- 1School of Food Science and Pharmaceutical Engineering, Nanjing Normal University, Nanjing, China
- 2Department of Chemical, Biochemical and Environmental Engineering, University of MD, Baltimore County, Baltimore, MD, United States
- 3College of Biological and Pharmaceutical Engineering, West Anhui University, Lu’an, Anhui, China
- 4Department of Chemical Engineering, Guangdong Technion-Israel Institute of Technology (GTIIT), Shantou, Guangdong, China
- 5The Wolfson Department of Chemical Engineering, Technion-Israel Institute of Technology, Haifa, Israel
Introduction: Systematic gene knockout studies may offer us novel insights on cell metabolism and physiology. Specifically, the lipid accumulation mechanism at the molecular or cellular level is yet to be determined in the oleaginous yeast Y. lipolytica.
Methods: Herein, we established ten engineered strains with the knockout of important genes involving in central carbon metabolism, NADPH generation, and fatty acid biosynthetic pathways.
Results: Our result showed that NADPH sources for lipogenesis include the OxPP pathway, POM cycle, and a trans-mitochondrial isocitrate-α-oxoglutarate NADPH shuttle in Y. lipolytica. Moreover, we found that knockout of mitochondrial NAD+ isocitrate dehydrogenase IDH2 and overexpression of cytosolic NADP+ isocitrate dehydrogenase IDP2 could facilitate lipid synthesis. Besides, we also demonstrated that acetate is a more favorable carbon source for lipid synthesis when glycolysis step is impaired, indicating the evolutionary robustness of Y. lipolytica.
Discussion: This systematic investigation of gene deletions and overexpression across various lipogenic pathways would help us better understand lipogenesis and engineer yeast factories to upgrade the lipid biomanufacturing platform.
1 Instruction
Currently, concerns about fossil fuel sustainability have driven researchers to develop renewable lipid-derived biofuels as an alternative (Gu et al., 2018; Lazar et al., 2018). Lipid synthesis using oleaginous yeasts, owing to their unique metabolism, can achieve high lipid titers, yields and productivity, thus creating the potential for cost-effective industrial-scale operations (Ma et al., 2020). Specifically, Yarrowia lipolytica is an oleaginous yeast with GRAS (generally regarded as safe) status and has several significant attributes, such as a broad substrate spectrum, relatively well-defined genome annotations, diversified genetic manipulation methods, and abundant pool size of metabolic precursors, including acetyl-CoA, malonyl-CoA, and tricarboxylic acid (TCA) cycle intermediates (Abdel-Mawgoud et al., 2018; Larroude et al., 2018; Markham and Alper, 2018). Therefore, this yeast has emerged as a popular non-model organism in the lipid biomanufacturing field (Liu et al., 2015; Wong et al., 2017; Liu et al., 2019a; Yang et al., 2019).
In Y. lipolytica, extracellular carbon feedstock, such as glucose, is firstly internalized and converted into cytosolic pyruvate by glycolytic pathway (Qiao et al., 2017). Then, pyruvate is transported into mitochondria to synthesize mitochondrial citrate. Next, citrate is excreted to the cytoplasm by mitochondrial citrate carrier YlYhm2p (Yuzbasheva et al., 2019) and further cleaved to acetyl-CoA for lipid synthesis by cytosolic ATP-citrate lyases (Qiao et al., 2017). However, lipids are the highly reduced metabolites that need to consume the NADPHs to reduce acetyl groups (CH3-CO-) to alkyl groups (-CH2-CH2-) for growing fatty acid carbon backbone (Wasylenko et al., 2015; Xu et al., 2017; Xue et al., 2017). Currently, optimizing the intracellular metabolic network of Y. lipolytica has become a trend to improve lipids production by metabolic engineering and synthetic biology methods (Qiao et al., 2015; Liu et al., 2016; Xu et al., 2016; Abdel-Mawgoud et al., 2018). For example, Lazar et al. knocked out POX1-6 (peroxidase) and TGL4 (triglyceride lipase) with overexpressing GDP1 (glycerol triphosphate dehydrogenase) and DGA2 (diacylglycerol acyltransferase) in Y. lipolytica, improving the titer of lipids to 34 g/L (Dulermo and Nicaud, 2011). Qiao et al. optimized the supply of NADPH, which maximized the utilization of electrons from alternative NADPH pathways to increase substrate-to-lipid yields, leading to a 25% improvement over the starting strain (Qiao et al., 2017). Notwithstanding, much effort has been investigated to improve lipid accumulation, such as deletion of degradation pathways, overexpression of lipid synthesis pathways, eliminating of regulatory bottlenecks, and strengthening the supply of precursor, such as acetyl-CoA and NADPH (Groenewald et al., 2014; Silverman et al., 2016; Liu et al., 2019b). However, the influences of the important gene deficiency on lipid accumulation, such as carbon feedstock degradation and NADPH metabolism, have not been explored in Y. lipolytica.
Herein, we systematically investigated the influence of deleting important lipogenesis genes on lipid synthesis and cell growth, involving in carbon metabolism, NADPH metabolism, and fatty acid biosynthetic pathways. Specifically, we demonstrated that the sources of NADPH for lipogenesis include the OxPPP pathway, pyruvate-oxaloacetate-malate (POM) cycle, and isocitrate-2-oxoglutarate shuttle in Y. lipolytica. Moreover, the knockout of mitochondrial NAD+ isocitrate dehydrogenase IDH2 and overexpression of cytosolic NADP+ isocitrate dehydrogenase IDP2 could facilitate lipid synthesis. Besides, we also concluded that acetate is a favorable carbon source for producing lipid in Y. lipolytica. This study may help us better understand lipogenesis mechanisms, and the insights obtained here will guide us to engineer efficient cell factories for oleochemical production.
2 Materials and methods
2.1 Strains, plasmid, primers, and chemicals
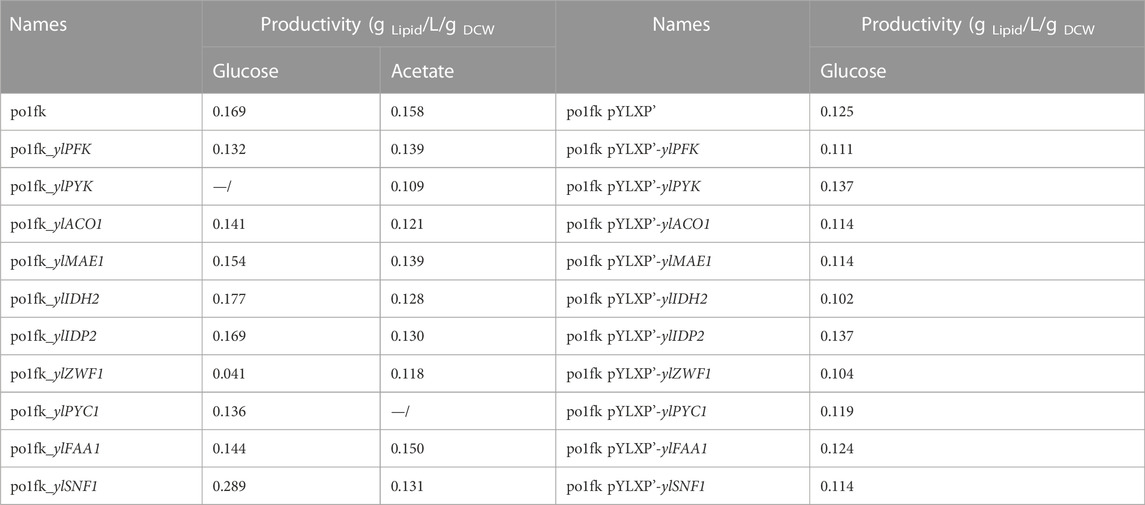
The strain po1fk obtained in our previous work was chosen as the starting strain in this study (Gu et al., 2020a). Moreover, strains, plasmids and primers have been listed in Table1 and Supplementary Table S1, respectively. Chemicals, including methyl tridecanoic acid and triglyceryl heptadecanoic acid were purchased from Sigma-Aldrich.
2.2 Gene knockout vectors construction
The marker-free Cre-loxP based gene knockout method was used as previously reported (Gu et al., 2020a; Gu et al., 2020b). In this study, constructed gene knockout plasmids included pHyloxP-ylPFK (encoding phosphofructose kinase), pHyloxP-ylPYK (encoding pyruvate kinase), pHyloxP-ylPYC1 (encoding pyruvate carboxylase), pHyloxP-ylMAE1 (encoding malic enzyme), pHyloxP-ylIDH2 (encoding mitochondrial isocitrate dehydrogenase), pHyloxP-ylIDP2 (encoding cytosolic isocitrate dehydrogenase), pHyloxP-ylFAA1 (encoding peroxisomal fatty acyl-CoA ligase), pHyloxP-ylACO1 (encoding aconitate hydratase), and pHyloxP-ylSNF1 (encoding AMP-activated protein kinase).
Here, we took the process of constructing the plasmid pHyloxP-ylPFK as an example. Firstly, the upstream and downstream sequences (both 1000 bp) of flanking gene ylPFK were obtained by the PCR-amplified reactions using primers ylPFK_UpF/ylPFK_UpR and ylPFK_DwF/ylPFK_DwR. Then, pYLXP’-loxP-Hyr containing loxP-Hyr-loxP cassette was digested by endonucleases AvrII and SalI to obtain loxP-Hyr-loxP cassette and plasmid backbone of pYLXP’-loxP-Hyr. Next, the upstream fragment ylPFK_Up, downstream fragments ylPFK_Dw, loxP-Hyr-loxP cassette and pYLXP’-loxP-Hyr backbone were joined by Gibson Assembly to generate the plasmid pHyloxP-ylPFK. The constructed plasmids were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China).
2.3 The expression plasmid construction and assembly
The pYLXP’, a YaliBrick plasmid, was used for gene expression (Gu et al., 2020a; Gu et al., 2020b). For example, to construct the recombinant plasmid pYLXP’-ylPFK, pYLXP’ was firstly digested by SnaBI and KpnI, obtaining the linearized pYLXP’. Then, the linearized pYLXP’ was ligase with the DNA fragment of gene PFK (that was obtained by the PCR-amplified reaction using primers PFK-F and PFK-R) by Gibson assembly method to generate plasmid pYLXP’-ylPFK, which was sequenced by Sangon Biotech Co., Ltd. (Shanghai, China).
2.4 Yeast transformation
The standard protocols of Y. lipolytica transformation have been reported (Gu et al., 2020b; Lv et al., 2020). In brief, 1-mL cells were harvested from YPD medium (yeast extract 10 g/L, peptone 20 g/L, and glucose 20 g/L) at 24 h. Then, cells were resuspended by using 105 uL transformation solution, containing 90 uL 50% PEG4000, 5 uL lithium acetate (2 M), 5 uL boiled single strand DNA (salmon sperm, denatured), and 5 uL DNA products. Next, the mixture was incubated at 39°C for 1 h before spreading on selected plates, which needed to be vortexed for 15 s every 15 min. In this study, the selected markers, including leucine and hygromycin, were used.
2.5 Shake flask cultivations
Shake flask cultivations of genetically modified Y. lipolytica were performed in 250 mL flasks with 25 mL fermentation medium at 220 r.p.m. and 30°C for 120 h. For this, 0.8 mL seed solution was inoculated, which was cultured in 14 mL shaking tubes under the conditions of 220 r.p.m. and 30°C for 48 h. One milliliter of fermentation broth was sampled every 24 h for OD600, lipid, glucose, and acetate measurements.
Seed culture medium of CSM (the yeast complete synthetic media) contains ammonium sulfate 5.0 g/L, YNB (yeast nitrogen base without ammonium sulfate) 1.7 g/L, glucose 20.0 g/L, CSM-Leu 0.74 g/L, and L-leucine 0.20 g/L. The CSM fermentation medium (C/N = 80) contains ammonium sulfate 1.1 g/L, YNB 1.7 g/L, glucose 40.0 g/L, CSM-Leu 0.74 g/L, and L-leucine 0.20 g/L. Specifically, phosphoric buffer solution (PBS), including 0.2 M Na2HPO4 and 0.2 M Na2HPO4, with pH 6.0 was used to replace water to make CSM-Acetate fermentation medium. Besides, CSM-acetate medium (C/N = 80) contains ammonium sulfate 1.1 g/L, YNB 1.7 g/L, acetate 41.0 g/L, CSM-Leu 0.74 g/L, and L-leucine 0.20 g/L.
2.6 Quantification of biomass, glucose, citrate and lipid
Cell densities were monitored by measuring the optical density at 600 nm (OD600). The OD600 value could be converted to dry cell weight (DCW) according to Eq. 1 OD600 = 0.35 g/L. The concentrations of glucose and acetate were measured under a flow rate of 0.6 mL/min of the mobile phase (5 mM of H2SO4) at 40°C by high-performance liquid chromatography (HPLC) equipped with a HPX-87H column (Bio-Rad, Hercules, CA, United States) and a refractive index detector through Shimadzu Prominence HPLC LC-20 A. To determine lipid composition, 4 OD units of cells were directly saponified with 0.5 M sodium methoxide with vortexing at 1,200 rpm for 2 h, and then neutralized with 40 μL 98% sulfuric acid to facilitate transesterification. Next, 400 μL hexane was added to extract fatty acid methyl ester for GC analysis. Moreover, to quantitatively determine lipid titer, 100 μL of 2 g/L of tridecanoate methyl ester and 2 g/L of glyceryl trihepta-decanoate were added as internal standard at the beginning, to benchmark the transesterification and saponification efficiency, respectively. Triplicate samples were taken and results were reported as average ± standard deviations. Lipid samples were taken at 120 h of shaking incubation. The method of GC analysis is consistent with previously reported method (Xu et al., 2016).
3 Results
3.1 Selecting the important genes for investigating the lipogenesis metabolism in Y. lipolytica
Noticeably, cytosolic acetyl-CoA is a direct precursor for lipid synthesis (Qiao et al., 2017), which is carboxylated to malonyl-CoA, further transacetylated to malonyl-ACPs, the true precursor to extending the growing chain of fatty acids, catalyzed by fatty acid synthases (FAS1 and FAS2). Thus, in acetyl-CoA metabolism, we chose three genes for investigation, including ylPFK (YALI0D16357g, encoding 6-phosphofructokinase), ylPYK (YALI0F09185g, encoding pyruvate kinase), and ylAC O 1 (YALI0D09361g, encoding aconitate hydratase), and further constructed three strain po1fk_ylPFK, po1fk_ylPYK, and po1fk_ylACO1.
On the other hand, acetyl groups (CH3-CO-) originating from acetyl-CoA is reduced to alkyl groups (-CH2-CH2-) when growing fatty acid carbon backbone, which needs to consume the reducing equivalent NADPH (Liu et al., 2019a). It has been reported that the sources of cytosolic NADPH include NADP+-dependent isocitrate dehydrogenase, the OxPP pathway, and POM cycle (Silverman et al., 2016), related to malate dehydrogenase (encoded by gene ylMAE1, YALI0E18634g), mitochondrial isocitrate dehydrogenase (encoded by gene ylIDH2, YALI0D06303g), cytosolic isocitrate dehydrogenase (encoded by gene ylIDP2, YALI0F04095g), and glucose-6-phosphate dehydrogenase (encoded by gene ylZWF1, YALI0E22649g). Particularly, the POM cycle (Figure 1) also involves the reductive carboxylation of pyruvate to oxaloacetate by pyruvate carboxylase (encoded by gene ylPYC1, YALI0C24101g). Thus, these five genes in the NADPH metabolism were deleted to obtained strains po1fk_ylMAE1, po1fk_ylIDH2, po1fk_ylIDP2, po1fk_ylZWF1, and po1fk_ylPYC1, respectively. Moreover, we also investigated the influences of deleting lipogenesis gene ylFAA1 (YALI0D17864g, long chain fatty acyl-CoA synthetase) on the lipid synthesis (Figure 1), which generated strain po1fk_ylFAA1. In addition, the regulator SNF1 (encoded by gene YALI0D02101g, carbon catabolite-derepressing protein kinase) was also knocked out, which is a cellular carbon metabolism regulator in Y. lipolytica to control the transition from growth phase to oleaginous production phase (Lazar et al., 2018), obtaining po1fk_ylSNF1.
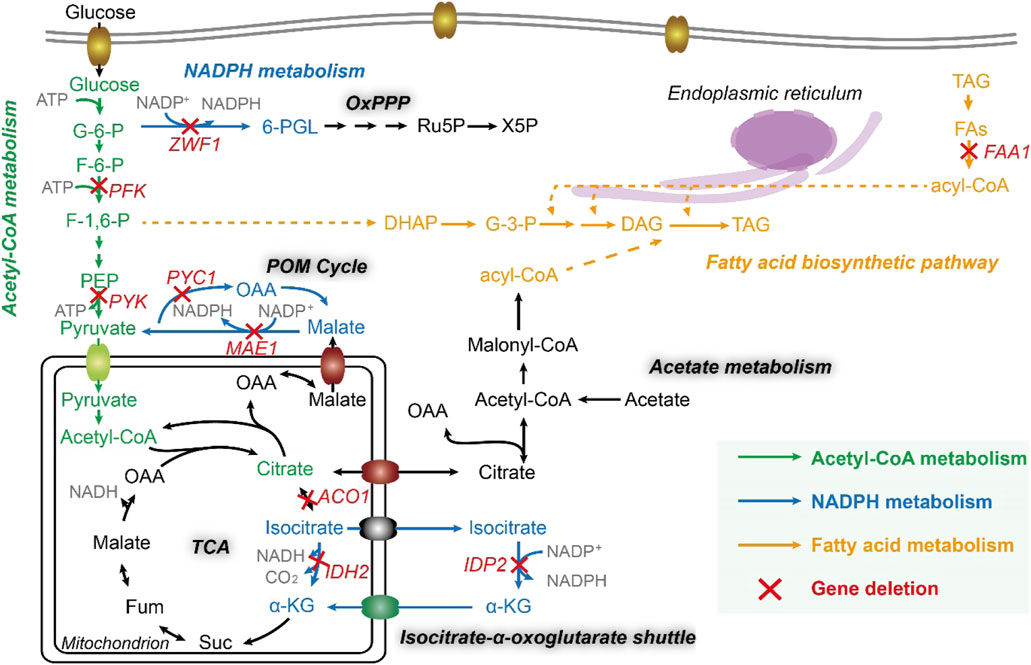
FIGURE 1. The fatty acid metabolic network of Y. lipolytica. FAA1, fatty acyl-CoA synthase; ZWF1, glucose-6-phosphate dehydrogenase; PFK, 6-phosphofructokinase; PYK, pyruvate kinase; PYC1, pyruvate carboxylase; MAE1, malic enzyme; ACO1, Aconitase; IDH2, mitochondrial isocitrate dehydrogenase; IDP2, cytosolic NADP-specific isocitrate dehydrogenase; G-6-P, Glucose-6-phosphate; F-6-P, fructose-6-phosphate; F-1,6-P, fructose-1,6-diphosphate; PEP, phosphoenolpyruvic acid; OAA, oxaloacetic acid; Fum, fumaric acid; Suc, succinyl coenzyme A; α-KG, 2-oxoglutarate; 6-PGL, 6-phosphogluconolactone; Ru5P, ribulose-5-phosphate; X5P, xylulose-5-phosphate; DHAP, dihydroxyacetone phosphate; G-3-P, glyceraldehyde-3-phosphate; DAG, diacylglycerol; TAG, triacylglycerol; FFA, free fatty acid; PL, phospholipid; Glx, glutamine; QH2, coenzyme QH2.
3.2 Characterization of engineered Y. lipolytica strains under the nitrogen-limited shaking cultivation
To confirm whether cell growth and lipid levels of engineered Y. lipolytica strains were changed, we performed the nitrogen-limited shaking cultivation using the CSM medium (C/N = 80) to measure the lipid titers and cell growth. As a result, the maximal biomasses of strains po1fk_ylPYC1, po1fk_ylACO1, and po1fk_ylMAE1 were slightly decreased compared with po1fk (Figure 2), and deletion of gene ylIDP2 and ylPFK almost does not influence on cell growth. However, cell growth of strains po1fk_ylPYK and po1fk_ylZWF is severely inhibited. The maximum biomass of po1fk_ylPYK and po1fk_ylZWF are 72.5% and 52.2% lower than that of the control strain po1fk, respectively, reaching 3.96 and 6.47 (OD600). Notably, gene ylPYK encodes pyruvate kinase, which converts the reaction of phosphoenolpyruvate to pyruvate in glycolysis. Therefore, deletion of gene ylPYK could block the metabolic flux from the glycolytic pathway to TCA cycles. On the other hand, glucose-6-phosphate dehydrogenase ZWF1 catalyzes glucose 6-phosphate to 6-phospho-glucono-1,5-lactone, providing the primary source of cytosolic NADPH in Y. lipolytica. It should be noted that NADPH functions as reducing equivalents to maintain cellular redox homeostasis and serves as important electron donors in the anabolic metabolism (Wang et al., 2017). In this study, we found that deletion of gene ylZWF gave a 76.9% reduction (0.18 ± 0.05 g/L with the yield of 13.01 mg/g Glucose) in the lipid synthesis, indicating that the OxPP pathway is the primary NADPH source for lipogenesis. However, an interesting phenomenon to note is that the surface wrinkle of po1fk_ylZWF1 is disappeared. We speculated that the reduced surface area of po1fk_ylZWF1 colony may be attributed to the less oxygen demand for the ZWF1 deficient strain.
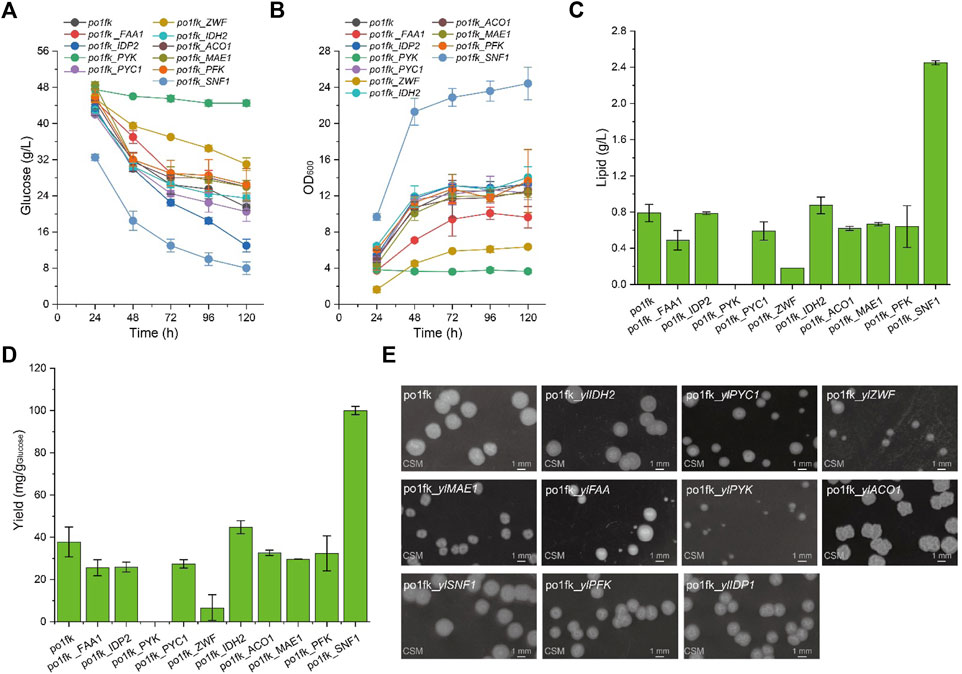
FIGURE 2. Time profiles of cell growth, glucose consumption, lipid titer and lipid yield in the CSM-leu medium and the morphology analysis of gene knockout strains. (A) Glucose consumption of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (B) Cell growth of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (C) Lipid titer of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (D) Lipid yield of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (E) the morphology analysis of gene knockout strains. For analyzing the morphology, cells were harvested from CSM-leu medium at 48 h and washed twice using .9% of NaCl solution. Then, cells were re-suspended and diluted to an appropriate concentration and spread on the CSM plates, cultivating at 30°C for 96 h.
Moreover, the lipid titer of po1fk_ylFAA1, po1fk_ylPYC1, po1fk_ylACO1, and po1fk_ylMAE1 are 0.49 ± 0.11, 0.59 ± 0.10, 0.62 ± 0.02, and 0.67 ± 0.02 g/L, decreased by 38.0%, 25.3%, 21.5%, and 15.2% relatively to po1fk (0.79 ± 0.10 g/L), respectively, suggesting that genes ylFAA1, ylPYC1, ylAC O 1, and ylMAE are necessary for the lipid synthesis in Y. lipolytica. However, the deletion of gene ylIDH2 increased the lipid titer, reaching .87 g/L with the yield of 44.8 mg/g Glucose, which were 1.10-fold and 1.19-fold of that in the strain po1fk, respectively. It is incomprehensible because the mitochondrial NAD+ isocitrate dehydrogenase IDH2 is important for TCA cycle, catalyzing isocitrate to 2-oxoglutarate. To explain this result, we performed a KEGG pathway analysis and found that the mitochondrial NAD+ isocitrate dehydrogenases have two isoenzymes in Y. lipolytica, encoded by genes YALI0E05137g and YALI0D06303g, respectively. Therefore, we speculate that deletion of IDH2 downregulated the activity of mitochondrial NAD+ isocitrate dehydrogenases, which strengthened the accumulation of citrate to promote the synthesis of lipids. Besides, the deletion of carbon catabolite repressor gene ylSNF1 significantly increased the lipid titer, reaching 2.45 ± 0.02 g/L with the yield of 100.1 mg/g Glucose, which was 3.10-fold and 2.65-fold of that of po1fk, respectively. This result is consistent with the previous research (Seip et al., 2013). The regulator SNF1 is involved in the transition from the growth phase to the oleaginous phase, and allows strains to accumulate lipids even in the nitrogen-abundant medium (Wang et al., 2020). However, how SNF1 regulates the lipid synthesis pathway has not been resolved (Lazar et al., 2018), possibly due to the SNF1-mediated MAP kinase, which phosphorylates acetyl-CoA carboxylase and downregulates lipid synthesis. In brief summary, knocking out mitochondrial IDH2 and SNF1 will improve lipid accumulation in Y. lipolytica under the culture condition of using the CSM medium.
3.3 Using acetate as carbon source for lipid synthesis by engineered Y. lipolytica strains
The acetate uptake pathway in Y. lipolytica could function as an acetyl-CoA shortcut, which has been demonstrated to achieve metabolic optimality in producing polyketides (Liu et al., 2019b). Therefore, we turned to use acetate as the carbon source for lipid synthesis. Specifically, extracellular acetate is first catalyzed by acetyl-coA synthase to generate acetyl-coA, which enters into cellular metabolic pathways for maintaining cell activities. However, it should be noted that partial acetyl-CoA would pass through gluconeogenesis into the OxPP pathway to provide NADPH for fatty acid synthesis.
Noticeably, the cultivation pH will keep increasing when sodium acetate is used as a carbon source. For this, an in situ pH indicator (bromocresol purple) was used to track the change of cultivation pH (Liu et al., 2019a). By performing the shaking flask, we found that po1fk_ylPYK, po1fk_ylZWF, and po1fk_ylFAA1 showed the remarkable recovery of cell growth (Figure 3) when acetate was used as sole carbon source, reaching 20.02 ± 1.18, 20.72 ± 1.56, and 21.38 ± 4.11 (the maximum biomass, OD600), respectively. These results indicate that the evolutionary robustness of cells that enable them adapt to alternate carbon sources when central metabolism is compromised, and most importantly, acetate could be used as a shortcut nutrient to support cell growth. However, the cell growth of po1fk_ylPYC1 was repressed in the CSM-acetate medium, and its maximum biomass (OD600) was 5.72 ± 0.50, which could be attributed to the fact that PYC is important for cell to replenish gluconeogenesis when the cells were grown on the acetate media. The maximal lipid titer was produced by po1fk_ylPFK, reaching 1.04 ± 0.23 g/L, which is 1.09-fold of that of po1fk (0.95 ± 0.04 g/L). Interestingly, the lipid titer of po1fk_ylSNF1 was close to that of po1fk using acetate as carbon source. Studies have reported that downregulation of Snf1 can activate the activity of acetyl-CoA carboxylase ACC1, which could facilitate the flux from acetyl-CoA to malonyl-CoA (Gross et al., 2019). Therefore, we speculated the increase flux from acetyl-CoA to malonyl-CoA in the engineering po1fk_ylSNF1 strain leads to a reduction of NADPH supply and limits the synthesis of lipid under the acetate cultivation.
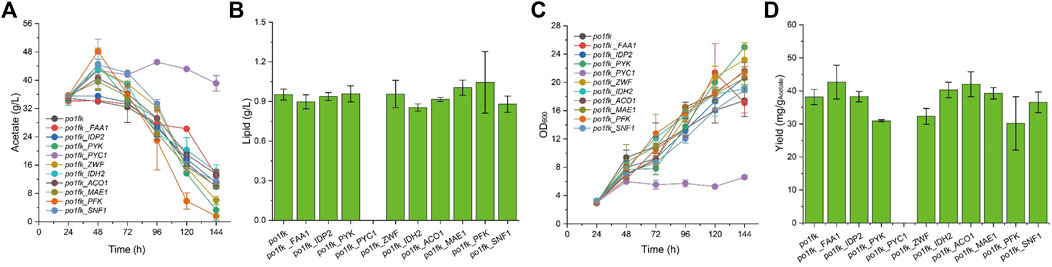
FIGURE 3. Time profiles of cell growth, acetate consumption, lipid titer and lipid yield in CSM-Acetate medium. (A) Glucose consumption of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (B) Lipid titer of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (C) Cell growth of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA; (D) Lipid yield of po1fk, po1fk_ylPYK, po1fk_ylACO1, po1fk_ylPFK, po1fk_ylSNF1, po1fk_ylIDP2, po1fk_ylZWF, po1fk_ylIDH2, po1fk_ylMAE, po1fk_ylPYC1, and po1fk_ylFAA.
3.4 The effect of overexpressing the selected genes on cell growth and lipid synthesis
To further identify the potential functions of the selected genes, we overexpressed these genes under the control of strong constitutive pTEF-intron promoter by the plasmid pYLXP’. Shake flask cultivation of these engineering strains, including po1kf pYLXP’-ylPFK, po1kf pYLXP’-ylPYK, po1kf pYLXP’-ylPYC1, po1kf pYLXP’-ylMAE1, po1kf pYLXP’-ylIDH2, po1kf pYLXP’-ylIDP2, po1kf pYLXP’-ylFAA1, po1kf pYLXP’-ylACO1, and po1kf pYLXP’-ylSNF1, show no significant differences in cell growth (Figure 4). Further, we analyzed the lipid titer of these engineering strains. As shown in Figure 4, individual overexpression of gene IDP2 result in a significant increase in the lipid titer, reaching 0.62 g/L with the yield of 29.58 mg/g Glucose. In Y. lipolytica, gene IDP2 encodes the cytosolic NADP+ isocitrate dehydrogenase IDP2, which catalyzes the reaction of isocitrate to 2-oxoglutarate with generating one molecule of NADPH. However, the cytoplasmic TCA cycle in Y. lipolytica is incomplete, which lacks cytoplasmic aconitate hydratase, succinyl-CoA ligase, fumarate hydratase, etc. Thus, we speculated that has a trans-mitochondrial isocitrate-α-oxoglutarate NADPH shuttle in Y. lipolytica, which can be responsible for maintaining the redox homeostasis and transporting the reducing equivalent from mitochondria NADH to cytoplasm NADPH.
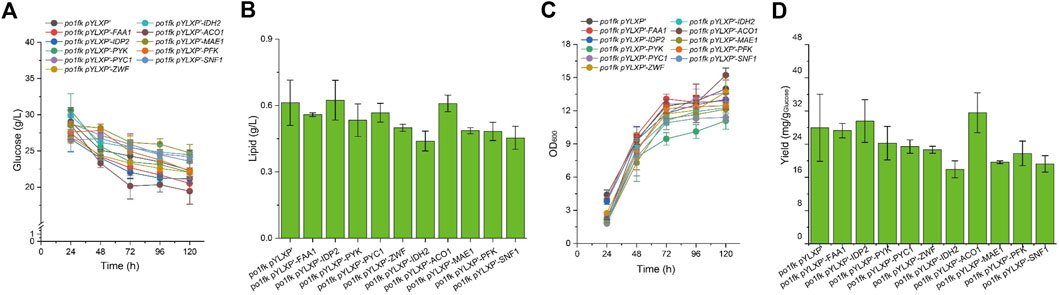
FIGURE 4. Time profiles of cell growth, glucose consumption, lipid titer and lipid yield in CSM-leu medium. (A) Glucose consumption of po1fk pYLXP’, po1fk pYLXP’-ylPYK, po1fk pYLXP’-ylACO1, po1fk pYLXP’-ylPFK, po1fk pYLXP’-ylSNF1, po1fk pYLXP’-ylIDP2, po1fk pYLXP’-ylZWF, po1fk pYLXP’-ylIDH2, po1fk pYLXP’-ylMAE, po1fk pYLXP’-ylPYC1, and po1fk pYLXP’-ylFAA; (B) The lipid titer of po1fk pYLXP’, po1fk pYLXP’-ylPYK, po1fk pYLXP’-ylACO1, po1fk pYLXP’-ylPFK, po1fk pYLXP’-ylSNF1, po1fk pYLXP’-ylIDP2, po1fk pYLXP’-ylZWF, po1fk pYLXP’-ylIDH2, po1fk pYLXP’-ylMAE, po1fk pYLXP’-ylPYC1, and po1fk pYLXP’-ylFAA; (C) Cell growth of po1fk pYLXP’, po1fk pYLXP’-ylPYK, po1fk pYLXP’-ylACO1, po1fk pYLXP’-ylPFK, po1fk pYLXP’-ylSNF1, po1fk pYLXP’-ylIDP2, po1fk pYLXP’-ylZWF, po1fk pYLXP’-ylIDH2, po1fk pYLXP’-ylMAE, po1fk pYLXP’-ylPYC1, and po1fk pYLXP’-ylFAA; (D) Lipid yield of po1fk pYLXP’, po1fk pYLXP’-ylPYK, po1fk pYLXP’-ylACO1, po1fk pYLXP’-ylPFK, po1fk pYLXP’-ylSNF1, po1fk pYLXP’-ylIDP2, po1fk pYLXP’-ylZWF, po1fk pYLXP’-ylIDH2, po1fk pYLXP’-ylMAE, po1fk pYLXP’-ylPYC1, and po1fk pYLXP’-ylFAA.
4 Discussions
In Y. lipolytica, lipogenic pathway is consisting of three parts: acetyl-CoA, NAPDH, and lipid metabolism pathways. Herein, we systematically investigated the knockout of ten important genes in these three parts, including ylPFK, ylPYK, ylAC O 1, ylMAE1, ylIDH2, ylIDP2, ylZWF1, ylPYC1, ylFAA1, and ylSNF1. Noticeably, pyruvate kinase PYK plays an important role in the glycolysis pathway, which provides the precursor pyruvate for TCA. Deleting gene ylPYK resulted in the block of the glycolytic pathway, and thus leaded to an impaired glucose uptake, retarded cell growth and compromised lipid synthesis. Specifically, the previous effort has showed that Y. lipolytica possesses strong acetate utilization pathway, which is equivalent or even superior to the hexose utilization pathway (Liu et al., 2019b). Therefore, it is believable that strain po1fk_ylPYK showed a remarkable recovery of cell growth using acetate as the carbon source. Specifically, we deduced the overall stoichiometry of glucose or acetate conversion to lipid (Supplementary Note 1). As suggested by the stoichiometry, the yield of lipid synthesized from acetate (0.294 g/g Acetate) is higher than that from glucose (0.271 g/g Glucose), indicating that acetate is a dominant carbon source for lipid synthesis. As a result, strain po1fk_ylPYK produced 0.96 ± 0.06 g/L of lipid using acetate as the carbon source.
The glucose-6-phosphate dehydrogenase ZWF is a key enzyme that catalyzes glucose 6-phosphate to 6-phosphogluconolactone with generating NADPH. NADPH provides reducing power for synthesizing biological macromolecules (such as lipids, proteins, and glycogen) and directly affects cell growth, protein expression, and other secondary metabolites synthesis (polyketides or triterpenoids) (Xu et al., 2018). We found that deletion of PYC1 or MAE1 both decreased the synthesis of lipid, suggesting the POM cycle participates in lipogenesis for NADPH supplementation in Y. lipolytica. However, in the CSM-acetate medium, the cell growth of po1fk_ylPYC1 was repressed, suggesting that the deficiency of pyruvate carboxylase PYC1 would lead to the disruption of gluconeogenesis. Moreover, it is reported that the distributions of the cellular reducing equivalent are highly compartmentalized in eukaryotes, which is attributed to the specific localization of metabolic pathways and impermeabilities of organelles membranes (Kory et al., 2020). Specifically, mitochondria are important organelles for the NADH metabolism due to its respiratory functions, the existence of electron transport chain for oxidative phosphorylation. Therefore, the dynamic balance of the reducing equivalent between mitochondria and cytoplasm are necessary for maintaining cellular redox homeostasis (Spinelli and Haigis, 2018). Our results of deletion of gene IDH2 and overexpression of gene IDP2 suggested the existence of a trans-mitochondrial isocitrate-α-oxoglutarate NADPH shuttle in Y. lipolytica. Specifically, according to the ways of NADPH supplement, we also deduced the overall stoichiometry of glucose or acetate conversion to lipid and calculated the yield of lipid (Supplementary Note 1), which could help us to understand the influences of NADPH supplement on lipid synthsis.
In addition, protein kinase ylSNF1 is a crucial regulator of glucose signal transduction, which is a negative regulator of the fatty acid biosynthesis pathway and acetyl-CoA carboxylase. It has been identified that SNF1 plays a vital role in the transition from growth to oil production in Y. lipolytica. Herein, deletion of genes ylSNF1 significantly increased the lipid titer in the CSM-leu medium, reaching 2.45 ± 0.02 g/L with the yield of 100.0 mg/g Glucose and the productivity of 0.289 g Lipid/L/g DCW (Table2), which is consistent with the previous research (Seip et al., 2013).
5 Conclusion
In this work, we established ten engineered strains with deleting the important gene involved in the central carbon, NADPH, and lipid metabolism, and systematically investigated the influence of such gene deletions on the lipid synthesis and cell growth. As a result, our study demonstrated that NADPH sources for lipogenesis include the OxPP pathway, POM cycle, and isocitrate-2-oxoglutarate shuttle in Y. lipolytica. Moreover, the knockout of mitochondrial isocitrate dehydrogenase IDH1 and the carbon catabolite repressor SNF1 could facilitate the lipid synthesis. Besides, acetate is a more favorable carbon source for the lipid synthesis when glycolysis step is impaired, indicating this could be alternate carbon source that reach metabolic optimality in lipid synthesis. This systematic investigation of gene deletions and overexpression across various lipogenic pathways would help us better understand lipogenesis and engineer yeast factories to upgrade the lipid biomanufacturing platform.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
Author contributions YG and PX conceived and designed research. YG, JZ, YY, and JM conducted experiments. YG, JZ, XS and PX analyzed data. YG, JZ, and PX wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was financially supported by the Natural Science Foundation of Jiangsu Province (BK20202002) and (BK20210572), and National Natural Science Foundation of China (22108126).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2023.1098116/full#supplementary-material
References
Abdel-Mawgoud, A. M., Markham, K. A., Palmer, C. M., Liu, N., Stephanopoulos, G., and Alper, H. S. (2018). Metabolic engineering in the host Yarrowia lipolytica. Metab. Eng. 50, 192–208. doi:10.1016/j.ymben.2018.07.016
Dulermo, T., and Nicaud, J. M. (2011). Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 13 (5), 482–491. doi:10.1016/j.ymben.2011.05.002
Groenewald, M., Boekhout, T., Neuvéglise, C., Gaillardin, C., van Dijck, P. W., and Wyss, M. (2014). Yarrowia lipolytica: Safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 40 (3), 187–206. doi:10.3109/1040841X.2013.770386
Gross, A. S., Zimmermann, A., Pendl, T., Schroeder, S., Schoenlechner, H., Knittelfelder, O., et al. (2019). Acetyl-CoA carboxylase 1-dependent lipogenesis promotes autophagy downstream of AMPK. J. Biol. Chem. 294 (32), 12020–12039. doi:10.1074/jbc.RA118.007020
Gu, Y., Ma, J., Zhu, Y., Ding, X., and Xu, P. (2020a). Engineering Yarrowia lipolytica as a chassis for de novo synthesis of five aromatic-derived natural products and chemicals. ACS Synth. Biol. 9 (8), 2096–2106. doi:10.1021/acssynbio.0c00185
Gu, Y., Ma, J., Zhu, Y., and Xu, P. (2020b). Refactoring ehrlich pathway for high-yield 2-phenylethanol production in Yarrowia lipolytica. ACS Synth. Biol. 9 (3), 623–633. doi:10.1021/acssynbio.9b00468
Gu, Y., Xu, X., Wu, Y., Niu, T., Liu, Y., Li, J., et al. (2018). Advances and prospects of Bacillus subtilis cellular factories: From rational design to industrial applications. Metab. Eng. 50, 109–121. doi:10.1016/j.ymben.2018.05.006
Kory, N., Uit de Bos, J., van der Rijt, S., Jankovic, N., Güra, M., Arp, N., et al. (2020). MCART1/SLC25A51 is required for mitochondrial NAD transport. Sci. Adv. 6 (43), eabe5310. doi:10.1126/sciadv.abe5310
Larroude, M., Rossignol, T., Nicaud, J. M., and Ledesma-Amaro, R. (2018). Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 36 (8), 2150–2164. doi:10.1016/j.biotechadv.2018.10.004
Lazar, Z., Liu, N., and Stephanopoulos, G. (2018). Holistic approaches in lipid production by Yarrowia lipolytica. Trends Biotechnol. 36 (11), 1157–1170. doi:10.1016/j.tibtech.2018.06.007
Liu, H., Marsafari, M., Deng, L., and Xu, P. (2019a). Understanding lipogenesis by dynamically profiling transcriptional activity of lipogenic promoters in Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 103 (7), 3167–3179. doi:10.1007/s00253-019-09664-8
Liu, H., Marsafari, M., Wang, F., Deng, L., and Xu, P. (2019b). Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab. Eng. 56, 60–68. doi:10.1016/j.ymben.2019.08.017
Liu, L., Pan, A., Spofford, C., Zhou, N., and Alper, H. S. (2015). An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab. Eng. 29, 36–45. doi:10.1016/j.ymben.2015.02.003
Liu, N., Qiao, K., and Stephanopoulos, G. (2016). (13)C Metabolic Flux Analysis of acetate conversion to lipids by Yarrowia lipolytica. Metab. Eng. 38, 86–97. doi:10.1016/j.ymben.2016.06.006
Lv, Y., Gu, Y., Xu, J., Zhou, J., and Xu, P. (2020). Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab. Eng. 61, 79–88. doi:10.1016/j.ymben.2020.05.005
Ma, J., Gu, Y., Marsafari, M., and Xu, P. (2020). Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J. Ind. Microbiol. Biotechnol. 47 (9-10), 845–862. doi:10.1007/s10295-020-02290-8
Markham, K. A., and Alper, H. S. (2018). Synthetic biology expands the industrial potential of Yarrowia lipolytica. Trends Biotechnol. 36 (10), 1085–1095. doi:10.1016/j.tibtech.2018.05.004
Qiao, K., Imam Abidi, S. H., Liu, H., Zhang, H., Chakraborty, S., Watson, N., et al. (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metab. Eng. 29, 56–65. doi:10.1016/j.ymben.2015.02.005
Qiao, K., Wasylenko, T. M., Zhou, K., Xu, P., and Stephanopoulos, G. (2017). Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat. Biotechnol. 35 (2), 173–177. doi:10.1038/nbt.3763
Seip, J., Jackson, R., He, H., Zhu, Q., and Hong, S. P. (2013). Snf1 is a regulator of lipid accumulation in Yarrowia lipolytica. Appl. Environ. Microbiol. 79 (23), 7360–7370. doi:10.1128/AEM.02079-13
Silverman, A. M., Qiao, K., Xu, P., and Stephanopoulos, G. (2016). Functional overexpression and characterization of lipogenesis-related genes in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 100 (8), 3781–3798. doi:10.1007/s00253-016-7376-0
Spinelli, J. B., and Haigis, M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell. Biol. 20 (7), 745–754. doi:10.1038/s41556-018-0124-1
Wang, J., Ledesma-Amaro, R., Wei, Y., Ji, B., and Ji, X. J. (2020). Metabolic engineering for increased lipid accumulation in Yarrowia lipolytica - a Review. Bioresour. Technol. 313 (123707), 123707. doi:10.1016/j.biortech.2020.123707
Wang, M., Chen, B., Fang, Y., and Tan, T. (2017). Cofactor engineering for more efficient production of chemicals and biofuels. Biotechnol. Adv. 35 (8), 1032–1039. doi:10.1016/j.biotechadv.2017.09.008
Wasylenko, T. M., Ahn, W. S., and Stephanopoulos, G. (2015). The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab. Eng. 30, 27–39. doi:10.1016/j.ymben.2015.02.007
Wong, L., Engel, J., Jin, E., Holdridge, B., and Xu, P. (2017). YaliBricks, a versatile genetic toolkit for streamlined and rapid pathway engineering in Yarrowia lipolytica. Metab. Eng. Commun. 5, 000999–e177. doi:10.1016/j.mec.2019.e00099
Xu, J. Z., Yang, H. K., and Zhang, W. G. (2018). NADPH metabolism: A survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Crit. Rev. Biotechnol. 38 (7), 1061–1076. doi:10.1080/07388551.2018.1437387
Xu, P., Qiao, K., Ahn, W. S., and Stephanopoulos, G. (2016). Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc. Natl. Acad. Sci. USA. 113 (39), 10848–10853. doi:10.1073/pnas.1607295113
Xu, P., Qiao, K., and Stephanopoulos, G. (2017). Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica. Biotechnol. Bioeng. 114 (7), 1521–1530. doi:10.1002/bit.26285
Xue, J., Balamurugan, S., Li, D. W., Liu, Y. H., Zeng, H., Wang, L., et al. (2017). Glucose-6-phosphate dehydrogenase as a target for highly efficient fatty acid biosynthesis in microalgae by enhancing NADPH supply. Metab. Eng. 41, 212–221. doi:10.1016/j.ymben.2017.04.008
Yang, Z., Edwards, H., and Xu, P. (2019). CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab. Eng. Commun. 22 (10), e00112. doi:10.1016/j.mec.2019.e00112
Yuzbasheva, E. Y., Agrimi, G., Yuzbashev, T. V., Scarcia, P., Vinogradova, E. B., Palmieri, L., et al. (2019). The mitochondrial citrate carrier in Yarrowia lipolytica: Its identification, characterization and functional significance for the production of citric acid. Metab. Eng. 54, 264–274. doi:10.1016/j.ymben.2019.05.002
Keywords: Yarrowia lipolytica, lipogenesis, central metabolism, lipid synthesis, metabolic engineering
Citation: Zhu J, Gu Y, Yan Y, Ma J, Sun X and Xu P (2023) Knocking out central metabolism genes to identify new targets and alternating substrates to improve lipid synthesis in Y. lipolytica. Front. Bioeng. Biotechnol. 11:1098116. doi: 10.3389/fbioe.2023.1098116
Received: 14 November 2022; Accepted: 03 January 2023;
Published: 13 January 2023.
Edited by:
Boyang Ji, BioInnovation Institute (BII), DenmarkReviewed by:
Dongming Xie, University of Massachusetts Lowell, United StatesJin Hou, Shandong University, China
Copyright © 2023 Zhu, Gu, Yan, Ma, Sun and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Gu, Z3V5YW5nQG5udS5lZHUuY24=; Peng Xu, cGVuZ3h1QGd0aWl0LmVkdS5jbg==
 Jiang Zhu1
Jiang Zhu1 Yang Gu
Yang Gu Yijing Yan
Yijing Yan