- 1Department of Mathematics and Statistics, Loyola University Chicago, Chicago, IL, United States
- 2School of Mathematical Sciences, University of Nottingham, Nottingham, United Kingdom
- 3Department of Statistics, The Ohio State University, Columbus, OH, United States
- 4Department of Geography and Anthropology, Louisiana State University, Baton Rouge, LA, United States
- 5Centre for the Exploration of the Deep Human Journey, University of the Witwatersrand, Johannesburg, South Africa
- 6Department of Computer Science, Loyola University Chicago, Chicago, IL, United States
- 7Department of Statistics, University of Connecticut, Mansfield, CT, United States
We consider the problem of classifying curves when they are observed only partially on their parameter domains. We propose computational methods for (i) completion of partially observed curves; (ii) assessment of completion variability through a nonparametric multiple imputation procedure; (iii) development of nearest neighbor classifiers compatible with the completion techniques. Our contributions are founded on exploiting the geometric notion of shape of a curve, defined as those aspects of a curve that remain unchanged under translations, rotations and reparameterizations. Explicit incorporation of shape information into the computational methods plays the dual role of limiting the set of all possible completions of a curve to those with similar shape while simultaneously enabling more efficient use of training data in the classifier through shape-informed neighborhoods. Our methods are then used for taxonomic classification of partially observed curves arising from images of fossilized Bovidae teeth, obtained from a novel anthropological application concerning paleoenvironmental reconstruction.
1 Introduction
Modern functional and curve data come in all shapes and sizes (pun intended!). A particular type of functional data that is starting to receive attention in recent years consists of univariate functions that are only observed in sub-intervals of their interval domains. Names for such data objects abound: censored functional data [1]; functional fragments [2,3]; functional snippets [4]; partially observed functional data [5]. Similar work with multivariate functional data or parametric curves in
The leitmotif of our approach lies in the explicit use of shapes of curves as a mechanism to not only counter the ill-posed nature of the problem of “sensibly” imputing or completing the missing piece of a partially observed curve, but also to use the metric geometry of the shape space of curves profitably when developing a suitable classifier. The rationale behind using shapes of curves can be explained quite simply. Fundamental to the routine task of comparing and identifying objects by humans or a computer is an implicit understanding of a set of symmetries or transformations pertaining to its shape: those properties or features of the object that are unaffected by nuisance information (e.g., orientation of the camera under which the object is imaged). Such an understanding assumes added importance when the object is only partially observed (e.g., identifying a chair hidden behind a table based on the backrest only) since it eliminates the need to consider a substantially large class of possible completions of the object. In the context of partially observed curves, working with their shapes leads to completions that are compatible with the shapes of fully observed curves in a training dataset. Relatedly, from an operational perspective, any formulation of completion of a missing piece of a curve based on an endpoints-constrained curve, either through deterministic or probabilistic model-based techniques, suffers from having too many degrees-of-freedom. As a result, the parameter space of missing pieces to search over needs to be constrained to obtain meaningful curve completions; we propose to impose such a shape-related constraint.
For example, in the anthropological application, any sensible completion of a bovid tooth should assume the shape of a tooth. We can constrain the parameter space comprising open curves, with endpoints constrained to match that of the partially observed curve, while determining a sensible completion based on the requirement that the completion should be tooth-like. Figure 1 shows an example of using shape information to complete a bovid tooth using Algorithm 1 (Section 3) and compares it to an arbitrary completion devoid of explicit shape information.
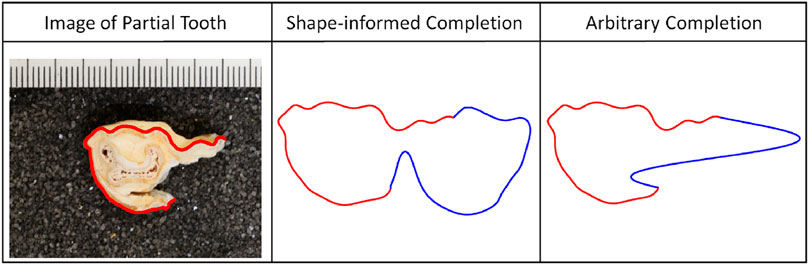
FIGURE 1. Anthropological application with bovid teeth. Left: Image of a partial tooth with the segmentation overlaid in red. Middle; Right: Shape-informed and arbitrary completions (blue) of the observed partial tooth (red), respectively.
An important consideration when considering shape of a curve is its scale. Strictly speaking, scaling a curve does not alter its shape, and it is hence a nuisance transformation. However, in our motivating application from anthropology, the size of bovid teeth is known to have important taxonomical information and can hence potentially improve discriminatory power in the classification problem [6]. We will therefore accord due consideration to scale information when comparing shapes of curves; in shape analysis vernacular, this is referred to as size-and-shape analysis. For simplicity, we will continue to chraracterize our approach as shape-informed.
Research in geometry-based statistical analysis of shapes of arbitrarily parameterized planar curves is quite mature; see, for example, [7,8] for foundational details and the R package fdasrvf for computational tools. Leveraging this, our main contributions are as follows.
1) We develop a gradient-based algorithm (Algorithm 1) for shape-informed partial matching and completion with respect to a complete template/donor curve.
2) In order to assess and visualize variability of completions from Algorithm 1, given a training dataset of fully observed curves, we propose an adaptation of the hot-deck imputation method used on traditional Euclidean data to generate several imputations or completions (Algorithm 2).
3) We propose two nearest neighbor classification procedures for partially observed curves based on shape distances by utlizing completions obtained from any of the above two algorithms.
1.1 Related Work
Partially observed curves arise as data in several applications. In medical imaging, the appearance of anatomies in images of various modalities is often summarized through the shapes of their outlines. Partial curves arise due to (i) low resolution and contrast of many medical imaging modalities (e.g., PET or CT); (ii) a boundary of an organ being obscured by other organs or hard to identify due to similar appearance of neighboring tissues [9]. In handwriting analysis, a key task is the segmentation of samples of handwriting (curves) into letters or syllables, followed by imputation of incomplete curves [10]. Shapes of occluded objects, such as tanks, are also routinely used in military applications, where only part of the object’s boundary is reliable and the rest must be imputed based on prior shape knowledge [9].
There is a substantial literature on missing data and shape analysis, however, most of the work is restricted to data obtained as multivariate morphological measurements. For example, [11] examines missing data in the morphology of crocodile skulls based on linear morphometric measurements of the skulls, in contrast to using landmarks or entire outlines (curves). Missing data methods for landmark-based shape data have been developed in [12, 13, 14, 15]. By defining landmarks on each shape, the problem can be framed in a more traditional statistical setting where each landmark can be thought of as a covariate and more traditional missing data techniques, such as the EM algorithm and multiple imputation, can be used. [16] look at four different methods for dealing with missing landmark data: Bayesian Principal Component Analysis (BPCA), least-squares regression, thin-plate splines (TPS), and mean substitution. Additional work on missing data can be found in [17], which focuses on missing data in Procrustes analysis, and [18], which considers occluded landmark data.
The work most closely related to this current study can be found in [19], which studies the problem of matching a partially observed shape to a full shape. [19] performs partial matching using the Square-Root Velocity framework, and this is the framework we use as well in the sequel. Our work in a certain sense goes beyond their work and incorporates missing data techniques into the completion procedure, and additionally is tailored towards the classification task. [9] incorporates prior shape information in Bayesian active contours that can be used to estimate object boundaries in images when the class of the object of interest is known; they demonstrate the usefulness of this approach when the object boundary is partially obscured. [20] considers the problem of identifying shapes in cluttered point clouds. They formulate a Bayesian classification model that also heavily relies on prior shape information. Finally, there is some recent work on missing data techniques for functional data analysis [21, 22, 23].
2 Shapes of Parameterized Curves
The main objects of interest in this work are parameterized curves and their shapes. Defining a suitable distance metric to compare their shapes is of fundamental importance in order to suitably formalize the notion of a “best completion of a partial curve”. From several available in the literature, we consider two distances that are suitable for our needs. We provide a description of the mathematical formulation for these two distances in the following, and refer the interested reader to [24] for more details. As discussed in the Introduction, the size of bovid teeth contains potentially taxon (class)-distinguishing information, and we hence consider the notion of size-and-shape of a curve. Throughout, for ease of exposition, we simply say shape to mean size-and-shape.
Denote by
This optimization problem can be solved in a straightforward fashion through Procrustes analysis [25]. The distance is termed non-elastic as it requires one to fix curve parameterizations to arc-length. Note that while dNE is defined on
If one desires to allow flexible parameterizations for shape analysis, the
Since re-parameterization completely preserves the image of a curve β, a distance based on a Riemannian metric that captures infinitesimal bending and stretching can be used. Several families of such metrics, termed as elastic have been considered [26, 27]; however, almost all of them are difficult to compute in practice and require non-trivial approximations.
A solution to this key issue was proposed in [7]. Specifically, a particular elastic metric is related to the usual
The corresponding elastic distance dE between two curves
where the equivalence class
In summary, if closed planar curves representing outlines of bovid teeth are assumed to be arc-length parameterized, we can use the non-elastic distance dNE on the shape space
3 Partial Shape Matching and Completion
We first focus on how a single partially observed curve can be completed. Indeed, this requires a template or donor curve that is fully observed, so that the partially observed one can be matched and compared to different pieces of the fully observed one. Once a match has been established, a completion can be subsequently determined. In principle, the two tasks can be carried out sequentially or in parallel; in this paper, we adopt the former approach and leave the latter for future work.
Accordingly, the key tasks are to (i) match the observed partial curve to a piece of the donor curve; (ii) impute or complete the observed curve by finding the closest match to the residual piece of the donor curve from a set of curves with fixed endpoints. These are non-trivial tasks since the set of curves
• A curve β is viewed as being composed of two pieces βo and βm, where the subscripts o and m identify the observed and missing portions of β, such that
• β = βo ⋆ βm denotes the concatenation of βo and βm, i.e., the complete curve. Throughout, βo will denote a partially observed curve, which conceptually is understood to be the observed portion of a curve β; in similar fashion, βm will throughout represent the missing piece of β.
• The restriction of a complete curve β to an open curve defined by parameter values [s1, s2] ⊂ [0, 1] is denoted as
• Denote the length of βo as
In line with our intention to use shape information of curves, we note that completion of βo with respect to a donor curve βdonor can be broken down into the following two steps.
(i) Determine the piece of βdonor that best matches the shape of βo.
(ii) Determine an open βm curve that then best matches the shape of the residual piece of the donor in (i); the required completion is then βo ⋆ βm.
Two points are worth considering here. First, the optimal βm is constrained to share the same endpoints as the determined piece of βdonor. Second, by virtue of its definition, the completion βo ⋆ βm exactly matches the partially observed curve βo when restricted to a suitable subset of the parameter domain. The latter is motivated by the quality of image data of bovid teeth, under which it is reasonable to assume that the partially observed curve is obtained under negligible measurement error.
The key consideration for developing an algorithm for the two steps is the choice of an objective function that quantifies the quality of matches, informed by either of the shape distances dNE and dE in Eqs 1, 2, respectively. Repeated computation of the elastic distance dE is computationally expensive (due to the additional optimization over Γ), and hence time-consuming inside an iterative algorithm (the potential donor set has large sample size). Since our main objective in this paper is classification of the partially observed curves, we use the more convenient non-elastic distance dNE in order to carry out partial matching and completion. However, we will employ the elastic distance dE when designing a classifier in order to better access pure shape features of curves that are potentially class-distinguishing.
We consider a two-step algorithm based on optimizing an objective function over two parameter spaces: ΩP for the partial match in step (i), and ΩC for the completion step (ii), defined as:
The parameter set ΩP consists of shape-preserving transformations for arc-length parameterized curves β and the length of βo, whereas ΩC represents the subset of endpoint constrained curves within
Note that
We propose to optimize over ΩC with a gradient-descent algorithm. First, rescale
where ⟨⋅, ⋅⟩ is the usual
where ϵ > 0 is a small step size. This is repeated until convergence. In practice, we reduce the dimension of the problem by truncating the basis at a finite number N; this additionally ensures that the optimal completion
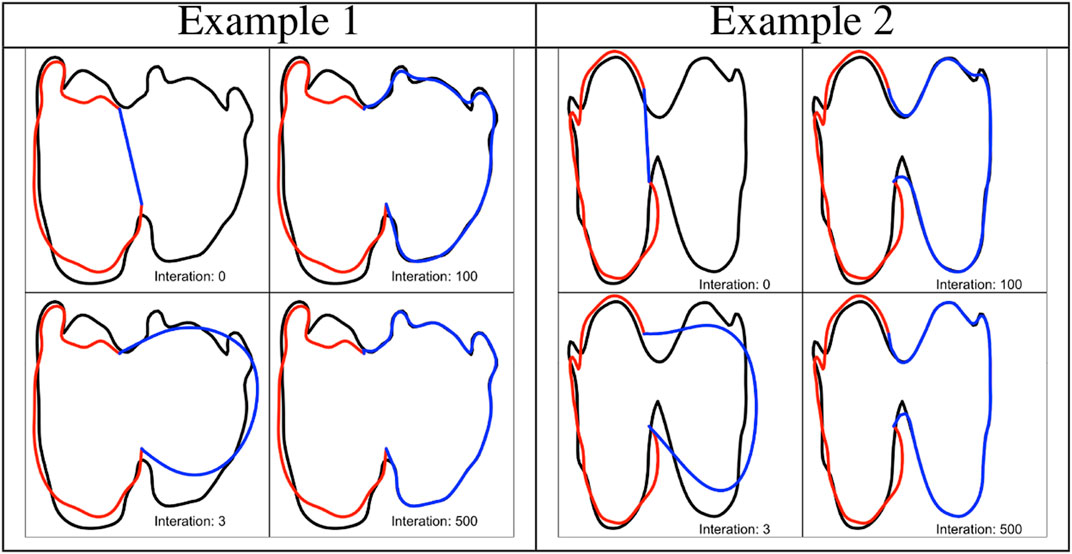
FIGURE 2. Two examples of shape imputation via Algorithm 1. The panels show the evolution of the completion (blue) at a few iterations of step 3, the observed partial curve (red) and the donor (black).
4 Assessing Variability in Completion Through Multiple Imputation
Algorithm 1 describes how a partially observed curve βo can be completed given a donor curve βdonor. The completion is deterministic and uncertainty estimates are unavailable. Further, the application of this procedure is only possible when a training dataset consisting of several curves is available. An attractive way to examine completion variability is to consider a multiple imputation framework for missing data. There are numerous multiple imputation methods to handle missing data in traditional multivariate settings; see [29, 30] for a broad overview and details on missing data techniques. Our choice is a nonparametric hot-deck multiple imputation procedure. We describe this technique in a regression setting with response
(i) Replace a missing value ymiss of y with randomly selected observed values in y, chosen from a donor pool of fixed size K < n comprising fully observed cases that are“similar” to the incomplete case.
(ii) Repeat step (i) M times to create M completed datasets.
(iii) Analyze the M completed datasets independently (e.g., mean estimation) and combine results using Rubin’s combining rules [29].
As a first step towards carrying out this program for partially observed curves, we propose an adaptation of the hot-deck imputation procedure for a classification task. However, the development of methods to combine classification results across M datasets, similar to Rubin’s rules, is an interesting research problem in its own right, and we leave that for future work (Section 7). Once steps (i) and (ii) are completed, it is possible to visualize variability associated with completion using Algorithm 1 by plotting the completions. Moreover, variability in classification results can also be computed as a function of a donor set of size K and number of completed datasets M. Algorithm 2 outlines our adapted hot-deck imputation procedure for generating M completions of a partially observed curve βo given a training dataset
Note that once
The key feature of Algorithm 2 is the use of the elastic distance, albeit not exactly in the form defined in (2) since an optimal rotation O* ∈ SO(2) has already been determined in line 5–we hence refer to this distance as partial elastic distance. The rationale for this is as follows. Once a piece of the donor βi of length L* corresponding to parameter values
Figure 3 provides an illustration of the hot-deck imputation procedure with M = 10 and K = 10 for two partially observed bovid teeth using Algorithm 2; see Section 6 for details on the bovid dataset.
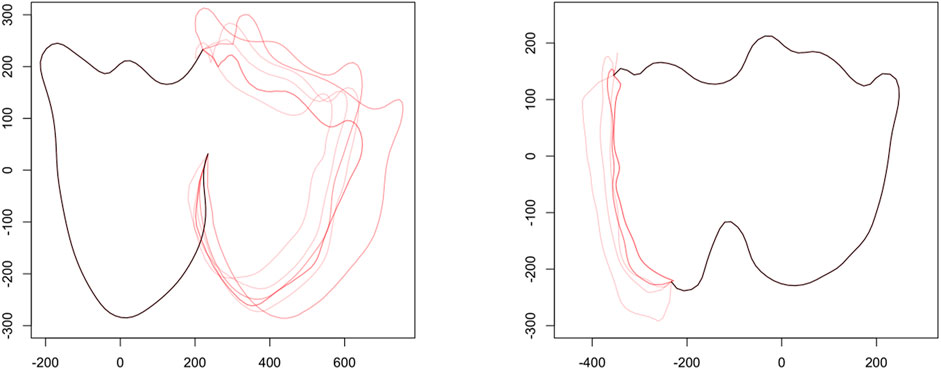
FIGURE 3. Examples of partial matches followed by imputation on partially observed teeth. Left: Approximately 50% of the tooth is observed. Right: Approximately 80% of the tooth is observed. The black curve denotes the fully observed portion of the tooth with each red curve being a single completion of the tooth. In both examples shown here M = 10 and K = 10.
5 Nearest Neighbor Classification
Consider a training dataset
A distance-based classification procedure is a natural choice, compatible with how completion and imputation is achieved. Accordingly, we consider the kn-nearest neighbor classification technique. A neighborhood of a curve in
The advantage of using the shape space lies in the fact that
In a kn-nearest neighbor setting, the radius r is distance, say
We will use the elastic distance dE again to accommodate for the possibility that curves in
•
and βo ⋆ βm is one completion obtained from Algorithm 1 with βm ∈ ΩC.
•
The class probability is thus obtained by averaging over all completions obtained by sampling M donor curves with replacement from the donor set
The
6 Tribe and Species Classification of Bovid Teeth
We examine performance of Algorithm 1 and Algorithm 2 with respect to classification of images of fully and partially observed bovid teeth using the two nearest neighbor methods under two settings.
(i) A simulated setting, where curves pertaining to partially observed teeth are created from fully observed ones with known class labels.
(ii) A real data setting comprising “true” partially observed teeth with unknown class labels.
There are numerous measures of classification performance. We use the log-loss measure to asses performance: let np denote the number of partially observed curves
where the class probability is defined as earlier depending on whether the
All computations are performed using routines available in the R [32] package fdasrvf [33] on a 16-node Intel Xeon-based computational cluster in the Computer Science Department at Loyola University Chicago. Full code for the analyses can be found on Github [34].
Our motivating application stems from anthropology, where fossil bovid teeth associated with our human ancestors are used to reconstruct past environments. Bovids are useful because they are ecologically sensitive to their environment and typically dominate the South African faunal assemblages [35, 36, 37, 38].
The tooth images for our study were obtained from four institutions in South Africa: National Museum, Bloemfontein; Ditsong Museum, Pretoria; and Amathole Museum, King William’s Town. Images were also taken at the Field Museum, Chicago, United States. The complete methodology as to how the teeth images were collected is outlined in [6]. Briefly, the occlusal surface of each of the three molars from the upper and lower dentitions for each extant bovid specimen were photographed separately. The specimen and the camera were levelled using a bubble level. A scale was placed next to the occlusal surface for every image.
Specifically, we consider images of teeth from 7 bovid tribes (Alcelaphini, Antilopini, Bovini, Hippotragini, Neotragini, Reduncini, and Tragelaphini) and 20 species (R. arundinum, A. buselaphus, S. caffer, R. campestris, P. capreolus, D. dorcas, K. ellipsiprymnus, H. equinus, R. fulvorufula, O. gazella, C. gnou, K. leche, A. marsupialis, H. niger, O. oreotragus, T. oryx, O. ourebi, T. scriptus, T. strepsiceros, C. taurinus). The dataset contains six tooth types: lower (i.e., mandibular) molars 1, 2 and 3 (LM1, LM2, LM3), and upper (i.e., maxillary) molars 1, 2 and 3 (UM1, UM2, UM3). Specific counts of the sample sizes of each tooth type and tribe are in Table 1. This dataset contains fully observed teeth of known taxa and will constitute the training data
Each tribe has unique dental characteristics that are shared by its members. Further, the complexity of the occlusal surface outline varies across the tribes. As such, considering shape for tribe classification is a natural endeavor in this application. Generally, classification at the species level is more difficult since the variability of shapes of occlusal surface outlines across species within the same tribe is not as large.
6.1 Simulated Setting
In this setting, a partially observed tooth was created from a fully observed one with known class label in
(i) K = 5, 10, 20 of size of donor set
(ii) M = 5, 10, 20 of number of imputations based on sampling with replacement from
(iii) kn = 1, 2, … , 20 of number of neighbors;
(iv) Tooth types LM1, LM2, LM3, UM1, UM2 and UM3;
(v) Side of tooth extracted, where Left is denoted as 1 and Right as 2.
A partially observed tooth was created in the following manner. The raw representation of each tooth in
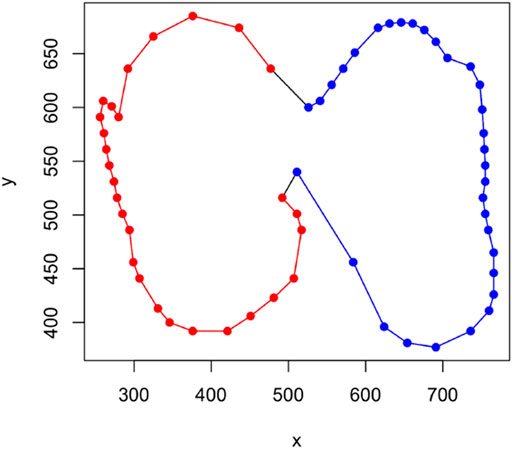
FIGURE 4. An example of how a partially observed tooth is obtained from a fully observed one in the simulated setting. The figure shows a lower molar 1 (LM1) from the tribe Alcelaphini with red points representing the extracted piece from the Left (1) side and blue points do the same for the Right (2) side.
6.1.1 Tribe classification
Figure 5 shows Log-loss curves associated with the
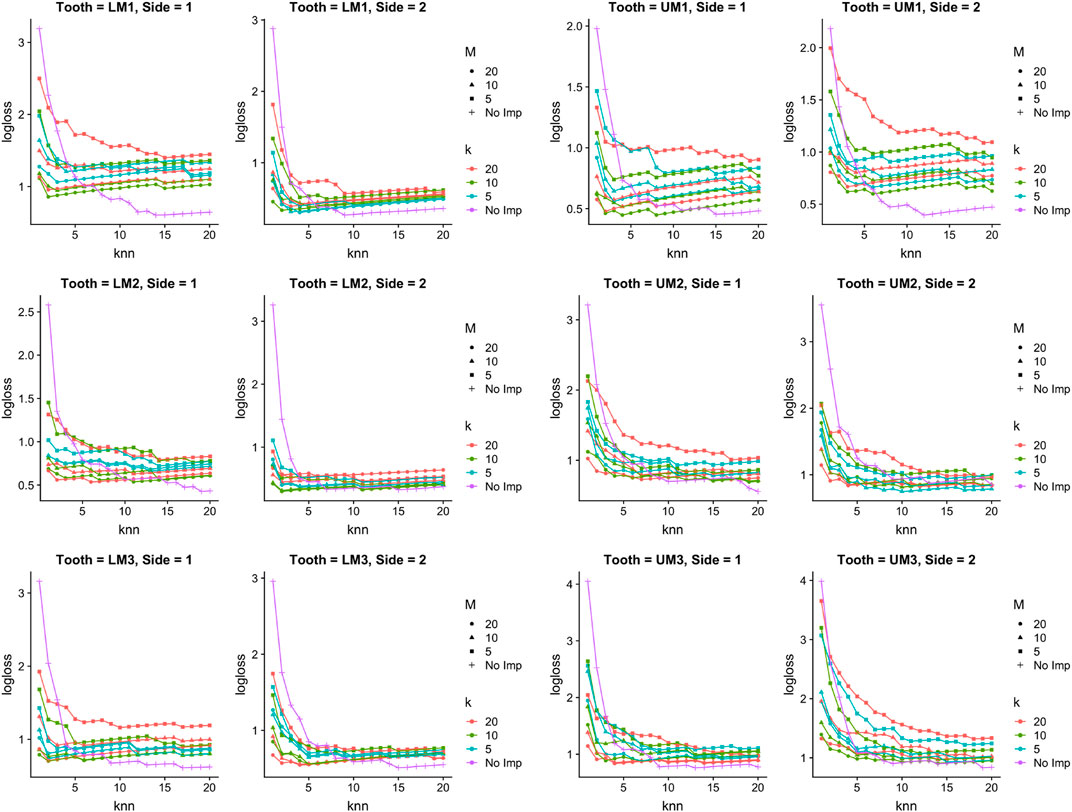
FIGURE 5. Tribe classification in simulated setting. Log-loss for the
6.1.2 Species classification
Figure 6 shows Log-loss curves for species classification and paints a fairly similar picture to Figure 5: when the number of nearest neighbors is chosen to be small (i.e., fewer than 5), there is always at least one imputation setting that has lower Log-loss than no imputation. However, in all cases as the number of nearest neighbors chosen gets close to 20, no imputation performs better in terms of Log-loss than doing imputation.
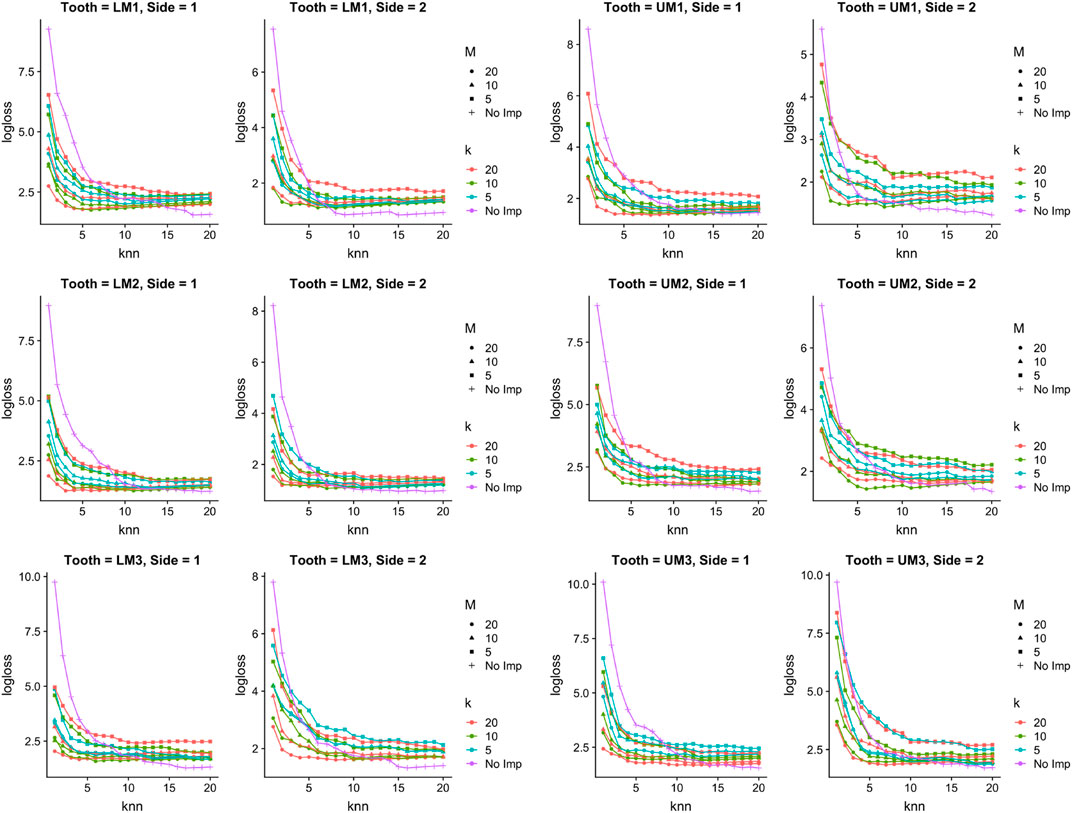
FIGURE 6. Species classification in simulated setting. Log-loss for the
6.2 Real Data Setting
A small set np = 7 of real, partially observed fossil bovid teeth from the site of Gladysvale, South Africa, extracted from images, with unknown class labels were used. We use the image numbers to label them: IMG4825, IMG4980, IMG4983, IMG4990, IMG5139, IMG9973 and IMG5514. Training data
For both
6.2.1 Classification at Tribe Level
Table 2 shows predicted class probabilities associated with the
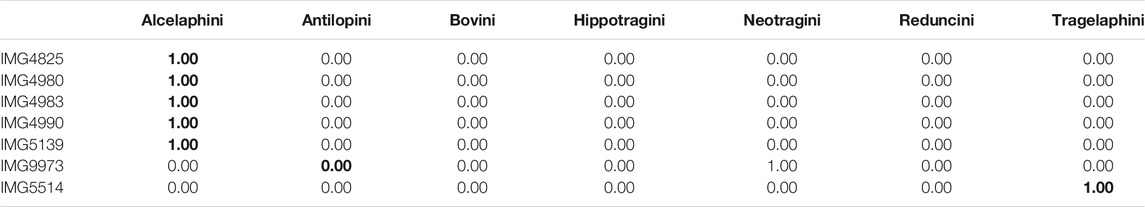
TABLE 2. Real data. Tribe level predicted class probabilities from the
Table 3 shows similar results for the
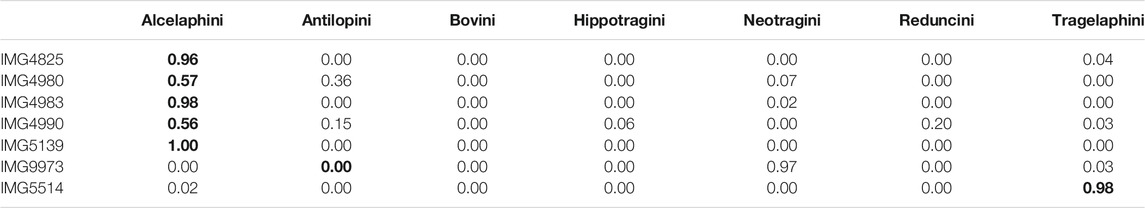
TABLE 3. Real data. Tribe level predicted class probabilities from the
6.2.2 Classification at the Species Level
Table 4 shows predicted class probabilities associated with the
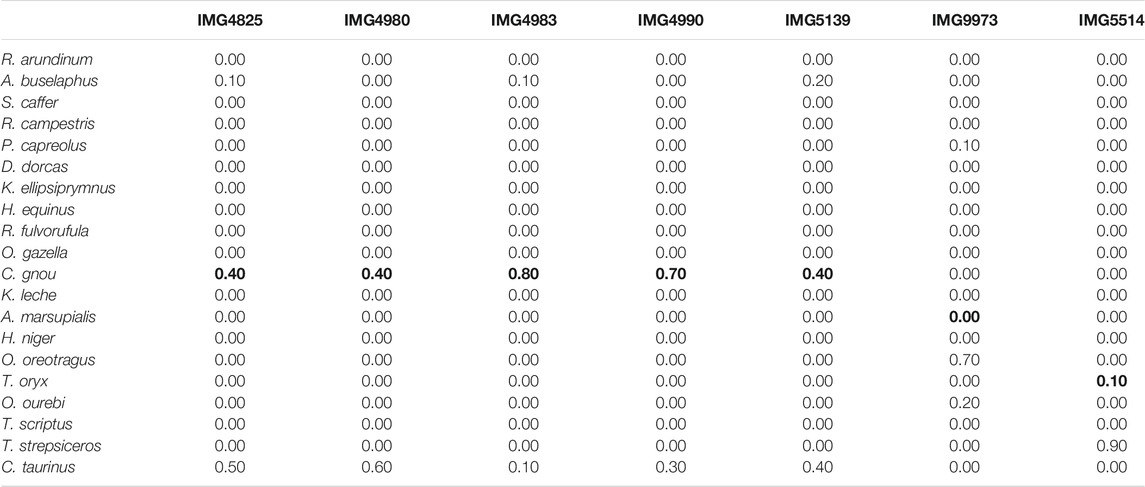
TABLE 4. Real data. Species level predicted class probabilities with
When classifying using the
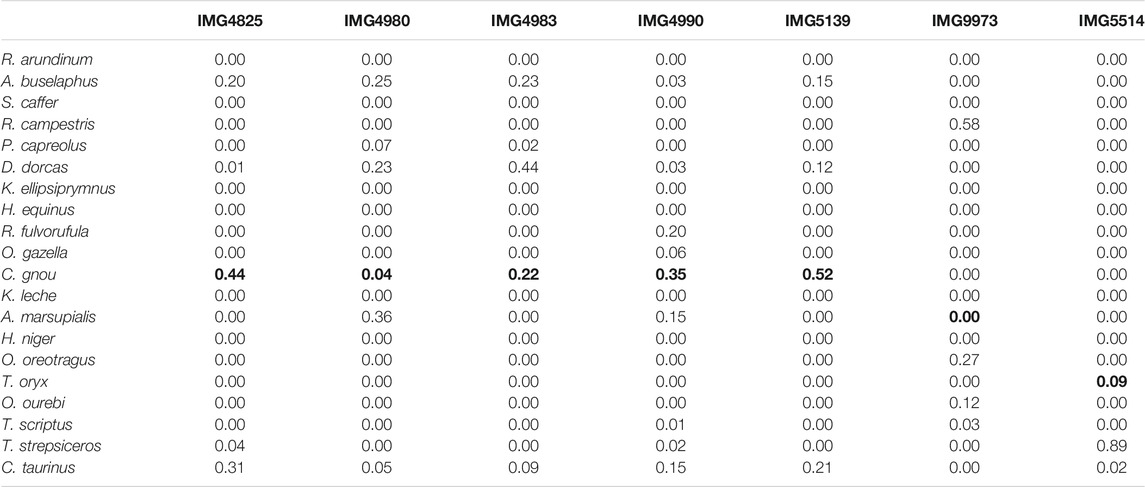
TABLE 5. Real data. Species-level predicted class probabilities with
7 Discussion
We have presented a computational approach for classifying partially observed curves. In particular, we presented two algorithms to complete and classify partially observed planar curves and simultaneously assess variability involved with the completion through a multiple imputation procedure. To our knowledge, this is the first work in literature to explicitly use the notion of shapes of parameterized curves in addressing the problem considered from the missing data perspective; coarsening the parameter space of suitable open curves, from which the partially observed curves are completed, through the notion of shape equivalence results in sensible completions. Moreover, shape-based distances used to define classifiers deliver satisfactory classification performance. The results from application of the algorithms to the dataset of images of bovid teeth are quite promising and are deserving of further, extensive, investigation involving several different classifiers.
Through the application of the proposed framework on real data, we have found that hot-deck imputation can sometimes deteriorate classification performance; there is an intuitive explanation for these findings. Classification performance is greatly affected by the “amount of information” contained in the observed partial curve. By “amount of information”, we specifically mean the ability to discriminate between different classes. In particular, if the observed partial curve contains a lot of information about its class membership compared to the missing portion, then imputation injects additional variability into the problem, which has a negative effect on classification performance. On the other hand, if the observed partial curve is not easily distinguishable across the different classes in the training data, then the variability coming from the imputation procedure provides valuable information, thus improving classification performance. Knowledge about information content in an observed partial curve for classification can be obtained either from a training dataset consisting of fully observed curves with class labels or from a subject matter expert. In such cases, a Bayesian classification model with a judicious choice of prior on class-specific templates can be developed; such an approach will extend the one recently proposed for univariate functional data [23] to the curve setting, and constitutes ongoing work.
As with any methodological development that represents a first foray into tackling a challenging problem, our approach suffers from a few shortcomings, which inevitably present many possible avenues for future research. Algorithm 1 can be improved. Ideally, the partial match and completion steps are carried out jointly. Moreover, assuming curves to be arc-length parameterized, while convenient, can sometimes be unrealistic in practice, especially when data curves are extracted as part of an elaborate pre-processing procedure. This points towards developing a version of Algorithm 1 based on the corresponding SRVFs qo and qm; the main challenge here is how to handle the interplay between points
In the current work, an explicit statistical model to handle the several sources of variation (e.g., measurement error in extracting curves from images) that can profoundly affect both completion and classification is conspicuous in its absence; without such a model, it is difficult to quantify uncertainty about the completions, which quite naturally percolates down to the classification task. An attractive model-based approach is to not just estimate the missing piece of the partially observed curve, but instead estimate an entire template that has a portion that is very similar in shape to the partially observed curve. Such an approach has recently been used for traditional univariate functional data under a Bayesian formulation [23] and appears promising.
Our primary task in this paper is classification. However, it is unclear how one can use the proposed algorithms if interest was in computing statistical summaries in the presence of partially observed curves, such as the mean shape or PCA on the space of shapes. For example, output of Algorithm 2 is a set of M closed curves
More generally, while the hot-deck imputation procedure worked reasonably well when combined with the completion task, there is a pressing need to systematically develop missing data concepts and imputation methods to better address the special structure of missingness in the context of shapes of curves. The following challenges naturally arise: (i) Is the notion of Missing Completely at Random (MCAR), so profitably used in traditional settings, ever a reasonable assumption for shapes of curves? It is almost impossible to disentangle measurement error from reasons for why a piece of a curve is missing. (ii) Conditional probability measures associated with random functions when conditioned on its values in a sub-domain are notoriously difficult, and rarely exist. Given this, how does one adapt, or perhaps circumvent, the traditional notion of sampling from the conditional distribution of the missing values conditioned on the observed values to the present setting? Much remains to be done in this direction.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
All authors participated in problem formulation. GM, KB, SK, and OH developed and implemented the methodology. GM analyzed the data. JB provided the data and interpreted the results. GT helped with computation.
Funding
This work is partially supported by the following grants: NSF DMS-2015320, NSF DMS-1811969 (OH); NSF DMS-1812065, NSF DMS-2015236 (JKB); NSF DMS-1812124 (GJM and GKT); NSF DMS- 2015374 (GJM and KB); NSF NIH R37-CA214955 (SK and KB); EPSRC EP/V048104/1 (KB); NSF CCF-1740761, NSF DMS-2015226, NSF CCF-1839252 (SK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Professor Ian Jermyn for valuable discussions on this topic. We would also like to thank Grady Flanary and Kajal Chokshi for their help in the early stages of this project.
Footnotes
1In practice, the basis is truncated to some finite number. We have found that between 40 and 80 total basis elements per coordinate function are sufficient, although this depends on the geometric complexity of the observed curves. In the application considered in Section 6, we used 80 basis elements.
References
1. Delaigle, A, and Hall, P. Classification using censored functional data. J Am Stat Assoc (2013) 108(504):1269–83. doi:10.1080/01621459.2013.824893
2. Delaigle, A, and Hall, P. Approximating fragmented functional data by segments of Markov chains. Biometrika (2016) 103:779–99. doi:10.1093/biomet/asw040
3. Descary, M-H, and Panaretos, VM. Recovering covariance from functional fragments. Biometrika (2018) 106(1):145–60. doi:10.1093/biomet/asy055
4. Lin, Z, Wang, J-L, and Zhong, Q. Basis expansions for functional snippets. Oxford: Biometrika (2020). In Press.
5. Kraus, D. Components and completion of partially observed functional data. J R Stat Soc B (2015) 77(5):777–801. doi:10.1111/rssb.12087
6. Brophy, JK, de Ruiter, DJ, Athreya, S, and DeWitt, TJ. Quantitative morphological analysis of bovid teeth and implications for paleoenvironmental reconstruction of Plovers Lake, Gauteng Province, South Africa. J Archaeological Sci (2014) 41:376–88. doi:10.1016/j.jas.2013.08.005
7. Srivastava, A, Klassen, E, Joshi, SH, and Jermyn, IH. Shape analysis of elastic curves in Euclidean spaces. IEEE Trans Pattern Anal Mach Intell (2011) 33:1415–28. doi:10.1109/tpami.2010.184
8. Kurtek, S, Srivastava, A, Klassen, E, and Ding, Z. Statistical modeling of curves using shapes and related features. J Am Stat Assoc (2012) 107(499):1152–65. doi:10.1080/01621459.2012.699770
9. Joshi, SH, and Srivastava, A. Intrinsic Bayesian active contours for extraction of object boundaries in images. Int J Comput Vis (2009) 81(3):331–55. doi:10.1007/s11263-008-0179-8
10. Kurtek, S, and Srivastava, A. Handwritten text segmentation using elastic shape analysis. In: International Conference on Pattern Recognition (2014). p. 2501–6. doi:10.1109/icpr.2014.432
11. Brown, CM, Arbour, JH, and Jackson, DA. Testing of the effect of missing data estimation and distribution in morphometric multivariate data analyses. Syst Biol (2012) 61(6):941–54. doi:10.1093/sysbio/sys047
12. Gunz, P, Mitteroecker, P, Neubauer, S, Weber, GW, and Bookstein, FL. Principles for the virtual reconstruction of hominin crania. J Hum Evol (2009) 57:48–62. doi:10.1016/j.jhevol.2009.04.004
13. Gunz, P, Mitteroecker, P, Bookstein, FL, and Weber, GW. Computer-aided reconstruction of incomplete human crania using statistical and geometrical estimation methods. In: Enter the past: the e-way into the four dimensions of cultural heritage; CAA 2003; computer applications and quantitative methods in archaeology. Oxford: Archaeopress (2004). p. 92–4.
14. Neeser, R, Ackermann, RR, and Gain, J. Comparing the accuracy and precision of three techniques used for estimating missing landmarks when reconstructing fossil Hominin Crania. Am J Phys Anthropol (2009) 140:1–18. doi:10.1002/ajpa.21023
15. Couette, S, and White, J. 3D geometric morphometrics and missing-data. Can extant taxa give clues for the analysis of fossil primates? Gen Palaeontology (2009) 9:423–33.
16. Arbour, JH, and Brown, CM. Incomplete specimens in geometric morphometric analyses. Methods Ecol Evol (2014) 5:16–26. doi:10.1111/2041-210x.12128
17. Albers, CJ, and Gower, JC. A general approach to handling missing values in procrustes analysis. Adv Data Anal Classif (2010) 4(4):223–37. doi:10.1007/s11634-010-0077-0
18. Mitchelson, JR. MOSHFIT: algorithms for occlusion-tolerant mean shape and rigid motion from 3D movement data. J Biomech (2013) 46:2326–9. doi:10.1016/j.jbiomech.2013.05.029
19. Robinson, DT. Functional Data Analysis and Partial Shape Matching in the Square Root Velocity Framework. PhD thesis. Florida: Florida State University (2012).
20. Srivastava, A, and Jermyn, IH. Looking for Shapes in Two-Dimensional Cluttered Point Clouds. IEEE Trans Pattern Anal Mach Intell (2009) 31(9):1616–29. doi:10.1109/tpami.2008.223
21. Liebl, D, and Rameseder, S. Partially observed functional data: The case of systematically missing parts. Comput Stat Data Anal (2019) 131:104–15. doi:10.1016/j.csda.2018.08.011
22. Ciarleglio, A, Petkova, E, and Harel, O. Elucidating age and sex-dependent association between frontal EEG asymmetry and depression: An application of multiple imputation in functional regression. J Am Stat Assoc (2021). In Press. doi:10.1080/01621459.2021.1942011
23. Matuk, J, Bharath, K, Chkrebtii, O, and Kurtek, S. Bayesian framework for simultaneous registration and estimation of noisy, sparse and fragmented functional data. J Am Stat Assoc (2021). In Press, doi:10.1080/01621459.2021.1893179
24. Srivastava, A, and Klassen, EP. Functional and Shape Data Analysis. New York, NY: Springer (2016).
25. Kendall, DG. Shape Manifolds, Procrustean Metrics, and Complex Projective Spaces. Bull Lond Math Soc (1984) 16:81–121. doi:10.1112/blms/16.2.81
26. Younes, L. Computable Elastic Distances between Shapes. SIAM J Appl Math (1998) 58(2):565–86. doi:10.1137/s0036139995287685
27. Mio, W, Srivastava, A, and Joshi, S. On shape of plane elastic curves. Int J Comput Vis (2007) 73(3):307–24. doi:10.1007/s11263-006-9968-0
29. Little, RJA, and Rubin, DB. Statistical Analysis with Missing Data. UK: John Wiley & Sons (1987).
30. Andridge, RR, and Little, RJA. A review of hot deck imputation for survey non-response. Int Stat Rev (2010) 78(1):40–64. doi:10.1111/j.1751-5823.2010.00103.x
32.R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing (2021).
34. Matthews, GJ. Shape Completion Matthews et al. US: GitHub repository (2021). Available at: https://github.com/gjm112/shape_completion_Matthews_et_al.
35. Bobe, R, and Eck, GG. Responses of African bovids to Pliocene climatic change. Paleobiology (2001) 27:1–48. doi:10.1666/0094-8373(2001)027<0001:roabtp>2.0.co;2
36. Bobe, R, Behrensmeyer, AK, and Chapman, RE. Faunal change, environmental variability and late Pliocene hominin evolution. J Hum Evol (2002) 42:475–97. doi:10.1006/jhev.2001.0535
37. Alemseged, Z. An integrated approach to taphonomy and faunal change in the Shungura Formation (Ethiopia) and its implication for hominid evolution. J Hum Evol (2003) 44:451–78. doi:10.1016/s0047-2484(03)00012-5
38. de Ruiter, DJ, Brophy, JK, Lewis, PJ, Churchill, SE, and Berger, LR. Faunal assemblage composition and paleoenvironment of Plovers Lake, a Middle Stone Age locality in Gauteng Province, South Africa. J Hum Evol (2008) 55:1102–17. doi:10.1016/j.jhevol.2008.07.011
Keywords: shapes of parameterized curves, curve completion, invariance, multiple imputation, classification
Citation: Matthews GJ, Bharath K, Kurtek S, Brophy JK, Thiruvathukal GK and Harel O (2021) Shape-Based Classification of Partially Observed Curves, With Applications to Anthropology. Front. Appl. Math. Stat. 7:759622. doi: 10.3389/fams.2021.759622
Received: 16 August 2021; Accepted: 28 September 2021;
Published: 26 October 2021.
Edited by:
Anuj Srivastava, Florida State University, United StatesReviewed by:
Mawardi Bahri, Hasanuddin University, IndonesiaChristian Gerhards, University of Vienna, Austria
Copyright © 2021 Matthews, Bharath, Kurtek, Brophy, Thiruvathukal and Harel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory J. Matthews, Z21hdHRoZXdzMUBsdWMuZWR1
 Gregory J. Matthews
Gregory J. Matthews Karthik Bharath2
Karthik Bharath2 Sebastian Kurtek
Sebastian Kurtek George K. Thiruvathukal
George K. Thiruvathukal