- 1National Institute for Basic Biology, National Institutes of Natural Sciences, Okazaki, Aichi, Japan
- 2Exploratory Research Center on Life and Living Systems (ExCELLS), National Institutes of Natural Sciences, Okazaki, Aichi, Japan
- 3The Graduate University for Advanced Studies (SOKENDAI), Okazaki, Aichi, Japan
- 4Shumei University, Yachiyo, Chiba, Japan
- 5Graduate School of Arts and Science, The university of Tokyo, Tokyo, Japan
To form tissues with unique functions and structures, it is important that the cells that comprise them maintain physical contact. On the other hand, with each mitosis, drastic changes in cell shapes, cell adhesion, and cytoskeletal architecture may cause such contacts to be temporarily weakened, risking improper development and maintenance of tissues. Despite such risks, tissues form properly during normal development. However, it is not well understood whether mitotic abnormalities affect tissue formation. Here, analysis of zebrafish embryos with aberrant mitosis shows that proper progression of mitosis is important to maintain cell contact in developing tissues. By screening mutants with abnormal trunk and tail development, we obtained a mutant with perturbed expression of some tissue-specific genes in embryonic caudal regions. The responsible gene is mastl/gwl, which is involved in progression of mitosis. Analysis focusing on the chordo-neural hinge (CNH), the primordium of axial tissues, shows that cell detachment from the CNH is increased in mastl mutant embryos. Time-lapse imaging reveals that this cell detachment occurs during mitosis. These results suggest that cells are unable to maintain contact due to abnormalities in progression of mitosis in mastl mutants.
Introduction
When animal cells enter mitosis, structural changes, such as nuclear envelope breakdown, chromosome condensation, and mitotic spindle formation occur in a coordinated manner. Once microtubule attachment is accomplished, cells undergo chromosome segregation, cytokinesis, and reassembly of interphase cell structures. Accompanying these changes, mitotic cells reduce adhesiveness and become more spherical to accommodate spindle formation to ensure equal DNA separation. These changes in cell shape, cell adhesiveness, and cytoskeletal architecture constitute risks for maintenance of tissue integrity during embryogenesis. Thus, strict control of mitotic progression may reduce such risks under physiological conditions. However, it still remains to be determined whether abnormalities in progression of mitosis impair proper maintenance of cell assembly in tissues during embryogenesis.
Mitotic events proceed through a combination of regulated phosphorylation, proteolysis, and dephosphorylation. The key factor initiating mitosis is cyclin-dependent kinase 1 (Cdk1)/cyclin B, which phosphorylates many mitotic substrates and triggers chromosome condensation, nuclear envelope breakdown, and spindle formation. After reaching the spindle assembly check point (SAC), degradation of Securin triggers chromosome separation, and degradation of CyclinB inactivates Cdk1. Protein phosphatase 2A (PP2A) in complex with B55 regulatory subunits reverses Cdk1 phosphorylation, and with other phosphatases, exercises many mitotic events to reenter interphase (Hein et al., 2017; Holder et al., 2019; Lacroix et al., 2022).
To ensure phosphorylation by Cdk1/CyclinB, phosphatase PP2A-B55 should be inactivated. This is accomplished by Microtubule-associated Ser/Thr kinase-like, Mastl, also known as Greatwall (Gwl), which is activated at the outset of mitosis and antagonizes PP2A-B55. Mastl, which is a member of the AGC family of serine/threonine protein kinases, phosphorylates cAMP-regulated phosphoprotein 19 (Arpp19) and α-endosulfine (ENSA) (Gharbi-Ayachi et al., 2010; Mochida et al., 2010). Phosphorylated forms of these proteins are substrates of PP2A-B55 with very high affinity, but very slow dephosphorylation kinetics, inhibiting the action of PP2A-B55 on other substrates (Williams et al., 2014). Thus, depletion of Mastl impairs inhibition of PP2A-B55 activity, resulting in defective entry into mitosis and maintenance of mitosis in Xenopus egg extracts (Yu et al., 2006; Vigneron et al., 2009). Defective or delayed commencement of mitosis was also observed in RNAi-mediated knockdown of mastl in human cells and Drosophila mastl mutant (Yu et al., 2004; Burgess et al., 2010; Voets and Wolthuis, 2010). In addition to these defects, delay in transitioning to anaphase and subsequent aberrant anaphase with lagging chromosomes or DNA bridges were observed in mastl-defective cells (Bettencourt-Dias et al., 2004; Yu et al., 2004; Burgess et al., 2010; Voets and Wolthuis, 2010; Álvarez-Fernández et al., 2013; Diril et al., 2016). These defects are thought to be caused by incomplete phosphorylation of mitotic substrates or precocious activation of PP2A-B55, resulting in failure of chromosome condensation (Yu et al., 2004), maintenance of SAC (Diril et al., 2016), inactivation of SAC (Burgess et al., 2010), destruction of CyclinB (Voets and Wolthuis, 2015) or orderly progression of mitotic exit (Cundell et al., 2013; 2016).
By screening of zebrafish mutants involved in early embryogenesis, we identified a mutant with abnormal tail elongation. This mutant exhibited disturbances in expression of several axial or paraxial marker genes in the tail bud region. Surprisingly, we found that the gene responsible for this mutant was mastl. Precise analysis focusing on cells in the chordo-neural hinge (CNH), which is the origin of axial cells, revealed that abnormalities in mitotic phase progression were related to cell detachment from the CNH.
Results
Abnormal gene expression is observed in the posterior region of zebrafish mutant kt441b
To better understand the molecular mechanism underlying axis elongation during embryogenesis, we performed ENU mutagenesis screening for mutations affecting morphogenesis of the trunk and tail in zebrafish (Koshida et al., 2005). Through this screening, we obtained a mutant in which tail elongation is defective due to a single recessive allele. In homozygotes for this mutation, kt441b, the tail elongation defect appeared after the 20-somite stage (Figures 1A–F). Homozygous embryos were less transparent than normal siblings, but most of them survived until the high-pec stage (42 h at 28°C, 3 days at 24°C).
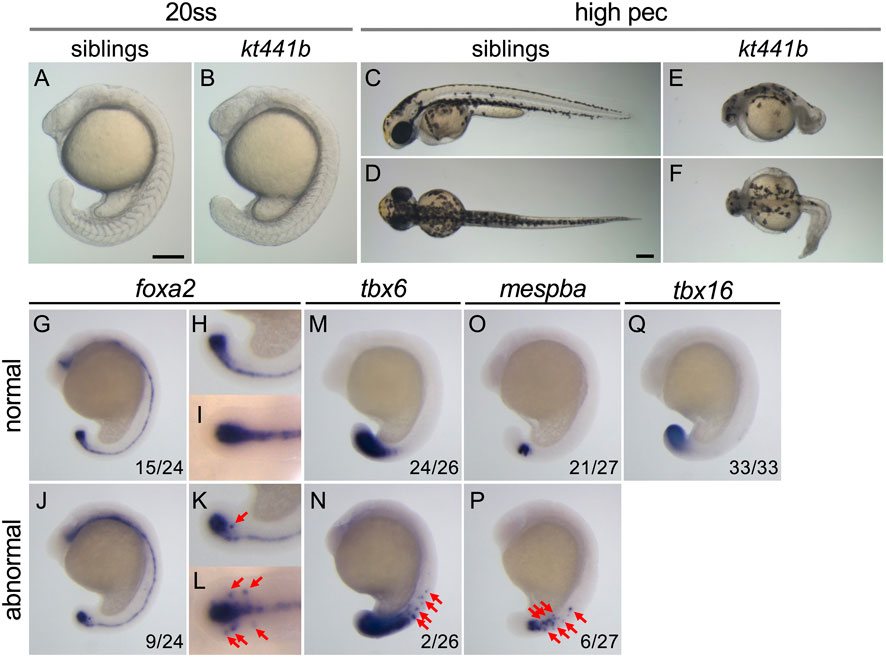
FIGURE 1. Phenotypes of kt441b homozygotes. (A–F) Morphology of siblings (A, C, D) and kt441b homozygous embryos (B, E, F) at the 20-somite stage; (A, B) and the high-pec stage (C–F). Side views (A, B, C, E) and dorsal views (D, F) are indicated. Stages were estimated by morphology according to the developmental stages of zebrafish defined by Kimmel (Kimmel C. B. (1995)). Estimated high-pec embryos had been grown at 24°C for 3 days. (G–Q) Expression patterns of foxa2 (G–L), tbx6 (M, N), mespba (O, P), and tbx16 (Q) in embryos at the 19-somite stage generated by crossing between kt441b heterozygous male and female fish. Embryos showing normal gene expression patterns (G–I, M, O, Q) and abnormal ones are shown (J–L, N, P). Side (G, H, J, K, M–Q) and dorsal (I, L) views are indicated. For foxa2 expression, magnified images in the tailbud region are also shown (H, I, K, L). In side-view images, anterior sides are located at the top. In embryos exhibiting abnormal gene expression, foxa2, tbx6, and mespba were additively expressed beside regions where these genes are expressed in normal embryos (red arrows). Embryos stained with foxa2, tbx6, and mespba probes were genotyped, and it was confirmed that penetrance of abnormal gene expression in kt441b homozygous embryos was 100%. Scale bars, 200 μm.
To examine morphogenesis and cell differentiation in kt441b homozygous embryos, expression of several tissue specific marker genes, foxa2 (the axial mesoderm, the floor plate, the hypochord, as well as their progenitors, the chordo-neural hinge (CNH); Figures 1G–L), tbx6 (posterior presomitic mesoderm; Figures 1M, N), and mespba (anterior presomitic mesoderm; Figures 1O, P), tbx16 (the tailbud; Figure 1Q) were analyzed. While no obvious difference was observed in the expression pattern of tbx16 at the 19-somite stage among embryos obtained by crossing between kt441b heterozygotes, misexpression of foxa2, tbx6, and mespba was found in some embryos (Embryos with misexpression were identified by genotyping as kt441b homozygotes after identification of the responsible gene. Penetrance of the phenotype of misexpression was 100%). This result suggests that a defect of the gene responsible for the kt441b mutation causes aberrant gene expression in the axial and paraxial mesoderm in developing zebrafish embryos.
mastl is the gene responsible for the kt441b phenotype
To better understand the phenotype of kt441b mutant embryos, we next identified the gene responsible for this mutation. By linkage analysis, we mapped kt441b to chromosome 24. Further fine mapping revealed that kt441b was located between two simple sequence length polymorphism (SSLP) markers that were generated close to the EST marker fi13d03 and on the BAC clone CR385039 (Figure 2A). Because the genome sequence was not completely assembled at that time, we manually aligned contigs and BAC clones between the two SSLP markers by Blast search and examined the expression pattern of several predicted genes on the contigs. Among them, expression of mastl was detectable in somite stages and reduced in kt441b embryos (Figure 2B). We suppose that this reduction is probably due to nonsense-mediated mRNA decay (Chang et al., 2007). Sequencing of mastl mRNA prepared from kt441b homozygous embryos showed a remnant of intron 4, and genomic sequencing revealed a one-base substitution of the splice donor of intron 4 (Figure 2C). RT-PCR analysis with primers designed on exons 3 and 5 of mastl showed two splicing variants, both of which contain the STOP codon just downstream of exon 4. These were specifically expressed in kt441b homozygous embryos (Figure 2D). Both mis-spliced products encode Mastl proteins truncated around the N-terminus of the kinase domain (Figure 2E). Furthermore, by CRISPR/Cas9-mediated genome editing, we generated a mutant containing an 11-bp deletion (mastlkt3002) just upstream of the conserved Mastl kinase domain (Supplementary Figure S1). This mutant showed the same live phenotype and did not complement kt441b (Figure 2F). Taken together, we concluded that mastl is the gene responsible for kt441b mutation.
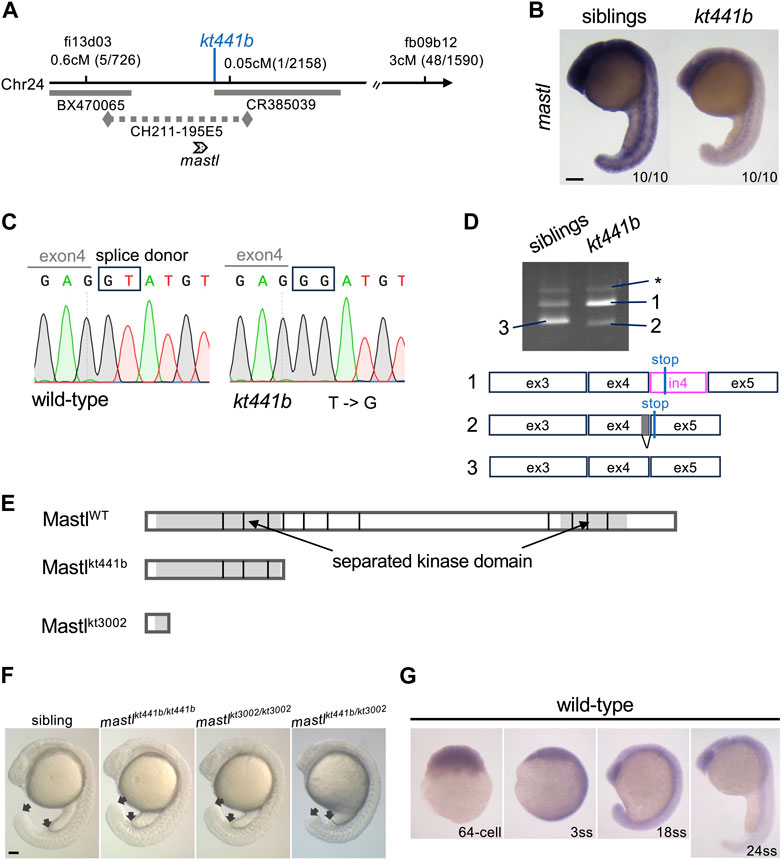
FIGURE 2. Identification of mastl as the gene responsible for the kt441b mutation. (A) A summary of genetic mapping of kt441b on chromosome 24. Numbers of recombinants and calculated distances for microsatellite markers are indicated. Gray bars indicate BAC clones, and a dotted line indicates a contig. (B) In situ hybridization showing expression of mastl in siblings and kt441b homozygous embryos at the 24-somite stage. For each, 10 embryos identified by their morphology, were stained at the same time. (C) Comparison of genomic sequences around a mutated nucleotide with the corresponding sequence of the wild-type allele. In kt441b mutation, a nucleotide in the splice donor site at the 3′ end of exon 4 of zebrafish mastl is changed from T to G. (D) PCR products of mastl cDNA prepared from 50 embryos of siblings and kt441b homozygous mutant embryos. cDNA was amplified with primers based on exons 3 and 5 of zebrafish mastl. Each PCR fragment from kt441b homozygotes was cloned and sequenced. The size of PCR fragment #3 is matched to the predicted one with proper splicing. Bands indicated with asterisks appear due to contamination of genomic DNA because they contain entire introns 3 and 4 in addition to exons 3,4 and 5 of zebrafish mastl. Note that siblings were a mixture of wild-type and heterozygote embryos. ex: exon, in: intron. (E) Schematic representation of zebrafish Mastl proteins with positions corresponding to exon-intron boundaries. Gray regions indicate the kinase domain, which is separated by a non-conserved region. One CRISPR mutant allele mastlkt3002 in which 11 bp are deleted in the mastl gene caused frameshifts upstream of the conserved kinase domain (Supplementary Figure S1). (F) Morphologies of mastlkt3002 homozygotes in comparison with mastlkt441b homozygotes. mastlkt3002 homozygous mutants exhibited similar morphology to mastlkt441b homozygotes and did not complement mastlkt441b. Arrows indicate the end of the yolk tube and the tip of the tailbud. (G) Expression of mastl mRNA at different stages of wild-type embryos. Expression of mastl mRNA was detectable even at the 64-cell stage, indicating that it is maternally expressed. Also, mastl mRNA was ubiquitously expressed from the 64-cell stage to the 24-somite stage. Scale bars, 100 μm.
In situ hybridization analysis showed that zebrafish mastl mRNA is expressed ubiquitously during early developmental stages, including the pre-MBT (midblastula transition) period (Figure 2G), indicating that maternally supplied mastl mRNA and protein can compensate for lack of zygotic products in early developmental stages (Kane et al., 1996). This is consistent with our observation that the morphological defect in kt441b homozygous embryos became evident after the 20-somite stage.
mastl is required for proper progression of mitosis in zebrafish embryos
Mastl is a kinase that functions in mitotic progression. Thus, we examined progression of mitosis in mastlkt441b homozygous embryos. Monitoring the number of cells positive for phosphorylated Histone H3 Ser10 (pHH3), a mitotic marker, showed an increase of mitotic cells in whole bodies of mastlkt441b homozygous embryos at the 14-somite stage, but not at the 6-somite stage (Figure 3A), perhaps because of depletion of maternal products.
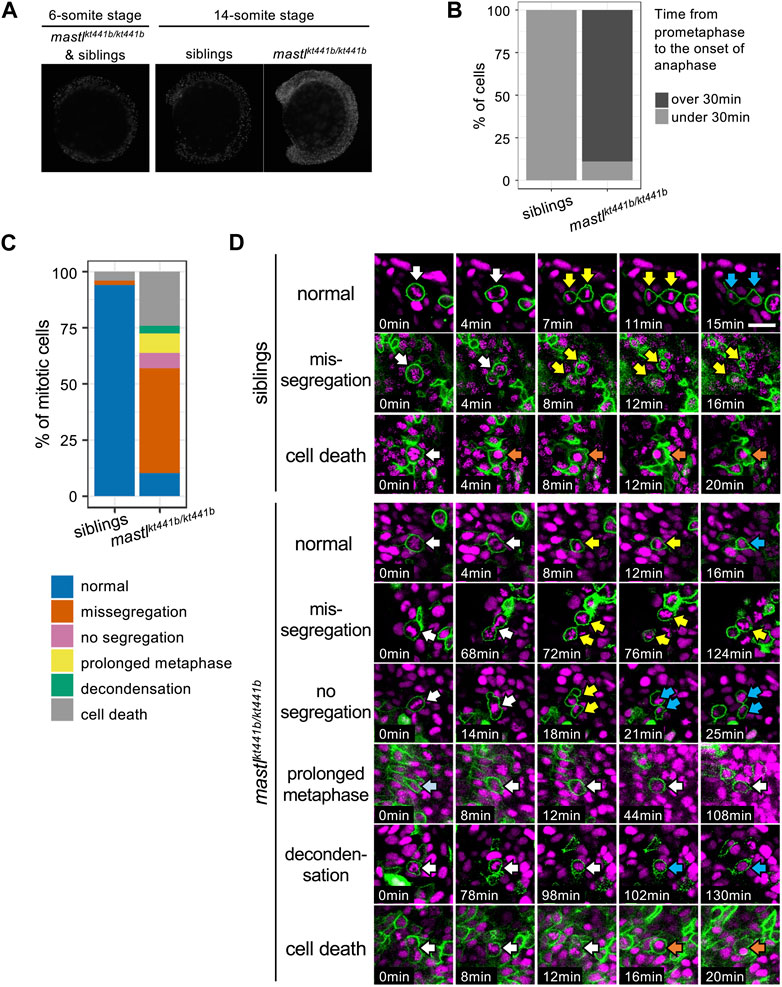
FIGURE 3. mastlkt441b homozygotes exhibit mitotic defects. (A) Phosphorylation of Histone H3 Ser10 (pHH3) in whole-mount embryos at the 6-somite stage (ss) and the 14 ss. At the 6 ss, no difference in phosphorylation was detectable among embryos containing siblings and mastlkt441b homozygotes. At the 14 ss, the number of pHH3-positive cells was increased in mastlkt441b homozygous embryos. For 14 ss embryos, 10 embryos of each phenotype were genotyped. (B) Time from prometaphase to the onset of anaphase of CNH cells. The duration was measured by time-lapse imaging of CNH cells with labeling of nuclei using Histone H2A-mChrery. The percentage of cells in which the duration of this process exceeded 30 min is indicated in dark gray. siblings: n = 10, mastlkt441b homozygotes: n = 18. (C) Classification of abnormalities of mitotic cells in the tail bud of siblings and mastlkt441b homozygous embryos. Embryos were labeled with Histone H2A-mCherry and sparsely labeled with membrane-Venus. All membrane-Venus labeled cells with condensed chromosomes within the first 30 min were tracked until the end of time-lapse imaging (2–3.5 h) or as long as we could track them (siblings: n = 50 cells, mastkt441b homozygous: n = 66 cells, from 3 embryos each). We classified tracked cells as follows. normal: normal cell division, missegregation: chromosomes were scattered around an elongated spindle, and cytokinesis was completed to pinch off some of the chromosomes, no segregation: cytokinesis was completed without obvious chromosome segregation, prolonged metaphase: metaphase was prolonged for more than 60 min without chromosome segregation or cytokinesis; decondensation: decondensation of chromosomes without segregation. cell death: change to hypercondensed nucleus. Time lapse images of representative cells for each type are shown in (D). (D) Time lapse images of representative cells for each type shown in (C). Green indicates membrane-Venus, while magenta indicates Histone H2A-mCherry. Mitotic cell types were classified as described in (C). White arrows: mother cells with condensed chromosomes; Yellow arrows: daughter cells with condensed chromosomes; Orange arrows: hypercondensed chromosomes. Since the state of chromosome condensation was sometimes unclear, we carefully judged it referring the orthogonal views. Scale bar, 20 μm.
To see whether the increase of mitotic cells was caused by the delay of mitosis, we sparsely labeled nuclei of CNH cells by injection of Histone H2A-mCherry mRNA into one blastomere at the 8-cell stage and performed time-lapse imaging. While the average duration from prometaphase to the onset of anaphase, i.e., from the start of chromosome condensation to chromosome segregation, was 15 min (SD 2.38, n = 10, from three embryos) for cells in siblings, it was more than 84 min (SD 40.83, n = 18, from three embryos) for cells in mastlkt441b homozygous embryos. Distribution of the duration in each cell is shown in Figure 3B and Supplementary Figure S2. The average duration in cells in mastlkt441b homozygous embryos should be longer than indicated because data obtained from mastlkt441b homozygous embryos includes duration in cells that were already in prometaphase or metaphase at the start of time-lapse imaging, or those that were still in metaphase at the end of time-lapse imaging. This prolonged duration from prometaphase to the onset of anaphase is consistent with previous reports (Yu et al., 2004; Burgess et al., 2010; Voets and Wolthuis, 2010; Álvarez-Fernández et al., 2013; Diril et al., 2016).
To investigate the mitotic defect more precisely, we sparsely labeled cell membranes with Venus, along with labeling of nuclei with Histone H2A-mCherry. In this analysis, we focused on tailbud cells that entered prometaphase within the first 30 min of our time-lapse imaging and tracked them until the end of imaging, or as long as they could be tracked. Interestingly, in mastlkt441b homozygous embryos, many prometaphase cells progressed to cytokinesis after a prolonged metaphase-like state (Figures 3C, D). Two types of aberrant cytokinesis were observed. One was that chromosomes were scattered in the elongated cell, and subsequent cytokinesis was completed to pinch off some of the chromosomes (missegregation). The other type was that cytokinesis was completed without obvious chromosome segregation (no segregation). Missegregation was the major mitotic defect in mastlkt441b homozygous embryos. Additionally, in missegregated cells, after completion of cytokinesis, daughter cells often maintained condensed chromosomes for a long time, even after daughter cells were well separated. Similar defects have not been reported in previous studies of cells or organisms with impaired mastl function, although failure of cytokinesis and appearance of polyploid nuclei or double nuclei were observed. Prolonged condensation of chromosomes in separated daughter cells observed in mastlkt441b homozygous zebrafish embryos might be an anomaly highlighted in the context of development of tailbuds in zebrafish. Taken together, we concluded that mastl is required for proper progression of mitosis in zebrafish embryos, as reported in other organisms, and that cytokinesis without completion of chromosome segregation and delay of chromosome decondensation in separated daughter cells is prominent, especially in zebrafish embryos.
Cell detachment from the CNH occurs in mastlkt441b homozygous embryos
How does perturbed gene expression occur in mastlkt441b homozygous embryos? Is it related to observed mitotic defects? To address these questions, we focused on aberrant expression of the foxa2 gene. This is because abnormal foxa2 expression was observed beyond CNH, which is specified at the gastrulation stage.
To explain how aberrant foxa2 gene expression occurs, we considered two hypotheses. One is that CNH cells aberrantly move out, maintaining their own gene expression profiles. The other is that cells outside of the CNH start to express foxa2 mRNA aberrantly. To distinguish these two cases, we generated transgenic fish that express a photoconvertible fluorescent protein, Kaede, under control of the foxa2 promoter (TgBac(foxa2:Kaede)). As shown in Figure 4A, Kaede expressed in CNH cells was photoconverted by UV irradiation and embryos were examined 5.5–6 h after photoconversion. Due to the stability of photoconverted Kaede protein, all cells in the CNH under irradiation should maintain the photoconverted form of Kaede, Kaede-red, throughout the experiment. Thus, if some CNH cells moved out, they should still preserve Kaede-red outside the CNH. In contrast, if cells outside the CNH newly start to express foxa2 after UV irradiation, the unconverted form of Kaede, Kaede-green, should be expressed. Note that Kaede fluorescence of TgBac(foxa2:Kaede) can be detected after the 3-somite stage only in axial tissue in our experiments, even though foxa2 mRNA starts to be expressed in the axial hypoblast and endodermal precursors from the early gastrulation stage.
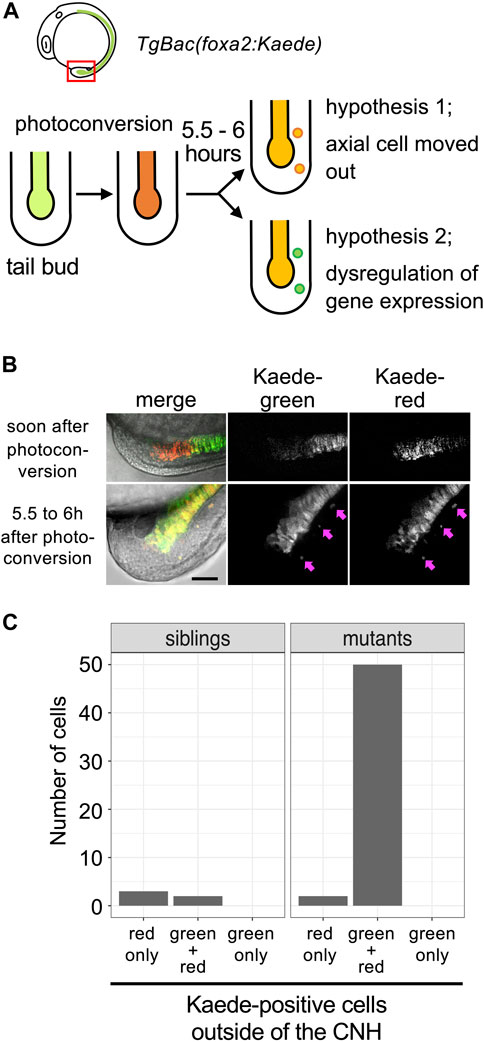
FIGURE 4. Detachment of CNH cells in mastlkt441b homozygous embryos. (A) The experimental design. mastlkt441b heterozygous male and female fish harboring transgene Bac(foxa2:Kaede) were crossed, and CNH cells of the progeny were photoconverted at the 4 to 5-somite stage. Five and half to 6 h after photoconversion, pictures of embryos were taken. Two hypotheses depicted in the figure can be distinguished by examining whether photoconverted (judged by red fluorescence) Kaede still remains in detached cells. (B) Examples of photoconverted tails. Pink arrows indicate Kaede-positive cells located outside the axial tissue observed at 5.5–6 h after photoconversion. Scale bar, 50 μm. (C) Kaede-positive cells outside of the CNH at 5.5–6 h after photoconversion were counted. The total number of cells of 3 embryos in the siblings or mastlkt441b homozygous mutant are shown.
All cells expressing Kaede fluorescence outside the CNH maintained Kaede-red in mastl homozygous embryos (n = 47 cells in 3 kt441b homozygous embryos, n = 5 in 11 sibling embryos; Figures 4B, C). Cells expressing only Kaede-green were not detected outside the CNH. These results support the former hypothesis, clearly showing that some CNH cells were abnormally detached in mastlkt441b homozygous embryos.
Cells are detached from the CNH during aberrant mitosis
To examine the process of cell detachment more precisely, we next performed time-lapse imaging of mastlkt441b homozygous embryos by labeling CNH cells with Kaede fluorescent protein using TgBac(foxa2:Kaede). By reverse-tracking of Kaede-positive cells located outside the CNH during time-lapse imaging, the process of detachment of these cells was examined. To make this tracking easier, nuclei were labeled sparsely by injection of Histone H2A-mCherry mRNA into one blastomere of the 8-cell stage.
Twenty-one cells in 3 individual mastlkt441b homozygous embryos were tracked and 12 cells were successfully tracked back to the point of detachment from the CNH (Figure 5A). The other 9 cells were already detached at the beginning of time-lapse imaging (Supplementary Figure S3). On the other hand, in 5 sibling embryos, only one detached cell, which died soon after detachment, was detected (Figure 5B, Supplementary Material S1). In mastlkt441b homozygous embryos, all 12 cells successfully traced from the CNH detached during mitosis, suggesting that cell detachment was related to mitosis. Importantly, all 12 cells exhibited aberrant chromosome segregation (Figures 5C, D, Supplementary Material S2–S6). While chromosome segregation appeared to have started, some chromosomes remained in the metaphase plane following division into two unequal clumps, which sometimes fused and then separated again. This aberrant chromosome segregation was similar to the missegregation shown in Figure 3D. These results suggests that aberrant mitosis caused by mastl depletion promotes cell detachment from the CNH.
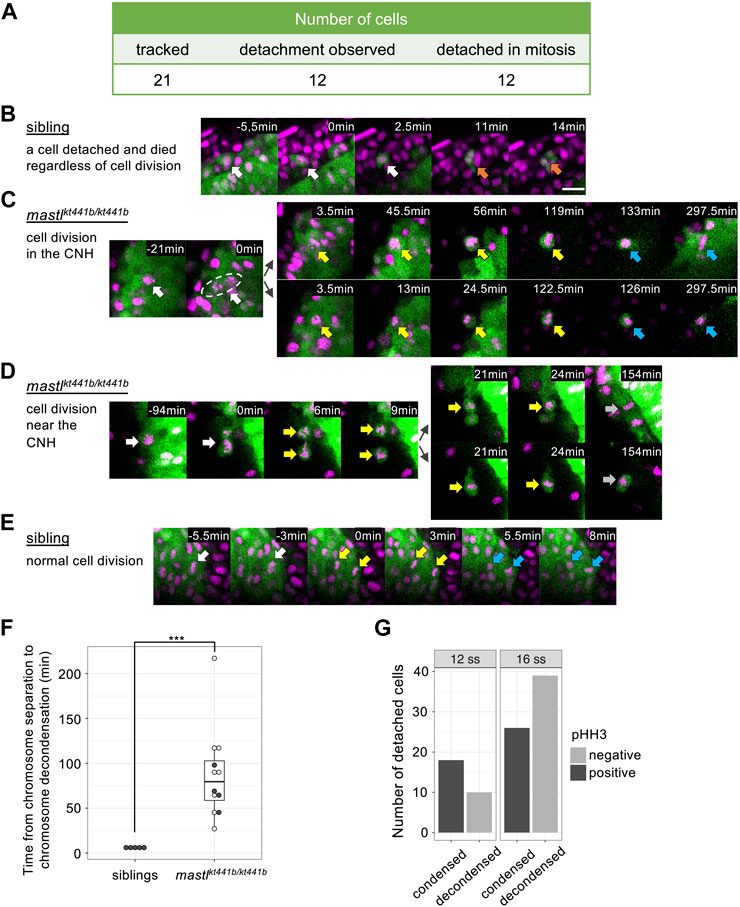
FIGURE 5. Cell detachment is related to aberrant mitosis in mastlkt441b homozygous embryos. Time-lapse imaging was carried out focusing on the tailbud region of mastlkt441b homozygous embryos carrying the Bac(foxa2:Kaede) transgene at the 8 to 16-somite stage. Kaede-positive cells that detached from the CNH were reverse tracked. (A) Summary of the tracking of detached cells in mastlkt441b homozygous embryos. Among 21 cells, 12 cells were successfully tracked back to the CNH, and all of them detached during aberrant mitosis. A schematic summary of tracked cells is shown in Supplementary Figure S3 (B) Time-lapse images of one detached cell in an embryo. Only one detached cell was detached among 5 sibling embryos. It was detached from the CNH at time 0 (white arrows) and died (orange arrows) soon thereafter. Chromosomes were not condensed. Scale bar, 20 μm. (C) A representative image of cells detaching from the CNH in mastlkt441b homozygous embryos. A mother cell in the CNH (white arrows) caused aberrant chromosome segregation at time 0. Both daughter cells detached from the CNH and maintained condensed chromosomes after segregation for around 2 h (yellow arrows). Their chromosomes appeared to decondense later (blue arrows). One of the daughter cells (upper panel) returned to the CNH. In this experiment, mastlkt441b homozygous embryos harboring the Bac(foxa2:Kaede) transgene were used. Nuclei were labeled by injection of Histone H2A-mCherry mRNA into 1 blastomere of the 8-cell stage. Green: CNH cells, magenta: Histone H2A-mCherry. (D) A representative cell that was detached and then divided near the CNH in mastlkt441b homozygous embryos. White arrows: the mother cell; Yellow arrows: condensed chromosomes; Gray arrows: chromosomes that could not be judged condensed or decondensed. (E) A representative image of normal cell division of CNH cells in sibling embryos. White arrows: metaphase; Yellow arrows: anaphase; Blue arrows: decondensed chromosomes in telophase. (F) Duration of the onset of chromosome segregation (anaphase) to chromosome decondensation. Detached cells were analyzed in mastlkt4441b homozygous embryos, comparing with CNH cells in siblings. In many cases, daughter cells in mastlkt4441b homozygotes could not be tracked until chromosome decondensation, because some daughter cells framed out, displayed reduced Kaede signals, or maintained condensed chromosomes until the time end of imaging (shown by open circles). On the other hand, examples in which time from chromosome segregation to chromosome decondensation was measured in the period of time-lapse imaging is shown by closed circles. Box plots of duration in each genotype show the first and third quartile, a line represents the median, and whiskers indicate the minimum and maximum inside 1.5 IQR (inter-quartile range). The time difference was statistically evaluated using the exact Wilcoxon-Mann-Whitney test. ***p = 0.00016 (G) The number of detached cells in 4 mastlkt441b homozygous embryos at the 12-somite stage and the 16-somite stage were classified by pHH3 staining and chromosome condensation. Note that all cells with condensed chromosomes were pHH3-positive.
Interestingly, all tracked cells after “missegregation” type cell division maintained round cell shapes and condensed chromosomes for prolonged periods (Figures 5C, D, F) compared to normal cell division in siblings (Figure 5E). Because some detached cells eventually decondensed their chromosomes (Figure 5C), completion of mitosis seems to be delayed in mastlkt441b mutant embryos. We confirmed the delay of mitotic exit in detached cells by staining other mastlkt441b homozygous embryos with an antibody that specifically recognizes phosphorylation at-Ser10 of Histone H3 (pHH3); all detached cells with condensed chromosomes were pHH3 positive, while all detached cells with decondensed chromosomes were pHH3 negative, as predicted (Figure 5G; Supplementary Figure S4). Of note, double staining of foxa2 mRNA and Kaede revealed that foxa2 mRNA in detached cells, i.e., Kaede-positive cells, was not detected in cells with condensed chromosomes, but was mostly detected in cells with decondensed chromosomes (Figure 6A; Supplementary Figure S5). This result suggests that foxa2 expression ceased during aberrant mitosis in detached cells, but restarted after mitotic exit. This is consistent with our observation that the proportion of foxa2 mRNA-positive cells among detached cells, increased as development progressed.
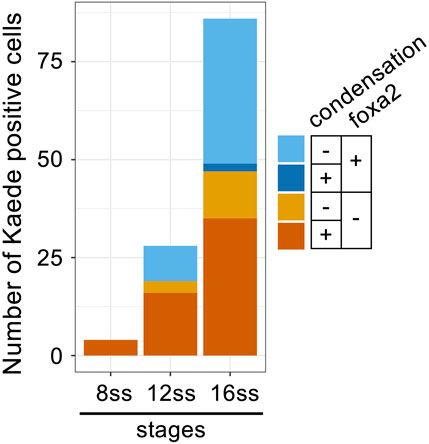
FIGURE 6. Decondensation of chromosomes and re-expression of foxa2 in detached cells in mastlkt441b homozygous embryos. Classification of cells detached from the CNH by foxa2 expression and chromosome condensation in mastlkt441b homozygous embryos carrying Bac(foxa2/Kaede) transgene at 8 ss, 12 ss and 16 ss. In situ hybridization of foxa2 mRNA and immunostaining with anti-Kaede antibody was carried out. Kaede-positive cells, i.e., cells detached from the CNH, in 4 mastlkt441b homozygous embryos are classified according to foxa2 mRNA expression and chromosome condensation. Note that most foxa2-expressing cells had decondensed chromosomes, suggesting that foxa2 expression was restarted after completion of mitosis.
Discussion
Defective mitotic progression observed in zebrafish mastl mutant embryos is similar to that reported in other organisms in regard to prolonged time from prometaphase to the onset of anaphase, and subsequent aberrant progression of anaphase with lagging chromosomes and a DNA bridge. Additionally, we found that mastl mutation caused forced separation of daughter cells and a continued mitotic state, such as chromosome condensation and phosphorylation at Ser 10 of Histone H3, even after separation of daughter cells, in zebrafish embryos. Judging from the observation of aberrant chromosome separation, aneuploid cells were likely to be dominant in zebrafish mastlkt441b mutants. In contrast, multinucleate cells or polyploid cells were prominent in mastl knockdown of HeLa cells (Burgess et al., 2010; Voets and Wolthuis, 2010) and mastl conditional knockout of MEF (Álvarez-Fernández et al., 2013; Diril et al., 2016). This difference may be due to the extent of Mastl inhibition, differences in regulatory mechanisms between species, or physical pressure in tissues.
Another highlight of this study is that cells were abnormally detached from tissues in mastlkt441b mutant zebrafish embryos. In this study, we found that loss of Mastl function results in cell detachment from the CNH. Our time-lapse imaging revealed that most cell detachment occurs in mitotic phase in mastl mutant embryos. In mitosis, cells reorganize the cytoskeleton, stiffen the cell surface, and increase intracellular pressure to become spherical, facilitating chromosome separation and partitioning of cellular contents without being disturbed by surrounding physical forces (Taubenberger et al., 2020). In addition, cell adhesion diminished in mitosis, and at the end of mitosis, rearrangement of adhesion molecules enables two daughter cells to associate with surrounding cells (Rizzelli et al., 2020). However, in spite of the biological significance of these mitotic changes, they also seem to be a potential risk for maintenance of tissue integrity. Our results show that some abnormal cell states caused by defective mitotic progression may promote cell detachment from the CNH in zebrafish embryos. For instance, impaired reorganization of the cytoskeleton may cause abnormal mechanical force, flicking cells out of the CNH. We also assume that prolonged reduction of cell adhesion may increase the risk of detachment of CNH cells in mastl mutant embryos. Therefore, we speculate that cytological changes in cell morphology, adhesion and mechanical force in mitosis should be tightly coordinated in maintaining integrity of the CNH.
It is uncertain whether cytological changes in mitosis may also affect the integrity of other tissues. As far as we observed, emergence of ectopic expression differs among marker genes. In contrast to foxa2 (a CNH marker gene), tbx6 (a marker gene for posterior presomitic mesoderm), and mespba (a marker gene for anterior presomitic mesoderm), no ectopic expression of tbx16 (a tailbud marker gene) was observed in kt441b homozygous embryos. These results suggest that the effect of prolonged mitosis on cell detachment may vary with tissue and cell differentiation status while it is also possible that the ease of restarting expression after prolonged mitosis may differ among these genes.
Details of the molecular mechanism by which mastl abnormalities cause prolonged mitosis and cell detachment from the CNH are currently unknown. Our results indicate that abnormalities in cell division caused by MASTL may indirectly promote cell detachment. On the other hand, it has been demonstrated that MASTL is involved in cell spreading and attachment to the ECM in mammary epithelial and breast cancer cells in a kinase-activity independent manner (Taskinen et al., 2020). MASTL induces cell contractility and cell migration by supporting expression of several proteins, including Rho guanine nucleotide exchange factor 2 (GEF-H1) and the SRF/MRTF target genes tropomyosin 4.2 (Tpm4.2), nonmuscle myosin IIB (NM-2B), and vinculin (VCL). Our results do not exclude the possibility that such abnormalities caused by defects in a kinase-independent role of mastl may be involved in detachment of cells from the CNH.
Materials and methods
Fish and embryos
Zebrafish with the TL2E background were used as the wild type, as described previously (Kishimoto et al., 2004). Zebrafish were maintained at 28 °C under a 14-h light/10-h dark cycle. Embryos were grown at 28.5 °C or 23.5 °C and their developmental stages were determined according to morphological criteria (Kimmel et al., 1995). mastlkt441b mutants were obtained with ENU-based mutant screening performed in our laboratory (Koshida et al., 2005).
Zebrafish lines generated in this study have been deposited in the National BioResource Project in Japan.
Gene mapping
For linkage analysis, kt441b heterozygous fish (TL2E background) were mated with wild-type India fish to generate F1 families. Homozygous kt441b mutant embryos were raised from the F1 cross and selected by morphological criteria (defect of tail elongation). Pools of genomic DNA from 10 homozygotes or siblings were subjected to segregation analyses using simple sequence polymorphism (SSLP) markers of the MGH or LN54 panels. BAC or contig sequences were obtained from the Sanger Institute Zebrafish Genome Browser (http://www.ensembl.org/) and UCSC Genome Browser (https://genome.ucsc.edu/). The GENSCAN web server at MIT (http://hollywood.mit.edu/GENSCAN.html) was used for prediction of genes near the mutation point.
Splicing valiant analysis of mastl mRNA expressed in mastlkt441b homozygous embryos
Total RNA was purified from 50 18-somite-stage embryos using Trizol (Thermo Fisher), and ssDNA was prepared by reverse transcription using SuperScript III (Thermo Fisher) with a mastl specific primer 5′-TTTTCCGATAACGGATGAGC-3′ either from the homozygotes or their siblings. Partial fragments of mastl cDNA were amplified using a primer pair, 5′-GATGGAGTATCTGATTGGAGG-3′, 5′- GACCAAGAGAGCTGATCAGTGA-3′, designed from exons 3 and 5, respectively. Amplified PCR fragments were separated by electrophoresis and each band of mastlkt441b homozygotes was subcloned using TOPO-TA cloning (Thermo Fisher). Four clones of each band were sequenced.
Bac transgenic zebrafish, TgBac(foxa2:Kaede)
To generate foxa2-promoter-driven Kaede transgenic fish, BAC clone DKEY-208C12, which contains the foxa2 genomic region, was used for bacteria-mediated homologous recombination, basically following standard protocols (Bussmann and Schulte-Merker, 2011), but using Escherichia coli strain SW102 for recombination. Briefly, two Tol2 long terminal repeats of Oryzias latipes Tol2 transposon in opposing directions flanking an ampicillin resistance cassette were inserted into the BAC vector backbone. A cassette containing the Kaede open reading frame (from pKaede-S1; MBL) with an SV40 polyA signal and a reversely oriented kanamycin-resistance gene was inserted at the start ATG of the foxa2 gene. (sequences of primers are in Supplementary Table S1). Successful recombinants were confirmed by PCR analysis. Recombinant foxa2:Kaede BAC was then injected with synthesized tol2 transposase capped-mRNA into wild-type embryos at the 1-cell stage, and transgenic lines were established by screening for Kaede expression. Even though foxa2 mRNA starts to express in axial hypoblast and endodermal precursors from the early gastrulation stage, Kaede fluorescence of the Bac(foxa2:Kaede) transgene can be detected after the 3-somite stage.
CRISPR/Cas9-mediated mastl mutation
Construction of guide RNA vectors and preparation of single-guide RNA (sgRNA) and Cas9 mRNA were performed as described previously (Cong et al., 2013; Hwang et al., 2013). The sgRNA sequence for targeting mastl was as follows: 5′-TGAAAGCTCCTTCCATCG-3’. Mutation efficiency was assessed by performing a T7 endonuclease assay (Chen et al., 2012) with the following primers: 5′-GACTTACCATCAGCCAGTTG’, 5′-TGTAACGTTAGCGGAGTACG-3′. Details on mastl mutagenesis and sequences are provided in Supplementary Figure S1. For microinjections, 1 nL of injection solution (25 ng/μL sgRNA, 0.4 μg/μL Cas9 mRNA, 0.2 M KCl and 0.05% Phenol Red) was injected into one-cell-stage zebrafish embryos using an IM300 micro injector (Narishige).
Genotyping primers
For genotyping of mastlkt441b embryos, genomic DNA fragments amplified by PCR using mastl-genoF: 5′- TGATTTGGAGTGTTTTGTTTGTC-3′ and mastl-genoR: 5′- TATTAATTATTAGGAAATAGAAAGAT-3′ were digested with MboI, which can digest DNA fragment from mastlkt441b, but not from mastlwt.
For genotyping of mastlkt3002, the following primers were used to amplify each allele of genomic DNA independently. For mastlkt3002: 5′-GTGGACTGAAAGGAAGACTC-3′ and 5′-AGCACGAAGTCCTCGCTTC-3′, for mastlwt: 5′-AACAGTGCAGTGAAAGCTCC -3′ and 5′- AACGTTAGCGGAGTACGTAC-3’.
Photoconversion of kaede-expressing fish
Photoconversion was carried out using the epifluorescence setup on a Zeiss Axio Imager Z1 microscope. Kaede-expressing embryos at the 6-somite stage were mounted on glass slides in 1/3 Ringer’s solution without removing the chorion. Directions of embryos were adjusted manually by moving the coverslip. A GFP filter set was first used to identify the region for photoconversion, and the size of the target field was adjusted to include the chordo-neural hinge region by adjusting the diameter of the diaphragm in the epifluorescence light path. Once target cells were in focus, a DAPI filter set was chosen, and UV light was switched on for 30 s at maximum intensity. After photoconversion, embryos were returned to the dishes with 1/3 Ringer’s solution and kept in the incubator at 24°C for 5.5–6 h. Embryos were mounted as described below and imaged on a LEICA SP8 confocal microscope.
Time-lapse imaging and processing
Male and female mastlkt441b/wt; TgBac(foxa2:Kaede)+/− fish were crossed and resulting embryos were labeled with Histone H2A-mCherry by injecting RNA into a blastomere at the 1-cell stage or into a blastomere at the 8-cell stage. Embryos that had Histone H2A-mCherry labeling in the tailbud region were selected for imaging. Embryos at the 6-somite stage were embedded in 0.5% low-melting-point agarose in 1/3 Ringer’s solution and imaged using a Leica SP8 confocal microscope with a 40x water immersion lens. Z-stacks were collected at 3–4 min intervals for 3–4 h. Cell movements were tracked manually, checking the orthogonal view with Las (Leica) or ImageJ. Images were compiled to movies with ImageJ.
In situ hybridization
Whole-mount in situ hybridization was performed as previously described (Koshida et al., 2005). Briefly, dechorionated embryos were fixed with a 2-h incubation at room temperature in 4% paraformaldehyde (PFA)/PBS. After fixation, embryos were dehydrated by treatment with methanol and kept at −20 °C until use. Hybridization was performed at 65 °C and signals were detected using anti-Digoxigenin-AP (Roche; 111093274910) and BM Purple AP Substrate, precipitating (Roche; 11442074001). For simultaneous detection of Kaede fluorescent protein with detection of foxa2 mRNA, anti-Digoxigenin-POD antibody (Roche; 11207733910) and a TSA Plus Cyanine3/Fluorescein System (Perkin Elmer; NEL753001KT) were used.
Plasmids for probes of foxa2, tbx6, and mespba were kindly donated by Drs. Phillip W. Ingham (Strähle et al., 1993), Masataka Nikaido (Nikaido et al., 2002), and Hiroyuki Takeda (Sawada et al., 2000). Primers for cloning zebrafish mastl probe were as follows: 5′-TATGCCATCAAGGTGGTGAA-3′, 5′-AGGCCATGAGGATGATCAAG-3′
Immunohistochemistry
Embryos were fixed with 4% paraformaldehyde (PFA)/PBS for 2 h at room temperature. Permeabilization was done with 0.5% Triton X-100 at room temperature for 1.5 h or acetone at −20°C for 5 min (for anti-phosphorylated Histone H3 Ser 10 antibody) and then incubated with 1/500 diluted rabbit anti-Kaede antibody (MBL; PM012) or 1/1000 diluted rabbit anti-phosphorylated Histone H3 Ser10 antibody (Millipore; 06–570) overnight at 4 °C. Signals were detected with 1/1,000 diluted anti-rabbit IgG Alexa 488 (Thermo Fisher; A11008) or 555 (Thermo Fisher; A21429). For simultaneous detection of Kaede and pHH3, Kaede-green fluorescence was detected without staining.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by The Animal Care and Use Committee of the National Institutes of Natural Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ST: Funding acquisition, Project administration, Supervision, Writing–original draft, Writing–review and editing, Conceptualization. HU: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing–original draft, Writing–review and editing. TY: Conceptualization, Methodology, Writing–review and editing. SK: Resources, Writing–review and editing. AY: Conceptualization, Methodology, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the following programs: Grants-in-Aid for Scientific Research (B), 18H02454 and 21H02498 to ST, Grants-in-aid for Scientific Research on Innovative Areas, 19H04797 to ST, Grants-in-aid for Transformative Research Areas, 22H05642 and 23H04930 to ST, and Grants-in-Aid for Encouragement of Scientists, 20H01001 to HU from the Japan Society for the Promotion of Science. This work was also supported by a grant from ExCELLS to ST (23EXC203 and 23EXC601).
Acknowledgments
We thank members of Optics and Imaging Facility, NIBB Trans-Scale Biology Center and personnel of the Center for Animal Resources and Collaborative Study for their technical support. Dr. Hisato Kondoh at JT Biohistory Research Hall, Dr. Masanori Mishima at the University of Warwick and all members of ST’s laboratory are gratefully acknowledged for helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1375655/full#supplementary-material
References
Álvarez-Fernández, M., Sánchez-Mart\’\inez, R., Sanz-Castillo, B., Gan, P. P., Sanz-Flores, M., Marianna, T., et al. (2013). Greatwall is essential to prevent mitotic collapse after nuclear envelope breakdown in mammals. Proc. Natl. Acad. Sci. U. S. A. 110, 17374–17379. doi:10.1073/pnas.1310745110
Bettencourt-Dias, M., Giet, R., Sinka, R., Mazumdar, A., Lock, W. G., Balloux, F., et al. (2004). Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980–987. doi:10.1038/nature03160
Burgess, A., Vigneron, S., Brioudes, E., Labbé, J.-C., Lorca, T., and Castro, A. (2010). Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. 107, 12564–12569. doi:10.1073/pnas.0914191107
Bussmann, J., and Schulte-Merker, S. (2011). Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development 138, 4327–4332. doi:10.1242/dev.068080
Chang, Y.-F., Imam, J. S., and Wilkinson, M. F. (2007). The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76, 51–74. doi:10.1146/annurev.biochem.76.050106.093909
Chen, J., Zhang, X., Wang, T., Li, Z., Guan, G., and Hong, Y. (2012). Efficient detection, quantification and enrichment of subtle allelic alterations. DNA Res. 19, 423–433. doi:10.1093/dnares/dss023
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi:10.1126/science.1231143
Cundell, M. J., Bastos, R. N., Zhang, T., Holder, J., Gruneberg, U., Novak, B., et al. (2013). The BEG (PP2A-B55/ENSA/Greatwall) pathway ensures cytokinesis follows chromosome separation. Mol. Cell 52, 393–405. doi:10.1016/j.molcel.2013.09.005
Cundell, M. J., Hutter, L. H., Nunes, B. R., Poser, E., Holder, J., Mohammed, S., et al. (2016). A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J. Cell Biol. 214, 539–554. doi:10.1083/jcb.201606033
Diril, M. K., Bisteau, X., Kitagawa, M., Caldez, M. J., Wee, S., Gunaratne, J., et al. (2016). Loss of the Greatwall kinase weakens the spindle assembly checkpoint. PLoS Genet. 12, e1006310. doi:10.1371/journal.pgen.1006310
Gharbi-Ayachi, A., Labbé, J.-C., Burgess, A., Vigneron, S., Strub, J.-M., Brioudes, E., et al. (2010). The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science 330, 1673–1677. doi:10.1126/science.1197048
Hein, J. B., Hertz, E. P. T., Garvanska, D. H., Kruse, T., and Nilsson, J. (2017). Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat. Cell Biol. 19, 1433–1440. doi:10.1038/ncb3634
Holder, J., Poser, E., and Barr, F. A. (2019). Getting out of mitosis: spatial and temporal control of mitotic exit and cytokinesis by PP1 and PP2A. FEBS Lett. 593, 2908–2924. doi:10.1002/1873-3468.13595
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31, 227–229. doi:10.1038/nbt.2501
Kane, D. A., Maischein, H. M., Brand, M., van Eeden, F. J., Furutani-Seiki, M., Granato, M., et al. (1996). The zebrafish early arrest mutants. Development 123, 57–66. doi:10.1242/dev.123.1.57
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi:10.1002/aja.1002030302
Kishimoto, Y., Koshida, S., Furutani-Seiki, M., and Kondoh, H. (2004). Zebrafish maternal-effect mutations causing cytokinesis defect without affecting mitosis or equatorial vasa deposition. Mech. Dev. 121, 79–89. doi:10.1016/j.mod.2003.10.001
Koshida, S., Kishimoto, Y., Ustumi, H., Shimizu, T., Furutani-Seiki, M., Hisato, K., et al. (2005). Integrinalpha5-dependent fibronectin accumulation for maintenance of somite boundaries in zebrafish embryos. Dev. Cell 8, 587–598. doi:10.1016/j.devcel.2005.03.006
Lacroix, B., Lorca, T., and Castro, A. (2022). Structural, enzymatic and spatiotemporal regulation of PP2A-B55 phosphatase in the control of mitosis. Front. Cell Dev. Biol. 10, 967909. doi:10.3389/fcell.2022.967909
Mochida, S., Maslen, S. L., Skehel, M., and Hunt, T. (2010). Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science 330, 1670–1673. doi:10.1126/science.1195689
Nikaido, M., Kawakami, A., Sawada, A., Furutani-Seiki, M., Takeda, H., and Araki, K. (2002). Tbx24, encoding a T-box protein, is mutated in the zebrafish somite-segmentation mutant fused somites. Nat. Genet. 31, 195–199. doi:10.1038/ng899
Rizzelli, F., Malabarba, M. G., Sigismund, S., and Mapelli, M. (2020). The crosstalk between microtubules, actin and membranes shapes cell division. Open Biol. 10, 190314. doi:10.1098/rsob.190314
Sawada, A., Fritz, A., Jiang, Y. J., Yamamoto, A., Yamasu, K., Kuroiwa, A., et al. (2000). Zebrafish Mesp family genes, mesp-a and mesp-b are segmentally expressed in the presomitic mesoderm, and Mesp-b confers the anterior identity to the developing somites. Development 127, 1691–1702. doi:10.1242/dev.127.8.1691
Strähle, U., Blader, P., Henrique, D., and Ingham, P. W. (1993). Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 7, 1436–1446. doi:10.1101/gad.7.7b.1436
Taskinen, M. E., Närvä, E., Conway, J. R. W., Hinojosa, L. S., Lilla, S., Mai, A., et al. (2020). MASTL promotes cell contractility and motility through kinase-independent signaling. J. Cell Biol. 219, e201906204. doi:10.1083/jcb.201906204
Taubenberger, A. V., Baum, B., and Matthews, H. K. (2020). The mechanics of mitotic cell rounding. Front. Cell Dev. Biol. 8, 687. doi:10.3389/fcell.2020.00687
Vigneron, S., Brioudes, E., Burgess, A., Labbé, J.-C., Lorca, T., and Castro, A. (2009). Greatwall maintains mitosis through regulation of PP2A. EMBO J. 28, 2786–2793. doi:10.1038/emboj.2009.228
Voets, E., and Wolthuis, R. (2015). MASTL promotes cyclin B1 destruction by enforcing Cdc20-independent binding of cyclin B1 to the APC/C. Biol. Open 4, 484–495. doi:10.1242/bio.201410793
Voets, E., and Wolthuis, R. M. F. (2010). MASTL is the human orthologue of Greatwall kinase that facilitates mitotic entry, anaphase and cytokinesis. Cell Cycle 9, 3591–3601. doi:10.4161/cc.9.17.12832
Williams, B. C., Filter, J. J., Blake-Hodek, K. A., Wadzinski, B. E., Fuda, N. J., Shalloway, D., et al. (2014). Greatwall-phosphorylated Endosulfine is both an inhibitor and a substrate of PP2A-B55 heterotrimers. Elife 3, e01695. doi:10.7554/eLife.01695
Yu, J., Fleming, S. L., Williams, B., Williams, E. V., Li, Z., Somma, P., et al. (2004). Greatwall kinase: a nuclear protein required for proper chromosome condensation and mitotic progression in Drosophila. J. Cell Biol. 164, 487–492. doi:10.1083/jcb.200310059
Keywords: MASTL, mitosis, zebrafish, axis elongation, chordo-neural hinge, cell detachment
Citation: Utsumi H, Yabe T, Koshida S, Yamashita A and Takada S (2024) Deficiency of mastl, a mitotic regulator, results in cell detachment from developing tissues of zebrafish embryos. Front. Cell Dev. Biol. 12:1375655. doi: 10.3389/fcell.2024.1375655
Received: 24 January 2024; Accepted: 22 February 2024;
Published: 11 March 2024.
Edited by:
Yoshiko Takahashi, Kyoto University, JapanCopyright © 2024 Utsumi, Yabe, Koshida, Yamashita and Takada. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shinji Takada, c3Rha2FkYUBuaWJiLmFjLmpw
 Hideko Utsumi
Hideko Utsumi Taijiro Yabe
Taijiro Yabe Sumito Koshida1,3,4
Sumito Koshida1,3,4 Akira Yamashita
Akira Yamashita Shinji Takada
Shinji Takada