Abstract
In the last decade, the CRISPR/Cas9 bacterial virus defense system has been adapted as a user-friendly, efficient, and precise method for targeted mutagenesis in eukaryotes. Though CRISPR/Cas9 has proven effective in a diverse range of organisms, it is still most often used to create mutant lines in lab-reared genetic model systems. However, one major advantage of CRISPR/Cas9 mutagenesis over previous gene targeting approaches is that its high efficiency allows the immediate generation of near-null mosaic mutants. This feature could potentially allow genotype to be linked to phenotype in organisms with life histories that preclude the establishment of purebred genetic lines; a group that includes the vast majority of vertebrate species. Of particular interest to scholars of early vertebrate evolution are several long-lived and slow-maturing fishes that diverged from two dominant modern lineages, teleosts and tetrapods, in the Ordovician, or before. These early-diverging or “basal” vertebrates include the jawless cyclostomes, cartilaginous fishes, and various non-teleost ray-finned fishes. In addition to occupying critical phylogenetic positions, these groups possess combinations of derived and ancestral features not seen in conventional model vertebrates, and thus provide an opportunity for understanding the genetic bases of such traits. Here we report successful use of CRISPR/Cas9 mutagenesis in one such non-teleost fish, sterlet Acipenser ruthenus, a small species of sturgeon. We introduced mutations into the genes Tyrosinase, which is needed for melanin production, and Sonic hedgehog, a pleiotropic developmental regulator with diverse roles in early embryonic patterning and organogenesis. We observed disruption of both loci and the production of consistent phenotypes, including both near-null mutants’ various hypomorphs. Based on these results, and previous work in lamprey and amphibians, we discuss how CRISPR/Cas9 F0 mutagenesis may be successfully adapted to other long-lived, slow-maturing aquatic vertebrates and identify the ease of obtaining and injecting eggs and/or zygotes as the main challenges.
Introduction
The central problem in modern biology is understanding how an organism’s one-dimensional genotype, i.e., its linear sequence of nucleotides, gives rise to its four-dimensional phenotype, i.e., its form and function through space and time. For decades, genotype was linked to phenotype by mapping genetic lesions in purebred lines of genetic model organisms. These organisms were carefully selected based on their rapid life cycles and ability to be easily maintained in the laboratory (Hedges, 2002). Mutations occurred either naturally, or were introduced randomly in the genome using radiation, chemicals, or transposable elements. In the 1980s and 1990s, various methods were developed that allowed targeted mutagenesis in select model organisms including Drosophila, Caenorhabditis elegans, and mouse (Robertson et al., 1988; Kaiser and Goodwin, 1990; Jansen et al., 1997; Faruqi et al., 1998). In the 2000s new “one-size-fits-all” gene targeting technologies, including TALENs and Zinc finger nucleases, allowed targeted mutagenesis in a greater variety of genetic models, such as zebrafish (Doyon et al., 2008; Meng et al., 2008; Huang et al., 2011; Wood et al., 2011). While effective, these methods still required the generation of purebred lines to determine the complete phenotype caused by the mutation. Thus, while substantially faster and more refined than previous methods, gene targeting was still largely limited to a handful of conventional genetic model systems. Furthermore, because these organisms were chosen specifically for their atypical life histories, our understanding of gene function was still largely limited to a few isolated twigs on the vast tree of life.
In the 2010s, these gene targeting strategies became largely supplanted by CRISPR/Cas9-mediated mutagenesis (Bassett et al., 2013; Chen et al., 2013; Fu et al., 2013; Hwang et al., 2013; Li-En et al., 2013; Nakayama et al., 2013; Waaijers et al., 2013; Port et al., 2014; Square et al., 2015; Véron et al., 2015; Martin et al., 2016; Suzuki et al., 2018; Rasys et al., 2019). The CRISPR/Cas9 method results in small, targeted lesions when the cell’s DNA repair mechanisms respond to double-stranded DNA breaks created by the Cas9 endonuclease (Barrangou et al., 2007; Jinek et al., 2012; Mali et al., 2013). Practically, CRISPR/Cas9 mutagenesis has several distinct advantages over TALENs and Zinc finger nucleases. Most significantly is its ease of use (Ran et al., 2013). Cas9 endonucleases are commercially available, and quickly and affordably programmed to cleave specific target sequences by binding short guide RNAs (sgRNAs) (Cong et al., 2013; Ran et al., 2013; Hsu et al., 2014). Another major advantage of CRISPR/Cas9 mutagenesis over previous gene targeting methods is its speed and efficiency (Mali et al., 2013; Ran et al., 2013; Fei et al., 2014). Target site cutting occurs minutes or hours after the Cas9-sgRNA complex enters the cytoplasm. In the case of zygotes, this means that the Cas9-sgRNA complex acts before and during early cell cleavage stages, creating mosaic mutant individuals (Mizuno et al., 2014; Yen et al., 2014). With highly efficient sgRNAs, most of the cells in these “F0” mosaic mutant individuals (sometimes called “crispants” (Burger et al., 2016)) will possess biallelic deletions in the targeted sequence (e.g. Blitz et al., 2013; Jao et al., 2013). CRISPR/Cas9 mutagenesis is also extremely versatile (Mali et al., 2013; Hsu et al., 2014). Despite evolving as a component of the prokaryotic adaptive immune system (Barrangou et al., 2007; Deveau et al., 2010; Garneau et al., 2010; Horvath and Barrangou, 2010; Makarova et al., 2011), Cas9 endonuclease appears to function efficiently and specifically in any cell type regardless of species. Thus, CRISPR/Cas9 allows the rapid creation of near-null mutants in virtually any organism whose eggs and/or zygotes are amenable to injection with proteins and RNA.
The efficiency and versatility of CRISPR/Cas9 means that genotype and phenotype can now be linked in organisms not suitable for the establishment of purebred lines. Thus, gene function can now be studied in organisms chosen for features aside from their ability to be lab-reared, including phylogenetic position, possession of derived phenotypes, or similarity to ancestral forms (Stolfi et al., 2014; Square et al., 2015; Trible et al., 2017; Rasys et al., 2019; Crawford et al., 2020; Kiyonari et al., 2021; Mori and Nakamura, 2021). This has opened up the possibility of side-by-side comparisons of gene function across diverse taxa, allowing researchers to more easily deduce ancestral gene functions, and identify the genes underlying novel phenotypes. CRISPR/Cas9 mutagenesis has become a powerful tool for understanding the evolution of genes, genomes, and phenotypes across both large and small evolutionary timescales (Komor et al., 2017).
CRISPR/Cas9 mutagenesis is having a large impact on the study of vertebrate evolution (e.g. Barske et al., 2020; Square et al., 2020; Hawkins et al., 2021). This is because the majority of vertebrates are large, long-lived, slow-maturing organisms, and do not survive well in small laboratory enclosures. This is especially true for taxa that diverged from the two dominant modern lineages, teleosts and tetrapods, in the Ordovician, or before. These so-called “basal” fish include the living jawless fish, hagfish and lamprey, and non-teleost jawed fish such as sturgeon, paddlefish, gar, bichir and bowfin. Unlike zebrafish and mouse, these vertebrates are typically large, with long generation times and extended predatory adulthoods.
Extant sturgeons are the few remaining representatives of once-diverse radiation of non-teleost ray-finned fish, the order acipenseriforms, that diverged from the lineage leading to modern teleosts about 345 million years ago (Hughes et al., 2018; Du et al., 2020). Because of their phylogenetic position, comparisons between sturgeons and teleost models, like zebrafish and medaka, can provide insights into the biology of early ray-finned fish (e.g. Minarik et al., 2017; Stundl et al., 2020). In addition, Acipenseriforms possess a combination of ancestral and derived vertebrate traits not seen in teleosts, including an endoskeleton lacking proper bone, a body armor made of bony scutes, a heterocercal caudal fin, and a lack of teeth in adulthood (Bemis et al., 1997). Several species are also polyploid (Havelka et al., 2013; Rajkov et al., 2014; Symonová et al., 2017) and roe of some is considered a delicacy (Bemis et al., 1997). Together, these characters make sturgeons an object of study for scientists from diverse specializations, from genetics and genomics to developmental and evolutionary biology, to aquaculture and food production. Despite this broad interest, understanding the genetic bases of sturgeon traits is difficult because they are large, long-lived, slow-maturing, and poorly suited for the establishment of purebred lines.
We recently adapted the CRISPR/Cas9 method to the sea lamprey, Petromyzon marinus, and the African clawed frog, Xenopus laevis (Square et al., 2015; Square et al., 2020), to better understand the ancestral functions of vertebrate developmental regulatory genes. Here we report the successful application of the same strategy to the non-teleost jawed fish sterlet, Acipenser ruthenus, a small species of sturgeon (Figure 1A). We then discuss specific variables and general considerations for workers seeking to apply the CRISPR/Cas9 mutagenesis method to other non-teleost fish, or any aquatic vertebrate for which establishing purebred lines is difficult or impossible.
FIGURE 1
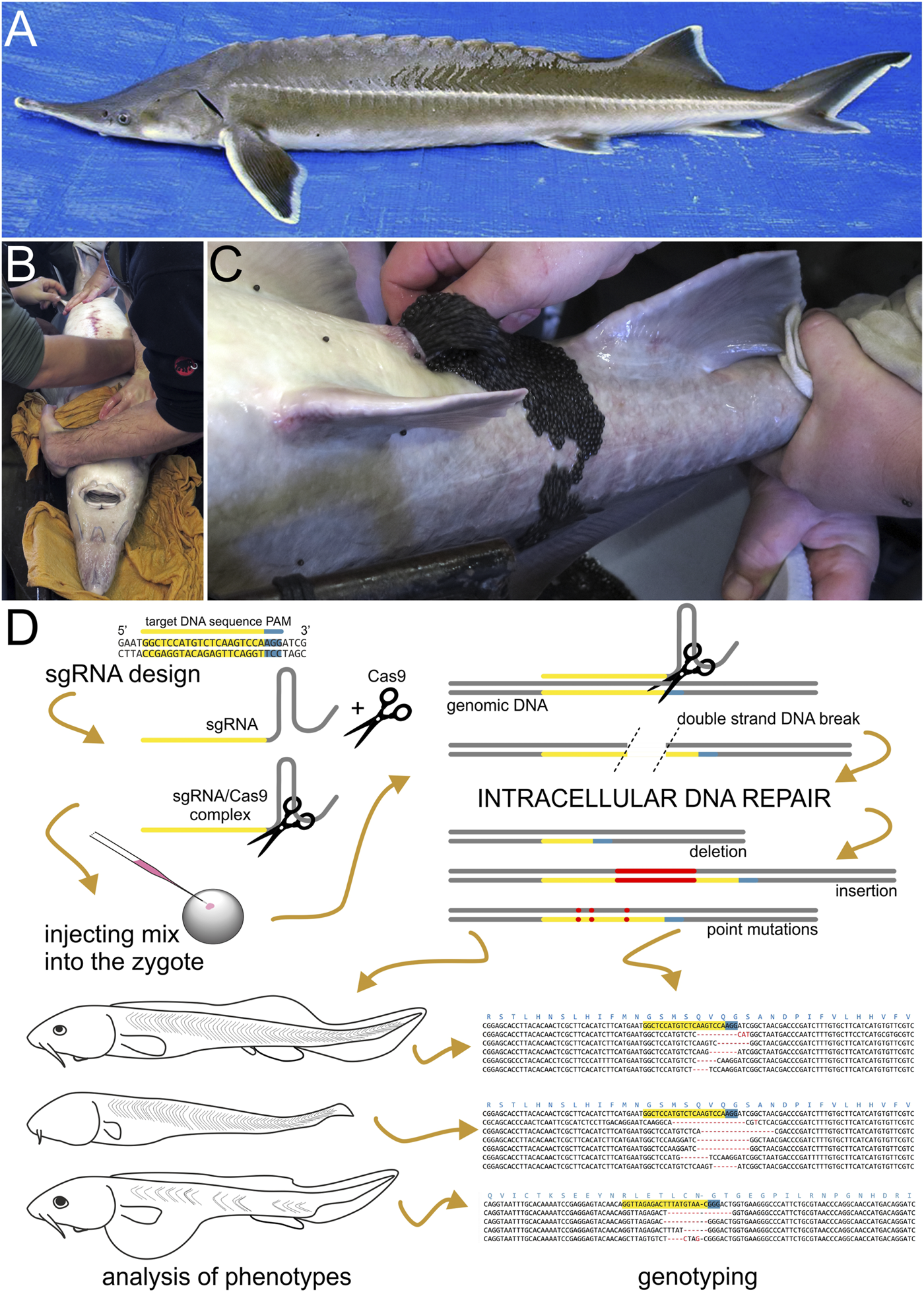
Sturgeon and CRISPR/Cas9 mutagenesis pipeline. (A) Dorsoleteral view of an adult individual of sterlet (Acipenser ruthenus). Abdomen of an adult anesthetized sturgeon female (pictured here is the Siberian sturgeon, Acipenser baerii) in dorsal recumbency (B) is manually massaged in anterior-to-posterior direction and ovulated eggs are collected into a bowl (C). (D) A scheme illustrating the sterlet CRISPR/Cas9 mutagenesis pipeline. First, the single guide RNA (sgRNA) is designed and synthesized based on the selected target DNA sequences and next to Protospacer Adjacent Motif (PAM). PAM is required for proper function of Cas9 nuclease and is not part of sgRNA. Next, the sgRNA is mixed with Cas9 protein (illustrated here as scissors) to form a sgRNA/Cas9 complex. This mix is injected into sterlet zygotes. Cas9 nuclease navigated to the target by sgRNA breaks the double strand of DNA, which is subsequently repaired inside the cleaving cells and nuclei. (mainly by non-homologous end joining) The imperfect DNA repairs introduce mutations into newly synthesized molecules of DNA resulting into a mosaic of cells bearing DNA with indels and/or point mutations at the targeted site. Embryos and/or raised larvae are later inspected for phenotypic effects and selected candidates are genotyped.
Materials and Methods
Animal Husbandry and in vitro Fertilization
We obtained the zygotes of sterlet (Acipenser ruthenus Linnaeus, 1758) from the adults kept and regularly bred at the Research Institute of Fish Culture and Hydrobiology, Faculty of Fisheries and Protection of Waters, University of South Bohemia in České Budějovice, Vodňany, Czech Republic (RIFCH). The husbandry, animal conditioning, gamete collection, and fertilization were described in detail by Chebanov and Galich (2011) and Saito et al. (2014). The adult breeding fish were handled under anesthesia in 0.05% tricaine. Briefly, both female and male adult sterlets, aged five to 9 years, were transferred from the outdoor ponds into 4,000 L indoor tanks with water temperature kept at constant 15°C. Spermiation in males was induced by intramuscular injection of carp pituitary extract at 4 mg/kg body weight in 0.9% NaCl solution. Sperm was collected 48 h later via a 0.6 mm catheter into cell culture flasks and kept on ice until in vitro fertilization. Female ovulation was induced in a similar way, but with two doses of the hormone instead of only one injected 12 h apart −0.5 and 4.5 mg/kg of body weight, respectively. Females ovulated 18–20 h after the hormone was administered. Ovulating females were placed in dorsal recumbency position and oviduct incision was performed using an eye microsurgery scalpel. This minimal invasive procedure allows accessing the ovulated eggs in sturgeon oviduct that is physiologically folded (see Chebanov and Galich, 2011). Then the female’s abdomen was massaged in anterior-to-posterior direction and eggs were collected into bowls (Figures 1B,C), sealed with aluminum foil and subsequently fertilized with sperm at 15°C in dechlorinated tap water. We only used sperm with the spermatozoa motility assessed at >80% (Chebanov and Galich, 2011). The zygotes were rinsed in 0.04% tannic acid to make them less adherent (Saito et al., 2014), and then kept in water at 15°C before they were used in experiments. After the injections, we transferred the embryos to 48-well plates, each in 1 ml volume of E2 zebrafish medium (Brand et al., 2002) containing antibiotics (120 ng/ml of penicillin and 200 ng/ml of streptomycin) kept at 15°C. The medium was changed daily or more often, if necessary. Embryos selected for raising to later developmental stages were transferred to well-oxygenated tanks with E2/antibiotics at 15–17°C shortly after they hatched, which typically happened 7 days post fertilization (st. 35). We staged the embryos and larvae using the staging system of Detlaff et al. (1993). When the embryos and larvae reached the desired developmental stage, they were anesthetized by tricaine (MS-222; Serva) and fixed in 4% PFA in PBS overnight at 4°C. After several washes in PBS, we gradually dehydrated the embryos and larvae through a series of PBS/methanol solutions and stored in 100% methanol at −20°C until further use.
Animal care and all experiments were approved by the Ministry of Agriculture of the Czech Republic (MSMT-12550/2016-3), followed the principles of the European Union Harmonized Animal Welfare Act of the Czech Republic, and Principles of Laboratory Animal Care and National Laws 246/1992 “Animal Welfare”, and were conducted in accordance with the Animal Research Committee of RIFCH. Authors of the study own the Certificate of professional competence for designing experiments and experimental projects under Section 15 d (3) of the Czech Republic Act no. 246/1992 Coll. on the Protection of Animals against Cruelty.
Identification of Endogenous Tyr and Shh Loci in Sterlet Genome and Design of the sgRNAs
We used similar F0 mutagenesis strategy as was described for sea lamprey (Petromyzon marinus) and African clawed frog (Xenopus laevis) (Square et al., 2015; Square et al., 2020) (Figure 1D). Using the spotted gar (Lepisosteus ocelatus) and zebrafish (Danio rerio) tyrosinase and sonic hedgehog protein sequences as queries we searched sterlet Tyr and Shh homologs in our de novo assembled transcriptomes of sterlet pharyngulae (available online at https://www.researchgate.net/profile/David-Jandzik/projects). The identity of recovered sequences was checked with BLAST (Altschul et al., 1990). We targeted the protein-coding exons at loci showing high evolutionary conservation across vertebrate taxa. In both tyr and shh we identified four putative exons. We selected the best CRISPR/Cas9 target sites with sequence 5′-GG (18N) NGG-3′ and with no off-target matches to our transcriptomes. The individual sequences were visually checked and identified as off-targets if they showed more than 85% similarity to our candidate sequence including PAM by BLAST (0–3 mismatches) and the mismatches were close to PAM site (more than one mismatch closer to PAM than 10 bp). The sequences of target sites were as follows: tyr sgRNA 3: GGTTAGAGACTTTATGTAAC (GGG), tyr sgRNA 4: GGCTCCATGTCTCAAGTCCA(AGG), shh gRNA1: (CCC)CAATGTGGCCGAGAAGACCC, shh gRNA2: GGGCCAGTGGCAGATATGAA(GGG) with PAM sites in parentheses and the PCR primers used to amplify the target sequences in Table 1. The amplified fragments were annealed and in vitro phosphorylated with T4 Polynuclease Kinase (NEB M0201S) at 37°C for 1h, and ligated into the DR274 plasmid (Hwang et al., 2013) pre-digested with BsaI (NEB R0535). Single guide RNAs were in vitro transcribed using T7 High Yield Kit (New England Biolabs) and purified by phenol-chloroform extraction followed by precipitation in 70% ethanol with 0.3M sodium acetate. The precipitate was resuspended in nuclease-free water.
TABLE 1
| Target | Forward primer | Reverse primer |
|---|---|---|
| A) | ||
| tyr sgRNA 3 | TAGGTTAGAGACTTTATGTAAC | AAACGTTACATAAAGTCTCTAA |
| tyr sgRNA 4 | TAGGCTCCATGTCTCAAGTCCA | AAACTGGACTTGAGACATGGAG |
| shh sgRNA 1 | TAGGGTCTTCTCGGCCACATTG | AAACCAATGTGGCCGAGAAGAC |
| shh sgRNA 2 | TAGGGCCAGTGGCAGATATGAA | AAACTTCATATCTGCCACTGGC |
| B) | ||
| tyr sgRNA 3 geno | GGAGGAAGCAAACAACATAAGCTACAG | CACGGATATGACTGGAGGTAACAGTC |
| tyr sgRNA 4 geno | GCAGTTTACTTTGCTGCATGTGTG | CCACGTGGCTGTCTATCGGTG |
| shh sgRNA 1and2 geno | CTTTGGTGTCCTCTGGGCTG | GAGCCTGTCAGCCCCAGTG |
| C) | ||
| foxD3 ISH probe | GAYGTGGAYATCGAYGTGGT | CTSARRAARCTVCCGTTGTC |
| ripply3 ISH probe | AGATGCAATCCACGGGCTAC | GTGGATTGTCGCTTGCACAG |
| twist1 ISH probe | GAAAWGWTGCARGANGAATC | TGVGATGYRGACATGGACCA |
Primers used A) to amplify the DNA templates for sgRNAs synthesis, B) for genotyping the putative mutants, and C) to amplify the DNA template for foxD3, ripply3, and twist1 RNA in situ hybridization probe.
Microinjection
We prepared a fresh injection mix on ice shortly before each injection session. The commercially produced recombinant Cas9 protein (PNA Bio Inc.) was resuspended per manufacturer’s instructions to the stock concentration of 1 mg/ml, aliquoted, and stored at −80°C. For a 6 μl injection mix we first incubated 1.6 μg of diluted Cas9 with 800 ng of total sgRNA for 10 min on ice, then brought the total volume up to 5.5 μl with nuclease-free water. Approximately 0.6 μl of 50 μg/μl lysinated Rhodamine-dextran (LRD; Invitrogen) in nuclease-free water was then added, resulting in a final LRD concentration of 5 μg/μl. One-cell-stage embryos were manually dechorionated using Dumont forceps and positioned in shallow holes in modeling clay in a Petri dish to facilitate their proper orientation and stability. The microinjection was performed either with mouth or manual injector (set to 100 hPa for 1 s) using microcapillary needles (Drummond Microcaps) pulled in a Narishige pc-10 puller (58°C with two weight elements; diameter 1.02 mm). The needle tip diameter was adjusted to allow to produce a ∼20 nl drop (approximately 1/7 of the sterlet zygote in diameter) containing around 2.67 ng of sgRNA and 5.3 ng Cas9 protein (in 1:2 wight ratio) in total. While injecting, we targeted the animal pole of the embryo at a ∼45° angle (Figure 1D). After injection the embryos were transferred to 48-well plates with E2/antibiotics at 15°C. We screened the embryos for LRD at the neurula stage (ca. st. 21). To control for mortality rates, we injected in parallel a few batches of embryos with a non-functional sgRNA at the same mix composition and concentrations as the experimental mixes. Each sgRNA was injected multiple times, in eggs from multiple clutches and by several authors of this study.
Analysis of Phenotypes
We assessed the efficiency of tyr mutagenesis in injected larvae raised approximately to 16 mm of total length, when pigmentation is visibly present and conspicuous on the head and body of the wild-type individuals. Based on the severity of pigmentation reduction we scored the observed phenotypes in four categories as 0–25% reduction, 25–50% reduction, 50–75% reduction, and 75–100% reduction. We also checked the larvae for any non-specific malformations and deformities. The embryos injected with shh sgRNAs with Cas9 protein were fixed at the pharyngula stage (st. 28), when several organs regulated by hedgehog signaling have formed. These include heart, pre-oral gut, olfactory and optic placodes, first and second pharyngeal pouches, fore-, mid-, and hindbrain, somites and pronephros. Rather than scoring the global phenotypes of the embryos and comparing the severity of the phenotypes among different characters, we scored each structure separately, recording whether it was present or reduced/deformed. We also used in situ hybridization to visualize changes in expression patterns of neural crest markers foxd3 and twist1 and pharyngeal pouch formation marker ripply3 in mutant embryos.
Genotyping
To confirm mutations of targeted loci, we genotyped selected sterlet embryos and larvae showing variable phenotypes; at least two individuals per sgRNA. Due to higher percentage of individuals showing no obvious reduction in pigmentation, i.e. wild-type-looking phenotype in tyr sgRNA injected larvae, we also genotyped a few of those to obtain a better picture of tyr sgRNA efficiency. In tyr sgRNA injected larvae we used tail clips obtained from freshly euthanized larvae, while in shh mutants we used the whole embryos after we photographed and analyzed their phenotypes or gene expression patterns. We digested the tissue with Proteinase K (80 IU/ml; Sigma-Aldrich) in 1X PCR buffer at 55°C for 12 h. The obtained genomic DNA extracts served as templates in PCR amplification reactions using GoTaq polymerase (Promega) with amplification primers listed in Table 1. The PCR program followed the manufacturer’s recommendations with the primer-specific annealing temperatures of 57.5° for both loci. We subcloned the resulting amplicons into pJet1.2 plasmid (Thermo Fisher) and fragments obtained from purified colony PCR reactions were sequenced using M13 forward and reverse primers. The alignments of mutant and wild-type sequences were prepared manually. We considered an individual to bear a mutant genotype if at least two sequences represented mutant alleles (see Table 4).
In situ Hybridization
Whole mount in situ hybridization (ISH) was carried out as described in detail by Minarik et al. (2017). We retrieved the putative sterlet homolog of ripply3 from our pharyngula transcriptomes. The 317-bp long DNA template sequence was PCR amplified from sterlet cDNA (amplification primers in Table 1) and subcloned into pGEM T-Easy vector (Promega) by standard procedures. Acquisition of foxd3 and twist1 sterlet sequences was described by Stundl et al. (2020). ISHs using injected and wild-type embryos were performed in separate tubes though in parallel and under the precisely same conditions to avoid variations in probe penetration and signal development.
Imaging
All photographs of sterlet embryos and larvae in PBS were taken with Olympus SZX12 stereoscopic microscope using z-stacking Deep Focus technology of QuickPhoto software (Promicra).
Results
We used CRISPR/Cas9 system to induce insertions and deletions (indels) Figure 1D into protein-coding sequences of two sterlet genes; tyr and shh. tyr encodes the enzyme tyrosinase involved in melanin synthesis in vertebrates. Successful mutagenesis is expected to reduce pigmentation of sterlet larvae but should not affect other aspects of their normal development. On the other hand, shh is a developmental regulator of several organ systems, and its mutation is expected to have dramatic effects on early embryonic development of tissues and organ systems derived from neural crest cells and all three germ layers.
Tyr Mutagenesis
We designed and synthesized two sgRNAs targeting two different exons of the sterlet homolog of human Tyr1 - sgRNA three and four against exons 2 and 3, respectively, sequences of which allowed us to design sgRNAs according to our criteria (see Methods and Square et al., 2015; Square et al., 2020). First, we co-injected 50 one-cell sterlet embryos with a mix containing both guides at a total amount of ∼2.67 ng of sgRNA and 5.3 ng of Cas9 protein (in 1:2 wight ratio) per single embryo. The total amount was calculated to match the amount of guide RNA and Cas9 protein relative to the egg size successfully used in Xenopus laevis CRISPR/Cas9 mutagenesis (Square et al., 2015; Square et al., 2020). In total, 32 injected embryos were LRD positive (i.e., with the lysinated Rhodamine-dextran of the injection mix glowing under the fluorescent light) at the neurula stage, however, no larva showed reduction in pigmentation or other developmental malformation. We suspected that this could have resulted from concentration of each individual sgRNA being too low in this combined sgRNA mix and therefore we used the same total amount of the injection mix but with each sgRNA separately in the next experiment. Both these experiments produced albinos with the variable extent of pigment reduction and normal morphology lacking any visible morphological abnormalities. We visually assessed the extent of pigment reduction and scored the phenotypes to four classes at increments of 25% pigment reduction (Table 2). Injection with sgRNA three resulted in >70% partial or total albinism occurrence, while treatment with sgRNA four produced >50% partially or completely albinotic larvae. Mutagenesis with sgRNA three was more efficient in producing phenotypes with stronger pigment reduction, although the numbers of complete or near-complete albinos were similar between the guides. We recorded 25–33% mortality, which was similar to the uninjected dechorionated wild-type larvae raised to the same stage (Table 2) and to the mortality of embryos injected with non-functional sgRNA (14/50 = 28%).
TABLE 2
| tyr sgRNA 3 | tyr sgRNA 4 | |
|---|---|---|
| 0–25% pigment reduction | 29% (6/21) | 50% (9/18) |
| 25–50% pigment reduction | 29% (6/21) | 26% (5/18) |
| 50–75% pigment reduction | 33% (7/21) | 11% (2/18) |
| 75–100% pigment reduction | 10% (2/21) | 11% (2/18) |
| Injected in total | 40 | 30 |
| LRD positive embryos | 21 | 18 |
| Mortality | 10 (25%) | 10 (33%) |
Summary of observed phenotypes of Δtyr sterlet larvae.
We verified the disruption of the target tyr loci by genotyping selected individuals from each phenotype class (Figure 2, Supplementary Figure S1). We found mutant alleles in 7/10 and 4/7 genotyped larvae generated with sgRNA 3 and 4, respectively, and confirmed mutants in all phenotype classes. Approximately half of all obtained sequences were mutant. We observed 27 unique mutant alleles (out of 36 sequences), 22 of which had only deletions, one had only insertion and four had both deletions and insertions (Supplementary Figure S1). While we did not observe a correlation between the relative occurrence of WT vs. mutant alleles and severity of the observed phenotypes, it appears that the stronger phenotypes have more alleles with indels that cause frameshifts (Figure 2, Supplementary Figure S1).
FIGURE 2
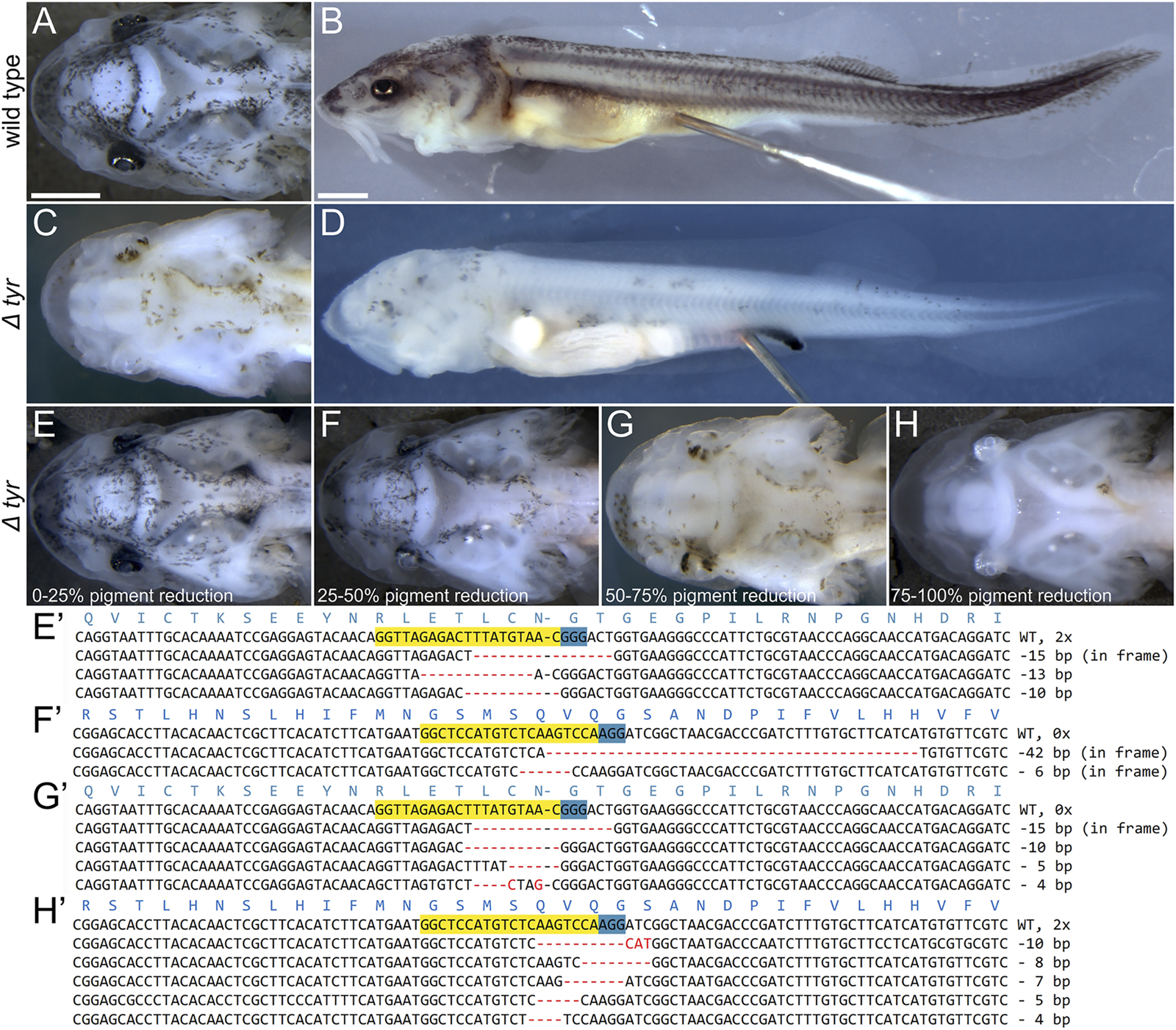
Phenotypes and genotypes of Δtyr sterlet larvae. Dorsal (A,C,E–H) and lateral (B–D) views of ∼16 mm long larvae with anterior to the left. Wild type [WT; (A,B)] and representative mutant (C,D) larvae and four mutant classes (E–H) and their genotypes (E′–H′). The CRISPR target sites and PAM on the forward sequences are highlighted in yellow and blue, respectively, and the red dashes and letters represent deletions, insertions, and polymorphisms relative to the WT sequence. Mutations in (E′,G′), and (F′,H′) were introduced by tyr sgRNA three and sgRNA 4, respectively. Mutant and WT sequences were identified in all phenotype classes (see also Supplementary Figure S1), while sequencing of larvae portrayed on panels (F,H) only returned mutant sequences (as was the case of 3/11 mutant larvae). Scale bars (same in A + C + E-H and B + D) represent 1 mm.
Shh Mutagenesis
Similar to our initial experiments with tyr mutagenesis, we first injected 50 single-cell sterlet embryos with a mix of both guides targeting shh locus with the total amount of ∼2.67 ng of sgRNA and 5.3 ng of Cas9 protein per embryo. We recorded relatively high mortality (36/50 = 72%), but the surviving LRD positive embryos (13; one was LRD negative) showed no discernable phenotypic effect. Next, we injected each sgRNA separately at the same total amount and sgRNA/Cas9 ratio. Due to high mortality in the initial experiment, we injected higher numbers with each guide in a separate mix - 191 with sgRNA 1 and 117 with sg RNA 2. We observed mortality of 34 and 38%, respectively (64/191 individuals in sgRNA one and 44/117 in sgRNA two injections) and obtained 93 and 57 LRD positive embryos that reached developmental st. 28. The embryos were severely affected with strong morphological deformations in almost all embryonic structures present at that stage (Table 3). The highest frequency phenotypes included missing pharyngeal pouches and pre-oral gut (up to 81%), followed by missing, reduced or malformed optic placode, not discernible heart, pronephros reduction and brain malformations. Such deformations were consistent with patterns of shh gene expression observed during the sterlet embryonic stages preceding the analyzed stages (Figures 3A–C). Only 7–10% of injected LRD positive individuals appeared to have no visible phenotype (9/93 and 4/57 for sgRNA 1 and 2, respectively). We did not observe any of the described phenotypes in the control embryos and their mortality was similar to the mortality in tyr mutagenesis experiment.
TABLE 3
| A) | shh sgRNA 1 | shh sgRNA 2 | ||
|---|---|---|---|---|
| Missing/not visible | Reduced/malformed | Missing | Reduced/malformed | |
| Heart | 23% (21/93) | — | 16% (9/57) | — |
| Pre-oral gut | 47% (44/93) | — | 46% (26/57) | — |
| Olfactory placode | 13% (12/93) | 8% (7/93) | 12% (7/57) | 9% (5/57) |
| Optic placode | 22% (20/93) | 34% (32/93) | 23% (13/57) | 30% (23/57) |
| Phar. pouch 1 | 73% (68/93) | — | 81% (46/57) | — |
| Phar. pouch 2 | 69% (64/93) | — | 68% (39/57) | — |
| Forebrain | 9% (8/93) | 24% (22/93) | 7% (4/57) | 21% (12/57) |
| Midbrain | 6% (6/93) | 17% (16/93) | 7% (4/57) | 12% (7/57) |
| Hindbrain | 6% (6/93) | 18% (17/93) | 4% (2/57) | 14% (8/57) |
| Somites | 9% (8/93) | — | 5% (3/57) | — |
| Pronefros | — | 33% (31/93) | — | 26% (15/57) |
| 100% Healthy embryos | 10% (9/93) | 7% (4/57) | ||
| B) | missing | Shh mutant | reduced | |
| foxd3 ISH | ||||
| -Trigeminal NC | 0/3 | 3/3 | ||
| -Hyoid NC | 0/3 | 3/3 | ||
| -Branchial NC | 0/3 | 3/3 | ||
| 100% Healthy embryos | 0/3 | |||
| twist1 ISH | ||||
| -Maxillary NC | 1/14 | 3/14 | ||
| -Mandibular NC | 4/14 | 3/14 | ||
| -Hyoid NC | 1/14 | 9/14 | ||
| -Branchial 1 NC | 1/14 | 7/14 | ||
| -Branchial 2 NC | 5/14 | 5/14 | ||
| 100% Healthy embryos | 3/14 | |||
| ripply3 ISH | ||||
| -Pharyngeal pouch 1 | 4/7 | 3/7 | ||
| -Pharyngeal pouch 2 | 0/7 | 7/7 | ||
| -Pharyngeal pouch 3 | 4/7 | 3/7 | ||
| 100% Healthy embryos | 0/7 | |||
Summary of observed phenotypic effects of Δshh sterlet embryos.
FIGURE 3
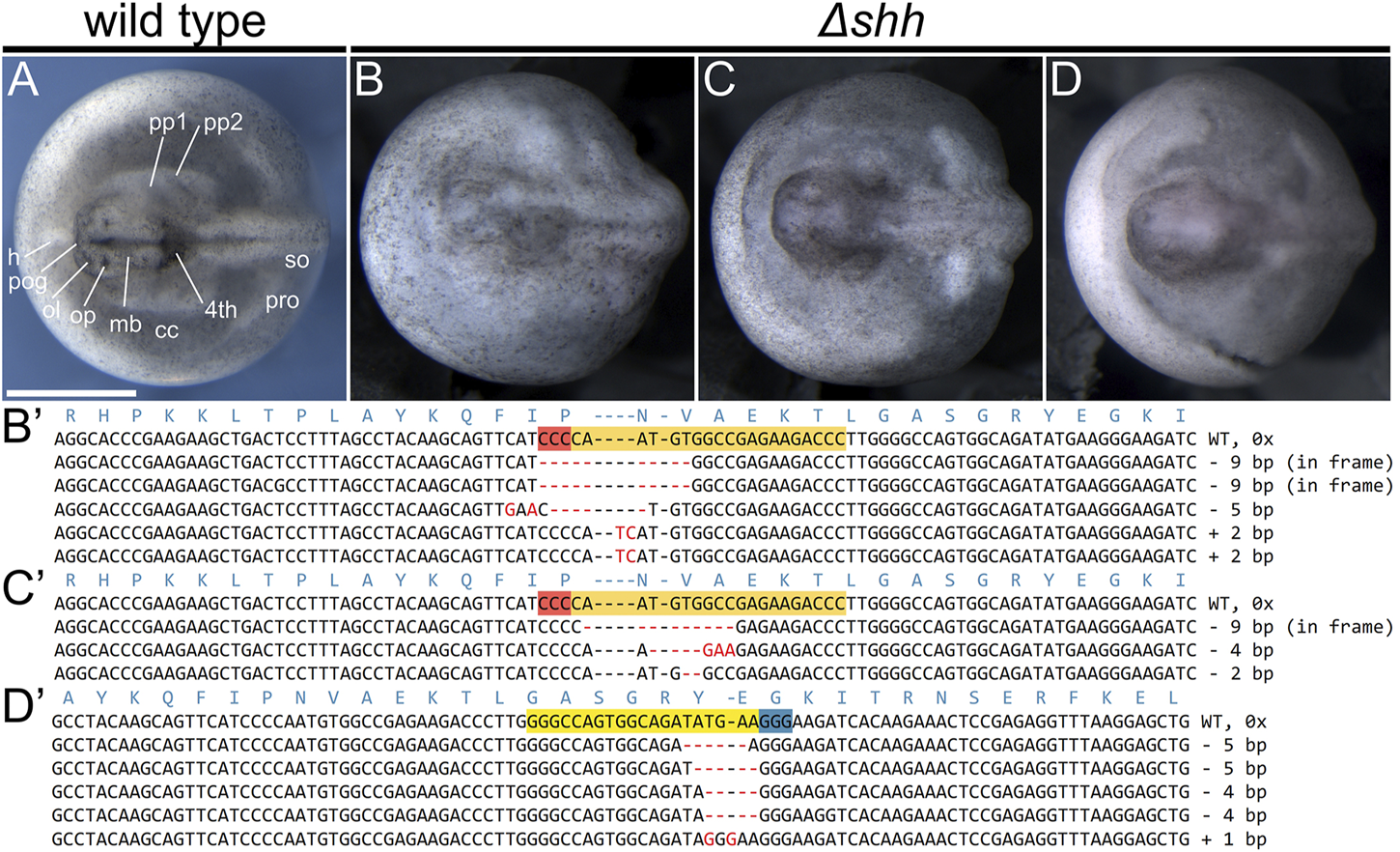
Phenotypes and genotypes of Δshh sterlet larvae. Dorsal views of st. 28 larvae with anterior to the left. Wild type [WT; (A)] and mutant (B–D) embryos and their mutant alleles (B′–D′). The CRISPR target sites are highlighted in orange with red PAM on the reverse strand sequences, while they are yellow with blue PAM on the forward strand. The red dashes and letters represent deletions, insertions, and polymorphisms relative to the WT sequence. Mutations in (B′,C′) were introduced using shh sgRNA 1, while the mutations in (D′) resulted from injections of shh sgRNA 2. No WT sequences were identified in any of the individuals showing shh phenotype (six genotyped individuals in total). The mutant embryo in (B) was scored as missing pre-oral gut, optic and olfactory placodes, forebrain, midbrain, part of the hindbrain, and pharyngeal pouches and with underdeveloped pronephros. The mutant embryo in (C) is missing pharyngeal pouches and has deformed forebrain and midbrain. The mutant embryo in (D) was identified as missing pharyngeal pouches and somites, the entire head is underdeveloped. Fourth—fourth brain chamber of hindbrain, cc—coelomic cavity, h—heart, mb—midbrain, ol—olfactory placode, op—optic placode, pog—pre-oral gut, pp—pharyngeal pouch, pro—pronephros, so—somites. Scale bar represents 1 mm.
Next, we analyzed effects of shh disruption on expression patterns in injected sterlet embryos by ISH. We examined three genes known to be involved in development of some of the affected structures at early embryonic stages: foxd3 marking NCCs at early migration stage, twist1 NCCs at later stages of migration, and ripply3 in forming pharyngeal pouches (Figure 4, Supplementary Figure S3). We found that all these markers were reduced in the injected sterlets at st. 28. foxd3 showed a reduction in the size of all three streams of migrating NCC (trigeminal, hyoid, and branchial), twist1 expression was reduced in all later NCC populations (maxillary, mandibular and branchial) and even entirely missing in some embryos. Only three out of 14 hybridized embryos appeared to have a wild-type expression pattern of the NCC markers. ripply3 ISH confirmed our observation of the strong effect of shh mutation on pharyngeal pouch formation in putative shh mutants; all seven analyzed embryos had either missing or reduced expression domains (Table 3).
FIGURE 4
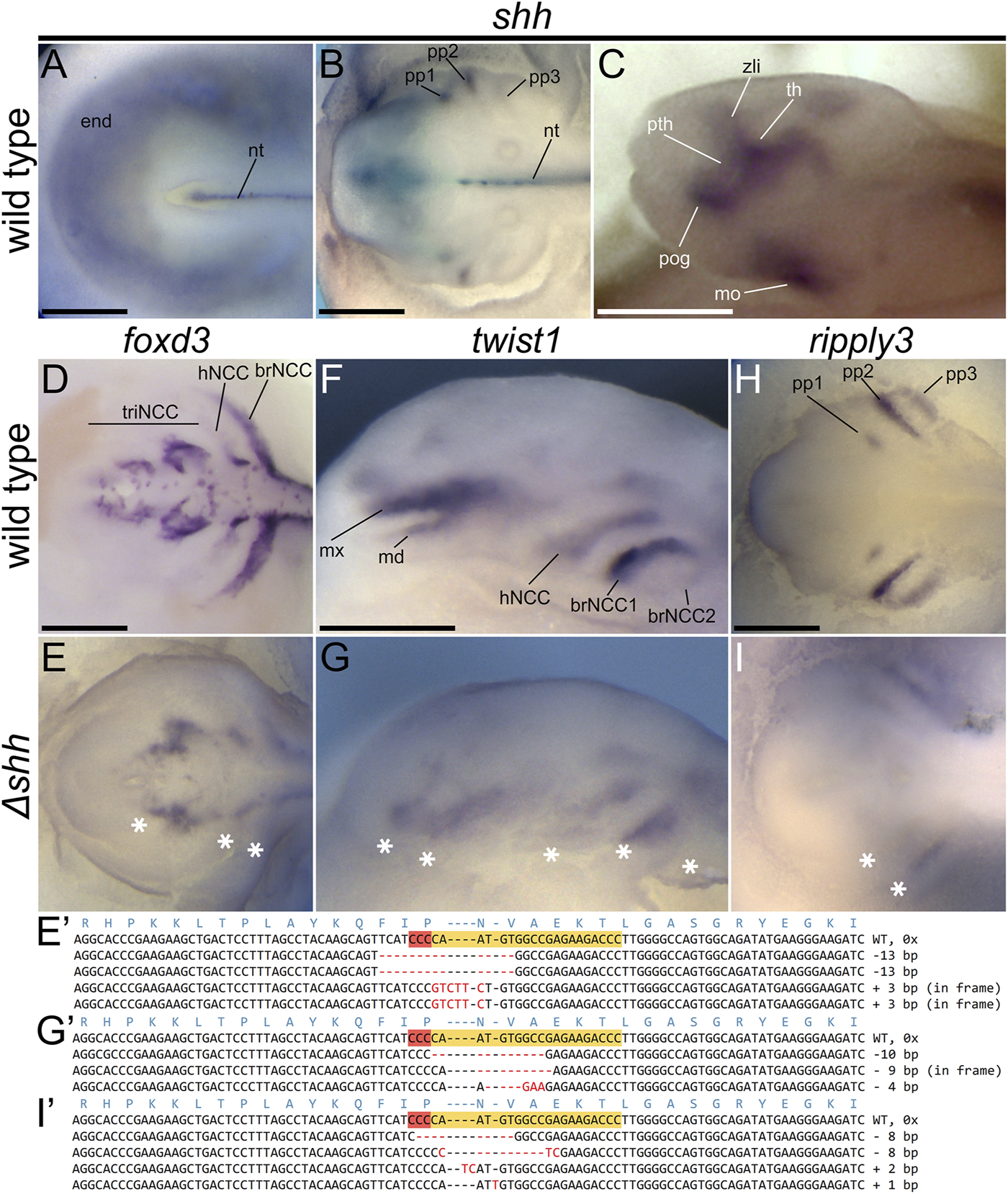
shh expression patterns and Δshh sterlet embryos at st. 28 showing defects in expression patterns of foxd3, twist1, and ripply3. Dorsal (A,B,D,E,H,I) and lateral (C,F,G) views with anterior to the left. Patterns of shh expression in various morphological structures of the wild-type sterlet embryos at stages 23 (A), 28 (B), and 30 (C). foxd3 marks neural crest cells (NCCs) at early stages of migration, twist1 marks NCC at later migration stages, and ripply3 is expressed in developing pharyngeal pouches. Compare with Supplementary Figure S3 to see the observed variation in expression patterns in both wild-type and mutant individuals. The sequences show the reverse DNA strands with shh sgRNA one CRISPR target sites highlighted in orange with red PAM site. The red dashes and letters show indels and polymorphisms relative to the WT sequence. Asterisks indicate missing or reduced expression in mutant embryos. brNCC—branchial stream of NCCs, hNCC—hyoid stream of NCCs, md—mandibular stream of trigeminal stream of NCCs, mo—mouth, mx—maxillary stream of trigeminal stream of NCCs, nt—notochord, pog—pre-oral gut, pth—prethalamus, pp—pharyngeal pouch, th—thalamus, triNCC—trigeminal stream of NCCs, zli—zona limitans intrathalamica. Scale bars represent 0.5 mm.
To verify mutagenesis in shh locus in analyzed sterlet embryos, we genotyped selected individuals injected by both sgRNAs, including those used for ISHs. All 33 obtained sequences had indels and consequently all nine genotyped individuals were confirmed mutants. We identified 21 unique alleles in total, five of which only showed insertions, 12 of them contained deletions and four had both insertions and deletions (Table 4). The majority of the observed mutations caused frameshifts; these were observed in at least half of all obtained sequences of each mutant individual. Only 8/26 sequences of mutations introduced by sgRNA one were in-frame (Figure 4, Figure 3, Supplementary Figure S2, Table 4).
TABLE 4
| Mutant | WT | Frameshift | Mutation rate | Frameshift rate (%) | |
|---|---|---|---|---|---|
| A) Sequences | |||||
| tyr sgRNA 3 | 21/46 | 25/46 | 11/21 | 46% | 52 |
| tyr sgRNA 4 | 15/30 | 15/30 | 8/15 | 50% | 53 |
| shh sgRNA 1 | 26/26 | 0/26 | 18/26 | 100% | 69 |
| shh sgRNA 2 | 7/7 | 0/7 | 7/7 | 100% | 100 |
| B) Unique alleles | |||||
| tyr sgRNA 3 | 14/21 | — | 7/14 | — | 50 |
| tyr sgRNA 4 | 13/15 | — | 8/13 | — | 62 |
| shh sgRNA 1 | 15/26 | 11/15 | 73 | ||
| shh sgRNA 2 | 6/7 | — | 6/6 | — | 100 |
| C) Individuals | |||||
| tyr sgRNA 3 | 7/10 | 3/10 | 6/7 | 70% | 86 |
| tyr sgRNA 4 | 4/7 | 3/7 | 2/4 | 57% | 50 |
| shh sgRNA 1 | 7/7 | 0/7 | 7/7 | 100% | 100 |
| shh sgRNA 2 | 2/2 | 0/2 | 2/2 | 100% | 100 |
Summary of sterlet Δtyr larvae and Δshh embryos genotyping. A) all obtained sequences from all genotyped individuals pooled together, B) unique alleles from all genotyped individuals pooled together, C) all genotyped individuals. The denominator numbers in A represent all obtained sequences from all individuals genotyped for a respective locus, and similarly in B, the denominators show the total number of mutant alleles obtained from sequencing all individuals. In C, the denominators indicate the total numbers of genotyped individuals for each respective locus. Mutation and frameshift rates are calculated for each row, i.e. they represent percentages of mutant sequences and unique alleles from all sequences obtained by genotyping all individuals pooled together in A and B, respectively, while they show percentages of genotyped individuals with mutant/frameshifted genotypes in C.
Discussion
Sterlet is Highly Amenable to CRISPR/Cas9 F0 Mutagenesis
Here we report the application of a CRISPR/Cas9 mutagenesis strategy previously optimized in the sea lamprey (Square et al., 2015) and African clawed frog (Square et al., 2020), to the sterlet. Previous reports using TALENs and CRISPR/Cas9 suggested sterlet zygotes will tolerate injection with proteins and nucleic acids and were amenable to gene targeting (Chen et al., 2018; Baloch et al., 2019). We found that our optimized protocol introduced biallelic mutations in the targeted loci at relatively high efficiency and yielded consistent and reproducible phenotypes. As expected, targeting of the gene tyr, resulted in larval albinism with no other discernible developmental defects. These mutants have similar mortality as their uninjected wild type siblings. The visual scoring of the larval phenotypes is straightforward, suggesting that tyr targeting could serve as a robust positive control in future studies. In contrast to tyr disruption, mutating the pleiotropic developmental regulator shh caused defects in a wide range of embryonic and larval tissues and structures. Interestingly, both sgRNAs used to target shh resulted in all injected individuals displaying defects consistent with the known roles of shh in vertebrate development. This demonstrates that determining the function of developmental regulators, even highly pleiotropic ones, is readily achievable in sterlet. Arguably, in CRISPR/Cas9 sterlet F0 mutants it could be more challenging to detect and score very subtle and less conspicuous phenotypes than in models with stable inbred lines.
Though sterlet is not well suited as a laboratory-propagated model organism, several features of its natural history make it a strong candidate for routine genetic analyses of “F0” mosaic mutants. Along with other sturgeon species, it is a frequently farmed fish, whose eggs can be seasonally obtained in large quantities. The eggs are relatively large and can be easily manipulated, dechorionated, and injected with just forceps and also simple hand-held and mouth operated injector. The embryos and larvae thrive in simple aquaria with no special care requirements beyond clean oxygenated water and a constant temperature of 15–17°C. Several common laboratory methods have also already been optimized in the species including histological sectioning, microCT scanning, RNA in situ hybridization, lineage tracing following injections, and immunohistochemistry (Minarik et al., 2017; Stundl et al., 2020).
Other features of sterlet, and acipenseriform fishes in general, were likely important for the successful application of our CRISPR/Cas9 mutagenesis strategy. Sterlet eggs are rich in yolk that is distributed throughout the egg (Dettlaff et al., 1993). The eggs of lamprey and frog, which are highly amenable to CRISPR/Cas9 mutagenesis (Square et al., 2015; Square et al., 2020), are similarly rich in yolk. Since as in frog, the sterlet egg yolk is slightly more concentrated on the vegetal pole, the animal pole with more yolk-free cytoplasm and nucleus are better accessible for the injected CRISPR/Cas9 mix. Therefore, we tried to target the animal pole, usually conveniently situated on the top of the embryo. However, we suspect that because acipenseriforms undergo holoblastic cleavage (Dettlaff et al., 1993; Ostaszewska and Dabrowski, 2009) injecting any part of the zygote’s cytoplasm would likely work well. Similar to frog, but unlike lamprey, sturgeon eggs have relatively thick envelope. While this does not prevent injection of the embryo with a capillary needle, we preferred to remove the thick outer layer with forceps to better see the injected droplet size and target the animal pole (frog eggs jelly membranes are removed by cysteine treatment; Sive et al., 2000). Due to the similarities between the zygotes of sterlet and those of lamprey with frog, we decided to use approximately the same concentration of injected RNA and protein, and approximately the same relative volume of injection solution. In practical terms, this means we injected a droplet of CRISPR/Cas9 injection mixture with a diameter approximately 1/7 that of the zygote diameter. Because this strategy works well in sterlet, lamprey, and African clawed frog, we suspect a 1/7 size droplet would yield good results in any vertebrate with isolecithal, mesolecithal, or polylecithal eggs displaying holoblastic cleavage. In contrast, zygotes of embryos with meroblastic cleavage (e.g., Takeuchi et al., 2009), such as zebrafish and other teleost fish, appear to tolerate lower amounts of RNA/protein (Hwang et al., 2013; Jao et al., 2013), and lower volumes of injection mix. For such embryos, the injected volume should likely be adjusted based on the size of the blastodisc rather than the entire embryo.
Recent publication of high-quality sterlet genome (Du et al., 2020) should allow the effective design of highly specific sgRNAs. We initially only searched our embryonic transcriptomes for potential off-target sequences. However, additional searches of the new sterlet genome, including non-coding sequences, did not identify any other off-target sequences. When designing sgRNAs, we found that approximately one third of the initially selected guide sequences conformed with off-target sequences according to the criteria described in Methods. When performing CRISPR/Cas9 mutagenesis in sea lamprey and African clawed frog we found that using at least two different sgRNAs per gene, and injecting them separately, is the best strategy for producing verifiable and consistent phenotypes and control for any defects caused by off-target lesions. We observed highly consistent phenotypes and no indication of off-target effects in sea lamprey after mutating more than 20 different genes (Square et al., 2020). The results presented here suggest a similar level of specificity and consistently can be expected in sterlet.
We only observed a moderate disparity in the efficiency of individual sgRNAs. While the mutation rates varied between tyr and shh loci, they were very similar between the guides targeting the same gene (Table 4), with subtle differences recorded among mutant categories. This contrasts with sea lamprey and other species, in which some degree of variation was observed among different guides targeting the same genes (Hsu et al., 2013; Square et al., 2015). We expect that injecting more eggs and guides would result in higher variation in sgRNA efficiency in sterlet as well. On the other hand, as we observed in sea lamprey, mutations causing frameshifts in coding DNA sequences correlated with more severe phenotypes in sterlet as well.
To confirm successful mutagenesis, we also genotyped representative individuals from each phenotypic class. While PCR fragment length is often used to tell whether an individual harbors mutant alleles, we find that this method often produces ambiguous results. We thus chose to clone and sequence genomic fragments including the target sequence and approximately ∼200 bp of flanking sequence on each side. Sequencing of phenotypic mutants and controls, confirmed highly efficient mutagenesis with all tested sgRNAs, with the frequency and type of alleles correlating with phenotype. While even two to three of sequences per individual are sufficient to confirm a baseline level of mutagenesis, we find sequencing ten or more clones gives a view of allelic composition that usually correlates with phenotypes, and also suggests the efficiency of the sgRNA. We also found that, as with lamprey embryos (York et al., 2017), fixed in situ hybridized sterlet embryos can be successfully genotyped if they are not post-fixed with formaldehyde for extended periods of time (see also Square et al., 2020).
Prospects for Adapting CRISPR/Cas9 F0 Mutagenesis to Other “Basal” Fishes
The efficiency, precision, and ease of use of CRISPR/Cas9 mutagenesis in sterlet, lamprey, and African clawed frog strongly suggest this method can be successfully applied to other non-teleost fish. Besides sterlet, several other sturgeon species are currently farmed. While sterlet is probably the most suitable model for developmental studies due to its smaller size and monoploid genome, several other species with different levels of polyploidy (Havelka et al., 2016) could be attractive for studies of genome evolution. While polyploidy necessarily complicates targeted gene mutagenesis by requiring simultaneous disruption of homeologs to yield a valid loss-of-function, the efficiency of CRISPR/Cas9 can be exploited to overcome this issue. We and others have found that targeting conserved homeolog sequences, and/or using multiple sgRNAs can yield strong phenotypes in the allotetraploid African clawed frog (Wang et al., 2015; Square et al., 2020). Gene manipulation in larger species of sturgeons can potentially be useful in sturgeon food production industry when targeting genes involved in growth, immunity, or egg production.
American paddlefish (Polyodon spathula) is morphologically unusual acipenseriform relative of sturgeons. Like sturgeon, it is also frequently farmed, and its embryos have been used in several evolutionary developmental studies (e.g. Davis et al., 2007; Modrell et al., 2011). There are currently no published reports of CRISPR/Cas9 mutagenesis in paddlefish. However, high similarity of paddlefish and sterlet embryos, and successful previous experiments involving dye injections at later embryonic stages suggest that CRISPR/Cas9 mutagenesis could be highly effective in paddlefish. Unfortunately, collecting of early paddlefish embryos appears to be more challenging than sterlet in part because of the shorter paddlefish spawning season (Modrell et al., 2017).
Besides acipenseriforms, three non-teleost fish lineages have extant members that have been utilized in comparative embryological studies and have published genomes - bichirs (Polypteriformes), gars (Lepisosteiformes), and bowfins (Amiiformes) (Askary et al., 2016; Braasch et al., 2016; Minarik et al., 2017; Stundl et al., 2019; Funk et al., 2020; Stundl et al., 2020; Thompson et al., 2021; Bi et al., 2021). While bichirs and bowfins are evolutionarily very attractive and informative species with unique morphologies, the main challenge in targeted mutagenesis in these organisms is obtaining zygotes for injection. Bichirs reproduce in aquaculture settings and can occasionally spawn in home aquaria. However, their mating is secretive and usually occurs at night. Thus, collection of zygotes requires constant observation, which could disturb the animals and prevent spawning. Once collected, though, bichir embryos are relatively sturdy, and can be manipulated and injected with vital dyes much like African clawed frog (Stundl et al., 2019). Bowfin eggs are typically only collected in the wild, and their zygotes are thus less accessible than those of bichir (Funk et al., 2020). Fortunately, decapsulating of later bowfin embryonic stages is relatively easy, so it is likely that early embryos could be injected if obtained (Brent Hawkins, personal communication). Cleavage of bowfin embryos is intermediate between the holoblastic mode of most non-teleost fishes and the meroblastic cleavage of teleosts, with reduced cleavage on the vegetal hemisphere (Cooper and Virta, 2007). This would suggest that if bowfin zygotes were collected, a smaller injection volume might be needed for embryos to survive. Besides sturgeons, perhaps the most promising non-teleost fish candidates for CRISPR/Cas9 mutagenesis are gars. Several species are kept and propagated at fish farms, and like in sturgeons, large quantities of eggs can be seasonally obtained in a controlled fashion in vitro fertilization (Braasch et al., 2014 in spotted gar; Minarik et al., 2017 in tropical gar). Subsequent manipulation with zygotes and raising the offspring is not complicated and the similarity of gar and sturgeon eggs should make injecting relatively straightforward. One potentially confounding factor worth mentioning is that gar embryos, like those of lamprey, lack maternal pigmentation, making it difficult to discern when cleavage begins (Comabella et al., 2014).
While other non-teleost ray-finned fish species are likely well suited to CRISPR/Cas9 F0 mutagenesis, two key groups of early-diverging vertebrates, the chondrichthyans, and hagfish, are unlikely to be amenable to this method, at least as described here. Representatives of both groups have been used in developmental studies in the last 2 decades and have provided some fundamental insights into the developmental bases of vertebrate evolution (Ota et al., 2008; Gillis et al., 2009; Oisi et al., 2013; Gillis and Tidswell, 2017). Embryos of both chondrichthyan clades; elasmobranchs (sharks, rays, and skates) and holocephalans (chimaeras) can be collected seasonally in the wild or in public aquaria, sometimes in the dozens (Gillis, 2011). However, zygotes that could potentially be injected are extremely difficult to obtain and, so far, have not been collected in the numbers needed for reproducible gene disruption. The logistics of hagfish embryology are even more challenging. The few classical reports of hagfish embryology have been based on the scarce embryonic material collected in late 19th century. Only recently have substantial numbers of live hagfish embryos been collected and analyzed (Ota and Kuratani, 2006; Kuratani and Ota, 2008; Ota and Kuratani, 2008). To our knowledge, none of these embryos have ever been successfully injected as zygotes.
Conclusion
Based on the results reported here, and previous experiences of ourselves and others, we posit that our CRISPR/Cas9 mutagenesis strategy will work well in any vertebrate with zygotes that are easily accessed and can tolerate microinjection. In such animals, we expect the main variable determining success will be injection droplet size, which will likely need to be adjusted according to the egg size and yolk distribution. Current limitations seen in chondrichthyans may be circumvented with techniques that allow mass transfection of cells at later embryonic stages, such as electroporation or viral vectors.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by The Animal Research Committee of Research Institute of Fish Culture and Hydrobiology, Faculty of Fisheries and Protection of Waters, University of South Bohemia in České Budějovice, Vodňany, Czech Republic and Ministry of Agriculture of the Czech Republic (MSMT-12550/2016-3).
Author contributions
DJ conceived and designed the project, JS, VS, RF, AP, VP, MP, and DJ performed the experiments, JS and DJ collected and interpreted the data, DJ, DM, and JS wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Funding
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 897949 (to JS). VS was supported by Charles University Research Centre program No. 204069, Charles University Grant SVV 260571/202, and the grant of the Czech Science Foundation GACR 18-04580S. The work of RF and MP was supported by the Ministry of Education, Youth and Sports of the Czech Republic, projects CENAKVA (LM2018099), Biodiversity (CZ.02.1.01/0.0/0.0/16_025/0007370) and Czech Science Foundation (20-23836S). RC was supported by the Czech Science Foundation GACR 19-18634S. MB was supported by National Institutes of Health grant R35NS111564. DM was supported by National Institutes of Health grant NIDCR R21 RDE025940A and National Science Foundation grant IOS 1656843. DJ was supported by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 751066 and by the Scientific Grant Agency of the Slovak Republic VEGA grant no. 1/0450/21.
Acknowledgments
We thank Marek Rodina, David Gela, Michaela Fučíková, and Martin Kahanec for their essential help with sterlet spawns; Vojtěch Miller and Štěpánka Novotná for technical assistance; Radek Šanda for allowing us to use the stereoscopic microscope with Z-stacking. Computational resources were supplied by the Ministry of Education, Youth and Sports of the Czech Republic under the Projects CESNET (Project No. LM2015042) and CERIT-Scientific Cloud (Project No. LM2015085) provided within the program Projects of Large Research, Development and Innovations Infrastructures. D.J. thanks Tyler Square for numerous discussions and priceless advice on everything CRISPR-related and Brent Hawkins for late night fish conversations and insightful comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.750833/full#supplementary-material
References
1
AltschulS. F.GishW.MillerW.MyersE. W.LipmanD. J. (1990). Basic Local Alignment Search Tool. J. Mol. Biol.215, 403–410. 10.1016/S0022-2836(05)80360-2
2
AskaryA.SmeetonJ.PaulS.SchindlerS.BraaschI.EllisN. A.et al (2016). Ancient Origin of Lubricated Joints in Bony Vertebrates. Elife5, e16415. 10.7554/eLife.16415
3
BalochA. R.FraněkR.TichopádT.FučíkováM.RodinaM.PšeničkaM. (2019). Dnd1 Knockout in Sturgeons by CRISPR/Cas9 Generates Germ Cell Free Host for Surrogate Production. Animals9, 174. 10.3390/ani9040174
4
BarrangouR.FremauxC.DeveauH.RichardsM.BoyavalP.MoineauS.et al (2007). CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science315, 1709–1712. 10.1126/science.1138140
5
BarskeL.FabianP.HirschbergerC.JandzikD.SquareT.XuP.et al (2020). Evolution of Vertebrate Gill Covers via Shifts in an Ancient Pou3f3 Enhancer. Proc. Natl. Acad. Sci. USA117, 24876–24884. 10.1073/pnas.2011531117
6
BassettA. R.TibbitC.PontingC. P.LiuJ.-L. (2013). Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cel Rep.4, 220–228. 10.1016/j.celrep.2013.06.020
7
BemisW. E.FindeisE. K.GrandeL. (1997). An Overview of Acipenseriformes. Environ. Biol. Fishes48, 25–71. 10.1023/A:1007370213924
8
BiX.WangK.YangL.PanH.JiangH.WeiQ.et al (2021). Tracing the Genetic Footprints of Vertebrate landing in Non-teleost ray-finned Fishes. Cell184, 1377–1391. 10.1016/j.cell.2021.01.046
9
BlitzI. L.BiesingerJ.XieX.ChoK. W. Y. (2013). Biallelic Genome Modification in F0Xenopus Tropicalisembryos Using the CRISPR/Cas System. Genesis51, 827–834. 10.1002/dvg.22719
10
BraaschI.GehrkeA. R.SmithJ. J.KawasakiK.ManousakiT.PasquierJ.et al (2016). The Spotted Gar Genome Illuminates Vertebrate Evolution and Facilitates Human-Teleost Comparisons. Nat. Genet.48 (4), 427–437. 10.1038/ng.3526
11
BraaschI.GuiguenY.LokerR.LetawJ. H.FerraraA.BobeJ.et al (2014). Connectivity of Vertebrate Genomes: Paired-Related Homeobox (Prrx) Genes in Spotted Gar, Basal Teleosts, and Tetrapods. Comp. Biochem. Physiol. C: Toxicol. PharmacologyCBP163, 24–36. 10.1016/j.cbpc.2014.01.005
12
BrandM.GranatoM.Nösslein-VolhardC. (2002). “Keeping and Raising Zebrafish,” in Zebrafish - A Practical Approach. Editors Nüsslein-VolhardC.DahmR. (Oxford, UK: Oxford University Press).
13
BurgerA.LindsayH.FelkerA.HessC.AndersC.ChiavacciE.et al (2016). Maximizing Mutagenesis with Solubilized CRISPR-Cas9 Ribonucleoprotein Complexes. Development143, 2025–2037. 10.1242/dev.134809
14
ChebanovM. S.GalichE. V. (2011). Sturgeon hatchery manual. FAO Fisheries and Aquaculture Technical Paper No. 558. Ankara: FAO, 303.
15
ChenB.GilbertL. A.CiminiB. A.SchnitzbauerJ.ZhangW.LiG.-W.et al (2013). Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell155, 1479–1491. 10.1016/j.cell.2013.12.001
16
ChenJ.WangW.TianZ.DongY.DongT.ZhuH.et al (2018). Efficient Gene Transfer and Gene Editing in Sterlet (Acipenser ruthenus). Front. Genet.9, 117. 10.3389/fgene.2018.00117
17
ComabellaY.CanabalJ.HurtadoA.García-GalanoT. (2014). Embryonic Development of Cuban Gar (Atractosteus Tristoechus) under Laboratory Conditions. Anat. Histol. Embryol.43, 495–502. 10.1111/ahe.12101
18
CongL.RanF. A.CoxD.LinS.BarrettoR.HabibN.et al (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science339, 819–823. 10.1126/science.1231143
19
CooperM. S.VirtaV. C. (2007). Evolution of Gastrulation in the ray-finned (Actinopterygian) Fishes. J. Exp. Zool.308B, 591–608. 10.1002/jez.b.21142
20
CrawfordK.Diaz QuirozJ. F.KoenigK. M.AhujaN.AlbertinC. B.RosenthalJ. J. C. (2020). Highly Efficient Knockout of a Squid Pigmentation Gene. Curr. Biol.30, 3484–3490. e4. 10.1016/j.cub.2020.06.099
21
DavisM. C.DahnR. D.ShubinN. H. (2007). An Autopodial-like Pattern of Hox Expression in the Fins of a Basal Actinopterygian Fish. Nature447, 473–476. 10.1038/nature05838
22
DettlaffT. A.GinsburgA. S.SchmalhausenO. I. (1993). Sturgeon Fishes—Developmental Biology and Aquaculture. New York: Springer.
23
DeveauH.GarneauJ. E.MoineauS. (2010). CRISPR/Cas System and its Role in Phage-Bacteria Interactions. Annu. Rev. Microbiol.64, 475–493. 10.1146/annurev.micro.112408.134123
24
DoyonY.McCammonJ. M.MillerJ. C.FarajiF.NgoC.KatibahG. E.et al (2008). Heritable Targeted Gene Disruption in Zebrafish Using Designed Zinc-finger Nucleases. Nat. Biotechnol.26, 702–708. 10.1038/nbt1409
25
DuK.StöckM.KneitzS.KloppC.WolteringJ. M.AdolfiM. C.et al (2020). The Sterlet sturgeon Genome Sequence and the Mechanisms of Segmental Rediploidization. Nat. Ecol. Evol.4, 841–852. 10.1038/s41559-020-1166-x
26
FaruqiA. F.EgholmM.GlazerP. M. (1998). Peptide Nucleic Acid-Targeted Mutagenesis of a Chromosomal Gene in Mouse Cells. Proc. Natl. Acad. Sci.95, 1398–1403. 10.1073/pnas.95.4.1398
27
FeiJ.-F.SchuezM.TazakiA.TaniguchiY.RoenschK.TanakaE. M. (2014). CRISPR-mediated Genomic Deletion of Sox2 in the Axolotl Shows a Requirement in Spinal Cord Neural Stem Cell Amplification during Tail Regeneration. Stem Cel Rep.3, 444–459. 10.1016/j.stemcr.2014.06.018
28
FuY.FodenJ. A.KhayterC.MaederM. L.ReyonD.JoungJ. K.et al (2013). High-frequency Off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol.31, 822–826. 10.1038/nbt.2623
29
FunkE. C.BreenC.SanketiB. D.KurpiosN.McCuneA. (2020). Changes in Nkx2.1, Sox2, Bmp4 , and Bmp16 Expression Underlying the Lung‐to‐gas Bladder Evolutionary Transition in ray‐finned Fishes. Evol. Develop.22, 384–402. 10.1111/ede.12354
30
GarneauJ. E.DupuisM.-È.VillionM.RomeroD. A.BarrangouR.BoyavalP.et al (2010). The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature468, 67–71. 10.1038/nature09523
31
GillisJ. A.DahnR. D.ShubinN. H. (2009). Shared Developmental Mechanisms Pattern the Vertebrate Gill Arch and Paired Fin Skeletons. Proc. Natl. Acad. Sci.106, 5720–5724. 10.1073/pnas.0810959106
32
GillisJ. A. (2011). Hard-to-find Fish Reveals Shared Developmental Toolbox of Evolution Elephant Fish. Available at: https://www.cam.ac.uk/research/news/hard-to-find-fish-reveals-shared-developmental-toolbox-of-evolution (Accessed July 15, 2021).
33
GillisJ. A.TidswellO. R. A. (2017). The Origin of Vertebrate Gills. Curr. Biol.27, 729–732. 10.1016/j.cub.2017.01.022
34
HavelkaM.BytyutskyyD.SymonováR.RábP.FlajšhansM. (2016). The Second Highest Chromosome Count Among Vertebrates Is Observed in Cultured sturgeon and Is Associated with Genome Plasticity. Genet. Sel Evol.48, 12. 10.1186/s12711-016-0194-0
35
HavelkaM.HulákM.BailieD. A.ProdöhlP. A.FlajšhansM. (2013). Extensive Genome Duplications in Sturgeons: New Evidence from Microsatellite Data. J. Appl. Ichthyol.29, 704–708. 10.1111/jai.12224
36
HawkinsM. B.HenkeK.HarrisM. P. (2021). Latent Developmental Potential to Form Limb-like Skeletal Structures in Zebrafish. Cell184, 899–911. e13. 10.1016/j.cell.2021.01.003
37
HedgesS. B. (2002). The Origin and Evolution of Model Organisms. Nat. Rev. Genet.3, 838–849. 10.1038/nrg929
38
HorvathP.BarrangouR. (2010). CRISPR/Cas, the Immune System of Bacteria and Archaea. Science327, 167–170. 10.1126/science.1179555
39
HsuP. D.LanderE. S.ZhangF. (2014). Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell157, 1262–1278. 10.1016/j.cell.2014.05.010
40
HsuP. D.ScottD. A.WeinsteinJ. A.RanF. A.KonermannS.AgarwalaV.et al (2013). DNA Targeting Specificity of RNA-Guided Cas9 Nucleases. Nat. Biotechnol.31, 827–832. 10.1038/nbt.2647
41
HuangP.XiaoA.ZhouM.ZhuZ.LinS.ZhangB. (2011). Heritable Gene Targeting in Zebrafish Using Customized TALENs. Nat. Biotechnol.29, 699–700. 10.1038/nbt.1939
42
HughesL. C.OrtíG.HuangY.SunY.BaldwinC. C.ThompsonA. W.et al (2018). Comprehensive Phylogeny of ray-finned Fishes (Actinopterygii) Based on Transcriptomic and Genomic Data. Proc. Natl. Acad. Sci. USA115, 6249–6254. 10.1073/pnas.1719358115
43
HwangW. Y.FuY.ReyonD.MaederM. L.TsaiS. Q.SanderJ. D.et al (2013). Efficient Genome Editing in Zebrafish Using a CRISPR-Cas System. Nat. Biotechnol.31 (3), 227–229. 10.1038/nbt.2501
44
JansenG.HazendonkE.ThijssenK. L.PlasterkR. H. A. (1997). Reverse Genetics by Chemical Mutagenesis in Caenorhabditis elegans. Nat. Genet.17, 119–121. 10.1038/ng0997-119
45
JaoL.-E.WenteS. R.ChenW. (2013). Efficient Multiplex Biallelic Zebrafish Genome Editing Using a CRISPR Nuclease System. Proc. Natl. Acad. Sci. USA110, 13904–13909. 10.1073/pnas.1308335110
46
JaoL.-E.WenteS. R.ChenW. (2013). Efficient Multiplex Biallelic Zebrafish Genome Editing Using a CRISPR Nuclease System. Proc. Natl. Acad. Sci. USA110, 13904–13909. 10.1073/pnas.1308335110
47
JinekM.ChylinskiK.FonfaraI.HauerM.DoudnaJ. A.CharpentierE. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science337, 816–821. 10.1126/science.1225829
48
KaiserK.GoodwinS. F. (1990). "Site-selected" Transposon Mutagenesis of Drosophila. Proc. Natl. Acad. Sci.87, 1686–1690. 10.1073/pnas.87.5.1686
49
KiyonariH.KanekoM.AbeT.ShiraishiA.YoshimiR.InoueK.-i.et al (2021). Targeted Gene Disruption in a Marsupial, Monodelphis Domestica, by CRISPR/Cas9 Genome Editing. Curr. Biol.31, 3956–3963. 10.1016/j.cub.2021.06.056
50
KomorA. C.BadranA. H.LiuD. R. (2017). CRISPR-based Technologies for the Manipulation of Eukaryotic Genomes. Cell168, 20–36. 10.1016/j.cell.2016.10.044
51
KurataniS.OtaK. G. (2008). Hagfish (Cyclostomata, Vertebrata): Searching for the Ancestral Developmental Plan of Vertebrates. Bioessays30, 167–172. 10.1002/bies.20701
52
MakarovaK. S.HaftD. H.BarrangouR.BrounsS. J. J.CharpentierE.HorvathP.et al (2011). Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol.9, 467–477. 10.1038/nrmicro2577
53
MaliP.YangL.EsveltK. M.AachJ.GuellM.DicarloJ. E.et al (2013). RNA-guided Human Genome Engineering via Cas9. Science339, 823–826. 10.1126/science.1232033
54
MartinA.SeranoJ. M.JarvisE.BruceH. S.WangJ.RayS.et al (2016). CRISPR/Cas9 Mutagenesis Reveals Versatile Roles of Hox Genes in Crustacean Limb Specification and Evolution. Curr. Biol.26, 14–26. 10.1016/j.cub.2015.11.021
55
MengX.NoyesM. B.ZhuL. J.LawsonN. D.WolfeS. A. (2008). Targeted Gene Inactivation in Zebrafish Using Engineered Zinc-finger Nucleases. Nat. Biotechnol.26, 695–701. 10.1038/nbt1398
56
MinarikM.StundlJ.FabianP.JandzikD.MetscherB. D.PsenickaM.et al (2017). Pre-oral Gut Contributes to Facial Structures in Non-teleost Fishes. Nature547 (7662), 209–212. 10.1038/nature23008
57
MizunoS.DinhT. T. H.KatoK.Mizuno-IijimaS.TanimotoY.DaitokuY.et al (2014). Simple Generation of Albino C57BL/6J Mice with G291T Mutation in the Tyrosinase Gene by the CRISPR/Cas9 System. Mamm. Genome25, 327–334. 10.1007/s00335-014-9524-0
58
ModrellM. S.BemisW. E.NorthcuttR. G.DavisM. C.BakerC. V. H. (2011). Electrosensory Ampullary Organs Are Derived from Lateral Line Placodes in Bony Fishes. Nat. Commun.2, 496. 10.1038/ncomms1502
59
ModrellM. S.LyneM.CarrA. R.ZakonH. H.BuckleyD.CampbellA. S.et al (2017). Insights into Electrosensory Organ Development, Physiology and Evolution from a Lateral Line-Enriched Transcriptome. eLife6, e24197. 10.7554/eLife.24197
60
MoriS.NakamuraT. (2021). An Evolutionarily Conserved Odontode Gene Regulatory Network Underlies Head Armor Formation in Suckermouth Armored Catfish. bioRxiv. 54. 10.1101/2021.06.21.449322
61
NakayamaT.FishM. B.FisherM.Oomen-HajagosJ.ThomsenG. H.GraingerR. M. (2013). Simple and Efficient CRISPR/Cas9-mediated Targeted Mutagenesis inXenopus Tropicalis. Genesis51, 835–843. 10.1002/dvg.22720
62
OisiY.OtaK. G.KurakuS.FujimotoS.KurataniS. (2013). Craniofacial Development of Hagfishes and the Evolution of Vertebrates. Nature493, 175–180. 10.1038/nature11794
63
OstaszewskaT.DabrowskiK. (2009). "Early Development of Acipenseriformes (Chondrostei", in Development of Non-teleost Fishes, ed by KunzY. W.LuerC. A.KapoorB. G. (New Hampshire: CRC Press), 170–229.
64
OtaK. G.KurakuS.KurataniS. (2008). Hagfish Embryology with Reference to the Evolution of the Neural Crest. Nature446, 672–675. 10.1038/nature05633
65
OtaK. G.KurataniS. (2008). Developmental Biology of Hagfishes, with a Report on Newly Obtained Embryos of the Japanese Inshore Hagfish, Eptatretus burgeri. Zoolog. Sci.25, 999–1011. 10.2108/zsj.25.999
66
OtaK. G.KurataniS. (2006). The History of Scientific Endeavors towards Understanding Hagfish Embryology. Zoolog. Sci.23, 403–418. 10.2108/zsj.23.403
67
PortF.ChenH.-M.LeeT.BullockS. L. (2014). Optimized CRISPR/Cas Tools for Efficient Germline and Somatic Genome Engineering in Drosophila. Proc. Natl. Acad. Sci.111, E2967–E2976. 10.1073/pnas.1405500111
68
RajkovJ.ShaoZ.BerrebiP. (2014). Evolution of Polyploidy and Functional Diploidization in Sturgeons: Microsatellite Analysis in 10 Sturgeon Species. J. Hered.105, 521–531. 10.1093/jhered/esu027
69
RanF. A.HsuP. D.WrightJ.AgarwalaV.ScottD. A.ZhangF. (2013). Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc.8, 2281–2308. 10.1038/nprot.2013.143
70
RasysA. M.ParkS.BallR. E.AlcalaA. J.LauderdaleJ. D.MenkeD. B. (2019). CRISPR-Cas9 Gene Editing in Lizards through Microinjection of Unfertilized Oocytes. Cel Rep.28, 2288–2292. e3. 10.1016/j.celrep.2019.07.089
71
RobertsonH. M.PrestonC. R.PhillisR. W.Johnson-SchlitzD. M.BenzW. K.EngelsW. R. (1988). A Stable Genomic Source of P Element Transposase in Drosophila melanogaster. Genetics118, 461–470. 10.1093/genetics/118.3.461
72
SaitoT.PšeničkaM.GotoR.AdachiS.InoueK.AraiK.et al (2014). The Origin and Migration of Primordial Germ Cells in Sturgeons. PLoS One9, e86861. 10.1371/journal.pone.0086861
73
SiveH. L.GraingerR. M.HarlandR. M. (2000). Early Development of Xenopus laevis: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor.
74
SquareT. A.JandzikD.MasseyJ. L.RomášekM.SteinH. P.HansenA. W.et al (2020). Evolution of the Endothelin Pathway Drove Neural Crest Cell Diversification. Nature585, 563–568. 10.1038/s41586-020-2720-z
75
SquareT.RomášekM.JandzikD.CattellM. V.KlymkowskyM.MedeirosD. M. (2015). CRISPR/Cas9-mediated Mutagenesis in the Sea Lamprey, Petromyzon marinus: a Powerful Tool for Understanding Ancestral Gene Functions in Vertebrates. Development (Cambridge, England)142 (23), 4180–4187. 10.1242/dev.125609
76
StolfiA.GandhiS.SalekF.ChristiaenL. (2014). Tissue-specific Genome Editing in Ciona Embryos by CRISPR/Cas9. Development141, 4115–4120. 10.1242/dev.114488
77
StundlJ.PospisilovaA.JandzikD.FabianP.DobiasovaB.MinarikM.et al (2019). Bichir External Gills Arise via Heterochronic Shift that Accelerates Hyoid Arch Development. Elife8, 1–13. 10.7554/eLife.43531
78
StundlJ.PospisilovaA.MatějkováT.PsenickaM.BronnerM. E.CernyR. (2020). Migratory Patterns and Evolutionary Plasticity of Cranial Neural Crest Cells in ray-finned Fishes. Develop. Biol.467 (1-2), 14–29. 10.1016/j.ydbio.2020.08.007
79
SuzukiM.HayashiT.InoueT.AgataK.HirayamaM.SuzukiM.et al (2018). Cas9 Ribonucleoprotein Complex Allows Direct and Rapid Analysis of Coding and Noncoding Regions of Target Genes in Pleurodeles waltl Development and Regeneration. Develop. Biol.443, 127–136. 10.1016/j.ydbio.2018.09.008
80
SymonováR.HavelkaM.AmemiyaC. T.HowellW. M.KořínkováT.FlajšhansM.et al (2017). Molecular Cytogenetic Differentiation of Paralogs of Hox Paralogs in Duplicated and Re-diploidized Genome of the North American Paddlefish (Polyodon spathula). BMC Genet.18, 1–12. 10.1186/s12863-017-0484-8
81
TakeuchiM.TakahashiM.OkabeM.AizawaS. (2009). Germ Layer Patterning in Bichir and Lamprey; an Insight into its Evolution in Vertebrates. Develop. Biol.332, 90–102. 10.1016/j.ydbio.2009.05.543
82
ThompsonA. W.HawkinsM. B.PareyE.WciselD. J.OtaT.KawasakiK.et al (2021). The Bowfin Genome Illuminates the Developmental Evolution of Ray-Finned Fishes. Nat. Genet.53 (9), 1373–1384. 10.1038/s41588-021-00914-y
83
TribleW.Olivos-CisnerosL.McKenzieS. K.SaragostiJ.ChangN.-C.MatthewsB. J.et al (2017). Orco Mutagenesis Causes Loss of Antennal Lobe Glomeruli and Impaired Social Behavior in Ants. Cell170, 727–735. e10. 10.1016/j.cell.2017.07.001
84
VéronN.QuZ.KipenP. A. S.HirstC. E.MarcelleC. (2015). CRISPR Mediated Somatic Cell Genome Engineering in the Chicken. Develop. Biol.407, 68–74. 10.1016/j.ydbio.2015.08.007
85
WaaijersS.PortegijsV.KerverJ.LemmensB. B. L. G.TijstermanM.van den HeuvelS.et al (2013). CRISPR/Cas9-targeted Mutagenesis in Caenorhabditis elegans. Genetics195, 1187–1191. 10.1534/genetics.113.156299
86
WangF.ShiZ.CuiY.GuoX.ShiY.-B.ChenY. (2015). Targeted Gene Disruption in Xenopus laevis Using CRISPR/Cas9. Cell Biosci5, 15. 10.1186/s13578-015-0006-1
87
WoodA. J.LoT.-W.ZeitlerB.PickleC. S.RalstonE. J.LeeA. H.et al (2011). Targeted Genome Editing across Species Using ZFNs and TALENs. Science333, 307. 10.1126/science.1207773
88
YenS.-T.ZhangM.DengJ. M.UsmanS. J.SmithC. N.Parker-thornburgJ.et al (2014). Somatic Mosaicism and Allele Complexity Induced by CRISPR/Cas9 RNA Injections in Mouse Zygotes. Develop. Biol.393, 3–9. 10.1016/j.ydbio.2014.06.017
89
YorkJ. R.YuanT.ZehnderK.McCauleyD. W. (2017). Lamprey Neural Crest Migration Is Snail-dependent and Occurs without a Differential Shift in Cadherin Expression. Develop. Biol.428 (1), 176–187. 10.1016/j.ydbio.2017.06.002
Summary
Keywords
CRISPR/Cas9, targeted mutagenesis, non-teleost fish, sturgeon, vertebrates, development, evolution, evo-devo
Citation
Stundl J, Soukup V, Franěk R, Pospisilova A, Psutkova V, Pšenička M, Cerny R, Bronner ME, Medeiros DM and Jandzik D (2022) Efficient CRISPR Mutagenesis in Sturgeon Demonstrates Its Utility in Large, Slow-Maturing Vertebrates. Front. Cell Dev. Biol. 10:750833. doi: 10.3389/fcell.2022.750833
Received
31 July 2021
Accepted
17 January 2022
Published
10 February 2022
Volume
10 - 2022
Edited by
Juan Pascual-Anaya, Malaga University, Spain
Reviewed by
Tetsuya Nakamura Rutgers, The State University of New Jersey, United States
Johanna Kowalko, Lehigh University, United States
Updates
Copyright
© 2022 Stundl, Soukup, Franěk, Pospisilova, Psutkova, Pšenička, Cerny, Bronner, Medeiros and Jandzik.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David Jandzik, David.Jandzik@uniba.sk
This article was submitted to Evolutionary Developmental Biology, a section of the journal Frontiers in Cell and Developmental Biology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.