- 1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Harbin Medical University, Harbin, China
- 2Department of Neurobiology, Heilongjiang Provincial Key Laboratory of Neurobiology, Harbin Medical University, Harbin, China
- 3Department of Pathology, Harbin Medical University, Harbin, China
Long non-coding RNAs (lncRNAs) have been reported to interact with BRCA1/2 to regulate homologous recombination (HR) by diverse mechanisms in ovarian cancers (OvCa). However, genome-wide screening of BRCA1/2-related lncRNAs and their clinical significance is still unexplored. In this study, we constructed a global BRCA1/2-directed lncRNA-associated ceRNA network by integrating paired lncRNA expression profiles, miRNA expression profiles, and BRCA1/2 expression profiles in BRCA1/2 wild-type patients and identified 111 BRCA1/2-related lncRNAs. Using the stepwise regression and Cox regression analysis, we developed a BRCA1/2-directed lncRNA signature (BRCALncSig), composing of three lncRNAs (LINC01619, DLX6-AS1, and AC004943.2) from the list of 111 BRCA1/2-related lncRNAs, which was an independent prognostic factor and was able to classify the patients into high- and low-risk groups with significantly different survival in the training dataset (HR = 2.73, 95 CI 1.65–4.51, p < 0.001). The prognostic performance of the BRCALncSig was further validated in the testing dataset (HR = 1.9, 95 CI 1.21–2.99, p = 0.005) and entire TCGA dataset (HR = 2.17, 95 CI 1.56–3.01, p < 0.001). Furthermore, the BRCALncSig is associated with chemo-response and was also capable of discriminating nonequivalent outcomes for patients achieving complete response (CR) (log-rank p = 0.003). Functional analyses suggested that mRNAs co-expressed with the BRCALncSig were enriched in cancer-related or cell proliferation-related biological processes and pathways. In summary, our study highlighted the clinical implication of BRCA1/2-directed lncRNAs in the prognosis and treatment response of BRCA1/2 wild-type patients.
Introduction
Ovarian cancer (OvCa), one of the most common cancers in the genital system, is the fifth leading cause of cancer death in females. It is estimated that there are 22,530 newly diagnosed cases, and 13,980 deaths occurred in the United States according to the cancer statistics 2019 (Siegel et al., 2019). The standard treatment of advanced OvCa is surgical tumor debulking, followed by platinum/taxane-based chemotherapy. However, most patients after therapy will suffer disease relapse and face a poor outcome with the overall 5-year survival rate of approximately 30% due to the lower complete response (CR) rate of 40–60% to first-line chemotherapy (Cancer Genome Atlas Research and Network, 2011; Kim et al., 2012; Sun J. et al., 2019). Therefore, identifying potential molecular biomarkers to predict patients’ outcomes and chemotherapy response will be critical for guiding individualized treatment of patients with poor outcomes and drug resistance.
BRCA1 and BRCA2 (abbreviated as BRCA1/2) are well-known tumor suppressor genes and play essential roles in DNA repair and damage response mainly through homologous recombination (HR) (Powell and Kachnic, 2003; Yoshida and Miki, 2004). Therefore, BRCA1/2 mutation is associated with HR deficiency and genomic instability, leading to favorable outcomes for OvCa patients harboring BRCA1/2 mutation when subjected to platinum-based treatment compared with BRCA1/2 wild-type patients (McCabe et al., 2006; Lu et al., 2014; da Cunha Colombo Bonadio et al., 2018). However, BRCA1/2 are essential for normal tissue cells, and therefore, even though cancer patients without BRCA mutation possibly have a better outcome after platinum-based treatment compared with those with BRCA mutation. Long non-coding RNAs (lncRNAs) is a newly discovered class of non-coding RNAs (ncRNAs) and play crucial roles in a wide range of biological processes by fulfilling regulatory roles at the transcriptional, post-transcriptional or epigenetic levels (Kornienko et al., 2013; Mercer and Mattick, 2013). A large number of studies have suggested that the aberrant expression of lncRNAs contribute to the development and progression of human cancers, and can serve as novel biomarkers in cancer diagnosis, prognosis and treatment response (Zhou et al., 2015a,b, 2016a,b, 2017, 2018a,b,c). Recent studies have reported that lncRNAs can interact with BRCA1/2 to regulate HR by diverse mechanisms (Sharma et al., 2015; Wu and Wang, 2017; Su et al., 2018; Bao et al., 2019). For example, Sharma et al. (2015) found that lncRNA (DNA damage-sensitive RNA1) can regulate HR and DNA damage response by interacting with BRCA1 and hnRNPUL1. Another lncRNA PCAT-1 was reported to have a functional deficiency in HR through its repression of the BRCA2 (Prensner et al., 2014). It is well known that lncRNAs can act as competing for endogenous RNA (ceRNA) to communicate with mRNAs by competing for binding to shared microRNAs (miRNAs) (Liu et al., 2018). However, BRCA1/2-directed lncRNA-associated ceRNA network and their implication in prognosis and treatment response of BRCA1/2 wild-type patients remain mostly unknown.
In this study, we tried to construct and analyze a global BRCA1/2-directed lncRNA-associated ceRNA network by integrating paired lncRNA expression profiles, miRNA expression profiles, and BRCA1/2 expression profiles in BRCA1/2 wild-type patients, and identified potential BRCA1/2-directed lncRNA biomarkers for predicting prognosis and chemotherapy response of BRCA1/2 wild-type patients.
Materials and Methods
OV Patient Datasets
A total of 459 OvCa patients and their clinical information, miRNA expression profiles, lncRNA expression profiles, mRNA expression profiles, somatic mutation and copy number variation information of BRCA1 and BRCA2 were downloaded from UCSC Xena1. After examining BRCA1/2 mutation information, these OvCa patients included 217 patients with BRCA1/2 mutation and 242 patients with wild-type BRCA1/2, which were randomly divided into a training dataset (n = 121) and a testing dataset (n = 121).
Construction of BRCA1/2-Directed ceRNA Network
The high-quality experimentally validated miRNA-BRCA1/2 target association data was obtained from Tarbase (version 8.0)2 (Karagkouni et al., 2018), miRTarBase (version 8.0)3 (Huang et al., 2020) databases. The high-quality experimentally validated miRNA-lncRNA target association data was obtained from ENCORI database4 (Li et al., 2014). The BRCA1/2-directed ceRNA network was built according to the “ceRNA hypothesis” as follows: (?) Pearson correlation coefficient (PCC) was calculated between BRCA1/2 and each lncRNA, and those BRCA1/2-lncRNA pairs with positive correlation and P-value < 0.05 was selected for further analysis; (?) A BRCA1/2-lncRNA pair in which both BRCA1/2 and lncRNA are regulated by more than one same miRNAs was selected as candidate BRCA1/2-lncRNA ceRNA pairs; (?) If both BRCA1/2 and lncRNA in this candidate BRCA1/2-lncRNA ceRNA pair were co-expressed negatively with a same miRNAs, this BRCA1/2-miRNA-lncRNA was defined as BRCA1/2-directed ceRNA triples; (?)These BRCA1/2-directed ceRNA triples was integrated to form a global BRCA1/2-directed ceRNA network (BRCA-CeNet).
Development of a BRCA1/2-Directed lncRNA Signature
The univariate Cox regression analysis was used to evaluate the association between lncRNA in a BRCA1/2-directed ceRNA network and survival. Then a stepwise regression was used to identify an optimal lncRNAs combination according to Akaike information criterion (AIC) as prognostic lncRNAs biomarkers. Finally, optimal prognostic lncRNAs were fitted in a multivariate Cox regression analysis to evaluate their relative contribution to survival prediction. Finally, a BRCA1/2-directed lncRNA signature (BRCALncSig) was developed by the linear combination of the expression value of optimal prognostic lncRNAs with multivariate Cox regression coefficient as the weight (Sun et al., 2020). The BRCALncSig classified patient into the high-risk group and low-risk group using the median value of the training dataset as risk cutoff.
Statistical Analysis
Kaplan–Meier survival curves and the log-rank test were used to compare survival differences between predicted high-risk group and low-risk group using the R package “survival.” Univariate and multivariate analyses were performed using Cox proportional hazards regression model. Chi-squared tests are used to test for differences between two groups. Functional enrichment analysis for GO and KEGG was performed using the R package “clusterProfiler” (Yu et al., 2012).
Results
Construction and Analysis of BRCA1/2-Directed ceRNA Network
We first performed an integrated analysis for paired BRCA1/2 expression profiles, miRNA expression profiles and lncRNA expression profiles of 242 patients with wild-type BRCA1/2 and experimentally validated target relationship among BRCA1/2, miRNA and lncRNAs. As described in the “Materials and Methods” section, we identified a total of 83 miRNA-directed BRCA1-lncRNA ceRNA triples (including 70 lncRNAs and three miRNAs) and 66 miRNA-directed BRCA2-lncRNA ceRNA triples (49 lncRNAs and three miRNAs). Then these miRNA-directed BRCA1/2-lncRNA ceRNA triples were integrated to build a global BRCA1/2-directed ceRNA network (BRCA-CeNet). The constructed BRCA-CeNet contained 119 nodes (including BRCA1/2, 111 lncRNAs, and six miRNAs) and 250 edges (Figure 1 and Supplementary Table S1).
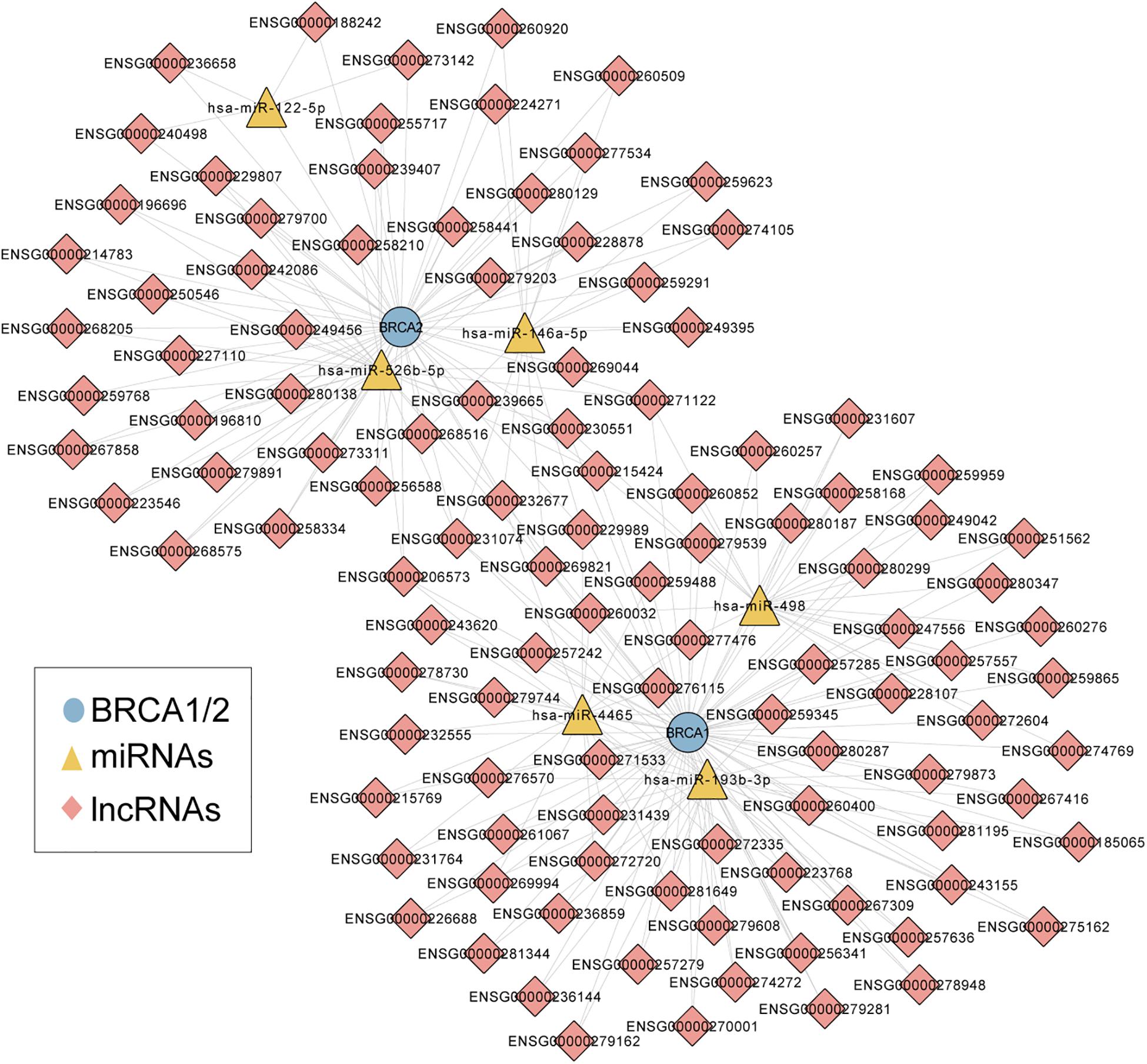
Figure 1. A global view of BRCA1/2-directed ceRNA networks comprised of 119 nodes (including BRCA1/2, 111 lncRNAs, and six miRNAs) and 250 edges.
Development and Validation of a BRCA1/2-Directed lncRNA Signature
We performed univariate Cox regression analysis to evaluate the association between lncRNAs in a BRCA1/2-directed ceRNA network and survival in the training dataset and identified five BRCA1/2-directed lncRNAs (LINC01619, DLX6-AS1, AC016747.4, AC027290.3, and AC004943.2) which are significantly associated with survival. Then we performed stepwise regression analysis for these five BRCA1/2-directed prognostic lncRNAs and identified an optimal combination of three lncRNAs (LINC01619, DLX6-AS1, and AC004943.2) according to AIC (Table 1). Three optimal prognostic lncRNAs were fitted in a multivariate Cox regression analysis to evaluate their relative contribution to survival prediction. Finally, a BRCA1/2-directed lncRNA signature (BRCALncSig) comprised of three lncRNAs were constructed as follows:BRCALncSig = (−0.571*EXLINC01619) + (−0.26*EXDLX6−AS1) + (−0.284*EXAC004943.2).
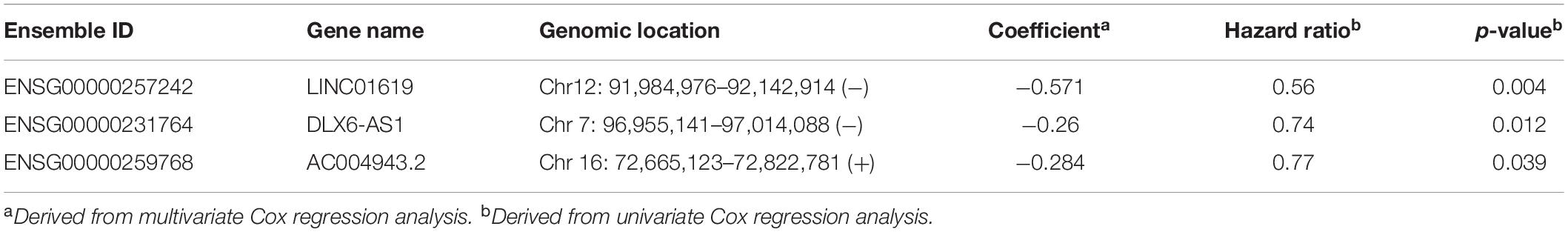
Table 1. The detailed information of three prognostic lncRNAs in the BRCA1/2-directed ceRNA network.
This BRCALncSig could stratify 121 patients of training dataset into two risk groups using the median value (0.175) as risk cutoff. As shown in Figure 2A, patients with high BRCALncSig tend to be high-risk, while patients with low BRCALncSig are low-risk. High-risk patients have significantly poor overall survival and disease-free survival compared with those with low-risk (log-rank p < 0.001 and p = 0.024) (Figures 2A,B). The median overall and disease-free survival time of patients in the high-risk group was 1102 days and 450 days, whereas the corresponding median survival time in the low-risk group was 1736 days and 616 days. The overall and disease-free survival rate of patients at 5 years in the high-risk group was 12% and 0%, respectively, which is significantly lower than those (45% and 14%) patients in the low-risk group. The expression pattern of BRCALncSig, scores distribution and survival time of 121 patients of training dataset were shown in Figure 2C. We found that three prognostic factors are protective factors that were significantly highly expressed in low-risk patients compared with high-risk patients (Figure 2C and Supplementary Figure S1).
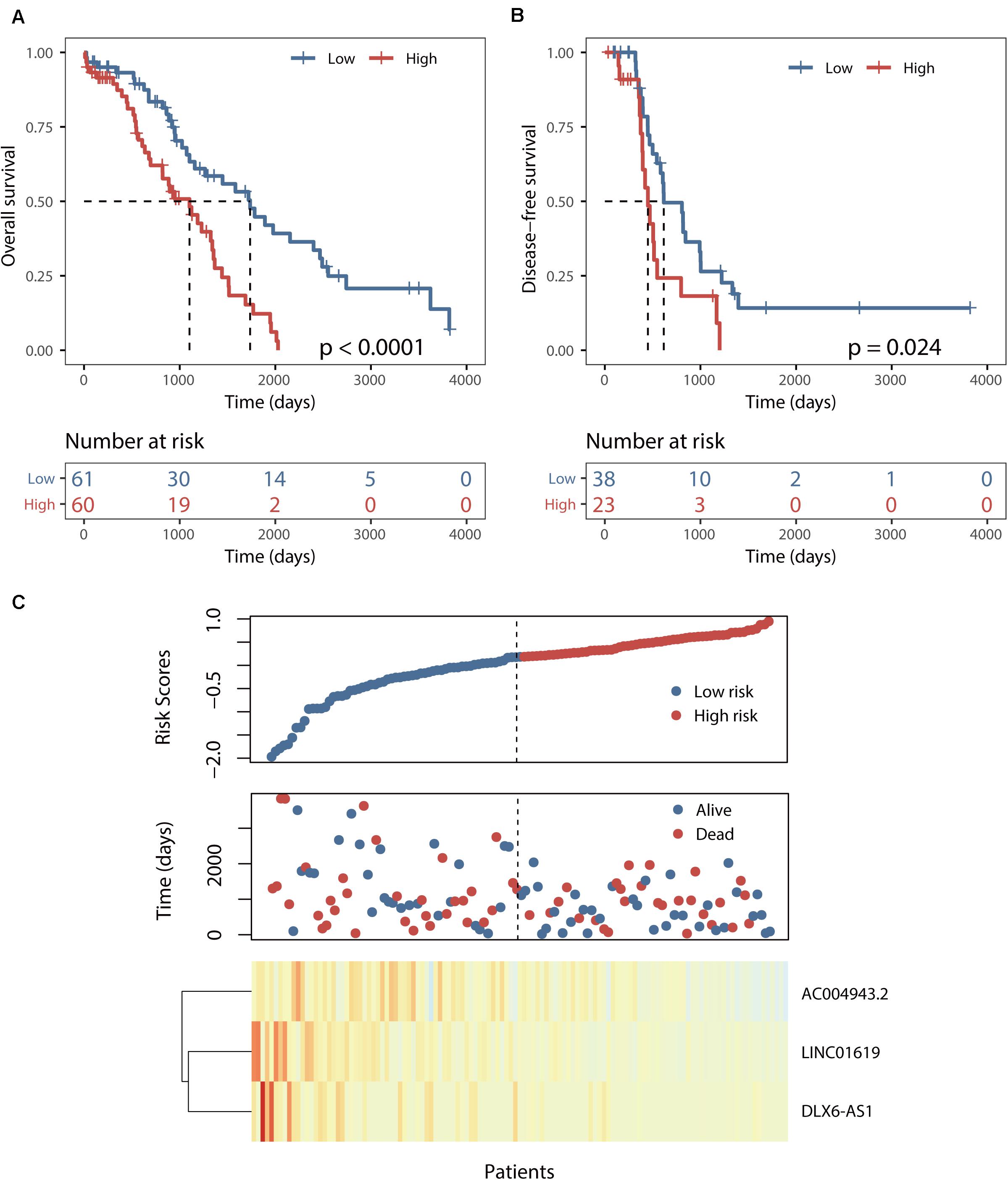
Figure 2. Identification of a BRCA1/2-directed lncRNA signature (BRCALncSig) in the training set. (A) Kaplan–Meier survival curves of overall survival between high-risk and low-risk patients. (B) Kaplan–Meier survival curves of disease-free survival between high-risk and low-risk patients. (C) Risk scores distribution and expression pattern of the BRCALncSig.
Further Validation of the BRCALncSig in the Independent Testing and TCGA Datasets
The predictive power of the BRCALncSig was further validated in another completely independent testing dataset. Using the same score model and risk cutoff derived from the training dataset without parameters re-estimation, all 121 patients of the testing dataset were divided into the high-risk group and low-risk group with significantly different overall survival (log-rank p = 0.005, Figure 3A) and disease-free survival (log-rank p = 0.001, Figure 3B). As shown in Figures 3A,B, the median overall and disease-free survival time of patients in the high-risk group was 1069 days and 422 days, which is significantly lower than that (1562 and 977 days) of patients in the low-risk group. The overall and disease-free survival rate of patients at 5 years in the high-risk group was 16% and 0%, respectively, whereas the corresponding rates are 35% and 24%, respectively. Similar results were observed for the TCGA dataset, with the median overall and disease-free survival time of patients in the high-risk group was 1069 days and 426 days, which is significantly lower than that (1620 and 817 days) of patients in the low-risk group. The overall and disease-free survival rate of patients at 5 years in the high-risk group was 14% and 0%, respectively, whereas the corresponding rates are 40% and 19% in the low-risk group, respectively (Figures 3C,D).
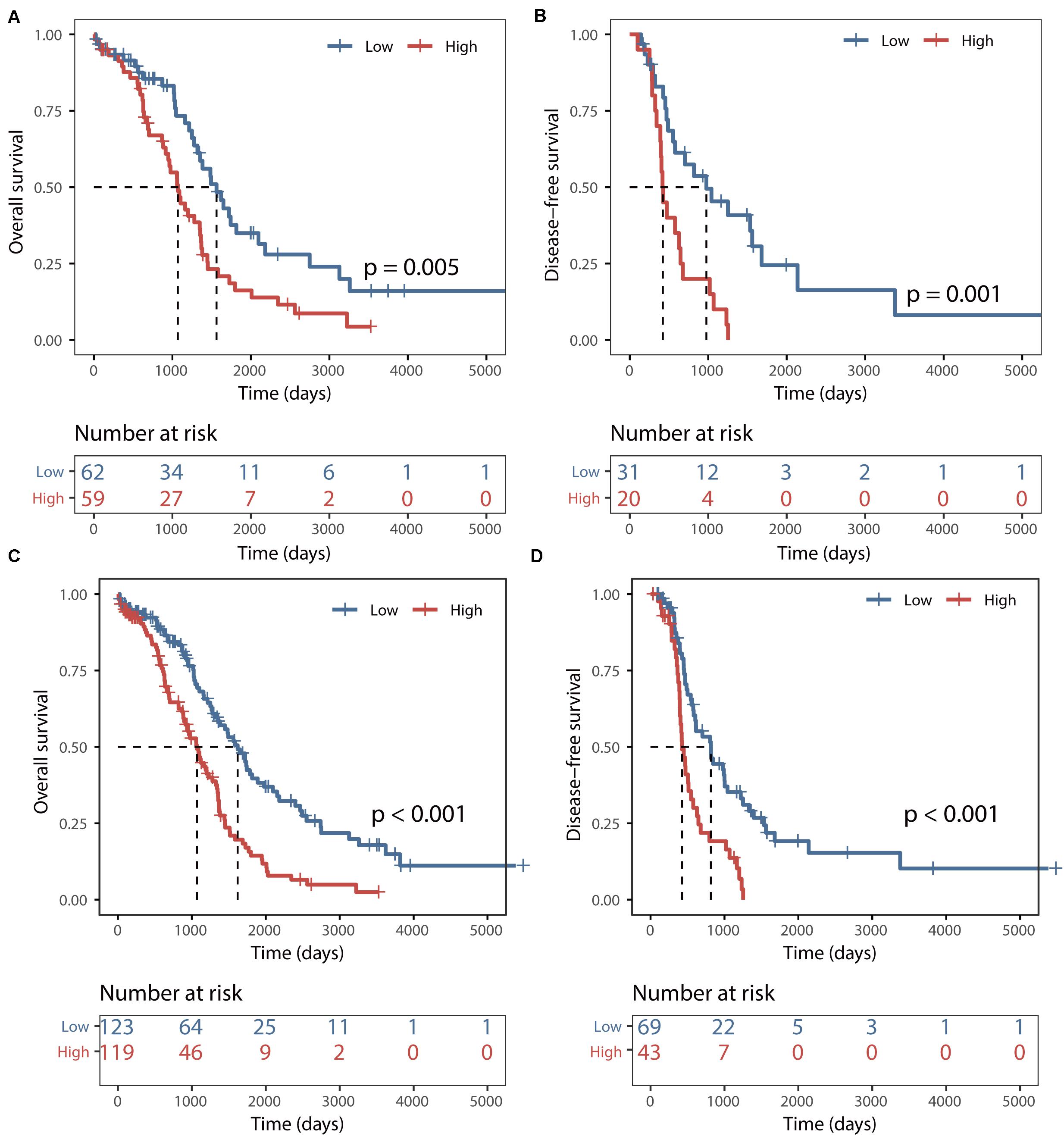
Figure 3. Validation of the BRCALncSig. Kaplan–Meier survival curves of overall survival between high-risk and low-risk patients in the testing set (A) and entire TCGA set (C). Kaplan–Meier survival curves of disease-free survival between high-risk and low-risk patients in the testing set (B) and entire TCGA set (D).
Prognostic Differences Among Patients With BRCA1/2 Mutation, BRCALncSig-Related High-Risk Patients and BRCALncSig-Related Low-Risk Patients
All 459 patients of the TCGA dataset were grouped into three categories: the BRCA1/2 mutation group (n = 217), the BRCALncSig-related high-risk wild-type group (n = 119) and the BRCALncSig-related low-risk wild-type group (n = 123). We found that there is a significant difference in survival time among these three patient groups (log-rank p < 0.001) (Figure 4A). The pairwise comparison among three patient groups showed that patients with BRCA1/2 mutation and patients in the BRCALncSig-related low-risk wild-type group both have significantly longer survival than patients in the BRCALncSig-related high-risk wild-type group (log-rank p < 0.001) (Figure 4C). The median survival time of patients with BRCA1/2 mutation and patients in the BRCALncSig-related low-risk wild-type group is 1579 days and 1620 days, respectively, while corresponding median survival time in the BRCALncSig-related high-risk wild-type group is 1069 days. However, patients with BRCA1/2 mutation showed no significant difference in survival from patients in the BRCALncSig-related low-risk wild-type group (Log-rank p = 0.95, median survival time 1620 vs. 1579 days; Figure 4B).
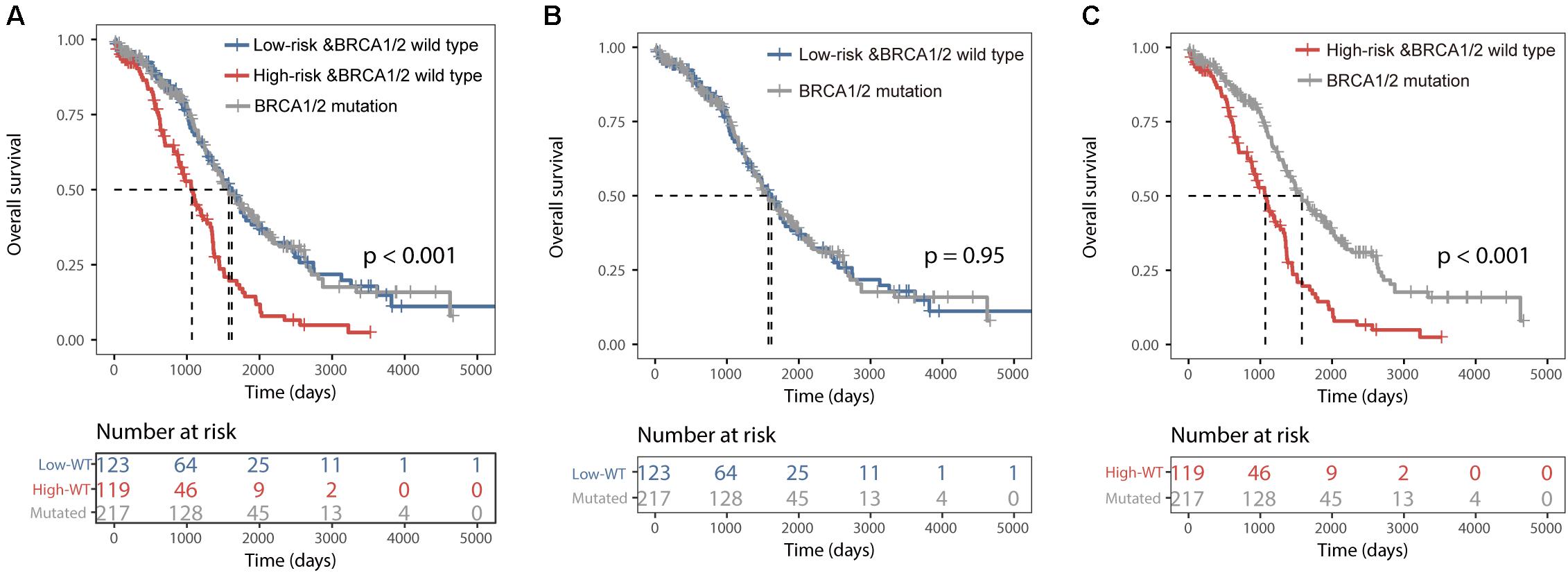
Figure 4. Prognostic differences among three group patients. (A) Kaplan–Meier survival curves of overall survival among BRCA1/2 mutation group, the BRCALncSig-related high-risk wild-type group and the BRCALncSig-related low-risk wild-type group. (B) Kaplan–Meier survival curves of overall survival between BRCA1/2 mutation group and BRCALncSig-related low-risk wild-type group. (C) Kaplan–Meier survival curves of overall survival between the BRCA1/2 mutation group and the BRCALncSig-related high-risk wild-type group.
Correlation Between the BRCALncSig and Chemo-Response
To examine whether the BRCALncSig correlated with the likelihood of CR, we plotted the percentage of patients achieving CR against the BRCALncSig and found that there is a marginally significant negative correlation between the BRCALncSig and CR (Pearson correlation coefficient r = 0.55, p = 0.097) (Figure 5A). Patients in the low-risk groups tended to have a higher likelihood of CR compared with those in the high-risk groups. In detail, 70% of patients in the low-risk group achieved CR to a platinum and taxane regimen, whereas the corresponding rate is 57% in the high-risk group (p = 0.078, Chi-squared test) (Figure 5B). Furthermore, the BRCALncSig was also capable of discriminating nonequivalent outcomes for all patients achieving CR. As shown in Figure 5C, patients in the high-risk group have significantly shorter survival time (median 1354 days) than that of patients in the low-risk group (median 1736 days) (log-rank p = 0.003; Figure 5C) even if these patients all received CR to a platinum and taxane regimen.
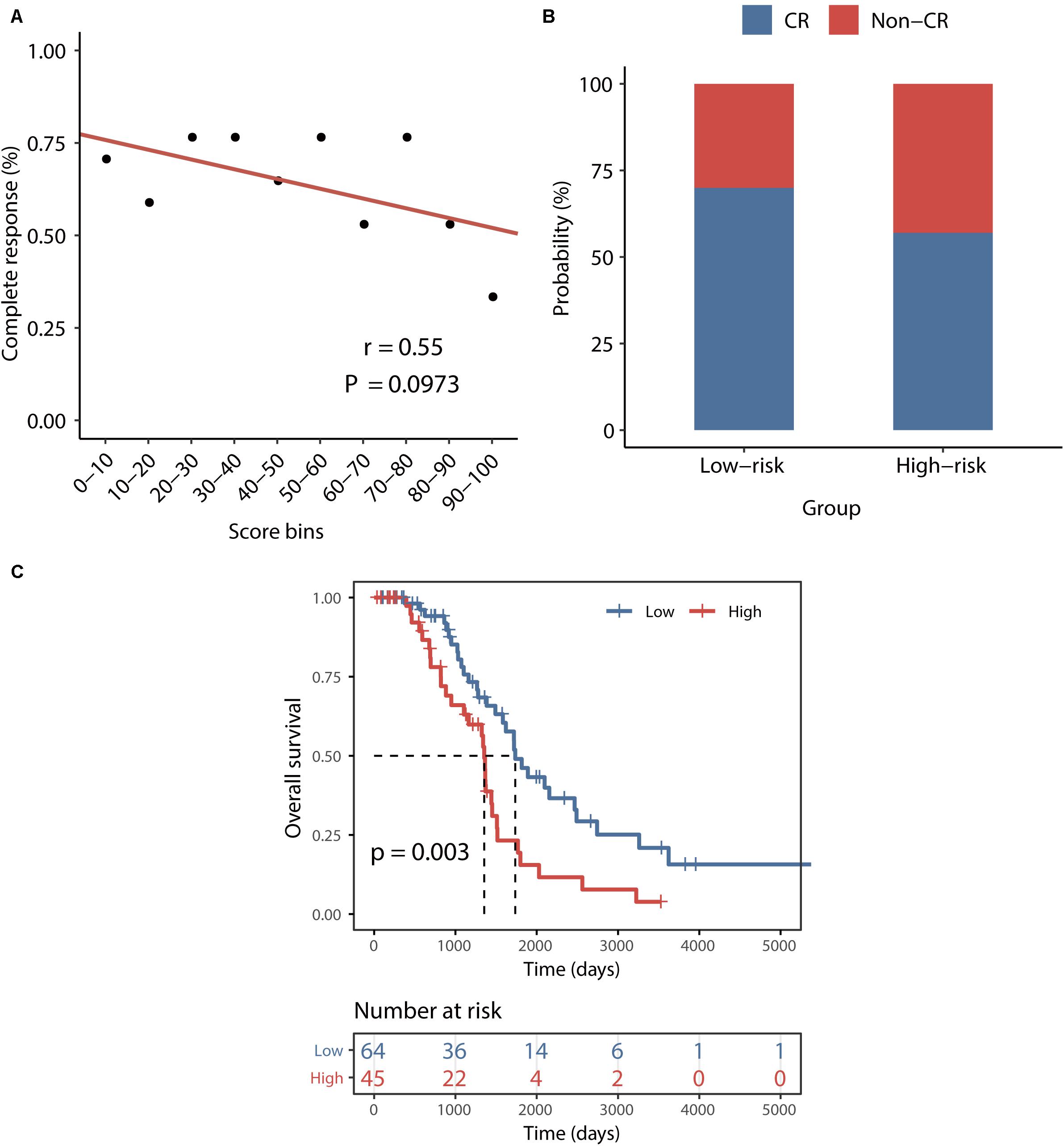
Figure 5. Association between the BRCALncSig and chemo-response. (A) The relationship between the BRCALncSig and the likelihood of complete response. (B) Differences in complete response ratios between the high-risk group and low-risk group. (C) Kaplan–Meier survival curves of overall survival between high-risk and low-risk patients for patients with CR.
Independence of the BRCALncSig From Other Clinicopathological Factors
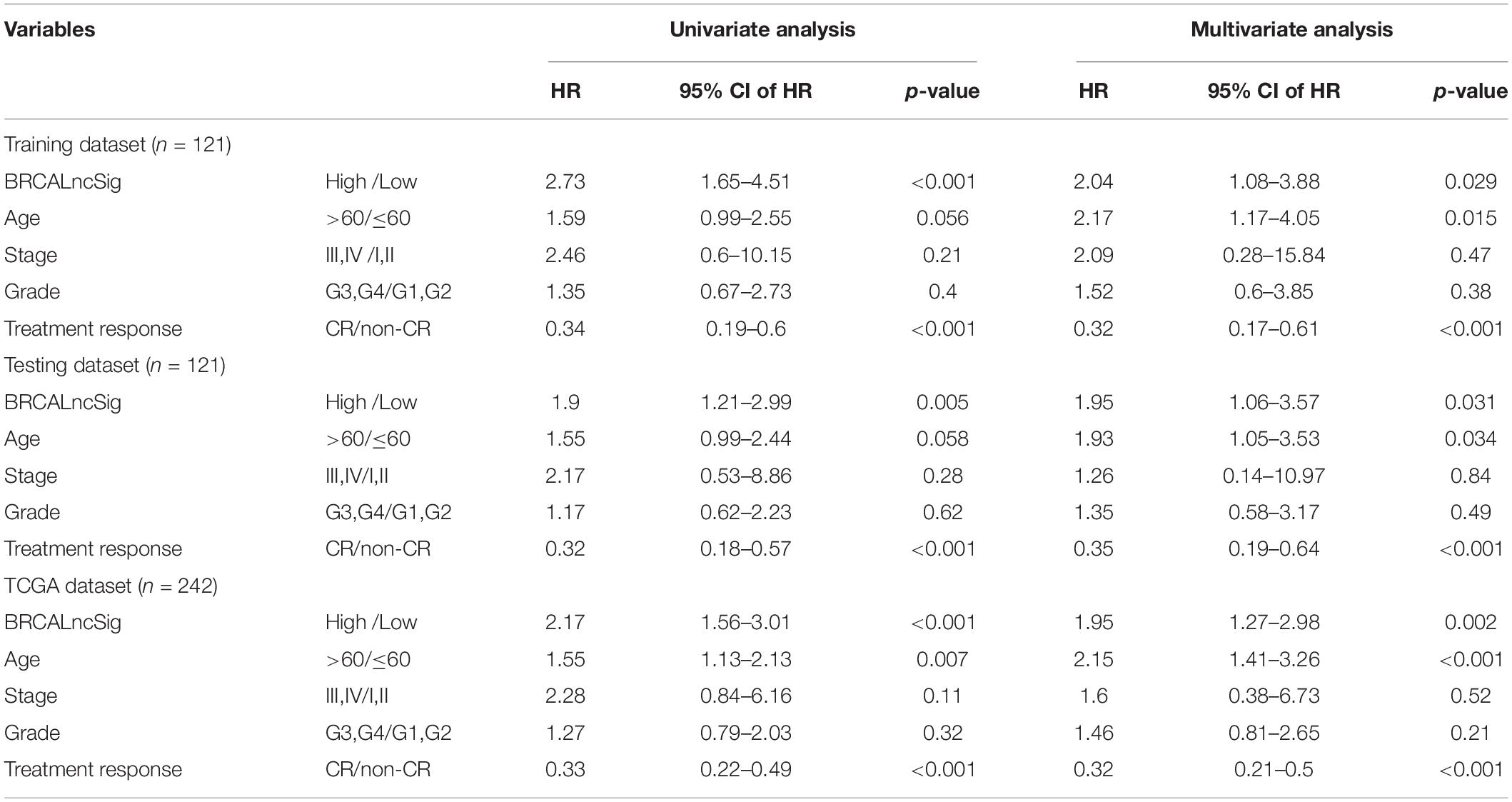
To further examine whether the BRCALncSig was independent of other clinicopathological factors, we performed multivariate Cox regression analysis for the BRCALncSig and other clinicopathological factors, including age, stage, grade and treatment response in each dataset. The results of the training set showed that the BRCALncSig (HR = 2.04, 95 CI 1.08–3.88, p = 0.029), age (HR = 2.17, 95% CI 1.17–4.05, p = 0.015), and treatment response (HR = 0.32, 95% CI 0.17–0.61, p < 0.001) was significantly correlated with overall survival (Table 2). Similar results also were observed in the testing dataset and entire TCGA dataset. The BRCALncSig still maintained a significant correlation with overall survival (HR = 1.95, 95% CI 1.06–3.57, p = 0.031 for testing dataset and HR = 1.95, 95% CI 1.27–2.98, p = 0.002 for entire TCGA dataset) after adjusted by age, stage, grade and treatment response in the testing and entire TCGA datasets (Table 2). The results of the multivariable Cox regression analysis thus indicated that the BRCALncSig was independent of other clinicopathological factors.
Discussion
Ovarian cancer is a complex disease characterized by heterogeneous molecular and clinical features (Cancer Genome Atlas Research and Network, 2011). The clinical outcome and chemotherapy response of OvCa are very different after standard treatment because of molecular heterogeneity. The clinical variables, such as stage, grade, and age, showed significant limitations in prognosis prediction and treatment decisions of the individual patient (Zhao et al., 2020). Many clinical studies have found that BRCA1/2 mutation was associated with higher rates of response to first-line platinum-based chemotherapy and improved outcome since deficiencies in BRCA1 and/or BRCA2 usually exhibit an impaired ability in HR-mediated double-strand break (DSB) repair which are highly prone to chromosomal damage induced by platinum-based chemotherapy (Pothuri, 2013; Lu et al., 2014; Rimar et al., 2017). However, recent studies indicated that BRCA1/2 wild type patients might have HR deficiency and favorable prognosis because the expression of BRCA1/2 could be affected not only by mutation but also by other molecular mechanisms, which perturbing DNA damage response and DNA repair pathways. For example, miRNAs have been reported to target BRCA1/2 and imped DNA damage repair in OvCa cells, which could impact chemotherapeutic sensitivity (Moskwa et al., 2011; Sun et al., 2013). Recent experimental studies found that several lncRNAs also participated in the DNA damage response and DNA repair pathways by interacting with BRCA1/2. However, genome-wide screening of BRCA1/2-related lncRNAs and their clinical significance is still unexplored.
In this study, we proposed a novel computational method to identify BRCA1/2-related lncRNAs based on the “ceRNA hypothesis.” Using this method, we constructed a global BRCA1/2-directed ceRNA network by integrating paired lncRNA expression profiles, miRNA expression profiles and BRCA1/2 expression profiles in BRCA1/2 wild-type patients and identified BRCA1/2-related 111 lncRNAs. These lncRNAs may be involved in DNA damage response and DNA repair pathways by interacting with BRCA1/2 via a ceRNA mechanism. Therefore, these BRCA1/2-related lncRNAs have the potential to serve as molecular biomarkers for predicting prognosis and chemotherapy response of BRCA1/2 wild-type OvCa patients. To clarify the clinical potential of these novel BRCA1/2-related lncRNAs, we examined the association of BRCA1/2-directed lncRNAs with survival and identified five BRCA1/2-directed lncRNAs (LINC01619, DLX6-AS1, AC016747.4, AC027290.3, and AC004943.2) significantly correlated with survival of BRCA1/2 wild-type patients in the training dataset. Then a BRCA1/2-directed lncRNA signature (BRCALncSig) comprised of three BRCA1-directed lncRNAs (LINC01619, DLX6-AS1, and AC004943.2) were constructed using the stepwise regression and multivariate regression methods. The BRCALncSig revealed robust prognostic value for predicting the overall survival of patients with wild-type BRCA1/2 in training, testing and TCGA datasets, which classified the patients into two groups with significantly different overall survival. Although a relatively high ratio of OvCa patients received CRs for platinum/taxane-based chemotherapy, a portion of patients receiving CR still has a poor five-survival rate because of recurrent disease (Lu et al., 2014; Gu et al., 2015; Sun J. et al., 2019). Therefore we further examined whether this BRCALncSig was able to distinguish patients with better outcomes from those with poor outcomes after platinum/taxane-based chemotherapy. Our results demonstrated the significant association between the BRCALncSig and CR, implying the potential application of the BRCALncSig in predicting CR and platinum-resistant patients. When the BRCALncSig was applied to patients with CR, we found that the BRCALncSig was able to identify those patients who will benefit from platinum-based chemotherapy. The BRCALncSig could stratify patients with CR into cases of significantly better outcomes and cases of poor outcome. Further, there are significantly different survival differences between high-risk BRCA1/2 wild-type patients and those with BRCA1/2 mutation, and no differences in survival time between low-risk BRCA1/2 wild-type patients and those with BRCA1/2 mutation.
To further explore the functional roles of the BRCALncSig in OvCa, we examined the correlation between mRNAs and the BRCALncSig, and selected the top 100 mRNAs as lncRNAs-related mRNAs. Then we performed GO and KEGG functional enrichment analysis of lncRNAs-related mRNAs, and found that these lncRNAs-related mRNAs were enriched in cancer-related or cell proliferation-related biological processes and pathways (Figure 6), provided the supporting evidence that the BRCALncSig are associated with OvCa and HR. Of three lncRNAs in the BRCALncSig, lncRNA DLX6-AS1 has been observed to be dysregulated in several cancers (Huang et al., 2019; Sun W. et al., 2019; Zhang et al., 2019). A recent study found that lncRNA DLX6-AS1 also plays an important role in the development and progression of OvCa (Zhao and Liu, 2019). The down-regulation of lncRNA DLX6-AS1 inhibits proliferation and metastasis via the Notch Signaling Pathway in human epithelial OvCa cells (Zhao and Liu, 2019). LncRNA LINC01619 has been reported to be involved in the regulation of Oxidative stress (Wang et al., 2019).
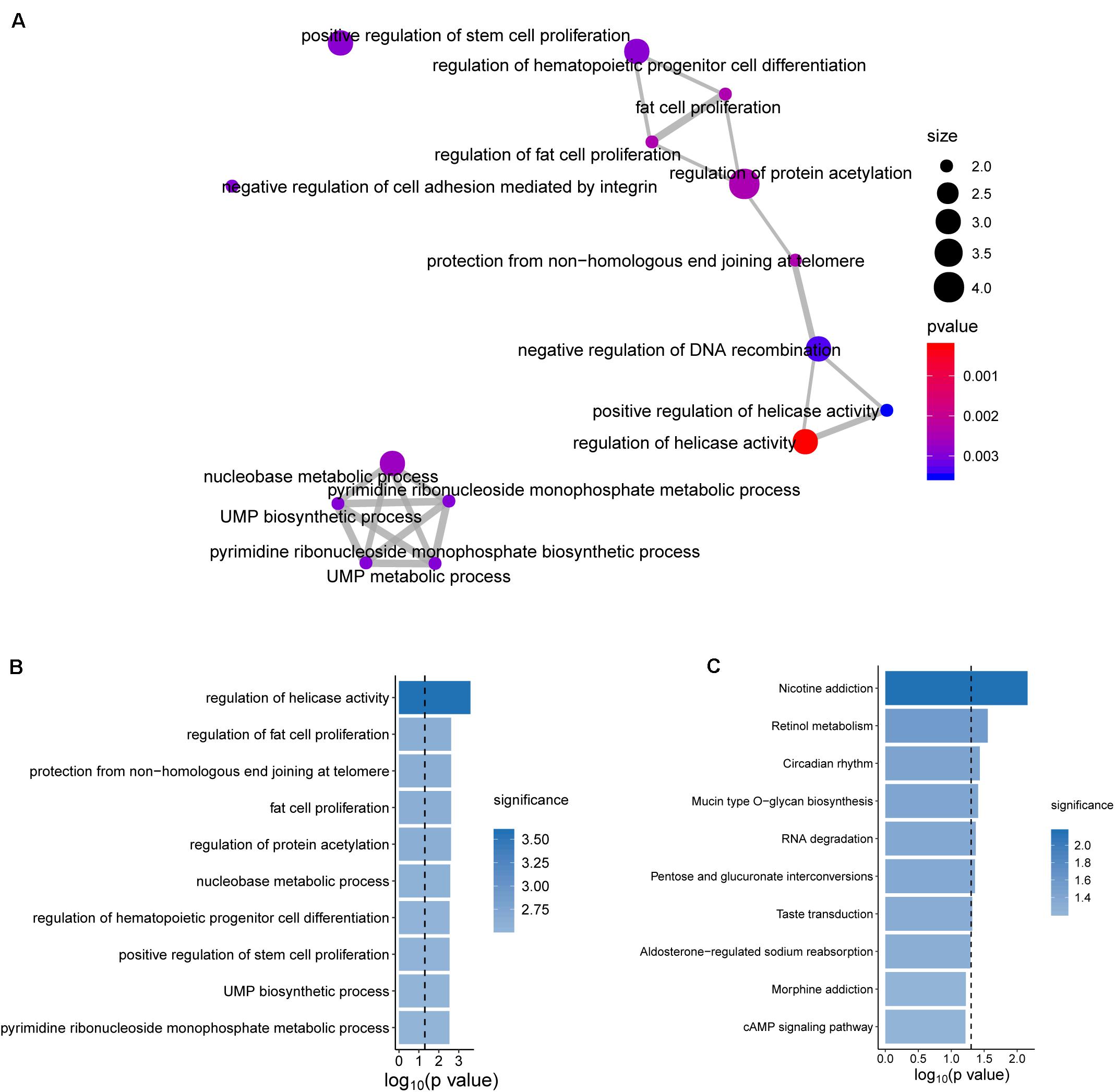
Figure 6. Functional enrichment analysis. (A) The functional map of enriched GO terms. (B) Enriched Go terms. (C) Enriched KEGG pathways.
In conclusion, we construed a BRCA1/2-directed lncRNA-associated ceRNA network by integrating paired lncRNA expression profiles, miRNA expression profiles and BRCA1/2 expression profiles in BRCA1/2 wild-type patients, and identified a BRCA1/2-directed lncRNA signature (BRCALncSig) that are significant with overall survival and chemotherapy response of BRCA1/2 wild-type patients. The BRCALncSig is independent prognostic factors which not only can robustly predict the survival of patients with wild-type BRCA1/2, but also identify OvCa patients who will benefit from platinum-based chemotherapy. However, further validation and investigation into the molecular mechanisms of how lncRNAs interact with BRCA1/2 to regulate HR and their impact on the prognosis and chemo-response should be done for in prospective studies.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: Clinical information, miRNA expression profiles, lncRNA expression profiles, mRNA expression profiles, somatic mutation, and copy number variation information of BRCA1 and BRCA2 of ovarian cancer patients were obtained from UCSC Xena (https://xena.ucsc.edu/).
Author Contributions
DW conceived and designed the experiments and wrote the manuscript. MZ, GW, and YZ analyzed the data. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 31671112).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00680/full#supplementary-material
FIGURE S1 | Expression pattern of three lncRNA biomarkers between low-risk patients and high-risk patients.
TABLE S1 | Global BRCA1/2-directed ceRNA network.
Footnotes
- ^ https://xena.ucsc.edu/
- ^ http://www.microrna.gr/tarbase
- ^ http://miRTarBase.cuhk.edu.cn/
- ^ http://starbase.sysu.edu.cn/
References
Bao, S., Zhao, H., Yuan, J., Fan, D., Zhang, Z., Su, J., et al. (2019). Computational identification of mutator-derived lncRNA signatures of genome instability for improving the clinical outcome of cancers: a case study in breast cancer. Brief. Bioinform. [Epub ahead of print]. doi: 10.1093/bib/bbz118
Cancer Genome Atlas Research and Network (2011). Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615. doi: 10.1038/nature10166
da Cunha Colombo Bonadio, R. R., Fogace, R. N., Miranda, V. C., and Diz, M. D. P. E. (2018). Homologous recombination deficiency in ovarian cancer: a review of its epidemiology and management. Clinics 73, (Suppl. 1): e450s.
Gu, Y., Zhang, M., Peng, F., Fang, L., Zhang, Y., Liang, H., et al. (2015). The BRCA1/2-directed miRNA signature predicts a good prognosis in ovarian cancer patients with wild-type BRCA1/2. Oncotarget 6, 2397–2406. doi: 10.18632/oncotarget.2963
Huang, H. Y., Lin, Y. C., Li, J., Huang, K. Y., Shrestha, S., Hong, H. C., et al. (2020). miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucl. Acids Res. 48, D148–D154. doi: 10.1093/nar/gkz896
Huang, Y., Ni, R., Wang, J., and Liu, Y. (2019). Knockdown of lncRNA DLX6-AS1 inhibits cell proliferation, migration and invasion while promotes apoptosis by downregulating PRR11 expression and upregulating miR-144 in non-small cell lung cancer. Biomed. Pharmacother. 109, 1851–1859. doi: 10.1016/j.biopha.2018.09.151
Karagkouni, D., Paraskevopoulou, M. D., Chatzopoulos, S., Vlachos, I. S., Tastsoglou, S., Kanellos, I., et al. (2018). DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucl. Acids Res. 46, D239–D245. doi: 10.1093/nar/gkx1141
Kim, A., Ueda, Y., Naka, T., and Enomoto, T. (2012). Therapeutic strategies in epithelial ovarian cancer. J. Exp. Clin. Cancer Res. 31:14. doi: 10.1186/1756-9966-31-14
Kornienko, A. E., Guenzl, P. M., Barlow, D. P., and Pauler, F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. doi: 10.1186/1741-7007-11-59
Li, J. H., Liu, S., Zhou, H., Qu, L. H., and Yang, J. H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucl. Acids Res. 42, D92–D97. doi: 10.1093/nar/gkt1248
Liu, G., Chen, Z., Danilova, I. G., Bolkov, M. A., Tuzankina, I. A., and Liu, G. (2018). Identification of miR-200c and miR141-Mediated lncRNA-mRNA crosstalks in muscle-invasive bladder cancer subtypes. Front. Genet. 9:422. doi: 10.3389/fgene.2018.00422
Lu, J., Wu, D., Li, C., Zhou, M., and Hao, D. (2014). Correlation between gene expression and mutator phenotype predicts homologous recombination deficiency and outcome in ovarian cancer. J. Mol. Med. 92, 1159–1168. doi: 10.1007/s00109-014-1191-9
McCabe, N., Turner, N. C., Lord, C. J., Kluzek, K., Białkowska, A., Swift, S., et al. (2006). Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly (ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109–8115. doi: 10.1158/0008-5472.can-06-0140
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Moskwa, P., Buffa, F. M., Pan, Y., Panchakshari, R., Gottipati, P., Muschel, R. J., et al. (2011). miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 41, 210–220. doi: 10.1016/j.molcel.2010.12.005
Pothuri, B. (2013). BRCA1- and BRCA2-related mutations: therapeutic implications in ovarian cancer. Ann. Oncol. 24, (Suppl. 8), viii22–viii27. doi: 10.1093/annonc/mdt307
Powell, S. N., and Kachnic, L. A. J. O. (2003). Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 22, 5784–5791. doi: 10.1038/sj.onc.1206678
Prensner, J. R., Chen, W., Iyer, M. K., Cao, Q., Ma, T., Han, S., et al. (2014). PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 74, 1651–1660. doi: 10.1158/0008-5472.CAN-13-3159
Rimar, K. J., Tran, P. T., Matulewicz, R. S., Hussain, M., and Meeks, J. J. (2017). The emerging role of homologous recombination repair and PARP inhibitors in genitourinary malignancies. Cancer 123, 1912–1924. doi: 10.1002/cncr.30631
Sharma, V., Khurana, S., Kubben, N., Abdelmohsen, K., Oberdoerffer, P., Gorospe, M., et al. (2015). A BRCA1-interacting lncRNA regulates homologous recombination. Embo Rep. 16, 1520–1534. doi: 10.15252/embr.201540437
Siegel, R. L., Miller, K. D., and Jemal, A. (2019). Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34. doi: 10.3322/caac.21551
Su, M., Wang, H., Wang, W., Wang, Y., Ouyang, L., Pan, C., et al. (2018). LncRNAs in DNA damage response and repair in cancer cells. Acta Biochim. Biophys. Sin. 50, 433–439. doi: 10.1093/abbs/gmy022
Sun, C., Li, N., Yang, Z., Zhou, B., He, Y., Weng, D., et al. (2013). miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J. Natl. Cancer Inst. 105, 1750–1758. doi: 10.1093/jnci/djt302
Sun, J., Bao, S., Xu, D., Zhang, Y., Su, J., Liu, J., et al. (2019). Large-scale integrated analysis of ovarian cancer tumors and cell lines identifies an individualized gene expression signature for predicting response to platinum-based chemotherapy. Cell Death Dis. 10:661. doi: 10.1038/s41419-019-1874-9
Sun, W., Zhang, L., Yan, R., Yang, Y., and Meng, X. (2019). LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/GSPT1 axis. Onco. Targets Ther. 12, 3945–3954. doi: 10.2147/OTT.S196865
Sun, J., Zhang, Z., Bao, S., Yan, C., Hou, P., Wu, N., et al. (2020). Identification of tumor immune infiltration-associated lncRNAs for improving prognosis and immunotherapy response of patients with non-small cell lung cancer. J. Immunother. Cancer 8:e000110. doi: 10.1136/jitc-2019-000110
Wang, X., Shen, C., Zhu, J., Shen, G., Li, Z., and Dong, J. (2019). Long noncoding RNAs in the regulation of oxidative stress. Oxid. Med. Cell Longev. 2019:1318795. doi: 10.1155/2019/1318795
Wu, Z., and Wang, Y. (2017). Studies of lncRNAs in DNA double strand break repair: what is new? Oncotarget 8, 102690–102704. doi: 10.18632/oncotarget.22090
Yoshida, K., and Miki, Y. (2004). Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci. 95, 866–871. doi: 10.1111/j.1349-7006.2004.tb02195.x
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, J. J., Xu, W. R., Chen, B., Wang, Y. Y., Yang, N., Wang, L. J., et al. (2019). The up-regulated lncRNA DLX6-AS1 in colorectal cancer promotes cell proliferation, invasion and migration via modulating PI3K/AKT/mTOR pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 8321–8331. doi: 10.26355/eurrev_201910_19143
Zhao, H., Gu, S., Bao, S., Yan, C., Zhang, Z., Hou, P., et al. (2020). Mechanistically derived patient-level framework for precision medicine identifies a personalized immune prognostic signature in high-grade serous ovarian cancer. Brief. Bioinform. bbaa069. doi: 10.1093/bib/bbaa069
Zhao, J., and Liu, H. R. (2019). Down-regulation of long noncoding RNA DLX6-AS1 defines good prognosis and inhibits proliferation and metastasis in human epithelial ovarian cancer cells via Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 3243–3252. doi: 10.26355/eurrev_201904_17684
Zhou, M., Diao, Z., Yue, X., Chen, Y., Zhao, H., Cheng, L., et al. (2016a). Construction and analysis of dysregulated lncRNA-associated ceRNA network identified novel lncRNA biomarkers for early diagnosis of human pancreatic cancer. Oncotarget 7, 56383–56394. doi: 10.18632/oncotarget.10891
Zhou, M., Sun, Y., Sun, Y., Xu, W., Zhang, Z., Zhao, H., et al. (2016b). Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget 7, 32433–32448. doi: 10.18632/oncotarget.8653
Zhou, M., Guo, M., He, D., Wang, X., Cui, Y., Yang, H., et al. (2015a). A potential signature of eight long non-coding RNAs predicts survival in patients with non-small cell lung cancer. J. Transl. Med. 13:231. doi: 10.1186/s12967-015-0556-3
Zhou, M., Zhao, H., Wang, Z., Cheng, L., Yang, L., Shi, H., et al. (2015b). Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J. Exp. Clin. Cancer Res. 34:102. doi: 10.1186/s13046-015-0219-5
Zhou, M., Hu, L., Zhang, Z., Wu, N., Sun, J., and Su, J. (2018a). Recurrence-associated long non-coding RNA signature for determining the risk of recurrence in patients with colon cancer. Mol. Ther. Nucl. Acids 12, 518–529. doi: 10.1016/j.omtn.2018.06.007
Zhou, M., Zhang, Z., Zhao, H., Bao, S., Cheng, L., and Sun, J. (2018b). An immune-related Six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol. Neurobiol. 55, 3684–3697. doi: 10.1007/s12035-017-0572-9
Zhou, M., Zhang, Z., Zhao, H., Bao, S., and Sun, J. (2018c). A novel lncRNA-focus expression signature for survival prediction in endometrial carcinoma. BMC Cancer 18:39. doi: 10.1186/s12885-017-3983-0
Keywords: ovarian cancers, long non-coding RNAs, BRCA1/2, ceRNA, signature
Citation: Zhang M, Wang G, Zhu Y and Wu D (2020) Characterization of BRCA1/2-Directed ceRNA Network Identifies a Novel Three-lncRNA Signature to Predict Prognosis and Chemo-Response in Ovarian Cancer Patients With Wild-Type BRCA1/2. Front. Cell Dev. Biol. 8:680. doi: 10.3389/fcell.2020.00680
Received: 24 May 2020; Accepted: 06 July 2020;
Published: 29 July 2020.
Edited by:
Lei Deng, Central South University, ChinaReviewed by:
Guoqing Liu, Inner Mongolia University of Science and Technology, ChinaLu Ao, Fujian Medical University, China
Copyright © 2020 Zhang, Wang, Zhu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Di Wu, bWFzdGVyd3U3MjhAMTYzLmNvbQ==
†These authors share first authorship
 Meiling Zhang1†
Meiling Zhang1† Di Wu
Di Wu