- Department of Pathology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, United States
PROteolysis-TArgeting Chimeras (PROTACs) is an emerging and promising approach to target intracellular proteins for ubiquitination-mediated degradation, including those so-called undruggable protein targets, such as transcriptional factors and scaffold proteins. To date, plenty of PROTACs have been developed to degrade various disease-relevant proteins, such as estrogen receptor (ER), androgen receptor (AR), RTK, and CDKs. However, the on-target off-tissue and off-target effect is one of the major limitation that prevents the usage of PROTACs in clinic. To this end, we and several other groups have recently developed light-controllable PROTACs, as the representative for the third generation controllable PROTACs, by using either photo-caging or photo-switch approaches. In this review, we summarize the emerging light-controllable PROTACs and the prospective for other potential ways to achieve temporospatial control of PROTACs.
Introduction
The ubiquitin-proteasome system (UPS) governs the degradation and turnover of protein, thus playing critical functions in many cellular processes including protein quality control, cell cycle progression, and cell signaling transduction (Komander and Rape, 2012; Pohl and Dikic, 2019). Catalyzed by the ubiquitin-activating enzyme (E1), the ubiquitin is transferred onto the ubiquitin-conjugating enzyme (E2), and eventually transferred onto protein target by the E3 ubiquitin ligase. The selectivity of the ubiquitination process on a protein substrate primarily relies on its recognition by a E3 ubiquitin ligase (Pickart, 2001; Bernassola et al., 2008; Zhou et al., 2013), through a short sequence motif on the protein substrate, known as degron (Mészáros et al., 2017; Kumar et al., 2020). For instance, the SCFβ–TrCP E3 ligase recognizes the phospho-degron of DpSGXXpS/pT (X represents any amino acid, and p represents phosphorylation modification), and the von Hippel-Lindau (VHL) E3 ligase binds to substrates with the proline-hydroxyl-degron of LAP-OH (P-OH represents the hydroxylation on the proline). Based on the growing understanding about biological function of E3 ligase and UPS, PROteolysis TArgeting Chimera (PROTAC) emerges as a new pharmaceutical approach since 2001 (Sakamoto et al., 2001). By hijacking the endogenous UPS to specifically degrade proteins of interest (POI), PROTACs are theoretically capable of targeting any proteins in cells (Sakamoto, 2010; Neklesa et al., 2017; Churcher, 2018; Guo et al., 2019; Paiva and Crews, 2019). Of the three functional moieties in the PROTAC molecule, the E3 ligase-ligand is designed for recruiting endogenous E3 ubiquitin ligase, and the warhead part (or called target-recruiting ligand) determines the specificity of protein targets, while the linker region between them should be optimized to achieve best efficiency and specificity to degrade individual substrate, in a case-by-case manner (Figure 1; Flanagan and Neklesa, 2019; Pettersson and Crews, 2019).
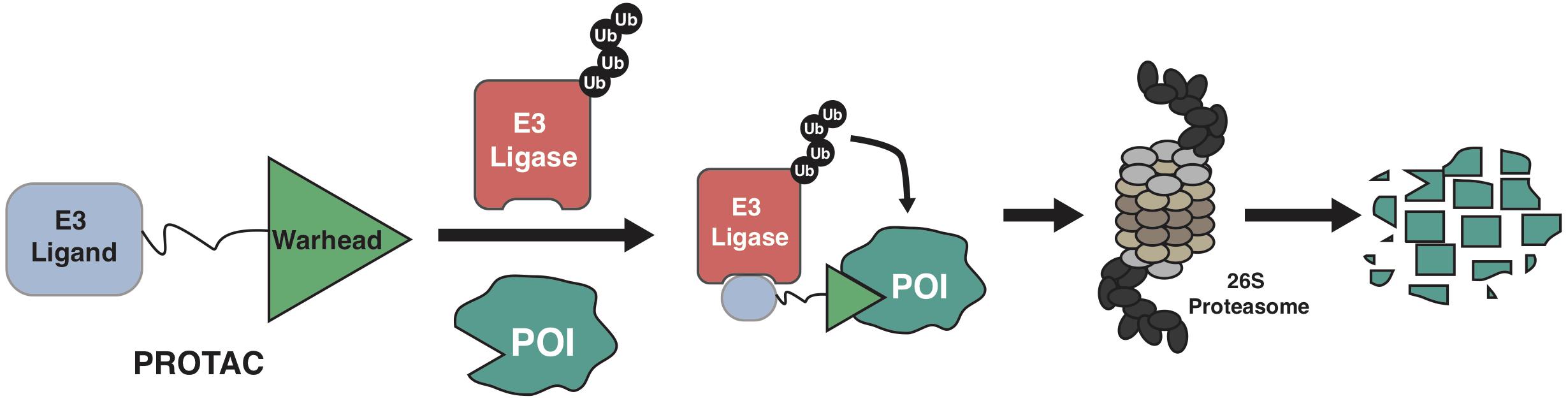
Figure 1. A schematic diagram for the action model of PROTAC. PROTAC recruits endogenous E3 ligase to ubiquitinate protein of interests (POIs), thus promoting the subsequent degradation of POI by the 26S proteasome.
The first generation of PROTACs take advantage of degron-derived peptides, such as phospho-peptides (Sakamoto et al., 2001, 2003) or hydroxyl-peptides (Schneekloth et al., 2004; Zhang et al., 2004; Rodriguez-Gonzalez et al., 2008), to recruit the endogenous β-TrCP or VHL E3 ubiquitin ligases, respectively. These peptide-based PROTACs have relatively high molecule weight, which limits their permeability into cells and their function as a bona fide drug. Moreover, peptide is unstable, and could only be injected into target cells, making them not practical in clinic. Recently, a modified version of peptide-based PROTAC, TD-PROTAC (Jiang et al., 2018), has been developed with better stability and cell-permeability, making it capable of degrading ERα in vitro and in vivo.
Besides these degron-derived peptides, small molecule inhibitors or binding partners have been developed for several E3 ligase, such as auxin for TIR E3 ligase (Dharmasiri et al., 2005), nutlin for mouse double minute 2 homolog (MDM2) E3 ligase (Vassilev et al., 2004). Based on these specific binding ligands of E3 ligases, the second generation small molecule PROTACs have been developed. In 2008, the first nutlin-based small molecule PROTAC has been developed to target androgen receptor (AR) for degradation in prostate cancer cells (Schneekloth et al., 2008). A recent study has shown that compared with VHL-based PROTACs, MDM2-based PROTACs might offer a synergistic anti-proliferative activity to cancer cells (Hines et al., 2019), in part due to the degradation of target protein bromodomain-containing protein 4 (BRD4), as well as the stabilization and accumulation of the tumor suppressor p53, a well-characterized endo-substrate of MDM2 (Chene, 2003). Several antagonists of cellular inhibitor of apoptosis protein 1 (cIAP1) E3 ligase, including bestatin (Sato et al., 2008), methyl bestatin (MeBS) (Sekine et al., 2008), MV1 (Varfolomeev et al., 2007) and LCL161 (Yang et al., 2016) have been reported to bind with cIAP1 and to promote its auto-ubiquitination and degradation. These small molecule antagonists have also been used in targeted protein degradation (TPD), also known as Specific and Non-genetic IAP-dependent Protein ERaser (SNIPER), to degrade many protein targets such as AR (Shibata et al., 2018), BCL-ABL (Demizu et al., 2016; Shibata et al., 2017; Shimokawa et al., 2017), BRDs (Ohoka et al., 2017b, 2019), Bruton’s tyrosine kinase (BTK) (Tinworth et al., 2019), cellular retinoic acid-binding protein 2 (CRABP2) (Okuhira et al., 2017), estrogen receptor (ER) (Okuhira et al., 2013), and transforming acidic coiled-coil containing protein 3 (TACC3) (Ohoka et al., 2014, 2017a).
In 2010, pomalidomide and its analogs immunomodulatory imide drugs (IMiDs) have been defined as molecule glues to bind with the endogenous cereblon (CRBN) E3 ligase (Ito et al., 2010; Fischer et al., 2014), subsequently causing the proteasomal degradation of several neo-substrates, including IKZFs (Kronke et al., 2014; Lu et al., 2014), CK1α (Kronke et al., 2015), GSPT1 (Matyskiela et al., 2016), SALL4 (Donovan et al., 2018), p63 (Asatsuma-Okumura et al., 2019) and ARID2 (Yamamoto et al., 2020). In 2015, IMiDs as ligands of the CRBN E3 ligase have been firstly used to develop CRBN-based PROTACs for the degradation of BRD4 and FKBP12 (Winter et al., 2015), and to date CRBN-based PROTACs have been applied to more than 30 different protein targets, for the treatment of cancer and inflammation disease (Supplementary Table 1; Mullard, 2021), among which ARV-110 (Neklesa et al., 2018) (NCT03888612) and ARV471 (Flanagan et al., 2019) (NCT04072952) are in Phase I/II clinical trials for the treatment of prostate cancer (Petrylak et al., 2020) and breast cancer (BRCA), respectively. In 2012, the small molecule VHL ligand (VHL ligand 1) has been developed to specifically interact with VHL without an inhibitory effect to the tumor suppressive function of the VHL E3 ligase (Buckley et al., 2012a,b; Galdeano et al., 2014). Furthermore, several other modified VHL ligands have been developed, including the 1, 3-fluoro-4-hydroxyprolines and methyl-VHL ligand 1 (Testa et al., 2018). Using these small molecule VHL ligands, dozens small molecule VHL-based PROTACs have been developed to target intracellular proteins, including AR (Salami et al., 2018; Han et al., 2019) and ER (Hu et al., 2019; Kargbo, 2019; Supplementary Table 1).
Compared with small molecule inhibitors, PROTACs have several advantages. First, unlike typical reversible enzymatic inhibitors, active center or allosteric site of protein targets is not necessary for PROTACs, making it possible to target those so-called undruggable proteins. Second, PROTACs function in a catalytic manner, and the drug could be recycled after the protein target being degraded, making it more potent than small molecule inhibitors. However, the catalytic feature of PROTACs might also introduce potential higher toxicity to cells in part due to the off-tissue on-target effects and off-target effects (Raina et al., 2016; Moreau et al., 2020), which is one of the major limitation for their application in practice. For example, thalidomide has been approved in 1950s for treating morning sickness in pregnant women in Europe, which caused a tragedy that affects thousands of children with severe birth defects (Rehman et al., 2011). Until recent, the teratogenic effects is defined for CRBN-mediated degradation of p63 (Asatsuma-Okumura et al., 2019) and SALL4 (Donovan et al., 2018). Besides, more and more CRBN neo-substrates of IMiDs have been reported, including IKZFs (Kronke et al., 2014; Lu et al., 2014), CK1α (Kronke et al., 2015), GSPT1 (Matyskiela et al., 2016), ARID2 (Yamamoto et al., 2020), RNF166 (You et al., 2020). ZNF827, and ZFP91 (Zorba et al., 2018). Furthermore, the subcutaneous injection of BRD4 degrader ARV-771 in xenograft tumor mice causes noticeable skin discoloration (Raina et al., 2016), which is consistent with the phenotype of Brd4 depleted mice (Bolden et al., 2014). Thus, next generation of PROTACs should at least have the property to distinguish target versus non-target tissues/cells to alleviate its toxicity issue.
The Third Generation PROTACs With Targeting Delivery And/Or Controllable Activation
One way to achieve targeted degradation of protein is to specifically deliver PROTACs into cancer cells, by taking advantage of the receptors expressed on the membrane of cancer cells, but not of normal cells. Recently, the antibody drug-conjugate (ADC) approach has been adopted for delivering PROTACs into cancer cells that expressing cancer-specific membrane-anchored receptors, such as HER2 (Dragovich et al., 2020, 2021a,b; Maneiro et al., 2020; Pillow et al., 2020). A major disadvantage of antibody-conjugated PROTAC is its relatively high molecule weight and weak stability. Thus, we have recently developed a small molecule version of targeting delivery platform for PROTACs, namely folate-PROTAC (Liu et al., 2021), by conjugating a folate group on the hydroxyl group of VHL ligand, to specific deliver PROTACs into cancer cells that express relatively high levels of folate receptor α (FOLR1) (Scaranti et al., 2020). Moreover, PROTACs that recruits cancer-specific E3 ligase might provide a way to achieve cancer-selective action of PROTACs (Nalawansha and Crews, 2020). For example, VHL-based PROTAC for BCL-xl is more tolerable than BCL-xl inhibitor ATB263, in part due to the relatively low expression of VHL in platelets than in cancer cells, thus reducing potential on-target toxicity (Khan et al., 2019). Several cancer specific or tissue specific E3 ligases have been recently identified (Schapira et al., 2019), however, none of these E3 ligase has ready-to-use small molecule binders yet, which prevents its further clinical development.
Another approach to achieve controllable protein degradation is to use an extraneous cellular signaling for the activation of PROTAC, such as by phosphoPROTACs (Hines et al., 2013). After stimulated with either nerve growth factor (NGF) or neuregulin, the two prototype phosphoPROTACs degraded fibroblast growth factor receptor substrate 2α (FRS2α) or PI3K, respectively (Hines et al., 2013). The phosphoPROTACs provide an option for controllable-PROTACs, but it still lacks tissue/cell specificity as these extraneous cues largely rely on universal receptors that are expressed in all cells regardless of normal or tumor cells. Recently, we and several other laboratories have independently developed light-controllable PROTACs, using either photo-cage or photo-switch approaches, which are widely used in photodynamic therapy (PDT) (Bethea et al., 1999; Moore et al., 2009; Agostinis et al., 2011; Shafirstein et al., 2016). Here, we summarize these light-controllable PROTACs and discuss for the advantages and limits for their applications in clinic.
Photo-Cage Enables Controllable PROTAC Activation to Degrade Proteins in Targeting Cells
Photo-Cage and Photo-Cage Chemical Group
Photo-cage groups, also known as photoremovable protecting groups, provide a standard approach to spatially and temporally control the release of chemicals in cells. To date, several types of photo-cage groups have been develop for the purpose of controlled release of organic molecules (Klan et al., 2013). However, only a few types of photo-cage groups are available for caging small molecule drugs, in part due to the strict release condition in water solution rather than other organic solution, such as methanol or ethanol (Klan et al., 2013). In the past few years, the development in biorthogonal chemistry prompts several photolabile groups for caging cellular molecules such as neurotransmitters, secondary messengers, and amino acids (Bardhan and Deiters, 2019), making it a powerful tool in biological studies. Taking advantage of these photo-cage groups, we and other groups have recently developed photo-caged PROTACs, which enable controllable activation of PROTACs in target cells (Xue et al., 2019; Liu et al., 2020; Naro et al., 2020; Figure 2).
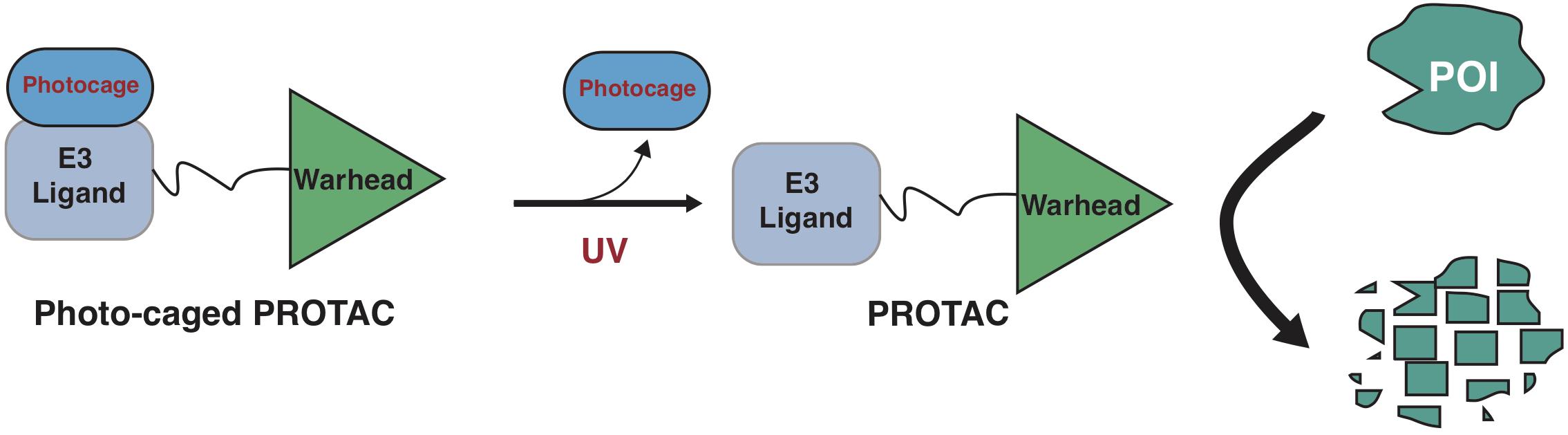
Figure 2. A schematic diagram for action model of photo-caged-PROTAC under control of UV illumination. The photo-caged-PROTAC is inert at beginning and activated by UV illumination, which leads to the release of the photocage group, thus enabling the degradation of POI in a controllable manner.
Photo-Cage Approach for CRBN-Based PROTACs
Further investigations on the crystal structure of CRBN and phthalimide complex indicate that the glutarimide NH in phthalidomide is critical for its binding with CRBN, particularly for the backbone carbonyl of the His380 residual (Petzold et al., 2016; Sievers et al., 2018; Matyskiela et al., 2020). Caging of glutarimide NH with methyl or other groups completely abolish the ability of pomalidomide to bind with the CRBN E3 ligase, and methyl-PROTACs are usually used as negative controls during the designation of PROTACs (Bondeson et al., 2018; Zhang et al., 2018). There are several photo-caged CRBN-based PROTACs that have been reported, including opto-PROTAC (Liu et al., 2020), pc-PROTAC (Xue et al., 2019), and others (Naro et al., 2020; Figure 3).
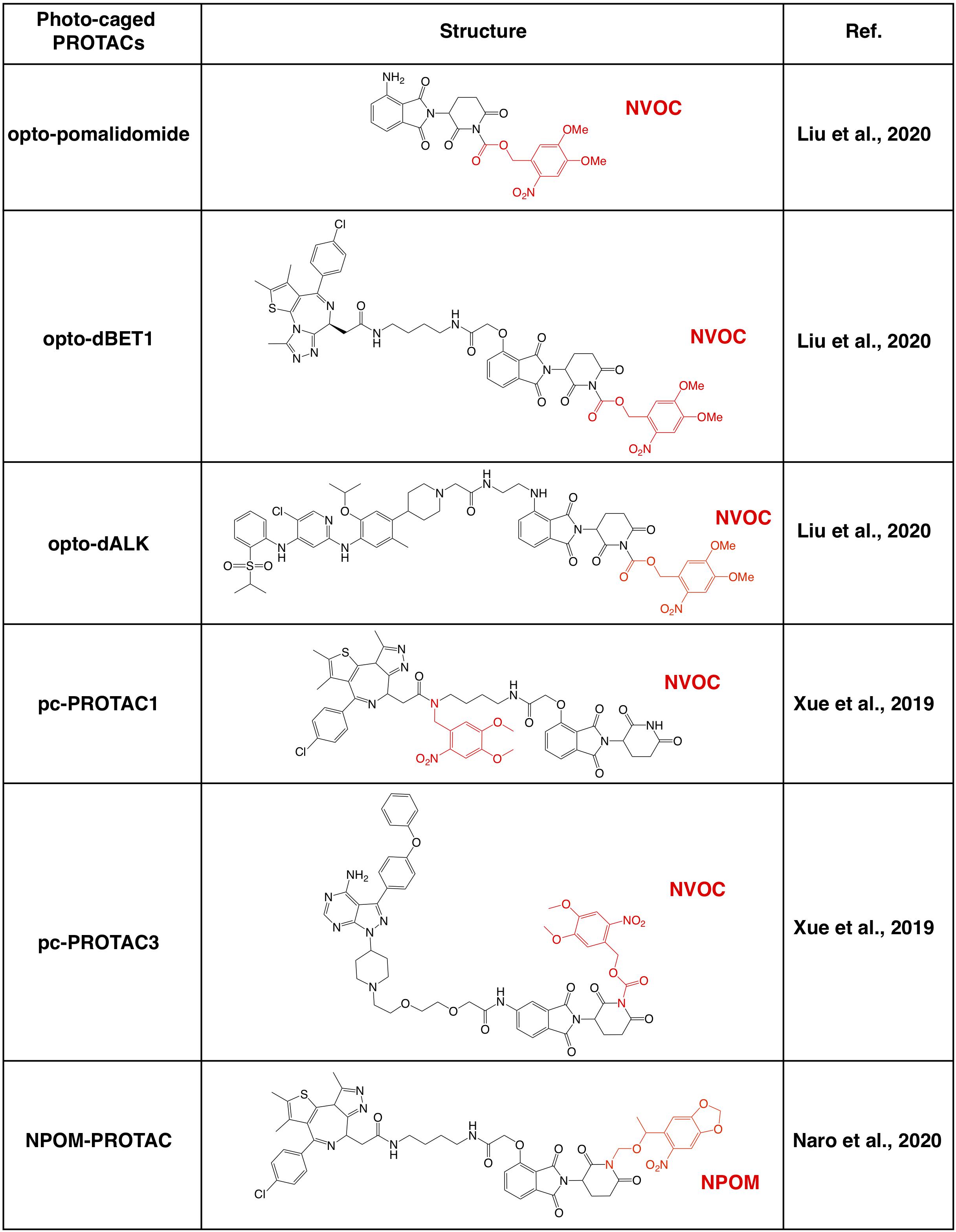
Figure 3. Summary of photo-caged CRBN-based PROTACs. The photo-cage groups are marked in red. NVOC, nitroveratryloxycarbonyl; NPOM, 6-nitropiperonyloxymethyl.
By incorporated a reversible photo-cage group, nitroveratryloxycarbonyl (NVOC) on the glutarimide NH of pomalidomide, opto-pomalidomide is inert and loss the capability in degrading IKZFs in cells (Liu et al., 2020), thus might be suitable to be applied to any other CRBN-based PROTACs. Two prototype opto-PROTACs, opto-dBET1 and opto-dALK, are inert and could be activated only after illuminated with UVA (λ = 365 nm) to degrade BRDs and ALK-fusion proteins, respectively (Liu et al., 2020). From another independent report, by using a similar photo-cage approach with NVOC, two pc-PROTACs prototypes, pc-PROTAC1 and pc-PROTAC3, degrade BRD4 and BTK, respectively, only after UVA illumination (Xue et al., 2019). Furthermore, by using a zebrafish model, they have validated the capability of pc-PROTAC1 in degrading endogenous BRDs under the control of UVA (λ = 365 nm) in vivo (Xue et al., 2019). Moreover, another photo-cage group, 6-nitropiperonyloxymethyl (NPOM) has also been used to cage the glutarimide NH in dBET1, and the resulting photo-caged PROTAC could degrade BRD4 after being illuminated with UVA (λ = 402 nm) (Naro et al., 2020). These studies together indicate that photo-cage on the glutarimide NH group could likely be an universal way for developing light-controllable PROTACs, and might be easily applied to other CRBN-based PROTACs in future studies.
Photo-Cage Approaches for VHL-Based PROTACs
Apart from CRBN-based PROTACs, VHL-based PROTACs represent another major class of second-generation small molecule PROTACs, and the photo-cage approach has also been used in VHL-based PROTACs (Figure 4). In a recent study, a photocleavable 4,5-dimethoxy-2-nitrobenzyl (DMNB) group has been incorporated onto the hydroxyl group of VHL ligand 1, and a prototype caged-PROTAC could degrade BRD4 after irradiation with UVA (λ = 365 nm) (Kounde et al., 2020). In another independent study, the photo-cage group diethylamino coumarin (DEACM) has been used to cage the VHL ligand in VHL-based PROTAC against ERRα, and the resulting caged-PROTAC is inert and regains the ability to degrade ERRα after activated by UVA (λ = 360 nm) (Naro et al., 2020). Given that the incorporation of photo-cage groups only affects the binding between PROTACs and the VHL E3 ligase, but not the protein substrate, those reported photo-cage methods could also be applied to other VHL-based PROTACs.
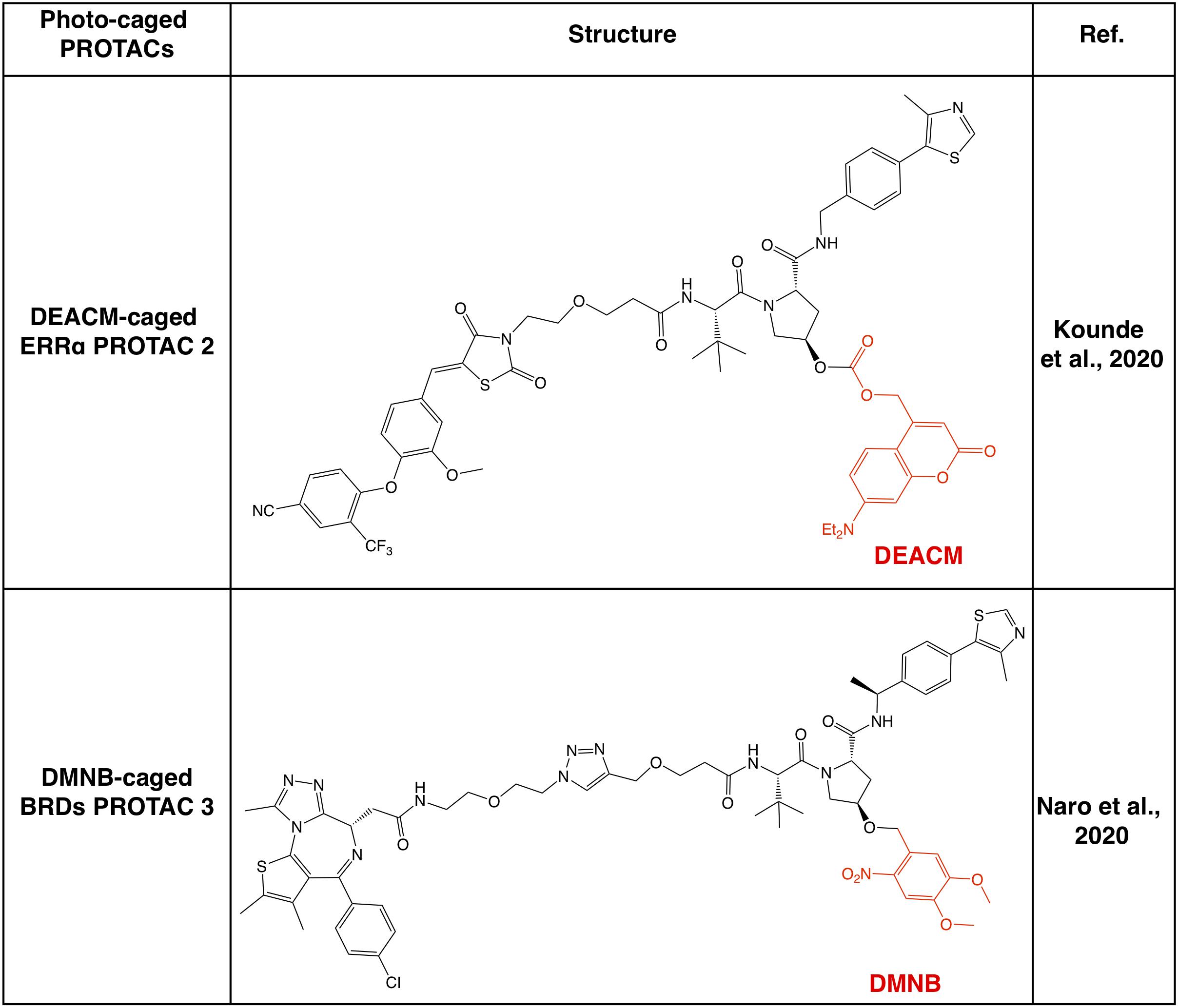
Figure 4. Summary of photo-caged VHL-based PROTACs. Photo-cage groups are marked in red. The photo-cage groups are in red. DMNB, 4,5-dimethoxy-2-nitrobenzyl; DEACM, diethylamino coumarin.
Photo-Switch Provide a Reversible On/Off Shift for PROTAC to Degrade Intracellular Proteins in Target Cells
Photo-Switchable Chemical Group in Biology
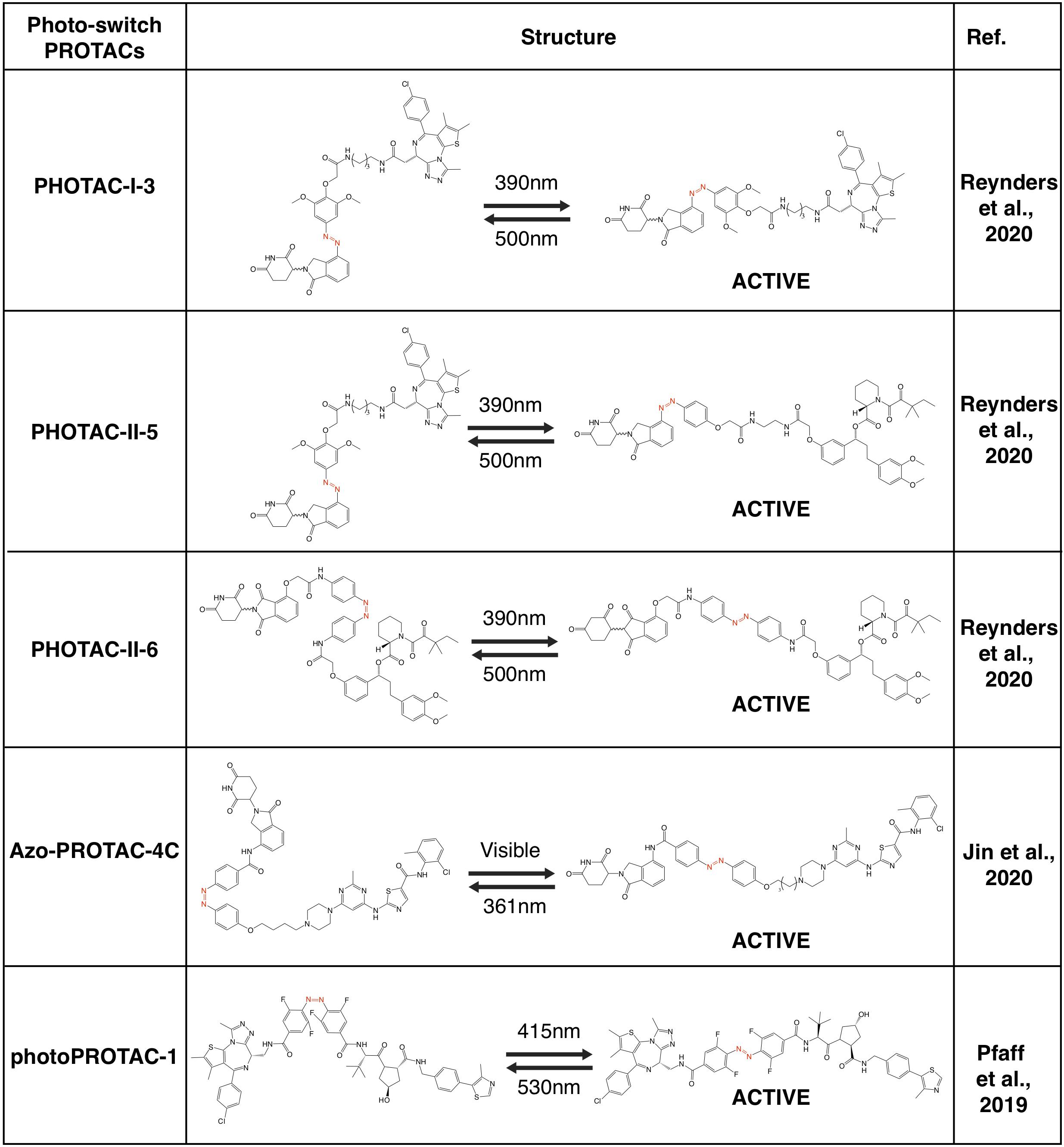
After entering target tissues/cells, focal UVA illumination leads to the release of activated PROTACs to be functional (Xue et al., 2019; Kounde et al., 2020; Liu et al., 2020; Naro et al., 2020). Activated PROTACs constantly degrade protein targets, and the degradation process will not stop before the clearance of PROTAC molecules. Thus, theoretically it should be better to add another OFF switch to inactivate the PROTACs, and the photo-switch provides a practical way. To this end, by taking advantage of the light-switchable azobenzene group or its analogs, several photo-switch PROTACs have been developed, including PHOTACs (Reynders et al., 2020), Azo-PROTACs (Jin et al., 2020) and photoPROTACs (Pfaff et al., 2019; Figures 5, 6).
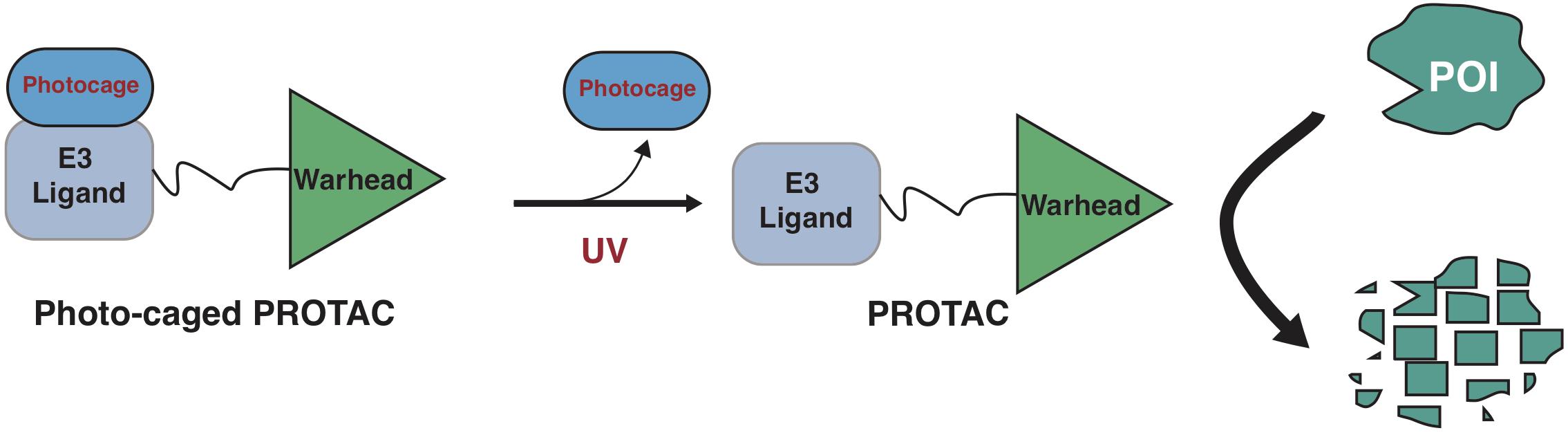
Figure 5. A schematic diagram for action model of photo-switch-PROTAC. The photo-switch PROTACs can be switched on and off by illumination with different wavelengths of light, which leads to the switch between the cis and trans forms of the photo-switch-PROTAC.
Photo-Switch PROTACs
Recently, several groups have utilized the photoswitch approach, i.e., azobenzene, to achieve photochemical isomerization of PROTAC molecules, and those photo-switch PROTACs could be reversibly turned on and off with light of different wave lengths (Reynders et al., 2020). By incorporating an azobenzene group in the linker region of pomalidomide-derived PROTACs, a type of light-inducible PROTACs, namely PHOTACs have developed. The two prototype PHOTACs remain in a trans inactive form in visible light (λ = 525 nm), and could be switched on with UVA illumination (λ = 390 nm), which leads to the conformation change to a cis active form, thus becoming capable of degrading BRDs and FKBP12, respectively. Furthermore, these PHOTACs could be turned off by visible light (λ = 525 nm), where PHOTACs return to the trans inactive form (Reynders et al., 2020). Furthermore, a similar photoswitchable azobenzene-based approach has been adopted in CRBN-based PROTACs to develop Azo-PROTACs. The prototype Azo-PROTAC could be switch between the trans active (ON) and the cis inactive (OFF) forms with either visible light or UV-C illumination, to ensure the light-controlled degradation of BCR-ABL fusion and ABL proteins in myelogenous leukemia K562 cells (Jin et al., 2020). Similarly, photo-switch could also be applied to VHL-based PROTAC. In another independent study, by using a similar photo-switch method to VHL-based PROTAC, photoPROTACs adopt the ortho-F4-azobenzene in the linker region between VHL ligand and warhead moiety against protein target (Pfaff et al., 2019). In contrast with PHOTACs, photoPROTACs remains as cis inactive form at beginning, and could be activated by UVA (λ = 415 nm) to change into a trans active form. Further illuminated by visible light (λ = 530 nm) could turn off the photoPROTAC, and the prototype photoPROTAC-1 could be switched on and off to degrade BRDs in cells in a light-controllable manner (Pfaff et al., 2019).
Limitations of Light-Controllable PROTACs and Perspective
The potential on-target off-tissue effects and off-target effects limit the application of PROTACs in clinic. These third-generation controllable PROTACs using light to activate or inactivate the PROTAC provide another layer of regulation on PROTACs, making it more practicable and controllable. However, those light-controllable PROTACs also have some disadvantages.
Notably, UVA light is used to activate or inactivate these light-controllable PROTACs, however, UVA light might trigger damage to DNA (Mouret et al., 2006; Cadet and Douki, 2011), especially when used in patients. Compared with UVB with shorter wavelength that causes DNA damage by triggering pyrimidine dimerization, UVA is less genotoxic (de Gruijl, 2002). However, UVA radiation is still thought to induce oxidant stress and DNA damage, which causes skin aging and possible skin cancers, including the deadly form of melanomas (de Gruijl, 2002). Moreover, UV light (used in both photo-caged PROTACs and photo-switch PROTACs) and visible light (used in photo-switch PROTACs) have limited penetration ability, thus making those light-controllable PROTACs only suitable for several types of cancer that can be accessed easily by light, such skin cancer or leukemia. To overcome such disadvantages, further effects should be focused on adopting other light source rather than UV light to trigger the photo-cage or photo-switch process. To this end, visible light or near-infrared light has longer wavelength and less energy than UV to trigger potential DNA damage (Mouret et al., 2006; Cadet and Douki, 2011), making them more suitable to be the cage group on PROTACs. More importantly, several photo-cage group with red and near-infrared light sensitivity have been developed recently (Vorobev and Moskalensky, 2020), including N-NO (Nakagawa, 2016) and benzoquinone-based photocage (Chen and Steinmetz, 2006; Wang and Kalow, 2018; Alabugin, 2019). Furthermore, other endogenous cues in cancers such as those cancer-specific antigens or receptors should be also useful for targeting delivery of PROTAC to cancer cells, thus eliminating possible toxic issue to normal tissues/cells (Liu et al., 2019; Saw and Song, 2019).
Another potential disadvantage of light-controllable PROTACs is due to their permeability. Compared with small molecule drug which is usually less than 500 Da, according to the Lipinski’s rule of five (Lipinski et al., 2001), standard PROTACs are usually more than 600 Da and these light-controllable PROTACs are usually near 1,000 Da. The relatively large molecule weight might compromise the pharmacokinetic and pharmacodynamic parameters of light-controllable PROTACs. To date, in most in vivo study, PROTACs are administrated by Raina et al. (2016); Ohoka et al. (2017b), Sun B. et al. (2018), intraperitoneally (Winter et al., 2015; Zhou et al., 2018; Gao et al., 2020) or intravenously (Mares et al., 2020) injection. Thus, it still warrants further in-depth investigation on optimization the pharmaceutical properties of PROTAC to make it possible for orally administered.
Finally, in clinic, there is lack of clear boundary between tumor tissues and adjunct normal tissues, making it hard to only activate these light-controllable PROTACs at the tumor tissues/cells. An alternative approach for controllable action of PROTACs in cancer cells could be taking advantage of cancer-specific receptors or transporters, such as HER2 and FOLR1 (Scaranti et al., 2020) for the guided delivery of PROTACs into cancer, but not normal cells. To this end, other types of third generation PROTACs, including antibody-conjugated PROTACs (Dragovich et al., 2020, 2021a,b; Maneiro et al., 2020; Pillow et al., 2020) and folate-PROTAC (Liu et al., 2021), have been recently developed, which specifically deliver PROTAC to cancer cells, thus avoiding potential toxicity to normal cells. Compared with the light-controllable PROTACs, folate-PROTAC (Liu et al., 2021) have relatively higher molecule weight of over 1,000 Da, and antibody-conjugated PROTACs (Dragovich et al., 2020, 2021a,b; Maneiro et al., 2020; Pillow et al., 2020) are macromolecule drug that could only be administrated by injection. Taken together, further studies are needed to make these third generation PROTACs (light-controllable PROTACs, antibody-conjugated PROTACs and folate-PROTAC) more practical in clinic.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported in part by funding from NIH (R01CA200651 and R35CA253027 to WW).
Conflict of Interest
WW is a co-founder and consultant for the ReKindle Therapeutics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize for not citing all relevant reports owing to space limitations.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.678077/full#supplementary-material
References
Adhikari, B., Bozilovic, J., Diebold, M., Schwarz, J. D., Hofstetter, J., Schroder, M., et al. (2020). PROTAC-mediated degradation reveals a non-catalytic function of AURORA-A kinase. Nat. Chem. Biol. 16, 1179–1188. doi: 10.1038/s41589-020-00652-y
Agostinis, P., Berg, K., Cengel, K. A., Foster, T. H., Girotti, A. W., Gollnick, S. O., et al. (2011). Photodynamic therapy of cancer: an update. CA Cancer J. Clin. 61, 250–281. doi: 10.3322/caac.20114
Alabugin, A. (2019). Near-IR photochemistry for biology: exploiting the optical window of tissue. Photochem. Photobiol. 95, 722–732. doi: 10.1111/php.13068
An, Z., Lv, W., Su, S., Wu, W., and Rao, Y. (2019). Developing potent PROTACs tools for selective degradation of HDAC6 protein. Protein Cell 10, 606–609. doi: 10.1007/s13238-018-0602-z
Asatsuma-Okumura, T., Ando, H., De Simone, M., Yamamoto, J., Sato, T., Shimizu, N., et al. (2019). p63 is a cereblon substrate involved in thalidomide teratogenicity. Nat. Chem. Biol. 15, 1077–1084. doi: 10.1038/s41589-019-0366-7
Bai, L., Zhou, H., Xu, R., Zhao, Y., Chinnaswamy, K., McEachern, D., et al. (2019). A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression In Vivo. Cancer Cell 36, 498.e17–511.e17. doi: 10.1016/j.ccell.2019.10.002
Bardhan, A., and Deiters, A. (2019). Development of photolabile protecting groups and their application to the optochemical control of cell signaling. Curr. Opin. Struct. Biol. 57, 164–175. doi: 10.1016/j.sbi.2019.03.028
Bassi, Z. I., Fillmore, M. C., Miah, A. H., Chapman, T. D., Maller, C., Roberts, E. J., et al. (2018). Modulating PCAF/GCN5 immune cell function through a PROTAC approach. ACS Chem. Biol. 13, 2862–2867. doi: 10.1021/acschembio.8b00705
Bernassola, F., Karin, M., Ciechanover, A., and Melino, G. (2008). The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21. doi: 10.1016/j.ccr.2008.06.001
Bethea, D., Fullmer, B., Syed, S., Seltzer, G., Tiano, J., Rischko, C., et al. (1999). Psoralen photobiology and photochemotherapy: 50 years of science and medicine. J. Dermatol. Sci. 19, 78–88. doi: 10.1016/S0923-1811(98)00064-4
Bian, J., Ren, J., Li, Y., Wang, J., Xu, X., Feng, Y., et al. (2018). Discovery of Wogonin-based PROTACs against CDK9 and capable of achieving antitumor activity. Bioorg. Chem. 81, 373–381. doi: 10.1016/j.bioorg.2018.08.028
Bolden, J. E., Tasdemir, N., Dow, L. E., van Es, J. H., Wilkinson, J. E., Zhao, Z., et al. (2014). Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition. Cell Rep. 8, 1919–1929. doi: 10.1016/j.celrep.2014.08.025
Bond, M. J., Chu, L., Nalawansha, D. A., Li, K., and Crews, C. M. (2020). Targeted degradation of oncogenic KRAS(G12C) by VHL-Recruiting PROTACs. ACS Cent. Sci. 6, 1367–1375. doi: 10.1021/acscentsci.0c00411
Bondeson, D. P., Smith, B. E., Burslem, G. M., Buhimschi, A. D., Hines, J., Jaime-Figueroa, S., et al. (2018). Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell Chem. Biol. 25, 78.e5–87.e5. doi: 10.1016/j.chembiol.2017.09.010
Brand, M., Jiang, B., Bauer, S., Donovan, K. A., Liang, Y., Wang, E. S., et al. (2019). Homolog-Selective degradation as a strategy to probe the function of CDK6 in AML. Cell Chem. Biol. 26, 300.e9–306.e9. doi: 10.1016/j.chembiol.2018.11.006
Buckley, D. L., Gustafson, J. L., Van Molle, I., Roth, A. G., Tae, H. S., Gareiss, P. C., et al. (2012a). Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF1alpha. Angew. Chem. Int. Ed. Engl. 51, 11463–11467. doi: 10.1002/anie.201206231
Buckley, D. L., Van Molle, I., Gareiss, P. C., Tae, H. S., Michel, J., Noblin, D. J., et al. (2012b). Targeting the von Hippel-Lindau E3 ubiquitin ligase using small molecules to disrupt the VHL/HIF-1alpha interaction. J. Am. Chem. Soc. 134, 4465–4468. doi: 10.1021/ja209924v
Burslem, G. M., Schultz, A. R., Bondeson, D. P., Eide, C. A., Savage Stevens, S. L., Druker, B. J., et al. (2019). Targeting BCR-ABL1 in chronic myeloid leukemia by PROTAC-Mediated targeted protein degradation. Cancer Res. 79, 4744–4753. doi: 10.1158/0008-5472.CAN-19-1236
Burslem, G. M., Smith, B. E., Lai, A. C., Jaime-Figueroa, S., McQuaid, D. C., Bondeson, D. P., et al. (2018). The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 25, 67.e3–77.e3. doi: 10.1016/j.chembiol.2017.09.009
Cadet, J., and Douki, T. (2011). Oxidatively generated damage to DNA by UVA radiation in cells and human skin. J. Invest. Dermatol. 131, 1005–1007. doi: 10.1038/jid.2011.51
Chen, H., Chen, F., Pei, S., and Gou, S. (2019). Pomalidomide hybrids act as proteolysis targeting chimeras: synthesis, anticancer activity and B-Raf degradation. Bioorg. Chem. 87, 191–199. doi: 10.1016/j.bioorg.2019.03.035
Chen, Y., and Steinmetz, M. G. (2006). Photoactivation of amino-substituted 1,4-benzoquinones for release of carboxylate and phenolate leaving groups using visible light. J. Org. Chem. 71, 6053–6060. doi: 10.1021/jo060790g
Chene, P. (2003). Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat. Rev. Cancer 3, 102–109. doi: 10.1038/nrc991
Cheng, M., Yu, X., Lu, K., Xie, L., Wang, L., Meng, F., et al. (2020). Discovery of potent and selective Epidermal Growth Factor Receptor (EGFR) bifunctional small-molecule degraders. J. Med. Chem. 63, 1216–1232. doi: 10.1021/acs.jmedchem.9b01566
Chi, J. J., Li, H., Zhou, Z., Izquierdo-Ferrer, J., Xue, Y., Wavelet, C. M., et al. (2019). A novel strategy to block mitotic progression for targeted therapy. EBioMedicine 49, 40–54. doi: 10.1016/j.ebiom.2019.10.013
Churcher, I. (2018). Protac-induced protein degradation in drug discovery: breaking the rules or just making new ones? J. Med. Chem. 61, 444–452. doi: 10.1021/acs.jmedchem.7b01272
Cromm, P. M., Samarasinghe, K. T. G., Hines, J., and Crews, C. M. (2018). Addressing kinase-independent functions of fak via PROTAC-Mediated degradation. J. Am. Chem. Soc. 140, 17019–17026. doi: 10.1021/jacs.8b08008
De Dominici, M., Porazzi, P., Xiao, Y., Chao, A., Tang, H. Y., Kumar, G., et al. (2020). Selective inhibition of Ph-positive ALL cell growth through kinase-dependent and -independent effects by CDK6-specific PROTACs. Blood 135, 1560–1573. doi: 10.1182/blood.2019003604
de Gruijl, F. R. (2002). Photocarcinogenesis: UVA vs. UVB radiation. Skin Pharmacol. Appl. Skin Physiol. 15, 316–320. doi: 10.1159/000064535
Demizu, Y., Shibata, N., Hattori, T., Ohoka, N., Motoi, H., Misawa, T., et al. (2016). Development of BCR-ABL degradation inducers via the conjugation of an imatinib derivative and a cIAP1 ligand. Bioorg. Med. Chem. Lett. 26, 4865–4869. doi: 10.1016/j.bmcl.2016.09.041
Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005). The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543
Donovan, K. A., An, J., Nowak, R. P., Yuan, J. C., Fink, E. C., Berry, B. C., et al. (2018). Thalidomide promotes degradation of SALL4, a transcription factor implicated in Duane Radial Ray syndrome. eLife 7:e38430. doi: 10.7554/eLife.38430
Dragovich, P. S., Adhikari, P., Blake, R. A., Blaquiere, N., Chen, J., Cheng, Y. X., et al. (2020). Antibody-mediated delivery of chimeric protein degraders which target estrogen receptor alpha (ERalpha). Bioorg. Med. Chem. Lett. 30:126907. doi: 10.1016/j.bmcl.2019.126907
Dragovich, P. S., Pillow, T. H., Blake, R. A., Sadowsky, J. D., Adaligil, E., Adhikari, P., et al. (2021a). Antibody-mediated delivery of chimeric BRD4 Degraders. Part 1: exploration of antibody linker, payload loading, and payload molecular properties. J. Med. Chem. 64, 2534–2575. doi: 10.1021/acs.jmedchem.0c01845
Dragovich, P. S., Pillow, T. H., Blake, R. A., Sadowsky, J. D., Adaligil, E., Adhikari, P., et al. (2021b). Antibody-mediated delivery of chimeric BRD4 Degraders. Part 2: improvement of in vitro antiproliferation activity and in vivo antitumor efficacy. J. Med. Chem. 64, 2576–2607. doi: 10.1021/acs.jmedchem.0c01846
Farnaby, W., Koegl, M., Roy, M. J., Whitworth, C., Diers, E., Trainor, N., et al. (2019). BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15, 672–680. doi: 10.1038/s41589-019-0294-6
Fischer, E. S., Bohm, K., Lydeard, J. R., Yang, H., Stadler, M. B., Cavadini, S., et al. (2014). Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53. doi: 10.1038/nature13527
Flanagan, J. J., and Neklesa, T. K. (2019). Targeting nuclear receptors with PROTAC degraders. Mol. Cell Endocrinol. 493:110452. doi: 10.1016/j.mce.2019.110452
Flanagan, J. J., Qian, Y., Gough, S. M., Andreoli, M., Bookbinder, M., Cadelina, G., et al. (2019). Abstract P5-04-18: ARV-471, an oral estrogen receptor PROTAC degrader for breast cancer. Cancer Res. 81(4 Suppl.), S17–S32.
Galdeano, C., Gadd, M. S., Soares, P., Scaffidi, S., Van Molle, I., Birced, I., et al. (2014). Structure-guided design and optimization of small molecules targeting the protein-protein interaction between the von Hippel-Lindau (VHL) E3 ubiquitin ligase and the hypoxia inducible factor (HIF) alpha subunit with in vitro nanomolar affinities. J. Med. Chem. 57, 8657–8663. doi: 10.1021/jm5011258
Gao, H., Zheng, C., Du, J., Wu, Y., Sun, Y., Han, C., et al. (2020). FAK-targeting PROTAC as a chemical tool for the investigation of non-enzymatic FAK function in mice. Protein Cell 11, 534–539. doi: 10.1007/s13238-020-00732-8
Gechijian, L. N., Buckley, D. L., Lawlor, M. A., Reyes, J. M., Paulk, J., Ott, C. J., et al. (2018). Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat. Chem. Biol. 14, 405–412. doi: 10.1038/s41589-018-0010-y
Guo, J., Liu, J., and Wei, W. (2019). Degrading proteins in animals: “PROTAC”tion goes in vivo. Cell Res. 29, 179–180. doi: 10.1038/s41422-019-0144-9
Han, X., Wang, C., Qin, C., Xiang, W., Fernandez-Salas, E., Yang, C. Y., et al. (2019). Discovery of ARD-69 as a highly potent proteolysis targeting chimera (PROTAC) degrader of androgen receptor (AR) for the treatment of prostate cancer. J. Med. Chem. 62, 941–964. doi: 10.1021/acs.jmedchem.8b01631
Han, X. R., Chen, L., Wei, Y., Yu, W., Chen, Y., Zhang, C., et al. (2020). Discovery of Selective Small Molecule Degraders of BRAF-V600E. J. Med. Chem. 63, 4069–4080. doi: 10.1021/acs.jmedchem.9b02083
He, Y., Zhang, X., Chang, J., Kim, H. N., Zhang, P., Wang, Y., et al. (2020). Using proteolysis-targeting chimera technology to reduce navitoclax platelet toxicity and improve its senolytic activity. Nat. Commun. 11:1996. doi: 10.1038/s41467-020-15838-0
Hines, J., Gough, J. D., Corson, T. W., and Crews, C. M. (2013). Posttranslational protein knockdown coupled to receptor tyrosine kinase activation with phosphoPROTACs. Proc. Natl. Acad. Sci. U.S.A. 110, 8942–8947. doi: 10.1073/pnas.1217206110
Hines, J., Lartigue, S., Dong, H., Qian, Y., and Crews, C. M. (2019). MDM2-Recruiting PROTAC offers superior, synergistic antiproliferative activity via simultaneous degradation of BRD4 and stabilization of p53. Cancer Res. 79, 251–262. doi: 10.1158/0008-5472.CAN-18-2918
Hu, J., Hu, B., Wang, M., Xu, F., Miao, B., Yang, C. Y., et al. (2019). Discovery of ERD-308 as a highly potent proteolysis targeting chimera (PROTAC) degrader of estrogen receptor (ER). J. Med. Chem. 62, 1420–1442. doi: 10.1021/acs.jmedchem.8b01572
Huang, H. T., Dobrovolsky, D., Paulk, J., Yang, G., Weisberg, E. L., Doctor, Z. M., et al. (2018). A chemoproteomic approach to query the degradable kinome using a multi-kinase degrader. Cell Chem. Biol. 25, 88.e6–99.e6. doi: 10.1016/j.chembiol.2017.10.005
Ito, T., Ando, H., Suzuki, T., Ogura, T., Hotta, K., Imamura, Y., et al. (2010). Identification of a primary target of thalidomide teratogenicity. Science 327, 1345–1350. doi: 10.1126/science.1177319
Jang, J., To, C., De Clercq, D. J. H., Park, E., Ponthier, C. M., Shin, B. H., et al. (2020). Mutant-selective allosteric EGFR degraders are effective against a broad range of drug-resistant mutations. Angew. Chem. Int. Ed. Engl. 59, 14481–14489. doi: 10.1002/anie.202003500
Jiang, B., Wang, E. S., Donovan, K. A., Liang, Y., Fischer, E. S., Zhang, T., et al. (2019). Development of dual and selective degraders of cyclin-dependent Kinases 4 and 6. Angew. Chem. Int. Ed. Engl. 58, 6321–6326. doi: 10.1002/anie.201901336
Jiang, Y., Deng, Q., Zhao, H., Xie, M., Chen, L., Yin, F., et al. (2018). Development of stabilized peptide-based PROTACs against estrogen receptor alpha. ACS Chem. Biol. 13, 628–635. doi: 10.1021/acschembio.7b00985
Jin, Y. H., Lu, M. C., Wang, Y., Shan, W. X., Wang, X. Y., You, Q. D., et al. (2020). Azo-PROTAC: novel light-controlled small-molecule tool for protein knockdown. J. Med. Chem. 63, 4644–4654. doi: 10.1021/acs.jmedchem.9b02058
Kang, C. H., Lee, D. H., Lee, C. O., Ha, J. Du, Park, C. H., and Hwang, J. Y. (2018). Induced protein degradation of anaplastic lymphoma kinase (ALK) by proteolysis targeting chimera (PROTAC). Biochem. Biophys. Res. Commun. 505, 542–547. doi: 10.1016/j.bbrc.2018.09.169
Kargbo, R. B. (2019). PROTAC-mediated degradation of estrogen receptor in the treatment of cancer. ACS Med. Chem. Lett. 10, 1367–1369. doi: 10.1021/acsmedchemlett.9b00397
Khan, S., Zhang, X., Lv, D., Zhang, Q., He, Y., Zhang, P., et al. (2019). A selective BCL-XL PROTAC degrader achieves safe and potent antitumor activity. Nat. Med. 25, 1938–1947. doi: 10.1038/s41591-019-0668-z
Klan, P., Solomek, T., Bochet, C. G., Blanc, A., Givens, R., Rubina, M., et al. (2013). Photoremovable protecting groups in chemistry and biology: reaction mechanisms and efficacy. Chem. Rev. 113, 119–191. doi: 10.1021/cr300177k
Komander, D., and Rape, M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81, 203–229. doi: 10.1146/annurev-biochem-060310-170328
Kounde, C. S., Shchepinova, M. M., Saunders, C. N., Muelbaier, M., Rackham, M. D., Harling, J. D., et al. (2020). A caged E3 ligase ligand for PROTAC-mediated protein degradation with light. Chem. Commun. 56, 5532–5535. doi: 10.1039/d0cc00523a
Kronke, J., Fink, E. C., Hollenbach, P. W., MacBeth, K. J., Hurst, S. N., Udeshi, N. D., et al. (2015). Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 523, 183–188. doi: 10.1038/nature14610
Kronke, J., Udeshi, N. D., Narla, A., Grauman, P., Hurst, S. N., McConkey, M., et al. (2014). Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305. doi: 10.1126/science.1244851
Kumar, M., Gouw, M., Michael, S., Samano-Sanchez, H., Pancsa, R., Glavina, J., et al. (2020). ELM-the eukaryotic linear motif resource in 2020. Nucleic Acids Res. 48, D296–D306. doi: 10.1093/nar/gkz1030
Lai, A. C., Toure, M., Hellerschmied, D., Salami, J., Jaime-Figueroa, S., Ko, E., et al. (2016). Modular PROTAC design for the degradation of oncogenic BCR-ABL. Angew. Chem. Int. Ed. Engl. 55, 807–810. doi: 10.1002/anie.201507634
Li, M. X., Yang, Y., Zhao, Q., Wu, Y., Song, L., Yang, H., et al. (2020). Degradation versus inhibition: development of proteolysis-targeting chimeras for overcoming statin-induced compensatory upregulation of 3-Hydroxy-3-methylglutaryl coenzyme a reductase. J. Med. Chem. 63, 4908–4928. doi: 10.1021/acs.jmedchem.0c00339
Li, W., Gao, C., Zhao, L., Yuan, Z., Chen, Y., and Jiang, Y. (2018). Phthalimide conjugations for the degradation of oncogenic PI3K. Eur. J. Med. Chem. 151, 237–247. doi: 10.1016/j.ejmech.2018.03.066
Li, Y., Yang, J., Aguilar, A., McEachern, D., Przybranowski, S., Liu, L., et al. (2019). Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double Minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem. 62, 448–466. doi: 10.1021/acs.jmedchem.8b00909
Li, Z., Pinch, B. J., Olson, C. M., Donovan, K. A., Nowak, R. P., Mills, C. E., et al. (2020). Development and characterization of a wee1 kinase degrader. Cell Chem. Biol. 27, 57.e9–65.e9. doi: 10.1016/j.chembiol.2019.10.013
Lipinski, C. A., Lombardo, F., Dominy, B. W., and Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26. doi: 10.1016/s0169-409x(00)00129-0
Liu, J., Chen, H., Liu, Y., Shen, Y., Meng, F., Kaniskan, H. U., et al. (2021). Cancer selective target degradation by folate-caged PROTACs. J. Am. Chem. Soc. 143, 7380–7387. doi: 10.1021/jacs.1c00451
Liu, J., Chen, H., Ma, L., He, Z., Wang, D., Liu, Y., et al. (2020). Light-induced control of protein destruction by opto-PROTAC. Sci. Adv. 6:eaay5154. doi: 10.1126/sciadv.aay5154
Liu, J., Lai, H., Xiong, Z., Chen, B., and Chen, T. (2019). Functionalization and cancer-targeting design of ruthenium complexes for precise cancer therapy. Chem. Commun. 55, 9904–9914. doi: 10.1039/c9cc04098f
Lu, G., Middleton, R. E., Sun, H., Naniong, M., Ott, C. J., Mitsiades, C. S., et al. (2014). The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 343, 305–309. doi: 10.1126/science.1244917
Lu, J., Qian, Y., Altieri, M., Dong, H., Wang, J., Raina, K., et al. (2015). Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem. Biol. 22, 755–763. doi: 10.1016/j.chembiol.2015.05.009
Maneiro, M. A., Forte, N., Shchepinova, M. M., Kounde, C. S., Chudasama, V., Baker, J. R., et al. (2020). Antibody-PROTAC conjugates enable HER2-dependent targeted protein degradation of BRD4. ACS Chem. Biol. 15, 1306–1312. doi: 10.1021/acschembio.0c00285
Mares, A., Miah, A. H. I, Smith, E. D., Rackham, M., Thawani, A. R., Cryan, J., et al. (2020). Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 3:140. doi: 10.1038/s42003-020-0868-6
Matyskiela, M. E., Clayton, T., Zheng, X., Mayne, C., Tran, E., Carpenter, A., et al. (2020). Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex. Nat. Struct. Mol. Biol. 27, 319–322. doi: 10.1038/s41594-020-0405-9
Matyskiela, M. E., Lu, G., Ito, T., Pagarigan, B., Lu, C. C., Miller, K., et al. (2016). A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature 535, 252–257. doi: 10.1038/nature18611
McCoull, W., Cheung, T., Anderson, E., Barton, P., Burgess, J., Byth, K., et al. (2018). Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC to provide insight into small molecule targeting of BCL6. ACS Chem. Biol. 13, 3131–3141. doi: 10.1021/acschembio.8b00698
Mészáros, B., Kumar, M., Gibson, T. J., Uyar, B., and Dosztányi, Z. (2017). Degrons in cancer. Sci. Signal. 10:eaak9982. doi: 10.1126/scisignal.aak9982
Moore, C. M., Pendse, D., and Emberton, M. (2009). Photodynamic therapy for prostate cancer–a review of current status and future promise. Nat. Clin. Pract. Urol. 6, 18–30. doi: 10.1038/ncpuro1274
Moreau, K., Coen, M., Zhang, A. X., Pachl, F., Castaldi, M. P., Dahl, G., et al. (2020). Proteolysis-targeting chimeras in drug development: a safety perspective. Br. J. Pharmacol. 177, 1709–1718. doi: 10.1111/bph.15014
Mouret, S., Baudouin, C., Charveron, M., Favier, A., Cadet, J., and Douki, T. (2006). Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. U.S.A. 103, 13765–13770. doi: 10.1073/pnas.0604213103
Mullard, A. (2021). Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discov. 20, 247–250. doi: 10.1038/d41573-021-00052-4
Nakagawa, H. (2016). Photocontrol of NO, H2S, and HNO release in biological systems by using specific caged compounds. Chem. Pharm. Bull. 64, 1249–1255. doi: 10.1248/cpb.c16-00403
Nalawansha, D. A., and Crews, C. M. (2020). PROTACs: an emerging therapeutic modality in precision medicine. Cell Chem. Biol. 27, 998–1014. doi: 10.1016/j.chembiol.2020.07.020
Naro, Y., Darrah, K., and Deiters, A. (2020). Optical control of small molecule-induced protein degradation. J. Am. Chem. Soc. 142, 2193–2197. doi: 10.1021/jacs.9b12718
Neklesa, T., Snyder, L. B., Willard, R. R., Vitale, N., Raina, K., Pizzano, J., et al. (2018). Abstract 5236: ARV-110: An androgen receptor PROTAC degrader for prostate cancer. Cancer Res. 78(13 Suppl.), 5236–5236.
Neklesa, T. K., Winkler, J. D., and Crews, C. M. (2017). Targeted protein degradation by PROTACs. Pharmacol. Ther. 174, 138–144. doi: 10.1016/j.pharmthera.2017.02.027
Nunes, J., McGonagle, G. A., Eden, J., Kiritharan, G., Touzet, M., Lewell, X., et al. (2019). Targeting IRAK4 for Degradation with PROTACs. ACS Med. Chem. Lett. 10, 1081–1085. doi: 10.1021/acsmedchemlett.9b00219
Ohoka, N., Nagai, K., Hattori, T., Okuhira, K., Shibata, N., Cho, N., et al. (2014). Cancer cell death induced by novel small molecules degrading the TACC3 protein via the ubiquitin-proteasome pathway. Cell Death Dis. 5:e1513. doi: 10.1038/cddis.2014.471
Ohoka, N., Nagai, K., Shibata, N., Hattori, T., Nara, H., Cho, N., et al. (2017a). SNIPER(TACC3) induces cytoplasmic vacuolization and sensitizes cancer cells to Bortezomib. Cancer Sci. 108, 1032–1041. doi: 10.1111/cas.13198
Ohoka, N., Okuhira, K., Ito, M., Nagai, K., Shibata, N., Hattori, T., et al. (2017b). In vivo knockdown of pathogenic proteins via specific and nongenetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs). J. Biol. Chem. 292, 4556–4570. doi: 10.1074/jbc.M116.768853
Ohoka, N., Ujikawa, O., Shimokawa, K., Sameshima, T., Shibata, N., Hattori, T., et al. (2019). Different degradation mechanisms of inhibitor of apoptosis proteins (IAPs) by the specific and nongenetic IAP-dependent protein eraser (SNIPER). Chem. Pharm. Bull. 67, 203–209. doi: 10.1248/cpb.c18-00567
Okuhira, K., Demizu, Y., Hattori, T., Ohoka, N., Shibata, N., Nishimaki-Mogami, T., et al. (2013). Development of hybrid small molecules that induce degradation of estrogen receptor-alpha and necrotic cell death in breast cancer cells. Cancer Sci. 104, 1492–1498. doi: 10.1111/cas.12272
Okuhira, K., Shoda, T., Omura, R., Ohoka, N., Hattori, T., Shibata, N., et al. (2017). Targeted degradation of proteins localized in subcellular compartments by hybrid small molecules. Mol. Pharmacol. 91, 159–166. doi: 10.1124/mol.116.105569
Paiva, S. L., and Crews, C. M. (2019). Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 50, 111–119. doi: 10.1016/j.cbpa.2019.02.022
Peng, L., Zhang, Z., Lei, C., Li, S., Ren, X., Chang, Y., et al. (2019). Identification of new small-molecule inducers of estrogen-related receptor alpha (ERRalpha) degradation. ACS Med. Chem. Lett. 10, 767–772. doi: 10.1021/acsmedchemlett.9b00025
Petrylak, P. D., Gao, X., Vogelzang, J. N., Garfield, H. M., Taylor, I., Moore, D. M., et al. (2020). First-in-human phase I study of ARV-110, an androgen receptor (AR) PROTAC degrader in patients (pts) with metastatic castrate-resistant prostate cancer (mCRPC) following enzalutamide (ENZ) and/or abiraterone (ABI). J. Clin. Oncol. 38, 3500–3500. doi: 10.1200/JCO.2020.38.15_suppl.3500
Pettersson, M., and Crews, C. M. (2019). PROteolysis TArgeting chimeras (PROTACs) - Past, present and future. Drug Discov. Today Technol. 31, 15–27. doi: 10.1016/j.ddtec.2019.01.002
Petzold, G., Fischer, E. S., and Thoma, N. H. (2016). Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature 532, 127–130. doi: 10.1038/nature16979
Pfaff, P., Samarasinghe, K. T. G., Crews, C. M., and Carreira, E. M. (2019). Reversible spatiotemporal control of induced protein degradation by bistable PhotoPROTACs. ACS Cent. Sci. 5, 1682–1690. doi: 10.1021/acscentsci.9b00713
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Pillow, T. H., Adhikari, P., Blake, R. A., Chen, J., Del Rosario, G., Deshmukh, G., et al. (2020). Antibody conjugation of a chimeric BET degrader enables in vivo activity. ChemMedChem 15, 17–25. doi: 10.1002/cmdc.201900497
Pohl, C., and Dikic, I. (2019). Cellular quality control by the ubiquitin-proteasome system and autophagy. Science 366, 818–822. doi: 10.1126/science.aax3769
Potjewyd, F., Turner, A. W., Beri, J., Rectenwald, J. M., Norris-Drouin, J. L., Cholensky, S. H., et al. (2020). Degradation of polycomb repressive complex 2 with an EED-Targeted bivalent chemical degrader. Cell Chem. Biol. 27, 47.e15–56.e15. doi: 10.1016/j.chembiol.2019.11.006
Powell, C. E., Gao, Y., Tan, L., Donovan, K. A., Nowak, R. P., Loehr, A., et al. (2018). Chemically induced degradation of anaplastic lymphoma kinase (ALK). J. Med. Chem. 61, 4249–4255. doi: 10.1021/acs.jmedchem.7b01655
Raina, K., Lu, J., Qian, Y., Altieri, M., Gordon, D., Rossi, A. M., et al. (2016). PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 7124–7129. doi: 10.1073/pnas.1521738113
Rana, S., Bendjennat, M., Kour, S., King, H. M., Kizhake, S., Zahid, M., et al. (2019). Selective degradation of CDK6 by a palbociclib based PROTAC. Bioorg. Med. Chem. Lett. 29, 1375–1379. doi: 10.1016/j.bmcl.2019.03.035
Rehman, W., Arfons, L. M., and Lazarus, H. M. (2011). The rise, fall and subsequent triumph of thalidomide: lessons learned in drug development. Ther. Adv. Hematol. 2, 291–308. doi: 10.1177/2040620711413165
Reynders, M., Matsuura, B. S., Berouti, M., Simoneschi, D., Marzio, A., Pagano, M., et al. (2020). PHOTACs enable optical control of protein degradation. Sci. Adv. 6:eaay5064. doi: 10.1126/sciadv.aay5064
Robb, C. M., Contreras, J. I., Kour, S., Taylor, M. A., Abid, M., Sonawane, Y. A., et al. (2017). Chemically induced degradation of CDK9 by a proteolysis targeting chimera (PROTAC). Chem. Commun. 53, 7577–7580. doi: 10.1039/c7cc03879h
Rodriguez-Gonzalez, A., Cyrus, K., Salcius, M., Kim, K., Crews, C. M., Deshaies, R. J., et al. (2008). Targeting steroid hormone receptors for ubiquitination and degradation in breast and prostate cancer. Oncogene 27, 7201–7211. doi: 10.1038/onc.2008.320
Sakamoto, K. M. (2010). Protacs for treatment of cancer. Pediatr. Res. 67, 505–508. doi: 10.1203/PDR.0b013e3181d35017
Sakamoto, K. M., Kim, K. B., Kumagai, A., Mercurio, F., Crews, C. M., and Deshaies, R. J. (2001). Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 98, 8554–8559. doi: 10.1073/pnas.141230798
Sakamoto, K. M., Kim, K. B., Verma, R., Ransick, A., Stein, B., Crews, C. M., et al. (2003). Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell Proteomics 2, 1350–1358. doi: 10.1074/mcp.T300009-MCP200
Salami, J., Alabi, S., Willard, R. R., Vitale, N. J., Wang, J., Dong, H., et al. (2018). Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun. Biol. 1:100. doi: 10.1038/s42003-018-0105-8
Sato, S., Aoyama, H., Miyachi, H., Naito, M., and Hashimoto, Y. (2008). Demonstration of direct binding of cIAP1 degradation-promoting bestatin analogs to BIR3 domain: synthesis and application of fluorescent bestatin ester analogs. Bioorg. Med. Chem. Lett. 18, 3354–3358. doi: 10.1016/j.bmcl.2008.04.031
Saw, P. E., and Song, E. W. (2019). Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell 10, 787–807. doi: 10.1007/s13238-019-0639-7
Scaranti, M., Cojocaru, E., Banerjee, S., and Banerji, U. (2020). Exploiting the folate receptor alpha in oncology. Nat. Rev. Clin. Oncol. 17, 349–359. doi: 10.1038/s41571-020-0339-5
Schapira, M., Calabrese, M. F., Bullock, A. N., and Crews, C. M. (2019). Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963. doi: 10.1038/s41573-019-0047-y
Schiedel, M., Herp, D., Hammelmann, S., Swyter, S., Lehotzky, A., Robaa, D., et al. (2018). Chemically induced degradation of sirtuin 2 (Sirt2) by a proteolysis targeting chimera (PROTAC) based on sirtuin rearranging ligands (SirReals). J. Med. Chem. 61, 482–491. doi: 10.1021/acs.jmedchem.6b01872
Schneekloth, A. R., Pucheault, M., Tae, H. S., and Crews, C. M. (2008). Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg. Med. Chem. Lett. 18, 5904–5908. doi: 10.1016/j.bmcl.2008.07.114
Schneekloth, J. S. Jr., Fonseca, F. N., Koldobskiy, M., Mandal, A., Deshaies, R., Sakamoto, K., et al. (2004). Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 126, 3748–3754. doi: 10.1021/ja039025z
Sekine, K., Takubo, K., Kikuchi, R., Nishimoto, M., Kitagawa, M., Abe, F., et al. (2008). Small molecules destabilize cIAP1 by activating auto-ubiquitylation. J. Biol. Chem. 283, 8961–8968. doi: 10.1074/jbc.M709525200
Shafirstein, G., Battoo, A., Harris, K., Baumann, H., Gollnick, S. O., Lindenmann, J., et al. (2016). Photodynamic therapy of non-small cell lung cancer. narrative review and future directions. Ann. Am. Thorac. Soc. 13, 265–275. doi: 10.1513/AnnalsATS.201509-650FR
Shibata, N., Miyamoto, N., Nagai, K., Shimokawa, K., Sameshima, T., Ohoka, N., et al. (2017). Development of protein degradation inducers of oncogenic BCR-ABL protein by conjugation of ABL kinase inhibitors and IAP ligands. Cancer Sci. 108, 1657–1666. doi: 10.1111/cas.13284
Shibata, N., Nagai, K., Morita, Y., Ujikawa, O., Ohoka, N., Hattori, T., et al. (2018). Development of protein degradation inducers of androgen receptor by conjugation of androgen receptor ligands and inhibitor of apoptosis protein ligands. J. Med. Chem. 61, 543–575. doi: 10.1021/acs.jmedchem.7b00168
Shimokawa, K., Shibata, N., Sameshima, T., Miyamoto, N., Ujikawa, O., Nara, H., et al. (2017). Targeting the allosteric site of oncoprotein BCR-ABL as an alternative strategy for effective target protein degradation. ACS Med. Chem. Lett. 8, 1042–1047. doi: 10.1021/acsmedchemlett.7b00247
Sievers, Q. L., Petzold, G., Bunker, R. D., Renneville, A., Slabicki, M., Liddicoat, B. J., et al. (2018). Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362:eaat0572. doi: 10.1126/science.aat0572
Smalley, J. P., Adams, G. E., Millard, C. J., Song, Y., Norris, J. K. S., Schwabe, J. W. R., et al. (2020). PROTAC-mediated degradation of class I histone deacetylase enzymes in corepressor complexes. Chem. Commun. 56, 4476–4479. doi: 10.1039/d0cc01485k
Smith, B. E., Wang, S. L., Jaime-Figueroa, S., Harbin, A., Wang, J., Hamman, B. D., et al. (2019). Differential PROTAC substrate specificity dictated by orientation of recruited E3 ligase. Nat. Commun. 10:131. doi: 10.1038/s41467-018-08027-7
Su, S., Yang, Z., Gao, H., Yang, H., Zhu, S., An, Z., et al. (2019). Potent and preferential degradation of CDK6 via proteolysis targeting chimera degraders. J. Med. Chem. 62, 7575–7582. doi: 10.1021/acs.jmedchem.9b00871
Sun, B., Fiskus, W., Qian, Y., Rajapakshe, K., Raina, K., Coleman, K. G., et al. (2018). BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia 32, 343–352. doi: 10.1038/leu.2017.207
Sun, N., Ren, C., Kong, Y., Zhong, H., Chen, J., Li, Y., et al. (2020). Development of a Brigatinib degrader (SIAIS117) as a potential treatment for ALK positive cancer resistance. Eur. J. Med. Chem. 193:112190. doi: 10.1016/j.ejmech.2020.112190
Sun, Y., Ding, N., Song, Y., Yang, Z., Liu, W., Zhu, J., et al. (2019). Degradation of Bruton’s tyrosine kinase mutants by PROTACs for potential treatment of ibrutinib-resistant non-Hodgkin lymphomas. Leukemia 33, 2105–2110. doi: 10.1038/s41375-019-0440-x
Sun, Y., Zhao, X., Ding, N., Gao, H., Wu, Y., Yang, Y., et al. (2018). PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res. 28, 779–781. doi: 10.1038/s41422-018-0055-1
Teng, M., Jiang, J., He, Z., Kwiatkowski, N. P., Donovan, K. A., Mills, C. E., et al. (2020). Development of CDK2 and CDK5 dual degrader TMX-2172. Angew. Chem. Int. Ed. Engl. 59, 13865–13870. doi: 10.1002/anie.202004087
Testa, A., Lucas, X., Castro, G. V., Chan, K. H., Wright, J. E., Runcie, A. C., et al. (2018). 3-Fluoro-4-hydroxyprolines: synthesis, conformational analysis, and stereoselective recognition by the VHL E3 Ubiquitin ligase for targeted protein degradation. J. Am. Chem. Soc. 140, 9299–9313. doi: 10.1021/jacs.8b05807
Tinworth, C. P., Lithgow, H., Dittus, L., Bassi, Z. I., Hughes, S. E., Muelbaier, M., et al. (2019). PROTAC-mediated degradation of bruton’s tyrosine kinase is inhibited by covalent binding. ACS Chem. Biol. 14, 342–347. doi: 10.1021/acschembio.8b01094
Tovell, H., Testa, A., Zhou, H., Shpiro, N., Crafter, C., Ciulli, A., et al. (2019). Design and characterization of SGK3-PROTAC1, an isoform specific SGK3 Kinase PROTAC Degrader. ACS Chem. Biol. 14, 2024–2034. doi: 10.1021/acschembio.9b00505
Varfolomeev, E., Blankenship, J. W., Wayson, S. M., Fedorova, A. V., Kayagaki, N., Garg, P., et al. (2007). IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131, 669–681. doi: 10.1016/j.cell.2007.10.030
Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848. doi: 10.1126/science.1092472
Vollmer, S., Cunoosamy, D., Lv, H., Feng, H., Li, X., Nan, Z., et al. (2020). Design, synthesis, and biological evaluation of MEK PROTACs. J. Med. Chem. 63, 157–162. doi: 10.1021/acs.jmedchem.9b00810
Vorobev, A. Y., and Moskalensky, A. E. (2020). Long-wavelength photoremovable protecting groups: on the way to in vivo application. Comput. Struct. Biotechnol. J. 18, 27–34. doi: 10.1016/j.csbj.2019.11.007
Wang, L., Shao, X., Zhong, T., Wu, Y., Xu, A., Sun, X., et al. (2021). Discovery of a first-in-class CDK2 selective degrader for AML differentiation therapy. Nat. Chem. Biol. 17, 567–575. doi: 10.1038/s41589-021-00742-5
Wang, M., Lu, J., Yang, C. Y., and Wang, S. (2020). Discovery of SHP2-D26 as a first, potent, and effective PROTAC Degrader of SHP2 Protein. J. Med. Chem. 63, 7510–7528. doi: 10.1021/acs.jmedchem.0c00471
Wang, S., Han, L., Han, J., Li, P., Ding, Q., Zhang, Q. J., et al. (2019). Uncoupling of PARP1 trapping and inhibition using selective PARP1 degradation. Nat. Chem. Biol. 15, 1223–1231. doi: 10.1038/s41589-019-0379-2
Wang, X., Feng, S., Fan, J., Li, X., Wen, Q., and Luo, N. (2016). New strategy for renal fibrosis: targeting Smad3 proteins for ubiquitination and degradation. Biochem. Pharmacol. 116, 200–209. doi: 10.1016/j.bcp.2016.07.017
Wang, X., and Kalow, J. A. (2018). Rapid aqueous photouncaging by red light. Org. Lett. 20, 1716–1719. doi: 10.1021/acs.orglett.8b00100
Wang, Z., He, N., Guo, Z., Niu, C., Song, T., Guo, Y., et al. (2019). Proteolysis targeting chimeras for the selective degradation of Mcl-1/Bcl-2 derived from nonselective target binding ligands. J. Med. Chem. 62, 8152–8163. doi: 10.1021/acs.jmedchem.9b00919
Wei, J., Hu, J., Wang, L., Xie, L., Jin, M. S., Chen, X., et al. (2019). Discovery of a first-in-class mitogen-activated protein kinase kinase 1/2 degrader. J. Med. Chem. 62, 10897–10911. doi: 10.1021/acs.jmedchem.9b01528
Winter, G. E., Buckley, D. L., Paulk, J., Roberts, J. M., Souza, A., Dhe-Paganon, S., et al. (2015). DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381. doi: 10.1126/science.aab1433
Winter, G. E., Mayer, A., Buckley, D. L., Erb, M. A., Roderick, J. E., Vittori, S., et al. (2017). BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5.e19–18.e19. doi: 10.1016/j.molcel.2017.06.004
Wu, H., Yang, K., Zhang, Z., Leisten, E. D., Li, Z., Xie, H., et al. (2019). Development of multifunctional histone deacetylase 6 degraders with potent antimyeloma activity. J. Med. Chem. 62, 7042–7057. doi: 10.1021/acs.jmedchem.9b00516
Wurz, R. P., and Cee, V. J. (2019). Targeted degradation of MDM2 as a new approach to improve the efficacy of MDM2-p53 inhibitors. J. Med. Chem. 62, 445–447. doi: 10.1021/acs.jmedchem.8b01945
Xue, G., Wang, K., Zhou, D., Zhong, H., and Pan, Z. (2019). Light-induced protein degradation with photocaged PROTACs. J. Am. Chem. Soc. 141, 18370–18374. doi: 10.1021/jacs.9b06422
Yamamoto, J., Suwa, T., Murase, Y., Tateno, S., Mizutome, H., Asatsuma-Okumura, T., et al. (2020). ARID2 is a pomalidomide-dependent CRL4(CRBN) substrate in multiple myeloma cells. Nat. Chem. Biol. 16, 1208–1217. doi: 10.1038/s41589-020-0645-3
Yang, C., Wang, H., Zhang, B., Chen, Y., Zhang, Y., Sun, X., et al. (2016). LCL161 increases paclitaxel-induced apoptosis by degrading cIAP1 and cIAP2 in NSCLC. J. Exp. Clin. Cancer Res. 35:158. doi: 10.1186/s13046-016-0435-7
Yang, K., Song, Y., Xie, H., Wu, H., Wu, Y. T., Leisten, E. D., et al. (2018). Development of the first small molecule histone deacetylase 6 (HDAC6) degraders. Bioorg. Med. Chem. Lett. 28, 2493–2497. doi: 10.1016/j.bmcl.2018.05.057
Yang, K., Wu, H., Zhang, Z., Leisten, E. D., Nie, X., Liu, B., et al. (2020). Development of selective histone deacetylase 6 (HDAC6) degraders recruiting von hippel-lindau (VHL) E3 ubiquitin ligase. ACS Med. Chem. Lett. 11, 575–581. doi: 10.1021/acsmedchemlett.0c00046
You, I., Erickson, E. C., Donovan, K. A., Eleuteri, N. A., Fischer, E. S., Gray, N. S., et al. (2020). Discovery of an AKT degrader with prolonged inhibition of downstream signaling. Cell Chem. Biol. 27, 66.e7–73.e7. doi: 10.1016/j.chembiol.2019.11.014
Zengerle, M., Chan, K. H., and Ciulli, A. (2015). Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777. doi: 10.1021/acschembio.5b00216
Zhang, C., Han, X. R., Yang, X., Jiang, B., Liu, J., Xiong, Y., et al. (2018). Proteolysis targeting chimeras (PROTACs) of Anaplastic Lymphoma Kinase (ALK). Eur. J. Med. Chem. 151, 304–314. doi: 10.1016/j.ejmech.2018.03.071
Zhang, D., Baek, S. H., Ho, A., Lee, H., Jeong, Y. S., and Kim, K. (2004). Targeted degradation of proteins by small molecules: a novel tool for functional proteomics. Comb. Chem. High Throughput Screen 7, 689–697. doi: 10.2174/1386207043328364
Zhang, H., Zhao, H. Y., Xi, X. X., Liu, Y. J., Xin, M., Mao, S., et al. (2020a). Discovery of potent epidermal growth factor receptor (EGFR) degraders by proteolysis targeting chimera (PROTAC). Eur. J. Med. Chem. 189:112061. doi: 10.1016/j.ejmech.2020.112061
Zhang, X., Thummuri, D., Liu, X., Hu, W., Zhang, P., Khan, S., et al. (2020b). Discovery of PROTAC BCL-XL degraders as potent anticancer agents with low on-target platelet toxicity. Eur. J. Med. Chem. 192:112186. doi: 10.1016/j.ejmech.2020.112186
Zhang, X., Xu, F., Tong, L., Zhang, T., Xie, H., Lu, X., et al. (2020b). Design and synthesis of selective degraders of EGFR(L858R/T790M) mutant. Eur. J. Med. Chem. 192:112199. doi: 10.1016/j.ejmech.2020.112199
Zhao, B., and Burgess, K. (2019). PROTACs suppression of CDK4/6, crucial kinases for cell cycle regulation in cancer. Chem. Commun. 55, 2704–2707. doi: 10.1039/c9cc00163h
Zhao, Q., Lan, T., Su, S., and Rao, Y. (2019). Induction of apoptosis in MDA-MB-231 breast cancer cells by a PARP1-targeting PROTAC small molecule. Chem. Commun. 55, 369–372. doi: 10.1039/c8cc07813k
Zhao, Q., Ren, C., Liu, L., Chen, J., Shao, Y., Sun, N., et al. (2019). Discovery of SIAIS178 as an effective BCR-ABL degrader by recruiting von hippel-lindau (VHL) E3 ubiquitin ligase. J. Med. Chem. 62, 9281–9298. doi: 10.1021/acs.jmedchem.9b01264
Zhou, B., Hu, J., Xu, F., Chen, Z., Bai, L., Fernandez-Salas, E., et al. (2018). Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J. Med. Chem. 61, 462–481. doi: 10.1021/acs.jmedchem.6b01816
Zhou, F., Chen, L., Cao, C., Yu, J., Luo, X., Zhou, P., et al. (2020). Development of selective mono or dual PROTAC degrader probe of CDK isoforms. Eur. J. Med. Chem. 187:111952. doi: 10.1016/j.ejmech.2019.111952
Zhou, H., Bai, L., Xu, R., Zhao, Y., Chen, J., McEachern, D., et al. (2019). Structure-based discovery of SD-36 as a potent, selective, and efficacious PROTAC degrader of STAT3 Protein. J. Med. Chem. 62, 11280–11300. doi: 10.1021/acs.jmedchem.9b01530
Zhou, W., Wei, W., and Sun, Y. (2013). Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases. Cell Res. 23, 599–619. doi: 10.1038/cr.2013.44
Zoppi, V., Hughes, S. J., Maniaci, C., Testa, A., Gmaschitz, T., Wieshofer, C., et al. (2019). Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) Identify VZ185 as a potent, fast, and selective von hippel-lindau (VHL) based dual degrader probe of BRD9 and BRD7. J. Med. Chem. 62, 699–726. doi: 10.1021/acs.jmedchem.8b01413
Keywords: ubiquitin, E3 ligase, PROTAC, tumorigenesis, light controllable
Citation: Liu J, Peng Y and Wei W (2021) Light-Controllable PROTACs for Temporospatial Control of Protein Degradation. Front. Cell Dev. Biol. 9:678077. doi: 10.3389/fcell.2021.678077
Received: 08 March 2021; Accepted: 14 June 2021;
Published: 19 July 2021.
Edited by:
Yu Rao, Tsinghua University, ChinaReviewed by:
Tae Ho Lee, Fujian Medical University, ChinaCatherine Lindon, University of Cambridge, United Kingdom
Copyright © 2021 Liu, Peng and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenyi Wei, d3dlaTJAYmlkbWMuaGFydmFyZC5lZHU=
†These authors have contributed equally to this work
 Jing Liu†
Jing Liu† Yunhua Peng
Yunhua Peng Wenyi Wei
Wenyi Wei