- 1Department of Clinical Laboratory, The First Affiliated Hospital of Guangxi Medical University, Nanning, China
- 2Key Laboratory of Clinical Laboratory Medicine of Guangxi Department of Education, Guangxi Medical University, Nanning, China
- 3Department of Gastroenterology, Meizhou People’s Hospital, Meizhou, China
- 4Department of Laboratory Medicine and Guangdong Engineering and Technology Research Center for Rapid Diagnostic Biosensors, Nanfang Hospital, Southern Medical University, Guangzhou, China
- 5Key Laboratory of Bioactive Materials, State Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Functional Polymer Ministry of Education, and College of Life Sciences, Nankai University, Tianjin, China
- 6Guangxi Key Laboratory of Thalassemia Research, Guangxi Medical University, Nanning, China
- 7Department of Laboratory Medicine, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, China
Rapid and accurate sensing of β-galactosidase (β-gal) activity is particularly critical for the early detection of many diseases and has become a topic of interest in recent years. However, most traditional probes for β-gal sensing often suffer from the disadvantages of narrow dynamic range, low reaction efficiency and are only employed with either colorimetric or fluorescence sensing. Furthermore, β-galactosidase sensing based assay for efficient detection and antibiotic resistance analysis of Escherichia coli (E.coli) is not available. Here, an enzyme-induced probe assay was reported for dual sensitive fluorescence and colorimetric measurement of β-gal activity, and was further employed for detection of Escherichia coli and their antibiotic resistance analysis. The DCM-βgal probe was virtually non-emissive in aqueous solution, while it could be activated by β-gal to produce bright emission. Under optimized conditions, DCM-βgal displayed high sensitivity, selectivity and rapid response to β-gal with a low detection limit of 1.5 × 10−3 U ml−1. Importantly, this assay was successfully applied to sensitive detection of E. coli cells with a fast detection process within 5 h and a low detection concentration of 1 × 103 CFU ml−1. Furthermore, the enzyme-activatable assay was also successfully applied for high throughput E. coli antibiotic resistance analysis. The DCM-βgal strategy is applied for the first time on the detection of E. coli cells and their antibiotic resistance analysis. It is provided with the advantages of high selectively, a simple operation, low cost and rapid detection. The detection platform can also be extended to analyze the level of β-gal in other types of cells or biological samples. Overall, the simple, effective and dual-readout assay holds promise for efficient sensing of β-gal activity and provides a potential tool for E. coli detection and their antibiotic resistance analysis.
Introduction
Enzymes play important roles in a variety of biological processes and have served as crucial biomarkers for disease diagnosis and monitoring. Rapid sensing of the specific enzymes activity is emerging as a critical strategy for accurate disease diagnostics and developing simple, effective sensing system has gained considerable attention in past decades. Among these specific enzymes, β-galactosidase (β-gal) is widely known as a common kind of hydrolase in cells and the main biological function of β-gal is to remove galactose residues from substrates (Komatsu and Urano, 2015). In recent years, β-gal has attracted extensive attention as a vital enzyme biomarker because it has been shown to be overexpressed in senescent cells, tumor cells as well as in Escherichia coli (E.coli) (Munoz-Espin and Serrano, 2014; Chen et al., 2016; Gu et al., 2016; Adkins et al., 2017; Nishihara et al., 2018). Therefore, developing efficient methods to detect β-gal with high efficiency and specificity is thus of great importance for the early diagnosis of the specific diseases and the identification of E.coli infection.
Although a number of approaches have been proposed for detecting the activity of β-gal, but most of them suffer from various disadvantages. For example, colorimetric methods have been suggested as convenient assays for visual detection of the β-gal concentration (Chen et al., 2015; Chen et al., 2016; Adkins et al., 2017; Jia et al., 2018). However, most of these enzyme-induced colorimetric strategies often do not have broad color changes enough for quantitative measurement of the enzyme activity. Electrochemical methods are sensitive enough to measure β-gal activity (Adkins et al., 2017; Wang et al., 2017). Unfortunately, they mostly rely upon advanced instrumentation and require cumbersome processes. Furthermore, the above methods are not suitable for the assessment of endogenous β-gal activity in vivo. In contrast to colorimetric and electrochemical methods, fluorescent sensors have attracted extensive attention because of their simplicity, high sensitivity, high signal-to-noise ratio, and their ability for imaging β-gal in vivo (Jiang et al., 2017; Fu et al., 2019; Gu et al., 2019; Zhang et al., 2019). However, most traditional fluorescent probes suffer from the disadvantages of background fluorescence interference, narrow dynamic range and long response time, which can greatly hamper their application in biological analysis. Thus, it is highly desirable to develop a fluorescent probe capable of detecting β-gal with an improved dynamic range and enhanced reaction efficiency.
E.coli contamination remain major public health challenge and substantial health burden worldwide, accounting for most urinary tract infections and over 90% of food and waterborne diseases (Oliver et al., 2005; Jones et al., 2008; Scharff, 2012). On the other hand, the increasing trend of antibiotics resistance poses another significant threat to public health. Therefore, the rapid and sensitive detection of E.coli concentration and E.coli antibiotic resistance has been a crucial research topic and is critical to the early diagnosis and prevention of E.coli infection. The current gold-standard method, culturing and plate counting, is accurate and reliable for E.coli detection. However, it is quite time-consuming and laborious, requiring at least 2 days from sampling to results and even longer for antibiotic resistance analysis (Rompre et al., 2002). Another widely used and promising method for E. coli detection is determination of β-gal activity. β-gal is a well-known intracellular enzyme that is encoded by the lacZ operon in E. coli cells and it has been widely used as an indicator for the determination of E. coli concentration in drinking water and food samples (Colquhoun et al., 1995; Derda et al., 2013; Burnham et al., 2014; Chen et al., 2015; Chen et al., 2016). A number of strategies and probes have been developed to detect β-gal in E. coli. Unfortunately, those strategies often take several hours for reaction and are only employed with either colorimetric or fluorescence sensing. Moreover, the requirement of multiple steps of T7 bacteriophage infection and complex chemical reaction in some assays further restrict their clinical application (Chen et al., 2015; Chen et al., 2016; Wang et al., 2017).
Here, we utilized a simple and effective probe, DCM-βgal, for dual sensitive fluorescence and colorimetric measurement of β-gal activity. For this sensing probe, dicyanomethylene-4H-pyran (DCM) chromophore is utilized as reporter, and a β-gal-responsive group (β-galactopyranoside) as the enzyme-active trigger (Figure 1) (Gu et al., 2016). DCM-βgal emits weak background fluorescence in aqueous buffer. In the presence of β-gal, the β-galactopyranoside unit is cleaved, releasing the DCM group, resulting in a remarkable fluorescent emission in the near-infrared region (NIR) and an obvious color shift from yellow to red. The probe enables sensitive and accurate detecting of β-gal activity with high-selectivity and simplicity. Unlike rhodamine derivates, there is only a single cleavage site in DCM-βgal, which makes the kinetic analysis much easier. The sensing strategy was employed to detect E. coli in aqueous solutions using β-gal activity as indicator. The bacterial cells were lysed by lysis solution to release β-gal for enzymatic reaction. Once released, β-gal catalyzed the β-gal cleavable unit in DCM-βgal to generate DCM and induced a colorimetric and fluorescence change (Figure 1). The concentration of E. coli cells was directly correlated with the colorimetric shift and the fluorescence intensity of emission spectra peak. DCM-βgal possesses several advantages, including a fast response speed, a high light-up ratio and a good sensitivity towards β-gal, making it a promising strategy for rapid sensing of β-gal.
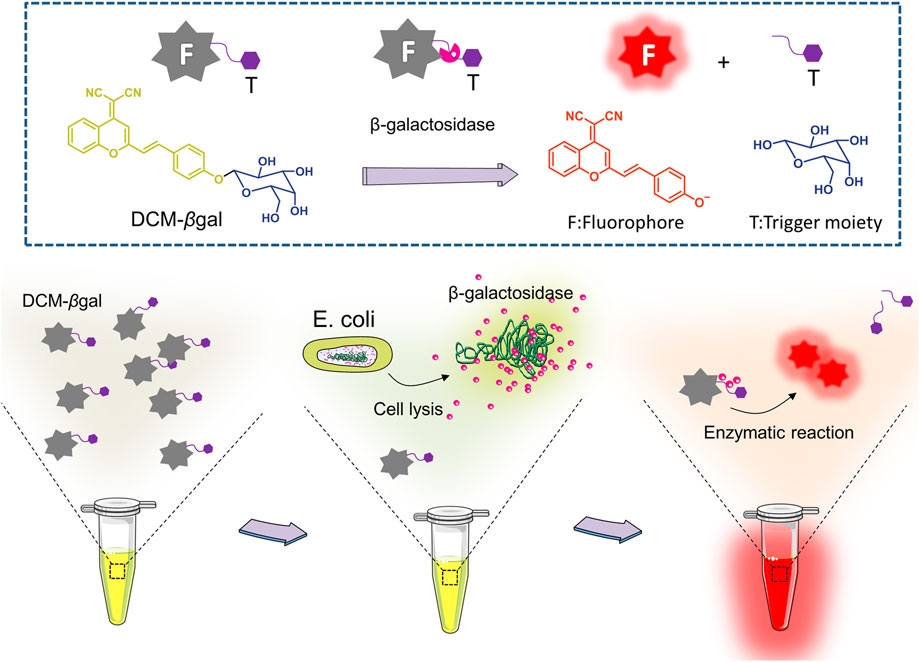
FIGURE 1. Schematic illustration of enzyme-induced probe assay for measuring of β-gal concentration and detection of Escherichia coli (E.coli). DCM-βgal probe consists of two parts: DCM-OH as the reporter unit and a β-gal-responsive unit. In the presence of β-gal, the responsive unit is cleaved and the DCM group shows enhance fluorescent emission and an obvious color shift from yellow to red. For E.coli detection, the bacterial cells are lysed to release β-gal for enzymatic reaction. The concentration of E. coli cells is correlated with the colorimetric shift and the fluorescence intensity of emission spectra peak.
Experimental section
Materials
β-Galactosidase (β-Gal), horseradish peroxidase, alkaline phosphatase, papain, pepsin, and α-mannosidase were purchased from Beyotime (Shanghai, China). Lysozyme, glucose, bovine serum albumin, γ-globulin, transferrin, mannitol tryptophan and proline were purchased from Solarbio (Beijing, China). Ciprofloxacin, ampicillin, kanamycin, dimethyl sulfoxide (DMSO), Polypropylene pyrrolidone K30 (PVP-K30, MW = 40,000) and all other chemicals and solvents were purchased from Sigma-Aldrich (St. Louis, United States). Luria-Bertani broth (LB) medium was purchased from BD Biosciences (San Jose, CA). All aqueous solutions were prepared with Milli-Q water (≥18 MΩ, Milli-Q, Millipore).
Synthesis of DCM-βgal
DCM-βgal was synthesized according to the processes described in Supplementary Scheme S1 in Supplementary Material. The substrate Ac4β-Gal-Ph-CHO (320 mg, 0.71 mmol, 1 equiv) was dissolved in 15 ml of acetonitrile followed by the addition of piperidine (70 μl, 0.71 mmol, 1 equiv) and dicyanomethyl chromone (DCMC) (221 mg, 1.06 mmol, 1.5 equiv). Reaction mixture was stirred at 70°C for 2 h and monitored by thin layer chromatography (Petroleum ether: Ethyl acetate = 2: 1). Upon completion, the reaction mixture diluted with Ethyl acetate (100 ml) was washed with saturated solution of 0.5 M HCl (100 ml). The organic layer was separated, washed with brine, dried over Na2SO4 and evaporated under reduced pressure. The orange residue was purified by column chromatography on silica gel (Petroleum ether: Ethyl acetate = 4: 1) affording Ac4β-Gal-DCM as an orange solid. The residue was dissolved in DCM and MeOH (15 ml, v/v = 1:2), and the MeONa (10 mg, 0.35 mmol, 0.5 equiv) was added into the solution. The mixture was stirred for 50 min, at room temperature and monitored by thin layer chromatography (DCM: MeOH = 10: 1). Upon completion, the reaction mixture was evaporated under reduced pressure. The orange residue was purified by column chromatography on silica gel (DCM: MeOH = 10: 1) affording the target product DCM-βgal as an orange solid (272 mg, 41% yield for two steps). And the NMR data is same as the literature reported (Gu et al., 2016).
UV-vis and fluorescence spectral measurements
DCM-βgal solution was prepared at a concentration of 100 µM in a 2 ml total volume of reaction buffer (pH 7.4, 30% DMSO and 70% PBS). 1 U ml−1 β-gal was added into DCM-βgal solution and the mixture was gently agitated and incubated at 37°C for 15 min. UV-vis absorption spectra for DCM-βgal and reaction product were respectively analyzed on a PerkinElmer Lambda 25 UV/vis Spectrometer. Photoluminescence (PL) spectra were analyzed on a PerkinElmer spectrfluorometer LS55 with a wavelength range of 550–800 nm. Fluorescence quantum yields were detected on a Hamamatsu absolute PL quantum yield spectrometer C11347 Quantaurus-QY.
Analytical procedures for detection of β-gal
The stock solution (100 mM) of DCM-βgal was prepared in DMSO and the work solution (100 μM, fDMSO% = 30 vol%) was diluted from the stock solution. Different concentrations of β-gal solution (10 µl) were added into work solution (1 ml) and incubated at 37°C for 15 min. The fluorescence intensity at 675 nm and colorimetric results were recorded for the enzymatic products using PerkinElmer spectrofluorometer LS55.
Photobleaching analysis
1 U ml−1 β-gal was added into DCM-βgal solution and the mixture was incubated at 37°C for 15 min. Then enzymatic products were treated with high density bright-light exposure (20 W). The fluorescence intensity of the solution was monitored from 0 to 45 min of exposure.
Bacterial culture
A single colony of E. coli was selected from a LB plate and added into LB liquid medium and incubated overnight at 37°C under in the orbital shaker of 200 rpm agitation. The E. coli cells was harvested by centrifugation at 6000 g for 3 min and re-suspended in PBS buffer. The centrifugation process was repeated for three times. The re-suspended bacterial solution was then serially diluted into various concentrations for further use. The diluted E. coli solution was plated on LB agar plate to confirm the visible counts (CFU ml−1).
Detection of E. coli using DCM-βgal based assay
Bacteria suspension with different concentrations was centrifugated by 6000 g for 3 min and discarded the supernatant. Subsequently, 0.7 ml lysis buffer containing 1 mg ml−1 lysozyme, 1% (w/v) sodium chloride, 0.2% (w/v) glycerin, 0.01% (w/v) PVP-K30 was added to the bacterial precipitate, mixed gently and incubated at 37°C for 30 min. The dissolved suspension was then added with 100 μM DCM-βgal and 0.3 ml DMSO and incubated at 37°C for 15 min after gently mixed. The fluorescence intensity at 675 nm and colorimetric results were recorded for the enzymatic solution using PerkinElmer spectrofluorometer LS55.
Two-step detection of E. coli using DCM-βgal based assay
Different concentrations of bacterial solutions (100 μL, 1×105, 5×104, 1×104, 5×103, and 1 × 103 CFU ml−1) were incubated in LB broth (900 µl) at 37°C for 1–3 h with 200 rpm agitation. PBS buffer without bacteria cells was taken as a negative control. After incubation, bacteria suspension was centrifugated and the bacterial precipitate was lysed by 0.7 ml lysis buffer as above. The dissolved suspension was added with 100 μM DCM-βgal and 0.3 ml DMSO and incubated at 37°C for 15 min. The fluorescence intensity and colorimetric results of solutions were recorded.
Bacteria antibiotic resistance analysis
For bacteria antibiotic resistance sensing, LB broth medium was added with different antibiotic drugs (ampicillin, kanamycin, or ciprofloxacin) with varying concentrations (0, 10, 20, 30, 40, and 50 μg ml−1). Then, E.coli solutions (105 CFU ml−1) were added in LB mixtures at 37°C for 3 h with 200 rpm agitation in a constant temperature shaker. After incubation, bacteria suspension was centrifugated and the bacterial precipitate was lysed by 0.7 ml lysis buffer as above. The dissolved suspension was then added with 100 μM DCM-βgal and 0.3 ml DMSO and incubated at 37°C for 15 min after gently mixed. Then, the supernatant was transferred to 96-well plate and the fluorescence intensity of solutions was analyzed by Cytation 5.
Statistical analysis
All continuous data was presented as means ± standard deviation. Data was statistically analyzed using t-test. p < 0.05 was considered statistically significant.
Results and discussion
Spectroscopic properties and optical response to β-gal
The basic chemical structure and the principle of DCM-βgal probe for β-gal activity sensing are shown in Figure 1. The synthetic route of DCM-βgal is display in Supplementary Scheme S1 in Electronic Supplementary Material. DCM derivatives are well-known as laser dyes that produce intense emission in the NIR region and DCM-OH is a commonly used signal reporter because of its special properties: it is a Donor–π–acceptor (D–π–A) molecule and it has a hydroxyl group that can be easily modified by other groups (Gu et al., 2016; Gu et al., 2019). Thus, DCM-βgal probe consists of two parts: DCM-OH as colorimetric and fluorescent reporter and a β-galactopyranoside unit as β-gal-responsive moiety. Firstly, to test the validity of DCM-βgal, its spectral properties were investigated by UV-vis-NIR spectroscopy and photoluminescence (PL) spectrum. As shown in Figure 2A, the absorption spectrum showed that DCM-βgal exhibited an obvious absorption peak at 440 nm. Upon addition of 1 U ml−1 β-gal, the absorption peak significantly decreased at 440 nm and a new absorption peak appeared at 535 nm. This new absorption was in accordance with that of DCM-O-, suggesting that DCM-βgal was cleaved by β-gal and resulting DCM-OH generation (Gu et al., 2016). Then, the emission profile of DCM-βgal was detected. Upon excitation at 535 nm, the fluorescence signal of DCM-βgal was very weak while the probe solution showed a remarkable NIR fluorescence at 675 nm in the presence of β-gal (Figure 2B). In particular, an obvious color shifting from yellow to red could be observed, allowing for the qualitative or semi-quantitative detection of β-gal by direct observation and rapid colorimetric analysis.
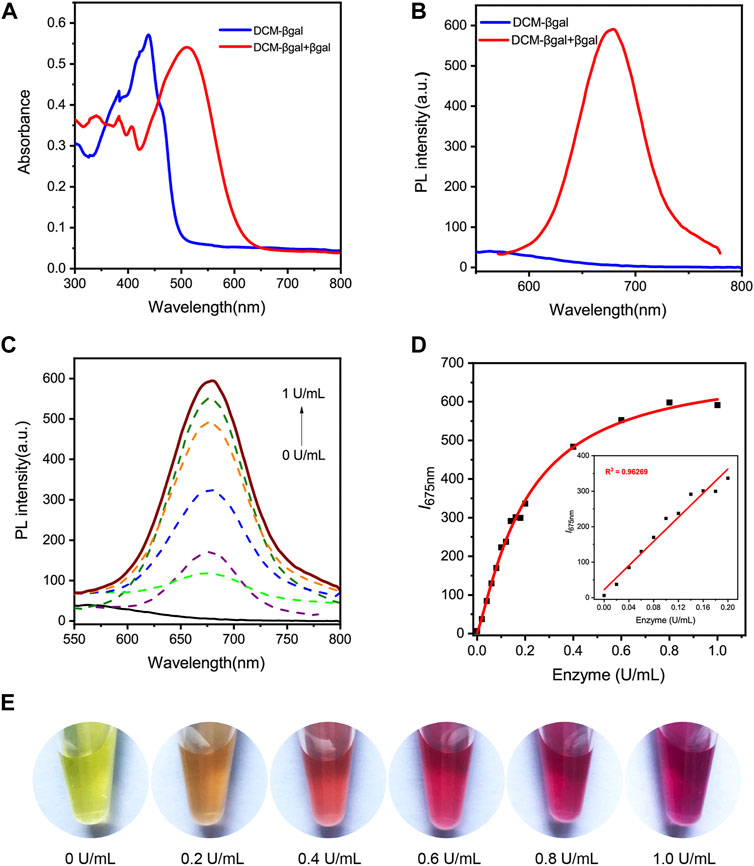
FIGURE 2. Spectral properties of DCM-βgal. (A) Normalized UV-vis absorption of DCM-βgal (100 μM) and DCM-βgal (100 μM) incubation with β-gal (1 U ml−1) in aqueous solution (PBS/DMSO = 7:3, v:v, pH = 7.4, 37°C). (B) Photoluminescence (PL) spectra of DCM-βgal (100 μM) and DCM-βgal (100 μM) incubation with β-gal (1 U ml−1) in aqueous solution (PBS/DMSO = 7:3, v:v, pH = 7.4, 37°C). (C) Emission spectra of DCM-βgal upon addition of different concentrations β-gal (0–1 U ml−1), λex = 535 nm. (D) The peak intensity of DCM-βgal at 675 nm toward various concentrations of β-gal (0–1 U ml−1). Inset: the linear relationship between I675 nm and β-gal concentration in a range from 0 U ml−1 to 0.2 U ml−1 (E) The photographs of DCM-βgal upon addition of various concentrations of β-gal.
Optimization of assay conditions
The reaction solution, temperature and pH are important factors for the sensing system. To obtain the best sensing performance, the optical properties of DCM-βgal were examined in different analytical parameters. Upon addition of 1 U ml−1 β-gal, the fluorescence intensity of DCM-βgal at 675 nm was tested in PBS/dimethyl sulfoxide (DMSO) mixtures with different DMSO volume fractions (fDMSO%). As shown in Supplementary Figure S1A,B (Electronic Supplementary Material), the fluorescence intensity of DCM-βgal solution increased quickly with fDMSO% and reached a maximum at fDMSO% = 30 vol%. Specifically, as the DMSO content continued to increase, the fluorescence intensity decreased sharply and DCM-βgal solution showed very weak fluorescence at fDMSO% = 50–100 vol%. Correspondingly, a similar colorimetric change could be observed with the change of fDMSO% (Supplementary Figure S1C in Supplementary Material). Since DCM-βgal is poor solvent in PBS, the addition of DMSO will increase its solubility and induces an increase in fluorescence emission. Thus, PBS solution with 30% DMSO was applied as the optimal reaction solution.
The effect of reaction temperature and pH was then evaluated. The fluorescence response of DCM-βgal to 1 U ml−1 β-gal was examined by incubation of the solution from 4°C to 56°C for 15 min. As expected, the fluorescence signal increased with temperature and reached the maximum at 37°C, and then decreased after 37°C (Supplementary Figure S2A in Supplementary Material). Hence, 37°C was selected as optimal reaction temperature. pH is another important factor affecting the rate of enzymatic reaction for the sensing probe. pH was adjusted to the desired value by using 0.1 M NaOH and 0.1 M HCl solutions. The pH was measured with a pH meter. The effect of pH (4.0–11.0) for the emission profiles of DCM-βgal was subsequently evaluated after 15 min of reaction. It can be observed that the fluorescence signal was very weak at pH lower than 6.12 and higher than 8.46, and the maximum fluorescence signal was achieved at pH = 7.59 (Supplementary Figure S2B in Supplementary Material). The result suggested that the enzymatic hydrolysis reaction for DCM-βgal could only be proceeding at the optimal pH range and pH = 7.59 was used as the optimal pH condition. In particular, the color of the solution varied from yellow to red corresponding to temperature and pH of the solution. The distinct color changes of the probe were therefore easily to identify visually (Supplementary Figures S2C,D in Supplementary Material).
Then, the fluorescence signal versus time was analyzed to investigate the enzyme response rate. As shown in Supplementary Figure S3A,B (Supplementary Material), as the incubation with β-gal, the fluorescence intensity of DCM-βgal gradually increased in the early stage and reached the maximum at around 15 min. Moreover, the fluorescence response of β-gal in different concentrations of DCM-βgal was investigated to determine the optimal probe concentration. Increasing concentrations of DCM-βgal (0–140 μM) were incubated with 1 U ml−1 β-gal at 37°C for 15 min. As depicted in Supplementary Figure S3C (Supplementary Material), the fluorescent signal increased gradually with increasing the concentration of DCM-βgal and reached a plateau after a concentration of 100 μM. Therefore, a DCM-βgal concentration of 100 μM was selected for further experiments to provide an optimal probe concentration. DCM-βgal displayed similar colorimetric response with the increasing of DCM-βgal concentration and incubation time, suggestive of its feasibility in dual-channel sensing for β-gal (Supplementary Figure S3D,E in Supplementary Material).
Analytical performance of DCM-βgal to β-gal
We next sought to investigate the analytical performance of DCM-βgal for β-gal detection. The fluorescence response of the probe to varying concentrations of β-gal was examined under optimized conditions. As expected, the fluorescence intensity of solution increased with the increasing of β-gal concentration and a 94.84-fold fluorescence enhancement was observed when incubated with 1 U ml−1 β-gal (Figures 2C,E). Notably, a good linear relationship between the fluorescence intensity at 675 nm and β-gal concentration in a range from 0 U ml−1 to 0.2 U ml−1 was observed (Figure 2D). The limit of detection (LOD) was calculated to be as low as 1.5 × 10–3 U ml−1 based on the signal of blank tests and the standard deviation, indicating the high sensitivity of the probe for the fast and quantitative detection of β-gal. Photo-stability is another important factor to evaluate the performance of the probe in long-term tracking and bioimaging of enzyme activity (Liu et al., 2016). The photo-stability of DCM-βgal was then assessed by time-dependent photobleaching measurements. 1 U ml−1 β-gal was added into DCM-βgal solution and the mixture was incubated at 37°C for 15 min. Then enzymatic products were treated with high density bright-light exposure (20 W). The results showed that rounded to 82% of DCM-βgal fluorescence intensity still remained after bright-light exposure for 30 min, indicating the high photo-stability of DCM fluorophore (Supplementary Figure S4 in Supplementary Material). These features make DCM-βgal a promising candidate for long-term tracking and imaging of β-gal in practical applications.
The selectivity of DCM-βgal
Subsequently, the selectivity of probe to β-gal sensing was then evaluated. Control experiments were conducted to investigate the selectivity of DCM-βgal towards of various biological species, including common enzymes (horseradish peroxidase, alkaline phosphatase, papain, pepsin, and α-mannosidase), bioactive molecules (glucose, bovine serum albumin, γ-globulin, transferrin, and mannitol) and amino acids (tryptophan and proline). As expected, an obvious fluorescence enhancement accompanying with an obvious color change from yellow to red was observed in the presence of β-gal (Figure 3). However, negligible fluorescence and color change was observed for other interferents. These results demonstrated the high selectivity of DCM-βgal for β-gal over other competitive analytes, suggesting its promising use as a bioprobe for detecting β-gal in biological systems.
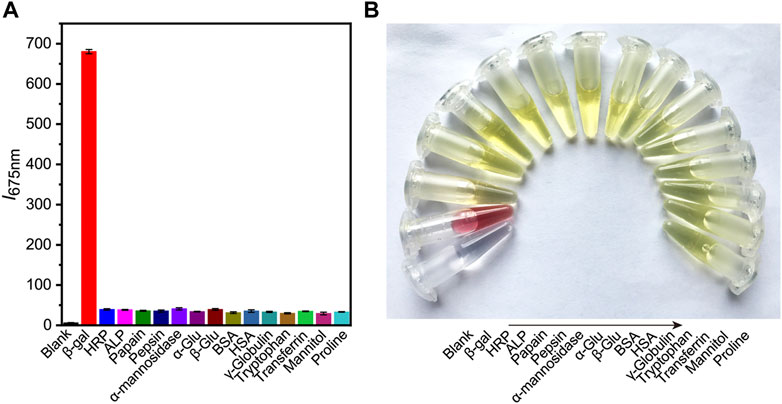
FIGURE 3. The selectivity of DCM-βgal towards of various biological species. Different types of biological samples are added into work solution of DCM-βgal and incubated at 37°C for 15 min. The fluorescence intensity at 675 nm and colorimetric results of solution are then recorded. (A) The fluorescence responses (I675 nm) of DCM-βgal to various analytes. Error bars represent the standard deviation of three replicates. (B) Corresponding color changes of DCM-βgal treated with different analytes.
Application for detection of Escherichia coli
Encouraged by the desirable results of DCM-βgal, we next sought to investigate the potential utility of DCM-βgal for detecting β-gal from E. coli cells. β-gal is an important bacteria enclosed enzyme encoded by the lacZ gene in E. coli cells, and can be used as an indicator for determination of the concentration of E. coli (Laczka et al., 2010; Derda et al., 2013; Burnham et al., 2014; Chen et al., 2016). E. coli ATCC 25926 was used as a model for investigating the performance of DCM-βgal in E. coli detection. Varying concentrations of bacterial cells were firstly lysed, resulting in the release of intracellular β-gal from cells into solution. Then, the solution containing the released β-gal was incubated with DCM-βgal to determine its concentration under optimized conditions. As a result, the E. coli concentration could be assessed using the fluorescence intensity of DCM, or directly by the naked eyes. As shown in Figure 4A, the fluorescence intensity and the red color of the enzymatic solution increased with the increasing of bacterial concentration, and distinct signal changes could be reproducibly detected or be distinguished visually for 1 × 105 CFU ml−1 of E. coli cells (Figure 4B). Compared with previously methods, the DCM-βgal based assay showed a low detection limit and provided a convenient colorimetric readout for visual detection, making it a sensitive and simple strategy for E. coli detection.
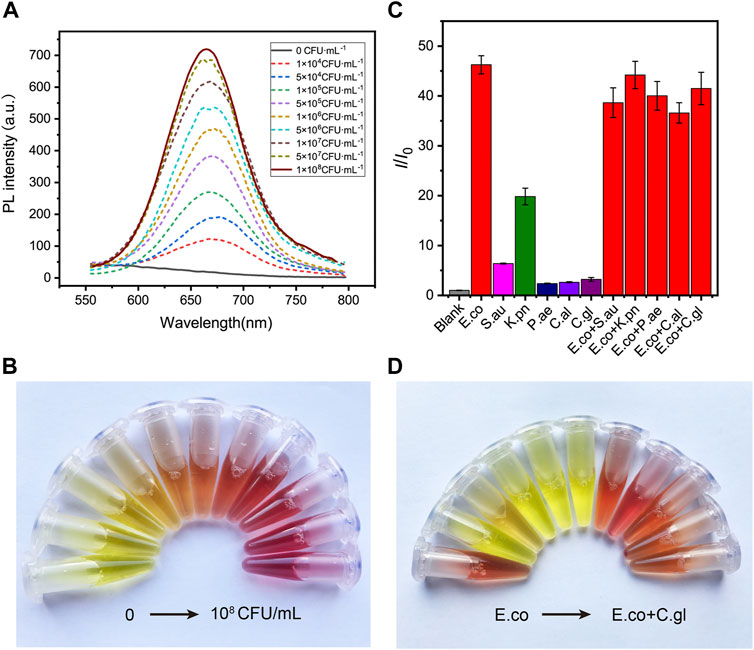
FIGURE 4. The sensitivity and specificity of DCM-βgal based assay to E. coli. (A) DCM-βgal based assay for the detection of E.coli in the concentration range of 0–1.0 × 108 CFU ml−1 and (B) its corresponding color changes. Error bars indicate ± SD of triplicate measurements. (C) Fluorescence ratio (I/I0) of specificity of the DCM-βgal probe against E.coli, S. aureus, K. pneumonia, P. aeruginosa, C.albicans, C.glabrata, E.coli + S. aureus, E.coli + K. pneumonia, E.coli + P. aeruginosa, E.coli + C.albicans and E.coli + C.glabrata at concentration of 1.0 × 106 CFU ml−1. (D) Its corresponding photographs.
To test the specificity of the assay to E. coli, the responses of the assay to common bacteria strains, including Staphylococcus aureus (S.aureus), Klebsiella pneumoniae (K. pneumoniae), Pseudomonas aeruginosa (P. aeruginosa), Candida albicans (C.albicans) and Candida glabrata (C.glabrata), as well as mixtures of different bacteria strains, were investigated. A concentration of 1 × 106 CFU ml−1 was used for solutions of each strain or their mixtures. Figures 4C,D shows the fluorescence and colorimetric detection results. No obvious fluorescence signal and color change were observed in S.aureus, P. aeruginosa, C.albicans and C.glabrata solutions. In contrast, a significant fluorescence response and distinct color change were detected in E. coli solution or in mixtures containing E. coli. In particular, the fluorescence intensity of K. pneumonia solution was slightly increased and the color of the solution changed from yellow to orange, correspondingly. This result was in accordance with the fact that a certain level of β-gal isoenzymes could be encoded by K. pneumonia (Wang et al., 2014). These results demonstrated that the dual-channel assay had a good selectively to β-gal for E. coli detection.
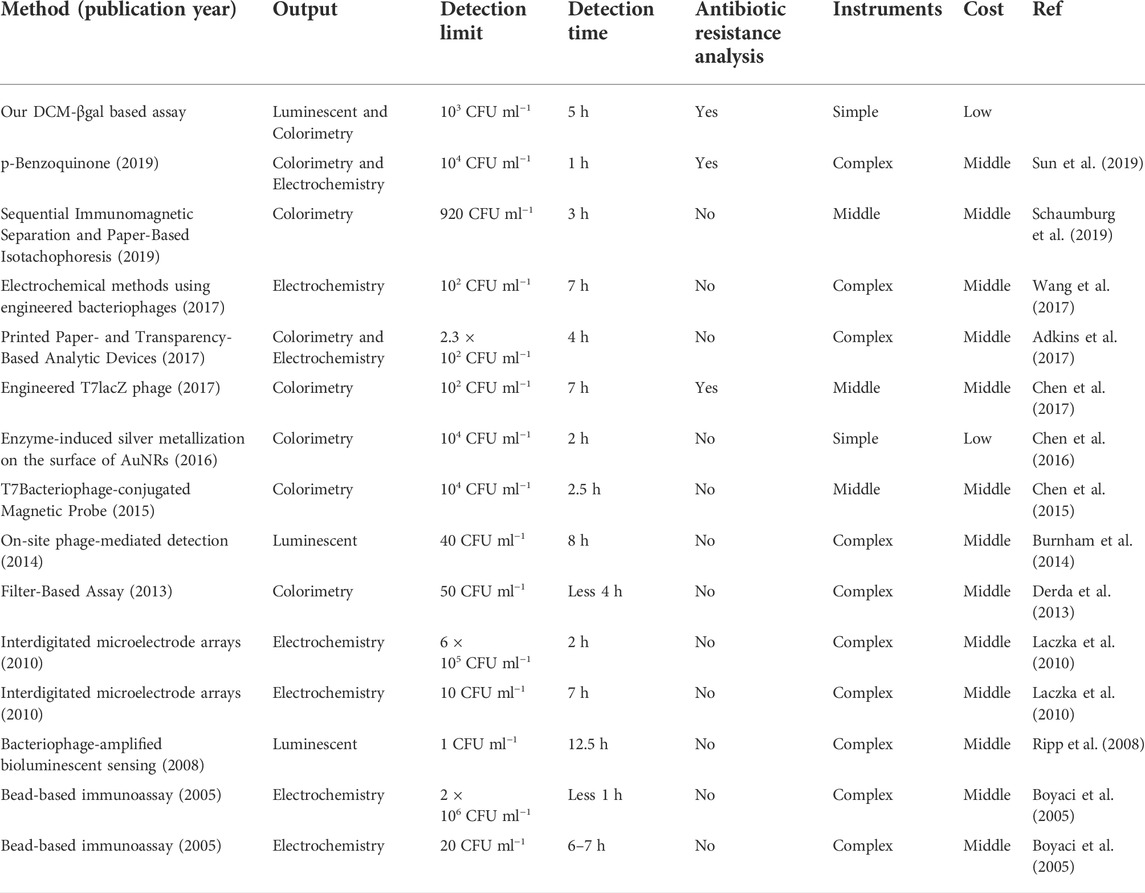
The concentration of E. coli cells is important for the enzymatic response, whereas E. coli in clinical urine samples is well below 1 × 104 CFU ml−1. To improve the clinical performance of the assay, two-step process was employed to detect E. coli cells at low concentrations. Pre-enrichment step was utilized to allow for bacterial growth, and the detection step was then used for determination of E. coli cells. Low concentrations of bacteria were incubated in LB liquid medium for 1, 2, and 3 h, respectively. The fluorescence intensity and the color change of solution were investigated after bacteria incubation and enzymatic reaction. As shown in Figures 5A,B, a significant fluorescence and obvious colorimetric response could be detected after 3 h pre-enrichment in all concentrations. E. coli cells at the concentration of 1 × 105 CFU ml−1, 5 × 104 CFU ml−1 and 1 × 104 CFU ml−1 could also be clearly detected after a pre-enrichment step of 2 h incubation. Compared with previous methods, the assay was a more efficient strategy with the whole detection time less than 5 h (Chen et al., 2017; Wang et al., 2017). As shown in Table 1, several similar assays for E. coli detection were summarized and compared. Sensitive methods often require advanced instrumentation and cumbersome processes, limiting their use in resource-limited settings. In contrast, the DCM-βgal assay would be a promising method for rapid detection of E. coli for their practicality, simplicity and low-cost. These results indicated that the DCM-βgal assay had a good performance for E. coli detection at low concentrations incorporating a pre-enrichment step.
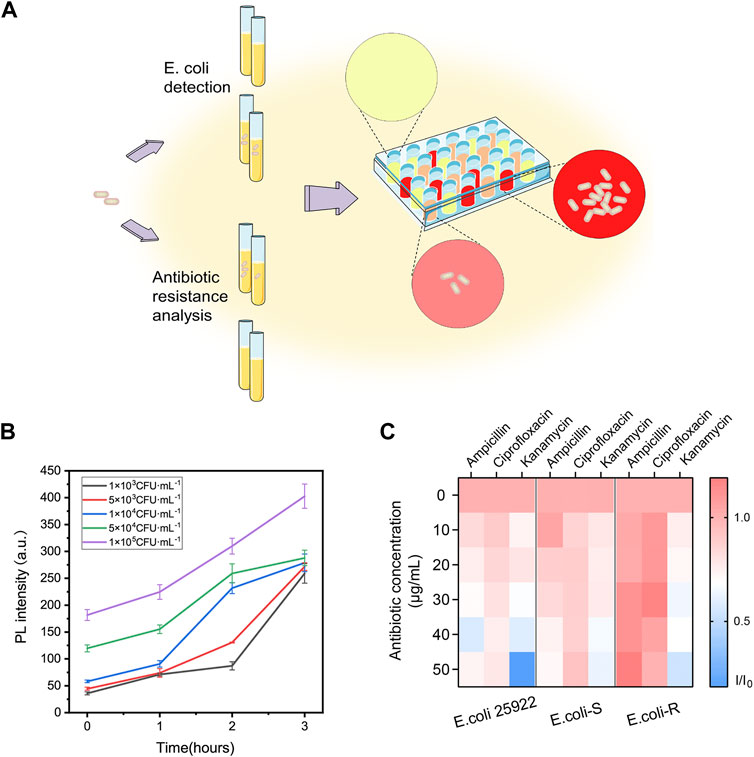
FIGURE 5. Two-step procedure for E. coli detection and high-throughput antibiotic screening using DCM-βgal based assay. (A) Schematic illustration of two-step procedure and high-throughput antibiotic analysis. E. coli cells are lysed to release intracellular β-gal and the signal generation based on enzyme-response is correlated with the concentration of E. coli cells. For antibiotic resistance analysis, E. coli cells can be treated with various antibiotic concentrations and fluorescence ratio of reaction products in 96-well plate can be high throughput monitored by Cytation. (B) Two-step detection of E. coli at low concentrations using DCM-βgal based assay. PL intensity (675 nm) with detection time (0–3 h) toward E. coli concentration of 1 × 103, 5 × 103, 1 × 104, 5 × 104, and 1 × 105 CFU ml−1. Error bars indicate ± SD of triplicate measurements. (C) Heatmap plot of fluorescence ratio (I/I0) of three types of E. coli cells (E. coli ATCC 25926, antibiotic-sensitive (E. coli-S) and resistant E. coli (E. coli-R) collected from clinical samples) toward various antibiotic concentrations.
Application for Escherichia coli antibiotic resistance determination
The prevalence of bacteria’s antibiotic resistance is a growing global health concern and it is in urgent need to develop a rapid, efficient strategy for the determination of E. coli antibiotic resistance (Walsh, 2000). Conventional test for bacteria antibiotic resistance detection requires cumbersome processes and often takes days to weeks for results. Inspired by the high sensing property of the probe, the ability of DCM-βgal based assay for rapidly detection of E. coli antibiotic resistance was then studied (Figure 5A). E. coli ATCC 25926, antibiotic-sensitive (E. coli-S) and resistant E. coli (E. coli-R) collected from clinical samples were used as bacterial model, and the response of the assay to the bacterial was examined in the presence of three common antibiotics, ampicillin, kanamycin, and ciprofloxacin. E. coli cells were incubated in LB liquid medium containing different concentrations of antibiotics at 37°C for 3 h. Bacterial cells were lysed and DCM-βgal probe was then added for enzymatic reaction to determine bacteria concentration. In the presence of effective antibiotic drugs, E. coli cells exhibited inhibited growth and a greatly reduced level of β-gal. However, E. coli cells showed exponential growth in the presence of antibiotic resistance. Varying levels of β-gal resulted in different levels of enzymatic response, thus providing a simple strategy for antibiotic resistance analysis. With the help of a high throughput fluorescence detection system, Cytation, the fluorescence information of 96-well plate could be easily quantified and monitored. As shown in Figure 5C, for E. coli ATCC 25926 and E. coli-S, the fluorescence intensity decreased with the increasing antibiotic concentration when incubated with ampicillin, kanamycin, and ciprofloxacin, indicating that these antibiotics were effective drugs for inhibiting cell growth of these strains. However, for E. coli-R, no decreasing trend for fluorescence intensity could be observed with the increasing concentration of ampicillin and ciprofloxacin, demonstrating that these antibiotics could not effectively inhibited cell growth of this strain. The resistance results were in consistent with those of Kirby-Bauer tests, a common conventional test. Thus, this enzyme-activatable assay has the potential to provide a simple and accessible tool for E. coli antibiotic resistance analysis.
Conclusion
In conclusion, we have proposed an enzyme-responsive NIR probe, DCM-βgal, for sensitive measuring and monitoring β-gal activity. DCM-βgal was almost non-emissive in the absence of β-gal, while it could be induced to produce bright emission upon the addition of β-gal. Particularly, the probe also provided a rapid and convenient colorimetric readout for visual assaying of the β-gal activity. The probe displayed high sensitivity, selectivity and rapid response to β-gal. Simultaneously, the probe also had a good photo-stability, and exhibited a good linear relationship with β-gal concentration from 0 to 0.2 U ml−1, with a low detection limit of 1.5 × 10−3 U ml−1. Importantly, this assay was successfully applied to sensitive detection of E. coli cells, as well as rapidly determination of the antibiotic resistance profile of E. coli via levels of the color response. The strategy provided several advantages for E. coli detection, including a fast detection process within 5 h, a low detection concentration of 1 × 103 CFU ml−1, a good selectively to E. coli sensing and dual fluorescent and colorimetric readout. Further developments include integrating this assay into point of care testing devices, detecting more real and complex samples would provide improved detection outcomes. Overall, the simple, low-cost and friendly-to-user approach provides an accessible tool for efficient quantification of β-gal activity and is promising for E. coli detection and antibiotic resistance analysis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
YH: Conceptualization, Methodology, Writing—original draft, Supervision. WF: Methodology, Data curation, Investigation. G-QZ: Synthesizing the probe. YQ, LL, and LP: Writing—review and editing. NC: Review and editing, Supervision.
Funding
This study was supported by the Open Project of NHC Key Laboratory of Thalassemia Medicine (GJWJWDP202203).
Acknowledgments
The authors thank Key Laboratory of Bioactive Materials, State Key Laboratory of Medicinal Chemical Biology, Key Laboratory of Functional Polymer Ministry of Education, and College of Life Sciences, Nankai University for their assistance in synthesizing of DCM-βgal probe. We thank Department of Clinical Laboratory, Guangzhou Twelfth People’s Hospital, Guangzhou, China for providing the bacterial cells.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1052801/full#supplementary-material
References
Adkins, J. A., Boehle, K., Friend, C., Chamberlain, B., Bisha, B., and Henry, C. S. (2017). Colorimetric and electrochemical bacteria detection using printed paper- and transparency-based analytic devices. Anal. Chem. 89 (6), 3613–3621. doi:10.1021/acs.analchem.6b05009
Boyaci, I. H., Aguilar, Z. P., Hossain, M., Halsall, H. B., Seliskar, C. J., and Heineman, W. R. (2005). Amperometric determination of live Escherichia coli using antibody-coated paramagnetic beads. Anal. Bioanal. Chem. 382 (5), 1234–1241. doi:10.1007/s00216-005-3263-8
Burnham, S., Hu, J., Anany, H., Brovko, L., Deiss, F., Derda, R., et al. (2014). Towards rapid on-site phage-mediated detection of generic Escherichia coli in water using luminescent and visual readout. Anal. Bioanal. Chem. 406 (23), 5685–5693. doi:10.1007/s00216-014-7985-3
Chen, J., Alcaine, S. D., Jackson, A. A., Rotello, V. M., and Nugen, S. R. (2017). Development of engineered bacteriophages for Escherichia coli detection and high-throughput antibiotic resistance determination. ACS Sens. 2 (4), 484–489. doi:10.1021/acssensors.7b00021
Chen, J., Alcaine, S. D., Jiang, Z., Rotello, V. M., and Nugen, S. R. (2015). Detection of Escherichia coli in drinking water using T7 bacteriophage-conjugated magnetic probe. Anal. Chem. 87 (17), 8977–8984. doi:10.1021/acs.analchem.5b02175
Chen, J., Jackson, A. A., Rotello, V. M., and Nugen, S. R. (2016). Colorimetric detection of Escherichia coli based on the enzyme-induced metallization of gold nanorods. Small 12 (18), 2469–2475. doi:10.1002/smll.201503682
Colquhoun, K. O., Timms, S., and Fricker, C. R. (1995). Detection of Escherichia coli in potable water using direct impedance technology. J. Appl. Bacteriol. 79 (6), 635–639. doi:10.1111/j.1365-2672.1995.tb00948.x
Derda, R., Lockett, M. R., Tang, S. K., Fuller, R. C., Maxwell, E. J., Breiten, B., et al. (2013). Filter-based assay for Escherichia coli in aqueous samples using bacteriophage-based amplification. Anal. Chem. 85 (15), 7213–7220. doi:10.1021/ac400961b
Fu, W., Yan, C., Zhang, Y., Ma, Y., Guo, Z., and Zhu, W. H. (2019). Near-infrared aggregation-induced emission-active probe enables in situ and long-term tracking of endogenous beta-galactosidase activity. Front. Chem. 7, 291. doi:10.3389/fchem.2019.00291
Gu, K., Qiu, W., Guo, Z., Yan, C., Zhu, S., Yao, D., et al. (2019). An enzyme-activatable probe liberating AIEgens: On-site sensing and long-term tracking of beta-galactosidase in ovarian cancer cells. Chem. Sci. 10 (2), 398–405. doi:10.1039/c8sc04266g
Gu, K., Xu, Y., Li, H., Guo, Z., Zhu, S., Shi, P., et al. (2016). Real-time tracking and in vivo visualization of beta-galactosidase activity in colorectal tumor with a ratiometric near-infrared fluorescent probe. J. Am. Chem. Soc. 138 (16), 5334–5340. doi:10.1021/jacs.6b01705
Jia, Z., Sukker, I., Muller, M., and Schonherr, H. (2018). Selective discrimination of Key enzymes of pathogenic and nonpathogenic bacteria on autonomously reporting shape-encoded hydrogel patterns. ACS Appl. Mat. Interfaces 10 (6), 5175–5184. doi:10.1021/acsami.7b15147
Jiang, G., Zeng, G., Zhu, W., Li, Y., Dong, X., Zhang, G., et al. (2017). A selective and light-up fluorescent probe for beta-galactosidase activity detection and imaging in living cells based on an AIE tetraphenylethylene derivative. Chem. Commun. 53 (32), 4505–4508. doi:10.1039/c7cc00249a
Jones, K. E., Patel, N. G., Levy, M. A., Storeygard, A., Balk, D., Gittleman, J. L., et al. (2008). Global trends in emerging infectious diseases. Nature 451, 990–993. doi:10.1038/nature06536
Komatsu, T., and Urano, Y. (2015). Evaluation of enzymatic activities in living systems with small-molecular fluorescent substrate probes. Anal. Sci. 31 (4), 257–265. doi:10.2116/analsci.31.257
Laczka, O., Garcia-Aljaro, C., del Campo, F. J., Munoz Pascual, F. X., Mas-Gordi, J., and Baldrich, E. (2010). Amperometric detection of Enterobacteriaceae in river water by measuring beta-galactosidase activity at interdigitated microelectrode arrays. Anal. Chim. Acta X. 677 (2), 156–161. doi:10.1016/j.aca.2010.08.001
Liu, X., Qiao, Q., Tian, W., Liu, W., Chen, J., Lang, M. J., et al. (2016). Aziridinyl fluorophores demonstrate bright fluorescence and superior photostability by effectively inhibiting twisted intramolecular charge transfer. J. Am. Chem. Soc. 138 (22), 6960–6963. doi:10.1021/jacs.6b03924
Munoz-Espin, D., and Serrano, M. (2014). Cellular senescence: From physiology to pathology. Nat. Rev. Mol. Cell Biol. 15 (7), 482–496. doi:10.1038/nrm3823
Nishihara, T., Kuno, S., Nonaka, H., Tabata, S., Saito, N., Fukuda, S., et al. (2018). Beta-galactosidase-responsive synthetic biomarker for targeted tumor detection. Chem. Commun. 54 (83), 11745–11748. doi:10.1039/c8cc06068a
Oliver, S. P., Jayarao, B. M., and Almeida, R. A. (2005). Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implications. Foodborne Pathog. Dis. 2 (2), 115–129. doi:10.1089/fpd.2005.2.115
Ripp, S., Jegier, P., Johnson, C. M., Brigati, J. R., and Sayler, G. S. (2008). Bacteriophage-amplified bioluminescent sensing of Escherichia coli O157:H7. Anal. Bioanal. Chem. 391 (2), 507–514. doi:10.1007/s00216-007-1812-z
Rompre, A., Servais, P., Baudart, J., de-Roubin, M. R., and Laurent, P. (2002). Detection and enumeration of coliforms in drinking water: Current methods and emerging approaches. J. Microbiol. Methods 49 (1), 31–54. doi:10.1016/s0167-7012(01)00351-7
Scharff, R. L. (2012). Economic burden from health losses due to foodborne illness in the United States. J. Food Prot. 75 (1), 123–131. doi:10.4315/0362-028X.JFP-11-058
Schaumburg, F., Carrell, C. S., and Henry, C. S. (2019). Rapid bacteria detection at low concentrations using sequential immunomagnetic separation and paper-based isotachophoresis. Anal. Chem. 91 (15), 9623–9630. doi:10.1021/acs.analchem.9b01002
Sun, J., Warden, A. R., Huang, J., Wang, W., and Ding, X. (2019). Colorimetric and electrochemical detection of Escherichia coli and antibiotic resistance based on a p-benzoquinone-mediated bioassay. Anal. Chem. 91 (12), 7524–7530. doi:10.1021/acs.analchem.8b04997
Walsh, C. (2000). Molecular mechanisms that confer antibacterial drug resistance. Nature 406 (6797), 775–781. doi:10.1038/35021219
Wang, D., Chen, J., and Nugen, S. R. (2017). Electrochemical detection of Escherichia coli from aqueous samples using engineered phages. Anal. Chem. 89 (3), 1650–1657. doi:10.1021/acs.analchem.6b03752
Wang, H., Yang, R., Jiang, X., Hua, X., Zhao, W., Zhang, W., et al. (2014). Expression and characterization of two beta-galactosidases from Klebsiella pneumoniae 285 in Escherichia coli and their application in the enzymatic synthesis of lactulose and 1-lactulose. Z Naturforsch C J. Biosci. 69 (11-12), 479–487. doi:10.5560/znc.2014-0061
Keywords: dual-readout probe, β-galactosidase sensing, Escherichia coli detection, antibiotic resistance analysis, enzyme reaction
Citation: Huang Y, Feng W, Zhang G-Q, Qiu Y, Li L, Pan L and Cao N (2022) An enzyme-activatable dual-readout probe for sensitive β-galactosidase sensing and Escherichia coli analysis. Front. Bioeng. Biotechnol. 10:1052801. doi: 10.3389/fbioe.2022.1052801
Received: 24 September 2022; Accepted: 17 October 2022;
Published: 31 October 2022.
Edited by:
Chang Li, Tongji University, ChinaReviewed by:
Xun Li, University of Maryland, United StatesYating Xiao, University of Chinese Academy of Sciences, China
Jiahong Shen, Northwestern University, United States
Copyright © 2022 Huang, Feng, Zhang, Qiu, Li, Pan and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yifang Huang, MDUwMWh5ZkAxNjMuY29t; Nannan Cao, Y2FvYmVpYmVpY25uQDE2My5jb20=
†These authors have contributed equally to this work
 Yifang Huang
Yifang Huang Weiwei Feng3,4†
Weiwei Feng3,4†