- 1Graduate School of Hebei Medical University, Shijiazhuang, China
- 2The Third Department of Surgery, The Fourth Hospital of Hebei Medical University, Shijiazhuang, China
- 3Department of Pathology, The Fourth Hospital of Hebei Medical University, Shijiazhuang, China
Gastric cancer (GC), as a common gastrointestinal tumor, is an important cause of death from cancer all around the world. Long non-coding RNAs (lncRNAs), a novel class of transcripts, have attracted great attention of researchers. However, the mechanisms of the clinical significance of most lncRNAs in human cancer are mainly undocumented. This research desires to explore the clinical significance, biological function, and mechanism of Lnc_ASNR (apoptosis suppressing-non-coding RNA) in GC. Cell proliferation, cell cycle, cell migration, and invasion abilities were respectively determined by 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT), flow cytometry, wound healing, and Transwell assay (Sigma-Aldrich, St. Louis, MO, United States). The association of Lnc_ASNR, miR-519e-5p, and fibroblast growth factor receptor 2 (FGFR2) was evaluated via luciferase reporter experiments. The tumor xenograft assay was conducted to confirm the results of cell experiments. High expressed Lnc_ASNR was detected in both GC cells and tissues using qRT-PCR. Downregulated Lnc_ASNR could reduce proliferation, migration, and invasion in GC cells, while upregulated Lnc_ASNR could promote the cell proliferation, migration, and invasion. Moreover, the effect of Lnc_ASNR on migration and invasion ability is closely related to epithelial-mesenchymal transition (EMT). The bioinformatics analysis, luciferase assay, and Western blot demonstrated that Lnc_ASNR inhibited miR-519e-5p expression but increased FGFR2 expression. Lnc_ASNR and FGFR2 were both targeted to miR-519e-5p, and they were negatively correlated with the expression of miR-519e-5p. All investigations indicated that Lnc_ASNR functioned as a ceRNA targeting miR-519e-5p and facilitated GC development by regulating the pathway of miR-519e-5p/FGFR2.
Introduction
Gastric cancer (GC) is a prevalent malignant tumor around the world with the sixth incidence rate and second mortality rate, which seriously affects human health (Bray et al., 2018). The advanced diagnosis and therapy of GC has achieved development in recent years. However, many limitations existed that hinder the effectiveness of GC treatment. The early symptoms of most GC patients are not obvious, and diagnosis delay becomes the main obstacle of GC treatment and prognosis. The combination of gastroscopy and biopsy is the universally recognized gold standard for GC diagnosis (Virgilio et al., 2017; den Hollander et al., 2019). However, its application in GC screening is still limited because it is not easy to be accepted by patients due to the invasiveness and high cost. At present, the most commonly used tumor biomarkers in clinical are insufficient to completely replace biopsy, so it is of great significance to explore more specific and sensitive biomarkers for early diagnosis of GC to improve the clinical treatment effect.
Long non-coding RNA (lncRNA) is an RNA molecule with a length between 200 and 100,000 nucleotides (nt), which can be transcribed independently. lncRNA is molecularly similar to mRNA, but there is no identifiable potential to encode functional proteins (Ulitsky, 2016). Researchers confirmed that lncRNA contains a significant biological function among several diseases (Montes et al., 2021; Zhang F. et al., 2021; Zhang Y. et al., 2021; Zhu et al., 2021). It is related to tumor development, chemotherapy sensitivity, and prognosis and especially plays a vital role in the resistance of multiple tumor chemotherapy drugs, especially in GC (Zhang and Wang, 2021), prostate cancer (Shang et al., 2019), and pancreatic cancer (Zhou et al., 2020). The expression of lncRNA is abnormal in a wide range of cancers. lncRNA demonstrates an essential influence in facilitating and inhibiting cancer occurrence and development, which proves its clinical potential as a biomarker and therapeutic target (Kim et al., 2018; Ramnarine et al., 2019; Wang Y. L. et al., 2019; Olivero et al., 2020). Recent findings indicated that lncRNA can interact with proteins, RNA, and lipids, thereby producing cancer signal transduction (Lin and Yang, 2018). In addition, lncRNAs, as competing endogenous RNAs (ceRNAs) or microRNA sponge, can regulate expression by competitively binding to microRNA (Song et al., 2017; Sun et al., 2017; Huang et al., 2019). Researchers have found that most lncRNAs are upregulated or downregulated expressly in tissues and serum plasma of patients with GC (Zhang et al., 2018; Wang C. J. et al., 2019; Guo et al., 2020; Yuan et al., 2020). Therefore, lncRNA can be used as a potential tumor marker for GC diagnosis.
Lnc_ASNR (non-coding RNA that inhibits apoptosis) is a nuclear-reserved lncRNA. Lnc_ASNR located in the chr14-q22.2 region is a newly discovered lncRNA associated with cancer and other diseases. Compared with normal tissues, Lnc_ASNR expression is significantly increased in four tumor tissues. Chen’s group (Chen et al., 2016) has found that Lnc_ASNR promotes the reduction of cytoplasmic AUF1 levels and inhibits AUF1-mediated degradation of Bcl-2 mRNA in colon cancer RKO cells, which leads to high expression of Bcl-2 and significantly inhibits cell apoptosis. However, the role and mechanism of Lnc_ASNR in tumorigenesis and development are unclear, which suggested that the study of the impact of Lnc_ASNR on tumors remains to be explored. Herein, we first evaluated whether Lnc_ASNR changes in the expression of lncRNA in GC and further clarified the biological function of Lnc_ASNR and the molecular mechanism that regulates the occurrence and development of GC. The research results help to identify important molecules that effectively regulate the occurrence and development of GC, which will provide a theoretical basis for clinical molecular diagnosis of GC and the development of targeted drugs. Our results exhibit important scientific significance and potential clinical application value for improving patient prognosis and enhancing survival rate.
Materials and Methods
Clinical and Histologic Evaluation of Human Tissues
In this study, 76 paired tumor tissues and adjacent non-tumor tissues were collected from GC patients at the Fourth Hospital of Hebei Medical University from 2017 to 2020. The GC diagnosis was histopathologically confirmed. Clinical pathological features, including gender, age, tumor-node-metastasis (TNM) staging, and tumor size, were summarized. None of them received preoperative treatment, including chemotherapy or radiotherapy. After obtaining the tissues, the samples were immediately placed in liquid nitrogen and then stored at −80°C. The research was approved by the Research Ethics Committee at the Fourth Hospital of Hebei Medical University (Shijiazhuang, China). All patients were informed and gave their consent.
Cell Culture
Four GC cell lines (MKN45, MKN28, AGS, and HGC27) and gastric mucosal epithelial cell (GES-1) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (GIBCO, Thermo Fisher Scientific, Waltham, MA, United States) containing 10% fetal bovine serum (FBS) (BI), 100 U/ml penicillin, and 100 mg/ml streptomycin (Invitrogen, Thermo Fisher Scientific) at 37°C with 5% CO2.
RNA Extraction and Quantitative Real-Time PCR
RNAiso Plus (TaKaRa, Tokyo, Japan) was utilized to extract total RNA from cells and tissue samples according to manufacturer’s information. The OD260/OD280 ratio ranging from 1.9 to 2.1 tested by UV spectrophotometer is qualified. Reverse transcription polymerase chain reaction was performed by LnRctue lncRNA First-Strand cDNA kit (TIANGEN, Beijing, China) and LnRctue lncRNA qPCR kit (SYBR Green) (TIANGEN) for quantitative real-time PCR (qRT-PCR) according to manufacturer’s information. ABI 7500 real-time PCR system (Applied Biosystems, Waltham, MA, United States) was utilized to conduct qRT-PCR and collect data. GAPDH was used as a loading control. Our qRT-PCR results were analyzed and expressed relative to threshold cycle (Ct) values, and then converted to fold changes. The specific primer sequences used in our study are provided in the Supplementary Table 1.
Vector Construction and Transfection and siRNA Transfection
Based on manufacturer’s information, Lipofectamine 2000 (Invitrogen) was used to transfect plasmid vectors and siRNAs into GC cells. The plasmid vector pcDNA3.1 used for Lnc_ASNR overexpression in MKN28, the siRNA (three individual ASNR siRNAs) used for Lnc_ASNR knockdown in MKN45 cells, miR-519e-5p inhibitor, and miR-519e-5p mimics, and the negative control (vector or NC) were obtained from Invitrogen. To upregulate ASNR, the coding sequence was increased based on the manufacturer’s instructions and subcloned into the pcDNA3.1 (+) vector (Invitrogen). Then Lipofectamine 2000 (Invitrogen) was used to transfect MKN28 cells with a negative control vector or a ASNR-expressed plasmid. In order to downregulate ASNR, Lipofectamine 2000 (Invitrogen) was utilized to transfect MKN28 cells with the target sequence of negative control siRNA. After being transfected for 48 h, cells were obtained for qRT-PCR and Western blot experiments. The siRNA sequences used in our study are provided in the Supplementary Table 2.
Lentiviral Infection
The MKN45 cell suspension was seeded onto a six-well plate at concentration of 3 × 104 cells/well, and the cells were grown to a density of 20%. We have created two different groups, sh-ASNR transfected with shASNR green fluorescent protein (GFP) lentivirus and shNC (GENECHEM) transfected with empty GFP lentivirus, respectively. An appropriate amount of lentivirus was added according to the differences in infection (MOI = 20). After 3 days of transfection, a fluorescence microscope was used to observe GFP-tagged gene expression. Cells with transfection efficiency > 80% were selected for further analyses.
Cell Proliferation Assays
The cell viability of MKN45 cells transfected with si-ASNR and MKN28 cells transfected with pcDNA3.1-ASNR was evaluated via MTT Cell Proliferation and Cytotoxicity Assay Kit (Solarbio, Beijing, China) based on the manufacturer’s guidance. Transfected MKN-45 cells and MKN28 cells (5 × 104/well) were incubated with MTT (20 μl/well) at 37°C for 4 h. Next, DMSO was added and mixed for 10 min to ensure the crystals were dissolved. The optical density value was measured at the absorbance 570 nm under Thermo Scientific Microplate Reader Multiskan MK3 (Thermo Fisher Scientific).
Flow Cytometry for Cell Cycle Analysis
si-ASNR- or si-NC-transfected MKN45 cells and pcDNA3.1-ASNR- or blank vector–transfected MKN28 cells were obtained after 48 h of transfection by Lipofectamine 2000 (Invitrogen). Next, the cells were dyed with propidium iodide (PI) utilizing the DNA content quantitative measurement method (cell cycle) (Solarbio) and detected via FACScan. Calculate and compare the percentage of cells in G0/G1, S, and G2/M phases.
Wound-Healing Assay
To conduct the wound healing experiment, 3 × 105 cells were seeded into six-well plates and cultured overnight. Cells were transfected with pcDNA3.1-ASNR, si-ASNR, and si-NC, respectively. When the cultured cells reached a density of 90%, a sterile 10-ml disposable pipette tip was used to produce uniform scratches. The cells were then grown in a medium containing 1% FBS for 48 h. At indicated time points, a microscope was used to obtain images of the plate (Olympus, Tokyo, Japan). Meanwhile, scratch area and wound-healing percentage were calculated. The experiment was repeated three times in parallel.
Cell Migration and Invasion Assays
si-ASNR- or si-NC transfected MKN45 cells and pcDNA3.1-ASNR-transfected MKN28 cells were harvested after 48 h of transfection and then collected. Twenty-four-well Transwell plates (8 μm pore size) (Corning Inc, Corning, NY, United States) were applied for cell migration and invasion assays. Briefly, 50 μl of Matrigel was placed into the Transwell upper chambers (Sigma-Aldrich). Cells (1 × 105) suspended in 100 μl of serum-free culture medium were seeded into the upper chambers. Added RPMI 1640 culture medium supplemented with 10% FBS into the wells under the chamber. The cells were cultured in the medium for 48 h at 37°C with 5% CO2. Then, a cotton swab was utilized to wipe away the remaining cells on the membrane. Cells migrated or invaded through the membrane were stained with methanol and 0.1% crystal violet. Cells were then imaged and counted using an IX71 inverted microscope (Olympus). Five random fields were counted in each well. The experiment was repeated three times in parallel.
Western Blot Assay and Antibodies
Radio immunoprecipitation assay (RIPA) protein lysis buffer (Solarbio) supplemented with protease inhibitor cocktail (Roche, Basel, Switzerland) and phenylmethylsulfonyl fluoride (Roche) was applied for protein extraction. The proteins concentration was evaluated through a BCA Protein Assay Kit (Solarbio). Ten percent sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was used to detach protein extracts (50 μg). The protein extracts were transferred to 0.22 mm polyvinylidene fluoride membranes (Sigma), then incubated with specific antibodies. ECL chromogenic substrate was applied to evaluate the specific bands. The specific bands were then quantified by densitometry (Quantity One software, Bio-Rad) with β-actin used as a control. All antibodies (1:2,000 dilutions) were purchased from Abcam (Cambridge, United Kingdom).
Luciferase Reporter Assay
The 3′-UTR of human FGFR2 or Lnc_ASNR was amplified from human genomic DNA and individually inserted into a pGL3-basic vector (Promega, Madison, WI, United States). The fragment of FGFR2 or lncRNA-ASNR 3′-UTR mutant was then inserted into the pGL3-basic vector (Promega) control vector at the same position. To perform the reporter experiments, transfect MKN45 cells with wild-type or mutant reporter plasmid and miR-519e-5p mimic by Lipofectamine 2000 (Invitrogen). After 36 h of transfection, the luciferase activities and the firefly luciferase vitalities were detected under Dual-Luciferase® Reporter Assay System (Promega). They were normalized based on Renilla luciferase activity.
Immunohistochemistry
Formalin and paraffin were used to fix and embed the isolated tumor tissues, respectively. The tissue examples were then cut into 5 mm thick. Immunochemical staining was performed following the standard protocol. Tumors from mice were immunostained for H&E, Ki67. Inverted microscope (Nikon, Tokyo, Japan) was applied to capture the staining intensities of Ki67, and ImageJ software was used to analyze the data. Anti-Ki67 (1:50) was purchased from Roche.
Xenograft Study
All experiments were approved by the Committee on the Ethics of Animal Experiments of Hebei Medical University. The investigations were conducted strictly on the basis of the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. MKN45 cells were stably infected with control shRNA or sh-ASNR using lentiviruses. GC cells were injected into either side of the armpit regions of the male BALB/c nude mice. The mice (4–5 weeks old) were randomly divided into two groups (n = 6/group). Tumor volumes and weights were measured every 3 days, and tumor volumes were measured regularly. Calculate as the equation: V = 0.5 × D × d2 (V, volume; D, longest diameter; d, diameter perpendicular to the longest diameter). After 16 days of treatment, mice were killed and tumors were excised. As mentioned earlier, H&E and IHC staining were performed on tumor tissues. The shRNA sequences used in our study were provided in the Supplementary Table 3.
Statistical Analysis
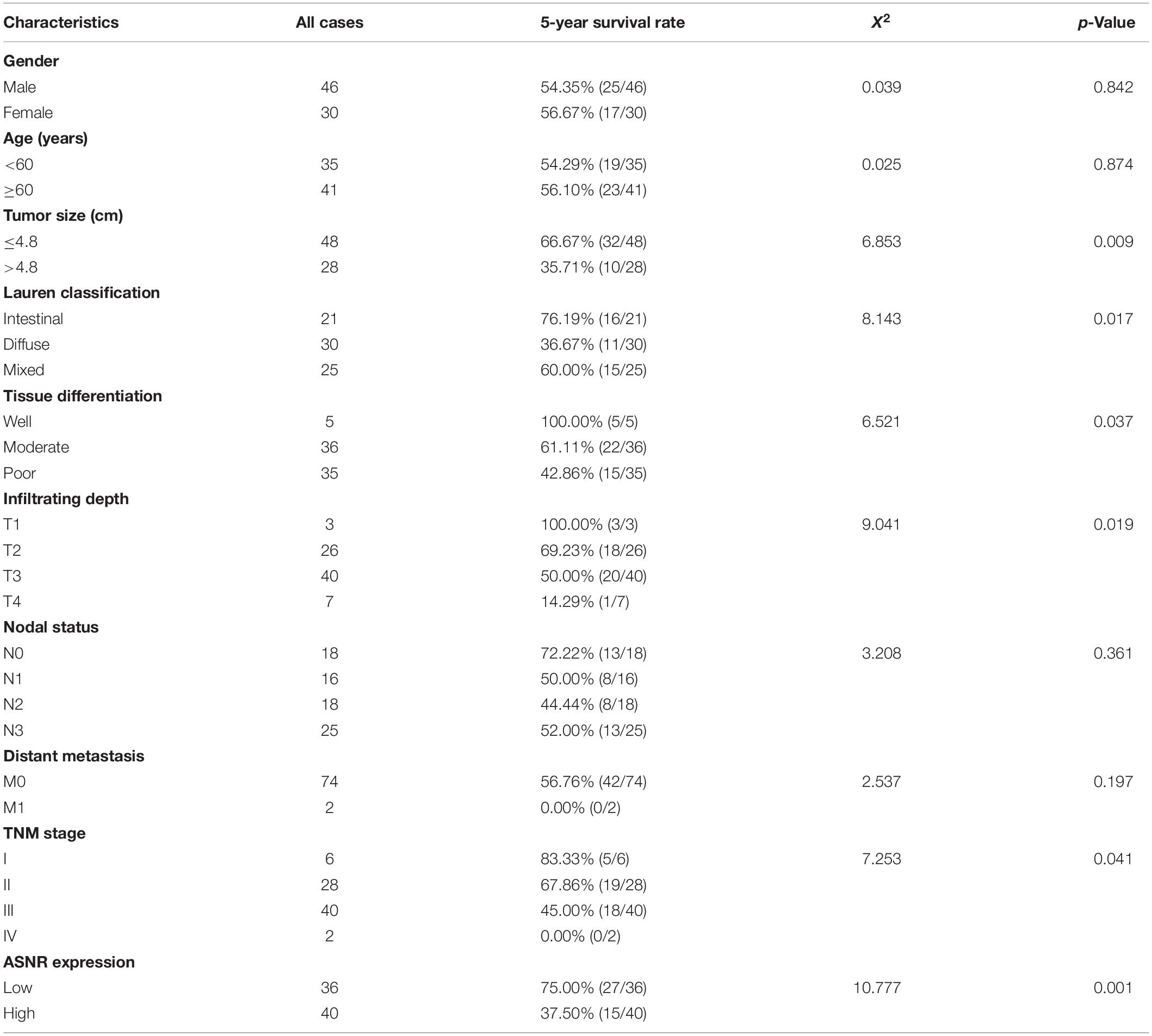
All experimental data analyses were carried out using Student’s t-test and one-way ANOVA (GraphPad Prism 8). All data were presented as mean ± SD. The clinical correlations between two genes were analyzed by linear regression. The interrelation between the expression of ASNR and the clinical features in GC was confirmed by Fisher’s exact test. Impact of variables on survival was evaluated by univariate and multivariate Cox proportional hazard models. Overall survival (OS) rates were determined via Kaplan–Meier method. P < 0.05 was regarded as statistical significance.
Results
Lnc_ASNR Is Highly Expressed in GC and High Level of Lnc_ASNR Predicts Poor Prognosis
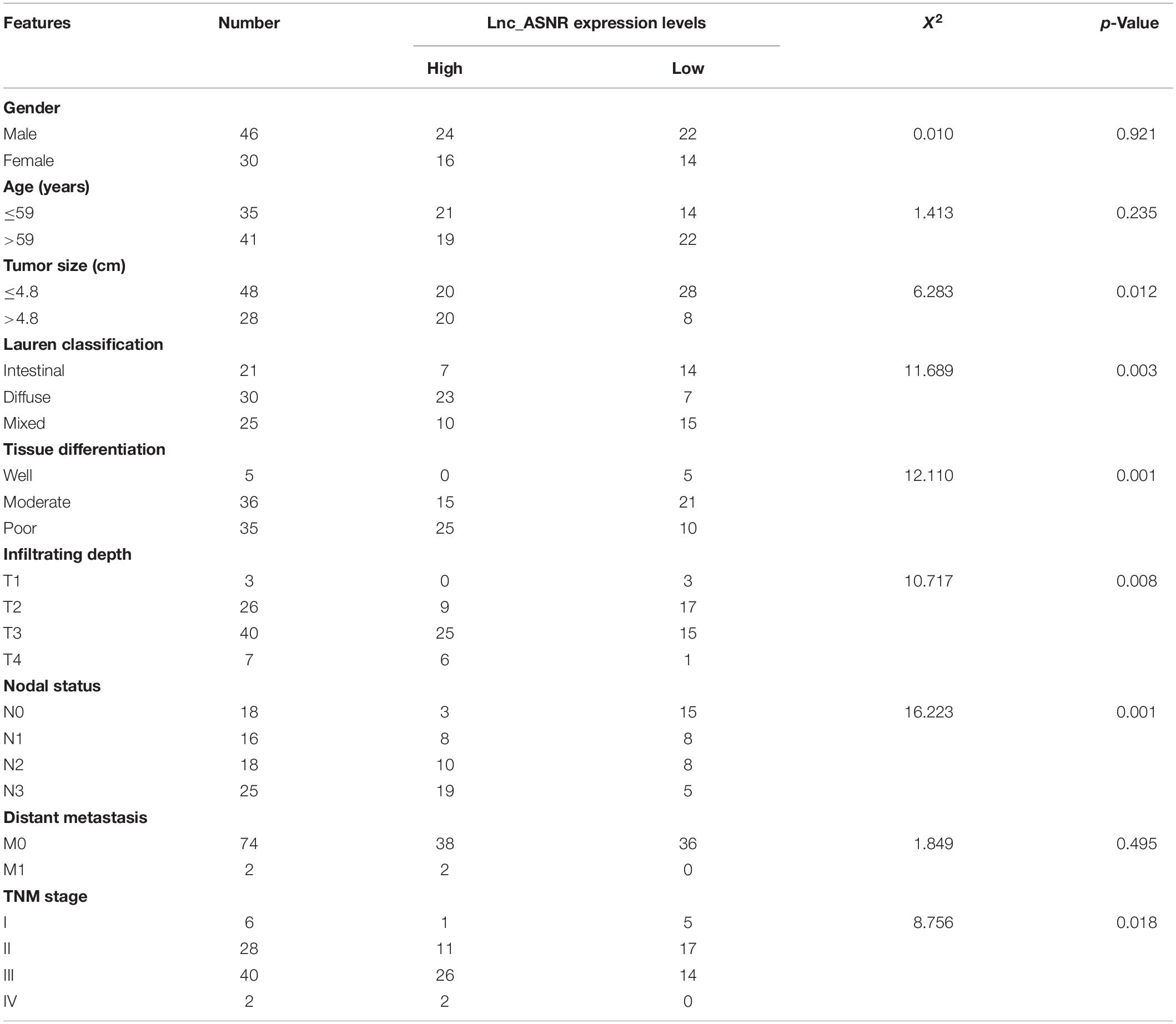
Lnc_ASNR was highly expressed in the GC tissues by analyzing their expression level in the TCGA datasets and GTEx datasets through using GEPIA2 online web tool1 (Figure 1A). To determine Lnc_ASNR expression in GC tissues, real-time PCR was carried out in GC tissues and adjacent normal tissues. The result exhibited that Lnc_ASNR expression was obviously increased in GC tissue (Figure 1B). Those results together indicated that Lnc_ASNR was highly expressed in GC tissue. In addition, we also measured Lnc_ASNR expression in GES-1, MKN28, AGS, MKN45, and HGC27 cells. As shown in Figure 1E, Lnc_ASNR expression in the GC cell lines was obviously higher than that in the GES-1 cell. Compared with the highly differentiated cell line MKN28, the Lnc_ASNR expression was higher in the lowly differentiated cell line MKN45 (Figure 1E).
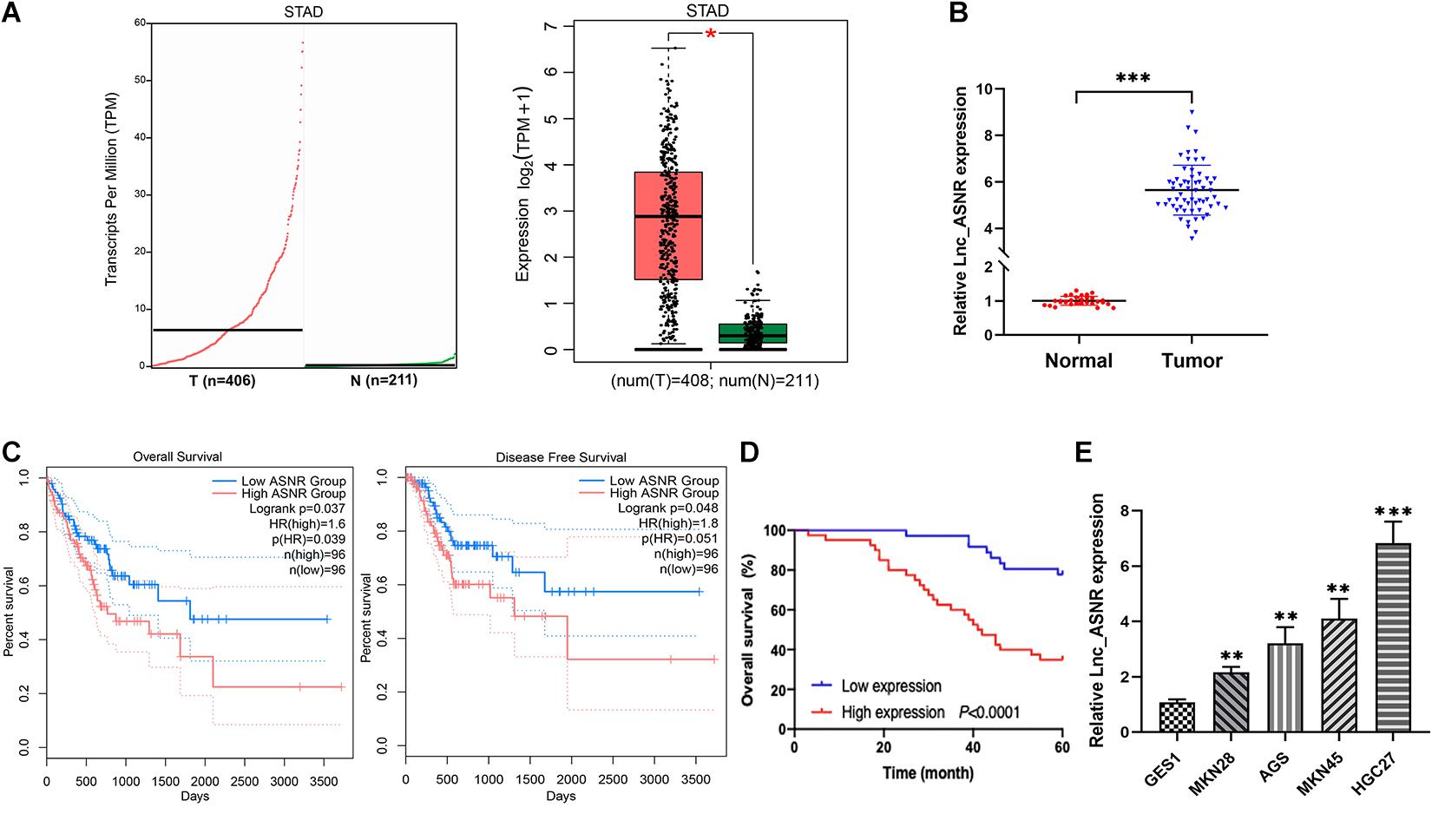
Figure 1. High expression of Lnc_ASNR in GC tissue samples and cell lines. (A) Lnc_ASNR expression was analyzed by using GEPIA2 in GC tissues and normal tissues. (B) qRT-PCR was applied to indicate the expression of Lnc_ASNR in GC tissue samples and adjacent non-tumor tissue samples (n = 76). (C) Overall survival and disease-free survival were analyzed using GEPIA2. (D) Kaplan–Meier analysis showed the relationship between Lnc_ASNR expression and overall survival time of GC patients. (E) qRT-PCR showed the expression of Lnc_ASNR in the normal cell line and GC cell lines. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
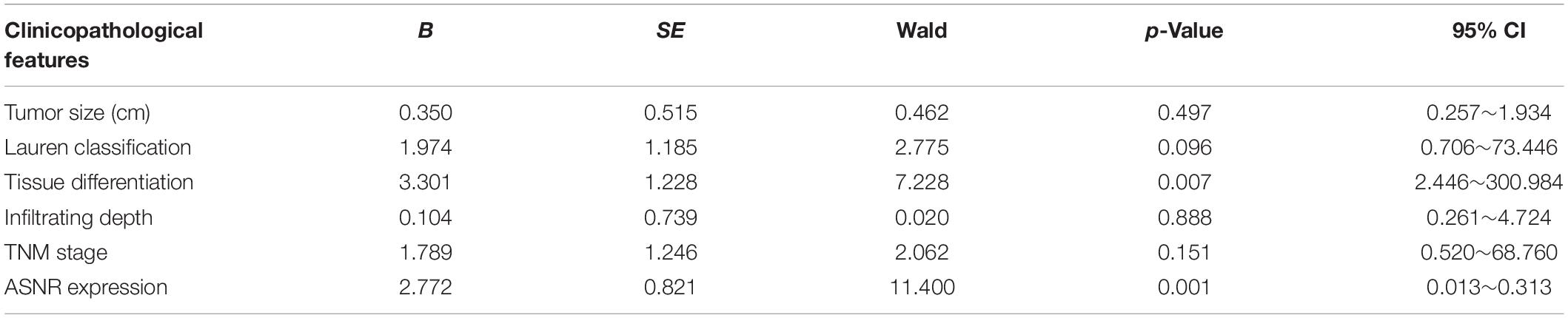
To evaluate the clinical significance of Lnc_ASNR overexpression in GC, we explored the expression of Lnc_ASNR in 76 patients with GC and evaluated their relationship with clinicopathological features. Median expression level was considered to be the key value. Tissues were grouped into low expression group with 36 samples and high expression group with 40 samples. As indicated in Table 1, high Lnc_ASNR levels was significantly associated with larger tumor size (p = 0.012), depth of invasion (p = 0.008), lymph node metastasis (p = 0.001), and TNM stage (p = 0.018). In contrast, no obvious association was found between Lnc_ASNR expression and other features including gender (p = 0.921) and age (p = 0.235). Correlation between Lnc_ASNR expression level and prognosis of GC patients was evaluated. According to the upper and lower quartiles of Lnc_ASNR expression, patients are divided into high-expression and low-expression groups. It was found that the expression of Lnc_ASNR affected the survival rate of the patient using GEPIA2 web tool. The overall survival (OS) and disease-free survival (DFS) of the low ASNR group were better than the high ASNR group (Figure 1C). It could be confirmed from Kaplan–Meier survival analysis that Lnc_ASNR highly expressed patients exhibited shorter OS, compared with Lnc_ASNR lowly expressed patients (Figure 1D). In addition, univariate survival analysis showed OS of patients were significantly correlated with that tumor size, infiltrating depth, Lauren classification, tissue differentiation, TNM stage, and Lnc_ASNR expression levels (Table 2). These factors were further evaluated by multifactor Cox regression analysis, indicating that Lnc_ASNR expression level and tissue differentiation are independent prognostic factors for patients (Table 3).
Knockdown of Lnc_ASNR Inhibits While ASNR Overexpression Promotes GC Cell Proliferation, Migration, and Invasion
We hypothesized that Lnc_ASNR may act as an oncogene, based on the increased expression of Lnc_ASNR in GC. Therefore, siRNA was used to downregulate the expression of Lnc_ASNR in MKN45 cells. qRT-PCR was used to verify knockdown efficiency of siRNA, and siRNA3 was chosen for subsequent experiments because of the highest knockdown efficiency (Figure 2A). Cell proliferation assay (MTT) results showed that silencing Lnc_ASNR inhibited the proliferation of GC cells (Figure 2B). It could be seen from the flow cytometry analysis that when Lnc_ASNR was silenced, the percentage of cells in G0/G1 phase would increase, while the percentage of cells in S phase would decrease (Figure 2D). The wound-healing assay and Transwell assay data suggested that compared with the control group, the si-Lnc_ASNR group significantly reduced the number of cells that migrated and invaded (Figure 2E).
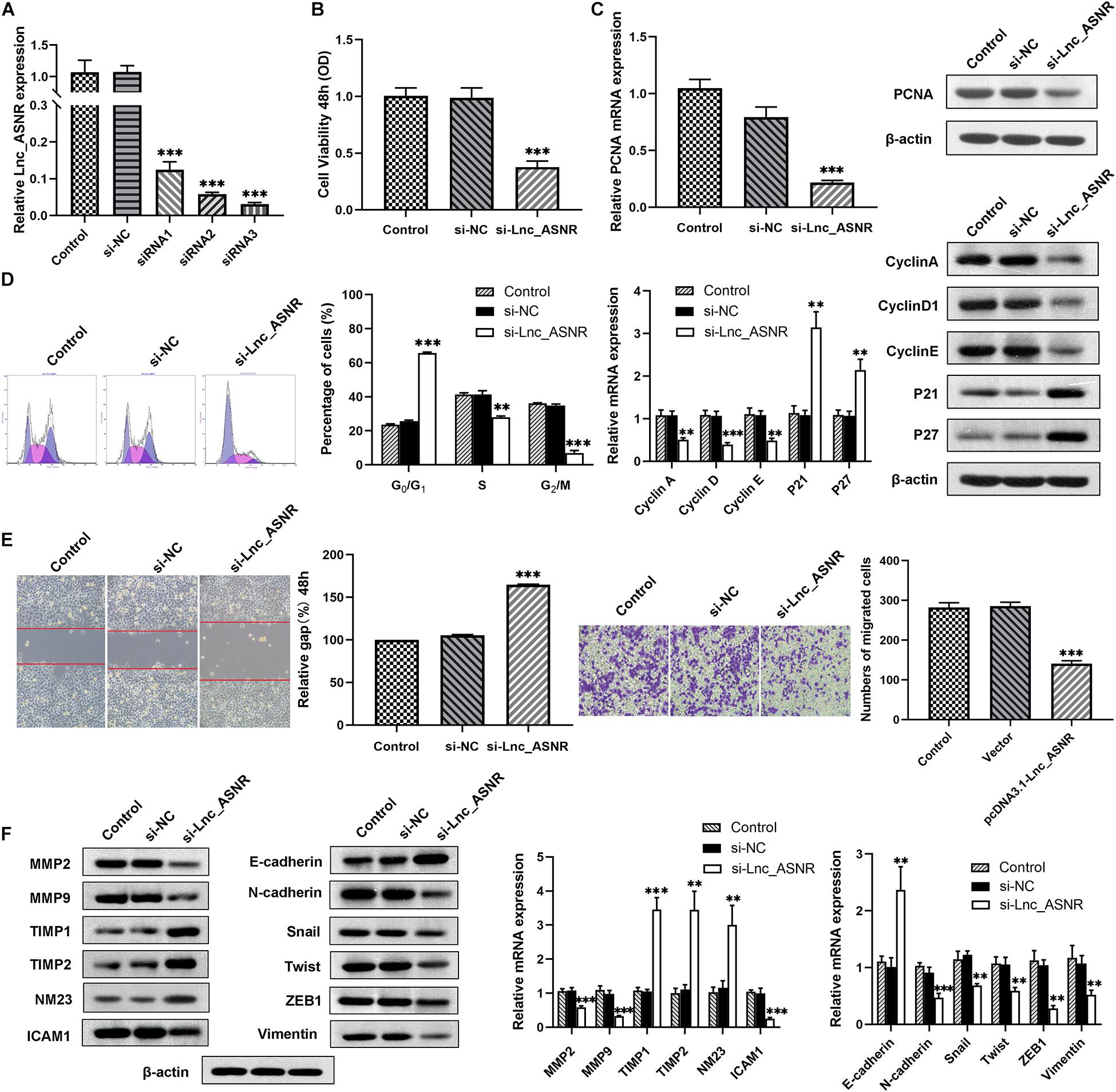
Figure 2. Silenced Lnc_ASNR inhibits GC cell proliferation, migration, and invasion. (A) qPCR showed the silence efficiency of Lnc_ASNR siRNAs. (B) MTT experiments detected the cell viability of MKN45 cells after treatment with Lnc_ASNR siRNA or control siRNAs. (C) qPCR and western blot assays showed PCNA mRNA and protein levels in si-Lnc_ASNR transfected MKN45 cells. (D) Flow cytometry was applied to verify cell cycle in MKN45 cell lines and qPCR and western blot assays were conducted to evaluate the cycle-related mRNA and protein levels. (E) The migration ability with Lnc_ASNR-silenced MKN45 cells was detected by wound-healing assay. The invasion ability was detected by invasion assay with Lnc_ASNR-silenced MKN45 cells. (F) The expression of metastasis-related mRNA and proteins with Lnc_ASNR-silenced MKN45 cells were investigated by performing qPCR and western blot assays. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To deeply verify whether Lnc_ASNR is related to GC, the expression of related mRNA and protein was evaluated by performing qRT-PCR and Western blot experiments. We found that both the mRNA and protein expression of PCNA, cell proliferation marker gene, was decreased (Figure 2C). Both mRNA and protein expression levels of cyclin A, cyclin D1, and cyclin E were decreased, while P21 and P27 expression levels were increased in the cells transfected with siRNA (Figure 2F). Furthermore, the mRNA and protein expression levels of MMP2, MMP9, and ICAM1, cell invasion- and migration-related gene, were decreased, while TIMP1, TIMP2, and NM23 were increased (Figure 2F).
Contrary to the results of silencing Lnc_ASNR, overexpression of Lnc_ASNR facilitated the proliferation, migration, and invasion of MKN28 cells (Figures 3A–F).
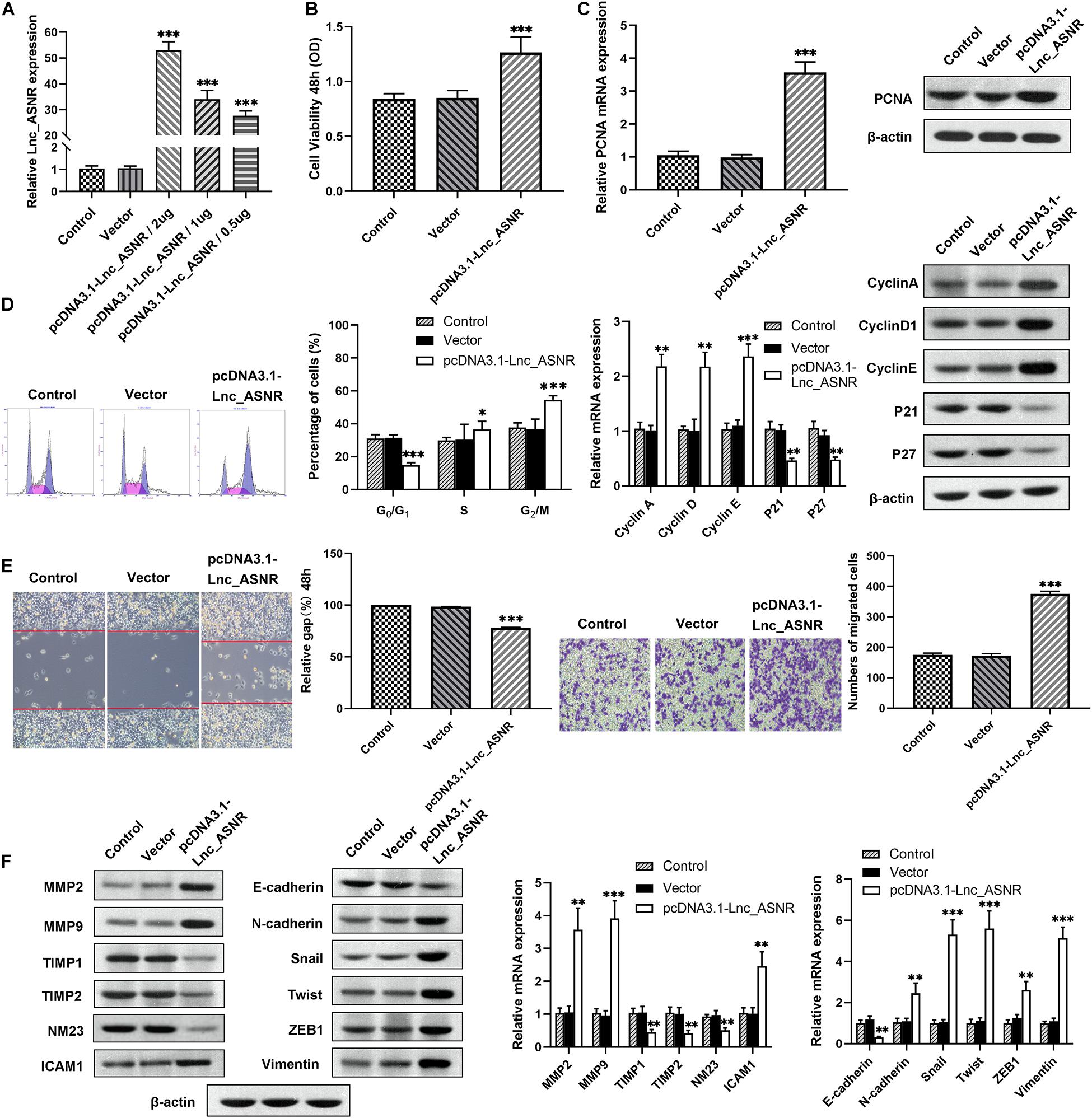
Figure 3. Lnc_ASNR overexpression promotes GC cell proliferation, migration, and invasion. (A) qPCR detected overexpress efficiency of pcDNA3.1-Lnc_ASNR. (B) MTT assays examined the cell viability of MKN28 cells after treatment with pcDNA3.1-Lnc_ASNR or vector. (C) PCNA mRNA and protein level in pcDNA3.1-Lnc_ASNR-transfected MKN28 cells were evaluated by carrying out qPCR and western blot assays. (D) Cell cycle was verified by using flow cytometry in MKN28 cell lines and cycle-related mRNA and protein level were evaluated by qPCR and western blot assays. (E) The migration ability with Lnc_ASNR-overexpressed MKN28 cells was detected by wound-healing assay. The invasion ability was detected by invasion assay with Lnc_ASNR-overexpressed MKN28 cells. (F) The expression of metastasis-related mRNA and proteins with Lnc_ASNR overexpressed MKN28 cells were investigated by performing qPCR and western blot assays. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Lnc_ASNR Influences GC Cell EMT
We found that cell migration and invasion are closely related to Lnc_ASNR levels. Since epithelial-mesenchymal transition (EMT) is a key step that contributes to tumor metastasis, we evaluated the effect of Lnc_ASNR on EMT. In order to further verify the abovementioned results, qRT-PCR and Western blot assay were conducted to evaluate the EMT-induced marker expression in cells downregulating or upregulating Lnc_ASNR. As shown in Figure 2F, high expressed epithelial markers E-cadherin, low expressed interstitial markers N-cadherin and vimentin were detected in the cells downregulated by Lnc_ASNR, indicating that the inhibition of Lnc_ASNR promoted the change of EMT to MET. Moreover, EMT transcription factor expression, including Snail, Twist, and ZEB1, was significantly downregulated (Figure 2F) in si-Lnc_ASNR-transfected cells. However, the results observed in the cells transfected with pcDNA3.1-Lnc_ASNR were just the opposite (Figure 3F). These results are consistent with our hypothesis that Lnc_ASNR affects the malignant phenotype by regulating EMT.
Lnc_ASNR Promotes GC Cell Tumorigenesis in vivo
In order to provide further evidence for the above findings, stably expressed sh-Lnc_ASNR or sh-NC AGS cells were constructed by infecting cells with shRNA vectors. Then, in order to determine the correlation between Lnc_ASNR and tumorigenesis of GC cells in vivo, AGS cells with stable knockdown of Lnc_ASNR were injected into nude mice. In contrary to the tumor formed by the control cells, the tumor formed by the Lnc_ASNR-silenced cells has taken significantly slower time (Figure 4A). In addition, compared with the control group, tumor volume of the sh-Lnc_ASNR group was significantly decreased (Figures 4B,C). We also found that the protein expression of cell proliferation marker gene, PCNA, and cell invasion and migration-related gene, MMP2, MMP9, and ICAM1, were decreased, while NM23 was increased in the sh-Lnc_ASNR group (Figure 4D). The above results demonstrated that the downregulated Lnc_ASNR could suppress tumor development in vivo. Furthermore, the results of H&E staining indicated that the sh-Lnc_ASNR group significantly suppressed tumorigenesis. Additionally, immunohistochemical staining of tumor tissues demonstrated that contrary to the sh-NC group, a decrease in Ki-67 was detected in the sh-Lnc_ASNR group (Figure 4E).
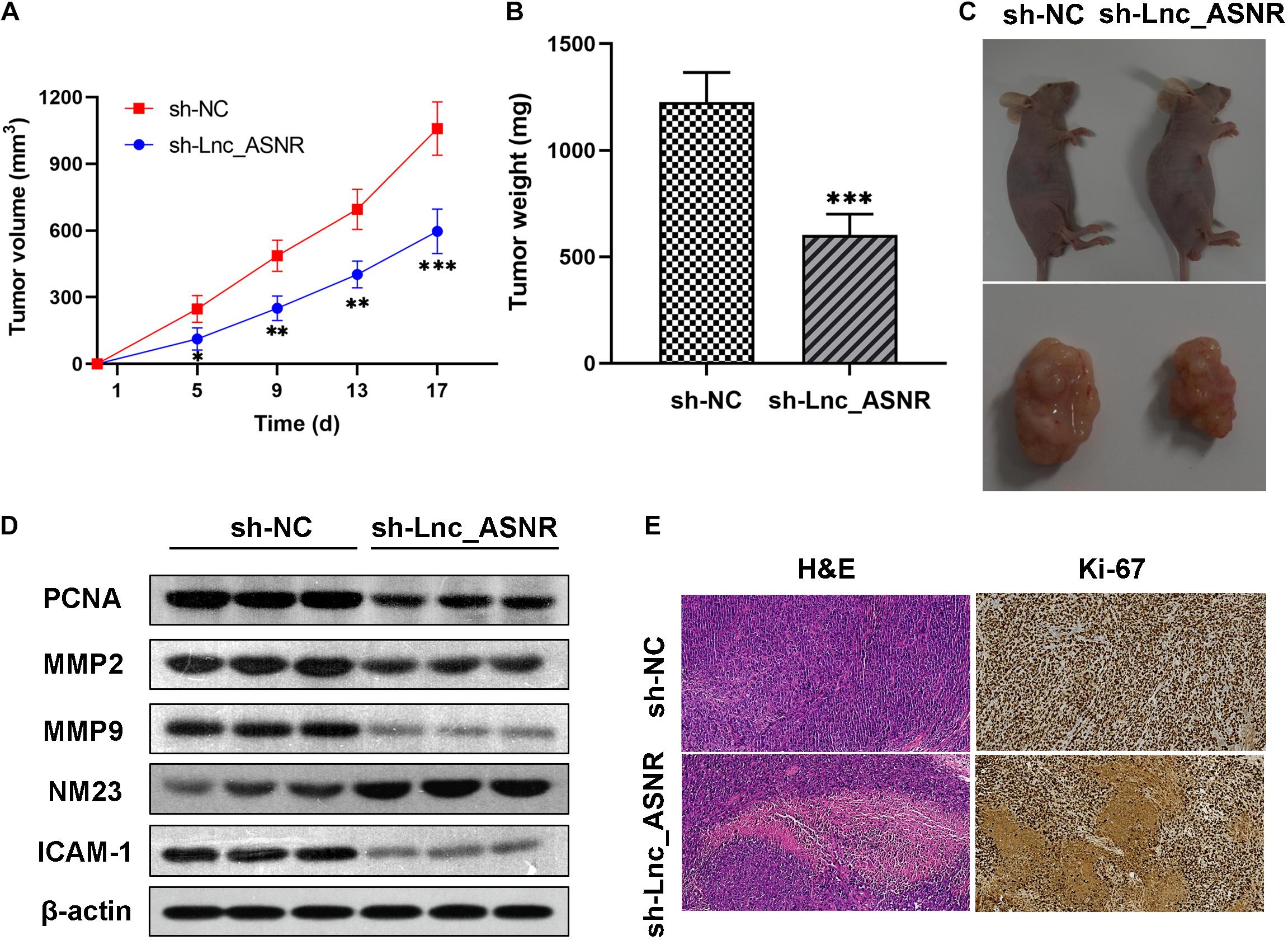
Figure 4. Silenced Lnc_ASNR suppresses the tumorigenesis ability in vivo. (A) The sizes of tumors were measured, and the volume was calculated every 4 days. (B) Tumor weights in different groups were shown. (C) Images of tumors formed from AGS cells in different groups were represented. (D) Related protein expression levels in tumor tissues were investigated via western blot assays. (E) Images of H&E staining and Ki67 immunostaining for nude mice tumor tissues in different groups. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
In vivo investigations above complement the evidence of in vitro functional studies involving Lnc_ASNR.
Lnc_ASNR Functions as ceRNA and Sponges miR-519e-5p in GC Cells
In addition to regulating the target in epigenetics, some lncRNAs can also act as competing endogenous RNA (ceRNA) of specific miRNA to affect the target gene expression. We predicted the potential target of Lnc_ASNR by searching the biological information database miRDB. We examined the Lnc_ASNR sequence and found the miR-519e-5p-binding site and identified miR-519e-5p as a candidate binding miRNA (Figure 5A). In order to detect the miR-519e-5p expression in GC tissues, real-time PCR was performed in GC tissues and matched adjacent normal tissues. It was clearly indicated that the expression of miR-519e-5p was markedly downregulated in GC samples (Figure 5B). Besides, Lnc_ASNR expression was negatively correlated with that of miR-519e-5p in GC samples (Figure 5C). We attempted to analyze the regulation of miR-519e-5p by Lnc_ASNR. Highly differentiated MKN28 cells were transfected pcDNA3.1(+) vector carrying the coding sequence of Lnc_ASNR to overexpress Lnc_ASNR, and downregulation of miR-519e-5p expression was detected by qPCR (Figure 5D). Whereas, lowly differentiated MKN45 cells were transfected with si-Lnc_ASNR to silence Lnc_ASNR and upregulated miR-519e-5p expression was detected by qPCR (Figure 5E). To further verify the targeting relationships between miR-519e-5p and Lnc_ASNR, luciferase reporter experiment was performed in MKN45 cells. As represented in Figure 5F, miR-519e-5p mimic apparently decreased the activity of luciferase in Lnc_ASNR-wild, while no significant change was detected in Lnc_ASNR-mut.
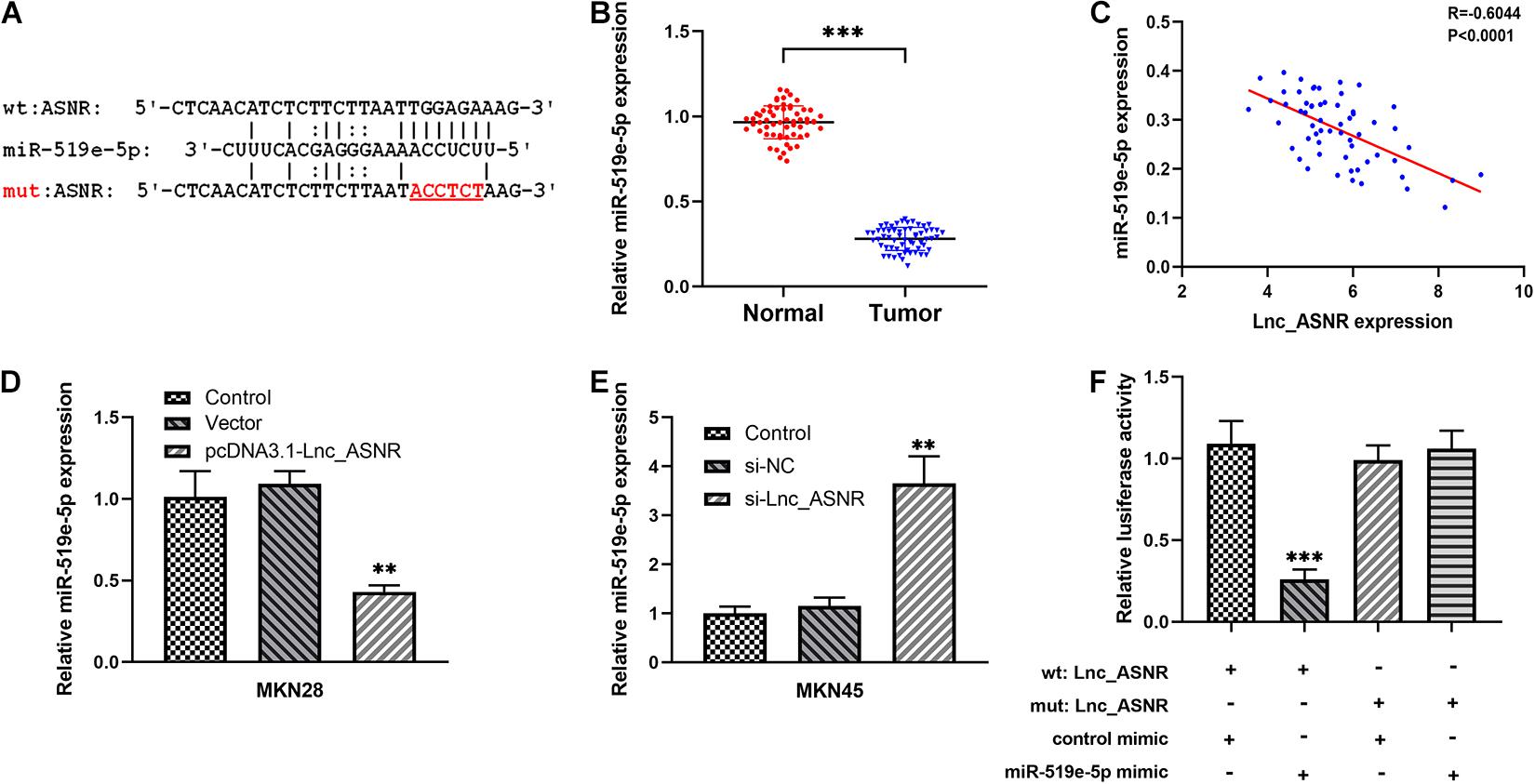
Figure 5. The relationship between Lnc_ASNR and miR-519e-5p. (A) The predicted combining positions between Lnc_ASNR and miR-519e-5p were predicted by using “miRDB” website (http://mirdb.org/). (B) qRT-PCR was applied to indicate the expression of miR-519e-5p in GC tissue samples and adjacent non-tumor tissue samples. (C) Correlation analysis was used to evaluate the relationship between miR-519e-5p expression and Lnc_ASNR expression. (D) qPCR detected the levels of miR-519e-5p in GC cells after Lnc_ASNR was overexpressed. (E) qPCR detected the levels of miR-519e-5p in GC cells after Lnc_ASNR was silenced. (F) Luciferase activity detection. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Therefore, these data suggested that Lnc_ASNR acts as a miR-519e-5p sponge in GC.
FGFR2 Is a Downstream Target of miR-519e-5p
Online miRNA target prediction databases (including Targetscan and miRDB) were applied to predict potential target genes of miR-519e-5p, and FGFR2 was determined as a potential downstream gene of miR-519e-5p (Figure 6A). Real-time PCR was carried out in GC tissues and matched adjacent normal tissues to evaluate the FGFR2 expression. An obvious upregulation of FGFR2 expression was detected in GC tissue samples (Figure 6B). Besides, FGFR2 expression was positively correlated with the expression of Lnc_ASNR (Figure 6C) and negatively related to miR-519e-5p expression in GC tissue samples (Figure 6D). In addition, FGFR2 expression in GES-1, MKN28, AGS, MKN45, and HGC27 cells was also detected. It was clearly indicated from Figure 6E that FGFR2 expression was markedly higher in GC cell lines than that of control cells.
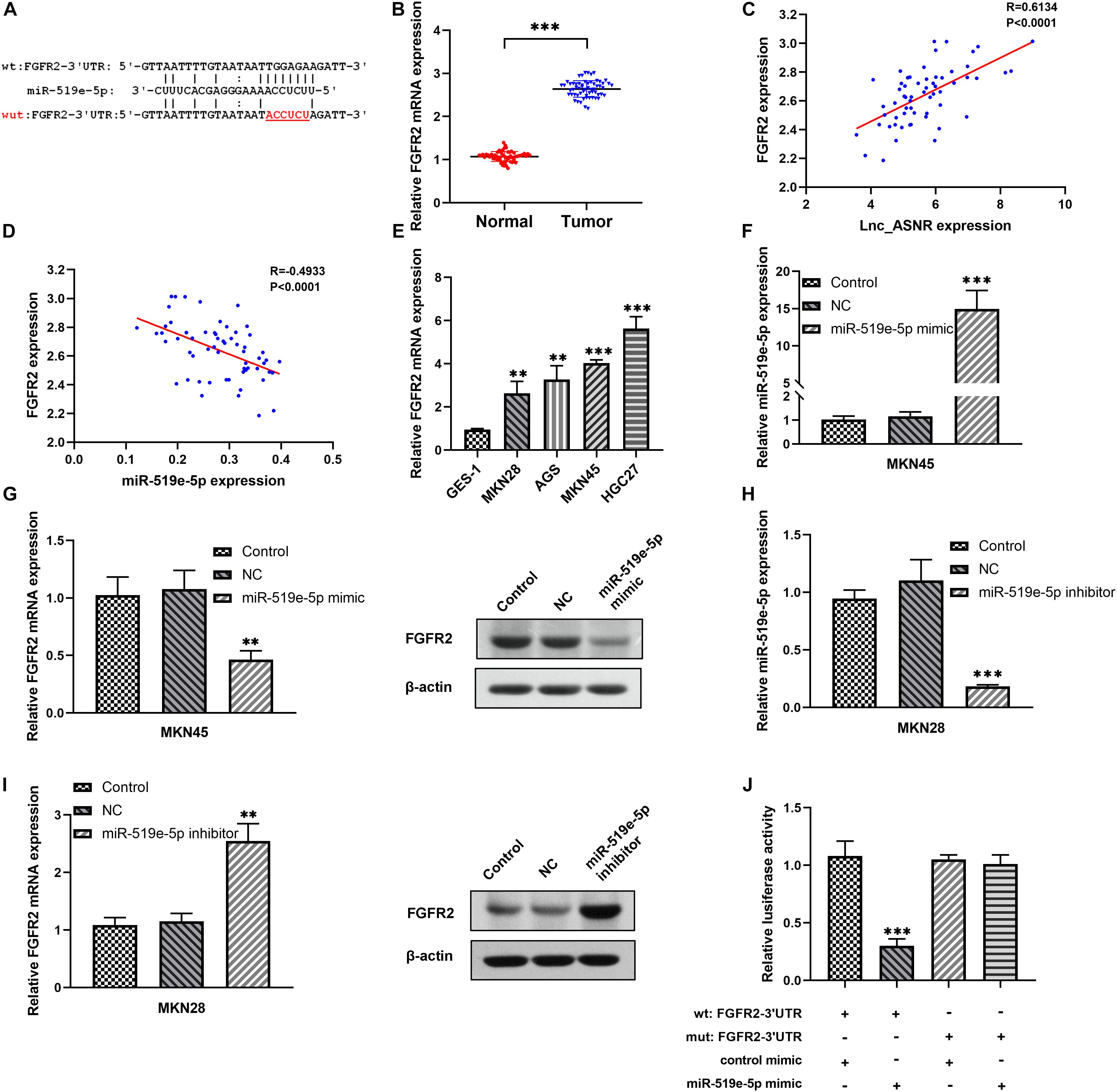
Figure 6. The relationship between FGFR2 and miR-519e-5p in GC cells. (A) The potential binding sites between miR-519e-5p and FGFR2 were predicted from “Targetscan” website (http://www.targetscan.org/) and “miRDB” website (http://mirdb.org/). (B) qRT-PCR was applied to indicate the expression of FGFR2 in GC tissue samples and adjacent non-tumor tissue samples. (C) Correlation analysis was used to evaluate the relationship between FGFR2 expression and Lnc_ASNR expression. (D) Correlation analysis was used to evaluate the relationship between FGFR2 expression and miR-519e-5p expression. (E) qRT-PCR showed the expression of FGFR2 in the normal cell line and GC cell lines. (F) qPCR detected the overexpress efficiency of miR-519e-5p mimic. (G) Relative FGFR2 expression and protein levels in GC cells after miR-519e-5p was overexpressed. (H) qPCR detected the silence efficiency of miR-519e-5p inhibitor. (I) Relative FGFR2 expression and protein levels in GC cells after miR-519e-5p was silence. (J) Luciferase activity examination. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, low differentiated MKN45 cells were transfected with miR-519e-5p mimic, and qRT-PCR was used to verify that miR-519e-5p expression was markedly increased (Figure 6F). In addition, the FGFR2 expression level was evaluated by qRT-PCR and Western blot experiments, and the data demonstrated that the FGFR2 expression level was markedly downregulated (Figure 6G). On the contrary, miR-519e-5p inhibitor was used to transfect highly differentiated MKN28 cells, and corresponding results demonstrated that the expression of miR-519e-5p was markedly decreased (Figure 6H). Moreover, FGFR2 expression was investigated by conducting qRT-PCR and Western blot experiments, and the results suggested that FGFR2 expression was obviously increased (Figure 6I). To further evaluate the targeting correlation between miR-519e-5p and FGFR2, luciferase reporter experiment was performed in the MKN45 cells. As exhibited in Figure 6J, miR-519e-5p mimic markedly suppressed the vitality of luciferase in FGFR2-wild, while no significant change was detected in FGFR2-mut.
Therefore, these data indicated that FGFR2 is an important downstream target of miR-519e-5p.
The Lnc_ASNR/miR-519e-5p/FGFR2 Axis Promotes Proliferation, Invasion, and Migration of GC
Mechanism research was conducted to clarify the relationship between Lnc_ASNR and FGFR2. Lnc_ASNR was silenced in lowly differentiated MKN45 cells. qRT-PCR and western blot assay inhibited that decreased Lnc_ASNR reduced the level of FGFR2 protein, which is consistent with the miR-519e-5p-induced FGFR2 protein downregulation. When transfected with si-ANSR + miR-519e-5p inhibitor in MKN45 cells, it was found that FGFR2 expression was restored (Figure 7A). In contrast, upregulated FGFR2 was detected in highly differentiated MKN28 cells who overexpressed Lnc_ASNR. When pcDNA3.1-ASNR + miR-519e-5p mimic was transfected in MKN45 cells, it was found that FGFR2 expression was reversed (Figure 7B). The effect of Lnc_ASNR/miR-519e-5p axis on GC was then evaluated. MKN28 cells were transfected with vectors, pcDNA3.1-ASNR, and pcDNA3.1-ASNR + miR-519e-5p minic, respectively. MKN45 cells were transfected with si-NC, si-ASNR, and si-ASNR + miR-519e-5p inhibitors, respectively. As shown in Figures 7C,D, miR-519e-5p mimics could counteract the enhancement effect of overexpression of Lnc_ASNR on cell migration and invasion ability, while miR-519e-5p inhibitor could reverse the inhibitory effect of knockdown of Lnc_ASNR on cell migration and invasion ability.
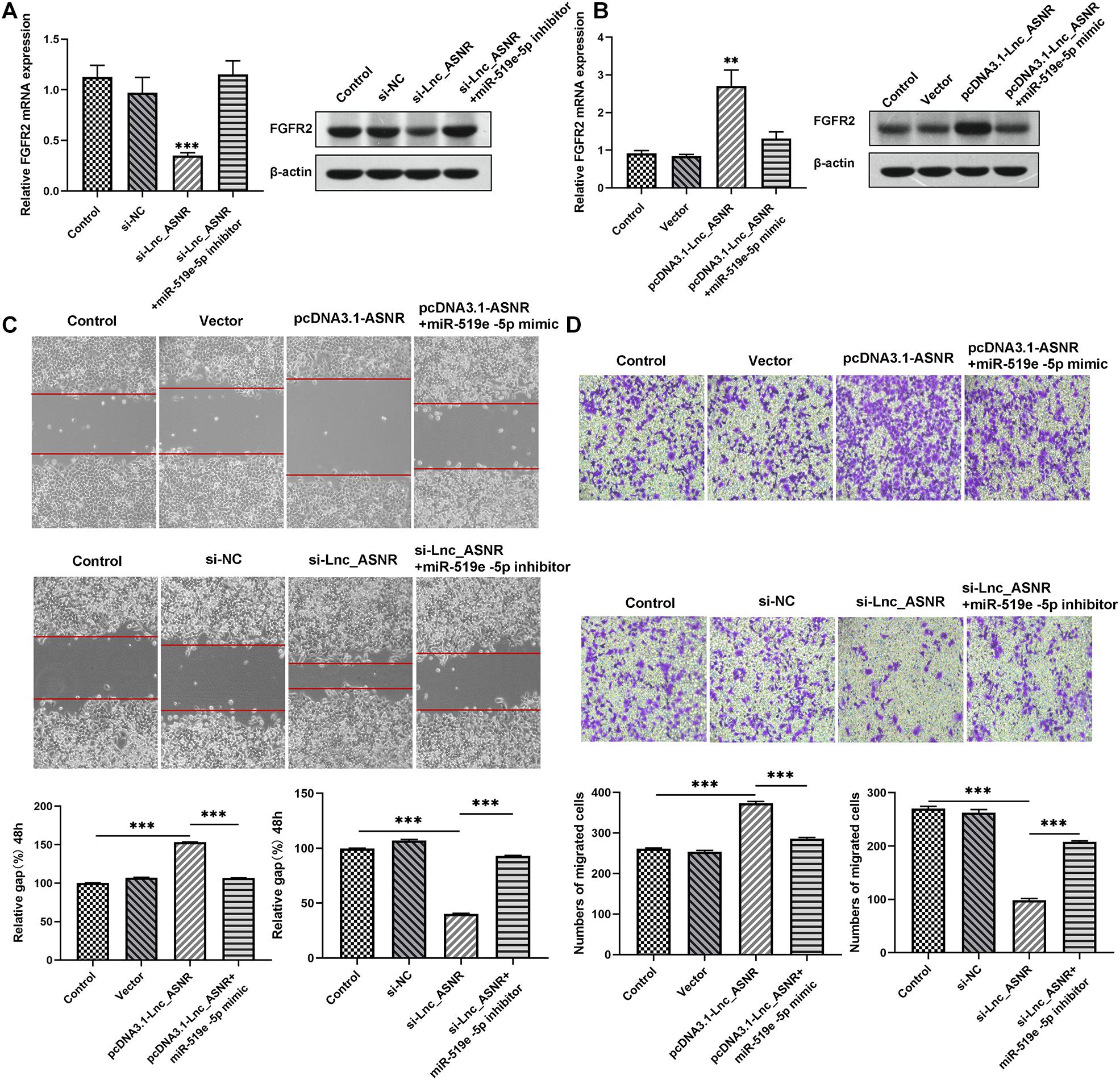
Figure 7. Lnc_ASNR/miR-519e-5p axis regulates GC progression. (A) The effects of Lnc_ASNR inhibition on the expression of FGFR2. (B) The effects of Lnc_ASNR overexpression on the expression of FGFR2. (C) The rescue experiments about si-Lnc_ASNR and pcDNA3.1-ASNR for cell migration. (D) The rescue experiments about si-Lnc_ASNR and pcDNA3.1-ASNR for cell invasion. Error bars, mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
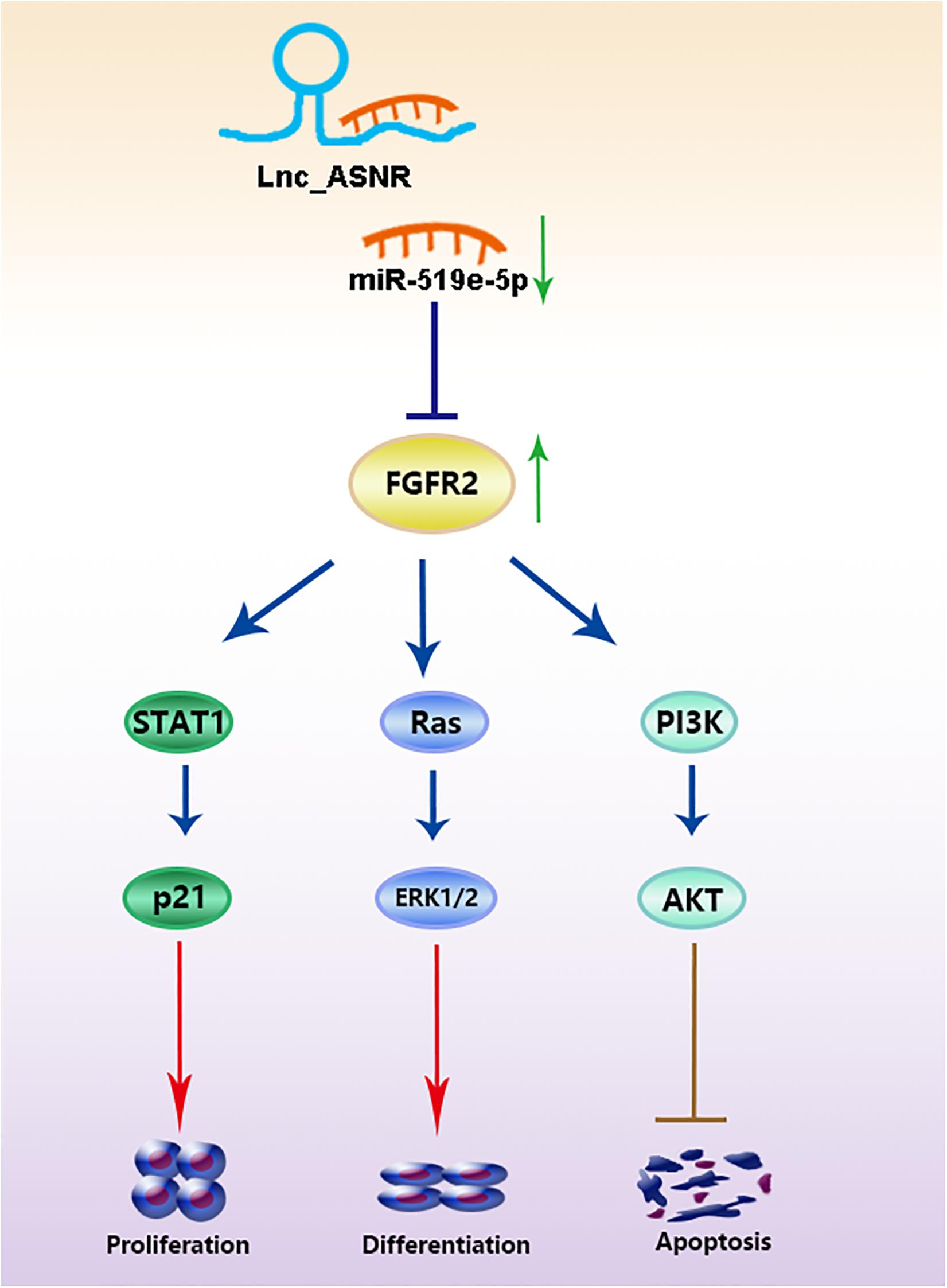
Ultimately, correlation analysis in GC showed that Lnc_ASNR expression and miR-519e-5p expression was negatively related, while Lnc_ASNR expression and FGFR2 expression was positively related. Overall, the above results demonstrated that Lnc_ASNR acted as a ceRNA for miR-519e-5p, thereby reducing FGFR2 expression and imposing posttranscriptional regulatory levels (Figure 8).
Discussion
Generally, explorations for cancer drivers are focused on protein-coding genes in cancer genomes before the discovery of non-coding RNAs. However, increasing research results verified that lncRNAs are crucial in cellular progression and diseases development (Han et al., 2019; Pradas-Juni et al., 2020), especially in cancer (Papaioannou et al., 2019; Schulte et al., 2020; Silva-Fisher et al., 2020). Abnormal expression of lncRNAs may lead to accelerated tumor growth (Geng et al., 2018; Lin and Yang, 2018; Sallam et al., 2018). The therapeutic methods available to advanced GC patients are limited, and there remains potential to improve the treatment effect. It is essential to investigate thoroughly the internal mechanisms of GC occurrence and development and explore novel prognostic markers. Large amounts of lncRNAs are verified to play crucial roles in GC development. Increasing researchers pay more attention to investigate the functions and regulation of lncRNAs to explore novel predictors for the diagnosis and therapeutics of GC (Fu et al., 2020; Qin et al., 2020; Ren et al., 2020). Herein, a GC-associated lncRNA ASNR was identified, which was markedly increased in GC cells and tissues. High Lnc_ASNR expression was related to advanced TNM staging, larger tumor size, and positive lymph node metastasis. In addition, the upregulated expression of Lnc_ASNR was related to the shortening of OS and DFS in GC patients. The above investigations showed that knocking down Lnc_ASNR suppressed cell proliferation, declined tumor growth, and facilitated cell apoptosis, while upregulated Lnc_ASNR accelerated cell proliferation, invasion, and metastasis. These functional evidences indicate that Lnc_ASNR plays a carcinogenic role in the occurrence of GC, which can be used as a potential prognostic indicator.
Increasing evidences indicate that there is a novel and extensive interaction network involving ceRNA. It was reported that ncRNAs could regulate target RNA through combining their linking position of protein coding messengers (Wang P. et al., 2020; Wang W. et al., 2020; Zhao et al., 2020). Emerging researches confirm the ceRNA hypothesis and indicate that ceRNA regulation is related to the carcinogenic effects of GC. For example, LncRNA MT1JP can suppress cell growth, migration, and invasion, promote cell apoptosis in vitro, and inhibit tumor growth and metastasis in vivo (Zhang et al., 2018). In our previous investigations, we suggested that lncRNA PWRN1 acts as a ceRNA-targeting miR-425-5p and inhibits the growth of GC by intervening p53 signaling pathway (Chen et al., 2018). Thus, we assumed that lncRNA may play the role of ceRNA and participate in occurrence of GC. It was confirmed by bioinformatics analysis and luciferase reporter gene analysis that miR-519e-5p was targeted by Lnc_ASNR. Recently, miR-519e-5p has only been reported in acute myocardial infarction (Wang F. et al., 2014) and pregnancy-related complications (Hromadnikova et al., 2015; Zhang et al., 2016), while few investigations on the relationship between miR-519e-5p and GC were reported. In our research, we confirmed that the endogenous level of Lnc_ASNR could influence the miR-519e-5p expression. The miR-519e-5p expression was increased in GC cells when Lnc_ASNR was silenced. Contrary to the results of silencing Lnc_ASNR, miR-519e-5p expression was decreased upon Lnc_ASNR overexpression, which confirmed our hypothesis. Above investigations revealed the importance of the correlation between Lnc_ASNR with miR-519e-5p in GC, which was contributed to that Lnc_ASNR exhibited a carcinogenic effect through causing miR-519e-5p to produce sponges in GC cells.
In general, lncRNAs exhibit function by relying on ceRNA to inhibit miRNA targets (Shen et al., 2018; Chen et al., 2019; Yang et al., 2019). Therefore, miRNA targets are important parts of ceRNA network. It is worth to note that FGFR2 is a potential miR-519e-5p target site which has never been reported via online-predicting database. In order to verify that miR-519e-5p was directly targeted by FGFR2, we carried out luciferase reporter experiments and confirmed that FGFR2 was targeted by miR-519e-5p at 3′-UTR. In addition, transfection of miR-519e-5p mimic inhibits FGFR2 mRNA and protein expression, while transfection of miR-519e-5p inhibitor promotes FGFR2 mRNA and protein expression. The protein encoded by the FGFR2 gene is one of the members of the fibroblast growth factor receptor (FGFR) family (Wu et al., 2013). At present, four types of FGFRs have been identified, namely FGFR1, FGFR2, FGFR3, and FGFR4. The main function of FGFR, a receptor for FGF, is to transduce FGF signal to RAS-ERK and PI3K-AKT signal cascade amplification. The missense mutations of the FGFR2 gene occur in endometrial cancer, cervical cancer, breast cancer, lung cancer, and GC. The amplified FGFR2 induces the activation of FGFR2 signaling. FGFR2 has been proposed as a target for targeted therapy of GC (Pearson et al., 2016; Kuboki et al., 2018; Kim et al., 2019). Yu et al. (2018) revealed that overexpressed FGFR2 accelerated the production of cancer-initiating cells (CIC) and enhanced the resistance to lapatinib in HER2-positive GC cells. We further determined that miR-519e-5p could reverse the effect of Lnc_ASNR on FGFR2 expression level, indicating that Lnc_ANSR acts on miR-519e-5p through sponge adsorption, which further affects the expression of downstream target gene FGFR2, thereby affecting cell proliferation, invasion, and migration activity.
In summary, our findings validated a novel lncRNA, called Lnc_ASNR, and found that Lnc_ASNR expression was related to poor prognosis in GC. The study found that Lnc_ASNR was a carcinogenic lncRNA exhibiting a targeting relationship with miR-519e-5p. GC patients with higher expression of Lnc_ASNR promoted tumor growth by regulating the miR-519e-5p/FGFR2 pathway. The current study reveals a further insight into the function of lncRNA–miRNA–mRNA–ceRNA network in GC progression. The promoter function of Lnc_ASNR in the GC progress was first reported. The potential molecular mechanism involved in the Lnc_ASNR/miR-519e-5p/FGFR2 axis could be a potentially useful approach for GC diagnosis and therapies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Research Ethics Committee at the Fourth Hospital of Hebei Medical University. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
ZC and YLo planned, designed, and coordinated this research and drafted the manuscript. BT and QZ participated in the conception and design of the manuscript and revised it critically for important intellectual content. FL and LF contributed to the conception and design and helped in drafting the manuscript. ZZ, XZ, and YLu contributed to analyzing and interpreting data and improved the crucial parts. DW helped in writing a part of the manuscript and analyzed the data. All the authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the members in our laboratory and department and all the patients and their families involved in the current study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.679176/full#supplementary-material
Abbreviations
GC, gastric cancer; lncRNAs, long non-coding RNAs; qRT-PCR, quantitative real-time PCR; MTT, 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide; EMT, epithelial-mesenchymal transition; FGFR, fibroblast growth factor receptor; nt, nucleotides; ceRNA, competitive endogenous RNA; TNM, tumor-node-metastasis; si-NC, negative control siRNA; GFP, green fluorescent protein; RIPA, radio immunoprecipitation assay; OS, overall survival; CIC, cancer-initiating cells.
Footnotes
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Chen, J., Liu, L., Wei, G., Wu, W., Luo, H., Yuan, J., et al. (2016). The long noncoding RNA ASNR regulates degradation of Bcl-2 mRNA through its interaction with AUF1. Sci. Rep. 6:32189. doi: 10.1038/srep32189
Chen, J., Yu, Y., Li, H., Hu, Q., Chen, X., He, Y., et al. (2019). Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 18:33. doi: 10.1186/s12943-019-0947-9
Chen, Z., Ju, H., Yu, S., Zhao, T., Jing, X., Li, P., et al. (2018). Prader-Willi region non-protein coding RNA 1 suppressed gastric cancer growth as a competing endogenous RNA of miR-425-5p. Clin. Sci. 132, 1003–1019. doi: 10.1042/CS20171588
den Hollander, W. J., Holster, I. L., den Hoed, C. M., Capelle, L. G., Tang, T. J., Anten, M. P., et al. (2019). Surveillance of premalignant gastric lesions: a multicentre prospective cohort study from low incidence regions. Gut 68, 585–593. doi: 10.1136/gutjnl-2017-314498
Fu, T., Ji, K., Jin, L., Zhang, J., Wu, X., Ji, X., et al. (2020). ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-kappaB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer 24, 45–59. doi: 10.1007/s10120-020-01096-y
Geng, H., Bu, H. F., Liu, F., Wu, L., Pfeifer, K., Chou, P. M., et al. (2018). In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology 155, 144–155. doi: 10.1053/j.gastro.2018.03.058
Guo, X., Lv, X., Ru, Y., Zhou, F., Wang, N., Xi, H., et al. (2020). Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: a multiphase study. JAMA Surg. 155, 572–579. doi: 10.1001/jamasurg.2020.1133
Han, X., Huang, S., Xue, P., Fu, J., Liu, L., Zhang, C., et al. (2019). LncRNA PTPRE-AS1 modulates M2 macrophage activation and inflammatory diseases by epigenetic promotion of PTPRE. Sci. Adv. 5:eaax9230. doi: 10.1126/sciadv.aax9230
Hromadnikova, I., Kotlabova, K., Ondrackova, M., Pirkova, P., Kestlerova, A., Novotna, V., et al. (2015). Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 34, 437–457. doi: 10.1089/dna.2014.2687
Huang, Y., Xu, Y., Lu, Y., Zhu, S., Guo, Y., Sun, C., et al. (2019). lncRNA Gm10451 regulates PTIP to facilitate iPSCs-derived beta-like cell differentiation by targeting miR-338-3p as a ceRNA. Biomaterials 216:119266. doi: 10.1016/j.biomaterials.2019.119266
Kim, H. S., Kim, J. H., and Jang, H. J. (2019). Pathologic and prognostic impacts of FGFR2 amplification in gastric cancer: a meta-analysis and systemic review. J. Cancer 10, 2560–2567. doi: 10.7150/jca.29184
Kim, J., Piao, H. L., Kim, B. J., Yao, F., Han, Z., Wang, Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715. doi: 10.1038/s41588-018-0252-3
Kuboki, Y., Schatz, C. A., Koechert, K., Schubert, S., Feng, J., Wittemer-Rump, S., et al. (2018). In situ analysis of FGFR2 mRNA and comparison with FGFR2 gene copy number by dual-color in situ hybridization in a large cohort of gastric cancer patients. Gastric Cancer 21, 401–412. doi: 10.1007/s10120-017-0758-x
Lin, C., and Yang, L. (2018). Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 28, 287–301. doi: 10.1016/j.tcb.2017.11.008
Montes, M., Lubas, M., Arendrup, F. S., Mentz, B., Rohatgi, N., Tumas, S., et al. (2021). The long non-coding RNA MIR31HG regulates the senescence associated secretory phenotype. Nat. Commun. 12:2459. doi: 10.1038/s41467-021-22746-4
Olivero, C. E., Martinez-Terroba, E., Zimmer, J., Liao, C., Tesfaye, E., Hooshdaran, N., et al. (2020). p53 activates the long noncoding RNA Pvt1b to inhibit Myc and suppress tumorigenesis. Mol. Cell 77, 761–774.e8. doi: 10.1016/j.molcel.2019.12.014
Papaioannou, D., Petri, A., Dovey, O. M., Terreri, S., Wang, E., Collins, F. A., et al. (2019). The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat. Commun. 10:5351. doi: 10.1038/s41467-019-13259-2
Pearson, A., Smyth, E., Babina, I. S., Herrera-Abreu, M. T., Tarazona, N., Peckitt, C., et al. (2016). High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 6, 838–851. doi: 10.1158/2159-8290.CD-15-1246
Pradas-Juni, M., Hansmeier, N. R., Link, J. C., Schmidt, E., Larsen, B. D., Klemm, P., et al. (2020). A MAFG-lncRNA axis links systemic nutrient abundance to hepatic glucose metabolism. Nat. Commun. 11:644. doi: 10.1038/s41467-020-14323-y
Qin, C., Jin, L., Li, J., Zha, W., Ding, H., Liu, X., et al. (2020). Long non-coding RNA LINC02163 accelerates malignant tumor behaviors in breast cancer by regulating the microRNA-511-3p/HMGA2 axis. Oncol. Res. 28, 483–495. doi: 10.3727/096504020X15928179818438
Ramnarine, V. R., Kobelev, M., Gibb, E. A., Nouri, M., Lin, D., Wang, Y., et al. (2019). The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur. Urol. 76, 546–559. doi: 10.1016/j.eururo.2019.07.040
Ren, Z., Liu, X., Si, Y., and Yang, D. (2020). Long non-coding RNA DDX11-AS1 facilitates gastric cancer progression by regulating miR-873-5p/SPC18 axis. Artif. Cells Nanomed. Biotechnol. 48, 572–583. doi: 10.1080/21691401.2020.1726937
Sallam, T., Sandhu, J., and Tontonoz, P. (2018). Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ. Res. 122, 155–166. doi: 10.1161/CIRCRESAHA.117.311802
Schulte, C., Barwari, T., Joshi, A., Zeller, T., and Mayr, M. (2020). Noncoding RNAs versus protein biomarkers in cardiovascular disease. Trends Mol. Med. 26, 583–596. doi: 10.1016/j.molmed.2020.02.001
Shang, Z., Yu, J., Sun, L., Tian, J., Zhu, S., Zhang, B., et al. (2019). LncRNA PCAT1 activates AKT and NF-kappaB signaling in castration-resistant prostate cancer by regulating the PHLPP/FKBP51/IKKalpha complex. Nucleic Acids Res. 47, 4211–4225. doi: 10.1093/nar/gkz108
Shen, L., Wang, Q., Liu, R., Chen, Z., Zhang, X., Zhou, P., et al. (2018). LncRNA lnc-RI regulates homologous recombination repair of DNA double-strand breaks by stabilizing RAD51 mRNA as a competitive endogenous RNA. Nucleic Acids Res. 46, 717–729. doi: 10.1093/nar/gkx1224
Silva-Fisher, J. M., Dang, H. X., White, N. M., Strand, M. S., Krasnick, B. A., Rozycki, E. B., et al. (2020). Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat. Commun. 11:2156. doi: 10.1038/s41467-020-15547-8
Song, Y. X., Sun, J. X., Zhao, J. H., Yang, Y. C., Shi, J. X., Wu, Z. H., et al. (2017). Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat. Commun. 8:289. doi: 10.1038/s41467-017-00304-1
Sun, Y., Zhou, Y., Bai, Y., Wang, Q., Bao, J., Luo, Y., et al. (2017). A long non-coding RNA HOTTIP expression is associated with disease progression and predicts outcome in small cell lung cancer patients. Mol. Cancer 16:162. doi: 10.1186/s12943-017-0729-1
Ulitsky, I. (2016). Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614. doi: 10.1038/nrg.2016.85
Virgilio, E., Proietti, A., D’Urso, R., Cardelli, P., Giarnieri, E., Montagnini, M., et al. (2017). Measuring intragastric tumor markers in gastric cancer patients: a systematic literature review on significance and reliability. Anticancer Res. 37, 2817–2821. doi: 10.21873/anticanres.11632
Wang, C. J., Zhu, C. C., Xu, J., Wang, M., Zhao, W. Y., Liu, Q., et al. (2019). The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol. Cancer 18:115. doi: 10.1186/s12943-019-1032-0
Wang, F., Long, G., Zhao, C., Li, H., Chaugai, S., Wang, Y., et al. (2014). Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One 9:e105734. doi: 10.1371/journal.pone.0105734
Wang, P., Li, X., Gao, Y., Guo, Q., Ning, S., Zhang, Y., et al. (2020). LnCeVar: a comprehensive database of genomic variations that disturb ceRNA network regulation. Nucleic Acids Res. 48, D111–D117. doi: 10.1093/nar/gkz887
Wang, W., Hu, W., Wang, Y., An, Y., Song, L., Shang, P., et al. (2020). Long non-coding RNA UCA1 promotes malignant phenotypes of renal cancer cells by modulating the miR-182-5p/DLL4 axis as a ceRNA. Mol. Cancer 19:18. doi: 10.1186/s12943-020-1132-x
Wang, Y. L., Liu, J. Y., Yang, J. E., Yu, X. M., Chen, Z. L., Chen, Y. J., et al. (2019). Lnc-UCID promotes G1/S transition and hepatoma growth by preventing DHX9-mediated CDK6 down-regulation. Hepatology 70, 259–275. doi: 10.1002/hep.30613
Wu, Y. M., Su, F., Kalyana-Sundaram, S., Khazanov, N., Ateeq, B., Cao, X., et al. (2013). Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 3, 636–647. doi: 10.1158/2159-8290.CD-13-0050
Yang, J., Qiu, Q., Qian, X., Yi, J., Jiao, Y., Yu, M., et al. (2019). Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol. Cancer 18:171. doi: 10.1186/s12943-019-1107-y
Yu, Y., Yu, X., Liu, H., Song, Q., and Yang, Y. (2018). miR494 inhibits cancerinitiating cell phenotypes and reverses resistance to lapatinib by downregulating FGFR2 in HER2positive gastric cancer. Int. J. Mol. Med. 42, 998–1007. doi: 10.3892/ijmm.2018.3680
Yuan, L., Xu, Z. Y., Ruan, S. M., Mo, S., Qin, J. J., and Cheng, X. D. (2020). Long non-coding RNAs towards precision medicine in gastric cancer: early diagnosis, treatment, and drug resistance. Mol. Cancer 19:96. doi: 10.1186/s12943-020-01219-0
Zhang, F., Wang, H., Yu, J., Yao, X., Yang, S., Li, W., et al. (2021). LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol. Cancer 20:6. doi: 10.1186/s12943-020-01299-y
Zhang, G., Li, S., Lu, J., Ge, Y., Wang, Q., Ma, G., et al. (2018). LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 17:87. doi: 10.1186/s12943-018-0829-6
Zhang, M., Muralimanoharan, S., Wortman, A. C., and Mendelson, C. R. (2016). Primate-specific miR-515 family members inhibit key genes in human trophoblast differentiation and are upregulated in preeclampsia. Proc. Natl. Acad. Sci. U.S.A. 113, E7069–E7076. doi: 10.1073/pnas.1607849113
Zhang, Y., Zhang, X., Cai, B., Li, Y., Jiang, Y., Fu, X., et al. (2021). The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia-reperfusion injury. Nat. Commun. 12:522. doi: 10.1038/s41467-020-20844-3
Zhang, Z., and Wang, H. (2021). HCP5 promotes proliferation and contributes to cisplatin resistance in gastric cancer through miR-519d/HMGA1 axis. Cancer Manag. Res. 13, 787–794. doi: 10.2147/CMAR.S289997
Zhao, X., Su, L., He, X., Zhao, B., and Miao, J. (2020). Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy 16, 70–85. doi: 10.1080/15548627.2019.1598750
Zhou, C., Yi, C., Yi, Y., Qin, W., Yan, Y., Dong, X., et al. (2020). LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. Cancer 19:118. doi: 10.1186/s12943-020-01237-y
Keywords: gastric cancer, long non-coding RNA, Lnc_ASNR, proliferation, invasion, migration
Citation: Chen Z, Li Y, Tan B, Li F, Zhao Q, Fan L, Zhang Z, Zhao X, Liu Y and Wang D (2021) Long Non-coding RNA ASNR Targeting miR-519e-5p Promotes Gastric Cancer Development by Regulating FGFR2. Front. Cell Dev. Biol. 9:679176. doi: 10.3389/fcell.2021.679176
Received: 11 March 2021; Accepted: 10 June 2021;
Published: 09 July 2021.
Edited by:
Yongbin Chen, Kunming Institute of Zoology (CAS), ChinaReviewed by:
Chaohong Liu, Huazhong University of Science and Technology, ChinaShuang Liu, Central South University, China
Shengtao Zhou, Sichuan University, China
Copyright © 2021 Chen, Li, Tan, Li, Zhao, Fan, Zhang, Zhao, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Li, bGlfeW9uZ19oYnRoQDEyNi5jb20=
 Zihao Chen
Zihao Chen Yong Li
Yong Li Bibo Tan2
Bibo Tan2