- 1Huzhou Key Laboratory of Molecular Medicine, Huzhou Central Hospital, The Affiliated Central Hospital of Huzhou University, Huzhou, China
- 2School of Basic Medicine College, Zhejiang Chinese Medical University, Hangzhou, China
- 3Department of Colorectal Surgery, Huzhou Central Hospital, The Fifth School of Clinical Medicine of Zhejiang Chinese Medical University, Huzhou, China
Gastrointestinal cancers account for approximately one-third of the total global cancer incidence and mortality with a poor prognosis. It is one of the leading causes of cancer-related deaths worldwide. Most of these diseases lack effective treatment, occurring as a result of inappropriate models to develop safe and potent therapies. As a novel preclinical model, tumor patient-derived organoids (PDOs), can be established from patients’ tumor tissue and cultured in the laboratory in 3D architectures. This 3D model can not only highly simulate and preserve key biological characteristics of the source tumor tissue in vitro but also reproduce the in vivo tumor microenvironment through co-culture. Our review provided an overview of the different in vitro models in current tumor research, the derivation of cells in PDO models, and the application of PDO model technology in gastrointestinal cancers, particularly the applications in combination with CRISPR/Cas9 gene editing technology, tumor microenvironment simulation, drug screening, drug development, and personalized medicine. It also elucidates the ethical status quo of organoid research and the current challenges encountered in clinical research, and offers a forward-looking assessment of the potential paths for clinical organoid research advancement.
1 Introduction
Gastrointestinal (GI) tumors are a common group of malignant tumors, including gastric, colorectal, esophageal, pancreatic, hepatocellular, and cholangiocellular cancers (Liu et al., 2022; Shi et al., 2022). Among them, gastric cancer (GC) and colorectal cancer (CRC) are the most prominent GI cancers (Weng et al., 2019; Huang et al., 2023). According to the global Cancer Statistics in 2020, there were about 19.3 million new cancer cases worldwide, including more than one million new cases of GC and 769,000 deaths. Globally, GC ranks fifth in incidence and fourth in mortality (Xia and Aadam, 2022). CRC is the third most common cause of cancer mortality worldwide, with more than 1.85 million cases and 850,000 deaths annually (Biller and Schrag, 2021). Furthermore, alterations in lifestyle behaviors, heightened levels of stress, and bacterial infections, etc., have contributed to the escalating prevalence of GI cancers, resulting in a younger age of onset and a substantial impact on individuals’ overall health and wellbeing (Tong et al., 2021).
The pathogenesis of GI tumors is a complex process caused by the interaction of genetic, environmental, and other factors (Tong et al., 2021). Genetic variations in key genes s such as tumor suppressors and oncogenes play a significant role in the transformation of normal cells into cancerous ones (Al-Ishaq et al., 2020). The accumulation of these mutations disrupts the body’s homeostasis, leading to the transformation of normal cells into cancerous ones (Liu et al., 2015). For example, mutations in the tumor suppressor genes MAD4, TP53, DCRC, and APC, as well as oncogenes KRAS, HER2, and C-MYC, are important factors for GI tumors development (Baugh et al., 2018; Wang and Fakih, 2021; Lee et al., 2022). The second oncogenic pathway involves epigenetic changes like aberrant DNA methylation, microRNA (miRNA) and noncoding RNA deregulation, and altered histone modifications (Grady, 2021). These alterations are present in virtually all cases of GI tumors (Okugawa et al., 2015). In addition, factors like cell proliferation, apoptosis, genetics, obesity, alcohol consumption, and Helicobacter pylori infection, have been implicated in the development of GI tumors (Narasimhan et al., 2020; Zhu and Li, 2023).
In the past half century, oncology treatment has evolved from chemotherapy to targeted therapy and immunotherapy (Liu and Langer, 2022). Recently, gene therapy has seen rapid development with various gene therapy drugs being used in clinical practice (Naldini, 2015). Gene therapy involves the introduction of exogenous normal genes into specific cells in order to correct or mitigate diseases resulting from genetic defects and abnormalities, with the ultimate goal of achieving therapeutic outcomes (Giamas, 2020). Gene editing technology is an important tool to study gene function. Among various gene editing techniques, the clustered regularly interspaced short palindromic repeats and associated protein 9 (CRISPR/Cas9) system, characterized by its superior efficiency and precision, is commonly preferred (Karimian et al., 2019). In 2015, scientists used CRISPR/Cas9 to create a mutation model in human intestinal organoids to study intestinal tumors for the first time (Matano et al., 2015). However, patient survival was not improved due to tumor heterogeneity and metastasis, highlighting the need for new treatment models (Zhao et al., 2022).
Organoid models, which are three-dimensional (3D) clusters originating from tumor tissue or tumor-specific stem cells, effectively replicate tumor characteristics and cellular diversity in vivo (Xu et al., 2022). These models have demonstrated significant advantages in elucidating the underlying mechanisms of tumorigenesis, progression, metastasis, and drug resistance, thereby offering valuable theoretical insights and practical tools for advancing precision medicine (Yoshida, 2020). In this review, we discuss the different in vitro models in current tumor research, the derivation of cells in patient-derived organoids (PDOs) models, and the application of PDO model technology in GI cancers, particularly the applications in combination with CRISPR/Cas9 gene editing technology, drug screening, drug development, personalized medicine, and tumor microenvironment simulation. Furthermore, we also highlight the existing constraints and obstacles that necessitate resolution for the progression of human-derived GI cancer organoids.
2 Models for tumor research
Tumor research relies on tumor models, which to some extent reflect the biological characteristics of human tumors and provide a fundamental guarantee for tumor mechanistic research and clinical transformation applications (Wang et al., 2022a). At present, commonly used tumor models in tumor research include cell lines, patient-derived xenograft (PDX) models, patient-derived tumor-like cell clusters (PTCs) models, organoids models, etc. They play important roles in the mechanisms of tumorigenesis, drug screening, drug development, and translation to clinical applications.
2.1 Cell line model
Cell line model is the most commonly used models in tumor research. A tumor cell line is a kind of immortal cell model established by using patient tumor tissues through a planar culture system, which has the advantages of easy construction, a short culture cycle, a high survival rate, and high throughput drug screening (Morgan et al., 2017). In the 1974s, studies established the first human immortal cell line, the HeLa cervical cancer cell line. With the continuous improvement of cytogenetics, 2D culture systems and other technologies, cell lines have been widely used as the most convenient and efficient tumor models (Nelson-Rees et al., 1974). With the research deepening, the shortcomings of cell line model have been gradually revealed. Under 2D culture conditions, cells are unable to maintain the genetic phenotype and genetic heterogeneity of the original tumor cells due to the lack of an immune microenvironment and vascular network system and are unable to recapitulate the morphology and function of the original tumor tissues (Nelson-Rees et al., 1974). In addition, cell line cells are homogeneous cells that have been stably passaged in vitro, with poor heterogeneity, making it impossible to simulate the types and interactions of tumor cells in vivo. As a result, many drugs that worked well in early cell line trials have proved ineffective in subsequent clinical trials (Habanjar et al., 2021).
2.2 PDX model
In recent years, researchers have been committed to developing experimental models that can approximately and realistically reflect the tumor characteristics of patients in vivo. Successfully, two of the most representative models, the PDX model and the PDOs model, have been constructed (Lannagan et al., 2021). The PDX model refers to xenografts obtained by directly transplanting tumor tissues derived from patients into an immunodeficient animal. Since transplantation directly using tumor tissue preserves the tumor cells and their cellular matrix to a large extent, PDX preserves the heterogeneity of the original tumor and the interaction between the tumor and the surrounding matrix. However, PDX cannot fully capture the genetic heterogeneity in the original tumor (Kemper et al., 2015; Morgan et al., 2017). Morgan et al. examined the number of mutations in the PDX model of patient with primary non-small cell lung cancer and found that only 43% of mutations were detected, whereas four mutations that were not present in the original tumor were found early in the PDX process, suggesting that clonal selection and mutation of tumor tissues may occur during the early stages of implantation in mice (Morgan et al., 2017). Moreover, during PDX model generation, mouse stroma gradually replaces human stromal components. Since the human tumor microenvironment cannot be replicated exactly in mice, the interaction between human tumor tissues and the mouse stromal microenvironment affects tumor secretion signaling, which is detrimental to the study of stroma-directed therapies (Yoshida, 2020). In addition to the above drawbacks, the PDX model has several other limitations, such as being cost-effective, time-consuming, and inadequate for high-flow drug screening (Praharaj et al., 2018). Although PDX has been extensively utilized in several fields, including preclinical drug evaluation and biomarker identification, it is still not the most ideal preclinical model in vitro (Wilding and Bodmer, 2014).
2.3 PTC model
PTCs arise from the self-assembly and proliferation of immune cells, fibroblast, and primary epithelial that physically and functionally mimic the original tumors. The PTC model was originally proposed as an in vitro tumor model by the Xi team in 2020 (Yin et al., 2020). The model emphasizes strategies for the maintenance and expansion of primary tumor cells under matrigel-free conditions. Suspended tumor cell clusters are established using patient-derived specimens (e.g., tumor tissue, puncture specimens, ascites specimens, etc.) through the use of specially optimized media and low-adherence culture plates. The PTC model has shown potential for clinical applications in the fields of tumor drug resistance, drug development, and precision medicine due to the short culture cycle, low requirement for specimen sources, and high throughput drug screening. Yin et al. established in vitro PTC models from patients with GC and CRC for preclinical drug testing (Yin et al., 2020). PTC tests of 59 patients with gastric, colorectal, or breast cancers revealed an overall accuracy of 93% in predicting their clinical outcomes. Additionally, Liu et al. generated PTC models from lung cancer patients that structurally and functionally recapitulated the original tumor features and exhibited a high degree of morphological similarities to surgically resected samples, which can be used to assess the efficacy of cisplatin-based chemotherapy (Liu et al., 2023a). Zhang et al. evaluated the responses of PTCs to specific drugs or combinations of drugs (Zhang et al., 2023a). In a study by Peng et al., PTC models were conducted to investigate PD-L1 mRNA expression in extracellular vesicles, and data showed that PTCs are significantly heterogeneous (Peng et al., 2023). However, the PTC model has just been proposed in recent years, later in time than the PDO model. Unlike the 3D matrix-embedded culture method, the tumor cell clusters obtained by the PTC technique cannot be used for gene editing and immortalization in culture. PTC models generated for clinical studies are still rare, so more reports on PTC models are needed for public acceptance.
2.4 Organoid model
Organoids have the ability to self-renewal and self-assembly, retain the physiological structure and function of the original tissue, and proliferate indefinitely under in vitro culture conditions (Vlachogiannis et al., 2018). In 2009, Sato et al. introduced organoid technology in the experiment for the first time and successfully cultured mouse small intestine cells dependent on stem cell culture (Sato et al., 2009). Subsequently, in 2013, Huch et al. generated a 3D organoid model from mouse pancreatic tissue (Huch et al., 2013). The successful attempts and applications of organoids in normal tissues have made organoid culture technology develop rapidly in the field of tumor research. In 2015, Clevers and Tuveson LABS collaborated to build organoid models of human and mouse ductal pancreatic cancer (Koikawa et al., 2021). These tumor organoids have a critical trait in that they can partially or totally replicate the heterogeneity of the original tumor, making them ideal for tumor research. Compared to cancer cell lines, organoid models reflect the phenotypic and genetic characteristics of the original tumor with higher fidelity (Li et al., 2023). At the same time, the culture success rate and culture cycle were significantly improved compared with PDX, suggesting that organoids may be a more effective and ideal model for detecting novel anticancer drugs (Neal et al., 2018). Additionally, the immortality characteristics non-possessed in the PTC model showed the organoid model has a broader application scenario. Table 1 summarizes the advantages and disadvantages of the cell lines, PDX, PTC, and organoid models.
3 GI tumor Organoid derivation and culture
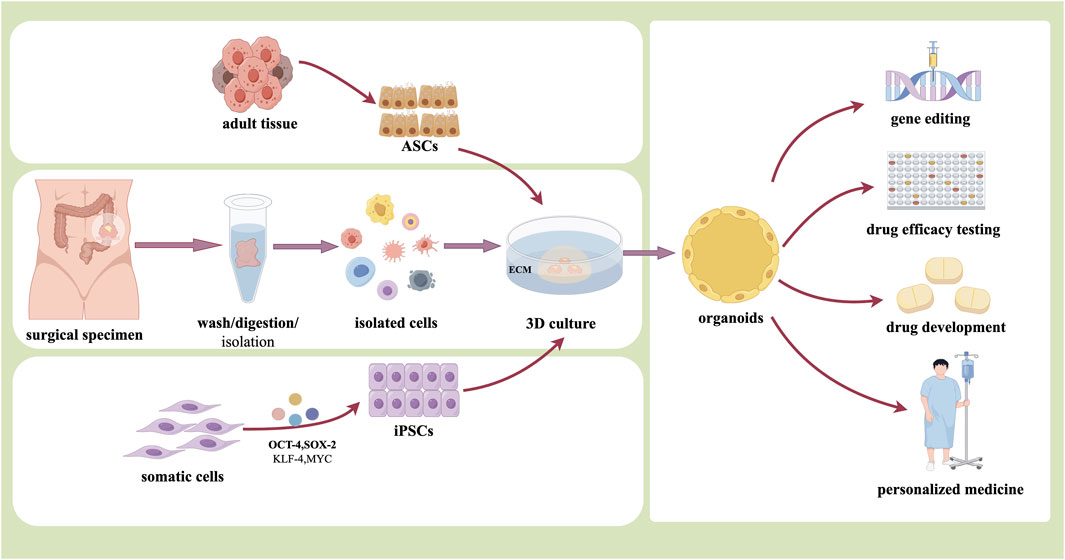
Organoids refer to cells obtained by surgical resection of tumor specimens, or biopsy tissues, or liquid biopsy samples cultured into 3D multicellular clusters with proliferation and spatial self-organization ability under specific culture conditions in vitro, which are also described as tiny tumors preserved in Petri-dishes (Artegiani and Clevers, 2018). Currently, the Matrigel is commonly used as a stereoscopic scaffold to support cell-cell and cell-matrix interactions in organoid culture (Onfroy-Roy et al., 2020; Xu et al., 2021). Matrigel is an ECM rich in laminin secreted by Engelbreth-Holm-Swarm tumor system, which contains a variety of growth factors and can regulate cell growth and metabolism (Kozlowski et al., 2021; Zhou et al., 2023). In the organoid culture system, the components that also need to be added mainly include activators, inhibitors and other specific cytokines such as epidermal growth factor and fibroblast growth factor, to ensure cell growth and differentiation (Almeqdadi et al., 2019; Seidlitz et al., 2019) (Figure 1).
3.1 Organoids from human pluripotent stem cells (PSCs)
Organoids can be differentiated from human PSCs or progenitor cells, which reserve certain functions and have similar cell types and tissue structures as internal organs (Reza et al., 2021). PSCs, including embryonic stem cells (ESCs) and induced PSCs (iPSCs), have been widely used in biomedical and biological sciences due to their potential to differentiate into all types of cells in the body (Yamanaka, 2020). The first pluripotent cell line was isolated and established from mouse blastocysts and reported in 1981 (Martin, 1981). After nearly 20 years of exploration, human ESCs were established by Thomson et al., in 1998 (Thomson et al., 1998). In 2006, Takahashi and Yamanaka reprogrammed mouse fibroblasts into PSCs by inserting four transcription factors encoding genes Oct4, Sox2, Klf4, and c-Myc. These cells, also known as iPSCs, have an intrinsic potential to develop into cells of all three germ layers in vivo, similar to those derived from ESCs (Takahashi and Yamanaka, 2006). Subsequently, human iPSC (hiPSC) technology was first introduced in 2007 and is now broadly used to generate human “dish disease” models. This hiPSC technology allows for personalized disease modeling and will be an important component of precision medicine (Takahashi et al., 2007).
Currently, hiPSC technology has been able to form intestinal, kidney, brain tissue, retina, liver, and other organoids in vitro (Taguchi and Nishinakamura, 2017; Workman et al., 2017; Nieto-Estévez and Hsieh, 2020; Zahmatkesh et al., 2021; Yamasaki et al., 2022). In 2011, Kyle W et al. developed a method to differentiate human embryos and PSC into intestinal organoids in vitro. In this study, hiPSC was first exposed to Activin A for definitive endoderm induction, then exposed to FGF3/Wnt4A to produce midgut/hindgut spheroids for 4 days, and then the spheroids were allowed to expand into intestinal tissues for 3–14 days (Spence et al., 2011). With the rapid development of regenerative medicine and precision therapy, it has been found that iPSC-derived organoids are more suitable for the study of reproducing the development process and internal changes of tissues or organs in time and space in human developmental biology. At the same time, the controversy surrounding its clinical application is highlighted by its tumorigenicity and heterogeneity.
3.2 Organoids from adult stem cells (ASCs)
Tissue-specific stem cells, also known as ASCs, exist in adult somatic tissues and contribute to tissue homeostasis and repair. ASCs have the capacity to self-renew and differentiate into specific cell types. The organoids formed by ASCs can well simulate the process of self-renewal or regeneration of tissues and organs after damage. Toshiro Sato and his colleagues established the first intestinal organoid model derived from ASCs in 2009 (Sato et al., 2009). The model entailed embedding mouse intestinal crypt leucine-rich repeat-containing G-protein-coupled receptor five-positive (LGR5+) cells with ECM and culturing with the necessary growth factors, including R-spondin-1, EGF, and BMP inhibitor Noggin. These organoids largely replicate the tissue architecture in vivo and contain relatively complete complements of stem cells, progenitor cells, and terminally differentiated cell types (Sato et al., 2009). Then, by changing the combination of growth factors and cell separation procedures, researchers can rapidly alter the organoid culture system to produce a variety of normal and cancerous human organoids, such as the colon, stomach, and liver (McCracken et al., 2014; Huch et al., 2015; Pleguezuelos-Manzano et al., 2020).
Compared with PSC-based organoids, ASC-based organoids are directly obtained from regenerated adult tissues, showing the advantages of simpler operation and shorter culture time. The maturity of ASC-derived organoids is closer to that of the source tissues, so ASC-derived organoids provide a better model for adult tissue repair (Tang et al., 2022). Unfortunately, ASC-derived organoids are not able to obtain stem cell-based tissues from adult organs such as the brain, heart, and islet. However, either ASC-derived organoids or PSC-derived organoids can maintain the genetic stability of source organs or tissues while expanding in vitro for long periods, making them an ideal choice for cell expansion and promoting their potential applications in new therapeutic strategies (Kimura et al., 2022).
PDOs, especially tumor-derived organoids, are a special type of ASC-based organoids. For PDOs, samples were mainly obtained from surgery, biopsy, ascites, or other means. The samples are mechanically minced, enzymatically digested, and erythrocytically lysed to obtain cell suspensions, which are then coated with special culture media and matrix gel to form 3D structures. In 2015, Wetering M et al. established tumor tissue-derived organoids in 20 patients with CRC. The tumor organoids model is suitable for high throughput drug screening and can detect gene-drug association (van de Wetering et al., 2015). Another study by Nuciforo et al. showed that hepatocellular carcinoma organoids could be successfully obtained by digesting tumor tissues obtained from needle biopsies of hepatocellular carcinoma with type IV collagenase and inoculating them with type II basement membrane extracts. The histomorphological and genetic characteristics the hepatocellular carcinoma organoids also retained those of the original tumor tissues (Nuciforo et al., 2018). Currently, the method of directly obtaining tumor cells from patients to establish tumor organoids is relatively mature and most widely used, through which a variety of PDO models such as CRC, cholangiocarcinoma (Saito et al., 2019), and squamous cell carcinoma of the head and neck have been successfully established (van de Wetering et al., 2015; Driehuis et al., 2019).
4 Application of organoids in GI tumors
4.1 Organoids and CRISPR/Cas9 technology
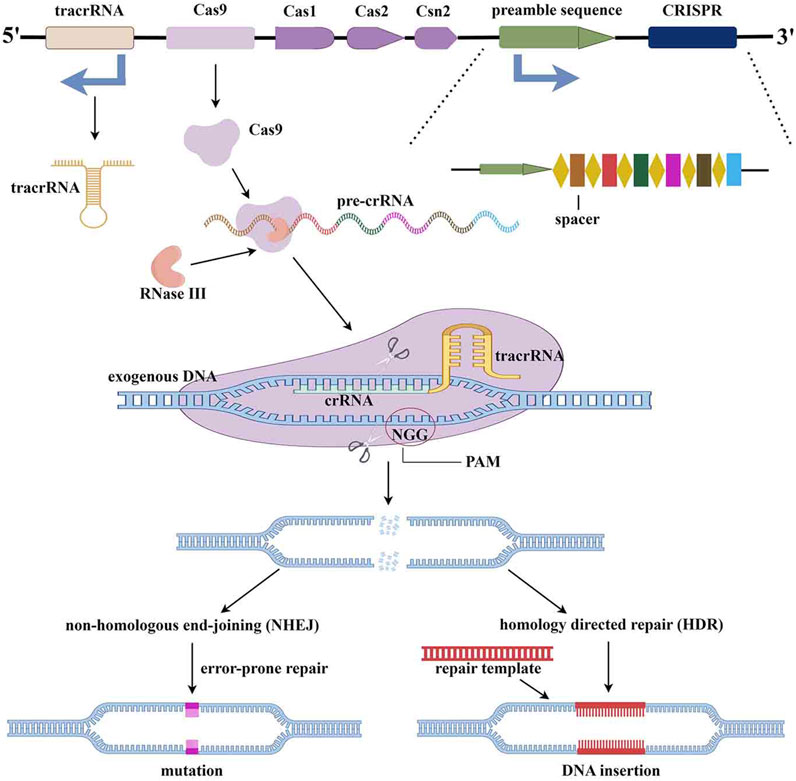
The CRISPR/Cas9 system is an adaptive immune defense developed by bacteria and archaea in the long-term evolution process to resist the invasion of viruses and exogenous DNA (Janik et al., 2020). CRISPR was first discovered and described in Escherichia coli bacteria DNA by Ishino et al. at Osaka University, Japan, in 1987. Subsequently, its powerful shearing and editing capabilities were definitively investigated in 2010 (Ishino et al., 1987). CRISPR/Cas9 consists primarily of the enzyme Cas9 and small guide RNA (sgRNA). SgRNA is a hairpin structure formed by complementary pairing of portions of crRNA and tracrRNA, and forms a complex with the Cas9 enzyme upon recruitment of sgRNA (Dominguez et al., 2016). Through the structure of Proto Spacer Adjacency Motif (PAM), the complex of CAS9 and sgRNA identifies and binds to the endogenous target genome. After the double-stranded DNA is unraveled, sgRNA will hybridize to complementary strand DNA, and then Cas9 enzymes cleave the complementary and non-complementary strands to produce flat ends. Finally, the target DNA of the double-strand break is destroyed during repair, leading to DNA silencing (Figure 2) (Dominguez et al., 2016; Janik et al., 2020).
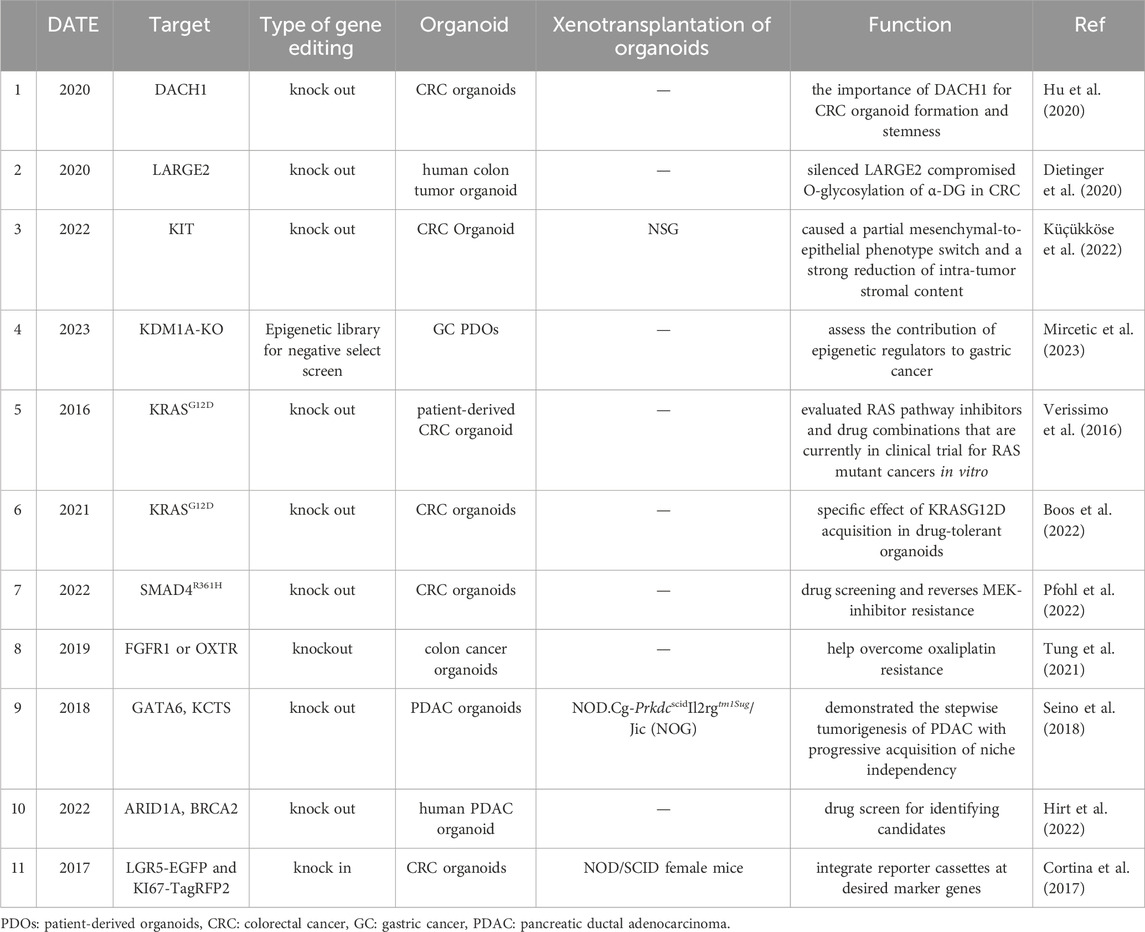
The CRISPR/Cas9 system has been widely used in basic medicine, translational medicine, and other fields due to its high efficiency and operability in gene editing. Here, we only focus on the study of target gene mutations introduced by CRISPR/Cas9 technology in the organoid platform and their xenograft or not, while other studies such as target gene mutations introduced in cells and xenografts have not been adopted (Table 2). As one of the sharpest and most promising gene editing tools, the CRISPR/Cas9 system has been applied to gene knockout in many types of organoid platforms. In CRC, driver genes such as DACH1 (Hu et al., 2020), LARGE2 (Dietinger et al., 2020), and KIT (Küçükköse et al., 2022) were knocked out by using CRIPSR/CAS9 gene editing technology to investigate the promoting effects of intestinal stemness, cell-matrix adhesion, and phenotype switch in carcinomatosis, respectively. In order to explore the epigenetic contribution in GC, the histone lysine demethylase-1A (KDM1A) gene was knocked out in PDOs, which in turn induced inhibition of Wnt signaling and a strong cell cycle arrest (Mircetic et al., 2023). For the drug susceptibility test, Verissimo CS et al. evaluated the effect of mutant KRAS on drug response and found that dual inhibition of the EGFR-MEK-ERK pathway in RAS mutant organoids induced a transient cell-cycle arrest rather than cell death (Verissimo et al., 2016). In 2021, Boos SL et al. further demonstrated that metastatic CRC PDOs introduced by KRASG12D are an ideal platform to model chemotherapy tolerance ex vivo and AURKA was found to be a therapeutic target in liver metastatic CRC (Boos et al., 2022). Besides, the SMAD4 (Pfohl et al., 2022) mutation and FGFR1 and OXTR (Tung et al., 2021) mutations were knocked out using CRIPSR/CAS9 technology in colon cancer PDOs for reversing drug resistance. Using an organoid platform based on CRISPR/Cas9 knockout technology, Seino T et al. found that GATA6 expression regulated Wnt niche independence, which was mainly acquired non-genetically through mutations of driver genes (KRASG12V, CDKN2A, TP53, and SMAD4) during pancreas tumorigenesis (Seino et al., 2018). In another report by Hirt CK et al., ARID1A and BRCA were knocked out in PDAC organoid by CRISPR/Cas9 technology for identifying drug repurposing candidates. Researchers have successfully screened 26 out of 1,172 FDA-approved compounds, showing effective killing of PDAC organoids, including 19 chemotherapy drugs currently approved for other cancer types (Hirt et al., 2022). As we all know, gene knock in of CRISPR/Cas9 technology is also another commonly used gene editing technology in tumor research. Cortina C et al. devised a strategy based on engineered human CRC organoids that carry EGFP and lineage-tracing cassettes knocked in the LGR5 locus to study cancer stem cells in human tumors (Cortina et al., 2017). This strategy described herein may have broad applications to study cell heterogeneity in human tumors. Overall, the combination of the organoid platform with the CRISPR/Cas9 system conquers the drawbacks of conventional cell line culture and provides a more accurate representation of the biological behavior of tumor cells, making this a potent platform for tumor research.
4.2 Organoids for drug screening
The research on tumor organoids in drug screening mainly focuses on the establishment of tumor organoid models and tumor organoid biobanks to evaluate the anti-tumor activity of drugs, and predict drug targets and drug sensitivities (Driehuis et al., 2020). Due to their suitability for high-throughput screening, PDOs are an efficient platform for tumor drug screening (Wang and Jeon, 2022). Compared with animal models and two-dimensional cell models, tumor organoids not only have a higher success rate, a shorter culture cycle, and more facility for large-scale culture and drug screening through pore plates, but they also retain the heterogeneity and physiological and pathological changes of tumors in patients. Therefore, tumor organoids are more suitable for evaluating the treatment effects, toxicity, and side effects of drugs in vivo (Mittal et al., 2019), and thus for improving the efficiency of drug development and the success rate of drug clinical trials.
4.2.1 CRC PDOs
Most CRCs are aggressive and lack effective strategies for selecting appropriate anticancer regimens. CRC PDOs have emerged as preclinical platforms for modeling clinical responses to drug screening. The two most common strategies for drug screening are targeted drug discovery (TDD) and phenotype drug discovery (PDD). The latter does not depend on an understanding of the target and mechanism. It can also be divided into drug screening based on a compound library or drug screening based on an antibody library. In recent years, fedratinib (Mao et al., 2023), vinorelbin (Mertens et al., 2023), metformin (Luo et al., 2023), etc., with special anti-cancer effects on CRC have been screened from the compound library. Besides, Soon-Chan Kim et al. demonstrated a platform of 12 sets of PDOs and cell lines isolated from multiple regions of single tumors from 12 patients that were used to clarify heterogeneous drug responses using a clinically relevant 24-compound library (Kim et al., 2022). MCLA-158, a bAb that specifically triggers epidermal growth factor receptor degradation in LGR5+ cancer stem cells but shows minimal toxicity toward healthy LGR5+ colon stem cells, was tested in a large-scale functional screen. Briefly, using the CRC organoid platform, MCLA-158 was identified as a therapeutic EGFR × Lgr5 bispecific antibody from 500 effective bAbs against epithelial tumors by Herpers B et al. (Herpers et al., 2022). Interestingly, De Angelis ML et al. generated organoids from CTCs isolated from an orthotopic CRC xenograft model and found that CTC-derived organoids (CTCDOs) have an increased sensitivity to YM155, a drug targeting Survivin, and to the HSP90 inhibitor AUY922 (Luminespib) (De Angelis et al., 2022). In contrast, a prospective experimental treatment of CRC patients based on organoid drug responses revealed that drugs that have been observed to be therapeutic on organoid models may not achieve the expected consistent clinical responses in patients (Ooft et al., 2021). However, the authors of this research also point out that limited drugs and limited number of patients do not allow firm conclusions on whether organoid in vitro sensitivity predicts clinical response in vivo.
4.2.2 GC PDOs
GC PDOs are widely heralded as a drug-screening platform to develop new anti-cancer therapies. Yan et al. established a primary GC organoids (GCO) biobank, with which 37 anticancer drugs were screened (Yan et al., 2018). Drugs recently approved or in clinical trials, such as Napabucasin, Abemaciclib, and the ATR inhibitor VE-822, were confirmed to be effective in large-scale screening. In other words, this GCO biobank, with linked genomic data, provided a useful resource for studying both cancer cell biology and precision cancer therapy. Li et al. established a 3D organoid culture derived from malignant ascites (MA) of GC for disease modeling and drug screening. Eleven MAPDOs were produced from MA tumor cells in GC cells (Li et al., 2019). In another study, different responses to 5-fluorouracil, oxaliplatin, docetaxel, and irinotecan were observed in sig-ring cell carcinoma (SRCC) and non-SRCC GC organoids, predicting a potential strategy for drug response testing (Puccini et al., 2022). SRCC is specific and heterogeneous in phenotype and drug resistance. Systemic therapy is often ineffective for patients with SRCC, so predictive biomarkers are urgently needed to guide treatment.
4.2.3 Other PDOs
The PDOs platform has also been wildly established for drug-screening in other GI tumors, such as pancreatic ductal adenocarcinoma, HCC, etc. For example, Li et al. generated an organoid-based tissue platform using patient tissues from pancreatic ductal adenocarcinoma and assessed the cellular response to standard-of-care chemotherapeutic compounds, demonstrating their usability for drug screening (Li et al., 2022). Recently, Zou et al. performed the drug sensitivity test of Atezolizumab on microchips based on PDO, MSC-PDO-PBMC, CAF-PDO-PBMC, and PDO-PBMC models (Zou et al., 2023). The later three models showed a more sensitive response, indicating that MSC (CAF)-PDO-PBMC models mimicking the original tumor microenvironment (TME) were more suitable than conventional PDO models for in vitro tests of immunotherapeutic drugs.
4.3 PDOs for anti-tumor drug development
Although a large number of innovative anticancer drug projects are declared every year, there are few ideal drugs that eventually enter clinical application. The reason is that in the pre-clinical research stage of drug development, there is no suitable drug development model to simulate the in vivo environment to evaluate the efficacy of drugs, resulting in most drugs showing good results in basic research, but in clinical trials, satisfactory results cannot be achieved. The organoid model can not only enable cells to be cultured in 3D, but also better evaluate the therapeutic effect of drugs by simulating the growth environment in vivo. It has been widely used in GI tumor research.
4.3.1 PDOs is an important in vitro model for mechanism research and targeted drug development
PDOs have been established for many types of cancer and have become a promising tool for personalized oncology research. These models can completely reproduce the pedigree of tumor development and can be used for large-scale analysis of therapeutic responses, that is, drug typing, biomarker discovery, and tumor-related pathway research (Piro et al., 2021). In CRC, Roulis M et al. described rare pericryptal Ptgs2-expressing fibroblasts exerting paracrine control over tumor-initiating stem cells via the druggable PGE2-Ptger4-Yap signaling axis and considered that the initiation of CRC correlates with the mesenchymal niche (Roulis et al., 2020). Besides, the signaling axis of CSN6-TRIM21 was reported to promote tumor stemness during tumorigenesis (Qin et al., 2020). TFEB-PGD was considered another signaling axis activated by ATP13A2 to promote tumor growth (Zhang et al., 2023b). In GC, signaling axis of KDM1A-NDRG1 and NF-κB-PIEZO1-YAP1-CTGF might serve as potential therapeutic options to block tumorigenesis (Chen et al., 2023; Mircetic et al., 2023). Besides, Chromosome eight open reading frame 76 (C8orf76) was for the first time identified as a novel therapeutic target for GC that directly binds to oncogenic lncRNA DUSP5P1 to induce its expression and activate MAPK signaling (Wang et al., 2019). Moreover, pyruvate kinase M2 isoform (PKM2) mediated by ARRB1 may be a promising therapeutic strategy in a subset of GC patients (Yu et al., 2022). Similarly, in hepatocarcinogenesis, Wong TL et al. found that CRAF methylation by PRMT6 regulates aerobic glycolysis-driven via ERK-dependent PKM2 nuclear relocalization and activation (Wong et al., 2020). In another ferroptosis study in hepatocellular carcinoma, WTAP was investigated as a key factor upregulating ATG5 in an m6A-YTHDC2-dependent manner and targeted inhibition of WTAP may be an innovative and effective treatment strategy for HCC (Li et al., 2024). Through a cohort study, the Zhou SL team evaluated broad differences among organoids with different BRAF variant subtypes in sensitivity to BRAF or MEK inhibitors, and suggested a precise treatment for patients with intrahepatic cholangiocarcinoma (ICC) (Xin et al., 2023).
4.3.2 PDOs is also an important in vitro model for drug development
In the era of precision medicine, clinical medical staff and oncologists are eager to find more realistic, economical, and timely in vitro tumor models to assist drug development. The emergence of the PDO platform seems to solve the urgent problem. Recently, Kim N et al. demonstrated that fisetin, a natural plant flavonoid, significantly delayed tumor growth and inhibited vascular endothelial growth factor (VEGF) and epithelial cell adhesion molecule (EpCAM) via upregulation of AKAP12 in CRC (Kim et al., 2023). Besides, ATP13A2 and CaCO3@Cur@QTX125@HA nanoparticles were also reported as potential targets for CRC therapy (Zhang et al., 2023b; Hu et al., 2023). Moreover, MCLA-158, as mentioned above, exhibits potential therapeutic properties in epithelial tumors (Herpers et al., 2022). In two HCC studies, SHP099, a Src homology two domain-containing phosphatase two inhibitor, and desloratadine were reported as novel anticancer drugs (Leung et al., 2020; Tan et al., 2023). In another pancreatic cancer research, Krukovine was found to be an effective therapy in Oxaliplatin-resistant pancreatic cancer cells (Lee et al., 2023). In addition, NTRC 0652-0, a novel LCK selective tyrosine kinase inhibitor (Conboy et al., 2023), and Heat Shock Protein 90 were reported as novel inhibitors in cholangiocarcinoma (Lampis et al., 2018). These findings reveal that this PDO model is consistent with the original tumor, which has great potential for personalized cancer therapy and is a prerequisite for subsequent drug screening.
4.3.3 PDO is also an important in vitro model for reversing drug resistance
Intrinsic and acquired resistance to therapeutic drugs occurs in virtually all types of cancer. For effective treatment, the development of new drugs or combination therapy plays an important role in eliminating extensive drug resistance. In CRC, it is reported that co-treatment with embedded proteasome inhibitors bortezomib and cisplatin in nanoparticles can improve efficacy and reduce the side effects caused by drug combination therapy (Shao et al., 2020). Over more, Mcl-1 inhibition and lnc-RP11-536 were reported as the critical contributors to reversing the resistance of regorafenib and oxaliplatin, respectively (Song et al., 2020; Li et al., 2021). Using the PDO model established from GC patients, BDNF was discovered as a therapy target to reverse anlotinib resistance, and MYOF was found to be a promising biomarker and therapeutic target for L-OHP-resistant patients (Harada et al., 2021; Jin et al., 2021). Additionally, in HCC, lysosomal protein transmembrane five and protein arginine N-methyltransferase 6 (PRMT6) were considered the critical contributors to sorafenib and lenvatinib resistance, separately (Pan et al., 2023; Li et al., 2024). For pancreatic cancer, Irbesartan overcomes gemcitabine resistance by suppressing stemness and iron metabolism via inhibition of the Hippo/YAP1/c-Jun axis. The combination of gemcitabine and mithramycin shows potential therapeutic efficacy in reversing the resistance of gemcitabine.
4.4 Clinical research and personalized treatment
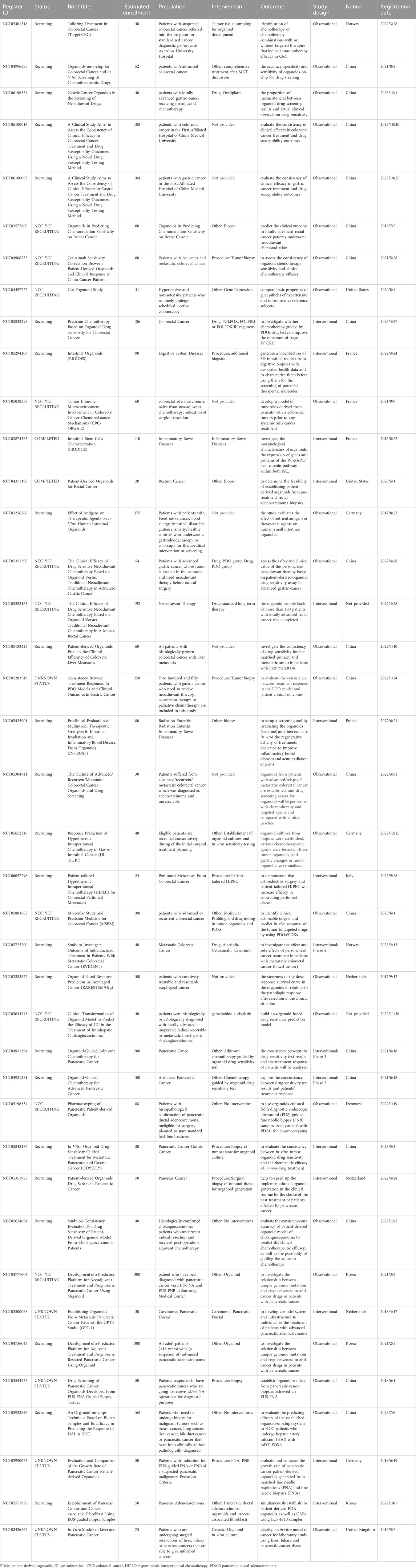
Recently, the PDOs model has been applied in clinical trials for personalized drug selection. The utility of PDOs in large-scale prospective clinical cohorts contributes to deeply integrating molecular biological characteristics and treatment responses in cancer patients, reducing the time of the clinic-laboratory-clinic cycle, and construct the platform for personalized oncology therapy (Shankaran et al., 2021). 40 case GI tumor PDO-related clinical trials have been registered in the Clinical Trials (Table 3).
4.5 Organoids and tumor microenvironment
The susceptibility of cancer to inflammation and immune deficiency demonstrates the critical role of immunity in tumorigenesis (Hibino et al., 2021). Cellular and drug immunotherapy can exert anti-cancer effects by enhancing the body’s immune monitoring function and local regulation of the tumor immune microenvironment, promising that the TME is a hot spot in the field of tumor research (Liu et al., 2023b). As we all know, the TME consists of immune cells, cancer-related fibroblasts (CAFs), interstitial cells, and ECM (Bejarano et al., 2021). The components in TME can maintain the differentiation activity of cancer stem cells and affect the occurrence, progress, metastasis, and drug resistance in tumorigenesis. At present, most tumor organoid models only contain tumor cells and lack TME components (Idris et al., 2021). Therefore, some research teams have constructed a co-culture system to explore the interaction between TME and PDOs (Mackenzie et al., 2022).
4.5.1 Co-culture system of CAFs and PDOs
CAFs are one of the main components of TME, which can secrete stimulating signals to support tumor development, suppress immunity, and promote drug resistance (Mao et al., 2021). Recently, organoid models of gastrointestinal tumors co-cultured with CAFs have been continuously established and developed. CRC PDO-CAF models developed by Luo et al., demonstrated that CAF can maintain the proliferation of CRC PDO culture in hydrogels even without the addition of traditional growth factors with which CRC-PDO can be cultured (Luo et al., 2021). In addition, the immune response-related pathways that exist in patient tissues but are missed in the non-co-culture model are reactivated. To compare the effects of different fibroblasts on PDO, primary fibroblasts from tumor adjacent tissues and CAFs were co-cultured with CRC organoids, respectively. The co-cultured organoids revealed greater heterogeneity in tumor cells than monocultured organoids, and crosstalk between tumor cells and fibroblasts leads to some imbalance in biological pathways (Atanasova et al., 2023). Similarly, Strating et al. co-cultured colon cancer organoids and CAFs in a highly standardized serum-free medium and optimized the ECM to investigate the interaction between cancer cells and CAFs and their effect on immune-related cytokines and T cell proliferation (Strating et al., 2023). In addition, Dang et al. studied the contribution of CAFs to the progress of early-stage CRC (T1CRC) through the co-culture model (Dang et al., 2023). Furthermore, Ramos Zapatero et al. developed a highly multiplexed mass cytometry platform and found that CAFs can regulate PDO plasticity-shifting proliferative colonic stem cells (proCSCs) to slow-cycling revival colonic stem cells (revCSCs) to protect cancer cells from chemotherapy (Ramos Zapatero et al., 2023). The PDO-CAFs model has also been established and applied to pancreatic cancer. Knoblauch M et al. established direct 3D co-cultures of primary PDAC organoids and patient-matched CAFs to investigate the influence of stromal components on chemosensitivity (Knoblauch et al., 2023). Similarly, Grützmeier SE et al. developed a co-culture model using PDTOs and CAFs derived from endoscopic ultrasound-guided fine-needle biopsies (EUS-FNBs) for potential use in drug screening and molecular mechanisms unraveling involved in the chemoresistance-supporting role of the tumor stroma (Grützmeier et al., 2023). In research reported by Zou et al., HCC-PDOs and PBMC are co-cultured with MSC and CAFs to construct HCC organoid-on-a-chip mimicking the original TME (Zou et al., 2023). This TME mimicked novel platform can be applied for high-throughput drug screening and to predict immunotherapy response of HCC patients. All these studies showed that the CRC PDO-CAFs model is suitable for evaluating standard care drugs, making it very useful for achieving personalized cancer drugs.
4.5.2 Co-culture system of PDOs with other immune cells
Co-culture of immune cells with PDO is helpful to further understand the interaction between tumor and immune system, which is of great significance to the research of tumor immunotherapy and can also promote personalized immunotherapy to adapt to reality. In 2018, Dijkstra et al. co-cultured PDO from mismatch repair deficient CRC patients with homologous peripheral blood lymphocytes and successfully enriched tumor-reactive T cells from peripheral blood. This not only realizes the dynamic evaluation of the curative effect of individualized immunotherapy under minimally invasive conditions, but also provides the possibility for adopting T cell therapy with peripheral blood (Dijkstra et al., 2018). Homologously, Cattaneo CM et al. co-cultured peripheral blood lymphocytes with PDO and assessed the generation and function of tumor-reactive T cells in CRC. T cells are evaluated for their capacity to carry out effector functions (IFN-γ secretion and degranulation) after recognition of tumor cells and their capacity to kill tumor organoids (Cattaneo et al., 2020). Besides, Jiang S et al. established a co-culture system with PDOs and macrophages, and Piro G et al. explored a co-culture system with PDOs and T-cells in pancreatic cancer (Piro et al., 2021; Jiang et al., 2023). An in vitro organoid/immune cell co-culture model reported by Holokai et al. was cultured to evaluate the specific target mechanisms that deplete MDSCs as a therapeutic strategy for PDAC (Wong et al., 2020). Schnalzger et al. reported a co-culture platform for CRC organoids and CAR-NK cells, which can dynamically and quantitatively monitor CAR-mediated cytotoxicity. The co-culture system showed the targeting effect of CAR cells on tumor-specific antigens and proved that CAR cells can still specifically kill CRC organoids without damaging normal organs under the condition of only a small amount of tumor antigen expression or complex microenvironment (Schnalzger et al., 2019). As described above, the co-culture of CRC organoids and immune cells can not only study the interaction between immune cells and tumors, but also the efficacy of tumor immunotherapy to develop new immunotherapy strategies. However, it has been a problem to be solved urgently: how to simulate the tumor immune microenvironment most realistically and quickly, evaluate the effect of immunotherapy, and screen out the target population that may benefit from itself clinically (Yuki et al., 2020). In addition, the interaction mode between immune cells and tumor cells in TME, the mechanism of directional metastasis of tumor-producing organoids, and the screening and identification of key cell subsets and regulatory molecules that regulate the formation of the metastatic microenvironment also need the support of a reliable co-culture platform in vitro (Yang and Yu, 2023).
5 Ethical consideration
Human organoids have been widely applied in medicine to extents such as drug development, disease modeling, developmental biology, disease pathology, cell biology, regeneration mechanisms, precision medicine and organ transplantation, which can efficiently simulate the physiological structure, function, development, and similarly maintenance processes of in situ tissues, showing great development potential (Tang et al., 2022). With the increasing development and maturity of organoid technology, various organoids, such as the stomach, intestine, liver, retina, brain, and other organoids have been established (Corrò et al., 2020). However, the ethical issues arising from the application are also worth pondering and noting.
5.1 Donor’s notification
It is well known that informed consent is necessary to be given to donors when their organs or specimens are donated. When informed consent is signed, the donors, whether have been informed, have the right to accept or reject it, or even have the right to revoke it. In addition, the donors may perceive the application scenarios and procedures for withdrawing informed consent post-sign (Munsie et al., 2017). All the above processes should be strictly implemented and need to be supervised and managed by specialized agencies.
5.2 Representativeness of the content of the notice
Organoids are derivatives of the specimens, which are derived from organs, tissues, biopsy specimens, pleural effusion, etc., in clinical assays (Rossi et al., 2018). In general, extensive informed consent was consulted and signed only for source specimens themselves, which is essentially different from the processing products of the source specimens (Boers et al., 2019). However, how to ensure that donors can inform the subsequent possible applications of such specimens in detail, such as successfully cultured organoids for scientific research or clinical utilization of specimen derivatives, remains unclear.
5.3 Privacy protection and information disclosure
Today, with the rapid development of various omics such as genomics, proteomics, metabolomics, etc., ethics in the era of big data have been challenged. It is legitimate that, on the premise of patient identity de-associated, human specimens utilized for scientific research after consent informed and confidentiality assurance However, the application of organoids seems to be limited for identity de-association because individual patients who might would be benefit from the study lost the opportunity to obtain experimental treatment. Moreover, the use value and application scenarios of human specimens will be greatly restricted if personal information is disconnected. Therefore, personal information privacy disclosures containing biological information related to biological samples of donors should be appropriately re-discussed.
5.4 Regenerative medicine and transplantation of organoids
The wide application of organoids in clinical trials or scientific research lies not only in their ability to highly simulate the physiological structure and function of in situ tissues or source organs but also in maintaining the stability and immortalization of genetic information through long-term passaging. Therefore, the application of organoids in regenerative medicine and organ transplantation poses emerging social and ethical challenges (Schneemann et al., 2020). As we all know, although the existing brain organoids have a very low probability of being conscious, how do people know if the brain organoids have developed consciousness (Lavazza and Chinaia, 2023). At present, human brain organoids or other organoids with consciousness or not have been cultivated either intentionally or unintentionally by scientists; the technology for detecting consciousness has not yet been broken through. Therefore, with the continuous development of in vitro culture conditions in organoids, people should be prepared for possible risks in brain organoids. When the emergence of stereotyped assembly and fusion technology provides a new probability to generate large-scale organoids, organ transplantation based on organoids will become a reality (Lavazza, 2021). In South Korea and Japan, intestinal organoids have been reported to be used in the clinical treatment of diseases such as ulcerative colitis or refractory Crohn’s disease. However, differences in genetic information between donors and recipients, immune rejection after transplantation, and the degree of benefit and risk to recipients also need to be accurately assessed after transplantation (Sugimoto et al., 2022). At the same time, organ transplantation is also a moral test for donors.
In conclusion, the current ethical norms and regulations need to be revised on a large scale to adapt to the ethical challenges brought by the rapid development of organoid technology and the implementation of clinical trial projects.
6 Challenges and prospects
6.1 The organoid cultivation system
Firstly, organoid culture technology depends largely on the establishment of the culture system, such as different growth factors and additives added to the medium. Although the organoid culture system of different organs and tissues has been established since the emergence of organoid culture technology, there is still no standard or universal training system or training program.
In the reported culture system, growth factors or additives will make the adapted cells grow preferentially, while the unadapted cells will die during the culture process. Studies have shown that intestinal tumor cells in the organoid culture system maintained the genomic, epigenetic, and transcriptomic characteristics of their in vivo-derived tumor tissues, but the organoid cells had different composition types and different proliferation rates (Wang et al., 2022b).
Secondly, matrigel is also a huge challenge for the stable construction of organoid technology. Matrigel, a basement membrane extract secreted by mouse sarcoma cells, is the most commonly used in organoid culture systems. Due to the unclear composition, heterogeneous characteristics, batch variation, and lack of reproducibility of matrigel, researchers have been committed to exploring alternatives to matrigel. For example, Takahashi et al. successfully used collagen gel instead of matrix gel and improved the medium composition, which was applied to human intestinal organoid culture for drug cytotoxicity screening (Takahashi et al., 2023). Alginate, hydrogel hyaluronic acid, and a mixture of hyaluronic acid and chitosan have also been reported to be good substitutes for matrigel (Sandilya and Singh, 2021; Hillion and Mahe, 2022; Chooi et al., 2023).
Finally, the tumor tissue itself has complex structures and tumor heterogeneity (Gao et al., 2023). In the process of in vitro organoid model construction, due to the limitation of the location of tumor tissue, which cannot represent the whole tumor, the limitation of culture conditions, the prolongation of culture time, and the increase of passages, only a part of the tumor cells adapted to the culture conditions, resulting in the surviving organoids losing tumor heterogeneity. In addition, in the process of culture and passage, the proliferation rate of tumor cells is lower than that of normal cells. Therefore, researchers have begun to focus on tumor organoid screening, which may be one of the next research directions (Wallaschek et al., 2019). During long-term in vitro culture, new mutations will also accumulate, resulting in increased differences between in vitro models and in vivo tumors (Yang et al., 2021). Therefore, in the future, more optimized training systems and conditions will be needed to solve these problems.
6.2 Culture microenvironment
Tumor stem cells, or epithelial stem cells, were adapted to grow in some specific medium, and matrigel finally formed the organoids. However, these stem cells lacked the in vivo tumor microenvironment during the culture process, such as fibroblasts, endothelial cells, immune cells, and other stromal cells and ECM components (Devarasetty et al., 2020). Even different individuals have different tumor microenvironments in vivo. However, in the matrigel-coated culture system, the same culture system is used, resulting in a partial lack of heterogeneity. The co-culture of organoids and immune cells is closer to the microenvironment in which cells grow in vivo, which is a new direction for the study of tumors and related immunotherapy (Schuth et al., 2022). In addition, the use of organoid microfluidic chip technology seems to be a breakthrough in the study of tumor microenvironment. However, the composition, concentration, and flow rate of multiple cytokines are the bottlenecks to the repeatability of organoid microenvironment reconstruction. In addition, the current organoid models cannot construct well-developed blood vessels, resulting in organoid models that cannot be cultured in vitro to a larger size, so the clinical application of organoids is also limited, and the development of engineered vascularization may overcome this problem (Garreta et al., 2021).
7 Conclusion
Over all, organoid, as one of the world’s top ten breakthrough technologies, is predicted to be the most promising preclinical disease transformation model in the 21st century. Although organoid technology currently faces considerable difficulties to be accomplished, its accurate simulation of human tumor characteristics still shows great potential for preclinical application. We all believed that, with the further precision of ethical recognition, the continuous maturity of organoid technology, and the solution of the microenvironment culture scheme, future efforts will undoubtedly bring this new technology closer to clinical practice.
Author contributions
SY: Software, Writing–original draft, Supervision. YuZ: Data curation, Formal Analysis, Writing–original draft. YH: Visualization, Writing–review and editing. WY: Visualization, Writing–review and editing. YC: Data curation, Writing–review and editing. CZ: Data curation, Investigation, Writing–original draft. FZ: Data curation, Visualization, Writing–review and editing. ZM: Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Zhejiang Provincial Basic Public Welfare Research Project (LGF20H16002) and the Huzhou Science and Technology Program Project (No.2019GZB06).
Acknowledgments
The authors would like to thank Hui Sun for supervision about the idea and all present and former lab members for helpful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2024.1384450/full#supplementary-material
References
Al-Ishaq, R. K., Overy, A. J., and Büsselberg, D. (2020). Phytochemicals and gastrointestinal cancer: cellular mechanisms and effects to change cancer progression. Biomolecules 10 (1), 105. doi:10.3390/biom10010105
Almeqdadi, M., Mana, M. D., Roper, J., and Yilmaz Ö, H. (2019). Gut organoids: mini-tissues in culture to study intestinal physiology and disease. Am. J. physiology. Cell physiology 317 (3), C405–C419. doi:10.1152/ajpcell.00300.2017
Artegiani, B., and Clevers, H. (2018). Use and application of 3D-organoid technology. Hum. Mol. Genet. 27 (R2), R99–R107. doi:10.1093/hmg/ddy187
Atanasova, V. S., de Jesus Cardona, C., Hejret, V., Tiefenbacher, A., Mair, T., Tran, L., et al. (2023). Mimicking tumor cell heterogeneity of colorectal cancer in a patient-derived organoid-fibroblast model. Cell. Mol. gastroenterology hepatology 15 (6), 1391–1419. doi:10.1016/j.jcmgh.2023.02.014
Baugh, E. H., Ke, H., Levine, A. J., Bonneau, R. A., and Chan, C. S. (2018). Why are there hotspot mutations in the TP53 gene in human cancers? Cell death Differ. 25 (1), 154–160. doi:10.1038/cdd.2017.180
Bejarano, L., Jordāo, M. J. C., and Joyce, J. A. (2021). Therapeutic targeting of the tumor microenvironment. Cancer Discov. 11 (4), 933–959. doi:10.1158/2159-8290.CD-20-1808
Biller, L. H., and Schrag, D. (2021). Diagnosis and treatment of metastatic colorectal cancer: a review. Jama 325 (7), 669–685. doi:10.1001/jama.2021.0106
Boers, S. N., van Delden, J. J. M., and Bredenoord, A. L. (2019). Organoids as hybrids: ethical implications for the exchange of human tissues. J. Med. ethics 45 (2), 131–139. doi:10.1136/medethics-2018-104846
Boos, S. L., Loevenich, L. P., Vosberg, S., Engleitner, T., Öllinger, R., Kumbrink, J., et al. (2022). Disease modeling on tumor organoids implicates AURKA as a therapeutic target in liver metastatic colorectal cancer. Cell. Mol. gastroenterology hepatology 13 (2), 517–540. doi:10.1016/j.jcmgh.2021.10.008
Cattaneo, C. M., Dijkstra, K. K., Fanchi, L. F., Kelderman, S., Kaing, S., van Rooij, N., et al. (2020). Tumor organoid-T-cell coculture systems. Nat. Protoc. 15 (1), 15–39. doi:10.1038/s41596-019-0232-9
Chen, B., Liu, X., Yu, P., Xie, F., Kwan, J. S. H., Chan, W. N., et al. (2023). H. pylori-induced NF-κB-PIEZO1-YAP1-CTGF axis drives gastric cancer progression and cancer-associated fibroblast-mediated tumour microenvironment remodelling. Clin. Transl. Med. 13 (11), e1481. doi:10.1002/ctm2.1481
Chooi, W. H., Ng, C. Y., Ow, V., Harley, J., Ng, W., Hor, J. H., et al. (2023). Defined alginate hydrogels support spinal cord organoid derivation, maturation, and modeling of spinal cord diseases. Adv. Healthc. Mater. 12 (9), e2202342. doi:10.1002/adhm.202202342
Conboy, C. B., Yonkus, J. A., Buckarma, E. H., Mun, D. G., Werneburg, N. W., Watkins, R. D., et al. (2023). LCK inhibition downregulates YAP activity and is therapeutic in patient-derived models of cholangiocarcinoma. J. hepatology 78 (1), 142–152. doi:10.1016/j.jhep.2022.09.014
Corrò, C., Novellasdemunt, L., and Li, V. S. W. (2020). A brief history of organoids. Am. J. physiology. Cell physiology 319 (1), C151–C165. doi:10.1152/ajpcell.00120.2020
Cortina, C., Turon, G., Stork, D., Hernando-Momblona, X., Sevillano, M., Aguilera, M., et al. (2017). A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 9 (7), 869–879. doi:10.15252/emmm.201707550
Dang, H., Harryvan, T. J., Liao, C. Y., Danen, E. H. J., Spalburg, V., Kielbasa, S. M., et al. (2023). Cancer-associated fibroblasts are key determinants of cancer cell invasion in the earliest stage of colorectal cancer. Cell. Mol. gastroenterology hepatology 16 (1), 107–131. doi:10.1016/j.jcmgh.2023.04.004
De Angelis, M. L., Francescangeli, F., Nicolazzo, C., Signore, M., Giuliani, A., Colace, L., et al. (2022). An organoid model of colorectal circulating tumor cells with stem cell features, hybrid EMT state and distinctive therapy response profile. J. Exp. Clin. cancer Res. CR 41 (1), 86. doi:10.1186/s13046-022-02263-y
Devarasetty, M., Forsythe, S. D., Shelkey, E., and Soker, S. (2020). In vitro modeling of the tumor microenvironment in tumor organoids. Tissue Eng. Regen. Med. 17 (6), 759–771. doi:10.1007/s13770-020-00258-4
Dietinger, V., García de Durango, C. R., Wiechmann, S., Boos, S. L., Michl, M., Neumann, J., et al. (2020). Wnt-driven LARGE2 mediates laminin-adhesive O-glycosylation in human colonic epithelial cells and colorectal cancer. Cell Commun. Signal. CCS 18 (1), 102. doi:10.1186/s12964-020-00561-6
Dijkstra, K. K., Cattaneo, C. M., Weeber, F., Chalabi, M., van de Haar, J., Fanchi, L. F., et al. (2018). Generation of tumor-reactive T cells by Co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174 (6), 1586–1598. doi:10.1016/j.cell.2018.07.009
Dominguez, A. A., Lim, W. A., and Qi, L. S. (2016). Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. cell Biol. 17 (1), 5–15. doi:10.1038/nrm.2015.2
Driehuis, E., Kolders, S., Spelier, S., Lõhmussaar, K., Willems, S. M., Devriese, L. A., et al. (2019). Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9 (7), 852–871. doi:10.1158/2159-8290.CD-18-1522
Driehuis, E., Kretzschmar, K., and Clevers, H. (2020). Establishment of patient-derived cancer organoids for drug-screening applications. Nat. Protoc. 15 (10), 3380–3409. doi:10.1038/s41596-020-0379-4
Gao, Q., Zhan, Y., Sun, L., and Zhu, W. (2023). Cancer stem cells and the tumor microenvironment in tumor drug resistance. Stem cell Rev. Rep. 19, 2141–2154. doi:10.1007/s12015-023-10593-3
Garreta, E., Kamm, R. D., Chuva de Sousa Lopes, S. M., Lancaster, M. A., Weiss, R., Trepat, X., et al. (2021). Rethinking organoid technology through bioengineering. Nat. Mater. 20 (2), 145–155. doi:10.1038/s41563-020-00804-4
Giamas, G. (2020). Cancer Gene Therapy: vision and strategy for the new decade. Cancer gene Ther. 27 (3-4), 115. doi:10.1038/s41417-020-0169-8
Grady, W. M. (2021). Epigenetic alterations in the gastrointestinal tract: current and emerging use for biomarkers of cancer. Adv. cancer Res. 151, 425–468. doi:10.1016/bs.acr.2021.02.006
Grützmeier, S. E., Kovacevic, B., Vilmann, P., Rift, C. V., Melchior, L. C., Holmström, M. O., et al. (2023). Validation of a novel EUS-FNB-derived organoid Co-culture system for drug screening in patients with pancreatic cancer. Cancers 15 (14), 3677. doi:10.3390/cancers15143677
Habanjar, O., Diab-Assaf, M., Caldefie-Chezet, F., and Delort, L. (2021). 3D cell culture systems: tumor application, advantages, and disadvantages. Int. J. Mol. Sci. 22 (22), 12200. doi:10.3390/ijms222212200
Harada, K., Sakamoto, N., Ukai, S., Yamamoto, Y., Pham, Q. T., Taniyama, D., et al. (2021). Establishment of oxaliplatin-resistant gastric cancer organoids: importance of myoferlin in the acquisition of oxaliplatin resistance. Gastric cancer 24 (6), 1264–1277. doi:10.1007/s10120-021-01206-4
Herpers, B., Eppink, B., James, M. I., Cortina, C., Cañellas-Socias, A., Boj, S. F., et al. (2022). Functional patient-derived organoid screenings identify MCLA-158 as a therapeutic EGFR × LGR5 bispecific antibody with efficacy in epithelial tumors. Nat. cancer 3 (4), 418–436. doi:10.1038/s43018-022-00359-0
Hibino, S., Kawazoe, T., Kasahara, H., Itoh, S., Ishimoto, T., Sakata-Yanagimoto, M., et al. (2021). Inflammation-induced tumorigenesis and metastasis. Int. J. Mol. Sci. 22 (11), 5421. doi:10.3390/ijms22115421
Hillion, K., and Mahe, M. M. (2022). Redesigning hydrogel geometry for enhanced organoids. Nat. methods 19 (11), 1347–1348. doi:10.1038/s41592-022-01656-3
Hirt, C. K., Booij, T. H., Grob, L., Simmler, P., Toussaint, N. C., Keller, D., et al. (2022). Drug screening and genome editing in human pancreatic cancer organoids identifies drug-gene interactions and candidates for off-label treatment. Cell genomics 2 (2), 100095. doi:10.1016/j.xgen.2022.100095
Hu, S., Xia, K., Huang, X., Zhao, Y., Zhang, Q., Huang, D., et al. (2023). Multifunctional CaCO(3)@Cur@QTX125@HA nanoparticles for effectively inhibiting growth of colorectal cancer cells. J. nanobiotechnology 21 (1), 353. doi:10.1186/s12951-023-02104-w
Hu, X., Zhang, L., Li, Y., Ma, X., Dai, W., Gao, X., et al. (2020). Organoid modelling identifies that DACH1 functions as a tumour promoter in colorectal cancer by modulating BMP signalling. EBioMedicine 56, 102800. doi:10.1016/j.ebiom.2020.102800
Huang, J., Lucero-Prisno, D. E., Zhang, L., Xu, W., Wong, S. H., Ng, S. C., et al. (2023). Updated epidemiology of gastrointestinal cancers in East Asia. Nat. Rev. Gastroenterology hepatology 20 (5), 271–287. doi:10.1038/s41575-022-00726-3
Huch, M., Bonfanti, P., Boj, S. F., Sato, T., Loomans, C. J., van de Wetering, M., et al. (2013). Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32 (20), 2708–2721. doi:10.1038/emboj.2013.204
Huch, M., Gehart, H., van Boxtel, R., Hamer, K., Blokzijl, F., Verstegen, M. M., et al. (2015). Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160 (1-2), 299–312. doi:10.1016/j.cell.2014.11.050
Idris, M., Alves, M. M., Hofstra, R. M. W., Mahe, M. M., and Melotte, V. (2021). Intestinal multicellular organoids to study colorectal cancer. Biochimica biophysica acta. Rev. cancer 1876 (2), 188586. doi:10.1016/j.bbcan.2021.188586
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 (12), 5429–5433. doi:10.1128/jb.169.12.5429-5433.1987
Janik, E., Niemcewicz, M., Ceremuga, M., Krzowski, L., Saluk-Bijak, J., Bijak, M., et al. (2020). Molecular aspects of mycotoxins-A serious problem for human health. Int. J. Mol. Sci. 21 (24), 8187. doi:10.3390/ijms21218187
Jiang, S., Deng, T., Cheng, H., Liu, W., Shi, D., Yuan, J., et al. (2023). Macrophage-organoid co-culture model for identifying treatment strategies against macrophage-related gemcitabine resistance. J. Exp. Clin. cancer Res. CR 42 (1), 199. doi:10.1186/s13046-023-02756-4
Jin, Z., Lu, Y., Wu, X., Pan, T., Yu, Z., Hou, J., et al. (2021). The cross-talk between tumor cells and activated fibroblasts mediated by lactate/BDNF/TrkB signaling promotes acquired resistance to anlotinib in human gastric cancer. Redox Biol. 46, 102076. doi:10.1016/j.redox.2021.102076
Karimian, A., Azizian, K., Parsian, H., Rafieian, S., Shafiei-Irannejad, V., Kheyrollah, M., et al. (2019). CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J. Cell. physiology 234 (8), 12267–12277. doi:10.1002/jcp.27972
Kemper, K., Krijgsman, O., Cornelissen-Steijger, P., Shahrabi, A., Weeber, F., Song, J. Y., et al. (2015). Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol. Med. 7 (9), 1104–1118. doi:10.15252/emmm.201404914
Kim, N., Kwon, J., Shin, U. S., and Jung, J. (2023). Fisetin induces the upregulation of AKAP12 mRNA and anti-angiogenesis in a patient-derived organoid xenograft model. Biomed. Pharmacother. = Biomedecine Pharmacother. 167, 115613. doi:10.1016/j.biopha.2023.115613
Kim, S. C., Park, J. W., Seo, H. Y., Kim, M., Park, J. H., Kim, G. H., et al. (2022). Multifocal organoid capturing of colon cancer reveals pervasive intratumoral heterogenous drug responses. Adv. Sci. 9 (5), e2103360. doi:10.1002/advs.202103360
Kimura, J. O., Bolaños, D. M., Ricci, L., and Srivastava, M. (2022). Embryonic origins of adult pluripotent stem cells. Cell 185 (25), 4756–4769.e13. doi:10.1016/j.cell.2022.11.008
Knoblauch, M., Ma, T., Beirith, I., Koch, D., Hofmann, F., Heinrich, K., et al. (2023). In-vitro model to mimic T cell subset change in human PDAC organoid co-culture. J. cancer Res. Clin. Oncol. 149 (14), 13051–13064. doi:10.1007/s00432-023-05100-7
Koikawa, K., Kibe, S., Suizu, F., Sekino, N., Kim, N., Manz, T. D., et al. (2021). Targeting Pin1 renders pancreatic cancer eradicable by synergizing with immunochemotherapy. Cell 184 (18), 4753–4771.e27. doi:10.1016/j.cell.2021.07.020
Kozlowski, M. T., Crook, C. J., and Ku, H. T. (2021). Towards organoid culture without Matrigel. Commun. Biol. 4 (1), 1387. doi:10.1038/s42003-021-02910-8
Küçükköse, E., Peters, N. A., Ubink, I., van Keulen, V. A. M., Daghighian, R., Verheem, A., et al. (2022). KIT promotes tumor stroma formation and counteracts tumor-suppressive TGFβ signaling in colorectal cancer. Cell death Dis. 13 (7), 617. doi:10.1038/s41419-022-05078-z
Lampis, A., Carotenuto, P., Vlachogiannis, G., Cascione, L., Hedayat, S., Burke, R., et al. (2018). MIR21 drives resistance to Heat Shock protein 90 inhibition in cholangiocarcinoma. Gastroenterology 154 (4), 1066–1079. doi:10.1053/j.gastro.2017.10.043
Lannagan, T. R., Jackstadt, R., Leedham, S. J., and Sansom, O. J. (2021). Advances in colon cancer research: in vitro and animal models. Curr. Opin. Genet. Dev. 66, 50–56. doi:10.1016/j.gde.2020.12.003
Lavazza, A. (2021). Potential ethical problems with human cerebral organoids: consciousness and moral status of future brains in a dish. Brain Res. 1750, 147146. doi:10.1016/j.brainres.2020.147146
Lavazza, A., and Chinaia, A. A. (2023). Human cerebral organoids: the ethical stance of scientists. Stem cell Res. Ther. 14 (1), 59. doi:10.1186/s13287-023-03291-x
Lee, J. H., Lee, S. H., Lee, S. K., Choi, J. H., Lim, S., Kim, M. S., et al. (2023). Antiproliferative activity of krukovine by regulating transmembrane protein 139 (TMEM139) in oxaliplatin-resistant pancreatic cancer cells. Cancers 15 (9), 2642. doi:10.3390/cancers15092642
Lee, R., Li, J., Li, J., Wu, C. J., Jiang, S., Hsu, W. H., et al. (2022). Synthetic essentiality of tryptophan 2,3-dioxygenase 2 in APC-mutated colorectal cancer. Cancer Discov. 12 (7), 1702–1717. doi:10.1158/2159-8290.CD-21-0680
Leung, C. O. N., Tong, M., Chung, K. P. S., Zhou, L., Che, N., Tang, K. H., et al. (2020). Overriding adaptive resistance to sorafenib through combination therapy with Src homology 2 domain-containing phosphatase 2 blockade in hepatocellular carcinoma. Hepatology 72 (1), 155–168. doi:10.1002/hep.30989
Li, J., Xu, H., Zhang, L., Song, L., Feng, D., Peng, X., et al. (2019). Malignant ascites-derived organoid (MADO) cultures for gastric cancer in vitro modelling and drug screening. J. cancer Res. Clin. Oncol. 145 (11), 2637–2647. doi:10.1007/s00432-019-03004-z
Li, N. T., Wu, N. C., Cao, R., Cadavid, J. L., Latour, S., Lu, X., et al. (2022). An off-the-shelf multi-well scaffold-supported platform for tumour organoid-based tissues. Biomaterials 291, 121883. doi:10.1016/j.biomaterials.2022.121883
Li, Q., Sun, H., Luo, D., Gan, L., Mo, S., Dai, W., et al. (2021). Lnc-RP11-536 K7.3/SOX2/HIF-1α signaling axis regulates oxaliplatin resistance in patient-derived colorectal cancer organoids. J. Exp. Clin. cancer Res. CR 40 (1), 348. doi:10.1186/s13046-021-02143-x
Li, Y., Gao, X., Ni, C., Zhao, B., and Cheng, X. (2023). The application of patient-derived organoid in the research of lung cancer. Cell. Oncol. Dordr. 46 (3), 503–519. doi:10.1007/s13402-023-00771-3
Li, Y., Guo, M., Qiu, Y., Li, M., Wu, Y., Shen, M., et al. (2024). Autophagy activation is required for N6-methyladenosine modification to regulate ferroptosis in hepatocellular carcinoma. Redox Biol. 69, 102971. doi:10.1016/j.redox.2023.102971
Liu, D., and Langer, R. (2022). Grading of tumor regression of gastrointestinal carcinomas after neoadjuvant therapy. Der Pathol. 43 (1), 51–56. doi:10.1007/s00292-021-01041-5
Liu, H., Su, H., Wang, F., Dang, Y., Ren, Y., Yin, S., et al. (2023a). Pharmacological boosting of cGAS activation sensitizes chemotherapy by enhancing antitumor immunity. Cell Rep. 42 (3), 112275. doi:10.1016/j.celrep.2023.112275
Liu, Y., Baba, Y., Ishimoto, T., Gu, X., Zhang, J., Nomoto, D., et al. (2022). Gut microbiome in gastrointestinal cancer: a friend or foe? Int. J. Biol. Sci. 18 (10), 4101–4117. doi:10.7150/ijbs.69331
Liu, Y., Hu, X., Han, C., Wang, L., Zhang, X., He, X., et al. (2015). Targeting tumor suppressor genes for cancer therapy. BioEssays news Rev. Mol. Cell. Dev. Biol. 37 (12), 1277–1286. doi:10.1002/bies.201500093
Liu, Y., Xun, Z., Ma, K., Liang, S., Li, X., Zhou, S., et al. (2023b). Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. hepatology 78 (4), 770–782. doi:10.1016/j.jhep.2023.01.011
Luo, X., Fong, E. L. S., Zhu, C., Lin, Q. X. X., Xiong, M., Li, A., et al. (2021). Hydrogel-based colorectal cancer organoid co-culture models. Acta biomater. 132, 461–472. doi:10.1016/j.actbio.2020.12.037
Luo, Z., Wang, B., Luo, F., Guo, Y., Jiang, N., Wei, J., et al. (2023). Establishment of a large-scale patient-derived high-risk colorectal adenoma organoid biobank for high-throughput and high-content drug screening. BMC Med. 21 (1), 336. doi:10.1186/s12916-023-03034-y
Mackenzie, N. J., Nicholls, C., Templeton, A. R., Perera, M. P., Jeffery, P. L., Zimmermann, K., et al. (2022). Modelling the tumor immune microenvironment for precision immunotherapy. Clin. Transl. Immunol. 11 (6), e1400. doi:10.1002/cti2.1400
Mao, X., Xu, J., Wang, W., Liang, C., Hua, J., Liu, J., et al. (2021). Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. cancer 20 (1), 131. doi:10.1186/s12943-021-01428-1
Mao, Y., Wang, W., Yang, J., Zhou, X., Lu, Y., Gao, J., et al. (2023). Drug repurposing screening and mechanism analysis based on human colorectal cancer organoids. Protein & cell, pwad038. doi:10.1093/procel/pwad038
Martin, G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U. S. A. 78 (12), 7634–7638. doi:10.1073/pnas.78.12.7634
Matano, M., Date, S., Shimokawa, M., Takano, A., Fujii, M., Ohta, Y., et al. (2015). Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 21 (3), 256–262. doi:10.1038/nm.3802
McCracken, K. W., Catá, E. M., Crawford, C. M., Sinagoga, K. L., Schumacher, M., Rockich, B. E., et al. (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516 (7531), 400–404. doi:10.1038/nature13863
Mertens, S., Huismans, M. A., Verissimo, C. S., Ponsioen, B., Overmeer, R., Proost, N., et al. (2023). Drug-repurposing screen on patient-derived organoids identifies therapy-induced vulnerability in KRAS-mutant colon cancer. Cell Rep. 42 (4), 112324. doi:10.1016/j.celrep.2023.112324
Mircetic, J., Camgöz, A., Abohawya, M., Ding, L., Dietzel, J., Tobar, S. G., et al. (2023). CRISPR/Cas9 screen in gastric cancer patient-derived organoids reveals kdm1a-NDRG1 Axis as a targetable vulnerability. Small methods 7 (6), e2201605. doi:10.1002/smtd.202201605
Mittal, R., Woo, F. W., Castro, C. S., Cohen, M. A., Karanxha, J., Mittal, J., et al. (2019). Organ-on-chip models: implications in drug discovery and clinical applications. J. Cell. physiology 234 (6), 8352–8380. doi:10.1002/jcp.27729
Morgan, K. M., Riedlinger, G. M., Rosenfeld, J., Ganesan, S., and Pine, S. R. (2017). Patient-derived xenograft models of non-small cell lung cancer and their potential utility in personalized medicine. Front. Oncol. 7, 2. doi:10.3389/fonc.2017.00002
Munsie, M., Hyun, I., and Sugarman, J. (2017). Ethical issues in human organoid and gastruloid research. Dev. Camb. Engl. 144 (6), 942–945. doi:10.1242/dev.140111
Naldini, L. (2015). Gene therapy returns to centre stage. Nature 526 (7573), 351–360. doi:10.1038/nature15818
Narasimhan, V., Wright, J. A., Churchill, M., Wang, T., Rosati, R., Lannagan, T. R. M., et al. (2020). Medium-throughput drug screening of patient-derived organoids from colorectal peritoneal metastases to direct personalized therapy. Clin. cancer Res. official J. Am. Assoc. Cancer Res. 26 (14), 3662–3670. doi:10.1158/1078-0432.CCR-20-0073
Neal, J. T., Li, X., Zhu, J., Giangarra, V., Grzeskowiak, C. L., Ju, J., et al. (2018). Organoid modeling of the tumor immune microenvironment. Cell 175 (7), 1972–1988. doi:10.1016/j.cell.2018.11.021
Nelson-Rees, W. A., Flandermeyer, R. R., and Hawthorne, P. K. (1974). Banded marker chromosomes as indicators of intraspecies cellular contamination. Sci. (New York, N.Y.) 184 (4141), 1093–1096. doi:10.1126/science.184.4141.1093
Nieto-Estévez, V., and Hsieh, J. (2020). Human brain organoid models of developmental epilepsies. Epilepsy Curr. 20 (5), 282–290. doi:10.1177/1535759720949254
Nuciforo, S., Fofana, I., Matter, M. S., Blumer, T., Calabrese, D., Boldanova, T., et al. (2018). Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep. 24 (5), 1363–1376. doi:10.1016/j.celrep.2018.07.001
Okugawa, Y., Grady, W. M., and Goel, A. (2015). Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 149 (5), 1204–1225. doi:10.1053/j.gastro.2015.07.011
Onfroy-Roy, L., Hamel, D., Foncy, J., Malaquin, L., and Ferrand, A. (2020). Extracellular matrix mechanical properties and regulation of the intestinal stem cells: when mechanics control fate. Cells 9 (12), 2629. doi:10.3390/cells9122629
Ooft, S. N., Weeber, F., Schipper, L., Dijkstra, K. K., McLean, C. M., Kaing, S., et al. (2021). Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO open 6 (3), 100103. doi:10.1016/j.esmoop.2021.100103
Pan, J., Zhang, M., Dong, L., Ji, S., Zhang, J., Zhang, S., et al. (2023). Genome-Scale CRISPR screen identifies LAPTM5 driving lenvatinib resistance in hepatocellular carcinoma. Autophagy 19 (4), 1184–1198. doi:10.1080/15548627.2022.2117893
Peng, W., Sun, D., Lu, W., Yin, S., Ye, B., Wang, X., et al. (2023). Comprehensive detection of PD-L1 protein and mRNA in tumor cells and extracellular vesicles through a real-time qPCR assay. Anal. Chem. 95 (28), 10625–10633. doi:10.1021/acs.analchem.3c00975
Pfohl, U., Loskutov, J., Bashir, S., Kühn, R., Herter, P., Templin, M., et al. (2022). A RAS-independent biomarker panel to reliably predict response to MEK inhibition in colorectal cancer. Cancers 14 (13), 3252. doi:10.3390/cancers14133252
Piro, G., Agostini, A., Larghi, A., Quero, G., Carbone, C., Esposito, A., et al. (2021). Pancreatic cancer patient-derived organoid platforms: a clinical tool to study cell- and non-cell-autonomous mechanisms of treatment response. Front. Med. 8, 793144. doi:10.3389/fmed.2021.793144
Pleguezuelos-Manzano, C., Puschhof, J., van den Brink, S., Geurts, V., Beumer, J., and Clevers, H. (2020). Establishment and culture of human intestinal organoids derived from adult stem cells. Curr. Protoc. Immunol. 130 (1), e106. doi:10.1002/cpim.106
Praharaj, P. P., Bhutia, S. K., Nagrath, S., Bitting, R. L., and Deep, G. (2018). Circulating tumor cell-derived organoids: current challenges and promises in medical research and precision medicine. Biochimica biophysica acta. Rev. cancer 1869 (2), 117–127. doi:10.1016/j.bbcan.2017.12.005
Puccini, A., Poorman, K., Catalano, F., Seeber, A., Goldberg, R. M., Salem, M. E., et al. (2022). Molecular profiling of signet-ring-cell carcinoma (SRCC) from the stomach and colon reveals potential new therapeutic targets. Oncogene 41 (26), 3455–3460. doi:10.1038/s41388-022-02350-6
Qin, B., Zou, S., Li, K., Wang, H., Wei, W., Zhang, B., et al. (2020). CSN6-TRIM21 axis instigates cancer stemness during tumorigenesis. Br. J. cancer 122 (11), 1673–1685. doi:10.1038/s41416-020-0779-9
Ramos Zapatero, M., Tong, A., Opzoomer, J. W., O'Sullivan, R., Cardoso Rodriguez, F., Sufi, J., et al. (2023). Trellis tree-based analysis reveals stromal regulation of patient-derived organoid drug responses. Cell 186 (25), 5606–5619.e24. doi:10.1016/j.cell.2023.11.005
Reza, H. A., Okabe, R., and Takebe, T. (2021). Organoid transplant approaches for the liver. Transpl. Int. official J. Eur. Soc. Organ Transplant. 34 (11), 2031–2045. doi:10.1111/tri.14128
Rossi, G., Manfrin, A., and Lutolf, M. P. (2018). Progress and potential in organoid research. Nat. Rev. Genet. 19 (11), 671–687. doi:10.1038/s41576-018-0051-9
Roulis, M., Kaklamanos, A., Schernthanner, M., Bielecki, P., Zhao, J., Kaffe, E., et al. (2020). Paracrine orchestration of intestinal tumorigenesis by a mesenchymal niche. Nature 580 (7804), 524–529. doi:10.1038/s41586-020-2166-3
Saito, Y., Muramatsu, T., Kanai, Y., Ojima, H., Sukeda, A., Hiraoka, N., et al. (2019). Establishment of patient-derived organoids and drug screening for biliary tract carcinoma. Cell Rep. 27 (4), 1265–1276. doi:10.1016/j.celrep.2019.03.088
Sandilya, S., and Singh, S. (2021). Development of islet organoids from human induced pluripotent stem cells in a cross-linked collagen scaffold. Cell Regen. Lond. Engl. 10 (1), 38. doi:10.1186/s13619-021-00099-z
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459 (7244), 262–265. doi:10.1038/nature07935
Schnalzger, T. E., de Groot, M. H., Zhang, C., Mosa, M. H., Michels, B. E., Röder, J., et al. (2019). 3D model for CAR-mediated cytotoxicity using patient-derived colorectal cancer organoids. EMBO J. 38 (12), e100928. doi:10.15252/embj.2018100928
Schneemann, S. A., Boers, S. N., van Delden, J. J. M., Nieuwenhuis, E. E. S., Fuchs, S. A., and Bredenoord, A. L. (2020). Ethical challenges for pediatric liver organoid transplantation. Sci. Transl. Med. 12 (552), eaau8471. doi:10.1126/scitranslmed.aau8471
Schuth, S., Le Blanc, S., Krieger, T. G., Jabs, J., Schenk, M., Giese, N. A., et al. (2022). Patient-specific modeling of stroma-mediated chemoresistance of pancreatic cancer using a three-dimensional organoid-fibroblast co-culture system. J. Exp. Clin. cancer Res. CR 41 (1), 312. doi:10.1186/s13046-022-02519-7
Seidlitz, T., Merker, S. R., Rothe, A., Zakrzewski, F., von Neubeck, C., Grützmann, K., et al. (2019). Human gastric cancer modelling using organoids. Gut 68 (2), 207–217. doi:10.1136/gutjnl-2017-314549
Seino, T., Kawasaki, S., Shimokawa, M., Tamagawa, H., Toshimitsu, K., Fujii, M., et al. (2018). Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22 (3), 454–467. doi:10.1016/j.stem.2017.12.009
Shankaran, A., Prasad, K., Chaudhari, S., Brand, A., and Satyamoorthy, K. (2021). Advances in development and application of human organoids. 3 Biotech. 11 (6), 257. doi:10.1007/s13205-021-02815-7
Shao, F., Lyu, X., Miao, K., Xie, L., Wang, H., Xiao, H., et al. (2020). Enhanced protein damage clearance induces broad drug resistance in multitype of cancers revealed by an evolution drug-resistant model and genome-wide siRNA screening. Adv. Sci. 7 (23), 2001914. doi:10.1002/advs.202001914
Shi, B., Liu, W. W., Yang, K., Jiang, G. M., and Wang, H. (2022). The role, mechanism, and application of RNA methyltransferase METTL14 in gastrointestinal cancer. Mol. cancer 21 (1), 163. doi:10.1186/s12943-022-01634-5
Song, X., Shen, L., Tong, J., Kuang, C., Zeng, S., Schoen, R. E., et al. (2020). Mcl-1 inhibition overcomes intrinsic and acquired regorafenib resistance in colorectal cancer. Theranostics 10 (18), 8098–8110. doi:10.7150/thno.45363
Spence, J. R., Mayhew, C. N., Rankin, S. A., Kuhar, M. F., Vallance, J. E., Tolle, K., et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470 (7332), 105–109. doi:10.1038/nature09691
Strating, E., Verhagen, M. P., Wensink, E., Dünnebach, E., Wijler, L., Aranguren, I., et al. (2023). Co-cultures of colon cancer cells and cancer-associated fibroblasts recapitulate the aggressive features of mesenchymal-like colon cancer. Front. Immunol. 14, 1053920. doi:10.3389/fimmu.2023.1053920
Sugimoto, S., Kobayashi, E., Kanai, T., and Sato, T. (2022). In vivo intestinal research using organoid transplantation. Keio J. Med. 71 (4), 73–81. doi:10.2302/kjm.2022-0019-IR
Taguchi, A., and Nishinakamura, R. (2017). Higher-order kidney organogenesis from pluripotent stem cells. Cell stem cell 21 (6), 730–746. doi:10.1016/j.stem.2017.10.011
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131 (5), 861–872. doi:10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126 (4), 663–676. doi:10.1016/j.cell.2006.07.024
Takahashi, Y., Inoue, Y., Sato, S., Okabe, T., Kojima, H., Kiyono, H., et al. (2023). Drug cytotoxicity screening using human intestinal organoids propagated with extensive cost-reduction strategies. Sci. Rep. 13 (1), 5407. doi:10.1038/s41598-023-32438-2
Tan, X. P., He, Y., Yang, J., Wei, X., Fan, Y. L., Zhang, G. G., et al. (2023). Blockade of NMT1 enzymatic activity inhibits N-myristoylation of VILIP3 protein and suppresses liver cancer progression. Signal Transduct. Target. Ther. 8 (1), 14. doi:10.1038/s41392-022-01248-9
Tang, X. Y., Wu, S., Wang, D., Chu, C., Hong, Y., Tao, M., et al. (2022). Human organoids in basic research and clinical applications. Signal Transduct. Target. Ther. 7 (1), 168. doi:10.1038/s41392-022-01024-9
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Sci. (New York, N.Y.) 282 (5391), 1145–1147. doi:10.1126/science.282.5391.1145
Tong, Y., Gao, H., Qi, Q., Liu, X., Li, J., Gao, J., et al. (2021). High fat diet, gut microbiome and gastrointestinal cancer. Theranostics 11 (12), 5889–5910. doi:10.7150/thno.56157
Tung, K. L., Chen, K. Y., Negrete, M., Chen, T., Safi, A., Aljamal, A. A., et al. (2021). Integrated chromatin and transcriptomic profiling of patient-derived colon cancer organoids identifies personalized drug targets to overcome oxaliplatin resistance. Genes & Dis. 8 (2), 203–214. doi:10.1016/j.gendis.2019.10.012
van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161 (4), 933–945. doi:10.1016/j.cell.2015.03.053
Verissimo, C. S., Overmeer, R. M., Ponsioen, B., Drost, J., Mertens, S., Verlaan-Klink, I., et al. (2016). Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 5, e18489. doi:10.7554/eLife.18489
Vlachogiannis, G., Hedayat, S., Vatsiou, A., Jamin, Y., Fernández-Mateos, J., Khan, K., et al. (2018). Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Sci. (New York, N.Y.) 359 (6378), 920–926. doi:10.1126/science.aao2774
Wallaschek, N., Niklas, C., Pompaiah, M., Wiegering, A., Germer, C. T., Kircher, S., et al. (2019). Establishing pure cancer organoid cultures: identification, selection and verification of cancer phenotypes and genotypes. J. Mol. Biol. 431 (15), 2884–2893. doi:10.1016/j.jmb.2019.05.031
Wang, C., and Fakih, M. (2021). Targeting KRAS in colorectal cancer. Curr. Oncol. Rep. 23 (3), 28. doi:10.1007/s11912-021-01022-0
Wang, J., Feng, X., Li, Z., Chen, Y., and Huang, W. (2022a). Patient-derived organoids as a model for tumor research. Prog. Mol. Biol. Transl. Sci. 189 (1), 259–326. doi:10.1016/bs.pmbts.2022.03.004
Wang, R., Mao, Y., Wang, W., Zhou, X., Wang, W., Gao, S., et al. (2022b). Systematic evaluation of colorectal cancer organoid system by single-cell RNA-Seq analysis. Genome Biol. 23 (1), 106. doi:10.1186/s13059-022-02673-3
Wang, X., Liang, Q., Zhang, L., Gou, H., Li, Z., Chen, H., et al. (2019). C8orf76 promotes gastric tumorigenicity and metastasis by directly inducing lncRNA DUSP5P1 and associates with patient outcomes. Clin. Cancer Res. 25 (10), 3128–3140. doi:10.1158/1078-0432.CCR-18-2804
Wang, Y., and Jeon, H. (2022). 3D cell cultures toward quantitative high-throughput drug screening. Trends Pharmacol. Sci. 43 (7), 569–581. doi:10.1016/j.tips.2022.03.014
Weng, M. T., Chiu, Y. T., Wei, P. Y., Chiang, C. W., Fang, H. L., and Wei, S. C. (2019). Microbiota and gastrointestinal cancer. J. Formos. Med. Assoc. = Taiwan yi zhi 118 (Suppl. 1), S32–S41. doi:10.1016/j.jfma.2019.01.002
Wilding, J. L., and Bodmer, W. F. (2014). Cancer cell lines for drug discovery and development. Cancer Res. 74 (9), 2377–2384. doi:10.1158/0008-5472.CAN-13-2971
Wong, T. L., Ng, K. Y., Tan, K. V., Chan, L. H., Zhou, L., Che, N., et al. (2020). CRAF methylation by PRMT6 regulates aerobic glycolysis-driven hepatocarcinogenesis via ERK-dependent PKM2 nuclear relocalization and activation. Hepatology 71 (4), 1279–1296. doi:10.1002/hep.30923
Workman, M. J., Mahe, M. M., Trisno, S., Poling, H. M., Watson, C. L., Sundaram, N., et al. (2017). Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23 (1), 49–59. doi:10.1038/nm.4233
Xia, J. Y., and Aadam, A. A. (2022). Advances in screening and detection of gastric cancer. J. Surg. Oncol. 125 (7), 1104–1109. doi:10.1002/jso.26844
Xin, H. Y., Sun, R. Q., Zou, J. X., Wang, P. C., Wang, J. Y., Ye, Y. H., et al. (2023). Association of BRAF variants with disease characteristics, prognosis, and targeted therapy response in intrahepatic cholangiocarcinoma. JAMA Netw. open 6 (3), e231476. doi:10.1001/jamanetworkopen.2023.1476
Xu, H., Jiao, D., Liu, A., and Wu, K. (2022). Tumor organoids: applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 15 (1), 58.
Xu, R., Zhou, X., Wang, S., and Trinkle, C. (2021). Tumor organoid models in precision medicine and investigating cancer-stromal interactions. Pharmacol. Ther. 218, 107668. doi:10.1016/j.pharmthera.2020.107668
Yamanaka, S. (2020). Pluripotent stem cell-based cell therapy-promise and challenges. Cell stem cell 27 (4), 523–531. doi:10.1016/j.stem.2020.09.014
Yamasaki, S., Kuwahara, A., Kishino, A., Kimura, T., Takahashi, M., and Mandai, M. (2022). Addition of Chk1 inhibitor and BMP4 cooperatively promotes retinal tissue formation in self-organizing human pluripotent stem cell differentiation culture. Regen. Ther. 19, 24–34. doi:10.1016/j.reth.2021.12.003
Yan, H. H. N., Siu, H. C., Law, S., Ho, S. L., Yue, S. S. K., Tsui, W. Y., et al. (2018). A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell stem cell 23 (6), 882–897. doi:10.1016/j.stem.2018.09.016
Yang, J., Huang, S., Cheng, S., Jin, Y., Zhang, N., and Wang, Y. (2021). Application of ovarian cancer organoids in precision medicine: key challenges and current opportunities. Front. cell Dev. Biol. 9, 701429. doi:10.3389/fcell.2021.701429
Yang, R., and Yu, Y. (2023). Patient-derived organoids in translational oncology and drug screening. Cancer Lett. 562, 216180. doi:10.1016/j.canlet.2023.216180
Yin, S., Xi, R., Wu, A., Wang, S., Li, Y., Wang, C., et al. (2020). Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Sci. Transl. Med. 12 (549), eaaz1723. doi:10.1126/scitranslmed.aaz1723
Yoshida, G. J. (2020). Applications of patient-derived tumor xenograft models and tumor organoids. J. Hematol. Oncol. 13 (1), 4. doi:10.1186/s13045-019-0829-z
Yu, H., Wang, M., Zhang, T., Cao, L., Li, Z., Du, Y., et al. (2022). Dual roles of β-arrestin 1 in mediating cell metabolism and proliferation in gastric cancer. Proc. Natl. Acad. Sci. U. S. A. 119 (40), e2123231119. doi:10.1073/pnas.2123231119
Yuki, K., Cheng, N., Nakano, M., and Kuo, C. J. (2020). Organoid models of tumor immunology. Trends Immunol. 41 (8), 652–664. doi:10.1016/j.it.2020.06.010
Zahmatkesh, E., Ghanian, M. H., Zarkesh, I., Farzaneh, Z., Halvaei, M., Heydari, Z., et al. (2021). Tissue-specific microparticles improve organoid microenvironment for efficient maturation of pluripotent stem-cell-derived hepatocytes. Cells 10 (6), 1274. doi:10.3390/cells10061274
Zhang, F., Wu, Z., Yu, B., Ning, Z., Lu, Z., Li, L., et al. (2023b). ATP13A2 activates the pentose phosphate pathway to promote colorectal cancer growth though TFEB-PGD axis. Clin. Transl. Med. 13 (5), e1272. doi:10.1002/ctm2.1272
Zhang, J., Hong, Z., Lu, W., Fang, T., Ren, Y., Yin, S., et al. (2023a). Assessment of drug susceptibility for patient-derived tumor models through lactate biosensing and machine learning. ACS sensors 8 (2), 803–810. doi:10.1021/acssensors.2c02381
Zhao, W., Jin, L., Chen, P., Li, D., Gao, W., and Dong, G. (2022). Colorectal cancer immunotherapy-Recent progress and future directions. Cancer Lett. 545, 215816. doi:10.1016/j.canlet.2022.215816
Zhou, C., Wu, Y., Wang, Z., Liu, Y., Yu, J., Wang, W., et al. (2023). Standardization of organoid culture in cancer research. Cancer Med. 12 (13), 14375–14386. doi:10.1002/cam4.5943
Zhu, X., and Li, S. (2023). Ferroptosis, necroptosis, and pyroptosis in gastrointestinal cancers: the chief culprits of tumor progression and drug resistance. Adv. Sci. 10 (26), e2300824. doi:10.1002/advs.202300824
Zou, Z., Lin, Z., Wu, C., Tan, J., Zhang, J., Peng, Y., et al. (2023). Micro-engineered organoid-on-a-chip based on mesenchymal stromal cells to predict immunotherapy responses of HCC patients. Adv. Sci. 10 (27), e2302640. doi:10.1002/advs.202302640
Glossary
Keywords: gastrointestinal cancers, organoid, 3D model, CRISPR/cas9, personalized medicine
Citation: Yan S, He Y, Zhu Y, Ye W, Chen Y, Zhu C, Zhan F and Ma Z (2024) Human patient derived organoids: an emerging precision medicine model for gastrointestinal cancer research. Front. Cell Dev. Biol. 12:1384450. doi: 10.3389/fcell.2024.1384450
Received: 09 February 2024; Accepted: 22 March 2024;
Published: 04 April 2024.
Edited by:
Océane C. B. Martin, Université de Bordeaux, FranceReviewed by:
Garima Kaushik, Champions Oncology, Inc., United StatesXiaolei Li, University of Pennsylvania, United States
Copyright © 2024 Yan, He, Zhu, Ye, Chen, Zhu, Zhan and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihong Ma, bWF6aGlob25nQGh6aG9zcGl0YWwuY29t
 Sicheng Yan
Sicheng Yan Yuxuan He1,2
Yuxuan He1,2 Zhihong Ma
Zhihong Ma