- 1Department of Periodontics, School of Stomatology, China Medical University, Shenyang, China
- 2State Key Laboratory of Oral Disease, School of Stomatology, Sichuan University, Chengdu, China
Recent investigations revealed the relationship between chronic periodontitis, Porphyromonas gingivalis and cancer. However, host genes that change in response to chronic infection with P. gingivalis and may contribute to oral cancer have remained largely unknown. In the present study, we aimed to comprehensively analyze microarray data obtained from the chronic infection model of immortalized oral epithelial cells that were persistently exposed to P. gingivalis for 15 weeks. Using protein-protein interaction (PPI) networks and Ingenuity Pathway Analysis (IPA), we identified hub genes, major biological processes, upstream regulators and genes potentially involved in tumor initiation and progression. We also validated gene expression and demonstrated genetic alteration of hub genes from clinical samples of head and neck cancer. Overall, we utilized bioinformatical methods to identify IL6, STAT1, LYN, BDNF, C3, CD274, PDCD1LG2, and CXCL10 as potential candidate genes that might facilitate the prevention and treatment of oral squamous cell carcinoma (OSCC), the most common type of head and neck squamous cell carcinoma (HNSCC).
Introduction
Periodontitis is a chronic infectious disease with high global prevalence (1). The persistent immune-inflammation response initiated by periodontal pathogens not only causes local damage in oral sites, but also promotes systemically chronic inflammation such as the pathogenesis of cancer (2). Based on the latest meta-analysis, periodontal disease is positively associated with the risk of pancreatic, lung, head and neck cancers (3). Furthermore, a cohort study with 10-year follow-up recently revealed a significant relationship between periodontitis and cancer mortality (4).
Porphyromonas gingivalis, one of the best characterized pathogens of periodontitis, is regarded as a keystone pathogen because of its ability to induce increased community biomass and manipulate subversion of host immune responses (2, 5). With a variety of virulence factors, P. gingivalis can interact with host cells for successful internalization and intracellular survival, which consequently results in chronic infection (6). Of note, P. gingivalis is now accepted as a risk factor of cancer such as oral squamous cell carcinoma (OSCC), the most common type of head and neck squamous cell carcinoma (HNSCC) (6–8). To date, several studies have shown that acute P. gingivalis infections (i.e., those lasting no more than 72 h) affected cell division, apoptosis, migration and invasion in gingival epithelial cells or OSCC cells (2). However, since cancer can be a complicated and gradual process (9), little is known about genes that contribute to OSCC in a microenvironment characterized by chronic P. gingivalis infection. To better mimic the infection mode in vivo, we established a novel model system using human immortalized oral epithelial cells (HIOECs) persistently exposed to P. gingivalis at a multiplicity of infection (MOI) of 1 (10). Furthermore, we reported the promoting effect of chronic infection by P. gingivalis on cellular morphology, cell proliferation, migration and invasion of HIOECs. Also, using microarray analysis and relevant validation of changes in expression, we identified genes that may be involved in the tumorigenic-like transformation of HIOECs induced by P. gingivalis (10). For further investigation, in this study, we aimed to systematically analyze the differentially expressed genes (DEGs) from previous microarray data. To our knowledge, this work is the first to show and analyze DEGs in response to P. gingivalis infections lasting up to 15 weeks. We identified IL6, STAT1, LYN, BDNF, C3, CD274, PDCD1LG2, and CXCL10 as candidate genes associated with the initiation and progression of oral cancer induced by P. gingivalis infection.
Methods
Establishment of Cellular Model and Microarray Analysis
The establishment of a novel cellular model of HIOECs persistently exposed to P. gingivalis was described previously (10). In short, P. gingivalis ATCC 33277 was routinely recovered on brain heart infusion (Difco Laboratories, MI, USA) agar plates and cultured in brain heart infusion broth medium supplemented with 0.5% yeast, 0.1% menadione, and 1% hemin at 37°C in an anaerobic environment (80% N2, 10% H2, and 10% CO2). HIOECs were kindly provided by Dr. Wantao Chen from Key Laboratory of Shanghai Oral Medicine, Ninth People's Hospital, Shanghai Jiao Tong University. HIOECs were cultured in Defined Keratinocyte-SFM (GibcoTM, Thermo Fisher Scientific Inc., MA, USA) with growth supplement in a humidified atmosphere (37°C, 5% CO2). To establish the infection model, actively growing HIOECs in six-well plates were infected with P. gingivalis at an MOI of 1 for 24 h each time. The infection was repeated at each passage. HIOECs without P. gingivalis infection were cultured at the same time as a control.
When the HIOECs infected by P. gingivalis showed significantly increased tumorigenic properties after 15 weeks, the total RNA of non-infected HIOECs and infected HIOECs was extracted with an RNAiso Plus kit (TaKaRa, Dalian, China). As described before (10), the microarray (Affymetrix) that could simultaneously detect coding and non-coding genes was conducted at the OEbiotech Corporation (Shanghai, China). An Affymetrix Scanner 3000 (Affymetrix) and Affymetrix GeneChip Command Console (version 4.0, Affymetrix) software was applied for array scan and raw data exaction, respectively. Expression Console (version 1.3.1, Affymetrix) software was used for robust multi-array average (RMA) normalization. DEGs were identified using GeneSpring software (version 13.1; Agilent Technologies). Student's t-test was used for statistical analysis. The threshold set for aberrantly regulated genes was a fold change ≥2.0 and a P < 0.05. The P-value was adjusted by multiple testing using the Benjamini-Hochberg false discovery rate (FDR) procedure and a standard threshold of 5% was selected for declaring significance (11). In the present study, we focused on the coding genes that were aberrantly expressed. The microarray dataset has been uploaded to NCBI GEO platform (GSE87539).
Data Analyses
To construct the protein-protein interaction (PPI) network, the 423 differentially expressed coding genes from the present dataset were imported into the STRING (version 10.5) online database (https://string-db.org) and filtered with the interaction score of high confidence (0.700) (12). The network was then visualized with Cytoscape software (version 3.6.0). Using cytoHubba, a plugin of Cytoscape, 10 hub genes were calculated and ranked by “Radiality.” Gene Ontology analysis was performed with BiNGO from Cytoscape software. The Ingenuity Pathway Analysis (IPA) was performed by Genery Bio Corporation (Shanghai, China). IPA platform provided regulator networks of biological functions and diseases based on a large-scale causal network derived from the Ingenuity Knowledge Base (13). In this study, the upstream regulators and the enriched gene functions of the present cellular model were analyzed by IPA. Fisher's Exact Test was used for statistical analysis.
To validate the expression of DEGs in clinical samples, we firstly screened the expression of the 10 hub genes by applying the OncomineTM platform (https://www.oncomine.org/resource/login.html). OncomineTM was a cancer microarray database which facilitated to explore genome-wide expression analyses in various types of cancer as well as respective normal tissues. Specifically, a variety of cancer subtypes were also available (14). We selected seven datasets of OSCC, one of the important types of HNSCC. The search term of each gene was consistent with the gene symbol shown in the Figure 1. A P-value < 1E-4 and a fold change > 2 were considered as the threshold with gene ranking in the top 10%. Further, we selected one of the datasets (Peng Head-Neck, 79 samples, 18148 measured genes) to find the specific fold change of hub genes. The P-value was provided by the program (OncomineTM). cBioPortal database was used to integrate multidimensional cancer genomic data, such as somatic mutations, DNA copy-number alterations (CNAs) and mRNA (15). The hub genes validated by OncomineTM were further analyzed for genetic alterations by using cBioPortal (http://www.cbioportal.org/). A network containing the hub genes and the most frequently altered neighbor genes was also produced by cBioPortal. Biological interactions were derived from public pathway databases including PANTHER, Reactome, pid, Phosphosite, HumanCyc, CancerRxGene and KEGG Drug.
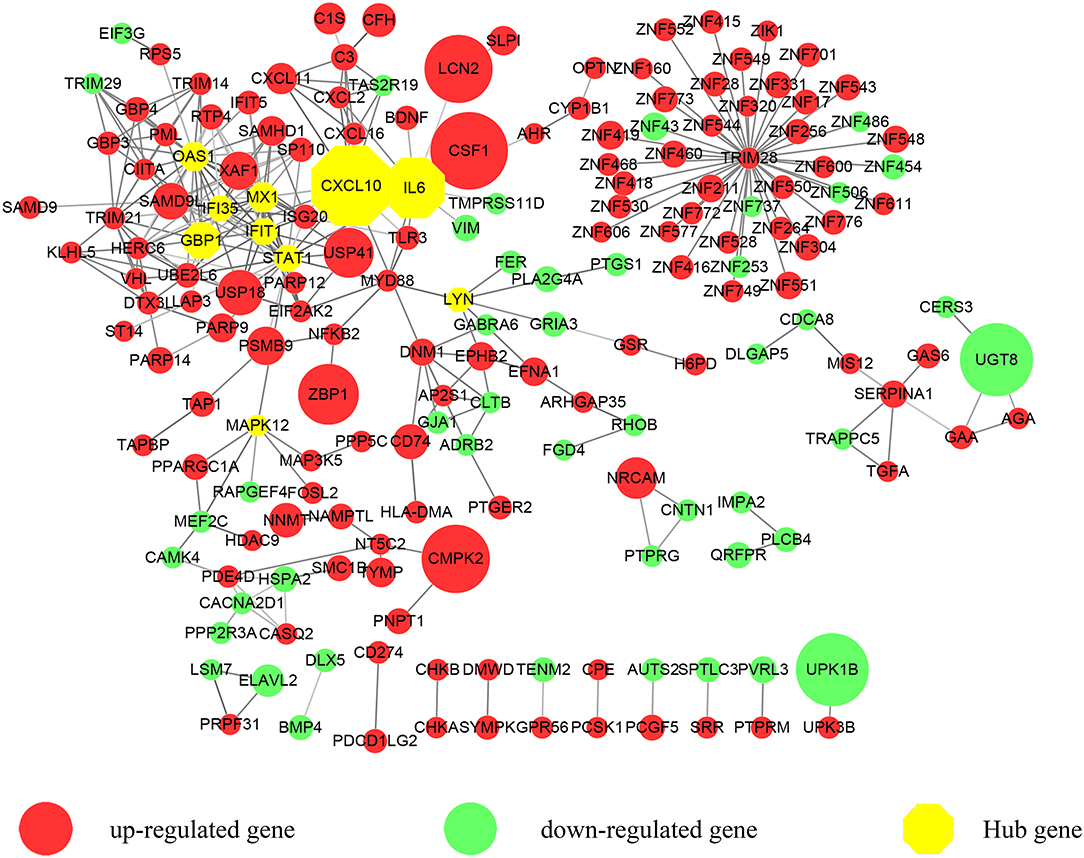
Figure 1. A protein-protein interaction (PPI) network exported from STRING was visualized with Cytoscape. 183 nodes and 309 edges were displayed. Up-regulated genes are shown in red while down-regulated genes are shown in green. Larger node sizes correspond to higher fold changes of DEGs. Edges were shown in gray from light to dark according to the combined score (from low to high). The hub genes were emphasized with octagon shapes in yellow.
Results
PPI Network Construction and Identification of Hub Genes
We focused on the coding genes in the present study. Among the 423 DEGs (fold change ≥ 2, P < 0.05), 296 genes were up-regulated while 127 genes were down-regulated. As shown in Figure 1, we first developed a PPI network. Three hundred and sixty-five of molecules were identified with STRING (PPI enrichment P < 1.0e−16). Disconnected nodes were hidden. As emphasized with a yellow octagon sign, STAT1, CXCL10, MX1, IFIT1, GBP1, IL6, OAS1, MAPK12, LYN, and IFI35 were identified as the hub genes of the network. STAT1, MX1, IFIT1, GBP1, OAS1, and IFI35 were associated with the most significant module. Hub gene ranks are shown in Supplementary Figure 1.
Gene Ontology Analysis
According to the analysis with Cytoscape software, statistically overrepresented biological processes are shown in Figure 2 as colored nodes, which range from yellow to orange based on P-value. Nodes such as “regulation of metabolic process,” “regulation of biological process” and “regulation of cellular process,” though presented in larger size and darker orange, were merely the results of the constitutive genes at nodes further down each branch (16); we chose to focus our analyses on these nodes. Nine enriched biological processes, highlighted in red in Figure 2, include: “regulation of transcription, DNA-dependent,” “negative regulation of collagen biosynthetic process,” “positive regulation of inflammation response,” “regulation of immune system process,” “positive regulation of development process,” “regulation of ossification,” “regulation of angiogenesis,” “regulation of cell proliferation” and “positive regulation of kinase activity.”
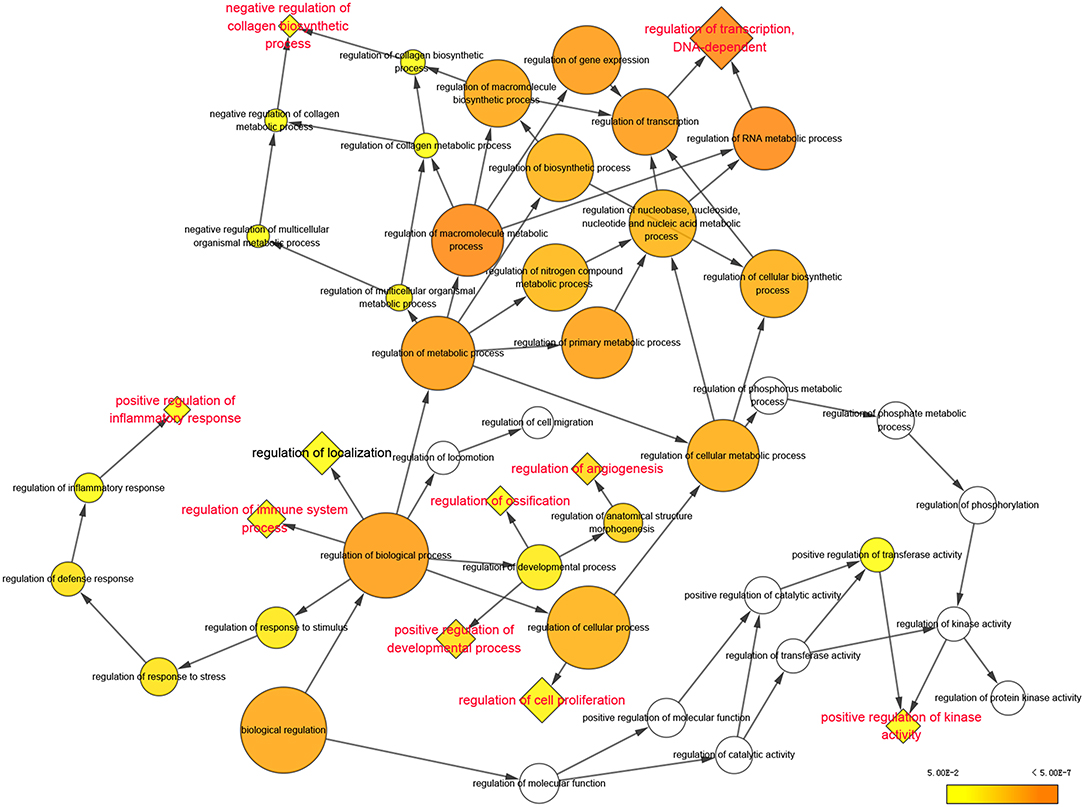
Figure 2. Significantly enriched biological processes of Gene Ontology were displayed in colored nodes. The nodes color got increasingly more orange for more significant P-values. The uncolored nodes that were not overrepresented were considered as the parents of the overrepresented categories further down. The size of the node depended on the number of genes annotated in the node.
Upstream Regulators
As shown in Figure 3A, STAT1, C3, IL6, and LCN2 were predicted to function as upstream regulators in the present model. STAT1 was predicted to directly up-regulate genes such as CXCL10, IFIT1, GBP1, OAS1, IFI35, CD274, and PDCD1LG2. IL6 was predicted to up-regulate downstream genes such as CD274, TGFA, SERPINA1, MYD88, and BDNF. C3 was predicted to up-regulate genes such as CSF1, CXCL16, and CXCL2. LCN2 was predicted to up-regulate CXCL10 and CXCL2 and to down-regulate VIM. Among the downstream regulators, CXCL10 was up-regulated by all four upstream regulators.
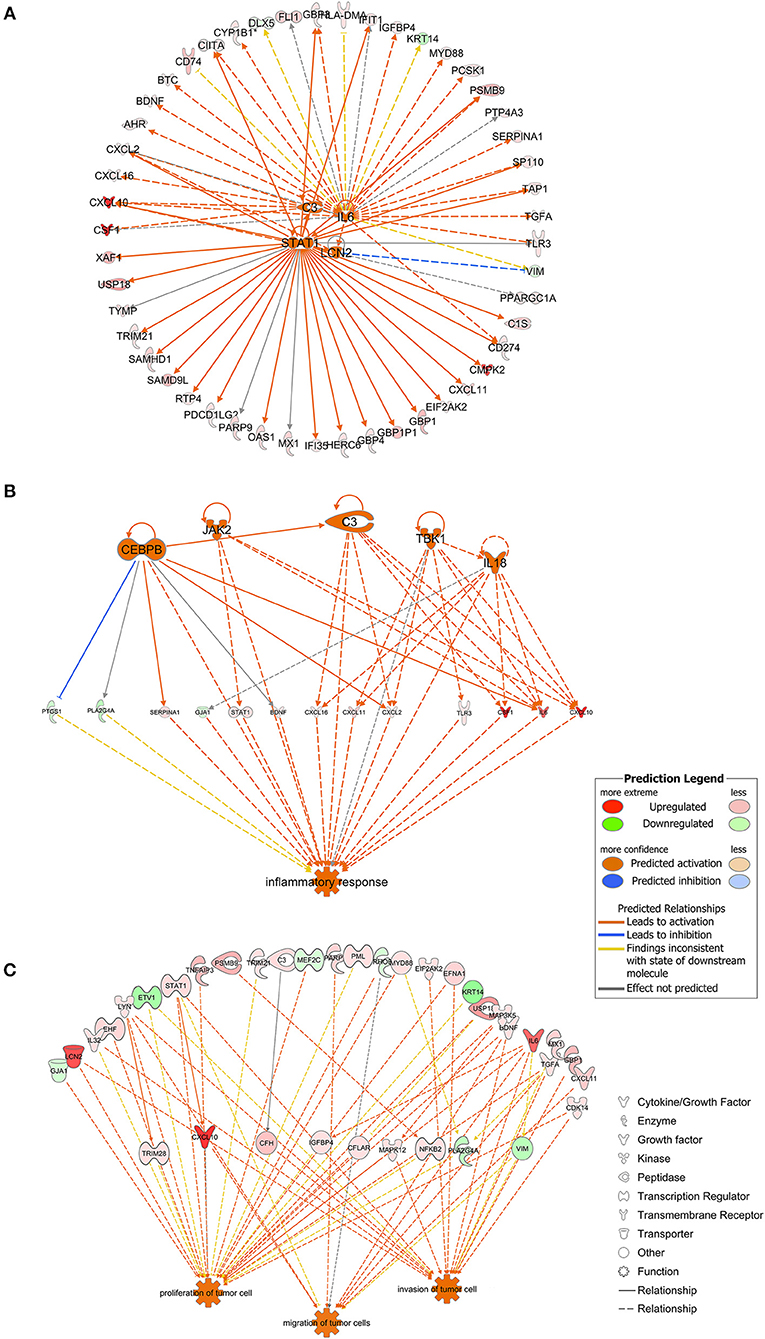
Figure 3. (A) The four molecules shown in the middle were considered as the upstream regulators and to regulate the surrounding genes. As shown in orange, STAT1, C3, IL6, and LCN2 were predicted to be activated in the present model. Full lines indicated the direct relationships while dashed lines indicated indirect relationships. (B) Inflammation-associated genes were highlighted. Genes containing CEBPB, JAK2, C3, TBK1, and IL18 were predicted to be activated and to regulate genes downstream. Full lines indicated direct relationships while dashed lines indicated indirect relationships. (C) Genes possibly involved in tumor cell proliferation, migration and invasion. Full lines indicated direct relationships while dashed lines indicated indirect relationships.
Analysis of Biological Functions
Genes associated with inflammatory response were analyzed using the IPA platform and are shown in Figure 3B. SERPINA1, STAT1, BDNF, CXCL16, CXCL11, CXCL2, TLR3, CSF1, IL6, and CXCL10 were up-regulated while PTGS1, PLA2G4A, and GJA1 were down-regulated based on the present dataset. CEBPB, JAK2, TBK1, and IL18, which did not belong to the dataset, were predicted to be activated according to the IPA platform. As shown in Figure 3B, CEBPB directly activated the expression of SERPINA1, CXCL2, IL6, and C3 and inhibited the expression of PTGS1.
Based on the results of the IPA platform, we discovered a series of genes that may be involved in regulation of tumorigenic properties of P. gingivalis-infected HIOECs. Most of the genes listed in Figure 3C were associated with cell proliferation and/or invasion, while genes associated with cell migration were relatively fewer in number. Genes such as LCN2, LYN, STAT1, BDNF, IL6, CXCL10, and CDK14 were related to cell proliferation and cell metastasis.
Validation of Hub Genes in Clinical Samples
To determine whether the DEGs identified in response to chronic infection by P. gingivalis were also aberrantly expressed in oral cancer, we chose to validate the expression of hub genes in clinical samples of OSCC from OncomineTM. With the exception of MAPK12, most genes were found to be generally over-expressed in OSCC samples compared with the normal samples (P < 0.05) (Figure 4). Notably, STAT1, GBP1, OAS1, IFI35, and LYN were overexpressed in more than 6 datasets. In one specific OSCC dataset, all hub genes were significantly up-regulated in oral cavity squamous cell carcinoma samples compared to control samples (P < 0.05) (Figure 5).
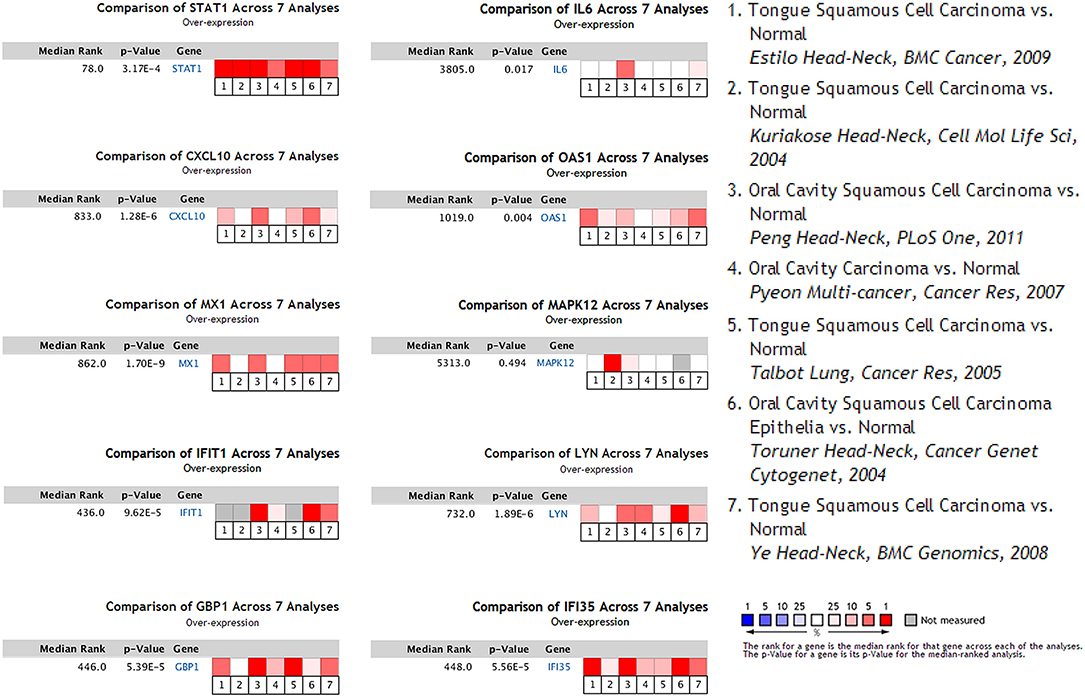
Figure 4. Seven OSCC datasets were selected and the expression of the hub genes was compared across the seven datasets. Among the datasets, three of them were obtained from general OSCC samples while the other four belonged to tongue squamous cell carcinoma, the most common subtype of OSCC. Normal tissues were selected as the control. Values above the average were considered over-expressed hub genes (red).
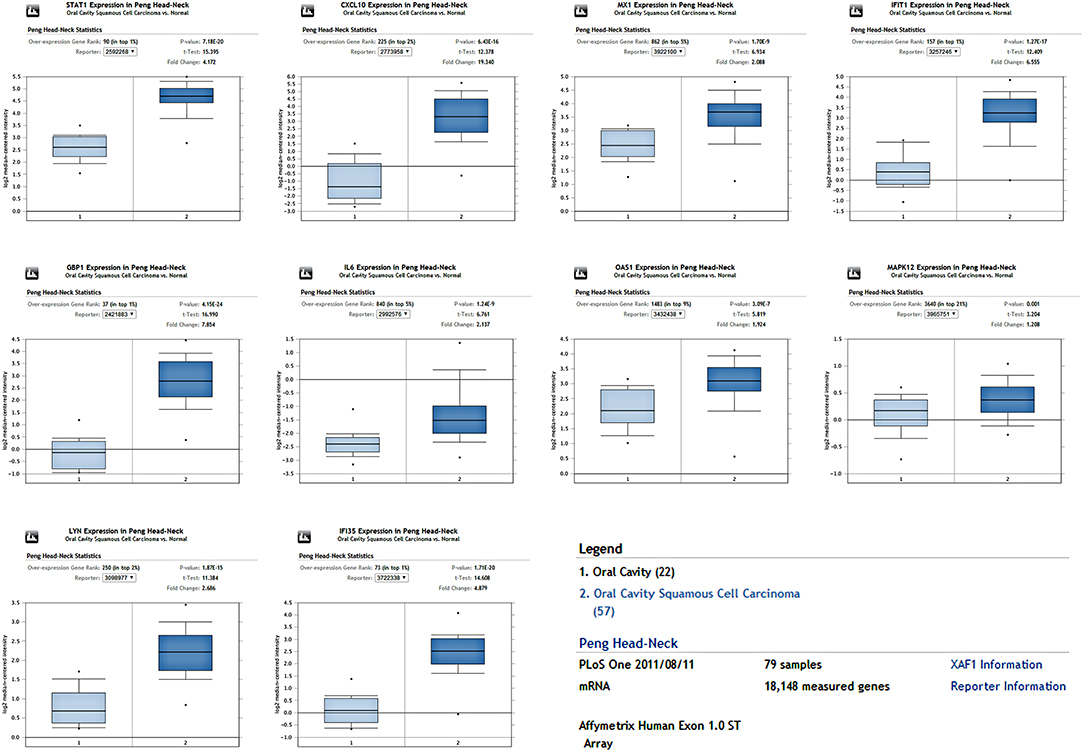
Figure 5. The expression of hub genes in one OSCC dataset containing 79 samples. All hub genes were overexpressed in the OSCC tissues compared to the control (P < 0.05).
Genetic Alterations of Validated Genes and the Relative Network
An HNSCC dataset provided by The Cancer Genome Atlas (TCGA) with 279 samples was used to analyze genetic alterations. As shown in Figure 6, the 10 hub genes were generally altered in 37% of the queried samples. Specifically, the gene set altered from 4 to 10% contained amplification, deep deletion, missense, mRNA up-regulation and truncation mutations. Using cBioPortal, we constructed a network that contained the hub genes and other frequently altered neighbor genes (Figure 7).
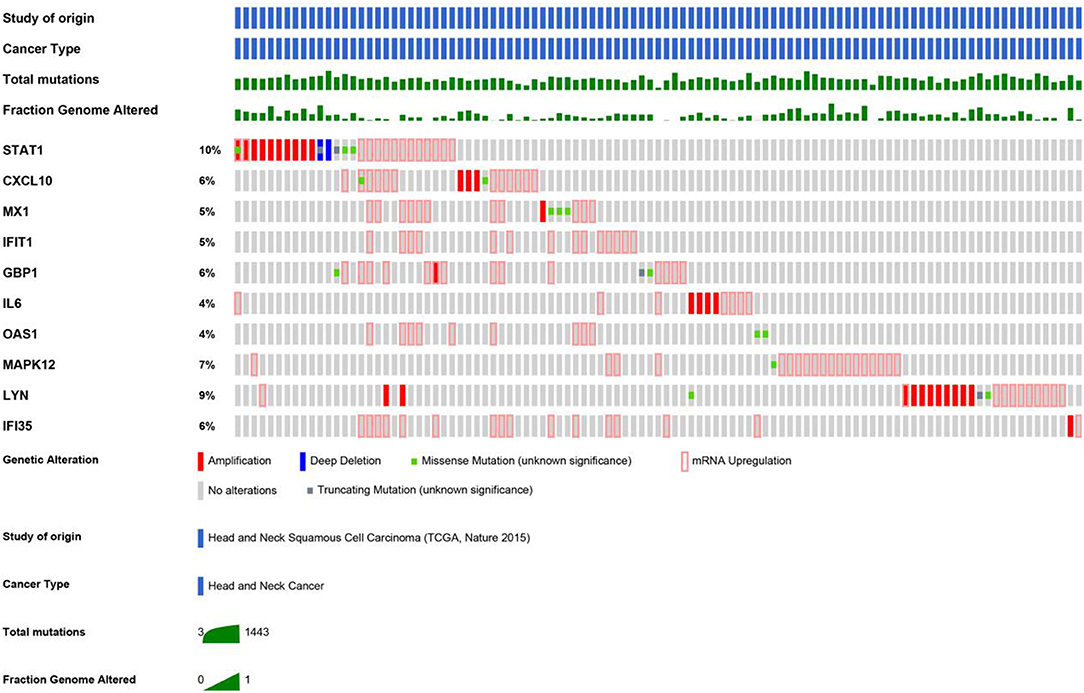
Figure 6. As shown, a study from HNSCC (TCGA, Nature 2015) was selected to analyze the genetic alterations of hub genes. The total mutations and fraction of the genome that was altered were also displayed, respectively.
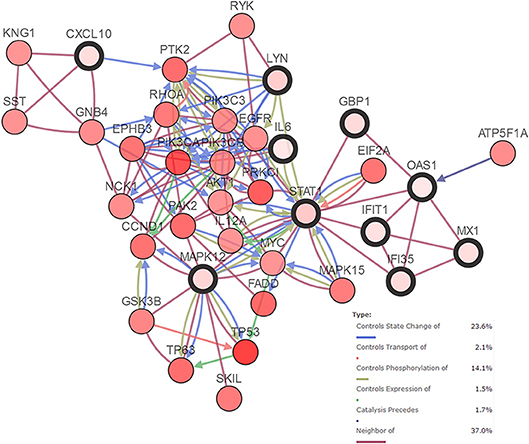
Figure 7. Network of hub genes (shown with thick border) and altered neighbor genes. Relationship among the genes included “Controls State Change of,” “Controls Transport of,” “Controls Phosphorylation of,” “Controls Expression of,” “Controls precedes,”and “Neighbor of”.
Potential Candidate Genes
By comprehensively analyzing the data shown in Figures 1, 3, 4, 7, genes including IL6, STAT1, LYN, BDNF, C3, CD274, PDCD1LG2, and CXCL10 were considered to be potential candidate genes and are presented in the proposed mode pattern in Figure 8. To be consistent with a previous study (2), CD274 and PDCD1LG2 are shown as B7H1 and B7DC, respectively, in Figure 8. In the proposed diagram, IL6, STAT1, LYN, and CXCL10 were hub genes while C3, IL6, and STAT1 were activated as the upstream regulators of the present dataset. CEBPB and JAK2, which were predicted as upstream regulators, are also shown in the network. Additionally, chemokines including CXCL2, CXCL11, CXCL16 were involved as possible downstream molecules. All candidate genes were associated with inflammation and/or tumors. Additional reasons for selecting these genes and their molecular functions in inflammation and tumors are discussed below.
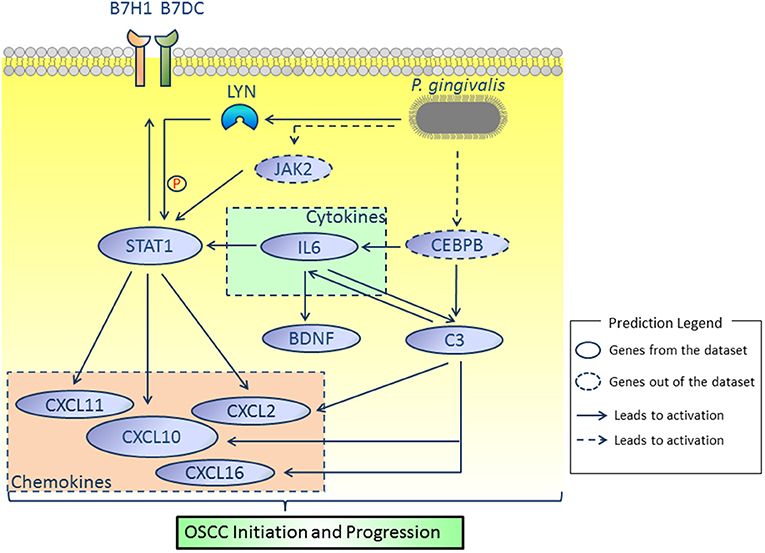
Figure 8. Proposed model. According to our results, IL6, STAT1, LYN, BDNF, C3, CD274, PDCD1LG2, and CXCL10 were considered as key molecules involved in OSCC initiation and progression induced by P. gingivalis. JAK2 and CEBPB, which were not identified in the present dataset, were predicted by the IPA platform and are shown in dotted lines. All relationships were based on IPA and cBioPortal database.
Discussion
It has been increasingly accepted that OSCC is the most relevant cancer type associated with oral bacterial infections (7). To date, several studies have reported the promoting effects of P. gingivalis on OSCC initiation and progression (10, 17–23). However, the mechanism of chronic infection by P. gingivalis in OSCC is not clearly understood. In the present study, we mined microarray data obtained from a cellular model of oral epithelial cells that were infected by P. gingivalis for 15 weeks. We first identified hub genes and found that DEGs were enriched in biological processes such as “positive regulation of inflammation response,” “regulation of immune system process,” “regulation of angiogenesis” and “regulation of cell proliferation.” Then, we specifically analyzed the upstream regulators and DEGs in tumor cell proliferation, migration, and invasion using the IPA platform. We previously validated some of the microarray data by qPCR (10). In the present study, using validation with datasets from clinical samples, we confirmed that some of the DEGs (hub genes) were also aberrantly expressed in OSCC clinical specimens.
Inflammation is well-accepted as a hallmark of tumorigenesis (24). Cytokines and chemokines play important roles in tumor initiation and progression. Cytokines such as IL6 promote tumor initiation by elevating intracellular reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI) as well as causing epigenetic alteration of certain genes. Furthermore, cytokines facilitate tumor progression by activating tumorigenic-related transcription factors (25). In turn, activated transcription factors induce the production of chemokines, which results in continuous tumor-associated inflammation (25). Among the genes listed in Figure 3, those such as IL6 (cytokine) and CXCL10 (chemokine) were shown to be involved in the inflammatory response (Figure 3B) and are positively related to cell proliferation and metastasis (Figure 3C). Interleukin (IL6), a tumor promoting cytokine in various types of cancer including OSCC (25–27), may function as a central molecule in the proposed network (Figure 8). Importantly, IL6 was identified as one of the most promising predictors for early diagnosis and prognosis of tongue squamous cell carcinoma, the most common type of OSCC with high risk of local invasion and recurrence (27, 28).
We paid specific attention to the relationship between CEBPB and IL6. It is well-known that IL6 is one of the major cytokines involved in P. gingivalis infection and periondontitis initiation. Based on previous studies of P. gingivalis infections, toll-like receptor 2/4 and nucleotide-binding oligomerization domain (NOD)-containing protein-like receptors were activated and mediated signaling pathways such as NF-κB and ERK1/2, which resulted in the overexpression of IL6 (29–31). In addition to these previously identified mechanisms, our results revealed that CEBPB was activated as an upstream regulator of IL6 (Figure 3B). CCAAT enhancer binding protein beta (CEBPB) is a transcription factor that plays an important role in immune and inflammation responses (32). CEBPB has been studied in the context of cellular transformation and cancer and was recently regarded as one of the master regulators in cancer biology, especially in mesenchymal glioblastoma (33). Importantly, a data mining study concluded that IL6 and CEBPB were up-regulated in late stage OSCC. Similar to our results, Bai et al. (34) reported that up-regulation of CEBPB in human periodontal ligament cells promoted the production of IL6 in response to the stimulation of lipopolysaccharide derived from P. gingivalis. However, direct regulation of IL6 expression by CEBPB and its effect in OSCC needs to be validated.
As reported previously, a carcinogen induced oral carcinoma model revealed that P. gingivalis and Fusobacterium nucleatum promoted OSCC progression with the activation of IL6-STAT3 (17). Interestingly, IL6 was also predicted to activate STAT1 in the present study (Figures 3A,B). Signal transducer and activator of transcription 1 (STAT1) was a hub gene with the highest median rank according to the expression validation with clinical samples. STAT1 is a critical transcription factor involved in regulating cellular responses to interferons (IFNs), cytokines, growth factors and hormones (35). The oncogenic role of STAT1 in certain types of cancer including HNSCC has been described (36–38). Phosphorylation of genes such as STAT1 was predicted to be regulated by LYN (Figure 7). LYN proto-oncogene (LYN), a hub gene that belongs to the Src family of kinases, was associated with various cellular functions such as cell proliferation, metabolism and differentiation. The presence of LYN kinase in the tumor microenvironment was regarded as a key component for tumor progression (39). Furthermore, LYN was considered as a target in both epithelial cells and stromal cells of HNSCC (40). Recently, LYN was found to be an independent biomarker of HNSCC and was correlated with poor survival. Upon validation using seven clinical datasets, we also confirmed the overexpression of LYN in OSCC (Figure 4). In this study, we speculated that phosphorylation of STAT1 might be one of the functions of LYN in OSCC in response to P. gingivalis infection.
An immunosuppressive microenvironment involving the activation of PD1/PDL1 signaling has been increasingly considered to be important for tumor initiation and progression. As a critical mediator that was overexpressed in various types of cancer, PD-L1 (programmed death-ligand 1, shown as CD274, also known as B7H1) was also observed to be up-regulated in OSCC cells by P. gingivalis infection, although the underlying mechanism was unclear (2, 41). In Figure 3A, CD274 and its paralog PDCD1LG2 (also known as B7DC) were regarded as targets of STAT1. Similarly, activation of STAT1 was shown to mediate the high expression of PD-L1 in HNSCC cells (36, 42). In addition, it was recently confirmed that IFNα, a cytokine belonging to the type I IFN family, induced overexpression of PDL1 through the activation of STAT1 (42). Interestingly, several type I IFN inducible genes such as OAS1 (2'-5'-oligoadenylate synthetase 1) were also identified in the present study as hub genes and were regulated by STAT1 (Figures 1, 3), which to some extent indicated the activation of endogenous IFNα (42). Taken together, we hypothesized that chronic infection by P. gingivalis contributed to an immunosuppressive microenvironment by targeting CD274 and PDCD1LG2 through the activation of STAT1.
IL6 was also predicted to activate BDNF (Figure 3A). Brain derived neurotrophic factor (BDNF) was of great interest as it was predicted to be associated with tumorigenic properties (tumor cell proliferation, migration and invasion) as well as inflammatory response (Figures 3B,C). Though BDNF belongs to the nerve growth factor family, it is also expressed in tissues outside the nerve system. BDNF has been shown to be overexpressed in neoplastic specimens in various types of tumors including HNSCC (43, 44). Stimulation of BDNF in vitro promoted the invasive properties of HNSCC cells (43), which was similar to our results. As previously reported, within an inflammatory microenvironment, production of BDNF was enhanced in response to pro-inflammatory cytokines such as IL6 (45). Correa et al. (46) were the first to report higher levels of BDNF in chronic periodontitis compared to control samples. However, it was recently suggested that recombinant human BDNF facilitated periodontal treatment by reducing excess inflammation (47). Thus, the functions of IL6 and BDNF in OSCC upon P. gingivalis infection require further exploration.
In addition to IL6, C3 was also a direct target of CEBPB, as predicted by IPA (Figure 3B). Complement system, the first defense against pathogens, is closely associated with tissue homeostasis. In the present study, Complement component 3 (C3) was considered as an upstream regulator and associated with inflammatory response. C3 was also predicted to activate genes such as IL6. The undesirable activation of complement results in pathogenesis of inflammatory diseases as well as cancer (24). Interestingly, apart from immune cells such as macrophages, epithelial cells also secrete complement proteins. In neoplastic tissues, C3 derived from tumor cells promoted tumor development by increasing cell proliferation (48). Several studies revealed the clinical role of C3 as a salivary or sera biomarker of OSCC (49–51). However, the relationship between P. gingivalis and C3 activation is rather complex and depends on the concentration of gingipains. C3 is specifically activated when the concentration of gingipains is low (52). In the present cellular model, C3 may be activated due to the low MOI of P. gingivalis, which is consistent with the low concentration of gingipains (52). Thus, we speculate that the constant activation of C3 which might be mediated by CEBPB during P. gingivalis infection at a low MOI may promote the tumorigenic properties of HIOECs. Furthermore, potential interaction between IL6 and C3 within the inflammatory microenvironment remain to be investigated. Finally, other targets of CEBPB, which might be associated with OSCC development under P. gingivalis infection, should be explored in future studies.
As shown in Figure 8, some chemokines were identified as downstream molecules in the network. Chemokines have complex roles in tumor biology. Indeed, certain chemokines that were overexpressed in a paracrine method were associated with tumor suppression and improved tumor prognosis owing to the recruitment of favorable immune cell populations (53). However, similar to the present cellular model, chemokines such as CXCL9, CXCL10, CXCL11 secreted in an autocrine manner by tumor cells were considered to contribute to a pro-tumoral microenvironment (25, 54). Thus, chemokines and their receptors are suggested to facilitate tumor initiation and progression and have been considered as key therapeutic targets for cancer (55, 56). CXCL10 was one of the hub genes with a high fold change (shown in dark red). From an integrated microarray analysis, CXCL10 was identified as a hub gene of periodontitis (57). As reported previously, CXCL10 was associated with tumor cell motility and metastasis in various type of cancer (56, 58). Meanwhile, other chemokines including CXCL2, CXCL11, and CXCL16 were also predicted to be activated downstream. A bioinformatics analysis suggested that CXCL10 and CXCL2 were members of the key molecules of epithelia in tongue squamous cell carcinoma (59). Overexpression of CXCL11 was observed in the premalignant stage of OSCC, which was thought to be involved in carcinogenesis of OSCC (60). Though few studies have focused on the role of CXCL16 in oral cancer, the promoting effect of CXCL16 in tumor progression was observed in other types of cancer (61, 62). Future work on malignant tumor properties associated with chemokines in response to P. gingivalis infection may shed light on the transition of periodontitis to OSCC.
We acknowledge some limitations of our present work. In this study, DEGs in response to repeated infection by P. gingivalis for 15 weeks were shown and candidate genes associated with inflammation and tumorigenic properties were analyzed. However, additional studies should be applied to explore the exact roles of the identified molecules in OSCC and to validate the proposed interactions. In addition, though we validated the expression of hub genes in some clinical datasets of OSCC, other datasets derived from larger-scale clinical samples which contain information on periodontal conditions and prevalence rates of P. gingivalis should be applied for further validation and evaluation.
In conclusion, we identified IL6, STAT1, LYN, BDNF, C3, CD274, PDCD1LG2, and CXCL10 as important candidates associated with OSCC in the present model and made attempts to illustrate the promoting role of P. gingivalis infection in OSCC initiation and progression. We suggested that chronic infection by P. gingivalis caused secretion of cytokines such as IL6 to initiate OSCC and to activate tumorigenic transcription factors such as STAT1 for tumor progression. During this complex process, overexpression of chemokines (such as CXCL10) may be involved in a positive loop of sustained tumor-associated inflammation. Immune escape of tumorigenic cells in response to P. gingivalis infection, regulated by CD274, should also be considered.
Author Contributions
YP, FG, and QW contributed conception and design of the study. FG, CL, and JL analyzed the data. FG and QW wrote the draft of the manuscript. YP, DZ, and SZ revised the draft manuscript. All authors read and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81670997 and 81870771).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2019.00091/full#supplementary-material
References
1. Frencken JE, Sharma P, Stenhouse L, Green D, Laverty D, Dietrich T. Global epidemiology of dental caries and severe periodontitis - a comprehensive review. J Clin Periodontol. (2017) 44 (Suppl. 18):S94–S105. doi: 10.1111/jcpe.12677
2. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. (2015) 15:30–44. doi: 10.1038/nri3785
3. Michaud DS, Fu Z, Shi J, Chung M. Periodontal disease, tooth loss, and cancer risk. Epidemiol Rev. (2017) 39:49–58. doi: 10.1093/epirev/mxx006
4. Heikkila P, But A, Sorsa T, Haukka J. Periodontitis and cancer mortality: register-based cohort study of 68,273 adults in 10-year follow-up. Int J Cancer (2018) 142:2244–53. doi: 10.1002/ijc.31254
5. Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. (2013) 7:1016–25. doi: 10.1038/ismej.2012.174
6. Miller DP, Hutcherson JA, Wang Y, Nowakowska ZM, Potempa J, Yoder-Himes DR, et al. Genes contributing to Porphyromonas gingivalis fitness in abscess and epithelial cell colonization environments. Front Cell Infect Microbiol. (2017) 7:378. doi: 10.3389/fcimb.2017.00378
7. Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. (2014) 10:e1003933. doi: 10.1371/journal.ppat.1003933
8. Sayehmiri F, Sayehmiri K, Asadollahi K, Soroush S, Bogdanovic L, Jalilian FA, et al. The prevalence rate of Porphyromonas gingivalis and its association with cancer: a systematic review and meta-analysis. Int J Immunopathol Pharmacol. (2015) 28:160–7. doi: 10.1177/0394632015586144
9. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi: 10.1016/j.cell.2011.02.013
10. Geng F, Liu J, Guo Y, Li C, Wang H, Wang H, et al. Persistent exposure to Porphyromonas gingivalis promotes proliferative and invasion capabilities, and tumorigenic properties of human immortalized oral epithelial cells. Front Cell Infect Microbiol. (2017) 7:57. doi: 10.3389/fcimb.2017.00057
11. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (1995) 57:289–300.
12. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. (2015) 43:D447–452. doi: 10.1093/nar/gku1003
13. Kramer A, Green J, Pollard JJr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics (2014) 30:523–30. doi: 10.1093/bioinformatics/btt703
14. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (2004) 6:1–6. doi: 10.1016/S1476-5586(04)80047-2
15. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. (2013) 6:pl1. doi: 10.1126/scisignal.2004088
16. Maere S, Heymans K, Kuiper M. BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics (2005) 21:3448–9. doi: 10.1093/bioinformatics/bti551
17. Binder Gallimidi A, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget (2015) 6:22613–23. doi: 10.18632/oncotarget.4209
18. Ha NH, Woo BH, Kim da J, Ha ES, Choi JI, Kim SJ, et al. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. (2015) 36:9947–60. doi: 10.1007/s13277-015-3764-9
19. Sztukowska MN, Ojo A, Ahmed S, Carenbauer AL, Wang Q, Shumway B, et al. Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell Microbiol. (2015) 18:844–58. doi: 10.1111/cmi.12554
20. Ha NH, Park DG, Woo BH, Kim da J, Choi JI, Park BS, et al. Porphyromonas gingivalis increases the invasiveness of oral cancer cells by upregulating IL-8 and MMPs. Cytokine (2016) 86:64–72. doi: 10.1016/j.cyto.2016.07.013
21. Lee J, Roberts JS, Atanasova KR, Chowdhury N, Han K, Yilmaz O. Human primary epithelial cells acquire an epithelial-mesenchymal-transition phenotype during long-term infection by the oral opportunistic pathogen, Porphyromonas gingivalis. Front Cell Infect Microbiol. (2017) 7:493. doi: 10.3389/fcimb.2017.00493
22. Utispan K, Pugdee K, Koontongkaew S. Porphyromonas gingivalis lipopolysaccharide-induced macrophages modulate proliferation and invasion of head and neck cancer cell lines. Biomed Pharmacother. (2018) 101:988–95. doi: 10.1016/j.biopha.2018.03.033
23. Wu JS, Zheng M, Zhang M, Pang X, Li L, Wang SS, et al. Porphyromonas gingivalis promotes 4-Nitroquinoline-1-Oxide-induced oral carcinogenesis with an alteration of fatty acid metabolism. Front Microbiol. (2018) 9:2081. doi: 10.3389/fmicb.2018.02081
24. Berraondo P, Minute L, Ajona D, Corrales L, Melero I, Pio R. Innate immune mediators in cancer: between defense and resistance. Immunol Rev. (2016) 274:290–306. doi: 10.1111/imr.12464
25. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell (2010) 140:883–99. doi: 10.1016/j.cell.2010.01.025
26. Gao J, Zhao S, Halstensen TS. Increased interleukin-6 expression is associated with poor prognosis and acquired cisplatin resistance in head and neck squamous cell carcinoma. Oncol Rep. (2016) 35:3265–74. doi: 10.3892/or.2016.4765
27. Hussein AA, Forouzanfar T, Bloemena E, de Visscher J, Brakenhoff RH, Leemans CR, et al. A review of the most promising biomarkers for early diagnosis and prognosis prediction of tongue squamous cell carcinoma. Br J Cancer (2018) 119:724–36. doi: 10.1038/s41416-018-0233-4
28. Domingues C, Serambeque BP, Laranjo Candido MS, Marto CMM, Veiga FJB, Sarmento Antunes Cruz Ribeiro AB, et al. Epithelial-mesenchymal transition and microRNAs: challenges and future perspectives in oral cancer. Head Neck (2018) 40:2304–13. doi: 10.1002/hed.25381
29. Herath TD, Darveau RP, Seneviratne CJ, Wang CY, Wang Y, Jin L. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-kappaB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS ONE (2013) 8:e58496. doi: 10.1371/journal.pone.0058496
30. Liu J, Wang Y, Ouyang X. Beyond toll-like receptors: Porphyromonas gingivalis induces IL-6, IL-8, and VCAM-1 expression through NOD-mediated NF-kappaB and ERK signaling pathways in periodontal fibroblasts. Inflammation (2014) 37:522–33. doi: 10.1007/s10753-013-9766-0
31. Ding PH, Darveau RP, Wang CY, Jin L. 3LPS-binding protein and its interactions with P. gingivalis LPS modulate pro-inflammatory response and Toll-like receptor signaling in human oral keratinocytes. PLoS ONE (2017) 12:e0173223. doi: 10.1371/journal.pone.0173223
32. Bundy LM, Sealy L. CCAAT/enhancer binding protein beta (C/EBPbeta)-2 transforms normal mammary epithelial cells and induces epithelial to mesenchymal transition in culture. Oncogene (2003) 22:869–83. doi: 10.1038/sj.onc.1206216
33. Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer (2017) 17:116–30. doi: 10.1038/nrc.2016.124
34. Bai Y, Wei Y, Wu L, Wei J, Wang X, Bai Y. C/EBP beta mediates endoplasmic reticulum stress regulated inflammatory response and extracellular matrix degradation in LPS-stimulated human periodontal ligament cells. Int J Mol Sci. (2016) 17:385. doi: 10.3390/ijms17030385
35. Olbrich P, Freeman AF. STAT1 and STAT3 mutations: important lessons for clinical immunologists. Expert Rev Clin Immunol. (2018) 14:1029–41. doi: 10.1080/1744666X.2018.1531704
36. Concha-Benavente F, Srivastava RM, Trivedi S, Lei Y, Chandran U, Seethala RR, et al. Identification of the cell-intrinsic and -extrinsic pathways downstream of EGFR and IFNgamma that induce PD-L1 expression in head and neck cancer. Cancer Res. (2016) 76:1031–43. doi: 10.1158/0008-5472.CAN-15-2001
38. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity (2018) 48:434–52. doi: 10.1016/j.immuni.2018.03.014
39. Nguyen PH, Fedorchenko O, Rosen N, Koch M, Barthel R, Winarski T, et al. LYN kinase in the tumor microenvironment is essential for the progression of chronic lymphocytic leukemia. Cancer Cell (2016) 30:610–22. doi: 10.1016/j.ccell.2016.09.007
40. Mao L, Deng WW, Yu GT, Bu LL, Liu JF, Ma SR, et al. Inhibition of SRC family kinases reduces myeloid-derived suppressor cells in head and neck cancer. Int J Cancer (2017) 140:1173–85. doi: 10.1002/ijc.30493
41. Groeger S, Domann E, Gonzales JR, Chakraborty T, Meyle J. B7-H1 and B7-DC receptors of oral squamous carcinoma cells are upregulated by Porphyromonas gingivalis. Immunobiology (2011) 216:1302–10. doi: 10.1016/j.imbio.2011.05.005
42. Ma H, Yang W, Zhang L, Liu S, Zhao M, Zhou G, et al. Interferon-alpha promotes immunosuppression through IFNAR1/STAT1 signalling in head and neck squamous cell carcinoma. Br J Cancer (2018) 120:317–30. doi: 10.1038/s41416-018-0352-y
43. Kupferman ME, Jiffar T, El-Naggar A, Yilmaz T, Zhou G, Xie T, et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene (2010) 29:2047–59. doi: 10.1038/onc.2009.486
44. de Moraes JK, Wagner VP, Fonseca FP, Vargas PA, de Farias CB, Roesler R, et al. Uncovering the role of brain-derived neurotrophic factor/tyrosine kinase receptor B signaling in head and neck malignancies. J Oral Pathol Med. (2018) 47:221–7. doi: 10.1111/jop.12611
45. Schulte-Herbruggen O, Nassenstein C, Lommatzsch M, Quarcoo D, Renz H, Braun A. Tumor necrosis factor-alpha and interleukin-6 regulate secretion of brain-derived neurotrophic factor in human monocytes. J Neuroimmunol. (2005) 160:204–9. doi: 10.1016/j.jneuroim.2004.10.026
46. Correa JD, Pereira DS, Madeira MF, Queiroz-Junior CM, Souza DG, Teixeira MM, et al. Brain-derived neurotrophic factor in chronic periodontitis. Mediators Inflamm. (2014) 2014:373765. doi: 10.1155/2014/373765
47. Sasaki S, Takeda K, Takewaki M, Ouhara K, Kajiya M, Mizuno N, et al. BDNF/HMW-HA complex as an adjunct to nonsurgical periodontal treatment of ligature-induced periodontitis in dogs. J Periodontol. (2018) 90:98–109. doi: 10.1002/JPER.18-0070
48. Cho MS, Vasquez HG, Rupaimoole R, Pradeep S, Wu S, Zand B, et al. Autocrine effects of tumor-derived complement. Cell Rep. (2014) 6:1085–95. doi: 10.1016/j.celrep.2014.02.014
49. Jessie K, Jayapalan JJ, Ong KC, Abdul Rahim ZH, Zain RM, Wong KT, et al. Aberrant proteins in the saliva of patients with oral squamous cell carcinoma. Electrophoresis (2013) 34:2495–502. doi: 10.1002/elps.201300107
50. Chen Y, Azman SN, Kerishnan JP, Zain RB, Chen YN, Wong YL, et al. Identification of host-immune response protein candidates in the sera of human oral squamous cell carcinoma patients. PLoS ONE (2014) 9:e109012. doi: 10.1371/journal.pone.0109012
51. Kawahara R, Bollinger JG, Rivera C, Ribeiro AC, Brandao TB, Paes Leme AF, et al. A targeted proteomic strategy for the measurement of oral cancer candidate biomarkers in human saliva. Proteomics (2016) 16:159–73. doi: 10.1002/pmic.201500224
52. Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. (2007) 178:7242–50. doi: 10.4049/jimmunol.178.11.7242
53. Cremonesi E, Governa V, Garzon JFG, Mele V, Amicarella F, Muraro MG, et al. Gut microbiota modulate T cell trafficking into human colorectal cancer. Gut (2018) 67:1984–94. doi: 10.1136/gutjnl-2016-313498
54. Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - a target for novel cancer therapy. Cancer Treat Rev. (2018) 63:40–7. doi: 10.1016/j.ctrv.2017.11.007
55. Mantovani A, Savino B, Locati M, Zammataro L, Allavena P, Bonecchi R. The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev. (2010) 21:27–39. doi: 10.1016/j.cytogfr.2009.11.007
56. Jin WJ, Kim B, Kim D, Park Choo HY, Kim HH, Ha H, et al. NF-kappaB signaling regulates cell-autonomous regulation of CXCL10 in breast cancer 4T1 cells. Exp Mol Med. (2017) 49:e295. doi: 10.1038/emm.2016.148
57. Guo X, Wang Y, Wang C, Chen J. Identification of several hub-genes associated with periodontitis using integrated microarray analysis. Mol Med Rep. (2015) 11:2541–7. doi: 10.3892/mmr.2014.3031
58. Zhou H, Wu J, Wang T, Zhang X, Liu D. CXCL10/CXCR3 axis promotes the invasion of gastric cancer via PI3K/AKT pathway-dependent MMPs production. Biomed Pharmacother. (2016) 82:479–88. doi: 10.1016/j.biopha.2016.04.069
59. Sun W, Qiu Z, Huang W, Cao M. Gene expression profiles and proteinprotein interaction networks during tongue carcinogenesis in the tumor microenvironment. Mol Med Rep. (2018) 17:165–71. doi: 10.3892/mmr.2017.7843
60. Xia J, Wang J, Chen N, Dai Y, Hong Y, Chen X, et al. Expressions of CXCR7/ligands may be involved in oral carcinogenesis. J Mol Histol. (2011) 42:175–80. doi: 10.1007/s10735-011-9322-x
61. Allaoui R, Bergenfelz C, Mohlin S, Hagerling C, Salari K, Werb Z, et al. Cancer-associated fibroblast-secreted CXCL16 attracts monocytes to promote stroma activation in triple-negative breast cancers. Nat Commun. (2016) 7:13050. doi: 10.1038/ncomms13050
Keywords: Porphyromonas gingivalis, chronic infection, inflammation, oral cancer, bioinformatics
Citation: Geng F, Wang Q, Li C, Liu J, Zhang D, Zhang S and Pan Y (2019) Identification of Potential Candidate Genes of Oral Cancer in Response to Chronic Infection With Porphyromonas gingivalis Using Bioinformatical Analyses. Front. Oncol. 9:91. doi: 10.3389/fonc.2019.00091
Received: 10 October 2018; Accepted: 31 January 2019;
Published: 21 February 2019.
Edited by:
Dietmar Thurnher, Medical University of Graz, AustriaReviewed by:
Soraya Castro Trindade, State University of Feira de Santana, BrazilWenhui Liu, First Affiliated Hospital of Zhengzhou University, China
Copyright © 2019 Geng, Wang, Li, Liu, Zhang, Zhang and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaping Pan, eXBwYW5AY211LmVkdS5jbg==
†These authors share co-first authorship
 Fengxue Geng
Fengxue Geng Qingxuan Wang2†
Qingxuan Wang2† Chen Li
Chen Li Dongmei Zhang
Dongmei Zhang Yaping Pan
Yaping Pan