Abstract
Resistance in Mycobacterium tuberculosis is a major obstacle for effective treatment of tuberculosis. Multiple studies have shown promising results for predicting drug resistance in M. tuberculosis based on whole genome sequencing (WGS) data, however, these tools are often limited to this single species. We have previously developed a common platform for resistance prediction in multiple species. This platform detects acquired resistance genes (ResFinder) and species-specific chromosomal mutations (PointFinder) associated with resistance, all based on WGS data. In this study, we present a new version of PointFinder together with an updated M. tuberculosis database. PointFinder now includes predictions based on insertions and deletions, and it explicitly reports frameshift mutations and premature stop codons. We found that premature stop codons in four resistance-associated genes (katG, ethA, pncA, and gidB) were over-represented in resistant strains, and we saw an increased prediction performance when including premature stop codons in these genes as resistance markers. Different M. tuberculosis resistance prediction tools vary in performance mostly due to the mutation library used. We found that a well-established mutation library included non-predictive linage markers, and through forward feature selection we eliminated those from the mutation library. Compared to other similar web-based tools, PointFinder performs equally good. The advantages of PointFinder is that together with ResFinder it serves as a common web-based and downloadable platform for resistance detection in multiple species. It is easy to use for clinicians and already widely used in the research community.
Introduction
Next generation sequencing is a rapidly evolving field and it is in the process of being adopted as the standard in many clinical and public health settings. Here, it replaces many traditional typing and phenotyping methods such as those for species determination and detection of antimicrobial resistance. Rapid and precise detection of antimicrobial resistance is important for correct treatment, surveillance and control efforts. Antimicrobial resistance occurs either through horizontal gene transfer or by de novo chromosomal mutations (Munita and Arias, 2016). In Mycobacterium tuberculosis all acquired resistance have been mediated by chromosomal mutations, and horizontal transfer have not been described (Smith et al., 2013). In addition to acquired resistance M. tuberculosis have a number of intrinsic resistance mechanisms including modification of drug targets, chemical modification of drugs, enzymatic degradation of drugs, molecular mimicry of drug targets, and drug deportation by efflux pumps (Smith et al., 2013). This is a serious obstacle for effective tuberculosis treatment and prevention of the disease worldwide (World Health Organization [WHO], 2017). Mutations and other genetic changes may lead to enzymatic inactivation of antibiotic molecules, overexpression of novel efflux pumps and porin alterations in the cell wall, trapping of drugs and overexpression of proteins involved in neutralizing the effect of drugs. Due to slow growth rates of M. tuberculosis, determining resistance by conventional drug susceptibility testing (DST) is highly time-consuming. Contrarily, next-generation sequencing rapidly yields accurate whole genome sequencing (WGS) data. Using prior knowledge on the genomic changes leading to resistance, WGS data can be used for rapid prediction of antimicrobial resistance (Koser et al., 2014). In fact, studies have already shown promising results for predicting resistance in M. tuberculosis based on WGS for first-line anti-tuberculosis drugs (Feuerriegel et al., 2015; The CRyPTIC Consortium and the 100, 000 Genomes Project et al., 2018). However, a challenge for applying this knowledge in a clinical setting is that resistance predictor tools are often limited to a single species. We have previously developed ResFinder (Zankari et al., 2012), an in silico method for detection of acquired genes associated with antimicrobial resistance in multiple species based on WGS data. ResFinder was recently expanded with PointFinder (Zankari et al., 2017), a species-specific tool detecting chromosomal mutations associated with drug resistance. PointFinder already includes five species. The rationale of this study is to expand PointFinder also to cover M. tuberculosis. In addition to point mutations insertions and deletions may also affect resistance. Especially if the insertion/deletion length is not a multiple of three they will cause the rest of the gene to be read out of frame, which have a high likelihood of introducing a stop codon leading to a truncated gene. We have therefore set out to do a thorough analysis of the correlation of premature stop codons with resistance. In this study we optimized and evaluated the performance of PointFinder’s prediction of resistance in a sixth species, M. tuberculosis. M. tuberculosis was chosen because of its importance for global health. It is also an organism for which many resistance mutations have been described. Here, we wanted to investigate is some of these are in fact non-informative when it comes to predicting resistance, and study in more detail how the presence of premature stop codons affects resistance. We used a data set of 3,528 M. tuberculosis isolates in the optimization which consisted of omitting non-predictive mutations from a well-established mutation library, and including premature stop codons as resistance markers. 2,480 isolates were used to validate the performed optimization.
Materials and Methods
PointFinder Database
The PointFinder database contains both a mutation library listing resistance-associated chromosomal mutation and a collection of reference sequences in which these mutations occur. All database files are available at bitbucket.org/genomicepidemiology/pointfinder_db.
The tuberculosis mutation library was obtained from pathogenseq.lshtm.ac.uk, under Tuberculosis and Rapid DR Study and described in Coll et al. (2015). Additional mutations were achieved from a genome wide association (GWA) study performed by the same group in Coll et al. (2018). Mutations, which were observed in the GWA study to be significantly associated with resistance and observed more than 10 times, were also included in the mutation library. All genes, RNA genes and promoter regions of interest for resistance prediction in M. tuberculosis are shown in Table 1. Reference sequences for genes and genomic regions listed in Table 1 were obtained from the H37Rv M. tuberculosis reference strain, NCBI-reference sequence: NC_000962.3.
TABLE 1
| Drug | Genes | RNA genes | Promoter regions |
| Rifampicin | rpoB, rpoC | ||
| Isoniazid | katG, inhA, kasA, ahpC | katG promoter, ahpC promoter, fabGl promoter | |
| Streptomycin | rpsL, gidB, embB, embC, | rrs (16SrRNA) | |
| Ethambutol | embA, embR, ubiA | embA promoter | |
| Amikacin | rrs (16SrRNA) | ||
| Capreomycin | tlyA | rrs (16SrRNA) | idsA2 promoter |
| Ethionamide | ethR, ethA, inhA | fabGl promoter, ethA promoter | |
| Kanamycin | rrs (16SrRNA) | eis promoter | |
| Pyrazinamide | pncA, panD, rpsA | pncA promoter | |
| Fluoroquinolone | gyrA, gyrB | ||
| Para-aminosalicylic acid | ridD, folC, thyA | thyX promoter | |
| Linezolid | rplC | rrl (23S rRNA) | |
| Bedaquiline | Rv0678 | ||
| Clofazimine | Rv0678 | ||
| d-Cycloserine | iniA, alr | ||
| XDR-TB | drrA | nuoA promoter |
Genes and genomic regions of interest for drug resistance in M. tuberculosis.
For each drug the genes and genomic regions of interest for resistance are listed. All gene names refer to a gene sequences in the H37Rv reference genome with the National Center for Biotechnology Information (NCBI) reference sequence: NC 000962.3. The loci in bold are novel resistance-associated genomic regions revealed by a genome wide association study from Coll et al. (2018).
PointFinder
PointFinder is both a web service and command line application for predicting resistance associated with chromosomal mutations based on WGS data. The web service is available on cge.cbs.dtu.dk/services/ResFinder/where users can specify to search “Chromosomal mutations” in six different species, including M. tuberculosis. The command line version of PointFinder is available on bitbucket.org/genomicepi-demiology/pointfinder.
PointFinder is a Python program that accepts both FastQ and Fasta files for resistance prediction. Initially, the genes of interest for resistance prediction (Table 1) are identified. KMA (Clausen et al., 2018) is used for mapping of raw reads, and BLASTN, RRID:SCR_001598 (Camacho et al., 2009) for aligning assembled genomes, to the genes of interest.
Mutations are detected by comparing the alignments between the reference sequences and the sequences found in the input file. The aligned sequences are compared nucleotide by nucleotide when the alignment represents a promoter region or an RNA gene and codon by codon when it represents a coding gene sequence. Effort has been put into detecting insertions and deletions and reporting any disruption or restoring of the reading frame. If a premature stop codon is detected, this will also be explicitly reported, and no further search for mutations will be performed after the detection of a stop codon. The observed mutations are looked up into the mutation library which holds information about mutations known to be predictive for resistance. If a found mutation exists in the mutation library, the resistance phenotype is returned together with the PubMed ID of the article linking the observed genotype with the predicted resistance phenotype.
M. tuberculosis Data Sets
All data sets used in this study exclusively consisted of paired-end WGS data associated with phenotype data. Phenotype data was given as Resistant or Susceptible based on laboratory determined DST results for multiple anti-tuberculosis drugs.
The first data set, called the ReSeq data set, was obtained from the Relational Sequencing TB Data Platform (Starks et al., 2015). It consisted of WGS data from 3,528 M. tuberculosis isolates. The second data set was obtained from the Supplementary Data in Coll et al. (2018) and was used as a validation data set. The validation data set contained 2,480 isolates. The ReSeq and validation data set contained sufficient phenotype data for 10 drugs namely; Rifampicin, Isoniazid, Streptomycin, Ethambutol, Amikacin, Capreomycin, Ethionamide, Kanamycin, Pyrazinamide, and Fluoroquinolones. The number of isolates with determined phenotype varied with each drug. Fluoroquinolones DSTs were determined for the specific compounds namely, Ciprofloxacin, Ofloxacin, Moxifloxacin, and Levofloxacin. However, in the analysis we considered Fluoroquinolones resistance as one, since the mutation library did not distinguish between different compounds. Thus, if an isolate was resistant to any of the Fluoroquinolone compounds it was considered Fluoroquinolones resistant. The data sets can be found in Supplementary Tables S1, S2.
A third data set was used to compare PointFinder to similar resistance predictor tools developed for M. tuberculosis. From a scientific report by Schleusener et al. (2017) we obtained 91 isolates that had been used to compare five existing M. tuberculosis resistance predictor tools. These 91 isolates were Illumina MiSeq paired end sequenced, and phenotype data existed for five drugs namely, Rifampicin, Isoniazid, Streptomycin, Ethambutol, and Pyrazinamide.
Measuring Prediction Performance
PointFinder’s detection of resistance-associated mutations was used for binary classification of resistance and susceptibility using the following rules. Isolates were predicted resistant to a drug if one or more mutations predictive of resistance to the drug were found. Isolates were predicted susceptible to a drug if all genes of interest for resistance to the drug were found with an identity above 90% and a sequence coverage above 60%, and no resistance-associated mutations were detected in the genes. We used default options and parameters when running PointFinder. To assess the quality of PointFinder’s binary classification we calculated the Matthew’s Correlation Coefficient (MCC) and the sensitivity and specificity of the prediction.
Forward Selection of Predictive Mutation
To detect non-predictive mutations, we applied forward feature selection optimized based the MCC over threefold cross-validation. We exclusively examined abundant mutations, defined as mutations found in the ReSeq data set 10 times or more. Mutations found less than 10 times were included in the initial state of the prediction model, whereas the abundant mutations were initially excluded. With each step of the forward selection one abundant mutation was added to the model. The mutation added was the one mutation that benefited the prediction the most based on the MCC. Mutations were added to the model one by one until adding any remaining mutations would decrease the quality of prediction. Examined mutations that were not selected in any of the threefold of the cross-validation were considered non-predictive for resistance.
Statistical Analyses
Significant over-representation of premature stop codons in resistant isolates was assessed with Pearson’s Chi-squared test on a 2 × 2 matrix using the statistical software R (Version 3.4.0). PointFinder was compared with a similar predictor called PhyResSE. We assessed if PhyResSE performed significantly better than PointFinder using bootstrapping.
Results
We created an updated method for predicting antimicrobial resistance from the genomic sequence. An overview of the method can be seen in Figure 1.
FIGURE 1
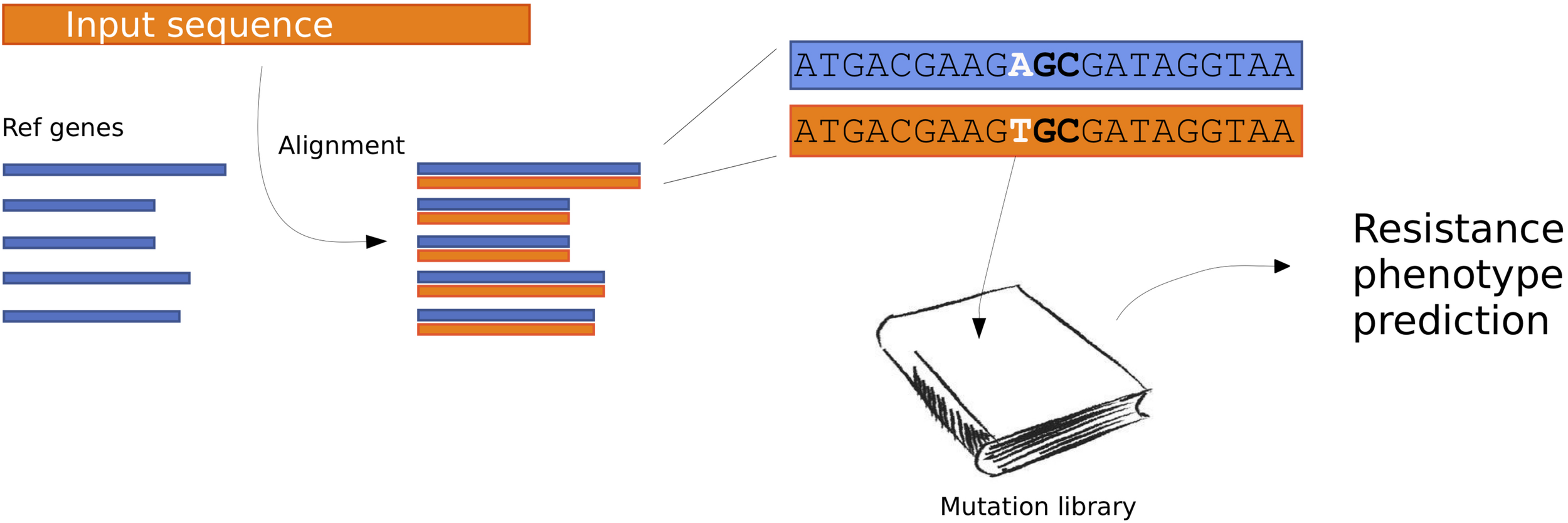
Flow chart describing the PointFinder workflow. The input sequences are aligned to a database of reference genes. The genetic differences observed in the alignments are compared to a mutation library, with annotated phenotypes. Based on this a resistance phenotype prediction is made.
Evaluating and Optimizing the Mutation Library
We calculated the sensitivity, specificity and MCC for predicting drug resistance using PointFinder compared to phenotypic DST results (Table 2). The resistance prediction was based on mutations from the mutation library detected in the 3,528 M. tuberculosis isolates from the ReSeq data set. The best prediction performances were obtained for the first-line drugs Rifampicin, Isoniazid and MDR (MCC of 0.85, 0.82, and 0.86, respectively). PointFinder’s prediction performance varied dependently on the drug with MCCs ranging from 0.386 to 0.848. Especially, the prediction of resistance to Ethambutol, Pyrazinamide, Amikacin, and Ethionamide was less successful, which indicated that the mutation library was not fully developed.
TABLE 2
| Drug | Res | Sus | Spec. | Sens. | MCC |
| RMP | 771 | 2710 | 0.965 | 0.887 | 0.848 |
| INH | 1093 | 2420 | 0.929 | 0.903 | 0.819 |
| STM | 728 | 1239 | 0.874 | 0.798 | 0.670 |
| EMB | 466 | 3040 | 0.796 | 0.850 | 0.484 |
| PZA | 325 | 2993 | 0.935 | 0.575 | 0.475 |
| KAN | 76 | 617 | 0.989 | 0.776 | 0.814 |
| FLQ | 240 | 1175 | 0.956 | 0.679 | 0.664 |
| AMK | 107 | 866 | 0.785 | 0.766 | 0.386 |
| ETH | 49 | 175 | 0.943 | 0.469 | 0.481 |
| CAP | 116 | 1024 | 0.971 | 0.474 | 0.512 |
PointFinder predicted resistance compared with phenotypic drug susceptibility testing on the ReSeq data set.
Res, number of resistant isolates determined by drug susceptibility testing; Sus, number of susceptible isolates determined by drug susceptibility testing; Spec, specificity; Sens, sensitivity; MCC, Mathew Correlation Coefficient; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide; KAN, Kanamycin; FLQ, Fluoroquinolones; AMK, Amikacin; ETH, Ethionamide; CAP, Capreomycin.
Table 3 shows the occurrence of PointFinder-detected premature stop codons in the resistance-associated genes found in resistant and susceptible isolates. Genes shown in bold in Table 3 were in the mutation library described with position-specific premature stop codons causing resistance. With the exception of the panD gene, these genes showed a significantly higher occurrence of premature stop codons among resistant strains. However, for many genes the analysis was only based on a few premature stop codons. Only for four genes, katG, pncA, ethA, and gidB premature stop codons occurred more than 10 times, and we used this as a threshold for a considerable frequency. Moreover, premature stop codons in these genes were significantly over-represented in strains resistant to the drug that the genes were associated with (see Table 1). For katG, pncA, and ethA the representative p-values were below 0.00001 and for gidB it was 0.006. PointFinder’s prediction performance given in Table 4 shows that considering premature stop codons in the four genes as resistance markers improved the MCC of the resistance prediction for drugs in question; Isoniazid, Streptomycin, Pyrazinamide, and Ethionamide. In the case of Streptomycin and Ethionamide, the performances were improved with a compromise of the specificity.
TABLE 3
| Drug | Res | Sus | Gene | Prem. stop codons in res. strains | Prem. stop codons in sus. strains | p-value |
| RMP | 771 | 2710 | rpoC | 3 | 1 | 0.011 |
| rpoB | 2 | 0 | 0.008∗ | |||
| inhA | 1 | 0 | 0.137 | |||
| INH | 1093 | 2420 | kasA | 3 | 0 | 0.01∗ |
| ahpC | 1 | 2 | 0.934 | |||
| katG | 13 | 0 | < 1.0e−5∗ | |||
| STM | 728 | 1239 | rpsL | 1 | 0 | 0.192 |
| gidB | 42 | 40 | 0.006∗ | |||
| embA | 1 | 4 | 0.658 | |||
| embR | 3 | 4 | 0.021 | |||
| EMB | 466 | 3040 | embC | 1 | 4 | 0.658 |
| embB | 1 | 2 | 0.306 | |||
| ubiA | 1 | 0 | 0.011 | |||
| PZA | 325 | 2993 | pncA | 372 | 7 | < 1.0e−5∗ |
| panD | 6 | 0.148 | ||||
| ETH | 49 | 175 | inhA | 1 | 0 | 0.058 |
| ethA | 21 | 24 | < 1.0e−5∗ | |||
| CAP | 116 | 1024 | tlyA | 2 | 2 | 0.008∗ |
Occurrence of resistance-associated genes with premature stop codons in resistant or susceptible strains in the ReSeq data set.
Genes annotated with premature stop codons in the mutation library are shown in bold. It was tested whether genes with premature stop codons were equally distributed between the two phenotypic groups. This was tested using a χ2 test on a 2 × 2 matrix, and the p-value is given. ∗p-value below the significance level 0.01. Res, population of resistant strains; Sus, population of susceptible strains. RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide; ETH, Ethionamide; CAP, Capreomycin.
TABLE 4
| Drug | Res | Sus | Spec. | Sens. | MCC |
| RMP | 771 | 2710 | 0.965 | 0.887 | 0.848 |
| INH | 1093 | 2420 | 0.929 | 0.909 | 0.823 |
| STM | 728 | 1239 | 0.847 | 0.839 | 0.674 |
| EMB | 466 | 3040 | 0.796 | 0.850 | 0.484 |
| PZA | 325 | 2993 | 0.935 | 0.612 | 0.502 |
| KAN | 76 | 617 | 0.989 | 0.776 | 0.814 |
| FLQ | 240 | 1175 | 0.956 | 0.679 | 0.664 |
| AMK | 107 | 866 | 0.785 | 0.766 | 0.386 |
| ETH | 50 | 175 | 0.829 | 0.800 | 0.564 |
| CAP | 116 | 1024 | 0.971 | 0.474 | 0.512 |
PointFinder predicted resistance compared with phenotypic drug susceptibility testing on the ReSeq data set when considering premature stop codons in katG, pncA, ethA, and gidB as resistance markers.
Green indicates an increase and Red a decrease in performance compared to the initial prediction in Table 2. Res, number of resistant strains determined by drug susceptibility testing; Sus, number of susceptible strains determined by drug susceptibility testing; Spec, specificity; Sens, sensitivity; MCC, Mathew Correlation Coefficient; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide; KAN, Kanamycin; FLQ, Fluoroquinolone; AMK, Amikacin; ETH, Ethionamide; CAP, Capreomycin.
Besides a possible lack of predictive premature stop codons, the mutation library also seemed to include mutations that were not predictive for resistance. For example, a low specificity was observed in the resistance prediction of Ethambutol and Amikacin, due to many false positive predictions. This indicated that the mutation library contained mutations, which should be omitted.
To detect such non-predictive mutations, we used a forward feature selection approach where the selection of mutations was optimized based the MCC over threefold cross-validation. Mutations not selected in any of the threefold of the cross-validation were considered non-predictive for resistance and shown in bold in Table 5. For 7 out of the 10 drugs, we found one or more mutations that were deselected in every fold and these mutations were omitted from the mutation library. The occurrence of the deselected mutations in resistant and susceptible isolates are shown in Supplementary Table S3.
TABLE 5
| Drug | CV-fold | Deselected mutations | MCC train | MCC test |
| 1 | rpoB I491F, rpoB H445N, rpoB L430P, rpoC L527V | 0.878 | 0.860 | |
| RMP | 2 | rpoB 149IF, rpoB H445N, rpoB L430P, rpoC L527V | 0.868 | 0.878 |
| 3 | rpoB 149 IF, rpoB H445N, rpoB L430P | 0.869 | 0.874 | |
| 1 | kasA G269S, kasA G312S, inhA V78A, fabGl promoter −8T > C | 0.880 | 0.884 | |
| INH | 2 | kasA G269S, kasA G312S, inhA V78A | 0.881 | 0.882 |
| 3 | kasA G269S, kasA G312S, inhA V78A | 0.883 | 0.877 | |
| 1 | rrs 1401A > G, rrs 492C > T | 0.743 | 0.744 | |
| STM | 2 | rrs 1401A > G, rrs 492C > T, gidB prem. stop codon | 0.733 | 0.762 |
| 3 | rrs 1401 A > G, rrs 492C > T, gidB prem. stop codon | 0.757 | 0.715 | |
| 1 | embB E378A, embC T270I, embB T1082A, | 0.648 | 0.590 | |
| embA promoter −12C > T, embA promoter −16C > T | ||||
| EMB | 2 | embB E378A, embC T270I, embB T1082A, embB G406D, embB D1024N, embB N296H | 0.643 | 0.616 |
| 3 | embB E378A, embC T270I, embB T1082A, embB G406D, embB N296H | 0.623 | 0.660 | |
| 1 | pncA I6L, pncA A146T, pncA W68G | 0.589 | 0.625 | |
| PZA | 2 | pncA I6L, pncA A146T | 0.590 | 0.636 |
| 3 | pncA I6L | 0.638 | 0.537 | |
| 1 | 0.789 | 0.864 | ||
| KAN | 2 | 0.829 | 0.785 | |
| 3 | 0.826 | 0.795 | ||
| 1 | gyrA T80A | 0.699 | 0.691 | |
| FLQ | 2 | gyrA T80A | 0.686 | 0.713 |
| 3 | gyrA T80A | 0.702 | 0.687 | |
| 1 | rrs 517C > T, rrs 514A > C | 0.702 | 0.744 | |
| AMK | 2 | rrs 517C > T, rrs 514A > C | 0.722 | 0.705 |
| 3 | rrs 517C > T, rrs 514A > C | 0.725 | 0.699 | |
| 1 | 0.652 | 0.393 | ||
| ETH | 2 | ethA prem. stop codon | 0.514 | 0.407 |
| 3 | 0.543 | 0.592 | ||
| 1 | 0.511 | 0.514 | ||
| CAP | 2 | 0.511 | 0.514 | |
| 3 | 0.514 | 0.509 |
Forward feature selection of resistance mutations on the ReSeq data set.
For all 10 drugs forward feature selection was performed over a threefold cross-validation to assess if the resistance performance could improve by omitting any mutations. CV-fold indicates for each fold of the cross-validation which mutations were deselected. Mutations shown in bold were deselected in every fold of the cross-validation and further considered non-predictive for resistance. MCC train, Mathew’s Correlation Coefficient obtained on the training data subset; MCC test, Mathew Correlation Coefficient obtained on the test data subset; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide; KAN, Kanamycin; FLQ, Fluoroquinolone; AMK, Amikacin; ETH, Ethionamide; CAP, Capreomycin.
Table 6 shows the prediction performance when excluding the mutations from the mutation library. Omitting the non-predictive mutations from the mutation library did compromises the sensitivity, yet since the forward feature selection was trained based on the MCCs, the MCC performance was improved for all seven drugs.
TABLE 6
| Drug | Res | Sus | Spec. | Sens. | MCC |
| RMP | 771 | 2710 | 0.978 | 0.878 | 0.871 |
| INH | 1093 | 2420 | 0.974 | 0.895 | 0.881 |
| STM | 728 | 1239 | 0.907 | 0.835 | 0.743 |
| EMB | 466 | 3040 | 0.898 | 0.848 | 0.631 |
| PZA | 325 | 2993 | 0.974 | 0.575 | 0.604 |
| KAN | 76 | 617 | 0.992 | 0.776 | 0.814 |
| FLQ | 240 | 1175 | 0.968 | 0.679 | 0.695 |
| AMK | 107 | 866 | 0.992 | 0.607 | 0.716 |
| ETH | 50 | 175 | 0.829 | 0.800 | 0.564 |
| CAP | 116 | 1024 | 0.971 | 0.474 | 0.512 |
PointFinder predicted resistance compared with phenotypic drug susceptibility testing on the ReSeq data set after including premature stop codons and exclusion of non-predictive mutations.
Green indicates an increase and red a decrease in performance compared to the prediction in Table 2. Res, number of resistant strains determined by drug susceptibility testing; Sus, number of susceptible strains determined by drug susceptibility testing; Spec, specificity; Sens, sensitivity; MCC, Mathew Correlation Coefficient; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide; KAN, Kanamycin; FLQ Fluoroquinolone. AMK, Amikacin; ETH, Ethionamide; CAP, Capreomycin.
Validating the Mutation Library Optimization
To validate the effects of including premature stop codons and excluding non-predictive mutations from the mutation library, we performed resistance predictions on a validation data set. This data set consisted of 2,480 isolates, and was independent of the ReSeq data set.
First, we examined occurrence of genes with premature stop codons in resistant and susceptible strains (Table 7). Like in the ReSeq data set premature stop codons occurred with a considerably frequency in the genes katG, pncA, ethA, and gibB. However, here only the katG and pncA premature stop codons were significantly over-represented in the resistant strains. gidB was close to the significant level of 0.01 (p-value: 0.015) whereas ethA premature stop codons seemed to be equally distributed between resistant and susceptible strains (p-value: 0.642).
TABLE 7
| Drug | Res | Sus | Loci | Prem. stop codons in resistant strains | Prem. stop codons in susceptible strains | p-value |
| RMP | 596 | 1814 | rpoC | 1 | 1 | 0.407 |
| INH | 768 | 1641 | ahpC | 0 | 1 | 0.566 |
| katG | 8 | 2 | 0.001∗ | |||
| STM | 379 | 687 | gidB | 28 | 27 | 0.015 |
| PZA | 248 | 420 | pncA | 25 | 0 | <1.0e−5∗ |
| ETH | 186 | 245 | ethR | 0 | 1 | 0.383 |
| ethA | 24 | 28 | 0.642 | |||
| CAP | 191 | 261 | tlyA | 0 | 1 | 0.392 |
Occurrence of resistance-associated genes with premature stop codons found in resistant or susceptible strains in the validation data set.
It was tested whether genes with premature stop codons were equally distributed between the two phenotypic groups. This was tested using a χ2 test on a 2 × 2 matrix, and the p-value is given. ∗p-value below the significance level of 0.01. Res, number of resistant strains determined by drug susceptibility testing; Sus, number of susceptible strains determined by drug susceptibility testing; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; PZA, Pyrazinamide; ETH, Ethionamide; CAP, Capreomycin.
Additionally, we looked at the occurrence of the mutations that were considered non-predictive in the forward feature selection analysis. Data is shown in Supplementary Table S3. Most of the mutations that were found to be non-predictive for resistance in the ReSeq data set were confirmed to be widely present in susceptible strains in the validation data set. The mutations, rpoB I491F, inhA V78A, pncA I6L, gyrA T80A, and rrs 517C > T, were present in none or in very few samples in the validation data set, and therefore, the positive effect of removing these mutations could not be validated.
Table 8 shows prediction performances on the validation data set using three different mutation libraries; first, the initial mutation library, secondly, the mutation library where premature stop codons in katG, pncA, ethA, and gidB were included as resistance markers, and thirdly, the mutation library containing both the four premature stop codon markers and excluding the non-predictive mutations. Table 8 shows an overall improved prediction performance when including the premature stop codons as resistance markers and when excluding the non-predictive mutations.
TABLE 8
| Drug | Res | Sus | Initial mutations | Including prem. stop codons | Excluding non-predictive muts | ||||||
| Spec. | Sens. | MCC | Spec. | Sens. | MCC | Spec. | Sens. | MCC | |||
| RMP | 596 | 1814 | 0.978 | 0.896 | 0.885 | 0.978 | 0.896 | 0.885 | 0.986 | 0.886 | 0.895 |
| INH | 768 | 1641 | 0.946 | 0.879 | 0.826 | 0.945 | 0.880 | 0.826 | 0.969 | 0.870 | 0.854 |
| STM | 379 | 687 | 0.85 | 0.744 | 0.592 | 0.817 | 0.805 | 0.606 | 0.902 | 0.778 | 0.688 |
| EMB | 304 | 880 | 0.739 | 0.914 | 0.576 | 0.739 | 0.914 | 0.576 | 0.825 | 0.908 | 0.666 |
| PZA | 239 | 420 | 0.971 | 0.799 | 0.802 | 0.971 | 0.808 | 0.809 | 0.971 | 0.808 | 0.809 |
| KAN | 211 | 176 | 0.920 | 0.896 | 0.814 | 0.920 | 0.896 | 0.814 | 0.920 | 0.896 | 0.814 |
| FLQ | 271 | 296 | 0.905 | 0.878 | 0.784 | 0.905 | 0.878 | 0.784 | 0.909 | 0.878 | 0.788 |
| AMK | 213 | 278 | 0.953 | 0.850 | 0.814 | 0.953 | 0.850 | 0.814 | 0.964 | 0.826 | 0.807 |
| ETH | 184 | 245 | 0.624 | 0.853 | 0.479 | 0.539 | 0.919 | 0.479 | 0.539 | 0.919 | 0.479 |
| CAP | 191 | 261 | 0.877 | 0.864 | 0.738 | 0.877 | 0.864 | 0.738 | 0.877 | 0.864 | 0.738 |
Validating the effect of including premature stop codons and excluding non-predictive mutations from the mutation library.
PointFinder-predicted resistance was compared to drug susceptibility test results on the validation data set. The quality of the prediction was evaluated with three different mutation libraries; the initial mutation library (initial mutations). The mutation library including premature stop codons in katG, pncA, ethA, and gidB (including prem. stop codons), and the mutation library both including those premature stop codons and excluding mutations considered non-predictive for resistance (excluding non-predictive muts). Green indicates an increase and red a decrease in the performance measurement compared to same measurement from the mutation library to the left. Res, number of resistant strains determined by drug susceptibility testing; Sus, number of susceptible strains determined by drug susceptibility testing; Spec, specificity; Sens, sensitivity; MCC, Mathew Correlation Coefficient; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide; KAN, Kanamycin; FLQ Fluoroquinolone; AMK, Amikacin; ETH, Ethionamide; CAP, Capreomycin.
Comparing PointFinder With Similar Tools
A scientific report from 2017 by Schleusener et al. PhyResSE generally showed the best performance, therefor we used the same data set to compare PointFinder to PhyResSE. We reran the data set through PhyResSE, to make a direct comparison. Table 9 show the prediction performance of PointFinder and PhyResSE based on WGS data and DST results from the 91 isolates. The mutation library used for PointFinder included premature stop codons in katG, pncA, ethA, and gidB and did not contain the non-predictive mutations. For Isoniazid, Streptomycin, and Ethambutol PhyResSE showed better performances. In the case of Streptomycin PheResSE performed significantly better than PointFinder which had a few false negative prediction, see Supplementary Table S4. For the drugs Rifampicin and Pyrazinamide PointFinder showed the best performance.
TABLE 9
| Drug | Res | Sus | PointFinder | PhyResSE | p-value | ||||
| Spec. | Sens. | MCC | Spec. | Sens. | MCC | ||||
| RMP | 14 | 77 | 0.961 | 1.000 | 0.890 | 0.935 | 1.000 | 0.830 | 0.751 |
| INH | 29 | 62 | 0.952 | 0.897 | 0.848 | 0.968 | 0.931 | 0.899 | 0.262 |
| STM | 37 | 54 | 0.981 | 0.649 | 0.693 | 0.981 | 0.838 | 0.843 | 0.047∗ |
| EMB | 14 | 77 | 0.961 | 0.857 | 0.796 | 0.974 | 0.857 | 0.831 | 0.395 |
| PZA | 8! | 83! | 0.964 | 0.750 | 0.677 | 0.964 | 0.625! | 0.589 | 0.666 |
Comparing PointFinder and PhyResSE prediction performance.
! Indicates that data varied from the previously published results, since the number of resistance and susceptible samples found in the published Supplementary Data did not correlate with amount assessed after rerunning the data through PheResSE. Res, number of resistant strains determined by drug susceptibility testing; Sus, number of susceptible strains determined by drug susceptibility testing; Spec, specificity; Sens, sensitivity; MCC, Mathew Correlation Coefficient; RMP, Rifampicin; INH, Isoniazid; STM, Streptomycin; EMB, Ethambutol; PZA, Pyrazinamide. It was tested weather PhyResSE was performing significantly better using bootstrapping, and the p-value is given, and the significance level was set at 0.05 indicated with ∗.
Discussion
In this study, we presented an improved version of PointFinder where the detection of insertion and deletion together with frameshift mutations were handled properly. As an effect of the improvements we were able to enhance PointFinder’s resistance prediction in M. tuberculosis by including premature stop codons as resistance markers. Additionally, we optimized the obtained M. tuberculosis mutation library by excluding mutations that through forward feature selection were considered non-predictive for resistance.
A scientific report from 2017 by Schleusener et al. compared five M. tuberculosis resistance prediction tools based on a data set of 91 isolates. These five tools were, CASTB (Iwai et al., 2015), PhyResSE (Feuerriegel et al., 2015), TBProfiler (Coll et al., 2015), KvarQ (Steiner et al., 2014), and Mykrobe Predictor TB (Bradley et al., 2015). To our knowledge it has not been studied thoroughly how the occurrence of premature stop codons in resistance-associated genes affect the resistance phenotype. The mutation library lists premature stop codons predictive for resistance, yet these premature stop codons are only considered as predictive markers if found at the specific position listed. However, the outcome of a premature stop codon – gene truncation – is, in most cases, independent of the position in the gene. The first version of PointFinder described in Zankari et al. (2017) did not consider insertions and deletions, and as a consequence of this, frameshift mutations and premature stop codons was not correctly detected. With this new version of PointFinder, efforts were put into detecting reading frame disruptions and premature stop codons caused by insertions and deletions, and the improved PointFinder version was used to assess the impact of premature stop codons on resistance emergence.
Among all genes annotated with predictive premature stop codons in the mutation library we found a significantly higher occurrence of premature stop codons among resistant strains in the ReSeq data set, with the exception of the panD gene (Table 3). A study from 2014, showed a M. tuberculosis panD deleted mutant still susceptible to Pyrazinamide (Dillon et al., 2014). The study postulated that panD is not a target for Pyrazinamide resistance, and our results support this hypothesis by indicating that loss of function of panD is not associated with Pyrazinamide resistance.
Our results suggest that katG and pncA premature stop codons are predictive for resistance, whereas the role of ethA and gidB premature stop codons was less clear. Isoniazid, Pyrazinamide, and Ethionamide are pro-drugs, and the proteins encoded by katG, pncA, and ethA are enzymes catalyzing the activation of these drugs, respectively (Zhang et al., 1992; Almeida Da Silva and Palomino, 2011). If the enzymatic activity is lost (e.g., by the occurrence of a premature stop codon), the drug cannot be converted to its active form, which can explain the emergence of drug resistance.
Surprisingly, premature stop codons in ethA also occurred with a high frequency in susceptible strains, and in the validation data set premature stop codons in ethA were not over-represented in resistant strains (Table 7). Since, ethA encodes the Ethionamide activating enzyme, we speculate whether this is not the only enzyme able to activate Ethionamide, or if Ethionamide also has antimicrobial effects as a pro-drug, or maybe premature stop codons can be neglected and not cause complete depletion of the ethA-encoded enzyme. Another explanation for the inconclusive effect of ethA premature stop codons, might be that the use of Ethionamide constitutes a selective pressure that favors premature stop codon in ethA leading to low-levels resistance close to the clinical breakpoint used in DST protocols.
Premature stop codons in gidB were slightly over-represented among the resistant strains both in the ReSeq (p-value = 0.006) and the validation data set (p-value = 0.015), yet, premature stop codons in gidB were also observed in many susceptible isolates (see Tables 3, 7). Like for ethA, this might reflect that depletion of the gidB-encoded protein causes resistance levels close to the clinical breakpoint. In fact, a functional study showed that knocking out gidB leads to low-level Streptomycin resistance (Wong et al., 2011). We observed an increased MCC when treating the mutation as a resistance marker, thereby, our study also indicates that loss of function of the gidB-encoded protein is associated with Streptomycin resistance.
The forward feature selection analysis implied that several mutations included in the obtained mutation library were misclassified as resistance markers, and the positive effects of removing these mutations were also seen in the MCC on the validation data set (Table 8), with the exception of predicting Amikacin resistance. The two mutation rrs 514A > C and rrs 517C > T that were removed in this case, were however also found in other studies to play no role in resistance to Amikacin (Maus et al., 2005; Jugheli et al., 2009).
Further investigation showed that the misclassification of many of the deselected mutations was also reported in other studies, for example for kasA G269S and kasA G312S (Sun et al., 2007), rrs 492C > T (Victor et al., 2001; Villellas et al., 2013), rrs 1401A > G (for Streptomycin resistance) (Via et al., 2010), gyrA T80A (Pantel et al., 2016), and embB E378A and embC T270I (Goude et al., 2009; Campbell et al., 2011; Koser et al., 2011). In the forward feature selection analysis, we chose to only include mutations that were observed 10 times or more, however, with more isolates or a lower threshold for including mutations, we might discover even more misclassified mutations. On the other hand, the mutation rpoB L430P, rpoB H445N, and rpoB I491F were considered non-predictive for resistance to Rifampicin based on the forward feature selection. However, studies have shown that DST performed on liquored-based mediums fails to detect resistance in strains with rpoB I491F and other rpoB mutations that were clinically associated with treatment failure (Rigouts et al., 2013; André et al., 2017). Thus, with forward feature selection we risk removing mutations that truly causes resistance but appears not to, due to erroneous DST results. This underlines a problem regarding using DST results as the standard for determining resistance. A well-established mutation library is important to avoid incorrect mutation interpretations.
When comparing PointFinder to PhyResSE we did see differences in variant interpretation. This was notable in the gidB gene associated with Streptomycin resistance. PointFinder only interpreted resistance based on premature stop codons in gidB, whereas PhyResSE included several gidB mutations in the interpretation, e.g., gidB A200E, V88A, and A138V (see Supplementary Table S4), and with the interpretation of these mutations as resistance markers PhyResSE showed a significantly better Streptomycin resistance prediction. A GWA study from 2018 did detect the same gidB mutations among 6,465 strains, but in this study this gidB mutations were either observed in less than 10 samples or not identified as being significantly associated with resistance (Coll et al., 2018). Based on this, we did not choose to include these gidB mutation in our mutation library. We have here evaluated the effect of genetic alterations on resistance. A limitation of this is that it is overlooked if mutations have an effect of for example fitness. Future studies may seek to clarify such correlations if large scale datasets with genomes and fitness estimations become available.
The predicting performance of PointFinder is comparable to other M. tuberculosis resistance prediction tools, like PhyResSE, and PointFinder has the advantage of being build into a larger platform for resistance prediction, that is not limited to a single species. Additionally, PointFinder is available on bitbucket.org/genomicepi-demiology/pointfinder, where all changes in the script are tracked. The databases are also available on bitbucket which gives the needed transparency. This creates a good foundation for future maintenance and improvements of the variant interpretation methods and the mutation library.
Conclusion
We have developed improved version of PointFinder with better detection of insertions and deletions as well as the possible associated frameshifts. We find that the accuracy of PointFinder’s resistance prediction in M. tuberculosis is improved as a result. We also optimized the M. tuberculosis mutation library by excluding mutations that through forward feature selection were found to be non-predictive for resistance. We think that these methods may also be applied to increase the antibiotic resistance prediction in other species. The method is flexible and can be updated if new genetic markers for resistance is identified. The method is freely available on the web as well as a stand alone version.
Statements
Data availability statement
The datasets analyzed in this study was obtained from platform.reseqtb.org and as Supplementary Data from Coll et al. (2018) and Schleusener et al. (2017). All accession numbers and phenotype data are also given as Supplementary Data (Supplementary Tables S1, S2, S4).
Author contributions
CJ implemented changes in the improved version of PointFinder, performed all analyses, and wrote the manuscript with inputs from all authors. PC provided help with statistical calculations and did proofreading. OL supervised the project and, together with FA, were in charge of overall direction and planning.
Funding
This study has received funding from the European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 643476 (COMPARE) and the Center for Genomic Epidemiology (Grant 09–067103/DSF). The funding body did not play any role in the design of the study, writing of the manuscript nor did they have any influence on the data collection, analysis or interpretation of the data and results.
Acknowledgments
We are grateful to Rosa Allesøe, Judit Szarvas, and Valeria Bortolaia for excellent technical support and theoretical guidance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02464/full#supplementary-material
TABLE S1ReSeq data set.
TABLE S2Validation data set.
TABLE S3Occurrence of non-predictive mutation in the ReSeq and validation data set.
TABLE S4Comparison of PointFinder and PhyResSE.
References
1
Almeida Da SilvaP. E.PalominoJ. C. (2011). Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs.J. Antimicrob. Chemother.661417–1430. 10.1093/jac/dkr173
2
AndréE.GoeminneL.ColmantA.BeckertP.NiemannS.DelmeeM. (2017). Novel rapid PCR for the detection of Ile491Phe rpoB mutation of Mycobacterium tuberculosis, a rifampicin-resistance-conferring mutation undetected by commercial assays.Clin. Microbiol. Infect.23267.e5–267.e7. 10.1016/j.cmi.2016.12.009
3
BradleyP.GordonN. C.WalkerT. M.DunnL.HeysS.HuangB.et al (2015). Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis.Nat. Commun.6:10063. 10.1038/ncomms10063
4
CamachoC.CoulourisG.AvagyanV.MaN.PapadopoulosJ.BealerK.et al (2009). BLAST+: architecture and applications.BMC Bioinformatics10:421. 10.1186/1471-2105-10-421
5
CampbellP. J.MorlockG. P.SikesR. D.DaltonT. L.MetchockB.StarksA. M. (2011). Molecular detection of mutations associated with firstand second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob.Agents Chemother.552032–2041. 10.1128/AAC.01550-10
6
ClausenP. T. L. C.AarestrupF. M.LundO. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA.BMC Bioinformatics19:307. 10.1186/s12859-018-2336-6
7
CollF.McNerneyR.PrestonM. D.Guerra-AssunçãoJ. A.WarryA.Hill-CawthorneG.et al (2015). Rapid determination of anti-tuberculosis drug resis-tance from whole-genome sequences.Genome Med.7:51. 10.1186/s13073-015-0164-0
8
CollF.PhelanJ.Hill-CawthorneG. A.NairM. B.MallardK.AliS.et al (2018). Genome-wide analysis of multiand extensively drug resistant Mycobacterium tuberculosis.Nat. Genet.50307–316.
9
DillonN. A.PetersonN. D.RosenB. C.BaughnA. D. (2014). Pantothenate and pantetheine antagonize the antitubercular activity of pyrazinamide.Antimicrob. Agents Chemother.587258–7263. 10.1128/AAC.04028-14
10
FeuerriegelS.SchleusenerV.BeckertP.KohlT. A.MiottoP.CirilloD. M.et al (2015). PhyResSE: a web tool delineating Mycobacterium tuberculosis Antibiotic Resistance and lineage from whole-genome sequencing data.J. Clin. Microbiol.531908–1914. 10.1128/JCM.00025-15
11
GoudeR.AminA. G.ChatterjeeD.ParishT. (2009). The arabinosyltransferase EmbC is inhibited by ethambutol in Mycobacterium tuberculosis.Antimicrob. Agents Chemother.534138–4146. 10.1128/AAC.00162-09
12
IwaiH.Kato-MiyazawaM.KirikaeT.Miyoshi-AkiyamaT. (2015). CASTB (the comprehensive analysis server for the Mycobacterium tuberculosis complex): A publicly accessible web server for epidemiological analyses, drug-resistance prediction and phylogenetic comparison of clinical isolates.Tuberculosis95843–844. 10.1016/j.tube.2015.09.002
13
JugheliL.BzekalavaN.de RijkP.FissetteK.PortaelsF.RigoutsL. (2009). High level of cross-resistance between kanamycin, amikacin, and capreomycin among Mycobacterium tuberculosis isolates from Georgia and a close relation with mutations in the rrs gene.Antimicrob. Agents Chemother.535064–5068. 10.1128/AAC.00851-09
14
KoserC. U.EllingtonM. J.PeacockS. J. (2014). Whole-genome sequencing to control antimicrobial resistance.Trends Genet.30401–407. 10.1016/j.tig.2014.07.003
15
KoserC. U.SummersD. K.ArcherJ. A. (2011). Thr270Ile in embC (Rv3793) is not a marker for ethambutol resistance in the Mycobacterium tuberculosis complex.Antimicrob. Agents Chemother.55:1825. 10.1128/aac.01607-10
16
MausC. E.PlikaytisB. B.ShinnickT. M. (2005). Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis.Antimicrob. Agents Chemother.493192–3197. 10.1128/aac.49.8.3192-3197.2005
17
MunitaJ. M.AriasC. A. (2016). Mechanisms of antibiotic resistance.Microbiol. Spectr.4:VMBF-0016-2015.
18
PantelA.PetrellaS.VezirisN.MatratS.BouigeA.FerrandH.et al (2016). Description of compensatory gyrA mutations restoring fluoroquinolone susceptibility in Mycobacterium tuberculosis.J. Antimicrob. Chemother.712428–2431. 10.1093/jac/dkw169
19
RigoutsL.GumusbogaM.de RijkW. B.NduwamahoroE.UwizeyeC.de JongB.et al (2013). Rifampin resistance missed in automated liquid culture system for Mycobacterium tuberculosis isolates with specific rpoB mutations.J. Clin. Microbiol.512641–2645. 10.1128/JCM.02741-12
20
SchleusenerV.KöserC. U.BeckertP.NiemannS.FeuerriegelS. (2017). Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools.Sci. Rep.7:46327. 10.1038/srep46327
21
SmithT.WolffK. A.NguyenL. (2013). Molecular biology of drug resistance in Mycobacterium tuberculosis.Curr. Top. Microbiol. Immunol.37453–80. 10.1007/82_2012_279
22
StarksA. M.AvilésE.CirilloD. M.DenkingerC. M.DolingerD. L.EmersonC.et al (2015). Collaborative effort for a centralized worldwide tuberculosis relational sequencing data platform.Clin. Infect. Dis.61S141–S146. 10.1093/cid/civ610
23
SteinerA.StuckiD.CoscollaM.BorrellS.GagneuxS. (2014). KvarQ: targeted and direct variant calling from fastq reads of bacterial genomes.BMC Genomics15:881. 10.1186/1471-2164-15-881
24
SunY. J.LeeA. S.WongS. Y.PatonN. I. (2007). Analysis of the role of Mycobacterium tuberculosis kasA gene mutations in isoniazid resistance.Clin. Microbiol. Infect.13833–835. 10.1111/j.1469-0691.2007.01752.x
25
The CRyPTIC Consortium and the 100,000 Genomes Project,Allix-BéguecC.ArandjelovicI.BiL.BeckertP.BonnetM.et al (2018). Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing.N. Engl. J. Med.3791403–1415. 10.1056/nejmoa1800474
26
ViaL. E.ChoS. N.HwangS.BangH.ParkS. K.KangH. S.et al (2010). Polymorphisms associated with resistance and cross-resistance to aminoglycosides and capreomycin in Mycobacterium tuberculosis isolates from South Korean Patients with drug-resistant tuberculosis.J. Clin. Microbiol.48402–411. 10.1128/JCM.01476-09
27
VictorT. C.van RieA.JordaanA. M.RichardsonM.van Der SpuyG. D.BeyersN.et al (2001). Sequence polymorphism in the rrs gene of Mycobacterium tuberculosis is deeply rooted within an evolutionary clade and is not associated with streptomycin resistance.J. Clin. Microbiol.394184–4186. 10.1128/jcm.39.11.4184-4186.2001
28
VillellasC.AristimuñoL.VitoriaM. A.PratC.BlancoS.García de ViedmaD.et al (2013). Analysis of mutations in streptomycin-resistant strains reveals a simple and reliable genetic marker for identification of the Mycobacterium tuberculosis Beijing genotype.J. Clin. Microbiol.512124–2130. 10.1128/JCM.01944-12
29
WongS. Y.LeeJ. S.KwakH. K.ViaL. E.BoshoffH. I.BarryC. E.III (2011). Mutations in gidB confer low-level streptomycin resistance in Mycobacterium tuberculosis.Antimicrob. Agents Chemother.552515–2522. 10.1128/AAC.01814-10
30
World Health Organization [WHO] (2017). Global Tuberculosis Report 2017.Geneva: WHO.
31
ZankariE.AllesøeR.JoensenK. G.CavacoL. M.LundO.AarestrupF. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens.J. Antimicrob. Chemother.722764–2768. 10.1093/jac/dkx217
32
ZankariE.HasmanH.CosentinoS.VestergaardM.RasmussenS.LundO.et al (2012). Identification of acquired antimicrobial resistance genes.J. Antimicrob. Chemother.672640–2644. 10.1093/jac/dks261
33
ZhangY.HeymB.AllenB.YoungD.ColeS. (1992). The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis.Nature358591–593. 10.1038/358591a0
Summary
Keywords
antimicrobial resistance (AMR), tuberculosis, whole genome sequencing, bioinformatics, resistance prediction
Citation
Johnsen CH, Clausen PTLC, Aarestrup FM and Lund O (2019) Improved Resistance Prediction in Mycobacterium tuberculosis by Better Handling of Insertions and Deletions, Premature Stop Codons, and Filtering of Non-informative Sites. Front. Microbiol. 10:2464. doi: 10.3389/fmicb.2019.02464
Received
04 January 2019
Accepted
15 October 2019
Published
31 October 2019
Volume
10 - 2019
Edited by
Benjamin Andrew Evans, University of East Anglia, United Kingdom
Reviewed by
Divakar Sharma, Indian Institute of Technology Delhi, India; James Sacchettini, Texas A&M University, United States
Updates
Copyright
© 2019 Johnsen, Clausen, Aarestrup and Lund.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ole Lund, olund@food.dtu.dk
This article was submitted to Antimicrobials, Resistance and Chemotherapy, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.