- Key Laboratory of Neuroregeneration of Jiangsu and Ministry of Education, Co-innovation Center of Neuroregeneration, Nantong University, Nantong, China
Previously, fidgetin (fign) and its family members fidgetin-like 1 (fignl1) and fidgetin-like 2 (fignl2) were found to be highly expressed during zebrafish brain development, suggesting their functions in the nervous system. In this study, we report the effects of loss-of-function of these genes on development. We designed and identified single-guide RNAs targeted to generate fign, fignl1, and fignl2 mutants and then observed the overall morphological and behavioral changes. Our findings showed that while fign and fignl1 null mutants displayed no significant defects, fignl2 null zebrafish mutants displayed pericardial edema, reduced heart rate, and smaller eyes; fignl2 null mutants responded to the light-darkness shift with a lower swimming velocity. fignl2 mRNAs were identified in vascular endothelial cells by in situ hybridization and re-analysis of an online dataset of single-cell RNAseq results. Finally, we used morpholino oligonucleotides to confirm that fignl2 knockdown resulted in severe heart edema, which was caused by abnormal vascular branching. The zebrafish fignl2 morphants also showed longer axonal length and more branches of caudal primary neurons. Taken together, we summarize that Fignl2 functions on cellular branches in endothelial cells and neurons. This study reported for the first time that the microtubule-severing protein Fignl2 contributes to cell branching during development.
Introduction
Morphology and motility of a cell are determined by the regulation of the cytoskeleton, especially microtubules. Microtubules form highly complex and dynamic arrays that play roles in various aspects of the development and function of cells. Microtubule dynamics are regulated by microtubule-associated proteins, of which microtubule severing proteins (MSPs) such as spastin, katanin, and fidgetin are key regulatory factors of microtubule dynamics (Karabay et al., 2004; Butler et al., 2010; Leo et al., 2015; Menon and Gupton, 2016). These MSPs are members of the AAA (ATPase family associated with various cellular activities) family, which is capable of severing microtubules into short fragments by forming a hexamer that consumes ATP (Sharp and Ross, 2012; McNally and Roll-Mecak, 2018).
Studies on katanin and spastin demonstrated that they primarily cut stable long microtubules into short ones for microtubule transport or elongation at new generating plus ends, while fidgetin (Fign) and its family members may have more functions. fign mutation resulted in mice “fidget” behavior (Yang et al., 2005); human FIGN suppressed microtubule growth via minus-end depolymerization during cell division (Mukherjee et al., 2012); FIGNL1 is regarded to regulate DNA homologous recombination repair (Yuan and Chen, 2013; Kumar et al., 2019) or meiotic crossovers (Girard et al., 2015); Charafeddine et al. reported that Fign12 modulates orientation of cell migration by shearing microtubules (Charafeddine et al., 2015), and O'Rourke el al. showed that Fign12 affected wound healing (O'Rourke et al., 2019). We previously studied Fign's function in rat brain astrocytes and found that Fign depletion resulted in a remarkable increase in tyrosine-modified microtubules, and thus changed the microtubule orientation in the cell cortical region (Hu et al., 2017). Leo et al. reported that Fign knockout in mice neurons increased the unacetylated microtubule mass (Leo et al., 2015). However, Drosophila Fign facilitates microtubule disassembly in dendrites but not in axons after neuron injury (Tao et al., 2016). These inconsistent functional features of Fign family members in different species or cell types have aroused our interest. Therefore, we attempted to comparatively study the functions of Fign family members in one animal model.
Similar to mice, rats and humans, there are three Fign paralogues, namely Fign, Fidgetin-like 1 (Fignl1), and Fidgetin-like 2 (Fignl2) in zebrafish. In the literature on Fign family members' functions, only Fignl1 was reported to be enriched in axons and growth cones of neurons and to play roles in the motor circuit in zebrafish larvae (Fassier et al., 2018), and Fignl1 overexpressed in zebrafish embryos inhibited ciliogenesis and decreased ciliary length (Zhao et al., 2016); however, the functions of Fign and Fignl2 remain elusive. Recently, we investigated the expression patterns of fign and its family members, fidgetin-like 1 (fignl1) and fidgetin-like 2 (fignl2), during zebrafish embryonic development, and fign genes were found highly expressed during brain development, suggesting their functions in the nervous system (Dong et al., 2021). In this study, we aim to compare the developmental features after loss-of-function of these genes to further clarify their functions and possible mechanism.
Results
Generation and Phenotype Analysis of Mutants of fign, fignl1, and fignl2
CRISPR/Cas9 was used to generate mutants harboring new alleles of Fign, Fignl1, and Fignl2 that contain frame-shift mutations. Selected target sites are shown in Table 1 as sense strand genomic sequences. Incross was performed in the F1 generation to produce homozygous mutant F2 and siblings of the other two genotypes shown in this study.
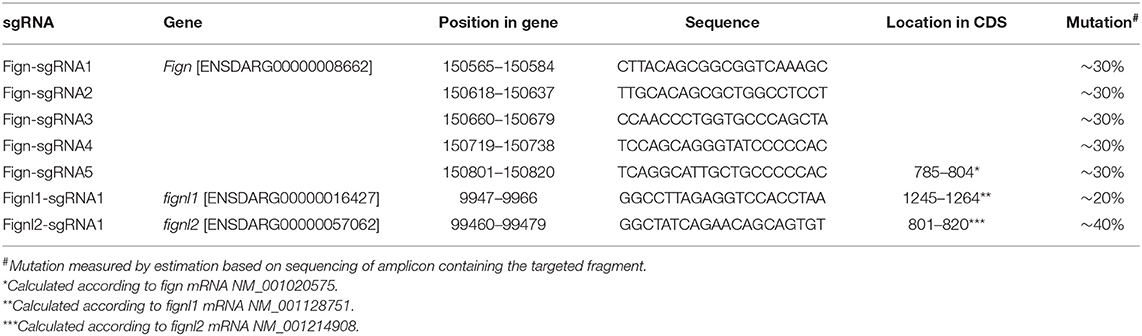
fign sgRNA1-5 were able to induce ~30% indel mutations in injected zebrafish embryos at 24 hpf. sgRNA5 was used to generate mutants for the following studies, producing mutated alleles ntu702 and ntu703 (Figure 1A), resulting in a protein completely missing the AAA domain (Figure 1A). fignl1 sgRNA1 induced a mutation level of 20% in 24 hpf embryos, generating the mutated allele ntu704 (Figure 1A), leading to a complete loss of the AAA-type ATPase domain. fignl2 sgRNA1 was found to be active with a somatic mutation rate of 30–40% in the founders at 24 hpf. One mutated allele was found to have a deletion of 3 bp and an insertion of 1 bp and was named ntu705 (Figure 1A), resulting in complete loss of AAA domain (Figure 1A).
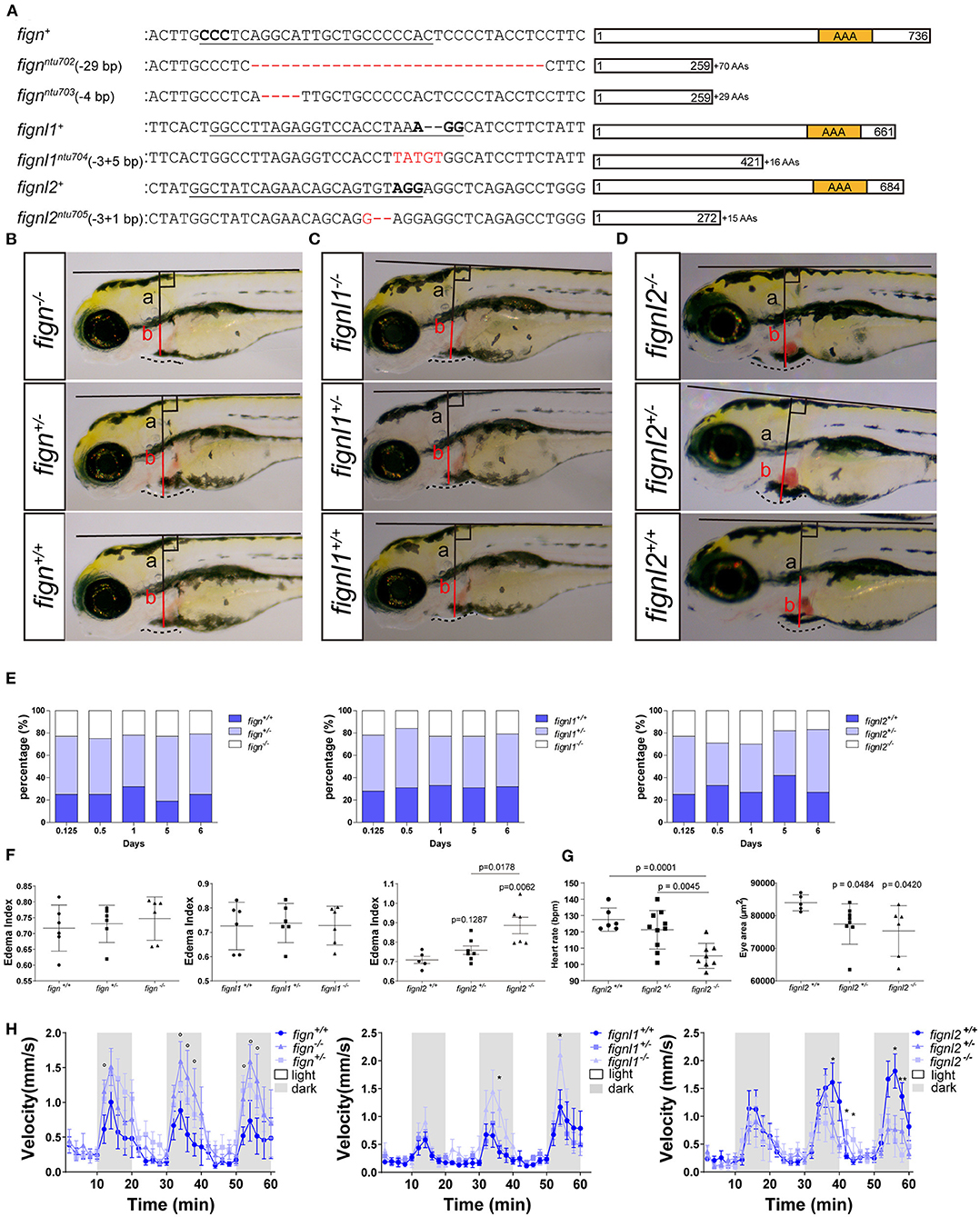
Figure 1. Loss of fignl2 leads to smaller eyes, pericardial edema, and reduced swimming velocity. (A) Mutated alleles generated using CRISPR/Cas9 lead to complete loss of the AAA-type ATPase domain. Underlined sequences are the CRISPR targets, wherein the bases in bold are the proto-spacer adjacent motifs (sequences are shown only on the sense strand). (B–D) Representative phenotype observation and edema index measurement in Fign mutants (B), Fignl1 mutants (C), and Fignl2 mutants (D) at 4 dpf. (E) Percentage of the filial embryos generated by crossing of fign+/ntu702 × fignl2+/ntu702, fignl1+/ntu704 × fignl2+/ntu704 and fignl2+/ntu705 × fignl2+/ntu705, showing partial embryonic lethality of fignl2 loss-of-function. Significance of differences compared to the wild type group is shown as p values on top of each dataset, and those between different mutant genotypes are shown on a horizontal line indicating groups used for comparison. (F) Statistics of pericardial edema index (PEI), n = (6,6,6) and (6,6,6) and (5,8,6); *p < 0.05, showing that the severity of edema was negatively correlated with functional fignl2 alleles. (G) Heart rate of the filial embryos generated by crossing of fignl2+/ntu705 × fignl2+/ntu705 showing abnormalities in the cardiovascular system in the fignl2 mutants, n = 6,10,8. Eyes of the filial embryos generated by crossing of fignl2+/ntu705 × fignl2+/ntu705 showing abnormalities in the cardiovascular system in the fignl2 mutants, n = 5,8,6. (H) Statistics of swimming velocity with 10-min light-dark cycle of fign, fignl1, and fignl2 mutants at 5 dpf, n = (12,25,9) and (14,22,11), and (20,19,9), showing weakened swimming ability of fignl2 mutants. Circles indicate a p < 0.1, asterisks (*) indicate a p < 0.05, and (**) indicates a p < 0.01, as indicated by the Student's t-test between the wild type and homozygous mutant groups.
F2 embryos were imaged for morphological changes at 4 dpf and behavioral analysis at 5 dpf and collected for genotyping to identify any phenotype caused by loss-of-function of fignl2. Embryos with fignl2 mutations were found to have pericardial edema and smaller eyes, and the phenotypes were dependent on gene dose because homozygous mutants showed more severe edema and heart congestion (Figure 1D), while Fign and Fignl1 mutants showed no significant phenotypes in the pericardial area and the heart (Figures 1B,C). Cardiovascular problems may lead to embryonic death; thus, we checked the survival of embryos with mutated fignl2. The ratios of genotypes at 5 and 6 dpf were far from the Mendelian segregation, where the percentage of homozygous mutants was significantly reduced, demonstrating that the survival ability of mutants was weakened by the mutation and suggesting developmental abnormalities caused by loss-of-function of fignl2 (Figure 1E). However, no similar reduction in the number of homozygous mutants during development was observed in fign or fignl1 mutants (Figure 1E). Homozygous mutants had greater pericardial edema index (PEI) compared to wild type and heterozygous mutant siblings (Figure 1F). fignl2 depletion leads to phenotypes with a lower heart rate and smaller eyes (Figure 1G), in addition to pericardial edema. Then, the mutants and wild type siblings were imaged for behavioral analysis using light stimuli. Larvae were treated with a 10-min light and 10-min darkness cycle after an initial 30-min adaptation in darkness. The fignl2 homozygous mutants responded to the light-dark shift with a lower swimming velocity than wild type siblings (Figure 1H), whereas fign and fignl1 knockout mutants were able to swim faster than wild type siblings.
fignl2 Is Highly Expressed in Vascular Endothelial Cells and Neuronal Cells in Zebrafish Compared With fign and fignl1
As the mutants had problems in the cardiovascular system, and the Fign homologs were not previously described to be expressed in the heart and vessels, we then examined the expression of fignl2 during zebrafish development using in situ hybridization. At 24 hpf, fignl2 was mainly observed in the midbrain-hindbrain boundary, hindbrain, and somites as well as in the eyes and pectoral fins. Interestingly, in the family of Fign homologs in zebrafish, only fignl2 was detected at a low level in the caudal vessels using in situ hybridization (Figure 2A), whereas fign and fignl1 were not detected.
Figure 2. Expression of fign, fignl1, and fignl2 in zebrafish embryos. (A) Expression analysis of fignl2 using in situ hybridization. (a1) fignl2 expression is observed in the nervous system and somites at 24 hpf, and weak staining is also observed in caudal blood vessels (arrowhead) (Enlarged in a1′). (a2) in situ hybridization using a fignl2 sense probe as control. (B) Single-cell expression analysis of fign, fignl1, and fignl2 during zebrafish development (a re-analysis of GEO GSE112294). (b1) Single-cells in the dataset (4–24 hpf) are mapped to different tissues. (b2–b4) Expression of fign, fignl1, and fignl2 in the single-cell dataset. (b5) Violin plot of fign, fignl1, and fignl2 expression in the single-cell expression dataset of zebrafish embryos (4–24 hpf). (b2′-b4′) Expression of fign, fignl1, and fignl2 in endothelial cells (18 and 24 hpf). (b5′) Violin plot of fign, fignl1, and fignl2 expression in endothelial cells (18 and 24 hpf). (b1′′) Neuronal cells were isolated by elavl3 expression. (b2′′-b4′′) Expression of fign, fignl1, and fignl2 in elavl3-positive cells. (b5′′) Violin plot of fign, fignl1, and fignl2 expression in elavl3-positive cells. (b5′′′) Violin plot of fign, fignl1, and fignl2 expression in mnx1 positive cells. Expression data for violin plots are normalized and presented as counts per 104.
Due to the limitation of sensitivity of in situ hybridization, we utilized the online resource of single-cell sequencing data of zebrafish embryos (Gene Expression Omnibus accession number GSE112294) (Wagner et al., 2018) to check the expression of fignl2. According to the original research, the single cells were mapped to the forebrain, midbrain, hindbrain, and other tissues. This dataset provided an abundance of resources containing the most systematic temporal expression information, and more importantly, the most in-depth sequencing, allowing analysis of genes expressed at low levels.
We then focused on verifying the expression of fignl2, especially in endothelial cells. We analyzed fignl2 expression during zebrafish development (along with its two paralogues, Figures 2Bb2–5) and labeled cells expressing fignl2 from the dataset (Figure 2Bb4). Cells expressing fignl2 were found in all brain regions and somites. Interestingly, when we highlighted endothelial cells at 18 and 24 hpf; many of these cells showed fignl2 expression (Figure 2Bb4′). According to the clustering results in the original research, cells expressing fignl2 include neural stem cells, neurons, epithelial cells, somite cells, and endothelial cells. Further analysis of the transcript levels of fign, fignl1, and fignl2 in the cluster of endothelial cells revealed that the transcript amount of fignl2 is remarkably more than those of fign and fignl1 (Figures 2Bb2′–b5′).
Using the same method, we distinguished the cluster of neuronal cells according to elavl3 expression and displayed the neuronal cells isolated from the dataset (Figure 2Bb1′′). The fign, fignl1, and fignl2 transcripts are shown in Figures 2Bb2′′–b4′′ and a violin plot (Figure 2Bb5′′), and the results clearly showed that fignl2 had significantly higher transcript levels in neuronal cells than did Fign or Fignl1. Given the fignl2 mutants with a lower swimming velocity, we also extracted the expression data of mnx1-positive motor neurons and analyzed the transcript levels of fign, fignl1, and fignl2 (Figure 2Bb5′′′), and the results showed that the level of fignl2 transcripts was notably higher than those of fign and fignl1.
The above results showed that fignl2 is expressed in more tissues with high levels than its paralogous genes, which may explain why fignl2 null mutant zebrafish displayed serious abnormal phenotypes.
Loss of fignl2 Caused Abnormalities in Branching in Endothelial Cells and Neurons
We then wanted to confirm whether the loss of Fignl2 indeed resulted in heart defects in zebrafish. Genomic gene defects may cause some secondary effects to alter developmental features; thus, we performed gene knockdown using morpholino oligonucleotides (MOs) to determine if similar phenotypes, that is, a phenocopy of the mutants, could be generated. Splicing-blocking MO was designed to target the splicing acceptor of intron 2 in fignl2 (Figure 3A), and a series of doses of the MO were tested. The fignl2-MO resulted in partial (0.3 mM) or complete (0.5 and 0.6 mM) deletion of a 34 bp fragment in the coding sequence due to activation of a cryptic splice site (Figures 3B,C). Then, 0.5 mM splice-blocking MO was injected into 1-cell stage Tübingen (Tu) zebrafish embryos. The phenotypes observed in the fignl2 mutants also appeared in these morphants. At 4 dpf, fignl2 morphants displayed pericardial edema and reduced eye size. fignl2 morphants with pericardial edema had a lower heart rate than did those injected with control MO (Figures 3D–D′′′). Morphants with higher levels of MO displayed more severe phenotypes (data not shown).
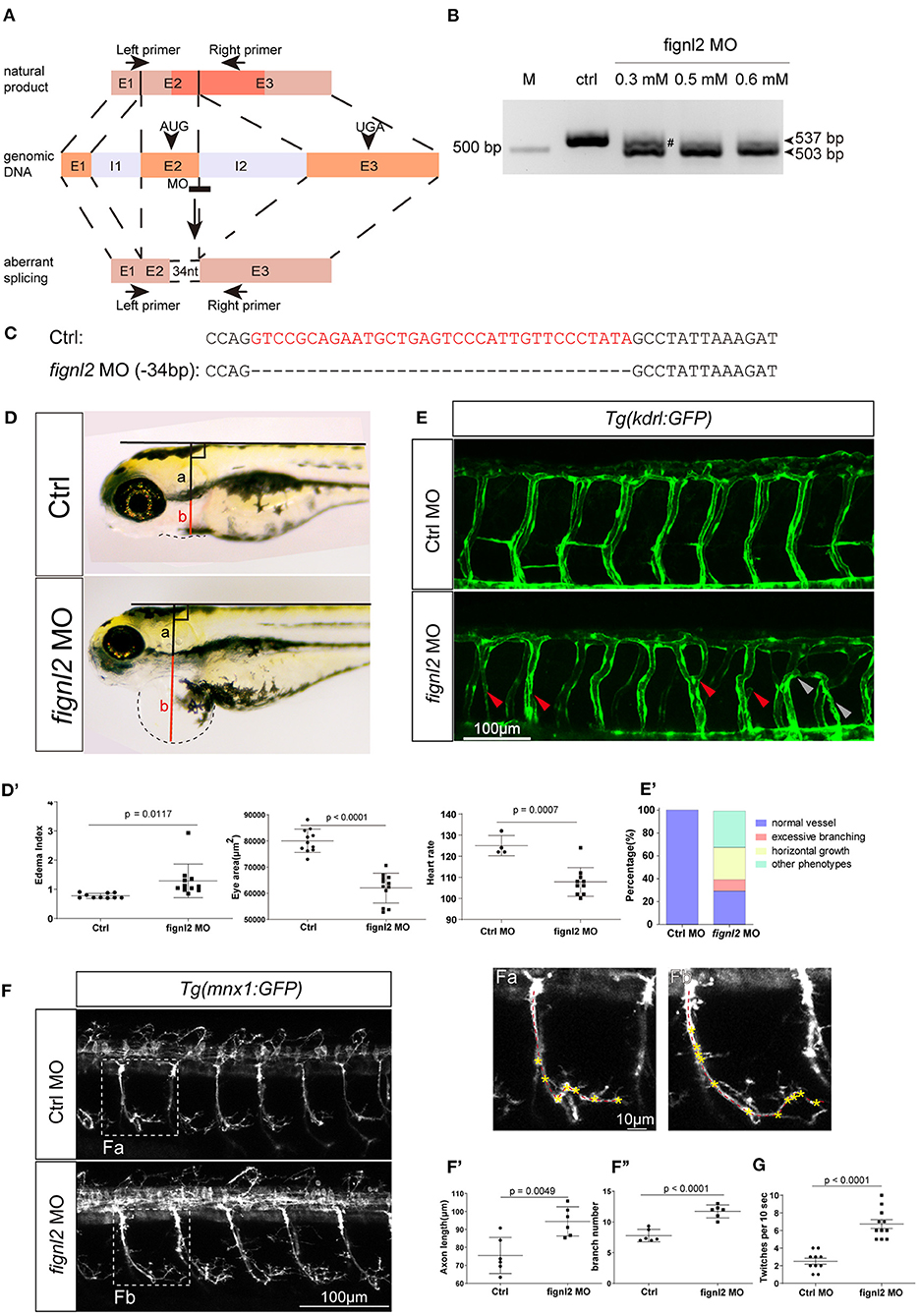
Figure 3. Depletion of fignl2 causes morphological changes in intersegmental vessels (ISVs) and caudal primary neurons. (A) Schematic diagram of morpholino oligonucleotide (MO) induced fignl2 knockdown causing aberrant splicing. (B) MO treatment results in mRNA size change in fignl2 RT-PCR amplicon. The pound sign (#) shows some amplicons of the original size detected after treatment with 0.3 mM MO. (C) MO treatment results in a 34-bp deletion in fignl2 mRNA. (D) fignl2 knockdown causes pericardial edema in developing zebrafish. Graphical representation of edema index (D′), eye size (D′′), heart rate (D′′′) in Ctrl and fignl2 knockdown zebrafish. Ctrl MO, n = 10; fignl2 MO, n = 11. (E) fignl2 knockdown causes morphological changes in ISVs, arrows showing malformations. Graphical representation (E′) of the percentage of different form of ISV malformations caused by fignl2 knockdown. Ctrl MO, n = 10; fignl2 MO, n = 11. (F) fignl2 knockdown causes morphological changes in caudal primary neurons, including greater axon length and increased branching. Representative axons were shown enlarged in (Fa) and (Fb). Graphical representation of axon length (F′), primary branch numbers (F′′) in Ctrl and fignl2 knockdown zebrafish. Ctrl MO, n = 10; fignl2 MO, n = 11. (G) fignl2 mutants shows more frequent twitching. Twitching was counted in 10 s. Ctrl MO, n = 10; fignl2 MO, n = 11.
Pericardial edema suggested abnormalities in the cardiovascular system during development. To further understand the function of fignl2 in zebrafish development, we generated fignl2 morphants using MO in transgenic zebrafish Tg(kdrl:GFP) labeling endothelial cells (Jin et al., 2005). fignl2 MO was injected into 1-cell stage Tg(kdrl:GFP) embryos, and the embryos were used for morphological observation and blood vessel imaging at 48 hpf.
Intersegmental blood vessels (ISVs) in fignl2 morphants showed more branching and an improper direction of growth. Some ISVs were linked to a neighboring ISV to form a Y shape or a circle (Figures 3E,E′), which was observed in both segmental arteries and veins, identified by whether they connected to the dorsal aorta or posterior cardinal vein (Ellertsdottir et al., 2010). Fignl2 is known to be a member of the AAA-type ATPase family with microtubule-severing function, and it is involved in regulating microtubule behaviors such as cellular pseudopodia extension and branching. We injected CRISPR/Cas9 in different combinations to determine whether the fign family members differ in their function in regulating endothelial branching. Co-injection of Cas9 mRNA and sgRNAs targeting fign and fignl1 caused minor defects in ISV branching, and in the embryos that received co-injection of CRISPR/Cas9 targeting fign, fignl1, and fignl2, the morphological changes seemed to be mainly contributed by mutation of fignl2 (Supplementary Figure 1A).
Microtubule dynamics are essential not only in endothelial branching, but also in the development of other cell types, especially in axon extension, and branching in neurons. Since the loss-of-function of Fignl2 affects branching in endothelial cells, it may cause changes in neurons where it is not only expressed but at a higher level. We performed similar treatments and observations at 28 hpf in Tg(mnx1:GFP) (Flanagan-Steet et al., 2005) (caudal primary neurons expressing GFP) for the effect of fignl2 knockdown, and the loss of fignl2 led to elongated axons and increased the number of branches in mnx1-expressing neurons (Figures 3F–F′′′) at 28 hpf and at 5 dpf (Supplementary Figure 2). Co-injection of Cas9 mRNA and sgRNAs targeting fign and fignl1 reduced neurite length, which was consistent with the reported observation with MO targeting fignl1 (Fassier et al., 2018). However, loss of fignl2 resulted in longer neurites and more branching, and a similar result was observed in the embryos that received injection of CRISPR/Cas9 targeting fign, fignl1, and fignl2 (Supplementary Figure 2).
Twitching of zebrafish embryos represents a type of spontaneous movement (Muto et al., 2011). We observed and recorded, and unexpectedly found that the spontaneous twitching at 28 hpf was more frequent in fignl2 morphants (Figure 3G and Supplementary Video 1).
Discussion and Conclusion
As members of the AAA ATPase family, fign paralogous genes play various roles, and their functions are relatively poorly understood for a short research history. We previously investigated and compared the expression patterns of fign, fignl1, and fignl2 during zebrafish development and found that Fignl2 is expressed at higher levels in the nervous system and other tissues than fign and fignl1 genes, which is consistent with the result of re-analyzed online single-cell RNAseq data (GEO GSE112294) (Wagner et al., 2018). Furthermore, fign, fignl1, and fignl2 were also shown to be expressed in zebrafish endothelial cells. However, knockout of fign or fignl1 did not yield obvious phenotypes, whereas depletion of fignl2 (at the genomic DNA level or mRNA level) caused serious zebrafish defects such as pericardial edema, reduced heart rate, and reduced swimming velocity. These hinted that Fignl2 may have some important functions, which Fign or Fignl1 fails to compensate, but not vice versa. We further used GFP-labeled vascular endothelial cells or motor neurons to reveal that fignl2 knockdown resulted in abnormal vascular branching as well as longer axonal length and more branches of caudal primary neurons, clarifying that Fignl2 regulates cellular branching. The pericardial edema or the weakened reaction to the lighting shifts in Fignl2-depleted zebrafish may be due to abnormal branching in endothelial cells or neurons. Twitching behavior, the first behavior in developing zebrafish, was used to evaluate the activity of the motor circuit (Muto et al., 2011). Twitching was more frequent in the fignl2 morphants (Figure 3G and Supplementary Video 1), suggesting that depletion of fignl2 impaired spontaneous movement, while the decreased response after light-to-darkness shift suggested changes in vision-related circuits in the fignl2 morphants.
Although there were some differences between the morphants and the mutants, which could have been caused by genetic compensations (Rossi et al., 2015), they exhibited similar changes after knock down or knockout. These results, together with those of the rescue experiment using fignl2 mRNA, indicate that the phenotypes seen in the endothelial cells and mnx1 neurons are fignl2-specific.
Cellular branch formation is a universal morphological change during development. In developmental processes, including angiogenesis and neurogenesis, microtubule dynamics regulates cell branching morphogenesis by mediating branch orientation or extension (Liu et al., 2010; Myers et al., 2011; Hu et al., 2012; Lyle et al., 2014; Dong et al., 2019). This study presented cellular branches alteration in morphology of ISVs and CaP neurons. Microtubule severing proteins like Fign homologs strongly affect microtubule length and direction in neurons (Leo et al., 2015; Tao et al., 2016; Fassier et al., 2018; Matamoros et al., 2019) and other cell types (Hu et al., 2017), and our study offered clear data that Fignl2 is involved in cell branching.
Our previous study showed that fign, fignl1, and fignl2 are similar in their high expression in the central nervous system during zebrafish early development, while fignl2 is also expressed in other tissues, e.g., pronephros, where fign and fignl1 are not detectable (Dong et al., 2021). The difference in expression patterns indicate figns function during cell branching may have differentiated. Although animals with ablated fign, fignl1, and/or fignl2 tend to swim fast under constant lighting, fignl2 null mutants responded to “light to darkness shift” more weakly than their wildtype or heterozygous siblings, indicating this change in vision-related behavior resulted from eye defect due to fignl2 loss of function.
In summary, this study provided comparative results of the preliminary functional analyses of fign, fignl1, and fignl2 in one zebrafish developmental system. Here, we report the finding that the microtubule-severing protein, Fignl2, contributes to proper cell branching during endothelial and neuronal development.
Materials and Methods
Zebrafish Husbandry
Zebrafish were housed in the Zebrafish Center at Nantong University. Zebrafish embryos of Tübingen and Tg(flk:GFP) were obtained through natural mating and maintained at 28.5°C. Embryos older than 24 h post-fertilization (hpf) were treated with 0.2 mM 1-phenyl-2-thio-urea (PTU, Sigma P7629, a tyrosinase inhibitor commonly used to block pigmentation and aid visualization of zebrafish development). Breeding of fign mutant zebrafish was performed by Nanjing YSY Biotech Company Ltd.
Bioinformatics
The zebrafish fignl2 genomic information was obtained from GenBank (Gene ID: 561837, mRNA NM_001214908.1, protein NP_001201837.1). Conserved domains of the fignl2 proteins were localized according to the Ensembl database (http://www.ensembl.org/). fignl2 sequences were aligned using Vector NTI software (http://www.thermofisher.com/). The phylogenetic tree was generated using MEGA X (https://www.megasoftware.net/) from aligned sequences generated with Clustal W (https://www.genome.jp/tools-bin/clustalw). The zebrafish embryo single-cell sequencing dataset (Wagner et al., 2018) was obtained from (https://kleintools.hms.harvard.edu/paper_websites/wagner_zebrafish_timecourse2018/mainpage.html) and visualized using SPRING (Weinreb et al., 2018). The run.py script for data preparation was modified to import original clustering results for visualization and is provided as Supplementary Document 1. Expression analyses were performed using Scanpy 1.5.1 (https://github.com/theislab/scanpy/) in a Python 3.8.3 environment, where neurons were extracted using the marker gene elavl3, and mnx1-positive neurons were extracted regarding mnx1 expression. Expression data of endothelial cells were extracted according to the original clustering results provided in the dataset. The Python script is provided in Supplementary Document 2.
Morpholino and Microinjection
The sgRNA templates were prepared by PCR with a forward primer composed of the first 17 bases of minimum T7 promoter followed by 20 bases identical to target proto-spacer and 20 bases identical to the first 20 bases of sgRNA scaffold, a reverse primer complementary to the last 25 bases of sgRNA scaffold (Table 2) and a template plasmid pT7-gRNA kindly provided by Prof. Bo Zhang at Peking University. The PCR products were used as templates for in vitro transcription using MAXIscript T7 Kit (Invitrogen, USA) to obtain sgRNAs. Capped Cas9 mRNA was prepared by in vitro transcription using mMessage mMachine T7 Kit (Invitrogen, USA) with a zebrafish optimized Cas9 template plasmid pGH-T7-zCas9 (Liu et al., 2014) kindly provided by Prof. Bo Zhang at Peking University. Cas9 mRNA and sgRNA were mixed and adjusted to a final concentration of 300 ng/μl:100 ng/μl. The MOs were synthesized by Gene Tools Company. MO antisense oligomers were prepared at a stock concentration of 1 mM according to the manufacturer's protocol. The sequence of zebrafish fignl2 splicing MO in this study was 5′-TCAGAAATGTAGCACTTACTATAGG-3′ and the standard control MO was 5′-CCTCTTACCTCAGTTACAATTTATA-3′. MOs or mixtures containing 300 ng/μl Cas9 mRNA and 100 ng/μL sgRNA were injected into Tg(flk:EGFP) or Tg(mnx1:EGFP) embryos at 1 nL of solution per embryo using borosilicate glass capillaries (Sutter, USA) pulled using a P-97 micropipette puller (Sutter, USA) and connected to an IM-400 pneumatic microinjector (Narishige, Japan).

Table 2. PCR primers for gRNA template preparation and genotyping of the mutants produced in this study.
RNA Extraction, Reverse Transcription, and RT-PCR
Tissue was homogenized and frozen in TRIzol UP (TransGen Biotech, Beijing, China) and stored at −80°C. Total RNA was extracted following the manufacturer's instructions. RNA (1 μg) was reverse-transcribed into cDNA using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions. Synthesized cDNA was stored at −20°C.
Whole Mount in situ Hybridizations
The 24 hpf cDNA served as templates for cloning fignl2 fragments to make antisense RNA probes for zebrafish fignl2. The fignl2 primers for RT-PCR were: Left primer, 5′-TCCTGCTATTTGGCCCTCAA-3′; Right primer, 5′-ACAACTCCCTTTCGCTGAGA-3′; the amplicon is a 441 bp fragment in the coding sequence for fignl2. Digoxigenin (DIG)-labeled RNA sense and antisense probes were made from the linearized plasmids using the DIG RNA Labeling Kit (SP6/T7) (Roche). Whole mount in situ hybridization was performed following the methods as previously described (Schulte-Merker et al., 1992). The concentration of probes was 2 ng/μl in fresh hyb+, and for blocking purpose, 2% blocking reagent (Boehringer blocking reagent, Roche), 10% sheep serum (Sigma), and 70% MAB (YSY, Nanjing, China) were used.
Imaging
For confocal imaging of blood vessel development in Tg(flk:GFP) zebrafish and the neurons in Tg(mnx1:GFP) zebrafish, embryos were anesthetized with egg water/0.16 mg/mL tricaine (Sigma E10521)/1% 1-phenyl-2-thiourea (Sigma P7629) and embedded in 0.6% agarose. Images were taken using a Leica TCS SP5 LSM confocal microscope (Leica, Wetzlar, Germany). The analysis was performed using Imaris (http://www.bitplane.com/). For behavioral studies, zebrafish juveniles were imaged using the Noldus DanioVision system and analyzed using EthoVision XT software (https://www.noldus.com/). Animals were placed in a 48-well plate, each juvenile occupying a well. Before the beginning of detection, the light was kept off for 30 min, followed by three tandem 10 min light/10 min darkness cycles with video recording for light/dark shift behavior studies, or 15 min with the light on for spontaneous behavior observation.
Measurement and Statistics
Axon length and branching were measured and counted as previously described (Dong et al., 2019). We measured the extent of pericardial edema by defining a PEI: from the center of the pericardium (P), a perpendicular to the extended back midline was made and let the intersection be A; let the distance from P to A be a and the radius of the pericardium be b; the pericardial edema index is defined as b/(b-a). All data analysis, statistical comparisons, and graphs were generated using GraphPad Prism 5 (http://www.graphpad.com/scientific-software/prism/). Significance of differences was analyzed using Student's t-test. Data are expressed as mean ± S.E.M. (standard error of the mean).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee on Animal Experimentation of Nantong University.
Author Contributions
ZD, XC, YL, RZ, and XL carried out the experiments, data collection, and analysis. ZD and ML prepared the manuscript. ZD and ML contributed to study design and management. All authors contributed to the article and approved the submitted version.
Funding
This study was supported from the National Natural Science Foundation of China (31701049 and 32070725), grants from Natural science foundation of Jiangsu Province (BK20150404 and BK20171253), funding from the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions, and the Large Instruments Open Foundation of Nantong University (KFJN2078).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.593234/full#supplementary-material
References
Butler, R., Wood, J. D., Landers, J. A., and Cunliffe, V. T. (2010). Genetic and chemical modulation of spastin-dependent axon outgrowth in zebrafish embryos indicates a role for impaired microtubule dynamics in hereditary spastic paraplegia. Dis. Model. Mech. 3, 743–751. doi: 10.1242/dmm.004002
Charafeddine, R. A., Makdisi, J., Schairer, D., O'Rourke, B. P., Diaz-Valencia, J. D., Chouake, J., et al. (2015). Fidgetin-Like 2: a microtubule-based regulator of wound healing. J. Invest. Dermatol. 135, 2309–2318. doi: 10.1038/jid.2015.94
Dong, Z., Li, Y., Chen, X., Lai, X., and Liu, M. (2021). A comparative study of the expression patterns of Fign family members in zebrafish embryonic development. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 251:110522. doi: 10.1016/j.cbpb.2020.110522
Dong, Z., Wu, S., Zhu, C., Wang, X., Li, Y., Chen, X., et al. (2019). Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9-mediated kif15 mutations accelerate axonal outgrowth during neuronal development and regeneration in zebrafish. Traffic 20, 71–81. doi: 10.1111/tra.12621
Ellertsdottir, E., Lenard, A., Blum, Y., Krudewig, A., Herwig, L., Affolter, M., et al. (2010). Vascular morphogenesis in the zebrafish embryo. Dev. Biol 341, 56–65. doi: 10.1016/j.ydbio.2009.10.035
Fassier, C., Freal, A., Gasmi, L., Delphin, C., Ten Martin, D., De Gois, S., et al. (2018). Motor axon navigation relies on fidgetin-like 1-driven microtubule plus end dynamics. J. Cell Biol. 217, 1719–1738. doi: 10.1083/jcb.201604108
Flanagan-Steet, H., Fox, M. A., Meyer, D., and Sanes, J. R. (2005). Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development 132, 4471–4481. doi: 10.1242/dev.02044
Girard, C., Chelysheva, L., Choinard, S., Froger, N., Macaisne, N., Lemhemdi, A., et al. (2015). AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 11:e1005369. doi: 10.1371/journal.pgen.1005369
Hu, J., Bai, X., Bowen, J. R., Dolat, L., Korobova, F., Yu, W., et al. (2012). Septin-driven coordination of actin and microtubule remodeling regulates the collateral branching of axons. Curr. Biol. 22, 1109–1115. doi: 10.1016/j.cub.2012.04.019
Hu, Z., Feng, J., Bo, W., Wu, R., Dong, Z., Liu, Y., et al. (2017). Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Mol. Bio. Cell 28, 545–553. doi: 10.1091/mbc.E16-09-0628
Jin, S. W., Beis, D., Mitchell, T., Chen, J. N., and Stainier, D. Y. (2005). Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209. doi: 10.1242/dev.02087
Karabay, A., Yu, W., Solowska, J. M., Baird, D. H., and Baas, P. W. (2004). Axonal growth is sensitive to the levels of katanin, a protein that severs microtubules. J. Neurosci. 24, 5778–5788. doi: 10.1523/JNEUROSCI.1382-04.2004
Kumar, R., Duhamel, M., Coutant, E., Ben-Nahia, E., and Mercier, R. (2019). Antagonism between BRCA2 and FIGL1 regulates homologous recombination. Nucleic Acids Res. 47, 5170–5180. doi: 10.1093/nar/gkz225
Leo, L., Yu, W., D'Rozario, M., Waddell, E. A., Marenda, D. R., Baird, M. A., et al. (2015). Vertebrate fidgetin restrains axonal growth by severing labile domains of microtubules. Cell Rep. 12, 1723–1730. doi: 10.1016/j.celrep.2015.08.017
Liu, D., Wang, Z., Xiao, A., Zhang, Y., Li, W., Zu, Y., et al. (2014). Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. J. Genet. Genomics 41, 43–46. doi: 10.1016/j.jgg.2013.11.004
Liu, M., Nadar, V. C., Kozielski, F., Kozlowska, M., Yu, W., and Baas, P. W. (2010). Kinesin-12, a mitotic microtubule-associated motor protein, impacts axonal growth, navigation, and branching. J. Neurosci. 30, 14896–14906. doi: 10.1523/JNEUROSCI.3739-10.2010
Lyle, K. S., Corleto, J. A., and Wittmann, T. (2014). Microtubule dynamics regulation contributes to endothelial morphogenesis. Bioarchitecture 2, 220–227. doi: 10.4161/bioa.22335
Matamoros, A. J., Tom, V. J., Wu, D., Rao, Y., Sharp, D. J., and Baas, P. W. (2019). Knockdown of fidgetin improves regeneration of injured axons by a microtubule-based mechanism. J. Neurosci. 39, 2011–2024. doi: 10.1523/JNEUROSCI.1888-18.2018
McNally, F. J., and Roll-Mecak, A. (2018). Microtubule-severing enzymes: From cellular functions to molecular mechanism. J. Cell Biol. 217, 4057–4069. doi: 10.1083/jcb.201612104
Menon, S., and Gupton, S. L. (2016). Building blocks of functioning brain: cytoskeletal dynamics in neuronal development. Int. Rev. Cell Mol. Biol. 322, 183–245. doi: 10.1016/bs.ircmb.2015.10.002
Mukherjee, S., Diaz Valencia, J. D., Stewman, S., Metz, J., Monnier, S., Rath, U., et al. (2012). Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle 11, 2359–2366. doi: 10.4161/cc.20849
Muto, A., Ohkura, M., Kotani, T., Higashijima, S., Nakai, J., and Kawakami, K. (2011). Genetic visualization with an improved GCaMP calcium indicator reveals spatiotemporal activation of the spinal motor neurons in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 108, 5425–5430. doi: 10.1073/pnas.1000887108
Myers, K. A., Applegate, K. T., Danuser, G., Fischer, R. S., and Waterman, C. M. (2011). Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J. Cell Biol. 192, 321–334. doi: 10.1083/jcb.201006009
O'Rourke, B. P., Kramer, A. H., Cao, L. L., Inayathullah, M., Guzik, H., Rajadas, J., et al. (2019). Fidgetin-Like 2 siRNA enhances the wound healing capability of a surfactant polymer dressing. Adv. Wound Care 8, 91–100. doi: 10.1089/wound.2018.0827
Rossi, A., Kontarakis, Z., Gerri, C., Nolte, H., Holper, S., Kruger, M., et al. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233. doi: 10.1038/nature14580
Schulte-Merker, S., Ho, R. K., Herrmann, B. G., and Nusslein-Volhard, C. (1992). The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development 116, 1021–1032.
Sharp, D. J., and Ross, J. L. (2012). Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125, 2561–2569. doi: 10.1242/jcs.101139
Tao, J., Feng, C., and Rolls, M. M. (2016). The microtubule-severing protein fidgetin acts after dendrite injury to promote their degeneration. J. Cell Sci. 129, 3274–3281. doi: 10.1242/jcs.188540
Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G., and Klein, A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. doi: 10.1126/science.aar4362
Weinreb, C., Wolock, S., and Klein, A. M. (2018). SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248. doi: 10.1093/bioinformatics/btx792
Yang, Y., Mahaffey, C. L., Berube, N., Nystuen, A., and Frankel, W. N. (2005). Functional characterization of fidgetin, an AAA-family protein mutated in fidget mice. Exp. Cell Res. 304, 50–58. doi: 10.1016/j.yexcr.2004.11.014
Yuan, J., and Chen, J. (2013). FIGNL1-containing protein complex is required for efficient homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 110, 10640–10645. doi: 10.1073/pnas.1220662110
Keywords: development, branching, neuron, vascular endothelial cells, fidgetin-like 2
Citation: Dong Z, Chen X, Li Y, Zhuo R, Lai X and Liu M (2021) Microtubule Severing Protein Fignl2 Contributes to Endothelial and Neuronal Branching in Zebrafish Development. Front. Cell Dev. Biol. 8:593234. doi: 10.3389/fcell.2020.593234
Received: 10 August 2020; Accepted: 21 December 2020;
Published: 18 January 2021.
Edited by:
Pedro Roda-Navarro, Universidad Complutense de Madrid, SpainReviewed by:
Srikala Raghavan, Institute for Stem Cell Science and Regenerative Medicine (inStem), IndiaAurnab Ghose, Indian Institute of Science Education and Research, Pune, India
Copyright © 2021 Dong, Chen, Li, Zhuo, Lai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Liu, bGl1bWVpQG50dS5lZHUuY24=
†These authors have contributed equally to this work
 Zhangji Dong
Zhangji Dong Xu Chen
Xu Chen Yuanyuan Li†
Yuanyuan Li† Mei Liu
Mei Liu