- 1Department of Nephrology, Shanghai Kidney Development and Pediatric Kidney Disease Research Center, Children’s Hospital of Fudan University, Shanghai, China
- 2State Key Laboratory of Genetic Engineering and National Center for International Research of Development and Disease, Institute of Developmental Biology and Molecular Medicine, Collaborative Innovation Center of Genetics and Development, School of Life Sciences, Fudan University, Shanghai, China
Congenital anomalies of the kidney and urinary tract (CAKUT) is a common birth defect and is the leading cause of end-stage renal disease in children. The etiology of CAKUT is complex and includes mainly genetic and environmental factors. However, these factors cannot fully explain the etiological mechanism of CAKUT. Recently, participation of long non-coding RNAs (lncRNAs) in the development of the circulatory and nervous systems was demonstrated; however, the role of lncRNAs in the development of the kidney and urinary tract system is unclear. In this study, we used the piggyBac (PB) transposon-based mutagenesis to construct a mouse with lncRNA 4933425B07Rik (Rik) PB insertion (RikPB/PB) and detected overexpression of Rik and a variety of developmental abnormalities in the urinary system after PB insertion, mainly including renal hypo/dysplasia. The number of ureteric bud (UB) branches in the RikPB/PB embryonic kidney was significantly decreased in embryonic kidney culture. Only bone morphogenetic protein 4 (Bmp4), a key molecule regulating UB branching, is significantly downregulated in RikPB/PB embryonic kidney, while the expression levels of other molecules involved in the regulation of UB branching were not significantly different according to the RNA-sequencing (RNA-seq) data, and the results were verified by quantitative real-time polymerase chain reaction (RT-PCR) and immunofluorescence assays. Besides, the expression of pSmad1/5/8, a downstream molecule of BMP4 signaling, decreased by immunofluorescence. These findings suggest that abnormal expression of Rik may cause a reduction in the UB branches by reducing the expression levels of the UB branching-related molecule Bmp4, thus leading to the development of CAKUT.
Introduction
Congenital anomalies of the kidney and urinary tract (CAKUT) are a group of developmental diseases with abnormal anatomical structure of the urinary system. The phenotype of CAKUT is broadly defined to include abnormalities of the kidneys, ureters, bladder, and urethra. CAKUT is a common developmental birth defect in children. On average, the prevalence of CAKUT varies from 3 to 6 per 1,000 newborns (Nicolaou et al., 2015), accounting for 20–30% of congenital birth defects (Murugapoopathy and Gupta, 2020). CAKUT is the leading cause of chronic kidney disease (CKD) in children, accounting for 40–50% of children receiving renal replacement therapy (Chesnaye et al., 2014), which is a heavy burden on the families of the children and on society.
The development of the mammalian urinary system includes three phases: pronephros, mesonephros, and metanephros. Metanephros eventually develops into a mature kidney. Metanephros development depends on the interaction between two types of cells: metanephric mesenchyme (MM) cells and ureteric bud (UB) cells. Metanephros development starts at embryonic day (E) 10.5. A signal from MM induces UB outgrowth from the nephric duct, and the UB then penetrates the MM. On E11.5, the UB forms the first T-shaped structure. After iterative branching, the UB eventually forms a renal collecting system; at the same time, a signal from the UB induces epithelial differentiation of MM cells with subsequent gradual nephron formation (Blake and Rosenblum, 2014; Short and Smyth, 2016). At this stage, abnormal UB outgrowth and branching leads to CAKUT. Insufficient UB outgrowth causes renal dysplasia, excessive UB outgrowth leads to a duplex renal collecting system, and reduced UB branching may result in renal hypoplasia or renal dysplasia (Kozlov and Schedl, 2020).
The etiological mechanism of CAKUT has not been fully elucidated. Currently, the classical etiological factors of CAKUT are considered to be genetic and environmental. Genetic factors included single gene mutation, repeat copy number variation, and abnormal chromosome structure, which account for 10, 15, and 20% of cases, respectively. However, etiological factors cannot be clearly defined in approximately half of the patients (Rosenblum et al., 2017). Thus, additional potential etiological factors of CAKUT have to be studied, including non-coding RNAs, histone modifications, and other epigenetic factors. Long non-coding RNAs (lncRNAs) may be one of these factors. lncRNAs are a class of non-coding RNAs with more than 200 nucleotides that lack an intact open reading frame and cannot encode proteins. In recent years, numerous studies have demonstrated that lncRNAs can regulate the developmental physiology of various tissues and organs, such as the circulatory and nervous systems. For instance, lateral mesoderm-specific lncRNA Fendrr anchors polycomb repressive complex 2 (PRC2), increasing PRC2 occupancy and the levels of histone H3 Lys27 tri-methylation (H3K27me3) in heart development (Grote et al., 2013). Loss of the lncRNA lncrps25 reduces locomotion behavior by regulating motor neuron progenitor development and expression of oligodendrocyte transcription factor (Olig2) (Gao et al., 2020). Expression of the MM-specific lncRNA GM29418 can regulate SIX homeobox 2 (Six2) in mouse MM cells in vitro, and Six2 has been demonstrated to regulate the development of the metanephros (Nishikawa et al., 2019). Expression of the lncRNA Hoxb3os (homeobox B3 and homeobox B2, opposite strand) is significantly decreased in the renal tissues of patients with autosomal dominant polycystic kidney disease (ADPKD) and in a mouse model of ADPKD (Aboudehen et al., 2018). Thus, we hypothesized that the presence of abnormal lncRNAs may lead to abnormal development of the urinary system and may induce CAKUT in a mouse model.
The intergenic lncRNA Rik is located at 14qC1: 46,602,806-46,637,423. A review of the literature found that abnormal expression of the lncRNA Rubie (14qC1: 46,568,263-46,587,983) located adjacent to Rik can cause vestibular malformations in the inner ear of mice. Thus, we hypothesized that Rik may participate in embryonic development including the urinary system development in mice. Therefore, in this study, we used the piggyBac (PB) transposon-based insertional mutagenesis to construct the lncRNA 4933425B07Rik (Rik) PB to insert in mice and generate a mutant mouse model (RikPB/PB) to investigate the phenotype and the possible mechanism.
Experimental Materials and Methods
Mice
RikPB/+ mice were obtained by using PB transposon-based insertional mutagenesis targeted to intron 3 of the Rik gene of FVB/NJ mice (Ding et al., 2005). RikPB/+ mice were inbred to obtain RikPB/PB and Rik+/+ mice. Hoxb7/myr-Venus is a previously successfully bred mouse strain that specifically expresses a fluorescent protein in the UB epithelium (Yu et al., 2020). RikPB/PB mice were mated with Hoxb7/myr-Venus mice to obtain Rik PB insertion mice specifically expressing fluorescent protein in the UB epithelium (RikPB/+; Hoxb7/myr-Venus abbreviated as RikPB/+; Hoxb7). RikPB/+; Hoxb7 were inbreed to obtain offspring namely RikPB/PB; Hoxb7/myr-Venus (RikPB/PB; Hoxb7), RikPB/+; Hoxb7, and control mice Rik+/+; Hoxb7/myr-Venus (Rik+/+; Hoxb7). We observed the mice urinary system phenotypes by selected the same nest of different genotypes. All mice used in the experiments were specific pathogen free (SPF) and were maintained at 18–22°C, 50–60% humidity, and 12 h day/night cycle. Animals were maintained and managed according to the animal welfare and usage management regulations of the School of Life Sciences of Fudan University [Protocol Approval No. SYXK (hu) 2020-0011].
RNA Extraction and Quantitative Real-Time PCR
The TRIzol (Life Technologies, United States) method was used to extract RNA from E12.5 whole embryos, from E14.5, E16.5, E18.5, and postnatal day (P) 0.5 kidneys, and from E14.5 heart, liver, brain, lung, ureter, and bladder. RNeasy mini kit (QIAGEN, Germany) was used to extract RNA from E12.5 embryonic kidney. Genomic DNA removal, reverse transcription, and quantitative real-time polymerase chain reaction (RT-PCR) were performed according to manufacturer instructions. Genomic DNA was removed using a gDNA eraser kit (Takara, Japan), and reverse transcription was performed using a reverse transcription kit (Takara, Japan). RT-PCR was performed using the AceQ qPCR SYBR Green Master Mix (Vazyme, China) and a real-time qPCR system (Agilent Mx3000P, United States). RT-PCR primers for Rik, bone morphogenetic protein 4 (Bmp4), Wnt9b, forkhead box C1 (Foxc1), Foxc2, GATA-binding protein 3 (Gata3), cyclin dependent kinase inhibitor 3 (Cdkn3), glia maturation factor beta (Gmfb), sterile alpha motif domain containing 4 (Samd4), cell growth regulator with ring finger domain 1 (Cgrrf1), cornichon family AMPA receptor auxiliary protein 1 (Cnih1), and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) are listed as follows. For Rik, 5′-TTGGGTGTGACTGGAGGAAA-3′ (forward) and 5′-CAAA CAGGATAAAGATGGGAGGT-3′ (reverse); for Bmp4, 5′-GCA AGTTTGTTCAAGATTGGCTCC-3′ (forward) and 5′-CCAT CAGCATTCGGTTACCAGG-3′ (reverse); for Wnt9b, 5′-TTC CCCGACACTCAGCAAAG-3′ (forward) and 5′-TTCCCCTT CTCAGTCTGTTCTCCG-3′(reverse); for Foxc1, 5′-GCCAAA TGGAATGGAACCCC-3′ (forward) and 5′-CGCTGGTGTGA GGAATCTTCTC-3′ (reverse); for Foxc2, 5′-CTTCTACCGCG AGAACAAGC-3′ (forward) and 5′-GACTTTCTTCTCGGCCT CCT-3′ (reverse); for Gata3, 5′-CCAGGCAAGATGAGAAA GAGTG-3′ (forward) and 5′-ATAGGGCGGATAGGTGGTA ATG-3′ (reverse); for Cdkn3, 5′-GAGCACATCTAGGGT CTCCA-3′ (forward) and 5′-ACACCGAATTCATGACTCA AGC-3′ (reverse); for Gmfb, 5′-CCAAGAGATCGCACGGC AAC-3′ (forward) and 5′-AGGCGTTCATCCTTGTCAATCT-3′ (reverse); for Cgrrf1, 5′-CTTCCCTAGCAACAGGCATGG-3′ (forward) and 5′-TCTTCTAACGCTTGCGGGG-3′ (reverse); for Cnih1, 5′-TCGCGGCCTTCTGCTATATG-3′ (forward) and 5′-GGAAGGACAAGAGGGTTCAGG-3′ (reverse); and for Gapdh, 5′-TGTTCCTACCCCCAATGTGTCC-3′ (forward) and 5′-GGAGTTGCTGTTGAAGTCGCAG-3′ (reverse). Gapdh was used as an internal control for relative quantification. The relative gene expression levels were calculated using the 2–Δ Δ CT method. Each group contained at least three samples, and each sample was assayed in triplicate.
Nuclear and Cytoplasmic Extraction
Mouse renal proximal tubular epithelial cells (mPTCs), an immortalized cell line, were acquired from ATCC and cultured in DMEM-F12 medium supplemented with 10% fetal bovine serum (FBS) (Yang et al., 2018). After collection of mPTCs, the cytoplasm and nuclei were isolated using the NE-PER Nuclear and Cytoplasmic Extraction kit (Thermo Fisher Scientific, United States). Cytoplasmic and nuclear RNA was extracted using the RNeasy kit (Qiagen, Germany). Gapdh mRNA, U1 spliceosomal RNA and metastasis associated lung adenocarcinoma transcript 1 (MALAT1) RNA were used as controls.
Gross Phenotypic Analysis
Phenotypes of newborn RikPB/PB, RikPB/+, and Rik+/+ mice were assessed. The animals were anesthetized with CO2 and the abdomen was dissected to expose the urinary system. The location, morphology, and number of kidneys, ureters, and bladders of mice in each group were observed under a stereoscope. The images were recorded under a microscope (Leica, Germany).
Hematoxylin-Eosin Staining
The kidneys of the mice were removed and fixed in 4% paraformaldehyde. After ethanol gradient dehydration, the tissues were embedded in paraffin and cut into 4 μm sections. Hematoxylin-eosin (H&E) staining was performed according to the standard protocols described in the literature (Xu et al., 1999).
Embryonic Kidney Culture
E11.5 embryonic kidneys from RikPB/PB; Hoxb7 and Rik+/+; Hoxb7 mice were isolated in phosphate-buffered saline (PBS) and placed on a 0.4 μm polyester membrane (Corning, United States) at 37°C and 5% CO2. Embryonic kidneys were cultured in DMEM-F12 (Gibco, United States) containing 10% FBS (Gibco, United States) and 1% penicillin/streptomycin (Gibco, United States) for 24, 48, and 72 h and then observed under a fluorescence microscope (Leica, Germany) to detect UB branches and count the UB tips (Kim et al., 2014).
RNA-Sequencing
The RNeasy Mini kit (QIAGEN, Germany) was used to extract the total RNA of E12.5 embryonic kidneys from the RikPB/PB; Hoxb7 and Rik+/+; Hoxb7 mice. The sequencing data was filtered with SOAPnuke (v1.5.2) (Li et al., 2008) by (1) removing reads containing sequencing adapter; (2) removing reads whose low-quality base ratio (base quality less than or equal to 5) is more than 20%; (3) removing reads whose unknown base (“N” base) ratio is more than 5%, afterward clean reads were obtained and stored in FASTQ format. The clean reads were mapped to the reference genome using HISAT2 (v2.0.4) (Kim et al., 2015), Bowtie2 (v2.2.5) (Li and Dewey, 2011) was applied to align the clean reads to the reference coding gene set, then expression level of gene was calculated by RSEM (v1.2.12) (Raivo, 2019). Using Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) algorithm for the expression of standardized, which counts by total exon fragments/[mapped reads (millions) × exon length (kb)], reflects the gene expression level. The heatmap was drawn by pheatmap (v1.0.8) (Love et al., 2014) according to the gene expression in different samples. Essentially, differentially expressed genes (DEGs) analysis was performed using the DESeq2 (v1.4.5) (Abdi, 2007) with fold change ≥2 and false discovery rate (FDR) ≤ 0.001. To take insight to the change of phenotype, GO1 enrichment analysis of annotated different expressed gene was performed by Phyper2 based on Hypergeometric test. The significant levels of terms and pathways were corrected by Q-value (=FDR) with a rigorous threshold (Q-value ≤ 0.05) by Bonferroni.
Immunofluorescence
Fresh kidney tissue was fixed with 4% paraformaldehyde overnight, dehydrated in 30% sucrose for 48 h, embedded in optimal cutting temperature compound (OCT compound), and stored at −80°C. The sections were frozen and sliced at a thickness of 15 μm. The sections were washed with PBS, permeabilized with 0.3% Triton X-100 for 15 min, and blocked with 5% donkey serum for 1 h at room temperature. The tissue slices were incubated with a primary antibody at 4°C overnight, washed several times with PBS, incubated with a secondary antibody at room temperature for 1 h, washed several times with PBS, and treated with an autofluorescence quencher. Slices were observed and imaged using a fluorescence confocal microscope (Olympus FV3000, Japan). The following antibodies were used: primary antibodies: anti-BMP4 (Abcam, ab39973, 1:200), anti-Smad1/Smad5/Smad8 (Affinity, AF-0614, 1:100), and anti-pSmad1/pSmad5/pSmad8 (Cell Signaling Technology, CST13820, 1:200); secondary antibodies: Alexa 647-conjugated anti-rabbit (Jackson ImmunoResearch, 1:400).
Statistical Analysis
All data were processed using the SPSS 24.0 statistical software. The data are presented as the mean ± standard deviation. The count data are presented as rates. Differences between the groups were analyzed using the Chi-squared test or unpaired t-test. P < 0.05 was defined as a statistically significant difference.
Results
Rik Overexpression in RikPB/PB Mice
We used the phylogenetic codon substitution frequency software (PhyloCSF) to identify an open reading frame and ORF Finder to assess the protein-coding potential of Rik; the results demonstrated that Rik is a lncRNA because of its low protein-coding potential (Supplementary Figure 1). Non-coding RNAs are able to accurately regulate the development of the metanephros in a specific spatiotemporal pattern due to unique tissue and embryonic expression characteristics. We detected the spatiotemporal expression distribution of Rik in wild-type mice by RT-PCR and found that the expression level was high during E12.5-E15.5; expression was highest on E12.5 and then gradually decreased (Figure 1A). At E14.5, high levels of Rik were expressed in the mouse urinary system (kidney, ureter, and bladder), while the expression levels in the brain, heart, lung, and liver were low (Figure 1B). The subcellular localization of Rik in mPTC was detected using the nuclear and cytoplasmic protein extraction assay; the data indicate that Rik is almost exclusively present in the nucleus (Figure 1C).
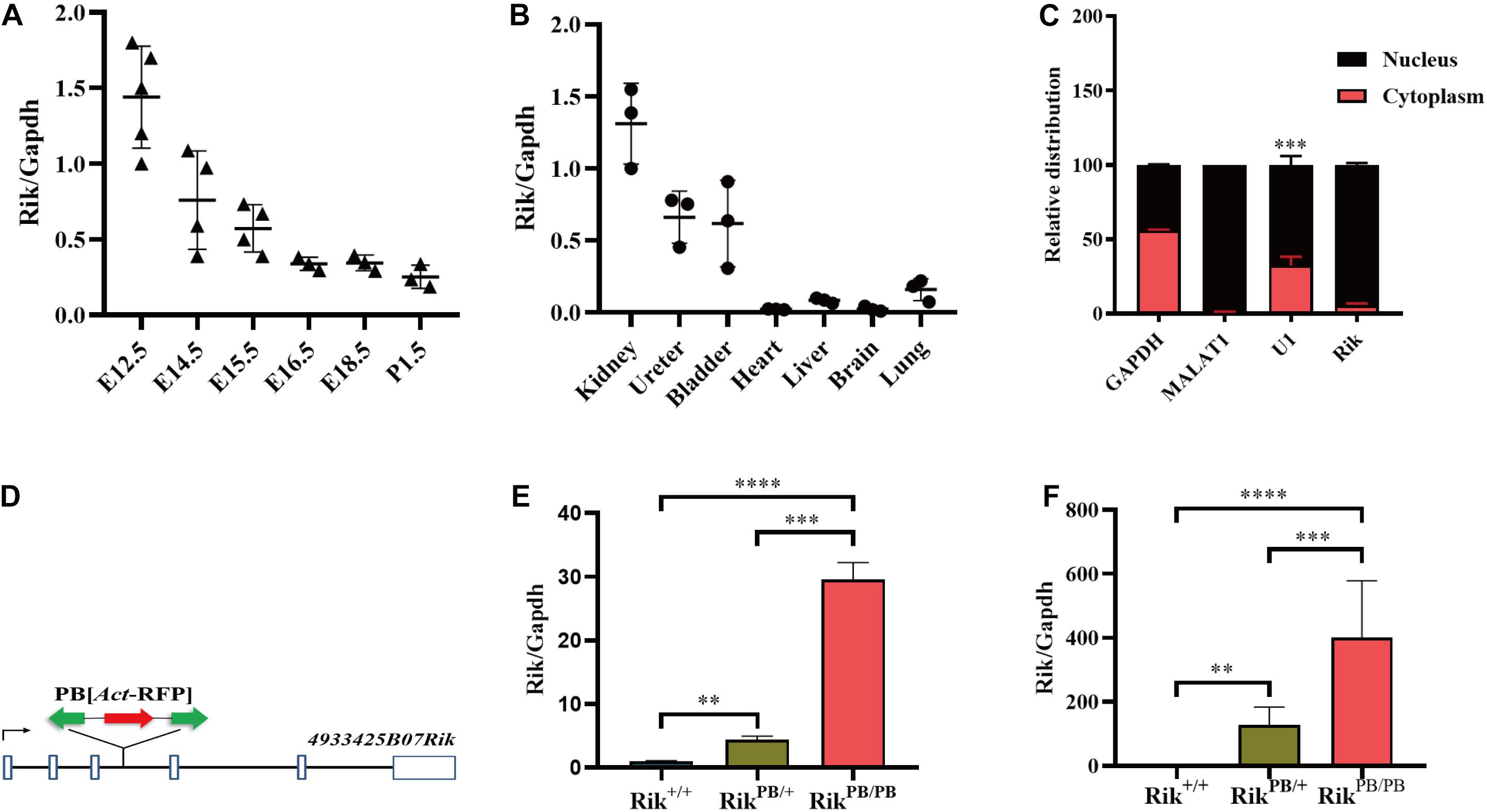
Figure 1. Rik overexpression in PB mice. (A) Rik expression during various kidney development stages in Rik+/+ mice. (B) Rik expression distribution in various tissues of Rik+/+ mice at E14.5. (C) Subcellular localization of Rik. (D) Genomic structure after PB insertion. PB (Act-RFP) was inserted into intron 3 of the Rik gene (solid line). Boxes, exons; arrows, transcription direction; green arrows, PB transposon; red arrows, RFP expression elements. (E) After PB insertion, the expression level of Rik was increased in E12.5 embryos, n = 3. (F) The expression level of Rik in E12.5 embryonic kidney was increased after PB insertion, n = 5. **P < 0.01; ***P < 0.001; ****P < 0.0001.
RikPB/PB mice were constructed using PB transposon-based insertional mutagenesis. PB was inserted into intron 3 of the mouse Rik gene (Figure 1D), resulting in Rik overexpression in the RikPB/PB mice. RT-PCR of whole embryos and embryonic kidneys of E12.5 RikPB/PB, RikPB/+, and Rik+/+ mice detected an increase in Rik expression levels in RikPB/PB and RikPB/+ mice compared to that in Rik+/+ mice (Figures 1E,F).
RikPB/PB Newborn Mice Develop Urinary Malformations
Gross phenotypes of the urinary system of RikPB/PB, RikPB/+, and Rik+/+ newborn mice were examined, and the results indicate a significantly higher incidence of CAKUT in RikPB/PB newborn mice than in RikPB/+ mice and Rik+/+ mice (66.7%, 34/51 vs 10.4%, 15/144, P < 0.001; 66.7%, 34/51 vs 9.3% (4/43), P < 0.001, respectively, Figure 2A). The CAKUT phenotype (Figures 2B–M) of the RikPB/PB mice included renal hypo/dysplasia (82.4%, 28/34, Figures 2C,G,K), duplex renal/collecting system (8.8%, 3/34, Figures 2D,H,L), hydronephrosis (5.9%, 2/34, Figures 2E,I,M), and vesicoureteral reflux (VUR) (2.9%, 1/34, Figure not shown) with renal hypo/dysplasia being the most common phenotype. The renal pathology of newborn mice in the RikPB/PB group was examined by H&E staining, which indicated that the structure and morphology of the glomerulus and renal tubules in animals with renal hypo/dysplasia and duplex kidneys were normal (Figures 2K,L). The numbers of glomeruli and tubules were reduced in the renal tissues in animals with hydronephrosis (Figure 2M). Gross anatomy and histopathology of the brain, heart, lung, and liver of RikPB/PB newborn mice were observed, and no significant abnormalities were detected (Supplementary Figure 2).
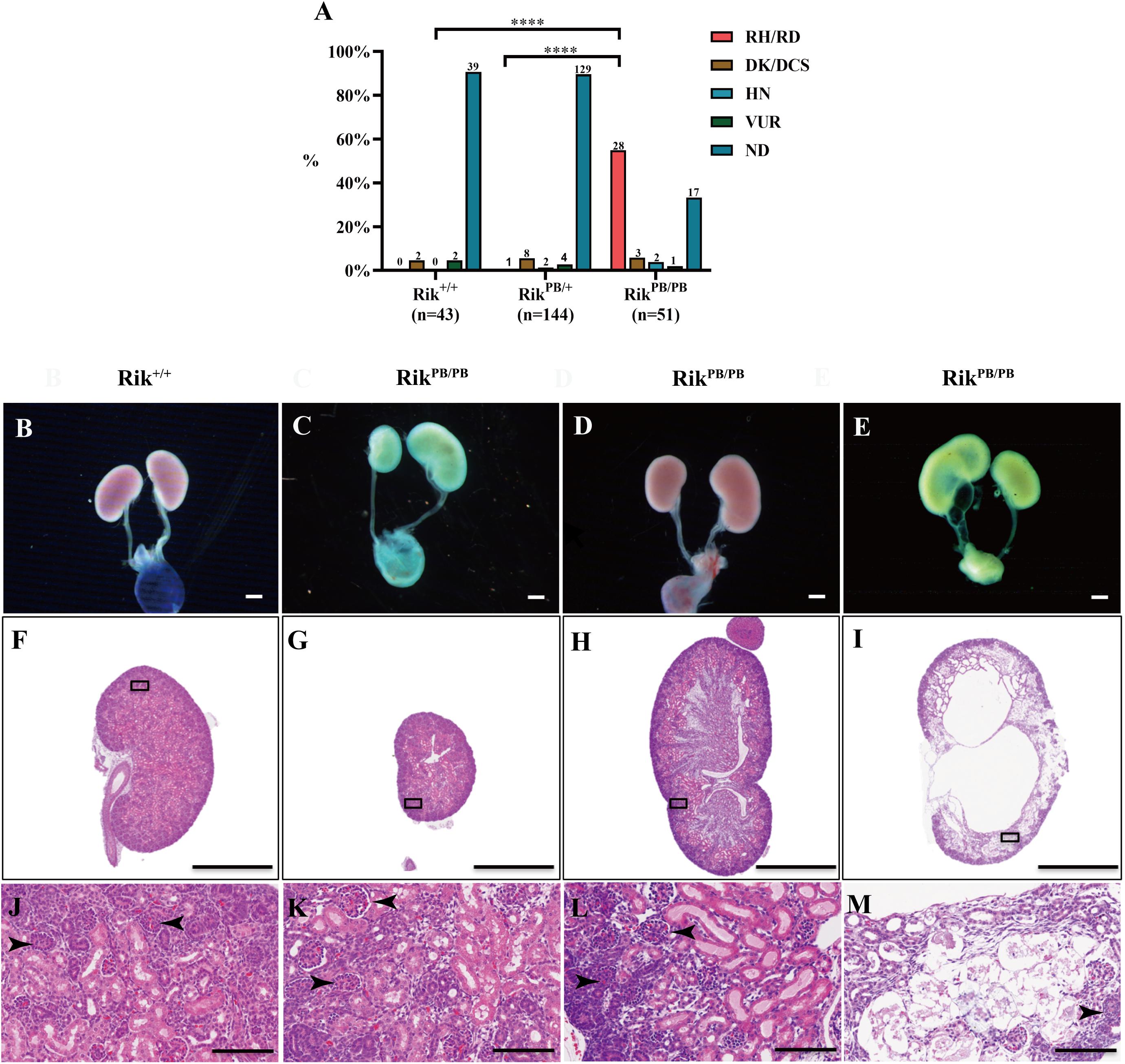
Figure 2. Various urinary malformations observed in RikPB/PB newborn mice predominantly manifested as renal hypo/dysplasia. (A) The percentage of urinary malformations in RikPB/PB, RikPB/+, and Rik+/+ newborn mice. The number n represents the number of cases. (B–E) Overview images of the urinary system of newborn mice. (F–M) H&E staining of the kidneys of newborn mice. (J–M) Local magnified images. (B,F,J) Normal kidney of Rik+/+ mice. (C,G,K) Renal dysplasia/hypoplasia in RikPB/PB mice. (D,H,I) Duplex kidney in RikPB/PB mice. (E,I,M) Hydronephrosis in RikPB/PB mice. RH/RD, renal hypo/dysplasia; DK/DCS, duplex kidneys/duplex collecting systems; HN, hydronephrosis; VUR, vesicoureteral reflux; ND, normal kidney. Arrowhead, nephron. ****P < 0.0001. Scar bars, 10 mm in (B–E); 1 mm in (F–I); 100 μm in (J–M).
UB Branches in the Early Stages of Metanephric Development of RikPB/PB Mice
The effect of Rik on UB branching in the early stages of metanephric development was observed. E11.5 embryonic kidneys of RikPB/PB and Rik+/+ mice were extracted for in vitro culture. The number of UB tips in these two groups was counted at 24, 48, and 72 h. The number of UB branches at these three time points was significantly lower in the RikPB/PB group than in the Rik+/+ group (9.0 ± 1.5 vs 6.5 ± 1.8, 25.1 ± 7.0 vs 12.5 ± 3.3, and 57.9 ± 10.8 vs 31.3 ± 11.2, respectively, Figures 3A–I).
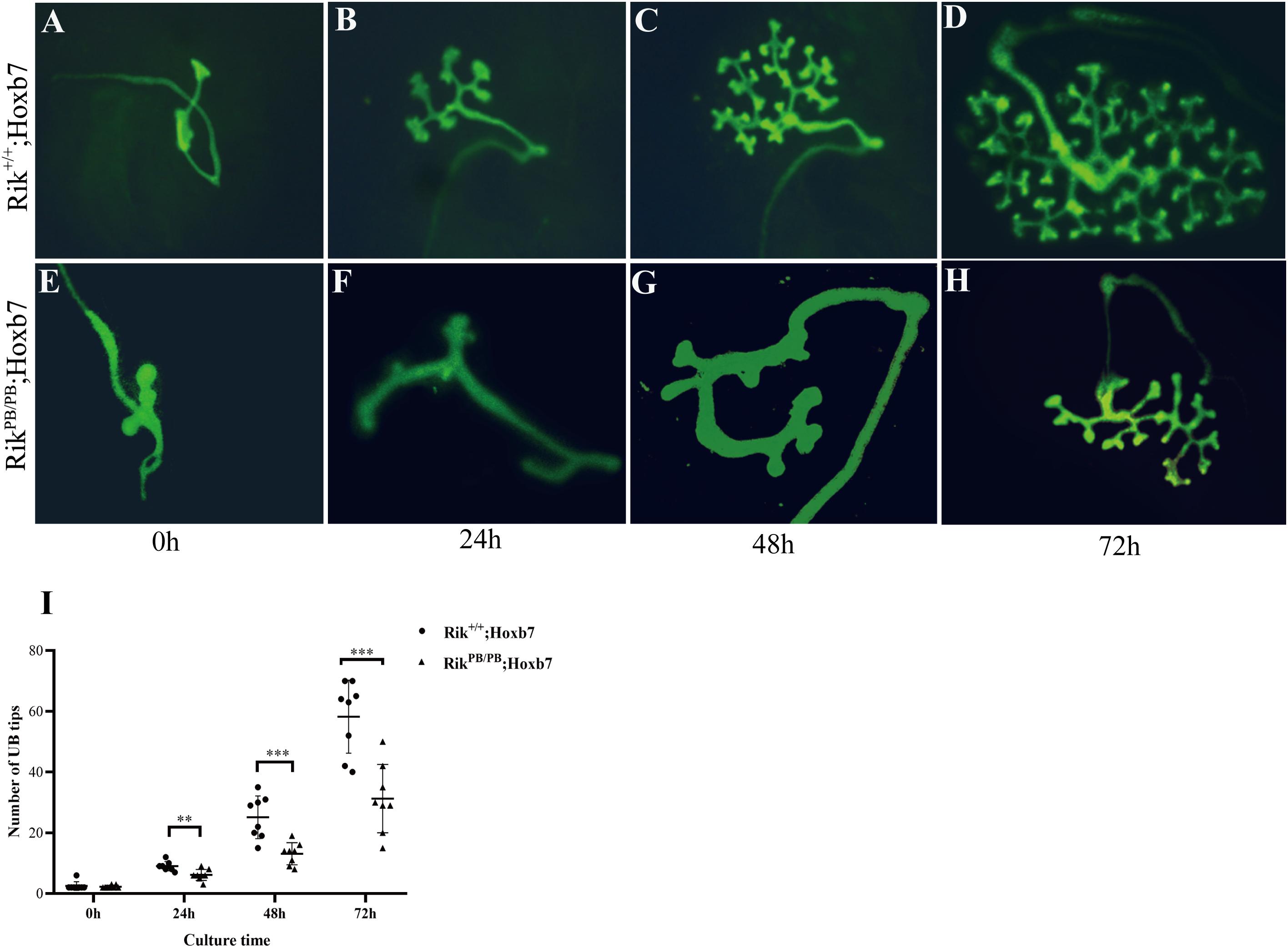
Figure 3. Decrease in UB branches in the embryonic kidney of RikPB/PB. (A,E) Embryonic kidney at E11.5. (B,F) Embryonic kidney after 24 h culture. (C,G) Embryonic kidney after 48 h culture. (D,H) Embryonic kidney after 72 h culture. (I) The UB tips count of UB branches in wild-type and homozygous embryonic kidneys, n = 8. **P < 0.01; ***P < 0.001.
Rik Overexpression Induces Abnormal Expression of Metanephric Development-Related Genes in Mice
To clarify the effect of Rik overexpression on the expression of genes related to metanephric development in mice, we extracted RNA from the kidneys of E12.5 Rik+/+ and RikPB/PB mice to perform RNA-sequencing (RNA-seq). The results showed that the number of DEGs in the wild-type vs homozygous mutant mice was 186, including 150 upregulated genes and 36 downregulated genes (Figures 4A,B). These 186 DEGs were classified and enriched, including the SMAD protein signal transduction pathway [Bmp4, transferrin (Trf), growth differentiation factor 2 (Gdf2), Gdf10, Gata4, hepatocyte nuclear factor 4 alpha (Hnf4a), and alpha-fetoprotein (Afp)], Figure 4C, which is related to the development of the urinary system. Subsequent analysis of the molecules involved in the regulation of the UB branching revealed differential expression: the upregulated genes (fold increase) included Foxc1 (1.20), Wnt9b (1.24), Foxc2 (1.18), and Gata3 (1.18), and the downregulated genes (fold decrease) included Bmp4 (0.48) (Supplementary Table 1).
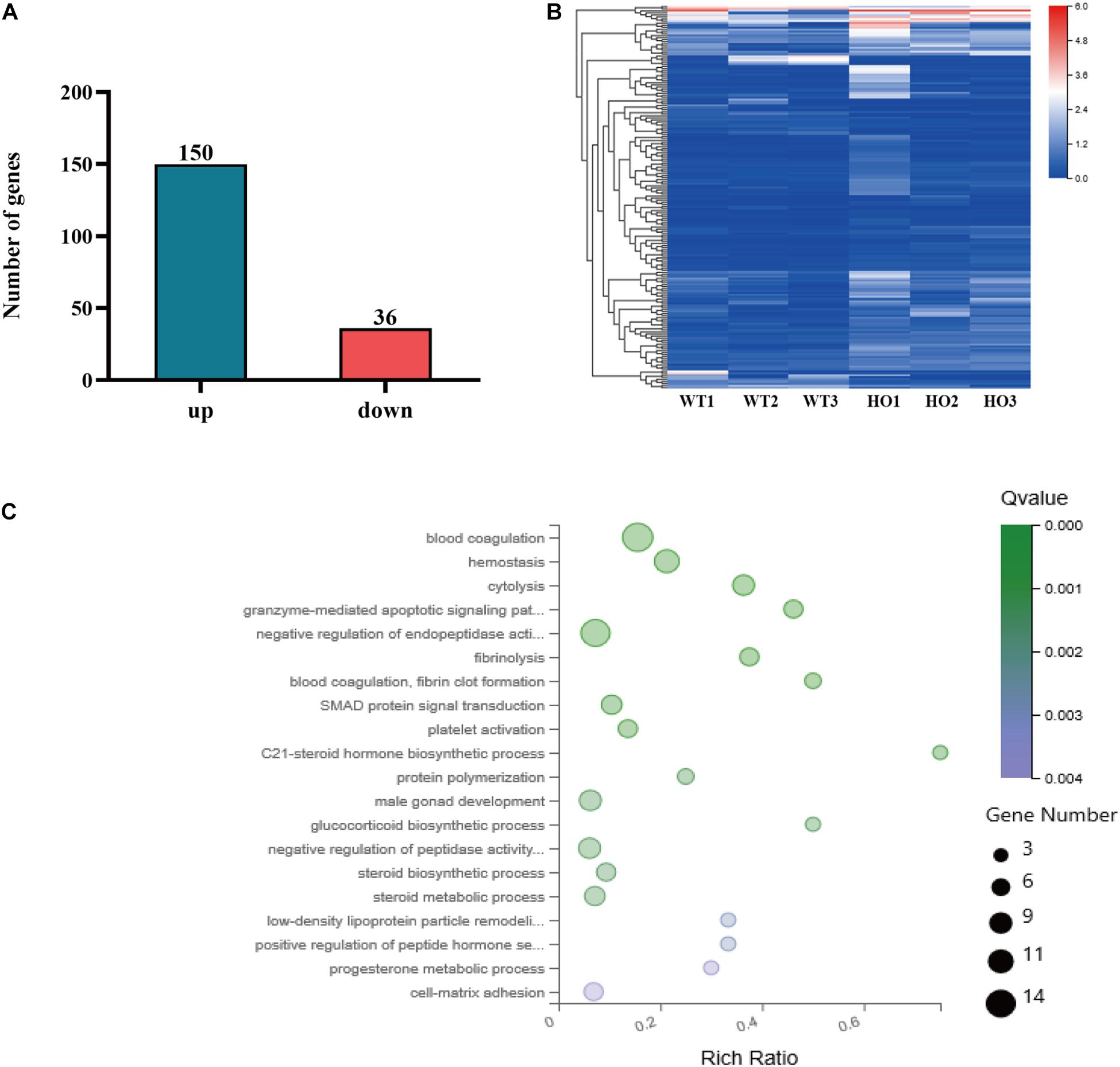
Figure 4. RNA-seq analysis. (A) Number of DEGs identified by RNA-seq. (B) Heat map of DEGs identified by RNA-seq. WT, wild-type namely Rik+/+; Hoxb7 mice; HO, homozygous namely RikPB/PB; Hoxb7 mice. (C) Bubble plots of the gene ontology (GO) enrichment of DEGs in the E12.5 embryonic kidney [Q-value means false discovery rate (FDR) or adjusted P-value. Rich ratio is the ratio of the signaling pathway of enrichment on the number of differentially expressed genes and signaling pathways of all the genes].
Then, RT-PCR was performed to validate the DEGs regulating UB branching. The expression of Bmp4 in the E12.5 RikPB/PB group was significantly decreased (Figure 5A), and the expression levels of other genes were not significantly different. RT-PCR detection revealed significantly decreased expression of Bmp4 in the E14.5 RikPB/PB group (Figure 5B).
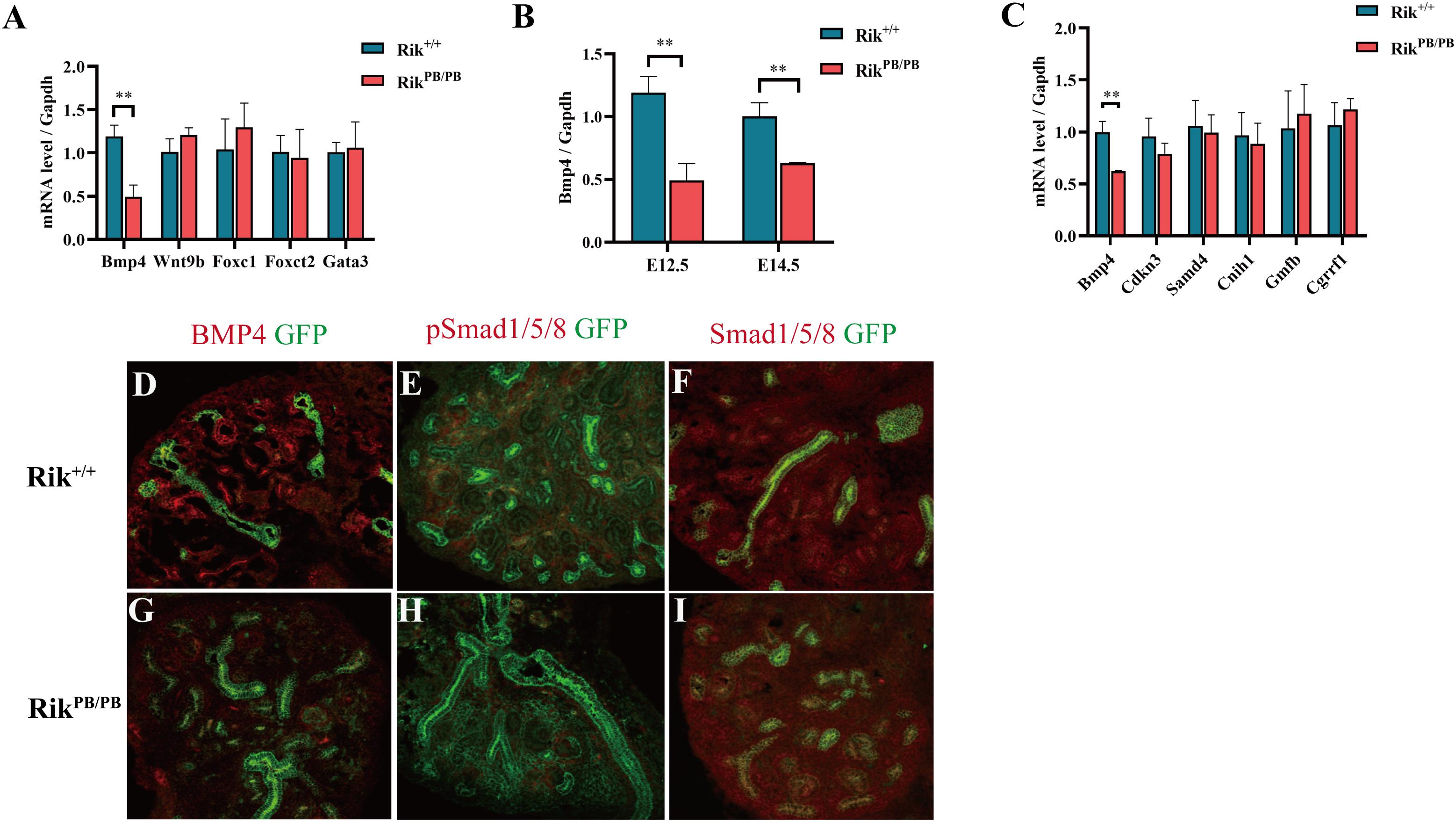
Figure 5. Rik overexpression-induced abnormal expression of genes related to mouse metanephric development. (A) Expression changes of the key genes in metanephric development at E12.5, n = 3. (B) Changes in Bmp4 expression at various embryonic ages, n = 4. (C) Expression of the genes adjacent to the Rik gene locus. (D–I) Immunofluorescence was used to detect the expression of BMP4, Smad1/Smad5/Smad8 and pSmad1/pSmad5/pSmad8 in the kidneys of Rik+/+ and RikPB/PB mice at E14.5. n = 3, original magnification ×20 (D–I).
Considering that Rik is mainly distributed in the nucleus, we identified the genes located adjacent to Rik in the same region of chromosome 14, including Bmp4, Cdkn3, Samd4, Cnih1, Gmfb, and Cgrrf1. The changes in the expression levels of these genes in the kidney of E14.5 RikPB/PB were analyzed using RT-PCR; the results indicated that Bmp4 was significantly downregulated (Figure 5C). Immunofluorescence analysis of Bmp4 and its downstream targets Smad1/Smad5/Smad8 and pSmad1/pSmad5/pSmad8 indicated that the expression levels of Smad1/Smad5/Smad8 were not significantly changed in E14.5 RikPB/PB homozygous kidney, while the expression levels of BMP4 and pSmad1/pSmad5/pSmad8 were decreased (Figures 5D–I).
Discussion
In this study, we successfully established RikPB/PB mice using PB transposon-based insertional mutagenesis and found that Rik is overexpressed after PB insertion. RikPB/PB mice had multiple developmental malformations of the urinary system predominantly manifested as renal hypo/dysplasia. Additionally, the PB insertion induced a decrease in the number of UB branches by influencing UB branching at the early stage of metanephric development. Analysis of the expression profiles of the molecules related to early UB branching in RikPB/PB mice revealed that the expression levels of Bmp4, a key gene regulating ureteral bud branching, were decreased in the RikPB/PB embryonic kidney. Therefore, we suggest that abnormal Rik expression may lead to the occurrence of CAKUT in mice and that the lesions may be mediated by altered expression of the key factor Bmp4 during the early UB branching stages.
Previous studies have shown that lncRNAs are related to the development of the mouse urinary system. Nishikawa et al. identified a lncRNA, Gm29418, that was specifically expressed in MM cells in mice. Gm29418 has an enhancer-like function on Six2 in vitro, which is the key regulatory gene in MM cells (Nishikawa et al., 2019). In another study, Hoxb3os, a kidney-specific, evolutionarily conserved lncRNA, was shown to be downregulated in the cyst epithelium in an ADPKD adult mouse model. The data indicate that Hoxb3os can regulate the signaling of mammalian target of rapamycin complex 1 (mTORC1) and mitochondrial respiration in vitro (Aboudehen et al., 2018). However, causal associations between abnormal lncRNA and CAKUT occurrence have not been confirmed in animal models. In this study, Rik-overexpressing mice were established and were shown to develop various phenotypes of urinary malformations predominantly manifested as renal hypo/dysplasia. The biological functions of lncRNAs are closely related to their tissue specificity and temporal and spatial specificity. This study demonstrates that Rik is specifically expressed in the mouse kidneys and that the expression level was increased in the early stages of metanephric development. These findings suggest that Rik regulates the development of the urinary system in mice, and abnormal Rik expression may lead to CAKUT.
The normal development of the mouse urinary system is closely associated with UB outgrowth and branching. UB outgrowth begins on E10.5, and the early branching stage occurs on E11.5–E15.5 (Short and Smyth, 2016). In this process, abnormal or ectopic UB outgrowth or abnormal position of the outgrowth may lead to renal agenesis, duplex kidney, hydronephrosis, or VUR; reduction in UB branches or early termination of the branching may result in renal hypo/dysplasia (Schedl, 2007). In this study, RikPB/PB mice predominantly manifested renal hypo/dysplasia. Dynamic observation of the UB branching process in in vitro culture showed that the number of UB branches in the RikPB/PB embryonic kidney was significantly reduced. Iterative UB branching is regulated by a complex molecular network, and abnormalities of certain key molecules may determine the lesions of the UB branches. Previous studies have shown that the abnormal expression of genes [Bmp4 (Chang et al., 2008), Foxc1 (Kume et al., 2000), Wnt9b (Carroll et al., 2005), Gata3 (Grote et al., 2008), Six2 (Self et al., 2006), Ret (Allison, 2012), Fgf7 (Qiao et al., 1999), Gremlin1 (Michos et al., 2004) etc.] can lead to a reduction in UB branches. This study used RNA-seq for the preliminary screening and RT-PCR and immunofluorescence to verify the changes in various molecules and found that only Bmp4 is significantly downregulated, while the expression levels of other molecules involved in the regulation of UB branching were not significantly different from Rik+/+ embryonic kidney. BMP4 belongs to the transforming growth factor beta (TGF-β) superfamily and participates in urinary system development by regulating cell proliferation, differentiation, and apoptosis (Hogan, 1996; von Bubnoff and Cho, 2001). Renal hypo/dysplasia has been observed in patients with BMP4 mutations (Bakrania et al., 2008; Weber et al., 2008; Davis et al., 2014). Moreover, the CAKUT phenotype predominantly manifested as renal hypo/dysplasia has been observed in Bmp4± mice (Miyazaki et al., 2003), similar to the CAKUT phenotype of RikPB/PB mice in the present study. Extracellular BMP4 can interact with BMP receptor type I or II (BMPR1/BMPR2), transmembrane serine/threonine kinase receptors, to form a complex, which triggers the signal transduction cascade of intracellular Smad1/Smad5/Smad8 protein phosphorylation, thereby regulating cellular processes (Bakrania et al., 2008). Studies of the mechanisms of renal hypo/dysplasia suggest that renal hypo/dysplasia may be associated with abnormal phosphorylation of the downstream molecules of BMP4 signaling (van der Ven et al., 2018). The present study also detected a significant reduction in the expression of pSmad1/pSmad5/pSmad8, which are located downstream of Bmp4, in RikPB/PB mice. These results suggest that abnormal Rik expression may influence Bmp4 signaling related to UB branching to reduce the number of UB branches in mice, thus leading to the development of CAKUT. Additional studies are needed to identify other molecules regulated by Rik.
Variability in the cellular localization of lncRNAs can regulate gene expression by various mechanisms. For example, cytoplasmic lncRNAs can act as a miRNA sponge, competitively inhibit miRNA, reduce degradation of mRNA by miRNA, and regulate mRNA stability and translation (Beermann et al., 2016; Yao et al., 2019). Nuclear lncRNAs mainly regulate the expression of their target genes at the transcriptional level and can bind to the promoter regions of the target genes or interact with transcription factors, thereby inhibiting or promoting expression of the adjacent target genes (Latos et al., 2012; Engreitz et al., 2016; Cho et al., 2018; Wang et al., 2019). The subcellular localization experiments performed in the present study suggest that Rik is located in the nucleus. We speculate that Rik may play a biological role by influencing the expression of the adjacent genes and found that Bmp4 expression is downregulated by Rik overexpression. Therefore, we suggest that Rik, which is located adjacent to Bmp4 in the genome, may inhibit the expression of Bmp4 via a certain transcription factor or by regulating the promoter of the Bmp4 gene. Specific mechanisms of the regulation of Bmp4 by Rik require additional investigation.
Conclusion
In summary, this study found that overexpression of the lncRNA Rik induced the development of CAKUT in mice, predominantly involving abnormal ureteral bud branching; abnormal urinary development may be mediated by Rik-dependent regulation of the expression of Bmp4, a key molecule in the UB branching. This study provides new evidence of the involvement of lncRNAs in the process of embryonic development and new insight into investigations of the etiological mechanisms of CAKUT.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the animal welfare and usage management regulations of the School of Life Sciences of Fudan University [Protocol Approval No. SYXK (hu) 2020-0011].
Author Contributions
LT, HX, and QS designed this study. LT completed the experiment, analyzed the data, and wrote the first draft. JC, YZ, XF, and XW interpreted the experimental results. JL, JJL, YL, and SX analyzed the images. MY and QS revised and refined the manuscript. QS edited and approved the final version. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (82070686), Science and Technology Commission of Shanghai (20ZR1408700), the National Children’s Medical Center “DengFeng” Cross creative team (No. EK112520180201) and the Establishment, Performance, and Quality Control of the Standardized Phenotype Analysis Process grant (No. 2018YFA0801102).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to Yufeng Zhou and Dashi Qi for their guidance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.594640/full#supplementary-material
Footnotes
References
Abdi, H. (2007). The bonferonni and sˇidák corrections for multiple comparisons. Encycl. Meas. Stat. 1, 1–9.
Aboudehen, K., Farahani, S., Kanchwala, M., Chan, S. C., Avdulov, S., Mickelson, A., et al. (2018). Long noncoding RNAHoxb3os is dysregulated in autosomal dominant polycystic kidney disease and regulates mTOR signaling. J. Biol. Chem. 293, 9388–9398. doi: 10.1074/jbc.RA118.001723
Allison, S. J. (2012). Basic research: ret signaling reveals insights into the pathogenesis of CAKUT. Nat. Rev. Nephrol. 8:432. doi: 10.1038/nrneph.2012.119
Bakrania, P., Efthymiou, M., Klein, J. C., Salt, A., Bunyan, D. J., Wyatt, A., et al. (2008). Mutations in BMP4 cause eye, brain, and digit developmental anomalies: overlap between the BMP4 and hedgehog signaling pathways. Am. J. Hum. Genet. 82, 304–319. doi: 10.1016/j.ajhg.2007.09.023
Beermann, J., Piccoli, M., Viereck, J., and Thum, T. (2016). Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol. Rev. 96, 1297–1325. doi: 10.1152/physrev.00041.2015
Blake, J., and Rosenblum, N. D. (2014). Renal branching morphogenesis: morphogenetic and signaling mechanisms. Semin Cell Dev. Biol. 36, 2–12. doi: 10.1016/j.semcdb.2014.07.011
Carroll, T. J., Park, J., Hayashi, S., Majumdar, A., and McMahon, A. P. (2005). Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. cell 9, 283–292. doi: 10.1016/j.devcel.2005.05.016
Chang, W., Lin, Z., Kulessa, H., Hebert, J., Hogan, B. L., and Wu, D. K. (2008). Bmp4 is essential for the formation of the vestibular apparatus that detects angular head movements. PLoS Genet. 4:e1000050. doi: 10.1371/journal.pgen.1000050
Chesnaye, N., Bonthuis, M., Schaefer, F., Groothoff, J. W., Verrina, E., Heaf, J. G., et al. (2014). Demographics of paediatric renal replacement therapy in Europe: a report of the ESPN/ERA-EDTA registry. Pediatr. Nephrol. 29, 2403–2410. doi: 10.1007/s00467-014-2884-2886
Cho, S. W., Xu, J., Sun, R., Mumbach, M. R., Carter, A. C., Chen, Y. G., et al. (2018). Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412. doi: 10.1016/j.cell.2018.03.068
Davis, T. K., Hoshi, M., and Jain, S. (2014). To bud or not to bud: the RET perspective in CAKUT. Pediatr. Nephrol. 29, 597–608. doi: 10.1007/s00467-013-2606-2605
Ding, S., Wu, X., Li, G., Han, M., Zhuang, Y., and Xu, T. (2005). Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483. doi: 10.1016/j.cell.2005.07.013
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi: 10.1038/nature20149
Gao, T., Li, J., Li, N., Gao, Y., Yu, L., Zhuang, S., et al. (2020). lncrps25 play an essential role in motor neuron development through controlling the expression of olig2 in zebrafish. J. Cell Physiol. 235, 3485–3496. doi: 10.1002/jcp.29237
Grote, D., Boualia, S. K., Souabni, A., Merkel, C., Chi, X., Costantini, F., et al. (2008). Gata3 acts downstream of beta-catenin signaling to prevent ectopic metanephric kidney induction. PLoS Genet. 4:e1000316. doi: 10.1371/journal.pgen.1000316
Grote, P., Wittler, L., Hendrix, D., Koch, F., Währisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. doi: 10.1016/j.devcel.2012.12.012
Hogan, B. L. (1996). Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 10, 1580–1594. doi: 10.1101/gad.10.13.1580
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, S. T., Ahn, S. Y., Swat, W., and Miner, J. H. (2014). DLG1 influences distal ureter maturation via a non-epithelial cell autonomous mechanism involving reduced retinoic acid signaling, ret expression, and apoptosis. Dev. Biol. 390, 160–169. doi: 10.1016/j.ydbio.2014.03.014
Kozlov, V. M., and Schedl, A. (2020). Duplex kidney formation: developmental mechanisms and genetic predisposition. F1000 Research 9:F1000FacultyRev-2.
Kume, T., Deng, K., and Hogan, B. L. (2000). Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127, 1387–1395.
Latos, P. A., Pauler, F. M., Koerner, M. V., Şenergin, H. B., Hudson, Q. J., Stocsits, R. R., et al. (2012). Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472. doi: 10.1126/science.1228110
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, R., Li, Y., Kristiansen, K., and Wang, J. (2008). SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714. doi: 10.1093/bioinformatics/btn025
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-558
Michos, O., Panman, L., Vintersten, K., Beier, K., Zeller, R., and Zuniga, A. (2004). Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development (Cambridge, England) 131, 3401–3410. doi: 10.1242/dev.01251
Miyazaki, Y., Oshima, K., Fogo, A., and Ichikawa, I. (2003). Evidence that bone morphogenetic protein 4 has multiple biological functions during kidney and urinary tract development. Kidney Int. 63, 835–844. doi: 10.1046/j.1523-1755.2003.00834.x
Murugapoopathy, V., and Gupta, I. R. (2020). A primer on Congenital Anomalies of the Kidneys and Urinary Tracts (CAKUT). Clin. J. Am. Soc. Nephrol. 15, 723–731. doi: 10.2215/CJN.12581019
Nicolaou, N., Renkema, K. Y., Bongers, E. M., Giles, R. H., and Knoers, N. V. (2015). Genetic, environmental, and epigenetic factors involved in CAKUT. Nat. Rev. Nephrol. 11, 720–731. doi: 10.1038/nrneph.2015.140
Nishikawa, M., Yuri, S., Kimura, H., Yanagawa, N., Hamon, M., Hauser, P., et al. (2019). Comprehensive analysis of chromatin signature and transcriptome uncovers functional lncRNAs expressed in nephron progenitor cells. Biochim. et Biophysica Acta. Gene Regul. Mechan. 1862, 58–70. doi: 10.1016/j.bbagrm.2018.09.006
Qiao, J., Uzzo, R., Obara-Ishihara, T., Degenstein, L., Fuchs, E., and Herzlinger, D. (1999). FGF-7 modulates ureteric bud growth and nephron number in the developing kidney. Development 126, 547–554.
Raivo, K. (2019). Package ‘Pheatmap’. 2019-01-04 13:50:12 UTC. Available online at: https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf
Rosenblum, S., Pal, A., and Reidy, K. (2017). Renal development in the fetus and premature infant. Semin. Fetal Neonatal Med. 22, 58–66. doi: 10.1016/j.siny.2017.01.001
Schedl, A. (2007). Renal abnormalities and their developmental origin. Nat. Rev. Genet. 8, 791–802. doi: 10.1038/nrg2205
Self, M., Lagutin, O. V., Bowling, B., Hendrix, J., Cai, Y., Dressler, G. R., et al. (2006). Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 25, 5214–5228. doi: 10.1038/sj.emboj.7601381
Short, K. M., and Smyth, I. M. (2016). The contribution of branching morphogenesis to kidney development and disease. Nat. Rev. Nephrol. 12, 754–767. doi: 10.1038/nrneph.2016.157
van der Ven, A. T., Vivante, A., and Hildebrandt, F. (2018). Novel insights into the pathogenesis of monogenic congenital anomalies of the kidney and urinary tract. J. Am. Soc. Nephrol. JASN 29, 36–50. doi: 10.1681/ASN.2017050561
von Bubnoff, A., and Cho, K. W. (2001). Intracellular BMP signaling regulation in vertebrates: pathway or network? Dev. Biol. 239, 1–14. doi: 10.1006/dbio.2001.0388
Wang, F., Ren, D., Liang, X., Ke, S., Zhang, B., Hu, B., et al. (2019). A long noncoding RNA cluster-based genomic locus maintains proper development and visual function. Nucleic Acids Res. 47, 6315–6329. doi: 10.1093/nar/gkz444
Weber, S., Taylor, J. C., Winyard, P., Baker, K. F., Sullivan-Brown, J., Schild, R., et al. (2008). SIX2 and BMP4 mutations associate with anomalous kidney development. J. Am. Soc. Nephrol. 19, 891–903. doi: 10.1681/ASN.2006111282
Xu, P. X., Adams, J., Peters, H., Brown, M. C., Heaney, S., and Maas, R. (1999). Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23, 113–117. doi: 10.1038/12722
Yang, Y., Yu, X., Zhang, Y., Ding, G., Zhu, C., Huang, S., et al. (2018). Hypoxia-inducible factor prolyl hydroxylase inhibitor roxadustat (FG-4592) protects against cisplatin-induced acute kidney injury. Clin. Sci. (Lond) 132, 825–838. doi: 10.1042/CS20171625
Yao, R. W., Wang, Y., and Chen, L. L. (2019). Cellular functions of long noncoding RNAs. Nat. Cell Biol. 21, 542–551. doi: 10.1038/s41556-019-0311-318
Keywords: long non-coding RNA, 4933425B07Rik, congenital anomalies of the kidney and urinary tract, kidney development, Bmp4
Citation: Tan L, Yu M, Li Y, Xue S, Chen J, Zhai Y, Fang X, Liu J, Liu J, Wu X, Xu H and Shen Q (2021) Overexpression of Long Non-coding RNA 4933425B07Rik Causes Urinary Malformations in Mice. Front. Cell Dev. Biol. 9:594640. doi: 10.3389/fcell.2021.594640
Received: 13 August 2020; Accepted: 01 February 2021;
Published: 19 February 2021.
Edited by:
Duan Ma, Fudan University, ChinaReviewed by:
Zong Wei, Mayo Clinic Arizona, United StatesJianhua Mao, Children’s Hospital of Zhejiang University, China
Copyright © 2021 Tan, Yu, Li, Xue, Chen, Zhai, Fang, Liu, Liu, Wu, Xu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Shen, c2hlbnFpYW5Ac2htdS5lZHUuY24=; Hong Xu, aHh1QHNobXUuZWR1LmNu
†These authors have contributed equally to this work
 Lihong Tan
Lihong Tan Minghui Yu
Minghui Yu Yaxin Li
Yaxin Li Shanshan Xue1
Shanshan Xue1 Yihui Zhai
Yihui Zhai Jialu Liu
Jialu Liu Qian Shen
Qian Shen