- 1Clinic of Pediatric Onco-Hematology, Department of Women's and Children's Health, University of Padova, Padova, Italy
- 2Istituto di Ricerca Pediatrica Città della Speranza, Padova, Italy
- 3Department of Molecular Medicine, University of Padova, Padova, Italy
- 4Gene Therapy Program, Dana Farber/Boston Children's Cancer and Blood Disorders Centers, Boston, MA, United States
- 5CRIBI Interdepartmental Research Center for Innovative Biotechnologies (CRIBI), University of Padova, Padova, Italy
Emerging evidence indicates that extracellular vesicles, particularly exosomes, play a role in several biological processes and actively contribute to cancer development and progression, by carrying and delivering proteins, transcripts and small RNAs (sRNAs). There is high interest in studying exosomes of cancer patients both to develop non-invasive liquid biopsy tests for risk stratification and to elucidate their possible involvement in disease mechanisms. We profiled by RNA-seq the sRNA content of circulating exosomes of 20 pediatric patients with Anaplastic Large Cell Lymphoma (ALCL) and five healthy controls. Our analysis disclosed that non-miRNA derived sRNAs constitute the prominent fraction of sRNA loaded in exosomes and identified 180 sRNAs significantly more abundant in exosomes of ALCL patients compared to controls. YRNA fragments, accounting for most of exosomal content and being significantly increased in ALCL patients, were prioritized for further investigation by qRT-PCR. Quantification of RNY4 fragments and full-length sequences disclosed that the latter are massively loaded into exosomes of ALCL patients with more advanced and aggressive disease. These results are discussed in light of recent findings on the role of RNY4 in the modulation of tumor microenvironment.
Introduction
Extracellular vesicles (EVs) are cell-derived membrane particles secreted from many cell types and circulating in body fluids, including plasma. Among different classes of EVs of different size and intracellular origin, exosomes are 40–150 nm endosome-derived EV originating from the inward budding of the limiting membrane of multivesicular bodies (1). In this process, exosomes are packed with proteins, lipids, DNAs, messenger RNAs (mRNAs) and non-coding RNAs, which can be transferred to recipient cells, and function both as paracrine and endocrine factors (2).
A large body of evidence collected in the last years proved the functional involvement of exosomes in cancer progression and spreading, induction of angiogenesis, as well as in chemoresistance and immune response evasion during tumor development (3). In this scenario, defining the peculiarities of exosomal cargo in cancer patients is a hot topic in biomedical research. The characterization of small non-coding RNAs (sRNAs) in plasmatic exosomes of cancer patients attracted interest for the identification of non-invasive disease biomarkers and, notably, in consideration of sRNA regulatory functions and their direct involvement in cancer mechanisms (4, 5).
Most functional studies on circulating sRNAs carried by tumor-derived exosomes were focused on microRNAs (miRNAs) because of their well-characterized regulatory roles in key signaling axes: exosome-delivered miRNAs have been shown to promote epithelial-mesenchymal transition (6, 7), induce angiogenesis and increase vascular leakage (8–10), prepare pre-metastatic niches to promote metastasis (11, 12) or induce tumor resistance to immune responses (13, 14).
Non-Hodgkin lymphoma (NHL) is a heterogeneous group of lymphoid malignancies and the fourth most common malignancy across the pediatric age spectrum. Considerable progress has been achieved in developing curative therapy for pediatric NHL, with an overall survival rate now exceeding 80% (15). There are three major categories of NHL: mature B-cell neoplasms, Lymphoblastic Lymphoma, and Anaplastic Large Cell Lymphoma (ALCL). Other NHL subtypes, including peripheral T-cell lymphomas, follicular lymphomas, and rare entities, represent <3% of the cases (16).
ALCL accounts for 10–15% of pediatric and adolescent NHL. Differently from ALCL in adults, ALCL in children is nearly universally ALK-positive and, in almost all of the cases, it is characterized by the t(2;5)(p23;q35) translocation, which leads to the constitutive expression of the NPM-ALK fusion protein (17). Although current treatment strategies achieve an event-free survival (EFS) of ~75% after 5 years, about 30% of the patients are resistant to therapy or experience a relapse (18). In this clinical context, new disease biomarkers are needed to enable the early identification of high-risk patients and a better tailoring of treatment. In ALCL, the identification of new active players in lymphomagenesis and in cancer cells dissemination mechanisms would have high potential for the design of innovative therapeutic interventions.
Currently, only a few, mainly descriptive, studies reported data regarding the sRNA cargo of lymphoma-derived exosomes and EVs. Moreover, only Diffuse Large B-cell Lymphoma (DLBCL), which is the most frequent histological subtype presenting in adults (19, 20), was investigated.
Studies aiming at the identification of clinically relevant sRNAs in plasmatic exosomes/EVs from lymphoma patients focused on miRNAs (21). The first evidence on EV miRNAs as a molecular diagnostic tool for disease monitoring in Hodgkin lymphoma (HL) patients was reported by Eijndhoven et al. (22). Specifically, lymphoma-associated miR-21-5p, miR-127-3p, miR-24-3p, let-7a-5p, and miR-155-5p were significantly increased in plasmatic EV from HL patients compared to healthy donors (HD). Exosome-derived miRNAs were also proposed as predictive biomarkers of chemotherapy resistance in DLBCL, where increased levels of miR-99a-5p and miR-125b-5p in patient plasmatic exosomes were associated with reduced progression-free survival (23). Data on exosomal sRNAs in pediatric lymphomas setting are currently missing.
Further, our appreciation of the small transcriptome complexity largely increased in the last years. Alternative processing of miRNA precursors (24–26) and housekeeping non-coding RNAs can generate miRNA-like sRNAs that can be functional and play roles in malignancies (27, 28). It's worth noting that several RNA-seq studies on exosomes derived from both tumoral and non-tumoral cell lines revealed that, differently from secreting cells, miRNAs constitute a minor percentage of EV-enclosed RNA. Besides protein coding mRNA, the EV fractions contain vault RNAs, YRNAs, small nuclear and nucleolar RNAs (snRNAs and snoRNAs), transfer RNAs (tRNA), as well as fragments deriving from long non-coding RNAs and transcribed pseudogenes (29–31).
In this perspective article, we present original data about the characterization of exosomal sRNAs in pediatric ALCL aiming attention on non-miRNA derived sRNAs, and discuss them in the frame of current literature, providing an original viewpoint of the possible translational relevance of these findings.
RNY4 Fragment Enrichment In Circulating Exosomes Of Alcl Patients Discovered By RNA-seq
To characterize the exosomal load of sRNAs in ALCL patients and disclose differences with healthy donors, sRNAs were examined by small RNA-seq of exosomes from 20 ALK-positive ALCL patients and five HD plasma samples, and from supernatant of five ALCL cell lines (Karpas299, SUDHL1, and SUP-M2, ALK-positive; FE-PD and MAC2A, ALK-negative). For patients, paired biopsy samples, all positive for the NPM-ALK fusion, were also sequenced. Exosomal RNA was extracted by using exoRNeasy Maxi/Midi kit (Qiagen) and assessed for proper amount and quality by Agilent 2100 Bioanalyzer (Agilent Technologies). RNA-seq libraries were prepared with NEBNext Multiplex Small RNA Library Prep Kit for Illumina (New England Biolabs), as previously reported (32), and sequenced on an Illumina HiSeq 4000 platform with single-end reads and average depth of 15 and 30 M for exosomal and biopsy samples, respectively.
After a preprocessing phase for adapter trimming and selection of high-quality reads (Qphred ≥ 30), data underwent analysis by miR&moRe software (24, 25). Briefly, miR&moRe maps filtered reads to genome assembly and the known hairpins sequences from miRBase extended in either directions by additional 30 bp and allows detection and prediction of miRNA hairpins and of the corresponding mature forms, as in Gaffo et al. (33). MiR&moRe allowed identification and quantification of 1,194 and 523 miRNA-derived sRNA species in biopsies and exosomal samples, including miRNAs and microRNA-offset RNAs derived from annotated and newly predicted miRNA precursors.
We observed that in biopsies ~59% of the reads derived from miRNAs, whereas the large majority of the reads from exosomal samples did not align to known nor predicted miRNA precursors, concordantly in exosomes isolated from ALCL patients (2.5%), cell lines (2.6%), and healthy donors (5.3%) (Figure 1A). A similarly large fraction of non-miRNA sRNAs in EVs was described by other studies on lymphoma, melanoma, breast cancer and immune cells (19, 20, 29–31).
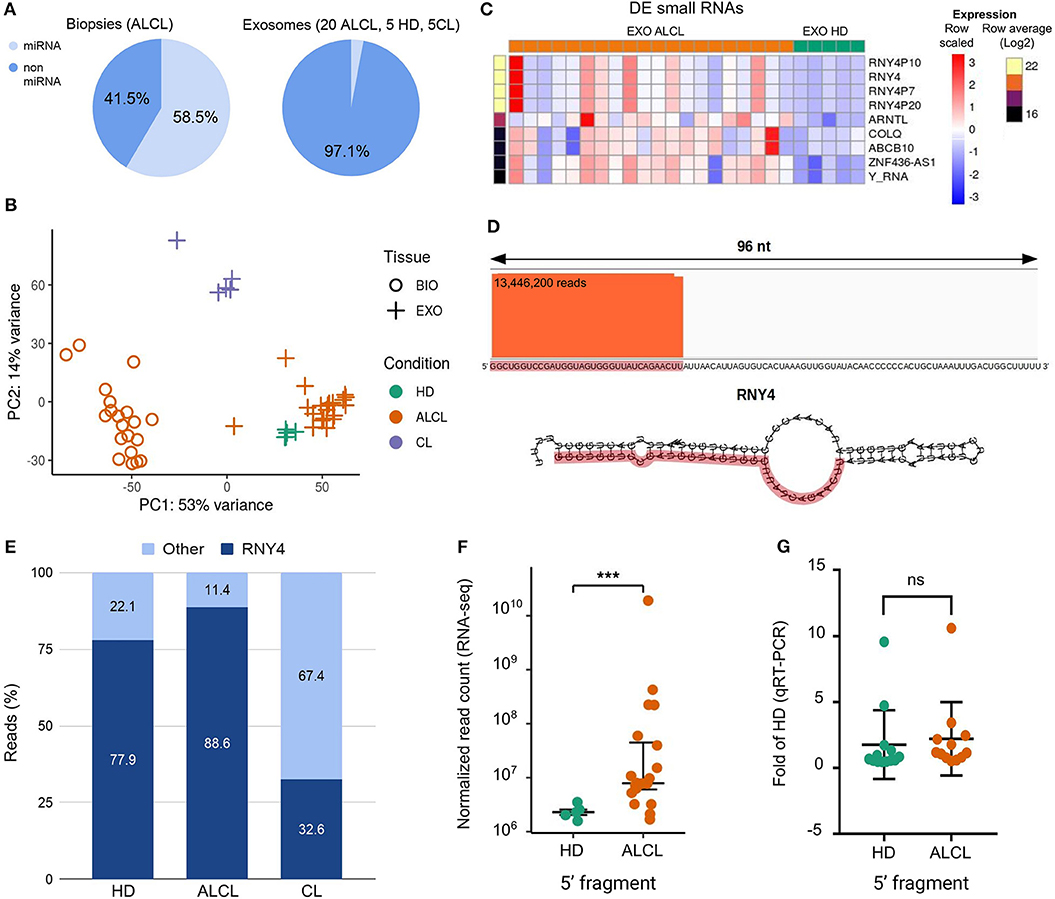
Figure 1. Characterization of exosomal small RNAs in ALCL patients. (A) Average proportion of sequence reads aligned to and outside miRNA precursors in anaplastic large cell lymphoma (ALCL) primary tumor biopsies, and in exosomes from ALCL patients, healthy donors (HD) and ALCL cell lines (CL). (B) Principal component analysis computed on expression profiles of the 1,007 most abundant (top 5% of expression in at least one sample) non miRNA-derived sRNAs. (C) Heatmap of expression profiles in exosome samples of the nine sRNAs most abundant (top 1%) and most varied comparing ALCL and HD (LFC>2). (D) Sequence reads alignment to RNY4 shows reads stacked on the RNY4 5′ end; RNY4 predicted secondary structure is displayed below, highlighting in red the 32 nt fragment identified through RNA-seq. (E) Average percentages of non-miRNA RNA-seq reads from exosomes samples mapping to RNY4 locus and to other putative sRNAs. (F) Box-plot of RNY4-5′F expression in HD (n = 5) and ALCL (n = 20) exosomes, according to RNA-seq data. (G) Box-plot of RNY4-5′F expression measured by qRT-PCR in HD (n = 13) and ALCL (n = 12) exosomes [***DESeq2 p.adj < 0.001 independent samples from those shown in (F), expression in ALCL relative to average in HD; comparative delta Ct method (2−ΔΔCt); miR-26a-5p used as endogenous control; Mann-Whitney].
The non-miRNA sRNAs fraction in exosomes of ALCL patients was further characterized. First, reads not aligned to known or predicted miRNA precursors were mapped with Bowtie v1.1.2 (34) to the reference genome, allowing no mismatches and up to 15 multiple alignments. The alignments were then analyzed with derfinder software tool (35) to identify expressed RNAs and the corresponding genomic regions. Among the 9,181 regions supported by at least 10 reads we selected those of 13–50 contiguous bases, consistently with the size of sequencing library fragments. After expression normalization [DESeq2 (36)], 1,007 most abundant (top 5% of expression) putative non-miRNA-derived sRNAs were considered for further characterization. Principal component analysis of expression profiles of these 1,007 sRNAs clearly distinguished tissue from exosome samples, separating as well as exosomes from healthy donors, patients and cell lines (Figure 1B).
Next, 180 sRNAs were identified with significantly different abundance between exosomes of ALCL patients and HD (DESeq2 p.adj < 0.001). The heatmap in Figure 1C shows the expression of the nine sRNAs with highest abundance and increase in ALCL exosomes. Apparently, the sRNA present in greater supply in ALCL exosomes was defined by reads aligned to the RNY4 gene, and to highly similar pseudogenes (RNY4P7, RNY4P10, and RNY4P20). Precisely, a fragment corresponding to the first 32 bases on the 5′ end of RNY4 (RNY4-5′F) (Figure 1D) accounted for at least 80% of the non-miRNA sRNA expression in exosomes, whereas it was less abundant in exosomes derived from the cell line supernatant, isolated with the same protocol (Figure 1E). Remarkably, RNY4-5′F was five times and significantly more abundant in ALCL than in HD exosomes (p.adj = 0.0003; Figure 1F). The expression of RNY4-5′F was assessed by quantitative real-time PCR (qRT-PCR; Custom TaqMan Small RNA assay designed on the 32 bases of the RNY4 sequence; ThermoFisher Scientific, Life Technologies) in 25 independent samples (12 ALCL and 13 HD). No difference was observed comparing exosomes from ALCL and HD (Figure 1G) not confirming RNA-seq result (Figure 1F).
YRNAs were first discovered in 1981 as 83–112 nt RNA components of circulating ribonucleoproteins, complexed to Ro60 and La autoantigens, in serum of patients with autoimmune diseases (37, 38). Evolutionary conserved in vertebrates (39), YRNAs fold in characteristic stem-loop secondary structures, with lower and upper stem loop sequences being the most conserved (40). Four different human YRNAs (RNY1, RNY3, RNY4, and RNY5) are transcribed in the nucleus by RNA polymerase III from genes clustered together at a single locus on chromosome 7q36 (41). Intracellularly, binding of the lower YRNA stem to Ro60 was shown to be involved in the maintenance of RNA stability and in cellular response to stress (42), whereas the upper stem was proven to be essential for the initiation of chromosomal DNA replication (43). In addition, YRNAs were also linked to alternative splicing and regulation of the translation of specific RNAs, since most YRNA-associated proteins are implicated in these processes (44).
In recent years, YRNAs and YRNA fragments derived from site-specific cleavage by RNase L were reported to be enriched in different types of EV compared to secreting cancer cells (19, 31, 45–47). Noteworthy, RNA fragments corresponding to the 5′ region of the RNY4, almost exactly corresponding to the RNY4-5′F detected in the present study, were shown to be the most abundant sRNA species in plasma samples from HD (30, 48–50) and melanoma patients (51), as well as in breast cancer patients' exosomes and plasma (30, 49), and plasmatic exosomes from non-small cell lung cancer (NSCLC) and chronic lymphocytic leukemia (CLL) patients (52, 53).
Since YRNA fragments derive from conserved ends of the YRNA hairpin, it was initially hypothesized that YRNAs could “conceal miRNAs” and be processed in miRNA-sized YRNA fragments that could function as miRNAs (54). However, this hypothesis was not supported by later studies. Since YRNA fragment biogenesis resulted to be Dicer-independent, they were found in complexes different from those associated with microRNAs and they did not co-immunoprecipitate with Ago2 (55). Moreover, they did not regulate targets tested by Thomson and colleagues in a miRNA-like manner (56).
Recently, since RNY5 fragments administration to human primary fibroblasts was shown to induce cell death (46) a role for RNY3 in enhancing “cleavage and polyadenylation specificity factor” (CPSF) recruitment to histone locus bodies has been proposed (57), thus associating YRNA fragments to functional activities different from those typical of miRNAs.
Genuine miRNA-Like RNY4 Fragments OR Full-Length RNY4?
The increased levels of RNY4-derived fragments or full-length transcripts circulating in plasma or exosomes of cancer patients compared to HD (30, 49, 53) triggered interest in the potential use of RNY4 as a cancer biomarker. However, whether RNY4-derived fragments detected by small RNA-seq are genuine fragments or reflect the presence of the full-length RNY4 is still a matter of debate.
By Northern blotting, Dhahbi et al. confirmed 5′ RNY4 fragments in plasma of HD (48), Haderk et al. validated the presence of both RNY4 full-length and 5′ fragments in CLL exosomes (53), whereas Driedonks et al. showed that EV released from dendritic cells mostly contain full-length RNY1 and only small amounts of 19–35 nt RNY1 fragments (58). In this regard, Driedonks and Nolte-'t Hoen suggested that YRNA secondary structures might impede full-length cDNA synthesis, leading to overestimation of fragmented non-coding RNA in sequencing data (59). Indeed, in a RNA-seq study, Godoy et al. detected fragments derived from both the 5′ and 3′ arms (60) whereas other works reported mostly full-length YRNAs (61, 62).
To verify if full-length RNY4 might be responsible for the differential expression detected by small RNA-seq, not validated by RT-PCR specific from RNY4-5′F, we quantified the full-length RNY4 in 12 ALCL and 12 HD plasmatic exosomes by qRT-PCR using primers from Tolkach et al. (63). The full-length RNY4 was significantly more abundant in ALCL than in HD (Mann Whitney, p = 0.017) (Figure 2A).
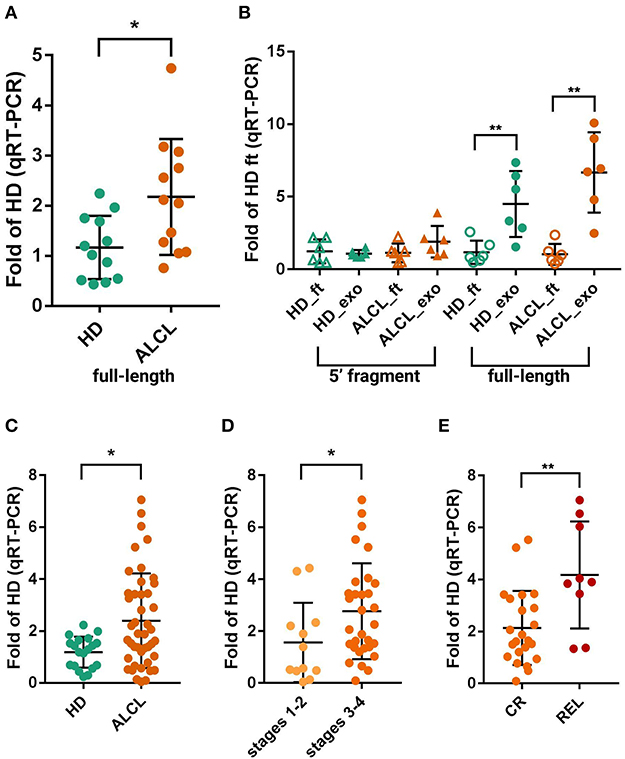
Figure 2. Quantification of RNY4-5′F and full-length RNY4 by qRT-PCR. (A) Expression of full-length RNY4 in 12 HD and 12 ALCL plasmatic exosomes measures by qRT-PCR. (B) Expression of RNY4-5′F (5′ fragment) and full-length RNY4 loaded into exosomes (exo) and freely circulating in plasma (flow-through, ft); expression in ALCL relative to average in HD_ft. Full-length RNY4 was significantly more abundant in both HD and ALCL exosomes than as free circulating RNA (6 HD, 6 ALCL), whereas the RNY4-5′F was almost equally distributed inside exosomes and as free circulating sRNA in both HD and ALCL. (C) Evaluation of full-length RNY4 in exosomes from an extended cohort of 44 ALCL and 19 HD confirmed a significantly increased expression of the full-length form in ALCL compared to HD. Full-length RNY4 was significantly more abundant in patients with stage 3–4 disease compared to those in stages 1–2 (D) and also among stage 3–4, in relapsed patients (REL) compared to those in stable complete remission (CR) (E). For panels (A,C,D,E), expression in ALCL has been calculated relative to average in HD; comparative delta Ct method (2−ΔΔCt), miR-26a-5p as endogenous control, Mann-Whitney test for sample comparison were used for all panels. (*0.01 < p < 0.05, **p < 0.01).
Further, we considered that full-length YRNAs were mainly reported as contained in exosomes (59), whereas fragments were previously validated by Northern blotting in total plasma and in exosomes (48, 53). We thus investigated the amount of full-length RNY4 and the RNY4-5′F, also distinguishing RNA loaded into exosomes and freely circulating in plasma (RNA of six HD and six ALCL from both exosomes and the column flow-through after the exosome binding step). Of note, most of the full-length RNY4 was inside exosomes (5.6 times more abundant inside, on average) of both HD (Mann-Whitney p = 0.009) and ALCL patient (p = 0.002) samples, whereas the fragment was present at a similar level in exosomes and as free circulating RNA (Figure 2B). Taken together, these results suggested that bona fide RNY4 fragments circulate in plasma both enclosed in membranes and as free RNAs, in amounts not discriminating ALCL and HD, whereas full-length RNY4 is mainly enclosed in exosomes, where it is significantly enriched in ALCL patients.
A further examination of an extended cohort of 44 ALCL and 19 HD plasmatic exosomes confirmed the upregulation of the full-length form in ALCL samples (Mann Whitney, p = 0.017) (Figure 2C).
Full-Length RNY4 Load In Exosomes Of ALCL Patients Correlates with Disease Aggressiveness
Next, the extended cohort was further examined considering clinical data. Of importance, RNY4 abundance correlated with ALCL patients' clinical characteristics. The full-length RNY4 was more abundant in exosomes of ALCL patients with advanced disease stages (32 ALCL in 3–4 stage vs. 12 ALCL in 1–2 stage; Mann Whitney, p = 0.049) (Figure 2D). Since most (9/10) of the relapsed patients were diagnosed in stages 3–4 (Figure 2D), we analyzed RNY4 amount at diagnosis in relation to relapse, considering only advanced stages. Compared to cases in stable complete remission (N = 23), relapsed patients (N = 9) presented at diagnosis with increased levels of exosomal full-length RNY4 (Mann Whitney, p = 0.0065) (Figure 2E).
These findings indicate exosomal RNY4 as a promising biomarker of disease aggressiveness in ALCL, to be quantified with a simple and non-invasive liquid biopsy. Moreover, our data and literature evidence collectively encourage further investigation to ascertain a possible functional role of RNY4 in ALCL disease aggressiveness, as well as in other lymphoproliferative diseases or different malignancies. Indeed, RNY4 delivery by CLL exosomes has been recently shown to induce key leukemia-associated phenotypes in monocytes, such as the release of pro-tumorigenic cytokines (CCL2, CCL4, and IL-6) and the expression of the immunosuppressive protein PD-L1, thus generating a tumor-supporting microenvironment (53). ALCL tumors are characterized by variable histological patterns, mostly depending on tumor cell size and the presence of a large number of reactive histiocytes in the background (64). The biological functions of YRNAs could be multidirectional and we speculate that the association between ALCL and changes in these circulating YRNA reflects some aspects of either the biology of the tumor or the immunosystem reaction of the individual to the tumor. In particular, our results pave the way for investigating the role of RNY4 as mediator of immunoescape in lymphoma patients. The treatment of monocytes ex vivo with tumor exosomes, the uptake as well as exosome-mediated responses by flow cytometry, or cytokine quantification can be used in the next future to elucidate this intriguing aspect.
In conclusion, RNY4 is a massively loaded molecule in exosomes of ALCL patients, with RNY4 significantly increased in patients compared to controls. Notably, significantly higher RNY4 levels were observed in patients diagnosed at advanced stages, and among them, in those that later relapsed. These findings, in the light of available functional data on exosomal RNY4, encourage further study of RNY4 involvement in ALCL tumor microenvironment and disease aggressiveness.
Data Availability Statement
Publicly available datasets were analyzed in this study, these can be found in the NCBI Gene Expression Omnibus (GSE144781).
Ethics Statement
The study was approved by the ethics committee for clinical experimentation of the Padova hospital CESC (Comitato Etico per la sperimentazione clinica azienda ospedaliera di Padova). Written informed consent was obtained from patients and/or their legal guardians in accordance with the institution's ethical review boards.
Author Contributions
FL designed the RNA-seq experiment and the experimental work, analyzed data, and wrote the manuscript. PD performed RNA-seq data analysis and qRT-PCR, and contributed to wrote the manuscript. EG contributed to RNA-seq data analysis and revised the manuscript. PD, FL, and EG prepared figures. CD, AG, and IG processed clinical samples and performed qRT-PCR. EC and MP collected clinical data and commented on manuscript. AB revised the manuscript. SB conceived the study, supervised the bioinformatics work, contributed to experimental results interpretation, and wrote the manuscript. LM conceived the study, supervised the experimental work, and revised the manuscript.
Funding
This work has been supported by Fondazione CA.RI.PA.RO, Padova, Italy (grant 17/03 to LM), Fondazione Roche, Roma, Italy (Roche per la Ricerca 2018 to FL), Fondazione Umberto Veronesi, Milano, Italy (fellowships to FL and EG), AIRC, Milano, Italy (Investigator Grant – IG 2018 #21385 to LM) and by Camera di Commercio Venezia, Venezia, Italy. This study was also partially supported by AIRC, Milano, Italy (Investigator Grant – IG 2017 #20052 to SB), by the Department of Molecular Medicine of the University of Padova, Italy (PRID 2017 to SB), and by Italian Ministry of Education, Universities and Research (PRIN 2017 #2017PPS2X4_003 to SB).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank all the AIEOP centers for clinical samples and data collection and Elisa Tosato for technical assistance.
References
1. Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. (2016) 36:301–12. doi: 10.1007/s10571-016-0366-z
2. Maia J, Caja S, StranoMoraes MC, Couto N, Costa-Silva B. Exosome-based cell-cell communication in the tumor microenvironment. Front Cell Dev Biol. (2018) 6:18. doi: 10.3389/fcell.2018.00018
3. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
4. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. (2011) 12:861–74. doi: 10.1038/nrg3074
5. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. (2018) 18:5–18. doi: 10.1038/nrc.2017.99
6. Xiao D, Barry S, Kmetz D, Egger M, Pan J, Rai SN, et al. Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. (2016) 376:318–27. doi: 10.1016/j.canlet.2016.03.050
7. Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: role of microRNA-210. Cancer BiolTher. (2016) 17:1062–9. doi: 10.1080/15384047.2016.1219815
8. Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. (2018) 37:2873–89. doi: 10.1038/s41388-018-0183-6
9. Fang J-H, Zhang Z-J, Shang L-R, Luo Y-W, Lin Y-F, Yuan Y, et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. (2018) 68:1459–75. doi: 10.1002/hep.29920
10. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. (2018) 9:5395. doi: 10.1038/s41467-018-07810-w
11. Ono M, Kosaka N, Tominaga N, Yoshioka Y, Takeshita F, Takahashi R-U, et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal. (2014) 7:ra63. doi: 10.1126/scisignal.2005231
12. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. (2015) 17:183–94. doi: 10.1038/ncb3094
13. Ye S-B, Li Z-L, Luo D-H, Huang B-J, Chen Y-S, Zhang X-S, et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. (2014) 5: 5439–52. doi: 10.18632/oncotarget.2118
14. Hsu Y-L, Hung J-Y, Chang W-A, Jian S-F, Lin Y-S, Pan Y-C, et al. Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol Ther. (2018) 26:568–81. doi: 10.1016/j.ymthe.2017.11.016
15. Minard-Colin V, Brugières L, Reiter A, Cairo MS, Gross TG, Woessmann W, et al. Non-Hodgkin lymphoma in children and adolescents: progress through effective collaboration, current knowledge, and challenges ahead. J Clin Oncol. (2015) 33:2963–74. doi: 10.1200/JCO.2014.59.5827
16. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. (2016) 127:2391–405. doi: 10.1182/blood-2016-03-643544
17. Turner SD, Lamant L, Kenner L, Brugières L. Anaplastic large cell lymphoma in paediatric and young adult patients. Br J Haematol. (2016) 173:560–72. doi: 10.1111/bjh.13958
18. Le Deley M-C, Reiter A, Williams D, Delsol G, Oschlies I, McCarthy K, et al. Prognostic factors in childhood anaplastic large cell lymphoma: results of a large European intergroup study. Blood. (2008) 111:1560–6. doi: 10.1182/blood-2007-07-100958
19. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. (2014) 8:1649–58. doi: 10.1016/j.celrep.2014.08.027
20. Rutherford SC, Fachel AA, Li S, Sawh S, Muley A, Ishii J, et al. Extracellular vesicles in DLBCL provide abundant clues to aberrant transcriptional programming and genomic alterations. Blood. (2018) 132:e13–23. doi: 10.1182/blood-2017-12-821843
21. Li J, Tian T, Zhou X. The role of exosomal shuttle RNA (esRNA) in lymphoma. Crit Rev Oncol Hematol. (2019) 137:27–34. doi: 10.1016/j.critrevonc.2019.01.013
22. van Eijndhoven MA, Zijlstra JM, Groenewegen NJ, Drees EE, van Niele S, Baglio SR, et al. Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients. JCI Insight. (2016) 1:e89631. doi: 10.1172/jci.insight.89631
23. Feng Y, Zhong M, Zeng S, Wang L, Liu P, Xiao X, et al. Exosome-derived miRNAs as predictive biomarkers for diffuse large B-cell lymphoma chemotherapy resistance. Epigenomics. (2019) 11:35–51. doi: 10.2217/epi-2018-0123
24. Bortoluzzi S, Bisognin A, Biasiolo M, Guglielmelli P, Biamonte F, Norfo R, et al. Characterization and discovery of novel miRNAs and moRNAs in JAK2V617F-mutated SET2 cells. Blood. (2012) 119:e120–30. doi: 10.1182/blood-2011-07-368001
25. Agnelli L, Bisognin A, Todoerti K, Manzoni M, Taiana E, Galletti S, et al. Expanding the repertoire of miRNAs and miRNA-offset RNAs expressed in multiple myeloma by small RNA deep sequencing. Blood Cancer J. (2019) 9:21. doi: 10.1038/s41408-019-0184-x
26. Bortoluzzi S, Biasiolo M, Bisognin A. MicroRNA–offset RNAs (moRNAs): by-product spectators or functional players? Trends Mol Med. (2011) 17:473–4. doi: 10.1016/j.molmed.2011.05.005
27. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. (2013) 110:1404–9. doi: 10.1073/pnas.1206761110
28. Asikainen S, Heikkinen L, Juhila J, Holm F, Weltner J, Trokovic R, et al. Selective microRNA-offset RNA expression in human embryonic stem cells. PLoS ONE. (2015) 10:e0116668. doi: 10.1371/journal.pone.0116668
29. Nolte-'t Hoen ENM, Buermans HPJ, Waasdorp M, Stoorvogel W, Wauben MHM, 't Hoen PAC. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. (2012) 40:9272–85. doi: 10.1093/nar/gks658
30. Tosar JP, Gámbaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. (2015) 43:5601–16. doi: 10.1093/nar/gkv432
31. Lunavat TR, Cheng L, Kim D-K, Bhadury J, Jang SC, Lässer C, et al. Small RNA deep sequencing discriminates subsets of extracellular vesicles released by melanoma cells–evidence of unique microRNA cargos. RNA Biol. (2015) 12:810–23. doi: 10.1080/15476286.2015.1056975
32. Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. (2016) 6:19413. doi: 10.1038/srep19413
33. Gaffo E, Zambonelli P, Bisognin A, Bortoluzzi S, Davoli R. miRNome of Italian Large White pig subcutaneous fat tissue: new miRNAs, isomiRs and moRNAs. Anim Genet. (2014) 45:685–98. doi: 10.1111/age.12192
34. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. (2009) 10:R25. doi: 10.1186/gb-2009-10-3-r25
35. Collado-Torres L, Nellore A, Frazee AC, Wilks C, Love MI, Langmead B, et al. Flexible expressed region analysis for RNA-seq with derfinder. Nucleic Acids Res. (2017) 45:e9. doi: 10.1093/nar/gkw852
36. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
37. Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA. Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol. (1981) 1:1138–49. doi: 10.1128/MCB.1.12.1138
38. Lerner MR, Boyle JA, Hardin JA, Steitz JA. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. (1981) 211:400–2. doi: 10.1126/science.6164096
39. Mosig A, Guofeng M, Stadler BMR, Stadler PF. Evolution of the vertebrate Y RNA cluster. Theory Biosci. (2007) 126:9–14. doi: 10.1007/s12064-007-0003-y
40. Teunissen SW, Kruithof MJ, Farris AD, Harley JB, Venrooij WJ, Pruijn GJ. Conserved features of Y RNAs: a comparison of experimentally derived secondary structures. Nucleic Acids Res. (2000) 28:610–9. doi: 10.1093/nar/28.2.610
41. Maraia RJ, Sasaki-Tozawa N, Driscoll CT, Green ED, Darlington GJ. The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hYscRNA genes. Nucleic Acids Res. (1994) 22:3045–52. doi: 10.1093/nar/22.15.3045
42. Sim S, Wolin SL. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscip Rev RNA. (2011) 2:686–99. doi: 10.1002/wrna.85
43. Christov CP, Gardiner TJ, Szüts D, Krude T. Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol. (2006) 26:6993–7004. doi: 10.1128/MCB.01060-06
44. Perreault J, Perreault J-P, Boire G. Ro-associated Y RNAs in metazoans: evolution and diversification. Mol Biol Evol. (2007) 24:1678–89. doi: 10.1093/molbev/msm084
45. Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. (2017) 8:1145. doi: 10.1038/s41467-017-01196-x
46. Chakrabortty SK, Prakash A, Nechooshtan G, Hearn S, Gingeras TR. Extracellular vesicle-mediated transfer of processed and functional RNY5 RNA. RNA. (2015) 21:1966–79. doi: 10.1261/rna.053629.115
47. Donovan J, Rath S, Kolet-Mandrikov D, Korennykh A. Rapid RNase L–driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA. (2017) 23:1660–71. doi: 10.1261/rna.062000.117
48. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Mote P, Martin DIK. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. (2013) 45:990–8. doi: 10.1152/physiolgenomics.00129.2013
49. Dhahbi JM, Spindler SR, Atamna H, Boffelli D, Martin DIK. Deep sequencing of serum small RNAs identifies patterns of 5′ tRNA half and YRNA fragment expression associated with breast cancer. Biomark Cancer. (2014) 6:BIC.S20764. doi: 10.4137/BIC.S20764
50. Yeri A, Courtright A, Reiman R, Carlson E, Beecroft T, Janss A, et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci Rep. (2017) 7:44061. doi: 10.1038/srep44061
51. Solé C, Tramonti D, Schramm M, Goicoechea I, Armesto M, Hernandez LI, et al. The circulating transcriptome as a source of biomarkers for melanoma. Cancers. (2019) 11:E70 doi: 10.3390/cancers11010070
52. Li C, Qin F, Hu F, Xu H, Sun G, Han G, et al. Characterization and selective incorporation of small non-coding RNAs in non-small cell lung cancer extracellular vesicles. Cell Biosci. (2018) 8:2. doi: 10.1186/s13578-018-0202-x
53. Haderk F, Schulz R, Iskar M, Cid LL, Worst T, Willmund KV, et al. Tumor-derived exosomes modulate PD-L1 expression in monocytes. Sci Immunol. (2017) 2:eaah5509. doi: 10.1126/sciimmunol.aah5509
54. Verhagen APM, Pruijn GJM. Are the Ro RNP-associated Y RNAs concealing microRNAs? Y RNA-derived miRNAs may be involved in autoimmunity. Bioessays. (2011) 33:674–82. doi: 10.1002/bies.201100048
55. Langenberger D, Çakir MV, Hoffmann S, Stadler PF. Dicer-processed small RNAs: rules and exceptions. J Exp Zool B Mol Dev Evol. (2013) 320:35–46. doi: 10.1002/jez.b.22481
56. Thomson DW, Pillman KA, Anderson ML. Assessing the gene regulatory properties of Argonaute-bound small RNAs of diverse genomic origin. Nucleic Acids Res. (2014) 43:470–81. doi: 10.1093/nar/gku1242
57. Köhn M, Ihling C, Sinz A, Krohn K, Hüttelmaier S. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. (2015) 29:1998–2003. doi: 10.1101/gad.266486.115
58. Driedonks TAP, van der Grein SG, Ariyurek Y, Buermans HPJ, Jekel H, Chow FWN, et al. Immune stimuli shape the small non-coding transcriptome of extracellular vesicles released by dendritic cells. Cell Mol Life Sci. (2018) 75:3857–75. doi: 10.1007/s00018-018-2842-8
59. Driedonks TAP, Nolte-'t Hoen ENM. Circulating Y-RNAs in extracellular vesicles and ribonucleoprotein complexes; implications for the immune system. Front Immunol. (2018) 9:3164. doi: 10.3389/fimmu.2018.03164
60. Godoy PM, Bhakta NR, Barczak AJ, Cakmak H, Fisher S, MacKenzie TC, et al. Large differences in small RNA composition between human biofluids. Cell Rep. (2018) 25:1346–58. doi: 10.1016/j.celrep.2018.10.014
61. Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci USA. (2017) 114:E8987–95. doi: 10.1073/pnas.1712108114
62. Qin Y, Yao J, Wu DC, Nottingham RM, Mohr S, Hunicke-Smith S, et al. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA. (2016) 22:111–28. doi: 10.1261/rna.054809.115
63. Tolkach Y, Stahl AF, Niehoff E-M, Zhao C, Kristiansen G, Müller SC, et al. YRNA expression predicts survival in bladder cancer patients. BMC Cancer. (2017) 17:749. doi: 10.1186/s12885-017-3746-y
Keywords: ALCL, liquidbiopsy, exosomes, YRNA, RNA-seq, small RNA
Citation: Lovisa F, Di Battista P, Gaffo E, Damanti CC, Garbin A, Gallingani I, Carraro E, Pillon M, Biffi A, Bortoluzzi S and Mussolin L (2020) RNY4 in Circulating Exosomes of Patients With Pediatric Anaplastic Large Cell Lymphoma: An Active Player? Front. Oncol. 10:238. doi: 10.3389/fonc.2020.00238
Received: 21 November 2019; Accepted: 12 February 2020;
Published: 27 February 2020.
Edited by:
Basem M. William, The Ohio State University, United StatesReviewed by:
Stefano Aldo Pileri, University of Bologna, ItalyRehan Khan, Mayo Clinic Arizona, United States
Copyright © 2020 Lovisa, Di Battista, Gaffo, Damanti, Garbin, Gallingani, Carraro, Pillon, Biffi, Bortoluzzi and Mussolin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefania Bortoluzzi, c3RlZmFuaWEuYm9ydG9sdXp6aUB1bmlwZC5pdA==
†These authors have contributed equally to this work
 Federica Lovisa
Federica Lovisa Piero Di Battista1,2†
Piero Di Battista1,2† Enrico Gaffo
Enrico Gaffo Stefania Bortoluzzi
Stefania Bortoluzzi Lara Mussolin
Lara Mussolin