- 1College of Animal Science and Technology, Northwest A&F University, Xianyang, China
- 2TUM School of Life Sciences, Technical University of Munich, Freising, Germany
- 3Medical College, Qinghai University, Xining, China
The health of mammary gland is essential for lactation. Epidermal growth factor (EGF) is reported to play an important role in lactation initiation and miR-223 is a conserved microRNA in anti-inflammation. In this study, EGF was found to induce a higher expression of miR-223 in goat mammary epithelial cell (gMEC). The downstream genes of miR-223 were screened by RNA sequencing, including Interferon-stimulated gene product 15 (ISG15), a pivotal immune responder, which was detected to be downregulated by EGF and miR-223. Due to the correlation between inflammation and apoptosis, the gMEC apoptosis modulated by EGF, miR-223, and ISG15 was investigated, and the protein expressions of Bcl-2/Bax, Caspase 3 and p53 were examined to evaluate the apoptosis of gMEC. The protein expressions of p-STAT3/STAT3, PR, FOXC1, and HOXA10, which had been shown to be related to inflammation, were detected to assess the inflammation of gMEC. This study provided a regulation axis, EGF/miR-223/ISG15, and illustrated its regulation to gMEC apoptosis and inflammation.
Introduction
Mammary gland is a complex tissue undergoing constant remodeling. It is characterized by highly branched tubular structures with luminal cells surrounded by myoepithelial cells during puberty. The mammary epithelium expands massively during pregnancy to form alveolar structures, which would differentiate into secretary units in late pregnancy to achieve lactation (Watson, 2006). At the onset of lactation, the expression of epidermal growth factor (EGF) surges compared to late pregnancy (Fu et al., 2015), indicating the importance of EGF in the lactation process.
MicroRNAs (miRNAs) are a class of endogenous non-coding RNAs about 22 nucleotides in length that function by repressing the expression of their target genes after transcription (Jonas and Izaurralde, 2015). MiRNAs play critical roles in animal development and physiology (Ambros, 2004), especially in mammary gland (Ji et al., 2019; Xuan et al., 2020; Zhang et al., 2020b,2021b). In this study, we found the expression of miR-223 could be induced by EGF in goat mammary epithelial cells (gMEC). Through RNA-sequencing and analysis of downstream differentially expressed genes (DEGs) of miR-223, the functions and pathways that miR-223 involved in were enriched. In the light of the enrichment, miR-223 was found to be highly associated with inflammation, which was consistent with previous studies (Haneklaus et al., 2013; Jeffries et al., 2019). Interferon-stimulated gene product 15 (ISG15) is one of the DEGs with the highest expression in negative control (NC) group and the second greatest fold-change between miR-223 and NC groups. As the first identified ubiquitin-like protein, ISG15 is a pivotal component of host responses to microbial infection that actively involves in inflammation (Zhang et al., 2015; Dzimianski et al., 2019), which is correlated with apoptosis (Vasilikos et al., 2017).
Here we explored the relationship among EGF, miR-223, and ISG15, and their involvement in the apoptosis and inflammation of gMEC. The gMEC apoptosis regulated by EGF, miR-223, and ISG15 was measured by flow cytometry and evaluated by the expression of classical apoptosis-related proteins (Zamzami and Kroemer, 2005; Ola et al., 2011) Bcl-2/Bax, p53, and Caspase 3. Then inflammation-related protein expressions (p-STAT3, PR, FOXC1, and HOXA10) were examined to evaluate the involvement of EGF, miR-223 and ISG15 in gMEC inflammation. Signal transducers and activators of transcription 3 (STAT3) is a cellular signal transcription factor involved in many cellular activities, and phosphorylated (p-) STAT3 on Ser727 is a crucial intermediary for inflammatory cytokine production (Hu et al., 2019a; Balic et al., 2020). Progesterone (P4) is one of the main hormones involved in mammary gland remodeling during lactation and implicated in host immune regulation (Xu et al., 2017). Previous studies revealed that progesterone receptor (PR) could protect cells from inflammation (Park et al., 2020). Forkhead box C1 (FOXC1) is reported to attenuate the inflammation in chronic obstructive pulmonary disease (Xia et al., 2019), and homeobox 10 (HOXA10) is demonstrated to contribute to the innate immune response and suppress inflammation (Wang et al., 2015). The pathways involved in the regulation of apoptosis and inflammation by EGF, miR-223 and ISG15 were assessed by detecting the expression of these proteins.
Materials and Methods
Animals
One cubic centimeter of mammary gland tissue was taken with a scalpel from 3-year-old Guanzhong dairy goats in peak lactation period after anesthetizing and stored in PBS with penicillin/streptomycin (100 U/mL, Harbin Pharmaceutical Group, China). The wound was sewed and sterilized immediately, and the goats recovered after a week. The mammary gland tissue was used to isolate gMECs as reported (Wang et al., 2010). Specifically, the tissue was cut into pieces around 1 mm3 and seeded in cell culture plates, and then gMECs would grow adherently. One week later, gMECs were obtained and purified. All the procedures conformed to the guidelines of the Animal Care and Use Committee of the Northwest A&F University (ethic code: #0726/2018).
Vector Construction
The full coding sequence of ISG15 was amplified with forward primer: 5′-cccaagcttgccaccATGGGCGGGGACCTGAAG-3′ and reverse primer: 5′-ccgctcgag CTACCCACCCCGCAG-3′ using 2 × HiFiTaq PCR StarMix with Loading Dye (Genstar, Beijing, China), and inserted into pcDNA3.1 (+) vector between the restriction sites Hind III and Xho I to achieve the overexpression of ISG15. The segment of ISG15 containing the seed site of miR-223 was amplified with forward primer: 5′-ccgctcgag ACCTTGACAGCAGGGAAGTG-3′ and reverse primer: 5′-ataagaatgcggccgcCAGAATTGGTCCGCTTGCAC-3′ using 2 × HiFiTaq PCR StarMix with Loading Dye (Genstar, Beijing, China), and inserted to a dual-luciferase reporter vector, psiCHECK2, between the restriction sites Xho I and Not I, for dual-luciferase reporter assay.
Cell Culture
Primary gMECs were cultured in DMEM/F12 medium (Hyclone, United States) with 10% fetal bovine serum and incubated at 37°C with 5% CO2 in a humid environment. The sRNAs and vectors were transfected into gMECs using Lipofectamine 2000 reagent (Invitrogen, United States), and the transfection was conducted when gMEC density reached about 80%. One hundred pmol sRNA (or 4 μg vector) and 5 μl Lipofectamine 2000 were diluted into 250 μl Opti-MEM (Gibco, United States), respectively, and sRNA or vector was gently mixed into Lipofectamine 2000 and incubated for 20 min at room temperature. The mixture was applied to gMECs in six-well plates. Four hours later, the medium was changed to the fresh medium. Forty-eight hours post-transfection, the gMECs were harvested. The sequences of sRNAs are given below. Negative control (NC): 5′-UUCUCCGAACGUGUCACGUTT-3′; inhibitor NC: 5′-CAGUACUUUUGUGUAGUACAA-3′; miR-223 mimic: 5′-UGUCAGUUUGUCAAAUACCCCA-3′; miR-223 inhibitor: 5′-UGGGGUAUUUGACAAACUGACA-3′; siISG15: 5′-GCGUGUGCAAGCGGACCAATT-3′. MiR-223 mimic was applied for the overexpression of miR-223, miR-223 inhibitor was for the inhibition of the expression of miR-223, and siISG15 was for the knockdown of the expression of ISG15.
Dual-Luciferase Reporter Assay
The plasmid of psiCHECK2-ISG15 vector was co-transfected with miR-223 or negative control (NC), respectively, into gMEC. After 48 h, cells were lysed with passive lysis buffer, and the Firefly luciferase (hluc+) activity and Renilla luciferase (hRluc) activity were measured by a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, United States) according to the manufacturers’ instruction. The ratio of hRluc and hluc+ was calculated to evaluate the relative luciferase activity.
RNA Isolation and Analysis
The gMECs were washed mildly by PBS and lysed using TRIzol Reagent (Invitrogen, United States) for RNA isolation. The total RNA was quality-controlled by Agilent bioanalyzer 2100 and applied for cDNA synthesis of mRNA by the PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). The cDNA of miRNA was acquired using the miRcute Plus miRNA First-Strand cDNA Kit (Tiangen, Beijing, China). RT-qPCR was conducted using SYBR Green qPCR Master Mix (Takara, Japan) with following primers: 5′-TTTGCAGCAGGTCAGAGAGA-3′ (MX1 forward); 5′-GTACG CCATCAGGTGTTGAA-3′ (MX1 reverse); 5′-CCAATCAGATC CCGTTCATC-3′ (MX2 forward); 5′-CCTGAAGCAGCCAGG AATAG-3′ (MX2 reverse); 5′-ACCTTGACAGCAGGGAAGTG-3′ (ISG15 forward); 5′-GTCGTTCCTCACCAGGATGT-3′ (ISG 15 reverse); 5′-GTGCATGGTGTTTCAGATGC-3′ (DDX58 for ward); 5′-TCCGTGCATCCTCACTGATA-3′ (DDX58 reverse); 5′-ACTCCCACCAGCGTCAATTA-3′ (RSAD2 forward); 5′-AG ACCTCTCTTGGCCTCCTC-3′ (RSAD2 reverse); 5′-CCAATT ACACCCAAGCATCC-3′ (IFI44L forward); 5′-GTTTGAGCTT CGCCATCATT-3′ (IFI44L reverse); 5′-CTGGCCATCGCAA TGTACTA-3′ (IFIT3 forward); 5′-AGGGCCAGGAGAACTTTG AT-3′ (IFIT3 reverse); 5′-GTGGCCAAGGACAACAAGAT-3′ (F HL1 forward); 5′-GTGCCCTTGTATTCCACGTT-3′ (FHL1 rev erse); 5′-GATCTGGCACCACACCTTCT-3′ (β-actin forward); 5′-GGGTCATCTTCTCACGGTTG-3′ (β-actin reverse); 5′-GCG TGTCAGTTTGTCAAATACCCCA-3′ (miR-223 forward); the reverse primer of miRNA was provided by Tiangen (Beijing, China). The Poly (A) mRNA Magnetic Isolation Module (NEBNext, United States) was used to capture mRNA from total RNA, and Ultra RNA Library Prep Kit for Illumina (NEBNext, United States) was applied for the library construction. Then the library was purified and quality-controlled for RNA-sequencing, which was performed by Illumina HiSeq 2500. The obtained raw reads were filtered for clean reads of high quality, which were mapped to the reference genome database1. The differentially expressed genes were screened, and GO and KEGG enrichments were conducted.
Flow Cytometry
The gMECs were digested by pancreatin free of EDTA and washed by PBS subsequently. The cells were centrifuged at 1,000 rpm for 5 min and resuspended with 1 × Binding Buffer (7Sea Biotech, Shanghai, China). The apoptosis was examined using the Annexin V-FITC/PI apoptosis kit (7Sea Biotech, Shanghai, China). Within 1 h after the staining, the apoptotic rate was measured by a flow cytometer (BD, United States). The apoptosis results were shown in a four-quadrant diagram: dots in lower left quadrant represented normal cells; dots in upper left quadrant represented necrotic cells; dots in lower right quadrant represented late apoptotic cells; dots in upper right represented early apoptotic cells. The total apoptotic rate was calculated to evaluate the apoptosis of gMEC.
Western Blot
The total protein of gMEC was harvested by using RIPA lysis buffer with protease inhibitor and phosphatase inhibitor (Thermo Fisher Scientific, United States). The concentration of the proteins was measured by a BCA protein assay kit (Solarbio, Beijing, China). Equal amount of protein was loaded to a 12% SDS-PAGE gel. Primary antibodies used were as follows: anti-Bcl-2 (BBI, D260117, Shanghai, China), anti-Bax (BBI, D220073, Shanghai, China), anti-p53 (Beyotime, AF7671, Shanghai, China), anti-caspase 3 (Cell Signaling Technology, 9665, United States), anti-p-STAT3 (BBI, Phospho-Ser727, D155018, Shanghai, China), anti-STAT3 (BBI, D320083, Shanghai, China), anti-PR (Beyotime, AF7737, Shanghai, China), anti-FOXC1 (BBI, D160347, Shanghai, China), anti-HOXA10 (BBI, D163629, Shanghai, China), anti-β-actin (Beyotime, AA128, Shanghai, China). Horseradish peroxidase conjugated anti-mouse (Beyotime, A0216, Shanghai, China) and anti-rabbit (Beyotime, A0208, Shanghai, China) antibodies were applied as secondary antibodies.
Statistics
The quality of sequencing data was evaluated using FastQC v0.10.1, and raw reads were filtered by Cutadapt 1.9.1. Short read alignment was performed by Hisat v2.0.14. DEGs were conditioned on a more than twofold difference and FDR < or = 0.05. GO function and KEGG pathway annotation analyses were based on the GO database2 and the KEGG database3, respectively. All the experiments were repeated at least three times independently. Statistical significance was calculated via Student’s t-test and one-way ANOVA (SPSS 22.0; SPSS Inc). Significance: ∗p < 0.05, ∗∗p < 0.01.
Results
MiR-223 Induced by EGF Inhibits gMEC Apoptosis
Epidermal growth factor was diluted into cell cultivator at concentrations of 0, 1, 5, 10, and 50 ng/ml to incubate gMEC for 48 h. Concentration of EGF at 0 ng/ml was set as the control group. The result revealed that 1 ng/ml of EGF was the most effective to induce a higher expression of miR-223 (Figure 1A). Therefore, 1 ng/ml of EGF was applied to the following experiments. The apoptotic rate of gMECs incubated with 1 ng/ml of EGF was measured, which illustrated an inhibitory effect of EGF on gMEC apoptosis (Figures 1B,D). To explore the effect of miR-223 on gMEC apoptosis, miR-223 and negative control (NC) was transfected into gMEC, and gMEC apoptotic rate was measured after 48 h. The result revealed that miR-223 suppressed the apoptosis of gMEC (Figures 1C,E).
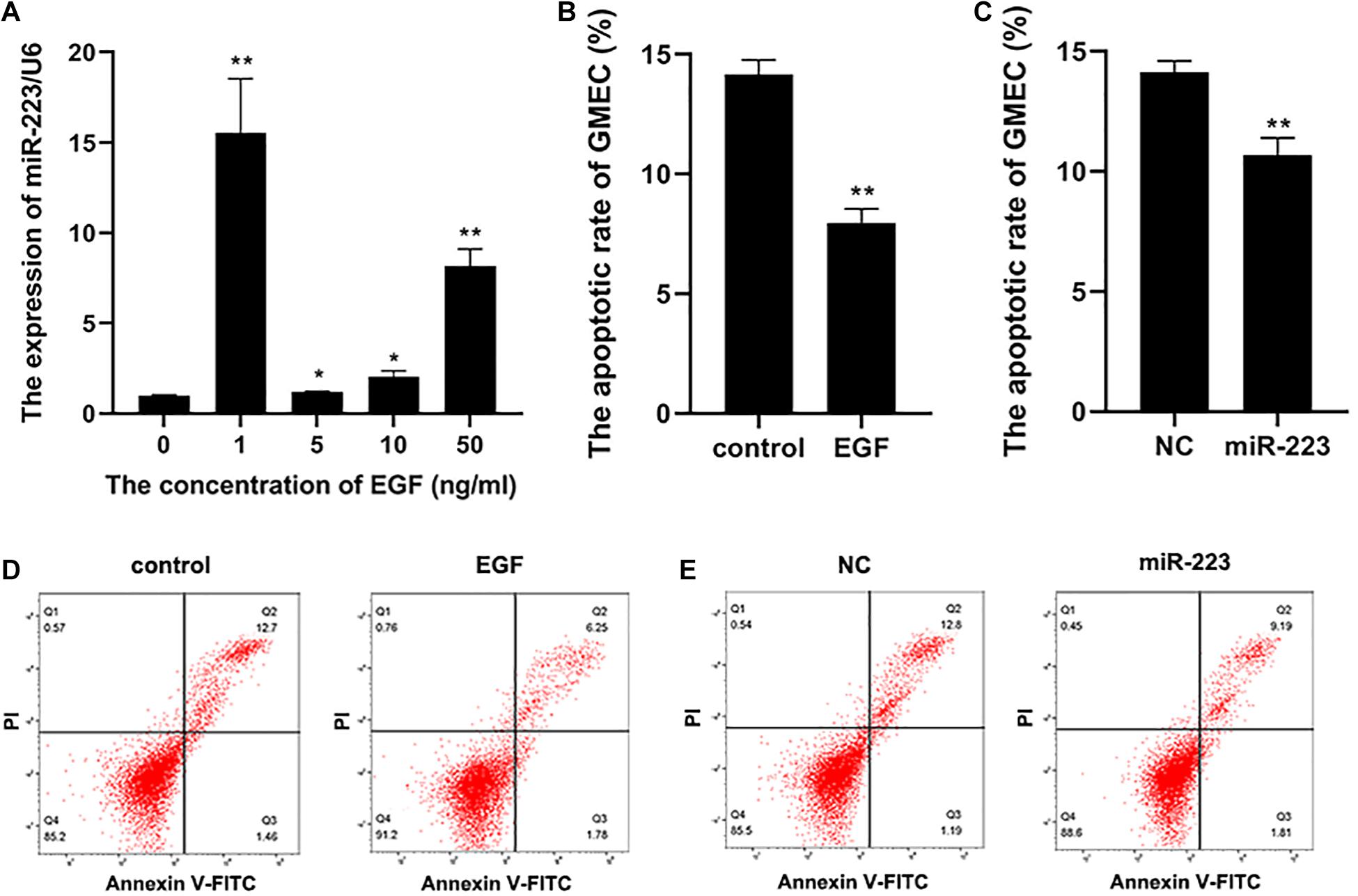
Figure 1. The apoptosis of gMEC regulated by EGF and miR-223. (A) A higher expression of miR-223 was induced by EGF. The gMECs were incubated for 48 h with EGF at concentrations of 0, 1, 5, 10, and 50 ng/ml, respectively, and the expression of miR-223 was then detected, which revealed that the expression of miR-223 was the highest in gMEC incubated with 1 ng/ml of EGF. (B,D) The regulation of gMEC apoptosis by EGF. The cell cultivators with or without EGF at 1 ng/ml were applied to incubate gMEC for 48 h, and a reduction of apoptotic rate in gMEC was found in EGF group. (C,E) The regulation of gMEC apoptosis by miR-223. MiR-223 and NC were transfected into gMEC, respectively, and 48 h post-transfection gMEC apoptotic rate was found reduced in miR-223 group. *p < 0.05; **p < 0.01.
Screening of miR-223 Downstream Genes
MiR-223 mimic and inhibitor were transfected into gMEC, respectively, and NC and inhibitor NC were transfected, respectively, as control. Total RNA of gMEC was isolated 48 h post-transfection. The efficiency of miR-223 mimics and miR-223 inhibitor was measured (Supplementary Figure 1). Groups miR-223 mimic and NC were used for RNA-seq (Sequence Read Archive: SRR13638390) to screen differentially expressed genes (DEGs). Thirty-six DEGs were found, including 24 downregulated genes and 12 upregulated genes. The heatmap of DEGs is shown in Figure 2A. To ensure the accuracy of RNA-sequencing, eight of the DEGs were randomly picked to perform RT-qPCR (Figure 2B), which ensured the reliability of the result of RNA-sequencing and demonstrated the down-regulation of ISG15 by miR-223. GO and KEGG enrichments of DEGs were conducted to predict the function of miR-223. GO enrichment showed that miR-223 might involve in 20 GO terms (Figure 2C), such as binding (GO:0003723), catalytic activity (GO:0003824), response to stimulus (GO:0050896) and immune system process (GO:0002376), and 18 KEGG pathways that miR-223 could participate in were found (Figure 2D), including RIG-I like receptor signaling pathway (ko04622).
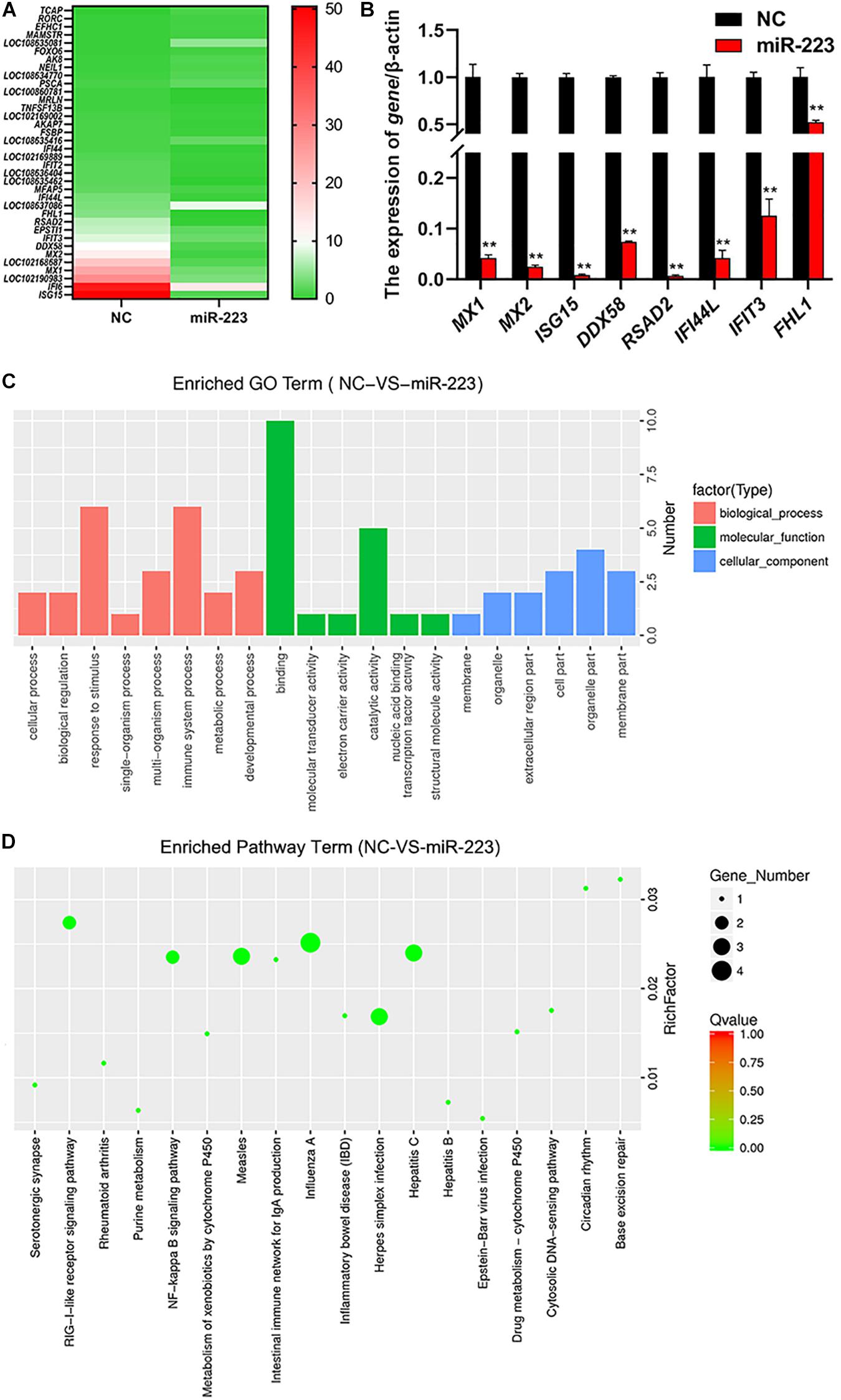
Figure 2. The analysis of differentially expressed genes induced by miR-223. (A) The downstream genes regulated by miR-223. MiR-223 and NC were transfected into gMECs, respectively, and gMECs were harvested after 48 h for RNA-seq to screen differentially expressed genes of miR-223, which were shown in the heatmap. (B) The accuracy test of RNA-seq by RT-qPCR. Eight differentially expressed genes were randomly chosen to ensure the accuracy of the RNA sequencing by RT-qPCR, and the result showed that the result of RNA-seq was reliable. (C,D) GO analysis and KEGG pathway analysis of differentially expressed genes of miR-223 were conducted to predict the function of miR-223. It showed that the differentially expressed genes involved in 20 GO terms (C), and 18 KEGG pathways (D). **p < 0.01.
MiR-223 Induces gMEC Apoptosis via ISG15
To investigate whether ISG15 could be targeted by miR-223, the sequence of ISG15 containing the seed site of miR-223 was amplified and inserted into psiCHECK2 (Figure 3A). The psiCHECK2-ISG15 vector was co-transfected with miR-223 and NC, respectively, into gMEC. The luciferase activities were detected after 48 h, and it showed a decreased relative luciferase activity in the miR-223 group (Figure 3B), which means ISG15 was targeted by miR-223. It is shown in Figure 3C that EGF reduced the expression of ISG15, especially at 1 ng/ml, and EGF at 1 ng/ml decreased the protein expression of ISG15 as well (Figure 3D). The miR-223 inhibitor and inhibitor NC were transfected into gMEC and the expression of ISG15 was measured after 48 h. The increase of ISG15 expression in miR-223 inhibitor group reassured the down-regulation of ISG15 by miR-223 (Figure 3E). It is shown in Figure 3F that miR-223 could restrain the protein expression of ISG15, and when miR-223 was inhibited, the protein expression of ISG15 was increased.
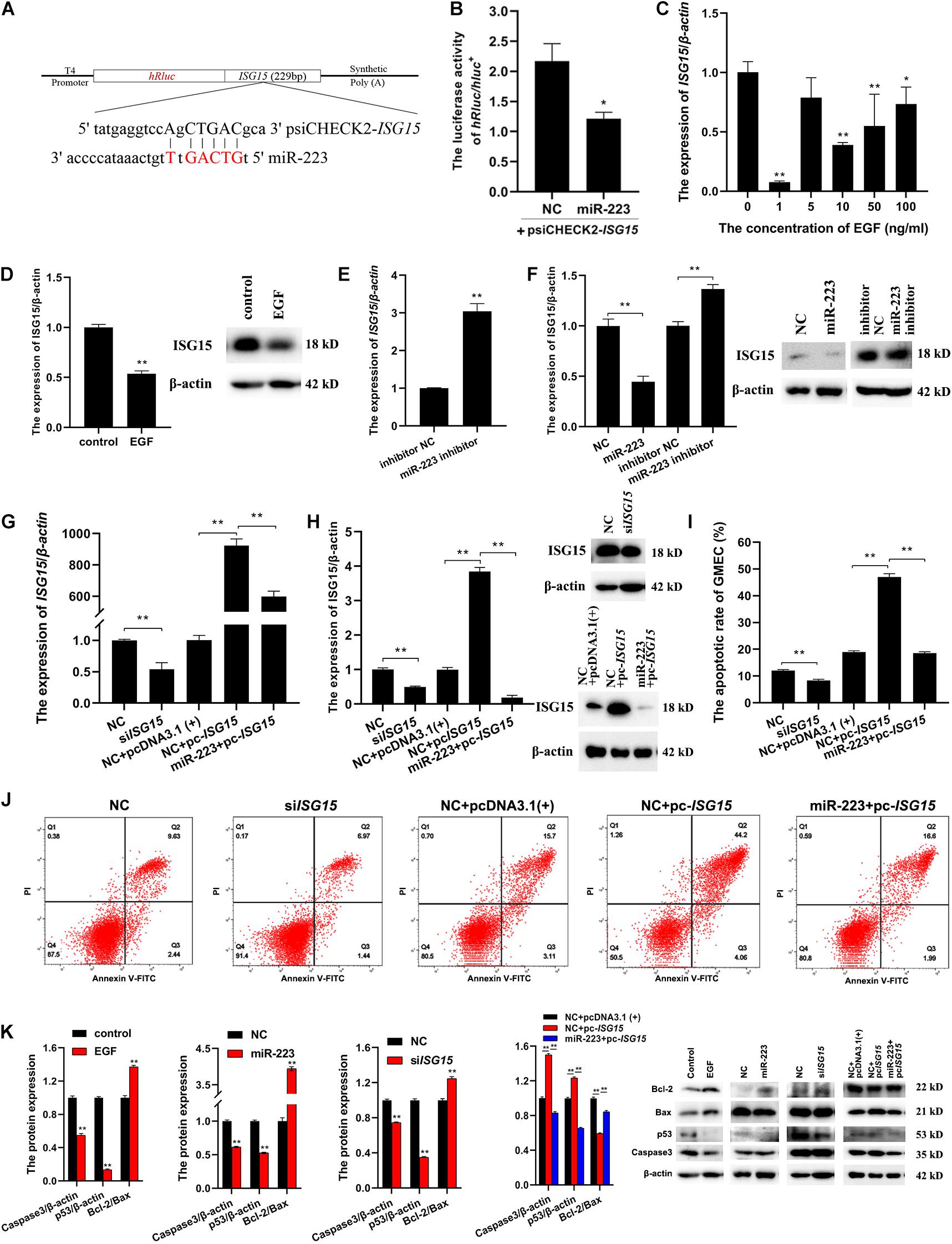
Figure 3. A further investigation of the relationships among EGF, miR-223 and ISG15, and their regulation to apoptosis. (A) The information of psiCHECK2-ISG15 vector for dual luciferase reporter assay. The sequence of ISG15 containing seed site of miR-223 was amplified and inserted into psiCHECK2 vector. (B) The relative luciferase activity regulated by miR-223. PsiCHECK2-ISG15 vector was co-transfected with miR-223 or NC, respectively, and 48 h later, a decreased relative luciferase activity was detected in miR-223 group. (C,D) The effect of EGF on the expression of ISG15 in gMEC. EGF at 0, 1, 5, 10, and 50 ng/ml was applied to incubate gMEC, and the expression of ISG15 was examined 48 h later. The group with 1 ng/ml of EGF witnessed the lowest expression of ISG15 (C), and the protein expression of ISG15 was also decreased by 1 ng/ml of EGF (D). (E,F) The regulation of ISG15 by miR-223. The expression of ISG15 regulated by miR-223 had been evaluated in the accuracy test of RNA-seq, and the inhibitor of miR-223 was applied to enhance the conclusion that miR-223 downregulated the expression of ISG15 (E). The measurement of protein expression of ISG15 modulated by miR-223 illustrated a reduction of ISG15 induced by miR-223 (F). (G,H) The efficiency of siISG15 and pc-ISG15. To explore the function of ISG15 in gMEC, the efficiency of siISG15 and pc-ISG15 was detected by RT-qPCR (G) and Western Blot (H), which showed the effectiveness of siISG15 and pc-ISG15. (I,J) The apoptotic rate of gMEC regulated by ISG15. Flow cytometry was used to detect gMEC apoptosis, and it showed that siISG15 suppressed gMEC apoptosis and pc-ISG15 enhanced gMEC apoptosis, while miR-223 impaired the promotion of apoptosis by pc-ISG15. (K) The protein expression of apoptosis-related biomarkers (Bcl-2/Bax, p53, caspase 3). EGF, miR-223, and siISG15 increased the expression of Bcl-2/Bax and reduced the expression of p53 and Caspase 3, and pc-ISG15 reduced the expression of Bcl-2/Bax and elevated the expression of p53 and Caspase 3, while miR-223 weakened its regulation. *p < 0.05; **p < 0.01.
To explore the effect of ISG15 on gMEC, the efficiency of ISG15 siRNA (siISG15) and pcDNA3.1-ISG15 vector (pc-ISG15) was ensured by RT-qPCR and Western Blot. Meanwhile, the efficiency of pc-ISG15 decreased when co-transfected with miR-223, which further confirmed the inhibitory effect of miR-223 on ISG15 expression (Figures 3G,H). The apoptotic rate of gMEC was measured by flow cytometry to investigate the regulation of gMEC apoptosis by ISG15. The results showed reduced apoptotic rates in siISG15 group and increased apoptotic rates in pc-ISG15 group. The apoptosis of gMEC induced by pc-ISG15 was alleviated by co-transfection with miR-223 (Figures 3I,J). The expression of apoptosis-related proteins, like p53, Caspase 3, and Bcl-2/Bax regulated by EGF/miR-223/ISG15 axis were detected to further explore the regulation of gMEC apoptosis. The results showed that the expressions of p53 and Caspase 3 were decreased, and the expression of Bcl-2/Bax was increased by EGF, miR-223 and siISG15. The overexpression of ISG15 enhanced the expressions of p53 and Caspase 3 and reduced the expression of Bcl-2/Bax, while miR-223 mitigated the regulation of apoptosis-related proteins by pc-ISG15 (Figure 3K).
The Regulation of EGF/miR-223/ISG15 Axis to Inflammation
To investigate the involvement of EGF, miR-223 and ISG15 in inflammation, the expressions of inflammation-related proteins (p-STAT3, PR, FOXC1, and HOXA10) were detected by Western Blot. The results showed that EGF, miR-223, and siISG15 significantly decreased the expression of p-STAT3 and elevated the expressions of PR, FOXC1, and HOXA10 in gMEC (Figure 4). When the ISG15 was overexpressed, the expression of p-STAT3 increased and the expressions of PR, FOXC1 and HOXA10 reduced, and miR-223 could impair the effect of ISG15 overexpression on the protein expressions in gMEC (Figure 4).
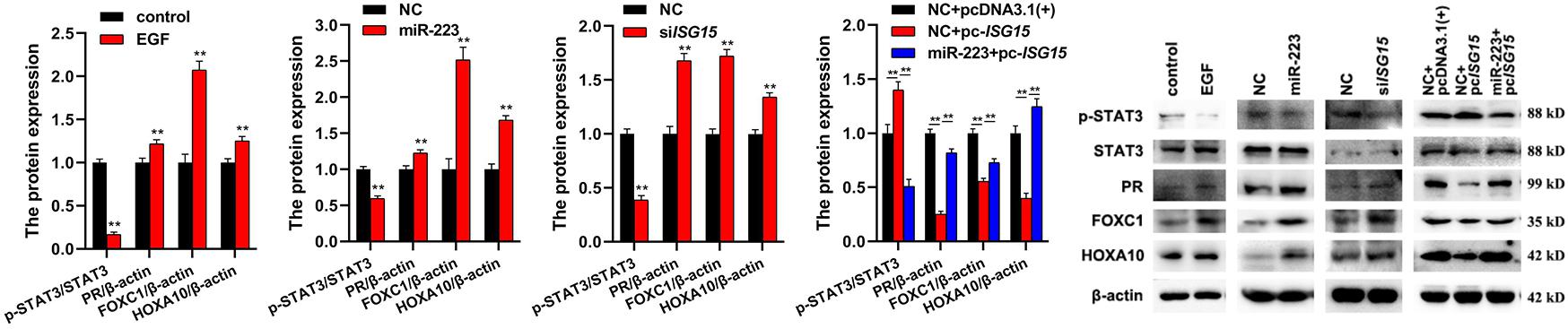
Figure 4. Effects of EGF, miR-223, and ISG15 on gMEC inflammation. The regulation of inflammation-related protein expressions (p-STAT3, PR, FOXC1, and HOXA10) by EGF, miR-223, and ISG15 was detected, and it showed that EGF, miR-223, and siISG15 significantly decreased the expression of p-STAT3 and elevated the expressions of PR, FOXC1, and HOXA10 in gMEC. Pc-ISG15 promoted the expression of p-STAT3 and reduced the expressions of PR, FOXC1, and HOXA10 in gMEC, while miR-223 weakened its regulation. EGF has been reported to surge at the onset of lactation, suggesting its importance in the lactation process. In this study, we found that the expression of miR-223, a conserved anti-inflammatory miRNA, could be induced by EGF. The downstream differentially expressed genes of miR-223 were screened by RNA-seq, and abundant interferon stimulated genes were found to be regulated by miR-223, including ISG15, a pivotal immune responder. Therefore, we explored the role of EGF, miR-223, and ISG15 in goat mammary epithelial cell apoptosis and inflammation to elaborate how mammary gland could be protected from mastitis during lactation. **p < 0.01.
Discussion
Our data showed that EGF could significantly elevate the expression of miR-223 in gMEC, especially at 1 ng/ml. Downstream differentially expressed genes (DEGs) between miR-223 and negative control (NC) groups were screened by RNA-seq. The innate immune response mediated by interferon is a robust first line of defense against pathogens (Schneider et al., 2014). Interferon signaling is important in immune response and inflammation (Chen et al., 2017; Zhang et al., 2017). The abundance of interferon-stimulated genes in the downregulated DEG list, such as interferon-stimulated gene product 15 (ISG15), IFI6, MX1, MX2, DDX58, IFIT3, RSAD2, IFI44L, IFIT2, and IFI44, indicates that miR-223 was likely to participate in immunity and inflammation. ISG15 was one of the DEGs with the highest expression in the NC group and was greatly downregulated by miR-223. The downregulation of ISG15 by miR-223 was confirmed by RT-qPCR and Western Blot. The targeting regulation of miR-223 to ISG15 was illustrated by a dual luciferase reporter assay. It was demonstrated that the expression of ISG15 was also reduced by epidermal growth factor (EGF), which suggested the expression of ISG15 could be reduced by EGF-induced miR-223. The apoptotic rate was measured by flow cytometry and the expressions of anti-apoptotic Bcl-2 (Bruckheimer et al., 1998), and pro-apoptotic caspase 3 (Choudhary et al., 2015) and p53 (Aubrey et al., 2018; Zhang et al., 2020a, 2021a; Niu et al., 2021a,b) were detected to evaluate the apoptosis of gMEC. The inflammation of gMEC was estimated by inflammation-related proteins. It showed that EGF and miR-223 reduced STAT3 phosphorylation levels at Ser727 and promoted the expression of PR, FOXC1, and HOXA10 by downregulating ISG15. The results indicated that inhibition of apoptosis and inflammation in gMECs by EGF can be achieved by increasing the expression of miR-223 that can downregulate ISG15.
Epidermal growth factor is a 53-amino acid polypeptide mitogen that transduces its signal to EGF receptor (EGFR). The surge of EGF expression at the onset of lactation indicates the importance of EGF in the lactation process (Hardy et al., 2010; Fu et al., 2015). However, studies show that EGF is likely to induce tumor cell invasion in breast cancer (Klijn et al., 1992; Chrysogelos and Dickson, 1994; Hardy et al., 2010), where the overexpression of EGFR is connected with the loss of estrogen receptor and poor prognosis (Chrysogelos and Dickson, 1994). The interesting thing is that EGFR expresses more frequently in normal mammary gland than in malignant tissue (Chrysogelos and Dickson, 1994). The role of EGFR might be different between normal breast and breast cancer. MiR-223 is a guard to prevent relapse of breast cancer (Fabris et al., 2016; Citron et al., 2020). In this study, the expression of miR-223 was raised about 15-fold by EGF, which might be a protection in lactating mammary tissues, and ISG15, a critical gene in inflammation (Hu et al., 2019b), was targeted and down-regulated by miR-223. The regulation of gMEC apoptosis and inflammation-related genes by EGF/miR-223/ISG15 indicated that EGF-induced miR-223 could prevent gMEC apoptosis and inflammation through targeting and down-regulating ISG15. In conclusion, our study provided a possible pathway for EGF to protect mammary gland during lactation.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject, Accession no: PRJNA699733 https://dataview.ncbi.nlm.nih.gov/object/PRJNA699733?reviewer=er592n5eopqpmo65ljj7983hh2 and NCBI SRR13638390.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the Northwest A&F University.
Author Contributions
YZ designed the project, performed the experiments, and wrote the manuscript. GLN, JDL, FJC and XPA helped with performing the experiments. BYC and QW applied for the funding for this study. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81860762), Scientific Research Guiding Plan Topic of Qinghai Hygiene Department (2018-wjzdx-131), Shaanxi Science and Technology Innovation Project Plan (2017ZDXM-NY-081 and 2018ZDCXL-NY-01-04), Shaanxi Key Research and Development Program (2020ZDLNY02-01 and 2020ZDLNY02-02), and Natural Science Foundation of Shaanxi Province (2020JQ-868).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.660933/full#supplementary-material
Footnotes
- ^ http://www.ensembl.org/
- ^ http://geneontology.org/page/go-database
- ^ http://www.kegg.jp/kegg/ko.html
References
Aubrey, B. J., Kelly, G. L., Janic, A., Herold, M. J., and Strasser, A. (2018). How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Different. 25, 104–113. doi: 10.1038/cdd.2017.169
Balic, J. J., Albargy, H., Luu, K., Kirby, F. J., Jayasekara, W. S. N., Mansell, F., et al. (2020). STAT3 serine phosphorylation is required for TLR4 metabolic reprogramming and IL-1β expression. Nat. Commun. 11:3816.
Bruckheimer, E. M., Cho, S. H., Sarkiss, M., Herrmann, J., and McDonnell, T. J. (1998). The Bcl-2 gene family and apoptosis. Adv. Biochem. Eng. Biotechnol. 62, 75–105. doi: 10.1007/bfb0102306
Chen, K., Liu, J., and Cao, X. (2017). Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J. Autoimmunity 83, 1–11. doi: 10.1016/j.jaut.2017.03.008
Choudhary, G. S., Al-Harbi, S., and Almasan, A. (2015). Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol. Biol. 1219, 1–9. doi: 10.1007/978-1-4939-1661-0_1
Chrysogelos, S. A., and Dickson, R. B. (1994). EGF receptor expression, regulation, and function in breast cancer. Br. Cancer Res. Treat. 29, 29–40. doi: 10.1007/bf00666179
Citron, F., Segatto, I., Vinciguerra, G. L. R., Musco, L., Russo, F., Mungo, G., et al. (2020). Downregulation of miR-223 expression is an early event during mammary transformation and confers resistance to cdk4/6 inhibitors in luminal breast cancer. Cancer Res. 80, 1064–1077. doi: 10.1158/0008-5472.can-19-1793
Dzimianski, J. V., Scholte, F. E. M., Bergeron, E., and Pegan, S. D. (2019). ISG15: It’s complicated. J. Mol. Biol. 431, 4203–4216. doi: 10.1016/j.jmb.2019.03.013
Fabris, L., Berton, S., Citron, F., D’Andrea, S., Segatto, I., Nicoloso, M. S., et al. (2016). Radiotherapy-induced miR-223 prevents relapse of breast cancer by targeting the EGF pathway. Oncogene 35, 4914–4926. doi: 10.1038/onc.2016.23
Fu, N. Y., Rios, A. C., Pal, B., Soetanto, R., Lun, A. T., Liu, K., et al. (2015). EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival. Nat. Cell Biol. 17, 365–375. doi: 10.1038/ncb3117
Haneklaus, M., Gerlic, M., O’Neill, L. A., and Masters, S. L. (2013). miR-223: infection, inflammation and cancer. J. Intern. Med. 274, 215–226. doi: 10.1111/joim.12099
Hardy, K. M., Booth, B. W., Hendrix, M. J., Salomon, D. S., and Strizzi, L. (2010). ErbB/EGF signaling and EMT in mammary development and breast cancer. J. Mammary Gland Biol. Neoplasia 15, 191–199. doi: 10.1007/s10911-010-9172-2
Hu, Y. S., Han, X., and Liu, X. H. (2019a). STAT3: a potential drug target for tumor and inflammation. Curr. Top. Med. Chem. 19, 1305–1317. doi: 10.2174/1568026619666190620145052
Hu, Y., Hong, X. Y., Yang, X. F., Ma, R. H., Wang, X., Zhang, J. F., et al. (2019b). Inflammation-dependent ISG15 upregulation mediates MIA-induced dendrite damages and depression by disrupting NEDD4/Rap2A signaling. Biochim. Biophys. Acta Mol. Basis. Dis. 1865, 1477–1489. doi: 10.1016/j.bbadis.2019.02.020
Jeffries, J., Zhou, W., Hsu, A. Y., and Deng, Q. (2019). miRNA-223 at the crossroads of inflammation and cancer. Cancer Lett. 451, 136–141. doi: 10.1016/j.canlet.2019.02.051
Ji, Z., He, R., Chao, T., Xuan, R., Liu, S., Wang, G., et al. (2019). chi-miR-143-3p promotes apoptosis of mammary gland epithelial cells from dairy goats by targeting Ndfip1. DNA Cell Biol. 38, 1188–1196. doi: 10.1089/dna.2019.4830
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi: 10.1038/nrg3965
Klijn, J. G., Berns, P. M., Schmitz, P. I., and Foekens, J. A. (1992). The clinical significance of epidermal growth factor receptor (EGF-R) in human breast cancer: a review on 5232 patients. Endocrine Reviews 13, 3–17. doi: 10.1210/er.13.1.3
Niu, G., Bak, A., Nusselt, M., Zhang, Y., Pausch, H., Flisikowska, T., et al. (2021a). Allelic expression imbalance analysis identified YAP1 amplification in p53- dependent osteosarcoma. Cancers 13:1364. doi: 10.3390/cancers13061364
Niu, G., Hellmuth, I., Flisikowska, T., Pausch, H., Rieblinger, B., Carrapeiro, A., et al. (2021b). Porcine model elucidates function of p53 isoform in carcinogenesis and reveals novel circTP53 RNA. Oncogene 40, 1896–1908. doi: 10.1038/s41388-021-01686-9
Ola, M. S., Nawaz, M., and Ahsan, H. (2011). Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol. Cell Biochem. 351, 41–58. doi: 10.1007/s11010-010-0709-x
Park, C. J., Lin, P. C., Zhou, S., Barakat, R., Bashir, S. T., Choi, J. M., et al. (2020). Progesterone receptor serves the ovary as a trigger of ovulation and a terminator of inflammation. Cell Rep. 31:107496. doi: 10.1016/j.celrep.2020.03.060
Schneider, W. M., Chevillotte, M. D., and Rice, C. M. (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Vasilikos, L., Spilgies, L. M., Knop, J., and Wong, W. W. (2017). Regulating the balance between necroptosis, apoptosis and inflammation by inhibitors of apoptosis proteins. Immunol. Cell Biol. 95, 160–165. doi: 10.1038/icb.2016.118
Wang, H., Bei, L., Shah, C. A., Hu, L., and Eklund, E. A. (2015). HoxA10 terminates emergency granulopoiesis by increasing expression of triad1. J. Immunol. 194, 5375–5387. doi: 10.4049/jimmunol.1401909
Wang, Z., Luo, J., Wang, W., Zhao, W., and Lin, X. (2010). [Characterization and culture of isolated primary dairy goat mammary gland epithelial cells]. Sheng Wu Gong Cheng Xue Bao 26, 1123–1127.
Watson, C. J. (2006). Involution: apoptosis and tissue remodelling that convert the mammary gland from milk factory to a quiescent organ. Br. Cancer Res. 8:203.
Xia, S., Qu, J., Jia, H., He, W., Li, J., Zhao, L., et al. (2019). Overexpression of Forkhead box C1 attenuates oxidative stress, inflammation and apoptosis in chronic obstructive pulmonary disease. Life Sci. 216, 75–84. doi: 10.1016/j.lfs.2018.11.023
Xu, D. D., Wang, G., He, X. J., Wang, J. F., Yang, B., Sun, Z. P., et al. (2017). 17β-Estradiol and progesterone decrease MDP induced NOD2 expression in bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 188, 59–64. doi: 10.1016/j.vetimm.2017.04.010
Xuan, R., Chao, T., Wang, A., Zhang, F., Sun, P., Liu, S., et al. (2020). Characterization of microRNA profiles in the mammary gland tissue of dairy goats at the late lactation, dry period and late gestation stages. PLoS One 15:e0234427. doi: 10.1371/journal.pone.0234427
Zamzami, N., and Kroemer, G. (2005). p53 in apoptosis control: an introduction. Biochem. Biophys. Res. Commun. 331, 685–687. doi: 10.1016/j.bbrc.2005.04.013
Zhang, X., Bogunovic, D., Payelle-Brogard, B., Francois-Newton, V., Francois-Newton, V., Speer, S. D., et al. (2015). Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature 517, 89–93. doi: 10.1038/nature13801
Zhang, Y., Guo, X., Wang, G., Liu, J., Liang, P., Wang, H., et al. (2020a). Effects of rhodioloside on the neurological functions of rats with total cerebral ischemia/reperfusion and cone neuron injury in the hippocampal CA1 region. PeerJ 8:e10056. doi: 10.7717/peerj.10056
Zhang, Y., Liu, J., Li, W., Cao, F., Niu, G., Ji, S., et al. (2020b). A regulatory circuit orchestrated by novel-miR-3880 modulates mammary gland development. Front. Cell Dev. Biol. 8:383. doi: 10.3389/fcell.2020.00383
Zhang, Y., Niu, G., Flisikowska, T., Schnieke, A., and Flisikowski, K. (2021a). A tissue- and gender-specific regulation of the SARS-CoV-2 receptor ACE2 by p53 in pigs. Biochem. Biophys. Res. Commun. 553, 25–29. doi: 10.1016/j.bbrc.2021.03.068
Zhang, Y., Wu, Q., Liu, J., An, X., and Cao, B. (2021b). Circ-140/chi-miR-8516/STC1-MMP1 regulates αs1-/β-casein secretion and lipid formation in goat mammary epithelial Cells. Genes 12:671. doi: 10.3390/genes12050671
Keywords: EGF, miR-223, ISG15, apoptosis, inflammation, mammary gland
Citation: Zhang Y, Wu Q, Niu G, Liu J, Cao F, An X and Cao B (2021) EGF-Induced miR-223 Modulates Goat Mammary Epithelial Cell Apoptosis and Inflammation via ISG15. Front. Cell Dev. Biol. 9:660933. doi: 10.3389/fcell.2021.660933
Received: 15 February 2021; Accepted: 08 June 2021;
Published: 30 June 2021.
Edited by:
Geeta Upadhyay, Uniformed Services University of the Health Sciences, United StatesReviewed by:
Mario Cioce, Campus Bio-Medico University, ItalyJiangjiang Zhu, Southwest Minzu University, China
Zhibin Ji, Shandong Agricultural University, China
Copyright © 2021 Zhang, Wu, Niu, Liu, Cao, An and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binyun Cao, Y2FvYmlueXVuQDEyNi5jb20=
†These authors have contributed equally to this work
 Yue Zhang
Yue Zhang Qiong Wu
Qiong Wu Guanglin Niu
Guanglin Niu Jidan Liu1
Jidan Liu1 Xiaopeng An
Xiaopeng An Binyun Cao
Binyun Cao