h. Migrated cells were collected from the lower migration chamber compartments and counted or analyzed by FACS for the combined expression of CD56 and CD16 markers. The specific chemotactic response of CD56brightCD16dim/neg NK cells to CCL19, CCL21 (B), and CXCL12 (E) was assessed as enrichment of this cell subset within migrated cells. The enrichment was calculated as fold increase of the CD56brightCD16dim/neg cell percentage within cells migrated to specific chemokines as compared to the CD56brightCD16dim/neg cell percentage within spontaneously migrated cells (see section Materials and Methods for details). (C,F) Representative experiments showing the enrichment of CD56brightCD16dim/neg NK cells within cells migrated in response to CCL19, CCL21 (C) or CXCL12 (F) as compared to cells that spontaneously migrated in the lower compartment in the absence of stimuli (CTR). (D) Specific chemotactic response to CXCL12 of the whole PB-NK cell population cultured under normoxic or hypoxic conditions. In (B,D,E) white and gray bars indicate data from NK cells cultured under normoxic or hypoxic conditions, respectively and represent the mean ± SEM of 6 independent experiments. * p < 0.05, ** p < 0.01.
The analysis of CXCR4-dependent chemotaxis indicated that, overall, NK cells exposed to hypoxia were responsive to CXCL12 more than NK cells cultured under normoxic conditions (Figure 6D). Remarkably, this difference was more pronounced when considering the CD56bright cell subset. Indeed, “hypoxic” (but not “normoxic”) NK cells gave rise to enrichment of the CD56bright cells within cells migrated to CXCL12 (Figures 6E,F). These results were in line with the observation that under hypoxia all CD56bright cells expressed CXCR4 at high levels while CD56dim NK cells included a variable fraction of CXCR4neg cells (Figure 5C).
Overall, these data demonstrate that hypoxia can sustain CXCR4- and CCR7-dependent chemotactic response of NK cells to specific chemokines, such as CXCL12, CCL19, or CCL21, and favor the recruitment of CD56bright cells, suggesting that a hypoxic environment may influence the extent and the nature of the NK cell infiltrate in different types of tumors.
Discussion
In the present study we analyze the global effects of hypoxia on NK cells, which are among the most potent immune effectors available to the host for the control of tumor development and progression (1, 2, 6). We first provide the transcriptional overview of the response of IL-2-primed NK cells to short-term (16 h) and prolonged (96 h) hypoxia, demonstrating that Hy-NK cells are functionally reprogrammed through the differential expression of a large number of genes implicated in various aspects of NK cell biology, including immunoregulation and migration. Then, we document hypoxia influence on the chemotactic properties of specific NK cell subsets and on NK cell ability to release cytokines and chemokines, providing important clues on the effective role of the O2 tension in determining the composition and the function of the NK cell infiltrate in tumor lesions.
So far, one transcriptional study describing how hypoxia could influence the cytokine-mediated activation of NK cells has been done for IL-15, while no data were available on IL-2, although this factor represents the most known and studied priming cytokine for NK cells. As assessed by GSEA, gene expression changes observed upon NK cell exposure to 1% O2 conditions follow a consensus hypoxia transcriptional profile. Several hypoxia-related and HIF-1α/HIF-2α target gene sets defined in previous studies are, in fact, significantly enriched in both the 16 and the 96 h Hy-NK transcriptomes. Moreover, we find enrichment of genes involved in glycolysis, gluconeogenesis, glucose transport, non-glycolytic metabolism, and ion transport, which is a common feature of hypoxic cells of different type, origin, and functional state being essential to compensate for the inhibition of oxidative metabolism and the malfunctioning of O2-dependent enzymes occurring under conditions of reduced oxygenation (32, 34, 47–49).
GO clustering of HMGs in NK cells indicates that hypoxia can modulate several biological processes and suggests that NK cells reaching hypoxic tumor areas may deeply change their mode to respond to stimuli or exert their functions. Processes related to metabolism and biosynthesis, response to stimuli, regulation of apoptosis and response to stress, cell proliferation, and signaling appear to be all affected by hypoxia suggesting that NK cells may modulate a wide range of functions in a hypoxic environment. In particular, the coordinated enrichment of down- or up-regulated genes in specific processes, such as cell cycle, DNA replication and repair, cellular component organization, regulation of gene transcription and expression, indicates that NK cells can moderate their biosynthetic and proliferative capabilities in response to decreased O2 tension.
Noteworthily, among HMGs we identified a significant cluster of immune-related genes, 43 of which coding for cytokines, chemokines, and their receptors. Several of these cytokine/chemokine-coding genes have not been previously reported to be affected by hypoxia in NK cells, although some of them are known from the literature to be modulated in other immune cells either exposed to short-term hypoxia (typically 8–24 h) (48, 49, 65, 66, 70, 71) or generated under conditions of long-term hypoxia (37, 38, 47, 67–69). On the other hand, some genes appear to be modulated uniquely in NK cells, as they have never been characterized in terms of responsiveness to hypoxia in other immune cells. These findings indicate that hypoxia regulates the expression of genes coding for cytokines/chemokines on different immune cell populations, but it can also activate a distinct transcriptional profile in NK cells.
The data on immune-related HMGs give some hints on how NK cells may be functionally skewed in a hypoxic microenvironment. The early downregulation of genes coding for IFNγ and for several members of the TNF family, such as TNFα, LTA, LTB, TNFSF14, TNFSF10, and TNFSF11, is of particular interest given the role of these molecules in triggering tumor immunogenicity, decreasing tumor proliferation and angiogenesis, and favoring apoptotic tumor cell killing. Likewise, the downregulation of genes coding for TNFRSF18 and IL1RL1 may be crucial, as these molecules act by enhancing IFNγ secretion and potentiating NK cell expansion and response to tumors (62, 63). Consistently, hypoxia also induces the up-regulation of genes coding for important proangiogenic, protumorigenic, prometastatic, and/or immune suppressive factors, namely VEGFA,B, SPP1,2, CXCL8, MIF, TGFβ2, and PDGFD (11, 34, 37, 48, 50–54). Finally, it is remarkable the modulation of genes coding for chemokines and chemokine receptors.
Collectively, our gene expression analysis suggests a major effect of hypoxia on the immunomodulatory functions and the chemotactic properties of NK cells. These aspects are not trivial, as the nature and the function of the NK cell infiltrate at the tumor site, as well as the tissue distribution of specific NK cell subsets, can influence the prognosis of different tumor types (11, 12, 28, 29). For this reason we decided to investigate in detail the effect of hypoxia on these specific functions of NK cells: cytokine/chemokine release and migration to specific stimuli.
Multiplex ELISA analysis of NK cell culture supernatants partly confirmed the indications obtained by the gene chip analysis, showing that, indeed, hypoxia can limit the ability of NK cells to release different factors involved in the host response to the tumor, such as IFNγ, TNFα, GM-CSF, CCL3, and CCL5. These factors are endowed with antitumor activity and/or can induce recruitment, differentiation, proliferation, and activation of APCs, Th1 lymphocytes, and NK cells (9, 11, 72, 73). Hypoxia appears to variably affect cytokine release, depending on the type of NK cell stimulation. Indeed, its inhibitory effect is particularly evident on NK cells exposed to the monokine combination, IL-15 + IL-18, while it does not reach statistical significance in case of IL-12 + IL18 stimulation. We couldn't find any transcriptional modulation of genes coding for IL-12, IL-15, IL-18, or IL-2 receptor subunits, suggesting that the differential effect of hypoxia on the various stimuli may involve other mechanisms such as the interference with specific signaling pathways.
That hypoxia could differentially modulate NK cells under different monokine stimuli is important both because stimulatory monokines can be released by immune cells in inflamed tissues and also in view of the recent lines of research aimed at the definition of effective monokine combinations in NK-based immunotherapy (2, 17, 74). Notably, the hypoxia-related factors CXCL8, VEGF, and MIF were not (or poorly) released by both “normoxic” and “hypoxic” NK cells, although they were induced by hypoxia at the mRNA level. This discrepancy suggests that target-specific translational regulation (49, 67) can shape NK cell response to hypoxia, giving rise to unique functional profiles. The fact that hypoxia fails to induce CXCL8, VEGF, and MIF secretion by NK cells has also been reported in a recent study by Velasquez et al. (41). In that study, however, in contrast to our present data, exposure to hypoxia could induce little, but significant, secretion of CCL3, CCL4, and CCL5. The discrepancy between our and their results may probably be ascribed to the different experimental protocols used, in particular with regard to the priming cytokines (IL-2, IL-12+IL-18, or IL-15+IL-18 in our study vs. IL-15 alone in that of Velasquez) and the time and duration of priming (the whole 20 h culture period in our study vs. the final 6 h culture in the Velasquez study).
Evaluation of chemokine receptor surface expression reveals that hypoxia can significantly increase the expression of CXCR4 receptor on a large fraction of PB-NK cells, suggesting that changes in the levels of O2 tension within tissues may significantly influence NK cell trafficking (11). The CXCL12-CXCR4 axis represents one of the mechanisms responsible for tumor spread, driven by pro-metastatic CXCR4+ tumor cells. In addition, CXCL12 expressed by Tumor Associated Fibroblasts (TAF) and tumor cells has been demonstrated to play an important role in favoring tumor growth and progression in primary lesions. Thus, sustained CXCR4 expression in NK cells may be important for reaching and infiltrating certain metastatic niches (for example in the bones) and also primary tumors. In this context, in a model of NKp46-targeted HIF1α KO mice, it has been recently shown that NK cells can reach hypoxic tumor tissues, influence angiogenesis, tumor growth, and metastasis spread in a HIF1α-dependent fashion (75). Remarkably, our data indicate that, in humans, hypoxia can differently affect two functionally distinct NK cell subsets. We observed that hypoxia-induced CXCR4 up-regulation involved the whole CD56bright NK cell population, while it affected only a fraction (even if large) of CD56dim cells. Accordingly, hypoxic NK cells that migrated to CXCL12 showed an enrichment of such CD56bright cell subset. Along this line, hypoxia also increased CCR7 expression on CD56bright cells, enhancing their selective migration in response to CCL19 and CCL21. The CCL19/21-CCR7 axis drives metastatic spread to Lymph Nodes but also promotes homing of specific leukocyte subsets. In addition, the CCL21-CCR7 interaction may be effective at the tumor site (76, 77). Overall, our data suggest that hypoxia can intervene in the recruitment of specific NK cell subsets at the site of both primary tumor and metastasis and offer new hints to explain the relative high frequencies of poorly cytotoxic CD56bright cells observed within the NK cell infiltrate of several tumors (29–31). Of course these findings, although suggestive, must be considered within the rich network of factors that regulates lymphocyte trafficking in different tumor sites. As an example, it has been recently described the complex correlation between the chemokine receptor pattern of different T lymphocyte subsets and the control of metastasis in specific sites (78). Also NK cells can respond to multiple chemokines that can be variably released in different tumor sites. In this context, it is worth-noting that NK cells can amplify their recruitment at the tumor site by killing tumor cells and inducing release of chemiotactic HMGB1 (79).
Various escape mechanisms induced by the tumor hypoxic environment have been documented in the last years, most relying on the suppression of different immune cell types, others involving the editing of the tumor cell targets or the tumor microenvironment (23, 33, 38, 47, 80–85). However, only few studies were focused on NK cells. This report represents the first comprehensive transcriptome analysis of human Hy-NK cells, which defines a wide array of hypoxia-modulated immunological genes. Remarkably, our study also describes how hypoxia can influence the type and function of NK cells reaching hypoxic tissues, thus providing new elements useful to design improved NK cell-based immunotherapeutic strategies.
Ethics Statement
This study was carried out following approved operational procedures of the Ethics Committee of the IRCCS Ospedale Policlinico San Martino (IOH78). Written informed consent was obtained from all the donors in adherence with the Declaration of Helsinki.
Author Contributions
MP and FR conducted the experiments, assembled and analyzed data, and participated in MS writing. DC provided statistical analysis of microarray data and performed GSEA analysis. CM, MB, and FB provided experimental support and helped in the analysis of data. AE revised the manuscript. LV, GP, LM, and MM revised the manuscript and provided financial support. MV and MCB conceived the study, designed experiments, interpreted the data, wrote the manuscript, and provided financial support.
Funding
This work was supported by Associazione Italiana Ricerca sul Cancro AIRC under grants: IG 2014 project n. 15428 (MV), IG 2014 project n. 15283 (LM), and IG 2017—Project n. 19920 (LM); PRA 2013 and FRA 2015, DIMES, University of Genoa (GP); 5 × 1000 Min. Sal. 2013 (MV and MM); by the Italian Ministry of Health (MCB); and by Ricerca Corrente 2018 (MCB). MP was recipient of the AIRC fellowship n. 18274, year 2016.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Marzia Ognibene for her valuable support in the carry out of NK cell culture under hypoxic conditions and Dt. Massimo Acquaviva for his support in the statistical analysis of data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2018.02358/full#supplementary-material
Abbreviations
PB, peripheral blood; pO2, low oxygen tension; GSEA, Gene Set Enrichment Analysis; qRT-PCR, Real time PCR; Hy-NK, hypoxic NK cells; HIF, hypoxia-inducible factor; NES, normalized enrichment score; FDR q-val, false discovery rate q-values; NOM p-val, nominal p-values; LEA, Leading Edge Analysis; GO, gene ontology; HMGs, hypoxia-modulated genes; MDMs, monocyte-derived macrophages; iDCs, immature DCs; mDCs, mature DCs; IONO, ionomycin.
References
1. Locatelli F, Pende D, Falco M, Della CM, Moretta A, Moretta L. NK cells mediate a crucial graft-versus-leukemia effect in haploidentical-HSCT to cure high-risk acute leukemia. Trends Immunol. (2018) 39:577–90. doi: 10.1016/j.it.2018.04.009
2. Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. (2016) 17:1025–36. doi: 10.1038/ni.3518
3. Lopez-Soto A, Gonzalez S, Smyth MJ, Galluzzi L. Control of metastasis by NK cells. Cancer Cell (2017) 32:135–54. doi: 10.1016/j.ccell.2017.06.009
4. Fend L, Rusakiewicz S, Adam J, Bastien B, Caignard A, Messaoudene M, et al. Prognostic impact of the expression of NCR1 and NCR3 NK cell receptors and PD-L1 on advanced non-small cell lung cancer. Oncoimmunology (2017) 6:e1163456. doi: 10.1080/2162402X.2016.1163456
5. Cantoni C, Huergo-Zapico L, Parodi M, Pedrazzi M, Mingari MC, Moretta A, et al. NK cells, tumor cell transition, and tumor progression in solid malignancies: new hints for NK-based immunotherapy? J Immunol Res. (2016) 2016:4684268. doi: 10.1155/2016/4684268
6. Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, et al. Innate or adaptive immunity? The example of natural killer cells. Science (2011) 331:44–9. doi: 10.1126/science.1198687
7. Barrow AD, Edeling MA, Trifonov V, Luo J, Goyal P, Bohl B, et al. Natural killer cells control tumor growth by sensing a growth factor. Cell (2018) 172:534–48. doi: 10.1016/j.cell.2017.11.037
8. Gaggero S, Bruschi M, Petretto A, Parodi M, Del Zotto G, Lavarello C, et al. Nidogen-1 is a novel extracellular ligand for the NKp44 activating receptor. Oncoimmunology (2018) 7:e1470730. doi: 10.1080/2162402X.2018.1470730
9. Maghazachi AA. Role of chemokines in the biology of natural killer cells. Curr Top Microbiol Immunol. (2010) 341:37–58. doi: 10.1007/82_2010_20
10. Fauriat C, Long EO, Ljunggren HG, Bryceson YT. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood (2010) 115:2167–76. doi: 10.1182/blood-2009-08-238469
11. Bernardini G, Santoni A. The pathophysiological role of chemokines in the regulation of NK cell tissue homing.Crit Rev Oncog. (2014) 19:77–90. doi: 10.1615/CritRevOncog.2014010386
12. Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The broad spectrum of human natural killer cell diversity. Immunity (2017) 47:820–33. doi: 10.1016/j.immuni.2017.10.008
13. Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood (2010) 116:3853–64. doi: 10.1182/blood-2010-04-281675
14. Moretta L. Dissecting CD56dim human NK cells. Blood (2010) 116:3689–91. doi: 10.1182/blood-2010-09-303057
15. Berahovich RD, Lai NL, Wei Z, Lanier LL, Schall TJ. Evidence for NK cell subsets based on chemokine receptor expression. J Immunol. (2006) 177:7833–40. doi: 10.4049/jimmunol.177.11.7833
16. Moretta A, Marcenaro E, Sivori S, Della CM, Vitale M, Moretta L. Early liaisons between cells of the innate immune system in inflamed peripheral tissues. Trends Immunol. (2005) 26:668–75. doi: 10.1016/j.it.2005.09.008
17. Vitale M, Cantoni C, Pietra G, Mingari MC, Moretta L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur J Immunol. (2014) 44:1582–92. doi: 10.1002/eji.201344272
18. Balsamo M, Scordamaglia F, Pietra G, Manzini C, Cantoni C, Boitano M, et al. Melanoma-associated fibroblasts modulate NK cell phenotype and antitumor cytotoxicity. Proc Natl Acad Sci USA. (2009) 106:20847–52. doi: 10.1073/pnas.0906481106
19. Pietra G, Manzini C, Rivara S, Vitale M, Cantoni C, Petretto A, et al. Melanoma cells inhibit natural killer cell function by modulating the expression of activating receptors and cytolytic activity. Cancer Res. (2012) 72:1407–15. doi: 10.1158/0008-5472.CAN-11-2544
20. Li H, Han Y, Guo Q, Zhang M, Cao X. Cancer-expanded myeloid-derived suppressor cells induce anergy of NK cells through membrane-bound TGF-beta 1. J Immunol. (2009) 182:240–9. doi: 10.4049/jimmunol.182.1.240
21. Trzonkowski P, Szmit E, Mysliwska J, Dobyszuk A, Mysliwski A. CD4+CD25+ T regulatory cells inhibit cytotoxic activity of T CD8+ and NK lymphocytes in the direct cell-to-cell interaction. Clin Immunol. (2004) 112:258–67. doi: 10.1016/j.clim.2004.04.003
22. Cekic C, Day YJ, Sag D, Linden J. Myeloid expression of adenosine A2A receptor suppresses T and NK cell responses in the solid tumor microenvironment. Cancer Res. (2014) 74:7250–9. doi: 10.1158/0008-5472.CAN-13-3583
23. Hasmim M, Messai Y, Ziani L, Thiery J, Bouhris JH, Noman MZ, et al. Critical role of tumor microenvironment in shaping NK cell functions: implication of hypoxic stress. Front Immunol. (2015) 6:482. doi: 10.3389/fimmu.2015.00482
24. El-Gazzar A, Groh V, Spies T. Immunobiology and conflicting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J Immunol. (2013) 191:1509–15. doi: 10.4049/jimmunol.1301071
25. Schlecker E, Fiegler N, Arnold A, Altevogt P, Rose-John S, Moldenhauer G, et al. Metalloprotease-mediated tumor cell shedding of B7-H6, the ligand of the natural killer cell-activating receptor NKp30. Cancer Res. (2014) 74:3429–40. doi: 10.1158/0008-5472.CAN-13-3017
26. Balsamo M, Vermi W, Parodi M, Pietra G, Manzini C, Queirolo P, et al. Melanoma cells become resistant to NK-cell-mediated killing when exposed to NK-cell numbers compatible with NK-cell infiltration in the tumor. Eur J Immunol. (2012) 42:1833–42. doi: 10.1002/eji.201142179
27. Elboim M, Gazit R, Gur C, Ghadially H, Betser-Cohen G, Mandelboim O. Tumor immunoediting by NKp46. J Immunol. (2010) 184:5637–44. doi: 10.4049/jimmunol.0901644
28. Remark R, Alifano M, Cremer I, Lupo A, Dieu-Nosjean MC, Riquet M, et al. Characteristics and clinical impacts of the immune environments in colorectal and renal cell carcinoma lung metastases: influence of tumor origin. Clin Cancer Res. (2013) 19:4079–91. doi: 10.1158/1078-0432.CCR-12-3847
29. Rusakiewicz S, Semeraro M, Sarabi M, Desbois M, Locher C, Mendez R, et al. Immune infiltrates are prognostic factors in localized gastrointestinal stromal tumors. Cancer Res. (2013) 73:3499–510. doi: 10.1158/0008-5472.CAN-13-0371
30. Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. (2011) 71:5412–22. doi: 10.1158/0008-5472.CAN-10-4179
31. Carrega P, Bonaccorsi I, Di CE, Morandi B, Paul P, Rizzello V, et al. CD56(bright)perforin(low) noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J Immunol. (2014) 192:3805–15. doi: 10.4049/jimmunol.1301889
32. Semenza GL. Oxygen sensing, homeostasis, and disease. N Engl J Med. (2011) 365:537–47. doi: 10.1056/NEJMra1011165
33. Noman MZ, Hasmim M, Messai Y, Terry S, Kieda C, Janji B, et al. Hypoxia: a key player in antitumor immune response. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol. (2015) 309:C569–79. doi: 10.1152/ajpcell.00207.2015
34. Bosco MC, Puppo M, Blengio F, Fraone T, Cappello P, Giovarelli M, et al. Monocytes and dendritic cells in a hypoxic environment: spotlights on chemotaxis and migration. Immunobiology (2008) 213:733–49. doi: 10.1016/j.imbio.2008.07.031
35. Palazon A, Aragones J, Morales-Kastresana A, de Landazuri MO, Melero I. Molecular pathways: hypoxia response in immune cells fighting or promoting cancer. Clin Cancer Res. (2012) 18:1207–13. doi: 10.1158/1078-0432.CCR-11-1591
36. Pierobon D, Bosco MC, Blengio F, Raggi F, Eva A, Filippi M, et al. Chronic hypoxia reprograms human immature dendritic cells by inducing a proinflammatory phenotype and TREM-1 expression. Eur J Immunol. (2013) 43:949–66. doi: 10.1002/eji.201242709
37. Blengio F, Raggi F, Pierobon D, Cappello P, Eva A, Giovarelli M, et al. The hypoxic environment reprograms the cytokine/chemokine expression profile of human mature dendritic cells. Immunobiology (2013) 21876–89. doi: 10.1016/j.imbio.2012.02.002
38. Raggi F, Pelassa S, Pierobon D, Penco F, Gattorno M, Novelli F, et al. Regulation of human macrophage M1-M2 polarization balance by hypoxia and the triggering receptor expressed on myeloid cells-1. Front Immunol. (2017) 8:1097. doi: 10.3389/fimmu.2017.01097
39. Balsamo M, Manzini C, Pietra G, Raggi F, Blengio F, Mingari MC, et al. Hypoxia downregulates the expression of activating receptors involved in NK-cell-mediated target cell killing without affecting ADCC. Eur J Immunol. (2013) 43:2756–64. doi: 10.1002/eji.201343448
40. Sarkar S, Germeraad WT, Rouschop KM, Steeghs EM, van GM, Bos GM, et al. Hypoxia induced impairment of NK cell cytotoxicity against multiple myeloma can be overcome by IL-2 activation of the NK cells. PLoS ONE (2013) 8:e64835. doi: 10.1371/journal.pone.0064835
41. Velasquez SY, Killian D, Schulte J, Sticht C, Thiel M, Lindner HA. Short term hypoxia synergizes with interleukin 15 priming in driving glycolytic gene transcription and supports human natural killer cell activities. J Biol Chem. (2016) 291:12960–77. doi: 10.1074/jbc.M116.721753
42. Bosco MC, Pierobon D, Blengio F, Raggi F, Vanni C, Gattorno M, et al. Hypoxia modulates the gene expression profile of immunoregulatory receptors in human mature dendritic cells: identification of TREM-1 as a novel hypoxic marker in vitro and in vivo. Blood (2011) 117:2625–39. doi: 10.1182/blood-2010-06-292136
43. Cangelosi D, Pelassa S, Morini M, Conte M, Bosco MC, Eva A, et al. Artificial neural network classifier predicts neuroblastoma patients' outcome. BMC Bioinformatics (2016) 17(Suppl. 12):83–93. doi: 10.1186/s12859-016-1194-3
44. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
45. Raggi F, Blengio F, Eva A, Pende D, Varesio L, Bosco MC. Identification of CD300a as a new hypoxia-inducible gene and a regulator of CCL20 and VEGF production by human monocytes and macrophages. Innate Immun. (2014) 20:721–34. doi: 10.1177/1753425913507095
46. Elvidge GP, Glenny L, Appelhoff RJ, Ratcliffe Pj, Ragoussis J, Gleadle JM. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem. (2006) 281:15215–26. doi: 10.1074/jbc.M511408200
47. Bosco MC, Varesio L. Dendritic cell reprogramming by the hypoxic environment. Immunobiology (2012) 217:1241–9. doi: 10.1016/j.imbio.2012.07.023
48. Bosco MC, Puppo M, Santangelo C, Anfosso L, Pfeffer U, Fardin P, et al. Hypoxia modifies the transcriptome of primary human monocytes: modulation of novel immune-related genes and identification of CC-chemokine ligand 20 as a new hypoxia-inducible gene. J Immunol. (2006) 177:1941–55. doi: 10.4049/jimmunol.177.3.1941
49. Gaber T, Haupl T, Sandig G, Tykwinska K, Fangradt M, Tschirschmann M, et al. Adaptation of human CD4+ T cells to pathophysiological hypoxia: a transcriptome analysis. J Rheumatol. (2009) 36:2655–69. doi: 10.3899/jrheum.090255
50. Shijubo N, Uede T, Kon S, Nagata M, Abe S. Vascular endothelial growth factor and osteopontin in tumor biology. Crit Rev Oncog. (2000) 11:135–46.
51. Krockenberger M, Dombrowski Y, Weidler C, Ossadnik M, Honig A, Hausler S, et al. Macrophage migration inhibitory factor contributes to the immune escape of ovarian cancer by down-regulating NKG2D. J Immunol. (2008) 180:7338–48. doi: 10.4049/jimmunol.180.11.7338
52. Ben Baruch A. Inflammation-associated immune suppression in cancer: the roles played by cytokines, chemokines and additional mediators. Semin Cancer Biol. (2006) 16:38–52. doi: 10.1016/j.semcancer.2005.07.006
53. Wu Q, Hou X, Xia J, Qian X, Miele L, Sarkar FH, et al. Emerging roles of PDGF-D in EMT progression during tumorigenesis. Cancer Treat Rev. (2013) 39:640–6. doi: 10.1016/j.ctrv.2012.11.006
54. Friese MA, Wischhusen J, Wick W, Weiler M, Eisele G, Steinle A, et al. RNA interference targeting transforming growth factor-beta enhances NKG2D-mediated antiglioma immune response, inhibits glioma cell migration and invasiveness, and abrogates tumorigenicity in vivo. Cancer Res. (2004) 64:7596–603. doi: 10.1158/0008-5472.CAN-04-1627
55. Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat Immunol. (2005) 6:600–7. doi: 10.1038/ni1197
56. Schoenborn JR, Wilson CB. Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol. (2007) 96:41–101. doi: 10.1016/S0065-2776(07)96002-2
57. Bertazza L, Mocellin S. The dual role of tumor necrosis factor (TNF) in cancer biology. Curr Med Chem. (2010) 17:3337–52. doi: 10.2174/092986710793176339
58. Croft M, Duan W, Choi H, Eun SY, Madireddi S, Mehta A. TNF superfamily in inflammatory disease: translating basic insights. Trends Immunol. (2012) 33:144–52. doi: 10.1016/j.it.2011.10.004
59. Fan Z, Yu P, Wang Y, Wang Y, Fu ML, Liu W, et al. NK-cell activation by LIGHT triggers tumor-specific CD8+ T-cell immunity to reject established tumors. Blood (2006) 107:1342–51. doi: 10.1182/blood-2005-08-3485
60. Holmes TD, Wilson EB, Black EV, Benest AV, Vaz C, Tan B, et al. Licensed human natural killer cells aid dendritic cell maturation via TNFSF14/LIGHT. Proc Natl Acad Sci USA. (2014) 111:E5688–96. doi: 10.1073/pnas.1411072112
61. Smyth MJ, Takeda K, Hayakawa Y, Peschon JJ, van den Brink MR, Yagita H. Nature's TRAIL–on a path to cancer immunotherapy. Immunity (2003) 18:1–6. doi: 10.1016/S1074-7613(02)00502-2
62. Nocentini G, Riccardi C. GITR: a modulator of immune response and inflammation. Adv Exp Med Biol. (2009) 647:156–73. doi: 10.1007/978-0-387-89520-8_11
63. Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R, Smith DE. IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int Immunol. (2008) 20:1019–30. doi: 10.1093/intimm/dxn060
64. Billottet C, Quemener C, Bikfalvi A. CXCR3, a double-edged sword in tumor progression and angiogenesis. Biochim Biophys Acta (2013) 1836:287–95. doi: 10.1016/j.bbcan.2013.08.002
65. Fang HY, Hughes R, Murdoch C, Coffelt SB, Biswas SK, Harris AL, et al. Hypoxia-inducible factors 1 and 2 are important transcriptional effectors in primary macrophages experiencing hypoxia. Blood (2009) 114:844–59. doi: 10.1182/blood-2008-12-195941
66. Bosco MC, Reffo G, Puppo M, Varesio L. Hypoxia inhibits the expression of the CCR5 chemokine receptor in macrophages. Cell Immunol. (2004) 228:1–7. doi: 10.1016/j.cellimm.2004.03.006
67. Ricciardi A, Elia AR, Cappello P, Puppo M, Vanni C, Fardin P, et al. Transcriptome of hypoxic immature dendritic cells: modulation of chemokine/receptor expression. Mol Cancer Res. (2008) 6:175–85. doi: 10.1158/1541-7786.MCR-07-0391
68. Yang M, Ma C, Liu S, Sun J, Shao Q, Gao W, et al. Hypoxia skews dendritic cells to a T helper type 2-stimulating phenotype and promotes tumour cell migration by dendritic cell-derived osteopontin. Immunology (2009) 128(1 Suppl.):e237–49. doi: 10.1111/j.1365-2567.2008.02954.x
69. Elia AR, Cappello P, Puppo M, Fraone T, Vanni C, Eva A, et al. Human dendritic cells differentiated in hypoxia down-modulate antigen uptake and change their chemokine expression profile. J Leukoc Biol. (2008) 84:1472–82. doi: 10.1189/jlb.0208082
70. Mancino A, Schioppa T, Larghi P, Pasqualini F, Nebuloni M, Chen IH, et al. Divergent effects of hypoxia on dendritic cell functions. Blood (2008) 112:3723–34. doi: 10.1182/blood-2008-02-142091
71. Caldwell CC, Kojima H, Lukashev D, Armstrong J, Farber M, Apasov SG, et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J Immunol. (2001) 167:6140–9. doi: 10.4049/jimmunol.167.11.6140
72. Kaufman HL, Ruby CE, Hughes T, Slingluff CL Jr. Current status of granulocyte-macrophage colony-stimulating factor in the immunotherapy of melanoma. J Immunother Cancer (2014) 2:11. doi: 10.1186/2051-1426-2-11
73. Lapteva N, Huang XF. CCL5 as an adjuvant for cancer immunotherapy. Expert Opin Biol Ther. (2010) 10:725–33. doi: 10.1517/14712591003657128
74. Ni J, Miller M, Stojanovic A, Garbi N, Cerwenka A. Sustained effector function of IL-12/15/18-preactivated NK cells against established tumors. J Exp Med. (2012) 209:2351–65 doi: 10.1084/jem.20120944
75. Krzywinska E, Kantari-Mimoun C, Kerdiles Y, Sobecki M, Isagawa T, Gotthardt D, et al. Loss of HIF-1alpha in natural killer cells inhibits tumour growth by stimulating non-productive angiogenesis. Nat Commun. (2017) 8:1597. doi: 10.1038/s41467-017-01599-w
76. Tutunea-Fatan E, Majumder M, Xin X, Lala PK. The role of CCL21/CCR7 chemokine axis in breast cancer-induced lymphangiogenesis. Mol Cancer (2015) 14:35. doi: 10.1186/s12943-015-0306-4
77. Sand LG, Berghuis D, Szuhai K, Hogendoorn PC. Expression of CCL21 in Ewing sarcoma shows an inverse correlation with metastases and is a candidate target for immunotherapy. Cancer Immunol Immunother. (2016) 65:995–1002. doi: 10.1007/s00262-016-1862-1
78. Jacquelot N, Enot DP, Flament C, Vimond N, Blattner C, Pitt JM, et al. Chemokine receptor patterns in lymphocytes mirror metastatic spreading in melanoma. J Clin Invest. (2016) 126:921–37. doi: 10.1172/JCI80071
79. Parodi M, Pedrazzi M, Cantoni C, Averna M, Patrone M, Cavaletto M, et al. Natural Killer (NK)/melanoma cell interaction induces NK-mediated release of chemotactic High Mobility Group Box-1 (HMGB1) capable of amplifying NK cell recruitment. Oncoimmunology (2015) 4:e1052353. doi: 10.1080/2162402X.2015.1052353
80. Zhang J, Cao J, Ma S, Dong R, Meng W, Ying M, et al. Tumor hypoxia enhances Non-Small Cell Lung Cancer metastasis by selectively promoting macrophage M2 polarization through the activation of ERK signaling. Oncotarget (2014) 5:9664–77. doi: 10.18632/oncotarget.1856
81. Tripathi C, Tewari BN, Kanchan RK, Baghel KS, Nautiyal N, Shrivastava R, et al. Macrophages are recruited to hypoxic tumor areas and acquire a pro-angiogenic M2-polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget (2014) 5:5350–68. doi: 10.18632/oncotarget.2110
82. Sitkovsky M, Lukashev D. Regulation of immune cells by local-tissue oxygen tension: HIF1 alpha and adenosine receptors. Nat Rev Immunol. (2008) 5:712–21. doi: 10.1038/nri1685
83. Westendorf AM, Skibbe K, Adamczyk A, Buer J, Geffers R, Hansen W, et al. Hypoxia enhances immunosuppression by inhibiting CD4+ effector T cell function and promoting treg activity. Cell Physiol Biochem. (2017) 41:1271–84. doi: 10.1159/000464429
84. Kumar V, Gabrilovich DI. Hypoxia-inducible factors in regulation of immune responses in tumour microenvironment. Immunology (2014) 143:512–9. doi: 10.1111/imm.12380
Keywords: NK cells, hypoxia, tumor immunology, cytokines/chemokines, chemokine receptors, CD56bright cells, tumor infiltration, transcriptome
Citation: Parodi M, Raggi F, Cangelosi D, Manzini C, Balsamo M, Blengio F, Eva A, Varesio L, Pietra G, Moretta L, Mingari MC, Vitale M and Bosco MC (2018) Hypoxia Modifies the Transcriptome of Human NK Cells, Modulates Their Immunoregulatory Profile, and Influences NK Cell Subset Migration. Front. Immunol. 9:2358. doi: 10.3389/fimmu.2018.02358
Received: 06 August 2018; Accepted: 24 September 2018;
Published: 16 October 2018.
Edited by:
Marina Cella, Washington University in St. Louis, United StatesReviewed by:
Kerry S. Campbell, Fox Chase Cancer Center, United StatesEmily Mace, Columbia University, United States
Copyright © 2018 Parodi, Raggi, Cangelosi, Manzini, Balsamo, Blengio, Eva, Varesio, Pietra, Moretta, Mingari, Vitale and Bosco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimo Vitale, bWFzc2ltby52aXRhbGVAaHNhbm1hcnRpbm8uaXQ=
†These authors have contributed equally to this work
‡These authors share senior authorship
§Deceased
 Monica Parodi
Monica Parodi Federica Raggi
Federica Raggi Davide Cangelosi
Davide Cangelosi Claudia Manzini
Claudia Manzini Mirna Balsamo
Mirna Balsamo Fabiola Blengio
Fabiola Blengio Alessandra Eva
Alessandra Eva Luigi Varesio
Luigi Varesio Gabriella Pietra1,4
Gabriella Pietra1,4 Lorenzo Moretta
Lorenzo Moretta Maria Cristina Mingari
Maria Cristina Mingari Massimo Vitale
Massimo Vitale Maria Carla Bosco
Maria Carla Bosco