- 1Department of Hepatobiliary Surgery II, Guangdong Provincial Research Center for Artificial Organ and Tissue Engineering, Guangzhou Clinical Research and Transformation Center for Artificial Liver, Institute of Regenerative Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 2The First Affiliated Hospital, Sun Yat-sen university, Guangzhou, China
- 3Guangdong Provincial Hospital of Traditional Chinese Medicine, Guangzhou, China
- 4Gaozhou People’s Hospital, Gaozhou, China
- 5State Key Laboratory of Organ Failure Research, Southern Medical University, Guangzhou, China
Pancreatic cancer is one of the major malignancies and causes of mortality worldwide. E3 ubiquitin–protein ligases transfer activated ubiquitin from ubiquitin-conjugating enzymes to protein substrates and confer substrate specificity in cancer. In this study, we first downloaded data from The Cancer Genome Atlas pancreatic adenocarcinoma dataset, acquired all 27 differentially expressed genes (DEGs), and identified genomic alterations. Then, the prognostic significance of DEGs was analyzed, and eight DEGs (MECOM, CBLC, MARCHF4, RNF166, TRIM46, LONRF3, RNF39, and RNF223) and two clinical parameters (pathological N stage and T stage) exhibited prognostic significance. RNF223 showed independent significance as an unfavorable prognostic marker and was chosen for subsequent analysis. Next, the function of RNF223 in the pancreatic cancer cell lines ASPC-1 and PANC-1 was investigated, and RNF223 silencing promoted pancreatic cancer growth and migration. To explore the potential targets and pathways of RNF223 in pancreatic cancer, quantitative proteomics was applied to analyze differentially expressed proteins, and metabolism-related pathways were primarily enriched. Finally, the reason for the elevated expression of RNF223 was analyzed, and KLF4 was shown to contribute to the increased expression of RNF233. In conclusion, this study comprehensively analyzed the clinical significance of E3 ligases. Functional assays revealed that RNF223 promotes cancer by regulating cell metabolism. Finally, the elevated expression of RNF223 was attributed to KLF4-mediated transcriptional activation. This study broadens our knowledge regarding E3 ubiquitin ligases and signal transduction and provides novel markers and therapeutic targets in pancreatic cancer.
Introduction
Pancreatic cancer (PC) is one of the major malignancies and causes of mortality worldwide (Cheng et al., 2019; Mizrahi et al., 2020; Siegel et al., 2020). The most common type of PC is adenocarcinoma, accounting for 95% of cases, which is classified as pancreatic ductal adenocarcinoma (He et al., 2014; Martens et al., 2019). The prognosis for PC remains poor, with only 4.4% of patients reaching a 5-year survival rate (Mizrahi et al., 2020). Risk factors for developing PC include family history, obesity, type 2 diabetes, and tobacco use (Mizrahi et al., 2020). Diagnosis of PC often occurs at a late stage, meaning that more than 80% of patients with PC are unsuitable for surgical resection (Buscail et al., 2020). Chemotherapy, targeted therapy, and immunotherapy are now the most widely used treatments for PC (Bear et al., 2020; Christenson et al., 2020; Hosein et al., 2020). However, because of delayed disease detection and the limited efficacy of systemic therapies, the prognosis for this disease remains very poor (Grossberg et al., 2020). Therefore, insights regarding the regulatory mechanisms underlying PC progression are required to identify novel diagnostic and/or prognostic markers.
Ubiquitination plays an essential role in protein posttranslational modification and is strongly linked to various diseases (Rape, 2018). E3 ubiquitin–protein ligases transfer activated ubiquitin from ubiquitin-conjugating enzymes to protein substrates and confer substrate specificity in cancer (Zheng and Shabek, 2017; Senft et al., 2018). The RING finger (RNF) protein family is a complex set of proteins containing an RNF domain with more than 200 members having been identified (Joazeiro and Weissman, 2000; Fang et al., 2003; Lipkowitz and Weissman, 2011). Many RNF family members have been reported to play key roles in carcinogenesis (Lipkowitz and Weissman, 2011), such as RNF45 (Tsai et al., 2007), RNF6 (Liang et al., 2018), RNF4 (Plechanovová et al., 2011), RNF7 (Sun and Li, 2013), RNF168 (Devgan et al., 2011), RNF183 (Geng et al., 2017), RNF20 (Dickson et al., 2016), and RNF180 (Deng et al., 2016). In PC, RNF13 is involved in tumorigenesis (Zhang et al., 2009). As a biomarker candidate of PC, RNF6 facilitates PC metastasis by enhancing the c-Myc–mediated Warburg effect (Qiu et al., 2021). RNF43 mutation might cause downregulation of the expression of ring finger protein 43 and synergistically associates with GNAS mutations during the development of PC (Sakamoto et al., 2015). In addition, mutational inactivation of RNF43 confers Wnt dependency and could be used as a predictive biomarker for the clinical development of Wnt inhibitors in PC (Jiang et al., 2013). Identification of additional RNF family members associated with PC will help to elucidate the process of carcinogenesis and to develop new therapeutic strategies.
In this study, we first downloaded gene expression data from The Cancer Genome Atlas (TCGA) pancreatic adenocarcinoma (PAAD) dataset and analyzed the expression differences in E3 ubiquitin ligases (Medvar et al., 2016). Then, the prognostic significance of differentially expressed genes (DEGs) was analyzed. Of the DEGs, RNF223 showed independent significance as an unfavorable prognostic marker and was chosen for subsequent analysis. Next, the function of RNF223 in the PC cell lines ASPC-1 and PANC-1 was investigated using shRNA-mediated RNA silencing. To explore the potential targets and pathways of RNF223 in PC, quantitative proteomics was applied to analyze differentially expressed proteins (DEPs) and their functions in RNF223-silenced ASPC-1 cells. Finally, the mechanism for the elevated expression of RNF223 in PC was analyzed, and the regulatory mechanism was validated using a luciferase assay. The flowchart was shown in Supplementary Figure 1. This study deepens our understanding of the clinical significance and role of the E3 ligase RNF223 in pancreatic cancer.
Materials and Methods
The Cancer Genome Atlas Pancreatic Adenocarcinoma Dataset and E3 Ligase Acquisition
TCGA PAAD transcriptome FPKM data were downloaded from the GDC Data Portal1. Clinical data, such as age, sex, clinical stage, and survival time, were also downloaded. The 377 E3 ligase genes were acquired from the online database2.
Differentially Expressed Genes and Functional Analysis
Gene expression in PC and control samples was compared using the LIMMA R package. A fold change ≥ 2 or ≤ 0.5 and p < 0.05 were set as the cutoff values for EG screening. The heatmap R package was used to draw heatmaps. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, Gene Ontology (GO) enrichment analysis, and protein–protein interaction enrichment analysis for DEGs were also performed using the EnrichR (Kuleshov et al., 2016) website tool3.
Genomic Alterations in Pancreatic Adenocarcinoma Samples
The genomic alteration (GA) information of DEGs was acquired using cBioPortal4. Two PAAD datasets were included in this study, the TCGA Pan-cancer atlas and the UTSW study (Witkiewicz et al., 2015). All 184 and 109 samples from the two datasets were included.
Cell Culture and Gene Silencing
The PANC-1 and ASPC-1 human PC cell lines were obtained from the American Type Culture Collection (United States) and cultured at 37°C with 95% air and 5% CO2. PANC-1 and ASPC-1 cells were cultured in Dulbecco modified Eagle medium (Gibco, Germany) supplemented with 10% fetal calf serum (Germany), 2 mM L-glutamine, 105 U/L penicillin, and 100 mg/L streptomycin. Short hairpin RNAs specifically targeting RNF223 and Kruppel-like factor 4 (KLF4) were designed and synthesized by Generay Biotech (Shanghai) Co., Ltd., RNF223 shRNAs were as follows: shRNA1 (5′–3′): GCACAGCAGCCACTGGAAGTC, shRNA2 (5′–3′): GCGAAAGGAGCCTGGCATCTC, and shRNA3 (5′–3′): GGAGCCTGGCATCTCTGAGGA.
KLF4 shRNAs were as follows: shRNA1 (5′–3′): GCTCC ATTACCAAGAGCTCAT, shRNA2 (5′–3′): CCAGCCAGAAA GCACTACAAT, and shRNA3 (5′–3′): GCCTTACACATGAAG AGGCAT.
Cell Counting Kit-8 Assay
The cell proliferation reagent WST-8 (Roche, Germany) was measured. Cell growth: 10 μL of cell counting kit-8 (CCK8) was added to each well at the time of harvest after plating cells in 96-well microtiter plates (Corning, NY) at 1.0 × 103/well, according to the manufacturer’s instructions. Cellular viability was determined by measuring the absorbance of the converted dye at 450 nm 2 h after adding CCK8.
Wound Healing Assay
PANC-1 and ASPC-1 cells were seeded into 6-well plates and incubated for 24 h, and a linear wound was created by dragging a 100-μL pipette tip through the monolayer prior to transfection. Cellular debris was removed by gentle washes with culture medium, following which transfection was immediately performed, and the cells were allowed to migrate for an additional 48 h. The healing process was dynamically imaged after the wound was introduced using a microscope (Olympus 600 Autobiochemical Analyzer, Tokyo, Japan). Migration distance was measured using images (five fields) taken at each indicated time point. The gap size was analyzed using Image Pro Plus 6.0 software. The residual gap between the migrating cells from the opposing wound edge is expressed as a percentage of the initial gap size.
Quantitative Real-Time Polymerase Chain Reaction
Total RNA was extracted from ASPC-1 and PANC-1 cells using TRIzol® RNA Isolation Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Reverse transcription was performed using the PrimeScriptTM RT reagent kit (Takara, Dalian, China). All mRNA levels were normalized to the housekeeping gene GAPDH. The following RNF223 and KLF4 primers were used in this study: RNF223: 5′-TGATGCTCTTCTGTGTGGCA-3′ (F) and 5′-TTATCAGTCAG AGGCCCGAG-3′ (R). KLF4: 5′-CCCACATGAAGCGACTTC CC-3′ (F) and 5′-CAGGTCCAGGAGATCGTTGAA-3′(R). The GAPDH primers were as follows: 5′-TGACTTCAACAGC GACACCCA-3′ (F) and 5′- CACCCTGTTGCTGTAGCCAAA-3′ (R). All samples were treated under the same conditions and analyzed by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) using SYBR Premix ExTaqTM (Takara, Dalian China) according to the manufacturer’s protocol.
Prognostic Significance of Genes
Univariate Cox regression analysis was performed to identify prognosis-associated E3 ligase genes. Genes with a hazard ratio (HR) < 1 were considered favorable for OS, whereas HR > 1 presented unfavorable for OS. Genes with p < 0.05 were considered significant markers. The Kaplan–Meier survival plot was constructed using the “survplot” R package. Pan-cancer survival analysis was conducted using the KM plotter online tool (Nagy et al., 2021)5.
Data-Independent Acquisition Quantitative Proteomics
Data-independent acquisition quantitative proteomics was conducted according to a previous study (Zhang et al., 2021). Briefly, cells underwent protein extraction and trypsin digestion into peptides, and then a spectral library was generated and quantified. Ten fractions were collected, and each fraction was dried in a vacuum concentrator. The fractions were redissolved in 0.1% formic acid and analyzed using nanospray liquid chromatography with tandem mass spectrometry (LC-MS/MS) on an Orbitrap Fusion Lumos Tribrid (Thermo Fisher Scientific, MA, United States) coupled to a Waters nanoACQUITY UPLC System (Waters, MA, United States). The mass spectrometer was run in DDA mode and automatically switched between MS and MS/MS modes. The DDA data were processed and analyzed using Spectronaut X (Biognosys, Schlieren, Switzerland) with default settings to generate an initial target list. A false discovery rate cutoff at the precursor and protein levels was applied at 1%. Finally, proteins were identified and quantified.
ChIP Sequencing Analysis of the Transcription Factors in RNF223
The ChIP sequencing peaks of RNF223 transcription factors were identified using the online tool ChIP-Atlas6. PC, including cell lines, datasets in bigwig format were downloaded and imported into the IGV browser. Transcription factors with binding peaks within the distance of ≤ 1 kb from the transcription start sites were considered candidate peaks.
Luciferase Reporter Assay
A luciferase reporter assay was performed according to a standard protocol as previously described (Gong et al., 2016). Briefly, ASPC-1 cells (3 × 104 cells/well) were seeded into 24-well plates in triplicate and allowed to attach for 24 h. The sequences (−28∼−54) of the wild-type (WT) and mutant ASPC-1 cells were WT: TATACCCTATGGGCCAAGGGTGTGGC and mutant (MUT): CGCCTTTACTGGGAACCGGGAGCAAA. The indicated plasmids and 1.5 ng pRL-TK Renilla plasmid were transfected using Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA, United States, cat. no. L3000008). Forty-eight hours posttransfection, luciferase and Renilla signals were assessed using a Dual Luciferase Reporter Assay Kit (Promega, cat. no. E1980) according to the manufacturer’s instructions as previously described (Hahn et al., 2002).
Statistical Analysis
The data are presented as means ± SD. SPSS 18.0 software was used to perform statistical analyses, and graphs were generated using GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, United States). Differences were examined using Student t-test or one-way analysis of variance. p < 0.05 was considered statistically significant.
Results
Comprehensive Analysis of the Clinical Significance of E3 Ligases in Pancreatic Cancer
To examine the expression differences in the E3 ligases in PC, 377 genes were applied for statistical analysis. By applying the cutoff criteria of fold change ≥ 2 or ≤ 0.5 and p < 0.05, 27 DEGs were acquired, including RNF166, MARCHF1, TRIM10, TRIM7, and so on (Figure 1A), and their expression in cancer and normal samples is shown in Figure 1B. Then, we examined their GAs in PC studies. As shown in Figure 1C, MEX3A exhibited the highest alteration frequency of 11%, followed by CBLC (10%), RNF43 (9%), MECOM (8%), TRIM58 (8%), and RNF39 (8%), mostly comprising amplification events. In another study (UTSW) by Witkiewicz et al. (2015), GA events displayed a comparatively different pattern. As shown in Figure 1D, RNF223 (26%), TRIM46 (24%), MEX3A (18%), CBLC (16%), TRIM58 (13%), and RNF43 (9%) showed the highest alteration frequencies. Notably, high occurrence of amplification events in MEX3A, CBLC, and MECOM and truncating mutations in RNF43 were observed in both datasets. In addition, RNF223 showed a distinct GA pattern in the two datasets, with only 2.2% in TCGA and 26% GA events (amplification and deep deletion events combined) in UTSW.
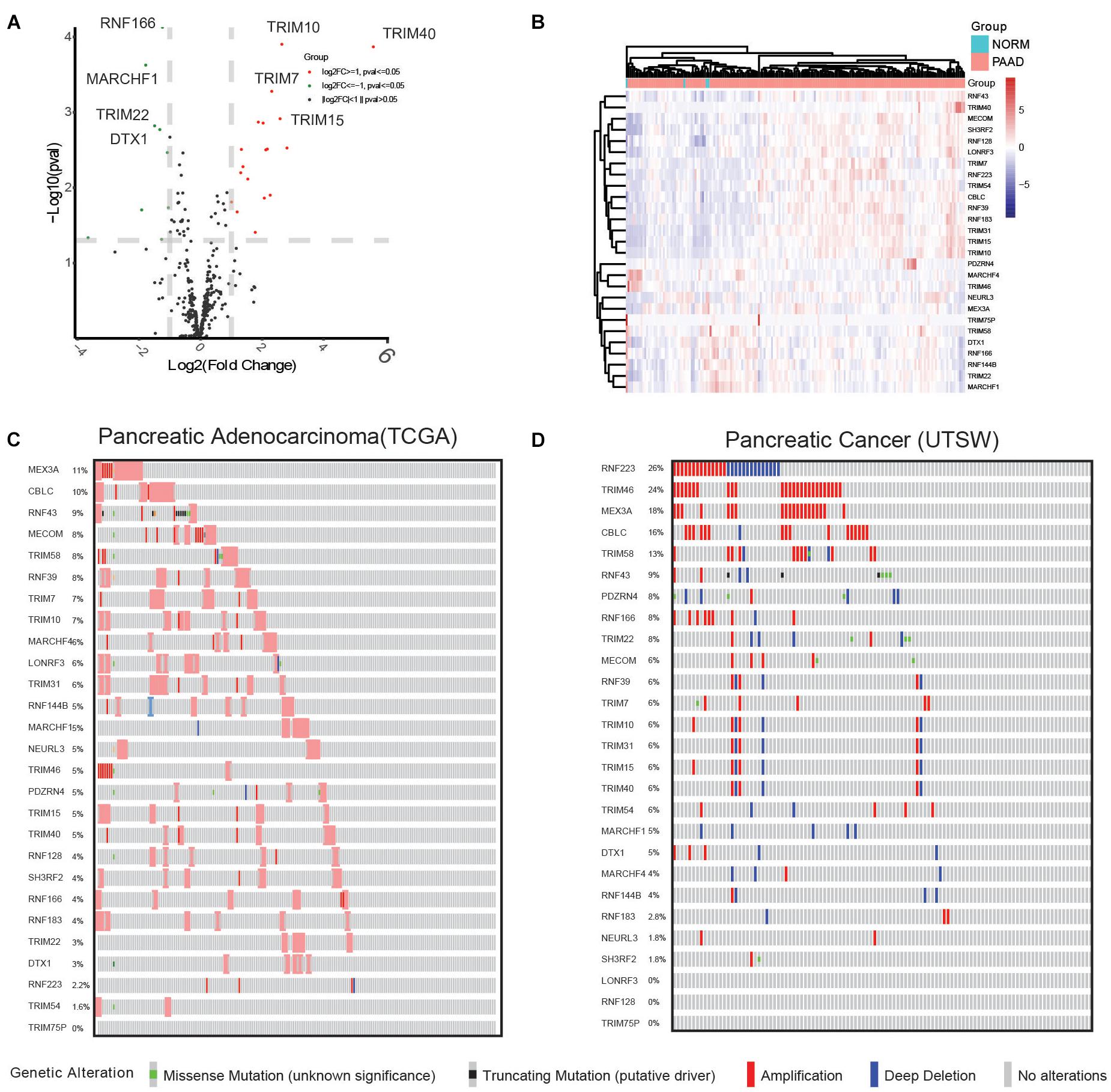
Figure 1. Comprehensive analysis reveals 27 differentially expressed E3 ubiquitin ligase genes and their GAs in the TCGA pancreatic cancer (PAAD) dataset. (A) Differential analysis of E3 ubiquitin ligases identified differentially expressed genes, shown in a volcano plot. Upregulated genes are shown as red dots, downregulated genes are shown as green dots, and genes with no significant differential expression are shown as gray dots. (B) Supervised hierarchical clustering of the DEGs in pancreatic cancer and normal samples. (C) Genetic alterations in pancreatic cancer samples included in the TCGA PAAD dataset. (D) Genetic alterations in pancreatic cancer samples included in the UTSW dataset.
Next, we conducted univariate survival analysis using the 27 DEGs and clinicopathological parameters of the TCGA dataset. As shown in Figure 2A, eight DEGs (MECOM, CBLC, MARCHF4, RNF166, TRIM46, LONRF3, RNF39, and RNF223) and two clinical parameters (pathological N stage and T stage) showed prognostic significance. The Kaplan–Meier survival plot of the eight DEGs is shown in Figure 2B. Of the eight genes, five genes, CBLC (HR = 1.7, p = 0.015), LONRF3 (HR = 2.1, p = 0.00092), RNF39 (HR = 1.6, p = 0.02), MECOM (HR = 1.8, p = 0.0059), and RNF223 (HR = 1.9, p = 0.0034), exhibited unfavorable prognostic significance, whereas TRIM46 (HR = 0.65, p = 0.046), MARCHF4 (HR = 0.63, p = 0.029), and RNF166 (HR = 0.65, p = 0.046) conveyed favorable survival significance. Finally, using the pan-cancer dataset, overall survival (OS) and relapse-free survival (RFS) analyses were conducted using the online tool Kaplan–Meier plotter. Consistent with the results in Figure 3, all eight genes showed prognostic significance, including the RFS data. Taken together, we acquired the expression and GA data of 27 DEGs, among which eight genes exhibited potential performance as prognostic indicators in PC.
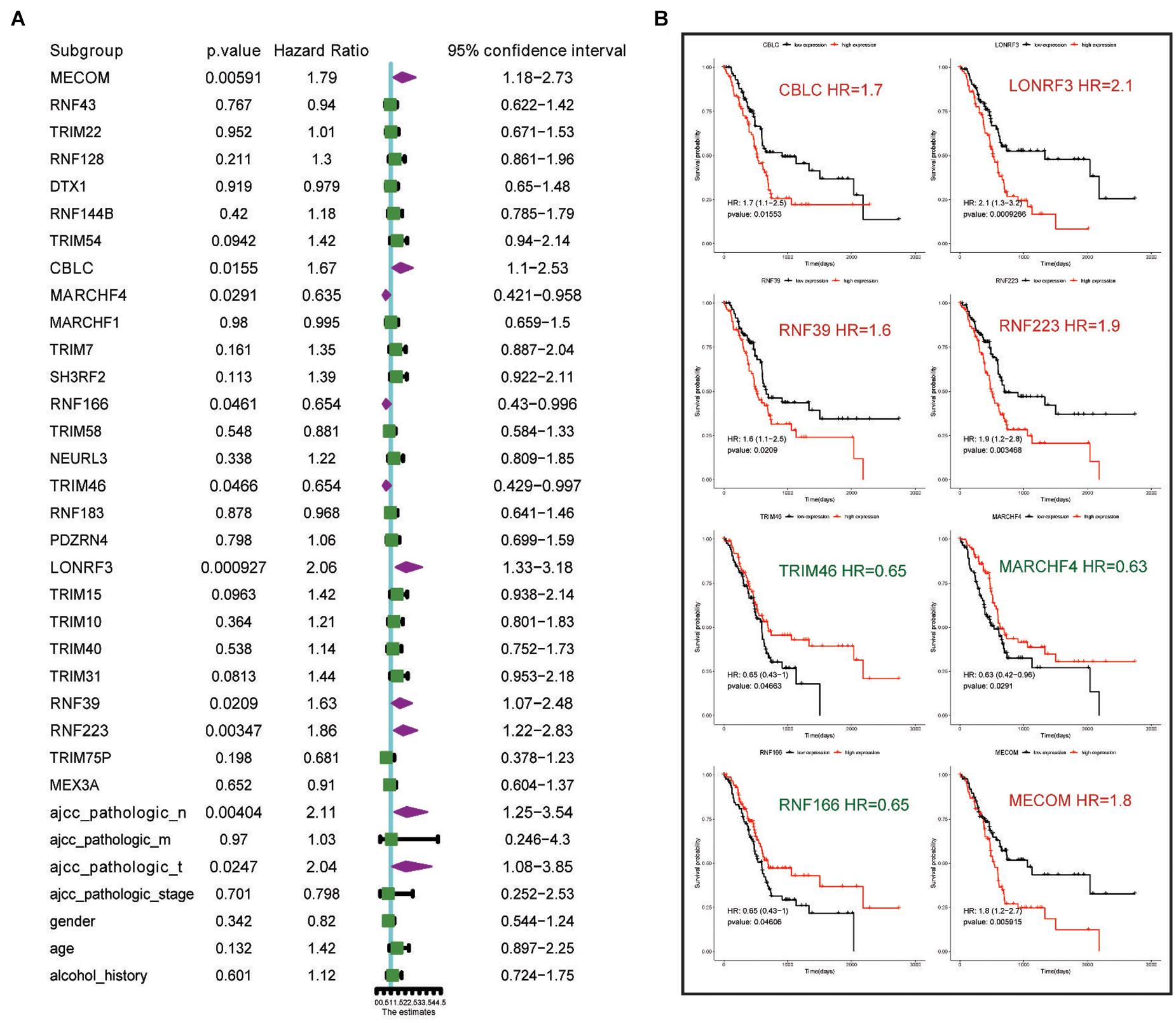
Figure 2. Univariate analysis highlights eight genes and clinical parameters with overall prognostic significance. (A) Univariate analysis of the 27 DEGs with prognostic significance in the TCGA PAAD dataset. (B) Kaplan–Meier survival analysis showing the HR of eight genes’ expression with OS rate in TCGA PAAD dataset. Favorable and unfavorable markers are labeled in green and red, respectively.
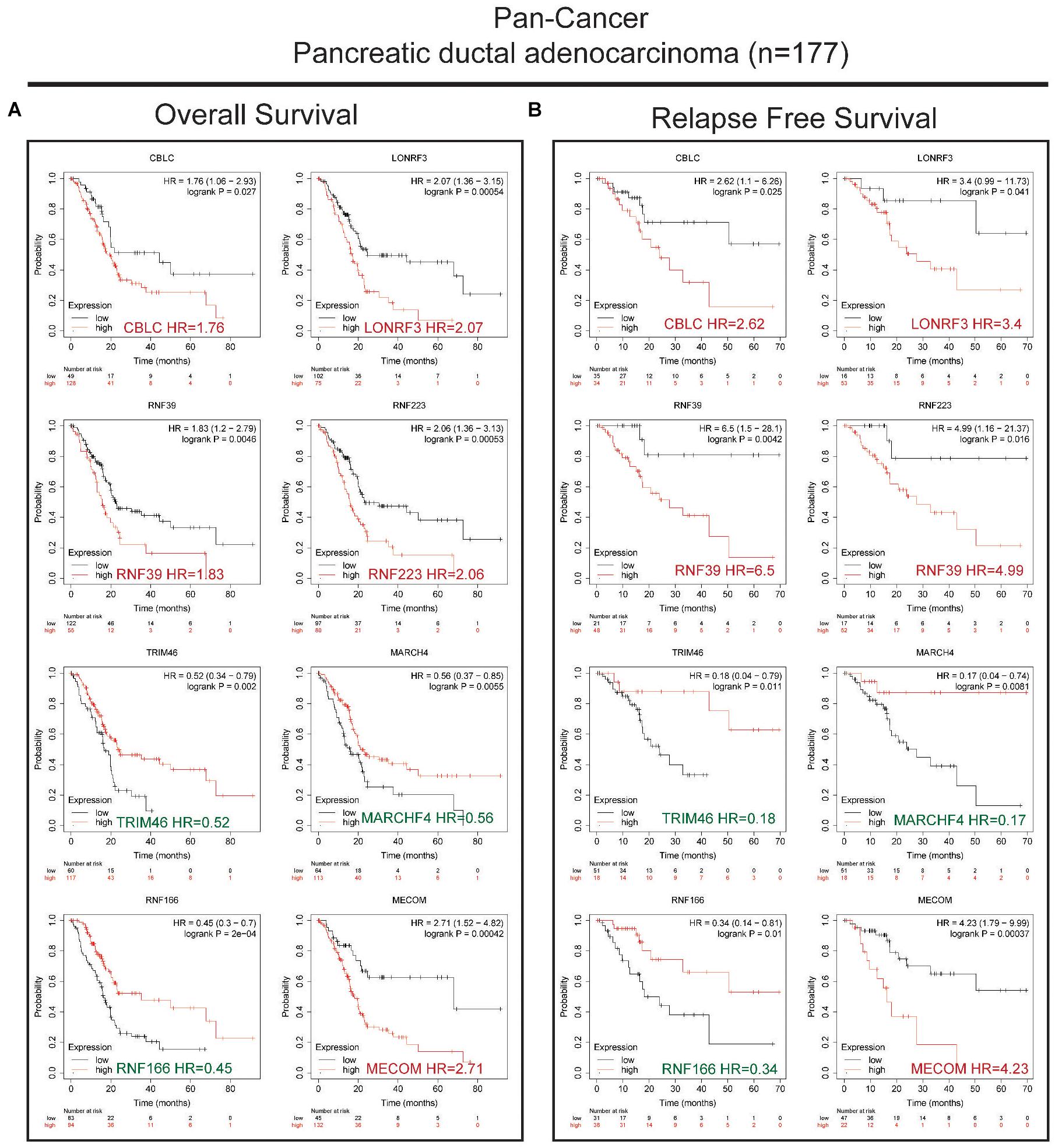
Figure 3. Validation of the OS and RFS rates using the PAAD dataset in a pan-cancer study. (A) Kaplan–Meier survival analysis showing the HR of eight genes’ expression with OS rate in the pan-cancer PAAD dataset. (B) Kaplan–Meier survival analysis showing the HR of eight genes’ expression with RFS rate in the pan-cancer PAAD dataset. Favorable and unfavorable markers were labeled in green and red, respectively.
Clinical and Functional Investigation of RNF223 in Pancreatic Cancer
Next, a multivariate analysis was conducted to analyze the prognostic significance of the 27 DEGs and the OS rate, and as a result, only RNF223 (among the eight prognostic genes) displayed prognostic significance, indicating that RNF223 may serve as an independent prognostic marker in PC (Figure 4A). Then, we compared the expression of RNF223 in groups of clinical parameters and as shown in Figure 4B, RNF223 in males (gender), high alcohol consumption, ductal/lobular neoplasms (disease type), pathological stage N0, tumor stage IIa, and vital status (dead) groups exhibited significantly higher expression compared to the other groups.
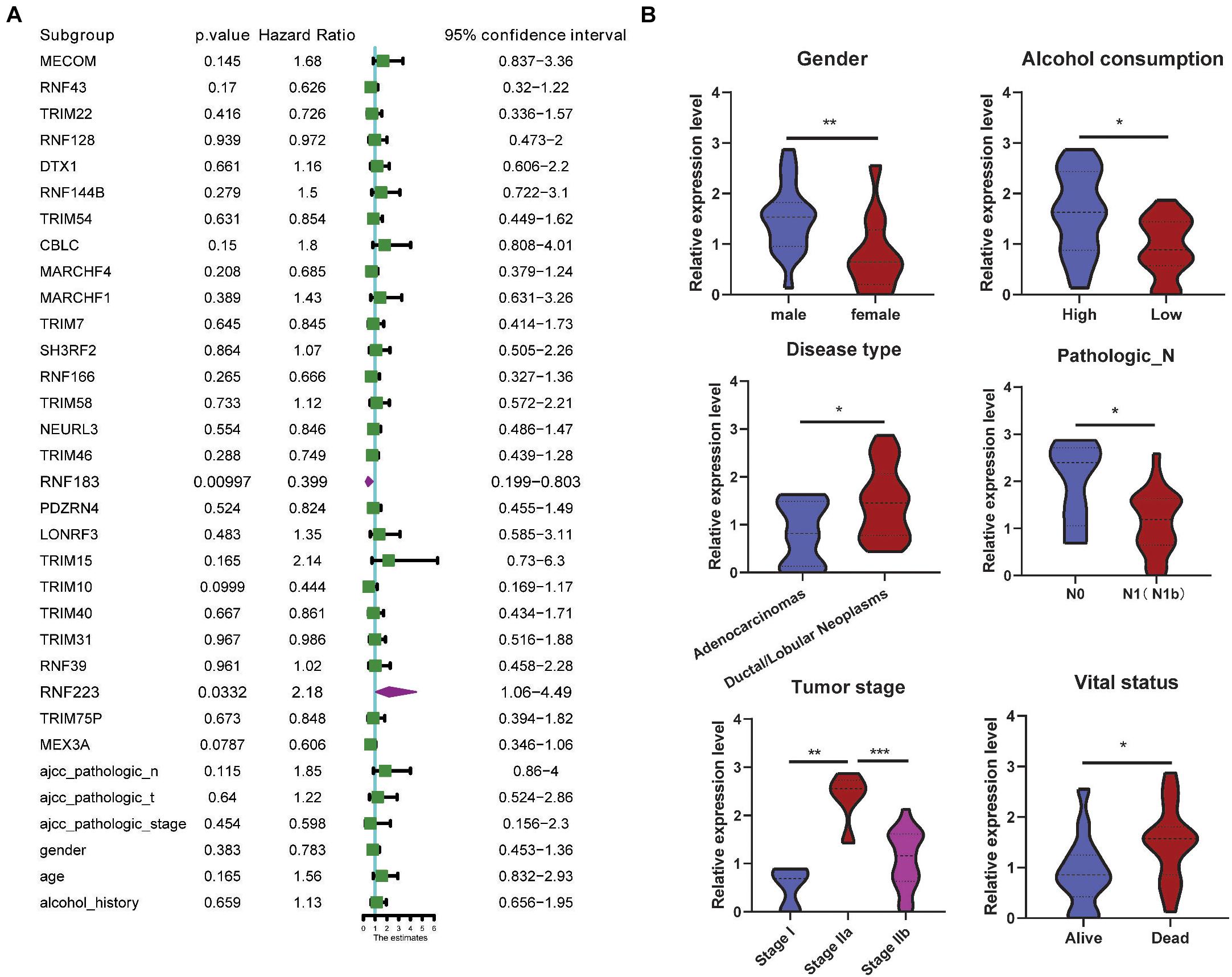
Figure 4. RNF223 serves as an independent prognostic marker and correlates with clinical parameters. (A) Multivariate analysis of the 27 DEGs with prognostic significance in the TCGA PAAD dataset. (B) Expression analysis of RNF23 with clinical parameters; *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we focused on RNF223 and explored the function of RNF223 silencing on PC cell line phenotypes (Figure 5A). After RNF223-targeting shRNA transfection, the RNF223 silencing efficiency was examined using qRT-PCR, and because shRNA2 exerted consistently higher (>50%) knockdown efficiency, we used shRNA2 as the subsequent shRNA (shRNF223) (Figure 5B). Then, CCK8 and wound healing assays were applied to study the impact of RNF223 silencing on the proliferation and migration capacity of ASPC-1 and PANC-1 cells. As shown in Figure 5C, RNF223 knockdown significantly reduced the cell number in both cell lines, indicating that RNF223 may promote PC growth. In addition, the wounds of shRNF223-transfected ASPC-1 and PANC-1 cell lines demonstrated reduced migration distance compared to the control group, indicating that RNF223 knockdown decreases the migration ability in both cell lines (Figure 5D). In summary, the above results revealed that RNF223 may represent an independent prognostic marker that promotes PC growth and migration.
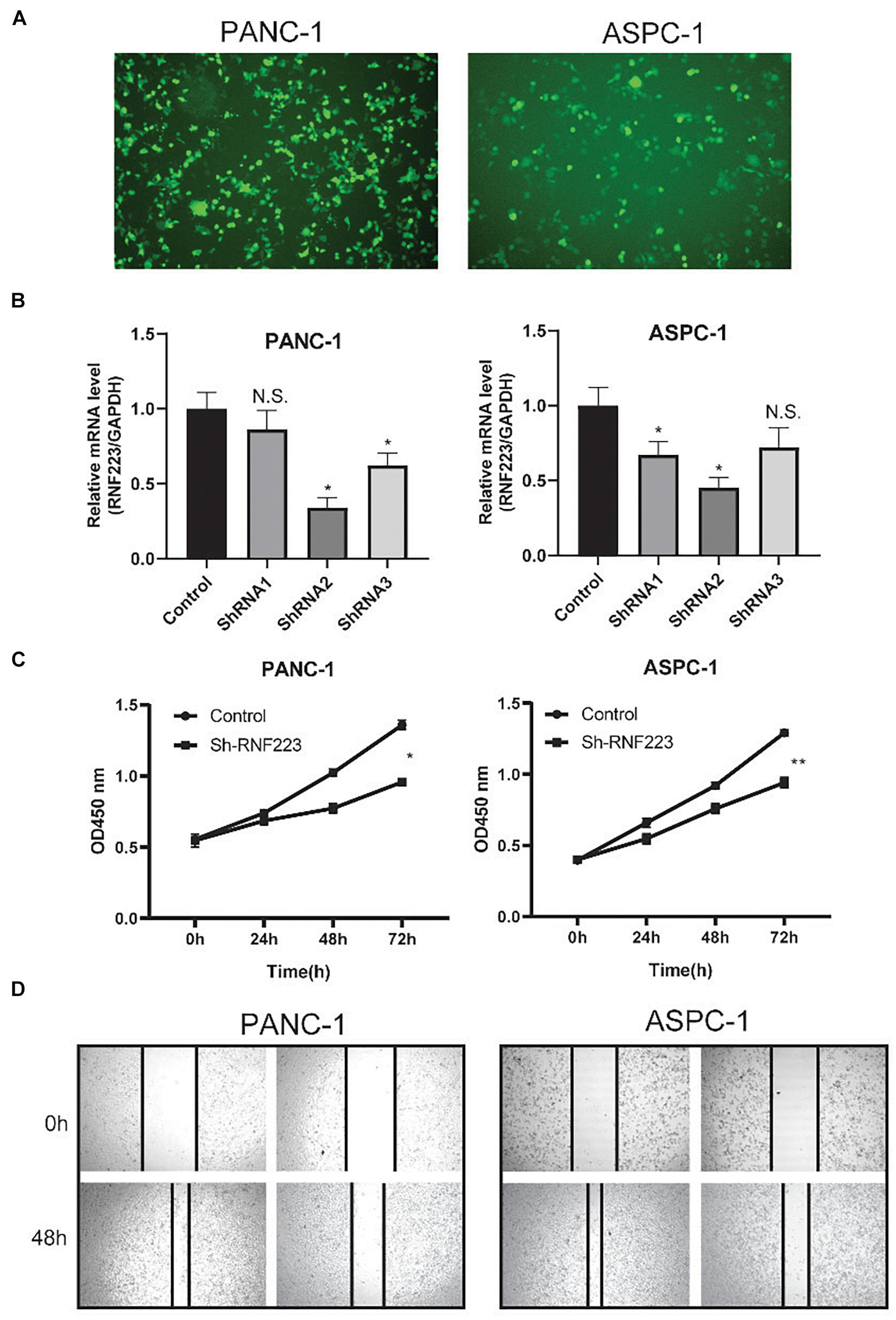
Figure 5. Functional investigation of RNF223 silencing in pancreatic cancer PANC-1 and ASPC-1 cell lines. (A) Representative cell morphology of transfection efficiency of RNF223 shRNAs in pancreatic cancer cells. (B) The silencing efficacy of three shRNAs was examined using qRT-PCR in pancreatic cancer PANC-1 and ASPC-1 cell lines. (C) The CCK8 assay was conducted to analyze the effect of RNF223 knockdown on the proliferation capability of pancreatic cancer PANC-1 and ASPC-1 cell lines. (D) The wound healing assay was conducted to analyze the effect of RNF223 knockdown on the migration capability of pancreatic cancer PANC-1 and ASPC-1 cell lines. All assays were conducted using three replicates and *p < 0.05, **p < 0.01.
The Molecular Mechanism of RNF223-Affected Pathways and Targets
Then, the downstream mechanism of RNF223 in PC was investigated using quantitative proteomics in RNF223 knockdown and control ASPC-1 cells. After protein quantification, all 885 DEPs were acquired (Figure 6A), and their expression is shown in Figure 6B. Then, the functions of these DEPs were annotated, and their enriched pathways and functions were identified using KEGG pathways and GO databases. Based on the enrichment score (−log10 p-value), the top 15 enriched pathways and GO biological processes (BPs), cellular components (CCs), and molecular functions (MFs) are shown in Figures 6C–F. The most enriched pathways were oxidative phosphorylation, and other items, such as regulation of cytoskeleton, pathways in cancer, metabolism pathways, and HIG1α signaling pathways, were also enriched. For GO-BPs, metabolism-related BPs were also enriched, such as cellular metabolic, primary metabolic, and nitrogen compound metabolic processes. In addition, the cell cycle process was also significantly enriched. For the GO-CC result, catalytic complex was the top enriched component, supporting the pathway and BPs of enrichment of metabolism-related factors. As expected, the GO-MM category identified enriched protein binding as the most significant item, consistent with the biochemical role of RNF223 as an E3 ligase. Finally, a functional network was created of RNF223 targets, and genes in the DNA synthesis and transforming growth factor β signaling pathways are shown. In summary, we identified potential protein targets and metabolism-related pathways of RNF223 in PC (Figure 6G).
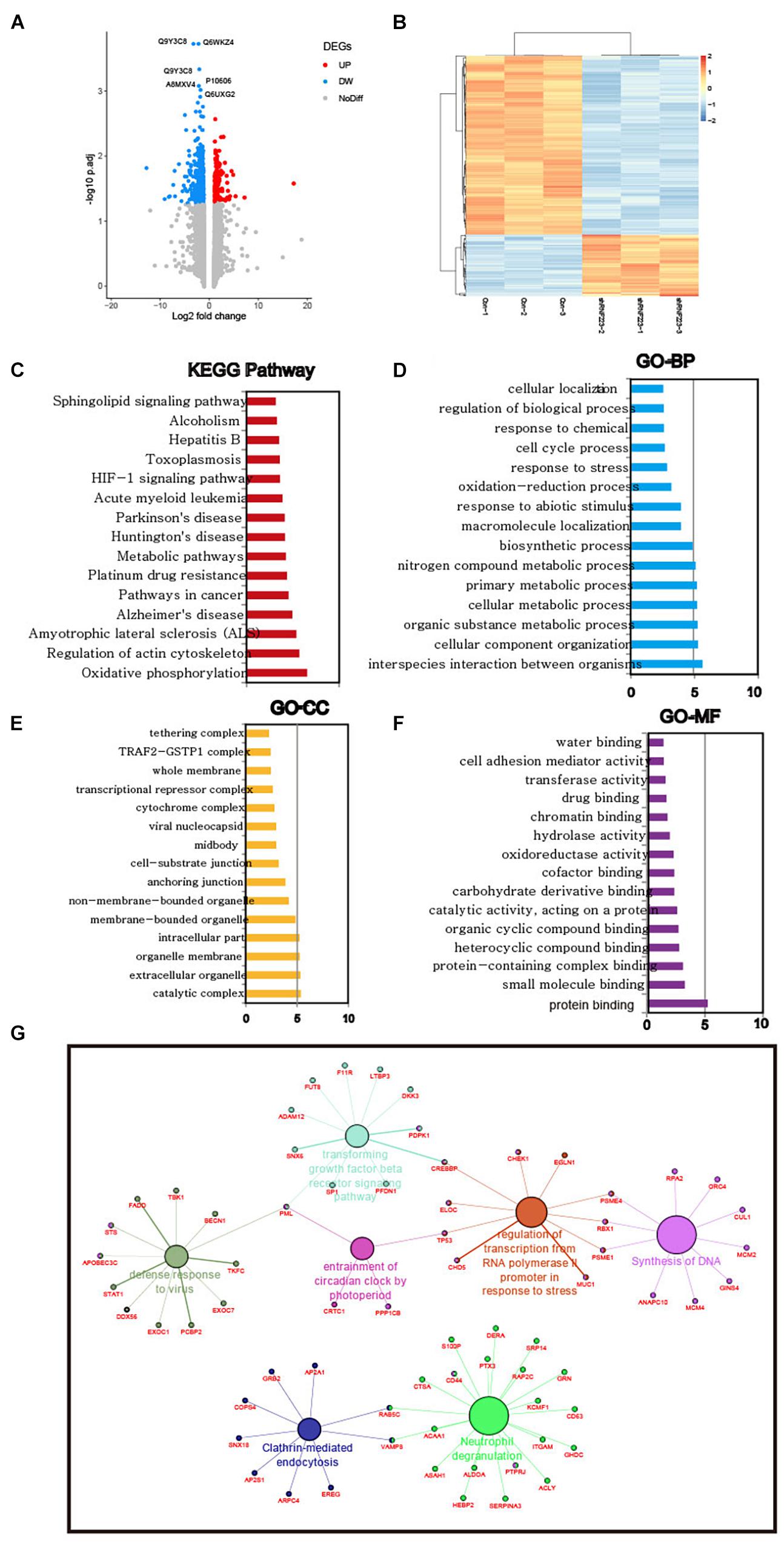
Figure 6. Quantitative proteomics analysis reveals pathways and BPs in the RNF223-silenced ASPC-1 cell line. (A) Analysis of DEGs in RNF223 knockdown ASPC-1 cells is shown using a volcano plot. Upregulated genes are shown as red dots, downregulated genes are shown as blue dots, and genes with no significant differential expression are shown as gray dots. (B) Supervised hierarchical clustering of the DEGs in RNF223 knockdown ASPC-1 cells. (C) The top 15 significantly enriched KEGG pathways of the DEGs in RNF223 knockdown ASPC-1 cells. (D) The top 15 significantly enriched Gene Ontology BPs of the DEGs in RNF223 knockdown ASPC-1 cells. (E) The top 15 significantly enriched Gene Ontology CCs of the DEGs in RNF223 knockdown ASPC-1 cells. (F) The top 15 significantly enriched Gene Ontology MFs of the DEGs in RNF223 knockdown ASPC-1 cells. (G) Functional network of the DEGs in pathways and BPs.
RNF223 Was Transactivated by Kruppel-Like Factor 4 in Pancreatic Cancer
Finally, the mechanism underlying the elevated expression of RNF223 was explored. As the genetic alteration frequency of RNF223 was not remarkably high in PC, we speculated that transcription factors may contribute to this process. First, we downloaded PC ChIP sequencing data from the recently published online tool ChIP-Atlas. As shown in Figure 7A, KLF4 exhibited a prominent peak in the promoter region of RNF223 DNA. In addition, coexpression analysis of KLF4 with RNF223 revealed a strong coefficient (R = 0.51) in TCGA PAAD datasets (Figure 7B). Then, to validate the role of KLF4 on the mRNA expression of RNF223, qRT-PCR was performed to examine the expression of RNF223 in the KLF4 knockdown cell line ASPC-1. As shown in Figure 7C, RNF223 exhibited significantly decreased expression in KLF4-silenced cells, indicating that KLF4 may upregulate RNF223 expression in ASPC-1 cells. Finally, a luciferase assay was conducted to validate the above results. As shown in Figure 7D, KLF4 silencing remarkably decreased luciferase intensity in the RNF223 WT group, whereas no significant difference was observed in the RNF223 MUT group. Altogether, these data show that KLF4 contributes to the increased expression of RNF233 in PC.
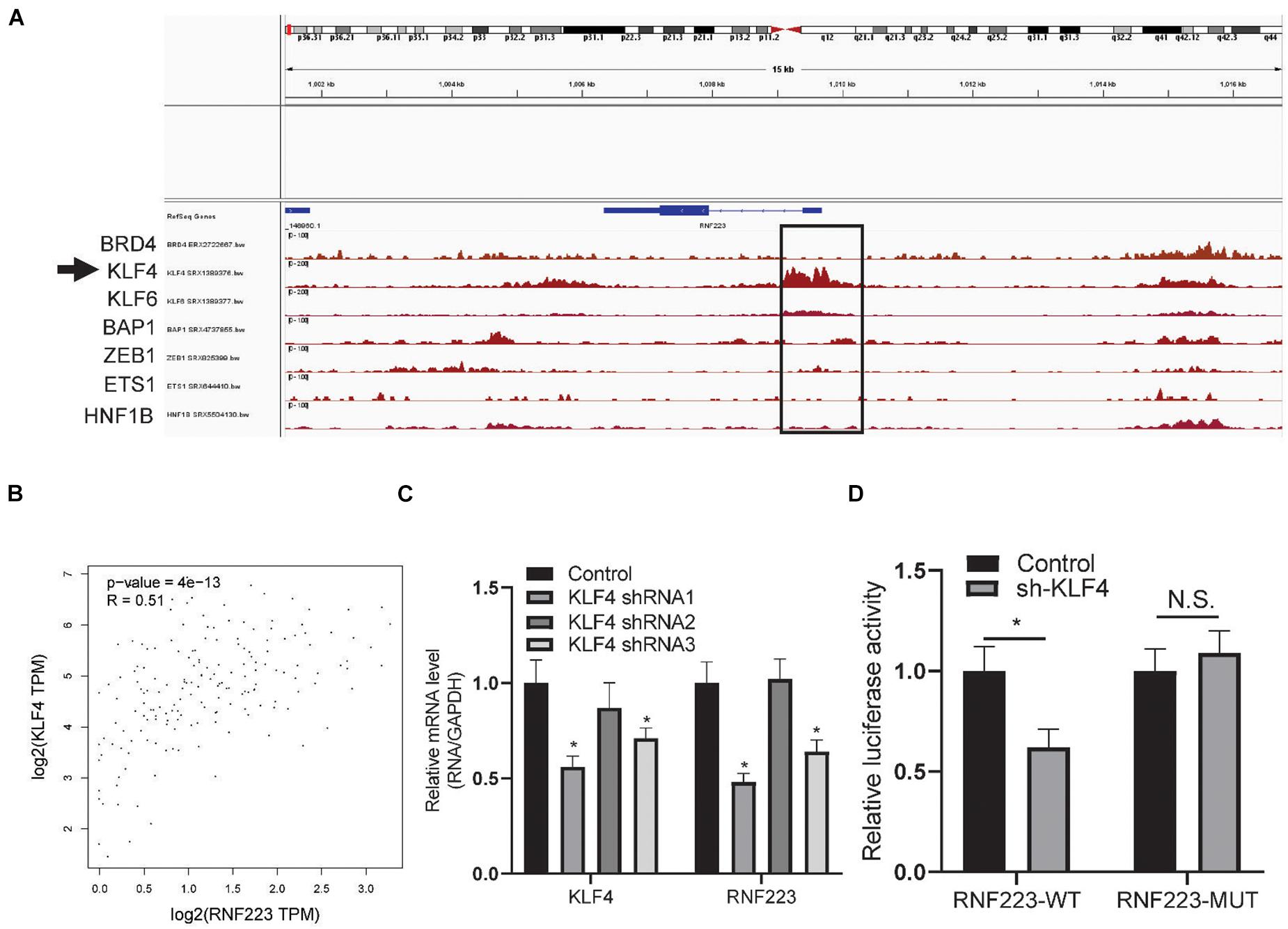
Figure 7. RNF223 is transcriptionally activated by KLF4 in pancreatic cancer. (A) Binding peaks of transcription factors in pancreatic cancer tissue and cell lines; arrow indicates that KLF4 shows remarkable binding affinity at the promoter of RNF223. (B) Expression correlation of KLF4 with RNF223 in the TCGA PAAD dataset. (C) Relative expression of KLF4 and RNF223 in KLF4 knockdown shRNA-transfected ASPC-1 cell lines. (D) Luciferase assay showing the binding affinity of KLF4 in RNF223 promoter WT and mutated (MUT) cells. All assays were conducted using three replicates and *p < 0.05.
Discussion
Mounting evidence indicates that E3 ubiquitin ligases play important roles in cancer onset, progression, and treatment response and serve as prognostic makers in cancer (Senft et al., 2018). Both genetic and epigenetic alterations account for the dysregulation of E3s in cancer (Qi and Ronai, 2015). Consequently, the stability and/or activity of E3 substrates are also altered, leading to downregulation of tumor-suppressor activities and upregulation of oncogenic activities (Senft et al., 2018; Fujita et al., 2019). Targeting E3 ligases has been previously proposed as a novel cancer therapeutic strategy (Micel et al., 2013; Sharp et al., 2021). A better understanding of the mechanisms underlying E3 regulation and function in tumorigenesis is expected to reveal novel prognostic markers and to enable the development of the next generation of anticancer therapies (Kumari et al., 2017; Wang et al., 2017; Khan et al., 2020). Here, by analyzing the clinical significance of E3 ligases in PC, we first identified 27 DEGs, of which eight DEGs showed prognostic performance. To the best of our knowledge, this is the first comprehensive study of E3 ligases in PC and provides an overall map of these E3 ligases in PC.
Among the eight prognostic markers, most have been reported in cancer. For instance, MDS1 and EVI1 complex loci (MECOM) interact with PAX8 and drive oncogenic functions in ovarian cancer (Bleu et al., 2021). Cbl Proto-Oncogene C (CBLC) was demonstrated to enhance epidermal growth factor receptor dysregulation and signaling in lung adenocarcinoma (Hong et al., 2018). MARCHF4, previously known as MARCH4, was identified as a potential therapeutic target in cutaneous squamous cell carcinoma (McHugh et al., 2020). The chimeric RNAs generated from tripartite motif containing 46 (TRIM46) with MUC1 and KRTCAP2 have been clinically implicated in high-grade serous ovarian carcinoma (Kannan et al., 2015). In PC, MECOM was shown to be a critical regulator that suppresses acinar cell death by permitting cellular dedifferentiation (Backx et al., 2021), but there are limited studies regarding the other seven genes in pancreatic cancer. Identification of these eight genes provides an alternative option for prognostic prediction in pancreatic cancer patients.
Subsequently, we identified RNF223 as an independent prognostic marker in pancreatic cancer, and further functional assays revealed that RNF223 may play an oncogenic role in pancreatic cancer progression. As a member of the ring finger proteins, most studies have focused on RNF43. RNF43 mutation might cause downregulation of the expression of ring finger protein 43 and associate synergistically with GNAS mutations during the development of PC (Sakamoto et al., 2015). In addition, mutational inactivation of RNF43 confers Wnt dependency and could be used as a predictive biomarker for the clinical development of Wnt inhibitors in PC (Jiang et al., 2013). For RNF223, mutation sites have been related to age, International Federation of Gynecology and Obstetrics stage, and histology in sporadic and Lynch syndrome–associated endometrial cancer (Sun et al., 2021). In addition, RNF223 was reported to serve as a prognostic marker for uterine sarcoma (Zhou et al., 2019). To date, no studies of RNF223 have been reported in PC. Moreover, we are conducting additional assays to reveal the role of RNF223 in PC, including the clinical significance of RNF223 in our collected PC samples, in vivo xenograft animal assays, and immunoprecipitation-coupled MS to identify targets of RNF223 in PC. In addition, as E3 ligases have been shown to function in both ubiquitin–proteasome-dependent and ubiquitin–proteasome-independent diseases (Cadena et al., 2019; Weinelt and van Wijk, 2021), the specific mechanism of RNF223 in PC remains to be uncovered in the future.
Finally, utilizing the ChIP data in ChIP-Atlas and further validation assays, such as luciferase assays, we identified KLF4 to be a hub regulon of RNF223 in PC. KLF4 has been widely reported as an oncogene in multiple cancer types (Rowland and Peeper, 2006; Hu et al., 2015; Hsieh et al., 2017; Murgai et al., 2017), including lung cancer (Yu et al., 2016), ovarian cancer (Zhang et al., 2019), esophageal squamous cell cancer (Tetreault et al., 2010), gastric cancer (Li et al., 2012), colorectal cancer (Li et al., 2011; Gamper et al., 2012), and leukemia (Faber et al., 2013; Li et al., 2015; Seipel et al., 2016; Park et al., 2019). In PC, KLF4 was demonstrated to contribute to carcinogenesis and progression by inducing acinar-to-ductal reprogramming (Wei et al., 2016), as well as the LDHA signaling pathway and aerobic glycolysis (Shi et al., 2014) and MSI2 signaling pathway–mediated cell growth (Guo et al., 2017). Moreover, increased expression of KLF4 is attributed to hypomethylation-mediated by DNA methyltransferase 1 (Xie et al., 2017). In this study, we first validated RNF223 as a novel target of KLF4, and the primary enriched metabolic pathways of RNF223 also corresponded to the function of KLF4 as a regulator of glycolysis.
Conclusion
This study comprehensively analyzed the expression difference in E3 ligases and identified eight prognostic markers among 27 DEGs. In addition, functional assays of RNA silencing revealed RNF223 as a tumor-promoting gene that may regulate cancer cell metabolism. Finally, the elevated expression of RNF223 was attributed to KLF4-mediated transcriptional activation. This study broadens our knowledge of E3 ubiquitin ligases and signal transduction and provides novel markers and therapeutic targets for PC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: iProX; PXD028446.
Ethics Statement
The study protocol was reviewed and approved by the Zhujiang Hospital Institutional Review Board.
Author Contributions
CZ, YG, and LF were responsible for the conception and design and study supervision. JW, JZ, JD, and LH were responsible for the development of the methodology, analysis, and experiments. CZ, LF, JW, JZ, CF, HL, and XX performed the statistical and bioinformatic analysis. All authors read and approved the final manuscript.
Funding
This study was co-supported by National Key R&D Program of China (Grant No. 2018YFC1106400), Guangdong Basic and Applied Basic Research Foundation (Grant No. 2020A1515111111), and Beijing iGandan Foundation (Grant No. RGGJJ-2021-008).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.738709/full#supplementary-material
Supplementary Figure 1 | The flowchart of this study.
Abbreviations
TCGA, The Cancer Genome Atlas; RNF, RING finger; DEGs, differentially expressed genes; GO, Gene Ontology; CCK8, cell counting kit-8; HR, hazard ratio; DIA, data-independent acquisition; FDR, false discovery rate; TSS, transcription start sites; GA, genomic alterations; OS, overall survival; RFS, relapse-free survival; DEPs, differentially expressed proteins; BP, biological processes; CC, cellular components; MF, molecular functions; WT, wild type; MUT, mutant; MECOM, MDS1 and EVI1 complex locus; TRIM46, tripartite motif containing 46; KLF4, Kruppel-like factor 4.
Footnotes
- ^ https://portal.gdc.cancer.gov/
- ^ https://hpcwebapps.cit.nih.gov/ESBL/Database/E3-ligases/
- ^ https://maayanlab.cloud/Enrichr/
- ^ http://cbioportal.org
- ^ www.kmplot.com
- ^ http://chip-atlas.org/
References
Backx, E., Wauters, E., Baldan, J., Van Bulck, M., Michiels, E., Heremans, Y., et al. (2021). Mecom permits pancreatic acinar cell dedifferentiation avoiding cell death under stress conditions. Cell Death Differ. 28, 2601–2615. doi: 10.1038/s41418-021-00771-6
Bear, A. S., Vonderheide, R. H., and O’Hara, M. H. (2020). Challenges and opportunities for pancreatic cancer immunotherapy. Cancer Cell 38, 788–802. doi: 10.1016/j.ccell.2020.08.004
Bleu, M., Mermet-Meillon, F., Apfel, V., Barys, L., Holzer, L., Bachmann, S. M., et al. (2021). Pax8 and mecom are interaction partners driving ovarian cancer. Nat. Commun. 12:2442. doi: 10.1038/s41467-021-22708-w
Buscail, L., Bournet, B., and Cordelier, P. (2020). Role of oncogenic kras in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 17, 153–168. doi: 10.1038/s41575-019-0245-4
Cadena, C., Ahmad, S., Xavier, A., Willemsen, J., Park, S., Park, J. W., et al. (2019). Ubiquitin-dependent and -independent roles of e3 ligase riplet in innate immunity. Cell 177, 1187–1200. doi: 10.1016/j.cell.2019.03.017
Cheng, Y., Wang, K., Geng, L., Sun, J., Xu, W., Liu, D., et al. (2019). Identification of candidate diagnostic and prognostic biomarkers for pancreatic carcinoma. EBioMedicine 40, 382–393. doi: 10.1016/j.ebiom.2019.01.003
Christenson, E. S., Jaffee, E., and Azad, N. S. (2020). Current and emerging therapies for patients with advanced pancreatic ductal adenocarcinoma: a bright future. Lancet Oncol. 21, e135–e145. doi: 10.1016/S1470-2045(19)30795-8
Deng, J., Liang, H., Zhang, R., Hou, Y., Liu, Y., Ying, G., et al. (2016). Clinical and experimental role of ring finger protein 180 on lymph node metastasis and survival in gastric cancer. Br. J. Surg. 103, 407–416. doi: 10.1002/bjs.10066
Devgan, S. S., Sanal, O., Doil, C., Nakamura, K., Nahas, S. A., Pettijohn, K., et al. (2011). Homozygous deficiency of ubiquitin-ligase ring-finger protein rnf168 mimics the radiosensitivity syndrome of ataxia-telangiectasia. Cell Death Differ. 18, 1500–1506. doi: 10.1038/cdd.2011.18
Dickson, K. A., Cole, A. J., Gill, A. J., Clarkson, A., Gard, G. B., Chou, A., et al. (2016). The ring finger domain e3 ubiquitin ligases brca1 and the rnf20/rnf40 complex in global loss of the chromatin mark histone h2b monoubiquitination (h2bub1) in cell line models and primary high-grade serous ovarian cancer. Hum. Mol. Genet. 25, 5460–5471. doi: 10.1093/hmg/ddw362
Faber, K., Bullinger, L., Ragu, C., Garding, A., Mertens, D., Miller, C., et al. (2013). Cdx2-driven leukemogenesis involves klf4 repression and deregulated pparγ signaling. J. Clin. Invest. 123, 299–314. doi: 10.1172/JCI64745
Fang, S., Lorick, K. L., Jensen, J. P., and Weissman, A. M. (2003). Ring finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 13, 5–14. doi: 10.1016/s1044-579x(02)00095-0
Fujita, Y., Tinoco, R., Li, Y., Senft, D., and Ronai, Z. A. (2019). Ubiquitin ligases in cancer immunotherapy - balancing antitumor and autoimmunity. Trends Mol. Med. 25, 428–443. doi: 10.1016/j.molmed.2019.02.002
Gamper, A. M., Qiao, X., Kim, J., Zhang, L., DeSimone, M. C., Rathmell, W. K., et al. (2012). Regulation of klf4 turnover reveals an unexpected tissue-specific role of pvhl in tumorigenesis. Mol. Cell. 45, 233–243. doi: 10.1016/j.molcel.2011.11.031
Geng, R., Tan, X., Zuo, Z., Wu, J., Pan, Z., Shi, W., et al. (2017). Synthetic lethal short hairpin rna screening reveals that ring finger protein 183 confers resistance to trametinib in colorectal cancer cells. Chin. J. Cancer. 36:63. doi: 10.1186/s40880-017-0228-1
Gong, L., Song, J., Lin, X., Wei, F., Zhang, C., Wang, Z., et al. (2016). Serine-arginine protein kinase 1 promotes a cancer stem cell-like phenotype through activation of wnt/β-catenin signalling in nsclc. J. Pathol. 240, 184–196. doi: 10.1002/path.4767
Grossberg, A. J., Chu, L. C., Deig, C. R., Fishman, E. K., Hwang, W. L., Maitra, A., et al. (2020). Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J. Clin. 70, 375–403. doi: 10.3322/caac.21626
Guo, K., Cui, J., Quan, M., Xie, D., Jia, Z., Wei, D., et al. (2017). The novel klf4/msi2 signaling pathway regulates growth and metastasis of pancreatic cancer. Clin. Cancer Res. 23, 687–696. doi: 10.1158/1078-0432.CCR-16-1064
Hahn, W. C., Dessain, S. K., Brooks, M. W., King, J. E., Elenbaas, B., Sabatini, D. M., et al. (2002). Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22, 2111–2123. doi: 10.1128/MCB.22.7.2111-2123.2002
He, D., Miao, H., Xu, Y., Xiong, L., Wang, Y., Xiang, H., et al. (2014). Mir-371-5p facilitates pancreatic cancer cell proliferation and decreases patient survival. PLoS One 9:e112930. doi: 10.1371/journal.pone.0112930
Hong, S. Y., Kao, Y. R., Lee, T. C., and Wu, C. W. (2018). Upregulation of e3 ubiquitin ligase cblc enhances egfr dysregulation and signaling in lung adenocarcinoma. Cancer Res. 78, 4984–4996. doi: 10.1158/0008-5472.CAN-17-3858
Hosein, A. N., Brekken, R. A., and Maitra, A. (2020). Pancreatic cancer stroma: an update on therapeutic targeting strategies. Nat. Rev. Gastroenterol. Hepatol. 17, 487–505. doi: 10.1038/s41575-020-0300-1
Hsieh, M. H., Chen, Y. T., Chen, Y. T., Lee, Y. H., Lu, J., Chien, C. L., et al. (2017). Parp1 controls klf4-mediated telomerase expression in stem cells and cancer cells. Nucleic Acids Res. 45, 10492–10503. doi: 10.1093/nar/gkx683
Hu, D., Gur, M., Zhou, Z., Gamper, A., Hung, M. C., Fujita, N., et al. (2015). Interplay between arginine methylation and ubiquitylation regulates klf4-mediated genome stability and carcinogenesis. Nat. Commun. 6:8419. doi: 10.1038/ncomms9419
Jiang, X., Hao, H. X., Growney, J. D., Woolfenden, S., Bottiglio, C., Ng, N., et al. (2013). Inactivating mutations of rnf43 confer wnt dependency in pancreatic ductal adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 110, 12649–12654. doi: 10.1073/pnas.1307218110
Joazeiro, C. A., and Weissman, A. M. (2000). Ring finger proteins: mediators of ubiquitin ligase activity. Cell 102, 549–552. doi: 10.1016/s0092-8674(00)00077-5
Kannan, K., Kordestani, G. K., Galagoda, A., Coarfa, C., and Yen, L. (2015). Aberrant muc1-trim46-krtcap2 chimeric rnas in high-grade serous ovarian carcinoma. Cancers (Basel) 7, 2083–2093. doi: 10.3390/cancers7040878
Khan, S., He, Y., Zhang, X., Yuan, Y., Pu, S., Kong, Q., et al. (2020). Proteolysis targeting chimeras (protacs) as emerging anticancer therapeutics. Oncogene 39, 4909–4924. doi: 10.1038/s41388-020-1336-y
Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97. doi: 10.1093/nar/gkw377
Kumari, N., Jaynes, P. W., Saei, A., Iyengar, P. V., Richard, J., and Eichhorn, P. (2017). The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim. Biophys. Acta Rev. Cancer 1868, 456–483. doi: 10.1016/j.bbcan.2017.09.002
Li, D., Peng, Z., Tang, H., Wei, P., Kong, X., Yan, D., et al. (2011). Klf4-mediated negative regulation of ifitm3 expression plays a critical role in colon cancer pathogenesis. Clin. Cancer Res. 17, 3558–3568. doi: 10.1158/1078-0432.CCR-10-2729
Li, Q., Jia, Z., Wang, L., Kong, X., Li, Q., Guo, K., et al. (2012). Disruption of klf4 in villin-positive gastric progenitor cells promotes formation and progression of tumors of the antrum in mice. Gastroenterology 142, 531–542. doi: 10.1053/j.gastro.2011.11.034
Li, W., Jiang, Z., Li, T., Wei, X., Zheng, Y., Wu, D., et al. (2015). Genome-wide analyses identify klf4 as an important negative regulator in t-cell acute lymphoblastic leukemia through directly inhibiting t-cell associated genes. Mol. Cancer 14:26. doi: 10.1186/s12943-014-0285-x
Liang, Q., Ma, D., Zhu, X., Wang, Z., Sun, T. T., Shen, C., et al. (2018). Ring-finger protein 6 amplification activates jak/stat3 pathway by modifying shp-1 ubiquitylation and associates with poor outcome in colorectal cancer. Clin. Cancer Res. 24, 1473–1485. doi: 10.1158/1078-0432.CCR-17-2133
Lipkowitz, S., and Weissman, A. M. (2011). Rings of good and evil: ring finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer. 11, 629–643. doi: 10.1038/nrc3120
Martens, S., Lefesvre, P., Nicolle, R., Biankin, A. V., Puleo, F., Van Laethem, J. L., et al. (2019). Different shades of pancreatic ductal adenocarcinoma, different paths towards precision therapeutic applications. Ann. Oncol. 30, 1428–1436. doi: 10.1093/annonc/mdz181
McHugh, A., Fernandes, K., Chinner, N., Ibrahim, A., Garg, A. K., Boag, G., et al. (2020). The identification of potential therapeutic targets for cutaneous squamous cell carcinoma. J. Invest. Dermatol. 140, 1154–1165. doi: 10.1016/j.jid.2019.09.024
Medvar, B., Raghuram, V., Pisitkun, T., Sarkar, A., and Knepper, M. A. (2016). Comprehensive database of human e3 ubiquitin ligases: application to aquaporin-2 regulation. Physiol. Genomics 48, 502–512. doi: 10.1152/physiolgenomics.00031.2016
Micel, L. N., Tentler, J. J., Smith, P. G., and Eckhardt, G. S. (2013). Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J. Clin. Oncol. 31, 1231–1238. doi: 10.1200/JCO.2012.44.0958
Mizrahi, J. D., Surana, R., Valle, J. W., and Shroff, R. T. (2020). Pancreatic cancer. Lancet 395, 2008–2020. doi: 10.1016/S0140-6736(20)30974-0
Murgai, M., Ju, W., Eason, M., Kline, J., Beury, D. W., Kaczanowska, S., et al. (2017). Klf4-dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nat. Med. 23, 1176–1190. doi: 10.1038/nm.4400
Nagy, Á, Munkácsy, G., and Győrffy, B. (2021). Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 11:6047. doi: 10.1038/s41598-021-84787-5
Park, C. S., Lewis, A. H., Chen, T. J., Bridges, C. S., Shen, Y., Suppipat, K., et al. (2019). A klf4-dyrk2-mediated pathway regulating self-renewal in cml stem cells. Blood 134, 1960–1972. doi: 10.1182/blood.2018875922
Plechanovová, A., Jaffray, E. G., McMahon, S. A., Johnson, K. A., Navrátilová, I., Naismith, J. H., et al. (2011). Mechanism of ubiquitylation by dimeric ring ligase rnf4. Nat. Struct. Mol. Biol. 18, 1052–1059. doi: 10.1038/nsmb.2108
Qi, J., and Ronai, Z. A. (2015). Dysregulation of ubiquitin ligases in cancer. Drug Resist. Updat. 23, 1–11. doi: 10.1016/j.drup.2015.09.001
Qiu, Y., Zhu, H., Xu, D., Feng, Q., Wen, C., Du, Y., et al. (2021). Ring-finger protein 6 enhances c-myc-mediated warburg effect by promoting mad1 degradation to facilitate pancreatic cancer metastasis. Am. J. Cancer Res. 11, 2025–2043.
Rape, M. (2018). Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70. doi: 10.1038/nrm.2017.83
Rowland, B. D., and Peeper, D. S. (2006). Klf4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 6, 11–23. doi: 10.1038/nrc1780
Sakamoto, H., Kuboki, Y., Hatori, T., Yamamoto, M., Sugiyama, M., Shibata, N., et al. (2015). Clinicopathological significance of somatic rnf43 mutation and aberrant expression of ring finger protein 43 in intraductal papillary mucinous neoplasms of the pancreas. Mod. Pathol. 28, 261–267. doi: 10.1038/modpathol.2014.98
Seipel, K., Marques, M. T., Bozzini, M. A., Meinken, C., Mueller, B. U., and Pabst, T. (2016). Inactivation of the p53-klf4-cebpa axis in acute myeloid leukemia. Clin. Cancer Res. 22, 746–756. doi: 10.1158/1078-0432.CCR-15-1054
Senft, D., Qi, J., and Ronai, Z. A. (2018). Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 18, 69–88. doi: 10.1038/nrc.2017.105
Sharp, M. F., Bythell-Douglas, R., Deans, A. J., and Crismani, W. (2021). The fanconi anemia ubiquitin e3 ligase complex as an anti-cancer target. Mol. Cell. 81, 2278–2289. doi: 10.1016/j.molcel.2021.04.023
Shi, M., Cui, J., Du, J., Wei, D., Jia, Z., Zhang, J., et al. (2014). A novel klf4/ldha signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin. Cancer Res. 20, 4370–4380. doi: 10.1158/1078-0432.CCR-14-0186
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30. doi: 10.3322/caac.21590
Sun, P., Shen, Y., Wang, T., He, Y., Zhang, Y., Tian, W., et al. (2021). Distinct clinical and genetic mutation characteristics in sporadic and lynch syndrome-associated endometrial cancer in a chinese population. Cancer Epidemiol. 73:101934. doi: 10.1016/j.canep.2021.101934
Sun, Y., and Li, H. (2013). Functional characterization of sag/rbx2/roc2/rnf7, an antioxidant protein and an e3 ubiquitin ligase. Protein Cell 4, 103–116. doi: 10.1007/s13238-012-2105-7
Tetreault, M. P., Wang, M. L., Yang, Y., Travis, J., Yu, Q. C., Klein-Szanto, A. J., et al. (2010). Klf4 overexpression activates epithelial cytokines and inflammation-mediated esophageal squamous cell cancer in mice. Gastroenterology 139, 2124–2134. doi: 10.1053/j.gastro.2010.08.048
Tsai, Y. C., Mendoza, A., Mariano, J. M., Zhou, M., Kostova, Z., Chen, B., et al. (2007). The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting kai1 for degradation. Nat. Med. 13, 1504–1509. doi: 10.1038/nm1686
Wang, D., Ma, L., Wang, B., Liu, J., and Wei, W. (2017). E3 ubiquitin ligases in cancer and implications for therapies. Cancer Metastasis Rev. 36, 683–702. doi: 10.1007/s10555-017-9703-z
Wei, D., Wang, L., Yan, Y., Jia, Z., Gagea, M., Li, Z., et al. (2016). Klf4 is essential for induction of cellular identity change and acinar-to-ductal reprogramming during early pancreatic carcinogenesis. Cancer Cell 29, 324–338. doi: 10.1016/j.ccell.2016.02.005
Weinelt, N., and van Wijk, S. (2021). Ubiquitin-dependent and -independent functions of otulin in cell fate control and beyond. Cell Death Differ. 28, 493–504. doi: 10.1038/s41418-020-00675-x
Witkiewicz, A. K., McMillan, E. A., Balaji, U., Baek, G., Lin, W. C., Mansour, J., et al. (2015). Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6:6744. doi: 10.1038/ncomms7744
Xie, V. K., Li, Z., Yan, Y., Jia, Z., Zuo, X., Ju, Z., et al. (2017). DNA-methyltransferase 1 induces dedifferentiation of pancreatic cancer cells through silencing of krüppel-like factor 4 expression. Clin. Cancer Res. 23, 5585–5597. doi: 10.1158/1078-0432.CCR-17-0387
Yu, T., Chen, X., Zhang, W., Liu, J., Avdiushko, R., Napier, D. L., et al. (2016). Klf4 regulates adult lung tumor-initiating cells and represses k-ras-mediated lung cancer. Cell Death Differ. 23, 207–215. doi: 10.1038/cdd.2015.85
Zhang, L., Zhou, Q., Qiu, Q., Hou, L., Wu, M., Li, J., et al. (2019). Circplekhm3 acts as a tumor suppressor through regulation of the mir-9/brca1/dnajb6/klf4/akt1 axis in ovarian cancer. Mol. Cancer 18:144. doi: 10.1186/s12943-019-1080-5
Zhang, Q., Meng, Y., Zhang, L., Chen, J., and Zhu, D. (2009). Rnf13: a novel ring-type ubiquitin ligase over-expressed in pancreatic cancer. Cell Res. 19, 348–357. doi: 10.1038/cr.2008.285
Zhang, Q., Zhang, Y., Sun, S., Wang, K., Qian, J., Cui, Z., et al. (2021). Acox2 is a prognostic marker and impedes the progression of hepatocellular carcinoma via pparα pathway. Cell Death Dis. 12:15. doi: 10.1038/s41419-020-03291-2
Zheng, N., and Shabek, N. (2017). Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157. doi: 10.1146/annurev-biochem-060815-014922
Keywords: pancreatic cancer, E3 ubiquitin ligase, prognosis, RNF223, KLF4, metabolism
Citation: Feng L, Wang J, Zhang J, Diao J, He L, Fu C, Liao H, Xu X, Gao Y and Zhou C (2021) Comprehensive Analysis of E3 Ubiquitin Ligases Reveals Ring Finger Protein 223 as a Novel Oncogene Activated by KLF4 in Pancreatic Cancer. Front. Cell Dev. Biol. 9:738709. doi: 10.3389/fcell.2021.738709
Received: 09 July 2021; Accepted: 15 September 2021;
Published: 14 October 2021.
Edited by:
Jian-ye Zhang, Guangzhou Medical University, ChinaReviewed by:
Omer Faruk Bayrak, Yeditepe University, TurkeyCamilla Abbehausen, State University of Campinas, Brazil
Copyright © 2021 Feng, Wang, Zhang, Diao, He, Fu, Liao, Xu, Gao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Gao, Z2FveWk2MTQ2QDE2My5jb20=; Chenjie Zhou, c2FudGNydXNAc211LmVkdS5jbg==
†ORCID: Yi Gao, orcid.org/0000-0002-4536-6191; Lei Feng, orcid.org/0000-0001-6053-8417; Chenjie Zhou, orcid.org/0000-0002-8774-6846
‡These authors have contributed equally to this work and share first authorship
 Lei Feng
Lei Feng Jieqing Wang2‡
Jieqing Wang2‡ Chaoyi Fu
Chaoyi Fu Xiaoping Xu
Xiaoping Xu Yi Gao
Yi Gao Chenjie Zhou
Chenjie Zhou