- 1Shanghai Key Laboratory of Maternal Fetal Medicine, Clinical and Translational Research Center, Shanghai First Maternity and Infant Hospital, School of Medicine, Tongji University, Shanghai, China
- 2Reproductive Medicine Center, Shanghai East Hospital, School of Medicine, Tongji University, Shanghai, China
- 3Liver Cancer Institute, Zhongshan Hospital, Key Laboratory of Carcinogenesis and Cancer Invasion, Ministry of Education, Fudan University, Shanghai, China
- 4Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, University of Chinese Academy of Sciences, Beijing, China
Early embryonic arrest denotes premature termination of development in preimplantation embryos, which is one of the major phenotypes of recurrent assisted reproduction failure. Padi6 is proven to be a member of the subcortical maternal complex (SCMC) in mice, which is essential in oocyte maturation and embryogenesis. We and other groups previously found that biallelic mutations in PADI6 caused female infertility manifesting as early embryonic arrest. In this study, we identified two novel homozygous variants (p.Cys163Arg, and p. Trp475*) of PADI6 in two infertile patients from a cohort of 75 females with the phenotype of early embryonic arrest. An in vitro expression study indicated severe decrease of PADI6, which might destruct the stability of SCMC. Our study expands the mutational spectrum of PADI6 and further supports the causality between PADI6 mutations and female infertility.
Introduction
The prevalence of infertility is estimated to be 8%–12% among reproductive-aged couples worldwide (Vander Borght and Wyns, 2018). The increasing number of infertile people accelerates the process of an aging population, which has become a serious social issue (Datta et al., 2016). With the help of assisted reproductive technology (ART), millions of couples with fertility problems deliver babies after 1–2 cycles of treatment. However, there is still a large number of people suffering from unexplained recurrent ART failure, among which early embryonic arrest accounts for a considerable proportion.
Early embryonic arrest is characterized by premature termination of development in preimplantation embryos. Its genetic basis was largely unknown for a long time. In the recent 5 years, with the advance of high-throughput sequencing technology, a group of pathogenic genes were identified, and biallelic mutations in PADI6 (610363), NLRP2 (609364), NLRP5 (609658), and FBXO43 (609110) were gradually reported to be responsible for the phenotype (Xu et al., 2016; Mu et al., 2019; Wang et al., 2021). PADI6, NLRP2, and NLRP5 are considered to form a subcortical maternal complex (SCMC), a multiprotein complex first identified in mice, which plays a critical role in essential events during the oocyte-to-embryo transition, including maternal RNA metabolism, mitochondrial rearrangement, and zygote genome activation (ZGA) (Lu et al., 2017). In 2016, we first reported biallelic mutations in PADI6 causing a lack of PADI6 in oocytes, which impaired the ZGA process of early embryonic development (Xu et al., 2016). Subsequently, studies from three independent groups strengthened the correlation between PADI6 variants and early embryonic arrest (Wang et al., 2018; Zheng et al., 2020; Liu et al., 2021). However, despite the previous findings, the pathogeny of the majority of patients was still unclear, and the genetic basis of human early embryonic development remained to be investigated.
Here, we recruited 75 Chinese patients diagnosed with early embryonic arrest in addition to the subjects studied in 2016 and identified two novel homozygous variants in PADI6 (p.Cys163Arg, and p. Trp475*). We investigated and compared the effects of the two variants with our previously identified variants (p.His211Gln, p. Gln324*, p. Gln381*, and p. Glu670Glyfs*48). Our study broadens the mutational spectrum of PADI6 and provides the further relationship between PADI6 variants and female infertility.
Materials and Methods
Patients
A total of 75 Han Chinese patients undergoing in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) treatment with the phenotype of recurrent early embryonic arrest were recruited from the Reproductive Medicine Center of Shanghai East Hospital and Shanghai First Maternity and Infant Hospital affiliated with the Tongji University School of Medicine. The cohort involved one consanguineous family and 74 nonconsanguineous families. All the patients were under 40 years of age, to whom written informed consent were provided. This study was approved by the ethical committee of Shanghai East Hospital and Shanghai First Maternity and Infant Hospital affiliated with Tongji University School of Medicine.
Genetic Analysis
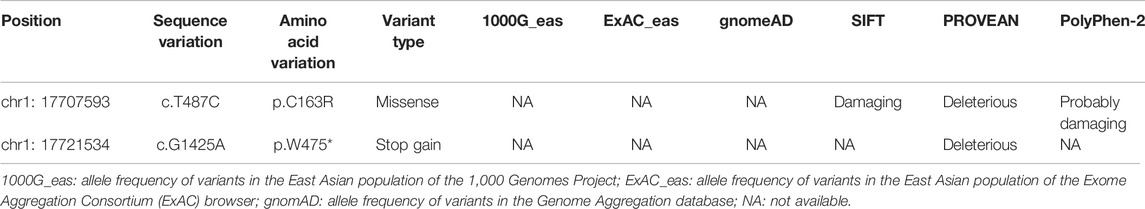
Genomic DNA of all participants was extracted from peripheral blood using the HiPure Blood DNA Mini Kit (Magen Biotechnology, China) and was sheared to acquire 150–200 bp fragments by Biorupter (Diagenode, Belgium). The ends repair and adaptor ligation was performed using the Fast Library Prep Kit (iGeneTech, China). Whole exons were captured with the AIExome Enrichment Kit V1 (iGeneTech, China), and sequencing was performed on the Illumina platform. FastQC filtered clean reads were mapped to the reference genome (hg19) using Bwa. After removing duplications, SNV and InDel were called and annotated using GATK. For the patient from the consanguineous family, homozygosity mapping was performed to identify homozygous regions using HomozygosityMapper (Seelow et al., 2009). Variants were filtered according to the following criteria: 1) not recorded or recorded to have a frequency less than 0.1% in four public databases (000 Genomes Project [1,000 g], ExAC, gnomAD, and the NHLBI Exome Sequencing Project Exome Variant Server [ESP6500]); 2) missense, nonsense, splice-site, and indel variants; 3) homozygous variants in the identified homozygous regions; 4) predicted to be damaging by at least two in silico prediction models (SIFT, PROVEAN, PolyPhen-2); 5) genes were highly or specifically expressed in oocytes and early embryos based on our in-house database; 6) genes previously reported to affect female fertility in human or mouse. Candidate genes were validated by Sanger sequencing. The co-segregation validated genes were then screened in the sporadic cases in our cohort.
Conservation Analysis and Molecular Modeling
Multiple sequence alignment of the nearby amino acid sequence of Cys163 and Trp475 was performed using ClustalW (https://www.genome.jp/tools-bin/clustalw). The result was displayed using ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) (Robert and Gouet, 2014). The wild-type model of PADI6 3D structure was generated based on the predicted result of AlphaFold Protein Structure database (https://www.alphafold.ebi.ac.uk/entry/Q6TGC4) (Jumper et al., 2021). The variant was mapped onto the atomic model using PyMol software.
Plasmid Construction and Mutagenesis
The wild-type PADI6 coding sequence was cloned and inserted into the pRK7 vector with the N-terminal Flag tag. The mutant plasmids were generated using the Mut Express MultiS Fast Mutagenesis Kit (Vazyme Biotech Co.,Ltd, China). All the constructs were confirmed by Sanger sequencing.
Cell Culture and Transfection
Human embryonic kidney 293T (HEK-293T) cells were cultured in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich) with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco). Transient transfections were performed using the PolyJet DNA Transfection Reagent (SignaGen Laboratories, United States) according to the manufacturers’ instructions. The pEGFP-C1 plasmid was co-transfected as controls. Three independent transfections were performed as the biological replicates.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated from 293T cells 36 h after transfection using TRIzol reagent, and 500 ng RNA was used for reverse transcription using the Evo M-MLV RT Mix Kit (Accurate Biotechnology, AG11728, China). Quantitative RT-PCR was performed using the SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, AG11718, China) and QuantStudio 5 Real-Time PCR Systems (Thermo Fisher Scientific). All reactions were performed in at least biological triplicates with technical duplicates. The primers used in the experiments are listed in Supplementary Table S1. The relative expression level was calculated by 2−ΔΔCt normalized by endogenous GAPDH expression.
Western Blot Analysis
Forty-eight hours after transfection, cells were harvested and lysed with RIPA lysis buffer containing 1% protease inhibitor cocktail (Bimake, United States). After 30 min incubating at 4°C, protein lysates were denatured in SDS protein loading buffer. Protein samples were separated in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes (Millipore, United States). The membranes were blocked in 5% skim milk (FUJIFILM Wako Pure Chemical Corporation, Japan) and incubated at 4°C overnight with primary antibodies against Flag (1:3,000, 14793, Cell Signaling Technology) or Vinculin (1:5,000, ab129002, Abcam). After 2 h incubation with secondary antibodies (1:5,000, Abmart), The membranes were visualized using the FluorChem E system (ProteinSimple, United States).
Results
Identification of Novel Variants in PADI6
Whole-exome sequencing (WES), combined with homozygosity mapping, was first performed on the patient from the consanguineous family. Variant filtering was then conducted following our criteria. The first three criteria excluded most of the variants, and only 11 variants were retained in the gene list. After the in silico prediction procedure, four homozygous variants remained: c. 487T > C [p.Cys163Arg] in PADI6 (NM_207421.4), c.524C > T [p.Pro175Leu] in EPHA8 (MIM: 176945, NM_020526.5), c.688A > C [p.Lys230Gln] in HAUS6 (MIM: 613433, NM_017645.5), and c.1559A > C [p.His520Pro] in SLCO1B3 (MIM: 605495, NM_019844.4). PADI6 is the only gene with high expression in oocytes and early stage embryos and was previously reported to associate with reproduction (Supplementary Figures S1A,B) (Yan et al., 2013). Sanger sequencing was subsequently performed to confirm the transmission pattern of the variant: The patient was homozygous, and her parents were heterozygous for the PADI6 variant (Figure 1A, left). The variant was located in a 3.2 Mb homozygous region (Chr1:17235196-20510690, Figure 1B, left), and was absent in ExAC, gnomAD, ESP6500, and 1000G database (Table 1). It was predicted to be damaging or deleterious by SIFT, PROVEAN, and PolyPhen-2 (Table 1). The mutant amino acid was located in the PAD middle domain of PADI6 and was highly conserved among different species (Figure 2A).
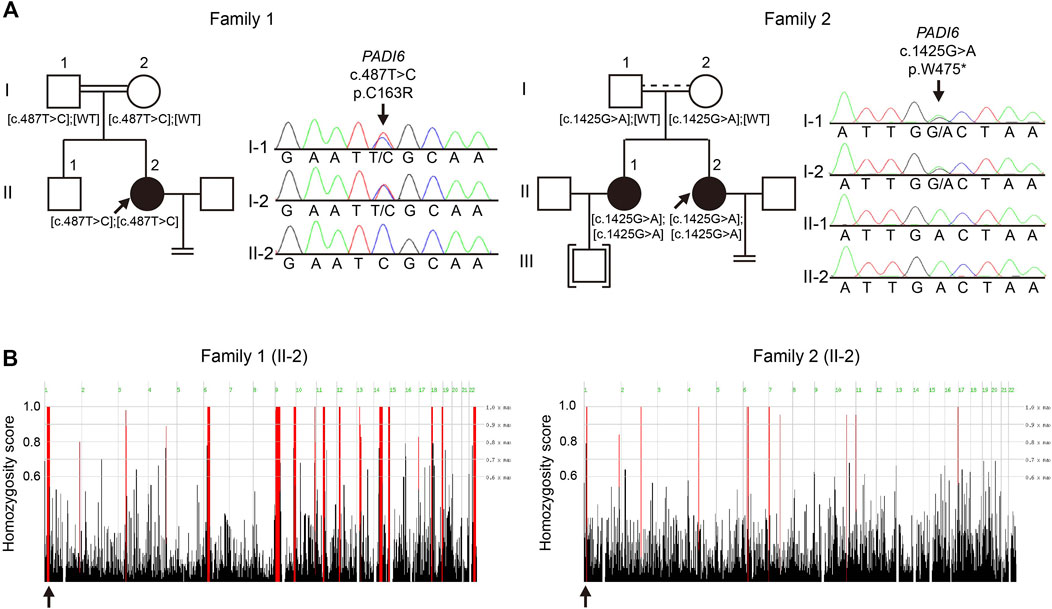
FIGURE 1. Identification of novel PADI6 variants in two Chinese families with early embryonic arrest. (A) Pedigrees and Sanger sequencing results of the families. Black solid circles denote the patients; black arrows denote probands of the families. Brackets denote adoption, equal signs indicate infertility. Parallel double solid lines represent consanguinity; parallel solid line with dash line represent suspected consanguinity. (B) Homozygosity mapping of the probands in two families. Homozygous regions are indicated in red, and the black arrows indicate the location of PADI6.
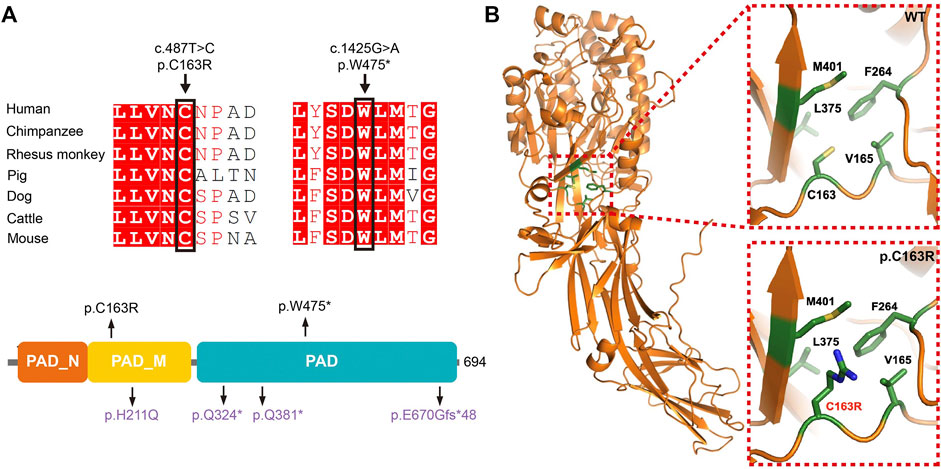
FIGURE 2. Location, conservation, and structure analysis of the mutant residues of PADI6. (A) Conservation analysis, and location of the mutant residues identified in our studies. Variants identified in this study are presented in black, and variants identified in our previous study are presented in purple. (B) Structure prediction of the p. Cys163Arg variant.
We subsequently screened exons of PADI6 in the cohort of 74 sporadic patients and identified one homozygous c.1425G > A [p.Trp475*] variant in PADI6 (Figure 1A, right). Surprisingly, the patient denied that she was from a consanguineous family when we provided genetic counseling before she was included in our study. To confirm the causality and clarify the inheritance pattern of the variant, we first performed Sanger sequencing and confirmed the heterozygosity of her parents (Figure 1A, right). Then, we performed WES and homozygosity mapping on the patient, and the result indicated that her parents might have a consanguineous marriage, and the variant was located in a 2 Mb homozygous region on chromosome 1 (Chr1:17517064-19452948, Figure 1B, right). None of the previously reported causative gene variants were identified in the WES data. The variant was absent in the four public databases and was predicted to be deleterious by PROVEAN (Table 1). The Trp475 and the nearby residues were highly conserved among species (Figure 2A).
In Silico Prediction of the Variant Effects on Protein Function
The p. Trp475* variant was located in the functional protein-arginine deiminase (PAD) domain, and the p. Cys163Arg variant was located in the PAD middle domain of PADI6 (Figure 2A). These two variants were located in the highly conserved regions among different species (Figure 2A). The p. Cys163Arg variant was predicted to be damaging or deleterious by SIFT, PROVEAN, and PolyPhen-2, and the p. Trp475* variant was predicted to be deleterious by PROVEAN (Table 1). According to the AlphaFold predicted PADI6 structure, the wild-type Cys163 was located in the core region of the protein. Arginine substitution produced a prolonged side chain with a positive charge, which might disrupt the surrounding environment and result in a conformational change and misfolding of protein (Figure 2B).
Clinical Phenotypes of the Patients
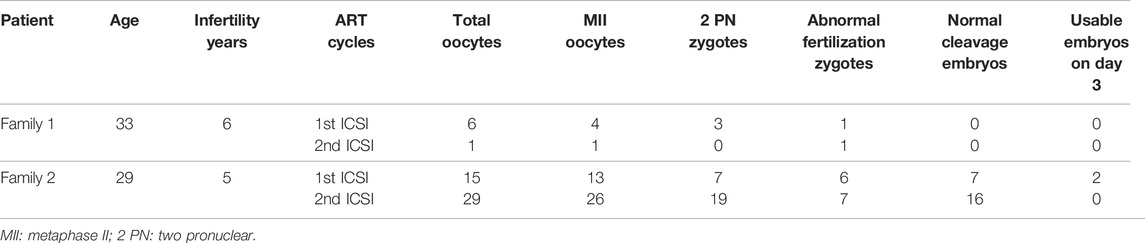
The patient from family 1 was from a consanguineous family. She was 33 years old and was diagnosed with primary infertility 6 years ago. She had experienced two failed ART attempts. In the first ART cycle, six oocytes were retrieved, and four matured oocytes were injected with sperm. Three oocytes were fertilized normally but were arrested at the zygote stage (Figure 3A). In the second cycle, one MII oocyte was retrieved and was inseminated by ICSI. However, only one pro-nuclear (PN) was observed on day 1, and the egg was discarded (Table 2).
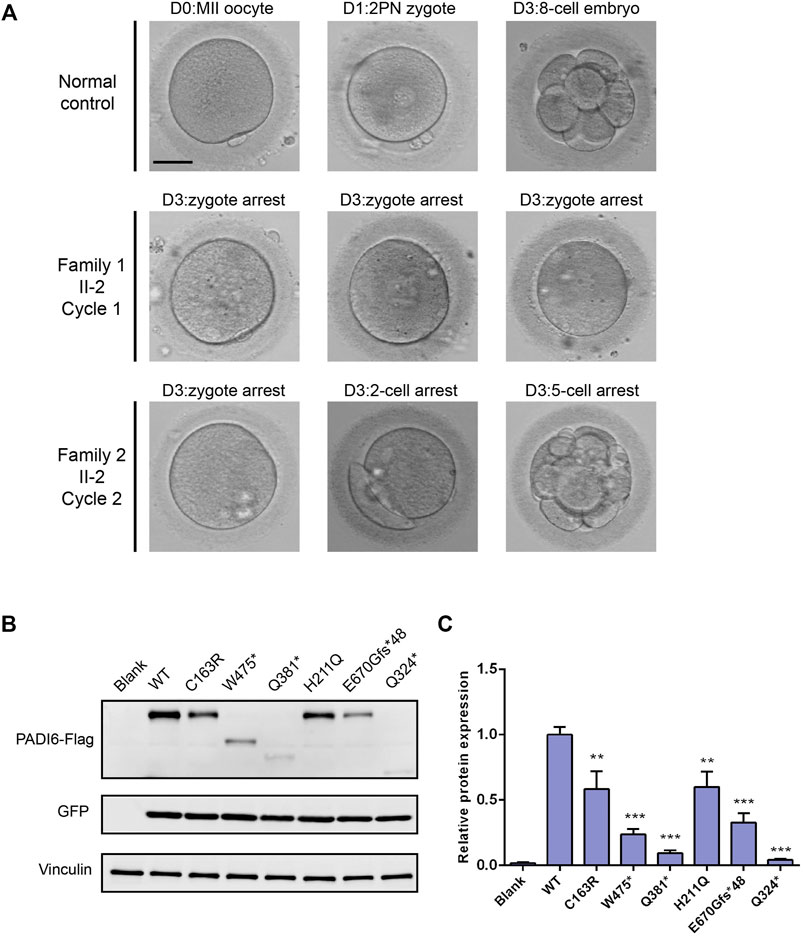
FIGURE 3. Effects of the variants on embryo development and protein expression. (A) Clinical phenotype of embryos from patients and control. Scale bar = 40 µm. (B) Western blot analysis of PADI6 protein expression in HEK-293T cells transfected with wild-type and mutant plasmids. (C) Quantitative analysis of wild-type and mutant PADI6 protein level. N = 3 independent transfections. **, p <.01; ***, p <.001.
The patient from family 2 was 29 years old and had not conceived with regular unprotected sexual intercourse for 5 years. During the ART treatment, a total of 44 oocytes were retrieved, and 39 MII oocytes were inseminated by ICSI. Among them, 26 oocytes were fertilized and cleaved normally. However, only two usable embryos were acquired on day 3, and most of the embryos were arrested before the eight-cell stage (Figure 3A). It is noteworthy that the patient has a sister diagnosed with primary infertility for several years, and the infertile couple eventually adopted a child after the failure of several years of pregnancy attempt. We performed Sanger sequencing on the sister and identified the homozygous c.1425G > A variant in PADI6 (Figure 1A, right, II-1).
PADI6 Variants Affected Protein Expression in HEK-293T Cells
We constructed PADI6 wild-type and mutant plasmids (two variants identified in this study and four variants identified in our previous study), and transiently transfected the constructs into HEK-293T cells to evaluate the functional influences of PADI6 variants in vitro. Immunoblot examination showed that, compared with wild-type PADI6, the expression level of p. Cys163Arg protein was significantly reduced. The p. Trp475* protein was observed at the expected smaller size with significantly reduced expression (Figures 3B,C). The variants identified in our previous study displayed the same reduction in protein expression level, which was consistent with our previous findings in patients’ oocytes (Figures 3B,C) (Xu et al., 2016). Quantitative RT-PCR showed a significant reduction in mRNA levels in p. Trp475*, p. Gln324*, and p. Gln381* variants (Supplementary Figure S2).
Discussion
In this study, we identified two novel homozygous variants (c.487T > C [p.Cys163Arg], and c.1425G > A [p.Trp475*]) in PADI6 in two infertile patients from a cohort of 75 females with the phenotype of early embryonic arrest, which caused recurrent ART failure. We transfected mutant constructs in HEK-293T cells to investigate the potential functional influences caused by the two variants. The result suggests that the two variants could cause significant reduction in protein expression in vitro, which is similar to the pathogenic variants identified in our previous study.
PADI6 is a member of the peptidylarginine deiminase (PADI) family, a family of calcium-dependent post-translational modify enzymes that can catalyze the process of citrullination by converting positively charged arginine residues to neutrally charged citrulline (Horibata et al., 2012). The PADI family includes five isozymes, PADI1, PADI2, PADI3, PADI4, and PADI6. Each PADI has biased expression among different tissues and has been proven to play critical roles in multiple diseases, including rheumatoid arthritis, Alzheimer’s disease, ulcerative colitis, and cancers (Horibata et al., 2012; Mondal and Thompson, 2019; Curran et al., 2020; Al-U’datt et al., 2021). PADI6 is uniquely and highly expressed in different stages of oocytes and cleavage stage embryos, which suggests its specific role in oocyte maturation and early embryonic development (Xu et al., 2016).
During mammalian oocyte and early embryonic development, a group of maternal effect genes were found to play critical roles in regulating oocyte maturation, fertilization, and early embryonic development by modifying multiple processes, including maternal mRNA clearance, chromatin remodeling, and zygote genome activation (Kim and Lee, 2014). Among them, a multiprotein complex called SCMC was recently identified in mice and was proven to be essential for mouse cleavage-stage embryogenesis (Lu et al., 2017). The knockout mouse models of SCMC members were infertile and displayed embryonic developmental attest at the two-cell stage (Tong et al., 2000; Li et al., 2008; Yurttas et al., 2008; Yu et al., 2014). Padi6 was considered to be a member of SCMC, and eggs ovulated by female mice lack of Padi6 could be fertilized but could not develop beyond the cleavage stage (Wright et al., 2003; Esposito et al., 2007).
In our previous study, we found that the PADI6 mutations could lead to the reduction of PADI6 expression in patients’ oocytes, which could impair the ZGA process and finally cause the developmental arrest of embryos (Xu et al., 2016). In this study, due to the limitation of clinical samples, we did not investigate the in vivo expression of PADI6 variants in patients’ oocytes. Instead, we conducted the in vitro expression assay and compared the expression level of p. Cys163Arg and p. Trp475* variants with the wild-type and the pathogenic variants identified in our previous study: p. His211Gln, p. Gln324*, p. Gln381*, and p. Glu670Glyfs*48. The two variants displayed a significant reduction in protein expression, which was similar to the previously identified variants. Additionally, the mRNA levels were significantly reduced in p. Trp475*, p. Gln324*, and p. Gln381* variants, suggesting that the premature termination codons might cause the degradation of mRNAs by nonsense-mediated mRNA decay (NMD), thus resulting in significantly reduced truncated proteins eventually. Because the p. Cys163Arg variant was a missense variant, we performed a 3-D structural prediction to provide supporting evidence of the pathogenicity of the variant. The wild-type Cys163 was located in the central core area of protein, and replacement of the noncharged cysteine residue by positive-charged arginine could alter the charge balance of the local area, which might lead to misfolding and degradation of the protein. We speculated that the variants could cause a significant reduction of PADI6 in oocytes, which might destroy the SCMC structural and functional stability, further affect the ZGA process of the early embryos, and finally lead to developmental arrest. Therefore, an effective supplement of PADI6 protein or mRNA in oocytes might rescue the phenotype, which is highly promising to investigate knockout mice in further study.
To summarize, we identified two novel homozygous variants (c.487T > C [p.Cys163Arg], and c.1425G > A [p.Trp475*]) in PADI6 in two females experiencing recurrent embryo developmental arrest. Our study expands the spectrum of genetic defects in female early embryonic arrest, and PADI6 is worth being screened in females with similar phenotypes.
Data Availability Statement
The data sets presented in this study can be found in online repositories. The name of the repository and accession number can be found below: China National GeneBank DataBase (CNGBdb), https://db.cngb.org/, CNP0002647.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethical committee of Shanghai East Hospital and Shanghai First Maternity and Infant Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YX, LJ, and SX conceived and designed this study. YX, LS, and HY recruited the patients. RW, YL, YZ, and SX contributed to embryo evaluation and imaging. YX, RW, and ZP collected the clinical samples and clinical data. YX analyzed the sequencing data and performed the experiments. YX, ZW, GW, SL, LJ, and SX drafted and revised the manuscript. All the authors contributed to the article and approved the submitted version.
Funding
This study was sponsored by National Natural Science Foundation of China (Nos 82001552, 81730039, 82071653, and 82171691), National Key Research and Development Program of China (2017YFC1001401), Shanghai Sailing Program (20YF1438300), and Natural Science Foundation of Shanghai (19ZR1440400).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the patients and their families for their participation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2022.819667/full#supplementary-material
Supplementary Figure S1 | Expression pattern of four candidate genes identified from family 1. (A) Expression dynamics of four candidate genes according to our in-house RNA-Seq data. GV: germinal vesicle oocyte; MI: metaphase I oocyte; MII: metaphase II oocyte. (B) Expression dynamics of four candidate genes according to single-cell RNA-Seq data published by Yan et al (Yan et al., 2013).
Supplementary Figure S2 | Quantitative RT-PCR analysis of PADI6 mRNA expression in HEK-293T cells transfected with wild-type or mutant plasmids. **, p < 0.01; ***, p < 0.001.
References
Al-U'datt, G. F. D., Allen, B. G., Hiram, R., and Alrabadi, N. (2021). Current Knowledge into the Role of the Peptidylarginine Deiminase (PAD) Enzyme Family in Cardiovascular Disease. Eur. J. Pharmacol. 891, 173765. doi:10.1016/j.ejphar.2020.173765
Curran, A. M., Naik, P., Giles, J. T., and Darrah, E. (2020). PAD Enzymes in Rheumatoid Arthritis: Pathogenic Effectors and Autoimmune Targets. Nat. Rev. Rheumatol. 16 (6), 301–315. doi:10.1038/s41584-020-0409-1
Datta, J., Palmer, M. J., Tanton, C., Gibson, L. J., Jones, K. G., Macdowall, W., et al. (2016). Prevalence of Infertility and Help Seeking Among 15 000 Women and Men. Hum. Reprod. 31 (9), 2108–2118. doi:10.1093/humrep/dew123
Esposito, G., Vitale, A. M., Leijten, F. P. J., Strik, A. M., Koonen-Reemst, A. M. C. B., Yurttas, P., et al. (2007). Peptidylarginine Deiminase (PAD) 6 Is Essential for Oocyte Cytoskeletal Sheet Formation and Female Fertility. Mol. Cell Endocrinol. 273 (1-2), 25–31. doi:10.1016/j.mce.2007.05.005
Horibata, S., Coonrod, S. A., and Cherrington, B. D. (2012). Role for Peptidylarginine Deiminase Enzymes in Disease and Female Reproduction. J. Reprod. Develop. 58 (3), 274–282. doi:10.1262/jrd.2011-040
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596 (7873), 583–589. doi:10.1038/s41586-021-03819-2
Kim, K.-H., and Lee, K.-A. (2014). Maternal Effect Genes: Findings and Effects on Mouse Embryo Development. Clin. Exp. Reprod. Med. 41 (2), 47–61. doi:10.5653/cerm.2014.41.2.47
Li, L., Baibakov, B., and Dean, J. (2008). A Subcortical Maternal Complex Essential for Preimplantation Mouse Embryogenesis. Develop. Cel 15 (3), 416–425. doi:10.1016/j.devcel.2008.07.010
Liu, J., Tan, Z., He, J., Jin, T., Han, Y., Hu, L., et al. (2021). Two Novel Mutations in PADI6 and TLE6 Genes Cause Female Infertility Due to Arrest in Embryonic Development. J. Assist. Reprod. Genet. 38 (6), 1551–1559. doi:10.1007/s10815-021-02194-1
Lu, X., Gao, Z., Qin, D., and Li, L. (2017). A Maternal Functional Module in the Mammalian Oocyte-To-Embryo Transition. Trends Mol. Med. 23 (11), 1014–1023. doi:10.1016/j.molmed.2017.09.004
Mondal, S., and Thompson, P. R. (2019). Protein Arginine Deiminases (PADs): Biochemistry and Chemical Biology of Protein Citrullination. Acc. Chem. Res. 52 (3), 818–832. doi:10.1021/acs.accounts.9b00024
Mu, J., Wang, W., Chen, B., Wu, L., Li, B., Mao, X., et al. (2019). Mutations in NLRP2 and NLRP5 Cause Female Infertility Characterised by Early Embryonic Arrest. J. Med. Genet. 56 (7), 471–480. doi:10.1136/jmedgenet-2018-105936
Robert, X., and Gouet, P. (2014). Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 42 (Web Server issue), W320–W324. doi:10.1093/nar/gku316
Seelow, D., Schuelke, M., Hildebrandt, F., and Nurnberg, P. (2009). HomozygosityMapper--an Interactive Approach to Homozygosity Mapping. Nucleic Acids Res. 37 (Web Server issue), W593–W599. doi:10.1093/nar/gkp369)
Tong, Z.-B., Gold, L., Pfeifer, K. E., Dorward, H., Lee, E., Bondy, C. A., et al. (2000). Mater, a Maternal Effect Gene Required for Early Embryonic Development in Mice. Nat. Genet. 26 (3), 267–268. doi:10.1038/81547
Vander Borght, M., and Wyns, C. (2018). Fertility and Infertility: Definition and Epidemiology. Clin. Biochem. 62, 2–10. doi:10.1016/j.clinbiochem.2018.03.012
Wang, W., Wang, W., Xu, Y., Shi, J., Fu, J., Chen, B., et al. (2021). FBXO43 Variants in Patients with Female Infertility Characterized by Early Embryonic Arrest. Hum. Reprod. 36 (8), 2392–2402. doi:10.1093/humrep/deab131
Wang, X., Song, D., Mykytenko, D., Kuang, Y., Lv, Q., Li, B., et al. (2018). Novel Mutations in Genes Encoding Subcortical Maternal Complex Proteins May Cause Human Embryonic Developmental Arrest. Reprod. BioMedicine Online 36 (6), 698–704. doi:10.1016/j.rbmo.2018.03.009
Wright, P. W., Bolling, L. C., Calvert, M. E., Sarmento, O. F., Berkeley, E. V., Shea, M. C., et al. (2003). ePAD, an Oocyte and Early Embryo-Abundant Peptidylarginine Deiminase-Like Protein that Localizes to Egg Cytoplasmic Sheets. Dev. Biol. 256 (1), 73–88. doi:10.1016/s0012-1606(02)00126-4
Xu, Y., Shi, Y., Fu, J., Yu, M., Feng, R., Sang, Q., et al. (2016). Mutations in PADI6 Cause Female Infertility Characterized by Early Embryonic Arrest. Am. J. Hum. Genet. 99 (3), 744–752. doi:10.1016/j.ajhg.2016.06.024
Yan, L., Yang, M., Guo, H., Yang, L., Wu, J., Li, R., et al. (2013). Single-cell RNA-Seq Profiling of Human Preimplantation Embryos and Embryonic Stem Cells. Nat. Struct. Mol. Biol. 20 (9), 1131–1139. doi:10.1038/nsmb.2660
Yu, X.-J., Yi, Z., Gao, Z., Qin, D., Zhai, Y., Chen, X., et al. (2014). The Subcortical Maternal Complex Controls Symmetric Division of Mouse Zygotes by Regulating F-Actin Dynamics. Nat. Commun. 5, 4887. doi:10.1038/ncomms5887
Yurttas, P., Vitale, A. M., Fitzhenry, R. J., Cohen-Gould, L., Wu, W., Gossen, J. A., et al. (2008). Role for PADI6 and the Cytoplasmic Lattices in Ribosomal Storage in Oocytes and Translational Control in the Early Mouse Embryo. Development 135 (15), 2627–2636. doi:10.1242/dev.016329
Keywords: female infertility, early embryonic arrest, PADI6, novel variant, subcortical maternal complex
Citation: Xu Y, Wang R, Pang Z, Wei Z, Sun L, Li S, Wang G, Liu Y, Zhou Y, Ye H, Jin L and Xue S (2022) Novel Homozygous PADI6 Variants in Infertile Females with Early Embryonic Arrest. Front. Cell Dev. Biol. 10:819667. doi: 10.3389/fcell.2022.819667
Received: 22 November 2021; Accepted: 16 February 2022;
Published: 01 April 2022.
Edited by:
Jennifer R. Wood, University of Nebraska System, United StatesReviewed by:
Margrit Urbanek, Northwestern University, United StatesJiaqiang Wang, Northeast Agricultural University, China
Copyright © 2022 Xu, Wang, Pang, Wei, Sun, Li, Wang, Liu, Zhou, Ye, Jin and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yao Xu, eHV5YW8xOTkxQGhvdG1haWwuY29t; Liping Jin, amlubHAwMUAxNjMuY29t; Songguo Xue, YWhhdXhzZ0AxNjMuY29t
†ORCID: Yao Xu, https://orcid.org/0000-0002-0792-1158
‡These authors have contributed equally to this work and share first authorship
 Yao Xu
Yao Xu Rongxiang Wang
Rongxiang Wang Zhi Pang
Zhi Pang Zhiyun Wei
Zhiyun Wei Lihua Sun
Lihua Sun Sa Li4
Sa Li4 Hongjuan Ye
Hongjuan Ye