- 1Water Research Institute, National Research Council (IRSA-CNR), Rome, Italy
- 2Institute of BioEconomy – National Research Council (IBE-CNR), Florence, Italy
- 3Institute of Geosciences and Earth Resources, National Research Council (IGG-CNR), Florence, Italy
- 4Department of Earth Sciences, University of Florence, Florence, Italy
- 5Department of Ecology and Evolutionary Biology, University of Colorado Boulder, Boulder, CO, United States
Direct and indirect effects of extremely high geogenic CO2 levels, commonly occurring in volcanic and hydrothermal environments, on biogeochemical processes in soil are poorly understood. This study investigated a sinkhole in Italy where long-term emissions of thermometamorphic-derived CO2 are associated with accumulation of carbon in the topsoil and removal of inorganic carbon in low pH environments at the bottom of the sinkhole. The comparison between interstitial soil gasses and those collected in an adjacent bubbling pool and the analysis of the carbon isotopic composition of CO2 and CH4 clearly indicated the occurrence of CH4 oxidation and negligible methanogenesis in soils at the bottom of the sinkhole. Extremely high CO2 concentrations resulted in higher microbial abundance (up to 4 × 109 cell g–1 DW) and a lower microbial diversity by favoring bacteria already reported to be involved in acetogenesis in mofette soils (i.e., Firmicutes, Chloroflexi, and Acidobacteria). Laboratory incubations to test the acetogenic and methanogenic potential clearly showed that all the mofette soil supplied with hydrogen gas displayed a remarkable CO2 fixation potential, primarily due to the activity of acetogenic microorganisms. By contrast, negligible production of acetate occurred in control tests incubated with the same soils, under identical conditions, without the addition of hydrogen. In this study, we report how changes in diversity and functions of the soil microbial community – induced by high CO2 concentration – create peculiar biogeochemical profile. CO2 emission affects carbon cycling through: (i) inhibition of the decomposition of the organic carbon and (ii) promotion of CO2-fixation via the acetyl-CoA pathway. Sites naturally exposed to extremely high CO2 levels could potentially represent an untapped source of microorganisms with unique capabilities to catalytically convert CO2 into valuable organic chemicals and fuels.
Introduction
Natural diffuse gas emitting areas, emanating almost pure volcanic or thermometamorphic CO2 to the atmosphere, commonly result in CO2 concentrations (>90% v/v) in the soil markedly higher than typical soil CO2 contents (ranging from near atmospheric levels to 100-fold higher values and generally <10% v/v; e.g., Amundson and Davidson, 1990; Oh et al., 2005; Schaetzl and Anderson, 2005), which are likely on par with CO2 concentrations in the Earth’s atmosphere when photosynthesis evolved (Beulig et al., 2016).
Soil biogeochemical processes in areas affected by direct and indirect influence of extremely high CO2 levels are poorly studied although they could potentially have significant ecological, environmental and biotechnological implications. Despite the fact that responses to high CO2 emissions were mainly studied on plants and on their capacity of carbon fixation, recent investigations clearly pointed out the pivotal role played by naturally occurring microbial communities on the utilization of volcanic CO2 and its incorporation into the soil organic matter (Beulig et al., 2016). Microbially driven CO2 utilization is a ubiquitous process in soils, carried out by different microbial metabolic pathways. Under chemical-physical conditions typical of venting spots (e.g., acidic pH and absence of oxygen), strictly anaerobic autotrophic prokaryotes, such as acetogenic bacteria (using hydrogen to reduce carbon dioxide into acetic acid) and hydrogenotrophic methanogenic archaea (using hydrogen to reduce carbon dioxide to methane), were reported to be the dominant members of the soil microbiome (Beulig et al., 2015). Both acetogens and methanogens use molecular hydrogen as electron donor and CO2 as terminal electron acceptor in their energy metabolism, producing acetate or methane as end-products. Several environmental parameters (e.g., temperature, pH, hydrogen partial pressure) influence competitive interactions between these two trophic groups, ultimately shaping the flow of carbon and electrons, since either acetate or methane can be produced. At low temperatures (i.e., <15°C) and acidic pH (i.e., <5), acetogenic bacteria can outcompete methanogens for hydrogen use, due to their higher growth rates (Cord-Ruwisch et al., 1988; Kotsyurbenko et al., 2001). By contrast, due to their higher affinity for hydrogen, documented by a lower half-saturation Michaelis-Menten constant and lower minimal hydrogen threshold concentration, methanogens can have a competitive advantage over acetogens in environments characterized by a low hydrogen availability (Cord-Ruwisch et al., 1988; Kotsyurbenko et al., 2001). To date, the competition between methanogens and acetogens was primarily investigated in laboratory-scale bioreactors typically operating under highly controlled, steady-state conditions, whereas only limited information is available on its relevance in natural ecosystems.
Sites naturally exposed to extremely high CO2 levels could potentially represent an untapped source of microorganisms with unique capabilities to catalytically convert CO2 (at levels as high as those typically occurring in flue gasses emissions of industries or even anaerobic digestion plants) into valuable organic chemicals and fuels. Such microorganisms have recently received attention in the context of microbial electrosynthesis, i.e., a novel technology in which electric current, ideally produced from renewable energy sources (e.g., solar, wind), is supplied to living microorganisms via a cathode to reduce CO2 to yield industrially relevant products, such as methane, volatile fatty acids, and alcohols (Rabaey and Rozendal, 2010). One of the most attractive aspects of microbial electrosynthesis is the remarkable conversion efficiency (> 80%) of electricity to chemicals (e.g., acetate). On the other hand, the relatively low production rates and costly product separation processes reported so far, still largely challenge the commercial exploitation of the technology.
Microbial processes at extremely high CO2 soil concentration levels are also relevant within the broader context of carbon capture and storage (CCS) technologies. The urgent need for large-scale solutions to reduce atmospheric levels of greenhouse gasses has indeed prompted the interest toward CO2 storage in deep saline aquifers or exhausted gas and oil reservoirs. However, before underground CO2 storage can be implemented at large scale, it is necessary to determine the potential environmental risks associated with the leakage of CO2 from the reservoir to the near surface environment, to minimize possible environmental impacts. In principle, leakage of large volumes of CO2 can remarkably affect structures and functions of soil microbial communities, and the overall biogeochemical processes they mediate (McFarland et al., 2013). Sites characterized by CO2-rich gas discharges represent unique natural analogs of engineered carbon storage sites experiencing steady CO2 leakage events. These sites could therefore provide the unique opportunity for studying the potential impacts of CO2 on near-surface ecosystems and groundwater and developing monitoring strategies and possibly preventing CO2 leakage events.
Natural CO2-rich reservoirs commonly occur worldwide (Pearce et al., 2004; Pearce, 2006) and their distribution is mainly controlled by Cenozoic rift systems, e.g., East African Rift System (Vaselli et al., 2002; Smets et al., 2010), Tertiary volcanism and hydrothermal and volcanic systems related to both Quaternary to recent volcanic activity and sedimentary basins.
The Bossoleto hydrothermal area at Rapolano (Tuscany, Italy), where fluxes of gas originated by both thermometamorphic processes on limestone and mantle degassing flow up to the surface in correspondence of deep fractures connected to the faults system (Minissale et al., 2002; Guerra and Raschi, 2004; Brogi et al., 2007), represents a typical example of natural CO2 storage site.
The objective of this study was to investigate the specific interactions between soil CO2 concentration changes and the microbial community dynamics in terms of structure and function to improve our understanding of microbially mediated C cycle, including the potential impact of CO2 on near-surface ecosystems. Onsite investigations in natural CO2 vents were integrated with laboratory experimental approaches in order to: (i) examine the diversity of soil microbial communities (both bacteria and archaea) in natural environments with extremely high CO2 concentration by 16S rRNA gene sequencing and by in situ hybridization approach (CARD-FISH) and (ii) use laboratory incubations to test the acetogenic and methanogenic potential of the microbial communities differently exposed to natural CO2 enrichment.
Materials and Methods
Site Description and Soil Sampling Strategy
The Bossoleto mofette belongs to the Rapolano hydrothermal area (Figure 1). It is a round shaped sinkhole characterized by gaseous emissions primarily consisting of CO2. In this sinkhole, a CO2 lake forms every night due to the combination of site topography and CO2 accumulation. Typically, CO2 concentrations range from 0.04 to 80% (v/v) over a 24-h period. During the night, the concentrations build up, reaching a maximum of about 80% at around 7:00 AM; afterward (typically after 9:00 AM), a rapid decrease occurs when direct radiation is incident on the bottom of the mofette (Van Gardingen et al., 1995; Kies et al., 2015). Due to infrared absorption by CO2, the temperature inside the sinkhole can be up to 30°C, i.e., higher than the corresponding air temperature (Kies et al., 2015). Inside the Bossoleto area, plant populations have therefore existed at elevated CO2 for hundreds of years (Miglietta et al., 1993; Körner and Miglietta, 1994; Raschi et al., 1997), an ample time for the occurrence of evolutionary changes in microorganisms with short generation times (Collins and Bell, 2006).
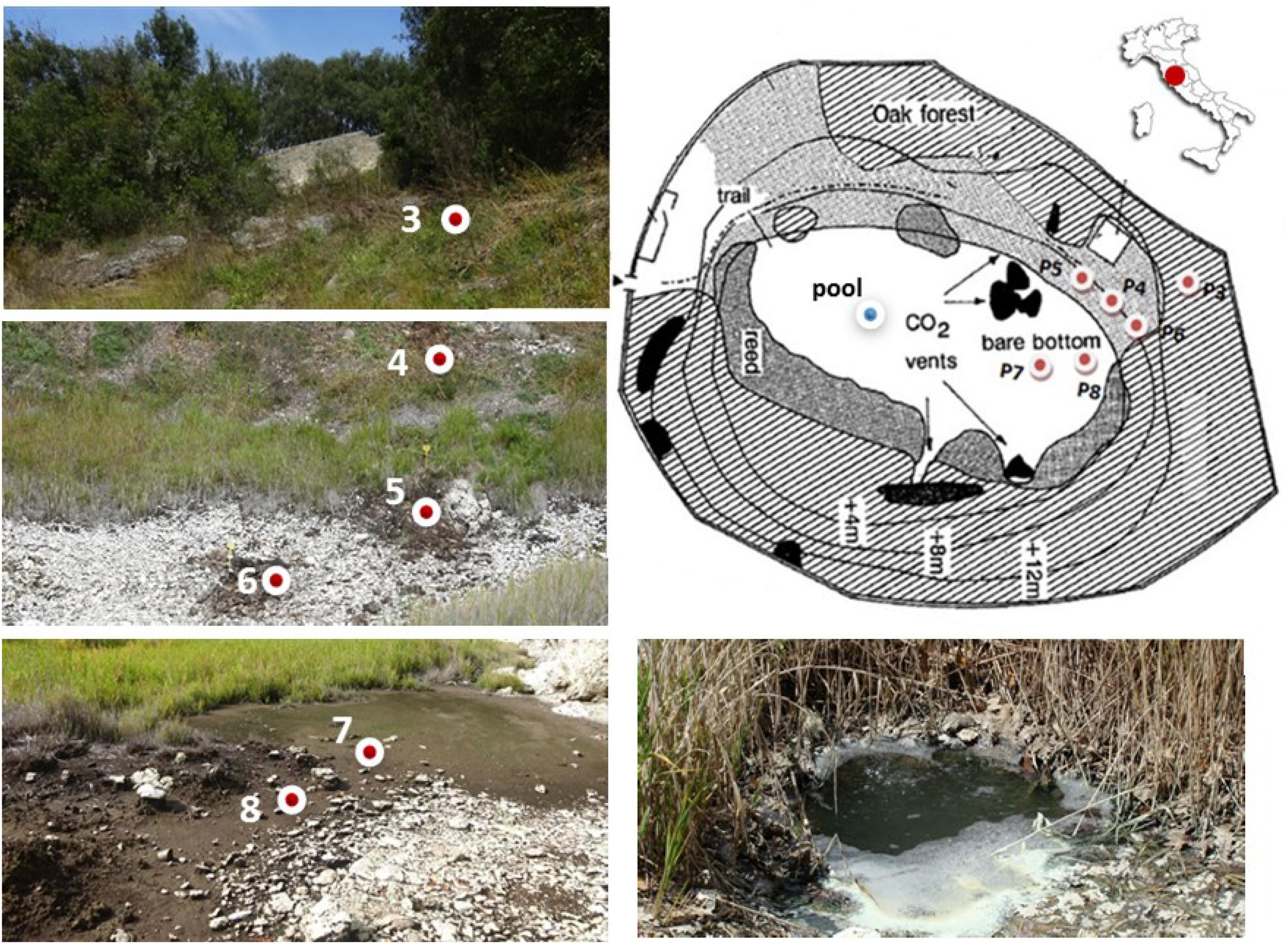
Figure 1. Sampling sites along the geomorphological gradient of the Bossoleto mofette (Rapolano, Tuscany, Italy) (Photos by S. Fazi, Site map modified after Körner and Miglietta, 1994).
Soil sampling was carried out at six locations along a geomorphological gradient spanning from the peak of the sink-rim (site 3), to the back-slope (sites 4-5-6) and to the toe-slope at the bottom of the sinkhole (sites 7-8) (Figure 1). At each sampling site in the sink-rim and at the back-slope, soil samples were collected from the topsoil (0–10 cm, named “Sur”) and the subsurface layers (35–45 cm, hereafter named “Sub”). At the bottom of the sinkhole in sites 7 and 8 the “Sub” samples were collected at 20–30 cm depth. Only at site 7 an additional subsurface sample was collected at −60 cm depth (hereafter named “Deep”). A free-gas sample was collected from one of the bubbling pools (named “BP”) located at the bottom of the Bossoleto sinkhole close to the soil gas sampling sites.
Soil Analyses
In three selected sites (site 3, site 4 and site 7 - Figure 1), soils were characterized in both the field and laboratory from the surface to a depth of approximately 60 cm depth, for a number of parameters including pH, HCl reaction, micro- and macro-nutrients and C-N content. Soil horizons were identified, described and sampled in the field following standard soil survey protocol (USDA, 2017). The pH was measured in a soil/water suspension (1:2.5 mass ratio). HCl reaction was observed in the field by using a 1M solution for determining effervescence class as a relative index of the carbonate amount in the soil matrix (USDA, 2017). Soil C and N contents were measured in laboratory using a CHN Elemental Analyzer (Carlo Erba Instruments, mod 1500 series 2). Dry sub-samples were also digested with a microwave oven (CEM, MARSXpress) according to the EPA method 3052 (U. S. Environmental Protection Agency, 2004). The solutions obtained after the mineralization were filtered (0.45 μm PTFE) and diluted. Micro- and macro-nutrients were determined by an ICP optical spectrometer (Varian Inc., Vista MPX) using scandium as internal standard.
Gas Sampling and Analysis
Interstitial soil gasses were sampled from 5 sites (i.e., sites 3, 4, 5, 6 and 8) along vertical profiles from −10 to −50 cm, at regular depth intervals of 10 cm, except at site 8 where the maximum sampling depth was 20 cm, due to the presence of a shallow water table (Figure 1). Gas sampling was carried out using a stainless-steel tube (internal diameter: 0.4 cm) as described in Tassi et al. (2015). The gas from the pool was collected in a pre-evacuated glass vial (60 cm3) equipped with a thorion© valve and containing 20 mL of 4 M NaOH (Giggenbach, 1975). During sampling, CO2 and H2S dissolved in the alkaline solution, whereas non-condensable gasses (N2, O2, Ar, CH4, C2H6) were stored in the flask headspace. A glass vial for the analysis of 13C/12C ratios in CO2 (δ13C-CO2) was also collected.
Inorganic gasses in both vials (CO2, H2S, N2 and O2 + Ar) and the flask headspace (N2 and O2 + Ar) were analyzed using a Shimadzu 15A gas chromatograph (GC) and a Thermo Focus gas chromatograph, was used to separate the Ar and O2 peaks, as described in Tassi et al. (2015). CH4 and C2H6 were analyzed using a Shimadzu 14A gas chromatograph equipped with flame ionization detector (FID) and a 10 m stainless steel column packed with 80/100 mesh Chromosorb PAW coated with 23% SP 1700 as described in Tassi et al. (2015). The δ13C-CO2 values were measured by a Finnigan MAT 252 mass spectrometer after the extraction and purification of the gas mixtures (Evans et al., 1998; Vaselli et al., 2006). The δ13C-CH4 were analyzed using MS (Varian MAT 250) according to the procedure reported by Schoell (1980).
Bacteria and Archaea Abundance and Diversity
The total bacteria and archaea cell abundances were assessed by Catalyzed Reported Deposition-Fluorescence in situ Hybridization (CARD-FISH) following extraction and detection procedures described elsewhere (Fazi et al., 2007; Amalfitano and Fazi, 2008). Subsamples of the purified cell suspension (1 mL) were filtered onto 0.2 μm-pore size polycarbonate membranes (47 mm diameter) and frozen at −20°C until analysis. Filter sections were then cut and hybridized in duplicate by rRNA-target HRP-labeled probes (Biomers, Ulm, Germany) targeting most Bacteria (EUB338, EUB338-II, EUB338- III) and Archaea (ARCH 915). The stained cells were quantified using an epifluorescence microscope (Leica, DM LB 30). At least 300 cells were counted in each filter section. Data are expressed as a percentage of total DAPI stained prokaryotes cells (%) and as abundance (cells/g of DW).
Sequencing approaches were employed to study microbial diversity across the environmental gradient. Samples of fresh soil from each site at two depths (three in site 7) were frozen directly in the field in dry ice and kept at −80°C until DNA extraction. DNA was extracted from each sample using the approach described in Fierer et al. (2012). To determine the diversity and composition of the bacterial communities, the protocol described in Leff et al. (2015) for amplicon 16S rRNA gene sequencing was used. Specifically, the 16S rRNA gene from isolated DNA was PCR-amplified using the 515f/806r primer pair. To prepare amplicons for sequencing, amplicon purification and normalization was done with Invitrogen SequalPrep Normalization Kit (Invitrogen Inc., CA, United States). Amplicons were combined into a single pool and sequenced using the Illumina MiSeq platform (BioFrontiers Institute, Boulder, CO, United States) using pair-end 2 × 150 bp chemistry.
Data Processing and Statistical Analysis
Forward-oriented sequences were demultiplexed, quality filtered and processed using the Quantitative Insights into Microbial Ecology (QIIME) v1.9.1 bioinformatics package (Caporaso et al., 2010b). 16S rRNA gene paired-end reads were joined and singletons were excluded from further analysis and sequences with >97% identity were clustered into an OTU via UCLUST (Edgar, 2010). Representative sequences for each OTU were chosen for classification and the Greengenes 13.5 database (DeSantis et al., 2006) was employed to assign taxonomy identification to each single OTU. Based on this classification, all mitochondrial and chloroplast OTUs based on this classification were removed from the bacterial data set. The taxonomic assignments of the top OTUs from each treatment were verified by using BLAST to search NCBI, and refined as needed. Sequences were aligned with the PyNAST (Caporaso et al., 2010a) and a phylogeny was built with the FastTree algorithm (Price et al., 2009). OTU tables were rarefied to the lowest number of sequences in the lowest populated sample to make more robust comparisons and were used to assess alpha diversity and relative abundance of all taxa. A community-level Bray-Curtis distance matrix was generated and analyzed with a permutational multivariate analysis of variance (PERMANOVA) using an ADONIS model (Oksanen et al., 2013) to partition the variance in community composition. Principal Coordinate Analysis (PCoA) ordination was constructed based on the basis of the Bray-Curtis distance matrix and Hellinger transformed in order to visualize differences in community composition between low and high CO2 concentrations. Similarity percentage analysis (SIMPER) was used to determine the OTUs that contributed most to the observed dissimilarity between communities of low and high CO2 concentrations. All statistical tests, unless otherwise stated, were performed using the “vegan” R package (Oksanen et al., 2013).
The chemical variables were incorporated into a Non-metric MultiDimensional Scaling (NMDS) ordination plot in order to graphically synthesize the Bray-Curtis dissimilarity among samples. Chemical and microbial data were then projected onto the NMDS ordination using a vector-fitting procedure, in which the length of the arrow is proportional to the correlation between NMDS axes and each variable. This method allowed determining the variation pattern of each projected variable discriminating the samples (Foulquier et al., 2013; Amalfitano et al., 2014).
The 16S rRNA gene sequences from Illumina MiSeq libraries from this study were deposited in the SRA (Short Read Archive) database under Bioproject ID PRJNA548940.
Assessment of the Acetogenic and Methanogenic Potential
To evaluate the acetogenic and methanogenic potential of the mofette microbial communities, a set of anoxic incubations was set up. Incubations were prepared in 120 mL (total volume) serum bottles, incubated statically, in the dark, at room temperature (20–25°C). Each bottle contained about 10 g (wet weight) of mofette soil (site 7sup, 7sub, 7deep, and site 3sub) and 40 mL of anaerobic mineral medium. The medium contained the following components: NH4Cl (0.5 g/L), MgCl2 × 6H2O (0.1 g/L), K2HPO4 (0.4 g/L), and CaCl2 × 2H2O (0.05 g/L). Upon preparation, all bottles were sealed with Teflon-faced butyl rubber stoppers, flushed with a N2/CO2 (70:30 v/v) gas mixture. H2 was added to half of the bottles to reach a final headspace concentration of 60:15 (v/v) H2:CO2. Upon setup, the pH value was in the range 5.5–6.0, hence close to typical pH values measured in the field.
Once all the bottles completely converted the initial dose of H2 (1st feeding cycle), they were flushed with the N2/CO2 gas mixture and then re-spiked with another dose of H2 (2nd feeding cycle). At the end of the first feeding cycle, the liquid volume cumulatively removed during incubation (i.e., for organic acids and pH analyses) was replaced with freshly prepared anaerobic medium. Each incubation experiment was set up in duplicate to ensure reproducibility.
Organic acids were analyzed by injecting 1 μL of filtered (0.22 μm porosity) liquid sample into a PerkinElmer Auto System gas-chromatograph equipped with a Flame Ionization Detector (FID). Gasses (H2, CH4 and CO2) were analyzed by injecting 50 μL of headspace sample into a Perkin-Elmer Auto System gas-chromatograph, equipped with a Thermal Conductivity Detector (TCD).
The percentage of reducing equivalents from the electron donor (i.e., H2) used in acetogenesis or methanogenesis was calculated at the end of each incubation from the measured levels of acetate and methane formed and the electron donor consumed. Molar equivalents factors used were: 8 eq/mol for acetate, 8 eq/mo1 for methane, and 2 eq/mol for hydrogen.
Results
Soil Characterization and Geochemical Analysis
Soils from the Bossoleto sinkhole represent a toposequence with some typical aspects of soil formation in semiarid Mediterranean environments developed on travertine parent materials. Shallow skeletal soils are more developed on the summit of the sinkhole (site 3; 270 m a.s.l.) under oak forest (Quercus ilex L.; Quercus pubescens L.; Fraxinus ornus L.) and scanty herbaceous vegetation cover (Cyclamen repandum Sibth & Sm.; Festuca inops De Not., Teucrium chamaedrys L.). Under these conditions, a thick litter develops and the soil is characterized by the presence of a Bk horizon (i.e., with accumulation of calcium carbonate). The horizons sequence observed in the field is O-A-Bk-BC-Cr (Figure 2) and the soil is classified as Typic Calcixerept according to Soil Taxonomy (USDA, 2010) or as Skeletic Haplic Calcisol (IUSS Working Group Wrb, 2015). Soils along the slope are shallower and less developed: in the back-slope position (site 4; 264 m a.s.l) with uneven grass cover (Sanguisorba minor Scop., Plantago lanceolata L., Centaurea deusta Ten. subsp. deusta), water erosion is more active and only a less developed transitional BC horizon is observed along the soil profile. The horizons sequence observed in the field is O-A-BC-Cr and the soil is classified as (Lithic) Typic Xerorthent (USDA, 2010) or as a Skeletic Regosol (humic) (IUSS Working Group Wrb, 2015). The bottom of the sinkhole (site 7; 258 m a.s.l.) has scant herbaceous cover (Agrostis stolonifera L., Phragmites australis (Cav.) Trin. ex Steud.). Soil profile is formed on colluvial materials whose development is strongly affected by: (i) the rate of delivery of organic and mineral materials from the slopes of the sinkhole, (ii) the presence of high CO2 concentrations and (iii) the shallow fluctuating groundwater. Under these conditions, organic matter accumulates at the top of the soil profile (Oa and O organic topsoil horizons) and strongly anaerobic conditions lead to the formation of deeper Cg horizons with undecomposed root materials. The soil is classified as Thapto-Histic Fluvaquents (USDA, 2010) or Histic Gleyic Fluvisol (IUSS Working Group Wrb, 2015). Soil pH (field, lab and HCl reaction) exhibited a clear decrease along the toposequence (range 4.68–7.60) due to increasing CO2 concentrations in the pedosphere (along soil depth and along the toposequence), which resulted in the complete exploitation of the soil buffer capacity despite the calcareous nature of the parent material (Table 1). In terms of macro-nutrients such as C and N, the soil in site 3 showed intermediate content of N with a positive trend toward the bottom of the sinkhole (range 0.08–1.46%). Sites 3 and 7 also recorded high content of total C (range 2.31–16.61%), with site 5 showing the highest C:N ratio (range 10.32–149.02).

Figure 2. Soil profiles for sites 3, 4 and 7. The sequence of soil horizons along each soil profile is reported. Capital letters designate the master horizons and layers (O, horizons or layers dominated by organic soil materials; A, mineral soil horizon or layers at the soil surface or below an O horizon; B, subsurface mineral horizons that typically formed below an A or O horizon; C, subsurface mineral horizons or layers, excluding strongly cemented and harder bedrock, that are little affected by pedogenic processes and lack properties of O, A, and B horizons; R, strongly cemented to indurated bedrock). Two capital-letter symbols are used for transitional horizons, dominated by properties of one master horizon but have subordinate properties of another. Lowercase letters are used as suffixes to indicate specific characteristics of master horizons and layer (a, highly decomposed organic material; k, accumulation of carbonates; g, strong gleying due to saturation with stagnant water). Arabic numbers as suffixes indicate vertical subdivisions within a horizon or layer.
In terms of micro- and macro-nutrients (Supplementary Table 1), a loss of Ca (−93%) and other bases (e.g., Mg, Li) was observed in site 7 with respect to both sites 3 and 4. This was coupled with an increase in total P (+ 604%) and K (+ 113%) and Cu, Pb, Fe, Al and other microelements, likely because of the effect of the pH on element mobility.
Chemical Composition of Gasses and δ13C-CO2 and δ13C-CH4 Values
The chemical composition (CO2, N2, H2S, O2, Ar in mmol/mol; CH4 and C2H6 in μmol/mol) and the δ13C-CO2 and δ13C-CH4 (in ‰ vs. V-PDB) of both soil gasses and bubbling pool is reported in Table 2. Interstitial soil gasses from profiles at sites 3, 4 and 5 were mainly consisting of N2 (from 744 to 821 mmol/mol) with variable concentrations of O2 (from 19 to 183 mmol/mol), CO2 (from 9.0 to 226 mmol/mol) and minor amounts of Ar (from 10 to 13 mmol/mol). Methane and C2H6 concentrations were up to 16 and 6.1 μmol/mol, respectively, whereas no H2S was detected (<0.05 mmol/mol). Sites 3, 4 and 5 were hereafter named LC: Low CO2 concentration sites.
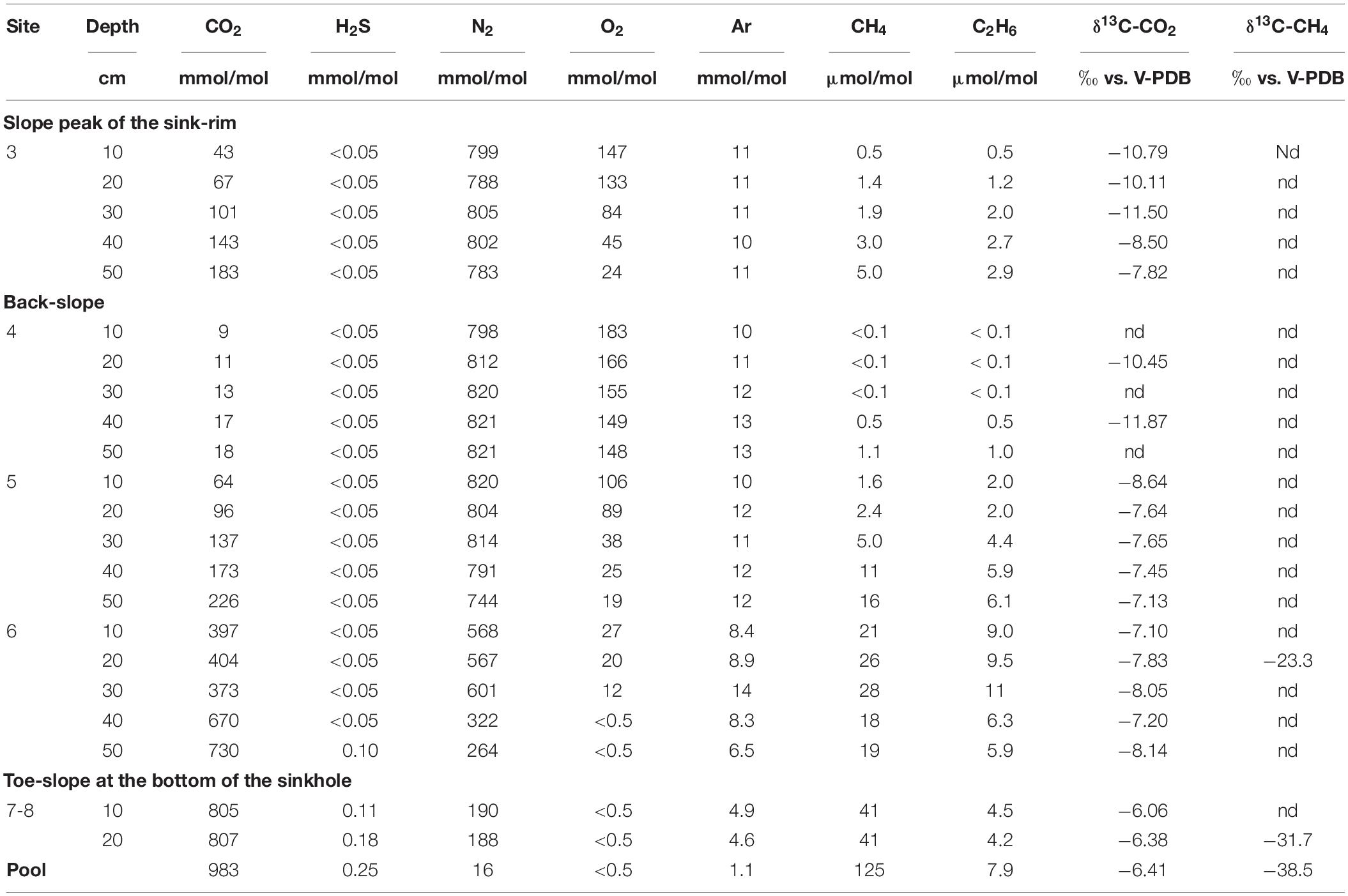
Table 2. Chemical and isotopic composition of interstitial soil gasses and bubbling gasses (pool) from Bossoleto mofette.
The soil gasses from profile at site 6 had CO2 and N2 at comparable concentrations (up to 601 and 730 mmol/mol, respectively), relatively low O2 (≤27 mmol/mol), with Ar from 6.5 to 14 mmol/mol, and CH4 and C2H6 concentrations (up to 28 and 11 μmol/mol, respectively) slightly higher than those measured at sites 3, 4 and 5. At the maximum depth of profile at site 6, CO2 reached a concentration of 730 mmol/mol and H2S was detected (0.10 mmol/mol). Soil gasses from profile at sites 7-8 were characterized by dominant CO2 (up to 807 mmol/mol), whereas N2 and Ar concentrations were up to 190 and 4.9 mmol/mol, respectively. Oxygen was below the detection limit (<0.5 mmol/mol), H2S (up to 0.18 mmol/mol) and CH4 and C2H6 were up to 41 and 4.5 μmol/mol. Sites 6sub, 7 and 8 were hereafter named HC: High CO2 concentration sites.
The bubbling gas was marked by the highest CO2 (983 mmol/mol), H2S (0.25 mmol/mol), CH4 (125μmol/mol) and C2H6 (7.9 μmol/mol) concentrations and the lowest N2 and Ar contents (16 and 1.1 mmol/mol, respectively). The chemical composition of the interstitial soil gasses collected at 10 and 20 cm depth at site 8 resembled that of the gas sample collected at the bubbling pool (BP) (Figure 3). The N2 vs. CH4 and CO2 binary diagrams (Figure 4) show that the interstitial gasses from sites 3 and 4 and those from 10 to 40 cm depth in site 5 were characterized by N2 concentrations higher than those expected for air-BP mixing. The interstitial gas sample from site 5 and those collected at 10–30 cm depth from site 6 showed CO2 content higher with respect to the air-BP mixing curves. The interstitial soil gasses from sites 6 at depth >30 cm and 8 were depleted in CH4 with respect to the air-BP mixing line (Figure 4).
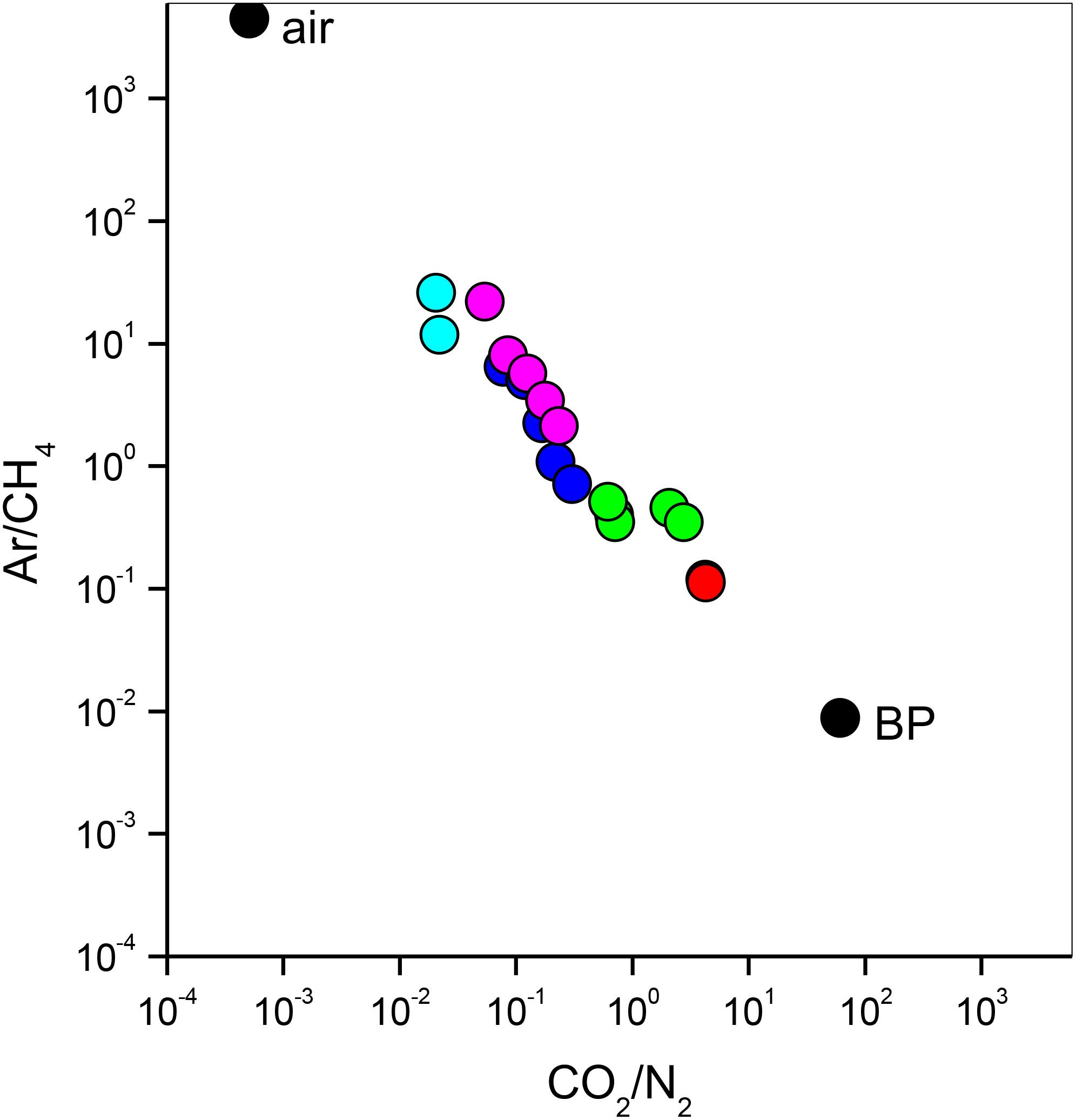
Figure 3. Ar/CH4 vs. CO2/N2 binary diagram for the interstitial soil gasses from site 3 (magenta circles), site 4 (cyan circles), site 5 (blue circles), site 6 (green circles) and site 8 (red circles). Air and gas from the bubbling pool (BP) are also reported (black circles).
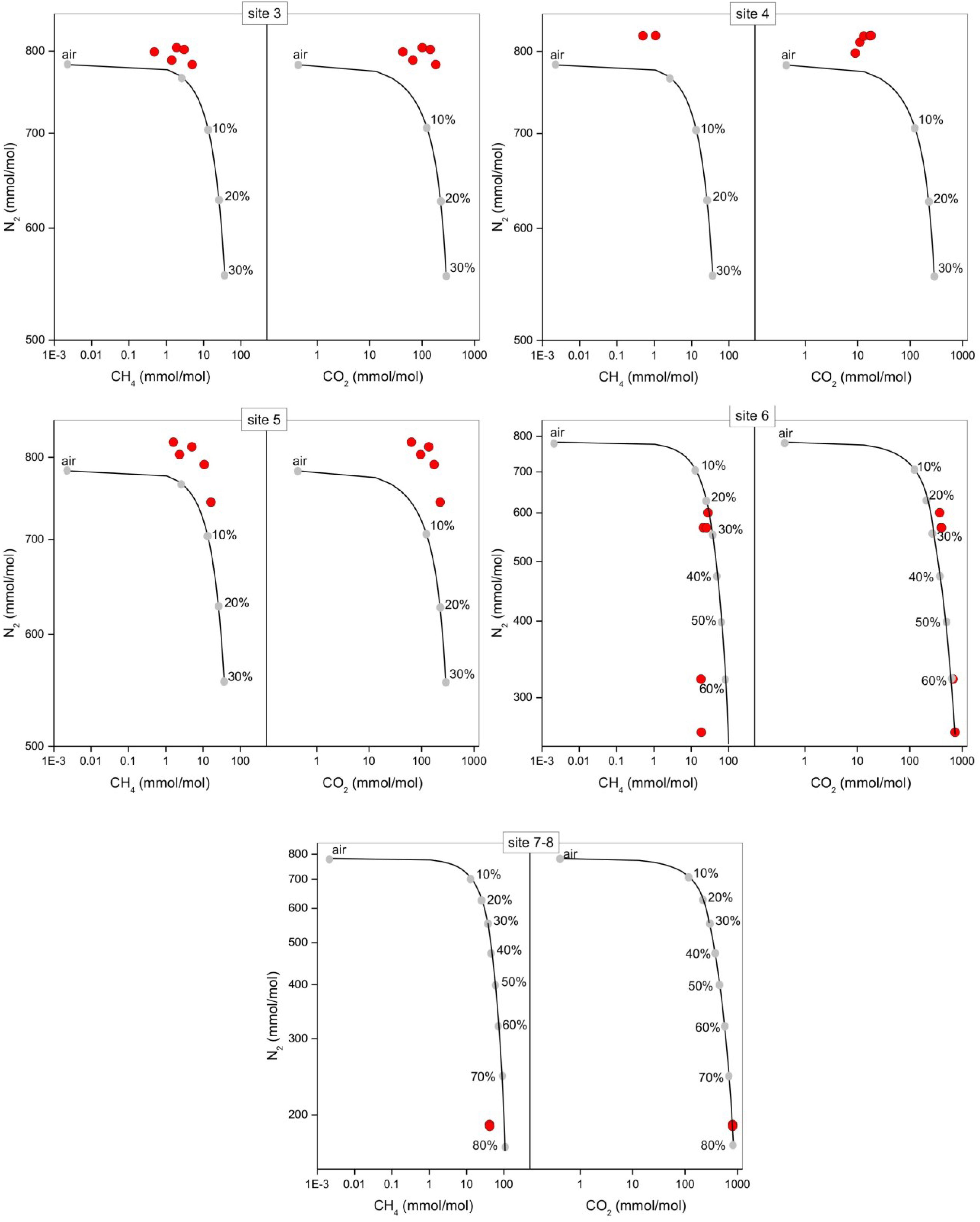
Figure 4. N2 vs. CH4 and N2 vs. CO2 binary diagrams for the interstitial soil gasses (red circles) from each sampling site at the different depths (data are shown in Table 2). The expected composition of soil gasses resulting from mixing of air with variable amounts of geogenic gas. The geogenic gas characteristics were obtained by analyzing a free-gas sample collected from one of the bubbling pools (named “BP”). The expected composition of soil gasses then obtained using a simple mass balance modeling approach, is shown (black line and gray dots). The fraction (in percentage) of the deep gas involved in the mixture is also reported.
The δ13C-CO2 values ranged from −11.5 to −6.06‰ vs. V-PDB, showing increasing trends from profiles at site 3 to site 8 and at increasing depth along each profile. The δ13C-CO2 value of the bubbling gas (−6.41‰ vs. V-PDB) was similar to those of profile at site 8. The δ13C-CH4 value measured in profile 6 at 20 cm depth was −23.3‰ vs. V-PDB, whereas those at site 8 (at 20 cm depth) and the bubbling gas were −31.7 and −38.5‰ vs. V-PDB, respectively.
Bacterial and Archaea Abundance and Diversity
The average prokaryotic abundance showed higher values in the surface (2.7 × 109 ± 1.3 × 109 cell/g DW) than in the subsurface (1.2 × 109 ± 3.7 × 108 cell/g DW) samples, with an overall increase passing from site 3 to sites 7 and 8. The highest abundances were observed at the surface from sites 7 and 8 (4.5 × 109 ± 1.2 × 109 cell/g DW and 3.9 × 109 ± 7.6 × 107 cell/g DW, respectively). The deep sample at site 7 showed an average abundance of 1.3 × 109 ± 1.3 × 108 cell/g DW (Figure 5). Overall, Bacteria (probe EUB338 I-III) represented about 80% of total DAPI stained cells. Archaea (probe ARCH 915) were on average 6% of total cells, with the highest values (12%) measured at site 8 subsurface samples.
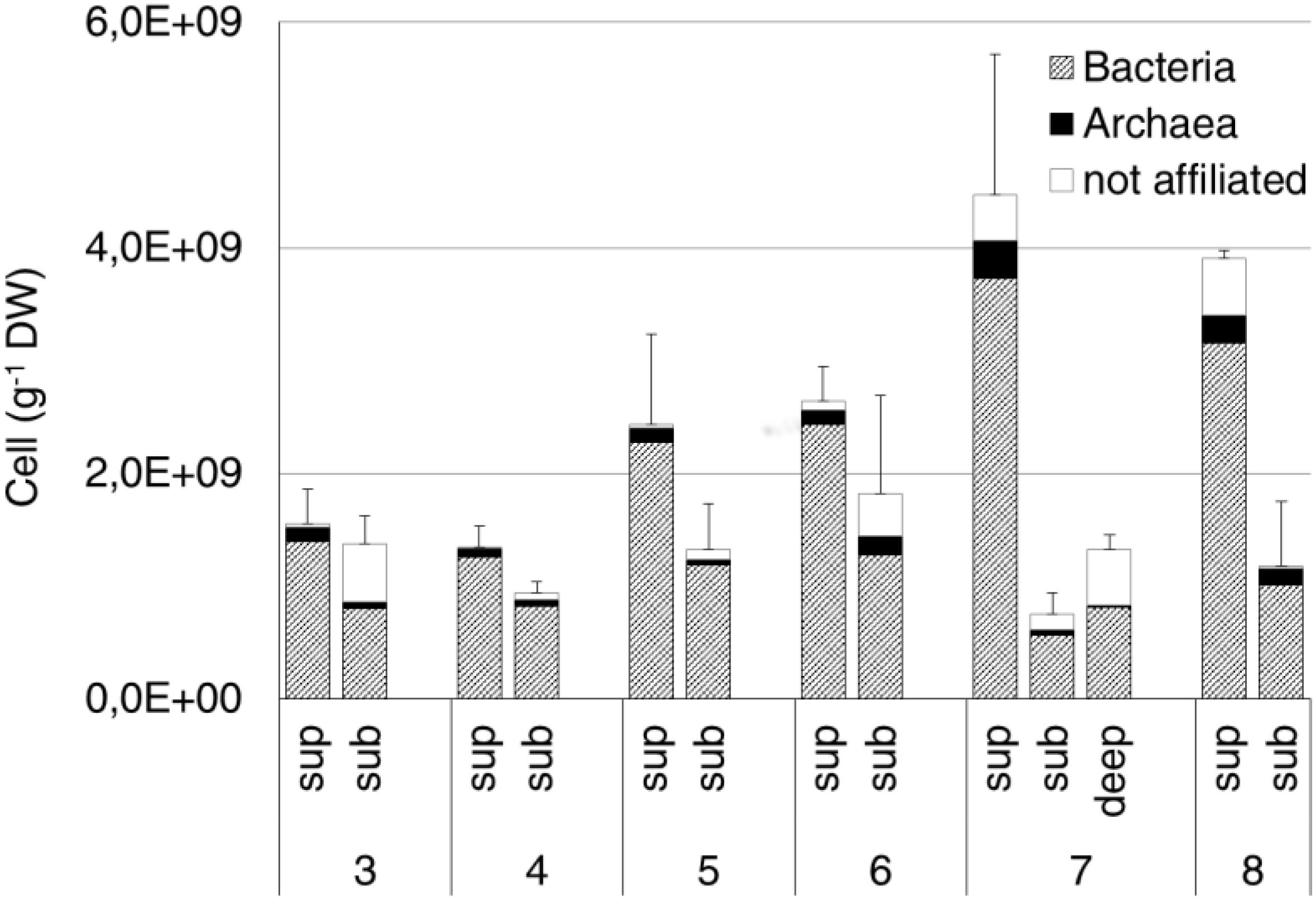
Figure 5. Bacteria and Archaea abundance estimated by CARD-FISH in the original soil samples. Data are expressed as number of cells per gram of dry weight (DW). Cells were hybridized by the specific probes EUB338 I-III and ARCH915 for Bacteria and Archaea, respectively. Error bars represent the standard deviation.
The number of OTUs retrieved in each sample was in the range of 1000–1700 and 2200–2600 at sites with high (HC) and low (LC) CO2 concentration, respectively. Rarefaction curves show that the LC samples harbor significantly higher diversity than HC samples, regardless of rarefaction depth and the two communities were significantly different from each other (PERMANOVA p = 0.002, R2 = 0.38) (Supplementary Figure 1).
The relative abundances of the 13 dominant phyla are shown in Figure 6, revealing the main differences between microbial communities inhabiting soils at the two CO2 concentration levels. Overall, the NMDS ordination plots, showing the variation patterns of the chemical and microbiological variables are reported in Figure 7. Proteobacteria dominated the community in all sampling sites with members within the Xanthomonadaceae (Gammaproteobacteria), being the most abundant family in HC soils. The closest un-cultured match for the most abundant Xanthomonadaceae phylotype is from a sludge reactor for toluene degradation (LC336110, 100% identity).
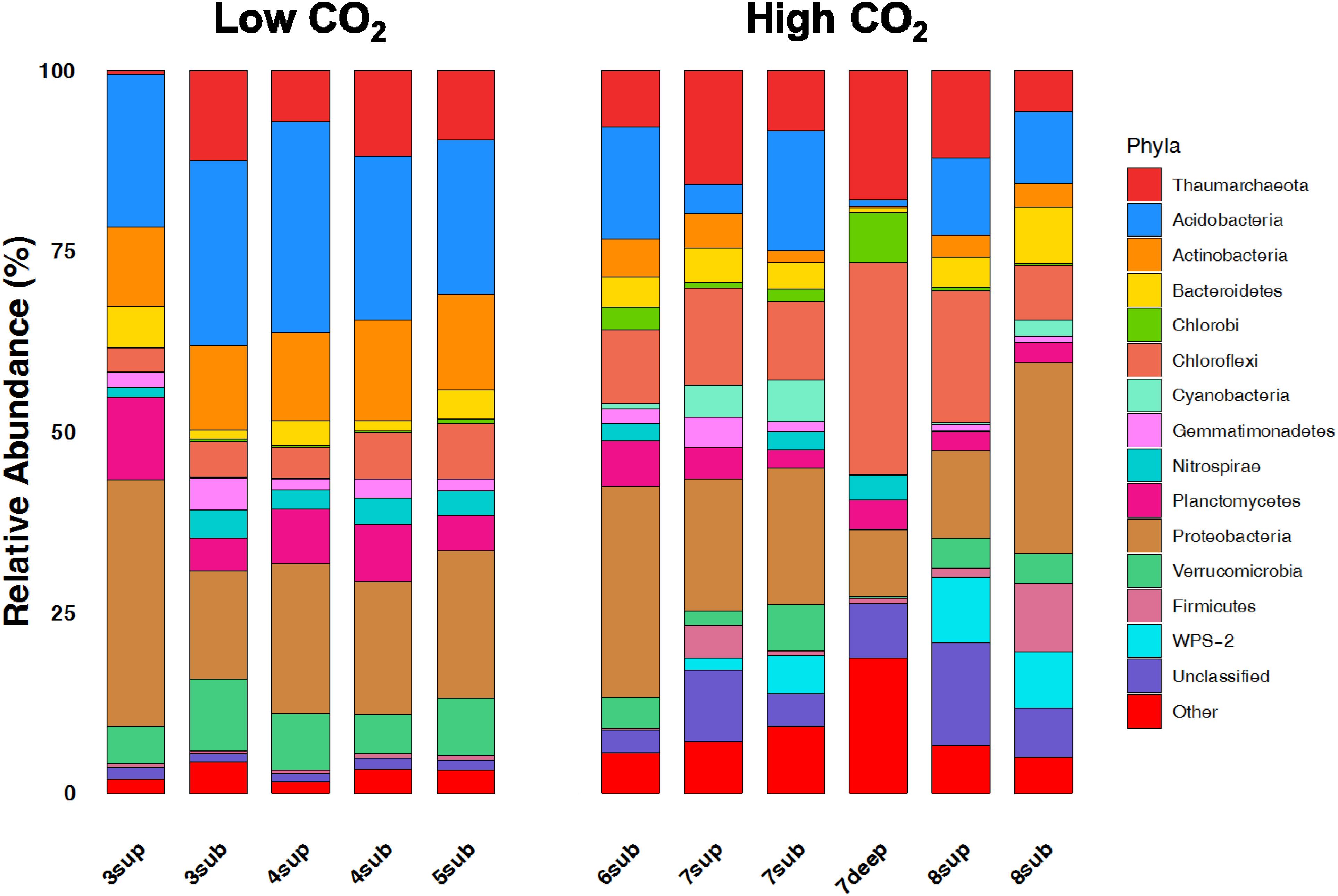
Figure 6. Broad taxonomic affiliation of 16S rRNA gene sequences obtained from environmental samples from high CO2 (HC) and low CO2 (LC) concentration sites. The relative abundances of the 13 dominant phyla of OTUs belonging to the domain of Bacteria and Archaea are shown as% of total OTUs.
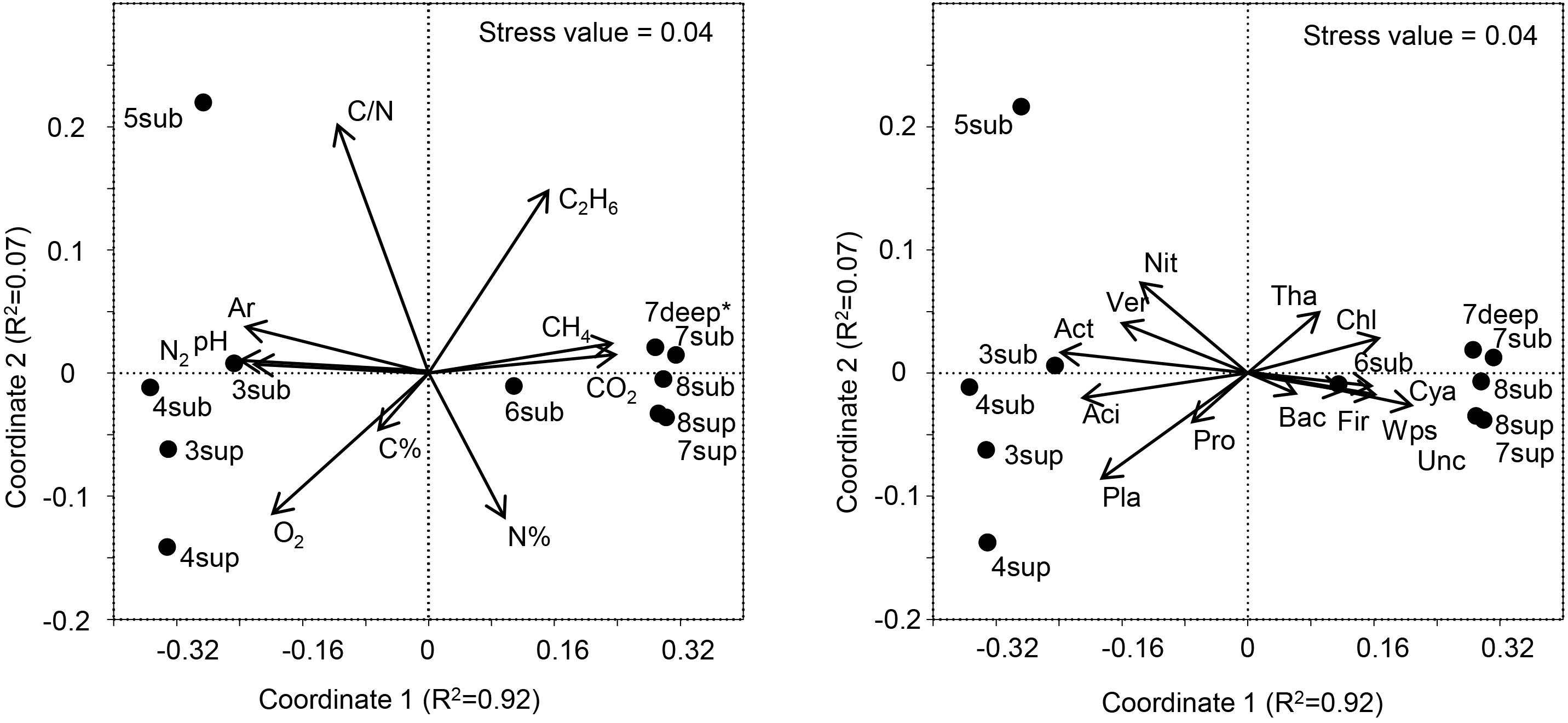
Figure 7. Left panel: NMDS ordination plot, based on the Bray Curtis distance matrix, showing the variation patterns of chemical variables. The vector length is proportional to the correlation between the NMDS axes and each chemical variable. Right panel: The typifying microbial composition revealed by 16S rRNA gene sequencing projected onto the NMDS ordination synthesizing the chemical dissimilarity between areas. The vector length is proportional to the correlation between the NMDS axes and relative abundance of each microbial phylum. The stress value (value 0.04) provides an accurate representation of the dissimilarity among areas affected or not affected by hydrothermal fluids. Wps, WPS; Chl, Chloroflexi; Ver, Verrucomicrobia; Cya, Cyanobacteria; Aci, Acidobacteria; Act, Actinobacteria; Pla, Planctomycetes; Tha, Thaumarchaeota; Pro, Proteobacteria; Bac, Bacteroidetes; Fir, Firmicutes; Nit, Nitrospira; Unc, Unclassified. ∗Gas profile for site 7deep was estimated from the values of sites 7-8sub.
The second most abundant Proteobacteria in HC samples was within the genus Geobacter, whose closest environmental match was from paddy soils (MG101255, 100% identity). This anaerobic genus was already reported in similar environments exposed to high CO2 fluxes (Oppermann et al., 2010). Acidobacteria, Actinobacteria, Planctomycetes and Verrucomicrobia showed significantly higher abundances in LC samples. It is worth noting that Chloroflexi, Firmicutes, WPS2 and Cyanobacteria were more abundant in HC samples. Moreover, HC samples showed a higher percentage of undefined sequences when compared to LC samples. The closest environmental match in the NCBI database of the most abundant Unclassified OTU in HC samples is an OTU retrieved in an acid mine drainage (HQ322903, 96% identity).
Chloroflexi classes, Ktedonobacteria and Anaerolineae were abundant only in HC samples. The most abundant Ktedonobacteria OTU, within the Thermogemmatisporaceae family, had a 97% match to an uncultured bacterium from Antarctic soils (EF221335, 97% identity), while the most abundant Anaerolineae phylotype retrieved from HC samples had closest matches with uncultured bacteria from marine sediments (MG637761, 97% identity) and from subfloor sediments in methane hydrate fields (AB540881, 97% identity). Chloroflexi class Dehalococcoidia was only retrieved in one of the HC samples (7deep) and had closest matches with uncultured bacteria from a groundwater aquifer (KC606861, 97% identity) and from sediment from high arsenic groundwater (KF632458, 97% identity).
Thaumarcheota were abundant in all sites except site 3. They made up ∼6% of total community in both LC and HC samples. Two different orders of Thaumarcheota were found in LC and HC samples, respectively: (i) Nitrosospherales (this group also showed to be more abundant in “reference soils (low CO2)” (Beulig et al., 2015) (N. gargensis and Candidatus Nitrososphaera) and (ii) Cenarcheales (SAGMA-X). The most abundant Nitrosospherales phylotype retrieved from LC samples had a closest match with uncultured bacteria from paddy soils (KP328055, 100% identity), while the closest environmental matches for the most abundant Thaumarchaeota phylotypes in HC samples were from bottled mineral water (JX458345, 99% identity), subtropical forest soil (MH016249, 100% identity) and landfill leachate (KM870444, 100% identity). Our results showed the nearly complete absence from soils of Methanogens that were found only in the most extreme conditions (1.5% only in one HC sample 7deep, data not shown).
Hierarchical clustering for the dataset was generated on the basis of a distance matrix calculated by using the Bray-Curtis distance (Supplementary Figure 2A). This clustering shows a clear profile of the core microbiome with soil samples at high (HC: sites 3-4-5) and low (LC: sites 6sub-7-8) CO2 concentrations in different groups. HC samples showed a more dissimilar composition (based on counts on each sample) among each other than the LC samples, as also shown by PCoA plot (Supplementary Figure 2B). The top 10 OTUs that explained the most variance between low and high CO2 concentrations according to SIMPER analysis are reported in Table 3 and the mean dissimilarity of the bacterial communities between them was 95%.
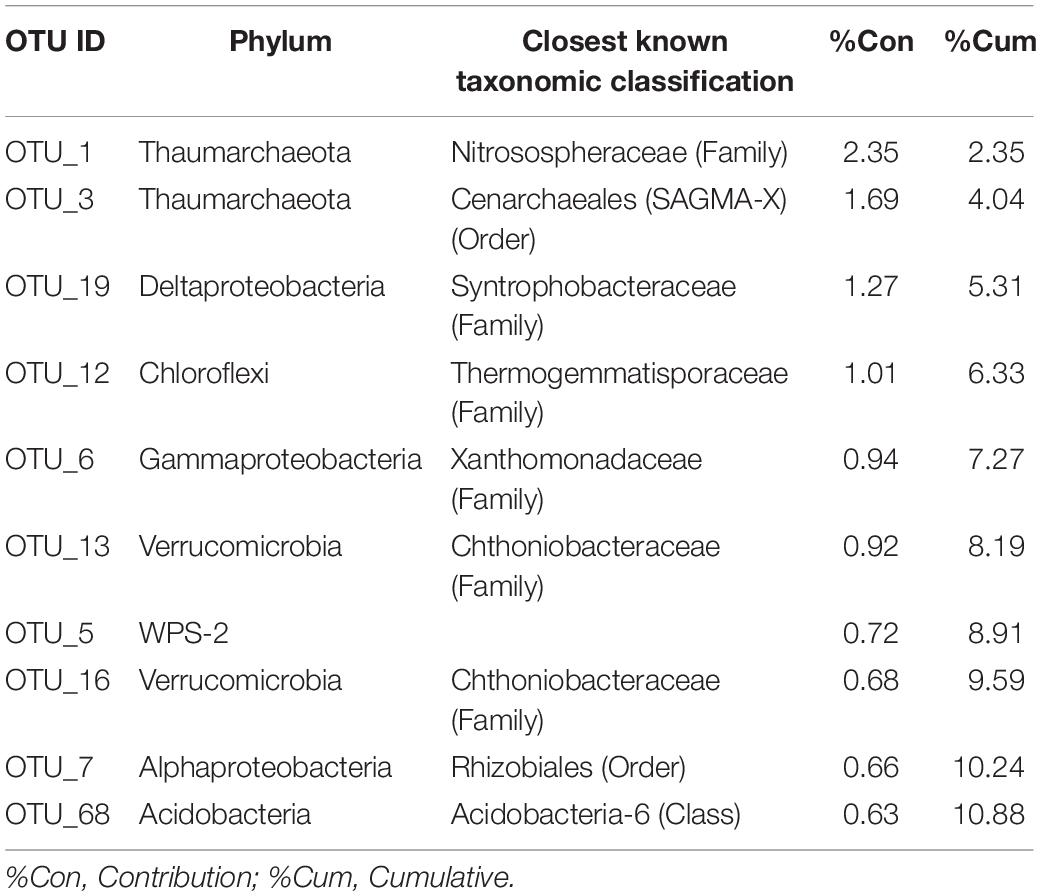
Table 3. Similarity percentage analysis (SIMPER) between environmental samples collected from high CO2 (HC) and low CO2 (LC) concentration sites.
Acetogenic Potential
All the mofette soil incubations supplied with hydrogen gas displayed a remarkable CO2 fixation potential, primarily due to the activity of acetogenic (i.e., acetate-producing) microorganisms originally present in the soil. By contrast, negligible production of acetate occurred in control tests incubated with the same soils, under identical conditions, without addition of hydrogen gas. As an example, Figure 8 shows the time course of hydrogen, carbon dioxide, and acetate in a representative incubation experiment setup with soil taken from site 7. Hydrogen and CO2 utilization, as well as acetate formation, commenced without any significant lag phase. Upon depletion of hydrogen (on day 28), both acetate production and CO2 consumption almost ceased. These latter processes, however, resumed as soon as H2 was re-spiked to the bottles (on day 40). Throughout the entire incubation period, the pH remained in the range of 5.5–6.5 and negligible formation of methane and/or of other reduced organic metabolites was detected. Figure 9A compares the observed H2-dependent specific acetate formation rates for the different mofette soils. Unexpectedly, the highest values, up to 6.7 ± 0.9 μmol of acetate produced per g of soil (dry weight) per day, were observed in the soil exposed to relatively lower CO2 levels. This finding may be due to the lower natural pH of soils exposed to higher CO2 levels, which may have resulted in a lower abundance of acetogenic microorganisms. By contrast, the mofette soils exposed to remarkably higher CO2 levels displayed a lower and similar acetogenic formation rate, ranging from 1.8 to 3.4 μmol/g ⋅ d. A mass balance indicated that acetate production accounted for 50–80% of consumed H2 (Figure 9B), possibly suggesting that other (still not identified) anaerobic H2 consuming processes such as sulfate reduction occurred during incubations.
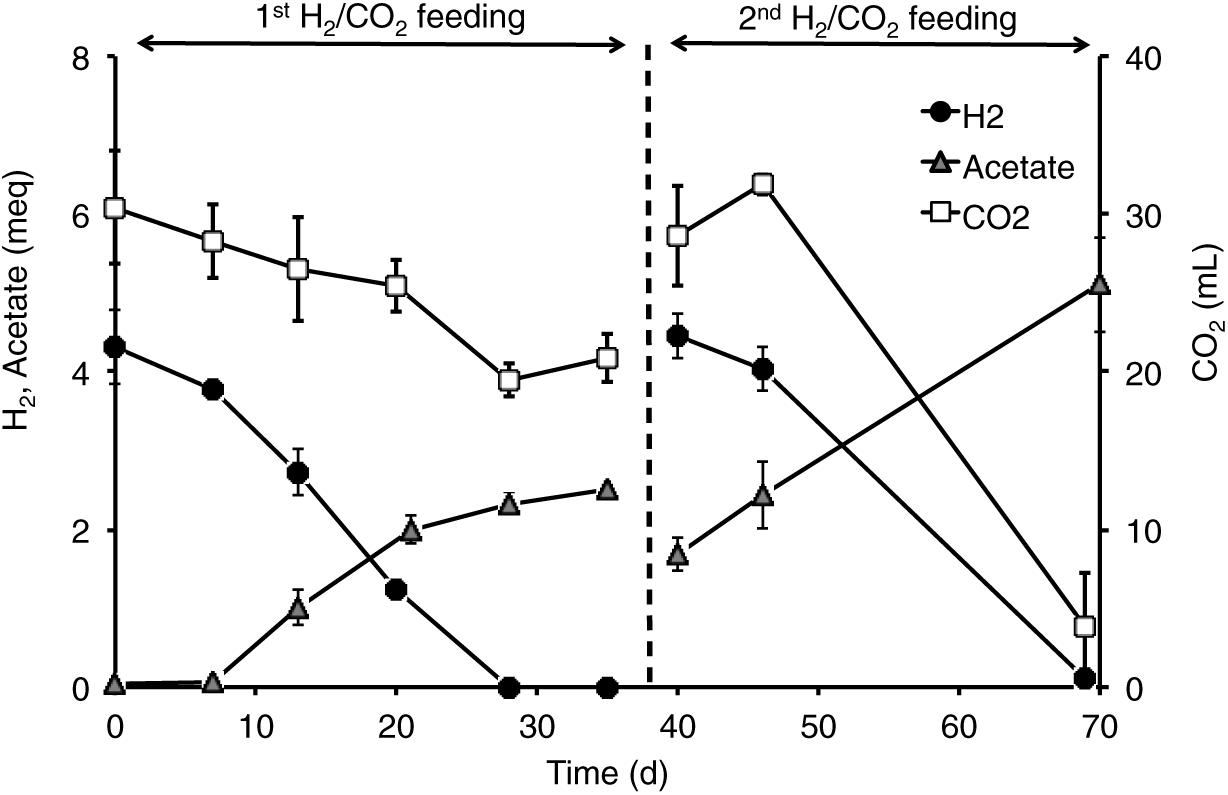
Figure 8. Time course of H2, acetate, and CO2 in the incubations setup with mofette soil (10 g wet weight in 40 mL anaerobic mineral medium buffered at pH 5.5-6-0) 7sup sampled from a high CO2 area. At the start of each feeding cycle, the headspace composition of the bottles consisted of H2 (60%, vol/vol), CO2 (15%, vol/vol), and N2 (25%, vol/vol). The total pressure was around 1 atm. Bottles were incubated statically, in the dark, at room temperature (20–25°C). Error bars represent the standard error of duplicate incubations.
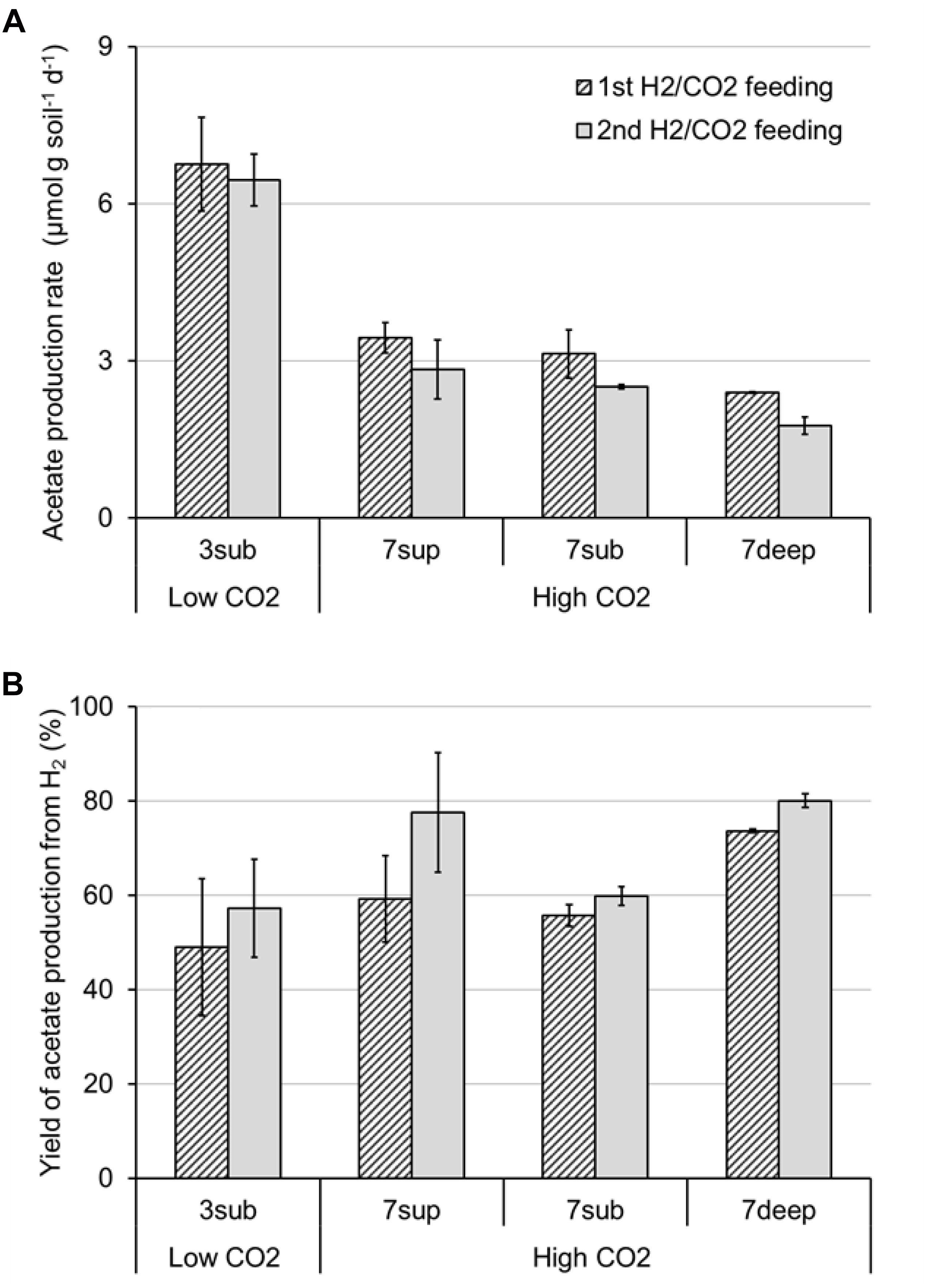
Figure 9. Maximum rate of acetate production (A) and yield of acetate production (B) from H2 in bottles incubated with the different mofette soils (3sub, 7sup, 7sub, and 7deep). Error bars represent the standard error of duplicate incubations.
Discussion
Origin of Interstitial Gases
The interstitial soil gasses collected at 10 and 20 cm depth at site 8 showed chemical composition similar to that recorded for the gas sample collected from the bubbling pool (BP). The latter, basically dominated by CO2 produced at depth by thermometamorphic reactions on carbonates and partially originating from mantle degassing, is to be considered typical of the thermal fluid emission of this area (e.g., Minissale et al., 2002). A moderate increase of the atmospheric inert gasses (N2 and Ar), characterizing the interstitial soil gasses with respect to the gas sample from the pool, can likely be related to soil permeability that allowed a significant air contamination of the deep-originated gasses. The atmospheric gas concentrations showed a significant decrease in interstitial gasses from the sites located at decreasing distance from the crater bottom, since air in the soil was counteracted by the flux of the deep-originated gasses, which achieved their highest concentrations in correspondence of the bubbling pool area (Figure 1). Hence, the chemical composition of the interstitial gasses was produced, at a first approximation, by air dilution of the BP-type gas (Figure 3). The consumption of O2, a process that typically occurs as fluids circulate underground where reducing conditions are dominating, may have produced an indirect N2 increase with respect to that in the air for those samples having a low deep gas contribution such as 3, 4 and 5 (10–40 cm) (Figure 4). The interstitial gas sample from site 5 and those collected at 10–30 cm depth from site 6 show CO2 concentrations higher with respect to the curves (Fig. 4) that were constructed considering a simple binary mixing between air and a gas phase having the BP composition. As suggested to explain the indirect N2 increase, such a CO2-excess was the result of O2 depletion affecting the air end-member during its diffusion within the soil, producing a relative enrichment in the other gasses. However, a minor contribution from CH4 oxidation cannot be ruled out, as apparently supported by the δ13C-CO2 values of the interstitial gasses, which were more negative than those of the CO2 from the pool, especially those collected at the shallower sampling depths (Table 3). The occurrence of CH4 oxidation was confirmed by (i) the composition of the interstitial soil gasses from sites 6 at depth >30 cm and 8 which were depleted in CH4 with respect to the air-BP mixing line, and (ii) the δ13C-CH4 values that were less negative than that of BP, as a result of isotopic fractionation during CH4 consumption that typically produces a significant 13C-enrichment in the residual CH4 (e.g., Tassi et al., 2015; Venturi et al., 2019).
Microbiome Profiling
The long-term abiotic selection pressure represented by high CO2 concentrations is known to drive compositional changes in soil microbial communities (Oppermann et al., 2010; Krüger et al., 2011; Frerichs et al., 2013; Šibanc et al., 2014). However, whether soil CO2 concentration itself directly impact soil microbes or whether microbial organisms respond indirectly to co-varying factors such as local hypoxia or elevated soil pH is less clear. O2 concentration and pH were among the main co-varying factors that differed between soils with different CO2 exposure, both being negatively correlated with CO2 (Figure 7). Since O2 and pH are major abiotic factors known to affect microbial communities, it is possible that these and not the direct exposure to CO2 levels would determine the microbial community composition at the mofette sites (Maček et al., 2011). Nevertheless, it must be considered that the observed variations in both the O2 contents and pH were ultimately controlled by the supply of geogenic CO2, the latter hindering diffusion of atmospheric O2 into the soil and enhancing soil acidification due to increased concentrations of H+ and H2CO3∗.
The variation patterns of major physicochemical parameters differentiated those samples positioned intermediately between the less affected by CO2 emission (sites 3–5) and the most affected ones (sites 6–8), as revealed by NMDS analysis (Figure 7, Left Panel). The high species richness in the CO2-rich soils (1000 to 2600 16S OTUs) puts them on a par with many soils from different environments (Solon et al., 2018). However, the presence of high CO2 concentration levels resulted in much lower species richness (16S rRNA gene) and significantly different communities than in sites characterized by lower CO2 content, regardless of the depth at which samples were taken. These results corroborate the findings of Beulig et al. (2015), where mofettes exhibited substantially lower prokaryotic diversity than the reference sites. In particular, Chloroflexi, Firmicutes, WPS2 and Cyanobacteria were associated to high CO2 conditions, whereas Acidobacteria, Actinobacteria, Planctomycetes and Verrucomicrobia showed higher percentages in Low CO2 samples (Figure 7, Right Panel). Most of OTUs retrieved were not identified at the genus level, suggesting that this environment harbors novel bacterial and archaeal diversity, which deserves further investigation to allow fine-scale phylogeny of these communities.
WPS2, initially described in a study on polluted soil in Germany, branch off from either Cyanobacteria or Deinococcus phylum (Nogales et al., 2001). Representatives of this group were already recorded in acidic environments (Grasby et al., 2013; Trexler et al., 2014; Bragina et al., 2015) and a recent study showed that WPS2 is likely an anoxygenic phototroph capable of carbon fixation (Holland-Moritz et al., 2018). Soils exposed to high CO2 concentrations could confer an advantage to this group, which would explain its higher abundance relative to lower CO2 concentrations.
Chloroflexi have been identified in many environments, including freshwater and marine sediments. Nonetheless, Chloroflexi remain a relatively understudied bacterial lineage. The phylum shows different metabolic lifestyles, including photoautotrophs (e.g., Chloroflexus aurantiacus), fermentative (e.g., Anaerolinea thermophila UNI-1), organohalide respiring organisms in the Dehalococcoidia, and aerobic thermophiles (e.g., Thermomicrobium) (Hug et al., 2013). Although members of Chloroflexi were not directly reported to grow acetogenically, previous studies (e.g., Chan et al., 2013; Hug et al., 2013; Wasmund et al., 2014) suggested that members of Chloroflexi can have the potential to utilize CO2 via the acetyl-CoA pathway. Beulig et al. (2015), by SIP analysis of mofette soil incubations, suggested that Chloroflexi perform acetogenesis. Moreover, two Chloroflexi genomes (RBG-2 and RBG-1351), closely related to the Dehalococcoidia, are described to be putative acetogens, utilizing a pathway for the formation of acetate less common in bacteria. For the first time, the complete acetyl-CoA pathway for carbon fixation was described in the Chloroflexi (Hug et al., 2013). In Dehalococcoidia, genes encoding enzymes of the reductive acetyl-CoA pathway were identified, which may enable the fixation of CO2 or complete oxidation of organics completely to CO2 (Wasmund et al., 2014).
Chloroflexi have been found in high abundance in similar sites exposed to high CO2 concentration (Fernández-Montiel et al., 2016; Crognale et al., 2018) and sequences retrieved in this study were affiliated with two classes associated with hypoxic and microhypoxic niches: Ktedonobacteria and Anaerolinea. Ktedonobacteria can thrive in microaerophilic conditions (Chang et al., 2011). Anaerolineae have been reported to grow under strictly anaerobic conditions with isolates found in anaerobic sludge and hot spring (Yamada et al., 2006). Previous studies reported that Ktedonobacteria are prominent in extreme environments such as volcanic, Antarctic, and cave ecosystems (Tebo et al., 2015; Yabe et al., 2017; Schmidt et al., 2018). They comprises only five described species and a large number of uncultured environmental clone sequences (Cavaletti et al., 2006; Yabe et al., 2011; King and King, 2014). Several other strains were isolated, but not formally described, from soil (genus Ktedonobacter), geothermal soils or compost (Stott et al., 2008; Yabe et al., 2011). Moreover, The genome of K. racemifer SOSP1-21 T was sequenced and contains a cox operon that confers the potential for carbon monoxide oxidation (Chang et al., 2011; King and King, 2014).
Anaerolineae class comprises only a few cultured strains and a large number of environmental 16S rRNA gene sequences. Since Anaerolineae were frequently found within various ecosystems, they were considered to be ubiquitous (Yamada and Sekiguchi, 2009). Two thermophilic, filamentous organisms (i.e., Anaerolinea thermophila and Caldilinea aerophila) were isolated from an anaerobic granular sludge and a hot spring (Sekiguchi et al., 2003). Moreover, thermophilic and mesophilic filamentous strains were isolated from anaerobic sludge blanket at a temperature range of 25–50°C (Yamada et al., 2005) and in hydrothermal vents in the Yaeyama Archipelago in Japan (Nunoura et al., 2013).
Acidobacteria were more abundant in low CO2 samples than in those exposed to high CO2 fluxes, a pattern previously reported in similar environments (Crognale et al., 2018). Members of the phylum Acidobacteria represent one of the predominant bacterial groups in soil but their ecological functions are still poorly understood (Jones et al., 2009; Foesel et al., 2014; Kielak et al., 2016). Acidobacteria were abundantly found in volcanic craters (Glamoclija and Garrel, 2004; Crognale et al., 2018), in marine vents (Sievert et al., 2000; López-García et al., 2003) and in the Yellowstone National Park geothermal areas (Norris et al., 2002).
In both LC and HC samples (except site 3), the archaeal OTUs were mainly affiliated to Thaumarchaeota, a deep-branching phylum within Archaea domain. An increase in Thaumarchaeota associated sequences was recorded in other mofettes with high CO2 concentrations (Frerichs et al., 2013; Šibanc et al., 2014; Crognale et al., 2018) and it was proposed that this could be an “indicator taxa” of high CO2 enriched soils (Frerichs et al., 2013), which could be connected to the CO2 soil acidification and the obligate acidophilic nature of these taxa (Lehtovirta-Morley et al., 2011). Thaumarchaeota are among the most abundant Archaea on Earth and are believed to significantly contribute to the global N-cycle and C-cycle. This phylum includes not only all known Ammonia-Oxidizing Archaea (AOA) but also several clusters with unknown energy metabolism (Pester et al., 2011; Stieglmeier et al., 2014). Previous studies suggested that ammonia-oxidizing Thaumarchaeota can be adapted to both low ammonia availability and an autotrophic or mixotrophic lifestyle (Zhang et al., 2008; Schleper and Nicol, 2010; Pratscher et al., 2011). It has also been suggested that members of this phylum may be involved in methane oxidation (Crognale et al., 2018) since ammonia monooxygenase and methane monooxygenase enzymes are evolutionarily linked (Holmes et al., 1995), enabling both methanotrophy and ammonia oxidation (O’Neill and Wilkinson, 1977; Jones and Morita, 1983). The presence of the two different clades in different CO2 concentrations, Nitrosospherales and Cenarcheales in LC and HC, respectively, suggests strong selection by local environmental conditions: Cenarcheales found at high CO2 may not be ammonia-oxidizers and have a different metabolic pathway for energy production or may have extremely high affinity for O2, which allows their survival as ammonia oxidizers at very high CO2 concentrations. At higher O2 concentrations they may be outcompeted by Nitrosospherales. The circadian Oxygen oscillation could also justify the presence of ammonia-oxidizing archaea. Interestingly, Methanobacteria class, belonging to the South African Gold Mine Euryarchaeotic Group (SAGMEG), was only found in the most extreme conditions (1.5% only in the sample 7deep). OTUs affiliated with archaeal groups that include methanogens were nearly absent from this site, as previously reported in similar acidic soils exposed to natural CO2 fluxes (Crognale et al., 2018). The nearly complete absence of methanogens in the study soils could be due to the fact that these microorganisms were outcompeted by either acetogens, as also discussed in the following paragraph, or the fluctuating oxygen concentration during daytime. This suggests that methanogenesis marginally contributes to the overall metabolic features of the microbial communities in such an environment.
Acetogenic Bacteria
Acetogenic bacteria are ubiquitous in nature and are a specialized group of strictly anaerobic bacteria.
Acetogens were isolated from diverse environments, including soils, hypersaline waters and sediments (Liu and Conrad, 2011). In these ecosystems, chemolithoautotrophic acetogenic bacteria can directly compete with methanogenic archaea (hydrogenotrophic) or syntrophically interact with acetoclastic methanogens (Chassard and Bernalier-Donadille, 2006; Liu and Conrad, 2011). Acetogens include Firmicutes, Spirochaetes, Delta-Proteobacteria and Acidobacteria (Drake et al., 2006). Recently Beulig et al. (2015) speculated that Chloroflexi might be involved in acetogenesis in wetland mofette. Comparing the community composition in the environmental samples from sites at low (LC) and at high CO2 concentration (HC), our results suggest a major role of Acidobacteria in LC sites whereas in HC sites Chloroflexi and Firmicutes are likely the predominant acetogens. This could also explain the difference in yield of acetate production observed in the incubation experiments with soil from low and at high CO2 concentration sites. Under standard conditions, H2-CO2-dependent methanogenesis (ΔGo’ = −130 kJ/reaction) is more energetically favorable than H2-CO2-dependent acetogenesis (ΔGo’ = −95 kJ/reaction). At low H2 concentrations, typically found in many anoxic environments, methanogenesis is energetically more favorable than acetogenesis, and hydrogenotrophic methanogens outcompete chemolithoautotrophic acetogens (Drake et al., 2006). However, acetogens have a kinetic advantage over methanogens at acidic pH and low temperature (Conrad et al., 1989; Kotsyurbenko et al., 1993), hence providing a likely explanation for the remarkable acetogenic potential observed in the incubation experiments, together with the lack of methanogenic activity, further confirmed by the absence of methanogens in sequencing analyses. In spite of that, it should be considered that the herein described incubation tests would have likely resulted in the selective enrichments of acetogens (e.g., Firmicutes), thereby overestimating the actual acetogenic potential of the natural communities.
Concluding Remarks
Gas discharges, characterized by extremely high CO2 concentrations, are significantly affecting soil formation processes interacting with the geomorphological gradient observed in the mofette. Consequently, low pH values at the bottom of the Bossoleto sinkhole, as well as increased organic carbon accumulation in the topsoil and removal of inorganic carbon, were recorded. Similarly, variations in macro and micro-elements concentrations were also observed, likely due to the effect of the pH on element mobility. The comparison between interstitial soil gasses and those collected in an adjacent bubbling pool and the isotopic carbon fractionation, clearly indicated an increase in CO2 due to CH4 oxidation at the bottom of the sinkhole. The extremely high CO2 concentrations resulted in higher microbial abundance and a lowered microbial diversity by favoring bacteria already reported to be involved in acetogenesis in mofette soils (i.e., Chloroflexi, Acidobacteria and Firmicutes). All the mofette soils supplied with hydrogen gas, in experimental incubations, displayed a remarkable CO2 fixation potential, primarily due to the activity of acetogenic (i.e., acetate-producing) microorganisms. By contrast, negligible production of acetate occurred in control tests incubated with the same soils, under identical conditions, without the addition of H2. Our results suggest that the composition of microbial communities at high CO2 concentrations affects carbon cycling through inhibition of organic carbon decomposition and increases CO2-fixation via the acetyl-CoA pathway. These acetogenic organisms, outcompeting methanogens, might determine considerable changes in carbon cycling leading to the accumulation of organic carbon in soils at the bottom of the mofette. Sites naturally exposed to extremely high CO2 levels could, therefore, potentially represent an untapped source of microorganisms with unique capabilities to catalytically convert CO2 into valuable organic chemicals for industrial applications.
Data Availability Statement
The 16S rRNA gene sequences from Illumina MiSeq libraries from this study were deposited in the SRA (Short Read Archive) database under Bioproject ID PRJNA548940.
Author Contributions
SF conceived the study. SF, FUn, FT, OV, AR, and FA contributed to the conception and design of the study. All authors performed the sampling campaign and the field and laboratory analysis, wrote the sections of the manuscript, contributed to manuscript revision, and read and approved the submitted version. LV and SV organized the database and performed the statistical analysis. SF wrote the first draft of the manuscript.
Funding
This research was supported by the CNR Short Term Mobility fellowship assigned to SF and by the Italian Ministry of Education, University and Research (MIUR) in the frame of the WE-MET Project (ERANETMED_NEXUS-14-035).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to deeply thank Noah Fierer of University of Colorado Boulder for the continuous support to the project and valuable suggestions and Tess Brewer, Teal Potter, and Smets Wenke for help with the analyses. Enrico Calvi (CNR-IGG Pisa) is warmly thanked for analyzing the carbon isotopic ratios of CO2 in the soil gasses, Ilaria Pizzetti and Adita Bandra for the assistance during sampling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02238/full#supplementary-material
References
Amalfitano, S., Del Bon, A., Zoppini, A., Ghergo, S., Fazi, S., Parrone, D., et al. (2014). Groundwater geochemistry and microbial community structure in the aquifer transition from volcanic to alluvial areas. Water Res. 65, 384–394. doi: 10.1016/j.watres.2014.08.004
Amalfitano, S., and Fazi, S. (2008). Recovery and quantification of bacterial cells associated with streambed sediments. J. Microbiol. Methods. 75, 237–243. doi: 10.1016/j.mimet.2008.06.004
Amundson, R. G., and Davidson, E. A. (1990). Carbon dioxide and nitrogenous gases in the soil atmosphere. J. Geochem. Explor. 38, 13–41. doi: 10.1016/0375-6742(90)90091-N
Beulig, F., Heuer, V. B., Akob, D. M., Viehweger, B., Elvert, M., Herrmann, M., et al. (2015). Carbon flow from volcanic CO2 into soil microbial communities of a wetland mofette. ISME J. 9, 746–759. doi: 10.1038/ismej.2014.148
Beulig, F., Urich, T., Nowak, M., Trumbore, S. E., Gleixner, G., Gilfillan, G. D., et al. (2016). Altered carbon turnover processes and microbiomes in soils under long-term extremely high CO2 exposure. Nat. Microbiol. 1:15025. doi: 10.1038/nmicrobiol.2015.25
Bragina, A., Berg, C., and Berg, G. (2015). The core microbiome bonds the Alpine bog vegetation to a transkingdom metacommunity. Mol. Ecol. 24, 4795–4807. doi: 10.1111/mec.13342
Brogi, A., Capezzuoli, E., and Gandin, A. (2007). I travertini delle terme di s. giovanni (rapolano terme, appennino settentrionale) e loro implicazione neotettonica. Italian J. Neurol. Sci. 20, 107–124.
Caporaso, J. G., Bittinger, K., Bushman, F. D., Desantis, T. Z., Andersen, G. L., and Knight, R. (2010a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267. doi: 10.1093/bioinformatics/btp636
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010b). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cavaletti, L., Monciardini, P., Bamonte, R., Schumann, P., Rohde, M., Sosio, M., et al. (2006). New lineage of filamentous, spore-forming, gram-positive bacteria from soil. Appl. Environ. Microbiol. 72, 4360–4369. doi: 10.1128/aem.00132-136
Chan, Y., Van Nostrand, J. D., Zhou, J., Pointing, S. B., and Farrell, R. L. (2013). Functional ecology of an antarctic dry valley. Proc. Natl. Acad. Sci. U.S.A. 110, 8990–8995. doi: 10.1073/pnas.1300643110
Chang, Y. J., Land, M., Hauser, L., Chertkov, O., del Rio, T. G., Nolan, M., et al. (2011). Non-contiguous finished genome sequence and contextual data of the filamentous soil bacterium Ktedonobacter racemifer type strain (SOSP1-21 T). Stand. Genomic Sci. 5, 97–111. doi: 10.4056/sigs.2114901
Chassard, C., and Bernalier-Donadille, A. (2006). H2 and acetate transfers during xylan fermentation between a butyrate-producing xylanolytic species and hydrogenotrophic microorganisms from the human gut. FEMS Microbiol. Lett. 254, 116–122. doi: 10.1111/j.1574-6968.2005.00016.x
Collins, S., and Bell, G. (2006). Evolution of natural algal populations at elevated CO2. Ecol. Lett. 9, 129–135. doi: 10.1111/j.1461-0248.2005.00854.x
Conrad, R., Bak, F., Seitz, H. J., Thebrath, B., Mayer, H. P., and Schütz, H. (1989). Hydrogen turnover by psychrotrophic homoacetogenic and mesophilic methanogenic bacteria in anoxic paddy soil and lake sediment. FEMS Microbiol. Lett. 62, 285–293. doi: 10.1016/0378-1097(89)90010-90014
Cord-Ruwisch, R., Seitz, H. J., and Conrad, R. (1988). The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch. Microbiol. 149, 350–357. doi: 10.1007/BF00411655
Crognale, S., Venturi, S., Tassi, F., Rossetti, S., Rashed, H., Cabassi, J., et al. (2018). Microbiome profiling in extremely acidic soils affected by hydrothermal fluids: the case of the solfatara crater (campi flegrei, southern italy). FEMS Microbiol. Ecol. 94, 1–17. doi: 10.1093/femsec/fiy190
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-3005
Drake, H. L., Küsel, K., and Matthies, C. (2006). “Acetogenic prokaryotes,” in The Prokaryotes Ecophysiology and Biochemistry, eds E. Dworkin, M. Rosenberg, E. Schleifer, and K. H. Stackebrandt, (New York, NY: Springer), 354–420. doi: 10.1007/0-387-30742-7_13
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Evans, W. C., White, L. D., and Rapp, J. B. (1998). Geochemistry of some gases in hydrothermal fluids from the southern Juan de Fuca ridge. J. Geophys. Res. 15, 305–313.
Fazi, S., Amalfitano, S., Pizzetti, I., and Pernthaler, J. (2007). Efficiency of fluorescence in situ hybridization for bacterial cell identification in temporary river sediments with contrasting water content. Syst. Appl. Microbiol. 30, 463–470. doi: 10.1016/j.syapm.2007.03.003
Fernández-Montiel, I., Sidrach-Cardona, R., Gabilondo, R., Pedescoll, A., Scheu, S., and Bécares, E. (2016). Soil communities are affected by CO2 belowground emissions at a natural vent in Spain. Soil Biol. Biochem. 97, 92–98. doi: 10.1016/j.soilbio.2016.02.007
Fierer, N., Lauber, C. L., Ramirez, K. S., Zaneveld, J., Bradford, M. A., and Knight, R. (2012). Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 6, 1007–1017. doi: 10.1038/ismej.2011.159
Foesel, B. U., Nägele, V., Naether, A., Wüst, P. K., Weinert, J., Bonkowski, M., et al. (2014). Determinants of Acidobacteria activity inferred from the relative abundances of 16S rRNA transcripts in German grassland and forest soils. Environ. Microbiol. 16, 658–675. doi: 10.1111/1462-2920.12162
Foulquier, A., Volat, B., Neyra, M., Bornette, G., and Montuelle, B. (2013). Long-term impact of hydrological regime on structure and functions of microbial communities in riverine wetland sediments. FEMS Microbiol. Ecol. 85, 211–226. doi: 10.1111/1574-6941.12112
Frerichs, J., Oppermann, B. I., Gwosdz, S., Möller, I., Herrmann, M., and Krüger, M. (2013). Microbial community changes at a terrestrial volcanic CO2 vent induced by soil acidification and anaerobic microhabitats within the soil column. FEMS Microbiol. Ecol. 84, 60–74. doi: 10.1111/1574-6941.12040
Giggenbach, W. F. (1975). A simple method for the collection and analysis of volcanic gas samples. Bull. Volcanol. 39, 132–145. doi: 10.1007/BF02596953
Glamoclija and Garrel, L.L.G.P. (2004). “The solfatara crater, italy: characterization of hydrothermal deposits, biosignatures and their astrobiological implication,” in Proceedings of the 5th Lunar and Planetary Science Conference, League, TX.
Grasby, S. E., Richards, B. C., Sharp, C. E., Brady, A. L., Jones, G. M., and Dunfield, P. F. (2013). The paint pots, kootenay national park, canada — a natural acid spring analogue for Mars. Can. J. Earth Sci. 50, 94–108. doi: 10.1139/e2012-060
Guerra, M., and Raschi, A. (2004). “Field trip guide book D03 field sight near rapolano terme (siena, tuscany). relationship between tectonics and fluid circulation,” in Proceedings of the 32nd International Geological Congress, Florence.
Holland-Moritz, H., Stuart, J., Lewis, L. R., Miller, S., Mack, M. C., McDaniel, S. F., et al. (2018). Novel bacterial lineages associated with boreal moss species. Environ. Microbiol. 20, 2625–2638. doi: 10.1111/1462-2920.14288
Holmes, A. J., Costello, A., Lidstrom, M. E., and Murrell, J. C. (1995). Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 132, 203–208. doi: 10.1016/0378-1097(95)00311-R
Hug, L. A., Castelle, C. J., Wrighton, K. C., Thomas, B. C., Sharon, I., Frischkorn, K. R., et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22. doi: 10.1186/2049-2618-1-22
IUSS Working Group Wrb, (2015). World Reference Base for Soil Resources 2014, Update 2015. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. Rome: FAO.
Jones, R. D., and Morita, R. Y. (1983). Methane oxidation by nitrosococcus oceanus and nitrosomonas europaea. Appl. Environ. Microbiol. 45, 401–410.
Jones, R. T., Robeson, M. S., Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3, 442–453. doi: 10.1038/ismej.2008.127
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016). The ecology of acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Kies, A., Hengesch, O., Tosheva, Z., Raschi, A., and Pfanz, H. (2015). Diurnal CO2-cycles and temperature regimes in a natural CO2 gas lake. Int. J. Greenh. Gas Control. 37, 142–145. doi: 10.1016/j.ijggc.2015.03.012
King, C. E., and King, G. M. (2014). Description of thermogemmatispora carboxidivorans sp. nov., a carbon-monoxideoxidizing member of the class ktedonobacteria isolated from a geothermally heated biofilm, and analysis of carbon monoxide oxidation by members of the class ktedonobacteria. Int. J. Syst. Evol. Microbiol. 64(Pt 4), 1244–1251. doi: 10.1099/ijs.0.059675-59670
Körner, C., and Miglietta, F. (1994). Long term effects of naturally elevated CO2 on mediterranean grassland and forest trees. Oecologia 99, 343–351. doi: 10.1007/BF00627748
Kotsyurbenko, O. R., Glagolev, M. V., Nozhevnikova, A. N., and Conrad, R. (2001). Competition between homoacetogenic bacteria and methanogenic archaea for hydrogen at low temperature. FEMS Microbiol. Ecol. 38, 153–159. doi: 10.1016/S0168-6496(01)00179-179
Kotsyurbenko, O. R., Nozhevnikova, A. N., and Zavarzin, G. A. (1993). Methanogenic degradation of organic matter by anaerobic bacteria at low temperature. Chemosphere 27, 1745–1761. doi: 10.1016/0045-6535(93)90155-X
Krüger, M., Jones, D., Frerichs, J., Oppermann, B. I., West, J., Coombs, P., et al. (2011). Effects of elevated CO 2 concentrations on the vegetation and microbial populations at a terrestrial CO 2 vent at Laacher See. Germany. Int. J. Greenh. Gas Control. 2, 1093–1098. doi: 10.1016/j.ijggc.2011.05.002
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Lehtovirta-Morley, L. E., Stoecker, K., Vilcinskas, A., Prosser, J. I., and Nicol, G. W. (2011). Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U.S.A. 108, 15892–15897. doi: 10.1073/pnas.1107196108
Liu, F., and Conrad, R. (2011). Chemolithotrophic acetogenic H2/CO2 utilization in Italian rice field soil. ISME J. 5, 1526–1539. doi: 10.1038/ismej.2011.17
López-García, P., Duperron, S., Philippot, P., Foriel, J., Susini, J., and Moreira, D. (2003). Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ. Microbiol. 5, 961–976. doi: 10.1046/j.1462-2920.2003.00495.x
Maček, I., Dumbrell, A. J., Nelson, M., Fitter, A. H., Vodnik, D., and Helgason, T. (2011). Local adaptation to soil hypoxia determines the structure of an arbuscular mycorrhizal fungal community in roots from natural CO 2 springs. Appl. Environ. Microbiol. 77, 4770–4777.
McFarland, J. W., Waldrop, M. P., and Haw, M. (2013). Extreme CO2 disturbance and the resilience of soil microbial communities. Soil Biol. Biochem. 65, 274–286. doi: 10.1016/j.soilbio.2013.04.019
Miglietta, F., Raschi, A., Bettarini, I., Resti, R., and Selvi, F. (1993). Natural CO2 springs in Italy: a resource for examining long-term response of vegetation to rising atmospheric CO2 concentrations. Plant. Cell Environ. 16, 873–878. doi: 10.1111/j.1365-3040.1993.tb00510.x
Minissale, A., Vaselli, O., Tassi, F., Magro, G., and Grechi, G. P. (2002). Fluid mixing in carbonate aquifers near Rapolano (central Italy): chemical and isotopic constraints. Appl. Geochem. 17, 1329–1342. doi: 10.1016/s0883-2927(02)00023-9
Nogales, B., Moore, E. R. B., Llobet-Brossa, E., Rossello-Mora, R., Amann, R., and Timmis, K. N. (2001). Combined use of 16S ribosomal DNA and 16S rRNA to Study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67, 1874–1884. doi: 10.1128/AEM.67.4.1874-1884.2001
Norris, T. B., Wraith, J. M., Castenholz, R. W., and McDermott, T. R. (2002). Soil microbial community structure across a thermal gradient following a geothermal heating event. Appl. Environ. Microbiol. 68, 6300–6309. doi: 10.1128/AEM.68.12.6300-6309.2002
Nunoura, T., Hirai, M., Miyazaki, M., Kazama, H., Makita, H., Hirayama, H., et al. (2013). Isolation and characterization of a thermophilic, obligately anaerobic and heterotrophic marine chloroflexi bacterium from a chloroflexi-dominated microbial community associated with a japanese shallow hydrothermal system, and proposal for thermomarinilin. Microbes Environ. 28, 228–235. doi: 10.1264/jsme2.me12193
Oh, N.-H., Kim, H.-S., and Richter, D. D. (2005). What regulates soil CO 2 concentrations? A modeling approach to CO 2 diffusion in deep soil profiles. Environ. Eng. Sci. 22, 38–45. doi: 10.1089/ees.2005.22.38
Oksanen, J., Blanchet, F. G., Kindt, R., Oksanen, M. J., and Suggests, M. (2013). Package ‘vegan.’ Community Ecol. Packag. Version.
O’Neill, J. G., and Wilkinson, J. F. (1977). Oxidation of ammonia by methane-oxidizing bacteria and the effects of ammonia on methane oxidation. J. Gen. Microbiol. 100, 407–412. doi: 10.1099/00221287-100-2-407
Oppermann, B. I., Michaelis, W., Blumenberg, M., Frerichs, J., Schulz, H. M., Schippers, A., et al. (2010). Soil microbial community changes as a result of long-term exposure to a natural CO2 vent. Geochim. Cosmochim. Acta. 74, 2697–2716. doi: 10.1016/j.gca.2010.02.006
Pearce, J. M. (2006). “What can we learn from natural analogues? an overview of how analogues can benefit the geological storage of CO2,” in Advances in the Geological Storage of Carbon Dioxide, eds S. Lombardi, L. K. Altunina, and S. E. Beaubien, (Netherlands: Springer), 129–139.
Pearce, J. M., Czernichowski-Lauriol, I., Lombardi, S., Brune, S., Nador, A., Baker, J., et al. (2004). “A review of natural CO2 accumulations in europe as analogues for geological sequestration,” in Geological Storage of Carbon Dioxide, eds R. H. Baines, and S. Worden, (Amsterdam: Elsevier), 29–42.
Pester, M., Schleper, C., and Wagner, M. (2011). The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr. Opin. Microbiol. 14, 300–306. doi: 10.1016/j.mib.2011.04.007
Pratscher, J., Dumont, M. G., and Conrad, R. (2011). Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U.S.A. 108, 4170–4175. doi: 10.1073/pnas.1010981108
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rabaey, K., and Rozendal, R. A. (2010). Microbial electrosynthesis - revisiting the electrical route for microbial production. Nat. Rev. Microbiol. 8, 706–716. doi: 10.1038/nrmicro2422
Raschi, A., Miglietta, F., Tognetti, R., and Van Gardingen, P. R. (1997). Plant Responses to Elevated CO2. Cambridge: Cambridge University Press.
Schaetzl, R. J., and Anderson, S. (2005). Soils: Genesis and Geomorphology. Cambridge: Cambridge University Press.
Schleper, C., and Nicol, G. W. (2010). Ammonia-oxidising archaea - physiology, ecology and evolution. Adv. Microb. Physiol. 57, 1–41. doi: 10.1016/B978-0-12-381045-8.00001-1
Schmidt, S. K., Gendron, E. M. S., Vincent, K., Solon, A. J., Sommers, P., Schubert, Z. R., et al. (2018). Life at extreme elevations on atacama volcanoes: the closest thing to mars on earth? antonie van leeuwenhoek. Int. J. Gen. Mol. Microbiol. 111, 1389–1401. doi: 10.1007/s10482-018-1066-1060
Schoell, M. (1980). The hydrogen and carbon isotopic composition of methane from natural gases of various origins. Geochim. Cosmochim. Acta. 26, 399–407. doi: 10.1016/0016-7037(80)90155-90156
Sekiguchi, Y., Yamada, T., Hanada, S., Ohashi, A., Harada, H., and Kamagata, Y. (2003). Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain bacteria at the subphylum level. Int. J. Syst. Evol. Microbiol. 53(Pt 6), 1843–1851. doi: 10.1099/ijs.0.02699-2690
Šibanc, N., Dumbrell, A. J., Mandić-Mulec, I., and Maček, I. (2014). Impacts of naturally elevated soil CO 2 concentrations on communities of soil archaea and bacteria. Soil Biol. Biochem. 68, 348–356. doi: 10.1016/j.soilbio.2013.10.018
Sievert, S. M., Kuever, J., and Muyzer, G. (2000). Identification of 16s ribosomal DNA-defined bacterial populations at a shallow submarine hydrothermal vent near Milos island (Greece). Appl. Environ. Microbiol. 66, 3102–3109. doi: 10.1128/AEM.66.7.3102-3109.2000
Smets, B., Tedesco, D., Kervin, F., Kies, A., Vaselli, O., and Yalire, M. M. (2010). Dry gas vents (“mazuku”) in goma region (north-kivu, democratic republic of congo): formation and risk assessment. J. African Earth Sci. 58, 787–798. doi: 10.1016/j.jafrearsci.2010.04.008
Solon, A. J., Vimercati, L., Darcy, J. L., Arán, P., Porazinska, D., Dorador, C., et al. (2018). Microbial communities of high-elevation fumaroles, penitentes, and dry tephra “Soils” of the puna de atacama volcanic zone. Microb. Ecol. 76, 340–351. doi: 10.1007/s00248-017-1129-1121
Stieglmeier, M., Klingl, A., Alves, R. J. E., Rittmann, S. K. M. R., Melcher, M., Leisch, N., et al. (2014). Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 64(Pt 8), 2738–2752. doi: 10.1099/ijs.0.063172-63170
Stott, M. B., Crowe, M. A., Mountain, B. W., Smirnova, A. V., Hou, S., Alam, M., et al. (2008). Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10, 2030–2041. doi: 10.1111/j.1462-2920.2008.01621.x
Tassi, F., Venturi, S., Cabassi, J., Vaselli, O., Gelli, I., Cinti, D., et al. (2015). Biodegradation of CO2, CH4 and volatile organic compounds (VOCs) in soil gas from the Vicano-Cimino hydrothermal system (central Italy). Org. Geochem. 86, 81–93.
Tebo, B. M., Davis, R. E., Anitori, R. P., Connell, L. B., Schiffman, P., and Staudigel, H. (2015). Microbial communities in dark oligotrophic volcanic ice cave ecosystems of Mt. Erebus, Antarctica. Front. Microbiol. 6:179. doi: 10.3389/fmicb.2015.00179
Trexler, R., Solomon, C., McClure, E. E., Brislawn, C. J., Grube, A. M., Wright, J. R., et al. (2014). Assessing impacts of unconventional natural gas extraction on microbial communities in headwater stream ecosystems in northwestern Pennsylvania. Front. Microbiol. 5:522. doi: 10.3389/fmicb.2014.00522
U. S. Environmental Protection Agency, (2004). Method 3052: Microwave Assisted Acid Digestion of Siliceous and Organically Based Matrices. Washington, D.C: U. S. Environmental Protection Agency.
USDA, (2010). Keys to Soil Taxonomy, 11th Edn. Washington DC: Soil Survey Staff, United States Department of Agriculture - Natural Resources Conservation Service.
USDA, (2017). Soil Survey Manual – Handbook No. 18. Washington DC: Soil Survey Staff, United States Department of Agriculture - Natural Resources Conservation Service.
Van Gardingen, P. R., Grace, J., Harkness, D. D., Miglietta, F., and Raschi, A. (1995). Carbon dioxide emission at an Italian mineral spring result in local greenhouse effect. Agric. For. Meteorol. 73, 17–27.
Vaselli, O., Capaccioni, B., Tedesco, D., Tassi, F., Yalire, M. M., and Kasareka, M. C. (2002). The “Evils winds” (Mazukus) at nyiragongo volcano (democratic republic of congo). Acta Vulcanol. 1, 123–128.
Vaselli, O., Tassi, F., Montegrossi, G., Capaccioni, B., and Giannini, L. (2006). Sampling and analysis of volcanic gases. Acta Vulcanol. 18, 65–76.
Venturi, S., Tassi, F., Magi, F., Cabassi, J., Ricci, A., Capecchiacci, F., et al. (2019). Carbon isotopic signature of interstitial soil gases reveals the potential role of ecosystems in mitigating geogenic greenhouse gas emissions: case studies from hydrothermal systems in Italy. Sci. Total Environ. 655, 887–898. doi: 10.1016/j.scitotenv.2018.11.293
Wasmund, K., Schreiber, L., Lloyd, K. G., Petersen, D. G., Schramm, A., Stepanauskas, R., et al. (2014). Genome sequencing of a single cell of the widely distributed marine subsurface dehalococcoidia, phylum chloroflexi. ISME J. 8, 383–397. doi: 10.1038/ismej.2013.143
Yabe, S., Aiba, Y., Sakai, Y., Hazaka, M., and Yokota, A. (2011). Thermogemmatispora onikobensis gen. nov., sp. nov. and Thermogemmatispora foliorum sp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. nov. and Thermogemmatisporales ord. nov. within the class Ktedonob. Int. J. Syst. Evol. Microbiol. 61(Pt 4), 903–910. doi: 10.1099/ijs.0.024877-24870
Yabe, S., Sakai, Y., Abe, K., and Yokota, A. (2017). Diversity of Ktedonobacteria with Actinomycetes-Like Morphology in Terrestrial Environments. Microbes Environ. Environ. 32, 61–70. doi: 10.1264/jsme2.me16144
Yamada, T., and Sekiguchi, Y. (2009). Cultivation of uncultured chloroflexi subphyla: significance and ecophysiology of formerly uncultured chloroflexi “Subphylum I” with natural and biotechnological relevance. Microbes Environ. 24, 205–216. doi: 10.1264/jsme2.me09151s
Yamada, T., Sekiguchi, Y., Hanada, S., Imachi, H., Ohashi, A., Harada, H., et al. (2006). Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. And Caldilineae classis nov. In the bacterial phylum chloroflexi. Int. J. Syst. Evol. Microbiol. 56(Pt 6), 1331–1340. doi: 10.1099/ijs.0.64169-64160
Yamada, T., Sekiguchi, Y., Imachi, H., Kamagata, Y., Ohashi, A., and Harada, H. (2005). Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl. Environ. Microbiol. 71, 7493–7503. doi: 10.1128/AEM.71.11.7493-7503.2005
Keywords: mofette, CO2, bacteria, soil, acetogenesis
Citation: Fazi S, Ungaro F, Venturi S, Vimercati L, Cruz Viggi C, Baronti S, Ugolini F, Calzolari C, Tassi F, Vaselli O, Raschi A and Aulenta F (2019) Microbiomes in Soils Exposed to Naturally High Concentrations of CO2 (Bossoleto Mofette Tuscany, Italy). Front. Microbiol. 10:2238. doi: 10.3389/fmicb.2019.02238
Received: 22 March 2019; Accepted: 12 September 2019;
Published: 04 October 2019.
Edited by:
Mirko Basen, University of Rostock, GermanyReviewed by:
Roey Angel, Academy of Sciences of the Czech Republic, CzechiaKristof Brenzinger, Netherlands Institute of Ecology (NIOO-KNAW), Netherlands
Copyright © 2019 Fazi, Ungaro, Venturi, Vimercati, Cruz Viggi, Baronti, Ugolini, Calzolari, Tassi, Vaselli, Raschi and Aulenta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefano Fazi, ZmF6aUBpcnNhLmNuci5pdA==
 Stefano Fazi
Stefano Fazi Fabrizio Ungaro2
Fabrizio Ungaro2 Stefania Venturi
Stefania Venturi Lara Vimercati
Lara Vimercati Carolina Cruz Viggi
Carolina Cruz Viggi Franco Tassi
Franco Tassi Orlando Vaselli
Orlando Vaselli Federico Aulenta
Federico Aulenta