- 1National Institution of Drug Clinical Trial, Xiangya Hospital, Central South University, Changsha, China
- 2Department of Clinical Pharmacology, Hunan Key Laboratory of Pharmacogenetics, Xiangya Hospital, Central South University, Changsha, China
- 3National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, China
- 4Department of Orthopaedics, The First Affiliated Hospital of the University of South China, Hengyang, China
- 5Department of Otolaryngology Head and Neck Surgery, Xiangya Hospital, Central South University, Changsha, China
- 6Otolaryngology Major Disease Research Key Laboratory of Hunan Province, Xiangya Hospital, Central South University, Changsha, China
- 7Department of Pharmacy, Xiangya Hospital, Central South University, Changsha, China
Objective: Lung cancer is one of the most prevalent cancers and the leading cause of cancer-related death in the world. Platinum-based chemotherapy plays an important role in lung cancer treatment, but the therapeutic effect varies from person to person. Heat shock proteins (HSPs) have been reported to be associated with the survival time of lung cancer patients, which may be a potential biomarker in lung cancer treatment. The aim of this study was to investigate the association between genetic polymorphisms and the prognosis in lung cancer patients treated with platinum-based chemotherapy.
Methods: We performed genotyping in 19 single nucleotide polymorphisms (SNPs) of HSP genes and Rho family genes of 346 lung cancer patients by SequenomMassARRAY. We used Cox proportional hazard models, state and plink to analyze the associations between SNPs and the prognosis of lung cancer patients.
Results: We found that the polymorphisms of HSPB1 rs2070804 and HSPA4 rs3088225 were significantly associated with lung cancer survival (p=0.015, p=0.049*, respectively). We also discovered the statistically significant differences between rs2070804 with age, gender, histology and stage, rs3088225 with gender and stage, which can affect lung cancer prognosis.
Conclusion: The results of our study suggest that HSPB1 rs2070804 (G>T) and HSPA4 rs3088225 (A>G) may be useful biomarkers for predicting the prognosis of lung cancer patients treated with platinum-based chemotherapy.
Introduction
Lung cancer is one of the deadliest malignancies in the world (Siegel et al., 2019). It can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC consists of adenocarcinoma, squamous cell cancer and large cell lung cancer (Granger, 2016; The Lancet Respiratory, 2017). The main treatments of lung cancer are surgery, radiotherapy, chemotherapy and immunotherapy, and platinum-based chemotherapy is the first-line chemotherapy regimens (Dunbar et al., 2018). However, most of the patients are at advanced stage when diagnosed (Woodman et al., 2020). The ongoing treatments are limited by drug-resistance and unpredictable adverse-drug-reactions (ADR) (Hong et al., 2020; Sun et al., 2020). They are big challenges preventing clinical therapeutic benefits, which lead to a very low five-year survival rate (Clausen & Langer, 2019). In recent years, more and more investigation found that the therapy effect varies with different individuals (Li et al., 2019; Zou et al., 2019). There are many gene polymorphisms that have been found to be associated with drug resistance or adverse drug reactions in lung cancer patients treated with platinum-based chemotherapy, such as translation initiation factor 3a (eIF3a) (Xu et al., 2013), Wnt-inducible signaling pathway protein 1 (WISP1) (Chen et al., 2015), DNA repair genes (XRCC5, RRM1) (Chen et al., 2016; Zheng et al., 2017), and the Ca2+-dependent C-type lectin (CLEC4M) (Tan et al., 2019).
Heat shock proteins (HSPs) are a large family of chaperones (Milani et al., 2019), which are classified by their molecular weights, such as HSP27, HSP40, HSP60, HSP70, and HSP90 (Yun et al., 2019). HSPs can regulate cellular proliferation and differentiation which are strongly implicated in cancer development and progression (Chatterjee and Burns, 2017). Intriguingly, studies have verified the abnormal expression levels of HSPs in different types of cancer, including prostate, bladder, breast, ovarian, colorectal, and lung cancers (Calderwood et al., 2006; Ledford, 2011). The overriding role of the HSPs is to stabilize the active functions of overexpressed and mutated cancer genes (Calderwood and Gong, 2016). HSPs are associated with the outcomes of anticancer therapies, such as radiotherapy and immunotherapy in lung cancer patients (Li et al., 2016; Das et al., 2019). They can facilitate protein folding and maintain protein structures that regulate cellular metabolisms, which are essential for cell survival and proliferation (Chatterjee and Burns, 2017). What’s more, HSPs have been reported to be associated with lung cancer patient’s prognosis (Liu et al., 2019). The HSPB (HSP27) can protect cells from damage induced by stress factors, and it can also regulate cell proliferation, differentiation, and apoptotic signal transduction (Wang et al., 2011). The expression of HSPB1 have distinct prognostic values in NSCLC patients, it can induce resistance to cisplatin in A549 cell through the regulation of Transforming growth factor β (TGF-β) (Huang et al., 2017; Huang et al., 2018). HSPA (HSP70) could be a valuable diagnostic and prognostic marker in lung cancer patients, and high serum HSPA level predicted unfavorable survival in SCLC patients (Balazs et al., 2017; Sojka et al., 2019). HSPB and HSPA are also very important in DNA damage and repair signaling pathway, they could be a general regulator of DNA repair, ensuring the turnover of nuclear proteins required for proper DNA repair (Dubrez et al., 2020). It has been reported that DNA damage and repair is closely relevant to cancer prognosis (Silva et al., 2019), and DNA repair genes’ polymorphisms are also found to be associated with lung cancer patient’s prognosis treated with platinum-based chemotherapy (Butkiewicz et al., 2012; Perez-Ramirez et al., 2019).
Rho GTPases, including RHO, RAC1, and CDC42, are molecular switches which can control a wide variety of signal transduction pathways in all eukaryotic cells (Reiner and Lundquist, 2018). Rho GTPases play important roles in the process of cell migration, adhesion, intracellular transport and cellular transformation (Guan et al., 2020). Rho GTPases can also regulate lung cancer cell migration and invasion by activating β-catenin signaling pathway (Wang et al., 2019). The overexpression of RAC1 was related to poor differentiation, high TNM stage, and lymph node metastasis in NSCLC patients, while down-regulation of RAC1 can reduce cell migration and invasion and sensitize cells to antitumor drugs (Chen et al., 2011). Moreover, the inhibition of RAC1 can also sensitize gefitinib-resistant NSCLC cells to gefitinib (Kaneto et al., 2014).
Genotype mutations in some key genes, including single nucleotide polymorphisms (SNPs), may cause disorder or over-activation of some specific signaling pathway, leading to tumor development and affect patients’ prognosis (Xu et al., 2019). We have found that RAC1 polymorphisms are associated with platinum-based chemotherapy toxicity in lung cancer patients in our previous study (Zou et al., 2016). In this study, we want to further explore the prognostic roles of the HSPs and Rho GTPases polymorphisms in lung cancer patients treated with platinum-based chemotherapy. The purpose of this investigation was to improve the prognosis of lung cancer patients and provide a basis for the development of lung cancer treatment measures.
Materials and Methods
Study Subjects and Treatment Procedures
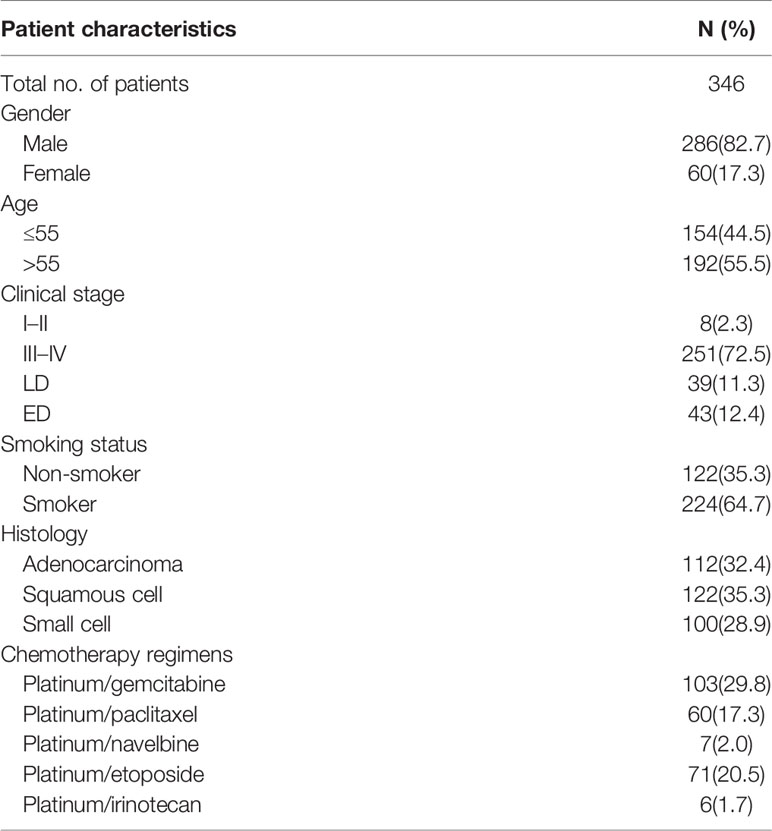
All patients were selected by the following inclusion criteria: (1) Patients newly diagnosed with lung cancer by histological examination at the Affiliated Cancer Hospital or Xiangya Hospital of Central South University (Changsha, Hunan, China) from August 2009 to January 2013; (2) Patients should receive at least 2 periods of platinum-based chemotherapy; (3) Patients with no history of surgery before chemotherapy. The clinical characteristics of these lung cancer patients enrolled are shown in Table 1. All patients were provided written informed consent before they participated in this study. The study protocol was approved by the Ethics Committee of Xiangya School of Medicine, Central South University.
Data Collection
The termination date for patient follow-up was July 15, 2019. Survival data were collected by telephone follow-up or residence registration. Overall survival (OS) time was defined as the time period between diagnosed of lung cancer and the date of the last follow-up or the death. Progression-free survival (PFS) time was calculated from the date diagnostic of lung cancer until the date of the first local recurrence or metastases in the last follow-up. Patients at the date of the last contact without progression were censored. As researchers, we were unaware of the presence of polymorphisms in the patients.
SNP Selecting, DNA Extraction, and Genotyping
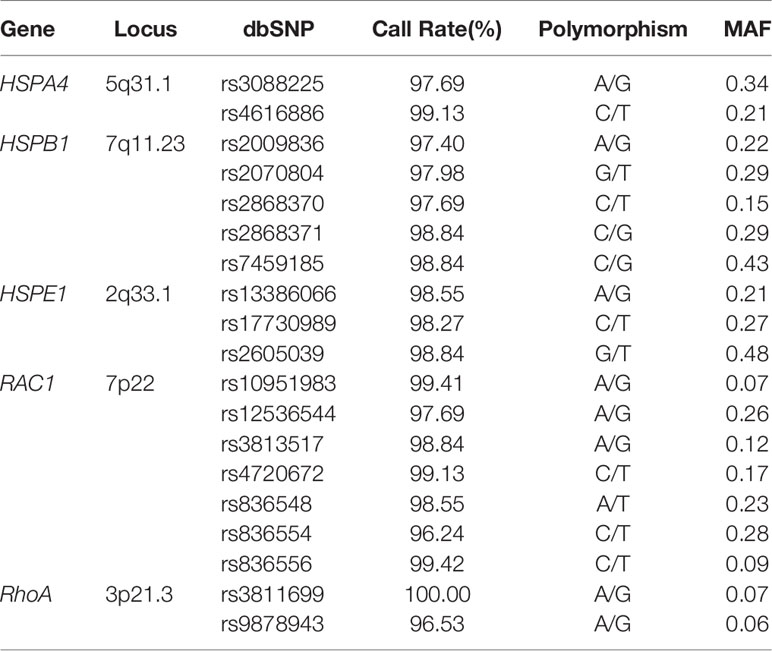
There were 19 common SNPs of HSPs and Rho GTPases selected in our study (Table 2). The candidate SNPs were located 5 kb upstream of the first exon and downstream of the last exon respectively. We used Haploview version 4.2 to choose the Haplotype tagging SNPs. They were chosen based on our previous research HSPs and Rho GTPases SNPs were related to lung cancer platinum-based chemotherapy toxicity (Zou et al., 2016), and they were associated with the outcome of cancers and involvement in multiple cancers (Li et al., 2016; Hung et al., 2020). And the selected SNPs must meet the condition that the minor allele frequency (MAF)>0.05 in the HapMap CHB population. The DNA we used for genotyping was separated from a 5ml peripheral blood sample using FlexiGene DNA Kit (Qiagen, Hilden, Germany). And all the samples were stored at 4°C before using. Genotyping were conducted by Sequenom’s MassARRAY system (Sequenom, San Diego, California, USA).
Statistical Analysis
We used Cox proportional hazard models to analyze the differences in age, gender, smoking status, histology, and clinical stage between the OS and PFS. We also screened the covariates used forward stepwise method of Cox proportional hazard models. Variables which were associated with OS or PFS significantly were considered as the covariates. And then, we fit these covariates into multivariate logistic regression model to adjust the covariates, through the command of –covar in PLINK. The p value was 2-sided and p<0.05 was considered statistically significant. All association analyses were conducted by three models including additive, dominant, and recessive. The additive model is for the additive effects of SNPs. It means that, if D is a minor allele and d is the major allele, the additive model means DD versus Dd versus dd. Dominant and recessive models are tests for the minor allele with two of the classes pooled. The dominant model means (DD, Dd) versus dd, and the recessive model means DD versus (Dd, dd). The aforementioned statistical analyses were performed using PLINK (ver 1.07, http://pngu.mgh.harvard.edu/purcell/plink/) and SPSS 18.0 (SPSS Inc, Chicago, Illinois, USA)
Results
Characteristics and Survival Status of the Lung Cancer Patients
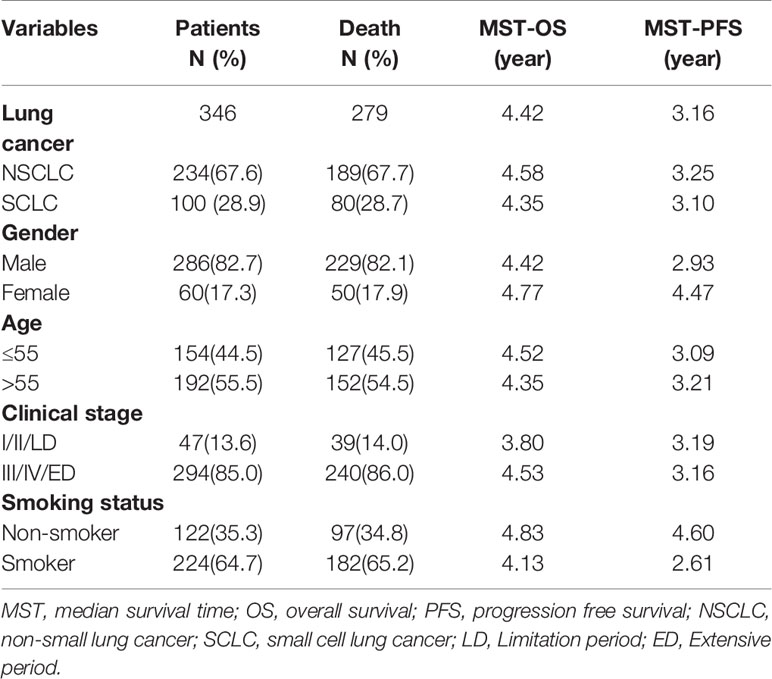
The demographic characteristics of the 346 lung cancer patients are provided in Table 3. Most patients were male (82.7%), and the median age at the time of the lung cancer patients diagnosed was 55 years (range 21–77 years). The median survival time of overall survival (MST-OS) is 4.42 year, and the median survival time of progression free survival (MST-PFS) is 3.16 year. The detailed information of the associations between clinical pathology characteristics and outcomes in lung cancer patients were also summarized in Table 3. And the Cox proportional hazard models were analyzed to find the covariates, and the results revealed that there are no significant differences between age, gender, smoking status, histology, and clinical stage with OS or PFS as shown in Table S1 (p>0.05).
Association Between the Polymorphisms and Prognosis in the Lung Cancer Patients
We found that the genetic polymorphism of HSPB1 rs2070804 (G>T) was significantly associated with the overall survival (OS) of lung cancer patients in additive and dominant models [Additive model: p=0.041, OR=0.66, 95%CI, (0.45–0.98); Dominant model: p=0.015, OR=0.55, 95%CI, (0.34–0.89)]. Patients who carry the HSPB1 rs2070804 GG genotype had a significantly shorter MST-OS than the patients who have the HSPB1 rs2070804 GT or TT variant genotypes (MST-OS: 3.047, 3.942, 4.971 years, respectively). What’s more, the genetic polymorphism of HSPA4 rs3088225 (A>G) was significantly associated with the progression free survival (PFS) of lung cancer patients in recessive model [p=0.049, OR=0.44; 95%CI, (0.19–1.00)]. Which means that the patients who carry the HSPA4 rs3088225 GG variant genotype have a shorter MST-PFS than the patients who have the HSPA4 rs3088225 AA or GA genotypes (MST-PFS: 2.504, 4.219, 3.103 year, respectively). In conclusion, those patients carrying the HSPB1 T allele rs2070804 and the allele A of HSPA4 rs3088225m are carriers of the protective allele in terms of the prognosis of lung cancer (Table 4, Figure 1).
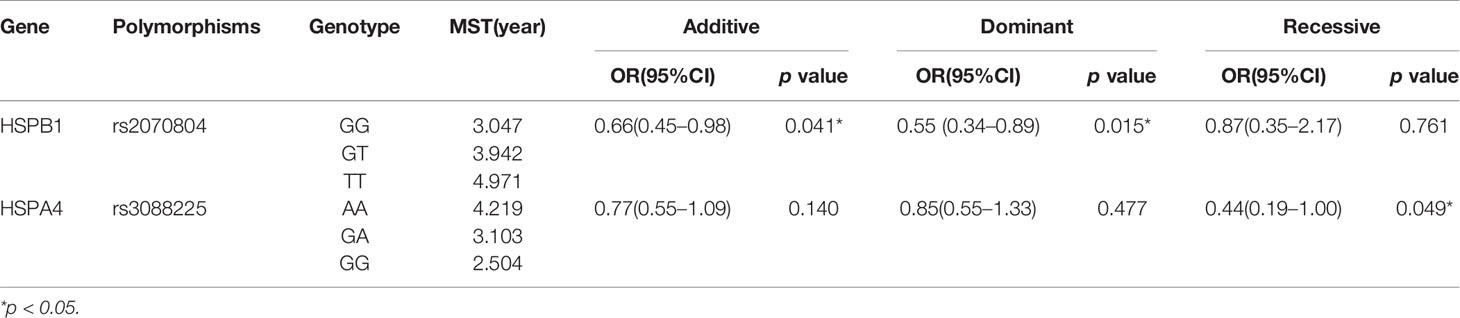
Table 4 Association of the HSPB1 rs2070804 polymorphisms and OS, HSPA4 rs3088225 polymorphisms and PFS in lung cancer patients.
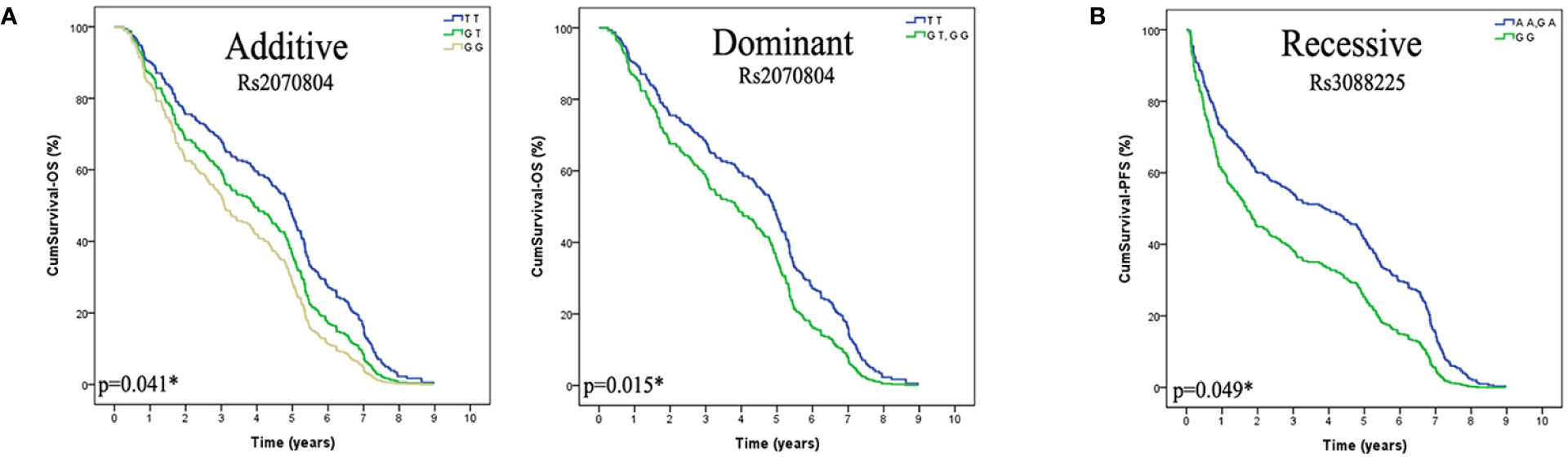
Figure 1 The HSPB1 rs2070804 and HSPA4 rs3088225 polymorphisms are significantly associcated with the prognosis in lung cancer patients treated with platinum-based chemotherapy, and the T variant allele of HSPB1 rs2070804, the A allele of HSPA4 rs3088225 are protective alleles. (A) Lung cancer patients that carried the genotype of HSPB1 rs2070804 TT have a longer overall survival time than GT or GG carries in Additive model (p=0.041*), and in the dominant model (p=0.015*). (B) Patients that carry the genotype of HSPA4 rs3088225 AA and GA have a longer progression free survival time than GG in Recessive model (p=0.049*).
To further investigate the association between these two SNPs and the prognosis in lung cancer patients, we performed subgroup analysis based on age, gender, smoking status, histology and stage. As shown in Figure 2, the polymorphisms of HSPB1 rs2070804 was significantly associated with the overall survival in dominant model in age more than 55 years old [p=0.027, OR=0.47, 95%CI, (0.24–0.92)] and male patients [p=0.042, OR=0.57, 95%CI, (0.33–0.98)]. It was also correlated to the overall survival in NSCLC patients in additive [p=0.048, OR=0.61, 95%CI, (0.37–0.99)] and dominant models [p=0.018, OR=0.48, 95%CI, (0.26–0.88)], and significant with patients whose clinical stage are at I/II/LD in additive model [p=0.034, OR=0.25, 95%CI, (0.07–0.90)]. The polymorphisms of HSPA4 rs3088225 was closely related to the progression free survival in NSCLC [p=0.033, OR=0.33, 95%CI, (0.12–0.92)] and patients whose clinical stage are in III/IV/ED [p=0.027, OR=0.37, 95%CI, (0.16–0.89)] in recessive model.
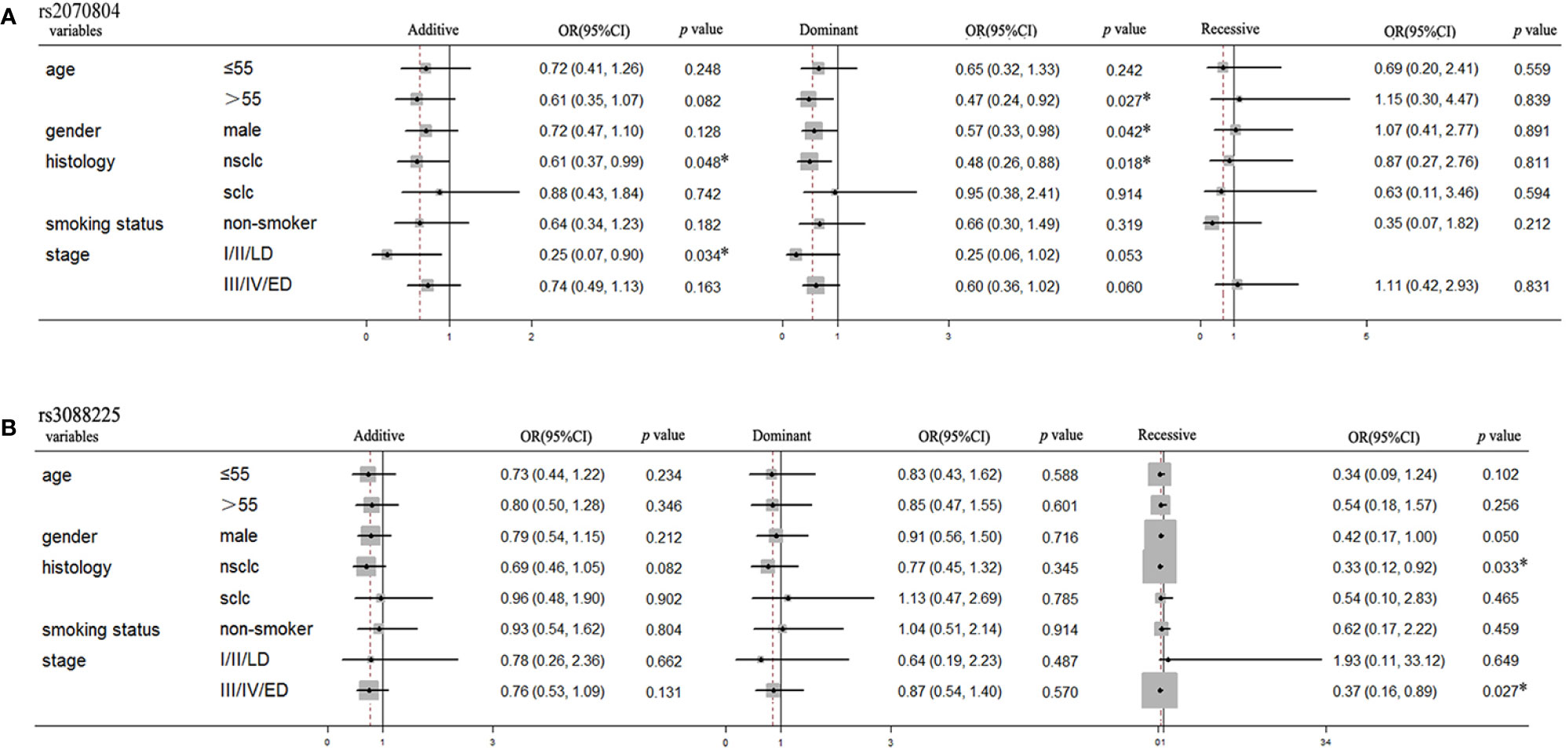
Figure 2 The HSPB1 rs2070804 and HSPA4 rs3088225 polymorphisms are significantly associated with the survival time in the subgroups of lung cancer patients treated with platinum-based chemotherapy. (A) HSPB1 rs2070804 polymorphism is significantly associated with the overall survival time in age more than 55 years, male, NSCLC, and patients whose clinical stage are I/II/LD. (B) HSPA4 rs3088225 polymorphism is significantly correlated with the progression free survival time in male, NSCLC, and patients whose clinical stage are III/IV/ED.
Stratification Analyses of Association Between Polymorphisms and Prognosis in Lung Cancer Patients
To further elucidate the association between the other 17 SNPs and the prognosis in lung cancer patients, we also performed subgroup analysis based on age, gender, smoking status, histology, and clinical stage. As shown in Table 5, the HSPE1 rs2605039 was related to overall survival in SCLC patients in dominant model [p=0.047, OR=0.39, 95%CI, (0.15–0.99)]. The RAC1 rs836548 [(Additive model: p=0.026, OR=2.01, 95%CI, (1.09–3.70); Dominant model: p=0.017, OR=2.46, 95%CI, (1.17–5.14)] and rs12536544 [Dominant model: p=0.039, OR=2.15, 95%CI, (1.04–4.43)] were significantly associated with overall survival in age less than 55-year old patients. The RAC1 rs3813517 was closely related to overall survival in patients whose clinical stage are at I/II/LD in dominant models [p=0.043, OR=5.25, 95%CI, (1.05–26.2)]. The HSPB1 rs2868370 was correlated with progression free survival in NSCLC [Additive model: p=0.012, OR=1.94, 95%CI, (1.16-3.25); Dominant model: p=0.033, OR=1.89, 95%CI, (1.05–3.37)], SCLC [Additive model: p=0.030, OR=0.30, 95%CI, (0.10–0.89); Dominant model: p=0.037, OR=0.28, 95%CI, (0.09–0.92)] and smoking patients [Additive model: p=0.021, OR=2.36, 95%CI, (1.14–4.89); Dominant model: p=0.021, OR=2.65, 95%CI, (1.16–6.06)]. The HSPB1 rs2009836 was closely associated with progression free survival in NSCLC patients in additive model [p=0.030, OR=1.61, 95%CI, (1.05–2.46)].
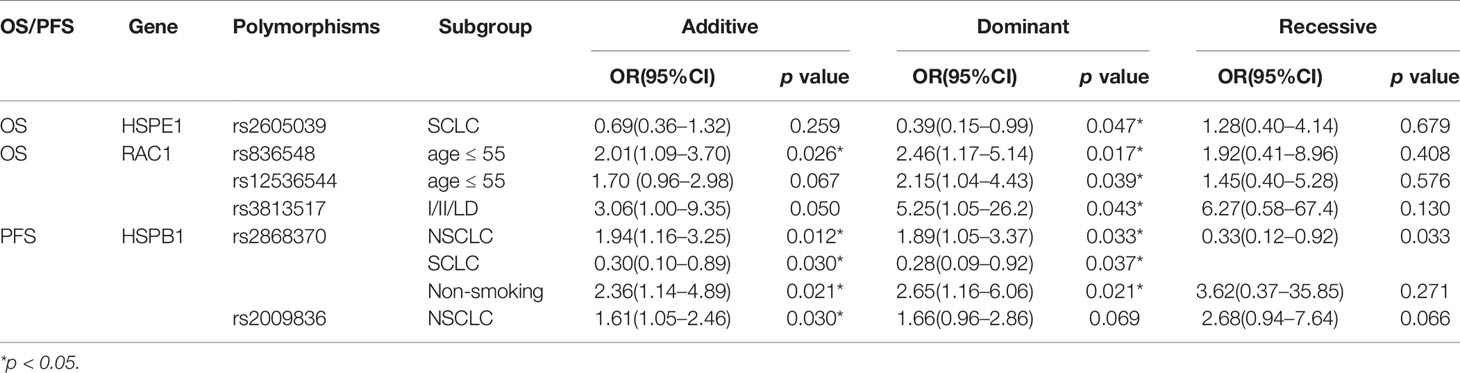
Table 5 Stratification analyses of Association between polymorphisms and OS or PFS in lung cancer patients.
Discussion
In this study, we found that the HSPB1 rs2070804 and HSPA4 rs3088225 polymorphisms are significantly associated with the prognosis in lung cancer patients with the platinum-based chemotherapy treatment. Patients who carry the rs2070804 T variant allele are more likely to have a longer overall survival (OS) time than patients with rs2070804 G allele. These relationship are mainly reflected in male, NSCLC, age more than 55-year-old patients and patients whose clinical stage are in I/II/LD. Moreover, the progression free survival (PFS) time in patients who carry the rs3088225 G variant allele are more likely to be shorter than patients with rs3088225 A allele. The rs3088225 polymorphisms in the subgroups of male, NSCLC, and clinical stage in III/IV/ED patients are related to the PFS time significantly in the stratified analysis. In conclusion, the HSPB1 rs2070804 T allele and HSPA4 rs3088225 A allele are the protective allele in the prognosis of lung cancer patients treated with platinum-based chemotherapy.
It has been reported that the expression of heat-shock proteins (HSPs) may be prognostic markers in several tumor types through the regulation of cell proliferation, invasion and metastasis (Saini and Sharma, 2018). And the inhibition of HSPs is currently an attractive potential therapeutic approach against cancer (Kaigorodova and Bogatyuk, 2014). HSPB suppression can result in the apoptotic death of MET-addicted EBC-1 in lung cancer cells, and oncogene-addicted cells require HSPB for survival (Konda et al., 2017). Abnormalities in the transforming growth factor b (TGF.B) pathway, is widely observed in drug resistance during lung cancer chemotherapy (Yokokura et al., 2016). HSPB can induce the resistance to cis-platinum in A549 cells through the regulation of TGF-β via decreasing cell viability and increasing cell apoptosis in A549 cell (Huang et al., 2017). HSPB1 can inhibit the endothelial-to-mesenchymal transition (EDMT) to suppress pulmonary fibrosis and lung tumorigenesis (Choi et al., 2016). It has been reported that the increased HSPB expression was correlated with malignant biological behavior of NSCLC and the increased HSPB expression was also related to the shorter survival of NSCLC patients (Sheng et al., 2017). There are some variants of HSPB found to be correlated with its expression level. The functional HSPB1 promoter -1271G>C variant may affect lung cancer susceptibility and survival time by modulating endogenous HSPB synthesis levels (Guo et al., 2010). The SNPs of HSPB1 rs7459185 was associated with radiation esophagitis in lung cancer through the regulation of HSPB1 expression level (Delgado et al., 2019). HSPB can regulate aggressive tumor behavior, metastasis, poor prognosis, and resistance to chemotherapy through the interference with theses effectiveness of targeted agents (Choi et al., 2017). There was one report about HSPB polymorphism found to be associated with NSCLC prognosis, in the US patients, it found that the CC genotype of HSPB1 rs2868371 was associated with poorer overall survival in US patients with NSCLC after radio(chemo)therapy (Xu et al., 2012). HSPA expression is a powerful and significant prognostic indicator in NSCLC patients, which is related to histopathological differentiation, lymph node metastasis, patients’ clinical stages, and smoking history (Huang et al., 2005). HSPA was reported to be a positive predictive factor in completely resected NSCLC treated with platinum-based adjuvant chemotherapy (Park et al., 2014). It was interestingly observed that HSPA1 was associated with good prognosis while HSPA2 correlated with bad prognosis in primary NSCLC (Sojka et al., 2019). HSPA1B A(1267)G polymorphism can influence the HSPA1B expression of SCLC cells, and the expression of HSPA1B was significantly decreased in GG as compared to cells of AA or AG genotype patients. The variant of GG genotype may be a negative prognostic factor for survival in SCLC patients, as the survival time of patients carry the GG genotype was significantly shorter as compared to carriers of the A allele (Szondy et al., 2012). The functional HSPA1B rs2763979 and rs6457452 variants are associated with lung cancer risk and survival (Guo et al., 2011). However, there are no reports about the relationship of HSPs polymorphisms and prognosis in platinum-based chemotherapy treatment of Chinese lung cancer patients.
The three GTPases, RHO, RAC, and Cdc42, play important roles in harmonizing many cellular activities among embryonic development, both in healthy cells and in disease conditions like cancers, RAC1 is significantly associated with the cell proliferation, metastasis-associated phenotypes, and drug-resistance especially on solid tumors (De et al., 2019). It has been reported that the overexpression of RAC1 is related to the poor prognosis and metastasis in NSCLC through the regulation of Epithelial Mesenchymal Transition, which means that RAC1 may be a potential therapeutic target in the treatment of NSCLC patients (Zhou et al., 2016). The Rho kinase pathway has an intimate relationship with cell growth, cell migration, and invasion in lung cancer, it has been found that RhoA knockdown can prevent cell proliferation and induces apoptosis in SPCA1 lung cancer cells (Liu et al., 2017). It has been reported that RAC1 or RhoA are mutated in cancers, and the expression levels of RAC1 or RhoA are aslo altered, which means that RhoA may play an crucial role in lung cancer treatment (Haga and Ridley, 2016).
Our study investigated the association between HSPs, RAC1 and RhoA and the survival time in Chinese lung cancer patients treated with platinum-based chemotherapy. All the patients we enrolled received the platinum-based chemotherapy regimens treatment for at least two periods, and we also performed stratified analysis in age, gender, smoking status, histology and clinical stage. Except for the polymorphisms of HSPB1 rs2070804 and HSPA4 rs3088225, we also found other polymorphisms which are associated with lung cancer prognosis in some specific subgroups. For instance, HSPE1 rs2605039 was correlated to the overall survival in SCLC patients (p=0.047). RAC1 rs836548 and rs12536544 were associated with the overall survival in age less than 55 years old patients (p=0.017, p=0.039, respectively), and RAC1 rs3813517 was related to overall survival in patients whose clinical stage were in I/II/LD (p=0.043). HSPB1 rs2868370 was associated with the progression free survival in NSCLC, CLCL and non-smoking patients (p=0.012, p=0.030, p=0.021, respectively). HSPB1 rs2009836 was also correlated to the progression free survival in NSCLC patients (p=0.030).
However, there were several limitations in our study. First, the sample size was not large enough, we just enrolled 346 patients in our study. Second, the biological function mechanisms of these SNPs need further study in vitro. Finally, the validation of our results needs replication studies with other independent subjects.
In conclusion, we found that the polymorphisms of HSPB1 rs2070804 and HSPA4 rs3088225 were significantly associated with the prognosis in lung cancer patients treated with platinum-based chemotherapy. Lung cancer patients who carry the HSPB1 rs2070804 T allele or HSPA4 rs3088225 A allele may have a better prognosis compared to the rs2070804 G allele or HSPA4 rs3088225 G allele. The genotypes of HSPB1 rs2070804 and HSPA4 rs3088225 may be an attractive biomarker used to predict the prognosis of platinum-based chemotherapy lung cancer patients.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Xiangya School of Medicine, Central South University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Study design was contributed by TZ, J-YL, JC, and Z-QL, with assistance from the rest of the authors. TZ and LS took the lead in sample collection and data analysis, assisted by J-YY, XL, X-PL, and H-HZ. Data interpretation was performed by TZ, with assistance from the other authors. The manuscript was written primarily by TZ and JC, with assistance from the other authors, and revised by Z-QL. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (81803640, 81874327), the Hunan Provincial Natural Science Foundation of China (2019JJ50946), and the Youth Science Foundation of Xiangya Hospital (2017Q02, 2018Q014).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2020.01029/full#supplementary-material
References
Balazs, M., Zsolt, H., Laszlo, G., Gabriella, G., Lilla, T., Gyula, O., et al. (2017). Serum Heat Shock Protein 70, as a Potential Biomarker for Small Cell Lung Cancer. Pathol. Oncol. Res. POR 23, 377–383. doi: 10.1007/s12253-016-0118-x
Butkiewicz, D., Drosik, A., Suwinski, R., Krzesniak, M., Rusin, M., Kosarewicz, A., et al. (2012). Influence of DNA repair gene polymorphisms on prognosis in inoperable non-small cell lung cancer patients treated with radiotherapy and platinum-based chemotherapy. Int. J. Cancer 131, E1100–E1108. doi: 10.1002/ijc.27596
Calderwood, S. K., Gong, J. (2016). Heat Shock Proteins Promote Cancer: It’s a Protection Racket. Trends Biochem. Sci. 41, 311–323. doi: 10.1016/j.tibs.2016.01.003
Calderwood, S. K., Khaleque, M. A., Sawyer, D. B., Ciocca, D. R. (2006). Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 31, 164–172. doi: 10.1016/j.tibs.2006.01.006
Chatterjee, S., Burns, T. F. (2017). Targeting Heat Shock Proteins in Cancer: A Promising Therapeutic Approach. Int. J. Mol. Sci. 18, 1978. doi: 10.3390/ijms18091978
Chen, Q. Y., Xu, L. Q., Jiao, D. M., Yao, Q. H., Wang, Y. Y., Hu, H. Z., et al. (2011). Silencing of Rac1 modifies lung cancer cell migration, invasion and actin cytoskeleton rearrangements and enhances chemosensitivity to antitumor drugs. Int. J. Mol. Med. 28, 769–776. doi: 10.3892/ijmm.2011.775
Chen, J., Yin, J. Y., Li, X. P., Wang, Y., Zheng, Y., Qian, C. Y., et al. (2015). Association of Wnt-Inducible Signaling Pathway Protein 1 Genetic Polymorphisms With Lung Cancer Susceptibility and Platinum-Based Chemotherapy Response. Clin. Lung Cancer 16298-304, e1–e2. doi: 10.1016/j.cllc.2014.12.008
Chen, J., Wu, L., Wang, Y., Yin, J., Li, X., Wang, Z., et al. (2016). Effect of transporter and DNA repair gene polymorphisms to lung cancer chemotherapy toxicity. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 37, 2275–2284. doi: 10.1007/s13277-015-4048-0
Choi, S. H., Nam, J. K., Kim, B. Y., Jang, J., Jin, Y. B., Lee, H. J., et al. (2016). HSPB1 Inhibits the Endothelial-to-Mesenchymal Transition to Suppress Pulmonary Fibrosis and Lung Tumorigenesis. Cancer Res. 76, 1019–1030. doi: 10.1158/0008-5472.CAN-15-0952
Choi, B., Choi, S. K., Park, Y. N., Kwak, S. Y., Lee, H. J., Kwon, Y., et al. (2017). Sensitization of lung cancer cells by altered dimerization of HSP27. Oncotarget 8, 105372–105382. doi: 10.18632/oncotarget.22192
Clausen, M. M., Langer, S. W. (2019). Improving the prognosis for lung cancer patients. Acta Oncol. 58, 1077–1078. doi: 10.1080/0284186X.2019.1632477
Das, J. K., Xiong, X., Ren, X., Yang, J. M., Song, J. (2019). Heat Shock Proteins in Cancer Immunotherapy. J. Oncol. 2019, 3267207. doi: 10.1155/2019/3267207
De, P., Aske, J. C., Dey, N. (2019). RAC1 Takes the Lead in Solid Tumors. Cells 8, 382. doi: 10.3390/cells8050382
Delgado, B. D., Enguix-Riego, M. V., Fernandez de Bobadilla, J. C., Herrero Rivera, D., Nieto-Guerrero Gomez, J. M., Praena-Fernandez, J. M., et al. (2019). Association of single nucleotide polymorphisms at HSPB1 rs7459185 and TGFB1 rs11466353 with radiation esophagitis in lung cancer. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 135, 161–169. doi: 10.1016/j.radonc.2019.03.005
Dubrez, L., Causse, S., Borges Bonan, N., Dumetier, B., Garrido, C. (2020). Heat-shock proteins: chaperoning DNA repair. Oncogene 39, 516–529. doi: 10.1038/s41388-019-1016-y
Dunbar, C. E., High, K. A., Joung, J. K., Kohn, D. B., Ozawa, K., Sadelain, M. (2018). Gene therapy comes of age. Science 359, 4672. doi: 10.1126/science.aan4672
Granger, C. L. (2016). Physiotherapy management of lung cancer. J. Physiother. 62, 60–67. doi: 10.1016/j.jphys.2016.02.010
Guan, X., Guan, X., Dong, C., Jiao, Z. (2020). Rho GTPases and related signaling complexes in cell migration and invasion. Exp. Cell Res. 388, 111824. doi: 10.1016/j.yexcr.2020.111824
Guo, H., Bai, Y., Xu, P., Hu, Z., Liu, L., Wang, F., et al. (2010). Functional promoter -1271G>C variant of HSPB1 predicts lung cancer risk and survival. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 28, 1928–1935. doi: 10.1200/JCO.2009.24.4954
Guo, H., Deng, Q., Wu, C., Hu, L., Wei, S., Xu, P., et al. (2011). Variations in HSPA1B at 6p21.3 are associated with lung cancer risk and prognosis in Chinese populations. Cancer Res. 71, 7576–7586. doi: 10.1158/0008-5472.CAN-11-1409
Haga, R. B., Ridley, A. J. (2016). Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 7, 207–221. doi: 10.1080/21541248.2016.1232583
Hong, S. H., Ku, J. M., Lim, Y. S., Lee, S. Y., Kim, J. H., Cheon, C., et al. (2020). Cucurbitacin D Overcomes Gefitinib Resistance by Blocking EGF Binding to EGFR and Inducing Cell Death in NSCLCs. Front. Oncol. 10, 62. doi: 10.3389/fonc.2020.00062
Huang, Q., Zu, Y., Fu, X., Wu, T. (2005). Expression of heat shock protein 70 and 27 in non-small cell lung cancer and its clinical significance. J. Huazhong Univ. Sci. Technol. Med. Sci. 25, 693–695. doi: 10.1007/BF02896173
Huang, Z., Yang, C., Sun, S., Nan, Y., Lang, Z., Wang, X., et al. (2017). Heat Shock Protein 27, a Novel Regulator of Transforming Growth Factor beta Induced Resistance to Cisplatin in A549 Cell. Pharmacology 100, 283–291. doi: 10.1159/000479320
Huang, Z. C., Li, H., Sun, Z. Q., Zheng, J., Zhao, R. K., Chen, J., et al. (2018). Distinct prognostic roles of HSPB1 expression in non-small cell lung cancer. Neoplasma 65, 161–166. doi: 10.4149/neo_2018_102
Hung, C. S., Huang, C. Y., Hsu, Y. W., Makondi, P. T., Chang, W. C., Chang, Y. J., et al. (2020). HSPB1 rs2070804 polymorphism is associated with the depth of primary tumor. J. Cell. Biochem. 121, 63–69. doi: 10.1002/jcb.28266
Kaigorodova, E. V., Bogatyuk, M. V. (2014). Heat shock proteins as prognostic markers of cancer. Curr. Cancer Drug Targets 14, 713–726. doi: 10.2174/1568009614666140926122846
Kaneto, N., Yokoyama, S., Hayakawa, Y., Kato, S., Sakurai, H., Saiki, I. (2014). RAC1 inhibition as a therapeutic target for gefitinib-resistant non-small-cell lung cancer. Cancer Sci. 105, 788–794. doi: 10.1111/cas.12425
Konda, J. D., Olivero, M., Musiani, D., Lamba, S., Di Renzo, M. F. (2017). Heat-shock protein 27 (HSP27, HSPB1) is synthetic lethal to cells with oncogenic activation of MET, EGFR and BRAF. Mol. Oncol. 11, 599–611. doi: 10.1002/1878-0261.12042
Li, X., Xu, S., Cheng, Y., Shu, J. (2016). HSPB1 polymorphisms might be associated with radiation-induced damage risk in lung cancer patients treated with radiotherapy. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 37, 5743–5749. doi: 10.1007/s13277-016-4959-4
Li, W., Zhang, M., Huang, C., Meng, J., Yin, X., Sun, G. (2019). Genetic variants of DNA repair pathway genes on lung cancer risk. Pathol. Res. Pract. 215, 152548. doi: 10.1016/j.prp.2019.152548
Liu, D., Mei, X., Wang, L., Yang, X. (2017). RhoA inhibits apoptosis and increases proliferation of cultured SPCA1 lung cancer cells. Mol. Med. Rep. 15, 3963–3968. doi: 10.3892/mmr.2017.6545
Liu, K., Kang, M., Li, J., Qin, W., Wang, R. (2019). Prognostic value of the mRNA expression of members of the HSP90 family in non-small cell lung cancer. Exp. Ther. Med. 17, 2657–2665. doi: 10.3892/etm.2019.7228
Milani, A., Basirnejad, M., Bolhassani, A. (2019). Heat-shock proteins in diagnosis and treatment: an overview of different biochemical and immunological functions. Immunotherapy 11, 215–239. doi: 10.2217/imt-2018-0105
Park, T. S., Kim, H. R., Koh, J. S., Jang, S. H., Hwang, Y. I., Yoon, H. I., et al. (2014). Heat shock protein 70 as a predictive marker for platinum-based adjuvant chemotherapy in patients with resected non-small cell lung cancer. Lung Cancer 86, 262–267. doi: 10.1016/j.lungcan.2014.08.009
Perez-Ramirez, C., Canadas-Garre, M., Alnatsha, A., Villar, E., Valdivia-Bautista, J., Faus-Dader, M. J., et al. (2019). Pharmacogenetics of platinum-based chemotherapy: impact of DNA repair and folate metabolism gene polymorphisms on prognosis of non-small cell lung cancer patients. Pharmacogenom. J. 19, 164–177. doi: 10.1038/s41397-018-0014-8
Reiner, D. J., Lundquist, E. A. (2018). Small GTPases. WormBook Online Rev. C. Elegans Biol. 2018, 1–65. doi: 10.1895/wormbook.1.67.2
Saini, J., Sharma, P. K. (2018). Clinical, Prognostic and Therapeutic Significance of Heat Shock Proteins in Cancer. Curr. Drug Targets 19, 1478–1490. doi: 10.2174/1389450118666170823121248
Sheng, B., Qi, C., Liu, B., Lin, Y., Fu, T., Zeng, Q. (2017). Increased HSP27 correlates with malignant biological behavior of non-small cell lung cancer and predicts patient’s survival. Sci. Rep. 7, 13807. doi: 10.1038/s41598-017-13956-2
Siegel, R. L., Miller, K. D., Jemal, A. (2019). Cancer statistics 2019. CA: Cancer J. Clin. 69, 7–34. doi: 10.3322/caac.21551
Silva, M. M., Rocha, C. R. R., Kinker, G. S., Pelegrini, A. L., Menck, C. F. M. (2019). The balance between NRF2/GSH antioxidant mediated pathway and DNA repair modulates cisplatin resistance in lung cancer cells. Sci. Rep. 9, 17639. doi: 10.1038/s41598-019-54065-6
Sojka, D. R., Gogler-Piglowska, A., Vydra, N., Cortez, A. J., Filipczak, P. T., Krawczyk, Z., et al. (2019). Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non-small cell lung carcinoma (NSCLC); an implication for NSCLC treatment. Sci. Rep. 9, 14394. doi: 10.1038/s41598-019-50840-7
Sun, J., Zhang, Z., Bao, S., Yan, C., Hou, P., Wu, N., et al. (2020). Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J. Immunother. Cancer 8, e000110. doi: 10.1136/jitc-2019-000110
Szondy, K., Rusai, K., Szabo, A. J., Nagy, A., Gal, K., Fekete, A., et al. (2012). Tumor cell expression of heat shock protein (HSP) 72 is influenced by HSP72 [HSPA1B A(1267)G] polymorphism and predicts survival in small Cell lung cancer (SCLC) patients. Cancer Invest. 30, 317–322. doi: 10.3109/07357907.2012.657815
Tan, L. M., Li, X., Qiu, C. F., Zhu, T., Hu, C. P., Yin, J. Y., et al. (2019). CLEC4M is associated with poor prognosis and promotes cisplatin resistance in NSCLC patients. J. Cancer 10, 6374–6383. doi: 10.7150/jca.30139
The Lancet Respiratory, M. (2017). Lung cancer-moving in the right direction. Lancet Respiratory Med. 5, 599. doi: 10.1016/S2213-2600(17)30271-0
Wang, W., Xu, X., Wang, W., Shao, W., Li, L., Yin, W., et al. (2011). The expression and clinical significance of CLIC1 and HSP27 in lung adenocarcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 32, 1199–1208. doi: 10.1007/s13277-011-0223-0
Wang, L., Shen, S., Wang, M., Ding, F., Xiao, H., Li, G., et al. (2019). Rho GTPase Activating Protein 24 (ARHGAP24) Silencing Promotes Lung Cancer Cell Migration and Invasion by Activating beta-Catenin Signaling. Med. Sci. Monitor Int. Med. J. Exp. Clin. Res. 25, 21–31. doi: 10.12659/MSM.911503
Woodman, C., Vundu, G., George, A., Wilson, C. M. (2020). Applications and strategies in nanodiagnosis and nanotherapy in lung cancer. Semin. Cancer Biol. doi: 10.1016/j.semcancer.2020.02.009
Xu, T., Wei, Q., Lopez Guerra, J. L., Wang, L. E., Liu, Z., Gomez, D., et al. (2012). HSPB1 gene polymorphisms predict risk of mortality for US patients after radio(chemo)therapy for non-small cell lung cancer. Int. J. Radiat. Oncol. Biol. Phys. 84, e229–e235. doi: 10.1016/j.ijrobp.2012.03.032
Xu, X., Han, L., Yang, H., Duan, L., Zhou, B., Zhao, Y., et al. (2013). The A/G allele of eIF3a rs3740556 predicts platinum-based chemotherapy resistance in lung cancer patients. Lung Cancer 79, 65–72. doi: 10.1016/j.lungcan.2012.10.005
Xu, Q., Lin, D., Li, X., Xiao, R., Liu, Z., Xiong, W., et al. (2019). Association between single nucleotide polymorphisms of NOTCH signaling pathway-related genes and the prognosis of NSCLC. Cancer Manage. Res. 11, 6895–6905. doi: 10.2147/CMAR.S197747
Yokokura, S., Kanaji, N., Tadokoro, A., Yokokura, S., Kadowaki, N., Bandoh, S. (2016). Confluence-dependent resistance to cisplatin in lung cancer cells is regulated by transforming growth factor-beta. Exp. Lung Res. 42, 175–181. doi: 10.3109/01902148.2016.1172370
Yun, C. W., Kim, H. J., Lim, J. H., Lee, S. H. (2019). Heat Shock Proteins: Agents of Cancer Development and Therapeutic Targets in Anti-Cancer Therapy. Cells 9, 60. doi: 10.3390/cells9010060
Zheng, Y., Deng, Z., Yin, J., Wang, S., Lu, D., Wen, X., et al. (2017). The association of genetic variations in DNA repair pathways with severe toxicities in NSCLC patients undergoing platinum-based chemotherapy. Int. J. Cancer 141, 2336–2347. doi: 10.1002/ijc.30921
Zhou, Y., Liao, Q., Han, Y., Chen, J., Liu, Z., Ling, H., et al. (2016). Rac1 overexpression is correlated with epithelial mesenchymal transition and predicts poor prognosis in non-small cell lung cancer. J. Cancer 7, 2100–2109. doi: 10.7150/jca.16198
Zou, T., Yin, J., Zheng, W., Xiao, L., Tan, L., Chen, J., et al. (2016). Rho GTPases: RAC1 polymorphisms affected platinum-based chemotherapy toxicity in lung cancer patients. Cancer Chemother. Pharmacol. 78, 249–258. doi: 10.1007/s00280-016-3072-0
Keywords: lung cancer, platinum-based chemotherapy, prognosis, genetic polymorphism, heat shock proteins
Citation: Zou T, Liu JY, She L, Yin JY, Li X, Li XP, Zhou HH, Chen J and Liu ZQ (2020) The Association Between Heat-Shock Protein Polymorphisms and Prognosis in Lung Cancer Patients Treated With Platinum-Based Chemotherapy. Front. Pharmacol. 11:1029. doi: 10.3389/fphar.2020.01029
Received: 29 April 2020; Accepted: 24 June 2020;
Published: 21 July 2020.
Edited by:
Amit V. Pandey, University of Bern, SwitzerlandReviewed by:
Huijuan Wang, Northwest University, ChinaLucia Taja-Chayeb, National Institute of Cancerology (INCAN), Mexico
Copyright © 2020 Zou, Liu, She, Yin, Li, Li, Zhou, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao-Qian Liu, enFsaXVAY3N1LmVkdS5jbg==; Juan Chen, Y2oxMDI4QGNzdS5lZHUuY24=
 Ting Zou1,2,3
Ting Zou1,2,3 Hong-Hao Zhou
Hong-Hao Zhou Juan Chen
Juan Chen Zhao-Qian Liu
Zhao-Qian Liu