- 1Department of Microbiology, Gauhati Medical College & Hospital, Guwahati, Assam, India
- 2Department of Microbiology, Dhubri Medical College & Hospital, Dhubri, Assam, India
- 3Department of Oncology (Head & Neck), State Cancer Institute, Gauhati Medical College, Guwahati, Assam, India
- 4Department of Gastroenterology, Gauhati Medical College and Hospital, Guwahati, Assam, India
Introduction: Helicobacter pylori (H. pylori), a globally prevalent bacterium is linked to various gastroduodenal diseases. Its genetic diversity arises from nucleotide variability, genome plasticity and inter-strain recombination. Genetic studies of H. pylori provide insights into human migration and regional disease risks. This study profiles the H. pylori gene pool in ethnically and geographically distinct population of Assam, Northeast India using multi-locus sequence typing (MLST) to explore its role in human migration and its potential role on the occurrence of gastroduodenal diseases in this region.
Methods: In this hospital-based study, gastric biopsy and serum samples were collected from 200 dyspeptic patients and H. pylori individual risk factors were assessed. Multivariate logistic regression was used to determine the statistical association between predictors and outcomes. Serum anti-H. pylori IgG levels were estimated and cagA and vacA virulence gene profiles were analysed by polymerase chain reaction. MLST analysis on the seven housekeeping genes and phylogenetic analysis were conducted to infer the genomic diversity and evolutionary relationship of the strains.
Results: H. pylori infection prevalence was 74.5% with significant associations between elevated serum IgG antibody levels, family history, virulent genotypes cagA, vacA s1m1 and gastric pathology, including cancer. MLST analysis identified 36 sequence types among 49 strains, including 8 new ones, with most strains clustering within the hpAsia2 and hpEurope populations. Phylogenetic analysis shows that H. pylori strains in the Assamese population cluster with native Thai strains, suggesting their introduction through Tai-Ahom migrants from Thailand, supporting gene flow into India. No clustering of gastric outcomes was observed among the strains. A strong familial link was noted, with 76.5% of gastric cancer cases having a family history, indicating possible intrafamilial transmission.
Discussion: Our results suggest that H. pylori strains from Assam share ancestry with Tai-Ahom migrants from Thailand. Further, along with virulent genotypes, a family history of gastric cancer and high IgG levels may indicate higher disease risk. Future research should analyse strain transmission and IgG levels to improve early detection and intervention strategies for gastric cancer. IgG is a key predictor of gastric cancer risk, highlighting the importance of regular monitoring for early diagnosis and follow-up of high-risk individuals.
1 Introduction
Helicobacter pylori (H. pylori) is a micro-aerophilic, spiral-shaped, Gram-negative, pathogenic bacterium capable of colonizing the human stomach for years. The prevalence of H. pylori infection varies globally, ranging from 30–50% in developed countries to over 80% in developing regions (Rothenbacher and Brenner, 2003; World Gastroenterology Organisation, 2011; Hunt et al., 2011). As a key member found in the human microbiome, H. pylori has co-evolved with Homo sapiens, accompanying human populations throughout their migration history (Linz et al., 2007; Falush et al., 2003). The bacterium is usually acquired during childhood and is well adapted to persist in the gastric mucosa for decades, with most infected individuals remaining asymptomatic.
Genomic analyses have revealed that the genetic landscape of H. pylori is highly diverse, which is due to its nucleotide sequence variability, genome plasticity and occurrence of various inter-strain recombination processes (Hanafiah and Lopes, 2020; Kersulyte et al., 2000; Blaser and Berg, 2001). While most recombination events are neutral, some influence the bacterium’s ability to colonize, persist, and contribute to disease development. H. pylori infection has been linked with various gastro-intestinal conditions, including peptic ulcer disease, chronic gastritis, gastric cancer and mucosa associated lymphoid tissue (MALT) lymphoma (Burkitt et al., 2017; Camilo and Henrique, 2018; Shirani et al., 2023).
Recent studies have examined the genetic diversity of H. pylori populations using MLST analysis on seven housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI and yphC) (Linz et al., 2007; Devi et al., 2007; Kumar et al., 2015; Kauser et al., 2005). These investigations have demonstrated distinct phylogeographic differentiation among H. pylori strains from various regions, supporting their use as a major landmark to track the migration patterns of human populations (Linz et al., 2007; Falush et al., 2003; Yamaoka, 2009). Additionally, the geographical diversity of H. pylori populations appears to correlate with clinical consequences, suggesting that bacterial genetic diversity may contribute to variations in disease risk (Correa and Piazuelo, 2012; Yamaoka et al., 2008).
Recent genetic analyses have been conducted on isolates obtained from various Indian populations (Devi et al., 2007; Kumar et al., 2011; Mukhopadhyay et al., 2000; Sarma et al., 2018; Ahmed et al., 2003; Datta et al., 2003; Katelaris et al., 1992; Sarma et al., 2017, 2024). Although extensive studies on the genetic diversity of H. pylori populations have been conducted from major cities such as Hyderabad (Kumar et al., 2011; Ahmed et al., 2003) and Kolkata (Mukhopadhyay et al., 2000; Patra et al., 2012) and as well as in the remote Ladakh region of northern India (Kauser et al., 2005), with molecular-level characterization, similar research is lacking for geographically isolated and ethnically diverse regions such as Assam in Northeast India.
H. pylori strains exhibit distinct phylogeographic differentiation across various geographic regions. As a result, genetic comparisons of H. pylori can provide valuable insights into human migration and population history. In fact, several studies have already employed H. pylori as a genetic marker to trace the origins of various ethnic groups (Linz et al., 2007; Falush et al., 2003; Devi et al., 2007; Achtman et al., 1999; Wirth et al., 2004). Furthermore, exploring the genetic variation of H. pylori across different regions may help explain variations in disease outcomes, as specific bacterial genotypes may be linked to varying risks of gastroduodenal diseases.
Gastric cancer is among the most prevalent malignancies in Northeast India and represents a significant public health concern in the region, particularly in Assam, where incidence rates surpass the national average (Shanker et al., 2021). Given the established association between H. pylori infection and gastric carcinogenesis, and considering the high prevalence of H. pylori in India, this study was undertaken to investigate the association of H. pylori in the development of gastric cancer in Assam. Understanding this association is crucial for developing targeted prevention and treatment strategies in the region. Moreover, a population study of H. pylori isolates from this region of India has not been conducted before. Although previous studies have mapped H. pylori in other parts of India, we undertook this study to compare our findings with those earlier reports.
The present study aims to profile the H. pylori gene pool in Assam, Northeast India, through MLST analysis of seven housekeeping genes. This study seeks to establish H. pylori as a surrogate marker for human migration and demographic research while also evaluating its potential role on the occurrence of gastroduodenal diseases in this region.
2 Materials and methods
2.1 Study design
The present study was a hospital based cross-sectional study which was conducted in the Microbiology laboratory of a tertiary care hospital situated in Guwahati, Assam, Northeast India from January 2022 to December 2024.
2.2 Calculation of sample size
Sample size determination was based on the formula for calculation of single population proportion. The formula used for calculating the sample size was n = Z2 × P(1 − P)/d2, where n is the sample size, Z is the statistic corresponding to the level of confidence (CI), P is the expected prevalence and d is the margin of relative error. As previous data was not available to us, a 50% of population proportion was selected to calculate the sample size. Corresponding to previous year, the population size was 350 patients. Considering Z=1.96 (95% confidence interval) and a marginal error of 5% (d= 0.05), the initial sample size for our study was 184. Due to degradation of some samples, 10% was added to it making the final sample size of 202.4, rounding off to 200 samples for this study.
2.3 Research participants
The research participants consisting of 200 consecutive patients were enrolled in the endoscopy units of two of the health facilities in our region: State Cancer Institute, Guwahati, India and Department of Gastroenterology of Gauhati Medical College & Hospital (GMCH), Guwahati, Assam, India and were referred for routine upper gastro-intestinal tract endoscopy during our study period for suspected gastritis, duodenitis, peptic ulcer disease or gastric cancer. These patients suffered from dyspeptic symptoms such as vomiting, stomach ache, nausea, abdominal discomfort, black tarry stool, bloating of the stomach. During the study period, patients who were previously treated with antibiotics or proton pump inhibitors within one month prior to endoscopy and patients with medical conditions such as chronic liver disease or with upper gastro-intestinal bleeding or anaemia, were excluded from the study. Before sample collection, a pre-formed structured questionnaire was designed to collect the patients’ clinical history and epidemiological information. Written informed consent was obtained from every patient participating in the study.
Among the 200 patients, 137 (68.5%) were male, while 63 (31.5%) were female patients. The patients’ ages ranged from 18 to 90 years, with a mean age of 50.81 ± 2.1 years. All patients were divided into two groups on the basis of the type of pathological lesions visible during endoscopy: Group 1 included patients without pathology- specifically those with non-ulcer dyspepsia (NUD) and Group 2 included patients with pathology, including those with chronic gastritis (CG), gastric cancer (GC) and peptic ulcer disease (PUD), which is further sub-divided into gastric ulcer (GU) and duodenal ulcer (DU). Among them, 55 (27.5%) patients had NUD, 19 (9.5%) had CG, 100 (50%) patients had GC and remaining 26 (13%) patients had PUD, out of which 15 (7.5%) had GU and 13 (6.5%) had DU.
2.4 Clinical sample collection
During endoscopic procedures, four gastric biopsy samples were collected: Two from corpus and two from the antrum area of the stomach from each patient suffering from gastrointestinal symptoms. One set of samples (one from corpus and one from antrum) was fixed in formalin solution (10%) for histopathological analysis and the other set was placed in a sterile micro centrifuge tube with 80 μl phosphate buffered saline, transported to our laboratory immediately and stored at minus 20°C until DNA extraction and polymerase chain reaction (PCR) for H. pylori detection, determination of virulence and MLST analysis. In addition, blood samples were drawn from the same patients before endoscopic procedures and transported to the laboratory, where the serum was separated and tested for determination of anti-H. pylori IgG antibodies.
2.5 Histopathological analysis
For histopathological analysis, the biopsy specimens subjected to formalin fixation were processed according to standard procedures (Lash and Genta, 2013) and embedded in paraffin wax and sliced into 3 µm thick sections. Out of each block, two slides were prepared: Haematoxylin-eosin (H and E) staining for assessment of inflammation and associated morphological changes in the gastric mucosa and Giemsa staining for visualization and identification of H pylori. All the slides were reviewed and reported by a consultant pathologist.
2.6 Determination of anti-H. pylori IgG antibodies in serum
Anti-H. pylori IgG antibodies in the patients’ serum samples were detected using the Vidas H. pylori IgG assay kit (BioMerieux, Marcy-l’Etoile, France) following the manufacturer’s instructions. According to the kit instructions, a sample is considered as positive if the test value is ≥1 and negative if the test value is <0.75. Values falling within these cutoff ranges were interpreted as equivocal.
2.7 Genomic DNA extraction
Genomic DNA was isolated from the gastric biopsy samples using QiAmp DNA mini kit (Qiagen, Hilden, Germany) following the methodology as mentioned in the manufacturer’s manual. The extracted DNA was then stored at −20°C until further use for PCR and MLST analysis.
2.8 PCR for H. pylori detection and identification of virulence genes
To validate the presence of H. pylori in the patient samples, gastric biopsy samples were subjected to PCR of ureA, ureC and 16S rRNA genes. For the H. pylori-positive samples, the virulent genes cagA and vacA with alleles s1, m1, s2 and m2 were amplified by multiplex PCR. Gene specific PCR primer sets used for all genes were selected on the basis of previously published sequences (Supplementary Table S1). PCR reactions were carried out in a 25 μl reaction volume containing PCR buffer, 1.5mM of MgCl2, 200 μM dNTPs (each), 2U Taq polymerase enzyme (Sigma Chemicals Co., St. Louis, MO, USA), 10 μM each of forward and reverse primers and 15 ng of genomic DNA as template. PCR amplification was performed with an initial denaturation at 95°C for 3 minutes, followed by 35 cycles consisting of denaturation at 94°C for 1 minute, annealing for 1 minute (at 45°C for ureA and 55°C for ureC, 16S rRNA, cagA, and vacA genes), extension at 72°C for 1 minute, and a final extension at 72°C for 7 minutes. Genomic DNA extracted from the H. pylori ATCC 26695 strain served as a positive control while nuclease-free water was used as the negative control for all PCR assays. The amplified PCR products were examined through agarose gel electrophoresis using a 1.5% agarose gel containing ethidium bromide and a 100 bp DNA ladder. he resulting bands were visualized with the Gel Doc XR+ system. (Bio-Rad laboratories, Milan, Italy).
2.9 Multi locus sequence typing analysis
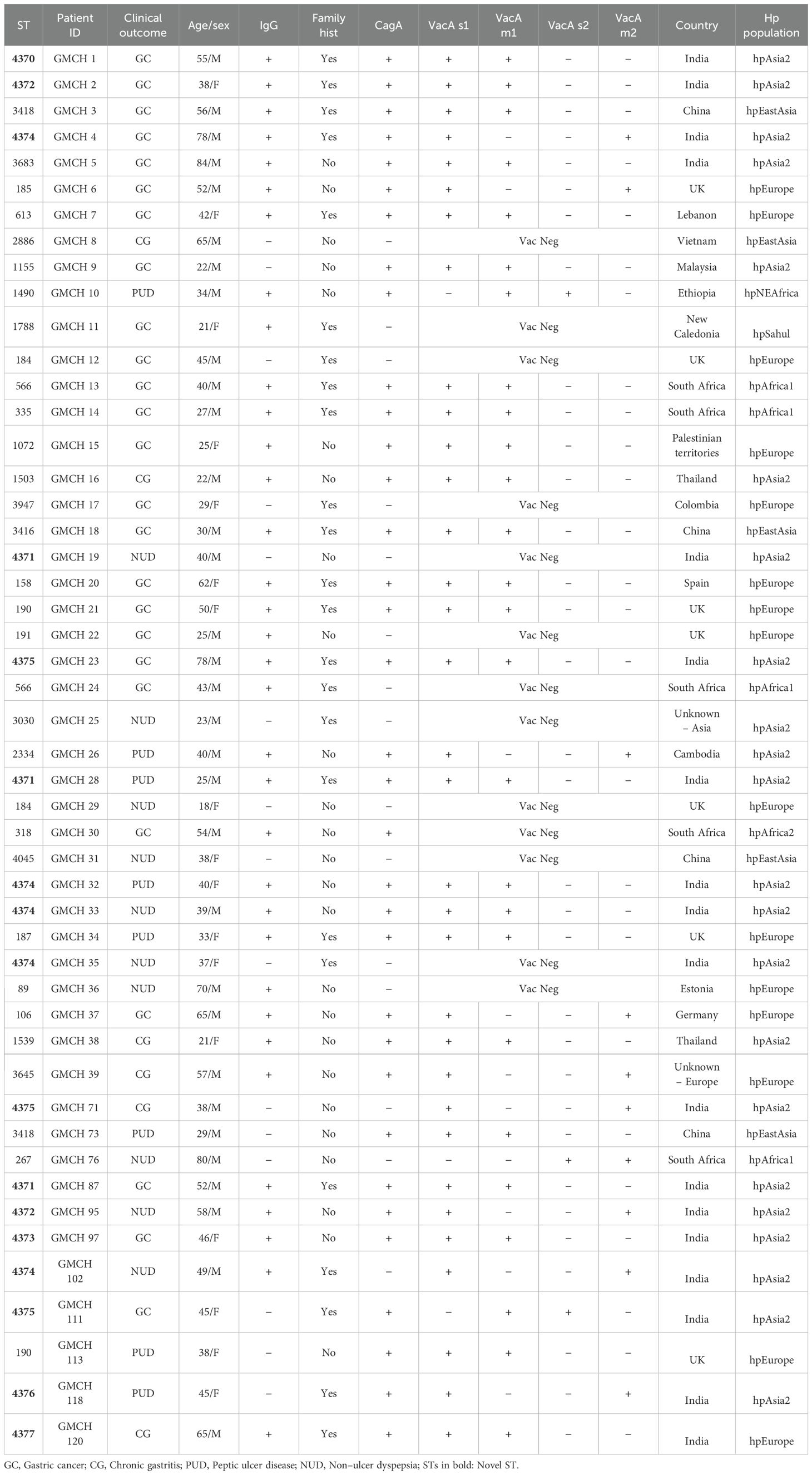
A total of 49 H. pylori positive cases were selected for MLST analysis on the basis of purposive sampling, which included 25 patients with GC, 6 patients with CG, 8 with PUD (GU: n=6, DU: n=2) and remaining 10 with NUD. The characteristics of the patients are shown in Table 1.
For MLST analysis, seven housekeeping genes of H. pylori: atpA, efp, mutY, ppa, trpC, ureI and yphC were amplified by PCR using the extracted DNA from each patient using gene-specific primers as given in Supplementary Table S1 as described on H. pylori PubMLST database (http://pubmlst.org/helicobacter/) (Jolley et al., 2018) and the PCR products were subjected to dideoxy nucleotide sequencing (Sanger sequencing method) using Genetic Analyzer 3500 (Applied Biosystems) in both directions as per the protocol detailed in PubMLST database. The 49 allele sequences were submitted to the sequence database for allele identification and sequence type (ST) assignment. Each unique sequence was assigned an allele number and STs were identified based on the combination of allele numbers of all the seven loci. DNA sequences that did not match existing PubMLST sequences were submitted to the database for new number assignment and sequence type.
2.10 Phylogenetic analysis
For phylogenetic analysis, partial DNA sequences of seven housekeeping genes were obtained from the 49 strains under study, concatenated end to end and aligned with the corresponding loci from 163 reference sequences downloaded from PubMLST database [http://pubmlst.org/helicobacter/] using MUSCLE in MEGA v11.0 (Tamura et al., 2021), comprising of the major bacterial populations of H. pylori: hpEurope, 22 sequences; hpEastAsia, 58 sequences; hpAfrica1, 10 sequences; hpAfrica2, 11 sequences; hspWestAfrica, 4 sequences; hpAsia2, 20 sequences; hpNEAfrica, 10 sequences; hpSahul, 10 sequences; hspMaori, 9 sequences and hspAmerind, 9 sequences. Additionally, concatenated sequences of 14 H. pylori strains obtained from our study belonging to hpAsia2 population were aligned with 103 South east Asian references strains comprising of Thailand, 34 sequences; Malaysia, 13 sequences and India, 56 sequences, deposited in PubMLST database [http://pubmlst.org/helicobacter/] to further elucidate the evolutionary relationship of the strains of our region. The phylogenetic trees were constructed using the Neighbour-joining method (Saitou and Nei, 1987) and Kimura 2-parameter model (Kimura, 1980) for nucleotide substitution in MEGA software v11.0. The evolutionary history of the analysed taxa was represented by the bootstrap consensus tree, inferred from 500 replicates.
2.11 Analysis of nucleotide diversity and polymorphism
The number of polymorphic sites among the house-keeping genes and the overall nucleotide diversity (π) were determined on the basis of allele sequences of the STs using the DnsSp programme v6.0. To assess the possibility of positive selection occurring on the house-keeping gene sequences, a codon-based Z-test of selection was conducted in MEGA v11.0 after aligning sequences by CLUSTAL W (codon-based) with the following criteria: Scope-Overall average; Test Hypothesis-Positive selection; variance estimation method: Bootstrap for 1000 replicates; Substitution type-Synonymous-Nonsynonymous; Model-Nei-Gojobori method; data subset-Pairwise deletion.
2.12 Nucleotide sequence accession numbers
The genome sequences of the 7 housekeeping genes for the 8 Indian strains from our study which is deposited in H. pylori PubMLST database: GMCH1, GMCH87, GMCH95, GMCH97, GMCH102, GMCH111, GMCH118 and GMCH120 have been submitted to NCBI GenBank with the following accession numbers: atpA: PV241101–PV241108, efp: PV241109–PV241116, mutY: PV241117–PV241124, ppa: PV241093–PV241100, trpC: PV241125–PV241132, ureI: PV241133–PV241140 and yphC: PV241141–PV241148.
2.13 Statistical analysis
The data was analyzed using the Epi-Info software (version 7.1.3; Atlanta, Georgia, USA). The Chi-square test and Fisher’s exact test were applied for evaluation, while a multivariate logistic regression model was employed for predicting the risk factors which were independently associated with prevalence of infection. A p-value of <0.05 was regarded as statistically significant.
3 Results
3.1 Demographic and clinical features of patients with dyspepsia
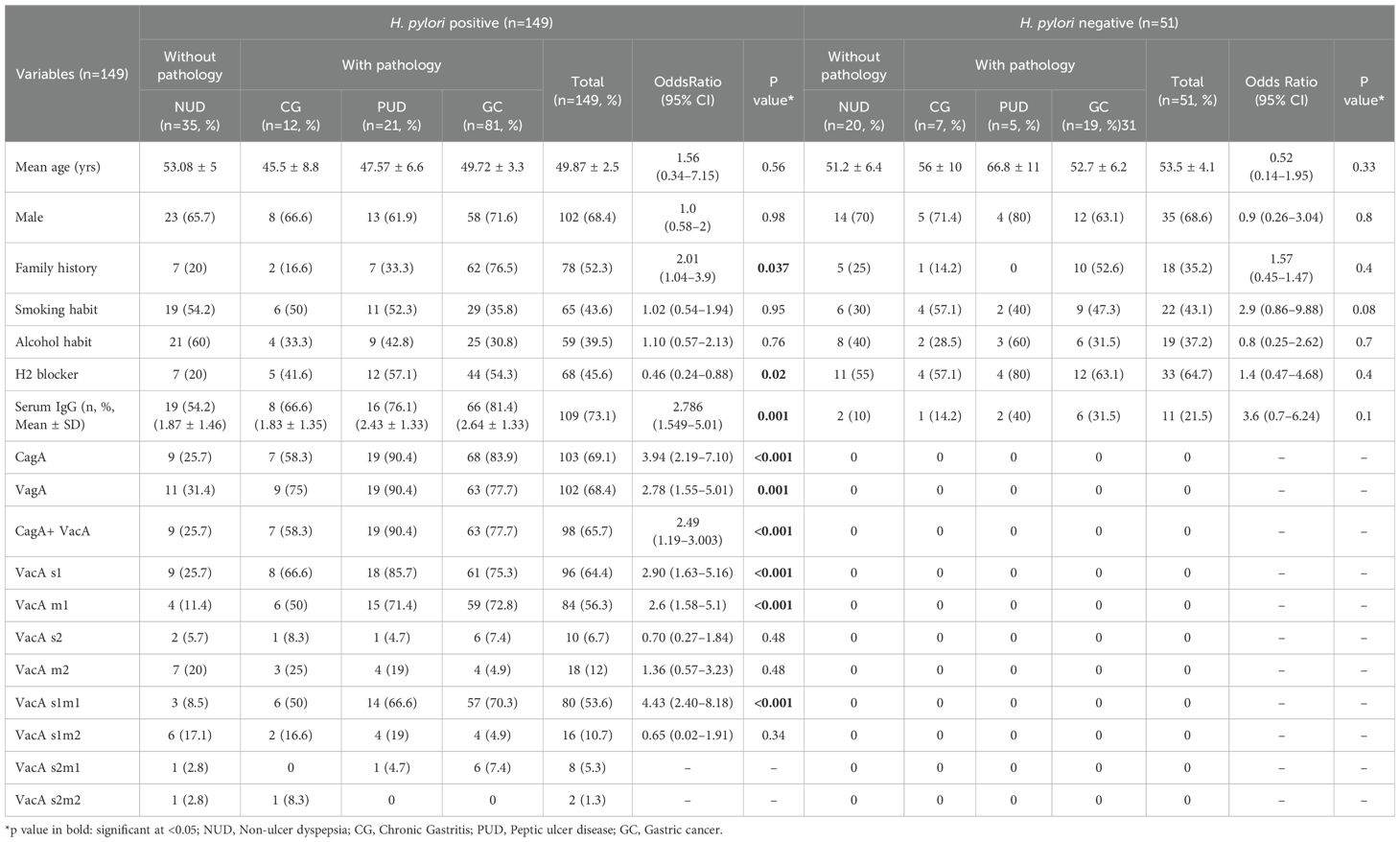
The overall positivity rate of H. pylori infection in our study by PCR of ureA, ureC and 16S rRNA was 74.5% (149/200), out of which 102 (68.4%) were male patients and 47 (31.5%) were female patients. Mean age of the infected patients was 49.87 ± 2.5 years. The male population showed an increase rate of H. pylori infection than females (102 vs 47). Out of 149 H. pylori positive samples, 35 (23.4%) were NUD cases, 12 (8%) were CG cases, 21 (14%) were PUD cases, out of which 13 (8.7%) were GU and 8 (5.3%) were DU patients and 81 (54.3%) were GC cases, out of which 63 (42.2%) were gastric adenocarcinoma cases, 12 (8%) were squamous cell carcinoma and remaining 5 (3.3%) were signet ring carcinoma cases (Table 2).
3.2 Distribution of virulent genotypes of H. pylori in the study population
Among the 149 H. pylori positive patients, overall prevalence of cagA, VacAs1m1, vacAs1m2, vacAs2m1, vacAs2m2 and cagA+vacA combined genotypes had a prevalence rate of 69.1% (103/149), 78.4% (80/149), 15.6% (16/149), 7.8% (8/149), 1.9% (2/149) and 65.7% (98/149) respectively (Table 2; Supplementary Figure S1).
3.3 Seroprevalence in H. pylori-positive patients
The percentage of H. pylori-positive patients who were seropositive for anti H. pylori IgG antibodies was 73.1% (109/149). Highest positivity of serum IgG antibodies against H. pylori was found in GC cases (66/149, 81.4%) with gastric adenocarcinoma cases exhibiting IgG positivity rate of 69.6% (46/66), followed by NUD (19/109, 17.4%), PUD (16/109, 14.6%) and CG (8/149, 5.3%).
3.4 Association between risk factors of H. pylori infection with gastro-intestinal pathology
Significant associations were detected between the pathological lesions (CG, PUD and GC) and several risk factors of H. pylori infection including serum IgG status, a family history of infection, H2 blocker use, H. pylori virulence factors cagA, vacA, cagA+vacA, vacAs1m1 and vacAs1m2 (p<0.05). The mean IgG antibody titre was significantly higher in GC (2.64 ± 1.33) and PUD (2.43 ± 1.33) compared to CG (1.83 ± 1.35) and NUD (1.87 ± 1.46), (p<0.05). Out of 81 GC cases, 62 (76.5%) had a family history of GC (Table 2).
The highest positivity of the cagA gene was found in PUD (19/21; 90.4%) and GC cases (68/81; 83.9%) and lowest in NUD (9/35; 25.7%) (p<0.001). The vacA gene positivity was highest in GC cases (63/81; 77.7%). The cagA+vacA gene and the vacA s1 gene was more prevalent in PUD cases (19/21; 90.4% and 18/21; 85.7%), whereas vacA m1 and vacA s1m1 was more prevalent in GC cases (59/81;72.8% and 57/81; 70.3%) than any other clinical lesion (p<0.001) (Table 2).
Multivariate logistic regression analysis identified risk factors that were independently associated with H. pylori infection included a family history of cancer (OR=2.014, 95% CI=1.043–3.89, p=0.037) and serum IgG antibody levels (OR= 2.786, 95% CI=1.549–5.01, p=0.001). Among the virulent genes, patients with vacA s1m1 (OR=4.438, 95% CI=2.408–8.178, p<0.001) and cagA (OR=3.946, 95% CI=2.193–7.103, p<0.001) were at a higher risk of H. pylori infection than patients with vacA s1 (OR=2.905, 95% CI=1.635–5.161, p<0.001) and combination of cagA+vacA genes (OR=2.49, 95% CI=1.19–3.003, p<0.001) (Table 2).
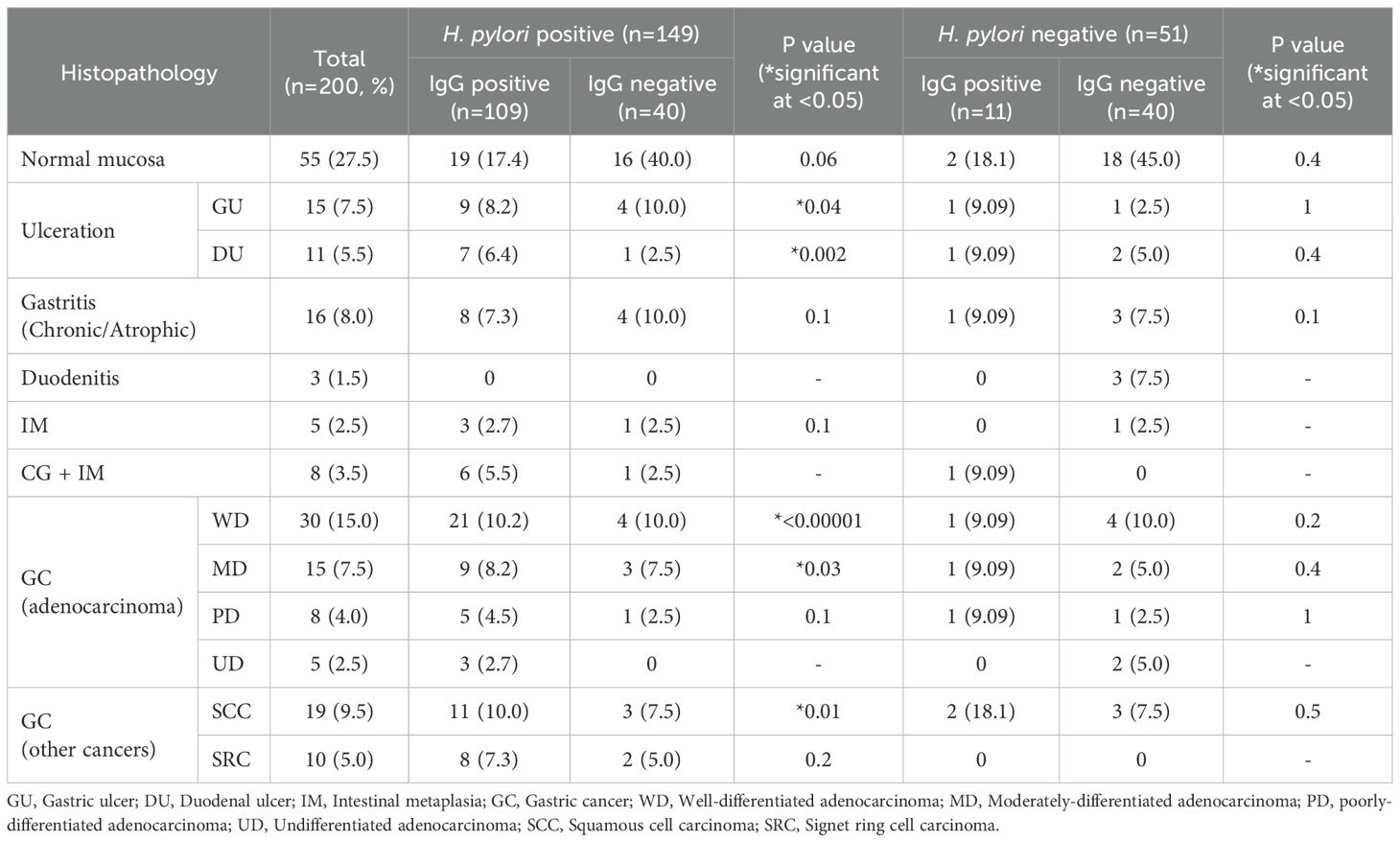
In determining the association between the histopathological variables and serum IgG status of the patients, patients with H. pylori infected peptic ulceration (gastric and duodenal ulcer), gastric adenocarcinoma (well-differentiated and moderately-differentiated) and squamous cell type carcinoma were significantly associated with serum IgG positivity status of the patients (p<0.05) (Table 3).
3.5 MLST and phylogenetic analysis of the 49 H. pylori strains
MLST analysis classified the 49 H. pylori strains from our study into 36 distinct sequence types (STs), out of which 28 were pre-defined STs and 8 were new STs (ST4370, ST4371, ST4372, ST4373, ST4374, ST4375, ST4376 and ST4377). Out of 49 strains, 34 (69.3%) were cagA positive and 36 strains (73.4%) were vacA positive. Majority of the strains were positive for serum IgG antibodies (34/49; 69.3%) (Table 1).
Out of the 36 STs, 8 STs were assigned to multiple clinical outcomes. ST4372 was assigned to one GC and one NUD case, ST3418 was assigned to one GC and one PUD case, ST4374 was assigned to one PUD, one GC and three NUD cases, ST184 was assigned to one GC and one NUD case, ST566 was assigned to two GC cases, ST4371 was assigned to one PUD, one NUD and one GC case, ST 190 was assigned to one PUD and one GC case and ST4375 was assigned to two GC cases and one case of CG (Table 1).
Phylogenetic analysis of the concatenated housekeeping genes based on MLST sequences of 49 H. pylori strains and 163 reference sequences deposited in PubMLST database revealed that all the strains were scattered in the MLST tree, irrespective of the gastric pathology. Out of the 36 STs derived from the 49 strains, 14 STs (38.8%) clustered within the hpAsia2 population, 12 (33.3%) clustered in the hpEurope population, 5 (13.8%) clustered in the hpAfrica population, 4 (11.1%) clustered within the hpEastAsia population and 1 ST (2.7%) clustered with hpSahul population, which indicated the geographical distribution of the diverse populations and sub-populations of H. pylori (Figure 1). All the new STs identified in this study (ST4370, ST4371, ST4372, ST4373, ST4374, ST4375, ST4376 and ST4377) clustered within the hpAsia2 population.

Figure 1. Phylogenetic relationship of the analysed STs from this study determined using the neighbour joining method conducted in MEGA v.11.0 and visualized in online software ITOL. The evolutionary distances were estimated using the Neighbour joining method and Kimura 2-parameter model and the scale bar denotes the number of nucleotide substitutions per site. The analysis included 36 STs from 49 analysed STs from this study and 163 reference sequences retrieved from the H. pylori PubMLST database [http://pubmlst.org/helicobacter/]. The positions and identification of the 36 STs identified in this study are shown in the tree. The major H. pylori populations are depicted in this tree according to the data available in the PubMLST.
The 14 STs clustering with the hpAsia2 population from our region (ST4370, ST4371, ST4372, ST4373, ST4374, ST4375, ST4376, ST4377, ST3683, ST1155, ST1503, ST3030, ST2334 and ST1539) were aligned with H. pylori strains from Southeast Asian countries including Thailand, Malaysia and India to construct a phylogenetic tree. The resulting analysis revealed the formation of 3 distinct clusters, on the basis of their genetic relatedness – Cluster 1 comprised strains from Thailand and Malaysia belonging to hspEastAsia population, Cluster 2 included strains from Ladakh, India, belonging to hpEurope population and Cluster 3 consisted of strains from Thailand, Malaysia and India classified under the hpAsia2 population. Notably, the 14 STs from our region exhibited genetic homology with Cluster 3 (Figure 2).
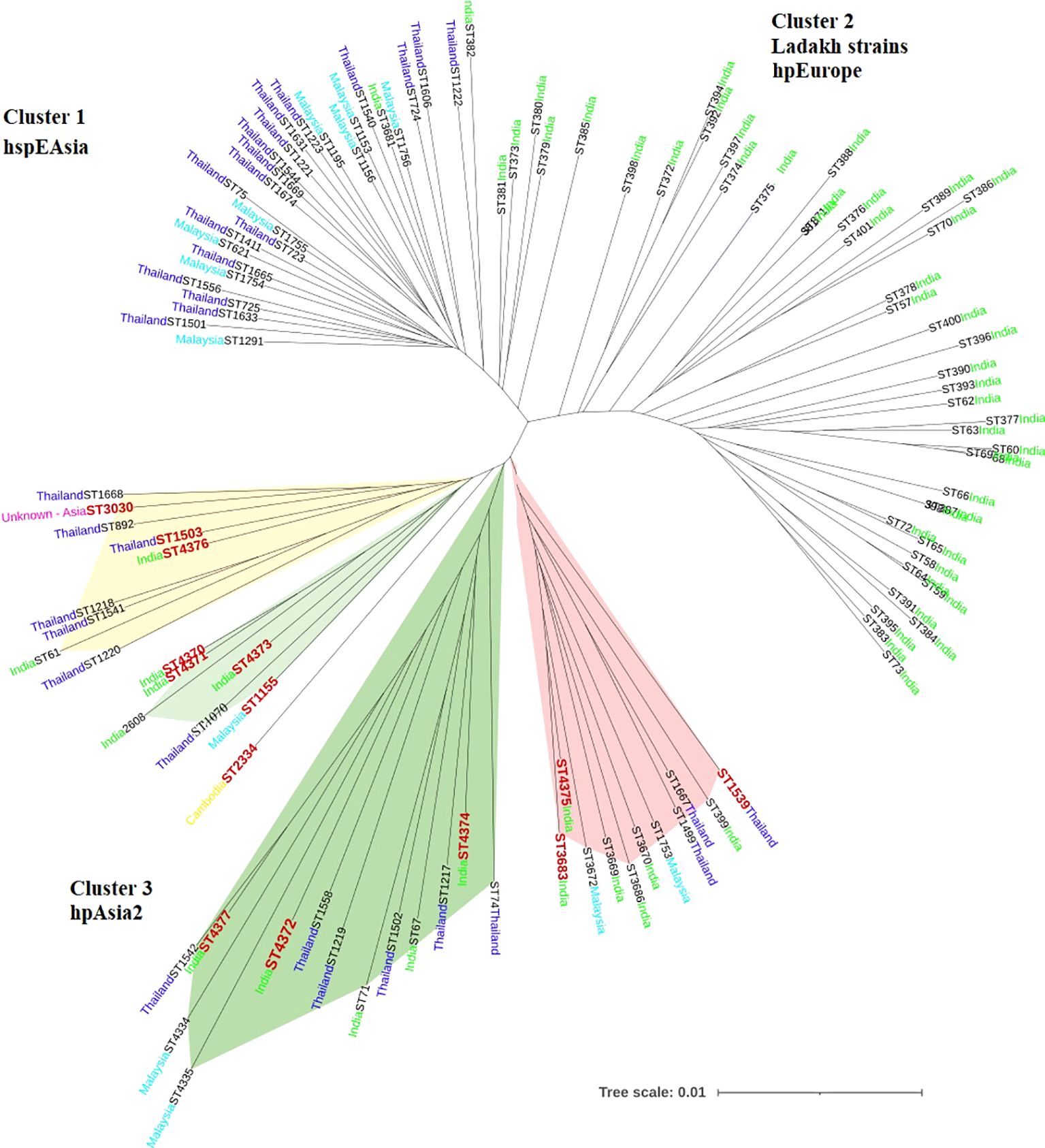
Figure 2. Phylogenetic tree showing the evolutionary relationship between H. pylori strains belonging to hpAsia2 from our region and 103 reference strains from Southeast Asia including Thailand, Malaysia and India, available from H. pylori PubMLST database [http://pubmlst.org/helicobacter/]. Sequence data from the seven house-keeping genes of the H. pylori strains were used to construct the tree. The STs depicted in red denote the strains from this study.
Additionally, H. pylori strains from Thailand were further categorized into 3 well-defined populations based on ethnicity- the hspEastAsian, the hpEurope and the indigenous hpAsia2 population of Thailand. All the 14 STs analysed in the present study clustered with the hpAsia2 population of Thailand, suggesting a strong genetic connection among the hpAsia2 strains from Assam, Northeast India and the hpAsia2 sub-population of Thailand (Figure 3).
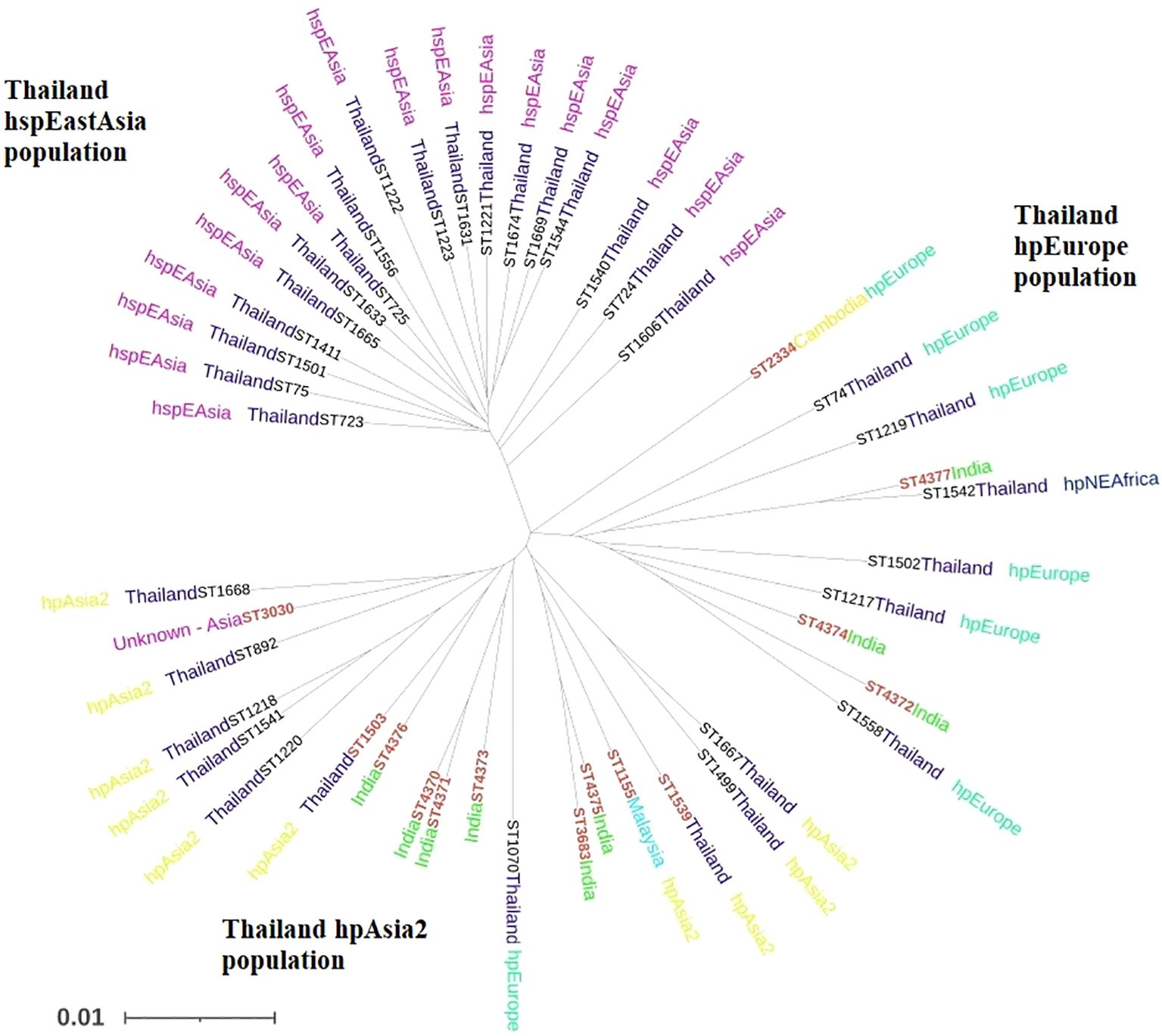
Figure 3. Phylogenetic tree showing the evolutionary relationship between H. pylori strains belonging to hpAsia2 from our region and 34 references strains from Thailand, Southeast Asia, available from H. pylori PubMLST database [http://pubmlst.org/helicobacter/]. Sequence data from the seven house-keeping genes of the H. pylori strains were used to construct the tree. The STs depicted in red denote the strains from this study.
3.6 Nucleotide diversity and polymorphism
The highest number of polymorphic sites were observed in trpC gene and the lowest in ureI gene. The order of such polymorphic sites from highest to lowest is trpC>mutY>yphC>efp>ppa> atpA>ureI. Singleton variable sites with two variants were observed in all the genes but none with three or four variants (Table 4).
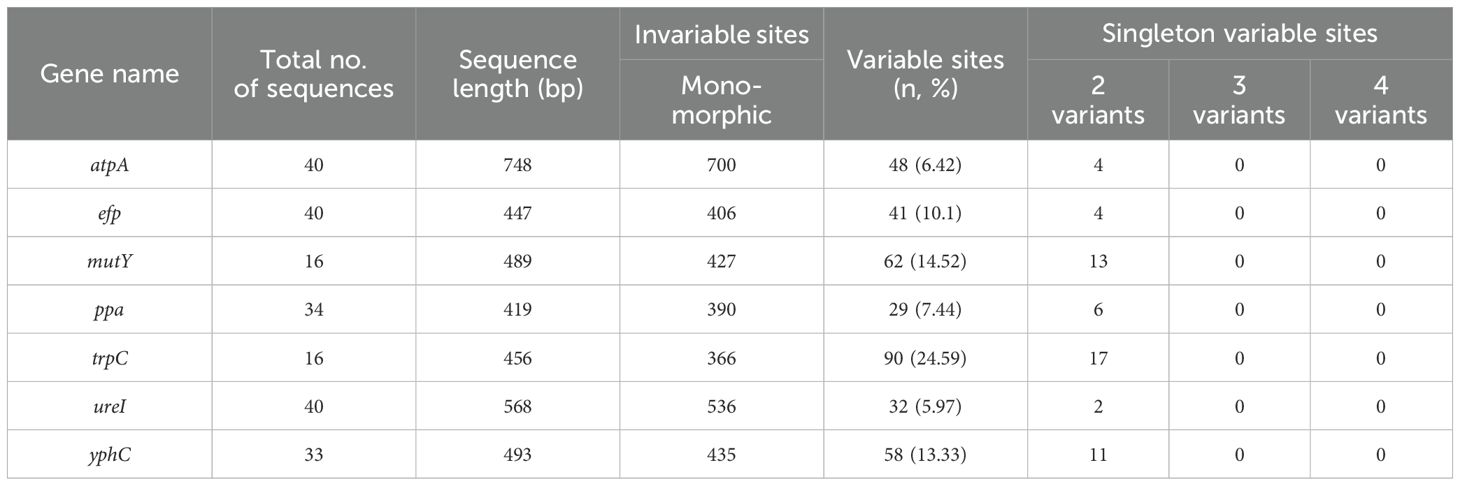
Table 4. Distribution of polymorphic sites in H. pylori housekeeping gene sequences available from the study.
The overall nucleotide diversity as determined by the DnaSP package was found to range from 0.01 (ppa) to 0.07 (trpC). The overall probability of the Z test ranged from 0 to 1 implying no positive selection of genes and purifying selection has been observed in 3 (atpA, mutY, ppa) out of 7 housekeeping genes (Table 5).
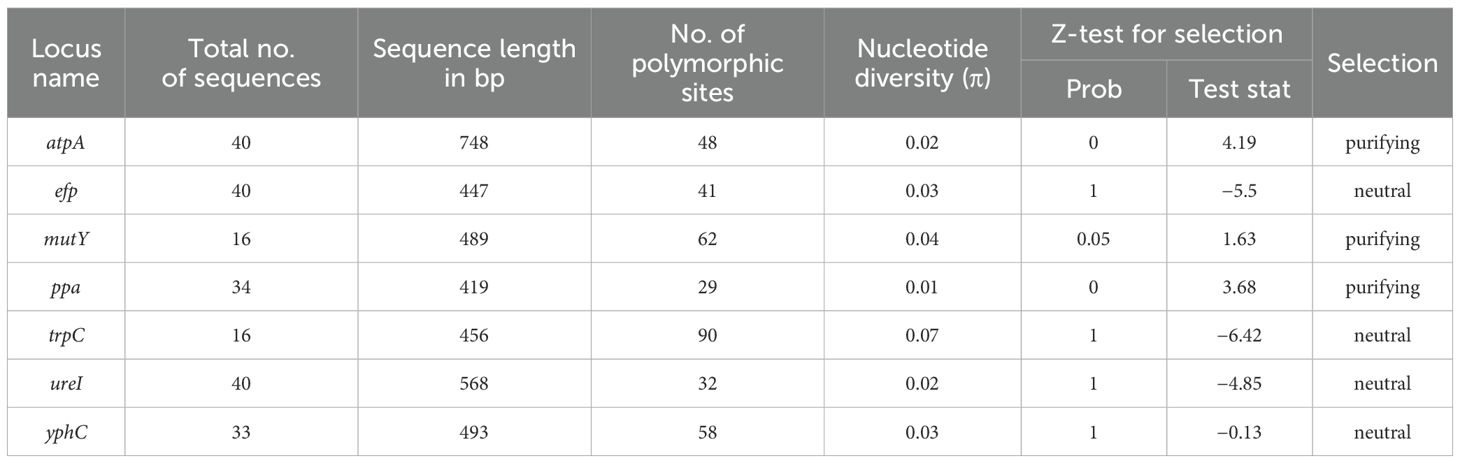
Table 5. Overall nucleotide diversity and codon-based positive selection of all the housekeeping gene sequences.
4 Discussion
In the present study, MLST was used to explore the occurrence of genetic diversity and evolutionary relationships among H. pylori strains in Assam, Northeast India to establish a possible link regarding the ancestral origin of the strains and correlate them with gastric outcome. Previous studies in India have used MLST tool to analyse the population structure and genetic variations among H. pylori strains to gain insight into the unique characteristics of the H. pylori lineages present within the Indian population and potentially link them to migration patterns or geographical variations (Devi et al., 2007; Kauser et al., 2005; Shetty et al., 2018).
Several studies have evaluated the genetic origins of H. pylori strains across different regions of India, revealing significant geographical diversity. In Varanasi (North India) and Hyderabad (South India), strains exhibit distinct genetic profiles, with Varanasi strains showing closer homology to Taiwanese strains and Hyderabad strains showing similarities to Brazilian strains (Kumar et al., 2011). In Ladakh, a high-altitude region in North India, H. pylori strains are genetically distinct, forming a subpopulation termed hpAsia2, which shares features with both Indo-European and East Asian gene pools (Kauser et al., 2005). Studies in West Bengal have shown that strains from tribal communities like the Santhals and Oraons possess virulence genes similar to those in mainstream Bengali populations, indicating gene flow between communities (Datta et al., 2003; Mukhopadhyay et al., 2000). Additionally, research from South India has identified strains belonging to both hpAsia2 and hpEurope populations, highlighting the complex evolutionary history of H. pylori in the region (Shetty et al., 2018). These findings underscore the importance of regional studies to understand the genetic diversity and evolution of H. pylori in India.
The northeastern region of India is geographically distinct from other parts of the country and is home to heterogeneous population groups consisting of more than 200 ethnic and tribal communities with around 80 tribes in Assam, each characterized by distinct cultural and ethnic diversity (Choudhury et al., 2017). Although a significant amount of genetic analyses studies on H. pylori genomes has been published from different parts of India, no comprehensive research has been specifically focused on the diversity of H. pylori strains and their associated clinical outcomes within Assam, North east India.
In the present study, a total of 49 strains out of 149 (25 patients with GC, 6 patients with CG, 8 with PUD and 10 with NUD) were subjected to MLST typing. A total of 36 STs were identified from 49 strains and among them, 8 were new STs. In the phylogenetic analysis, most strains from Assam clustered in two previously described population in India – hpEurope (n=12) and hpAsia2 (n=14) (Linz et al., 2007; Devi et al., 2007; Shetty et al., 2018).
However, phylogenetic reconstruction revealed that hpAsia2 strains from Assamese population are mostly clustered within native Thailand population (hpAsia2) commonly known as ‘Thais’. Northeast India, including Assam, has long been a hub for human migration, with many ethnic groups tracing their ancestry to Southeast Asia (Cordaux et al., 2004; Das, 2015; Tagore et al., 2022; Lusome and Bhagat, 2020). Internationally, Northeast India holds a strategic position, connecting South and Southeast Asia. The migration of Thai people to Assam primarily refers to the historical movement of the Ahom or Tai-Ahom tribe, an ethnic group of Assam, whose members were admixed descendants of the “Tai” people, who migrated from their ancestral homeland in Thailand to the Brahmaputra Valley in Assam, India via the Patkai mountains during the 12th century and established the historic Ahom dynasty that ruled Assam for nearly 600 years (Kumar et al., 2024; Sheikh, 2021). While the Ahoms are considered to be of Tai origin, which is associated with modern-day Thailand, their migration route is generally traced back to the Mong Mao region in southern China (present-day Yunnan province) (Sheikh, 2021). This suggests that H. pylori was likely introduced to Assam through these early migrants, thus supporting the idea of gene flow into India. However, strains also clustered within hpEastAsia (n=4), hpAfrica (n=5) and hpSahul (n=1) which confirms the presence of heterogeneity in H. pylori strains within the Assamese population.
Generally, the incidence of gastric cancer is closely associated with H. pylori groups classified through population analysis using MLST (Yamaoka et al., 2008). According to reports, H. pylori strains from hpEastAsia (Yamaoka et al., 2008; Zhu et al., 2024; Zaidi, 2016; Cho et al., 2010; Fock et al., 2008) and hpEurope strains (de Sablet et al., 2011) are linked to a higher risk of gastric cancer. Studies revealed that hpEurope strains were prevalent in high-risk stomach cancer regions, associated with advanced gastric lesions and increased DNA damage (de Sablet et al., 2011). However, in this study, we did not detect any variations in the phylogenetic distribution between various PUD and GC cases.
Although STs with a history of GC were mostly scattered within the hpAsia2 population, previous studies indicate that hpAsia2 strains are generally less virulent than East Asian strains (Misra et al., 2014). However, in the present study, 9 out of 14 hpAsia2 strains were associated with GC with some STs linked to both GC and other conditions such as PUD and NUD. This signifies that there is no specific clustering among H. pylori strains from PUD and GC cases present within hpAsia2 strains, which was consistent with findings from previous studies (Shiota et al., 2014).
Differences in the outcomes of H. pylori infection are shaped by individual host factors, such as family history, genetics and immune response, environmental and dietary influences, as well as differences in H. pylori strains (Misra et al., 2014; Yamaoka, 2010; Kao et al., 2016). However, several studies indicate that presence of virulent genes like cagA and vacA, particularly vacAs1m1 genotype strongly correlate with gastric pathology, primarily gastric cancer (Atherton et al., 1995; Atrisco-Morales et al., 2018; Carlosama-Rosero et al., 2022; Njenga et al., 2023; Mahant et al., 2025; Shetty et al., 2021; Chattopadhyay et al., 2002). Yet, in the present study, 6 out of 49 GC patients tested negative for the cagA and vacA genes. Among them, 4 patients had a family history of cancer, which signifies that beyond virulent genes, host genetic factors also play a predominant role in gastric pathology.
Epidemiological studies suggest that intra-familial transmission of infection, particularly among first-degree relatives, such as having a family member or sibling affected by cancer, is associated with an increased risk of gastrointestinal tract cancer (Song et al., 2018; Choi et al., 2022; Yaghoobi et al., 2010). In the present study, 62 out of 81 GC cases (76.5%) had a family history of GC, indicating a potential familial transmission of H. pylori strains. However, additional research is required to confirm whether the same strain is present among family members. The possibility of intrafamilial transmission is supported by existing research, which links a familial history of gastric cancer to an increased likelihood of transmission (Brenner et al., 2000; Bernini et al., 2006; Choi and Kim, 2016).
In addition to family history, elevated serum IgG antibody levels are also linked to an increased risk of H. pylori infection (Li et al., 2003; Tu et al., 2014). Therefore, future research should not only analyse H. pylori strains responsible for intrafamilial transmission but also assess IgG antibody levels. Since H. pylori infection often remains asymptomatic, detecting IgG levels could serve as an early predictive marker for gastric pathology, including cancer. By combining strain analysis, antibody profiling and longitudinal follow-ups, early diagnosis and intervention strategies for gastric cancer may be improved.
5 Conclusion
This study offers important insights into the genetic variation and evolutionary connections of H. pylori strains in Northeast India, suggesting that H. pylori was likely introduced to Assam through Tai-Ahom migrants from their ancestral homeland in Thailand, supporting the idea of gene flow into India, which may play a crucial role in shaping gastric disease patterns by influencing strain diversity, virulence and host interactions. Although our study did not reveal clustering of gastric outcomes, the findings suggest that patients with a familial history of gastric cancer and increased IgG antibody levels may be more vulnerable to the disease. Further research is needed to analyze both H. pylori strains and IgG levels to determine whether additional virulence factors are associated with gastric cancer. Moreover, IgG serves as a valuable predictor of gastric cancer risk. Therefore, regular monitoring of IgG levels could aid in the early detection and follow-up of high-risk individuals.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by the Institutional Ethics Committee, Gauhati Medical College & Hospital, Guwahati, Assam, India. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
AS: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Resources, Software, Validation, Writing – original draft, Writing – review & editing. DM: Writing – review & editing. MS: Investigation, Methodology, Resources, Writing – review & editing. LS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Resources, Supervision, Validation, Visualization, Writing – review & editing. BC: Investigation, Methodology, Resources, Writing – review & editing. MB: Investigation, Methodology, Resources, Writing – review & editing. VS: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was financially supported by the Department of Health Research, Ministry of Health and Family Welfare, Government of India under the HRD scheme “Fellowship Programme to Young Scientists” vide Sanction letter No.12014/32/2020-HR/E-Office:8047051, dated 07/08/2020.
Acknowledgments
The authors acknowledge the Department of Health Research, Ministry of Health and Family Welfare, Government of India, for providing financial support for the study. The authors are thankful to Dr. Nabanita Das (MD), Pathologist, Guwahati, India, for reviewing and reporting the histopathological analysis of our study and Dr. Kaushal Yadav (MSc, PhD) Dibrugarh, India, for his help with the Bioinformatics analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbrio.2025.1589230/full#supplementary-material
Supplementary Table S1 | PCR primers for amplification of UreA, UreC, 16S rRNA, CagA, VacA and MLST housekeeping genes.
Supplementary Figure S1 | PCR amplification of ureA, ureC, 16S rRNA, vacA (alleles: s1, s2, m1, m2) and the cagA gene of H. pylori. (a) Amplification of ureA gene at 411 bp. (b) Amplification of ureC gene at 294 bp. (c) Amplification of 16S rRNA gene at 522 bp. (d) Amplification of virulent genes vacA (alleles: s1, s2, m1, m2) and the cagA gene: Lane 1 – 100 bp DNA ladder. Lane 2 – positive control. Lanes 3, 4 and 5 – VacA s1m1-CagA genotype at 259 bp, 567 bp and 350 bp, respectively. Lane 6 – VacA s1m2-CagA genotype at 259 bp, 642 bp and 350 bp, respectively. Lane 7 – VacA s2m1-CagA genotype at 286 bp, 567 bp and 350 bp, respectively. Lane 8 – VacA s2m2-CagA genotype at 286 bp, 642 bp and 350 bp, respectively.
Abbreviations
MLST, Multi-locus sequence typing; PCR, Polymerase chain reaction; ureA, Urease A gene; ureC, Urease C gene; 16S rRNA, 16S Ribosomal RNA gene, cagA, Cytotoxin-associated gene A; vacA, Vacuolating cytotoxin A; ST, Sequence type; SD, Standard deviation; NUD, Non-ulcer dyspepsia; PUD, Peptic ulcer disease; CG, Chronic gastritis; GC, Gastric cancer; H and E, Haematoxylin eosin stain; dNTP, Deoxynucleotide Triphosphate; GMCH, Gauhati Medical College & Hospital; ATCC, American Type Culture Collection; atpA, gene encoding an ATP synthase subunit alpha; efp, gene encoding an elongation factor P; mutY, gene encoding a DNA glycosylase; ppa, gene encoding an inorganic diphosphatase protein; trpC, gene encoding an anthranilate isomerase; ureI, gene encoding a urease subunit I; yphC, gene encoding a GTP binding protein.
References
Achtman M., Azuma T., Berg D. E., Ito Y., Morelli G., Pan Z.-J., et al. (1999). Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32, 459–470. doi: 10.1046/j.1365-2958.1999.01382.x
Ahmed N., Khan A. A., Alvi A., Tiwari S., Jyothirmayee C. S., Kauser F., et al. (2003). Genomic analysis of Helicobacter pylori from Andhra Pradesh, South India: molecular evidence for three major genetic clusters. Curr. Sci. 85, 101–108.
Atherton J. C., Cao P., Peek R. M. Jr., Tummuru M. K., Blaser M. J., and Cover T. L. (1995). Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270, 17771–17777. doi: 10.1074/jbc.270.30.17771
Atrisco-Morales J., Martínez-Santos V. I., Román-Román A., Alarcón-Millán J., De Sampedro-Reyes J., Cruz-Del Carmen I., et al. (2018). vacA s1m1 genotype and cagA EPIYA-ABC pattern are predominant among Helicobacter pylori strains isolated from Mexican patients with chronic gastritis. J. Med. Microbiol. 67, 314–324. doi: 10.1099/jmm.0.000660
Bernini M., Barbi S., Roviello F., Scarpa A., Moore P., Pedrazzani C., et al. (2006). Family history of gastric cancer: a correlation between epidemiologic findings and clinical data. Gastric Cancer 9, 9–13. doi: 10.1007/s10120-005-0350-7
Blaser M. J. and Berg D. E. (2001). Helicobacter pylori genetic diversity and risk of human disease. J. Clin. Invest. 107, 767–773. doi: 10.1172/JCI12672
Brenner H., Bode G., and Boeing H. (2000). Helicobacter pylori infection among offspring of patients with stomach cancer. Gastroenterology 118, 31–35. doi: 10.1016/s0016-5085(00)70411-2
Burkitt M. D., Duckworth C. A., Williams J. M., and Pritchard D. M. (2017). Helicobacter pylori-induced gastric pathology: insights from in vivo and ex vivo models. Dis. Model Mech. 10, 89–104. doi: 10.1242/dmm.027649
Camilo V. and Henrique R. (2018). Oncogenic potential of CHAF1A in gastric cancer: A novel link with Helicobacter pylori-driven carcinogenesis? EBioMedicine 38, 3–4. doi: 10.1016/j.ebiom.2018.11.033
Carlosama-Rosero Y. H., Acosta-Astaiza C. P., Sierra-Torres C. H., and Bolaños-Bravo H. J. (2022). Helicobacter pylori genotypes associated with gastric cancer and dysplasia in Colombian patients. Rev. Gastroenterol. Mex. (Engl. Ed) 87, 181–187. doi: 10.1016/j.rgmxen.2021.09.003
Chattopadhyay S., Datta S., Chowdhury A., Chowdhury S., Mukhopadhyay A. K., Rajendran K., et al. (2002). Virulence genes in Helicobacter pylori strains from West Bengal residents with overt H. pylori-associated disease and healthy volunteers. J. Clin. Microbiol. 40, 2622–2625. doi: 10.1128/JCM.40.7.2622-2625.2002
Cho S. J., Choi I. J., Kim C. G., Lee J. Y., Kook M. C., Seong M. W., et al. (2010). Helicobacter pylori seropositivity is associated with gastric cancer regardless of tumor subtype in Korea. Gut Liver 4, 466–474. doi: 10.5009/gnl.2010.4.4.466
Choi H. G., Chun W., and Jung K. H. (2022). Association between gastric cancer and the family history of gastric cancer: a cross-sectional study using Korean Genome and Epidemiology Study data. Eur. J. Cancer Prev. 31, 408–414. doi: 10.1097/CEJ.0000000000000724
Choi Y. J. and Kim N. (2016). Gastric cancer and family history. Korean J. Intern Med. 31, 1042–1053. doi: 10.3904/kjim.2016.147
Choudhury S., Bahadur B., Krishnamurthy K. V., and Adams S. J. (2017). “Ethnic diversity of North-East India,” in Ethnobotany of India: Volume 3: North-East India and Andaman and Nicobar Islands. Eds. Pullaiah T., Krishnamurthy K. V., and Bahadur B. (Florida, USA: Apple Academic Press), 15–34.
Cordaux R., Weiss G., Saha N., and Stoneking M. (2004). The northeast Indian passageway: a barrier or corridor for human migrations? Mol. Biol. Evol. 21, 1525–1533. doi: 10.1093/molbev/msh151
Correa P. and Piazuelo M. B. (2012). Evolutionary history of the helicobacter pylori genome: implications for gastric carcinogenesis. Gut Liver 6, 21–28. doi: 10.5009/gnl.2012.6.1.21
Das S. D. (2015). Ethnic and cultural ties between northeast India and China: insights from the past. Int. Res. J. Soc. Sci. 4, 44–47.
Datta S., Chattopadhyay S., Balakrish Nair G., Mukhopadhyay A. K., Hembram J., Berg D. E., et al. (2003). Virulence genes and neutral DNA markers of Helicobacter pylori isolates from different ethnic communities of West Bengal, India. J. Clin. Microbiol. 41, 3737–3743. doi: 10.1128/JCM.41.8.3737-3743.2003
de Sablet T., Piazuelo M. B., Shaffer C. L., Schneider B. G., Asim M., Chaturvedi R., et al. (2011). Phylogeographic origin of Helicobacter pylori is a determinant of gastric cancer risk. Gut 60, 1189–1195. doi: 10.1136/gut.2010.234468
Devi S. M., Ahmed I., Francalacci P., Hussain M. A., Akhter Y., Alvi A., et al. (2007). Ancestral European roots of Helicobacter pylori in India. BMC Genomics 8, 184. doi: 10.1186/1471-2164-8-184
Falush D., Wirth T., Linz B., Pritchard J. K., Stephens M., Kidd M., et al. (2003). Traces of human migrations in Helicobacter pylori populations. Science 299, 1582–1585. doi: 10.1126/science.1080857
Fock K. M., Talley N., Moayyedi P., Hunt R., Azuma T., Sugano K., et al. (2008). Asia-Pacific consensus guidelines on gastric cancer prevention. J. Gastroenterol. Hepatol. 23, 351–365. doi: 10.1111/j.1440-1746.2008.05314.x
Hanafiah A. and Lopes B. S. (2020). Genetic diversity and virulence characteristics of Helicobacter pylori isolates in different human ethnic groups. Infect. Genet. Evol. 78, 104135. doi: 10.1016/j.meegid.2019.104135
Hunt R. H., Xiao S. D., Megraud F., Leon-Barua R., Bazzoli F., van der Merwe S., et al. (2011). Helicobacter pylori in developing countries: World Gastroenterology Organisation global guideline. J. Gastrointestin. Liver Dis. 20, 299–304.
Jolley K. A., Bray J. E., and Maiden M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124. doi: 10.12688/wellcomeopenres.14826.1
Kao C. Y., Sheu B. S., and Wu J. J. (2016). Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. BioMed. J. 39, 14–23. doi: 10.1016/j.bj.2015.06.002
Katelaris P. H., Tippett G. H., Norbu P., Lowe D. G., Brennan R., and Farthing M. J. (1992). Dyspepsia, Helicobacter pylori, and peptic ulcer in a randomly selected population in India. Gut 33, 1462–1466. doi: 10.1136/gut.33.11.1462
Kauser F., Hussain M. A., Ahmed I., Ahmad N., Habeeb A., Khan A. A., et al. (2005). Comparing genomes of Helicobacter pylori strains from the high-altitude desert of Ladakh, India. J. Clin. Microbiol. 43, 1538–1545. doi: 10.1128/JCM.43.4.1538-1545.2005
Kersulyte D., Mukhopadhyay A. K., Velapatiño B., Su W., Pan Z., Garcia C., et al. (2000). Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182, 3210–3218. doi: 10.1128/JB.182.11.3210-3218.2000
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kumar S., Kumar A., and Dixit V. K. (2011). Genetic diversity in strains of Helicobacter pylori from India and their relatedness to strains from other parts of the world. Infect. Genet. Evol. 11, 242–247. doi: 10.1016/j.meegid.2010.09.001
Kumar N., Mariappan V., Baddam R., Lankapalli A. K., Shaik S., Goh K. L., et al. (2015). Comparative genomic analysis of Helicobacter pylori from Malaysia identifies three distinct lineages suggestive of differential evolution. Nucleic Acids Res. 43, 324–335. doi: 10.1093/nar/gku1271
Kumar S., Singh P. P., Pasupuleti N., Tripathy V. M., Chauley M. K., Chaubey G., et al. (2024). The genetic admixture and assimilation of Ahom: a historic migrant from Thailand to India. Hum. Mol. Genet. 33, 1015–1019. doi: 10.1093/hmg/ddae054
Lash J. G. and Genta R. M. (2013). Adherence to the Sydney System guidelines increases the detection of Helicobacter gastritis and intestinal metaplasia in 400738 sets of gastric biopsies. Aliment Pharmacol. Ther. 38, 424–431. doi: 10.1111/apt.12383
Li S., Lu A. P., Zhang L., and Li Y. D. (2003). Anti-Helicobacter pylori immunoglobulin G (IgG) and IgA antibody responses and the value of clinical presentations in diagnosis of H. pylori infection in patients with precancerous lesions. World J. Gastroenterol. 9, 755–758. doi: 10.3748/wjg.v9.i4.755
Linz B., Balloux F., Moodley Y., Manica A., Liu H., Roumagnac P., et al. (2007). An African origin for the intimate association between humans and Helicobacter pylori. Nature 445, 915–918. doi: 10.1038/nature05562
Lusome R. and Bhagat R. B. (2020). Migration in northeast India: inflows, outflows and reverse flows during pandemic. Ind. J. Labour Econ. 63, 1125–1141. doi: 10.1007/s41027-020-00278-7
Mahant S., Singh S., Dutta S., Sharma N., Das P., Mukhopadhyay A. K., et al. (2024). High prevalence of cagA positive Vac A s1m1 Helicobacter pylori strains isolated from patients suffering from various gastroduodenal diseases in Guwahati, Assam, India. Indian J. Pathol. Microbiol. 68, 51–60. doi: 10.4103/ijpm.ijpm_1002_23
Misra V., Pandey R., Misra S. P., and Dwivedi M. (2014). Helicobacter pylori and gastric cancer: Indian enigma. World J. Gastroenterol. 20, 1503–1509. doi: 10.3748/wjg.v20.i6.1503
Mukhopadhyay A. K., Kersulyte D., Jeong J. Y., Datta S., Ito Y., Chowdhury A., et al. (2000). Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182, 3219–3227. doi: 10.1128/jb.182.11.3219-3227.2000
Njenga P., Njau A., Moloo Z., Revathi G., Tshibangu E., and Yamaoka Y. (2023). Pattern and trends of Helicobacter pylori genotypes in gastric cancer: A Kenyan 8-year study. Front. Med. (Lausanne) 10. doi: 10.3389/fmed.2023.1119513
Patra R., Chattopadhyay S., De R., Ghosh P., Ganguly M., Chowdhury A., et al. (2012). Multiple infection and microdiversity among Helicobacter pylori isolates in a single host in India. PloS One 7, e43370. doi: 10.1371/journal.pone.0043370
Rothenbacher D. and Brenner H. (2003). Burden of Helicobacter pylori and H. pylori-related diseases in developed countries: recent developments and future implications. Microbes Infect. 5, 693–703. doi: 10.1016/s1286-4579(03)00111-4
Saitou N. and Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sarma A., Borkakoti J., Sarma M. K., and Saikia L. (2024). Serum IgG Antibody against Helicobacter pylori and the CagA-VacA s1 Genotype as a Predictor of Gastric Cancer Development: A Case-control Study. J. Pure Appl. Microbiol. 18, 2336–2345. doi: 10.22207/JPAM.18.4.06
Sarma A., Saikia L., Bhuyan R. K., and Hussain E. (2017). Molecular identification and detection of virulent factors in Helicobacter pylori from gastric biopsy samples of patients attended at Assam Medical College and Hospital, Dibrugarh, Assam, India. Indian J. Med. Microbiol. 35, 600–603. doi: 10.4103/ijmm.IJMM_17_232
Sarma A., Saikia L., Gogoi M., and Yadav K. (2018). Molecular characterisation of virulent gene vacA in Helicobacter pylori clinical isolates from patients with gastroduodenal diseases in Assam, India. Indian J. Med. Microbiol. 36, 178–185. doi: 10.4103/ijmm.IJMM_18_67
Shanker N., Mathur P., Das P., Sathishkumar K., Martina Shalini A. J., and Chaturvedi M. (2021). Cancer scenario in North-East India & need for an appropriate research agenda. Indian J. Med. Res. 154, 27–35. doi: 10.4103/ijmr.IJMR_347_20
Sheikh M. R. A. (2021). The ahom migration to assam and the establishment of the Ahom kingdom. IJNRD - Int. J. Novel Res. Dev. 6, 58–65. Available at: https://ijnrd.org/papers/IJNRD2106008.pdf.
Shetty V., Lamichhane B., Chua E. G., Ballal M., and Tay C. Y. (2018). Draft genome sequences of 42 helicobacter pylori isolates from rural regions of south India. Genome Announc. 6, e01486–e01417. doi: 10.1128/genomeA.01486-17
Shetty V., Lingadakai R., Pai G. C., and Ballal M. (2021). Profile of Helicobacter pylori cagA &vacA genotypes and its association with the spectrum of gastroduodenal disease. Indian J. Med. Microbiol. 39, 495–499. doi: 10.1016/j.ijmmb.2021.06.001
Shiota S., Suzuki R., Matsuo Y., Miftahussurur M., Tran T. T., Binh T. T., et al. (2014). Helicobacter pylori from gastric cancer and duodenal ulcer show same phylogeographic origin in the Andean region in Colombia. PloS One 9, e105392. doi: 10.1371/journal.pone.0105392
Shirani M., Pakzad R., Haddadi M. H., Akrami S., Asadi A., Kazemian H., et al. (2023). The global prevalence of gastric cancer in Helicobacter pylori-infected individuals: a systematic review and meta-analysis. BMC Infect. Dis. 23, 543. doi: 10.1186/s12879-023-08504-5
Song M., Camargo M. C., Weinstein S. J., Best A. F., Männistö S., Albanes D., et al. (2018). Family history of cancer in first-degree relatives and risk of gastric cancer and its precursors in a Western population. Gastric Cancer 21, 729–737. doi: 10.1007/s10120-018-0807-0
Tagore D., Majumder P. P., Chatterjee A., and Basu A. (2022). Multiple migrations from East Asia led to linguistic transformation in NorthEast India and mainland Southeast Asia. Front. Genet. 13. doi: 10.3389/fgene.2022.1023870
Tamura K., Stecher G., and Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tu H., Sun L., Dong X., Gong Y., Xu Q., Jing J., et al. (2014). Serum anti-Helicobacter pylori immunoglobulin G titer correlates with grade of histological gastritis, mucosal bacterial density, and levels of serum biomarkers. Scand. J. Gastroenterol. 49, 259–266. doi: 10.3109/00365521.2013.869352
Wirth T., Wang X., Linz B., Novick R. P., Lum J. K., Blaser M., et al. (2004). Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: Lessons from Ladakh. Proc. Natl. Acad. Sci. U S A 101, 4746–4751. doi: 10.1073/pnas.0306629101
World Gastroenterology Organisation (2011). World Gastroenterology Organisation Global Guideline: Helicobacter pylori in developing countries. J. Clin. Gastroenterol. 45, 383–388. doi: 10.1097/MCG.0b013e31820fb8f6
Yaghoobi M., Bijarchi R., and Narod S. A. (2010). Family history and the risk of gastric cancer. Br. J. Cancer 102, 237–242. doi: 10.1038/sj.bjc.6605380
Yamaoka Y. (2009). Helicobacter pylori typing as a tool for tracking human migration. Clin. Microbiol. Infect. 15, 829–834. doi: 10.1111/j.1469-0691.2009.02967.x
Yamaoka Y. (2010). Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 7, 629–641. doi: 10.1038/nrgastro.2010.154
Yamaoka Y., Kato M., and Asaka M. (2008). Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 47, 1077–1083. doi: 10.2169/internalmedicine.47.0975
Zaidi S. F. (2016). Helicobacter pylori associated Asian enigma: Does diet deserve distinction? World J. Gastrointest. Oncol. 8, 341–350. doi: 10.4251/wjgo.v8.i4.341
Keywords: Helicobacter pylori, gastric cancer, genetic diversity, serum IgG, intrafamilial transmission, multi-locus sequence typing, phylogenetic relationship, virulence genes
Citation: Sarma A, Medhi D, Sarma MK, Saikia L, Choudhury BN, Bhattacharya M and Sarma V (2025) Helicobacter pylori genetic landscape in Northeast India and its impact in peptic ulcer disease and gastric cancer. Front. Bacteriol. 4:1589230. doi: 10.3389/fbrio.2025.1589230
Received: 07 March 2025; Accepted: 09 May 2025;
Published: 30 May 2025.
Edited by:
Darina Cejkova, Brno University of Technology, CzechiaReviewed by:
Padhmanand Sudhakar, Kumaraguru College of Technology, IndiaSukhithasri Vijayrajratnam, Washington University in St. Louis, United States
Copyright © 2025 Sarma, Medhi, Sarma, Saikia, Choudhury, Bhattacharya and Sarma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lahari Saikia, bGFoYXJpLnNhaWtpYUB5YWhvby5jb20=; Anisha Sarma, YW5pc2hhc2FybWExMUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Anisha Sarma
Anisha Sarma Devyashree Medhi2
Devyashree Medhi2 Lahari Saikia
Lahari Saikia