- 1Department of Mathematical Sciences, University of Bath, Bath, United Kingdom
- 2School of Computing, Engineering and The Built Environment, Edinburgh Napier University, Edinburgh, United Kingdom
There exist several methods for simulating biological and physical systems as represented by chemical reaction networks. Systems with low numbers of particles are frequently modeled as discrete-state Markov jump processes and are typically simulated via a stochastic simulation algorithm (SSA). An SSA, while accurate, is often unsuitable for systems with large numbers of individuals, and can become prohibitively expensive with increasing reaction frequency. Large systems are often modeled deterministically using ordinary differential equations, sacrificing accuracy and stochasticity for computational efficiency and analytical tractability. In this paper, we present a novel hybrid technique for the accurate and efficient simulation of large chemical reaction networks. This technique, which we name the regime-conversion method, couples a discrete-state Markov jump process to a system of ordinary differential equations by simulating a reaction network using both techniques simultaneously. Individual molecules in the network are represented by exactly one regime at any given time, and may switch their governing regime depending on particle density. In this manner, we model high copy-number species using the cheaper continuum method and low copy-number species using the more expensive, discrete-state stochastic method to preserve the impact of stochastic fluctuations at low copy number. The motivation, as with similar methods, is to retain the advantages while mitigating the shortfalls of each method. We demonstrate the performance and accuracy of our method for several test problems that exhibit varying degrees of inter-connectivity and complexity by comparing averaged trajectories obtained from both our method and from exact stochastic simulation.
1. Introduction
A chemical reaction network (CRN) is a representation of a reacting (bio)chemical system of several species interacting via some number of reaction channels. CRNs, such as those found in biological systems, are often represented by continuous time, discrete-state Markov processes [1]. This modeling regime is appropriate when the described system has a small number of interacting particles and provides an exact description of reaction dynamics under appropriate assumptions; specifically, that the inter-event times between the “firing” of reaction channels are independent and exponentially distributed. Such Markov processes are most often simulated via a stochastic simulation algorithm (SSA), the prototypical example of which is the Gillespie direct method [2]. Several improvements to the Gillespie direct method have been proposed for reaction networks with particular structural characteristics. For example, the next reaction method [3] and the optimized direct method [4] are exact and efficient SSAs for systems with a large number of loosely-coupled reaction channels. Further extensions also exist, such as the modified next reaction method [5], that facilitate the simulation of systems with time-dependent reaction rates.
For any reaction network, and under mild differentiability assumptions, one can derive a system of ordinary differential equations called the chemical master equation (CME) that describes the time-evolution of the probability density of the system existing in any given state [6]. The CME, as a single equation that encapsulates all stochastic information of a system, is neither solvable analytically nor practicable to solve numerically in all but the most straightforward of systems. Rather, the practical utility of the CME lies in the ease with which one can derive time-evolution equations for the raw moments of the system. These moment equations take the form of a system of ordinary differential equations (ODEs) that govern the moments of each constituent species. In cases where the CRN contains reactions of at least second-order, these moment equations do not form a closed system; in particular, the equations governing the nth moments will, in general, depend on the (n + 1)th or higher-order moments. These systems are not solvable analytically. As such, one generally applies a so-called “moment-closure” that closes the system of moment equations at a given order by making explicit assumptions about the relationships between lower- and higher-order moments. Commmon moment-closures (or, simply, closures) include the mean-field closure, wherein all moments above the first are set to zero, and the Poisson closure, where diagonal cumulants are assumed equal to their corresponding mean and all mixed cumulants are set to zero [7].
In general, determining the most appropriate closure assumptions for a given system can be challenging and higher-order closures often yield systems of moment equations that can be difficult to solve; as such, straightforward closures like the mean-field see the widest application. In the case of the mean-field closure, the resulting system of mean-field ODEs provides an approximate, continuous, and deterministic description of the time evolution of the mean of the underlying Markov process, and can be solved either analytically or numerically.
The primary downside of SSAs is that they may become computationally intractable for large systems of interacting particles. Even for systems with favorable network structures, large systems can quickly become infeasible to simulate exactly. This is contrasted with deterministic modeling techniques that sacrifice accuracy in exchange for computational efficiency where, notably, the efficiency of numerical simulation methods (i.e., those for ODEs and PDEs) is typically independent of copy number. The various advantages and disadvantages of each modeling regime discussed have motivated the development of so-called hybrid methods that combine regimes to leverage their advantages and mitigate their limitations [8]. Several general hybrid approaches have been developed to tackle these issues.
One such approach is to model certain species under a continuous regime (such as an ODE or SDE) and others under a discrete regime (via a SSA). Typically, this extension of the system is accomplished by categorizing reactions as either being “fast” or “slow”, applying a continuous representation to the former and using a discrete method for the latter. Cao, Gillespie, and Petzold [9] pioneered this technique in the development of the ‘slow-scale SSA', a method for simulating dynamically stiff chemical reaction networks. Their method separates reactions and reactant species into fast and slow categories in a manner that allows for only the slow-scale reactions and species to be simulated stochastically, subject to certain stability criteria of the fast system. The fast-slow paradigm was also applied by Cotter et al. [10] for simulating chemical reaction networks that can be extended into fast and slow ‘variables', which may be reactant species or combinations thereof. They define a “conditional stochastic simulation algorithm” that can draw sample values of fast variables conditioned on the values of the slow variables. These samples are then used to approximate the drift and diffusion terms in a Fokker-Planck equation that describes the overall state of the system.
There are a number of other hybrid-type methods in the literature for simplifying the computation of SSAs that do not necessarily partition species into fast/slow reactions. Hellander and Löstedt [11] present a hybrid method for simulating chemical systems with disparities in species copy number or reaction rates that would render pure stochastic simulation extremely expensive. Those species which exhibit both small variance and take part in fast reactions are simulated using approximate reaction rate equations, while the evolution of the probability density function of those species which are involved in slow reactions or have large variance are estimated using a modified SSA to preserve accuracy. Smith et al. [12] take an approach based not on the separation of species by reaction time-scale but on the separation of species by their abundance. This involves forming a “reduced” CME from the non-abundant species by taking a limit of the CME as the number of abundant species tends to infinity. This reduced CME can then be sampled using an SSA. Jahnke [13] contributes to a much-studied line of enquiry investigating approximations of the chemical master equation. Particularly, it provides error bounds for the modeling error of two reduced models from the literature and proposes another, called the model reduction by conditional expectations (MRCE). Roughly, these reduced models partition the species into two subsets: those deemed of interest and the remaining variables. Approximations of the CME occur as different assumptions are made about the overall probability distribution in terms of these two subsets, for example, that it decomposes into a product of probability distributions (the product approximation) and the so-called Hellander-Lötstedt model from [11], which approximates a marginal probability distribution of one subset and the expectation with respect to the other.
In this paper we detail the development of a novel hybrid simulation technique for well-mixed CRNs; that is, systems of interacting (bio)chemical species distributed homogeneously within a reactor vessel of fixed volume. As discussed, continuum methods are advantageous when copy numbers are high and the effects of stochasticity can be safely assumed to be small. Discrete methods, on the other hand, are best applied in low copy number systems and where stochasticity is a critical driver of the dynamics. It is this fundamental tension between computational efficiency and model accuracy that our method seeks to address. Where other, similar methods aim to subdivide species and/or the reactions between them into categories based on reaction rates, we take a simpler approach that is instead based on particle density. Our objective is to create a method that is simple to implement, computationally efficient, accurate, and flexible enough to handle not only reaction networks with fast/slow reactions, but also more uniform reaction networks where no such fast/slow distinctions can be leveraged. Further, the method offers additional flexibility by permitting species to transition between regimes during run-time, as opposed to being fixed in a predetermined regime.
Our method, which we term the regime-conversion method (RCM), consists of a system of ODEs and a discrete-state Markov jump process that, taken together, form an inexact yet computationally amenable representation of a well-mixed CRN. The key idea behind the method is to run, simultaneously, a numerical method for solving the system of ODEs alongside a SSA for simulating stochastic trajectories. Individuals in the system are represented by exactly one of the two regimes at any given time, but are permitted to switch back and forth between each modeling regime in response to the current concentration of their species. To accomplish this, we describe a “network extension” procedure by which one can convert a CRN into a larger network that is probabilistically equivalent to the original in a manner that we describe. The extended network is larger than the original in three specific ways. First, each species in the original corresponds to two species in the extended network, where one species is to be governed by the discrete regime and the other by the continuous. Second, to satisfy the combinatorial requirements that give rise to the probabilistic equivalence of each network, the extension requires that we add additional reactions that allow the continuous and discrete species to interact. The final ingredient in the extended network are first-order conversion reactions that allow discrete species to enter the continuous regime and vice versa, adaptively redistributing species concentrations between regimes to maximize computational efficiency and accuracy.
From the extended network we construct an augmented reaction network (ARN) that governs the same species as the extended network. The critical difference is that we represent the species marked as “continuous” (and the reactions between them) in the extended network by a system of ODEs. This system of ODEs is derived by forming the CME that would govern the continuous species (were they discrete) from the set of reactions that act exclusively on continuous species, deriving the moment equations for these species, and taking an appropriate moment closure. Under this representation, reactions between continuous species are governed exclusively by the continuum approximation, and reactions between discrete species are governed exclusively by the discrete simulation regime. To retain accuracy in bimolecular reactions, and to mitigate the impact of moment closure, reactions that have both a continuous and a discrete reactant are governed by the discrete simulation regime. Given that mass is converted back-and-forth between discrete and continuous representations depending on copy-number, we can reasonably view the ARN as a mechanism for representing “low copy-number reactions” under the discrete simulation regime, and “high copy-number reactions” under the continuum approximation. This new structure, the ARN, provides an intermediate description of a CRN that is both continuous and discrete. The RCM, then, is a method for simulating the trajectories of an ARN. We find that the RCM can indeed strike a balance between efficiency and accuracy.
The remainder of this work is divided into three sections. In Section 2, we outline the construction of an ARN from a CRN alongside the mathematical prerequisites, the theoretical justification, and the specific algorithmic formulation of the RCM. In Section 3, we present numerical results that evaluate the accuracy and bias of our method for a series of test problems of increasing complexity. We conduct this evaluation by comparing the results from the RCM against results from an exact SSA. Finally, in Section 4 we give remarks on the relative advantages and limitations of our method vs. traditional stochastic or numerical methods, and signpost future potential avenues of development and application for the method.
2. Method
In this section we describe the regime-conversion method (RCM) which couples a CRN described by a discrete-state Markov jump process with a system of ordinary differential equations representing the mean dynamics of the same CRN. We begin our discussion of the method with some preliminary information regarding stochastic simulation and continuum modeling before presenting the theoretical justification and implementation of our proposed coupling scheme.
2.1. Stochastic simulation and stoichiometry
We consider a CRN, , with K chemical species that interact via a set R of reaction channels within a reaction vessel of unit volume. Denote by Xk(t) ∈ ℕ, for k = 1, …, K, the number of individuals of the kth species at continuous time t, and denote the overall state of the system by X(t): = (X1(t), …, XK(t)). We make the assumption that reaction r ∈ R fires with an exponentially distributed waiting time with rate λr. The reaction rate coefficient λr is typically taken to be constant over time; however, we note that the results in the remainder of this paper hold in the case that λr is piecewise constant in time, with the caveat that there are only finitely many such discontinuities. Reactions in the network take the form
where μr = (μrk)k = 1, …, K and ηr = (ηrk)k = 1, …, K. We can thus, for each reaction, define the stoichiometric vector
which represents the change in state upon the firing of reaction r. These vectors are often collected into a single stoichiometric matrix, which we denote S, where each column in S corresponds to a stoichiometric vector νr. To form this matrix, one must decide on an ordering of the reactions in R- we note that this choice is arbitrary and bears no impact on the dynamics of the system.
The most common method for drawing sample trajectories of X(t) is the aforementioned Gillespie direct method (GDM). Whilst the coupling technique for our hybrid method, which we will discuss later, is strictly independent of the choice of SSA, we will describe its implementation under the Gillespie direct method.
2.2. Continuum modeling
Given a CRN, , we can derive the associated CME as follows. Define for each reaction a propensity function αr(X(t)), defined such that αr(X(t))dt is the probability that said reaction occurs within the infinitesimally small time interval [t, t + dt). Under the law of mass-action, the propensity functions are given by
where for brevity we have subsumed any combinatorial coefficients into the rate coefficient λr [14]. Standard techniques [6] reveal that the corresponding CME for this system is given by
where p(x, t) is the probability that X(t) = x at time t. Multiplying equation (1) by xk and summing over the state space xk, yields the evolution equation for the mean concentration of each species. Denoting by 〈f(x)〉 the expectation of f(x) with respect to p(x, t) for some function f, we have
Defining the vector of propensity functions α(x) = (αr(x))r ∈ R, this can be written in matrix form,
assuming that the enumeration of reactions in the vector α corresponds to the column order of the stoichimetric matrix S. One can likewise, albeit through a somewhat laborious calculation, obtain higher-order moments of the system. These equations, however, do not in general admit closed-form solutions. Indeed, for CRNs with reactions of at least second-order, the system of moment equations itself is not closed; for example, for species which are reactants in a second-order reaction, the equation governing the evolution of the first moment of that species depends on the equations for the second moments, the equations for the second moments depend on the equations for the third moments, and so on.
Making a moment-closure approximation requires the explicit adoption of some set of assumptions about the moments of a system. As such, these closures are necessarily ad hoc and it is, in general, impossible to quantify a given closure's accuracy a priori. Nevertheless, there are several closures that see wide application. The simplest and possibly most common closure is the so-called “mean-field” closure [15, p. 82]. Under the mean-field closure, all variances and covariances are assumed to be zero, yielding
for all i, j = 1, …, K. Another common closure is the Poisson closure [16], which assumes that variances are equal to their corresponding means and that all covariances are zero, i.e.:
for all i = 1, …, K, and
for all i, j = 1, …, K where i ≠ j. Both the mean-field and Poisson closures close the system of moment equations at first-order. While there exist several higher-order closures [7], they are generally unsuitable for use in hybrid methods, as there is currently no clear method for coupling higher-order moment equations to SSAs.
2.3. Reaction network extension
We begin our discussion of the RCM by noting that we will henceforth only consider reactions of at most second-order. These are reactions for which at most two individual reactant molecules are present. While a simultaneous interaction of three or more individuals is, in principle, possible, collision theory suggests that the probability of three or more distinct molecules interacting simultaneously is vanishingly small [17]. Accordingly, a more realistic description of interactions of this type involves the formation of a highly reactive intermediary complex that subsequently reacts with the remaining reactants—such a system is of at most second order [18].
The RCM partitions each chemical species Xk into two “partition species”, Ck and Dk, each of which is governed by a different modeling regime, termed continuous and discrete, respectively. On these extension species we define a new reaction network that is both equivalent to the original network and computationally amenable. Further, this new “extended” reaction network contains additional “conversion” reactions that permit individuals to switch their partition at a rate proportional to the species-wise density. To do so, for each reaction in the network, we generate a new extended set of reactions for each possible combination of reactant regimes. In each reaction r, at most two species appear as reactants, which we label without loss of generality Xi and Xj, where i, j ∈ {1, …, K}, and where we may have that i = j. We require that this extended set of reactions obeys the following criteria:
C1 To maximize efficiency, we wish to minimize unnecessary conversion back-and-forth between regimes. We thus determine that all molecules produced by reaction r belonging to the ith species (resp. jth) are allocated to the same regime as reactant Xi (resp. Xj).
C2 To maximize accuracy, we aim to retain much of the stochasticity in the system. In particular, for each reaction r, we allocate all product molecules from non-reactant species (i.e. species other than Xi and Xj) to the discrete regime.
C3 Applying C2 without further restriction could yield a “trivial” reaction network wherein all continuous molecules are gradually converted to discrete molecules over time. As such, for reactions r where all reactant molecules are in the continuous regime, we assign all the reaction's products to the continuous regime also.
We begin our exposition of the RCM with reactions of order zero; that is, reactions of the form
for some set of reaction products P. The choice of whether to place these reaction products into the discrete or continuous regime may be problem dependent; specifically, it may be the case that all products in P belong to species that are known a priori to be of high copy number, and as such might best be placed in the continuous regime. Nevertheless, in light of C2, we place any such products into the discrete regime.
First-order reactions are dealt with trivially when applying the criteria above. Specifically, reactions of the form
are extended into
Any second-order reaction r ∈ R can be written uniquely in the form
for some i, j ∈ {1, …, K} with i ≤ j. To extend such a reaction we consider the four possible combinations of reactant regimes and apply C1–C3, yielding
Note that in the case of a homodimerisation, where i = j, reactions (7) and (8) are identical. Nevertheless, both must be included in the resultant network—this is explained in detail in Section 2.4. Applying this extension procedure to each reaction in the original network yields a new extended reaction network with chemical species Ck and Dk for k = 1, …, K.
Remaining are the regime conversion reactions that facilitate the conversion of species at high- and low-copy numbers to the continuous and discrete regimes, respectively. To this end, we append to the extended network reactions of the form
where κf,k and κb,k are non-constant rates of the form
for pre-determined regime-conversion rates γf,k and γb,k, conversion thresholds Tk, and where the subscript characters f and b indicate the “forward” and “backward” conversions, respectively.
The collection of the species Ck and Dk for k = 1, …, K alongside the set of reactions obtained from the procedures detailed above form the extended version of the network . In completing our description of this network, it is useful at this point to introduce notational conventions that reflect both its structure and its provenance. For a CRN , we denote its extended version by . We denote the state vector of by Y(t), taking without loss of generality , where C(t) = (C1, …, CK), D(t) = (D1, …DK), and the operator ⊕ denotes vector concatenation. Finally, we denote the collection of reactions in by .
2.4. Network equivalence
We claim that the evolution of the quantity Xk in the CRN is the same as the evolution of the quantity Ck + Dk in the partitioned version , for all i = 1, …, K, provided that the species Ck are treated as discrete and simulated using the stochastic simulation algorithm. Before embarking on the derivation of this equivalence, we must first specify what, precisely, we are aiming to demonstrate. Define p(x, t) to be the probability that {X(t) = x} and q(x, t) to be the probability that {C(t)+D(t) = x}. Our aim, therefore, is to demonstrate that for any choice of x ∈ ℕK and t > 0 we have q(x, t) = p(x, t), provided that the initial conditions for Ck + Dk are the same as those for Xk.
To this end, consider a CRN with K species and |R| = |R0| + |R1| + |R2| reactions, where R0, R1, and R2 are the sets of zeroth-, first-, and second-order reactions in the network, respectively. Recalling that the CME for this network is given by equation (1), we rewrite the CME for in the form
The extension procedure from Section 2.3 gives a CRN with 2K species and a set of reactions , where . We associate each reaction in (excluding the 2K regime conversion reactions) with the original reactions from which they were extended. Each zeroth-order reaction in is associated with a zeroth order reaction in . Similarly, first- and second-order reactions in are associated with two first- and four second-order reactions in , respectively. To track these relationships, we must introduce some new notation. We denote by , where r ∈ Rd, ℓ = 1, …, 2d, and d = 0, 1, 2, the stoichiometric vectors for the 2d reactions in associated with reaction r in . In particular, notice that our extension procedure guarantees that
for all reactions r ∈ R, ℓ = 1, …, 2d, d = 0, 1, 2, and where vn:m = (vn, ..., vm) for n ≤ m. Additionally, we define the extended set of propensity functions for each reaction r ∈ R via the usual mass-action kinetics, denoted by for ℓ = 1, …, 2d. Note that in both cases, there is an implied ordering on the stoichiometric vectors and propensity functions associated with each reaction that is induced by ℓ - any such enumeration is arbitrary and exists only for notational utility; the only restriction is that the enumerations of stoichiometric vectors and propensity functions match for any given r.
The propensity functions for the forward and backward regime conversion reactions (10) are not strictly governed by mass-action kinetics by virtue of their rates' dependence on the concentration of non-reactant species. Specifically, we choose the propensity functions for the forward and backward reactions for each species k = 1, …, K to take the forms
respectively, with associated stoichiometric vectors given by
again respectively, and where ek denotes the kth standard basis vector in ℝ2K. The CME for the network can thus be expressed as
where denotes the probability that {Y(t) = y} at time t, where y = c ⊕ d. Recalling the definition of q(x, t), we can additionally write the master equation governing q(c + d, t),
noticing that the regime conversion reactions contribute nothing to the evolution of q, since each conserves the quantity c(t)+d(t). Comparing equations (11) and (13), the critical step in our proof of equivalence is demonstrating that
for all r ∈ R, and for any c, d ∈ ℕK where c + d = x. To prove this, we will consider how the sum (14) behaves for each reaction order. To begin, fix c, d ∈ ℕK and x = c + d. Consider the case z = 0, where z denotes the reaction order we are considering. For any zeroth order reaction under the law of mass-action, we trivially have that for all r ∈ R0. Since each reaction r ∈ R0 corresponds with exactly one reaction in , equation (14) holds for d = 0.
We now consider the case z = 1 and consider a reaction r ∈ R1 of the form (2), taking without loss of generality ℓ = 1 to denote the reaction (3) and ℓ = 2 to denote the reaction (4). Notice that we have and . Further, under mass-action, the functions αr are linear for any first-order reaction r. Therefore, we have
for all r ∈ R1 and Equation (14) holds for first-order reactions.
Next, consider z = 2 and consider a second-order reaction r of the form (5). Similarly to the first-order case, we enumerate without loss of generality the propensity functions by setting ℓ = 1, …, 4 to correspond with reactions (6) through (9), respectively. Note that there are two distinct classes of second-order reaction; namely, homodimerizations, where both reactants are of the same species, and heterodimerizations, where both reactants are of different species. Each class yields propensity functions of a different functional form and must, therefore, be considered separately. For a homodimerization r of reactant species Xk, we have that
under mass-action kinetics. Summing these four equations yields
and therefore Equation (14) holds for homodimerizations. Likewise, for a heterodimerization r and reactant species of reactant species Xi and Xj, we have
under mass-action kinetics. Summing these four equations yields
and therefore Equation (14) holds for heterodimerizations. The final step of the proof is to observe that the innermost summand in the master equation (13) can be written
where the second step follows from equivalence (14) and the third follows from relationship (12). Taken together, Equations (14) and (15) allow us to rewrite (13) as
which, upon inspection, is identical to the evolution equation that governs p; namely, Equation (11).
2.5. The augmented reaction network
In this subsection, we use the extended network to construct an augmented reaction network (ARN), which we denote , that consists of both a chemical reaction network (simulated stochastically) and a set of ODEs (simulated deterministically) that, taken together, provide an approximation of the original network and that can be simulated at lower computational expense. Indeed, simply simulating the network using an SSA would be at least as computationally expensive as simply simulating . Specifically, the ARN contains all 2K species of — the key difference is that in forming the ARN we separate out all reactions that contain only continuous species. These “continuous-only” reactions are not simulated using the discrete method; rather, we derive from the continuous-only reactions a system of approximate time-evolution equations that govern (in part) the means of the continuous species Ck. It is this system of equations that we simulate using the continuous method. Note that not all reactions in which the Ck participate are continuous-only; indeed, many of the first- and second-order reactions in contain both continuous and discrete species. These reactions that involve both continuous and discrete species are of “mixed-type', and are simulated using the discrete method. In this manner, the discrete species are governed exclusively by the discrete method; on the other hand, the continuous species are governed by the continuous method for all high copy-number reactions (the continuous-only reactions) and by the discrete method for low copy-number reactions (the mixed-type reactions).
We now detail the construction of the ARN. Beginning with a CRN, , we apply the extension procedure set out in Section 2.3 to produce the extended network . As before, we denote by C(t) and D(t) the number of individuals in the continuous and discrete regimes at time t, respectively, which we combine into a single state vector Y(t) = C(t)⊕D(t). The complete set of reactions in the extended network numbers |R0| + 2|R1| + 4|R2| + 2K, of which a total of |R1| + |R2| are continuously-only — one for each first-order reaction and one for each second-order reaction in the original network. We denote the sets of continuous-only first- and second-order reactions by and , respectively. From we derive a master equation governing the evolution of ℙ(C(t) = c(t)) under this set of reactions. Finally, we derive mean time-evolution equations and close the system at first-order (via the mean-field or Poisson moment closures, for example). This procedure yields a system of ODEs that will ultimately be simulated by the continuous method. The remaining |R0| + |R1| + 3|R2| + 2K reactions are those aforementioned mixed-type and discrete-only reactions, which will be simulated by the discrete method.
Following this procedure, we find that the mean of the kth continuous species under the action of the reactions in the set obeys the following evolution equation,
Given that this description contains only first and second-order reactions, it is straightforward to derive mean time-evolution equations for each of the Ci under the mean-field and Poisson closures. Define for a reaction r the function πr(n) that returns the nth reactant species of said reaction, where n = 1, …, d. For example, for a reaction r of the form (6), the function takes the values πr(1) = Ci and πr(2) = Cj. Denote by and the sets of hetero and homodimerizations, respectively, such that . Note that the definition of a homodimerization guarantees that for any such reaction r, πr(1) = πr(2). We can now write the mean time-evolution equations for each of the Ck. Under the mean-field closure, Equation (16) becomes
Similarly, under the Poisson closure, Equation (16) becomes
To complete our description of the ARN, we also must specify the stoichiometry matrix, denoted M, that represents the set of reactions that will be simulated using the discrete method. This matrix may be written in block form,
where MR is the stoichiometric matrix obtained all remaining |R0| + |R1| + 3|R2| discrete reactions in , and MK is the stoichiometric matrix representing the regime conversion reactions. Notice that without loss of generality we can write
where IK is the K × K-dimensional identity matrix.
The ARN corresponding to the CRN is thus defined to be the tuple of the set of 2K species Ci, Di (i = 1, …, K), the stoichiometry matrix M and associated propensity functions (), and the system of ODEs given by either (17) or (18), depending on the chosen closure. We call these the mean-field ARN (M-ARN) and the Poisson ARN (P-ARN) associated with the CRN , respectively.
2.6. The regime-conversion method
We now describe in detail our proposed algorithm for the efficient simulation of an ARN : the regime-conversion method. The method itself resembles that of other hybrid methods based on the Gillespie direct method, and its implementation is straightforward—the mathematical machinery that gives the method its computational efficiency is implicit in the structure of the ARN.
The only strictly numerical parameters in the method are Δt, the ODE update step size, which should be chosen according to the numerical method used for solving the system of ODEs, and; the regime conversion rates γf,k and γb,k and thresholds Tk, which can be iteratively refined for a given problem of interest over the course of several shorter test runs. In the present description of the method, we take the step size Δt to be fixed; however, we note that all instances of fixed Δt may be replaced with a suitable value to accommodate, for example, adaptive time-stepping methods. We further comment that in all test problems presented here, the forward and backward regime conversion rates γf,k and γb,k are taken to be equal for each k = 1, ..., K.
The method is initialized by specifying the initial conditions Y(0) = C(0)⊕D(0), the first ODE update time, td = Δt, and the initial and final simulation times t0 and tf, respectively. We next calculate the value of each propensity function at the initial time t = t0 and calculate their sum α0(t). As in the Gillespie direct method, the sum α0(t) is used to determine the time until the next discrete-regime reaction τ using the formula
where u ~ U(0, 1) is a uniformly distributed random number.
If, at time t, the time of the next reaction is before that of the next ODE update (i.e. t + τ < td) then a regular stochastic event is executed. Notice, however, that since the state C is partially governed by the system of ODEs, the mass of any given species Ck is not necessarily integer-valued. It is possible then that the firing of an event in the usual manner may result in Ck < 0 for some k = 1, …, K. To avoid this unphysical occurrence we perform a rejection sampling step when a reaction attempts to destroy or convert a continuous mass molecule of species k when Ck ∈ (0, 1). Specifically, we sample u ~ U(0, 1) – if u < Ck, we execute the reaction and set Ck = 0; otherwise, the reaction does not occur.
If t + τ > td we set t = t + τ. Then, we enumerate without loss of generality all reactions by the order in which they appear in the stoichiometry matrix M of , denoting by the value of the propensity function at time t associated with the pth reaction under said enumeration. The reaction to be executed is then sampled by selecting r ~ U(0, 1) uniformly at random and finding j such that
In the case that the next reaction would occur after that of the next ODE update (i.e. t + τ > td), an ODE update is performed to calculate the concentrations of the continuous species C. This may be achieved using any suitable numerical method. After this, the time is set to be equal to the current ODE update time t = td, the time of the next ODE update is set td = td + Δt, and the process of sampling a new stochastic event is begun anew at time t. This procedure continues until the final time tf is reached, and forms the entirety of the RCM. An algorithmic description of the RCM is given in Algorithm 1.
3. Results
In this section we demonstrate the accuracy of the RCM for three example problems of increasing complexity. We choose to use the classical fourth-order Runge Kutta method [19, p. 352] for solving the systems of ODEs, and the GDM for simulating stochastic trajectories. We make special note that the validity of our coupling is independent of the chosen numerical method for simulating the system of ODEs. Nevertheless, the accuracy of the method as a whole will naturally depend to a large extent on the accuracy of the underlying numerical techniques; a phenomenon that we explore in Test Case 3.2. To measure the error in a simulation run, we define the relative error between the SSA and the RCM by
where fk,SSA is the computed density of the kth species at time t as approximated by the SSA (resp. by the RCM). Likewise, we define the relative error between the system of ODEs and the SSA by
where fk,ODE is the computed density of the kth species at time t according to the system of ODEs as simulated by the numerical method.
3.1. Test case 1—Alternating exponential growth
Our first test case aims to demonstrate the accuracy of the method in the case of network with a single species, where continuous mass is degraded by a first-order degradation reaction to induce a continuous-to-discrete regime conversion, and discrete mass is produced by a zeroth-order production reaction to induce a discrete-to-continuous regime conversion. We thus consider the following simple reaction network consisting of a single species X and two reactions,
where the rates are of the form
where ki > 0 and Ii is some finite, non-empty union of time intervals. In our specific example, we choose these intervals such that the degradation reaction is “on” precisely when the production reaction is “off”, and vice-versa. This network has stoichiometry matrix
and propensity functions
From this, we form the corresponding M-ARN with two species C and D. This network has stoichiometry matrix
and propensity functions
corresponding to the zeroth-order production and the first-order degradation of discrete mass, respectively. The first-order degradation of continuous mass is modeled via the ODE
We present the results of this test case in Figure 1 using the parameter values given in Table 1. This proof-of-concept example demonstrates the key behavior of the RCM—the conversion between discrete- and continuum-governed mass. As expected, when overall density falls below the threshold value we observe the conversion of continuum to discrete mass, and vice versa when density again becomes sufficiently high. We observe no evidence of bias in the RCM, with the fluctuations away from zero in Figure 1A not persisting between simulation runs.
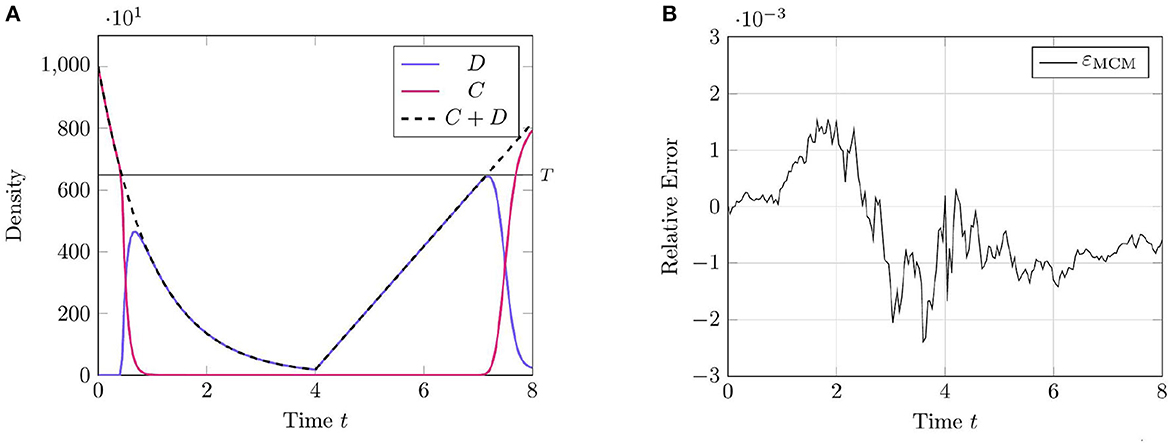
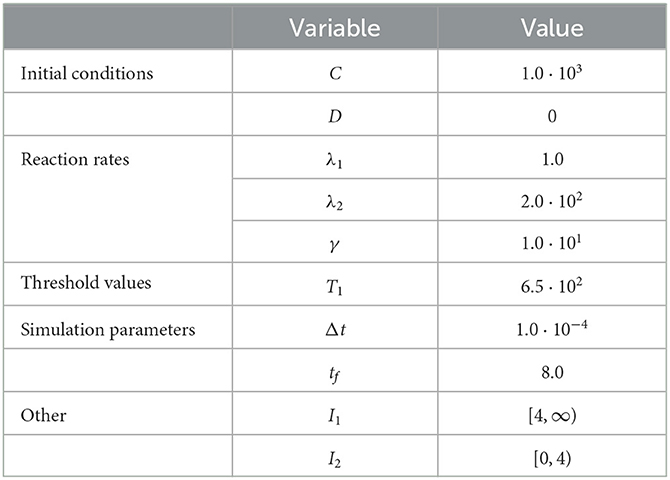
Figure 1. Results of Test Case 1 (Section 3.1) with parameters as specified in Table 1 with conversion threshold T = 650. Simulation results averaged over 105 repeats. (A) Plot of the density of D + C as simulated by the RCM. (B) Relative error in D + C between the RCM and the SSA.
3.2. Test case 2—Alternating logistic growth
Our second test case aims to demonstrate the accuracy of the method in the case of a network with a single species, this time where continuous mass is degraded by a second-order degradation reaction to induce a continuous-to-discrete regime conversion, and discrete mass is produced by a first-order production reaction to induce a discrete-to-continuous regime conversion. As in Test Case 1, we take consisting of a single species X, this time with reactions
where the rate λ1 is constant over time and λ2 is governed by
where k2 > 0 and I is some finite, non-empty union of time intervals. Again, we select these intervals such that the production reaction is “on” precisely when the degradation reaction is “off”, and vice-versa. This network has stoichiometry matrix
this time with propensity functions
Following extension, we obtain an ARN with two species C and D. This network has stoichiometry matrix
and propensity functions
representing the production of a discrete molecule from a discrete molecule, the degradation of a discrete molecule by a discrete molecule, the degradation of a continuous molecule by a discrete molecule, and the degradation of a discrete molecule by a continuous molecule, respectively. We form the equation governing the second-order degradation of continuous mass by continuous mass and the production of continuous mass from continuous mass using the Poisson closure; this equation is given by the ODE
We present the results of this test case in Figure 2 using the parameter values given in Table 2. The results of this test case demonstrate a particular limitation of the RCM; namely, that the error in the RCM is, in some sense, “tethered” to the error in the solution to the system of ODEs in the associated ARN. We see this most clearly at the parameter transition point t = 20, when the second-order reaction degradation activates.
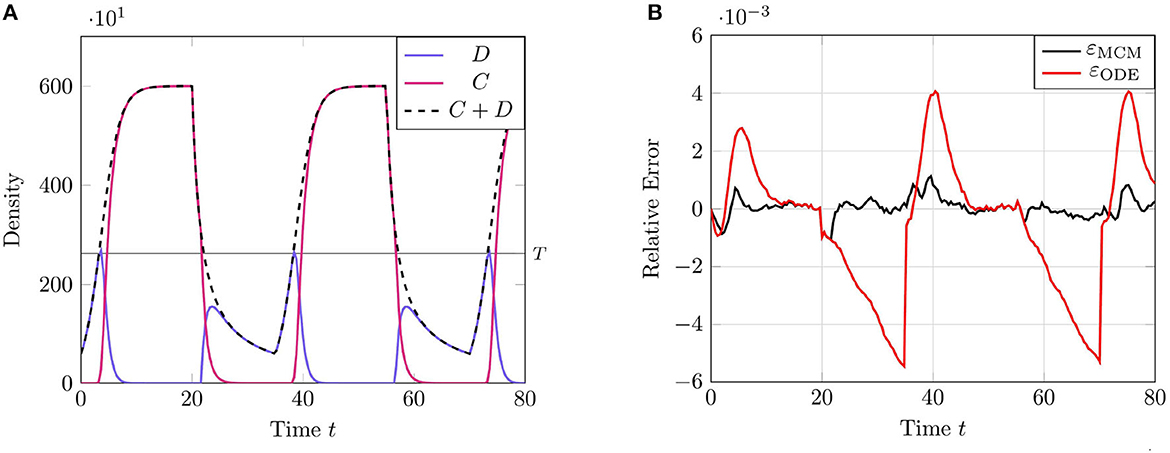
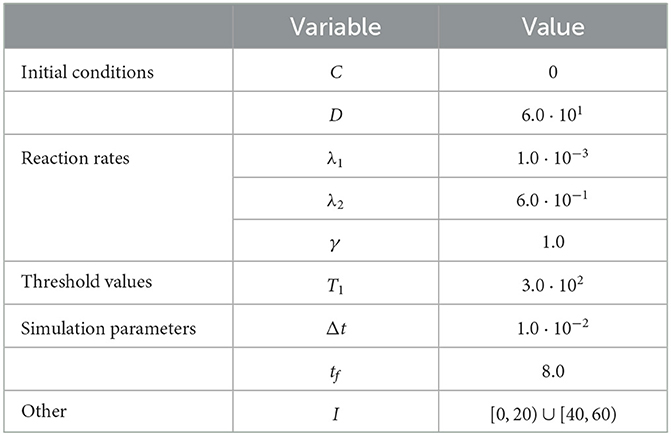
Figure 2. Results of Test Case 2 (Section 3.2) with parameters as specified in Table 2 with conversion threshold T = 300. Simulation results averaged over 105 repeats. (A) Plot of the density of D + C as simulated by the RCM. (B) Relative error in D + C between the RCM and the SSA and between the system of ODEs and the SSA.
3.3. Test case 3—Chemical signaling
For our third test case, consider a CRN, , consisting of three chemical species X1, X3, and X2, which we refer to as the signal, intermediate, and product species respectively, within a reactor vessel of unit volume. The product X2 is produced via the intermediate X3 and is degraded via a first-order sink reaction. The intermediate is produced via a zeroth-order source reaction.
The signal species X1 is coupled indirectly with X2 via the following reaction,
in which the signal degrades the intermediate X3. Finally, the signal species itself is produced and degraded according to the same reaction system we used in Test Case 1,
This CRN, , has stoichiometry matrix
with propensity functions given by
Under the mean-field closure, the means of X1, X2, and X3 are governed by the following system of ODEs
As demonstrated in Paulsson et al. [20], the steady-state behavior of is determined to a substantial degree by the stochastic fluctuations of X3. This system, therefore, benefits greatly from a hybrid modeling approach, where the low-copy-number X1 and X3 can be modeled discretely. From the CRN we form the M-ARN , which has stoichiometry matrix
where M3 is defined in equation (19); reaction propensities
and; the following system of ODEs,
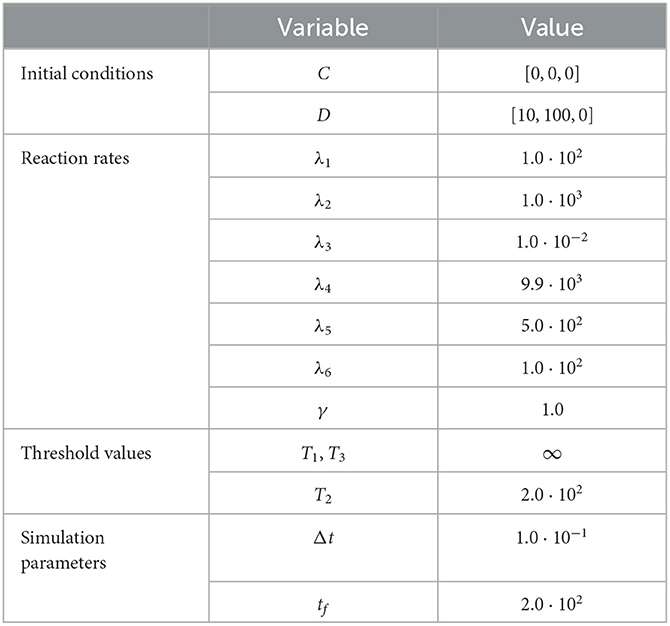
To demonstrate the utility of the RCM in this case, we compare the mean densities of as approximated by both the Gillespie SSA and by the mean-field equation (20) with the mean density of as approximated by the RCM. For this problem, we wish to simulate the species X1 and X3 purely via the discrete regime and the product species X2 will be permitted to switch regimes dependent on density. The model parameters used for our test case are listed in Table 3. We present the results of this test case in Figure 3. Notice that the density of D2 appears to decrease before reaching the threshold value. This is to be expected since, as the system is governed wholly by the discrete regime until the threshold is reached, a non-zero number of simulation trajectories reach threshold before the mean trajectory. This manifests as the mean trajectory beginning regime transition before the threshold is actually reached.
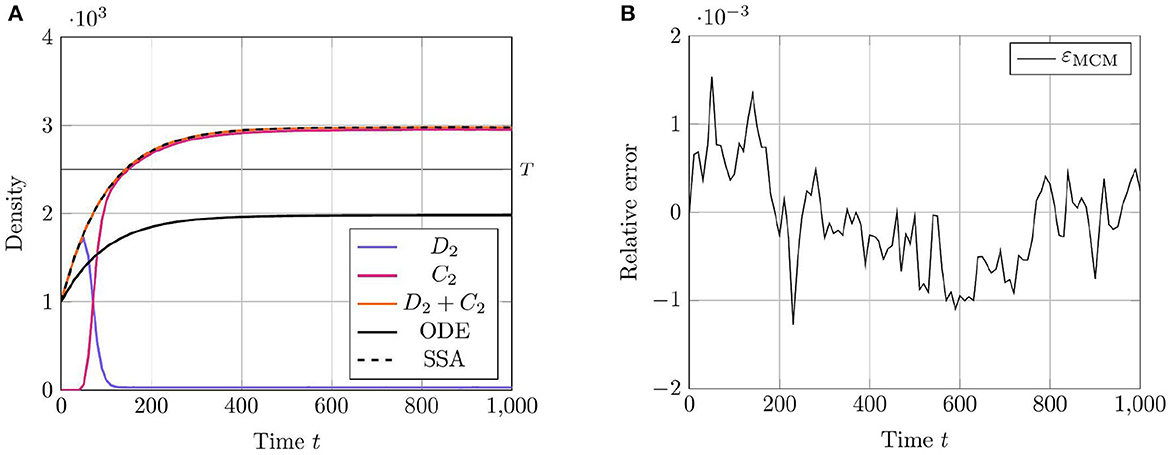
Figure 3. Results of Test Case 3 (Section 3.3) with parameter values given in Table 2. (A) Plot of the density of C2 + D2 as simulated by the RCM, SSA, and ODEs. Note that the density as determined by the SSA is indistinguishable at this scale from the density determined by the RCM—as such the trajectory of the RCM obscures that of the SSA in the plot. (B) Relative error in C2 + D2 between the RCM and the SSA. Simulation results averaged over 1.6 · 104 repeats.
Evidently, the RCM substantially outperforms the mean-field ODEs at approximating the true trajectory of this reaction network. The reason for this is that the RCM guarantees the simulation of the X3 species exclusively via the discrete regime by setting the relevant regime conversion threshold values to infinity. As such, the method retains information of the stochastic fluctuations in X3 where the system of mean-field ODEs does not. We further note the lack of bias in the error of the RCM.
3.4. Test case 4—Michaelis-Menten enzyme kinetics
Here we apply the RCM to the well-studied Michaelis-Menten model of enzyme kinetics [21, 22]. We consider a slight generalization of the classical model wherein the substrate species is continuously supplied to the system. The model can be represented as a CRN with the following reactions:
This network models the conversion of a substrate species S into a product species P via catalysis with some enzyme E. This conversion occurs when a member of the substrate species binds with the enzyme to form an intermediate enzyme-substrate complex M. The complex M can then unbind either into its original constituents E + S or into a new product P, freeing the enzyme E to bind with further substrate. Note that, since E acts only as a catalyst in the above network, the quantity ET = E + M is conserved over time.
For the purposes of our demonstration, tracking the growth in copy number of the species P is unimportant. Thus, we henceforth neglect to include this species in the network, though we retain the reaction channel to leave the dynamics of the remaining species unchanged. Taking the mean-field closure of the master equation formed from the system of reactions (21), we obtain the following system of ODEs:
which can be shown to have a steady-state solution given by
We now form the M-ARN for system (21), which has stoichiometry matrix
reaction propensities
and the following system of ODEs,
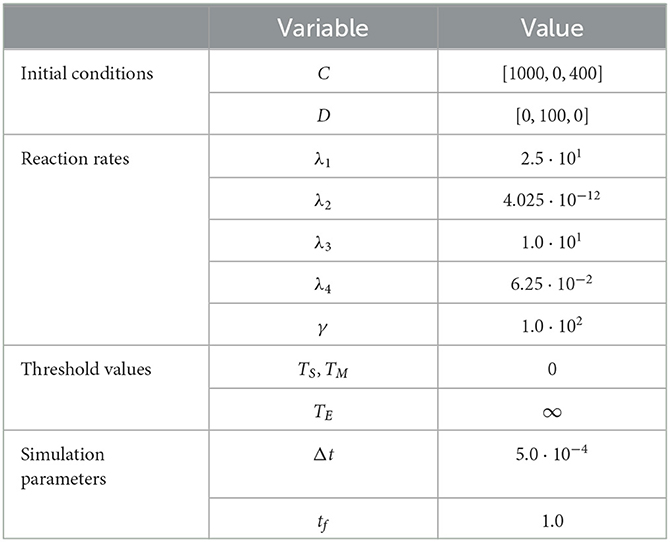
As in prior test cases, we again evaluate the accuracy of the RCM against the Gillespie SSA. For this system, we select parameters such that all species except for the enzyme species E are simulated using the continuous regime in order to evaluate how well the RCM performs at estimating both the mean and the variance of E at steady state. We present the results for the mean estimate in Figure 4. The model parameters used for this test case are listed in Table 4. This case highlights a key feature of the RCM. Notice that, despite E having a threshold of ∞ (and therefore suggesting that all mass should be governed by the discrete regime), a small proportion of the mass is nevertheless represented by the continuous regime at steady state. This proportion can be tweaked to different values depending on the specific enzymatic reaction being modeled by increasing or decreasing the regime transition rates γf,E and γb,E. This non-zero mass CE, as well as the fact that all other species are governed primarily by the continuous regime, manifests as a slight positive bias in the RCM vs. the SSA (Figure 4B). A parameter sweep demonstrates that this bias is reduced by decreasing the step size used in the numerical method; however, this naturally comes at greater computational cost.
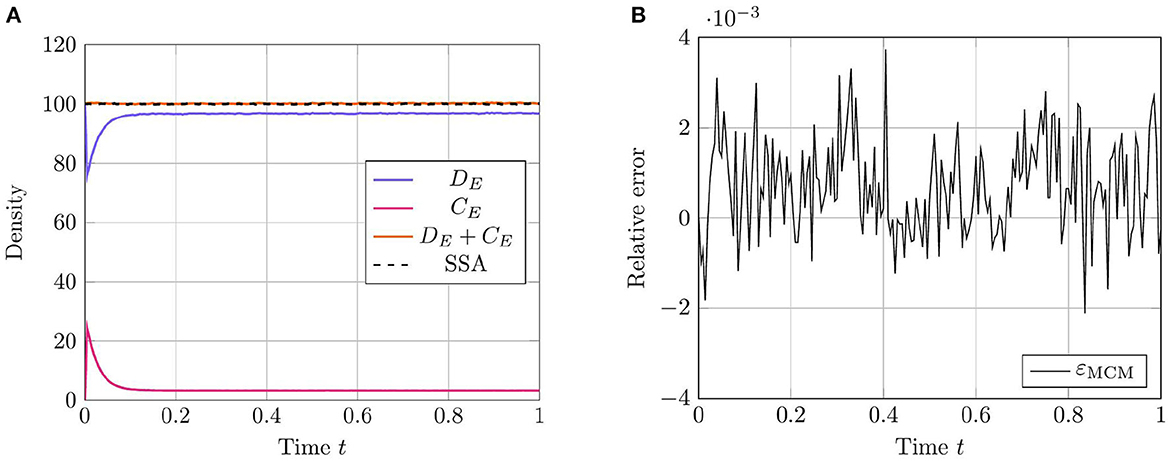
Figure 4. Results of Test Case 4 (Section 3.4) with parameter values given in Table 3. Simulation results averaged over 1 · 105 repeats. (A) Plot of the density of DE + CE as simulated by the RCM. Shown is the amount of mass DE in the discrete regime and the amount of mass CE in the continuous regime, alongside the total mass DE + CE. Results from the SSA over the same time period are overlaid. (B) Relative error in the density of DE + CE as predicted by the RCM and vs. the SSA over time.
Figure 5 compares the steady-state distribution of the mass of E as estimated by the RCM vs. the SSA. In both cases, we consider the system to have reached steady state by t = 5, and sample the distribution of E at this time. Here, we observe that while the overall shape of the distribution is largely preserved, the variance predicted by the RCM is slightly lower than that predicted by the SSA, as evidenced by the thinner tails of the distribution. Unlike the bias in the mean, this damping of the variance is not dependent on the step size used. This discrepancy in the variance is perhaps unsurprising, given that a proportion of E is governed by the (approximate and deterministic) continuous regime.
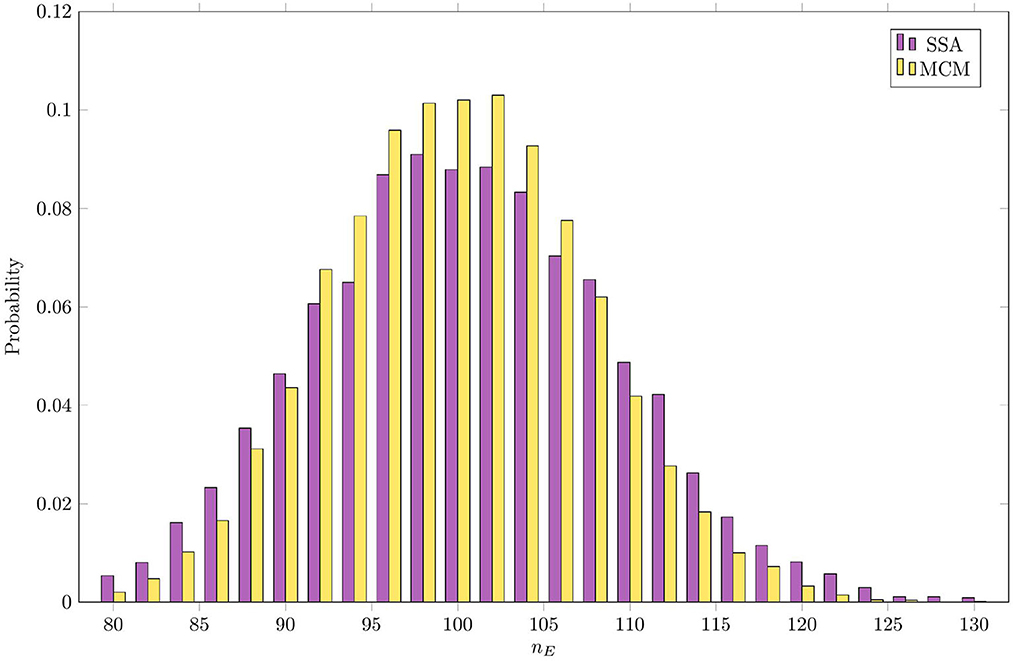
Figure 5. Steady-state distribution of the amount of mass in species E as estimated by the RCM vs. the SSA.
4. Discussion
In this work we introduced a novel hybrid method for simulating well-mixed chemical reaction networks. This method couples a system of ODEs with a Markov process representation of a chemical reaction network by constructing a so-called augmented reaction network that combines both representations. The continuous and discrete components of the augmented network can be simulated simultaneously using different techniques to maximize computational efficiency and minimize the loss of accuracy resultant from taking continuum approximations. We demonstrated the accuracy of the method in three separate test problems of increasing complexity, evidencing in the final test case a substantial improvement in accuracy using our method vs. the standard continuum approximation technique.
While our method demonstrates substantially better accuracy vs. the continuum-only models in the test cases we present, its advantage vs. a traditional SSA is, in general, dependent on network structure. Specifically, in systems where the majority of computation time (when simulated via a SSA) is spent on the simulation of low copy-number species interacting with high copy-number species via bimolecular reaction channels, there is little computational benefit to our approach. The reason for this is that such reactions are (assuming each species is below and above the transition thresholds, respectively) necessarily simulated using the SSA, and therefore may impart no computational benefit in the RCM vs. the SSA alone. In cases where both reactant species in a bimolecular reaction are of sufficiently high concentration to be above their respective transition thresholds, it may be the case that the RCM yields similar accuracy to that of a continuum-based approach. Nevertheless, in neither case is there reason to expect a priori that use of the RCM is necessarily disadvantageous. With these caveats in mind, there are clear instances where the RCM may be suitable to use over traditional methods. In loosely-coupled networks where the majority of interactions are of first-order (networks of this type frequently arise when modeling cellular populations [23–26]), the RCM demonstrates a clear computational advantage.
Another limitation of the RCM is that it may estimate moments of order two and above with some inaccuracy. This is a limitation shared by several other hybrid methods [11, 12]. Indeed, we see that by simulating a significant proportion of the dynamics of a system via the continuous method (as seen in Test Case 3.4, one induces a damping effect on the variance in species numbers. Nevertheless, the results demonstrate that the RCM allows for partial recovery of the distributions of constituent species. A possible solution to this problem would be to replace the system of deterministic, mean-field ODEs governing the continuous regime with appropriate stochastic differential equations. This approach has been used to solve the variance damping problem in spatial hybrid methods [27]. Additional work is required to conduct a full examination of the evolution of higher-order moments in the RCM and to quantify how such evolution is related to model parameters.
Our method differs from similar hybrid methods [11–13] in two crucial ways. First, our method allows for mass to transition dynamically between regimes. While it is possible to set thresholds (by setting threshold values to 0 or ∞) and transition rates (by setting transition rates to particularly small or particularly large values) such that mass is preferentially represented by one of the two regimes, the intended use case of the RCM is for systems where there is significant variability in the copy number of one of more species over the course of a simulation run. Second, in many cases, species simulated by the RCM have both a discrete and a continuous component. This allows for the partial recovery of these species' distributions, which would not be possible with a continuum-only approximation of the first moments of a network.
There are several ways in which the RCM might be extended to accommodate a wider variety of problems and to increase its computational efficiency. The first and most obvious direction is to extend its dimensionality; for example, to a spatial setting. The RCM, being an effective simulation technique for well-mixed reaction networks, might be extended to a spatial reaction-diffusion setting in several ways. Under a mesoscopic modeling regime [8], where individual system components are collected into well-mixed spatial “bins” of fixed size, the RCM could be used to simulate reactions by treating individuals in each bin as distinct species that do not interact with neighboring bins. In this framework, diffusive jumps between bins are simply reactions that convert individuals in one “bin species” to another. A spatial model consisting of binned particles and ordinary differential equations associated with each bin is thereby easily treated via the RCM. Nevertheless, this representation of a reaction-diffusion process is limited - for spatial domains with many bins, simulating large systems of (potentially) non-linear ODEs may be prohibitively expensive. A more sensible choice would be to represent the continuous approximation as a system of partial differential equations on an explicitly spatial domain; indeed, contemporary spatially-extended hybrid methods that couple continuous and mesoscopic regimes generally use this representation [8]. In this case, the matter of coupling the stochastic and diffusive reactions in each bin is not so straightforward, requiring numerical integration of the partial differential equation over relevant spatial regions. Extending the RCM in this manner to a spatially-extended mesoscopic-to-continuous hybrid method will form the basis of an upcoming investigation.
The RCM may also be extended to incorporate additional dimensionality along non-spatial lines. An important class of demographic and biological models are those with size- or age-structure, or a combination thereof. These model systems of interacting individuals (either eukaryotic or prokaryotic cells) undergoing some variant of the classical cell-cycle [28], and for which an individual's size (or age) is an important contributor to overall population dynamics. These systems are often modeled as either discrete-state stochastic processes [26, 29–31] or as continuous partial differential equations via the McKendrick-von Foerster equation [32, 33]. Despite their ubiquity, to the best of the authors' knowledge there exist no hybrid simulation techniques that can accommodate, without modification, size- or age-structure. Depending on the specific functional form of any size- or age-mediated reactions, a method of “spatial” numerical integration over intervals of age or size similar to that proposed for spatial extension may prove fruitful for coupling these two modeling regimes.
An important area of research in numerical methods in general is the development of so-called “adaptive” methods. These are methods for which certain numerical parameters can be changed mid-way through a simulation run to adapt to situations that might otherwise prove numerically challenging or computationally infeasible. The prototypical example of this is in adaptive time-stepping methods for solving systems of ordinary differential equations, wherein the usual fixed time step of a numerical solver is replaced with a variable time step that is recalculated at each update step to ensure stability even when the derivatives of the system undergo large variations [34]. As noted in our description of the RCM, one could apply such an adaptive time-stepping method for computing approximations to the continuum regime description without needing to modify the algorithm. Nevertheless, it is not difficult to conceive of a more specialized form of time-stepping that would take into consideration more than just the mass in the continuous regime and instead consider both the discrete mass and the calculated propensity functions at the time of an update step. For example, large numbers of individuals either entering or leaving the continuous regime may, by affecting the gradient of the continuum approximation, inject undesirable numerical instabilities into the RCM in extreme cases—something that traditional adaptive time-stepping methods are not designed to handle. Adaptivity in terms of time-stepping is not the only potential improvement, however. Presently, the RCM requires that the threshold values for the regime conversion reactions are set and fixed a priori. One can envisage modifications to the RCM where the conversion thresholds vary in response to changes in density, computational cost, rates of density change, and the stochasticity present in the system at any given time.
Finally, the method may be extended to incorporate reactions of arbitrary molecular order. While any reactions of molecular order of at least three can be decomposed into sequences of reactions of molecular order of at most two, these decompositions can be difficult to compute in practice. We conjecture that the same techniques used to demonstrate equivalence between the CRN and its associated ARN apply to higher-order reactions; however, proving this in generality is likely to be cumbersome. Further, one needs 2d ODEs to satisfy the coupling requirements C1, C2 for a reaction of order d which, while not necessarily impacting computation time, may quickly become impractical to implement for large networks.
To summarize, our method provides a novel and computationally efficient technique for simulating well-mixed chemical reactions networks using a hybrid discrete/continuous methodology. Unlike similar existing methods, ours does not depend on the system of interest possessing certain properties; i.e., a particular decomposability of reactions or species into “fast” and “slow” categories. Further, it represents a promising coupling mechanism between the mesoscopic and macroscopic regimes that may permit for the development of new spatially-extended hybrid techniques that have a particular intrinsic adaptivity; namely, the ability to simulate spatial density distributions with significant and dynamic heterogeneity.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
AH performed preliminary investigations. JK performed the numerical and mathematical analysis and took lead in writing and preparing the manuscript. CY and CG provided feedback and suggestions during the analysis. All authors contributed to the design and conception of the method, and read and approved the submitted version.
Funding
JK is supported by a scholarship from the EPSRC Centre for Doctoral Training in Statistical Applied Mathematics at Bath (SAMBa), under the project EP/L015684/1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
2. Gillespie DT. Exact stochastic simulation of coupled chemical reactions. J Phys Chem. (1977) 81:2340–61. doi: 10.1021/j100540a008
3. Gibson MA, Bruck J. Efficient exact stochastic simulation of chemical systems with many species and many channels. J Phys Chem A. (2000) 104:1876–89. doi: 10.1021/jp993732q
4. Cao Y, Li H, Petzold L. Efficient formulation of the stochastic simulation algorithm for chemically reacting systems. J Chem Phys. (2004) 121:4059–67. doi: 10.1063/1.1778376
5. Anderson DF. A Modified next reaction method for simulating chemical systems with time dependent propensities and delays. J Chem Phys. (2007) 127:214107. doi: 10.1063/1.2799998
6. Gillespie DT. A rigorous derivation of the chemical master equation. Physica A. (1992) 188:404–25. doi: 10.1016/0378-4371(92)90283-V
7. Schnoerr D, Sanguinetti G, Grima R. Comparison of different moment-closure approximations for stochastic chemical kinetics. J Chem Phys. (2015) 143:185101. doi: 10.1063/1.4934990
8. Smith CA, Yates CA. Spatially extended hybrid methods: a review. J R Soc Interface. (2018) 15:20170931. doi: 10.1098/rsif.2017.0931
9. Cao Y, Gillespie DT, Petzold LR. The slow-scale stochastic simulation algorithm. J Chem Phys. (2005) 122:014116. doi: 10.1063/1.1824902
10. Cotter SL, Zygalakis KC, Kevrekidis IG, Erban R. A constrained approach to multiscale stochastic simulation of chemically reacting systems. J Chem Phys. (2011) 135:094102. doi: 10.1063/1.3624333
11. Hellander A, Lötstedt P. Hybrid method for the chemical master equation. J Comp Phys. (2007) 227:100–22. doi: 10.1016/j.jcp.2007.07.020
12. Smith S, Cianci C, Grima R. Model reduction for stochastic chemical systems with abundant species. J Chem Phys. (2015) 143:214105. doi: 10.1063/1.4936394
13. Jahnke T. On reduced models for the chemical master equation. Multiscale Model Simul. (2011) 9:1646–76. doi: 10.1137/110821500
14. Van Kampen NG. Stochastic Processes in Physics and Chemistry. 3rd ed London: Elsevier. (2007). doi: 10.1016/B978-044452965-7/50006-4
15. Barrat A, Barthélemy M, Vespignani A. Dynamical Processes on Complex Networks. 1st ed. Cambridge: Cambridge University Press (2008). doi: 10.1017/CBO9780511791383
16. Nåsell I. An extension of the moment closure method. Theor Popul Biol. (2003) 64:233–239. doi: 10.1016/S0040-5809(03)00074-1
17. Laidler KJ. Reaction Kinetics. 1st ed Pergamon: Elsevier. (1963). doi: 10.1016/B978-1-4831-9738-8.50005-4
18. Aris R, Gray P, Scott SK. Modelling cubic autocatalysis by successive bimolecular steps. Chem Eng Sci. (1988) 43:207–11. doi: 10.1016/0009-2509(88)85032-2
19. Suli E, Mayers DF. An Introduction to Numerical Analysis. Cambridge: Cambridge University Press. (2003).
20. Paulsson J, Berg OG, Ehrenberg M. Stochastic Focusing: Fluctuation-Enhanced Sensitivity of Intracellular Regulation. PNAS. (2000) 97:7148–53. doi: 10.1073/pnas.110057697
22. Murray JD. Reaction Kinetics. In: Murray JD, editor. Mathematical Biology. New York, NY: Springer New York (1993). p. 175–217. doi: 10.1007/978-0-387-22437-4_6
23. Simpson MJ, Towne C, McElwain DS, Upton Z. Migration of breast cancer cells: understanding the roles of volume exclusion and cell-to-cell adhesion. Phys Rev E. (2010) 82:041901. doi: 10.1103/PhysRevE.82.041901
24. Simpson MJ, Landman KA, Hughes BD. Cell invasion with proliferation mechanisms motivated by time-lapse data. Physica A. (2010) 389:3779–90. doi: 10.1016/j.physa.2010.05.020
25. Yates CA, Ford MJ, Mort RL. A multi-stage representation of cell proliferation as a Markov process. Bull Math Biol. (2017) 79:2905–28. doi: 10.1007/s11538-017-0356-4
26. Kynaston JC, Guiver C, Yates CA. Equivalence framework for an age-structured multistage representation of the cell cycle. Physical Review E. (2022) 105:064411. doi: 10.1103/PhysRevE.105.064411
27. Alexander FJ, Garcia AL, Tartakovsky DM. Algorithm refinement for stochastic partial differential equations: I. Linear Diffusion. J Comp Phys. (2002) 182:47–66. doi: 10.1006/jcph.2002.7149
28. Schafer KA. The cell cycle: a review. Vet Pathol. (1998) 35:461–78. doi: 10.1177/030098589803500601
29. Stukalin EB, Aifuwa I, Kim JS, Wirtz D, Sun SX. Age-dependent stochastic models for understanding population fluctuations in continuously cultured cells. J R Soc Interface. (2013) 10:325. doi: 10.1098/rsif.2013.0325
30. Greenman CD, Chou T. A kinetic theory for age-structured stochastic birth-death processes. Phys Rev E. (2016) 93:012112. doi: 10.1103/PhysRevE.93.012112
31. Chou T, Greenman CD. A hierarchical kinetic theory of birth, death and fission in age-structured interacting populations. J Stat Phys. (2016) 164:49–76. doi: 10.1007/s10955-016-1524-x
32. Trucco E. Mathematical models for cellular systems. The von Foerster equation Part I B. Math Biophys. (1965) 27:285–304. doi: 10.1007/BF02478406
33. Rossini L, Contarini M, Speranza S. A Novel version of the von Foerster equation to describe poikilothermic organisms including physiological age and reproduction rate. Ric Mat. (2021) 70:489–503. doi: 10.1007/s11587-020-00489-6
Keywords: population dynamics, stochastic simulation, chemical reaction network simulation, hybrid method, continuum model
Citation: Kynaston JC, Yates CA, Hekkink AVF and Guiver C (2023) The regime-conversion method: a hybrid technique for simulating well-mixed chemical reaction networks. Front. Appl. Math. Stat. 9:1107441. doi: 10.3389/fams.2023.1107441
Received: 24 November 2022; Accepted: 03 July 2023;
Published: 07 September 2023.
Edited by:
Ivo Siekmann, Liverpool John Moores University, United KingdomReviewed by:
Ramon Grima, University of Edinburgh, United KingdomLutz Brusch, Technical University Dresden, Germany
Copyright © 2023 Kynaston, Yates, Hekkink and Guiver. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joshua C. Kynaston, am9zaC5reW5hc3RvbkBiYXRoLmVkdQ==
 Joshua C. Kynaston
Joshua C. Kynaston Christian A. Yates
Christian A. Yates Anna V. F. Hekkink1
Anna V. F. Hekkink1 Chris Guiver
Chris Guiver