- Department of Endocrinology, Endocrinology Research Center, Xiangya Hospital of Central South University, Changsha, China
Osteogenesis and senescence of BMSCs play great roles in age-related bone loss. However, the causes of these dysfunctions remain unclear. In this study, we identified a differentially expressed ASPH gene in middle-aged and elderly aged groups which were obtained from GSE35955. Subsequent analysis in various databases, such as TCGA, GTEx, and CCLE, revealed that ASPH had positive correlations with several osteogenic markers. The depletion of mouse Asph suppressed the capacity of osteogenic differentiation in bone marrow mesenchymal stem cells (BMSCs). Notably, the expression of ASPH in vitro decreased during aging and senescence. The deficiency of Asph accelerated cellular senescence in BMSCs. Conversely, the overexpression of Asph enhanced the capacity of osteogenic differentiation and inhibited cellular senescence. Mechanistically, ASPH regulated Wnt signaling mediated by Gsk3β. Taken together, our data established that ASPH was potentially involved in the pathogenesis of age-related bone loss through regulating cellular senescence and osteogenic differentiation, which provides some new insights to treat age-related bone loss.
Introduction
Osteoporosis is characterized by a reduction of bone mass and dysfunction of bone micro-architecture (Al Anouti et al., 2019; Yang et al., 2019). With the progressive aging of the general population, age-related osteoporosis is becoming a prevalent chronic disease in various countries. Elderly individuals are susceptible to suffer from the age-related osteoporosis because of the loss of bone mass and strength caused by skeletal aging (Li et al., 2015, 2018; Yu et al., 2018; Xiao et al., 2020). Skeletal aging-associated bone loss can be traced back to bone marrow mesenchymal stem cells (BMSCs). BMSCs play a great role in bone formation because of their potential of self-renewal and multi-lineage differentiation including osteogenic lineage (Li et al., 2018; Peng et al., 2019). During aging, BMSCs present with a reduced capacity of self-renewal and osteogenic lineage commitment, which thereby results in the aberrant age-dependent bone formation (Childs et al., 2015; Li H. et al., 2017).
Cellular senescence refers to a state of irreversible cell cycle arrest and distinct cellular alterations in response to various stress (Childs et al., 2015). This process is controlled by various types of tumor suppressors, such as p16INK4A, p15INK4b, Rb, and p21CIP1 (Campisi and d’Adda di Fagagna, 2007; Childs et al., 2015; Li C. et al., 2017). Aging has been reported to be associated with increased cellular senescence. Accumulation of cellular senescence, in turn, can accelerate organismal aging and contribute to the phenotype of aging (Childs et al., 2015). BMSCs will start the aging process early, that is, premature aging, upon stimulated by the stress. Previous studies also revealed that BMSCs underwent cellular senescence during skeletal aging and age-related modulators can cause or prevent premature aging (Lin et al., 2014; Li H. et al., 2017; Zhou et al., 2020).
Aspartate β-hydroxylase encoded by ASPH is a type II membrane protein, which includes several domains majorly including N-terminal cytoplasmic domain, transmembrane (TM) domain, Ca2+ binding domain and C-terminal catalytic domains (Treves et al., 2000; Finotti et al., 2008). ASPH has various isoforms because of its extensive alternative splicing. The longest ASPH isoform a containing C-terminal Aspartyl/Asparaginyl beta-hydroxylase catalytic domain can regulate some proteins with the epidermal growth factor (EGF)-like domains (Dinchuk et al., 2000). Based on the current studies, ASPH has been reported to be involved in the regulation of tumorigenesis via cell proliferation, colony formation and cellular senescence (Iwagami et al., 2016; Hou et al., 2018). Notably, a Genome-Wide Association Study (GWAS) conducted by Koller et al. (2010) described a SNP located closely with 3’ region of ASPH. They found this SNP was conceivably associated with the lumbar spine bone mineral density in the premenopausal European-American women (Koller et al., 2010). In addition, some studies reported that ASPH was involved in the regulation of various signaling pathways, such as Wnt signaling, Notch signaling, IGF signaling, IRS signaling and so on (Cantarini et al., 2006; Tomimaru et al., 2013; Iwagami et al., 2016; Hou et al., 2018). It is noted that these signaling pathways are the canonical pathways during bone modeling and remodeling (Malaguarnera and Belfiore, 2014; Tu et al., 2015; Yang et al., 2017). However, the molecular network orchestrating bone formation and cellular senescence of BMSCs mediated by ASPH still remain unclear.
In this study, we reported that the level of ASPH decreased with aging or senescence. Intriguingly, ASPH longest isoform a facilitating the osteogenic differentiation of BMSCs, whereas preventing the process of cellular senescence. Conversely, the knockdown of ASPH accelerated the cellular senescence while suppressed the osteogenic differentiation. Mechanistically, these observations might be associated with aberrant Wnt signaling. Taken together, our study revealed ASPH promoting the process of osteogenesis while inhibiting cellular senescence through regulating Gsk3β-mediated Wnt signaling, which potentially provided new insights for aged-related bone loss.
Materials and Methods
Differentially Expressed mRNAs Filtering and Bioinformatics Analysis
Human mRNA expression data were downloaded from Gene Expression Omnibus (GEO Accession: GEO35955)1. The differentially expressed mRNAs between middle-aged and elderly aged groups have been filtered using t-test. The R package “limma” (Ritchie et al., 2015) applied to normalize the data and identify DEGs. The DEGs was kept when adjust p = 0.01 and | logFC| = 2. Then, kept genes have been assessed the functional enrichments, including GO (Gene Ontology) Biological Processes term and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway using the R package “Clusterprofiler”(Yu et al., 2012). All probes of the interested ASPH gene have been extracted from the raw data. We calculated ASPH expression at different coding region according to the recognition sites of probes. Its expressions at different regions then were output the violin figures using t-test.
Co-expression of ASPH With Other Genes
The raw expression data of ASPH and RUNX2, COL1A1, GSK3B, and CTNNB1 genes were downloaded from The Cancer Genome Atlas (TCGA) project2, the Genotype Tissue Expression (GTEx) program3 and CCLE (Cancer Cell Line Encyclopedia) project4. The Pearson correlation (r) and p-value have been calculated using R and R Studio. The p-value has been marked as “0” when it is less than 1∗10–8.
BMSCs Culture
Mouse BMSCs (MUBMX-01001; Cyagen Biosciences) were cultured in Mouse Mesenchymal Stem Cell Growth Medium (MUBMX-90011; Cyagen Biosciences). For human BMSCs, we purified from bone marrow of participants underwent hip replacement. The gender of these subjects is male. The ages of young groups were 24, 28, 31, 27, and 32, respectively The ages in old groups were 75, 75, 78, 79, and 85, respectively.
qRT-PCR Analysis
For analysis of mRNA expression, total RNA from cultured cells was extracted using Trizol reagent (Takara). One-thousand nanograms of RNA was reverse-transcribed into first-strand cDNA using the Reverse Transcription Kit (Takara). For Human BMSCs RNA and cDNA have collected before. qPCR was performed using SYBR Green PCR Master Mix (Takara) and mRNA expression was normalized to reference genes GAPDH.
Western Blot
The protein was lysed using the mixture of RIPA, protease inhibitor (1:100) and phosphatase inhibitor (1:100). Western Blot was performed according to the previous described method (Li et al., 2015). The primary antibodies, GSK-3β Rabbit mAb (#9315; CST), Non-phospho (Active) β-Catenin (Ser33/37/Thr41) Rabbit mAb (#8814;CST), β-Catenin Antibody (#9562;CST), beta Actin mouse monoclonal antibody (#TA811000, ORIGENE) Phospho-GSK-3β (Ser9) Antibody (#9336; CST), and alpha-tubulin (11224-1-AP; Proteintech), were incubated overnight at 4°C, then incubated with appropriate HRP-conjugated secondary antibodies for 1 h at room temperature. The blots were visualized using ECL detection reagents.
Immunofluorescence
For immunofluorescence, cells were washed by phosphate-buffered saline (PBS) three times. Then, cells were fixed in 4% paraformaldehyde for 15 min and followed by permeabilization and blocking with 7% FCS, 1% TritonX100 in PBS for 30 min at room temperature. Primary antibodies of total β-Catenin (#9562; CST, 1:200) and Non-phospho (Active) β-Catenin (Ser33/37/Thr41) (#8814; CST, 1:800) were incubated overnight in 7% BSA in PBS overnight at 4°C and followed by Alexa Fluor R 488 Goat Anti– rabbit (Thermo Fisher Scientific, United States) at a dilution of 1:400, and finally stained and mounted by mounting medium. Images were visualized on Olympus microscope using cellSense Dimension software.
Cell Transfection
The Asph siRNA, Gsk3b siRNA and the negative control (NC) were purchased from Ribibio (Guangzhou, China). The siRNAs were transfected at the concentration of 100 nM using lipofectamine RNAiMAX (Invitrogen, United States) according to manufacturer’s recommendations. mAsph pcDNA3.1-3xFlag-C construct (GenBank:NM_004318.4) was purchased from YouBao Biology. One microgram mAsph (AAH) construct and negative control were transfected into BMCSs per well of 6-well-plate using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s recommendations.
Osteogenic Differentiation and Mineralization Assay
To induce osteogenic differentiation of BMSCs, BMSCs were cultured in 6-well plates at 2.5 × 106 cells per well with the mesenchymal stem cell osteogenic differentiation (MUBMX-90021; Cyagen Biosciences). At the 2rd day of osteogenic differentiation, the cell lysates were homogenized for ALP activity assay by spectrophotometric measurement of p-nitrophenol release using an Alkaline Phosphatase Assay Kit (P0321S, Beyotime). At the 7th day of osteogenic differentiation, alkaline phosphatase staining (ALP staining) was performed to evaluate the cell-matrix mineralization. We firstly washed the cells using PBS three times followed by 10% paraformaldehyde for 5 min. Then, cells were incubated in ALP incubation buffer (0.2 g barbital sodium, 0.4 g magnesium sulfate, 0.2 g calcium chloride and 0.3 g beta-glycerophosphate) at 37°C for 2 h. Next, washed cells with 2% calcium chloride and incubated with 2% cobaltous nitrate for 5 min. Then, cells were incubated in 1:80 ammonium sulfate for 10 s. At the 21st day of osteogenic differentiation, Alizarin Red staining was performed to evaluate the cell-matrix mineralization according to the manufacturer’s instructions (MUBMX-90021; Cyagen Biosciences). Briefly, cells were washed using PBS three times followed by 4% paraformaldehyde for 30 min. After washed by PBS for three times, cells were stained in Alizarin red solution at 37°C for 5 min. The stained wells were imaged using the camera (ILCE-5100, SONY). Alizarin Red S released from the cell matrix into the cetyl-pyridinium chloride solution was quantified by spectrophotometry at 540 nm.
β-Galactosidase Staining
BMSCs with Asph siRNA interference were detected by senescence β-Galactosidase staining kit according to the manufacturer’s instructions (G1580, Solarbio Life Science). Briefly, we firstly prepared the staining working solution which comprised of 10 μl β -galactosidase stain A, 10 μl β -galactosidase stain B, 930 μl β -galactosidase stain C and 50 μl X-Gal solution for each well. Then, we rinsed the cell plates with PBS followed by the fixation by 1 ml fix solution of β-galactosidase staining at room temperature for 15 min. After removal of fix solution, the plates were rinsed for three times. Then, cells were incubated with 1 ml staining working solution in incubator at 37°C overnight. Finally, the plates were visualized through Olympus microscope.
Giemsa Staining
BMSCs with Asph siRNA interference was detected by Giemsa staining kit according to the manufacturer’s instructions (G4640, Solarbio Life Science). Briefly, we washed the cell plates with PBS followed by fixation using 1 ml methanol for 2 min. After removal of fix solution, the plates were washed three times using PBS. Then, cells were incubated in 1 ml staining solution for 15 min. Finally, the CFUs were recorded using the camera (ILCE-5100, SONY).
Statistical Analysis
Data was imported into Excel and scaled and normalized to appropriate controls. Unpaired, two-tailed Student’s t-tests were performed for the comparisons of two groups and one-way ANOVA for comparison within multiple groups. Critical P-values were Bonferroni corrected and expressed as follows: ns: No significance, ∗P < 0.05, ∗∗P < 0.01, ∗∗P < 0.001, ###P < 0.001.
Results
ASPH Is an Age-Dependent Gene in BMSCs
To identify the possible causes of age-related bone loss, we analyzed the differentially expressed genes of GSE35955 using R and R studio software (version 3.5) (Raw data was obtained from a public repository of GEO which includes human middle-aged individuals and elderly individuals) (Benisch et al., 2012). Firstly, we normalized the expression of all samples (Figure 1A). Secondly, we kept the deferentially expressed probes through setting the cutoff of adjust p-value as 0.01 and LogFC (Log FoldChange) as 2.0, respectively. A total of 152 probes (including 127 genes) have been kept. Of the differentially expressed 152 probes, the expression of 116 probes decreased while the expression of 36 probes increased in elderly aged groups in comparison to the middle-aged group (Figures 1B,C). Thirdly, we analyzed the top 10 enriched signaling using three sub-ontologies (BP for Biological Process, MF for Molecular Function, and CC for Cellular Component) of GO biological processes and KEGG pathway analysis (Figures 1D,E). KEGG analysis showed the differentially expressed genes were majorly enriched in cellular senescence, EGFR tyrosine kinase inhibitor resistance and some cancer development (Figure 1E). Fourthly, we searched potential roles of all genes in bone formation via setting “Senescence OR Aging,” “Bone,” and “Gene symbol (from the differentially expressed genes)” as keywords in Pubmed database. Surprisingly, we found ASPH was associated with cellular senescence in cancer and possibly involved in bone health. Thus, we selected “ASPH” as our interested gene to conduct further studies.
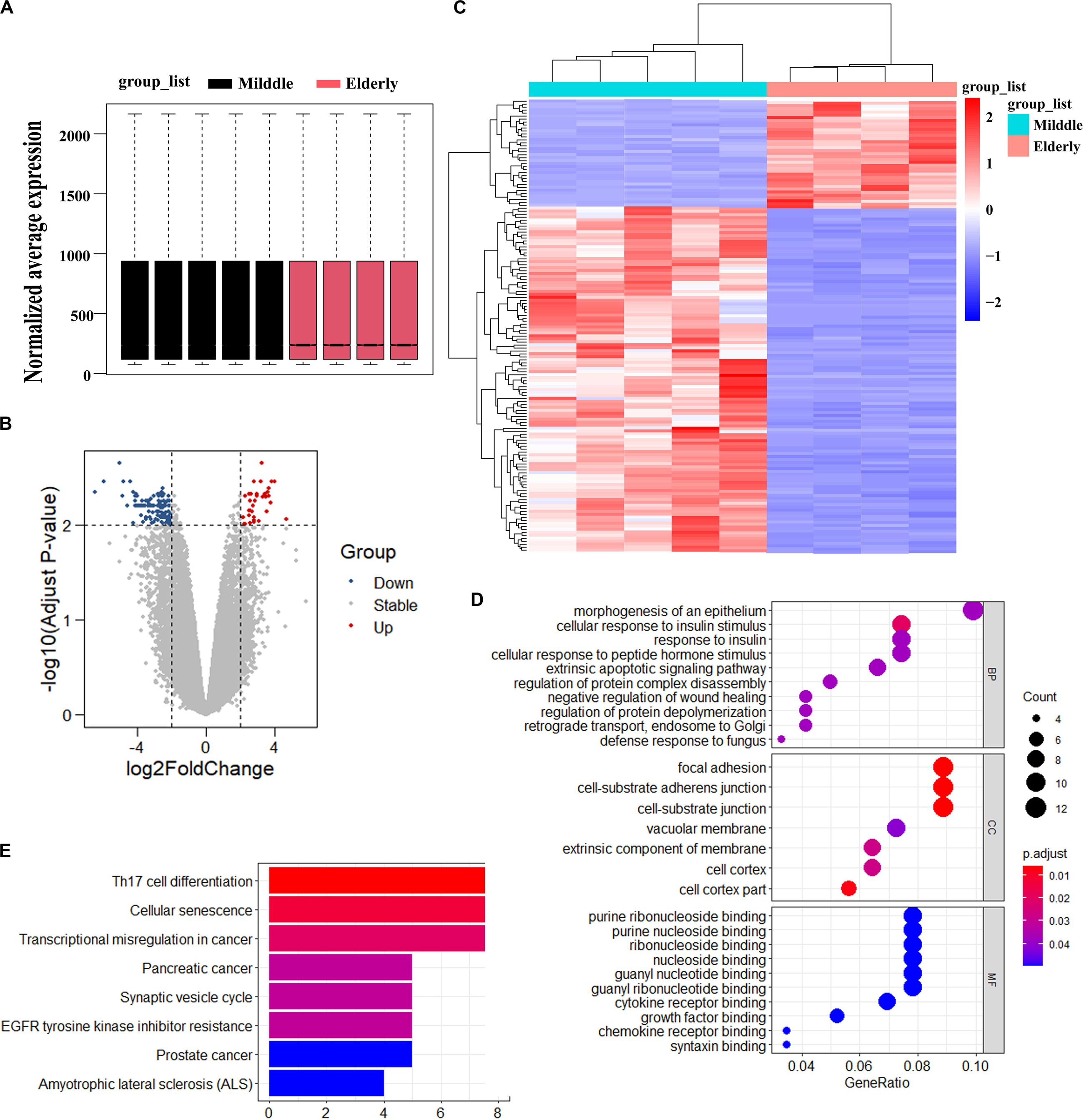
Figure 1. Analysis of differentially expressed genes in BMSCs from middle-aged individuals and elderly aged individuals mined from GSE35955. (A) Boxplot of RNA-seq profiling of normalized mRNA expression in BMSCs. (B) Volcano Plot of normalized mRNA expression in BMSCs. (C) Heatmap of mRNA expression in BMSC. | Fold change| > 2, adjust p < 0.01. (D) GO (Gene Ontology) analysis of differentially expressed gene in the two groups. (E) KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of differentially expressed gene in the two groups. p < 0.05.
ASPH Promotes Osteogenic Differentiation
Previous studies indicated ASPH might be associated with bone mineral density (Koller et al., 2010; Mantila Roosa et al., 2011; Benisch et al., 2012). To investigate if ASPH plays a role in bone formation, we analyzed the co-expression of ASPH with osteogenic markers such as RUNX2 and COL1A1 in various databases. In most of normal tissues from GTEX, the correlations of ASPH with RUNX2 and COL1A1 were positive, which presented in the upper right region of Figures 2A,E. ASPH had a moderate positive correlation with RUNX2 both in 7858 tissues (Pearson r = 0.4, p = 0) (Figure 2B) and specifically in 70 tissues of bone marrow (Pearson r = 0.51, p = 0) (Figure 2C). Similarly, ASPH had a positive correlation with COL1A1 both in 7858 tissues (Pearson r = 0.48, p = 0) (Figure 2F) even though didn’t show significant correlation with RUNX2 in bone marrow (Figure 2G). In 7801 tumor tissues, ASPH also showed a positive correlation with RUNX2 as well as COL1A1 (Figures 2D,H). Moreover, in 1019 cancer cell lines, there were positive correlation between ASPH and RUNX2 (Figure 2I) or COL1A1 (Figure 2J). Notably, the 26 bone-related cell lines did not show significant co-expression between ASPH and RUNX2 (Figure 2K), but showed positive correlation between ASPH and COL1A1 (Pearson r = 0.74, p = 0) (Figure 2L). Collectively, these results partially supported that ASPH was positively correlated with these two genes (Figures 2A–L).
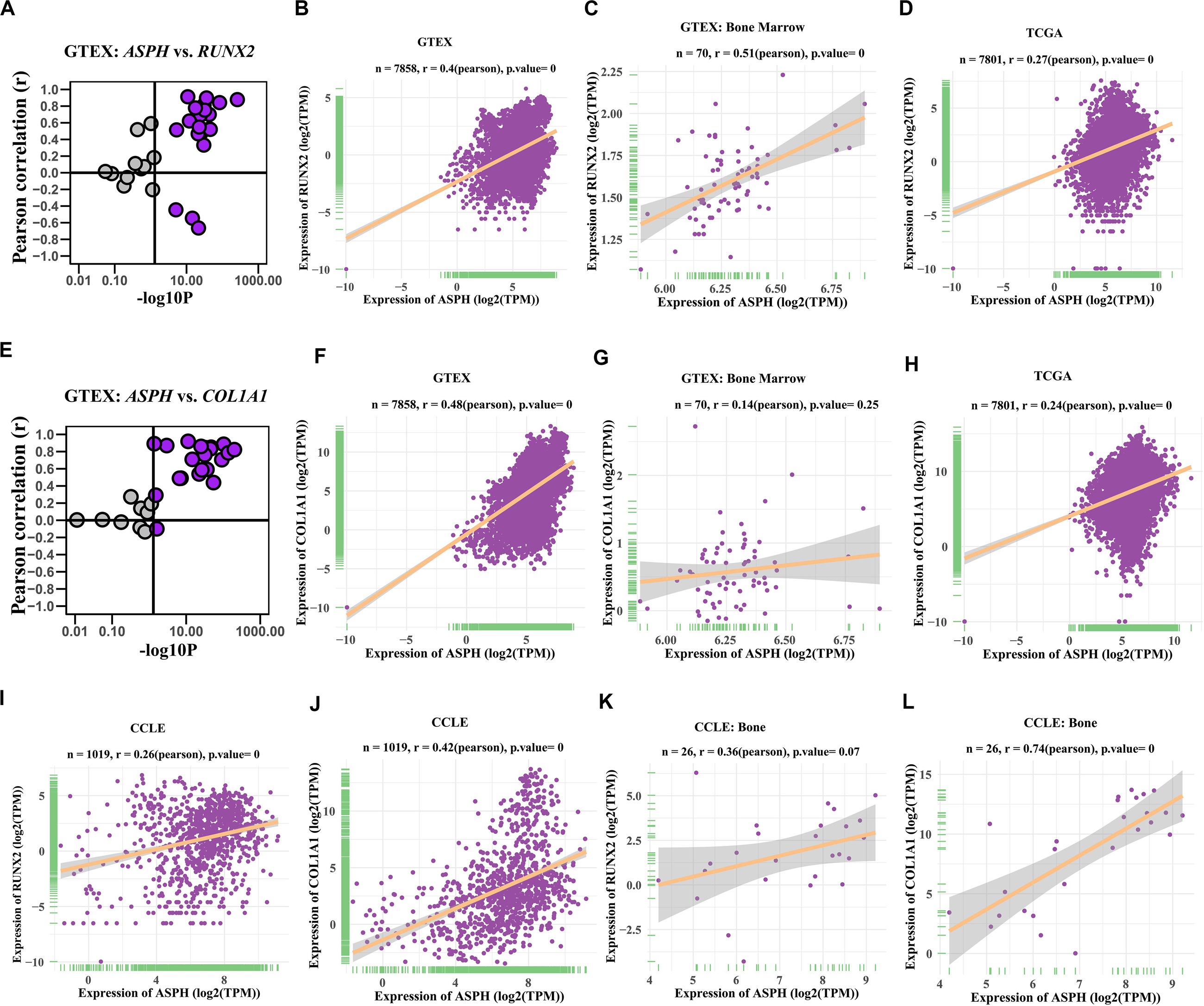
Figure 2. Co-expression of ASPH with osteogenic markers. (A–D) Correlation of ASPH with RUNX2 in expression in normal tissues (A,B), bone marrow from normal tissues (C), and cancer samples (D), based on the data from Genotype Tissue Expression (GTEx) and The Cancer Genome Atlas (TCGA) databases, respectively. (E–H) Correlation of ASPH with COL1A1 in expression in normal tissues (E,F), bone marrow from normal tissues (G) and cancer samples (H), based on the data from GTEx and databases, respectively. (I,J) Correlation of ASPH with RUNX2 in expression in cancer cell lines (I) and in bone related cancer cell lines (J), based on the data from Cancer Cell Line Encyclopedia (CCLE). (K,L) Correlation of ASPH with COL1A1 in expression in cancer cell lines (K) and in bone related cancer cell lines (L), based on the data from CCLE. The correlation coefficient (r) and P-value were calculated by Pearson’s Correlation analysis. Note that every dot represents one tissue type (A–C,E–G) or one cancer type (D,H) or one cell line (I–L).
ASPH is an 85KD type II transmembrane protein which can generate various splicing variants including aspartyl beta-hydroxylase (AAH), junctin, and junctate (Figure 3A). The longest isoform a (GenBank:NM_004318.4) is comprised of several domains, majorly including N-terminal cytoplasmic domain, transmembrane (TM) domain, Ca2+ binding domain and C-terminal catalytic domains. We extracted the expression of all “ASPH” probes and classified them into three types according to the probe-targeting positions (e.g., Exon 14–25 with C-terminal catalytic domain, Exon 4–13 with Ca2+ binding domain, and Exon 1–3 with a positively charged domain) (Figure 3A). Next, BMSCs were induced by osteogenic differentiation medium and tested its expression of different domains of ASPH. The qRT-PCR data showed increased expression of mouse Asph during osteogenesis of BMSCs (Figure 3B). Subsequently, BMSCs were transfected with mouse Asph siRNA targeting the catalytic domain (Figure 3C) followed by induction by osteogenic differentiation medium. qRT-PCR data suggested that downregulation of Asph expression was accompanied by a decline of osteogenic markers at the 7th day, including Runx2 and Col1a1 (Figure 3E). Furthermore, the matrix memorization measured by Alkaline phosphatase (ALP) activity, ALP staining and Alizarin Red staining showed a decline of AAH expression resulted in the impairment of osteogenesis in BMSCs (Figures 3F–I). Conversely, BMSCs were transfected with the construct of longest mouse Asph isoform variant 1 (GenBank:NM_023066.3) (Figure 3D) followed by induction by osteogenic differentiation medium. qRT-PCR suggested that up-regulation Asph expression was correlated with the increase of osteogenic markers, Runx2 and Col1a1 (Figure 3J). Similarly, matrix memorization measured by ALP activity, ALP staining and Alizarin Red staining showed over-expression of Asph resulted in the enhancement of osteogenesis in BMSCs (Figures 3K–N).
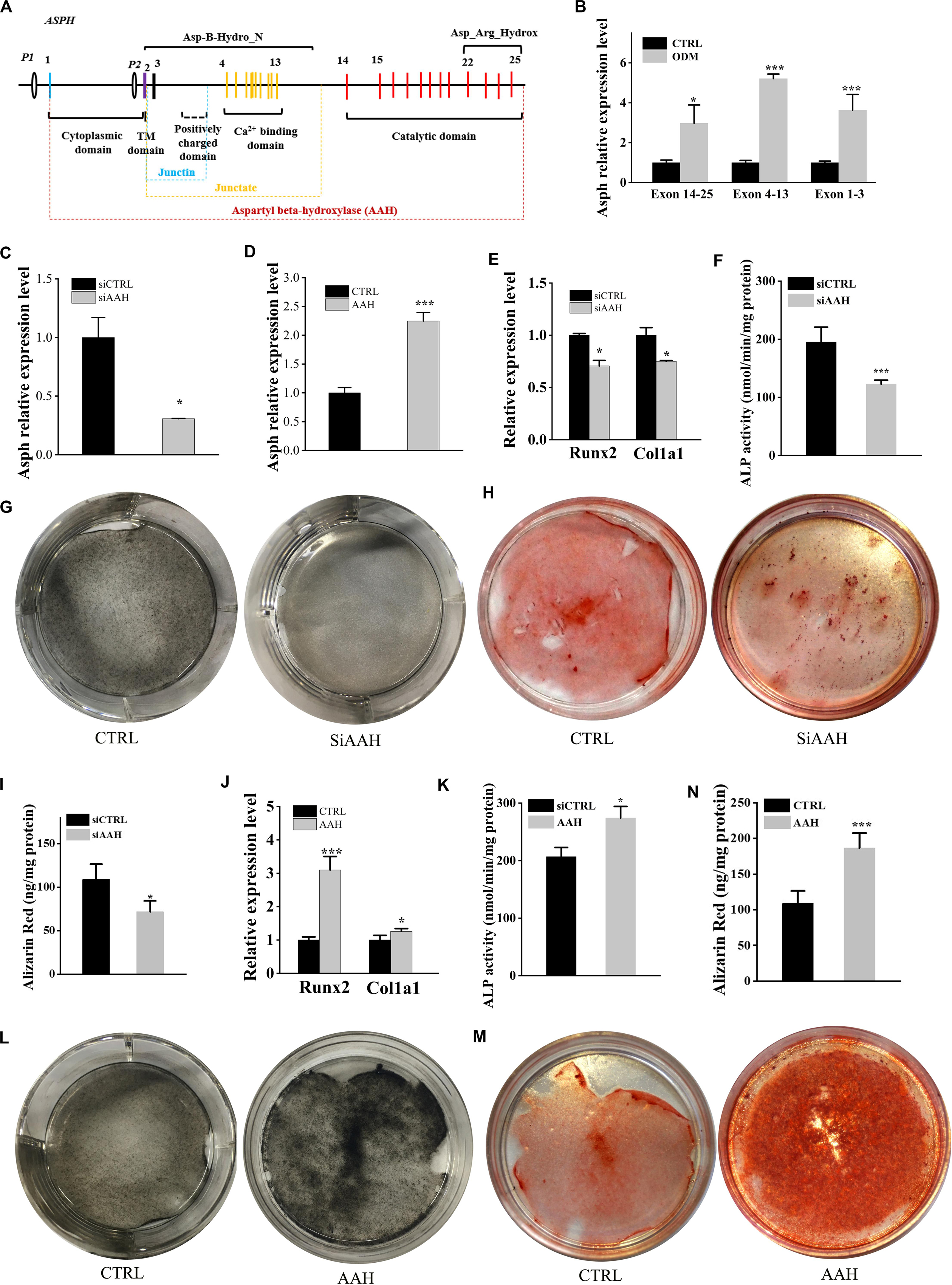
Figure 3. ASPH promotes osteogenic differentiation. (A) schematic graph for ASPH isoforms. (B) qRT-PCR analysis of ASPH regions’ expression during osteogenic differentiation. (C) qRT-PCR analysis of depletion of AAH. (D) qRT-PCR analysis of overexpression of AAH. (E) qRT-PCR analysis of the relative levels of Runx2 and Col1a1 in BMSCs with depletion of AAH. (F) Analysis of ALP activity in BMSCs with depletion of AAH. (G,H) Representative images of ALkaline Phosphatase (ALP) staining (G) and Alizarin red S staining (H,I) in BMSCs with depletion of AAH. (J) qRT-PCR analysis of the relative levels of Runx2 and Col1a1 in BMSCs with overexpression of AAH. (K) Analysis of ALP activity in BMSCs with overexpression of AAH. (L,M) Representative images of ALP staining (L) and Alizarin Red S staining (M,N) in BMSCs with overexpression of AAH. These experiments were replicated three times. Error bars show standard deviation. *P < 0.05, ***P < 0.001.
Ablation of ASPH Results in Premature Senescence of BMSCs
ASPH is known to be involved in the cellular senescence in hepatocellular carcinoma and gliomas (Iwagami et al., 2016; Sturla et al., 2016). In order to understand if ASPH is a cellular senescence-related gene in BMSCs, we calculated the mean ASPH expression and expression of different ASPH region (Supplementary Table 1; Peng et al., 2020a). The result suggested ASPH was lower in elderly aged individuals than middle-aged individuals (Figure 4A). The exon 4–13 and exon 14–25 of ASPH both decreased with aging in the two groups. However, the exon 1–3 expression locating at the common region of different ASPH isoforms was stable (Figure 4B). Then, we collected human BMSCs (hBMSCs) with different ages and tested their ASPH expression at different regions. As expected, compared to the aged group (age from 75 to 90), the young group (age from 24 to 32) showed a higher expression of ASPH longer isoforms (Figure 4C). Additionally, we tested the Asph expression in young mouse BMSCs (passage 0) and senescent mouse BMSCs (passage 5). Consistently, the expression of Asph longer isoforms were lower in senescent BMSCs than in young BMSCs (Figure 4D). These observations suggested that senescence suppressed Asph expression. To further test this hypothesis, we performed a series of functional analysis. qRT-PCR results demonstrated that the Asph expression was remarkably decreased along with cellular senescence (Figure 4E). Conversely, the senescence-associated p16 expression was positively correlated with cellular senescence (Figure 4E). Additionally, an impairment of the replicative capacity was observed in Asph siRNA transfected group (Figure 4F). CFU colonies obtained from Giemsa staining further showed the inhibition of cell proliferation in BMSCs interfered with Asph siRNA (Figure 4G). qRT-PCR results revealed that the depletion of Asph promoted markers of senescence, such as p15, p16, and p21 expression in BMSC (Figures 4H–J). Consistently, the senescence-associated β-galactosidase staining revealed that the deficiency of Asph promoted the cellular senescence (Figure 4K).
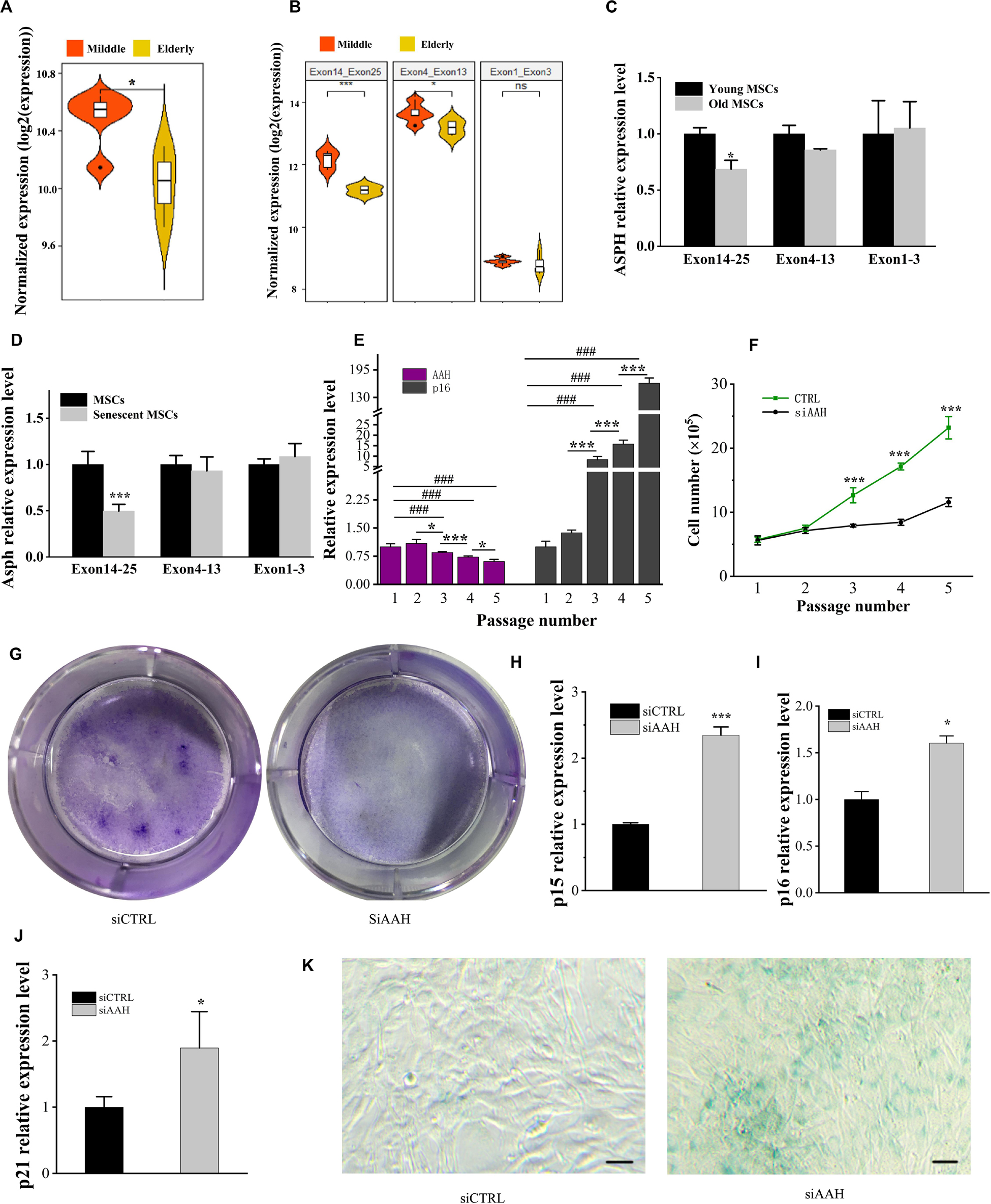
Figure 4. Deficiency of ASPH accelerates cellular senescence in BMSCs. (A) Mean expression of ASPH in the BMSCs from middle-aged individuals and elderly aged individuals (Raw data was obtained from GSE35955). (B) Mean expression of different ASPH regions in the BMSCs from middle-aged individuals and elderly aged individuals (Raw data was obtained from GSE35955). (C) The expression of ASPH isoforms in the BMSCs from young and old individuals. (D) The expression of Asph isoforms in the young or senescent BMSCs. (E) The negative correlation of AAH and p16 expression during passaging (P1–P5) of BMSCs. (F) Doublings of BMSCs interfered with AAH siRNA in vitro. (G) Giemsa staining for the CFU-F colonies of BMSCs from BMSCs interfered with AAH siRNA in vitro. (H–J) Expression of p15 (H), p16 (I), and p21 (J) in BMSCs with depletion of AAH. (K) Representative images of SA-β-Gal staining of BMSCs in Asph siRNA transfected and control group. Scale Bar = 100 μm. These experiments were replicated three times. Error bars show standard deviation. *P < 0.05, ***P < 0.001, ###P < 0.001.
ASPH Regulates Wnt Signal Mediated by Gsk3β
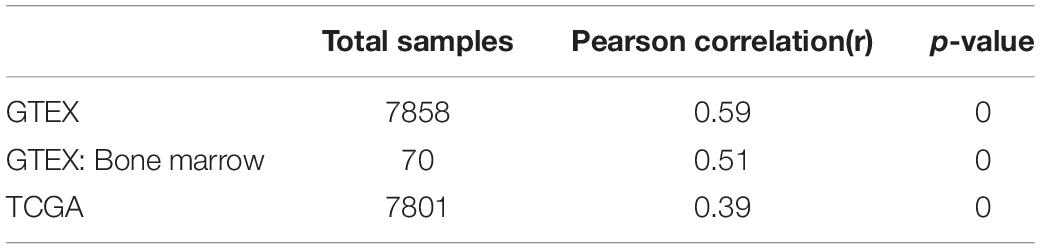
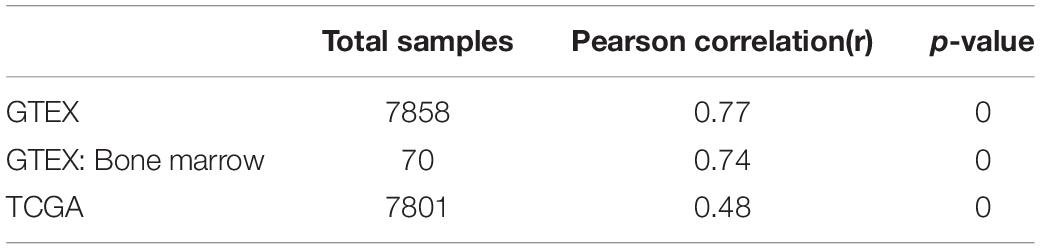
It is well established that Gsk3β and β-catenin have important roles in bone formation. Based on the involvement of ASPH in Wnt signaling and cellular senescence in various cancers, we hypothesized that ASPH was involved in the bone formation and cellular senescence through regulating Gsk3β and β-catenin (Iwagami et al., 2016; Peng et al., 2020b). Thus, we analyzed the co-expression of ASPH with GSK3B and CTNNB1 in GTEX, TACC and CCLE databases. Intriguingly, the analysis showed that ASPH has considerable positive correlations in mRNA expression with GSK3B and CTNNB1 in most of normal/tumor tissues (Figures 5A,B,D,E). Furthermore, ASPH has a moderate or strong correlation with GSK3B and CTNNB1 in the normal tissues including bone marrow from GTEX database (Tables 1, 2). In bone-related cancer cell lines, ASPH has significant correlations with GSK3B (Pearson r = 0.50, p = 0.01) (Figure 5C) and CTNNB1 (Pearson r = 0.54, p = 0) (Figure 5F). Consistent with these correlations, the expression of GSK3B and CTNNB1 showed significant decrease in BMSCs with depletion of Asph (Figures 5G–O). The ratio of phos-Ser9 GSK3β to total Gsk3β has increased in the Asph siRNA transfected BMSCs (Figure 5J). Asph inhibition lead to the downregulation of total β-Catenin as well as active β-Catenin (Figures 5K–O). Consistently, cells transfected with Asph siRNA showed predominantly decreased distribution of the β-Catenin protein both in cytoplasm and nucleus (Figure 6A). The immunofluorescence analysis also suggested Asph inhibition ameliorated distribution of active β-Catenin in cytoplasm (Figure 6B). Taken together, these results suggested that Asph possibly regulated Gsk3β/β-catenin signaling, contributing to bone formation and preventing cellular senescence (Figure 7).
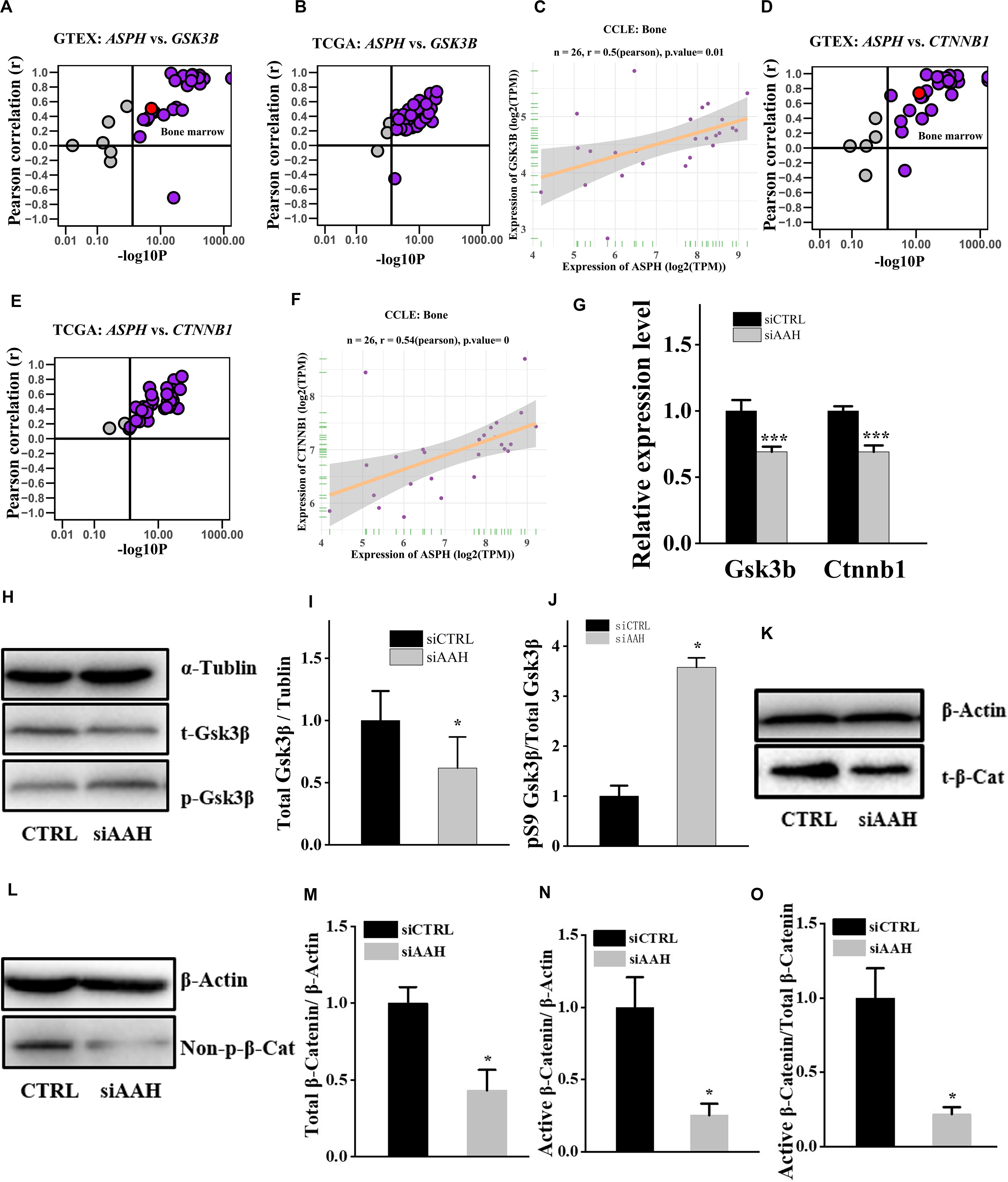
Figure 5. ASPH regulates Gsk3β and β-Catenin. (A–C) Co-expression of ASPH with GSK3B in expression in normal tissues (A), cancer samples (B), and bone-related cell lines (C), based on the data from GTEx, TCGA, and CCLE databases, respectively. (D–F) Co-expression of ASPH with CTNNB1 in expression in normal tissues (D), cancer samples (E), and bone-related cell lines (F), based on the data from GTEx, TCGA, and CCLE databases, respectively. The correlation coefficient (r) and P-value were calculated by Pearson’s Correlation analysis. Note that every dot represents one tissue type (A,D) or one cancer type (B,E). The red dots indicate tissue of bone marrow. (G) qRT-PCR analysis of GSK3B and CTNNB1 in BMSCs interfered with Asph siRNA in vitro. (H) Western Blot analysis of the levels of total GSK3β and phos-Ser9 GSK3β in BMSCs transfected with Asph siRNA. (I) The ratio of total Gsk3β to α-Tublin in BMSCs transfected with Asph siRNA. (J) The ratio of phos-Ser9 GSK3β (pS9 GSK3β) to total GSK3β in BMSCs transfected with Asph siRNA. (K) Western Blot analysis of the levels of total β-Catenin (t-β-Cat) in BMSCs transfected with Asph siRNA. (L) Western Blot analysis of the levels of active β-Catenin [Non-phospho β-Catenin (Ser33/37/Thr41): Non-p-β-Cat] in BMSCs transfected with Asph siRNA. (M) The ratio of total β-Catenin to β-Actin in BMSCs transfected with Asph siRNA. (N) The ratio of active β-Catenin to β-Actin in BMSCs transfected with Asph siRNA. (O) The ratio of active β-Catenin to total β-Catenin in BMSCs transfected with Asph siRNA. These experiments were replicated 3 times. Error bars show standard deviation. *P < 0.05, ***P < 0.001.
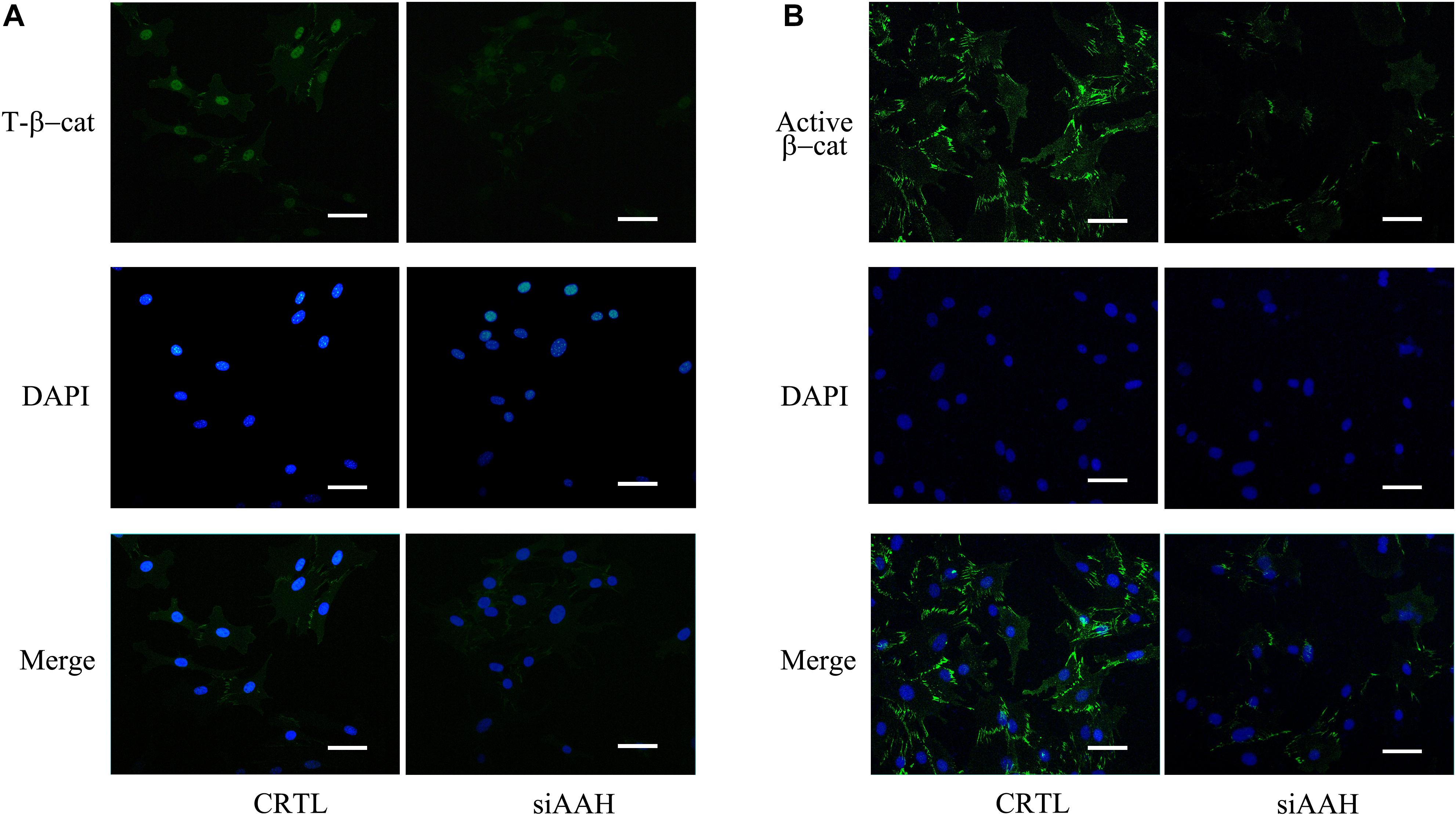
Figure 6. Subcellular localization of total and active β-Catenin. (A) Subcellular localization of total β-Catenin protein in BMSCs transfected with Asph siRNA. Nuclei, DAPI (blue), total β-Catenin (green). (B) Subcellular localization of active β-Catenin [Non-phosph-β-Catenin (Ser33/37/Thr41)] protein in BMSCs transfected with Asph siRNA. Nuclei, DAPI (blue), active β-Catenin (green). Scale bar, 250 μm.
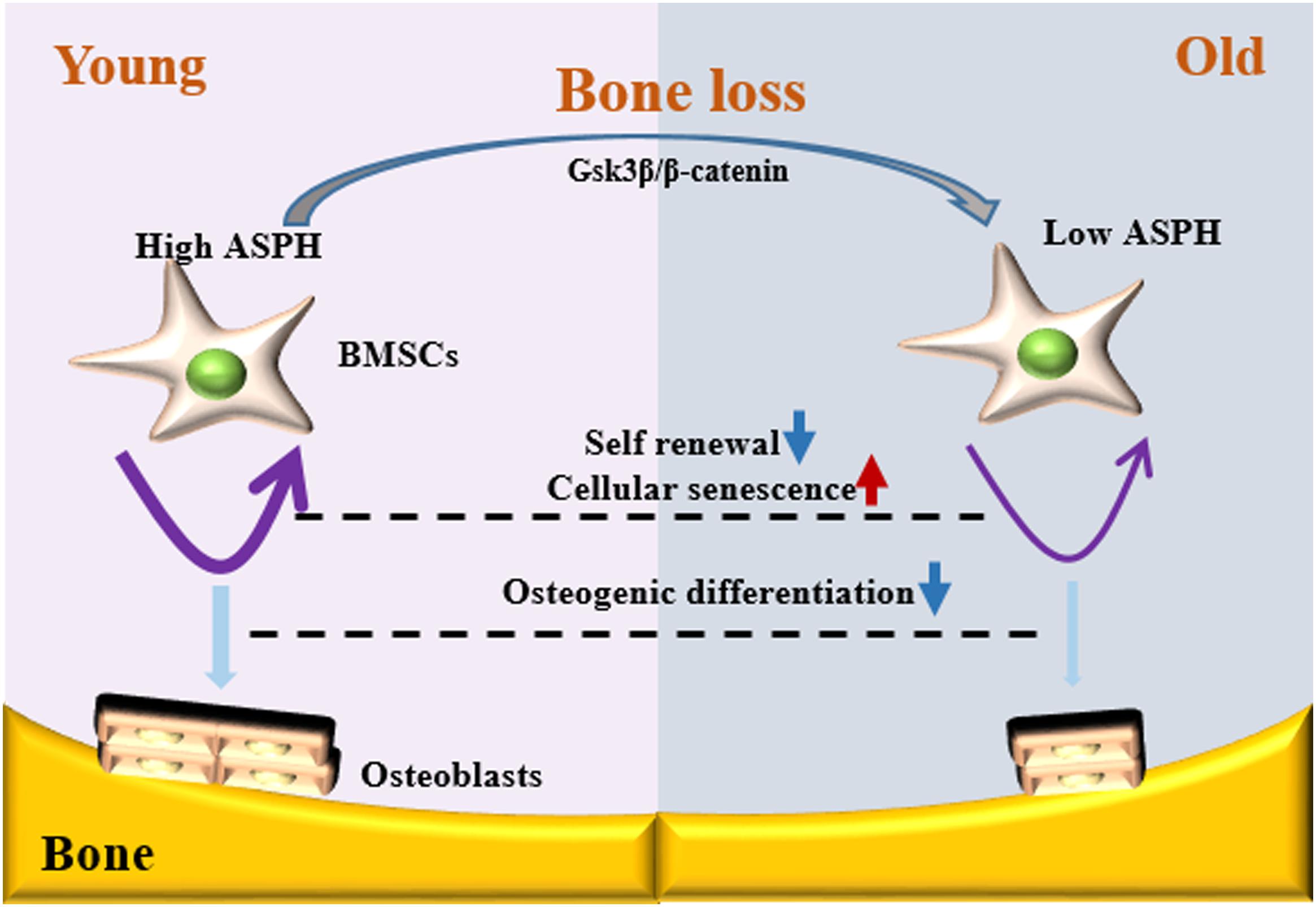
Figure 7. Schematic representation of ASPH regulating cellular senescence and osteogenic differentiation of BMSCs during aging. Low expression of ASPH restrains the accumulation and activation of Gsk3β and β-Catenin to maintain the capacity of self-renewal and osteogenic differentiation. In the old individuals, the decrease of ASPH is companied with the cellular senescence and impaired osteogenic differentiation of BMSCs.
Discussion
Osteogenesis and cellular senescence of BMSCs play great roles in bone formation (Qadir et al., 2020). In this study, our data showed ASPH longest isoform promoting the osteogenesis while inhibiting cellular senescence, indicating it potentially results in an elevated capacity of bone formation. Mechanistically, ASPH modulated the accumulation and activation of Gsk3β and β-catenin. In general, our study revealed ASPH was an aged-dependent modulator to regulate osteogenic differentiation and senescence in BMSCs (Figure 6).
Aspartate β-hydroxylase (ASPH) is a type II membrane protein, which is comprised of several domains, such as N-terminal cytoplasmic domain, transmembrane domain, and C-terminal catalytic domain (Finotti et al., 2008). Through data mining in GEO database, we found some genes differentially expressed in middle-aged subjects in comparison with elderly aged subjects. Through a set of filtering criteria as described in “Results” section, we finally hypothesized ASPH gene was partially involved in bone formation. ASPH is associated with the regulation of protein complex disassembly and regulation of protein depolymerization (Lee et al., 2012), which has been presented in the top 10 signaling through Go analysis (Figure 1D). Furthermore, KEGG analysis showed that the differentially expressed genes enriched in cellular senescence, EGFR tyrosine kinase inhibitor resistance and some cancer development. These observations gave us some clues that ASPH might be involved in bone modeling through regulating cellular senescence.
Accumulated evidence demonstrated that Asph was highly expressed during embryogenesis, which is crucial to maintain cell migration and organ development (Treves et al., 2000; Mahmood et al., 2010; Hou et al., 2018). Patel et al. (2014) performed the whole-mount in situ hybridization of Asph at embryonic day 11.5 (E11.5) and E12.5 mouse embryos. They found a strong expression of Asph in snout, limbs, and eyes (Patel et al., 2014). To date, ASPH mutations identified in patients were established to be associated with Traboulsi syndrome which is characterized by facial dimorphism, lens dislocation, anterior segment abnormalities, and spontaneous filtering blebs (OMIM:601552) (Patel et al., 2014; Chandran et al., 2019), but does not refer to the abnormal bone mass. The knock-out Asph mice also replicated similar phenotypes with these patients (Dinchuk et al., 2002; Yuan et al., 2007; Patel et al., 2014). However, Peggy et al. demonstrated that ASPH expressed differentially in hBMSCs and was thought to be associated with the BMD and risk of bone fracture (Benisch et al., 2012). GWAS results suggested that a mutation occurred in 3’ region of ASPH might be related to bone mass (Koller et al., 2010). In addition, the previous study suggested that ASPH played great roles in the other key signaling pathways like Wnt signaling, Notch signaling and so on, which are all closely involved in bone formation (Tomimaru et al., 2013; Hou et al., 2018). As expected, our study found that ASPH was an important modulator during osteogenesis. The correlation analysis suggested that ASPH had positive correlation with osteogenic markers. The expression of Asph was also increasing during osteogenic differentiation. Moreover, the depletion of Asph inhibited osteogenic differentiation while the overexpression of Asph promoted osteogenic differentiation. Nevertheless, Mantila Roosa et al. (2011) suggested that Asph was downregulated during matrix formation, which is not consistent with our results. In this study, stable expression at common region of ASPH (exon 1–3) and decreased expression at exon 4–25 have been showed in Figure 3B, which indicated that junctin has increased during aging and senescence. Thus, we suspected that ASPH different isoforms might be involved in the same process with opposite role.
Further study revealed that the deficiency of Asph in BMSCs resulting in the dysfunction of osteogenesis and cellular senescence. Conversely, overexpression of longest Asph variant 1 in BMSCs restored the capacity of osteogenic differentiation and prevented the cellular senescence. Gsk3β has been reported to be regulated by ASPH inhibitor in hepatocellular carcinoma (Iwagami et al., 2016). In addition, Gsk3β can phosphorylates the sites of β-catenin at the sites of Thr41/Ser37/Ser33 followed by the degradation of β-catenin (Wu and Pan, 2010; Peng et al., 2020b). Gsk3β-mediated Wnt signaling can promote bone formation and prevent cellular senescence (Ye et al., 2007; Seo et al., 2008; Gillespie et al., 2013). Thus, we investigated if ASPH regulated osteogenic differentiation and cellular senescence in BMSCs through regulating Gsk3β or β-catenin. Of note, positive correlations of ASPH with GSK3B and CTNNB1 have been found in the normal tissues, cancer tissues as well as cancer cell lines. Furthermore, we confirmed these positive correlations in vitro. The deficiency of Asph inhibited the accumulation of Gsk3β. Nevertheless, the non-phosphorylation of β-catenin at Thr41/Ser37/Ser33 has decreased which explained the downregulation of total β-catenin. The immunofluorescence analysis supported the downregulated Wnt signaling as well.
Of note, previous studies suggested that a relatively low level of ASPH expression in normal mature tissues, while abundant expression of ASPH in a variety of malignant tumors (Lavaissiere et al., 1996; Lin et al., 2019). Thus, ASPH has been thought to be a potential therapeutic target for different cancers (Shimoda et al., 2012; Dong et al., 2015; Sturla et al., 2016). However, according to our current data in vitro, it suggested that the treatment of ASPH inhibitor in patients with cancer need to be concerned because of their potential risks of bone loss or bone fracture.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: the GEO database (Accession number: GSE35955; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc~=~GSE35955).
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Xiangya Hospital of Central South University. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Xiangya Hospital of Central South University Ethics Committee.
Author Contributions
HP designed this study, carried out most of the experiments, and wrote the manuscript. QG, TS, YX, T-JJ, and L-JG helped to collect samples. MW supervised the experiments, analyzed results, and proofread the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Grant No. 81873670 from the National Natural Science Foundation of China (NSFC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Prof. Stephen P. Robertson and Dr. Zandra A. Jenkins at the University of Otago for helping to form the hypothesis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00872/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/geo/
- ^ http://cancergenome.nih.gov/
- ^ https://www.gtexportal.org/
- ^ https://portals.broadinstitute.org/ccle
References
Al Anouti, F., Taha, Z., Shamim, S., Khalaf, K., Al Kaabi, L., and Alsafar, H. (2019). An insight into the paradigms of osteoporosis: from genetics to biomechanics. Bone Rep. 11:100216. doi: 10.1016/j.bonr.2019.100216
Benisch, P., Schilling, T., Klein-Hitpass, L., Frey, S. P., Seefried, L., Raaijmakers, N., et al. (2012). The transcriptional profile of mesenchymal stem cell populations in primary osteoporosis is distinct and shows overexpression of osteogenic inhibitors. PLoS One 7:e45142. doi: 10.1371/journal.pone.0045142
Campisi, J., and d’Adda di Fagagna, F. (2007). Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740. doi: 10.1038/nrm2233
Cantarini, M. C., de la Monte, S. M., Pang, M., Tong, M., D’Errico, A., Trevisani, F., et al. (2006). Aspartyl-asparagyl β hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology 44, 446–457. doi: 10.1002/hep.21272
Chandran, P., Chermakani, P., Venkataraman, P., Thilagar, S. P., Raman, G. V., and Sundaresan, P. (2019). A novel 5 bp homozygous deletion mutation in ASPH gene associates with Traboulsi syndrome. Ophthalm. Genet. 40, 185–187. doi: 10.1080/13816810.2019.1605390
Childs, B. G., Durik, M., Baker, D. J., and van Deursen, J. M. (2015). Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat. Med. 21, 1424–1435. doi: 10.1038/nm.4000
Dinchuk, J. E., Focht, R. J., Kelley, J. A., Henderson, N. L., Zolotarjova, N. I., Wynn, R., et al. (2002). Absence of post-translational aspartyl β-hydroxylation of epidermal growth factor domains in mice leads to developmental defects and an increased incidence of intestinal neoplasia. J. Biol. Chem. 277, 12970–12977. doi: 10.1074/jbc.M110389200
Dinchuk, J. E., Henderson, N. L., Burn, T. C., Huber, R., Ho, S. P., Link, J., et al. (2000). Aspartyl β-Hydroxylase (Asph) and an evolutionarily conserved isoform of asph missing the catalytic domain share exons with junctin. J. Biol. Chem. 275, 39543–39554. doi: 10.1074/jbc.M006753200
Dong, X., Lin, Q., Aihara, A., Li, Y., Huang, C.-K., Chung, W., et al. (2015). Aspartate β-hydroxylase expression promotes a malignant pancreatic cellular phenotype. Oncotarget 6, 1231–1248. doi: 10.18632/oncotarget.2840
Finotti, A., Treves, S., Zorzato, F., Gambari, R., and Feriotto, G. (2008). Upstream stimulatory factors are involved in the P1 promoter directed transcription of the AbetaH-J-J locus. BMC Mol. Biol. 9:110. doi: 10.1186/1471-2199-9-110
Gillespie, J. R., Bush, J. R., Bell, G. I., Aubrey, L. A., Dupuis, H., Ferron, M., et al. (2013). GSK-3β function in bone regulates skeletal development, whole-body metabolism, and male life span. Endocrinology 154, 3702–3718. doi: 10.1210/en.2013-1155
Hou, G., Xu, B., Bi, Y., Wu, C., Ru, B., Sun, B., et al. (2018). Recent advances in research on aspartate β-hydroxylase (ASPH) in pancreatic cancer: a brief update. Bosn. J. Basic Med. Sci. 18, 297–304. doi: 10.17305/bjbms.2018.3539
Iwagami, Y., Huang, C.-K., Olsen, M. J., Thomas, J.-M., Jang, G., Kim, M., et al. (2016). Aspartate β-hydroxylase modulates cellular senescence through glycogen synthase kinase 3β in hepatocellular carcinoma. Hepatology 63, 1213–1226. doi: 10.1002/hep.28411
Koller, D. L., Ichikawa, S., Lai, D., Padgett, L. R., Doheny, K. F., Pugh, E., et al. (2010). Genome-Wide association study of bone mineral density in premenopausal european-american women and replication in african-american women. J. Clin. Endocrinol. Metab. 95, 1802–1809. doi: 10.1210/jc.2009-1903
Lavaissiere, L., Jia, S., Nishiyama, M., de la Monte, S., Stern, A. M., Wands, J. R., et al. (1996). Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. J. Clin. Invest. 98, 1313–1323. doi: 10.1172/JCI118918
Lee, K. W., Maeng, J.-S., Choi, J. Y., Lee, Y. R., Hwang, C. Y., Park, S. S., et al. (2012). Role of junctin protein interactions in cellular dynamics of calsequestrin polymer upon calcium perturbation. J. Biol. Chem. 287, 1679–1687. doi: 10.1074/jbc.M111.254045
Li, C., Chai, Y., Wang, L., Gao, B., Chen, H., Gao, P., et al. (2017). Programmed cell senescence in skeleton during late puberty. Nat. Commun. 8:1312. doi: 10.1038/s41467-017-01509-0
Li, H., Liu, P., Xu, S., Li, Y., Dekker, J. D., Li, B., et al. (2017). FOXP1 controls mesenchymal stem cell commitment and senescence during skeletal aging. J. Clin. Invest. 127, 1241–1253. doi: 10.1172/JCI89511
Li, C., Cheng, P., Liang, M., Chen, Y., Lu, Q., Wang, J., et al. (2015). MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Invest. 125, 1509–1522. doi: 10.1172/JCI77716
Li, C., Xiao, Y., Yang, M., Su, T., Sun, X., Guo, Q., et al. (2018). Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. J. Clin. Invest. 128, 5251–5266. doi: 10.1172/JCI99044
Lin, Q., Chen, X., Meng, F., Ogawa, K., Li, M., Song, R., et al. (2019). ASPH-notch axis guided exosomal delivery of prometastatic secretome renders breast cancer multi-organ metastasis. Mol. Cancer 18:156. doi: 10.1186/s12943-019-1077-0
Lin, S. P., Chiu, F. Y., Wang, Y., Yen, M. L., Kao, S. Y., and Hung, S. C. (2014). RB maintains quiescence and prevents premature senescence through upregulation of DNMT1 in mesenchymal stromal cells. Stem Cell Rep. 3, 975–986. doi: 10.1016/j.stemcr.2014.10.002
Mahmood, A., Harkness, L., Schrøder, H. D., Abdallah, B. M., and Kassem, M. (2010). Enhanced differentiation of human embryonic stem cells to mesenchymal progenitors by inhibition of TGF-β/activin/nodal signaling using SB-431542. J. Bone Miner. Res. 25, 1216–1233. doi: 10.1002/jbmr.34
Malaguarnera, R., and Belfiore, A. (2014). The emerging role of insulin and insulin-like growth factor signaling in cancer stem cells. Front. Endocrinol. 5:10. doi: 10.3389/fendo.2014.00010
Mantila Roosa, S. M., Liu, Y., and Turner, C. H. (2011). Gene expression patterns in bone following mechanical loading. J. Bone Miner. Res. 26, 100–112. doi: 10.1002/jbmr.193
Patel, N., Khan, A. O., Mansour, A., Mohamed, J. Y., Al-Assiri, A., Haddad, R., et al. (2014). Mutations in ASPH cause facial dysmorphism, lens dislocation, anterior-segment abnormalities, and spontaneous filtering blebs, or traboulsi syndrome. Am. J. Hum. Genet. 94, 755–759. doi: 10.1016/j.ajhg.2014.04.002
Peng, H., Guo, Q., Xiao, Y., Su, T., Jiang, T., Guo, L., et al. (2020a). ASPH regulates osteogenic differentiation and cellular senescence of BMSCs. Figshare Dataset. doi: 10.6084/m9.figshare.12436757.v1
Peng, H., Jenkins, Z. A., White, R., Connors, S., Hunter, M. F., Ronan, A., et al. (2020b). An activating variant in CTNNB1 is associated with a sclerosing bone dysplasia and adrenocortical neoplasia. J. Clin. Endocrinol. Metab. 105, 688–695. doi: 10.1210/clinem/dgaa034
Peng, H., Yang, M., Guo, Q., Su, T., Xiao, Y., and Xia, Z.-Y. (2019). Dendrobium officinale polysaccharides regulate age-related lineage commitment between osteogenic and adipogenic differentiation. Cell Prolif. 52, 1–10. doi: 10.1111/cpr.12624
Qadir, A., Liang, S., Wu, Z., Chen, Z., Hu, L., and Qian, A. (2020). Senile osteoporosis: the involvement of differentiation and senescence of bone marrow stromal cells. Int. J. Mol. Sci. 21:349. doi: 10.3390/ijms21010349
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. doi: 10.1093/nar/gkv007
Seo, Y.-H., Jung, H.-J., Shin, H.-T., Kim, Y.-M., Yim, H., Chung, H.-Y., et al. (2008). Enhanced glycogenesis is involved in cellular senescence via GSK3/GS modulation. Aging Cell 7, 894–907. doi: 10.1111/j.1474-9726.2008.00436.x
Shimoda, M., Tomimaru, Y., Charpentier, K. P., Safran, H., Carlson, R. I., and Wands, J. (2012). Tumor progression-related transmembrane protein aspartate-β-hydroxylase is a target for immunotherapy of hepatocellular carcinoma. J. Hepatol. 56, 1129–1135. doi: 10.1016/j.jhep.2011.12.016
Sturla, L.-M., Tong, M., Hebda, N., Gao, J., Thomas, J.-M., Olsen, M., et al. (2016). Aspartate-β-hydroxylase (ASPH): a potential therapeutic target in human malignant gliomas. Heliyon 2:e00203. doi: 10.1016/j.heliyon.2016.e00203
Tomimaru, Y., Koga, H., Yano, H., de la Monte, S., Wands, J. R., and Kim, M. (2013). Upregulation of T-cell factor-4 isoform-responsive target genes in hepatocellular carcinoma. Liver Int. 33, 1100–1112. doi: 10.1111/liv.12188
Treves, S., Feriotto, G., Moccagatta, L., Gambari, R., and Zorzato, F. (2000). Molecular cloning, expression, functional characterization, chromosomal localization, and gene structure of junctate, a novel integral calcium binding protein of Sarco(endo)plasmic reticulum membrane. J. Biol. Chem. 275, 39555–39568. doi: 10.1074/jbc.M005473200
Tu, X., Delgado-Calle, J., Condon, K. W., Maycas, M., Zhang, H., Carlesso, N., et al. (2015). Osteocytes mediate the anabolic actions of canonical Wnt/β-catenin signaling in bone. Proc. Natl. Acad. Sci. U.S.A. 112, E478–E486. doi: 10.1073/pnas.1409857112
Wu, D., and Pan, W. (2010). GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem. Sci. 35, 161–168. doi: 10.1016/j.tibs.2009.10.002
Xiao, Y.-Z., Yang, M., Xiao, Y., Guo, Q., Huang, Y., Li, C.-J., et al. (2020). Reducing hypothalamic stem cell senescence protects against aging-associated physiological decline. Cell Metab. 31, 534–548.e5. doi: 10.1016/j.cmet.2020.01.002
Yang, M., Guo, Q., Peng, H., Xiao, Y.-Z., Xiao, Y., Huang, Y., et al. (2019). Krüppel-like factor 3 inhibition by mutated lncRNA Reg1cp results in human high bone mass syndrome. J. Exp. Med. 216, 1944–1964. doi: 10.1084/jem.20181554
Yang, M., Li, C.-J., Sun, X., Guo, Q., Xiao, Y., Su, T., et al. (2017). MiR-497 195 cluster regulates angiogenesis during coupling with osteogenesis by maintaining endothelial Notch and HIF-1α activity. Nat. Commun. 8, 1–11. doi: 10.1038/ncomms16003
Ye, X., Zerlanko, B., Kennedy, A., Banumathy, G., Zhang, R., and Adams, P. D. (2007). Downregulation of Wnt signaling is a trigger for formation of facultative heterochromatin and onset of cell senescence in primary human cells. Mol. Cell 27, 183–196. doi: 10.1016/j.molcel.2007.05.034
Yu, B., Huo, L., Liu, Y., Deng, P., Szymanski, J., Li, J., et al. (2018). PGC-1α controls skeletal stem cell fate and bone-fat balance in osteoporosis and skeletal aging by inducing TAZ. Cell Stem Cell 23, 193–209.e5. doi: 10.1016/j.stem.2018.06.009
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Yuan, Q., Fan, G.-C., Dong, M., Altschafl, B., Diwan, A., Ren, X., et al. (2007). Sarcoplasmic reticulum calcium overloading in junctin deficiency enhances cardiac contractility but increases ventricular automaticity. Circulation 115, 300–309. doi: 10.1161/CIRCULATIONAHA.106.654699
Keywords: ASPH, BMSCs, cellular senescence, osteogenesis, aging
Citation: Peng H, Guo Q, Xiao Y, Su T, Jiang T-J, Guo L-J and Wang M (2020) ASPH Regulates Osteogenic Differentiation and Cellular Senescence of BMSCs. Front. Cell Dev. Biol. 8:872. doi: 10.3389/fcell.2020.00872
Received: 07 June 2020; Accepted: 11 August 2020;
Published: 03 September 2020.
Edited by:
Chao Liang, Hong Kong Baptist University, Hong KongCopyright © 2020 Peng, Guo, Xiao, Su, Jiang, Guo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Wang, bWlud2FuZzExQDEyNi5jb20=
 Hui Peng
Hui Peng Ye Xiao
Ye Xiao Min Wang
Min Wang