- 1Institutes of Biomedical Sciences, Fudan University, Shanghai, China
- 2ENT Institute, Department of Facial Plastic and Reconstructive Surgery, Eye & ENT Hospital, Fudan University, Shanghai, China
- 3Center for Reproductive Medicine, Department of Obstetrics and Gynecology, National Clinical Research Center for Obstetrics and Gynecology, Ministry of Education, Beijing, China
- 4Beijing Key Laboratory of Reproductive Endocrinology and Assisted Reproductive Technology, Peking University Third Hospital, Beijing, China
- 5Key Laboratory of Assisted Reproduction, Peking University, Beijing, China
- 6State Key Laboratory of Oral Diseases and National Clinical Research Center for Oral Diseases and Department of Cleft Lip and Palate, West China Hospital of Stomatology, Sichuan University, Chengdu, China
- 7Department of Oral and Cranio-Maxillofacial Surgery, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 8School of Basic Medical Sciences, Fudan University, Shanghai, China
- 9Children’s Hospital of Fudan University, Shanghai, China
- 10Medical Laboratory of Nantong ZhongKe, Nantong, China
Non-syndromic cleft lip and palate (NSCLP) is one of the most common congenital malformations with multifactorial etiology. Although long non-coding RNAs (lncRNAs) have been implicated in the development of lip and palate, their roles in NSCLP are not fully elucidated. This study aimed to investigate how dysregulated lncRNAs contribute to NSCLP. Using lncRNA sequencing, bioinformatics analysis, and clinical tissue sample detection, we identified that lncRNA ZFAS1 was significantly upregulated in NSCLP. The upregulation of ZFAS1 mediated by SP1 transcription factor (SP1) inhibited expression levels of Wnt family member 4 (WNT4) through the binding with CCCTC-binding factor (CTCF), subsequently inactivating the WNT/β-catenin signaling pathway, which has been reported to play a significant role on the development of lip and palate. Moreover, in vitro, the overexpression of ZFAS1 inhibited cell proliferation and migration in human oral keratinocytes and human umbilical cord mesenchymal stem cells (HUC-MSCs) and also repressed chondrogenic differentiation of HUC-MSCs. In vivo, ZFAS1 suppressed cell proliferation and numbers of chondrocyte in the zebrafish ethmoid plate. In summary, these results indicated that ZFAS1 may be involved in NSCLP by affecting cell proliferation, migration, and chondrogenic differentiation through inactivating the WNT/β-catenin signaling pathway.
Introduction
Cleft lip and palate (CLP) is one of the most common congenital craniofacial malformations around the world, resulting from failure in fusion of the facial primordia. Even though surgical treatments are doing well, children with CLP still face a variety of postoperative complications and psychosocial problems (Dixon et al., 2011).
There are two forms of CLP, syndromic and non-syndromic CLP (NSCLP). Syndromic CLP was often caused by single-gene mutations and chromosomal abnormalities. NSCLP, accounting for 70% of CLP, is a disorder without additional symptoms of atypical conditions (Jiang et al., 2006; Leslie and Marazita, 2013). It arises in about one per 500–1,000 live births (Mohamad Shah et al., 2019). NSCLP is a complex disease with multiple factors including genetic, epigenetic, and environmental factors. A number of susceptibility genes for NSCLP have been identified in previous studies (Dixon et al., 2011). However, only a small minority of NSCLP results from single-gene mutations. Because proper development of the lip and palate requires a coordination of a series of complex events, including cell growth, migration, differentiation, and apoptosis (Leslie and Marazita, 2013), which are regulated by precise gene expression, NSCLP could also be associated with the disturbances in this process due to dysregulation of genes.
Epigenetic modifications play an important role in the regulation of gene expression. As an important component of epigenetic modifications, long non-coding RNAs (lncRNAs), consisting of over 200 nucleotides, are non-coding transcripts and numerous in eukaryotes (Ma et al., 2020a; Ye et al., 2020b). They were thought to be “noise” in genome and a byproduct of transcription. Nevertheless, recent studies have revealed that lncRNAs are involved in several biological processes (BPs) through regulating gene expression at epigenetic, genomic transcription, and post-transcriptional levels (Ye et al., 2019; Ma et al., 2020b). Moreover, dysregulation of lncRNAs could be related to the progression of many disorders (Peng et al., 2017; Chi et al., 2019; Ye et al., 2020a). They have also been shown to participate in the development of lip and palate and the pathogenesis of related diseases (Gerrard et al., 2016; Gao et al., 2019; Shu et al., 2019). There have been a few of reports on the role of lncRNAs in CLP (Uslu et al., 2014; Gao et al., 2017; Yun et al., 2019), let alone NSCLP. RNA sequencing to identify transcriptome profiles including mRNAs, lncRNAs, and microRNAs (miRNAs) in NSCLP was performed in 2019 for the first time, and the potential role of lncRNAs in NSCLP has been suggested (Gao et al., 2019). So far, only several lncRNAs have been identified to be associated with NSCLP in previous studies, including lncRNA RP11-462G12.2, CCDC26 lncRNA, and a long non-coding interval at 8q24 (Uslu et al., 2014; Wu et al., 2019; Yun et al., 2019). Therefore, the underlying mechanism and function of lncRNAs in the involvement of NSCLP remain largely unclear.
In the present study, we identified the lncRNA profiles of tissue samples from patients with NSCLP using lncRNA sequencing followed by a series of bioinformatics analysis, and then we focused on ZFAS1 with unknown function in CLP. ZFAS1 was significantly upregulated in patients with NSCLP. In vitro, the overexpression of ZFAS1 significantly suppressed cell proliferation, cell migration, and chondrogenic differentiation. In vivo, upregulation of ZFAS1 in zebrafish embryos decreased cell proliferation and numbers of chondrocyte in the ethmoid plate. Exploration for further molecular mechanism suggested that ZFAS1 mediated by transcription factor SP1 inactivated the WNT/β-catenin signaling pathway through the binding with CCCTC-binding factor (CTCF).
Materials and Methods
Study Subjects and Samples
This study was reviewed and approved by the Institutional Research Ethics Committee of the Children’s Hospital of Fudan University (2016-121). A total of 40 tissues from patients with NSCLP and 15 normal control (NC) tissues, which were the extra tissues from surgery, were obtained from the Children’s Hospital of Fudan University. None of the patients with NSCLP had been diagnosed with extra disorders. NC samples were from patients with a traumatism for surgery and without any congenital disorders. All samples for RNA extraction were maintained in RNAlater RNA Stabilization Solution (Thermo Fisher Scientific, Waltham, MA, United States) after surgery and were stored at −80°C.
Long Non-coding RNA Sequencing and Functional Enrichment Analysis
Long non-coding RNAs sequencing of tissue samples from four NSCLP and two NC was performed by Precisiongenes (Nantong, China) to identify the differentially expressed lncRNAs. In brief, ribosomal RNA (rRNA) in total RNA was removed, and the remaining RNAs were subjected to construct sequencing library. The library was then sequenced on an Illumina platform (Illumina, Inc., San Diego, CA, United States), and the trimmed reads were mapped to the human genome Hg38. Finally, the transcript read count was calculated. The DAVIDdatabase1 was applied to perform gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses on the differentially expressed lncRNAs. P < 0.05 was considered to be statistically significant.
RNA Extraction, Reverse Transcription, and Quantitative Polymerase Chain Reaction
Total RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. Reverse transcription of 1,000 ng of RNA was performed to synthesize cDNA using a Primer Script RT Reagent Kit (Takara, Tokyo, Japan), followed by quantitative polymerase chain reaction (qPCR) using SYBR Premix ExTM Taq (Takara) on a StepOnePlusTM Real-Time PCR system (Thermo Fisher Scientific). Gene expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and calculated by the relative expression method (2–ΔΔCt). The primers used were as follows:
ZFAS1-Q-F: TCTCCTAGTTGCAGTCAGGC,
ZFAS1-Q-R: ATGCGGGTGTTGGAAGTAGA;
GAPDH-Q-F: GGGAGCCAAAAGGGTCAT,
GAPDH-Q-R: GAGTCCTTCCACGATACCAA;
U1-Q-F: GACGGGAAAAGATTGAGCGG,
U1-Q-R: CCACGAAGAGAGTCTTGAAGG;
Actin-Q-F: CATGTACGTTGCTATCCAGGC,
Actin-Q-R: CTCCTTAATGTCACGCACGAT;
Sox9-Q-F: ATGAAGATGACCGACGAGCA,
Sox9-Q-R: AACTTGTCCTCCTCGCTCTC;
ACAN-Q-F: CTTCTGAGTTCGTGGAGGGT,
ACAN-Q-R: CGGACGTCTCACTGCTAGAT;
SP1-Q-F: TCCAGACCATTAACCTCAGTGC,
SP1-Q-R: ACCACCAGATCCATGAAGACC;
WNT4-Q-F: GCTGTGACAGGACAGTGCAT,
WNT4-Q-R: GCCTCATTGTTGTGGAGGTT.
Cell Culture
Human oral keratinocyte (HOK) cells were cultured in Dulbecco’s modified Eagle’s medium (Biological Industries, Beit HaEmek, Israel) with 10% fetal bovine serum (FBS) (Biological Industries) and 1% penicillin–streptomycin (Biological Industries) at 37°C in 5% CO2.
Human umbilical cord mesenchymal stem cells (HUC-MSCs) were cultured in Dulbecco’s modified Eagle’s medium/F12 (Gibco, Carlsbad, CA, United States) with 10% FBS (Gibco) and 1% penicillin–streptomycin (Biological Industries) at 37°C in 5% CO2. The expanded cells at passages 3–7 were used for experiments.
All cell culture dishes and culture plates were purchased from Hangzhou Xinyou Biotechnology Co., Ltd. (Hangzhou, China).
Plasmid Construction
The plasmids of ZFAS1-PCDH and SP1-PCDH were obtained from GeneRay (Shanghai, China). The plasmids were sequenced and shown to be consistent with the sequence in the National Center for Biotechnology Information database2. The recombinant luciferase plasmids (pGL3-basic-ZFAS1-E1-WT; pGL3-basic-ZFAS1-E2-WT; and pGL3-basic-ZFAS1-E1-Del1/pGL3-basic-ZFAS1-E1-Del2) were constructed with the corresponding sequences amplified from human genomic DNA. Short hairpin RNAs (shRNAs) targeting ZFAS1 and SP1 were designed and cloned into the lentiviral vector pGreen. Empty vector was used as a negative control.
The shRNA sequences used were as follows:
ZFAS1-shRNA: GATCCGCTATTGTCCTGCCCGTTAGAG TTCAAGAGACTCTAACGGGCAGGACAA TAGCTTTTTTG;
SP1-shRNA: CCGGGCTGGTGGTGATGGAATACATCT CGAGATGTATTCCATCACCACCAGCTTTTT.
Cell Proliferation Assay
Cell proliferation assay was detected using Cell Counting Kit-8 (CCK8; Dojindo, Tokyo, Japan) according to the manufacturer’s instructions. Cells were seeded into 96-well plates at 1 × 104 cells/well. After cell culture for 0, 24, 48, and 72 h, 10 μl of CCK8 solution was added to each well and incubated at 37°C for 3 h. The absorbance was measured at a wavelength of 450 nm for each well. All samples were prepared in triplicate and normalized to blank controls.
Cell Cycle Analysis
Cells were harvested and fixed with 75% ethanol at 4°C overnight. And cells were washed with phosphate-buffered saline (PBS) for three times, followed by incubation with RNA enzyme containing propidium iodide (PI; 40%) (Sigma-Aldrich, St. Louis, MO, United States) for 30 min at room temperature. Cell cycle was detected by FACS Calibur (BD Biosciences, San Jose, CA, United States), and data analysis was performed by FACS Diva (BD Biosciences).
Transwell Assay
Cells were starved in serum-free medium overnight, and then a cell suspension of 200 μl of serum-free medium was seeded into each well of the upper transwell chamber (Corning, New York, NY, United States). In the lower chamber, 750 μl of medium with 25% FBS was added. Through incubation at 37°C for 48 h, cells on the upper surface were removed by a cotton tip. Cells on the lower surface were fixed with 4% paraformaldehyde (PFA) and stained with 0.1% crystal violet for 30 min. The results were observed by EVOSTM Microscope M5000 Imaging System (Invitrogen).
Nuclear–Cytoplasmic Separation
Nuclear and cytoplasmic fractions were isolated from cells using PARIS kit (Thermo Fisher Scientific) according to the manufacturer’s protocols. Actin was used as a cytoplasmic control, and U1 was used as a nuclear control. RNA levels of ZFAS1, actin, and U1 in the cytoplasm and nuclear fractions were quantified by qPCR.
RNA Fluorescence in situ Hybridization Assay
Cells were seeded in a 24-well plate for 24 h and fixed with 4% PFA for 20 min. The cells in the plate were pre-hybridized with a hybridization buffer at 42°C for 1 h and overnight with hybridization reaction solution containing the fluorescence in situ hybridization (FISH) probe in the dark. The nonspecific probe was removed with 2× saline sodium citrate (SSC) containing 50% formamide for 10 min at 37°C, 1× SSC for 2 × 5 min at 37°C, and 0.5× SSC for 10 min at room temperature. The anti-biotin monoclonal antibody and secondary antibody were used for detecting biotin-labeled ZFAS1. Cells in the plates were incubated with DAPI (Cell Signalling Technology, Boston, MA, United States). Finally, the cells were subjected to fluorescence signal detection under an EVOSTM Microscope M5000 Imaging System (Invitrogen). The probe of ZFAS1 was synthesized in Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
In vitro Chondrogenic Differentiation
High-density pellet cultures were performed with HUC-MSCs at passages 3–7. To form a cell pellet, 2 × 105 HUC-MSCs were centrifuged in a 15-ml polypropylene conical tube at 1,000 rpm for 5 min. And pellets were incubated in the StemPro chondrogenesis differentiation medium (Gibco) at 37°C in 5% CO2 for 7/14 days. The differentiation medium was changed every 3 days.
Histology Staining and Immunohistochemistry
The cell pellets were fixed in 4% PFA for 24 h, dehydrated in graded series of ethanol (70–100%), processed with xylene (50–100%), and then embedded in paraffin. Paraffin blocks were cut into consecutive sections using a microtome and placed onto glass slides. Following deparaffinization, the sections were used for subsequent experiments.
For Alcian blue staining, the pellets sections were treated with 1% Alcian blue (Sigma-Aldrich) dissolved in acetic acid for 1 h. For toluidine blue staining, the sections were incubated with toluidine blue solution (Sigma-Aldrich) for 1 h. These staining sections were observed in EVOSTM Microscope M5000 Imaging System (Invitrogen).
The pellet sections were subjected to immunohistochemistry (IHC). The antibody used was Collagen type II (1:100; Affinity Biosciences, Cincinnati, OH, United States). Antigen was retrieved by pepsin (Sigma-Aldrich) for 10 min, and 5% goat serum was applied to block the unspecific background antigen. Sections were incubated with the primary antibodies overnight at 4°C. According to the manufacturer’s instructions, a streptavidin–horseradish peroxidase (HRP) detection system (MxBiotech, Eugene, OR, United States) was applied, followed by hematoxylin staining and neutral balsam sealing. These sections were observed in EVOSTM Microscope M5000 Imaging System (Invitrogen).
Zebrafish Breeding and Husbandry
The Institutional Research Ethics Committee of the Children’s Hospital of Fudan University, China, approved and monitored all zebrafish procedures following the guidelines and recommendations outlined by the Guide for the Care and Use of Laboratory Animals (2016-121). Zebrafish were raised at 28°C under a condition of 14-h light/10-h dark with twice-a-day feeding at the Fudan University. The Tg(col2a1a-mcherry) zebrafish, which is cartilage specific and labeled with red fluorescence was purchased from Nanjing YSY Biotech Company Ltd (Nanjing, China). Adult zebrafish were acclimated to laboratory conditions for 4 weeks prior to breeding for further experiments.
Microinjection
Full-length lncRNAs were in vitro transcribed using a mMESSAGE mMACHINE Kit (Ambion, Austin, TX, United States) with the dsDNA templates coupled with SP6 promoter at 37°C overnight. After in vitro transcription, the DNA templates were removed by RNase-free DNase at 37°C for 15 min, followed by purification with RNAclean Kit (Tiangen, Beijing, China). At the night before mating, the males and females were separated by a transparent wall. The following morning, the transparent wall was removed, and the water was changed with warm water at temperature of 29°C. LncRNAs were diluted to 300 ng/μl using RNase-free water for microinjection of experimental group. RNase-free water was for the control group. Embryos were collected 20 min after spawning. Then microinjection into single-cell regions of embryos was performed with a Narishige IM-300 microinjector (Narishige Laboratory Instruments Ltd., Tokyo, Japan). Later, embryos were incubated in the E3 medium at 28°C. Unfertilized eggs and dead embryos were removed immediately, and E3 medium was refreshed every day. And then these embryos were used for further experiments and observation.
Bromodeoxyuridine Assay
Zebrafish embryos were incubated in E3 medium containing 10 mM of bromodeoxyuridine (BrdU) for 6 h, and then embryos were fixed with 4% PFA for 4 h, followed by 0.2 N of HCl for 1 h at room temperature. Anti-BrdU antibody (1:100; Santa Cruz Biotechnology, Paso Robles, CA, United States) and DAPI (1:100) were incubated overnight at 4°C. Embryos were washed with PBS and incubated with Goat Anti-Mouse IgG488 (1:200; Yisheng, Shanghai, China) overnight at 4°C. Histochemical detection was performed by Olympus MVX10 microscope (Olympus, Tokyo, Japan).
RNA Pull-Down Assay and Mass Spectrometry Analysis
Biotinylated ZFAS1 sense and antisense RNA were transcribed using Large Scale RNA Production System-T7 (Promega, Madison, WI, United States) and RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific) in vitro. About 1 × 107 cells were dissolved in cell lysis buffer with RNasin (Promega). RNA pull-down assay was conducted through PierceTM Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific). In brief, the biotinylated ZFAS1 sense and antisense RNA were incubated with Pierce Nucleic-Acid Compatible Streptavidin Magnetic Beads and total cell lysates at room temperature for 2 h. Finally, the retrieved proteins were detected by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels for mass spectrometry analysis. Primers used are as follow:
ZFAS1-sense-F: TAATACGACTCACTATAGGGCCACGC TGTTTTGACCTCCA,
ZFAS1-sense-R: GAGCGATCGCAGATCCTTCG;
ZFAS1-antisense-F: CCACGCTGTTTTGACCTCCA,
ZFAS1-antisense-R: TAATACGACTCACTATAGGGGAGC GATCGCAGATCCTTCG.
RNA Immunoprecipitation Assay
About 1 × 107 cells were harvested and lysed with RNA immunoprecipitation (RIP) lysis buffer at −80°C overnight. Five micrograms of human anti-CTCF (Abcam, Cambridge, United Kingdom) or NC rabbit anti-IgG antibody (ProteinTech, Chicago, IL, United States) were incubated with magnetic beads at room temperature for 1 h and then incubated with cell lysates at 4°C overnight. RNase-free DNase I (Promega) and proteinase K (Yeasen, Shanghai, China) were used for digesting extra DNA and proteins, respectively. Finally, the retrieved RNA was subjected to qRT-PCR analysis of ZFAS1. Total RNA (input controls) and rabbit IgG controls were measured simultaneously to normalize the binding capacity between RNA and protein.
Western Blotting and Antibodies
Cells were harvested and lysed with radioimmunoprecipitation assay (RIPA) buffer (Yeasen) containing a protease inhibitor cocktail (Sigma-Aldrich). Proteins were quantified by a Bradford Protein Assay Kit (Abcam), and the same amounts of proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes (Pall, Port Washington, NY, United States), followed by a block with 8% milk dissolved in Tris-buffered saline Tween for 1 h at room temperature. And then the membranes were incubated with specific primary antibodies at 4°C overnight, followed by secondary antibodies at room temperature for 1 h. Antibodies used were as follows: CTCF (1:5,000, Abcam), GAPDH (1:5,000, ProteinTech), WNT4 (1:1,000, Santa Cruz), β-catenin (1:1,000, Cell Signalling Technology), and c-Myc (1:5,000, ProteinTech).
Data Mining of Chromatin Immunoprecipitation Sequencing Data
Chromatin immunoprecipitation sequencing (ChIP seq) data of CTCF were mined and selected in Cistrome Data Browser3 based on the following requirements: (1) species, Homo sapiens; (2) cell type, HEK293T cells; (3) factors, CTCF; and (4) quality control meets the basic requirements. GSM2026782 was selected for further analysis.
RNA Sequencing
A group of HOK cells overexpressing ZFAS1 and a group of NC cells were subjected to RNA sequencing by GENEWIZ (Suzhou, China). In brief, RNA sequencing processes included RNA extraction, RNA sample quality detection, library construction/purification/detection/quantification, generation of sequencing cluster, and sequencing. And then the results were subjected to perform GO/KEGG analysis.
Chromatin Immunoprecipitation Assay
Cells were crosslinked with 1% formaldehyde and quenched in glycine solution. ChIP assay was performed by Pierce Magnetic Chip Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Two micrograms of anti-CTCF antibody (Abcam) and normal rabbit IgG (ProteinTech) were used for immunoprecipitation. ChIP-enriched DNA samples were quantified by qPCR to determine the CTCF binding sites of WNT4 genome. The value was shown as relative enrichment normalized to IgG. Primers used for ChIP-qPCR were as follows:
WNT4-peak-Q-F: TGACAGGTCGCTGAGGAAG,
WNT4-peak-Q-R: CCCCAAGCAGAAACAGGTC.
Dual-Luciferase Reporter Assay
The recombinant plasmid pGL3-basic-ZFAS1-E1-WT/del1/del2 was constructed. Cells were seeded in 96-well plates at 1 × 104 cells per well and incubated at 37°C overnight. SP1 was co-transfected into cells with pGL3-basic-ZFAS1-E1-WT/del1/del2 using Lipofectamine 3000 (Invitrogen). Cells were collected 48 h after transfection. Both firefly and Renilla luciferase activities were measured by a Dual-Luciferase Reporter Assay (Promega). And the firefly luciferase activities were normalized to Renilla luciferase activities.
Statistical Analysis
All experiments were repeated three times. All statistical analyses were conducted with paired two-tailed Student’s t-test with GraphPad Software Inc. (La Jolla, CA, United States). P < 0.05 was considered significant.
Results
ZFAS1 Was Screened Through Long Non-coding RNA Sequencing and Further Bioinformatics Analysis
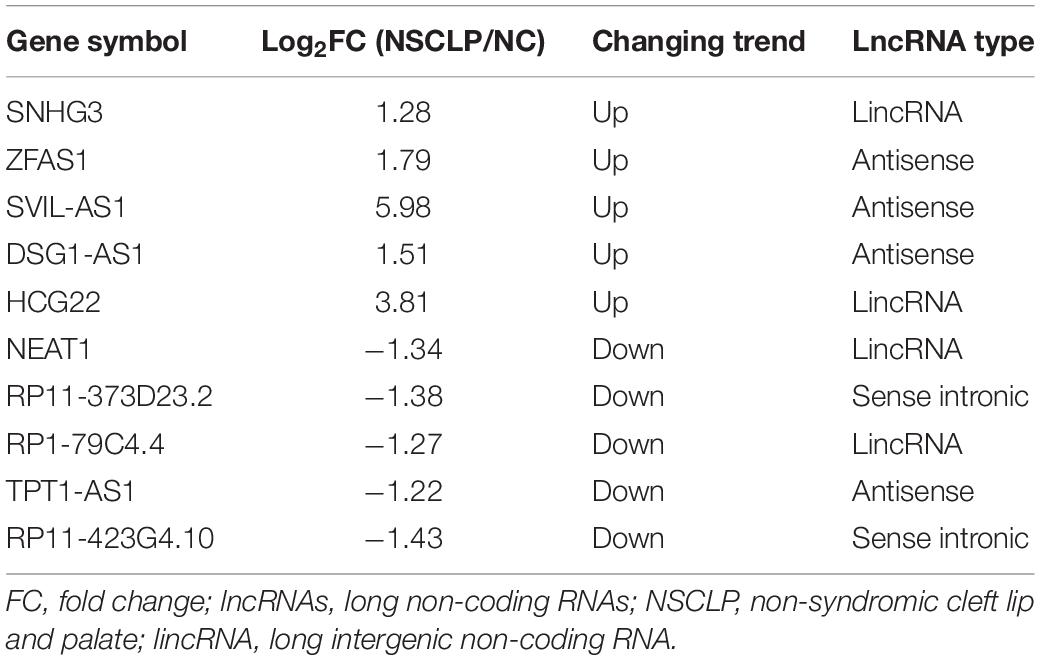
A total of 349 lncRNAs were significantly dysregulated, with 59 upregulated lncRNAs and 290 downregulated lncRNAs in NSCLP by lncRNA sequencing and bioinformatics analysis (Figures 1A,B). The GO pathway analysis revealed these differentially expressed lncRNAs were distinctly enriched in BP terms such as “epidermis development,” “skin development,” and “protein targeting” (Figure 1C). And the KEGG pathway analysis showed these lncRNAs are enriched in “Ribosome,” “Focal adhesion,” and so on (Figure 1D). Among the differentially expressed lncRNAs, we then selected 10 lncRNAs for expanded sample qPCR validation (Table 1), according to the following conditions: (1) log2 (FoldChange) > 1.5, P < 0.05; (2) the pathways that the lncRNA may be involved in are related to CLP based on enrichment analysis; and (3) the function of the lncRNA is associated with CLP by literature retrieval.
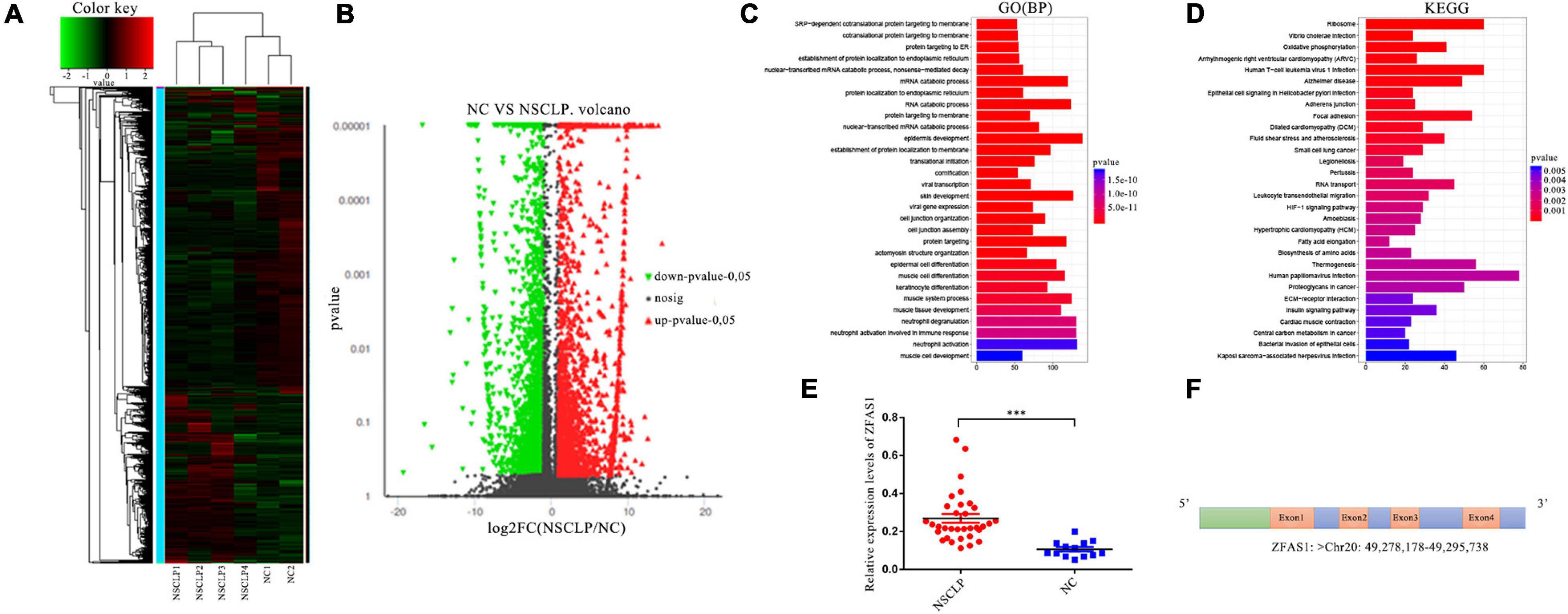
Figure 1. ZFAS1 was screened through long non-coding RNA (lncRNA) sequencing and further qPCR assay. (A) Heatmap of dysregulated lncRNAs identified from lncRNA sequencing by hierarchical clustering analysis between the non-syndromic cleft lip and palate (NSCLP) (NSCLP1/2/3/4) and normal control (NC) (NC1/2) samples. Expression values are represented by red and green shades, indicating expressions above and below the median expression level across all samples, respectively. (B) The volcano plot of differentially expressed lncRNAs. The fold change threshold is 1.5, and P-value ≤ 0.05. The red dots indicate upregulated genes, the green dots indicate downregulated genes, and the black dots indicate non-differentially expressed genes. (C,D) The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways significantly enriched in the differentially expressed lncRNAs. The length and color of the bar represent the amount of differentially expressed lncRNAs enriched in the pathway and enrichment significance, respectively. BP, biological processes. (E) ZFAS1 was upregulated in tissues from NSCLP compared with NC tissues by qPCR assay. (F) The schema of ZFAS1 in chromosome. Values are mean ± SEM, n = 3, ***P < 0.0001.
ZFAS1 Was Upregulated in Tissue Samples From Patients With Non-syndromic Cleft Lip and Palate
We detected expression levels of these 10 lncRNAs in 36 tissue samples from patients with NSCLP and 13 NC tissue samples using qPCR assay. In view of the non-conservation of lncRNAs and feasibility of animal experiment, we chose the upregulated lncRNA as the research target in principle. Finally, we selected ZFAS1, which was the most significantly upregulated lncRNA in NSCLP compared with NC (Figure 1E and Supplementary Figure 1). ZFAS1 was an lncRNA located at chromosome 20q13, with four exons (Figure 1F).
Overexpression of ZFAS1 Inhibited Cell Proliferation and Cell Migration in Human Oral Keratinocyte Cells and Human Umbilical Cord Mesenchymal Stem Cells
To investigate the effects of ZFAS1 on cell phenotypes related to NSCLP, ZFAS1 was successfully upregulated and downregulated in HOK cells and HUC-MSCs (Figures 2A,E). HUC-MSCs have been identified by their special markers on a flow cytometry platform (Supplementary Figure 2). CCK8 assay showed that ZFAS1 overexpression significantly inhibited cell proliferation, consistent with the tendency in cells downregulating ZFAS1 (Figures 2B,F). Then cell cycle distribution of HOK cells presented that the G1 phase was much higher in cells overexpressing ZFAS1, indicating that the overexpression of ZFAS1 contributed to G1 phase arrest (Figures 2C,D). Transwell assay revealed that cell migration of HOK and HUC-MSCs was both significantly inhibited with the overexpression of ZFAS1, in accordance with the tendency in cells to downregulate ZFAS1 (Figures 2G,H). However, flow cytometry analysis showed that ZFAS1 dysregulation did not affect apoptosis in HOK cells (Supplementary Figure 3).
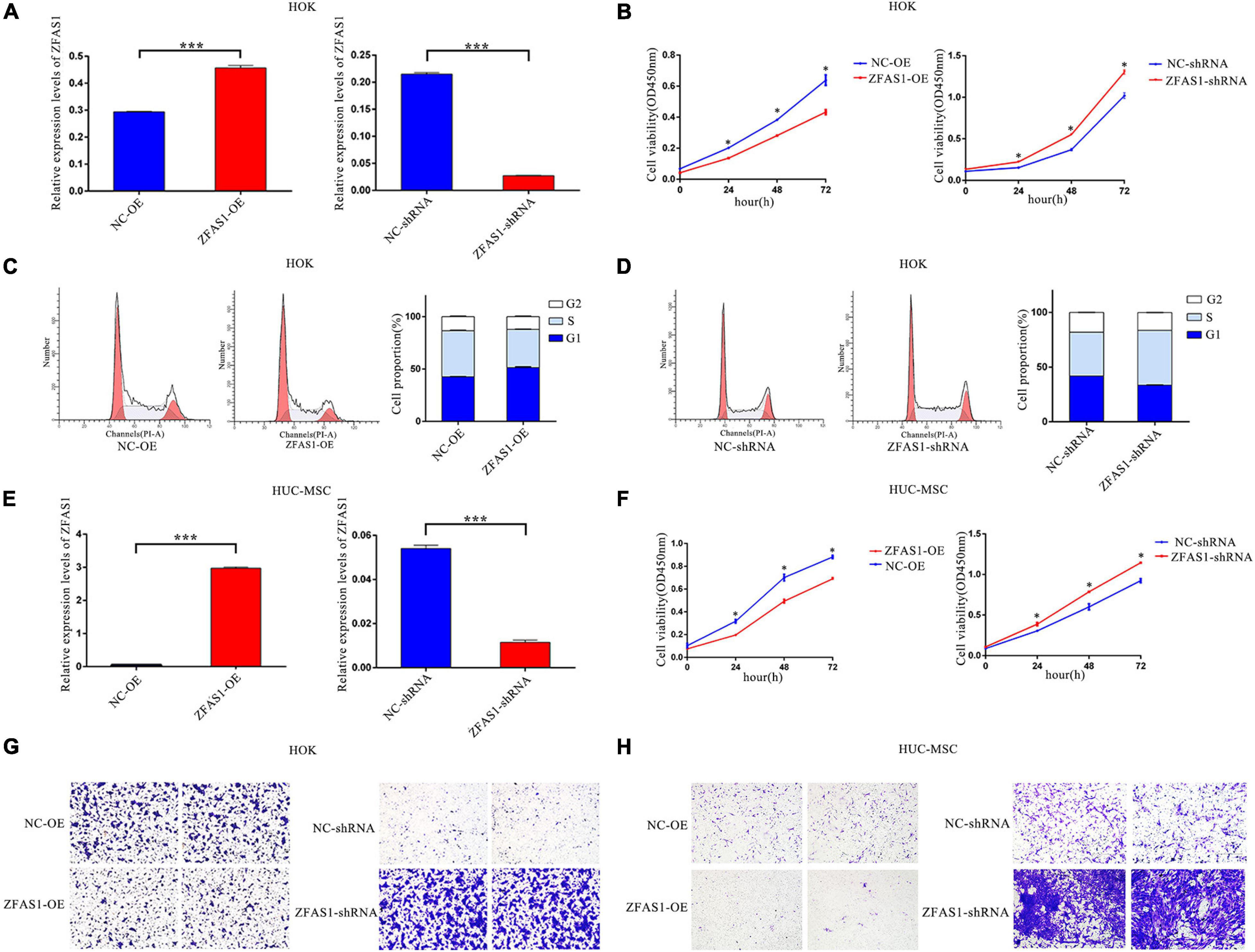
Figure 2. Overexpression of ZFAS1 inhibited cell proliferation and migration in human oral keratinocyte (HOK) cells and human umbilical cord mesenchymal stem cells (HUC-MSCs). (A) ZFAS1 was upregulated and downregulated in HOK cells using overexpression vector of ZFAS1 and shRNA targeting to ZFAS1, respectively. (B) Negative regulation of cell proliferation by ZFAS1 was revealed by Cell Counting Kit-8 (CCK8) assay in HOK cells. (C,D) ZFAS1 overexpression and knockdown in HOK cells increased and decreased G1/S arrest by cell cycle assay, respectively. The two left panels indicate the results from fluorescence-activated cell sorting (FACS), and the right panel indicates the statistical result of the two left panels. (E) ZFAS1 was upregulated and downregulated in HUC-MSCs using overexpression vector of ZFAS1 and shRNA targeting to ZFAS1, respectively. (F) Negative regulation of cell proliferation by ZFAS1 was revealed by CCK8 assay in HUC-MSCs. (G,H) Transwell assays revealed a negative regulation of cell migration by ZFAS1 in HOK cells and HUC-MSCs. Cells were stained with crystal violet. Values are mean ± SEM, n = 3, *P < 0.05, ***P < 0.0001. ShRNA, short hairpin RNAs; OE, overexpression.
ZFAS1 Overexpression Suppressed the Chondrogenic Differentiation Potential in Human Umbilical Cord Mesenchymal Stem Cells
Abnormal chondrogenic differentiation during palatogenesis may lead to NSCLP. To determine the biological effects of ZFAS1 on the chondrogenic ability, traditional high-density pellet cultures of HUC-MSCs were used for the induction of differentiation. At day 14 after cartilage pellet culture, both Alcian blue and toluidine blue staining showed that upregulating ZFAS1 significantly interfered with chondrogenesis (Figures 3A,C), while HUC-MSCs with ZFAS1 knockdown presented higher chondrogenesis potential (Figures 3B,D). By IHC in cartilage pellets, the expression of type II collagen, marker of chondrogenic matrix, was consistent with the results of staining (Figures 3E,F). The mRNA levels of chondrogenic markers SRY-box transcription factor 9 (Sox9) and aggrecan (ACAN) were significantly decreased in pellets overexpressing ZFAS1 at days 7 and 14, compared with NC (Figures 3G,H).
Figure 3. ZFAS1 overexpression suppressed the chondrogenic differentiation potential in human umbilical cord mesenchymal stem cells (HUC-MSCs). (A,B) Alcian blue staining of paraffin sections from chondrocyte pellets revealed a negative regulation of chondrogenic differentiation by ZFAS1. The color depth of Alcian blue indicates the levels of chondrogenic differentiation. (C,D) Toluidine blue staining of paraffin sections from chondrocyte pellets revealed a negative regulation of chondrogenic differentiation by ZFAS1. The color depth of toluidine blue indicates the levels of chondrogenic differentiation. (E,F) The immunohistochemistry (IHC) results of collagen type II in the paraffin sections from chondrocyte pellets. The brown regions represent the signal of collagen type II; the blue dots represent the signal of nuclei. (G,H) The qPCR assay showed that ZFAS1 overexpression downregulated the expression of Sox9 and ACAN in the chondrogenesis-induced pellets at 0/7/14 days. Values are mean ± SEM, n = 3, ***P < 0.0001. d, days.
ZFAS1 Overexpression in Zebrafish Embryos Significantly Decreased Cell Proliferation and Numbers of Chondrocyte in the Ethmoid Plate
To investigate the role of ZFAS1 in the palatogenesis, ZFAS1 was microinjected into zebrafish embryos to establish zebrafish overexpressing ZFAS1, and the efficiency was validated by qPCR at 48 h post-fertilization (hpf) (Figure 4A). The observation on the whole zebrafish indicated that microinjection did not affect the overall development of zebrafish, and BrdU assay demonstrated that the overexpression of ZFAS1 in zebrafish at 48 hpf can inhibit cell proliferation in the ethmoid plate (Figures 4B–D). To understand well the chondrocyte in the zebrafish ethmoid, ZFAS1 was overexpressed in the Tg(col2a1a-mcherry) zebrafish (Figure 4E), and the numbers of positive cells for col2a1a-mcherry were significantly decreased in the ethmoid plate at 72 hpf (Figures 4F,G), indicating a suppression of palatogenesis.
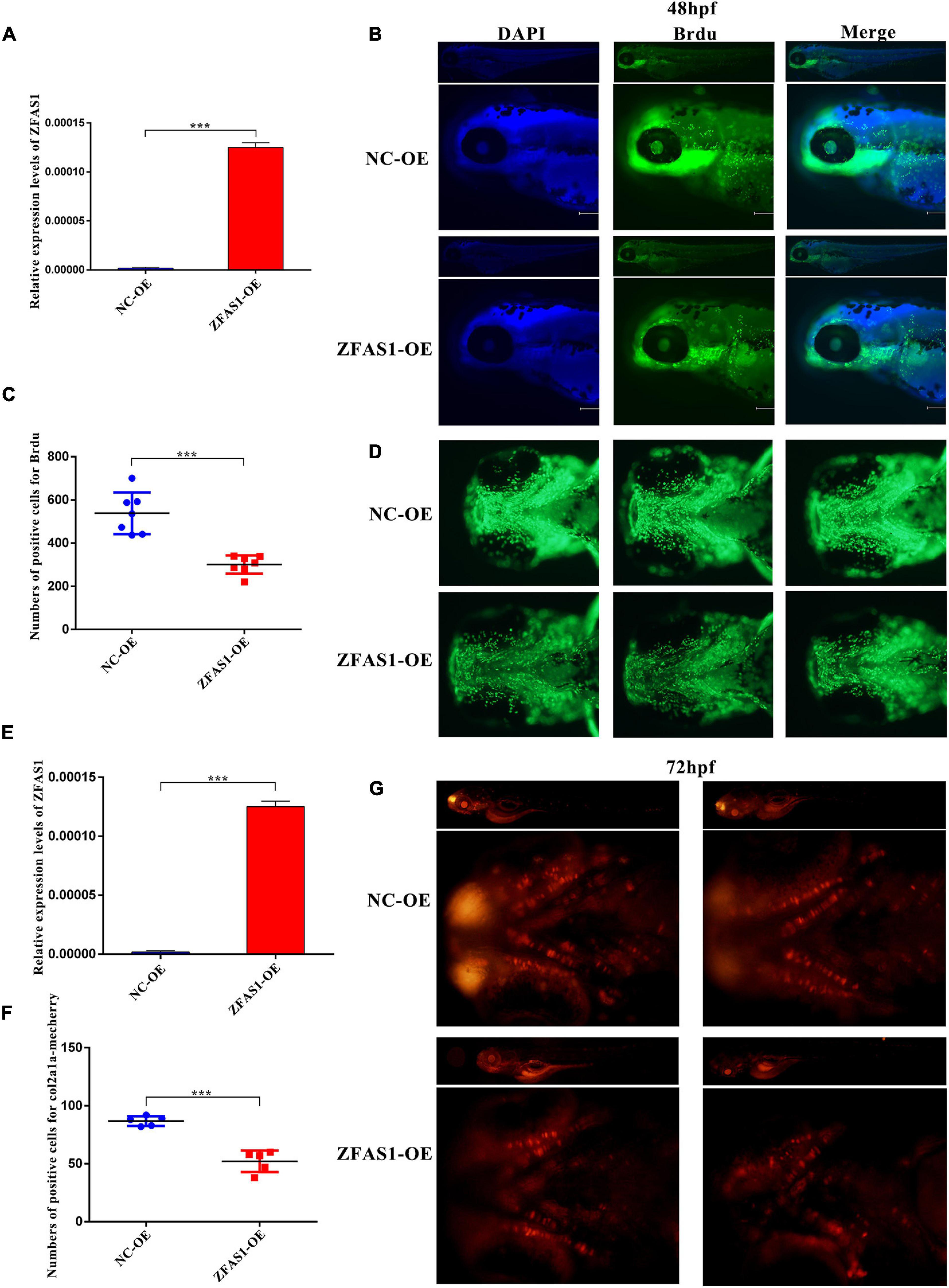
Figure 4. ZFAS1 overexpression in zebrafish embryos significantly decreased cell proliferation and numbers of chondrocyte in the ethmoid plate. (A) The efficiency of overexpressing ZFAS1 in wild-type zebrafish embryos by microinjection using the qPCR assay. (B) The side view of bromodeoxyuridine (BrdU)-labeling cells in zebrafish at 48 hpf detected by fluorescence microscope. Left, blue fluorescence for DAPI; middle, green fluorescence for BrdU; right, the merged image. (C) The statistical results of BrdU-positive cells in the zebrafish ethmoid plate. (D) The ventral view of BrdU-positive cells in the zebrafish ethmoid plate. (E) The efficiency of overexpressing ZFAS1 in Tg(col2a1a-mcherry) zebrafish embryos by microinjection using the qPCR assay. (F) The statistical results of col2a1a-mcherry-positive cells in the zebrafish ethmoid plate. (G) The ventral view of col2a1a-mcherry-positive cells in the zebrafish ethmoid plate at 72 hpf. Red fluorescence, col2a1a-mcherry. Values are mean ± SEM, n = 3, ***P < 0.0001.
ZFAS1 Was Distributed in Both the Nucleus and Cytoplasm of Human Oral Keratinocyte Cells and Human Umbilical Cord Mesenchymal Stem Cells
The subcellular localization of ZFAS1 determines its action mode; therefore, to further explore the underlying mechanism of ZFAS1 affecting phenotypes above, cell localization testing is necessary. Nuclear–cytoplasmic separation assay indicated that ZFAS1 was distributed in both the nucleus and cytoplasm in HOK and HUC-MSCs (Figures 5A,C), which was verified by RNA-FISH using ZFAS1-probe (Figures 5B,D).
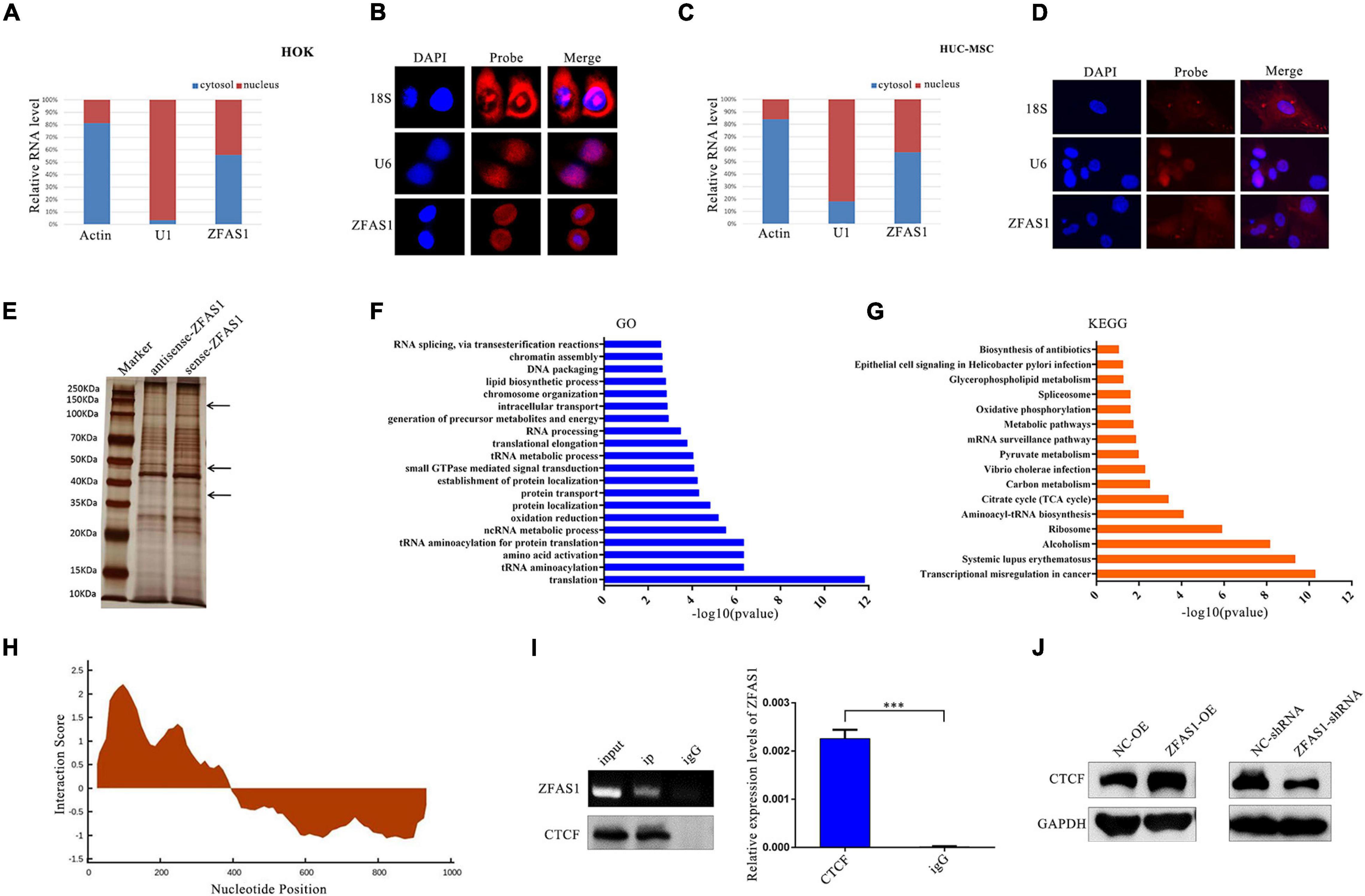
Figure 5. ZFAS1 regulated the protein level of CCCTC-binding factor (CTCF) through binding with CCCTC-binding factor (CTCF) protein. (A,C) Nucleus cytoplasm separation indicated that ZFAS1 was distributed in both the nucleus and cytoplasm of human oral keratinocyte (HOK) cells and human umbilical cord mesenchymal stem cells (HUC-MSCs). (B,D) RNA fluorescence in situ hybridization (FISH) assays indicated that ZFAS1 was distributed in both the nucleus and cytoplasm of HOK cells and HUC-MSCs. Left, DAPI was used to stain the nuclei (blue); middle, red fluorescence was from the biotin fusions; right, the merged image. (E) Proteins retrieved from the ZFAS1 pull-down assay were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The arrows indicate the unique proteins in the sense-ZFAS1 group compared with antisense-ZFAS1 group. (F,G) The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in these proteins from RNA pull-down assay. (H) The predicted binding sites of ZFAS1 with CTCF by catRAPID. The interaction score indicated the possibility of binding. (I) RNA immunoprecipitation (RIP) assay of ZFAS1 with anti-CTCF antibody. ZFAS1 and CTCF interaction in CTCF-RNA precipitates was revealed by nucleic acid gel (top left panel) and qPCR assays (right panel). Bottom left panel indicates that the CTCF protein was pulled down by anti-CTCF antibody. (J) ZFAS1 positively regulated CTCF protein levels by western blotting (WB). Values are mean ± SEM, n = 3, ***P < 0.0001.
ZFAS1 Affected the Protein Level of CCCTC-Binding Factor Through Binding to CCCTC-Binding Factor Protein
Growing evidence indicated that lncRNAs often exert their functions by binding to protein partners. To identify potential proteins that interact with ZFAS1, in vitro RNA pull-down assay of ZFAS1 was performed in HOK cells, followed by mass spectrometry (Figure 5E). Then GO and KEGG pathway analyses indicated that the proteins in mass spectrometry were enriched in the pathways related to chromosome organization, DNA packaging, transcription, translation, and so on (Figures 5F,G). One of these proteins was CTCF, a transcriptional regulator with diverse functionality in disease through transcriptional activation or repression. ZFAS1 was predicted to have a high probability to directly interact with CTCF at 101–152 nucleotides of ZFAS1 using an online tool catRAPID (Figure 5H). RIP assay was used to verify the binding ability of ZFAS1 to CTCF. In comparison with the IgG group, ZFAS1 enrichment was significantly upregulated in the CTCF group (Figure 5I). Moreover, the protein level of CTCF was significantly upregulated after ZFAS1 overexpression and downregulated after ZFAS1 knockdown (Figure 5J).
ZFAS1 Inhibited WNT/β-Catenin Signaling Pathway via Suppressing WNT4 Expression Levels by Binding With CCCTC-Binding Factor
To further investigate the downstream genes of CTCF, RNA seq and ChIP seq analyses were performed in HOK cells. Numerous potential target genes of CTCF were identified from the ChIP seq data (GSM2026782) in Cistrome Data Browser (Figures 6A,B). RNA seq showed that there were 1,414 differentially expressed genes in HOK cells overexpressing ZFAS1 compared with NC, and then GO and KEGG pathway analyses revealed that these genes were enriched in “Regulation of transcription,” “DNA binding” and “Focal adhesion,” “Signaling pathways regulating pluripotency of stem cells,” “RNA degradation,” and so on (Figures 6C,D).
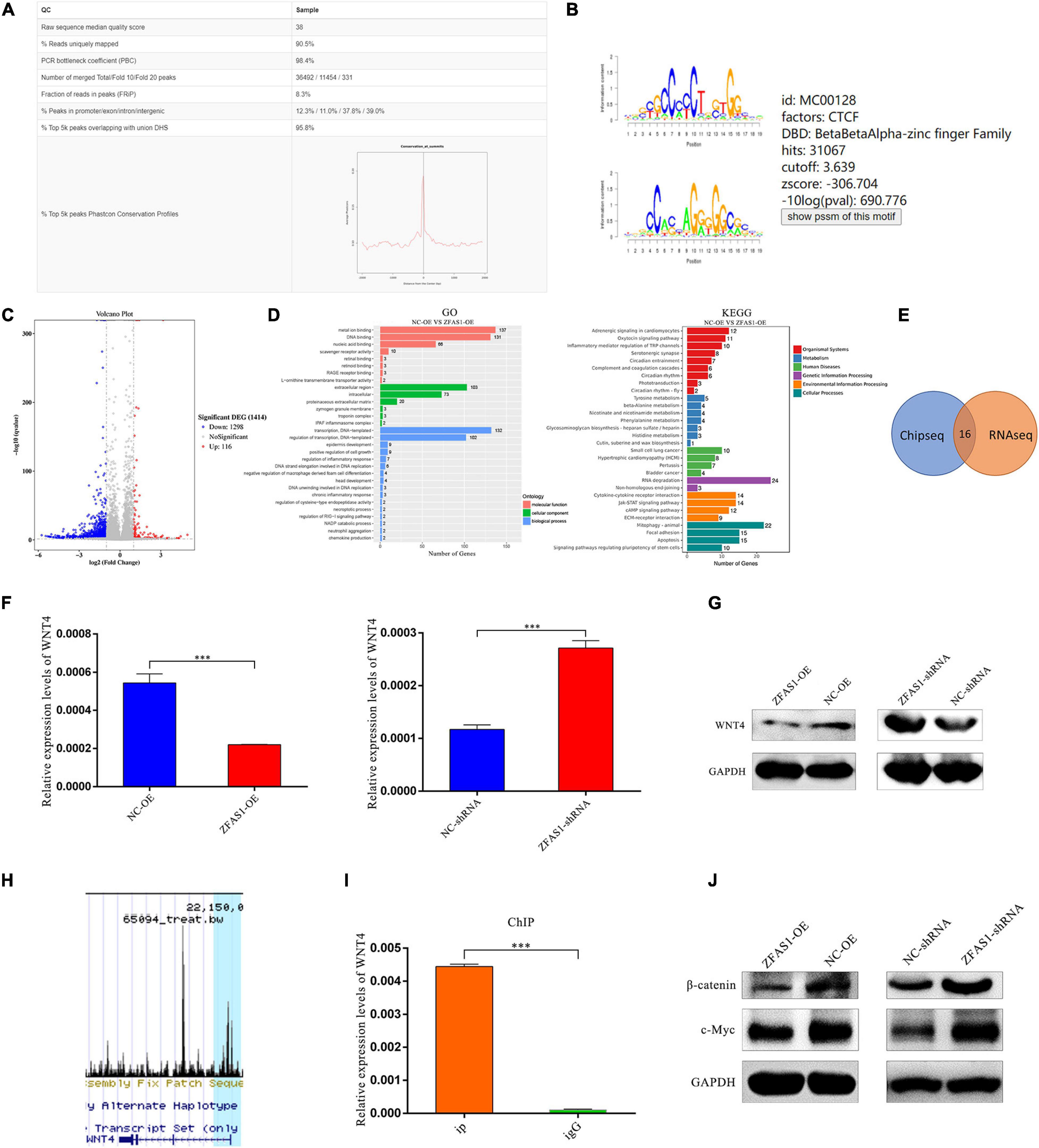
Figure 6. ZFAS1 inhibited WNT/β-catenin signaling pathway via suppressing WNT4 expression levels by binding with CCCTC-binding factor (CTCF). (A) The quality report of chromatin immunoprecipitation sequencing (ChIP seq) of CTCF mined in the Cistrome Data Browser. (B) The annotated binding motif of CTCF from the mined ChIP seq. (C) The volcano plot of differentially expressed genes from the RNA seq of human oral keratinocyte (HOK) cells overexpressing ZFAS1. The fold change threshold is 1.5, and P-value ≤ 0.05. The red dots indicate upregulated genes, the blue dots indicate downregulated genes, and the gray dots indicate non-differentially expressed genes. (D) The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in differentially expressed genes. Length of the bar represents the amount of differentially expressed genes enriched in the pathway. (E) The integrated analysis of ChIP seq and RNA seq. The intersection region indicates the merged genes of them. (F) ZFAS1 negatively regulated expression levels of WNT4 by qPCR assay. (G) ZFAS1 negatively regulated protein levels of WNT4 by western blotting (WB) assay. (H) The peak of WNT4 from the ChIP seq of CTCF. The blue highlight indicates the peak. (I) The binding capacity of CTCF to WNT4 was measured by ChIP qPCR assay. (J) ZFAS1 negatively regulated protein levels of β-catenin and c-Myc by WB assay. Values are mean ± SEM, n = 3, ***P < 0.0001.
Among the 16 possible target genes by integrated analysis of RNA seq and ChIP seq (Figure 6E), WNT4, which belongs to the WNT/β-catenin signaling pathway related with CLP, was selected for further research. The qPCR assay revealed that overexpressing ZFAS1 downregulated mRNA levels of WNT4, and knockdown of ZFAS1 upregulated mRNA levels of WNT4 (Figure 6F), which was validated in protein levels by western blotting (WB) (Figure 6G). Based on the ChIP seq of CTCF collected by the Cistrome Data Browser and its visualization window, there was a peak near the regulatory region of WNT4 (Figure 6H). ChIP qPCR results revealed that CTCF directly bound to the predicted binding sites of WNT4 (Figure 6I). Moreover, WB results indicated that upregulating ZFAS1 significantly also decreased β-catenin and c-myc expression levels, which were important molecules in the WNT/β-catenin signaling pathway, consistent with the tendency in cells downregulating ZFAS1 (Figure 6J). In summary, we suggested that ZFAS1 overexpression inhibited the WNT/β-catenin signaling pathway through suppressing WNT4 by interacting with CTCF.
SP1 Regulated Expression Levels of ZFAS1 via Binding to the E1 Region of ZFAS1 Promoter
Transcription factors binding to motifs located at genes’ promoter region plays a critical role in their transcription. To ascertain the underlying cause for the elevated expression of ZFAS1 in NSCLP, PROMO database was used to predict the potential transcription factors that possibly bound to the promoter region of ZFAS1. SP1 was selected, and qPCR results revealed that SP1 was upregulated in NSCLP (Figure 7A). And the binding motif in promoters of ZFAS1 was predicted by JASPAR database (Figure 7B). In order to make clear the regulatory relationship between SP1 and ZFAS1, SP1 was successfully upregulated and downregulated in HOK cells, respectively (Figures 7C,D). Moreover, the overexpression of SP1 significantly upregulated the expression of ZFAS1 in HOK cells (Figure 7E). Subsequent bioinformatics analyses in JASPAR database also predicted two binding domains (E1 and E2) of ZFAS1 with SP1 (Figure 7F). To determine the transcriptional activation of SP1 on the ZFAS1, HOK cells were co-transfected with the ZFAS1 promoter luciferase reporter (pGL3-basic-ZFAS1-E1/E2) and SP1. Dual-luciferase assay revealed that SP1 markedly increased ZFAS1 promoter activity via binding to the E1 region instead of the E2 region (Figure 7G). Whether depleting the motif1 or motif2 in E1 region, they both destroyed the binding of SP1 and E1 region, thus leading to the suppression of ZFAS1 transcription (Figure 7H).
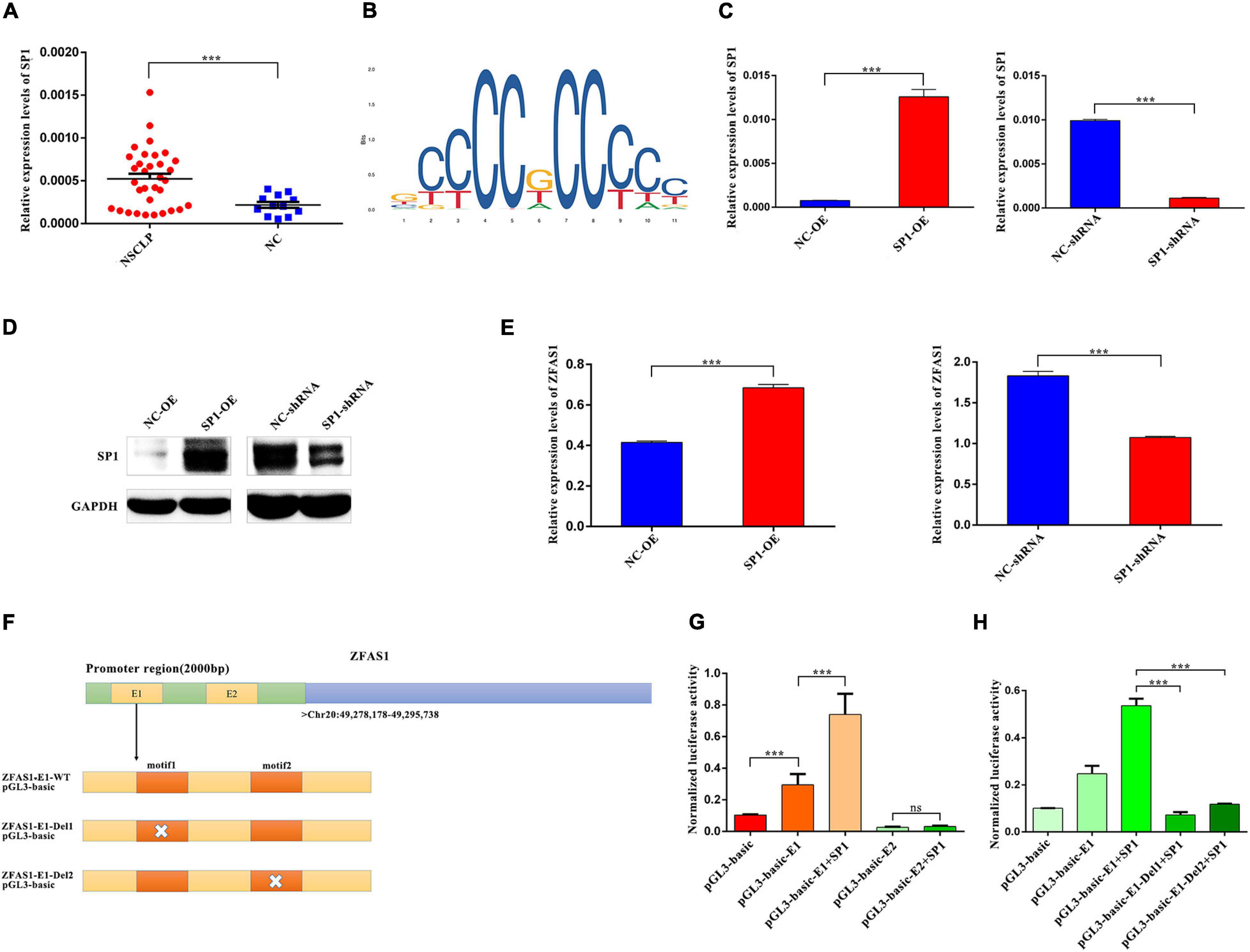
Figure 7. SP1 regulated expression levels of ZFAS1 via binding to the E1 region in the promoter of ZFAS1. (A) SP1 was upregulated in tissues from non-syndromic cleft lip and palate (NSCLP) compared with NC tissues by qPCR assay. (B) The annotated DNA binding motif of ZFAS1 promoter with SP1 in JASPAR database. (C,D) The qPCR and western blotting (WB) assays revealed that SP1 was significantly upregulated and downregulated in human oral keratinocyte (HOK) cells. (E) SP1 positively regulated expression levels of ZFAS1 by qPCR assay. (F) The schema of ZFAS1 for the construction of luciferase reporter plasmids. The orange regions indicate the motif of E1 region in the promoter of ZFAS1; the white cross indicates the deletion of the motif. (G) Dual luciferase reporter assay revealed that SP1 significantly increased ZFAS1 promoter activity via binding to E1 region instead of E2 region. Compared with the “pGL3-basic-E1” group, the “SP1+pGL3-basic-E1” group showed a higher luciferase activity. (H) Dual luciferase reporter assays revealed that deletion of motif1/motif2 in E1 region destroyed the binding of SP1 and E1 region. Compared with the “pGL3-basic-E1 group+SP1 group,” both the “pGL3-basic-E1-Del1+SP1” group and “pGL3-basic-E1-Del2+SP1” showed a significant decrease in the luciferase activity. Values are mean ± SEM, n = 3, ***P < 0.0001.
Discussion
Long non-coding RNAs have been reported to play a critical role in embryonic development and related disorders (Grote et al., 2013; Alvarez-Dominguez and Lodish, 2017). However, little was known about the function of lncRNA on lip and the palate morphogenesis or CLP. For a better understanding of lncRNA expression profiles in NSCLP, we performed lncRNA sequencing in tissues from NSCLP and NC and then identified many lncRNAs that were aberrantly expressed in NSCLP, which were enriched in “epidermis development,” “skin development,” “protein targeting,” “Ribosome,” “Focal adhesion,” and so on by GO and KEGG analyses. Among these differentially expressed lncRNAs, we focused on ZFAS1, which showed a significant upregulation in NSCLP. ZFAS1 has been reported to be involved in several cancers, including hepatocellular carcinoma (Li et al., 2015), osteosarcoma (Liu et al., 2017), breast cancer (Askarian-Amiri et al., 2011), and colorectal cancer (Zeng et al., 2018), by modulating cell proliferation and migration. ZFAS1 has also been shown to be dysregulated in the cartilage of osteoarthritis (Xiao et al., 2019). Nevertheless, none of the reports declared the role of ZFAS1 on the lip and palate morphogenesis or CLP. Lip and palate morphogenesis is involved in proliferation, migration, apoptosis of a variety of cells, and chondrogenic differentiation of precursor cells, such as MSCs.
To figure out the relationship of upregulated ZFAS1 and NSCLP, two cell models and zebrafish model were selected for further experiments in vitro and in vivo. One precursor cell was HUC-MSCs, which can differentiate into chondrocytes (Brown et al., 2019). Another mature cell line was HOK cells from oral mucosa, which was used to study the pathogenesis of NSCLP in previous studies (Zhou et al., 2018; Li et al., 2020). The zebrafish palate consists of cells derived from the frontonasal and maxillary domain, which is similar to the palatogenesis of mammals (Duncan et al., 2017). So zebrafish is also a proper study model for mammalian palate and CLP.
In vitro, the overexpression of ZFAS1 significantly inhibited cell proliferation and migration in HUC-MSCs and HOK cells and also suppressed chondrogenic differentiation of HUC-MSCs. In vivo, upregulating ZFAS1 significantly decreased cell proliferation and numbers of chondrocyte in the zebrafish ethmoid plate. These abnormal cell and zebrafish phenotypes were all associated with NSCLP (Kang and Svoboda, 2005; Jiang et al., 2006; Shull et al., 2020). Therefore, we speculated that upregulation of ZFAS1 may contribute to NSCLP through disturbances in these cell processes, which was consistent with the function of ZFAS1 in previous studies (Askarian-Amiri et al., 2011; Li et al., 2015; Liu et al., 2017). In order to find out the molecular pathways underlying ZFAS1 affecting the phenotypes above, we searched for its molecular chaperones. Increasing studies have reported that lncRNAs could recruit proteins and regulate the downstream pathway (Wang et al., 2019). CTCF was identified as one of the binding proteins of ZFAS1 by RNA pull-down and RIP. What is more, there was a positive regulation of ZFAS1 on CTCF protein level. The change of protein level was mainly due to the changes in the process of its synthesis or degradation. LncRNA could be involved in the protein stability through ubiquitylation (Zhang et al., 2015). CTCF was also reported to be interacted with ubiquitin (Qi et al., 2012). Moreover, among the proteins pulled down by ZFAS1, there was an E3 ubiquitin-protein ligase. So we speculated that ZFAS1 might regulate the protein level of CTCF through ubiquitin proteasome pathway. CTCF is a highly conserved 11 zinc finger DNA-binding domain protein (Franco et al., 2014) and is involved in diverse regulatory functions, including transcriptional activation or repression, modulation of histone modifications, and blocking of enhancer–promoter communication (Huang et al., 2013; Roy et al., 2018). Given the characteristics of CTCF, its target gene WNT4 was determined by ChIP seq and RNA seq. Further assays revealed that upregulated ZFAS1 inhibited the expression levels of WNT4 through the binding with CTCF. WNT4 is a member of WNT protein family, and the WNT molecules activate intracellular signals through specific receptors, subsequently modulating various cell processes, including proliferation, migration, differentiation, and apoptosis (Hu et al., 2013a). They exert activations through three WNT signaling pathway, including canonical WNT/β-catenin, WNT/calcium, and planar cell polarity pathways (Song et al., 2009). The WNT/β-catenin pathway is a major signaling pathway that modulates facial morphogenesis especially the lip and palate (Hu et al., 2013b; Jin et al., 2020). A series of phenotype about craniofacial development were observed in β-catenin conditional knockout and Tcf/Lef knockout embryos (Brault et al., 2001; Brugmann et al., 2007). Canonical WNT signaling in oral epithelium was proved to play a dynamic role in tongue and palate development (Lin et al., 2011). In our present study, β-catenin and c-Myc, key components in the canonical WNT/β-catenin pathway, were inhibited when ZFAS1 was overexpressed. So we speculated that the WNT/β-catenin pathway was the underlying mechanism of ZFAS1 regulating cell proliferation, migration, and chondrogenic differentiation in NSCLP.
To further explore the reason of ZFAS1 upregulation in NSCLP, the transcription factors that ZFAS1 may bind to were predicted, and we focused on SP1. It was reported to be a key transcription factor regulating expression levels of lncRNAs (Xu et al., 2015; Wang et al., 2017). Moreover, SP1 could combine with the promoter of ZFAS1 to regulate ZFAS1 in colorectal cancer (Chen et al., 2018). Following luciferase reporter assays, SP1 markedly increased ZFAS1 promoter activity via binding to E1 region in the promoter of ZFAS1 instead of E2 region. Moreover, SP1 was upregulated in tissue samples from patients with NSCLP, which was in accordance with ZFAS1. Therefore, we have made a conclusion that SP1 could promote the transcription of ZFAS1 by binding to the E1 region in the promoter of ZFAS1 in NSCLP.
Conclusion
In conclusion, we have demonstrated that lncRNA ZFAS1 was involved in NSCLP for the first time. The upregulation of ZFAS1 in NSCLP mediated by SP1 inhibited cell proliferation, migration, and chondrogenic differentiation through inactivation of the WNT/β-catenin signaling pathway, which was due to the interaction of ZFAS1 and CTCF to regulate the expression of WNT4. However, further studies are needed to declare the detailed mechanism. Moreover, if the primary cell lines from lip and palate are used for in vitro study, the conclusion will be more credible.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: SRA data database, accession: PRJNA700692; PRJNA700786.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Research Ethics Committee of the Children’s Hospital of Fudan University, Shanghai, China (2016-121). Written informed consent from the participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements. The animal study was reviewed and approved by the Research Ethics Committee of the Children’s Hospital of Fudan University, China, approved and monitored all zebrafish procedures following the guidelines and recommendations outlined by the Guide for the Care and Use of Laboratory Animals (2016-121).
Author Contributions
JM, DM, and CT were responsible for the idea, project design, and concept of the manuscript. SC and YG performed bioinformatics analysis. SC, ZJ, MC, DW, and TW collected the clinical samples and information. SC, MY, BZ, PW, and YX performed the clinical sample detection and experiments. JM, SC, DM, and CT wrote, edited, and revised the manuscript. All authors read and approved the manuscript.
Funding
This study was supported by grants from the National Key Research and Development Program of China (Nos. 2016YFC1000501 and 2016YFC1000503) and Shanghai Science and Technology Innovation Action Plan (No. 18411953300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.662780/full#supplementary-material
Supplementary Figure 1 | The qPCR results of another 9 lncRNAs in tissues from NSCLP compared with NC tissues. Values are mean ± SEM, n = 3, ns: no significance, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.0001.
Supplementary Figure 2 | HUC-MSCs were identified with a specific panel of cell surface markers of MSC by FACS. (A) The FACS results of HUC-MSCs stained with nothing; (B) the FACS results of HUC-MSCs identified by negative markers(CD34; CD11b; CD19; CD45; HLA-DR); (C–E) the FACS results of HUC-MSCs identified by positive marker(CD90; CD105; CD73); (F) The statistical results of A–E.
Supplementary Figure 3 | ZFAS1 did not affect cell apoptosis in HOK cells. (A,B) The cell apoptosis results in HOK cells with upregulating ZFAS1. (C) The statistical results of A,B. (D,E). The cell apoptosis results in HOK cells with downregulating ZFAS1. (F) The statistical results of D,E.
Footnotes
References
Alvarez-Dominguez, J. R., and Lodish, H. F. (2017). Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood 130, 1965–1975. doi: 10.1182/blood-2017-06-788695
Askarian-Amiri, M. E., Crawford, J., French, J. D., Smart, C. E., Smith, M. A., Clark, M. B., et al. (2011). SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. Rna 17, 878–891. doi: 10.1261/rna.2528811
Brault, V., Moore, R., Kutsch, S., Ishibashi, M., Rowitch, D. H., McMahon, A. P., et al. (2001). Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128, 1253–1264.
Brown, C., McKee, C., Bakshi, S., Walker, K., Hakman, E., Halassy, S., et al. (2019). Mesenchymal stem cells: cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 13, 1738–1755. doi: 10.1002/term.2914
Brugmann, S. A., Goodnough, L. H., Gregorieff, A., Leucht, P., Ten Berge, D., Fuerer, C., et al. (2007). Wnt signaling mediates regional specification in the vertebrate face. Development 134, 3283–3295. doi: 10.1242/dev.005132
Chen, X., Zeng, K., Xu, M., Hu, X., Liu, X., Xu, T., et al. (2018). SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 9:982. doi: 10.1038/s41419-018-0962-6
Chi, Y., Wang, D., Wang, J., Yu, W., and Yang, J. (2019). Long non-coding rna in the pathogenesis of cancers. Cells 8:1015. doi: 10.3390/cells8091015
Dixon, M. J., Marazita, M. L., Beaty, T. H., and Murray, J. C. (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat. Rev. Genet. 12, 167–178. doi: 10.1038/nrg2933
Duncan, K. M., Mukherjee, K., Cornell, R. A., and Liao, E. C. (2017). Zebrafish models of orofacial clefts. Dev. Dyn. 246, 897–914. doi: 10.1002/dvdy.24566
Franco, M. M., Prickett, A. R., and Oakey, R. J. (2014). The role of CCCTC-binding factor (CTCF) in genomic imprinting, development, and reproduction. Biol Rep. 91:125. doi: 10.1095/biolreprod.114.122945
Gao, L., Liu, Y., Wen, Y., and Wu, W. (2017). LncRNA H19-mediated mouse cleft palate induced by all-trans retinoic acid. Hum. Exp. Toxicol. 36, 395–401. doi: 10.1177/0960327116651121
Gao, Y., Zang, Q., Song, H., Fu, S., Sun, W., Zhang, W., et al. (2019). Comprehensive analysis of differentially expressed profiles of non-coding RNAs in peripheral blood and ceRNA regulatory networks in non-syndromic orofacial clefts. Mol. Med. Rep. 20, 513–528. doi: 10.3892/mmr.2019.10261
Gerrard, D. T., Berry, A. A., Jennings, R. E., Piper Hanley, K., Bobola, N., and Hanley, N. A. (2016). An integrative transcriptomic atlas of organogenesis in human embryos. Elife 5:e15657. doi: 10.7554/eLife.15657
Grote, P., Wittler, L., Hendrix, D., Koch, F., Währisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. doi: 10.1016/j.devcel.2012.12.012
Hu, X., Gao, J. H., Liao, Y. J., Tang, S. J., and Lu, F. (2013a). Dexamethasone alters epithelium proliferation and survival and suppresses Wnt/β-catenin signaling in developing cleft palate. Food Chem. Toxicol. 56, 67–74. doi: 10.1016/j.fct.2013.02.003
Hu, X., Gao, J., Liao, Y., Tang, S., and Lu, F. (2013b). Retinoic acid alters the proliferation and survival of the epithelium and mesenchyme and suppresses Wnt/β-catenin signaling in developing cleft palate. Cell Death Dis. 4:e898. doi: 10.1038/cddis.2013.424
Huang, K., Jia, J., Wu, C., Yao, M., Li, M., Jin, J., et al. (2013). Ribosomal RNA gene transcription mediated by the master genome regulator protein CCCTC-binding factor (CTCF) is negatively regulated by the condensin complex. J. Biol. Chem. 288, 26067–26077. doi: 10.1074/jbc.M113.486175
Jiang, R., Bush, J. O., and Lidral, A. C. (2006). Development of the upper lip: morphogenetic and molecular mechanisms. Dev. Dyn. 235, 1152–1166. doi: 10.1002/dvdy.20646
Jin, Y. R., Han, X. H., Nishimori, K., Ben-Avraham, D., Oh, Y. J., Shim, J. W., et al. (2020). Canonical WNT/β-Catenin Signaling Activated by WNT9b and RSPO2 cooperation regulates facial morphogenesis in mice. Front. Cell Dev. Biol. 8:264. doi: 10.3389/fcell.2020.00264
Kang, P., and Svoboda, K. K. (2005). Epithelial-mesenchymal transformation during craniofacial development. J. Dent. Res. 84, 678–690. doi: 10.1177/154405910508400801
Leslie, E. J., and Marazita, M. L. (2013). Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C Semin. Med. Genet. 163C, 246–258. doi: 10.1002/ajmg.c.31381
Li, M. X., Li, Z., Zhang, R., Yu, Y., Wang, L. S., Wang, Q., et al. (2020). Effects of small interfering RNA-mediated silencing of susceptibility genes of non-syndromic cleft lip with or without cleft palate on cell proliferation and migration. Int. J. Pediatr. Otorhinolaryngol. 138:110382. doi: 10.1016/j.ijporl.2020.110382
Li, T., Xie, J., Shen, C., Cheng, D., Shi, Y., Wu, Z., et al. (2015). Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 75, 3181–3191. doi: 10.1158/0008-5472.Can-14-3721
Lin, C., Fisher, A. V., Yin, Y., Maruyama, T., Veith, G. M., Dhandha, M., et al. (2011). The inductive role of Wnt-β-Catenin signaling in the formation of oral apparatus. Dev. Biol. 356, 40–50. doi: 10.1016/j.ydbio.2011.05.002
Liu, G., Wang, L., Han, H., Li, Y., Lu, S., Li, T., et al. (2017). LncRNA ZFAS1 promotes growth and metastasis by regulating BMI1 and ZEB2 in osteosarcoma. Am. J. Cancer Res. 7, 1450–1462.
Ma, J., Chen, S., Hao, L., Sheng, W., Chen, W., Ma, X., et al. (2020a). Hypermethylation-mediated down-regulation of lncRNA TBX5-AS1:2 in Tetralogy of Fallot inhibits cell proliferation by reducing TBX5 expression. J. Cell Mol. Med. 24, 6472–6484. doi: 10.1111/jcmm.15298
Ma, J., Chen, S., Hao, L., Sheng, W., Chen, W., Ma, X., et al. (2020b). Long non-coding RNA SAP30-2:1 is downregulated in congenital heart disease and regulates cell proliferation by targeting HAND2. Front. Med. 15:91–100. doi: 10.1007/s11684-020-0778-5
Mohamad Shah, N. S., Sulong, S., Wan Sulaiman, W. A., and Halim, A. S. (2019). Two novel genes TOX3 and COL21A1 in large extended Malay families with nonsyndromic cleft lip and/or palate. Mol. Genet. Genomic. Med. 7:e635. doi: 10.1002/mgg3.635
Peng, W. X., Koirala, P., and Mo, Y. Y. (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667. doi: 10.1038/onc.2017.184
Qi, C. F., Kim, Y. S., Xiang, S., Abdullaev, Z., Torrey, T. A., Janz, S., et al. (2012). Characterization of ARF-BP1/HUWE1 interactions with CTCF, MYC, ARF and p53 in MYC-driven B cell neoplasms. Int. J. Mol. Sci. 13, 6204–6219. doi: 10.3390/ijms13056204
Roy, A. R., Ahmed, A., DiStefano, P. V., Chi, L., Khyzha, N., Galjart, N., et al. (2018). The transcriptional regulator CCCTC-binding factor limits oxidative stress in endothelial cells. J. Biol. Chem. 293, 8449–8461. doi: 10.1074/jbc.M117.814699
Shu, X., Dong, Z., Zhang, M., and Shu, S. (2019). Integrated analysis identifying long non-coding RNAs (lncRNAs) for competing endogenous RNAs (ceRNAs) network-regulated palatal shelf fusion in the development of mouse cleft palate. Ann. Transl. Med. 7:762. doi: 10.21037/atm.2019.11.93
Shull, L. C., Sen, R., Menzel, J., Goyama, S., Kurokawa, M., and Artinger, K. B. (2020). The conserved and divergent roles of Prdm3 and Prdm16 in zebrafish and mouse craniofacial development. Dev. Biol. 461, 132–144. doi: 10.1016/j.ydbio.2020.02.006
Song, L., Li, Y., Wang, K., Wang, Y. Z., Molotkov, A., Gao, L., et al. (2009). Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development 136, 3161–3171. doi: 10.1242/dev.037440
Uslu, V. V., Petretich, M., Ruf, S., Langenfeld, K., Fonseca, N. A., Marioni, J. C., et al. (2014). Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat. Genet. 46, 753–758. doi: 10.1038/ng.2971
Wang, Y., Lu, J. H., Wu, Q. N., Jin, Y., Wang, D. S., Chen, Y. X., et al. (2019). LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 18:174. doi: 10.1186/s12943-019-1105-0
Wang, Z. Q., Cai, Q., Hu, L., He, C. Y., Li, J. F., Quan, Z. W., et al. (2017). Long noncoding RNA UCA1 induced by SP1 promotes cell proliferation via recruiting EZH2 and activating AKT pathway in gastric cancer. Cell Death Dis. 8:e2839. doi: 10.1038/cddis.2017.143
Wu, D., Wang, M., Wang, X., Zhang, Y. B., Song, T., Yin, N., et al. (2019). Interaction between interferon regulatory factor 6 and glycine receptor beta shows a protective effect on developing nonsyndromic cleft lip with or without cleft palate in the Han Chinese population. Eur. J. Oral. Sci. 127, 27–32. doi: 10.1111/eos.12587
Xiao, K., Yang, Y., Bian, Y., Feng, B., Li, Z., Wu, Z., et al. (2019). Identification of differentially expressed long noncoding RNAs in human knee osteoarthritis. J. Cell Biochem. 120, 4620–4633. doi: 10.1002/jcb.27750
Xu, T. P., Liu, X. X., Xia, R., Yin, L., Kong, R., Chen, W. M., et al. (2015). SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene 34, 5648–5661. doi: 10.1038/onc.2015.18
Ye, M., Liu, B., Zhang, J., and Dong, K. (2020b). Role of long noncoding RNA in neuroblastoma. Discov. Med. 30, 71–82.
Ye, M., Lu, H., Tang, W., Jing, T., Chen, S., Wei, M., et al. (2020a). Downregulation of MEG3 promotes neuroblastoma development through FOXO1-mediated autophagy and mTOR-mediated epithelial-mesenchymal transition. Int. J. Biol. Sci. 16, 3050–3061. doi: 10.7150/ijbs.48126
Ye, M., Ma, J., Liu, B., Liu, X., Ma, D., and Dong, K. (2019). Linc01105 acts as an oncogene in the development of neuroblastoma. Oncol. Rep. 42, 1527–1538. doi: 10.3892/or.2019.7257
Yun, L., Ma, L., Wang, M., Yang, F., Kan, S., Zhang, C., et al. (2019). Rs2262251 in lncRNA RP11-462G12.2 is associated with nonsyndromic cleft lip with/without cleft palate. Hum. Mutat. 40, 2057–2067. doi: 10.1002/humu.23859
Zeng, K., Chen, X., Xu, M., Liu, X., Hu, X., Xu, T., et al. (2018). CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 9:417. doi: 10.1038/s41419-018-0454-8
Zhang, A., Zhao, J. C., Kim, J., Fong, K. W., Yang, Y. A., Chakravarti, D., et al. (2015). LncRNA HOTAIR enhances the androgen-receptor-mediated transcriptional program and drives castration-resistant prostate cancer. Cell Rep. 13, 209–221. doi: 10.1016/j.celrep.2015.08.069
Zhou, X. L., Chen, G., Li, M. X., Wang, H. X., Hong, J. W., Shen, J. Y., et al. (2018). Targeting YOD1 by RNA interference inhibits proliferation and migration of human oral keratinocytes through transforming growth factor-β3 signaling pathway. Biomed. Res. Int. 2018:6254308. doi: 10.1155/2018/6254308
Keywords: NSCLP, ZFAS1, Sp1, CTCF, Wnt/β-catenin pathway
Citation: Chen S, Jia Z, Cai M, Ye M, Wu D, Wan T, Zhang B, Wu P, Xu Y, Guo Y, Tian C, Ma D and Ma J (2021) SP1-Mediated Upregulation of Long Noncoding RNA ZFAS1 Involved in Non-syndromic Cleft Lip and Palate via Inactivating WNT/β-Catenin Signaling Pathway. Front. Cell Dev. Biol. 9:662780. doi: 10.3389/fcell.2021.662780
Received: 01 February 2021; Accepted: 27 May 2021;
Published: 29 June 2021.
Edited by:
Lingqian Wu, Central South University, ChinaReviewed by:
Tong Cao, National University of Singapore, SingaporeZhengwei Yuan, ShengJing Hospital of China Medical University, China
Copyright © 2021 Chen, Jia, Cai, Ye, Wu, Wan, Zhang, Wu, Xu, Guo, Tian, Ma and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chan Tian, VGlhbmNoYW5fY2RjQDEyNi5jb20=; Duan Ma, ZHVhbm1hQGZ1ZGFuLmVkdS5jbg==; Jing Ma, bWoxOTgxNTIwOEB5ZWFoLm5ldA==
†These authors have contributed equally to this work and share first authorship
 Shiyu Chen1†
Shiyu Chen1† Zhonglin Jia
Zhonglin Jia Ming Cai
Ming Cai Mujie Ye
Mujie Ye Bowen Zhang
Bowen Zhang Yuntao Guo
Yuntao Guo Duan Ma
Duan Ma Jing Ma
Jing Ma