- Center for Bio/Molecular Science and Engineering, Naval Research Laboratory, Washington, DC, United States
Over the past two decades, various scaffolds have been designed and synthesized to organize enzyme cascades spatially for enhanced enzyme activity based on the concepts of substrate channeling and enhanced stability. The most bio-compatible synthetic scaffolds known for enzyme immobilization are protein and DNA nanostructures. Herein, we examined the utility of the T4 phage capsid to serve as a naturally occurring protein scaffold for the immobilization of a three-enzyme cascade: Amylase, Maltase, and Glucokinase. Covalent constructs between each of the enzymes and the outer capsid protein Hoc were prepared through SpyTag–SpyCatcher pairing and assembled onto phage capsids in vitro with an estimated average of 90 copies per capsid. The capsid-immobilized Maltase has a fourfold higher initial rate relative to Maltase free in solution. Kinetic analysis also revealed that the immobilized three-enzyme cascade has an 18-fold higher converted number of NAD+ to NADH relative to the mixtures in solution. Our results demonstrate that the T4 phage capsid can act as a naturally occurring scaffold with substantial potential to enhance enzyme activity by spatially organizing enzymes on the capsid Hoc.
Introduction
Catalytic properties of enzymes are greatly affected by their surrounding microenvironment, particularly enzymes retained in a small area either by limited surface or restricted volume (Kuchler et al., 2016). Specifically, high concentration of enzymes in a confined environment are more stable than those free in solution. As a result, there is a preference for a folded state over an unfolded state and their close proximity allows them to execute a series of biocatalytic events more efficiently through substrate channeling (Miles et al., 1999; Zhang, 2011). Based on these concepts, various synthetic scaffolds aimed to spatially organize enzymes were designed and used for immobilizing enzymes to enhance their activity (Conrado et al., 2008; Dueber et al., 2009; Fu et al., 2014). Artificial multi-enzyme scaffolds have been utilized both in vivo and in vitro to improve product production (Siu et al., 2015; Ellis et al., 2019). Among these synthetic scaffolds, protein arrays and DNA nanostructures are the most biocompatible and have the potential to form the basis of a powerful platform to enhance multi-enzyme catalysis for biotechnology applications (Klein et al., 2019; Lim et al., 2019). Our team is researching naturally occurring scaffolds possessing the ability to spatially organize enzymes.
One benefit that naturally occurring phage scaffolds possess is that they are monodisperse and can be produced economically from bacteria hosts. In addition, most of the icosahedral phage capsids are composed of arrays of hexamers formed by capsid proteins, which can serve as a platform for immobilizing enzymes spatially. T4 capsids are composed of the major capsid protein, gp23, and minor capsid protein, gp24, along with two accessory proteins, Hoc (highly antigenic outer capsid) and Soc (small outer capsid), and the portal protein, gp20 (Rao and Black, 2010). The capsids are homogeneous in size and structure and can only assemble inside the host bacteria with the expression of phage chaperone proteins and proteases. During the early infection of phage in bacteria, gp23 and gp24 assemble into a prohead shell wrapping around the core structure, later removed by protease. The vacant proheads allow DNA packaged inside through a packaging machinery, followed by the attachment of tail and tail fibers. Without DNA packaging, tail and tail fibers do not attach to the capsids.
The biggest hurdle for using phage scaffolds for displaying protein is that most capsids are quite rigid and can only display short peptides or few numbers of large proteins in vivo (Cardinale et al., 2012; Patel et al., 2017). However, T4 phage capsids allow one to overcome this difficulty by providing the needed flexibility to enable the display of large proteins through fusion with either of two outer capsid proteins, Hoc and Soc, without disrupting the capsid structure (Ren and Black, 1998). Moreover, both Hoc and Soc are dispensable for T4 phage propagation, their absence having no impact on T4 production. Soc proteins are closely associated with capsid proteins and are assembled next to each other, while Hoc is located at the center of hexamers, separated by both capsid proteins and Soc, based on a cryo-electron microscopy (EM) model (Fokine et al., 2004). Another advantage is that both Hoc and Soc fusions can be assembled onto Hoc and Soc deletion phage capsids as a scaffold either in vitro or in vivo (Rao and Black, 2010).
Our previously established work on characterization of T4 phage capsid structure using atomic force microscopy (AFM) showed that purified capsids are intact and stable (Archer and Liu, 2009; Robertson and Liu, 2012), and based on this work, we developed a new strategy to display a multi-enzyme cascade on phage capsids through SpyTag(St)/SpyCatcher(SC) pairing (Reddington and Howarth, 2015). Amylase (Aml), Maltase (Mal), and Glucokinase (GK) were selected to assemble onto phage scaffolds for catalytic analysis. These three enzymes are part of a four-enzyme biocatalytic pathway, which converts maltoheptaose into NADH and 6-phosphogluconolactone and the bio-catalytic assays for analyzing the enzyme activity have been well-established (Klein et al., 2019). The assembly of Hoc fusions onto phage scaffolds was conducted in vitro for better characterization. Enzyme–SC–St–Hoc fusions were purified and incubated in controlled ratios with T4 capsid. As observed in other scaffold systems, when assembled on the T4 capsid, the spatially organized assembled enzymes exhibit enhanced enzyme catalytic activity. The immobilized enzyme fusions on the phage scaffold showed enhanced biocatalytic activity for the number of NAD+ converted to NADH per second up to 18-fold higher than the enzyme fusions free in solution. We have successfully demonstrated T4 icosahedral phage as a naturally occurring scaffold adaptable for multi-enzyme cascade assembly.
Materials and Methods
Construction of Enzyme–SC Fusions and St–Hoc Plasmids
All the primer sequences used for cloning are listed in Table S1 and gene maps and sequences are described in the Supplementary Information. The enzyme–SC fusions were cloned into pET28, while St–Hoc was inserted into pACYCduet according to Anderson et al. (2010) and Goldman et al. (2017). Details are provided in the Supplementary Information.
Preparation of Fusion Proteins
Protein was produced using a protocol similar to one that had been previously described with several modifications (Walper et al., 2014). The protocol for protein production and purification is provided in the Supplementary Information. Protein concentrations were determined using UV-Visible spectroscopy and the molar extinction coefficient predicted from the protein sequence.
Preparation of Tailless ΔHoc T4 Phage Scaffolds
E. coli, CR63 (F− supD60 lamB63), or B40I (F−supD), containing tRNA suppressor E, was used for growing del Hoc T4 phage (ΔHoc T4), a gift from Dr. Lindsay W. Black, Professor of Biochemistry Department, UMB. The phage capsid preparation was performed according to Liu et al. (2014). Details are provided in the Supplementary Information. The concentration of phage capsids was estimated based on 1 mg = 1013 capsids by taking into account the copy numbers of capsid proteins, Hoc, Soc, and portal proteins in one capsid (Fokine et al., 2004; Liu et al., 2014). The protein concentration was determined by the absorbance 280 nm and Bradford assay (Bio-Rad, Hercules, CA).
Assembly of Hoc–Enzyme Fusions Onto ΔHoc T4 Phage Capsids
The three model enzyme–Hoc fusions (Aml–Hoc; Mal–Hoc; and GK–Hoc) were first mixed at various molar ratios, 1:1:1; 1:4:1; 1:16:1. Enzyme–Hoc mixtures were subsequently added to phage capsids at ratios of total enzyme to phage capsid of 60:1 or 40:1. The same amount of enzyme–Hoc fusions was also prepared without added phage capsids. The mixtures were incubated at 22°C for 2 h or overnight (16 h). The assembly buffer consisted of PBS supplemented with 10 mM MgCl2. To assess assembly, after incubation, the reactions were loaded onto 1% agarose and passed through 100 V for 2 h in 1 × Tris–Acetate–EDTA buffer. The scaffolds and proteins were then visualized with GelCode blue safe protein stain (Thermo Fisher Scientific).
Measurements of Enzymatic Activity
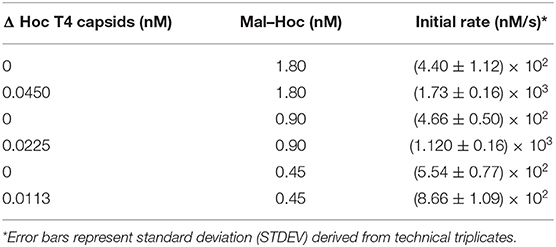
For assessing Maltase activity, the production of 4-Nitrophenol (absorbance at 405 nm) as a function of time was recorded for 3–4 h using a Tecan Infinite M1000 plate reader immediately after the addition of the substrate, 10 mM 4-Nitrophenyl-α-D-glucopyranoside to a mixture of Maltase or Mal-Hoc fusions in 250 mM HEPES (pH 7.4) buffer. For measuring the three-enzyme cascade activity, glucose-6-phopsphate dehydrogenase was included in excess in the reaction mixture and the rate was measured as NAD+ conversion to NADH (absorbance at 340 nm) as a function of time after the addition of Maltoheptaose. The reaction mixture included 5 mM ATP, 1 mM NAD+, and 10 U of glucose-6-phosphate dehydrogenase (G8404-2KU) in 250 mM HEPES (pH 7.4). The reaction schemes are described in more detail in Figure S1. The initial rate was measured as the slope after conversion of absorbance to nM/s, and Vmax, Kcat, and Km were calculated using Michaelis-Menton kinetics. Measurements were performed in triplicate. Chemicals were purchased from Millipore Sigma (St. Louis, MO), unless otherwise mentioned. Error bars were standard deviation derived from either biological replicates or technical triplicates.
Results
Demonstration of Purified Enzyme–Hoc Assembled on Phage Capsids
Wild-type (WT) T4 capsids are composed of mainly gp23, T4 capsid protein, which forms an array of hexamers, and two accessory outer capsid proteins (Hoc and Soc) as indicated in Figure 1A along with the minor capsid protein and portal protein (not shown in Figure 1A; Fokine et al., 2004). Although proteins can be displayed onto the capsids through both Hoc and Soc fusions, we choose Hoc for this work, as there is more intermolecular space for enzyme fusions immobilized onto phage capsids. Future studies could also examine Soc–enzyme fusions to evaluate what effects on the enzymatic activity are obtained when the intermolecular spacing of the fusion enzymes is lessened.
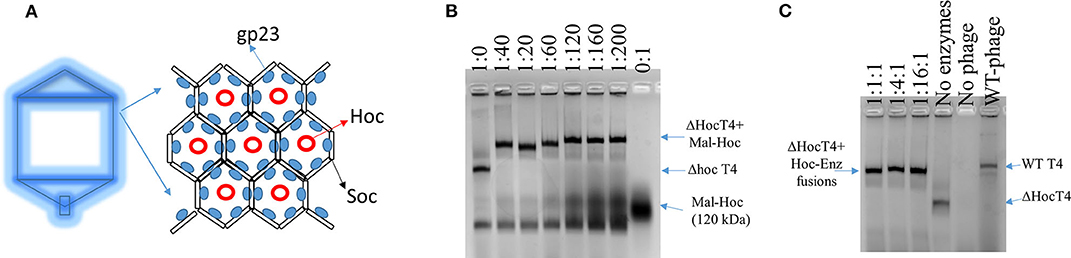
Figure 1. Scheme of T4 capsid hexamers and the assembly of Hoc–enzyme fusions onto ΔHoc T4 phages. (A) T4 capsid protein, gp23, forms hexamers and Hoc is located at the center of the hexamers. (B) The agarose image of mixtures of phage scaffolds and Maltase-Hoc fusions at a series of ratios. Ratios are given above each lane as phage scaffold: Mal–Hoc. (C) The agarose image of mixtures of the phage scaffolds and three Hoc–enzyme fusions, Aml–Hoc, Mal–Hoc, and GK–Hoc, with the total sum concentration 60 nM, which have a 60:1 ratio for the total enzyme fusions to phage scaffolds. The stoichiometric ratios for Aml–Hoc, Mal–Hoc, and GK–Hoc are 1:1:1, 1:4:1, and 1:16:1 as indicated. The no enzyme lane contains only the ΔHoc phage, the no phage lane contains only Hoc–enzyme fusions (1:16:1), and wild-type (WT) phage is also shown. Protein bands were visualized by staining gels with GelCode blue safe protein dye.
The enzyme–SC and St–Hoc were covalently attached in vivo to produce enzyme–SC–St–Hoc (a.k.a. enzyme–Hoc) fusions with a 6 × His tag by co-expressing enzyme–SC and St–Hoc in the same E. coli host. The fusion was purified via Ni-Sepharose resins, and separated from un-fused enzyme–SC through size exclusion chromatography (Goldman et al., 2017). Larger enzyme–Hoc fusions eluted earlier than smaller enzyme–SC fusions from the size-exclusion column (Figure S2). This purified material migrated slower on SDS-PAGE gel since the enzyme–Hoc was 40 kDa larger than enzyme–SC (Figure S3). Tailless ΔHoc T4 capsids were produced from ΔHoc T4 phage with a yield of 1–3 mg/L in the presence of 9-aminoacridine, which blocks genomic DNA packaged into the capsids and thus results in no tail attachment to the capsids (Schaerli and Kellenberger, 1980). They show faster electrophoretic mobility than WT T4 capsids as indicated in Figure 1C (lanes 4 vs. 6). The addition of Mal–Hoc onto ΔHoc T4 capsids at various molar ratios slows down their mobility and the Mal–Hoc assembled T4 capsids at ratios over 60–1 (Mal–Hoc: capsid) all maintain the same mobility (Figure 1B), suggesting that the numbers of Mal–Hoc assembled on the phage capsids reached maximum between 60 and 120 (~90 in average; Lanes 4 and 5 in Figure 1B). A darker band with the same position of free Mal–Hoc appears in the ratio of 120–1 (Lane 5 in Figure 1B), indicating that free Mal–Hoc exists following the binding reaction, while there is much less free Mal–Hoc in the ratio of 60–1 in Figure 1B. Likewise, the samples with the ratio of 60–1 for the total enzymes to phage capsids showed the same effect on the electrophoretic mobility as indicated in Figure 1C. Based on these results, we constructed the enzyme assembly onto phage scaffolds at ratios of 40–1 for Maltase alone, while 60–1 was used for the three enzyme mixtures in Tables 2, 3 to ensure better basal Kcat from free enzyme–Hoc fusions for comparison with the assembled ones. Both ratios exhibited minimal background free enzyme–Hoc activity.
Higher Initial Rate for the Immobilized Mal–Hoc on Phage Capsids
Since Maltase is the rate-limiting step in the enzyme cascade, we measured the initial rate of Mal–Hoc (Klein et al., 2019). The enzyme activity of immobilized Mal–Hoc vs. free Mal–Hoc in solution was measured using 4-Nitrophenyl-α-D-glucopyranoside as a substrate and the yield of the end product of 4-Nitrophenol (4-NP) was monitored by measuring its absorbance at 405 nm. The assembly of Mal–Hoc and phage capsids was conducted at 22°C for 16 h before measuring Mal activity. After prolonged incubation, the immobilized Mal–Hoc on phage capsids still retains its activity and exhibit fourfold higher initial rate relative to the free Mal–Hoc in solution (Figure S4 and Table 1).
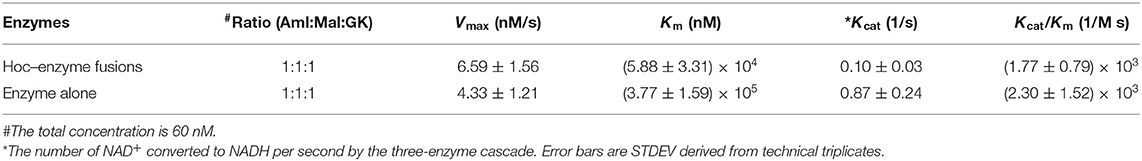
Kinetic Analysis for Enzyme–Hoc Fusions vs. Unfused Enzymes
The kinetics for the three enzyme–Hoc fusions and the three unfused enzymes was measured at a mixing ratio of 1:1:1 of the three enzymes with the total concentration of 0.06 μM. The Vmax was slightly higher for enzyme–Hoc, while Km and Kcat were smaller than enzyme alone (Table 2). Although the Kcat was slower in enzyme–Hoc, a lower concentration of substrate was needed for enzyme–Hoc to reach 1/2 Vmax, resulting in similar catalytic efficiency (Kcat/Km) for unfused enzymes.
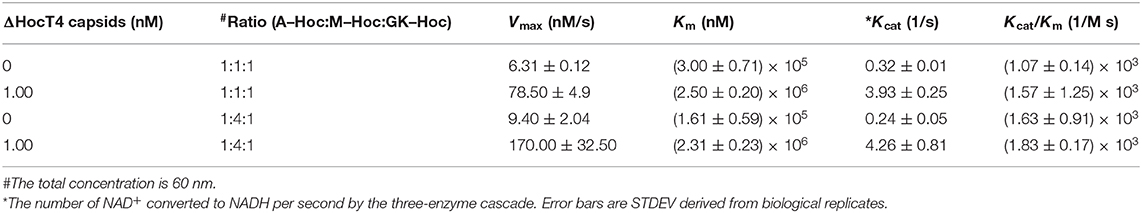
Kinetic Analysis for Immobilized Enzyme–Hoc vs. Enzyme–Hoc Free in Solution
The catalysis of a three-enzyme cascade immobilized vs. the mixture of three enzyme fusions free in solution was measured and compared. The immobilized enzymes on phage scaffolds consistently showed higher Kcat (Table 3) with more enzymatic efficiency (Kcat/Km) for the conversion of NAD+ to NADH. Specifically, assembled enzyme–Hoc fusions with the mixture of three enzymes at a ratio of 1:1:1 appears to have at least 12-fold higher numbers of conversion of NAD+ to NADH relative to corresponding enzyme fusions free in solution (Table 3). Likewise, assembled enzyme–Hoc fusions at a mixing ratio of 1:4:1 without changing the total enzyme–Hoc concentration also exhibit an increase in Kcat of 18-fold relative to the corresponding ones off the scaffolds in solution. However, the sample with a 1:4:1 molar ratio of Aml:Mal:GK did not show better catalytic efficiency than the one with a 1:1:1 ratio of the three enzymes.
Discussion
A Naturally Occurring Phage Scaffold for Assembly of Spatially Organized Enzymes
Reconstructed T4 capsid from cryo-electron microscopy (EM) images revealed that Hoc was dumbbell-shaped with a distance, 14 nm, apart from each other, while Soc was rod-shaped closely associated with each other with a few nanometers distance apart (Fokine et al., 2004). Therefore, the distance between two enzyme–Hoc fusions is within the range where substrate channeling can occur between two co-immobilized enzymes within a cascade (Fu et al., 2014; Lim et al., 2019). Consistent with this prediction, our results showed that the immobilized enzyme–Hoc fusions enhanced catalytic activity in comparison to enzymes free in bulk solution. The assembly of Hoc onto phage capsid is mainly driven by the interaction of its C-terminus with T4 capsid protein through electrostatic, Van der Waals, hydrogen bond forces (Sathaliyawala et al., 2010). According to the cryo-EM model, there are ~155 positions for Hoc–enzyme fusions on each ΔHoc capsid; however, the decorated numbers on the capsids depend on the size or charges of the Hoc fusions (Fokine et al., 2004). The addition of enzyme–Hoc fusions significantly changes the electrophoretic mobility of the assembled phage scaffolds, shifting to the same position as WT T4 (lanes 4 and 6 in Figure 1C). Our previous study showed that the loss of Hoc on the capsids (ΔHoc T4) resulted in a lower pI and more negative zeta potential, but no significant change in size (Robertson et al., 2012). Consistently, our results show that ΔHoc T4 migrated faster toward the positive electrode. Based on the estimate from the band density, ~90 copies on average of enzyme–Hoc were decorated on the capsid; therefore, every immobilized enzyme should have at least two neighboring enzymes on the capsids. Having neighboring enzymes likely facilitates the catalysis via substrate channeling. Enhanced stability could also contribute to higher catalytic activity. Thus, we have demonstrated that T4 phage capsids provide a naturally occurring scaffold to immobilize enzymes for efficient biocatalysis.
Comparable Enzymatic Activity of Enzyme–Hoc Fusion vs. Enzyme Alone
St and SC are two partners of the same domain of Streptococcus pyogenes fibronectin-binding protein (FbaB) and the pair is covalently attached through spontaneous amide bond formation between Asp and Lys in the respective partner (Zakeri et al., 2012; Reddington and Howarth, 2015). We individually fused St onto the N-terminus of Hoc and SC to the C-terminus of enzyme to produce enzyme–SC–St–Hoc fusions in vivo for subsequent one-step purification. The advantage of St–SC pairing allows the separate expression of enzyme and Hoc in the host and avoids the direct expression of large Hoc fusions, which often have the propensity for improper folding and low yields during the production. By this method, we were able to obtain reasonable yields (~5–10 mg/L) for the enzyme–Hoc fusions with the enzymatic activity comparable to enzymes alone.
Demonstration of Enhanced Activity for Immobilized Single Enzyme and the Three-Enzyme Cascade
The kinetics were measured after 2 h of assembly and the measurements were finished within 4 h to ensure that the majority of the enzyme fusions were still active. Although the specific molecular mechanism for the three-enzyme cascade is not clearly identified, we attribute the enhancement of the immobilized enzymes mainly to substrate channeling (Miles et al., 1999; Klein et al., 2019). Mal is the rate-limiting step of the three-enzyme cascade (Klein et al., 2019); therefore, we also specifically measured the initial rate for immobilized Mal–Hoc after 16 h of assembly. Our results show that immobilization on phage scaffolds stabilizes the Mal–Hoc after prolonged incubation (Figure S4). Therefore, the enhancement of the immobilized enzyme activity on phage scaffolds is the sum of substrate channeling and improved stability. Although the scaffolded enzyme–Hoc fusions have higher Km with reduced binding affinity possibly due to the interference of enzyme active sites while attaching to the scaffolds, the greater benefit of substrate channeling compensates for the disadvantage and results in a higher conversion rate of NAD+ to NADH.
In our experiments, the three enzyme–Hoc constructs were incorporated into ΔHoc T4 in vitro. This type of in vitro synthetic biology has been exploited to circumvent drawbacks to cell-based approaches, such as toxicity and the presence of competing pathways (Koch et al., 2018; Sperl and Sieber, 2018). In addition, in vitro assembly allows better control over the stoichiometry of the enzymes in the multi-enzyme-scaffolded complex. However, the T4 system could also be adapted for in vivo display of scaffolded enzymes.
Conclusion
T4 phage scaffolds are composed of prolated protein shells containing arrays of hexamers of capsid protein decorated with Hoc at the center of the hexamers. In this work, we prepared enzyme–Hoc fusions through St–SC pairing and successfully immobilized a three-enzyme cascade, Aml, Mal, and GK, onto naturally occurring T4 phage scaffolds. The immobilized enzymes exhibit enhanced catalytic activity up to 18-fold relative to enzymes free in solution. Thus, we have successfully demonstrated an icosahedral T4 phage scaffold as a new platform for enzyme assembly to enhance biocatalysis applicable to variety of biotechnology applications.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
JL designed and conducted the experiments and wrote the manuscript. DZ, JB, and GA performed the experiments and edited the manuscript. EG designed the experiments and wrote the manuscript All authors contributed to the article and approved the submitted version.
Funding
This work was supported by office of Naval Research through NRL 6.1 base fund.
Disclaimer
Distribution a from Naval Research Laboratory: approved for public release, distribution is unlimited.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly thank Drs. Lindsay W. Black at University of Maryland at Baltimore for the generous gift of T4 phage and Igor Medintz's insightful suggestions. We also thank Drs. Scott A. Walper and Kendrick D. Turner for the gift of plasmids containing the WT maltase, amylase, and glucokinase sequences.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00571/full#supplementary-material
References
Anderson, J., Dueber, J. E., Leguia, M., Wu, G. C., Goler, J. A., Arkin, A. P., et al. (2010). BglBricks: a flexible standard for biological part assembly. J. Biol. Eng. 4:1. doi: 10.1186/1754-1611-4-1
Archer, M. J., and Liu, J. L. (2009). Bacteriophage t4 nanoparticles as materials in sensor applications: variables that influence their organization and assembly on surfaces. Sensors (Basel) 9, 6298–6311. doi: 10.3390/s90806298
Cardinale, D., Carette, N., and Michon, T. (2012). Virus scaffolds as enzyme nano-carriers. Trends Biotech. 30, 369–376. doi: 10.1016/j.tibtech.2012.04.001
Conrado, R. J., Varner, J. D., and DeLisa, M. P. (2008). Engineering the spatial organization of metabolic enzymes: mimicking nature's synergy. Curr. Opin. Biotechnol. 19, 492–499. doi: 10.1016/j.copbio.2008.07.006
Dueber, J. E., Wu, G. C., Malmirchegini, G. R., Moon, T. S., Petzold, C. J., Ullal, A. V., et al. (2009). Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759. doi: 10.1038/nbt.1557
Ellis, G. A., Klein, W. P., Lasarte-Aragonés, G., Thakur, M., Walper, S. A., and Medintz, I. L. (2019). Artificial multienzyme scaffolds: pursuing in vitro substrate channeling with an overview of current progress. ACS Catal. 9, 10812–10869. doi: 10.1021/acscatal.9b02413
Fokine, A., Chipman, P. R., Leiman, P. G., Mesyanzhinov, V. V., Rao, V. B., and Rossmann, M. G. (2004). Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 101, 6003–6008. doi: 10.1073/pnas.0400444101
Fu, J., Yang, Y. R., Johnson-Buck, A., Liu, M., Liu, Y., Walter, N. G., et al. (2014). Multi-enzyme complexes on DNA scaffolds capable of substrate channelling with an artificial swinging arm. Nat. Nanotechnol. 9, 531–536. doi: 10.1038/nnano.2014.100
Goldman, E. R., Broussard, A., Anderson, G. P., and Liu, J. L. (2017). Bglbrick strategy for the construction of single domain antibody fusions. Heliyon 3:e00474. doi: 10.1016/j.heliyon.2017.e00474
Klein, W. P., Thomsen, R. P., Turner, K. B., Walper, S. A., Vranish, J., Kjems, J., et al. (2019). Enhanced catalysis from multienzyme cascades assembled on a DNA origami triangle. ACS Nano 13, 13677–13689. doi: 10.1021/acsnano.9b05746
Koch, M., Faulon, J.-L., and Borkowski, O. (2018). Models for cell-free synthetic biology: make prototyping easier, better, and faster. Front. Bioeng. Biotechnol. 6:182. doi: 10.3389/fbioe.2018.00182
Kuchler, A., Yoshimoto, M., Luginbuhl, S., Mavelli, F., and Walde, P. (2016). Enzymatic reactions in confined environments. Nat. Nanotechnol. 11, 409–420. doi: 10.1038/nnano.2016.54
Lim, S., Jung, G. A., Glover, D. J., and Clark, D. S. (2019). Enhanced enzyme activity through scaffolding on customizable self-assembling protein filaments. Small 15:e1805558. doi: 10.1002/smll.201805558
Liu, J. L., Dixit, A. B., Robertson, K. L., Qiao, E., and Black, L. W. (2014). Viral nanoparticle-encapsidated enzyme and restructured DNA for cell delivery and gene expression. Proc. Natl. Acad. Sci. U.S.A. 111, 13319–13324. doi: 10.1073/pnas.1321940111
Miles, E. W., Rhee, S., and Davies, D. R. (1999). The molecular basis of substrate channeling. J. Biol. Chem. 274, 12193–12196. doi: 10.1074/jbc.274.18.12193
Patel, A. N., Anne, A., Chovin, A., Demaille, C., Grelet, E., Michon, T., et al. (2017). Scaffolding of enzymes on virus nanoarrays: effects of confinement and virus organization on biocatalysis. Small 13:1603163. doi: 10.1002/smll.201603163
Rao, V. B., and Black, L. W. (2010). Structure and assembly of bacteriophage T4 head. Virol. J. 7:356. doi: 10.1186/1743-422X-7-356
Reddington, S. C., and Howarth, M. (2015). Secrets of a covalent interaction for biomaterials and biotechnology: SpyTag and SpyCatcher. Curr. Opin. Chem. Biol. 29, 94–99. doi: 10.1016/j.cbpa.2015.10.002
Ren, Z., and Black, L. W. (1998). Phage T4 SOC and HOC display of biologically active, full-length proteins on the viral capsid. Gene 215, 439–444. doi: 10.1016/S0378-1119(98)00298-4
Robertson, K., Furukawa, Y., Underwood, A., Black, L., and Liu, J. L. (2012). Deletion of the Hoc and Soc capsid proteins affects the surface and cellular uptake properties of bacteriophage T4 derived nanoparticles. Biochem. Biophys. Res. Commun. 418, 537–540. doi: 10.1016/j.bbrc.2012.01.061
Robertson, K. L., and Liu, J. L. (2012). Engineered viral nanoparticles for flow cytometry and fluorescence microscopy applications. Wiley Interdiscipl. Rev. Nanomed. Nanobiotechnol. 4, 511–524. doi: 10.1002/wnan.1177
Sathaliyawala, T., Islam, M. Z., Li, Q., Fokine, A., Rossmann, M. G., and Rao, V. B. (2010). Functional analysis of the highly antigenic outer capsid protein, Hoc, a virus decoration protein from T4-like bacteriophages. Mol. Microbiol. 77, 444–455. doi: 10.1111/j.1365-2958.2010.07219.x
Schaerli, C., and Kellenberger (1980). Head maturation pathway of bacteriophages T4 and T2. V. Maturable epsilon-particle accumulating an acridine-treated bacteriophage T4-infected cells. J. Virol. 33, 830–844. doi: 10.1128/JVI.33.2.830-844.1980
Siu, K. H., Chen, R. P., Sun, Q., Chen, L., Tsai, S. L., and Chen, W. (2015). Synthetic scaffolds for pathway enhancement. Curr. Opin. Biotechnol. 36, 98–106. doi: 10.1016/j.copbio.2015.08.009
Sperl, J. M., and Sieber, V. (2018). Multienzyme cascade reactions—status and recent advances. ACS Catal. 8, 2385–2396. doi: 10.1021/acscatal.7b03440
Walper, S. A., Battle, S. R., Lee, P. A. B., Zabetakis, D., Turner, K. B., Buckley, P. E., et al. (2014). Thermostable single domain antibody-maltose binding protein fusion for Bacillus anthracis spore protein BclA detection. Anal. Biochem. 447, 64–73. doi: 10.1016/j.ab.2013.10.031
Zakeri, B., Fierer, J. O., Celik, E., Chittock, E. C., Schwarz-Linek, U., Moy, V. T., et al. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–697. doi: 10.1073/pnas.1115485109
Keywords: phage scaffold, Hoc, SpyTagSpyCatcher, amylase, maltase, glucokinase
Citation: Liu JL, Zabetakis D, Breger JC, Anderson GP and Goldman ER (2020) Multi-Enzyme Assembly on T4 Phage Scaffold. Front. Bioeng. Biotechnol. 8:571. doi: 10.3389/fbioe.2020.00571
Received: 04 March 2020; Accepted: 12 May 2020;
Published: 24 June 2020.
Edited by:
Simon J. Moore, University of Kent, United KingdomReviewed by:
Hung-En Lai, Victoria University of Wellington, New ZealandShen-Long Tsai, National Taiwan University of Science and Technology, Taiwan
Copyright © 2020 Liu, Zabetakis, Breger, Anderson and Goldman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinny L. Liu, amlubnkubGl1QG5ybC5uYXZ5Lm1pbA==
 Jinny L. Liu
Jinny L. Liu Daniel Zabetakis
Daniel Zabetakis Joyce C. Breger
Joyce C. Breger George P. Anderson
George P. Anderson Ellen R. Goldman
Ellen R. Goldman