- Department of Microbiology, University of Manitoba, Winnipeg, MB, Canada
The phenylacetic acid (PA) degradative pathway is the central pathway by which various aromatic compounds (e.g., styrene) are degraded. Upper pathways for different aromatic compounds converge at common intermediate phenylacetyl-CoA (PA-CoA), which is then metabolized to succinyl-CoA and acetyl-CoA. We previously made a link in Burkholderia cenocepacia between PA degradation and virulence by showing that insertional mutagenesis of paaA and paaE genes, that encode part of a multicomponent oxidase of PA-CoA, results in PA-conditional growth and an attenuated killing phenotype in the Caenorhabditis elegans model of infection. However, insertional mutagenesis of paaK1, which encodes a phenylacetate-CoA ligase, did not result in a PA-conditional growth probably due to the presence of a putative paralog gene paaK2. Recently published crystallographic and enzyme kinetics data comparing the two PaaK ligases showed that PaaK1 is more active than PaaK2 and that the larger binding pocket of PaaK1 can accommodate hydroxylated PA derived molecules such as 3-hydroxyphenylacetic (3-OHPA) acid and 4-hydroxyphenylacetic acid (4-OHPA). The higher activity and broader substrate specificity suggested a more active role in pathogenesis. In this work, we aimed to determine the relevance of PaaK1 activity to the killing ability of B. cenocepacia to C. elegans. Using reporter activity assays, we demonstrate that 3-OHPA activated PA degradation gene promoters of Burkholderia cenocepacia K56-2 in a paaK1-dependent manner, while 4-OHPA had no effect. We compared the pathogenicity of a paaK1 deletion mutant with that of the wild type in C. elegans and observed no differences in the killing ability of the strains. Taken together, these studies suggest that 3-OHPA, but not 4-OHPA, can induce the PA pathway and that this induction is dependent on the paaK1 gene. However, the more active PaaK1 does not play a distinct role in pathogenesis of B. cenocepacia as previously suggested.
Introduction
The phenylacetic acid (PA) degradative pathway is a well-conserved, central pathway in bacteria utilized in the breakdown of many aromatic compounds such as styrene and phenylalanine (Luengo et al., 2001). A unique feature of the PA degradation pathway is that it exploits CoA activation of PA in aerobic conditions. Thus, intermediates are processed as CoA thioesters that are finally broken down into the TCA intermediates, acetyl-CoA, and succinyl-CoA.
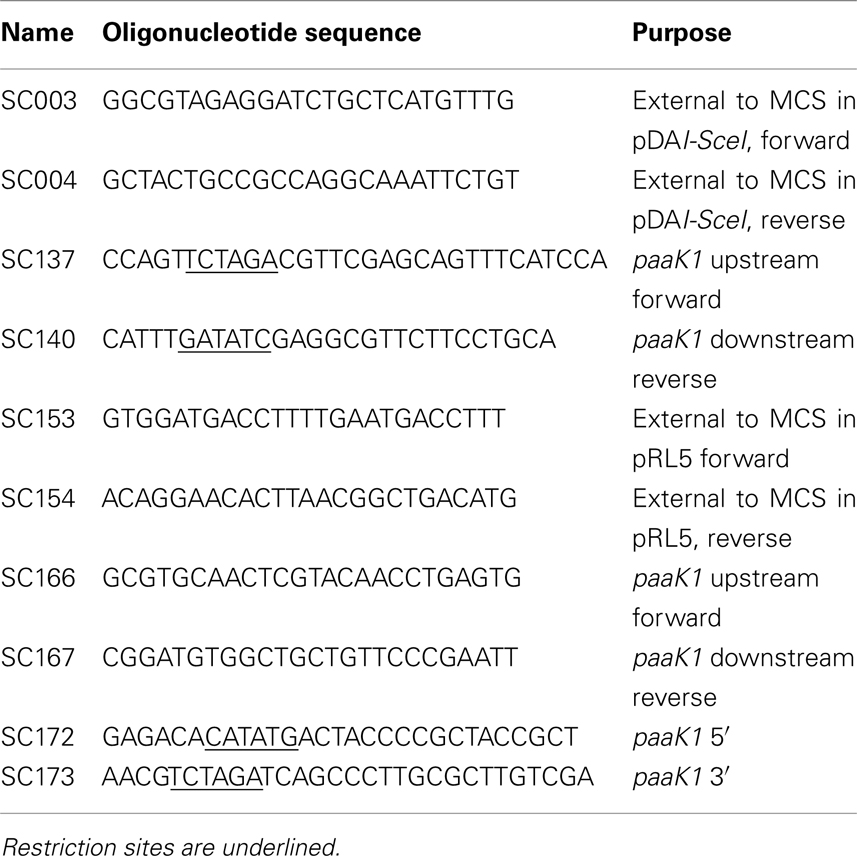
The detailed mechanism of the PA degradation pathway was recently elucidated for E. coli K12 and Pseudomonas putida Y2 (Teufel et al., 2010; Figure 1A). The initial step is crucial as it consists of the activation of PA by CoA mediated by the phenylacetate-CoA ligase PaaK (Martinez-Blanco et al., 1990; El-Said Mohamed, 2000). In Burkholderia cenocepacia, the PA degradation genes are organized in three clusters across two of three chromosomes (Figure 1B). There are two similar yet non-identical genes that encode phenylacetate-CoA ligases (Law et al., 2008). Each gene (BCAL0404 or paaK1 and BCAM1711 or paaK2) is located on chromosome 1 and chromosome 2 respectively. Recently, Law and Boulanger (2011) provided a rationale for the presence of the two-paralog PA-CoA ligases of B. cenocepacia by in vitro studies, showing that PaaK1 has a much higher affinity for PA than PaaK2 with Km values of 62 and 150 μM, respectively. A striking difference was that PaaK1 was able to accommodate 3′ and 4′ substituted PA due to a larger binding pocket. Although both enzymes were able to accept 3-OHPA, PaaK1 showed 6.5-fold greater affinity for 3-OHPA than PaaK2. PaaK1 could also use 4-OHPA as substrate, while the activity of PaaK2 with 4-OHPA was undetectable (Law and Boulanger, 2011).
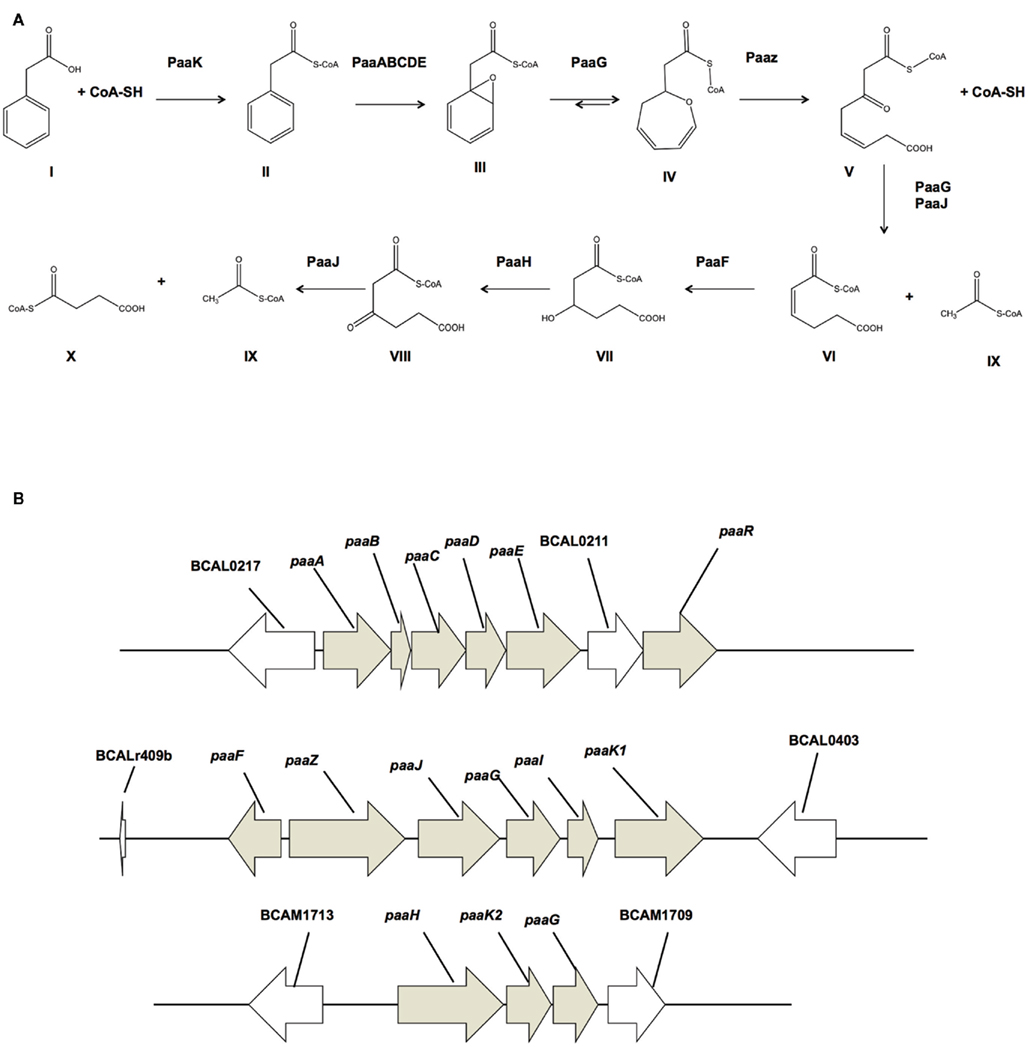
Figure 1. Proposed PA degradation pathway for B. cenocepacia J2315. (A) Detailed mechanism of the PA degradation pathway for E. coli K12 and Pseudomonas putida Y2 (Teufel et al., 2010). (I) Phenylacetic acid; (II) Phenylacetyl-CoA ligase; (III) ring 1,2-epoxyphenylacetyl-CoA; (IV) 2-oxepin-2(3H)-ylideneacetyl-CoA; (V) 3-oxo-5,6-dehydrosuberyl-CoA; (VI) 2,3-dehydroadipyl-CoA; (VII) 3-hydroxyadipyl-CoA; (VIII) 3-oxoadipyl-CoA; (IX) Acetyl-CoA; (X) succinyl-CoA. Enzymes: PaaK, phenylacetyl-CoA ligase; PaaABCDE, Monooxygenase complex; PaaG, ring 1,2-epoxyphenylacetyl-CoA isomerase/3,4-dehydroadipyl-CoA isomerase; PaaZ, oxepin-CoA hydrolase/3-oxo-5,6-dehydrosuberyl-CoA semialdehyde dehydrogenase; PaaJ, 3-oxoadipyl-CoA/3-oxo-5,6-dehydrosuberyl-CoA thiolase; PaaF, 2,3-dehydroadipyl-CoA hydratase; PaaH, 3-hydroxyadipyl-CoA dehydrogenase. (B) PA degradation gene clusters of B. cenocepacia J2315. paaABCDE and paaFZJGIK1 are located on the chromosome 1 and paaHK2paaG is located on chromosome 2.
We previously demonstrated that the B. cenocepacia K56-2 catabolic pathway for phenylacetic acid (PA) degradation is a requirement for full pathogenesis of B. cenocepacia (Law et al., 2008) in the Caenorhabditis elegans model of infection (Aballay and Ausubel, 2002) but the role of the PaaK enzymes in pathogenicity and the biological relevance of 3-OHPA and 4-OHPA as substrates was not addressed. With the benefit of reporter activity assays and C. elegans as a host model organism, we aimed to address the relevance of the more active and substrate-promiscuous PaaK1 for pathogenesis of B. cenocepacia against C. elegans. We demonstrate that a hydroxylated PA derived molecule such as 3-OHPA can induce the PA degradative pathway and that this induction is paaK1-dependent. A clean deletion of paaK1 resulted however, in wild type-like pathogenicity of B. cenocepacia in C. elegans.
Materials and Methods
Bacterial Strains, Nematodes, and Growth Conditions
Bacterial strains and plasmids are listed in Table 1. B. cenocepacia strains were grown at 37°C in Luria Bertani (LB), M9 media, or modified nematode growth media (mNGM; 3 g L−1 NaCl, 17 g L−1 bacto-agar, 3.5 g L−1 peptone, 1 mL 5 mg mL−1 cholesterol, 1 mL 1 M CaCl2, 1 mL 1 M MgSO4, 25 mL phosphate buffer) supplemented, as required, with 100 μg/mL trimethoprim (Tp), and 200 μg/mL chloramphenicol (Cm). Unless indicated otherwise, glycerol and phenylacetic acid were added at 0.1% w/v and 5 mM, respectively. The nematode C. elegans, strain DH26 and E. coli OP50 were obtained from the Caenorhabditis Genetics Center (CGC), University of Minnesota, Minneapolis. C. elegans strains were maintained on nematode growth media (NGM) according to standard practices at the CGC.
Molecular Biology Techniques
E. coli DH5α cells were transformed using the calcium chloride protocol (Cohen et al., 1972) and electroporation was used for transformation of E. coli SY327 cells (Miller and Mekalanos, 1988). Conjugation into B. cenocepacia K56-2, and IAI1 was accomplished by tri-parental mating (Craig et al., 1989) with E. coli DH5α carrying the helper plasmid pRK2013 (Figurski and Helinski, 1979). DNA was amplified using an Eppendorf Mastercycler ep gradient S thermal cycler with Taq DNA polymerase PCR Kit (Qiagen). Amplification conditions were optimized for each primer pair. PCR products and plasmids were purified with QIAquick purification kit (Qiagen) and QIAprep Miniprep kit (Qiagen), respectively.
Construction of paak1 Deletion Mutant IAI1
IAI1 (paaK1) was created using the techniques outlined in Flannagan et al. (2008) using B. cenocepacia wild type K56-2. Briefly, introduction of plasmid pRL5 (Table 1), which contains the flanking regions of paaK1 as well as a yeast I-SceI restriction site (TAGGGATAACAGGGTAAT) was performed by tri-parental mating. The first homologous recombination event and the recombination site were confirmed by colony PCR with primer pairs 154/165 or 153/164 (Table 2), which anneal to the plasmid and the genome, respectively. Following homologous recombination a second plasmid pDAI-SceI, which constitutively expresses the yeast homing endonuclease I-SceI, was introduced by tri-parental mating. Resulting clones were screened for trimethoprim sensitivity and tetracycline resistance by replica plating on LB containing 100 μg/mL of tetracycline with and without 100 μg/mL of trimethoprim. Subsequently, trimethoprim sensitive clones were screened for the deletion of paaK1 with primers 166 and 167 (Table 2). Following confirmation by colony PCR that the gene had been deleted, pDAI-SceI was cured by serial dilution in LB broth. Loss of the plasmid was confirmed by colony PCR using primers 3 and 4, which anneal external to the multi-cloning site of pDAI-SceI. Deletion of the paaK1 in B. cenocepacia IAI1 gene was further confirmed by sequencing.
Bacterial Growth
One-milliliter aliquots of M9 plus different carbon sources were inoculated with 100 μL from overnight culture grown in LB, washed with PBS, and adjusted to an optical density at 600 nm (OD600) of 2.0. One hundred fifty microliters aliquots were added in triplicate to 96-well format plates and the microplates were incubated 24 h at 37°C with shaking at 210 rpm. Growth was measured as the OD600 was measured with a BioTek Synergy 2 plate reader at hourly intervals.
Nematode Infection Assays
Caenorhabditis elegans slow killing assays were performed as previously described (Law et al., 2008). Thirty 5 mm plates filled with mNGM were seeded with 50 μL of stationary phase culture adjusted to an OD600 of 1.7. In these growth conditions, the PA degradation pathway is activated. Plates were incubated overnight at 37°C to allow growth of a bacterial lawn before 30–40 hypochlorite-synchronized L4 larvae of C. elegans DH26 were inoculated to each plate and plates were then incubated at 25°C for the duration of the experiment. Plates were scored for live worms at the time of inoculation and every 24 h subsequently for 7 days using a stereo-master dissecting microscope. Worms were considered dead when unresponsive to touch with a sterile wire pick. Assays were performed in triplicate and analyzed using survival curves generated by the Kaplan–Meier statistical method. The log rank test was used to compare survival differences for statistical significance using GraphPad Prism, version 5.02. A P value <0.05 was considered to be significant.
Reporter Activity Assays
Green Fluorescent Protein fluorescence was measured according to its excitation/emission wavelengths of 488/509 (Cormack et al., 1996). Relative fluorescence, defined as the ratio between arbitrary fluorescence and OD600 was measured with a BioTek Synergy 2 plate reader, using excitation 485/20 and emission 528/20 filter sets.
Results
Burkholderia cenocepacia Utilizes 3-OHPA and 4-OHPA as Sole Carbon Sources, but not Exclusively Through PA Degradation
To address the biological relevance of 3-OHPA and 4-OHPA as substrates of the PaaK enzymes, we started out by asking if B. cenocepacia could use 3-OHPA and 4-OHPA as sole carbon sources. Given the evidence that 3-OHPA and 4-OHPA can be substrates of the PaaK enzymes (Law and Boulanger, 2011) we also considered that these compounds could be funneled exclusively through the PA catabolic pathway. B. cenocepacia K56-2 was grown in 3-OHPA or 4-OHPA as sole carbon sources and growth was recorded as final optical density. The hydroxylated phenolic acids supported 80 and 54% growth related to growth in glucose, respectively while 3-OHPA and 4-OHPA supported growth to 94 and 75% respectively, in comparison to growth in PA (Figure 2). Similarly, the paaK1 deletion mutant IAI1 did not show any growth-defective phenotype in PA according to the presence of a second paaK gene (paaK2). Moreover, deletion of the paaK1 gene did not affect the ability of this strain to use 3-OHPA or 4-OHPA (87% and 80% growth related to glucose, respectively). Concomitantly, when the PA degradation defective mutants PaaA and PaaE were grown with 3-OHPA, 4-OHPA, or PA the growth-defective phenotype was observed only in the presence of PA (3.4 and 4% growth, respectively) while both mutants exhibited wild type-like growth in the presence of the hydroxylated phenolic acids. This suggests that 3-OHPA and 4-OHPA are degraded through other routes, possibly the homogentisate and homoprotocatechuate pathways (Méndez et al., 2011). Taken together, these results suggest that 3-OHPA and 4-OHPA can serve as carbon sources for B. cenocepacia and that these compounds do not seem to be degraded exclusively through the PA degradative pathway.
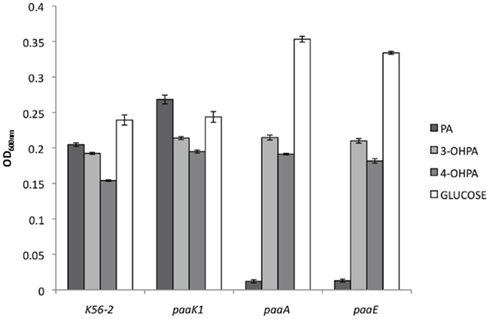
Figure 2. Growth of B. cenocepacia strains with 3-OHPA, 4-OHPA as sole carbon sources. B. cenocepacia K56-2 (wild type), deletion mutant IAI1 (paaK1), and insertional mutants STC179, (paaA) and STC155 (paaE) were grown for 24 h in M9 media supplemented with 5 mM PA, 3-OHPA, 4-OHPA, or 0.2% glucose as sole carbon sources. Error bars represent the SD of three replicates.
3-OHPA but not 4-OHPA Induces the PA Degradative Pathway
We reasoned that 3-OHPA and 4-OHPA, as substrates for PaaK1, could induce the PA degradative pathway in Burkholderia cenocepacia. In order to test this, we utilized a PA pathway induction reporter assay previously developed in our laboratory (Hamlin et al., 2009). The translational reporter consists of a plasmid with the enhanced green fluorescent protein gene (e-gfp) controlled by the paa promoter for each of the three paa gene clusters of the B. cenocepacia paa operons that start with the paaZ, paaA, and paaH genes. Growth of a strain carrying the reporter plasmid in minimal media with PA results in expression of eGFP, which can be measured by fluorescence as an indication of the induction of the PA pathway. Plasmids pJH1 and pJH2 carry e-gfp under control of the constitutive expression promoter DHFR and a promoterless e-gfp, respectively.
Burkholderia cenocepacia K56-2 reporter strains were grown in M9 minimal media with glycerol supplemented with 5 mM PA, 3-OHPA, or 4-OHPA (Figure 3). After 24 h of incubation at 37°C the relative fluorescence of wild type and reporter strains was analyzed. Low levels of background fluorescence were observed for B. cenocepacia K56-2 wild type and the reporter strain carrying a promoterless e-gfp gene. On the contrary, when B. cenocepacia/pJH1 was grown with glycerol or glycerol plus phenolic acids, increased fluorescence was observed. The levels of relative fluorescence due to PA, 3-OHPA, or 4-OHPA were comparable suggesting that there are no metabolic effects of these compounds on plasmid copy number. When PA pathway reporter strains were grown in the presence of PA, an increase in relative fluorescence was observed as previously described. However, only 3-OHPA (but not 4-OHPA) was capable of inducing the paaZ, paaA, and paaH promoters (Figure 2). Across the board, we noted an average of five times less relative fluorescence in media containing 4-OHPA than in media containing PA. Interestingly, B. cenocepacia K56-2/pJH6 registered a twofold increase in relative fluorescence for 3-OHPA in comparison with PA (7389 and 4281, respectively). However, this was not the case for B. cenocepacia K56-2 harboring pJH7 or pJH8. These strains showed slightly higher relative fluorescence in response to PA in comparison with 3-OHPA (11206 and 9966 units, respectively for pJH7 and 4454 and 3502 units, respectively for pJH8). For all carbon sources tested, pJH7 registered a phenotype characterized with strongest levels of relative fluorescence compared to the other reporter strains, as described previously (Hamlin et al., 2009).
Figure 3. B. cenocepacia K56-2 reporter activity assays. PA pathway induction in B. cenocepacia K56-2 (wild type) measured as relative fluorescence at 12 h. Strains were grown in M9 minimal media with PA, 3-OHPA, or 4-OHPA (plus 0.1% glycerol) or 0.2% glycerol. pJH7, paaA promoter reporter; pJH8, paaH promoter reporter; pJH6, paaZ promoter reporter; pJH1, positive control; pJH2, negative control. Error bars represent the SD of three replicates.
Induction of PA Degradation by 3-OHPA is Dependent on the paak1 Gene
To shed light on the role of paaK1 gene in the induction of PA degradation by 3-OHPA, we constructed a B. cenocepacia K56-2 paaK1 deletion mutant, IAI1 and introduced the reporter plasmid pJH7 in this strain. Next, we compared the relative fluorescence of the reporter system in the paaK1 genetic background to the one in the wild type upon induction by 3-OHPA. Figure 4 shows an increase in relative fluorescence in the presence of PA in both wild type and IAI1 reporter strains. This is expected due to the presence of at least one functioning paaK gene. It is probable then, that the PaaK2 enzyme is able to synthesize PA-CoA, which is necessary to activate the paa promoters (Yudistira et al., 2011). Nevertheless, compared to K56-2/pJH7, IAI/pJH7 exhibited 10% less fluorescence possibly due to the presence of two functioning ligases in the wild type variant compared to one in the mutant. K56-2/pJH7 registered induction of the PA degradative pathway in 3-OHPA, (but not 4-OHPA) converse to IAI1/pJH7, which did not for either carbon source. We can therefore deduce that induction of the PA degradative pathway by 3-OHPA is dependent on the presence of PaaK1 and not PaaK2.
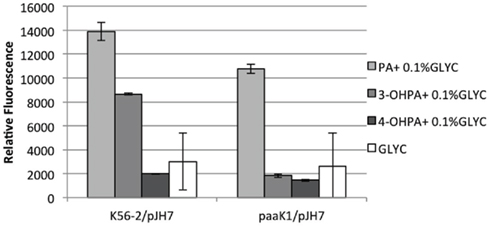
Figure 4. paaK1 reporter activity assays. paaA promoter induction in B. cenocepacia K56-2 (wild type) and B. cenocepacia deletion mutant IAI1 (paaK1) measured as relative fluorescence at 12 h. Strains were grown in M9 minimal media with PA, 3-OHPA, or 4-OHPA (plus 0.1% glycerol) or 0.2% glycerol. pJH7, paaA promoter reporter plasmid. Error bars represent the SD of three replicates.
PaaK1 does not Play a Role in B. cenocepacia Pathogenicity
Previous preliminary work done by Law (2009) in our laboratory had suggested that a paaK1 deletion mutant in B. cenocepacia K56-2 (RJL1) resulted in an attenuated phenotype in C. elegans. However, introduction of a paaK1 gene in trans in this strain did not restore the phenotype to wild type levels (data not shown) suggesting that the attenuated phenotype was not related to paaK1 but to a secondary site mutation.
In order to finally elucidate the role of PaaK1 in pathogenicity, we conducted slow killing assays with the paaA strain as a control of attenuated pathogenicity (Figure 5A) and with the newly constructed deletion mutant IAI1 (Figure 5B). Survival of the worms exposed to the wild type strain, K56-2, registered a sharp drop by more than 75% on day 2 of the assay with complete loss of survival at day 4. For worms infected with IAI1, a 50% drop in survival occurred on day 1 followed by a wild type-like decline in survival in subsequent days. Overall, we found no significant difference (P value = 0.063) in pathogenicity between wild type and paaK1 mutant. This finding supports a strong likelihood that PaaK1 may not play a role in PA pathway related killing.
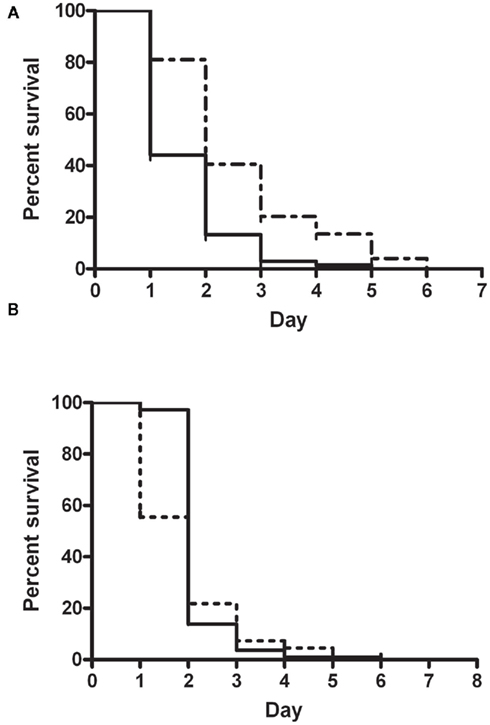
Figure 5. Nematode slow killing assays. Kaplan–Meier Survival curve comparing killing phenotypes of (A) B. cenocepacia wild type strain K56-2 (n = 68; solid line) and insertional mutant, paaA (n = 74; broken line); P value < 0.0001 (S) and (B) B. cenocepacia wild type strain K56-2 (n = 113; solid line) and paaK1 deletion mutant, IAI1 (n = 110; broken line); P value = 0.063 (NS).
Discussion
The premise that PaaK1, the activating enzyme of the PA pathway, can also bind 3-OHPA and 4-OHPA (Law and Boulanger, 2011) suggests that these phenolic acids may also be involved in inducing the pathway particularly 3-OHPA which binds PaaK1 with a lower Km than 4-OHPA. We supported this assertion with the results from our reporter activity assays showing PA pathway induction in minimal media containing 3-OHPA. Of particularly note is the observation that the strain harboring reporter plasmid (pJH6), featuring the paaZ promoter, displays double the strength of induction in 3-OHPA than in PA unlike the other two reporter plasmids (pJH7 and pJH8), which register only slightly stronger induction in PA. This is noteworthy because paaK1 is downstream of and under the control of the paaZ promoter. More interestingly, the gene upstream of paaK1 is the paaI gene, which encodes a hotdog thioesterase PaaI that preferentially binds 3-OHPA-CoA and 4-OHPA-CoA as substrates compared to PA-CoA (Song and Ko, 2007).
In toto, our results show that 3-OHPA induces the PA degradative pathway whereas 4-OHPA does not and that this induction is dependent on the presence of a functional paaK1 gene. Thus, we provide in vivo evidence for the role of 3-OHPA as a substrate of PaaK1 in Burkholderia cenocepacia, while 4-OHPA is likely not physiologically relevant as a substrate of PaaK1.
The basis of this study was preliminary data showing that a deletion of the paaK1 gene in B. cenocepacia RJL1 resulted in an attenuated killing phenotype in the nematode host model (Law, 2009). According to this finding, Law and Boulanger (2011) suggested that the more active, and substrate-versatile PaaK1 might be more beneficial during infection. However, our results show that PaaK1 (and possibly 3-OHPA as a PA degradation inducer) does not play a significant role in pathogenicity.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Ann Karen Brassinga for helping to improve our C. elegans protocols and for critically reading this manuscript and Marie Speare, Liaison Librarian for Microbiology and Chemistry at University of Manitoba, for help with referencing style. This study was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) to Silvia T. Cardona.
References
Aballay, A., and Ausubel, F. M. (2002). Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr. Opin. Microbiol. 5, 97–101.
Aubert, D. F., Flannagan, R. S., and Valvano, M. A. (2008). A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect. Immun. 76, 1979–1991.
Cohen, S. N., Chang, A. C., and Hsu, L. (1972). Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U.S.A. 69, 2110–2114.
Cormack, B. P., Valdivia, R. H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38.
Craig, F. F., Coote, J. G., Parton, R., Freer, J. H., and Gilmour, N. J. (1989). A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135, 2885–2890.
El-Said Mohamed, M. (2000). Biochemical and molecular characterization of phenylacetate-coenzyme A ligase, an enzyme catalyzing the first step in aerobic metabolism of phenylacetic acid in Azoarcus evansii. J. Bacteriol. 182, 286–294.
Figurski, D. H., and Helinski, D. R. (1979). Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652.
Flannagan, R. S., Linn, T., and Valvano, M. A. (2008). A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10, 1652–1660.
Hamlin, J. N., Bloodworth, R. A., and Cardona, S. T. (2009). Regulation of phenylacetic acid degradation genes of Burkholderia cenocepacia K56-2. BMC Microbiol. 9, 222. doi:10.1186/1471-2180-9-222
Izard, N. C., Hächler, H., Grehn, M., and Kayser, F. H. (1992). Ribotyping of coagulase-negative staphylococci with special emphasis on intraspecific typing of Staphylococcus epidermidis. J. Clin. Microbiol. 30, 817–823.
Law, A., and Boulanger, M. J. (2011). Defining a structural and kinetic rationale for paralogous copies of phenylacetate-CoA ligases from the cystic fibrosis pathogen Burkholderia cenocepacia J2315. J. Biol. Chem. 286, 15577–15585.
Law, R. J. (2009). The Relationship between Phenylacetic acid Catabolism and Pathogenicity of Burkholderia cenocepacia K56-2 in the Caenorhabditis elegans Host Model. Dissertation, University of Manitoba.
Law, R. J., Hamlin, J. N. R., Sivro, A., McCorrister, S. J., Cardama, G. A., and Cardona, S. T. (2008). A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J. Bacteriol. 190, 7209–7218.
Luengo, J. M., Garcia, J. L., and Olivera, E. R. (2001). The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39, 1434–1442.
Mahenthiralingam, E., Coenye, T., Chung, J. W., Speert, D. P., Govan, J. R., Taylor, P., and Vandamme, P. (2000). Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38, 910–913.
Martinez-Blanco, H., Reglero, A., Rodriguez-Aparicio, L. B., and Luengo, J. M. (1990). Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J. Biol. Chem. 265, 7084–7090.
Méndez, V., Agulló, L., González, M., and Seeger, M. (2011). The homogentisate and homoprotocatechuate central pathways are involved in 3- and 4-hydroxyphenylacetate degradation by Burkholderia xenovorans LB400. PLoS ONE 6, e17583. doi:10.1371/journal.pone.0017583
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583.
Song, J. H., and Ko, K. S. (2007). Detection of essential genes in Streptococcus pneumoniae using bioinformatics and allelic replacement mutagenesis. Methods Mol. Biol. 416, 401–408.
Teufel, R., Mascaraque, V., Ismail, W., Voss, M., Perera, J., Eisenreich, W., Haehnel, W., and Fuchs, G. (2010). Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U.S.A. 107, 14390–14395.
Yudistira, H., McClarty, L., Bloodworth, R. A., Hammond, S. A., Butcher, H., Mark, B. L., and Cardona, S. T. (2011). Phenylalanine induces Burkholderia cenocepacia phenylacetic acid catabolism through degradation to phenylacetyl-CoA in synthetic cystic fibrosis sputum medium. Microb. Pathog. 51, 183–196.
Keywords: Burkholderia cepacia, 3-hydroxyphenylacetic, phenylacetic acid, PA-CoA, CoA ligase, gene reporter
Citation: Imolorhe IA and Cardona ST (2011) 3-Hydroxyphenylacetic acid induces the Burkholderia cenocepacia phenylacetic acid degradation pathway – toward understanding the contribution of aromatic catabolism to pathogenesis. Front. Cell. Inf. Microbio. 1:14. doi: 10.3389/fcimb.2011.00014
Received: 31 August 2011;
Accepted: 20 November 2011;
Published online: 14 December 2011.
Edited by:
Joanna Goldberg, University of Virginia Health System, USAReviewed by:
Vincent Joseph Starai, The University of Georgia, USAGeorg Fuchs, University of Freiburg, Germany
Douglas Gordon Storey, University of Calgary, Canada
Copyright: © 2011 Imolorhe and Cardona. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Silvia T. Cardona, Department of Microbiology, University of Manitoba, Buller Building, Room 418, Winnipeg, MB, Canada R3T 2N2. e-mail:Y2FyZG9uYUBjYy51bWFuaXRvYmEuY2E=
 Ijeme A. Imolorhe
Ijeme A. Imolorhe