- Georgia Institute of Technology, Atlanta, GA, United States
Testing for independence plays a fundamental role in many statistical techniques. Among the nonparametric approaches, the distance-based methods (such as the distance correlation-based hypotheses testing for independence) have many advantages, compared with many other alternatives. A known limitation of the distance-based method is that its computational complexity can be high. In general, when the sample size is n, the order of computational complexity of a distance-based method, which typically requires computing of all pairwise distances, can be O(n2). Recent advances have discovered that in the univariate cases, a fast method with O(n log n) computational complexity and O(n) memory requirement exists. In this paper, we introduce a test of independence method based on random projection and distance correlation, which achieves nearly the same power as the state-of-the-art distance-based approach, works in the multivariate cases, and enjoys the O(nK log n) computational complexity and O( max{n, K}) memory requirement, where K is the number of random projections. Note that saving is achieved when K < n/ log n. We name our method a Randomly Projected Distance Covariance (RPDC). The statistical theoretical analysis takes advantage of some techniques on the random projection which are rooted in contemporary machine learning. Numerical experiments demonstrate the efficiency of the proposed method, relative to numerous competitors.
1 Introduction
Test of independence is a fundamental problem in statistics, with many existing work including the maximal information coefficient (MIC) [1], the copula based measures [2,3], the kernel based criterion [4] and the distance correlation [5,6], which motivated our current work. Note that the above works as well as ours focus on the testing for independence, which can be formulated as statistical hypotheses testing problems. On the other hand, interesting developments (e.g., [7]) aim at a more general framework for interpretable statistical dependence, which is not the goal of this paper.
Distance correlation proposed by [6] is an important method in the test of independence. The direct implementation of distance correlation takes O(n2) time, where n is the sample size. The time cost of distance correlation could be substantial when the sample size is just a few thousand. When the random variables are univariate, there exist efficient numerical algorithms of time complexity O(n log n) [8]. However, for the multivariate random variables, we have not found any efficient algorithms in existing papers after an extensive literature survey.
Independence tests of multivariate random variables could have a wide range of applications. In many problem settings, as mentioned in [9], each experimental unit will be measured multiple times, resulting in multivariate data. Researchers are often interested in exploring potential relationships among subsets of these measurements. For example, some measurements may represent attributes of physical characteristics while others represent attributes of psychological characteristics. It may be of interest to determine whether there exists a relationship between the physical and psychological characteristics. A test of independence between pairs of vectors, where the vectors may have different dimensions and scales, becomes crucial. Moreover, the number of experimental units, or equivalently, sample size, could be massive, which requires the test to be computationally efficient. This work will meet the demands for numerically efficient independence tests of multivariate random variables.
The newly proposed test of independence between two (potentially multivariate) random variables X and Y works as follows. Firstly, both X and Y are randomly projected to one-dimensional spaces. Then the fast computing method for distance covariances between a pair of univariate random variables is adopted to compute for a surrogate distance covariance. The above two steps are repeated numerous times. The final estimate of the distance covariance is the average of all aforementioned surrogate distance covariances.
For numerical efficiency, we will show (in Theorem 3.1) that the newly proposed algorithm enjoys the O(Kn log n) computational complexity and O( max{n, K}) memory requirement, where K is the number of random projections and n is the sample size. On the statistical efficiency, we will show (in Theorem 4.19) that the asymptotic power of the test of independence by utilizing the newly proposed statistics is as efficient as its original multivariate counterpart, which achieves the state-of-the-art rates.
The rest of this paper is organized as follows. In Section 2, we review the definition of distance covariance, its fast algorithm in univariate cases, and related distance-based independence tests. Section 3 gives the detailed algorithm for distance covariance of random vectors and corresponding independence tests. In Section 4, we present some theoretical properties on distance covariance and the asymptotic distribution of the proposed estimator. In Section 5, we conduct numerical examples to compare our method against others in the existing literature. Some discussions are presented in Section 6. We conclude in Section 7. All technical proofs, as well as the formal presentation of algorithms, are relegated to the appendix when appropriate.
Throughout this paper, we adopt the following notations. We denote
2 Review of Distance Covariance: Definition, Fast Algorithm, and Related Independence Tests
In this section, we review some related existing works. In Section 2.1, we recall the concept of distance variances and correlations, as well as some of their properties. In Section 2.2, we discuss the estimators of distance covariances and correlations, as well as their computation. We present their applications in the test of independence in Section 2.3.
2.1 Definition of Distance Covariances
Measuring and testing the dependency between two random variables is a fundamental problem in statistics. The classical Pearson’s correlation coefficient can be inaccurate and even misleading when nonlinear dependency exists [6]. propose the novel measure–distance correlation–which is exactly zero if and only if two random variables are independent. A limitation is that if the distance correlation is implemented based on its original definition, the corresponding computational complexity can be as high as O(n2), which is not desirable when n is large.
We review the definition of the distance correlation in [6]. Let us consider two random variables
where two constants cp and cq have been defined at the end of Section 1. The distance correlation is defined as
The following property has been established in the aforementioned paper.
Theorem 2.1 nSuppose
1) X is independent of Y;
2) ϕX,Y(t, s) = ϕX(t)ϕY(s), for any
3)
4)
Given sample (X1, Y1), …, (Xn, Yn), we can estimate the distance covariance by replacing the population characteristic function with the sample characteristic function: for
Consequently one can have the following estimator for
Note that the above formula is convenient to define a quantity, however, is not convenient for computation, due to the integration on the right-hand side. In the literature, other estimates have been introduced and will be presented in the following.
2.2 Fast Algorithm in the Univariate Cases
The paper [10] gives an equivalent definition for the distance covariance between random variables X and Y:
where the double centered distance d(⋅, ⋅) is defined as
where
Motivated by the above definition, one can give an unbiased estimator for
It has been proven [8, 28] that
is an unbiased estimator of
Theorem 2.2 (Theorem 3.2 & Corollary 4.1 in [8]). Suppose X1, …, Xn and
Corollary 2.3 The quantity
can be computed by an O(n log n) algorithm.We will use the above result in our test of independence. However, as far as we know, in the multivariate cases, there does not exist any work on the fast algorithm of the order of complexity O(n log n). This paper will fill in this gap by introducing an order O(nK log n) complexity algorithm in multivariate cases.
2.3 Distance Based Independence Tests
Ref. [6] proposed an independence test using the distance covariance. We summarize it below as a theorem, which serves as a benchmark. Our test will be aligned with the following one, except that we introduced a new test statistic, which can be more efficiently computed, and it has comparable asymptotic properties with the test statistic that is used below.
Theorem 2.4 ([6], Theorem 6). For potentially multivariate random variables X and Y, a prescribed level αs, and sample size n, one rejects the independence if and only if
where
Moreover, let α(X, Y, n) denote the achieved significance level of the above test. If
Note that the quantity
3 Numerically Efficient Method for Random Vectors
This section is made of two components. We present a random-projection-based distance covariance estimator that will be proven to be unbiased with a computational complexity that is O(Kn log n) in Section 3.1. In Section 3.2, we describe how the test of independence can be done by utilizing the above estimator. For users’ convenience, stand-alone algorithms are furnished in the Supplementary Appendix.
3.1 Random Projection Based Methods for Approximating Distance Covariance
We consider how to use a fast algorithm for univariate random variables to compute or approximate the sample distance covariance of random vectors. The main idea works as follows: first, projecting the multivariate observations on some random directions; then, using the fast algorithm to compute the distance covariance of the projections; at last, averaging distance covariances from different projecting directions.
More specifically, our estimator can be computed as follows. For potentially multivariate
1) For each k (1 ≤ k ≤ K), randomly generate uk and vk from Uniform
2) Let
Note that samples
3) Utilize the fast (i.e., order O(n log n)) algorithm that was mentioned in Theorem 2.2 to compute for the unbiased estimator in Eq. 2.5 with respect to
where Cp and Cq have been defined at the end of Section 1.
(4) The above three steps are repeated for K times. The final estimator is
To emphasize the dependency of the above quantity with K, we sometimes use a notation
See Algorithm 1 in the Supplementary Appendix for a stand-alone presentation of the above method. In the light of Theorem 2.2, we can handily declare the following.
Theorem 3.1 For potentially multivariate
3.2 Test of Independence
By a later result (cf. Theorem 4.19), we can apply
1) For each ℓ, 1 ≤ ℓ ≤ L, generate a random permutation of Y:
2) Using the algorithm in Section 3.1, one can compute the estimator
3) The above two steps are executed for all ℓ = 1, …, L. One rejects
See Algorithm 2 in the Supplementary Appendix for a stand-alone description.
It is notified that one can use the approximate asymptotic distribution information to estimate a threshold in the independence test. The following describes such an approach. Recall that random vectors
1) For each k (1 ≤ k ≤ K), randomly generate uk and vk from uniform
2) Use the fast algorithm in Theorem 2.2 to compute the following quantities:
where Cp and Cq have been defined at the end of Section 1 and in the last equation, the
3) For the aforementioned k, randomly generate
where Cp and Cq have been defined at the end of Section 1.
4) Repeat the previous steps for all k = 1, …, K. Then we compute the following quantities:
5) Reject
The above procedure is motivated by the observation that the asymptotic distribution of the test statistic
4 Theoretical Properties
In this section, we establish the theoretical foundation of the proposed method. In Section 4.1, we study some properties of the random projections and the subsequent average estimator. These properties will be needed in studying the properties of the proposed estimator. We study the properties of the proposed distance covariance estimator (Ωn) in Section 4.2, taking advantage of the fact that Ωn is a U-statistic. It turns out that the properties of eigenvalues of a particular operator play an important role. We present the relevant results in Section 4.3. The main properties of the proposed estimator
4.1 Using Random Projections in Distance-Based Methods
In this section, we will study some properties of distance covariances of randomly projected random vectors. We begin with a necessary and sufficient condition of independence.
Lemma 4.1 Suppose u and v are points on the hyper-spheres:
if and only if
The proof is relatively straightforward. We relegate a formal proof to the appendix. This lemma indicates that the independence is somewhat preserved under projections. The main contribution of the above result is to motivate us to think of using random projection, to reduce the multivariate random vectors into univariate random variables. As mentioned earlier, there exist fast algorithms of distance-based methods for univariate random variables.The following result allows us to regard the distance covariance of random vectors of any dimension as an integral of distance covariance of univariate random variables, which are the projections of the aforementioned random vectors. The formulas in the following lemma provide the foundation for our proposed method: the distance covariances in the multivariate cases can be written as integrations of distance covariances in the univariate cases. our proposed method essentially adopts the principle of Monte Carlo to approximate such integrals. We again relegate the proof to the Supplementary Appendix.
Lemma 4.2 Suppose u and v are points on unit hyper-spheres:
where Cp and Cq are two constants that are defined at the end of Section 1. Moreover, a similar result holds for the sample distance covariance:
Besides the integral equations in the above lemma, we can also establish the following result for the unbiased estimator. Such a result provides the direct foundation of our proposed method. Recall that Ωn, which is in Eq. 2.5, is an unbiased estimator of the distance covariance
Lemma 4.3 Suppose u and v are points on the hyper-spheres:
where Cp and Cq are constants that were mentioned at the end of Section 1.From the above lemma, recalling the design of our proposed estimator
Corollary 4.4 The proposed estimator
Lemma 4.5 Suppose E[|X|2] < ∞ and E[|Y|2] < ∞. For any ϵ > 0, we have
where ΣX and ΣY are the covariance matrices of X and Y, respectively, Tr[ΣX] and Tr[ΣY] are their matrix traces, and
4.2 Asymptotic Properties of the Sample Distance Covariance Ωn
The asymptotic behavior of the sample distance covariance Ωn in Eq. 2.5 of this paper, has been studied in many places, seeing [5,8,10,12]. We found that it is still worthwhile to present them here, as we will use them to establish the statistical properties of our proposed estimator. The asymptotic distributions of Ωn will be studied under two situations: 1) a general case and 2) when X and Y are assumed to be independent. We will see that the asymptotic distributions are different in these two situations.
It has been showed in ([8], Theorem 3.2) that Ωn is a U-statistic. In the following, we state the result without formal proof. We will need the following function, denoted by h4, which takes four pairs of input variables:
Note that the definition of h4 coincides with Ωn when the number of observations n = 4.
Lemma 4.6 (U-statistics). Let Ψ4 denote all distinct 4-subset of {1, …, n} and let us define Xψ = {Xi|i ∈ ψ} and Yψ = {Yi|i ∈ ψ}, then Ωn is a U-statistic and can be expressed as
From the literature of the U-statistics, we know that the following quantities play critical roles. We state them here:
where
Lemma 4.7 (Variance of the U-statistic). The variance of Ωn could be written as
where O(⋅) is the standard big O notation in mathematics.From the above lemma, we can see that Var(h1) and Var(h2) play indispensable roles in determining the variance of Ωn. The following lemma shows that under some conditions, we can ensure that Var(h1) and Var(h2) are bounded. A proof has been relegated to the appendix.
Lemma 4.8 If we have
Lemma 4.9 (Generic h1 and h2). In the general case, assuming (X1, Y1), (X, Y), (X′, Y′), and (X″, Y″) are independent and identically distributed, we have
We have a similar formula for h2((X1, Y1), (X2, Y2)) in (B.7). Due to its length, we do not display it here.If one assumes that X and Y are independent, we can have a simpler formula for h1, h2, as well as their corresponding variances. We list the results below, with detailed calculations relegated to the appendix. One can see that under independence, the corresponding formulas are much simpler.
Lemma 4.10 When X and Y are independent, we have the following. For (X, Y) and (X′, Y′) that are independent and identically distributed as (X1, Y1) and (X2, Y2), we have
where
Theorem 4.11 Suppose 0 < Var(h1) < ∞ and Var(h4) < ∞, then we have
moreover, we have
When X and Y are independent, the asymptotic distribution of
The following theorem, which applies a result in ([13], Chapter 5.5.2), indicates that nΩn converges to a weighted sum of (possibly infinitely many) independent
Theorem 4.12 If X and Y are independent, the asymptotic distribution of Ωn is
where
where function h2((⋅, ⋅), (⋅, ⋅)) was defined in (4.3).
Proof The asymptotic distribution of Ωn is from the result in ([13], Chapter 5.5.2).See Section 4.3 for more details on methods for computing the value of λi’s. In particular, we will show that we have
4.3 Properties of Eigenvalues λi’s
From Theorem 4.12, we see that the eigenvalues λi’s play important role in determining the asymptotic distribution of Ωn. We study its properties here. Throughout this subsection, we assume that X and Y are independent. Let us recall that the asymptotic distribution of sample distance covariance Ωn,
where λi’s are the eigenvalues of the operator G that is defined as
where function h2((⋅, ⋅), (⋅, ⋅)) was defined in Eq. 4.3. By definition, eigenvalues λ1, λ2, … corresponding to distinct solutions of the following equation
We now study the properties of λi’s. Utilizing Lemma 12 and Eq. 4.4 in [12], we can verify the following result. We give details of verifications in the Supplementary Appendix.
Lemma 4.13 Both of the following two functions are positive definite kernels:
and
The above result gives us a foundation to apply the equivalence result that has been articulated thoroughly in [12]. Equipped with the above lemma, we have the following result, which characterizes a property of λi’s. The detailed proof can be found in the Supplementary Appendix.
Lemma 4.14 Suppose {λ1, λ2, …} are the set of eigenvalues of kernel 6h2((x1, y1), (x2, y2)),
that is, each λi satisfying (4.5) can be written as, for some j, j′,
where
Corollary 4.15 The aforementioned eigenvalues
As a result, we have
and
4.4 Asymptotic Properties of Averaged Projected Sample Distance Covariance
We have reviewed the properties of the statistics Ωn in a previous section (Section 4.2). The disadvantage of directly applying Ωn (which is defined in Eq. 2.5) is that for multivariate X and Y, the implementation may require at least O(n2) operations. Recall that for univariate X and Y, an O(n log n) algorithm exists, cf. Theorem 2.2. The proposed estimator (
As preparation for presenting the main result, we recall and introduce some notations. Recall the definition of
where
and constants Cp, Cq have been defined at the end of Section 1. By Corollary 4.4, we have
where
We have seen that quantities h1 and h2 play significant roles in the asymptotic behavior of statistic Ωn. Let us define the counterpart notations as follows:
where
In the general case, we do not assume that X and Y are independent. Let U = (u1, …, uK) and V = (v1, …, vK) denote the collection of random projections. We can write the variance of
Lemma 4.16 Suppose
Equipped with the above lemma, we can summarize the asymptotic properties in the following theorem. We state it without proof as it is an immediate result from Lemma 4.16 as well as the contents in ([13], Chapter 5.5.1 Theorem A).
Theorem 4.17 Suppose
And, the asymptotic distribution of
1) If K → ∞ and K/n → 0, then
2) If n → ∞ and K/n → ∞, then
3) If n → ∞ and K/n → C, where C is some constant, then
Since our main idea is to utilize
Remark 4.18 Let us recall the asymptotic properties of Ωn,
Then, we make the comparison in the following different scenarios.
1) If K → ∞ and K/n → 0, then the convergence rate of
2) If n → ∞ and K/n → ∞, then the convergence rate of
3) If n → ∞ and K/n → C, where C is some constant, then the convergence rate of
Generally, when X is not independent of Y,
And, by Lemma 4.1, we know that
which implies
Therefore, we only need to consider
Theorem 4.19 If X and Y are independent, given the value of U = (u1, …, uK) and V = (v1, …, vK), the asymptotic distribution of
where
Remark 4.20 Let us recall that if X and Y are independent, the asymptotic distribution of Ωn is
Theorem 4.19. shows that under the null hypotheses,
where
See [14] Section 3 for an empirical justification on this Gamma approximation. See [15] for a survey on different approximation methods of the weighted sum of the chi-square distribution.The following result shows that both
Proposition 4.21 We can approximate
5 Simulations
Our numerical studies follow the works of [4,6,12]. In Section 5.1, we study how the performance of the proposed estimator is influenced by some parameters, including the sample size, the dimensionalities of the data, as well as the number of random projections in our algorithm. We also study and compare the computational efficiency of the direct method and the proposed method in Section 5.2. The comparison of the corresponding independence test with other existing methods will be included in Section 5.3.
5.1 Impact of Sample Size, Data Dimensions and the Number of Monte Carlo Iterations
In this part, we will use some synthetic data to study the impact of sample size n, data dimensions (p, q) and the number of the Monte Carlo iterations K on the convergence and test power of our proposed test statistic
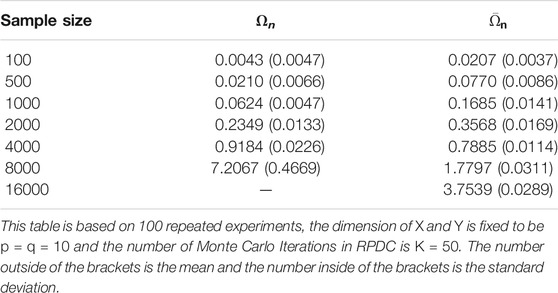
In first two examples, we fix data dimensions p = q = 10 and let the sample size n vary in 100, 500, 1000, 5000, 10000 and let the number of the Monte Carlo iterations K vary in 10, 50, 100, 500, and 1000. The data generation mechanism is described as follows, and it generates independent variables.
Example 5.1 We generate random vectors
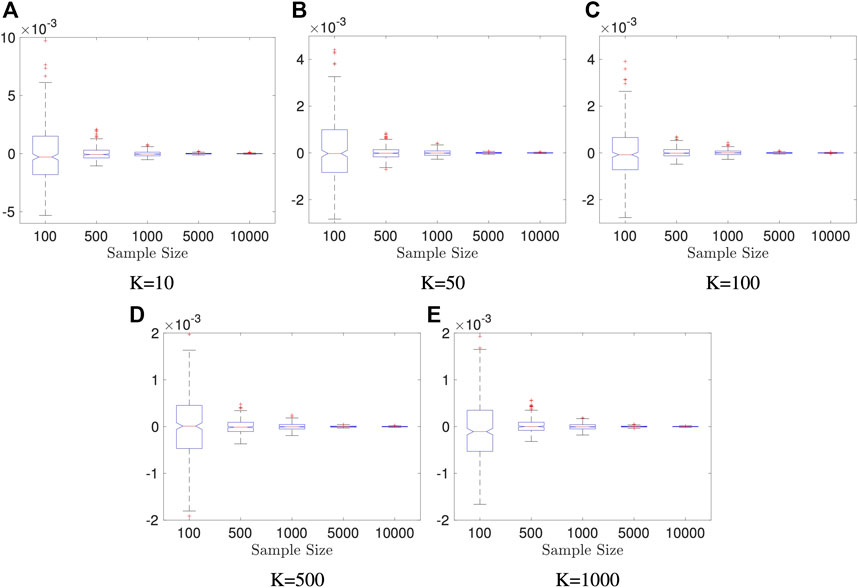
FIGURE 1. Boxplots of estimators in Example 5.1: dimension of X and Y is fixed to be p = q = 10; the result is based on 400 repeated experiments. In each subplot, y-axis represents the value of distance covariance estimators.
Example 5.2 We generate random vectors
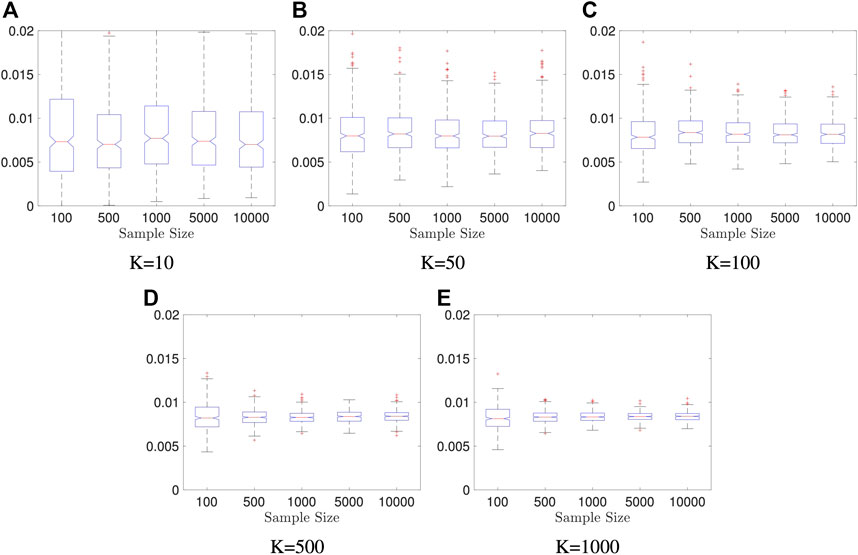
FIGURE 2. Boxplots of our estimators in Example 5.2: dimension of X and Y is fixed to be p = q = 10; the result is based on 400 repeated experiments. In each subplot, y-axis represents the value of distance covariance estimators.
Example 5.3 We generate random vectors
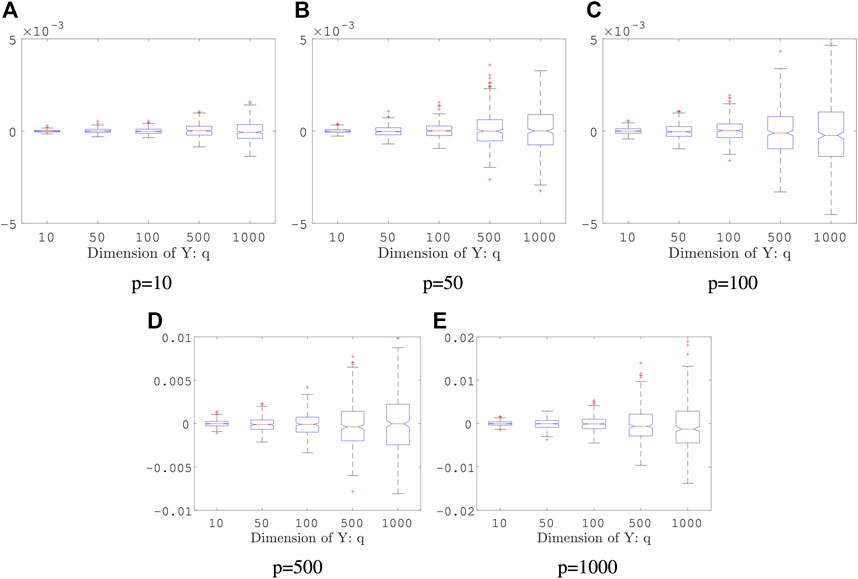
FIGURE 3. Boxplot of Estimators in Example 5.3: both sample size and the number of Monte Carlo iterations is fixed, n = 2000, K = 50; the result is based on 400 repeated experiments. In each subplot, y-axis represents the value of distance covariance estimators.
Example 5.4 We generate random vectors
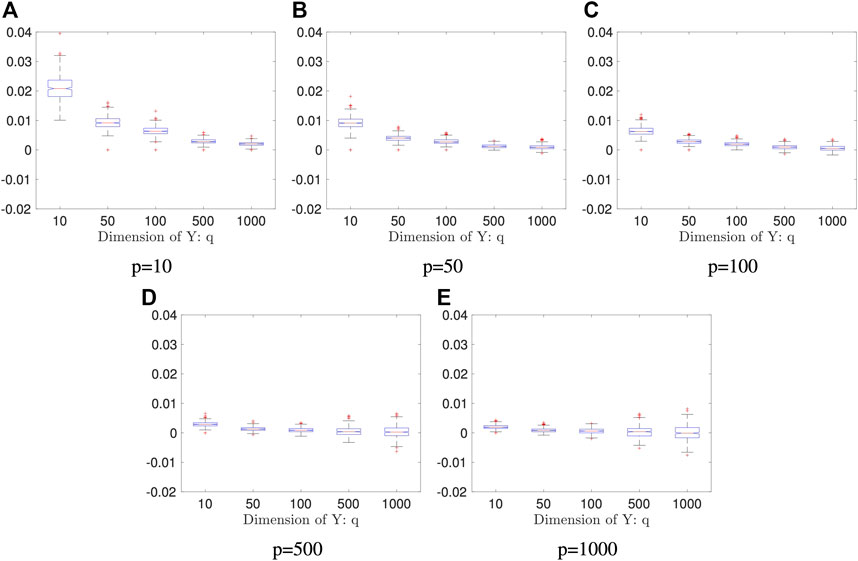
FIGURE 4. Boxplots of the proposed estimators in Example 5.4: both sample size and the number of the Monte Carlo iterations are fixed: n = 2000 and K = 50; the result is based on 400 repeated experiments. In each subplot, y-axis represents the value of distance covariance estimators.
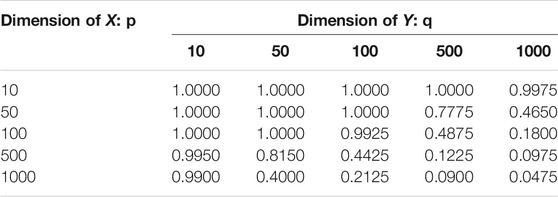
TABLE 1. Test Power in Example 5.4: this result is based 400 repeated experiments; the significance level is 0.05.
5.2 Comparison With Direct Method
In this section, we would like to illustrate the computational and space efficiency of the proposed method (RPDC). RPDC is much faster than the direct method (DDC, Eq. 2.5) when the sample size is large. It is worth noting that DDC is infeasible when the sample size is too large as its space complexity is O(n2). See Table 2 for a comparison of computing time (unit: second) against the sample size n. This experiment is run on a laptop (MacBook Pro Retina, 13-inch, Early 2015, 2.7 GHz Intel Core i5, 8 GB 1867 MHz DDR3) with MATLAB R2016b (9.1.0.441655).
5.3 Comparison With Other Independence Tests
In this part, we compare the statistical test power of the proposed test (RPDC) with Hilbert-Schmidt Independence Criterion (HSIC) [4] as HSIC is gaining attention in machine learning and statistics communities. In our experiments, a Gaussian kernel with standard deviation σ = 1 is used for HSIC. We also compare with Randomized Dependence Coefficient (RDC) [16], which utilizes the technique of random projection as we do. Two classical tests for multivariate independence, which are described below, are included in the comparison as well as Direct Distance Covariance (DDC) defined in Eq. 2.5.
• Wilks Lambda (WL): the likelihood ratio test of hypotheses Σ12 = 0 with μ unknown is based on
where det(⋅) is the determinant, S, S11 and S22 denote the sample covariances of (X, Y), X and Y, respectively, and S12 is the sample covariance
has the Wilks Lambda distribution Λ(q, n − 1 − p, p), see [17].
• Puri-Sen (PS) statistics: [18], Chapter 8, proposed similar tests based on more general sample dispersion matrices T. In that test S, S11, S12 and S22 are replaced by T, T11, T12 and T22, where T could be a matrix of Spearman’s rank correlation statistics. Then, the test statistic becomes
The critical values of the Wilks Lambda (WL) and Puri-Sen (PS) statistics are given by Bartlett’s approximation ([19], Section 5.3.2b): if n is large and p, q > 2, then
has an approximation χ2(pq) distribution.
The reference distributions of RDC and HSIC are approximated by 200 permutations. And the reference distributions of DDC and RPDC are approximated by Gamma Distribution. The significance level is set to be αs = 0.05 and each experiment is repeated for N = 400 times to get reliable type-I error/test power.
We start with an example that (X, Y) is multivariate normal. In this case, WL and PS are expected to be optimal as the assumptions of these two classical tests are satisfied. Surprisingly, DDC has comparable performance with the aforementioned two methods. RPDC can achieve satisfactory performance when the sample size is reasonably large.
Example 5.5 We set the dimension of the data to be p = q = 10. We generate random vectors
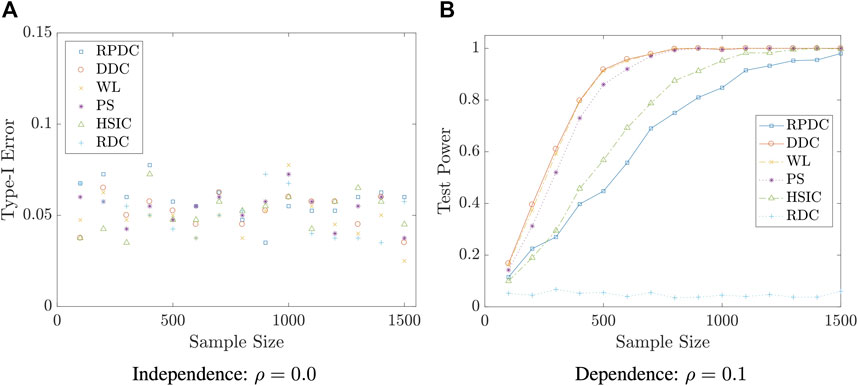
FIGURE 5. Type-I Error/Test Power vs. Sample Size n in Example 5.5: the result is based on 400 repeated experiments.
Example 5.6 We set the dimension of data to be p = q = 10. We generate random vector
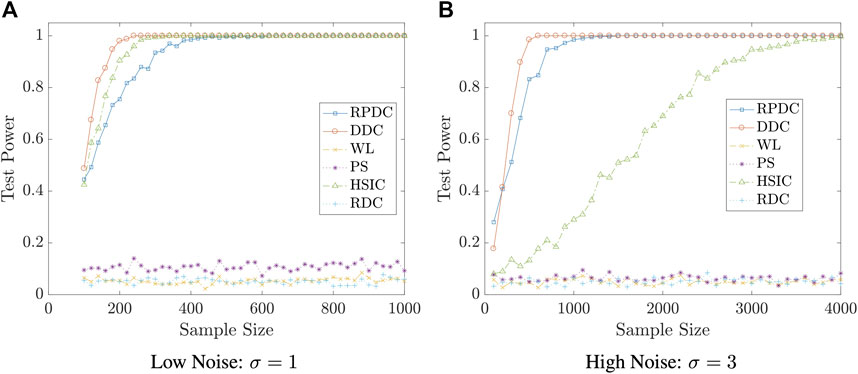
FIGURE 6. Test Power vs. Sample Size n in Example 5.6: significance level is αs = 0.05; the result is based on N = 400 repeated experiments.
Example 5.7 We set the dimension of data to be p = q = 10. We generate random vector
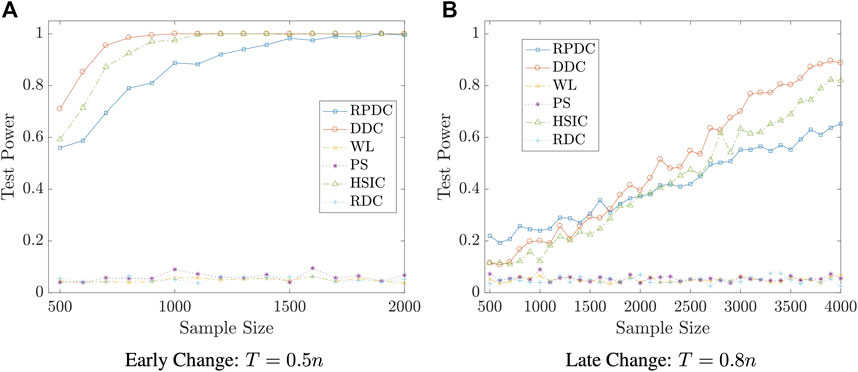
FIGURE 7. Test Power vs. Sample Size n in Example 5.7: significance level is αs = 0.05; the result is based on N = 400 repeated experiments.
Remark 5.8 The examples in this subsection show that though RPDC underperforms DDC when the sample size is relatively small, RPDC could achieve the same test power with DDC when the sample size is sufficiently large. Thus, when the sample size is large enough, RPDC is superior to DDC because of its computational efficiency in both time and space.
6 Discussions
6.1 A Discussion on the Computational Efficiency
We compare the computational efficiency of the proposed method (RPDC) and the direct method (DDC) in Section 5.2. We will discuss this issue here.
As
There is no doubt that O2 will eventually be much less than O1 as sample size n grows. Due to the complexity of the fast algorithm, we expect L2 > L1, which means the computing time of RPDC is even larger than DDC when the sample size is relatively small. Then, we need to study an interesting problem: what is the break-even point in terms of sample size n when RPDC and DDC have the same computing time?
Let n0 = n0(p + q, K) denote the break-even point, which is a function of p + q and number of Monte Carlo iterations K. For simplicity, we fix K = 50 since 50 iterations could achieve satisfactory test power as we showed in Example 5.4. Then, n0 becomes a function solely depending on p + q. Since it is hard to derive the close form of n0, we derive it numerically instead. For fixed p + q, we let the sample size vary and record the difference between the running time of the two methods. Then, we fit the difference of running time against sample size with a smoothing spline. The root of this spline is the numerical value of n0 at p + q.
We plot the n0 against p + q in Figure 8. As the figure shows, the break-even sample size decreases as the data dimension increases, which implies that our proposed method is more advantageous than the direct method when random variables are of high dimension. However, as shown in Example 5.4, the random projection-based method does not perform well when high dimensional data have a low dimensional dependency structure. We should be cautious to use the proposed method when the dimension is high.
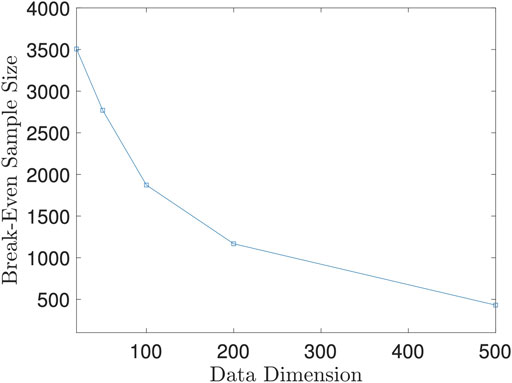
FIGURE 8. Break-Even Sample Size n0 against Data Dimension p + q. This figure is based on 100 repeated experiments.
6.2 Connections With Existing Literature
It turns out that distance-based methods are not the only choices in independence tests. See [20] and the references therein to see alternatives.
Our proposed method utilizes random projections, which bears a similarity with the randomized feature mapping strategy [21] that was developed in the machine learning community. Such an approach has been proven to be effective in kernel-related methods [22–26]. However, a closer examination will reveal the following difference: most of the aforementioned work is rooted in the Bochner’s theorem [27] from harmonic analysis, which states that a continuous kernel in the Euclidean space is positive definite if and only if the kernel function is the Fourier transform of a non-negative measure. In this paper, we will deal with the distance function which is not a positive definite kernel. We will manage to derive a counterpart to the randomized feature mapping, which was the influential idea that has been used in [21].
Random projections have been used in [28] to develop a powerful two-sample test in high dimensions. They derived an asymptotic power function for their proposed test, and then provide sufficient conditions for their test to achieve greater power than other state-of-the-art tests. They then used the receiver operating characteristic (ROC) curves (that are generated from simulated data) to evaluate its performance against competing tests. The derivation of the asymptotic relative efficiency (ARE) is of its own interests. Despite the usage of random projection, the details of their methodology are very different from the one that is studied in the present paper.
Several distribution-free tests that are based on sample space partitions were suggested in [29] for univariate random variables. They proved that all suggested tests are consistent and showed the connection between their tests and the mutual information (MI). Most importantly, they derived fast (polynomial-time) algorithms, which are essential for large sample size, since the computational complexity of the naive algorithm is exponential in sample size. Efficient implementations of all statistics and tests described in the aforementioned paper are available in the R package HHG, which can be freely downloaded from the Comprehensive R Archive Network, http://cran.r-project.org/. Null tables can be downloaded from the first author’s website.
Distance-based independence/dependence measurements sometimes have been utilized in performing a greedy feature selection, often via dependence maximization [8,30,31], and it has been effective on some real-world datasets. This paper simply mentions such a potential research line, without pursuing it.
Paper [32] derives an efficient approach to compute for the conditional distance correlations. We noted that there are strong resemblances between the distance covariances and its conditional version. The search for a potential extension of the work in this paper to conditional distance correlation can be a meaningful future topic of research.
Paper [33] provides some important insights into the power of distance covariance for multivariate data. In particular, they discover that distance-based independence tests have limiting power under some less common circumstances. As a remedy, they propose tests based on an aggregation of marginal sample distance and extend their approach to those based on Hilbert-Schmidt covariance and marginal distance/Hilbert-Schmidt covariance. It could be another interesting research direction but beyond the scope of this paper.
7 Conclusion
A significant contribution of this paper is we demonstrated that the multivariate variables in the independence tests need not imply the higher-order computational desideratum of the distance-based methods.
Distance-based methods are important in statistics, particularly in the test of independence. When the random variables are univariate, efficient numerical algorithms exist. It is an open question when the random variables are multivariate. This paper studies the random projection approach to tackle the above problem. It first turns the multivariate calculation problem into univariate calculation one via a random projection. Then they study how the average of those statistics out of the projected (therefore univariate) samples can approximate the distance-based statistics that were intended to use. Theoretical analysis was carried out, which shows that the loss of asymptotic efficiency (in the form of the asymptotic variance of the test statistics) is likely insignificant. The new method can be numerically much more efficient, when the sample size is large, which is well-expected under this information (or big-date) era. Simulation studies validate the theoretical statements. The theoretical analysis takes advantage of some newly available results, such as the equivalence of the distance-based methods with the reproducible kernel Hilbert spaces [12]. The numerical methods utilize a recently appeared algorithm in [8].
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This material was based upon work partially supported by the National Science Foundation under Grant DMS-1127914 to the Statistical and Applied Mathematical Sciences Institute. Any opinions, findings, and conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. This work has also been partially supported by NSF grant DMS-1613152.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fams.2021.779841/full#supplementary-material
References
1. David, N, Yakir, A, Hilary, K, Sharon, R, Peter, J, Eric, S, et al. Detecting Novel Associations in Large Data Sets. Science (2011) 334(6062):1518–24.
2. Schweizer, B, and Wolff, EF. On Nonparametric Measures of Dependence for Random Variables. Ann Stat (1981) 879–85. doi:10.1214/aos/1176345528
3. Siburg, KF, and Stoimenov, PA. A Measure of Mutual Complete Dependence. Metrika (2010) 71(2):239–51. doi:10.1007/s00184-008-0229-9
4. Gretton, A, Bousquet, O, Smola, A, and Schölkopf, B. Measuring Statistical Dependence with hilbert-schmidt Norms. International Conference on Algorithmic Learning Theory. Springer (2005). p. 63–77. doi:10.1007/11564089_7
5. Székely, GJ, and Rizzo, ML. Brownian Distance Covariance. Ann Appl Stat (2009) 3(4):1236–65. doi:10.1214/09-aoas312
6. GáborSzékely, J, MariaRizzo, L, and Bakirov, NK. Measuring and Testing Dependence by Correlation of Distances. Ann Stat (2007) 35(6):2769–94. doi:10.1214/009053607000000505
7. Matthew, R, and Nicolae, DL. On Quantifying Dependence: a Framework for Developing Interpretable Measures. Stat Sci (2013) 28(1):116–30.
8. Huo, X, and Székely, GJ. Fast Computing for Distance Covariance. Technometrics (2016) 58(4):435–47. doi:10.1080/00401706.2015.1054435
9. Taskinen, S, Oja, H, and Randles, RH. Multivariate Nonparametric Tests of independence. J Am Stat Assoc (2005) 100(471):916–25. doi:10.1198/016214505000000097
10. Lyons, R. Distance Covariance in Metric Spaces. Ann Probab (2013) 41(5):3284–305. doi:10.1214/12-aop803
11. Hoeffding, W. Probability Inequalities for Sums of Bounded Random Variables. J Am Stat Assoc (1963) 58(301):13–30. doi:10.1080/01621459.1963.10500830
12. Sejdinovic, D, Sriperumbudur, B, Gretton, A, and Fukumizu, K. Equivalence of Distance-Based and RKHS-Based Statistics in Hypothesis Testing. Ann Stat (2013) 41(5):2263–91. doi:10.1214/13-aos1140
13. Serfling, RJ. Approximation Theorems of Mathematical Statistics (Wiley Series in Probability and Statistics). Wiley-Interscience (1980).
14. George, EP, Some Theorems on Quadratic Forms Applied in the Study of Analysis of Variance Problems, I. Effect of Inequality of Variance in the One-Way Classification. Ann Math Stat (1954) 25(2):290–302.
15. DeanBodenham, A, and Adams, NM. A Comparison of Efficient Approximations for a Weighted Sum of Chi-Squared Random Variables. Stat Comput pages (2014) 1–12.
16. Lopez-Paz, D, Hennig, P, and Schölkopf, B. The Randomized Dependence Coefficient. In Advances in Neural Information Processing Systems, 1–9. 2013.
17. Wilks, SS. On the Independence of K Sets of Normally Distributed Statistical Variables. Econometrica (1935) 3:309–26. doi:10.2307/1905324
18. Puri, ML, and Sen, PK. Nonparametric Methods in Multivariate Analysis. Wiley Series in Probability and Mathematical Statistics. Probability and mathematical statistics. Wiley (1971).
19. Mardia, KV, Bibby, JM, and Kent, JT. Multivariate Analysis. New York, NY: Probability and Mathematical Statistics. Acad. Press (1982).
20. Lee, K-Y, Li, B, and Zhao, H. Variable Selection via Additive Conditional independence. J R Stat Soc Ser B (Statistical Methodology) (2016) 78(Part 5):1037–55. doi:10.1111/rssb.12150
21. Rahimi, A, and Recht, B. Random Features for Large-Scale Kernel Machines. In Advances in Neural Information Processing Systems, 1177–84. 2007.
22. Achlioptas, D, McSherry, F, and Schölkopf, B. Sampling Techniques for Kernel Methods. In Annual Advances In Neural Information Processing Systems 14: Proceedings Of The 2001 Conference (2001).
23. Avrim, B. Random Projection, Margins, Kernels, and Feature-Selection. In Subspace, Latent Structure and Feature Selection, 52–68. Springer, 2006.
24. Cai, T, Fan, J, and Jiang, T. Distributions of Angles in Random Packing on Spheres. J Mach Learn Res (2013) 14(1):1837–64.
25. Drineas, P, and Mahoney, MW. On the Nyström Method for Approximating a Gram Matrix for Improved Kernel-Based Learning. J Machine Learn Res (2005) 6(Dec):2153–75.
26. Frieze, A, Kannan, R, and Vempala, S. Fast Monte-Carlo Algorithms for Finding Low-Rank Approximations. J Acm (2004) 51(6):1025–41. doi:10.1145/1039488.1039494
28. Miles, L, Laurent, J, and Wainwright, J. A More Powerful Two-Sample Test in High Dimensions Using Random Projection. In Advances in Neural Information Processing Systems, 1206–14. 2011.
29. Heller, R, Heller, Y, Kaufman, S, Brill, B, and Gorfine, M. Consistent Distribution-free K-Sample and independence Tests for Univariate Random Variables. J Machine Learn Res (2016) 17(29):1–54.
30. Li, R, Zhong, W, and Zhu, L. Feature Screening via Distance Correlation Learning. J Am Stat Assoc (2012) 107(499):1129–39. doi:10.1080/01621459.2012.695654
31. Zhu, L-P, Li, L, Li, R, and Zhu, L-X. Model-free Feature Screening for Ultrahigh-Dimensional Data. J Am Stat Assoc (2012).
32. Wang, X, Pan, W, Hu, W, Tian, Y, and Zhang, H. Conditional Distance Correlation. J Am Stat Assoc (2015) 110(512):1726–34. doi:10.1080/01621459.2014.993081
Keywords: independence test, distance covariance, random projection, hypotheses test, multivariate hypothesis test
Citation: Huang C and Huo X (2022) A Statistically and Numerically Efficient Independence Test Based on Random Projections and Distance Covariance. Front. Appl. Math. Stat. 7:779841. doi: 10.3389/fams.2021.779841
Received: 19 September 2021; Accepted: 16 November 2021;
Published: 13 January 2022.
Edited by:
David Banks, Duke University, United StatesReviewed by:
Dominic Edelmann, German Cancer Research Center, GermanyGabor J. Szekely, National Science Foundation (NSF), United States
Copyright © 2022 Huang and Huo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoming Huo, aHVvQGdhdGVjaC5lZHU=
 Cheng Huang
Cheng Huang Xiaoming Huo
Xiaoming Huo