- 1Cell Signal Transduction Laboratory, Bioinformatics Department of Predictive Medicine, Bioinformatics Center, Henan Provincial Engineering Center for Tumor Molecular Medicine, School of Software, School of Basic Medical Sciences, Institute of Biomedical Informatics, Henan University, Kaifeng, China
- 2Department of Anesthesia, Stanford University, Stanford, CA, United States
- 3Henan Bioengineering Research Center, Zhengzhou, China
- 4Department of Thoracic Surgery, The Affiliated Nanshi Hospital of Henan University, Nanyang, China
Esophageal Adenocarcinoma (EAC) is one of the most common gastrointestinal tumors in the world. However, molecular prognostic systems are still lacking for EAC. Hence, we developed an Online consensus Survival analysis web server for Esophageal Adenocarcinoma (OSeac), to centralize published gene expression data and clinical follow up data of EAC patients from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). OSeac includes 198 EAC cases with gene expression profiling and relevant clinical long-term follow-up data, and employs the Kaplan Meier (KM) survival plot with hazard ratio (HR) and log rank test to estimate the prognostic potency of genes of interests for EAC patients. Moreover, we have determined the reliability of OSeac by using previously reported prognostic biomarkers such as DKK3, CTO1, and TXNIP. OSeac is free and publicly accessible at http://bioinfo.henu.edu.cn/EAC/EACList.jsp.
Introduction
Esophageal cancer is a common malignant tumor of digestive tract. The incidence of esophageal cancer is eighth in all tumors and sixth in fatal cancer (1). EAC is one of the most common histological types of esophageal cancer (2, 3) and has increased markedly in Western countries in recent decades (4, 5). Esophagectomy with the addition of perioperative chemotherapy or chemoradiotherapy had improved prognosis of EAC (6–8), however not all EAC patients got cured. The use of molecularly targeted agents are not satisfied so far for EAC and it has lagged behind other cancers (9). Therefore, it is necessary to identify new predictive and prognostic biomarkers for EAC patients to improve clinical outcome.
The expression of genes has been shown to guide the prognosis of cancer patients (10). Three genes DKK3, TXNIP, and CTO1 have been reported as prognosis biomarkers in EAC patients (11–13). However, these biomarkers need independent further validation to increase their sensitivity and specificity before clinical application. The advanced bioinformatic methods and resources have been developed for breast cancer, bladder cancer, esophageal squamous cell carcinoma, leiomyosarcoma, and lung cancer to analyze the prognostic abilities of genes (14–19), and greatly facilitate the development of cancer prognostic biomarkers. However, there is a lack of prognostic analysis system for EAC.
In this study, an online prognostic analysis tool for EAC was developed. It can not only rapidly evaluate the value of prognostic molecular biomarkers, but also provide the opportunities to identify the potential new therapeutic targets for EAC patients.
Materials and Methods
Data Collection
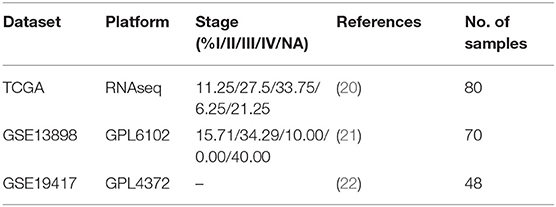
Three EAC datasets were collected from GEO (http://www.ncbi.nlm.nih.gov/geo/) and TCGA (http://cancergenome.nih.gov), these datasets include gene expression profiles and clinical follow-up information of EAC (Table 1).
Development of OSeac
The OSeac server is developed as we previously described (17–19), and hosted in a windows server and adopts Appache Tomcat as web application server. Use HTML and JSP for the front end page and server side code is compiled to Java. The R package “Rserve” as a middleware enables Java to call programs written in R language. The SQL Server database is used as backend database which stores data of the gene expression profiles and clinical information. The central server for OSeac can be accessed at http://bioinfo.henu.edu.cn/EAC/EACList.jsp.
Results
OSeac Establishment and Usage
We collected gene expression profiles and clinical follow-up information of 198 EAC patients from TCGA and GEO databases, and established OSeac to measure the association between a queried gene and patient outcome by Kaplan Meier plot. Before performing analysis, users can set up filter conditions by confounding clinical factors such as stage and gender. Four outcome terms including overall survival (OS), disease specific survival (DSS), disease-free interval (DFI) (23), and progression-free interval (PFI) (23) can be determined in OSeac.
OSeac provides the main functions that evaluate the prognostic value of gene of interests. When users input official gene symbol in the textbox, select an individual dataset or combined datasets [the combined datasets mean that all the patients from individual datasets were combined as a pool after they were stratified into subgroups (high vs. low) based on the expression level of inputted gene in each dataset], then choose an appropriate cutoff value for gene expression (including median, quartile or 30%) and click the “Kaplan-Meier plot” button, the server will generate survival curves and display HR with 95% confidence intervals and P-value on the output web page. Currently, five clinical factors were set as optional factors to limit the analysis in a subgroup of EAC patients for special needs from different researchers, these clinical factors include TNM stage, gender, race, grade and treatment response.
Validation of Previously Published EAC Biomarkers in OSeac
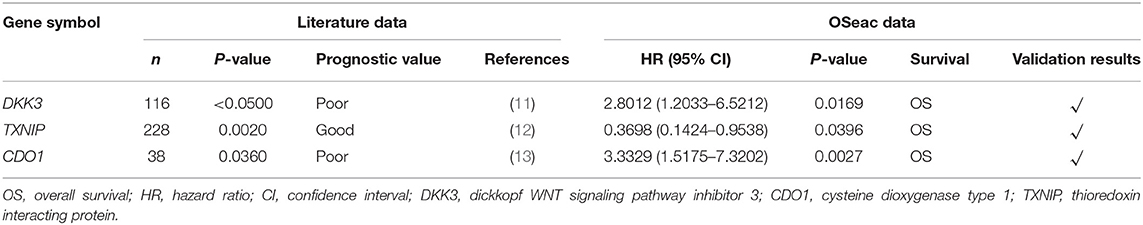
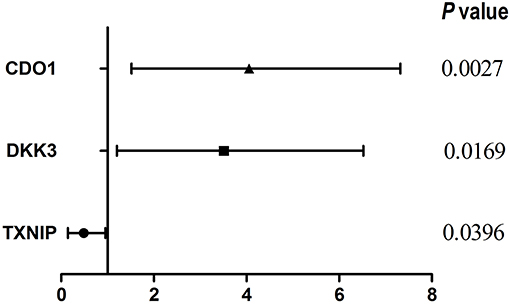
We evaluated three published prognostic biomarkers, including DKK3, CDO1, and TXNIP (Table 2, Figure 1) in OSeac. As shown in Table 2 and reported in original literatures, the higher expression of gene TXNIP in EAC patients implies a significant better overall survival rate, whereas higher expression of other two genes predict a significant worse overall survival. This result shows the validity and reliability of OSeac in determining the prognostic potency of genes of interests.
Discovering Potential Biomarkers for EAC in OSeac
In addition, to further test the prediction power of OSeac, we choose a known prognostic marker of gastric cancer (GC) (24) to analyze it in OSeac. We examined prognostic potency of RAP1B for EAC patients by OSeac, and found that RAP1B was significantly associated with unfavorable OS in TCGA (P = 0.0464, HR: 2.0045, 95% CI: 1.0111–3.9741), GSE13898 (P = 0.0241, HR: 3.0920, 95% CI: 1.1596–8.2446) and GSE19417 (P = 0.0138, HR: 2.4119, 95% CI: 1.1965–4.8618). This suggests that RAP1B could be a potential prognostic indicator of poor OS for EAC (Figure 2).
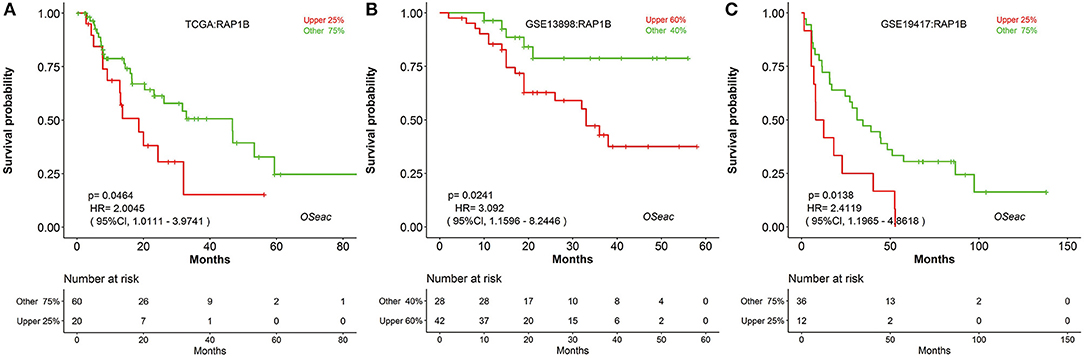
Figure 2. Kaplan Meier plots of a potential prognosis biomarker RAP1B in TCGA (A) GSE13898 (B) and GSE19417 (C) “upper 25%” and “other 75%”: sub-categorizing approach. After the expression level of inputted gene is sorted, take the patients with top 25% high expression level as “upper 25%” subgroup and remaining patients as “other 75%” subgroup.
Discussion
In this study, we have developed an online tool OSeac to analyze prognostic biomarkers in EAC. As most researchers may concern the accuracy rate and potential error from OSeac, for example, we performed the prognosis analysis for one gene each time in OSeac, the gene will be regarded significant for prognosis when P-value is < 0.05, while there are more than 20,000 genes in human genome, which means we may get 1,000 significant prognostic genes by chance when we do genome-wide repeated measurements. In addition, the EAC expression datasets may come from different analyzing platforms/technologies, or from different ethnicities, all these may influence the results of prognosis analysis. Nevertheless, in order to increase the specificity of prognosis biomarkers, we did our best to collect EAC expression datasets from GEO, TCGA and literatures as many as possible, and offered the opportunities to do independent validation across different EAC cohorts, which as we know is most important for biomarker development. Finally, to determine the performance of OSeac, three published EAC prognostic biomarkers were assessed in OSeac, and all of them reach statistical significance for prognosis in OSeac as expected, indicating the good performance of OSeac in prognostic biomarker screening. OSeac could also be used to screen novel prognosis biomarker for EAC, for example, RAP1B contributes to tumor malignant behavior and poor prognosis in GC (24). Using OSeac to assess the prognostic value of RAP1B in EAC, we found that RAP1B is a potential unfavorable prognostic indicator for EAC patients.
In conclusion, OSeac could help clinicians, biologists, and researchers to easily evaluate prognostic significance of genes of interests in EAC. The limitation of OSeac is that currently OSeac contains only 3 datasets and 198 samples, the sample number is relatively low, therefore, we will keep update and expand OSeac when new EAC datasets are available.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: http://bioinfo.henu.edu.cn/EAC/EACList.jsp.
Ethics Statement
Written informed consent was obtained from the individual(s), and minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author Contributions
XG: study concept and design. ZY, LX, XG, LG, NL, WZ, YW, and XL: acquisition of data. QW, LX, XG, MY, XS, WZ, YW, and XL: analysis and interpretation of data. QW, LX, GZ, YL, XG, and XL: draft of the manuscript. QW, ZY, GZ, LX, WZ, and XG: critical revision of the manuscript for intellectual content.
Funding
This study was supported by National Natural Science Foundation of China (Nos. 81602362 and 81801569), Supporting grants of Henan University (Nos. 2015YBZR048 and B2015151), Yellow River Scholar Program (No. H2016012), and Program for Innovative Talents of Science and Technology in Henan Province (No. 18HASTIT048), Program for Science and Technology Development in Henan Province (No. 162102310391), Program for Young Key Teacher of Henan Province (2016GGJS-214), Kaifeng Science and Technology Major Project (18ZD008), Supporting grant of Bioinformatics Center of Henan University (No. 2018YLJC01).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Hongo M, Nagasaki Y, Shoji T. Epidemiology of esophageal cancer: orient to occident. Effects of chronology, geography and ethnicity. J Gastroenterol. Hepatol. (2009) 24:729–35. doi: 10.1111/j.1440-1746.2009.05824.x
2. Parkin DM, Bray F, Ferlay J, Pisani P. Estimating the world cancer burden: Globocan 2000. Int J Cancer. (2001) 94:153–6. doi: 10.1002/ijc.1440
3. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. (2015) 65:87–108. doi: 10.3322/caac.21262
4. Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Semin Radiation Oncol. (2007) 17:2–9. doi: 10.1016/j.semradonc.2006.09.003
5. Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. (2005) 97:142–6. doi: 10.1093/jnci/dji024
6. Allum WH, Stenning SP, Bancewicz J, Clark PI, Langley RE. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. (2009) 27:5062–7. doi: 10.1200/JCO.2009.22.2083
7. Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, et al. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. (2006) 355:11–20. doi: 10.1056/NEJMoa055531
8. van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. (2012) 366:2074–84. doi: 10.1056/NEJMoa1112088
9. Secrier M, Li X, De Silva N, Eldridge MD, Contino G, Bornschein J, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. (2016) 48:1131. doi: 10.1038/ng.3659
10. Onda M, Emi M, Nagai H, Nagahata T, Tsumagari K, Fujimoto T, et al. Gene expression patterns as marker for 5-year postoperative prognosis of primary breast cancers. J Cancer Res Clin Oncol. (2004) 130:537–45. doi: 10.1007/s00432-004-0574-7
11. Wang Z, Lin L, Thomas DG, Nadal E, Chang AC, Beer DG, et al. The role of Dickkopf-3 overexpression in esophageal adenocarcinoma. J Thoracic Cardiovasc Surg. (2015) 150:377–85. e2. doi: 10.1016/j.jtcvs.2015.05.006
12. Woolston CM, Madhusudan S, Soomro IN, Lobo DN, Reece-Smith AM, Parsons SL, et al. Thioredoxin interacting protein and its association with clinical outcome in gastro-oesophageal adenocarcinoma. Redox Biol. (2013) 1:285–91. doi: 10.1016/j.redox.2013.04.006
13. Kojima K, Yamashita K, Ushiku H, Katoh H, Ishii S, Tanaka T, et al. The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Dis Esophagus. (2017) 30:1–9. doi: 10.1093/dote/dow001
14. Gyorffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. (2013) 8:e82241. doi: 10.1371/journal.pone.0082241
15. Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. (2012) 19:197–208. doi: 10.1530/ERC-11-0329
16. Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. (2010) 123:725–31. doi: 10.1007/s10549-009-0674-9
17. Zhang G, Wang Q, Yang M, Yuan Q, Dang Y, Sun X, et al. OSblca: a web server for investigating prognostic biomarkers of bladder cancer patients. Front Oncol. (2019) 9:466. doi: 10.3389/fonc.2019.00466
18. Wang Q, Wang F, Lv J, Xin J, Xie L, Zhu W, et al. Interactive online consensus survival tool for esophageal squamous cell carcinoma prognosis analysis. Oncol Lett. (2019) 18:1199–206. doi: 10.3892/ol.2019.10440
19. Wang Q, Xie L, Dang Y, Sun X, Xie T, Guo J, et al. OSlms: a web server to evaluate the prognostic value of genes in leiomyosarcoma. Front Oncol. (2019) 9:190. doi: 10.3389/fonc.2019.00190
20. Analysis WG, Asan U, Network CGAR. Integrated genomic characterization of oesophageal carcinoma. Nature. (2017) 541:169. doi: 10.1038/nature20805
21. Kim SM, Park Y-Y, Park ES, Cho JY, Izzo JG, Zhang D, et al. Prognostic biomarkers for esophageal adenocarcinoma identified by analysis of tumor transcriptome. PLoS ONE. (2010) 5:e15074. doi: 10.1371/journal.pone.0015074
22. Peters CJ, Rees JR, Hardwick RH, Hardwick JS, Vowler SL, Ong CAJ, et al. A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology. (2010) 139:1995–2004. e15. doi: 10.1053/j.gastro.2010.05.080
23. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. (2018) 173:400–16. e11. doi: 10.1158/1538-7445.AM2018-3287
Keywords: EAC, prognostic, survival analysis, biomarker, web server
Citation: Wang Q, Yan Z, Ge L, Li N, Yang M, Sun X, Xie L, Zhang G, Zhu W, Wang Y, Li Y, Li X and Guo X (2020) OSeac: An Online Survival Analysis Tool for Esophageal Adenocarcinoma. Front. Oncol. 10:315. doi: 10.3389/fonc.2020.00315
Received: 09 August 2019; Accepted: 21 February 2020;
Published: 06 March 2020.
Edited by:
Jorge A. R. Salvador, University of Coimbra, PortugalReviewed by:
Chengqi Xu, Huazhong University of Science and Technology, ChinaChunbo Zhang, Nanchang University, China
Copyright © 2020 Wang, Yan, Ge, Li, Yang, Sun, Xie, Zhang, Zhu, Wang, Li, Li and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianzhe Li, bnNseHoyMDA3QDE2My5jb20=; Xiangqian Guo, eHFndW9AaGVudS5lZHUuY24=
†These authors have contributed equally to this work
 Qiang Wang
Qiang Wang Zhongyi Yan1†
Zhongyi Yan1† Xiaoxiao Sun
Xiaoxiao Sun Longxiang Xie
Longxiang Xie Guosen Zhang
Guosen Zhang Wan Zhu
Wan Zhu Yongqiang Li
Yongqiang Li Xiangqian Guo
Xiangqian Guo