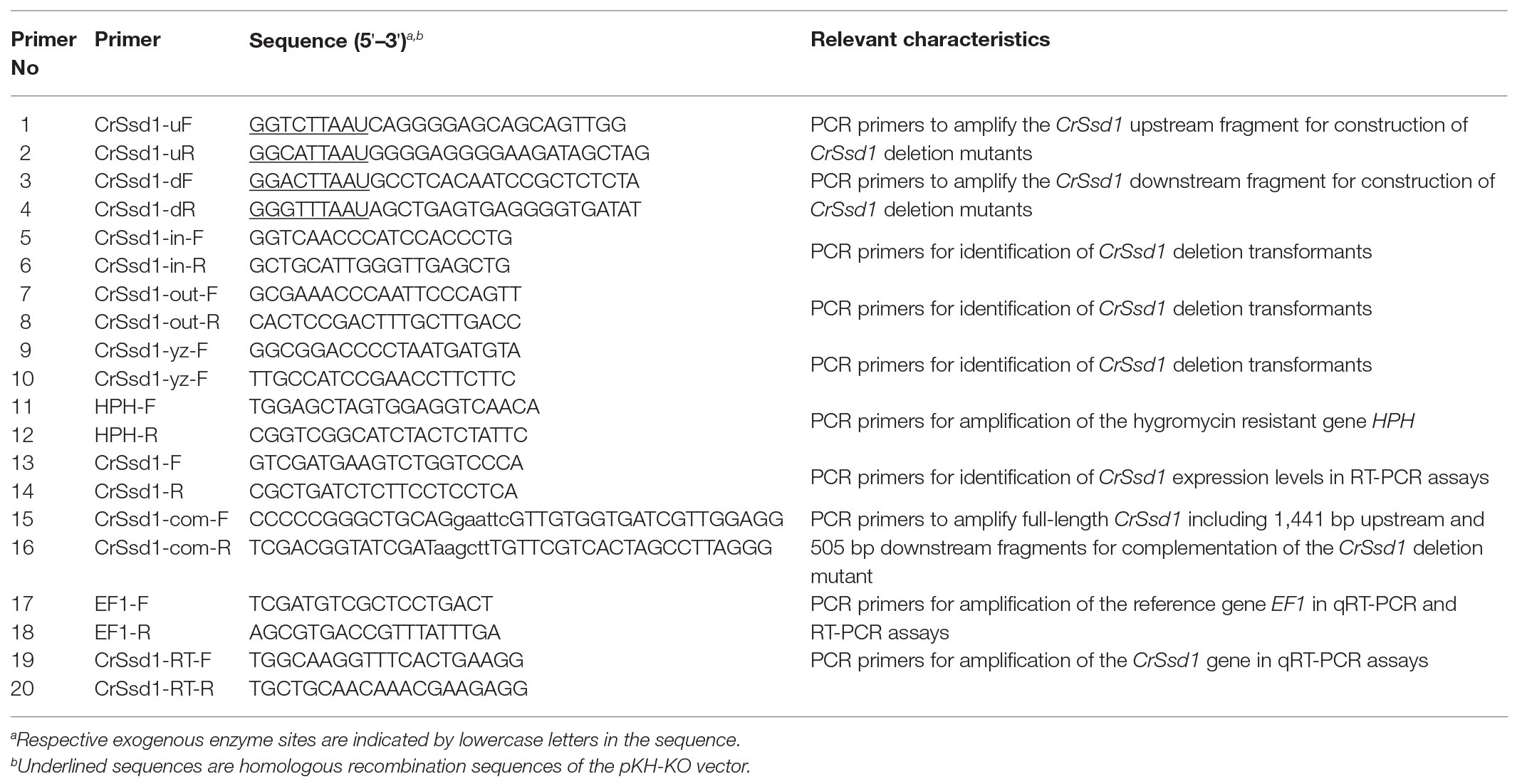
- Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Cell wall biogenesis protein phosphatases play important roles in various cellular processes in fungi. However, their functions in the widely distributed mycoparasitic fungus Clonostachys rosea remain unclear, as do their potential for controlling plant fungal diseases. Herein, the function of cell wall biogenesis protein phosphatase CrSsd1 in C. rosea 67-1 was investigated using gene disruption and complementation approaches. The gene-deficient mutant ΔCrSsd1 exhibited much lower conidiation, hyphal growth, mycoparasitic ability, and biocontrol efficacy than the wild-type (WT) strain, and it was more sensitive to sorbitol and Congo red. The results indicate that CrSsd1 is involved in fungal conidiation, osmotic stress adaptation, cell wall integrity, and mycoparasitism in C. rosea.
Introduction
Cell walls are essential structures that help organisms resist environmental stresses and protect a variety of natural cellular processes. In fungi, the biogenesis and integrity of cell walls are vital for fungal pathogenesis and survival (Scrimale et al., 2009; Wei et al., 2016), and their remodeling and expansion determine cell growth and reproduction (Fuchs and Mylonakis, 2009; Wang et al., 2013; Feng et al., 2017). Several cell wall building-related proteins have been identified in fungi, including the RNA-binding protein Ssd1 that was first cloned from Saccharomyces cerevisiae and is involved in various cellular processes and pathways such as cell wall integrity, signal transduction, and the cell cycle (Kaeberlein and Guarente, 2002; Reinke et al., 2004; Jansen et al., 2009). Ssd1 is highly conserved in fungi and contains a RNase II (RNB) domain (SMART No. SM00955), which is the catalytic domain of ribonuclease II, suggesting that it may be involved in post-transcriptional regulation by directly interacting with messenger RNAs (mRNAs; Mir et al., 2009; Kurischko et al., 2011b). The protein was also found to be remarkably enriched in message transmission-related proteins during cell wall biogenesis (Jansen et al., 2009).
In S. cerevisiae, Ssd1 regulates cell wall remodeling by inhibiting the translation of related proteins (Luukkonen and Séraphin, 1999; Wanless et al., 2014), and its inactivation is regulated by the nuclear Dbf2-related (Ndr)/large tumour suppressor (LATS) family protein kinase Cbk1 through phosphorylation of the N-terminal region of Ssd1 (Bidlingmaier et al., 2001; Du and Novick, 2002; Kurischko et al., 2011a). Lack of this regulation significantly impairs bud expansion and causes severe aberrant cell wall organization (Moriya and Isono, 1999). Three independent pathways separately mediated by Mpt5, Ssd1, and Pkc1 regulate cellular integrity; Mpt5 and Ssd1 act post-transcriptionally during cell wall biosynthesis and maintain cell structure as upstream regulators (Kaeberlein and Guarente, 2002), while Pkc1 activates a mitogen-activated protein kinase (MAPK) cascade that controls the transcription and expression of genes involved in cell wall formation (Gerik et al., 2005, 2008; Mir et al., 2009).
Orthologs of Ssd1 have been characterized in several pathogenic fungi, and they exhibit various functions. In Candida albicans, Ssd1 impedes the mutation of other genes and weakens the effects of mutations on cellular processes (Gank et al., 2008; Ohyama et al., 2010; Avrahami-Moyal et al., 2012). In Colletotrichum orbiculare, Ssd1 is essential for penetration of appressoria into the epidermal cells of susceptible plants, and the Ssd1-deficient mutant displays enhanced basal resistance to Nicotiana benthamiana (Tanaka et al., 2007, 2009). Moreover, deletion of Ssd1 in Colletotrichum higginsianum and Magnaporthe grisea leads to weakened penetration and virulence (Schmidpeter et al., 2017; Yu et al., 2019). However, the functions of orthologs of Ssd1 in biocontrol fungi remain poorly understood.
C. rosea (syn. Gliocladium roseum) is a widely distributed mycoparasite associated with a range of pathogenic fungi, such as Sclerotinia sclerotiorum, Rhizoctonia solani, and Botrytis cinerea (Xue, 2003; Zhang et al., 2008; Kosawang et al., 2014). This species has great potential for controlling various plant fungal diseases and promoting crop growth (Atanasova et al., 2018), and functional genes in particular have attracted much attention. Fatema et al. (2018) demonstrated that two polyketide synthase genes, PKS22 and PKS29, play important roles in the synthesis of antifungal agents, clonorosein A–D that are effective against B. cinerea. Dubey et al. (2016) found that the ABC transporter gene ABCG29 is involved in fungal adaptation to oxidative stress in the early stages of mycelial development and biocontrol of B. cinerea and Fusarium graminearum. Sun et al. (2019a) indicated that the heat shock protein 70 gene, crhsp, had a remarkable effect on C. rosea morphological characteristics and significantly reduced its ability to parasitize S. sclerotiorum sclerotia. However, there have been no studies on the functions of Ssd1, and whether Ssd1 is related to cell wall formation in C. rosea remains unknown.
In the present study, we identified and characterized the CrSsd1 gene, which is orthologous to S. cerevisiae Ssd1 and markedly upregulated during C. rosea parasitizing S. sclerotiorum (Sun et al., 2015b). Our results indicate that CrSsd1 is involved in conidiation, responses to osmotic stress, cell wall integrity, and mycoparasitism in C. rosea. This knowledge reinforces our understanding of the mechanisms underlying C. rosea mycoparasitism and lays a foundation for developing new potent biocontrol agents. To the best of our knowledge, this is the first report of CrSsd1 as a mycoparasitism-associated protein involved in C. rosea against fungal plant pathogens.
Materials and Methods
Fungal Strains
C. rosea 67-1 (ACCC 39160) was originally isolated from a vegetable yard in Hainan Province, China, using the sclerotia-baiting method (Zhang et al., 2004). S. sclerotiorum Ss-H (ACCC 39161) was separated from sclerotia-infected soybean stems in a field in Heilongjiang Province, China. B. cinerea TC-B1 was isolated from infected tomato fruits in a greenhouse (Sun et al., 2015b). All strains were maintained at 4°C in the Biocontrol of Soilborne Diseases Lab of the Institute of Plant Protection, Chinese Academy of Agricultural Sciences.
Bioinformatics Analysis
The DNA sequence of CrSsd1 was obtained from the draft genome sequence of C. rosea 67-1. NCBI1 and UniProt2 were used for BLASTp analysis. Functional domains of CrSsd1 were predicted using SMART3. The Clustal X program was used for amino acid alignments. The phylogenetic tree was constructed by MEGA 7.0 using the maximum likelihood method with 1,000 bootstrap replicates.
Quantitative Reverse Transcription PCR of CrSsd1
Strain 67-1 genomic DNA was extracted using a Biospin Fungus Genomic DNA Extraction Kit (Bioer Technology Co. Ltd., Hangzhou, China) according to the manufacturer’s instructions. Plasmid DNA was isolated using a Plasmid Miniprep Purification Kit (BioDev Co., Beijing, China).
We analyzed the expression levels of CrSsd1 in strain 67-1 during different stages of mycoparasitizing sclerotia. Strain 67-1 was incubated on potato dextrose agar (PDA) at 26°C for 10 days, spores were washed with sterile water and adjusted to 1 × 107 spores/ml, and spore suspensions were smeared evenly on a PDA plate and covered with cellophane. Uniformly sized sclerotia were placed onto the surface of 67-1 plates evenly after culturing for 48 h, and C. rosea 67-1 mycelia were collected at 8, 24, and 48 h and placed immediately in liquid nitrogen. Each treatment included five replicates. Total RNA was extracted using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. RNase-free DNase I (Invitrogen) was used to eliminate DNA contamination. Reverse transcription was performed using a cDNA FastQuant RT Kit (Tiangen, Beijing, China). Gene expression was analyzed by quantitative reverse transcription PCR (qRT-PCR) using a Bio-Rad IQ 5 Real-Time System (Bio-Rad, CA, USA) and SYBR Premix Ex Taq (Takara, Dalian, China) with primers listed in Table 1. Elongation factor gene EF1 (GenBank accession number: KP274074) was used as a reference gene to normalize gene expression in C. rosea 67-1 under sclerotia induction (Sun et al., 2015a,b), and mycelial samples without added sclerotia acted as a control. The relative expression levels of CrSsd1 were calculated using the 2−∆∆Ct method, and three replicates were included for each sample.
Generation of Gene Deletion and Complementation Mutants
The plasmid pKH-KO containing two uracil-specific excision reagent (USER) cloning sites (USC1 and USC2) on either side of the hygromycin resistance gene hph was used to construct a CrSsd1 disruption vector (Frandsen et al., 2008). Upstream and downstream flanking sequences of CrSsd1 were amplified using primer pairs CrSsd1-uF/CrSsd1-uR and CrSsd1-dF/CrSsd1-dR, respectively, and cloned into two USC sites using the USER-friendly cloning method to generate CrSsd1-deletion vector pKH-KO-CrSsd1.
For construction of the gene complementation vector, the full-length sequence of CrSsd1, including the promoter, protein-coding, and terminator regions was amplified from 67-1 genomic DNA and cloned into the pKN vector (carrying the G418 resistance gene neo; Kong et al., 2019). The resulting gene deletion and complementation vectors were transformed into protoplasts of 67-1 and ΔCrSsd1, respectively, to generate gene deletion and complementation mutants using protoplast formation and transformation of C. rosea (Sun et al., 2017). Primers (Table 1) were designed and mutants were verified by PCR and DNA sequencing. Furthermore, the expression levels of CrSsd1 in wild-type (WT), deletion and complementation strains were tested using reverse transcription PCR (RT-PCR) with primers CrSsd1-F and CrSsd1-R and reference gene EF-1 gene (Table 1; Sun et al., 2015a,b).
Fungal Growth, Conidiation, and Stress Tolerance
To analyze differences in vegetative growth among C. rosea 67-1, ΔCrSsd1, and ΔCrSsd1-C strains, agar blocks (3 mm) of strains were inoculated onto the center of a PDA plate and cultured at 26°C. The size and morphology of colonies were measured daily. After 15 days, fungal spores were collected by adding 5 ml sterile distilled water, and spores were counted under a BX41 microscope (Olympus, Tokyo, Japan). The conidial germination rates of all strains were determined. Spore suspensions of WT, ΔCrSsd1 and ΔCrSsd1-C strains with a concentration of 1 × 107 spores/ml were prepared and inoculated in potato dextrose (PD) broth on a rotary shaker at a speed of 180 rpm. Samples were incubated at 26°C, and the germinated conidia were counted at 8 and 16 h post inoculation. To evaluate the stress response, cultures were grown on PDA plates amended with different stress agents [1 M NaCl, 1 M KCl, 1 M glycerin, 1 M sorbitol, 20 mM H2O2, 0.03% SDS, and 0.3 mg/ml Congo red (CR)] for 10 days. Diameters of colonies were counted, and microscopic observation of the hypha under different stress conditions was performed with a fluorescence microscope system (DM6 B, Leica, Germany). All assays were repeated three times.
Antagonistic Activity Against Botrytis cinerea
Antagonistic activity of C. rosea 67-1, ΔCrSsd1, and ΔCrSsd1-C strains against B. cinerea was tested on 9 cm PDA plates. A 3-mm agar plug of strains was inoculated 2 cm from the edge of the plate and cultured at 26°C for 5 days. A plug of B. cinerea was then placed equidistant from the other side of the plate and cultured at 26°C for 20 days. The distance of hyphal extension for each strain was measured (Dubey et al., 2014; Tzelepis et al., 2015; Filizola et al., 2019).
Mycoparasitic Ability Against Sclerotinia sclerotiorum Sclerotia
Sclerotia of uniform size were surface-sterilized with 1% NaClO for 3 min, rinsed three times with sterile water, and then immersed in spore suspensions of wild-type (WT) 67-1, ΔCrSsd1, and ΔCrSsd1-C strains at a concentration of 1 × 107 spores/ml for 10 min. Sclerotia were picked and placed onto a piece of wet sterile filter paper in a Petri dish (diameter 9 cm) and incubated at 26°C. Treatment with sterile water was used as a control. The number of sclerotia infected by the transformants was counted under a stereo microscope (SMZ-10, Nikon, Tokyo, Japan) at 8, 16 and 24 h. Sclerotia covered with C. roses mycelia were regarded as parasitized, and parasitic rates of all strains were calculated (Sun et al., 2017). After 7 days, we investigated parasitic severity of sclerotia using a BX41 inverted microscope (Olympus) based on a four-grade scale (0 = no C. rosea hyphae were detected on the surface of sclerotia; 1 = loose C. rosea hyphae extended to the sclerotia; 2 = sclerotia were covered with C. rosea hyphae but not softened; and 3 = sclerotia were covered with C. rosea hyphae and exhibited soft rot; Sun et al., 2019a). A total of 30 sclerotia were tested for each treatment, and three replicates were performed.
Control Efficacy Against Soybean Sclerotinia Rot
Pot experiments were carried out to test the ability of C. rosea 67-1, ΔCrSsd1, and ΔCrSsd1-C strains to control S. sclerotiorum on soybean in the greenhouse. Soybean seeds (Zhonghuang 13; Institute of Crop Sciences, CAAS, China) were sown in sterile soil in plastic pots (diameter 11 cm). When nine compound leaves had grown, seedlings were sprayed with 100 ml spore suspension (1 × 107 spores/ml) from each strain. After drying for 2 h, an equivalent amount of S. sclerotiorum mycelial suspension was inoculated onto leaves. Plants treated with sterile water followed by the pathogen served as controls, and 12 pots were tested for each isolate. The greenhouse was maintained at 26–28°C and 60% relative humidity, and all pots were arranged randomly. After 7 days, disease severity of Sclerotinia rot was scored using grades 0–4 according to the percentage of lesions on soybean leaves (0 = no symptoms on soybean leaves; 1 = less than 10% lesions on soybean leaves, 2 = 10–30% lesions on soybean leaves; 3 = 30–50% lesions on soybean leaves; and 4 = over 50% lesions on soybean leaves). All unfolded compound leaves were checked and three replicates were performed for each treatment.
Statistical Analysis
Statistical software SPSS 2.0 (Chicago, IL, USA) was used for ANOVA. Statistical tests were carried out using Tukey’s test for multiple comparisons and a p < 0.05 was considered statistically significant.
Results
Identification and Expression Levels of CrSsd1
Gene cloning and bioinformatics analysis showed that CrSsd1 (GenBank accession number: MN816008) is 3,894 bp in length with no introns and encodes a 1,298-amino-acid polypeptide that contains the RNB domain (Figure 1A), which is the catalytic domain of ribonuclease II. CrSsd1 shares 45.8% identity with S. cerevisiae Ssd1, which is involved in a range of cellular processes, including cell wall integrity, signal transduction, and RNA deterioration. Phylogenetic analysis and sequence alignment of CrSsd1 with other fungal species revealed close homology with homologs of Ssd1 in Fusarium oxysporum and Trichoderma arundinaceum, and it is highly conserved among various fungi (Figure 1B).
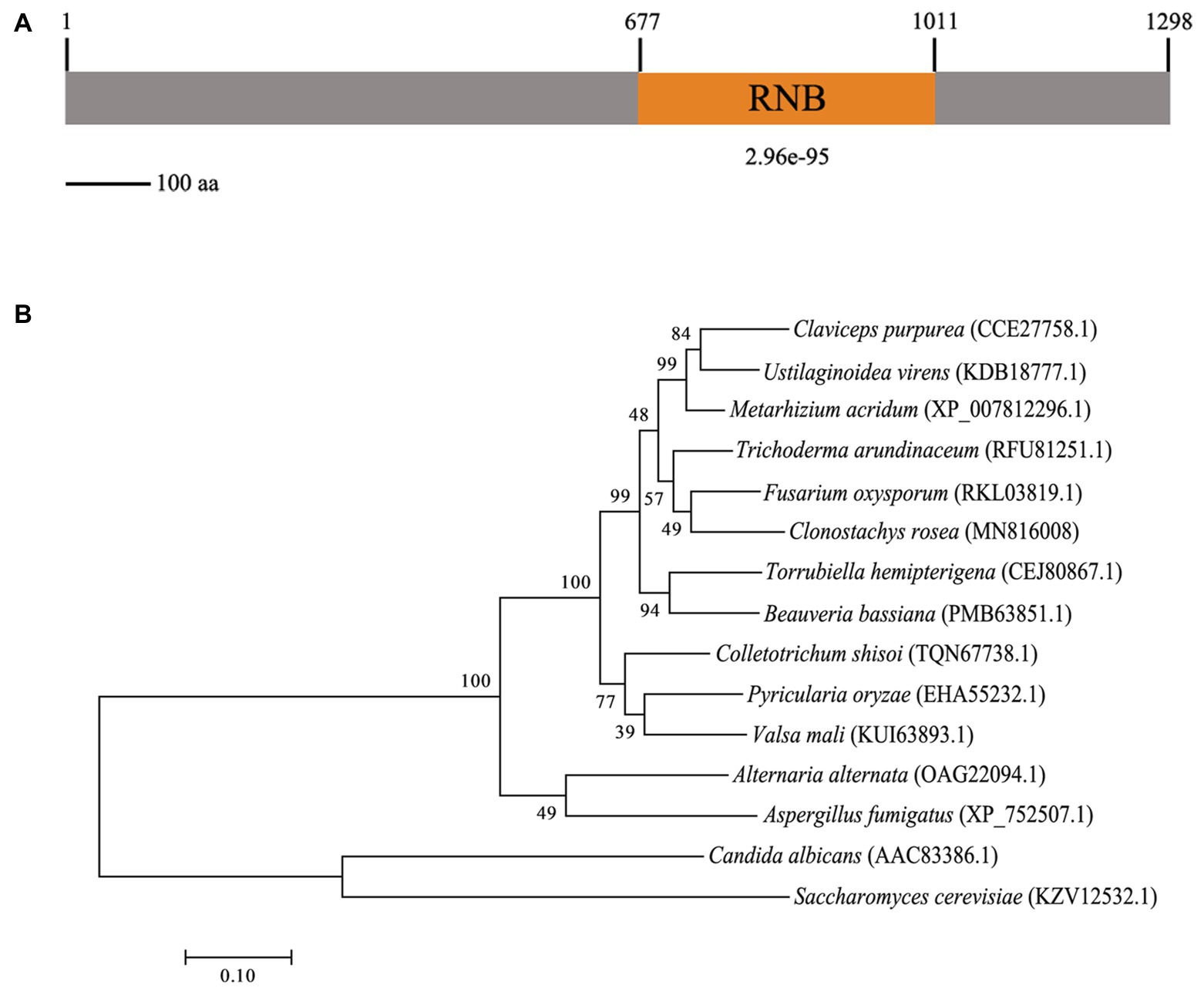
Figure 1. Characterization of the Clonostachys rosea CrSsd1 protein. (A) The domain structure of C. rosea CrSsd1 as annotated by SMART Mode (http://smart.embl.de/). (B) Phylogenetic analysis of CrSsd1 of C. rosea and its homologs from other fungi. Amino acid sequences were aligned by Clustal X and analyzed by MEGA 7.0 using the maximum likelihood method. Numbers in parentheses indicate GenBank accession numbers. Numbers at the nodes represent the bootstrap values of 1,000 bootstrap replicates. Bars = 0.10 and represent sequence divergence.
The expression levels of CrSsd1 in 67-1 were also investigated during different stages of mycoparasitizing sclerotia by qRT-PCR. Analysis of gene expression indicated that CrSsd1 was upregulated in C. rosea throughout mycoparasitism, particularly at 24 h, and expression levels were more than four-fold higher than the control (Figure 2), which is consistent with the transcriptome data from C. rosea parasitizing S. sclorotiorum (Sun et al., 2015b).
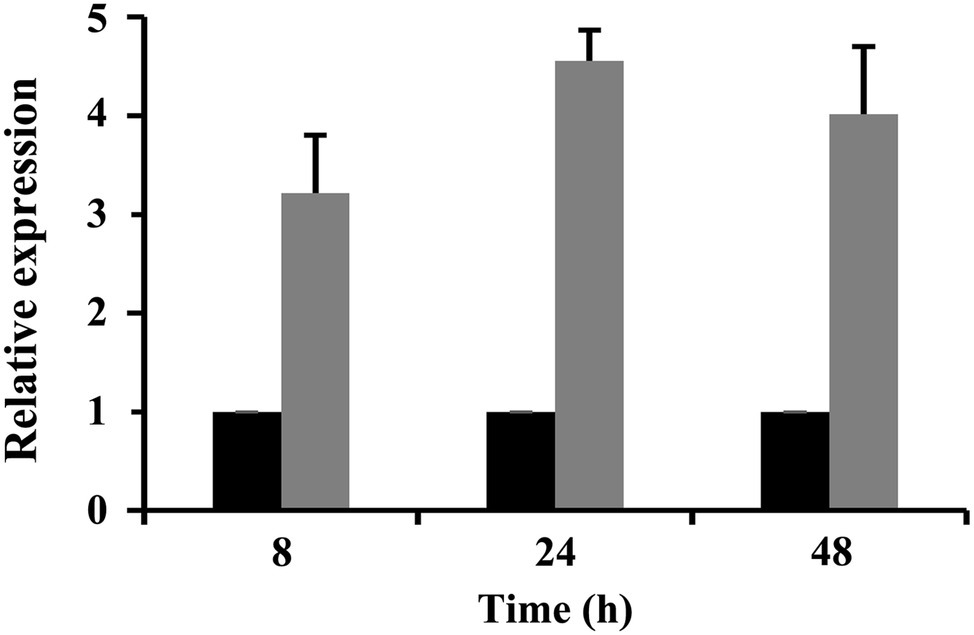
Figure 2. Expression levels of CrSsd1 in C. rosea under sclerotia mycoparasitism conditions. Black columns represent samples added with fresh sclerotia. Gray columns represent controls (samples without sclerotia). The relative expression levels of CrSsd1 were calculated using the 2−∆∆Ct method. Error bars show the standard deviation (SD) of three replicates.
CrSsd1 Disruption and Complementation
To identify the role of CrSsd1 in C. rosea, single gene deletion mutants were generated using a homologous recombination strategy (Figure 3A). Among 187 hygromycin-resistant transformants, three ΔCrSsd1 strains with identical phenotypic characteristics were confirmed by PCR analysis with primers CrSsd1-in-F/R (inside of the target gene), CrSsd1-yz-F/R (outside of the homologous fragment), HPH-F/R (on both ends of the hph gene), and CrSsd1-yz-F/HPH-R (Figure 3C). Moreover, fragments amplified by primer pair CrSsd1-yz-F/R were sequenced, and the results showed that the CrSsd1 gene was successfully replaced with a hygromycin B resistance cassette as expected. For complementation of CrSsd1, the vector pKN-CrSsd1-C was transformed into the ΔCrSsd1 strain and 11 complementation strains were finally obtained. RT-PCR verification demonstrated a complete loss of CrSsd1 transcript in ΔCrSsd1 mutants, whereas specific products were detected in the WT and complementation strains. In addition, the expression of EF-1 gene was detected in all strains (Figure 3D).
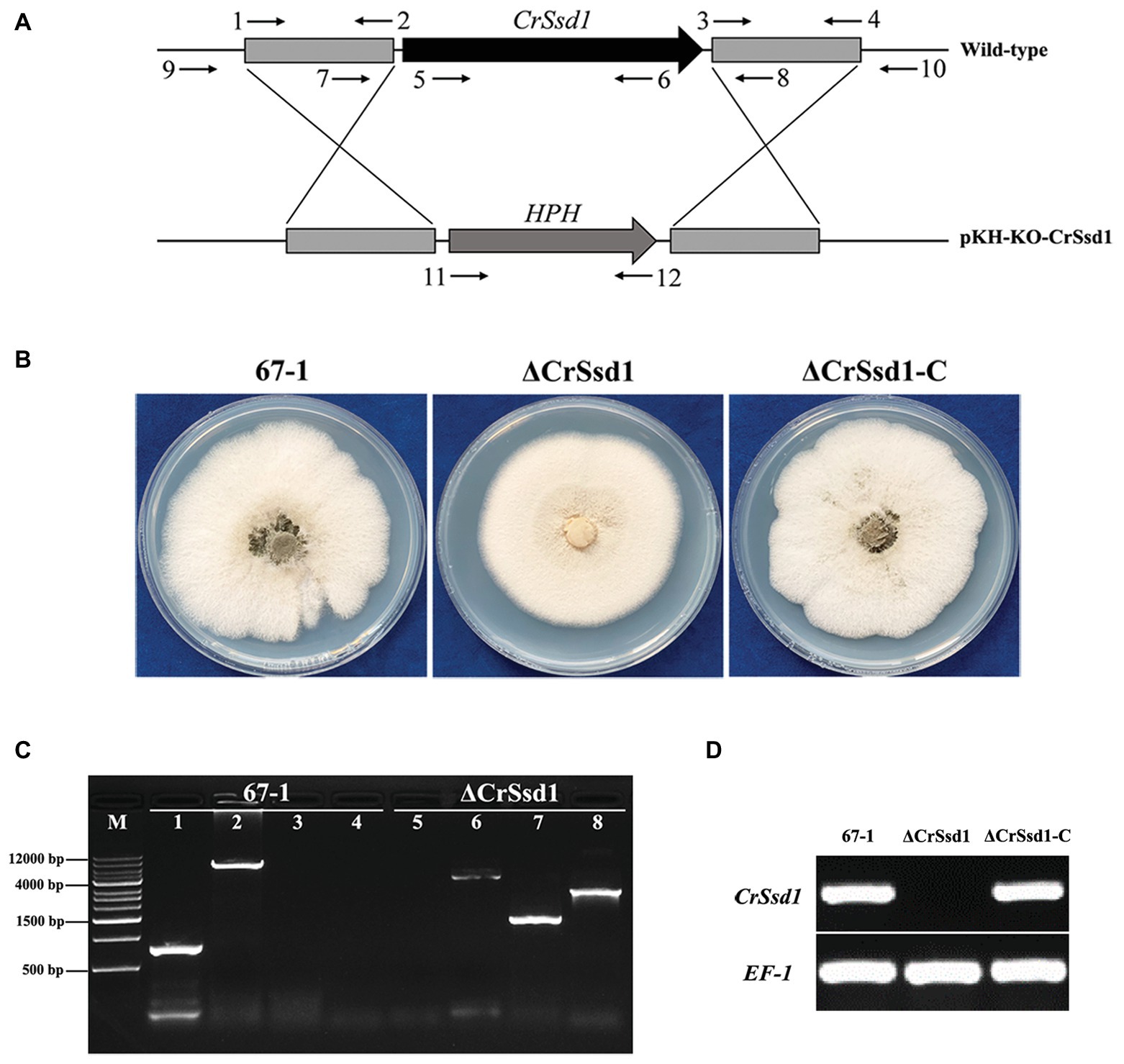
Figure 3. Construction of replacement vectors and confirmation of CrSsd1 deletion mutants. (A) Schematic representation of the gene disruption strategy. The hygromycin resistance cassette (hph) was cloned into the corresponding sites of vector pKH-KO-CrSsd1 to replace the 3,894 bp CrSsd1 open reading frame. The annealing sites of primers are indicated by small black arrows. (B) Colony morphologies of 67-1, ΔCrSsd1, and ΔCrSsd1-C grown on potato dextrose agar (PDA) plates at 26°C for 10 days. (C) PCR analysis of 67-1 and CrSsd1 deletion mutant strains using primers 5/6 (CrSsd1-in-F/R), 9/10 (CrSsd1-yz-F/R), 11/12 (HPH-F/R), and 9/12 (CrSsd1-yz-F/HPH-R). Lanes 1–4, PCR products amplified with the above primers using 67-1 as template; lanes 5–8, PCR products amplified with the above primers using ΔCrSsd1 as template. Primer numbers refer to binding sites shown in Table 1. (D) Reverse transcription PCR (RT-PCR) analysis of CrSsd1 gene expressions in 67-1, ΔCrSsd1, and ΔCrSsd1-C strains, using CrSsd1 specific CrSsd1-F/R primers (Table 1). The RT-PCR product of 247 bp was expected from 67-1 and ΔCrSsd1-C but not in the CrSsd1 deletion mutants.
Effects of CrSsd1 on Fungal Growth, Conidiation and Sensitivity to Stresses
Three ΔCrSsd1 and ΔCrSsd1-C mutants were selected to analyze the functions of CrSsd1 gene. The colony morphology showed that ΔCrSsd1 mutants had flatter and thinner mycelia than those of the WT 67-1 and the complemented transformant ΔCrSsd1-C (Figure 3B). Moreover, the mycelial growth rates of mutants were slightly slower than that of the WT and ΔCrSsd1-C strains. When grown on PDA for 9 days, the colony diameter of 67-1 reached 5.95 cm, while that of ΔCrSsd1 was 5.29 cm, and the difference was significant (p < 0.05; Figures 4A,B). Surprisingly, gene-deficient strains lost almost all ability to undergo conidiation. After incubation on PDA for 15 days, only 1 × 106 spores/plate were harvested for ΔCrSsd1, compared with 4.9 × 107 spores/plate for the WT strain (p < 0.01; Figures 4A,C). Conidial germination rate of the ΔCrSsd1 mutants was 46.9%, significantly lower than that of the WT strain (68.7%) at 8 h (p < 0.05); however, both strains increased to approximately 100% at 16 h (Supplementary Figure S1). The complemented transformants showed similar results with WT.
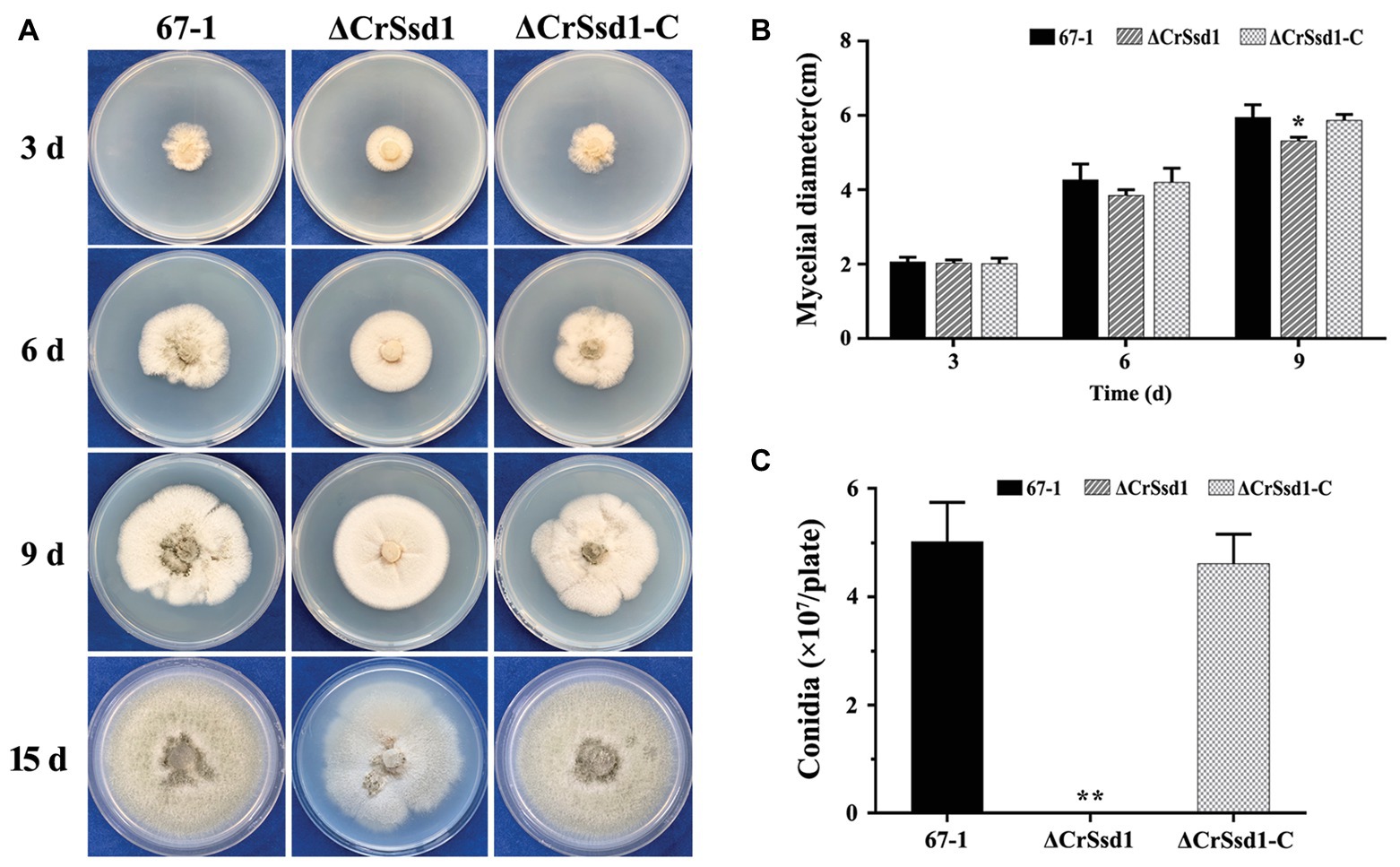
Figure 4. Impact of CrSsd1 deletion on mycelial growth and conidia formation. (A) Mycelial growth of 67-1, ΔCrSsd1, and ΔCrSsd1-C on PDA medium after 3, 6, and 9 days of incubation, and conidiation of these strains on PDA after 15 days. (B) Statistical analysis of colony diameters in (A). (C) Number of conidia produced by each strain on PDA plates. The data are the means of the three ΔCrSsd1 and ΔCrSsd1-C mutants and the means and standard errors are calculated from three independent replicates. Statistical tests were carried out using Tukey’s test for multiple comparisons. Asterisks indicate statistically significant differences (p < 0.05).
The sensitivity of mutants to a variety of environmental stresses, including osmotic stress, oxidative stress, cell membrane stress, and cell wall stress, was investigated. The results showed no significant differences among strains under treatment with NaCl (1 M), KCl (1 M), glycerin (1 M), H2O2 (20 mM), or SDS (0.03%). However, interestingly, ΔCrSsd1 grew much slower in media containing sorbitol (1 M) or CR (0.3 mg/ml) compared with WT and complemented strains, indicating that ΔCrSsd1 deletion mutants were more sensitive to osmotic and cell wall stresses (Figures 5A,B). To further investigate the stress sensitivity of ΔCrSsd1 and 67-1, the hyphal phenotypes under different stress conditions were observed. Our findings demonstrated that the loss of CrSsd1 impaired hyphae branching under NaCl, KCl, sorbitol, and CR, indicating that the CrSsd1 gene played an important role in C. rosea response to osmotic and cell wall stresses (Supplementary Figure S2).
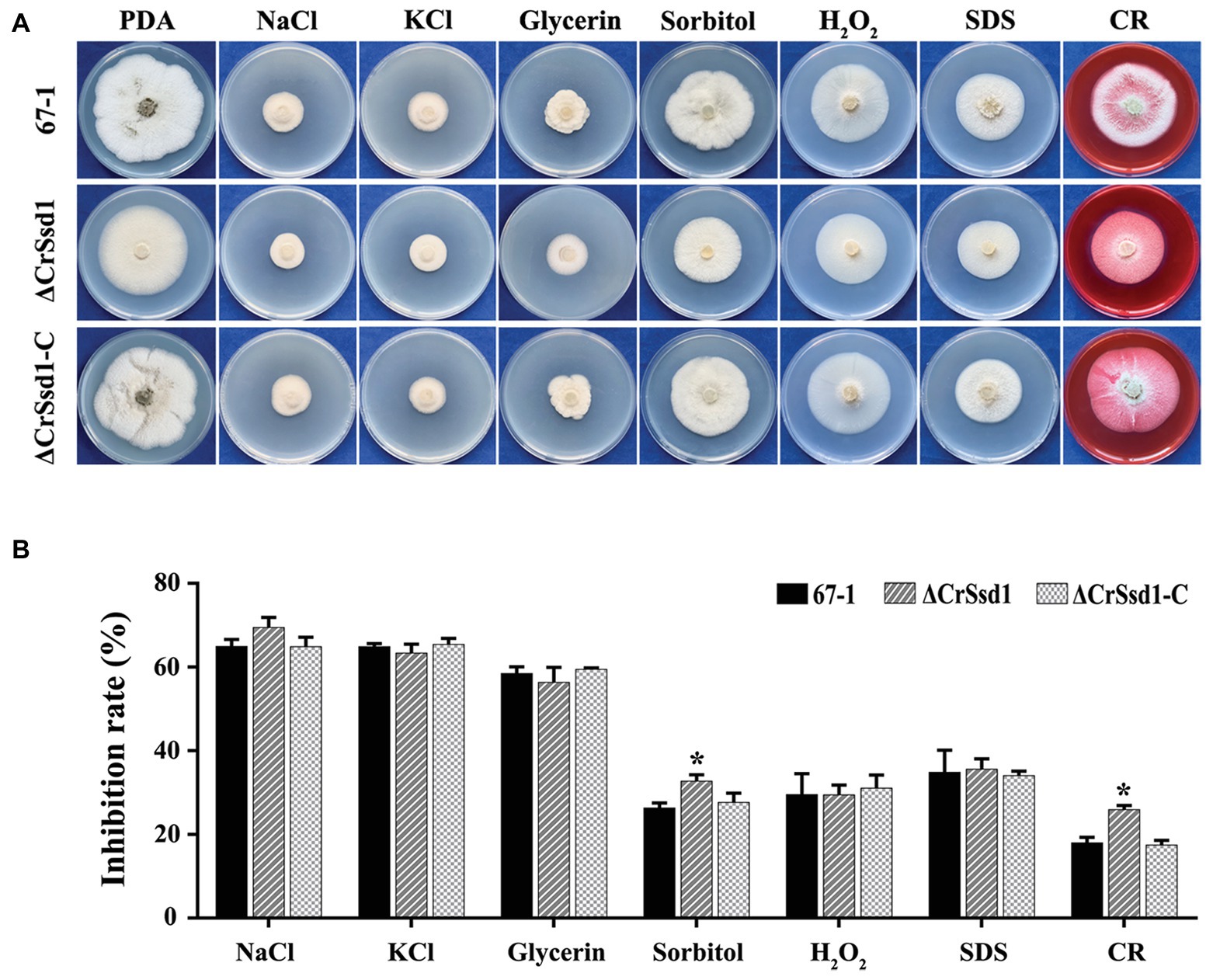
Figure 5. Sensitivity of 67-1, ΔCrSsd1, and ΔCrSsd1-C to diverse stresses. (A) Sensitivity of strains grown on PDA plates containing different stress agents; 1 M NaCl, 1 M KCl, 1 M glycerin, 1 M sorbitol, 20 mM H2O2, 0.03% SDS, and 0.3 mg/ml Congo red (CR). Images were captured after 10 days of incubation at 26°C. (B) Inhibition of mycelial growth compared with non-treated controls. The data are the means of three mutants, and the means and standard errors are calculated from three independent replicates. Statistical tests were carried out using Tukey’s test for multiple comparisons. Asterisks indicate statistically significant differences (p < 0.05).
Effects of CrSsd1 on Antagonistic Activity, Mycoparasitism and Control Efficacy
In vitro antagonistic activity tests showed that C. rosea 67-1, ΔCrSsd1, and ΔCrSsd1-C strains could all overgrow a colony of B. cinerea after culturing for 20 days. However, the hyphal extension ability was decreased by 41.3% for ΔCrSsd1 mutants compared with the WT strain (p < 0.05), and the complemented strain ΔCrSsd1-C recovered this ability almost to the WT level (a decrease of only 3.6%; Figure 6).
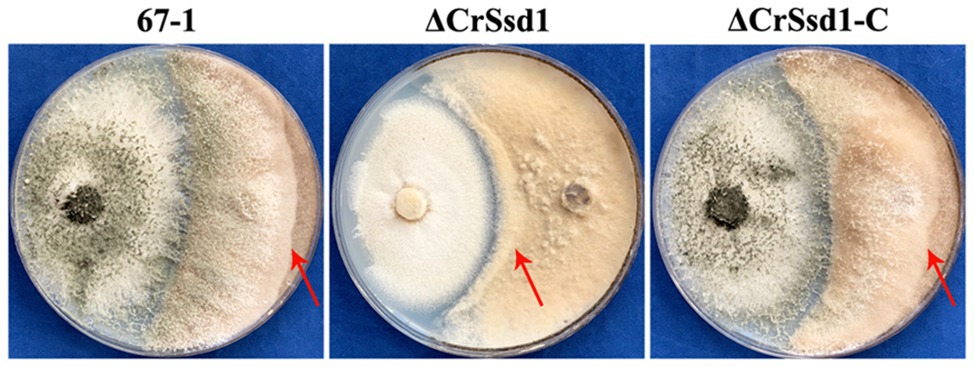
Figure 6. Impact of CrSsd1 deletion on the antagonistic activity of C. rosea. Plate confrontation assay of 67-1, ΔCrSsd1, and ΔCrSsd1-C against Botrytis cinerea at 20 days post-inoculation. Red arrows indicate the hyphal extension distance of each strain toward B. cinerea.
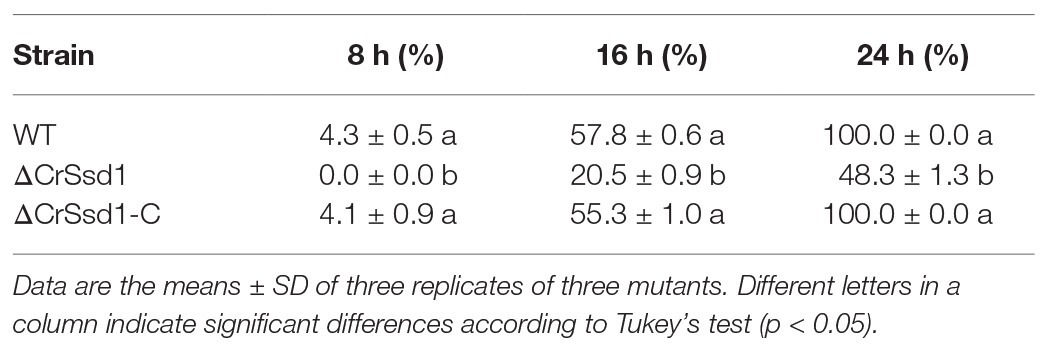
No hyphae of ΔCrSsd1 mutants were detected on the surface of the sclerotia at 8 h after inoculation, and the parasitic rate was 20.5% at 16 h, which was remarkably lower than the WT (57.8%, p < 0.05). By 24 h, hypha of 67-1 and ΔCrSsd1-C covered the whole sclerotia surface, while only 48.3% were parasitized by the ΔCrSsd1 mutants (Table 2). After 7 days of cultivation in a moist environment, the mycoparasitism level of ΔCrSsd1 on sclerotia was markedly reduced compared with that of WT 67-1 and complemented strain ΔCrSsd1-C. From the external phenotype and the inner structure of the sclerotia, we could see that infected sclerotia were completely softened and rotten, resulting in high parasitic severity (grade 4), whereas those treated with the ΔCrSsd1 deletion mutant were covered only sparsely in hyphae and remained relatively firm, equating to mycoparasitism grade 1, indicating that deletion of the CrSsd1 gene substantially weakened the mycoparasitism of C. rosea. Additionally, mycoparasitic ability was recovered in the complemented strain (Figures 7A,B).
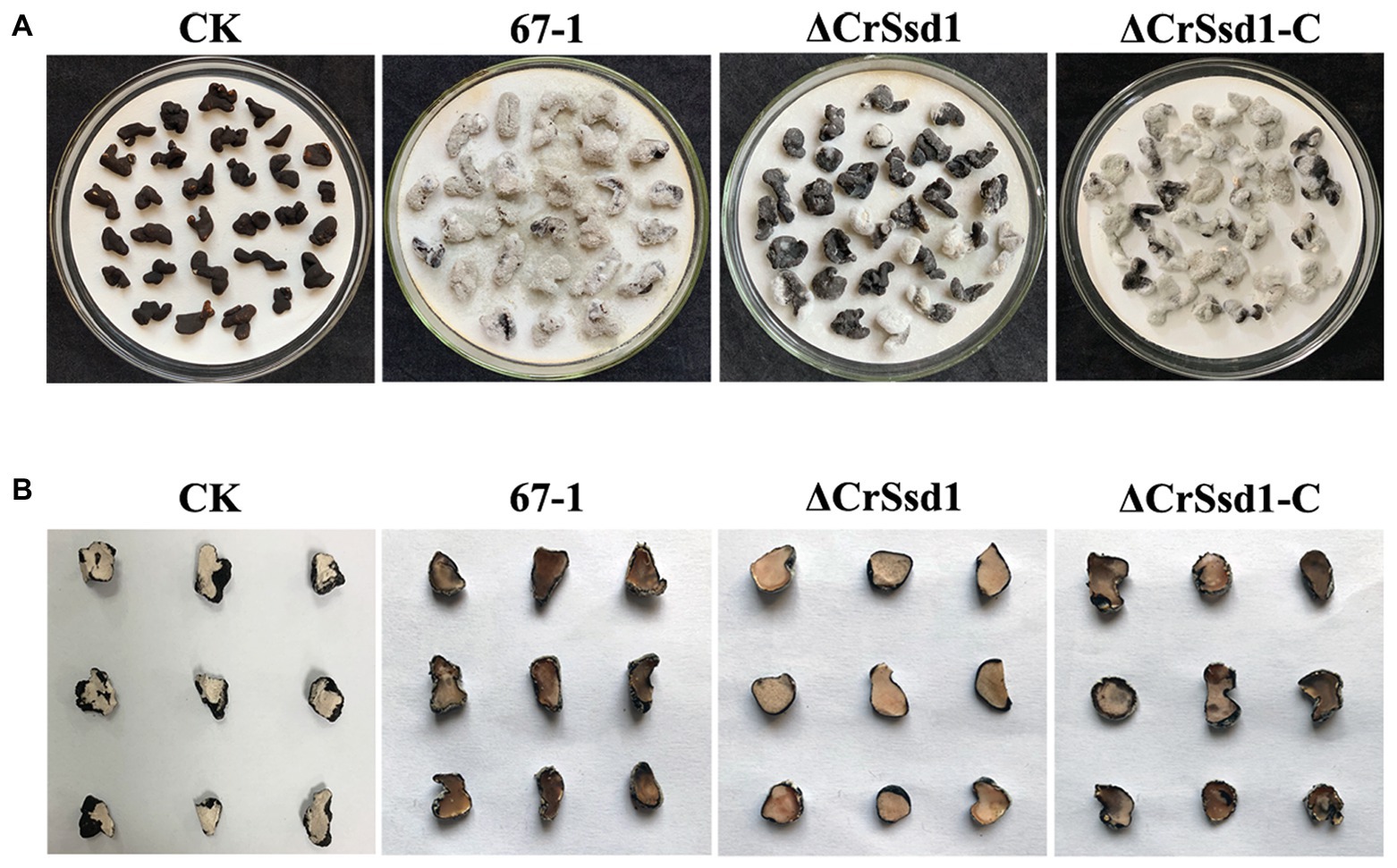
Figure 7. Mycoparasitism of C. rosea strains against Sclerotinia sclerotiorum sclerotia. (A) External phenotypes of healthy and infected sclerotia. (B) Transection of infected and uninfected sclerotia. Images were captured after 7 days incubation at 26°C.
After inoculation with S. sclerotiorum for 7 days, severe leaf lesions were observed in control soybean seedlings. However, soybean seedlings treated with the biocontrol fungus 67-1 were much healthier and displayed less damage, consistent with excellent control efficacy against soybean Sclerotinia rot. Interestingly, when the CrSsd1 gene was deleted, the control efficacy of the mutant was markedly reduced, while the efficiency was regained in the complemented strain (Figure 8 and Table 3), demonstrating that CrSsd1 could dramatically affect the biocontrol efficacy of C. rosea.

Figure 8. Impact of CrSsd1 deletion on efficacy against soybean Sclerotinia rot. Soybean Sclerotinia rot by C. rosea 67-1, ΔCrSsd1, and ΔCrSsd1-C after 7 days in a greenhouse. Plants treated with sterile water followed by the S. sclerotiorum pathogen served as controls (CK), and 12 pots were tested for each isolate. Disease severity of Sclerotinia rot was investigated after 7 days.
Discussion
The fungal cell wall is the first line of defense for protecting against environmental stresses, and any action to remodel it is tightly controlled to maintain balance with stress resistance and osmotic stability. Ssd1 is known to be involved in cell wall integrity and biosynthesis in several pathogenic fungi. To explore the functions of the cell wall biogenesis protein phosphatase CrSsd1 in mycoparasites, we investigated CrSsd1 in C. rosea using gene deletion and complementation methods and found that loss of CrSsd1 led to defects in conidiation, stress responses, mycoparasitism, and biocontrol efficacy. To the best of our knowledge, this is the first demonstration that CrSsd1 plays an important role in conidiation and mycoparasitism in C. rosea, which is of great value for the development of biocontrol fungal agents.
Normal mycelial growth is crucial to ensure fungal vitality and other activities (Cota et al., 2008; Liu et al., 2016). In our current study, ΔCrSsd1 deletion mutants exhibited reduced hyphal growth, suggesting diverse functions for CrSsd1 in different fungal pathogens. The ΔCrSsd1 strain also showed a significant reduction in conidiation, which plays important roles in the completion of the fungal life cycle, and the ability to produce conidia is essential for successful colonization and mycoparasitism (Iqbal et al., 2019; Kong et al., 2019; Sun et al., 2019b). These phenotypes indicate that CrSsd1 is required for vegetative growth and asexual reproduction. The exploration of CrSsd1 regulating fungal sporulation will be very interesting and worth to be further investigated.
Fungi respond in various ways to exogenous stresses in order to maintain cell shape and normal physiological processes (Leng and Zhong, 2015; Wang et al., 2019). The fungal cell wall is a highly dynamic structure and is the first barrier that interacts with diverse environmental stresses. In addition, cell well integrity is vital for survival and pathogenesis but the regulatory mechanisms are complicated (Reinke et al., 2004). In the present study, CrSsd1 deletion mutants displayed greater sensitivity to the cell wall inhibitor CR, consistent with previous observations for S. cerevisiae and C. albicans (Moriya and Isono, 1999; Gank et al., 2008; Yanamoto et al., 2011). Ssd1 has been implicated in the maintenance of cell wall integrity in C. albicans, and deletion of Ssd1 can render cells more susceptible to cell wall-perturbing agents such as Calcofluor white (Ram and Klis, 2006; Gank et al., 2008). Additionally, the loss of CrSsd1 leads to increased sensitivity to sorbitol and osmotic stress agent, and overexpression of Ssd1 rescues the sorbitol dependence of cell wall-defective mutants in C. albicans (Avrahami-Moyal et al., 2012). It was also noticed that the hyphal branching of the ΔCrSsd1 strains markedly lessened under the stresses of NaCl, KCl, sorbitol, and CR, which might be a possible explanation for CrSsd1 regulating C. rosea response to osmotic and cell wall stresses. From these observations, we deduced that CrSsd1 may perform a different regulatory mechanism in stress responses in different fungi. Nevertheless, until recently, there was no experimental evidence for the contribution of CrSsd1 to cell wall integrity in mycoparasites.
During mycoparasitism, a host fungus is parasitized by and provides a nutrient source for another biocontrol fungus, such as species of the genus Trichoderma, and C. rosea (Karlsson et al., 2017; Nygren et al., 2018). Mycoparasitism comprises several steps; when encountering a fungal host, mycoparasites trigger gene expression associated with recognition, penetration, and parasitism, through various mechanisms related to mycoparasitism, antifungal activity, competition, and production of cell wall-degrading enzymes (Qualhato et al., 2013; Lysoe et al., 2017; Gomez-Rodriguez et al., 2018; Ramirez-Valdespino et al., 2018). Our findings confirmed that the deletion of CrSsd1 severely impaired C. rosea antagonistic activity and mycoparasitic ability to S. sclerotiorum and B. cinerea and dramatically decreased the control efficacy against soybean Sclerotinia rot. These observations were further supported by analysis of CrSsd1 gene expression during different stages of C. rosea parasitizing S. sclerotiorum sclerotia, which showed that CrSsd1 was highly expressed throughout mycoparasitism, particularly in the first stage of infection. It has been reported that Ssd1 is an important component of the regulation of Ace2p activity and morphogenesis (RAM) pathway comprised of two kinases and four associated proteins and a conserved Cbk1 target involving phosphorylation in S. cerevisiae (Bidlingmaier et al., 2001). Current researches suggest that Cbk1 and RAM regulate polarized growth, mating efficiency, and cell wall morphogenesis (Schmidpeter et al., 2017). We hypothesize that CrSsd1 influences the mycoparasitic activity and cell wall integrity of C. rosea by regulating the expressions of genes related to RAM pathway.
Herein, we analyzed the functions of CrSsd1 in hyphal growth, conidiation, and stress responses in C. rosea and found that it is involved in cell wall integrity and osmotic stress. Additionally, we found that CrSsd1 is involved in mycoparasitism and biocontrol efficacy. The results provide new insight into the mycoparasitism-associated mechanisms of C. rosea and may assist the development of new biocontrol agents for controlling fungal plant pathogens. In-depth studies will be needed to further clarify the exact regulatory mechanism, such as comparative analysis of transcription profiles.
Conclusion
In summary, the CrSsd1 protein of C. rosea was demonstrated to be essential for conidiation and responses to sorbitol and CR. Furthermore, CrSsd1 was found to be involved in mycoparasitism and biocontrol efficacy, indicating that it plays diverse and essential roles in this fungus.
Data Availability Statement
The datasets generated for this study can be found in the GenBank accession number: MN816008.
Author Contributions
MS, SL, and BL conceived and designed the study. BL performed the experiments, analyzed the data, and wrote the manuscript. NJ, RH, and YC prepared the figures and tables. MS and SL provided funding and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Key Research and Development Program of China (2019YFD1002003), the Science and Technology Major Project of Inner Mongolia (zdzx2018009), the Science and Technology Program of Qinghai (2019-NK-116), and the Agricultural Science and Technology Innovation Program (CAAS-ZDXT2018005).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01640/full#supplementary-material.
Supplementary FIGURE S1Impact of CrSsd1 deletion on conidial germination. Spore suspensions of WT, ΔCrSsd1, and ΔCrSsd1-C strains were inoculated in PD broth at 26°C, and the germinated conidia were counted at 8 and 16 h. The data are the means of three mutants, and the means and standard errors are calculated from three independent replicates. Statistical tests were carried out using Tukey’s test for multiple comparisons. Asterisks indicate statistically significant differences (p < 0.05).
Supplementary FIGURE S2Impact of CrSsd1 deletion on hyphae branching under diverse stresses. Sensitivity of 67-1, ΔCrSsd1, and ΔCrSsd1-C strains was determined on PDA plates containing different stress agents, 1 M NaCl, 1 M KCl, 1 M glycerin, 1 M sorbitol, 20 mM H2O2, 0.03% SDS, and 0.3 mg/ml Congo red (CR). Microscopic photos of the hypha under stresses were captured after 10 days of incubation at 26°C with a fluorescence microscope system.
Footnotes
References
Atanasova, L., Dubey, M., Grujic, M., Gudmundsson, M., Lorenz, C., Sandgren, M., et al. (2018). Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbiol. 18:178. doi: 10.1186/s12866-018-1310-9
Avrahami-Moyal, L., Braun, S., and Engelberg, D. (2012). Overexpression of PDE2 or SSD1-V in Saccharomyces cerevisiae W303-1A strain renders it ethanol-tolerant. FEMS Yeast Res. 12, 447–455. doi: 10.1111/j.1567-1364.2012.00795.x
Bidlingmaier, S., Weiss, E. L., Seidel, C., Drubin, D. G., and Snyder, M. (2001). The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 2449–2462. doi: 10.1128/MCB.21.7.2449-2462.2001
Cota, L. V., Maffia, L. A., and Mizubuti, E. S. (2008). Brazilian isolates of Clonostachys rosea: colonization under different temperature and moisture conditions and temporal dynamics on strawberry leaves. Lett. Appl. Microbiol. 46, 312–317. doi: 10.1111/j.1472-765X.2007.02312.x
Du, L., and Novick, P. (2002). Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 503–514. doi: 10.1091/mbc.01-07-0365
Dubey, M. K., Jensen, D. F., and Karlsson, M. (2014). An ATP-binding cassette pleiotropic drug transporter protein is required for xenobiotic tolerance and antagonism in the fungal biocontrol agent Clonostachys rosea. Mol. Plant-Microbe Interact. 27, 725–732. doi: 10.1094/MPMI-12-13-0365-R
Dubey, M., Jensen, D. F., and Karlsson, M. (2016). The ABC transporter ABCG29 is involved in H2O2 tolerance and biocontrol traits in the fungus Clonostachys rosea. Mol. Gen. Genomics 291, 677–686. doi: 10.1007/s00438-015-1139-y
Fatema, U., Broberg, A., Jensen, D. F., Karlsson, M., and Dubey, M. (2018). Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea. Sci. Rep. 8:15009. doi: 10.1038/s41598-018-33391-1
Feng, X., Ramamoorthy, V., Pandit, S. S., Prieto, A., Espeso, E. A., and Calvo, A. M. (2017). cpsA regulates mycotoxin production, morphogenesis and cell wall biosynthesis in the fungus Aspergillus nidulans. Mol. Microbiol. 105, 1–24. doi: 10.1111/mmi.13682
Filizola, P. R. B., Luna, M. A. C., de Souza, A. F., Coelho, I. L., Laranjeira, D., and Campos-Takaki, G. M. (2019). Biodiversity and phylogeny of novel Trichoderma isolates from mangrove sediments and potential of biocontrol against Fusarium strains. Microb. Cell Factories 18:89. doi: 10.1186/s12934-019-1108-y
Frandsen, R. J., Andersson, J. A., Kristensen, M. B., and Giese, H. (2008). Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 9:70. doi: 10.1186/1471-2199-9-70
Fuchs, B. B., and Mylonakis, E. (2009). Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8, 1616–1625. doi: 10.1128/EC.00193-09
Gank, K. D., Yeaman, M. R., Kojima, S., Yount, N. Y., Park, H., Edwards, J. E. Jr., et al. (2008). SSD1 is integral to host defense peptide resistance in Candida albicans. Eukaryot. Cell 7, 1318–1327. doi: 10.1128/EC.00402-07
Gerik, K., Bhimireddy, S., Ryerse, J., Specht, C., and Lodge, J. (2008). PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 10, 1685–1698. doi: 10.1128/EC.00146-08
Gerik, K., Donlin, M., Soto, C., Banks, A., Banks, I., Maligie, M., et al. (2005). Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58, 393–408. doi: 10.1111/j.1365-2958.2005.04843.x
Gomez-Rodriguez, E. Y., Uresti-Rivera, E. E., Patron-Soberano, O. A., Islas-Osuna, M. A., Flores-Martinez, A., Riego-Ruiz, L., et al. (2018). Histone acetyltransferase TGF-1 regulates Trichoderma atroviride secondary metabolism and mycoparasitism. PLoS One 13:e0193872. doi: 10.1371/journal.pone.0193872
Iqbal, M., Dubey, M., Broberg, A., Viketoft, M., Jensen, D. F., and Karlsson, M. (2019). Deletion of the nonribosomal peptide synthetase gene nps1 in the fungus Clonostachys rosea attenuates antagonism and biocontrol of plant pathogenic Fusarium and nematodes. Phytopathology 109, 1698–1709. doi: 10.1094/PHYTO-02-19-0042-R
Jansen, J. M., Wanless, A. G., Seidel, C. W., and Weiss, E. L. (2009). Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr. Biol. 19, 2114–2120. doi: 10.1016/j.cub.2009.10.071
Kaeberlein, M., and Guarente, L. (2002). Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics 160, 83–95.
Karlsson, M., Atanasova, L., Jensen, D. F., and Zeilinger, S. (2017). Necrotrophic mycoparasites and their genomes. Microbiol. Spectr. 5:FUNK-0016-2016. doi: 10.1128/microbiolspec.FUNK-0016-2016
Kong, X., Zhang, H., Wang, X., van der Lee, T., Waalwijk, C., van Diepeningen, A., et al. (2019). FgPex3, a peroxisome biogenesis factor, is involved in regulating vegetative growth, conidiation, sexual development, and virulence in Fusarium graminearum. Front. Microbiol. 10:2088. doi: 10.3389/fmicb.2019.02088
Kosawang, C., Karlsson, M., Velez, H., Rasmussen, P. H., Collinge, D. B., Jensen, B., et al. (2014). Zearalenone detoxification by zearalenone hydrolase is important for the antagonistic ability of Clonostachys rosea against mycotoxigenic Fusarium graminearum. Fungal Biol. 118, 364–373. doi: 10.1016/j.funbio.2014.01.005
Kurischko, C., Kim, H. K., Kuravi, V. K., Pratzka, J., and Luca, F. C. (2011a). The yeast Cbk1 kinase regulates mRNA localization via the mRNA-binding protein Ssd1. J. Cell Biol. 192, 583–598. doi: 10.1083/jcb.201011061
Kurischko, C., Kuravi, V. K., Herbert, C. J., and Luca, F. C. (2011b). Nucleocytoplasmic shuttling of Ssd1 defines the destiny of its bound mRNAs. Mol. Microbiol. 81, 831–849. doi: 10.1111/j.1365-2958.2011.07731.x
Leng, Y., and Zhong, S. (2015). The role of mitogen-activated protein (MAP) kinase signaling components in the fungal development, stress response and virulence of the fungal cereal pathogen Bipolaris sorokiniana. PLoS One 10:e0128291. doi: 10.1371/journal.pone.0128291
Liu, Z., Zhang, X., Liu, X., Fu, C., Han, X., Yin, Y., et al. (2016). The chitin synthase FgChs2 and other FgChss co-regulate vegetative development and virulence in F. graminearum. Sci. Rep. 6:34975. doi: 10.1038/srep34975
Luukkonen, B. G., and Séraphin, B. (1999). A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicingmutant suppressor. Nucleic Acids Res. 27, 3455–3465. doi: 10.1093/nar/27.17.3455
Lysoe, E., Dees, M. W., and Brurberg, M. B. (2017). A three-way transcriptomic interaction study of a biocontrol agent (Clonostachys rosea), a fungal pathogen (Helminthosporium solani), and a potato host (Solanum tuberosum). Mol. Plant-Microbe Interact. 30, 646–655. doi: 10.1094/MPMI-03-17-0062-R
Mir, S. S., Fiedler, D., and Cashikar, A. G. (2009). Ssd1 is required for thermotolerance and Hsp104-mediated protein disaggregation in Saccharomyces cerevisiae. Mol. Cell. Biol. 29, 187–200. doi: 10.1128/MCB.02271-07
Moriya, H., and Isono, K. (1999). Analysis of genetic interactions between DHH1, SSD1 and ELM1 indicates their involvement in cellular morphology determination in Saccharomyces cerevisiae. Yeast 15, 481–496. doi: 10.1002/(SICI)1097-0061(199904)15:6<481::AID-YEA391>3.0.CO;2-M
Nygren, K., Dubey, M., Zapparata, A., Iqbal, M., Tzelepis, G. D., Durling, M. B., et al. (2018). The mycoparasitic fungus Clonostachys rosea responds with both common and specific gene expression during interspecific interactions with fungal prey. Evol. Appl. 11, 931–949. doi: 10.1111/eva.12609
Ohyama, Y., Kasahara, K., and Kokubo, T. (2010). Saccharomyces cerevisiae Ssd1p promotes CLN2 expression by binding to the 59-untranslated region of CLN2 mRNA. Genes Cells 15, 1169–1188. doi: 10.1111/j.1365-2443.2010.01452.x
Qualhato, T. F., Lopes, F. A., Steindorff, A. S., Brandao, R. S., Jesuino, R. S., and Ulhoa, C. J. (2013). Mycoparasitism studies of Trichoderma species against three phytopathogenic fungi: evaluation of antagonism and hydrolytic enzyme production. Biotechnol. Lett. 35, 1461–1468. doi: 10.1007/s10529-013-1225-3
Ram, A. F., and Klis, F. M. (2006). Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 1, 2253–2256. doi: 10.1038/nprot.2006.397
Ramirez-Valdespino, C. A., Porras-Troncoso, M. D., Corrales-Escobosa, A. R., Wrobel, K., Martinez-Hernandez, P., and Olmedo-Monfil, V. (2018). Functional characterization of TvCyt2, a member of the p450 monooxygenases from Trichoderma virens relevant during the association with plants and mycoparasitism. Mol. Plant-Microbe Interact. 31, 289–298. doi: 10.1094/MPMI-01-17-0015-R
Reinke, A., Anderson, S., McCaffery, J. M., Yates, J. 3rd, Aronova, S., Chu, S., et al. (2004). TOR complex 1 includes a novel component, Tco89p (YPL180w), and cooperates with Ssd1p to maintain cellular integrity in Saccharomyces cerevisiae. J. Biol. Chem. 279, 14752–14762. doi: 10.1074/jbc.M313062200
Schmidpeter, J., Dahl, M., Hofmann, J., and Koch, C. (2017). ChMob2 binds to ChCbk1 and promotes virulence and conidiation of the fungal pathogen Colletotrichum higginsianum. BMC Microbiol. 17:22. doi: 10.1186/s12866-017-0932-7
Scrimale, T., Didone, L., de Mesy Bentley, K. L., and Krysan, D. J. (2009). The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 164–175. doi: 10.1091/mbc.E08-08-0809
Sun, Z. B., Li, S. D., and Sun, M. H. (2015a). Selection of reliable reference genes for gene expression studies in Clonostachys rosea 67-1 under sclerotial induction. J. Microbiol. Methods 114, 62–65. doi: 10.1016/j.mimet.2015.05.009
Sun, Z. B., Sun, M. H., and Li, S. D. (2015b). Identification of mycoparasitism-related genes in Clonostachys rosea 67-1 active against Sclerotinia sclerotiorum. Sci. Rep. 5:18169. doi: 10.1038/srep18169
Sun, Z. B., Sun, M. H., Zhou, M., and Li, S. D. (2017). Transformation of the endochitinase gene Chi67-1 in Clonostachys rosea 67-1 increases its biocontrol activity against Sclerotinia sclerotiorum. AMB Express 7:1. doi: 10.1186/s13568-016-0313-x
Sun, Z. B., Wang, Q., Sun, M. H., and Li, S. D. (2019a). The heat shock protein 70 gene is involved for colony morphology, sporulation and mycoparasitism of Clonostachys rosea. FEMS Microbiol. Lett. 366:fnz188. doi: 10.1093/femsle/fnz188
Sun, Z. B., Zhang, J., Sun, M. H., and Li, S. D. (2019b). Identification of genes related to chlamydospore formation in Clonostachys rosea 67-1. Microbiology 8:e00624. doi: 10.1002/mbo3.624
Tanaka, S., Ishihama, N., Yoshioka, H., Huser, A., O’Connell, R., Tsuji, G., et al. (2009). The Colletotrichum orbiculare SSD1 mutant enhances Nicotiana benthamiana basal resistance by activating a mitogen-activated protein kinase pathway. Plant Cell 21, 2517–2526. doi: 10.1105/tpc.109.068023
Tanaka, S., Yamada, K., Yabumoto, K., Fujii, S., Huser, A., Tsuji, G., et al. (2007). Saccharomyces cerevisiae SSD1 orthologs are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol. Microbiol. 64, 1332–1349. doi: 10.1111/j.1365-2958.2007.05742.x
Tzelepis, G., Dubey, M., Jensen, D. F., and Karlsson, M. (2015). Identifying glycoside hydrolase family 18 genes in the mycoparasitic fungal species Clonostachys rosea. Microbiology 161, 1407–1419. doi: 10.1099/mic.0.000096
Wang, Q., Chen, X., Chai, X., Xue, D., Zheng, W., Shi, Y., et al. (2019). The involvement of Jasmonic acid, ethylene, and salicylic acid in the signaling pathway of Clonostachys rosea-induced resistance to gray mold disease in tomato. Phytopathology 109, 1102–1114. doi: 10.1094/PHYTO-01-19-0025-R
Wang, M., Zhao, Q., Yang, J., Jiang, B., Wang, F., Liu, K., et al. (2013). A mitogen-activated protein kinase Tmk3 participates in high osmolarity resistance, cell wall integrity maintenance and cellulase production regulation in Trichoderma reesei. PLoS One 8:e72189. doi: 10.1371/journal.pone.0072189
Wanless, A. G., Lin, Y., and Weiss, E. L. (2014). Cell morphogenesis proteins are translationally controlled through UTRs by the Ndr/LATS target Ssd1. PLoS One 9:e85212. doi: 10.1371/journal.pone.0085212
Wei, W., Xiong, Y., Zhu, W., Wang, N., Yang, G., and Peng, F. (2016). Colletotrichum higginsianum mitogen-activated protein kinase ChMK1: role in growth, cell wall integrity, colony melanization, and pathogenicity. Front. Microbiol. 7:1212. doi: 10.3389/fmicb.2016.01212
Xue, A. G. (2003). Biological control of pathogens causing root rot complex in field pea using Clonostachys rosea strain ACM941. Phytopathology 93, 329–335. doi: 10.1094/PHYTO.2003.93.3.329
Yanamoto, T., Miyamoto, A., Ikeda, K., Hatano, T., and Matsuzaki, H. (2011). The relationship between chromosomal positioning within the nucleus and the SSD1 gene in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 75, 1713–1721. doi: 10.1271/bbb.110242
Yu, L., Xiong, D., Han, Z., Liang, Y., and Tian, C. (2019). The mitogen-activated protein kinase gene CcPmk1 is required for fungal growth, cell wall integrity and pathogenicity in Cytospora chrysosperma. Fungal Genet. Biol. 128, 1–13. doi: 10.1016/j.fgb.2019.03.005
Zhang, Y. H., Gao, H. L., Ma, G. Z., and Li, S. D. (2004). Mycoparasitism of Gliocladium roseum 67-1 on Sclerotinia sclerotiorum. Acta Phytopathol. Sin. 34, 211–214.
Keywords: Clonostachys rosea, Ssd1, cell wall integrity, mycoparasitism, biocontrol efficacy, conidiation
Citation: Lv B, Jiang N, Hasan R, Chen Y, Sun M and Li S (2020) Cell Wall Biogenesis Protein Phosphatase CrSsd1 Is Required for Conidiation, Cell Wall Integrity, and Mycoparasitism in Clonostachys rosea. Front. Microbiol. 11:1640. doi: 10.3389/fmicb.2020.01640
Edited by:
Hector Mora Montes, University of Guanajuato, MexicoReviewed by:
Georgios Tzelepis, Swedish University of Agricultural Sciences, SwedenDaohong Jiang, Huazhong Agricultural University, China
Copyright © 2020 Lv, Jiang, Hasan, Chen, Sun and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manhong Sun, c3VubWFuaG9uZzIwMTNAMTYzLmNvbQ==; Shidong Li, c2RsaUBpcHBjYWFzLmNu
 Binna Lv
Binna Lv Na Jiang
Na Jiang Rakibul Hasan
Rakibul Hasan Manhong Sun
Manhong Sun