- 1College of Medicine, Southwest Jiaotong University, Chengdu, Sichuan, China
- 2Clinical Biobank Center and Laboratory Animal Center, The General Hospital of Western Theater Command, Chengdu, Sichuan, China
- 3Department of General Surgery, The General Hospital of Western Theater Command, Chengdu, China
- 4College of Life Science and Engineering, Southwest Jiaotong University, Chengdu, Sichuan, China
Long non-coding RNAs (LncRNA), exceeding 200 nucleotides in size, have emerged as important regulators of genes involved in multiple biological functions including cell growth, migration, invasion, drug resistance and apoptosis. They are increasingly being explored in human diseases. Notably, the recently identified LncRNA Cardiac hypertrophy-related factor (CHRF) has gained attention for its involvement in the molecular mechanisms of various diseases. CHRF was originally identified as a contributive LncRNA in cardiovascular diseases. Subsequent studies also revealed that it exerts an important role in promoting fibrosis and drug resistance. However, CHRF exhibits oncogenic functions in numerous cancers, including Non-small cell lung cancer (NSCLC), Colorectal cancer (CRC), Ovarian cancer (OC), Gastric cancer (GC), indicating its crucial roles in cancer progression. CHRF exhibits tremendous potential as both therapeutic target and diagnostic biomarker, particularly in cardiomyopathy, fibrosis, and cancer. To enhance our comprehensive understanding, this review synthesizes the pathophysiological mechanisms associated with CHRF and discusses its biological significance and clinical implication. Additionally, This review provides a comprehensive discussion on therapeutic strategies based on Non-coding RNA targets and discuss the potential of targeting CHRF, which is expected to offer readers a research approach for identifying the correct target strategies.
1 Introduction
Long non-coding RNAs (LncRNAs) are RNA molecules longer than 200 base pairs that are not translated into proteins (Dahariya et al., 2019; Quinn and Chang, 2016). Initially, as a by-products of RNA polymerase II transcription, LncRNAs have no biological function (Schier and Taatjes, 2020; Mattick et al., 2023). However, an increasing body of evidence have demonstrated that LncRNAs regulate various cellular processes (Ransohoff et al., 2018), including chromosomal and genomic modification, transcriptional activation and interference, and nuclear transport (Dykes and Emanueli, 2017; Ferrer and Dimitrova, 2024). This has spurred further research into the role of LncRNA in human biology (Yan and Bu, 2021; Chi et al., 2019). LncRNA can be classified by their length, function, location, and targeting mechanisms, though a unified classification system has yet to be established (Matsui and Corey, 2017). According to their position relative to protein-coding genes, LncRNAs are categorized as sense, antisense, bidirectional, intronic, intergenic, or enhancer-associated (Sanchez Calle et al., 2018). Mechanistically, LncRNA are typically divided into four categories: bait, scaffold, signal, and guide (Schmitz et al., 2016). Recent advancements have highlighted that certain LncRNAs play a crucial role in encoding small peptides and modulating general biological processes in a tissue-specific manner. These findings further emphasize the intricate nature and significant biological relevance of LncRNAs (Nelson et al., 2016; Choi et al., 2019; Chen et al., 2021).
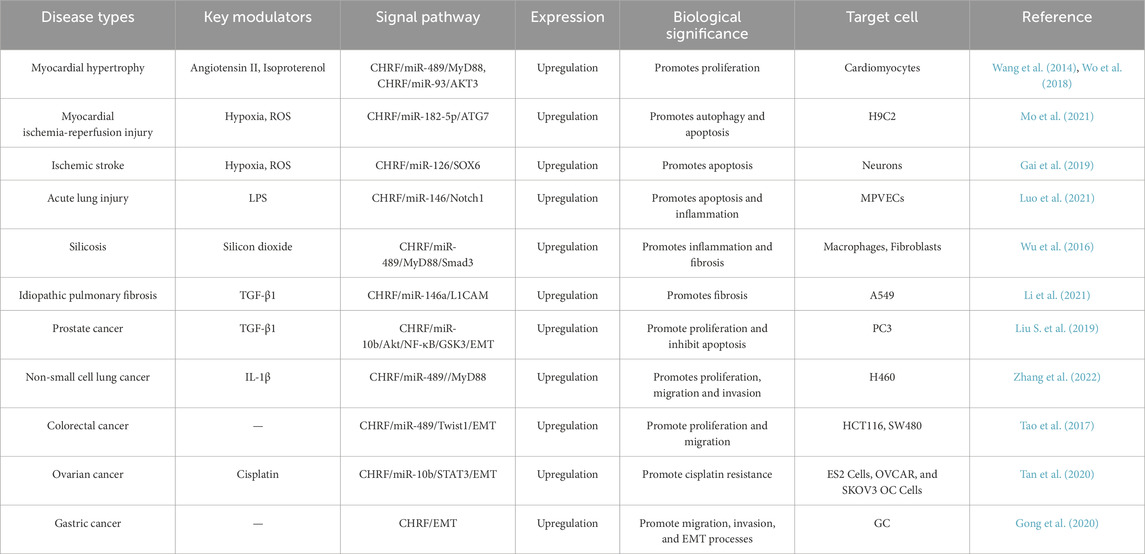
Cardiac hypertrophy-related factor (CHRF) is a recently identified LncRNA that functions as a central regulatory role in numerous cancers and diseases. It is a LncRNA with a length of 1,843 nucleotides that lacks protein-coding potential. It is genomically located on chromosome 18. While the full-length sequence of CHRF exhibits poor conservation across species, its binding site for miRNA demonstrates cross-species conservation (Wang et al., 2014). As an oncogene, CHRF is upregulated in a variety of pathological conditions, including Myocardial hypertrophy (Wang et al., 2014; Wo et al., 2018), Myocardial ischemia-reperfusion injury (Mo et al., 2021), Ischemic stroke (Gai et al., 2019), Acute lung injury (Luo et al., 2021), Silicosis (Wu et al., 2016), Idiopathic pulmonary fibrosis (Li et al., 2021), Prostate cancer (Liu S. et al., 2019), Non-small cell lung cancer (Zhang et al., 2022), Colorectal cancer (Tao et al., 2017), Ovarian cancer (Tan et al., 2020), Gastric cancer (Gong et al., 2020) and others. Given its critical role in these diseases, scientists are actively exploring the potential of CHRF as a biomarker for disease screening and working to develop technologies and drugs that target this LncRNA. However, despite CHRF’s significant impact on various diseases, the development of therapeutics targeting CHRF has lagged behind, largely due to the incomplete understanding of its underlying molecular network.
We provide an overview of the mechanisms by which CHRF contributes to disease progression, and discuss CHRF’s crucial role in cardiovascular diseases, fibrotic conditions, and cancer. Additionally, we summarize the recent advances based on non-coding RNA (ncRNA) and explore the potential and challenges of targeting LncRNA CHRF as a future therapeutic strategy. In conclusion, CHRF has garnered significant attention as a highly promising therapeutic target for clinically relevant diseases. Elucidating its precise mechanisms and functions in these conditions is likely to offer critical insights that could inform the development of more effective, targeted treatment strategies.
2 Performance of LncRNA CHRF in human diseases
CHRF is closely associated with the development and progression of cardiovascular diseases, cancer and fibrotic conditions. CHRF expression is typically closely associated with disease severity or progression. As a potential therapeutic and diagnostic target, CHRF regulates multiple key processes, which have cell proliferation, migration, invasion, epithelial-mesenchymal transition (EMT), and drug resistance. The mechanisms by which CHRF mainly involve its interactions with different microRNA (miRNA) and signaling pathways. Understanding the role of CHRF can aid in the development of novel therapies targeting CHRF. Association of CHRF with diseases is summarized in Table 1, and the specific mechanisms of CHRF are detailed in the following sections.
2.1 LncRNA CHRF in cardiovascular diseases
Myocardial hypertrophy and myocardial ischemia-reperfusion (I/R) injury are prevalent pathological conditions in cardiovascular diseases.
Myocardial hypertrophy is a compensatory response to various cardiac stresses, but if it persists, it can lead to heart failure (Heinzel et al., 1985). Emerging evidence has demonstrated that the long non-coding RNA CHRF is implicated in the regulation of cardiac hypertrophy (Wang et al., 2014; Wo et al., 2018; Zhou et al., 2019; Shen et al., 2017). Isoproterenol (Iso), which stimulates the sympathetic nervous system, induces changes in cardiomyocyte proteins, glycogen, collagen, and lipids, leading to myocardial fibrosis and hypertrophy, exacerbating the condition. Research indicates that CHRF is upregulated in Iso-induced myocardial hypertrophy models and promotes hypertrophy through the miR-93/AKT3 axis in stimulated cardiomyocytes (Wo et al., 2018). Additionally, CHRF is upregulated in hypertrophic mouse and human heart failure samples. Recent in vitro studies have demonstrated that miR-489 functions as an anti-hypertrophic microRNA (Wang et al., 2014). CHRF has been shown to directly bind to miR-489, thereby downregulating its expression. MyD88, a validated target of miR-489, is known to promote hypertrophic responses. The anti-hypertrophic effect of miR-489 is mediated through the inhibition of MyD88 expression. Thus, CHRF functions as an endogenous sponge to inhibit miR-489 activity and affect MyD88 levels, contributing to myocardial hypertrophy (Wang et al., 2014; Zhou et al., 2019).
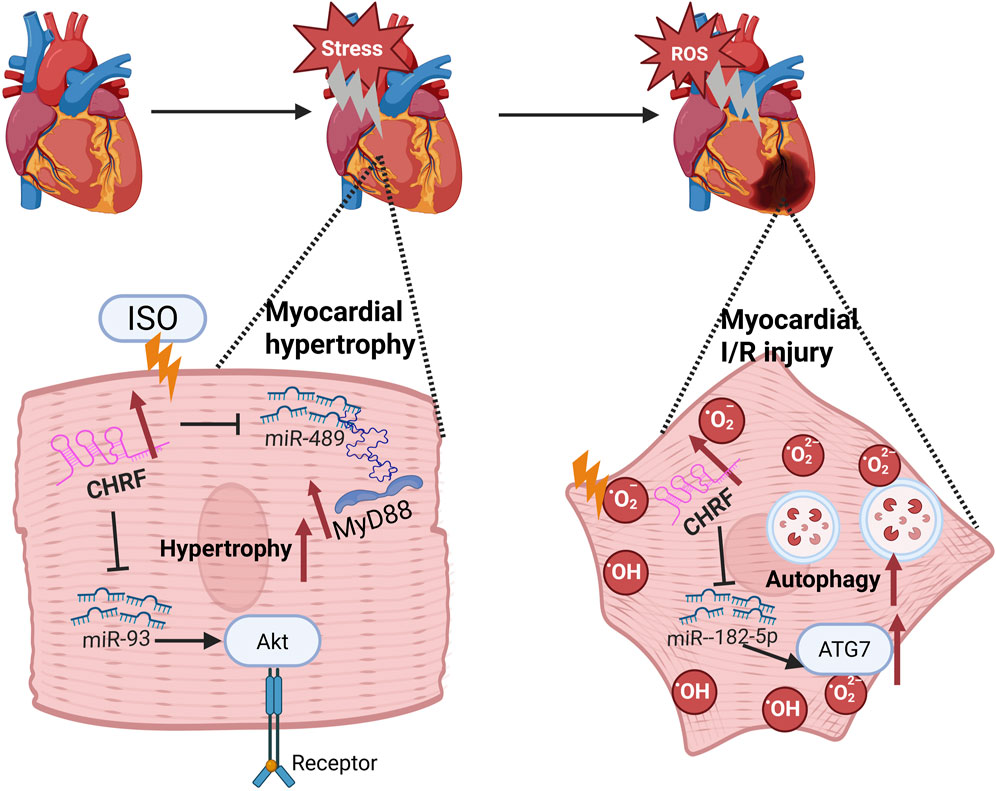
I/R injury, often caused by the restoration of blood flow after ischemia, is currently managed most effectively by early restoration of myocardial perfusion through thrombolysis (Giannakakis et al., 2017) or percutaneous coronary intervention (PCI) (Al-Lam et al., 2019), which are strategies proven to reduce infarct size and improve clinical outcomes (Li et al., 2022a). Paradoxically, the production of reactive oxygen species (ROS) can cause further damage (Hausenloy and Yellon, 2013; Davidson et al., 2019). Therefore, identifying new biomarkers for these conditions is crucial for diagnosis and treatment (Li Y. et al., 2018; Kumari et al., 2022). In the context of I/R injury, CHRF knockout significantly reduces myocardial damage, and its downregulation inhibits hypoxia/reoxygenation (H/R)-induced autophagy in cardiomyocytes. Autophagy is a vital process during myocardial ischemia-reperfusion (Popov et al., 2023; Ding et al., 2024). Bioinformatics analysis has identified miR-182-5p as a direct target of CHRF. Silencing miR-182-5p significantly enhances the effects of CHRF on cell viability, lactate dehydrogenase (LDH) levels, apoptosis, and autophagy, thereby revealing an inverse relationship between CHRF and miR-182-5p. Furthermore, autophagy-related gene 7 (ATG7), a downstream target of miR-182-5p, is positively regulated by CHRF. Overexpression of ATG7 reverses the reduction in apoptosis caused by CHRF silencing and exacerbates cardiomyocyte autophagy. This indicates that CHRF promotes myocardial ischemia-reperfusion injury by positively regulating ATG7 through the negative regulation of miR-182-5p, highlighting the therapeutic potential of targeting CHRF in treating myocardial hypertrophy and I/R injury (Solevag et al., 2020). These findings underscore the therapeutic potential of targeting CHRF in the treatment of both myocardial hypertrophy and I/R injury (Figure 1).
2.2 LncRNA CHRF in cancers
2.2.1 Prostate cancer (PC)
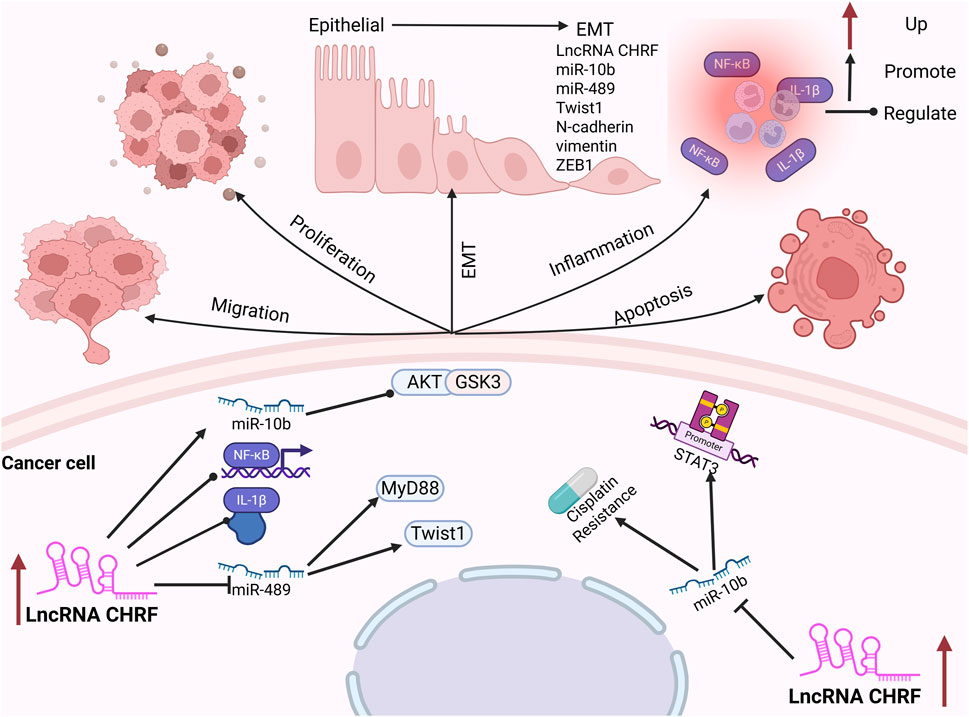
Prostate cancer (PC) is the sixth most prevalent malignant tumor in men globally, with its incidence rising progressively with age (Siegel et al., 2020; Robinson et al., 2015). Elevated expression levels of CHRF have been observed in prostate cancer tissues. Notably, silencing of CHRF effectively inhibits PC cell proliferation while promoting cell apoptosis. Epithelial-mesenchymal transition (EMT) is a critical process in the development and progression of tumors, playing a significant role in tumorigenesis (Lamouille et al., 2014; Pastushenko and Blanpain, 2019). The upregulation of CHRF leads to a marked decrease in E-cadherin expression, while the levels of N-cadherin, vimentin, and ZEB1 (an EMT activator) are notably increased, thereby promoting the EMT process in prostate cancer. Furthermore, CHRF positively regulates the high expression of miR-10b in PC tissues, and inhibiting miR-10b can repress the proliferation and EMT induced by CHRF, indicating that CHRF acts in conjunction with miR-10b as an oncogene in the progression of prostate cancer. Additionally, the miR-10b/AKT/GSK3β and NF-κB pathways may be activated by CHRF, further promoting the malignant progression of PC (Liu S. et al., 2019).
2.2.2 Non-small cell lung cancer (NSCLC)
Non-Small Cell Lung Cancer (NSCLC) originates from the bronchial mucosa, bronchial glands, and alveolar epithelium (Bade and Dela Cruz, 2020; Alexander et al., 2020). A recent study has underscored the crucial role of CHRF in the progression of NSCLC. Compared with adjacent normal tissues, both CHRF and interleukin-1β (IL-1β) are significantly upregulated in NSCLC tissues. IL-1β is a well-known pro-inflammatory and tumor-promoting cytokine (Weber et al., 2010), which can enhance the expression of CHRF and promote the proliferation and metastasis of NSCLC cells. Silencing CHRF can reduce the inflammatory response induced by IL-1β in NSCLC tissues. CHRF functions as a molecular sponge for microRNA-489 (miR-489), exhibiting a negative correlation with miR-489 expression. Inhabiting of miR-489 promotes the proliferation and migration of NSCLC cells, are facilitated by CHRF. Additionally, myeloid differentiation factor 88 (MyD88) is a downstream target of miR-489. MyD88 is negatively regulated by miR-489 and positively regulated by CHRF. This regulatory axis (CHRF/miR-489/MyD88) has been shown to play a significant role in the progression of NSCLC (Zhang et al., 2022).
2.2.3 Colorectal cancer (CRC)
Colorectal cancer (CRC) ranks as the fourth leading cause of cancer-related mortality globally and is among the three most prevalent malignant tumors worldwide (Haggar and Boushey, 2009; Si and mon, 2016). Despite significant advancements in CRC treatment, early diagnosis remains challenging (Van Cutsem et al., 2010; Fan et al., 2021; Dekker et al., 2019). The expression of CHRF is significantly upregulated in CRC tissues, and its knockdown in CRC cells leads to reduced migration and invasion capabilities. miR-489, a target of CHRF, shows a notable decrease in expression in CRC cells, indicating a negative regulatory relationship between CHRF and miR-489 in CRC. The overexpression of miR-489 inhibits the epithelial-mesenchymal transition (EMT) process, consequently suppressing the migration and invasion of CRC cells. Emerging evidence indicates that Twist1 is a downstream target of miR-489. CHRF positively regulates Twist1 expression while negatively modulating miR-489 levels. Overexpression of Twist1 effectively counteracts the inhibitory effects of miR-489 on colorectal cancer (CRC) progression. Specifically, Twist1 promotes epithelial-mesenchymal transition (EMT), thereby facilitating CRC cell migration and invasion (Tao et al., 2017).
2.2.4 Ovarian cancer (OC)
Ovarian Cancer (OC) ranks third in incidence among gynecological malignancies but has the highest mortality rate (Kujawa et al., 2015; Penny, 2020). Early diagnosis remains challenging, with the majority of patients being diagnosed at advanced stages (Penny, 2020; Orr and Edwards, 2018). Cisplatin is one of the primary chemotherapeutic agents used for the treatment of OC (Samuel et al., 2016). Studies have shown that the expression of CHRF is significantly elevated in patients with cisplatin-resistant OC, particularly in the liver, which is the first organ for OC metastasis, indicating that CHRF may play a role in cisplatin resistance in OC. It is further indicated that CHRF can directly bind to miR-10b, with a negative regulatory relationship. Silencing CHRF significantly enhances the inhibitory effect of cisplatin on OC cell proliferation, suggesting that CHRF contributes to cisplatin resistance. Moreover, this resistance is reversed when miR-10b is knocked out, reinforcing the regulatory role of the CHRF-miR-10b axis in OC. Additionally, the downregulation of CHRF leads to a significant decrease in STAT3 phosphorylation in OC cells, and this effect is similarly weakened by the upregulation of miR-10b. The CHRF-miR-10b axis exerts a significant influence on cisplatin resistance in ovarian cancer (OC) through the regulation of the epithelial-mesenchymal transition (EMT) process and the STAT3 signaling pathway (Tan et al., 2020).
2.2.5 Gastric cancer (GC)
Gastric Cancer (GC) originates from the gastric mucosal epithelium and is increasingly affecting younger individuals due to changes in dietary habits, increased work-related stress, and Helicobacter pylori infection (Thrift et al., 2023). A recent study has demonstrated that the expression of CHRF is significantly upregulated in gastric cancer (GC), particularly in cases with lymph node metastasis, where its levels are notably higher compared to non-metastatic cases. This finding indicates that CHRF plays a crucial role in promoting the progression of GC. Knocking out CHRF significantly inhibits the invasion and migration of GC cells, thereby highlighting its potential role in tumor aggressiveness. The role of CHRF in epithelial-mesenchymal transition (EMT) is of particular interest. Overexpression of CHRF suppresses the expression of the epithelial cell marker E-cadherin while promoting the expression of mesenchymal markers such as vimentin and N-cadherin. This dual effect facilitates the EMT process and contributes to the progression of GC (Gong et al., 2020) (Figure 2).
2.3 LncRNA CHRF in fibrotic diseases
2.3.1 Silicosis
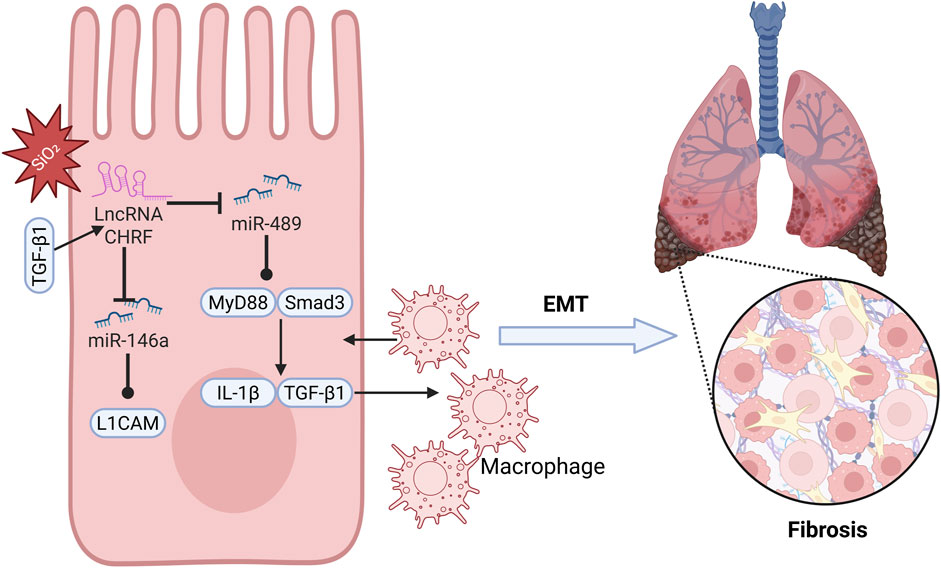
Silicosis is an incurable occupational disease caused by long-term inhalation of dust containing high concentrations of free crystalline silicon dioxide, leading to diffuse nodular pulmonary fibrosis (Chen Y. et al., 2018; Greenberg et al., 2007; Li et al., 2022b). Although silicosis is relatively common, its underlying mechanisms have not been fully explored (Tan and Chen, 2021), necessitating further research into its pathophysiological processes (Adamcakova and Mokra, 2021; Pollard, 2016). Studies indicate that miR-489 plays a role in regulating inflammation and pulmonary fibrosis associated with silicosis. Specifically, miR-489 expression is decreased in silica-induced pulmonary fibrosis, and increasing miR-489 expression significantly reduces the level of α-SMA and Vimentin proteins, suggesting that miR-489 inhibits the development of silicosis. Bioinformatics analysis identifies MyD88 and Smad3 as downstream targets of miR-489 and observes their direct interaction. Additionally, the key fibrotic factor Transforming Growth Factor-beta 1 (TGF-β1), which is produced through the activation of inflammatory macrophages and damage, is a downstream effector gene of MyD88. miR-489 may indirectly inhibit IL-1β and TGF-β1 by regulating MyD88, alleviating the inflammatory process of silicosis. Notably, CHRF exerts negative regulation on the expression of miR-489. Silencing CHRF significantly enhances the inhibitory effect of miR-489 on inflammation and fibrosis induced by silica exposure in silicosis (Figure 3). In summary, CHRF may exacerbate inflammation and fibrosis associated with silicosis through the miR-489/MyD88/Smad3 pathway (Wu et al., 2016).
2.3.2 Idiopathic pulmonary fibrosis (IPF)
Idiopathic pulmonary fibrosis (IPF) is a chronic, progressive fibrosing interstitial lung disease that predominantly affects individuals in middle and older age groups (Stowasser and Hallmann, 2015; Cho and Stout-Delgado, 2020). With the increasing annual incidence of IPF and the limited survival period for patients, the situation is particularly urgent (Glass et al., 2022; Lynch et al., 2018), highlighting the pressing need to identify potential therapeutic targets for IPF (King et al., 2011). Epithelial-mesenchymal transition (EMT) is a critical process in the pathogenesis of idiopathic pulmonary fibrosis (IPF), and CHRF is a significant contributor to this process. Studies have shown that in the process of alveolar epithelial cell transformation induced by TGF-β1, CHRF is significantly upregulated in IPF cells, indicating its close relationship with the progression of IPF. Furthermore, miR-146a has been identified as a direct target of CHRF. In A549 cells treated with TGF-β1, silencing miR-146a suppresses the mRNA level of E-cadherin while enhancing the expression of Vimentin, Slug, and N-cadherin, thereby promoting EMT in lung epithelial cells and accelerating the progression of IPF. Bioinformatics analysis reveals that L1 cell adhesion molecule (L1CAM) is a downstream target of miR-146a, positively regulated by CHRF. High levels of L1CAM can reverse the inhibition of EMT observed in IPF cells when CHRF expression is reduced. In summary, as a competing endogenous RNA (ceRNA), CHRF drives the EMT process and promotes the progression of IPF by negatively regulating miR-146a and positively regulating L1CAM (Figure 3). This suggests that CHRF may become an important target for the diagnosis and treatment of IPF in the future (Li et al., 2021).
2.4 LncRNA CHRF in other diseases
2.4.1 Ischemic stroke
Ischemic stroke, also known as ischemic apoplexy, is a condition caused by the narrowing or occlusion of cerebral arteries, leading to brain tissue necrosis and insufficient blood supply, often involving the carotid and vertebral arteries (Xu et al., 2016; Schmidt-Kastner, 2015). It progresses rapidly and is among the leading causes of disability and mortality worldwide (Green and Shuaib, 2006; Zhao et al., 2022). Research has found that after middle cerebral artery occlusion (MACO), CHRF is significantly upregulated in ischemic brain tissue. Knocking down CHRF can alleviate ischemia-reperfusion injury and improve neurological function. Bioinformatics analysis has revealed a significant correlation between CHRF and miR-126, with the upregulation of miR-126 counteracting the pro-apoptotic effects of CHRF overexpression on neurons, indicating that CHRF negatively regulates miR-126 to promote neuronal death. SOX6, acting as a downstream target of miR-126, competitively binds with CHRF, and silencing CHRF leads to reduced SOX6 levels, which in turn decreases ischemia-reperfusion injury, infarct size, and neuronal death, improving neurological function and behavioral outcomes. Therefore, the CHRF/miR-126/SOX6 axis may represent a promising therapeutic target for mitigating the progression of ischemic stroke (Gai et al., 2019).
2.4.2 Acute lung injury (ALI)
Acute Lung Injury (ALI) is a severe clinical condition characterized by diffuse alveolar damage and caused by various injurious factors. These factors lead to damage of alveolar epithelial cells and capillary endothelial cells, ultimately resulting in pulmonary interstitial and alveolar edema (Parsons et al., 2005; Mokra, 2020). Sepsis is closely associated with the occurrence of ALI, triggering inflammatory and apoptotic pathways that cause alveolar epithelial injury and leakage of edema fluid (Chen Y. et al., 2018; Kumar, 2020). The long non-coding RNA (LncRNA) CHRF plays a significant role in the pathogenesis of ALI, with increased expression in septic mice, and the knockdown of CHRF can suppress inflammation and apoptosis in microvascular pulmonary endothelial cells. In septic models, silencing CHRF reduces the risk of ALI. CHRF promotes pneumonia by modulating pro-inflammatory and anti-inflammatory cytokines. Lipopolysaccharide (LPS) increases CHRF levels in ALI, suggesting that CHRF may promote ALI by inducing LPS production (Chen et al., 2010; Ehrentraut et al., 2019). Bioinformatics analysis reveals a negative correlation between CHRF and miR-146a, and the upregulation of miR-146a can counteract the adverse effects of CHRF. CHRF competitively binds with miR-146a to regulate Notch1, playing an active role in Notch1 expression. Therefore, CHRF exacerbates inflammation and apoptosis in microvascular pulmonary endothelial cells, promotes LPS production, and advances the progression of ALI by modulating the miR-146/Notch1 axis (Luo et al., 2021).
3 Multifunctional regulatory roles of LncRNA CHRF in diseases and therapeutics
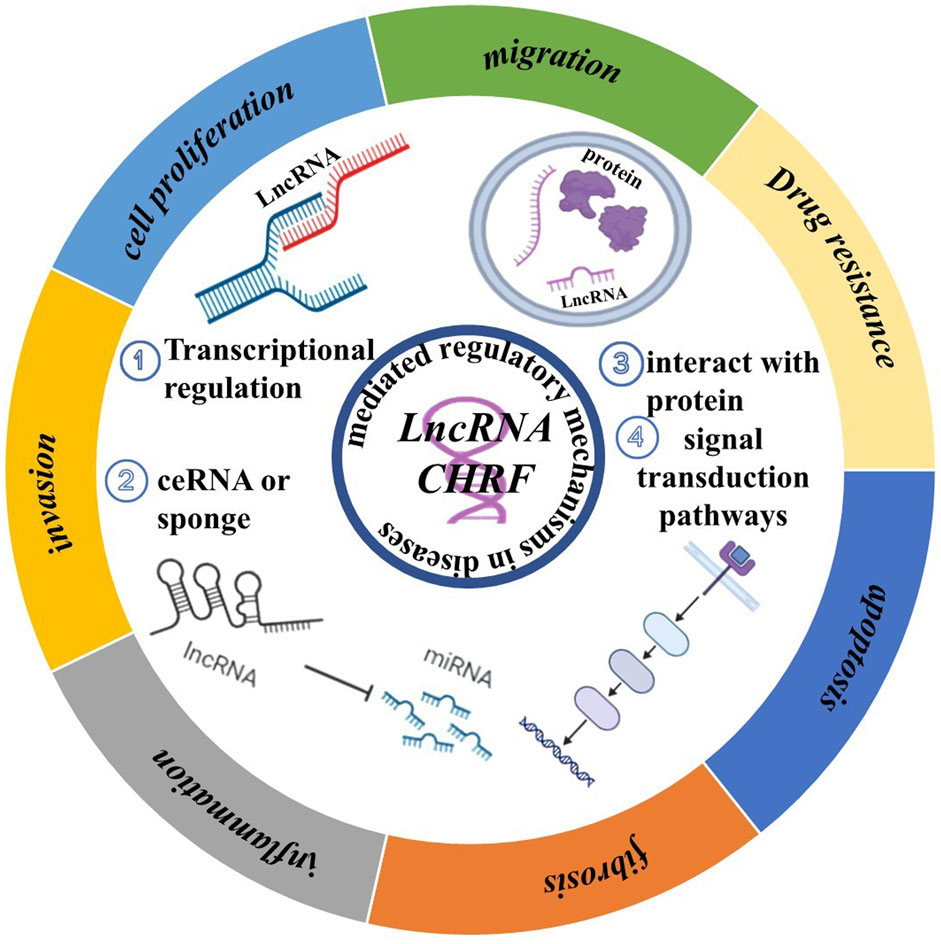
CHRF is increasingly recognized for its multifaceted roles in the progression and development of diseases. As shown in Figure 4, we review the common mechanisms of CHRF across multiple diseases, aiming to highlight its significance as a drug target.
In various diseases, CHRF acts as a Competing Endogenous RNA (ceRNA) or Transcripts by binding to specific miRNAs, thereby inhibiting their function and regulating downstream gene expression. For instance, in cardiac hypertrophy, CHRF interacts with miR-93 and miR-489 to regulate AKT3 and MyD88, promoting myocardial hypertrophy. In non-small cell lung cancer and silicosis, CHRF exacerbates inflammation and fibrosis via the miR-489/MyD88/Smad3 axis. Additionally, CHRF is a crucial factor in tumors and fibrosis-related diseases by interacting with proteins, which promotes cell migration and invasion. In prostate cancer, colorectal cancer, and gastric cancer, CHRF influences Twist1 or other EMT-related factors through miR-489 or miR-10b, facilitating the EMT of cancer cells. In pulmonary fibrosis, CHRF promotes EMT and fibrosis by regulating miR-146a and L1CAM. Interestingly, CHRF contributes to cell apoptosis and tumor drug resistance. For example, in ovarian cancer, CHRF enhances cisplatin resistance through the regulation of the miR-10b and STAT3 pathways. In myocardial ischemia-reperfusion injury, CHRF modulates autophagy and apoptosis via the miR-182-5p/ATG7 pathway. Besides, CHRF exhibits significant regulatory roles in inflammatory and fibrotic diseases, particularly in acute lung injury, silicosis, and idiopathic pulmonary fibrosis. In these conditions, CHRF promotes inflammatory responses and fibrotic processes by regulating factors such as miR-489, MyD88, and Smad3.
CHRF also exerts its effects in various diseases through similar miRNA regulatory axes, including the CHRF/miR-489/MyD88 Axis, which is involved in cardiac hypertrophy, non-small cell lung cancer, and silicosis and is often associated with inflammatory responses, cell proliferation, and fibrosis. In the CHRF/miR-93/AKT3 Axis, CHRF promotes myocardial hypertrophy in cardiac hypertrophy through this regulatory pathway (Figure 4).
4 Targeting non-coding RNA for drug development
Non-coding RNAs, including long LncRNAs and MicroRNAs, have emerged as critical regulators in the pathogenesis and progression of various diseases. We have summarized the current popular methods for targeting Non-coding RNA (ncRNA), and how to select them, which include the following three approaches. Additionally, Based on LncRNA CHRF, we discuss the potential strategies targeted LncRNA CHRF.
4.1 Strategies for targeting non-coding RNA
4.1.1 Small interfering RNA (siRNA)
Small interfering RNA (siRNA) is a double-stranded RNA molecule typically comprising 20 to 25 nucleotides. It specifically targets ncRNA and subsequently recruits the RNA-induced silencing complex (RISC) to mediate the degradation of the target ncRNA (Figure 5a). Studies have shown that siRNA has been used to degrade related ncRNA in various disease models (Khorkova and Wahlestedt, 2017), demonstrating significant therapeutic potential. In esophageal squamous cell carcinoma, the use of siRNA targeting LncRNA CASC9 significantly inhibited cancer cell invasion and migration (Liang et al., 2018). With the development of drug delivery systems, the first siRNA drug, Onpattro (patisiran), for the treatment of patients with hereditary transthyretin-mediated amyloidosis and another siRNA drug, Givlaari (givosiran), approved for the treatment of adults with acute hepatic porphyria, have been launched.
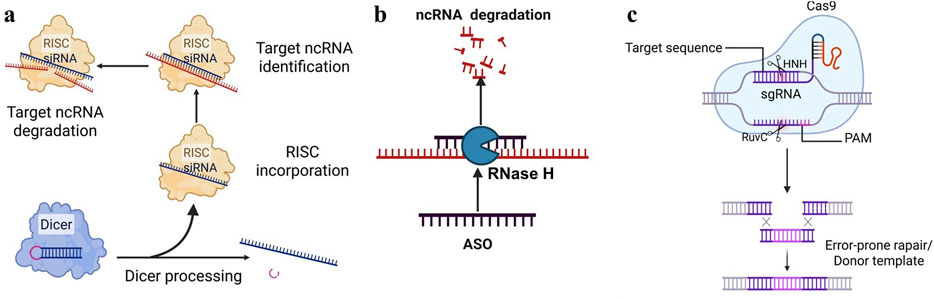
Figure 5. Prevalent targeted strategies of Non-coding RNA. (a) siRNA: siRNA specifically binds to and degrades the corresponding ncRNA, thereby preventing the continued translation of mRNA. (b) ASO: Forming a complementary strand with a specific ncRNA sequence, an heteroduplex is created, which is then recognized and degraded by endogenous cellular RNase H, thereby inducing gene silencing. (c) CRISPR/Cas9: Utilizing site-specific Cas nucleases to induce double-strand breaks (DSBs) at specific genomic loci, which are subsequently repaired via the cell’s intrinsic non-homologous end joining (NHEJ) or homologous recombination repair (HDR) pathways, ultimately enables precise genomic modifications, including gene knockout and base editing.
4.1.2 Antisense oligonucleotides (ASOs)
Antisense oligonucleotides (ASOs) are single-stranded DNA or RNA molecules that are 15–22 nucleotides in length. ASOs can bind RNA target, forming RNA/DNA heteroduplexes that are recognized and degraded by endogenous cellular RNase H, thereby inducing gene silencing (Crooke, 2017; Viney et al., 2016). It has been reported that ASOs have shown good application prospects in various diseases, especially in neurodegenerative diseases. Studies have shown that ASOs targeting MALAT1 have shown good effects in the treatment of breast cancer and multiple myeloma (Rinaldi and Wood, 2017; Arun et al., 2016). Additionally, ASO technology is characterized by its high targeting specificity, allowing for the selective inhibition of CHRF expression and preventing its pathological roles in diseases. It is also noteworthy that ASOs can inhibit transcription independently of RNase H cleavage, for example, by spatially blocking ribosome binding (Crooke, 2017). Moreover, ASOs can be chemically modified to enhance their stability and cell penetration, thereby facilitating their effective delivery into diseased tissues to inhibit ncRNA (Figure 5b). Recently, ASO-based therapies have achieved significant breakthroughs in clinical applications. To date, the U.S. Food and Drug Administration (FDA) has approved three ASO-based drugs (Rinaldi and Wood, 2017).
4.1.3 CRISPR/Cas9
CRISPR/Cas9 is a widely used gene-editing tool that can target and cut specific LncRNAs (Tsai and Joung, 2014; Kopp et al., 2019; Liu et al., 2016). Single-guide RNA (sgRNA) and Cas9 enzyme are key factors in the CRISPR/Cas9 system. The CRISPR/Cas system employs site-specific Cas nucleases to introduce double-strand breaks (DSBs) at specific genomic target sites. Subsequently, the cell’s intrinsic repair mechanisms, namely, non-homologous end joining (NHEJ) or homologous recombination repair (HDR), are harnessed to repair these DSBs. This process ultimately enables precise modifications to the target gene, including gene knockout and base editing (Kopp et al., 2019; Esposito et al., 2019). Studies have shown that CRISPR/Cas9 has been used to explore the function of LncRNA (Figure 5c). For example, in prostate cancer, researchers have used CRISPR/Cas9 to knock out TTTY15, which can significantly inhibit the growth of cancer cells (Xiao et al., 2019; Chen Q. et al., 2018). This indicates that by designing a single-guide RNA (sgRNA) (Slaymaker et al., 2016; Chen et al., 2017; Koonin et al., 2023) targeting ncRNA, the Cas9 protein can recognize and cut the gene, thereby permanently blocking its expression.
However, it is important to note that the choice of targeting CHRF in preclinical models should be based on its subcellular localization within cells. ASOs are primarily employed for targeting LncRNAs located in the nucleus, whereas small interfering RNA (siRNA) is mainly used for LncRNAs in the cytoplasm. In contrast, the CRISPR/Cas9 system can be applied to LncRNAs regardless of their subcellular localization, whether in the cytoplasm or the nucleus. Importantly, the precise subcellular localization of LncRNA CHRF remains unclear, highlighting the need for further investigation. Recent studies have demonstrated the successful application of the Smart Silence method, an improved approach that combines ASOs and siRNA reagents. This method includes three siRNAs and three ASOs, and has been effectively utilized in numerous in vivo and in vitro experiments involving LncRNA knockdown (Li H. et al., 2018; Li et al., 2019; Tao et al., 2018; Liu J. et al., 2019). Therefore, for LncRNA CHRF with unclear subcellular localization, a mixed method of ASOs and siRNA should be chosen for experimentation.
4.2 Small molecule inhibitors
Non-coding RNAs, like proteins, possess multi-dimensional structures. Small molecule inhibitors can bind to the secondary or tertiary structures of ncRNAs, thereby blocking their interactions with proteins or other molecules (Pedram Fatemi et al., 2015). Numerous studies have demonstrated that various types of RNAs possess functional sites, including Drosha and Dicer processing sites in microRNA (miRNA) precursors, internal ribosome entry sites (IRES) in viral RNA and certain human mRNAs, riboswitches in bacteria, splicing enhancers and silencers in pre-mRNA, and regulatory structures within the 5′ and 3′ untranslated regions (UTRs). Research has further shown that small molecule inhibitors, such as ellipticine, can effectively inhibit the interaction between ncRNAs and proteins (Engreitz et al., 2014). Designing small molecule inhibitors for ncRNA involves a series of intricate steps. Initially, computational methods are employed to predict the secondary and tertiary structures of the RNA, which are then experimentally validated using techniques such as chemical modifiers and high-throughput sequencing. Following this, functional RNA structures are identified, either through computational or experimental approaches.
Subsequently, affinity mass spectrometry, fluorescence-based assays, and microarray screening are utilized to screen and identify small molecules capable of binding to specific RNA structures. The lead molecules obtained are then optimized through medicinal chemistry methods to enhance their pharmacological properties, including affinity, selectivity, cell permeability, and pharmacokinetics. Furthermore, the direct binding of small molecules to RNA and their biological activity must be verified, potentially involving methods such as covalent bond formation, RNA degradation, and resistance analysis. Finally, the safety and efficacy of the small molecules are assessed through preclinical and clinical studies. Throughout this process, structure-guided methods and modular assembly approaches may also be necessary to optimize the small molecules, ensuring they possess the desired pharmaceutical properties. This meticulous process ensures the development of effective small molecule inhibitors tailored for LncRNA (Childs-Disney et al., 2022) (Figure 6). Inforna is a strategy for identifying lead compounds that involves comparing the structural features present in cellular RNAs with those in experimentally determined RNA-small molecule interaction databases. The overlap identified through this comparison provides potential lead targets and lead small molecules for further investigation (Disney, 2019). Additionally, structure-based small molecule design relying on RNA or RNA-ligand complex structural models can guide the modification of interactions between RNA and small molecules (Batool et al., 2019). This offers the possibility for small molecule drugs to target ncRNAs.
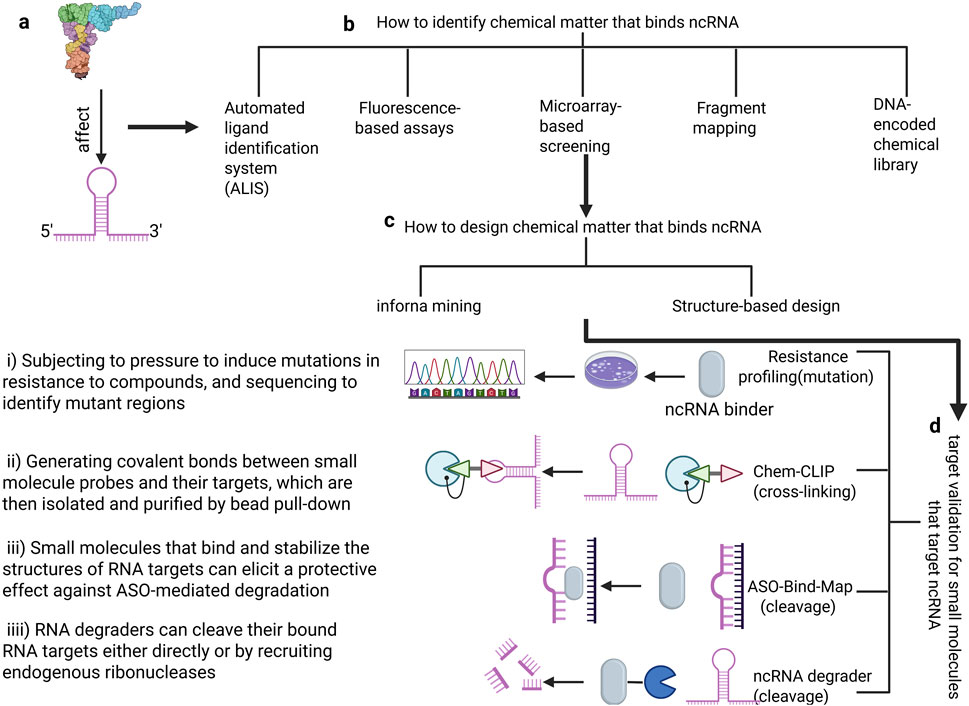
Figure 6. The process of designing small molecule inhibitors of Non-coding RNA. (a) Defining RNA structures for small-molecule targeting (b). How to identify chemical matter that binds ncRNA: Including five methods (ALIS, Fluorescence-based assays, Microarray-based screening, Fragment mapping, DNA-encoded chemical library) (c). How to design chemical matter that binds ncRNA: There are two prevalent strategies- infoma mining and structure-based design (d). Target validation for small molecules that target ncRNA: mutation, crossing-linking, ASO-bind-Map, ncRNA degrader.
4.3 Natural compounds
Natural compounds, such as resveratrol and curcumin, have shown potential in regulating ncRNA (Mishra et al., 2019; Xia et al., 2018; Zhang et al., 2013). For example, resveratrol alleviated Parkinson’s disease pathology by modulating MALAT1 (Xiao et al., 2019). Theoretically, these natural compounds could also regulate ncRNA’s expression or activity, reducing its promotion of cardiac hypertrophy, thus offering a natural therapeutic option. By screening and validating natural compounds that can regulate ncRNA expression or function, safe and effective drug candidates for treating diseases could be discovered.
4.4 The potential strategies targeted LncRNA CHRF
Increasing research has shown that LncRNA CHRF exhibits great potential in cancer, cardiac diseases, and fibrosis. While therapeutic strategies targeting it remain underdeveloped. Based on our ncRNA-targeted development strategy, we propose potential therapeutic approaches targeting LncRNA CHRF. As summarized in Figure 4, current research indicates that LncRNA CHRF influences cellular biological function in diseases, primarily through transcriptional regulation, molecular sponging, protein interactions, and signaling pathway modulation. Among these mechanisms, CHRF predominantly functions as a competitive endogenous RNA (ceRNA). In both cardiac hypertrophy and myocardial ischemia-reperfusion injury, in vivo and in vitro knockout experiments consistently demonstrate the detrimental role of CHRF in cardiac pathologies. Similarly, cancer studies reveal CHRF’s oncogenic function in patient samples and animal models, with its silencing in cellular experiments confirming tumor-promoting activities. Furthermore, CHRF silencing experiments in fibrotic and other diseases substantiate its pathogenic contributions. These collective findings provide a compelling rationale for developing CHRF-targeted therapeutics using strategies outlined in Section 4.1 (Strategies for Targeting Non-coding RNAs). Critically, CHRF’s functional impact depends on its interactions with proteins and other biomolecules. This presents opportunities to develop small-molecule inhibitors via high-throughput screening or computer-aided drug design to specifically disrupt CHRF’s interaction networks. Besides, Exploit CHRF’s structural features within disease-specific microenvironments to screen naturally derived compounds targeting this LncRNA. For instance, in cancer, CHRF accelerates tumor progression by regulating epithelial-mesenchymal transition (EMT)-related protein expression. This mechanistic insight establishes a solid foundation for designing CHRF-directed small-molecule inhibitors.
5 Conclusion and discussion
Research has found that CHRF (Cardiac Hypertrophy-Related Factor) is significantly upregulated in various diseases, including cardiac disorders, cancers, and fibrotic diseases. Mechanistically, CHRF acts as a competing endogenous RNA (ceRNA) in multiple pathologies, binding to specific miRNAs and inhibiting their activity. This competitive binding represents a key mechanism through which CHRF influences tumorigenesis and other pathological processes. Ultimately, CHRF impacts critical biological processes such as cell proliferation, migration, apoptosis, autophagy, drug resistance, and inflammatory responses, which are closely linked to patient prognosis.
Notably, the CHRF/miR-489/Myd88 axis is dysregulated in multiple diseases. Furthermore, epithelial-mesenchymal transition (EMT), a crucial biological process in tumor progression and fibrotic diseases, is also regulated by CHRF. However, the key modulators and biological significance of CHRF vary across different diseases (as summarized in Table 1). These findings underscore the potential of CHRF as a pivotal molecule for future research and highlight its promise as a valuable therapeutic target.
However, therapeutic strategies specifically targeting LncRNA CHRF remain largely unexplored. While the functional diversity of LncRNAs offers multiple avenues for therapeutic development, targeting strategies must be tailored to the specific mechanism of action of the individual LncRNA. Current approaches for LncRNA targeting include siRNA, ASO, CRISPR-Cas9, and the screening of small molecule inhibitors or natural compounds based on unique LncRNA structures. Studies demonstrating that CHRF knockout or silencing reverses disease progression provide a solid theoretical foundation for designing CHRF-targeted drugs. Despite this promise, the clinical translation of RNA-based therapies faces significant challenges, including insufficient specificity, inefficient delivery, and tolerability issues. Specificity problems stem primarily from off-target effects, which can arise from unintended activity in non-target cells, variations in the target sequence, or non-specific effects caused by dosing exceeding endogenous levels. Delivery challenges include the inherent instability of unmodified RNA molecules, the need to overcome the endosomal escape barrier for effective intracellular release, and the lack of carriers capable of precise targeting to specific organs or cell types. Furthermore, tolerability concerns and the combined impact of these challenges often lead to the discontinuation of RNA therapies in clinical trials due to insufficient efficacy.
Therefore, based on the current progress in LncRNA CHRF research, we propose rational hypotheses for its targeting strategies. However, despite our comprehensive review of CHRF’s mechanisms across different diseases, its specific pathophysiological roles—including its subcellular localization and upstream regulatory signals—require further elucidation.
Author contributions
JM: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review and editing. CL: Investigation, Supervision, Validation, Writing – original draft. WZ: Conceptualization, Investigation, Methodology, Validation, Writing – original draft. YS: Investigation, Methodology, Validation, Writing – original draft. JP: Investigation, Methodology, Supervision, Validation, Writing – original draft. YC: Investigation, Methodology, Validation, Writing – original draft. XG: Conceptualization, Investigation, Methodology, Supervision, Validation, Writing – original draft. YF: Conceptualization, Investigation, Methodology, Supervision, Validation, Writing – review and editing. HS: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work is funded by Joint Research Project of General Hospital of Western Theater Command (2019LH04)and General Program of General Hospital of Western Theater Command (2021-XZYG-B09), awarded to HS
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2025.1573723/full#supplementary-material
References
Adamcakova, J., and Mokra, D. (2021). New insights into pathomechanisms and treatment possibilities for lung silicosis. Int. J. Mol. Sci. 22 (8), 4162. doi:10.3390/ijms22084162
Alexander, M., Kim, S. Y., and Cheng, H. (2020). Update 2020: management of non-small cell lung cancer. Lung 198 (6), 897–907. doi:10.1007/s00408-020-00407-5
Al-Lamee, R. K., Nowbar, A. N., and Francis, D. P. (2019). Percutaneous coronary intervention for stable coronary artery disease. Heart 105 (1), 11–19. doi:10.1136/heartjnl-2017-312755
Arun, G., Diermeier, S., Akerman, M., Chang, K. C., Wilkinson, J. E., Hearn, S., et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes and Dev. 30 (1), 34–51. doi:10.1101/gad.270959.115
Bade, B. C., and Dela Cruz, C. S. (2020). Lung cancer 2020: epidemiology, etiology, and prevention. Clin. Chest Med. 41 (1), 1–24. doi:10.1016/j.ccm.2019.10.001
Batool, M., Ahmad, B., and Choi, S. (2019). A structure-based drug discovery paradigm. Int. J. Mol. Sci. 20 (11), 2783. doi:10.3390/ijms20112783
Chen, H., Bai, C., and Wang, X. (2010). The value of the lipopolysaccharide-induced acute lung injury model in respiratory medicine. Expert Rev. Respir. Med. 4 (6), 773–783. doi:10.1586/ers.10.71
Chen, J. S., Dagdas, Y. S., Kleinstiver, B. P., Welch, M. M., Sousa, A. A., Harrington, L. B., et al. (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550 (7676), 407–410. doi:10.1038/nature24268
Chen, Q., Cai, J., Wang, Q., Wang, Y., Liu, M., Yang, J., et al. (2018). Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β-Catenin pathway by scaffolding EZH2. Clin. Cancer Res. 24 (3), 684–695. doi:10.1158/1078-0432.CCR-17-0605
Chen, Y., Guo, L., Lang, H., Hu, X., Jing, S., Luo, M., et al. (2018). Effect of a stellate ganglion block on acute lung injury in septic rats. Inflammation 41 (5), 1601–1609. doi:10.1007/s10753-018-0803-x
Chen, Y., Li, Z., Chen, X., and Zhang, S. (2021). Long non-coding RNAs: from disease code to drug role. Acta Pharm. Sin. B 11 (2), 340–354. doi:10.1016/j.apsb.2020.10.001
Chi, Y., Wang, D., Wang, J., Yu, W., and Yang, J. (2019). Long non-coding RNA in the pathogenesis of cancers. Cells 8 (9), 1015. doi:10.3390/cells8091015
Childs-Disney, J. L., Yang, X., Gibaut, Q. M. R., Tong, Y., Batey, R. T., and Disney, M. D. (2022). Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 21 (10), 736–762. doi:10.1038/s41573-022-00521-4
Cho, S. J., and Stout-Delgado, H. W. (2020). Aging and lung disease. Annu. Rev. Physiol. 82, 433–459. doi:10.1146/annurev-physiol-021119-034610
Choi, S. W., Kim, H. W., and Nam, J. W. (2019). The small peptide world in long noncoding RNAs. Brief. Bioinform 20 (5), 1853–1864. doi:10.1093/bib/bby055
Crooke, S. T. (2017). Molecular mechanisms of antisense oligonucleotides. Nucleic Acid. Ther. 27 (2), 70–77. doi:10.1089/nat.2016.0656
Dahariya, S., Paddibhatla, I., Kumar, S., Raghuwanshi, S., Pallepati, A., and Gutti, R. K. (2019). Long non-coding RNA: classification, biogenesis and functions in blood cells. Mol. Immunol. 112, 82–92. doi:10.1016/j.molimm.2019.04.011
Davidson, S. M., Ferdinandy, P., Andreadou, I., Bøtker, H. E., Heusch, G., Ibáñez, B., et al. (2019). Multitarget strategies to reduce myocardial ischemia/reperfusion injury: JACC review topic of the week. J. Am. Coll. Cardiol. 73 (1), 89–99. doi:10.1016/j.jacc.2018.09.086
Dekker, E., Tanis, P. J., Vleugels, J. L. A., Kasi, P. M., and Wallace, M. B. (2019). Colorectal cancer. Lancet 394 (10207), 1467–1480. doi:10.1016/S0140-6736(19)32319-0
Ding, X., Zhu, C., Wang, W., and Gao, B. (2024). SIRT1 is a regulator of autophagy: implications for the progression and treatment of myocardial ischemia-reperfusion. Pharmacol. Res. 199, 106957. doi:10.1016/j.phrs.2023.106957
Disney, M. D. (2019). Targeting RNA with small molecules to capture opportunities at the intersection of chemistry, biology, and medicine. J. Am. Chem. Soc. 141 (17), 6776–6790. doi:10.1021/jacs.8b13419
Dykes, I. M., and Emanueli, C. (2017). Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinforma. 15 (3), 177–186. doi:10.1016/j.gpb.2016.12.005
Ehrentraut, H., Weisheit, C. K., Frede, S., and Hilbert, T. (2019). Inducing acute lung injury in mice by direct intratracheal lipopolysaccharide instillation. J. Vis. Exp. 149. doi:10.3791/59999
Engreitz, J. M., Sirokman, K., McDonel, P., Shishkin, A. A., Surka, C., Russell, P., et al. (2014). RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159 (1), 188–199. doi:10.1016/j.cell.2014.08.018
Esposito, R., Bosch, N., Lanzós, A., Polidori, T., Pulido-Quetglas, C., and Johnson, R. (2019). Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell 35 (4), 545–557. doi:10.1016/j.ccell.2019.01.019
Fan, A., Wang, B., Wang, X., Nie, Y., Fan, D., Zhao, X., et al. (2021). Immunotherapy in colorectal cancer: current achievements and future perspective. Int. J. Biol. Sci. 17 (14), 3837–3849. doi:10.7150/ijbs.64077
Ferrer, J., and Dimitrova, N. (2024). Transcription regulation by long non-coding RNAs: mechanisms and disease relevance. Nat. Rev. Mol. Cell Biol. 25 (5), 396–415. doi:10.1038/s41580-023-00694-9
Gai, H. Y., Wu, C., Zhang, Y., and Wang, D. (2019). Long non-coding RNA CHRF modulates the progression of cerebral ischemia/reperfusion injury via miR-126/SOX6 signaling pathway. Biochem. Biophys. Res. Commun. 514 (2), 550–557. doi:10.1016/j.bbrc.2019.04.161
Giannakakis, S., Galyfos, G., Sachmpazidis, I., Kapasas, K., Kerasidis, S., Stamatatos, I., et al. (2017). Thrombolysis in peripheral artery disease. Ther. Adv. Cardiovasc. Dis. 11 (4), 125–132. doi:10.1177/1753944716687517
Glass, D. S., Grossfeld, D., Renna, H. A., Agarwala, P., Spiegler, P., DeLeon, J., et al. (2022). Idiopathic pulmonary fibrosis: current and future treatment. Clin. Respir. J. 16 (2), 84–96. doi:10.1111/crj.13466
Gong, J., Wang, Y., and Shu, C. (2020). LncRNA CHRF promotes cell invasion and migration via EMT in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 24 (3), 1168–1176. doi:10.26355/eurrev_202002_20168
Green, A. R., and Shuaib, A. (2006). Therapeutic strategies for the treatment of stroke. Drug Discov. Today 11 (15-16), 681–693. doi:10.1016/j.drudis.2006.06.001
Greenberg, M. I., Waksman, J., and Curtis, J. (2007). Silicosis: a review. Dis. Mon. 53 (8), 394–416. doi:10.1016/j.disamonth.2007.09.020
Haggar, F. A., and Boushey, R. P. (2009). Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin. Colon Rectal Surg. 22 (4), 191–197. doi:10.1055/s-0029-1242458
Hausenloy, D. J., and Yellon, D. M. (2013). Myocardial ischemia-reperfusion injury: a neglected therapeutic target. J. Clin. Invest 123 (1), 92–100. doi:10.1172/JCI62874
Heinzel, F. R., Hohendanner, F., Jin, G., Sedej, S., and Edelmann, F. (1985). Myocardial hypertrophy and its role in heart failure with preserved ejection fraction. J. Appl. Physiol. 119 (10), 1233–1242. doi:10.1152/japplphysiol.00374.2015
Khorkova, O., and Wahlestedt, C. (2017). Oligonucleotide therapies for disorders of the nervous system. Nat. Biotechnol. 35 (3), 249–263. doi:10.1038/nbt.3784
King, T. E., Pardo, A., and Selman, M. (2011). Idiopathic pulmonary fibrosis. Lancet 378 (9807), 1949–1961. doi:10.1016/S0140-6736(11)60052-4
Koonin, E. V., Gootenberg, J. S., and Abudayyeh, O. O. (2023). Discovery of diverse CRISPR-Cas systems and expansion of the genome engineering toolbox. Biochemistry 62 (24), 3465–3487. doi:10.1021/acs.biochem.3c00159
Kopp, F., Elguindy, M. M., Yalvac, M. E., Zhang, H., Chen, B., Gillett, F. A., et al. (2019). PUMILIO hyperactivity drives premature aging of Norad-deficient mice. Elife 8, e42650. doi:10.7554/eLife.42650
Kujawa, K. A., and Lisowska, K. M. (2015). Ovarian cancer--from biology to clinic. Postepy Hig. Med. Dosw (Online) 69, 1275–1290. doi:10.5604/17322693.1184451
Kumar, V. (2020). Pulmonary innate immune response determines the outcome of inflammation during pneumonia and sepsis-associated acute lung injury. Front. Immunol. 11, 1722. doi:10.3389/fimmu.2020.01722
Kumari, R., Ranjan, P., Suleiman, Z. G., Goswami, S. K., Li, J., Prasad, R., et al. (2022). mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc Res. 118 (7), 1680–1692. doi:10.1093/cvr/cvab160
Lamouille, S., Xu, J., and Derynck, R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15 (3), 178–196. doi:10.1038/nrm3758
Li, H., Chen, C., Fan, J., Yin, Z., Ni, L., Cianflone, K., et al. (2018). Identification of cardiac long non-coding RNA profile in human dilated cardiomyopathy. Cardiovasc Res. 114 (5), 747–758. doi:10.1093/cvr/cvy012
Li, J., Jiang, Z. Z., Li, Y. Y., Tang, W. T., Yin, J., and Long, X. P. (2021). LncRNA CHRF promotes TGF-β1 induced EMT in alveolar epithelial cells by inhibiting miR-146a up-regulating L1CAM expression. Exp. Lung Res. 47 (4), 198–209. doi:10.1080/01902148.2021.1891354
Li, T., Yang, X., Xu, H., and Liu, H. (2022b). Early identification, accurate diagnosis, and treatment of silicosis. Can. Respir. J. 2022, 3769134. doi:10.1155/2022/3769134
Li, W., Dong, X., He, C., Tan, G., Li, Z., Zhai, B., et al. (2019). LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 38 (1), 183. doi:10.1186/s13046-019-1177-0
Li, Y., Gao, Y., and Li, G. (2022a). Preclinical multi-target strategies for myocardial ischemia-reperfusion injury. Front. Cardiovasc Med. 9, 967115. doi:10.3389/fcvm.2022.967115
Li, Y., Liang, Y., Zhu, Y., Zhang, Y., and Bei, Y. (2018). Noncoding RNAs in cardiac hypertrophy. J. Cardiovasc Transl. Res. 11 (6), 439–449. doi:10.1007/s12265-018-9797-x
Liang, Y., Chen, X., Wu, Y., Li, J., Zhang, S., Wang, K., et al. (2018). LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 25 (11), 1980–1995. doi:10.1038/s41418-018-0084-9
Liu, J., Zhang, X., Chen, K., et al. (2019). CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-Mediated glycolysis. Immunity 50 (3), 600–615.e15. doi:10.1016/j.immuni.2019.01.021
Liu, S., Wang, L., Li, Y., Cui, Y., Wang, Y., and Liu, C. (2019). Long non-coding RNA CHRF promotes proliferation and mesenchymal transition (EMT) in prostate cancer cell line PC3 requiring up-regulating microRNA-10b. Biol. Chem. 400, 1035–1045. doi:10.1515/hsz-2018-0380
Liu, X., Homma, A., Sayadi, J., Yang, S., Ohashi, J., and Takumi, T. (2016). Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci. Rep. 6 (1), 19675. doi:10.1038/srep19675
Luo, S., Ding, X., Zhao, S., Mou, T., Li, R., and Cao, X. (2021). Long non-coding RNA CHRF accelerates LPS-induced acute lung injury through microRNA-146a/Notch1 axis. Ann. Transl. Med. 9 (16), 1299. doi:10.21037/atm-21-3064
Lynch, D. A., Sverzellati, N., Travis, W. D., Brown, K. K., Colby, T. V., Galvin, J. R., et al. (2018). Diagnostic criteria for idiopathic pulmonary fibrosis: a fleischner society white paper. Lancet Respir. Med. 6 (2), 138–153. doi:10.1016/S2213-2600(17)30433-2
Matsui, M., and Corey, D. R. (2017). Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 16 (3), 167–179. doi:10.1038/nrd.2016.117
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24 (6), 430–447. doi:10.1038/s41580-022-00566-8
Mishra, S., Verma, S. S., Rai, V., Awasthee, N., Chava, S., Hui, K. M., et al. (2019). Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell. Mol. Life Sci. 76 (10), 1947–1966. doi:10.1007/s00018-019-03053-0
Mo, Y., Wu, H., Zheng, X., Xu, L., Liu, L., and Liu, Z. (2021). LncRNA CHRF aggravates myocardial ischemia/reperfusion injury by enhancing autophagy via modulation of the miR-182-5p/ATG7 pathway. J. Biochem. Mol. Toxicol. 35 (4), e22709. doi:10.1002/jbt.22709
Mokra, D. (2020). Acute lung injury - from pathophysiology to treatment. Physiol. Res. 69 (Suppl. 3), S353–S366. doi:10.33549/physiolres.934602
Nelson, B. R., Makarewich, C. A., Anderson, D. M., Winders, B. R., Troupes, C. D., Wu, F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351 (6270), 271–275. doi:10.1126/science.aad4076
Orr, B., and Edwards, R. P. (2018). Diagnosis and treatment of ovarian cancer. Hematol. Oncol. Clin. North Am. 32 (6), 943–964. doi:10.1016/j.hoc.2018.07.010
Parsons, P. E., Eisner, M. D., Thompson, B. T., Matthay, M. A., Ancukiewicz, M., Bernard, G. R., et al. (2005). Lower tidal volume ventilation and plasma cytokine markers of inflammation in patients with acute lung injury. Crit. Care Med. 33 (1), 1–232. doi:10.1097/01.ccm.0000149854.61192.dc
Pastushenko, I., and Blanpain, C. (2019). EMT transition states during tumor progression and metastasis. Trends Cell Biol. 29 (3), 212–226. doi:10.1016/j.tcb.2018.12.001
Pedram Fatemi, R., Salah-Uddin, S., Modarresi, F., Khoury, N., Wahlestedt, C., and Faghihi, M. A. (2015). Screening for small-molecule modulators of long noncoding RNA-protein interactions using AlphaScreen. SLAS Discov. 20 (9), 1132–1141. doi:10.1177/1087057115594187
Pollard, K. M. (2016). Silica, silicosis, and autoimmunity. Front. Immunol. 7, 97. doi:10.3389/fimmu.2016.00097
Popov, S. V., Mukhomedzyanov, A. V., Voronkov, N. S., Derkachev, I. A., Boshchenko, A. A., Fu, F., et al. (2023). Regulation of autophagy of the heart in ischemia and reperfusion. Apoptosis 28 (1-2), 55–80. doi:10.1007/s10495-022-01786-1
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17 (1), 47–62. doi:10.1038/nrg.2015.10
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19 (3), 143–157. doi:10.1038/nrm.2017.104
Rinaldi, C., and Wood, M. J. A. (2017). Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat. Rev. Neurol. 14 (1), 9–21. doi:10.1038/nrneurol.2017.148
Robinson, D., Van Allen, E. M., Wu, Y. M., Schultz, N., Lonigro, R. J., Mosquera, J. M., et al. (2015). Integrative clinical genomics of advanced prostate cancer. Cell 161 (5), 1215–1228. doi:10.1016/j.cell.2015.05.001
Samuel, P., Pink, R. C., Brooks, S. A., and Carter, D. R. (2016). miRNAs and ovarian cancer: a miRiad of mechanisms to induce cisplatin drug resistance. Expert Rev. Anticancer Ther. 16 (1), 57–70. doi:10.1586/14737140.2016.1121107
Sanchez Calle, A., Kawamura, Y., Yamamoto, Y., Takeshita, F., and Ochiya, T. (2018). Emerging roles of long non-coding RNA in cancer. Cancer Sci. 109 (7), 2093–2100. doi:10.1111/cas.13642
Schier, A. C., and Taatjes, D. J. (2020). Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34 (7-8), 465–488. doi:10.1101/gad.335679.119
Schmidt-Kastner, R. (2015). Genomic approach to selective vulnerability of the hippocampus in brain ischemia-hypoxia. Neuroscience 309, 259–279. doi:10.1016/j.neuroscience.2015.08.034
Schmitz, S. U., Grote, P., and Herrmann, B. G. (2016). Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 73 (13), 2491–2509. doi:10.1007/s00018-016-2174-5
Shen, S., Jiang, H., Bei, Y., Xiao, J., and Li, X. (2017). Long non-coding RNAs in cardiac remodeling. Cell Physiol. Biochem. 41 (5), 1830–1837. doi:10.1159/000471913
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics. CA Cancer J. Clin. 70 (1), 7–30. doi:10.3322/caac.21590
Simon, K. (2016). Colorectal cancer development and advances in screening. Clin. Interv. Aging 11, 967–976. doi:10.2147/CIA.S109285
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., and Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351 (6268), 84–88. doi:10.1126/science.aad5227
Solevag, A. L., Schmolzer, G. M., and Cheung, P. Y. (2020). Hypoxia - reoxygenation in neonatal cardiac arrest: results from experimental models. Semin. Fetal Neonatal Med. 25 (2), 101085. doi:10.1016/j.siny.2020.101085
Stowasser, S., and Hallmann, C. (2015). New guideline for idiopathic pulmonary fibrosis. Lancet 386 (10006), 1823–1824. doi:10.1016/S0140-6736(15)00765-5
Tan, S., and Chen, S. (2021). The mechanism and effect of autophagy, apoptosis, and pyroptosis on the progression of silicosis. Int. J. Mol. Sci. 22 (15), 8110. doi:10.3390/ijms22158110
Tan, W. X., Sun, G., Shangguan, M. Y., Gui, Z., Bao, Y., Li, Y. F., et al. (2020). Novel role of lncRNA CHRF in cisplatin resistance of ovarian cancer is mediated by miR-10b induced EMT and STAT3 signaling. Sci. Rep. 10 (1), 14768. doi:10.1038/s41598-020-71153-0
Tao, S. C., Rui, B. Y., Wang, Q. Y., Zhou, D., Zhang, Y., and Guo, S. C. (2018). Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 25 (1), 241–255. doi:10.1080/10717544.2018.1425774
Tao, Y., Han, T., Zhang, T., Ma, C., and Sun, C. (2017). LncRNA CHRF-induced miR-489 loss promotes metastasis of colorectal cancer via TWIST1/EMT signaling pathway. Oncotarget 8 (22), 36410–36422. doi:10.18632/oncotarget.16850
Thrift, A. P., Wenker, T. N., and El-Serag, H. B. (2023). Global burden of gastric cancer: epidemiological trends, risk factors, screening and prevention. Nat. Rev. Clin. Oncol. 20 (5), 338–349. doi:10.1038/s41571-023-00747-0
Tsai, S. Q., and Joung, J. K. (2014). What’s changed with genome editing? Cell Stem Cell 15 (1), 3–4. doi:10.1016/j.stem.2014.06.017
Van Cutsem, E., Nordlinger, B., and Cervantes, A.ESMO Guidelines Working Group (2010). Advanced colorectal cancer: ESMO clinical practice guidelines for treatment. Ann. Oncol. 21 (Suppl. 5), v93–v97. doi:10.1093/annonc/mdq222
Viney, N. J., van Capelleveen, J. C., Geary, R. S., Xia, S., Tami, J. A., Yu, R. Z., et al. (2016). Antisense oligonucleotides targeting apolipoprotein(a) in people with raised lipoprotein(a): two randomised, double-blind, placebo-controlled, dose-ranging trials. Lancet 388 (10057), 2239–2253. doi:10.1016/S0140-6736(16)31009-1
Wang, K., Liu, F., Zhou, L. Y., Long, B., Yuan, S. M., Wang, Y., et al. (2014). The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 114 (9), 1377–1388. doi:10.1161/CIRCRESAHA.114.302476
Weber, A., Wasiliew, P., and Kracht, M. (2010). Interleukin-1 (IL-1) pathway. Sci. Signal 3 (105), cm1. doi:10.1126/scisignal.3105cm1
Wo, Y., Guo, J., Li, P., Yang, H., and Wo, J. (2018). Long non-coding RNA CHRF facilitates cardiac hypertrophy through regulating Akt3 via miR-93. Cardiovasc Pathol. 35, 29–36. doi:10.1016/j.carpath.2018.04.003
Wu, Q., Han, L., Yan, W., Ji, X., Han, R., Yang, J., et al. (2016). miR-489 inhibits silica-induced pulmonary fibrosis by targeting MyD88 and Smad3 and is negatively regulated by lncRNA CHRF. Sci. Rep. 6, 30921. doi:10.1038/srep30921
Xia, D., Sui, R., and Zhang, Z. (2018). Administration of resveratrol improved Parkinson’s disease-like phenotype by suppressing apoptosis of neurons via modulating the MALAT1/miR-129/SNCA signaling pathway. J. Cell. Biochem. 120 (4), 4942–4951. doi:10.1002/jcb.27769
Xiao, G. A., Yao, J., Kong, D., Ye, C., Chen, R., Li, L., et al. (2019). The long noncoding RNA TTTY15, which is located on the Y chromosome, promotes prostate cancer progression by sponging let-7. Eur. Urol. 76 (3), 315–326. doi:10.1016/j.eururo.2018.11.012
Xu, Q., Deng, F., Xing, Z., Wu, Z., Cen, B., Xu, S., et al. (2016). Long non-coding RNA C2dat1 regulates CaMKIIδ expression to promote neuronal survival through the NF-κB signaling pathway following cerebral ischemia. Cell Death Dis. 7, e2173. doi:10.1038/cddis.2016.57
Yan, H., and Bu, P. (2021). Non-coding RNA in cancer. Essays Biochem. 65 (4), 625–639. doi:10.1042/EBC20200032
Zhang, S., Xin, H., Li, Y., Zhang, D., Shi, J., Yang, J., et al. (2013). Skimmin, a coumarin fromHydrangea paniculata, slows down the progression of membranous glomerulonephritis by anti-inflammatory effects and inhibiting immune complex deposition. Evidence-Based Complementary Altern. Med. 2013, 819296–10. doi:10.1155/2013/819296
Zhang, Y., Zeng, Q., Li, C., Zhou, H., Liu, J., et al. (2022). IL-1β-Triggered long non-coding RNA CHRF induces non-small cell lung cancer by modulating the microRNA-489/Myd88 Axis. J. Cancer 13 (8), 2620–2630. doi:10.7150/jca.63256
Zhao, Y., Zhang, X., Chen, X., and Wei, Y. (2022). Neuronal injuries in cerebral infarction and ischemic stroke: from mechanisms to treatment (Review). Int. J. Mol. Med. 49 (2), 15. doi:10.3892/ijmm.2021.5070
Keywords: cancer, cardiovascular diseases, fibrosis, long non-coding RNA CHRF, molecular mechanism, target strategies
Citation: Mou J, Luo C, Zhang W, Shao Y, Pei J, Chen Y, Guo X, Fan Y and Sun H (2025) LncRNA CHRF: molecular mechanisms and therapeutic potentials in cardiovascular diseases, cancers and fibrosis. Front. Cell Dev. Biol. 13:1573723. doi: 10.3389/fcell.2025.1573723
Received: 11 February 2025; Accepted: 10 June 2025;
Published: 19 June 2025.
Edited by:
Roland Wohlgemuth, Lodz University of Technology, PolandReviewed by:
Xiang Nie, Huazhong University of Science and Technology, ChinaXiaorong Hu, Wuhan University, China
Copyright © 2025 Mou, Luo, Zhang, Shao, Pei, Chen, Guo, Fan and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Sun, c2hvbmd5dTIwMDhAMTYzLmNvbQ==
†ORCID: Hongyu Sun, orcid.org/0000-0002-8587-0499
 Jie Mou1,2
Jie Mou1,2 Hongyu Sun
Hongyu Sun