Abstract
The formation of biofilms by Staphylococcus aureus and Staphylococcus epidermidis is an important aspect of many staphylococcal infections, most notably endocarditis, osteomyelitis and infections associated with indwelling medical devices. The major constituents of staphylococcal biofilms are polysaccharides, such as poly N-acetyl glucosamine (PIA/PNAG), cell surface and secreted bacterial proteins, and extracellular DNA. The exact composition of biofilms often varies considerably between different strains of staphylococci and between different sites of infection by the same strain. PIA/PNAG is synthesized by the products of four genes, icaADBC, that are encoded in a single operon. A fifth gene, icaR, is a negative regulator of icaADBC. Expression of icaADBC is tightly regulated, but can often be induced in vitro by growing staphylococci in the presence of high salt, high glucose, or ethanol. Regulation of icaADBC is complex and numerous regulatory factors have been implicated in control of icaADBC. Many of these are well known global transcriptional regulatory factors like SarA and sigmaB, whereas other regulators, such as IcaR, seem to affect expression of relatively few genes. Here, we will summarize how various regulatory factors affect the production of PIA/PNAG in staphylococci.
Staphylococcus aureus is a major nosocomial and community acquired pathogen causing a diverse array of infections ranging from superficial infections of the skin and mucosa to highly invasive and potentially lethal infections. Perhaps not surprisingly, S. aureus encodes a large array of virulence factors that enable the organism to infect different tissues within its host. Despite the potential of S. aureus to cause disease, the organism asymptomatically colonizes approximately one third of the adult population with the nares being the most common niche (Iwase et al., 2010). A number of S. aureus infections are associated with the formation of biofilms, including endocarditis, septic arthritis and osteomyelitis, and infections associated with implanted medical devices such as prosthetic heart valves, skeletal prostheses, and catheters. The formation of biofilms not only facilitates bacterial colonization of a host, but also provides resistance to antibiotics and the host immune system. Biofilms can also serve as foci of infection for metastatic spread of bacteria and release of toxins into the bloodstream (Gotz, 2002; Fitzpatrick et al., 2005; O'Gara, 2007; Otto, 2008; Boles and Horswill, 2011).
S. epidermidis is a human commensal and an opportunistic pathogen capable of causing disease in immunocompromised individuals. In healthy individuals, S. epidermidis typically causes infections only if introduced into subcutaneous tissues by some form of trauma especially in the presence of foreign bodies. S. epidermidis is also a common cause of biofilm-associated infections. Because it is present on skin and mucosal surfaces, the organism has the potential to be introduced into deeper tissues during the implantation of medical devices. S. epidermidis is much less virulent than S. aureus and the capacity to form biofilms is considered the most important virulence trait of the organism (O'Gara, 2007; Otto, 2009; Fey and Olson, 2010).
Formation and composition of biofilms
Bacterial biofilms are complex communities of organisms containing layers of bacteria within a glycoccalyx. A mature biofilm contains specific three dimensional structures referred to as towers or mushrooms separated by fluid filled channels (Costerton et al., 1999; Stoodley et al., 2002). The formation of biofilms occurs in multiple stages, initial attachment, microcolony and macrocolony formation, and detachment or disassembly (Otto, 2008; Fey and Olson, 2010; Boles and Horswill, 2011). The initial attachment of staphylococci is often mediated by cell surface proteins that bind to mammalian extracellular matrix/plasma proteins such as fibrinogen, fibronectin, collagen, vitronectin, or laminin. Collectively these bacterial proteins are frequently referred to as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) (Patti et al., 1994). Staphylococci have dozens of MSCRAMMs which can be covalently or noncovalently bound to the cell surface. Many staphylococci are capable of binding directly to plastic surfaces and researchers have often measured attachment to plastic as an in vitro model of attachment in vivo. Implanted medical devices are usually coated by plasma proteins, however, possibly obviating a need to bind directly to abiotic surfaces (Tsang et al., 2008; Beenken et al., 2010).
Non-MSCRAMM, surface localized proteins can also mediate attachment. The major cell wall autolysins, AtlA, and AtlE, (Heilmann et al., 1997; Houston et al., 2011) promote binding to hydrophobic surfaces for initial attachment and possibly biofilm accumulation (Heilmann et al., 1997; Hirschhausen et al., 2010; Houston et al., 2011). Teichoic (TA) and lipoteichoic (LTA) acids can also aid in initial attachment (Qin et al., 2007). TAs and LTAs are common components of the cell envelopes of Gram-positive bacteria that often play a role in bacterial adherence to host cells. S. aureus strains with a mutation in the dlt operon or tagO, both involved in TA/LTA synthesis, exhibit reduced binding to polystyrene and glass or other abiotic surfaces (Gross et al., 2001; Vergara-Irigaray et al., 2008).
The formation of microcolonies and biofilm accumulation require mechanisms for intercellular aggregation of bacteria. The production of exopolysaccharides is a common and important factor in biofilm accumulation. In both S. aureus and S. epidermidis, the major exopolysaccharide produced is termed polysaccharide intercellular adhesion (PIA), also known as poly-N-acetyl-glucosamine (PNAG) (Mack et al., 1996). PIA/PNAG, which has a net positive charge, may promote intercellular interactions by binding to the negatively charged surfaces of bacterial cells. PIA/PNAG may or may not interact with TAs and LTAs to foster intercellular interactions (O'Gara, 2007; Vergara-Irigaray et al., 2008). PNAG has been found to be essential for biofilm formation by many strains of S. aureus and S. epidermidis. In addition to PIA/PNAG, biofilms contain bacterial proteins and DNA as essential components with the ratios of these components being variable.
A number of staphylococcal strains exhibit PIA/PNAG-independent biofilm formation. In the latter strains, secreted proteins and extracellular DNA appear to substitute for PIA/PNAG. The fibronectin-fibrinogen binding MSCRAMMs, FnbA, and FnbB (O'Neill et al., 2008) the IgG binding Spa protein (Merino et al., 2009), and the adhesin SasG (Geoghegan et al., 2010) all contribute to biofilm formation in S. aureus. The biofilm-associated protein (Bap) encoded by some S. aureus strains that cause bovine mastitis, appears absent in human isolates. Bap is important for both initial attachment and biofilm accumulation (Cucarella et al., 2004). The accumulation-associated protein (Aap) is commonly found in S.epidermidis isolates. Aap forms fibrillar structures on the cell surface and may facilitate intercellular interactions (Rohde et al., 2005). Bhp, a homolog of Bap, is another accumulation-associated protein produced by some S. epidermidis strains (Cucarella et al., 2001). At least some strains of staphylococci appear able to switch from PIA-dependent to PIA/PNAG-independent biofilm formation (Hennig et al., 2007).
A number of soluble extracellular proteins can also affect biofilm formation. Beta toxin is a S. aureus sphingomyelinase capable of lysing sheep erythrocytes under the appropriate assay conditions, and killing lymphocytes (Marshall et al., 2000; Huseby et al., 2007). Huseby et al. (2010) demonstrated that beta toxin promotes biofilm accumulation by forming crosslinks with itself in the presence of extracellular DNA, producing an insoluble nucleoprotein matrix. Alpha hemolysin, a small pore forming toxin, has also been shown to be required for biofilm production in the 8325-4 strain of S. aureus. Inactivation of the hla gene, encoding alpha hemolysin, resulted in a strain capable of initial attachment but incapable of the cell to cell interactions required for biofilm accumulation (Caiazza and O'Toole, 2003).
Detachment of biofilms is widely regarded as a mechanism for bacterial spread in an infected host, probably initiated by changes in pH, nutrient depletion, and waste accumulation within the biofilm. Detachment involves the degradation of the biofilm matrix by proteases and nucleases (Otto, 2008; Beenken et al., 2010; Boles and Horswill, 2011). Degradation of PNAG apparently does not occur in staphylococcal biofilms, as staphylococci do not seem to have a PNAG hydrolytic enzyme (Otto, 2009). A group of small amphiphilic α-helical peptides, known as phenol-soluble modulins seem to function as surfactants, disrupting cell-to-cell interactions within the biofilm. It has been proposed that phenol-soluble modulins may play a more important role in detachment of biofilms than do proteases (Otto, 2008; Boles and Horswill, 2011).
Regulation of PIA/PNAG production and icaADBC expression
Production of PIA/PNAG is tightly regulated and, at least in vitro, seems to occur primarily at the transcriptional level. Although the signals controlling PIA/PNAG production in vivo are not clearly defined, a number of environmental conditions affect production in vitro. High temperature, anaerobiosis, high osmolarity, glucose, and ethanol can all induce PIA/PNAG production although there is strain-to strain variation in regard to which conditions result in increased PIA/PNAG production. Subinhibitory concentrations of specific antibiotics, including tetracycline, gentamicin, and the streptogramins, quinopristin and dalfopristin, can also increase PNAG (Rachid et al., 2000b; Nuryastuti et al., 2011).
PIA/PNAG is synthesized by four proteins, IcaA, IcaD, IcaB, and IcaC, encoded by the ica operon (Figure 1A). The transmembrane proteins, IcaA, and IcaD, work in concert as an N-acetylglucosaminyltransferase to synthesize PNAG oligomers that are less than 20 residues in length. IcaC is a membrane protein believed to transport IcaAD-synthesized oligomers across the cell membrane. IcaC is also involved in the formation of long oligomers of PIA/PNAG. The IcaB protein, which can be found in association with the bacterial cell surface and culture supernatants, deacetylates PIA/PNAG resulting in a positively charged polymer. Deacetylation is believed to promote the interaction of PIA/PNAG with the negatively charged cell surface.
Figure 1
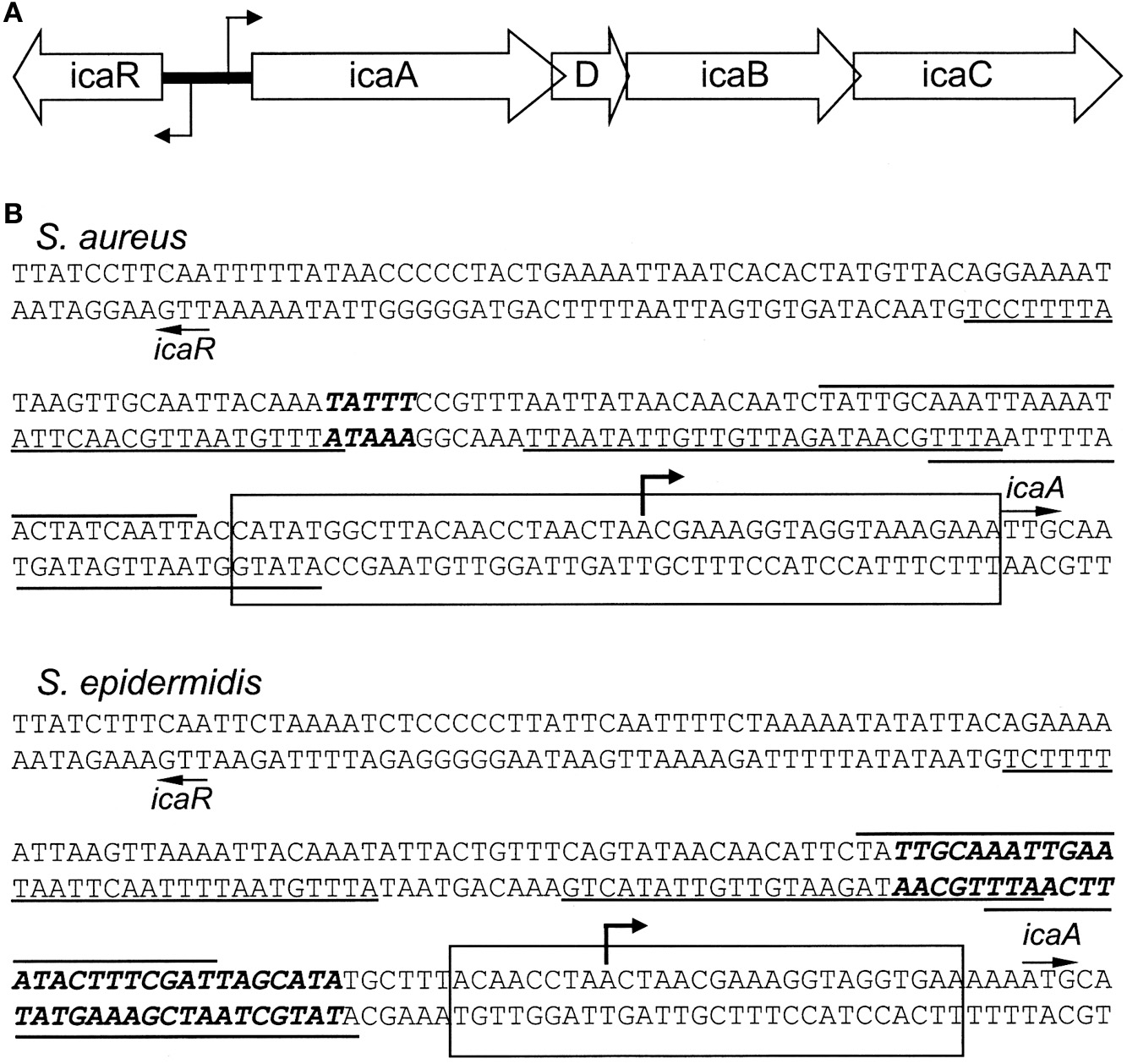
(A) Organization of the intercellular adhesion (ica) operon in S. aureus and S. epidermidis. The bent arrows indicate the transcriptional start sites. (B) The icaR-to-icaA intergenic regions. The start sites of icaR and icaA are indicated by arrows. The putative binding sites for the SarA protein are underlined or overlined (Tormo et al., 2005). The bent arrow indicates the icaADBC transcription start site determined for S. epidermidis RP62A; (Heilmann et al., 1996b; Mack et al., 2000). Top: Sequence of the S. aureus NCTC 8325 ica locus (Gillaspy et al., 2006). The bold, italicized nucleotides indicate base pairs deleted in S. aureus MN8m that resulted in PIA/PNAG overproduction (Jefferson et al., 2003). The rectangle indicates the region bound by IcaR in DNase I protection experiments (Jefferson et al., 2003). Bottom: Sequence of S. epidermidis RP62A ica locus (Heilmann et al., 1996b). The bold, italicized nucleotides represent the highest affinity TcaR binding site (Chang et al., 2010). The rectangle indicates the IcaR binding site (Jeng et al., 2008).
The ica locus was originally identified by screening a library of S. epidermidis transposon insertion mutants for isolates with defects in biofilm formation. A mutant with an insertion in the ica locus exhibited defects in biofilm formation, intercellular aggregation, and PIA synthesis (Heilmann et al., 1996a,b). The transposon insertion could be complemented by a plasmid carrying the icaADBC genes. Moreover, the icaADBC plasmid could confer a biofilm positive, aggregation, and PIA producing phenotype on the heterologous host species, S. carnosus (Heilmann et al., 1996b; McKenney et al., 1998). Not every isolate of S. epidermidis carries the ica locus, but ica genes seem to be more common in nosocomial and invasive isolates than in skin isolates (Rogers et al., 2008; Fey and Olson, 2010). It has been argued that carriage of icaADBC may actually be detrimental for the survival of skin isolates of S. epidermidis (Rogers et al., 2008).
Subsequent to its discovery in S. epidermidis, the ica locus was found in S. aureus and appears to be present in nearly all isolates of the latter (Cramton et al., 1999). The S. aureusica genes are organized as in S. epidermidis and the encoded proteins share from 79% to 89% similarity and 62–78% identity. The cloned S. aureus genes could confer biofilm production, PNAG synthesis and N-acetylglucosaminyltransferase activity to an S. aureusica mutant and to S. carnosus.
The regulation of icaADBC expression is mediated by a number of regulatory factors (Table 1). These factors include global regulatory proteins such as SarA and σB, as well as factors like IcaR and TcaR which seem to regulate relatively few genes. Some factors regulate icaADBC expression directly (e.g., IcaR) whereas regulation by other proteins seems to be indirect (e.g., σB). Notably, mechanisms governing ica expression often vary not only between different species of staphylococci, but also between different strains of the same species. It is also worth noting that different laboratories induce ica expression and measure biofilm formation under a variety of different conditions. For example, most studies utilize standard 96 well microtiter plates to assay biofilm production in vitro. Some researchers use untreated plates whereas others coat their plates with serum prior to adding bacteria to the wells. Proponents of the latter method argue that serum coating more closely approximates in vivo conditions. These variations in assay conditions can complicate comparisons of results from different laboratories. Moreover, no in vitro conditions or animal model can replicate the environment of an infected human host. Despite these limitations, the studies cited below have established the importance of the ica genes in biofilm formation by staphylococci. Here, we will summarize what is known about various regulators of the ica locus.
Table 1
| Regulatory factor | Overall effect on icaADBC transcription | Probable mechanism | Reference |
|---|---|---|---|
| Sa IcaR | Negative | Direct binding to icaADBC promoter | Conlon et al. (2002) |
| Se IcaR | Jefferson et al. (2003) | ||
| Sa SarA | Positive | Direct binding to icaADBC promoter | Valle et al. (2003) |
| Se SarA | Tormo et al. (2005) | ||
| Se SarX | Positive | Direct binding to icaADBC promoter | Rowe et al. (2010) |
| Se SarZ | Positive | Unknown | Wang et al. (2008) |
| Sa TcaR | Negative | Direct binding to icaADBC promoter | Jefferson et al. (2004) |
| Se TcaR | Chang et al. (2010) | ||
| Sa σB | Variable depending on study | Indirect | See text |
| Se σB | |||
| Sa Rbf | Positive | Repression of icaR through a hypothetical regulator | Cue et al. (2009) |
| Se Rbf | Rowe (2010) | ||
| Se LuxS | Negative | Unknown | Xu et al. (2006) |
| Sa Spx | Negative | Upregulation of icaR | Pamp et al. (2006) |
| Se Spx | Negative | Unknown, but not through icaR | Wang et al. (2010) |
| Sa SrrAB | Positive | Direct binding to icaADBC promoter | Ulrich et al. (2007) |
| Se Ygs | Positive | Unknown | Wang et al. (2011) |
| Sa GdpS | Positive | Unknown | Holland et al. (2008) |
| Se Gdps | Tu Quoc et al. (2007) | ||
| Sa CcpA | Positive | Indirect, see text | Seidl et al. (2008) |
| Se CcpA | Sadykov et al. (2011) |
Regulatory proteins affecting icaADBC expression in staphylococci.
Regulatory factors affecting ica expression
IcaR
The ica locus contains the same five known genes in both S. aureus and S. epidermidis (Figure 1A). IcaADBC are encoded by a single transcript that initiates 29 bp upstream of the icaA start codon in S. epidermidis strain RP62A (Accession number U43366) (Heilmann et al., 1996b; Mack et al., 2000). The fifth gene, icaR, is transcribed divergently from the other ica genes. The start codons of icaA and icaR are separated by approximately 163 bp of DNA (Conlon et al., 2002).
IcaR is an approximately 22 kDa protein of the TetR family of transcriptional regulators, (Conlon et al., 2002; Jeng et al., 2008). Amino acid sequence alignments first suggested that icaR might encode a transcriptional regulator (Ziebuhr et al., 1999; Rachid et al., 2000a). Conlon et al. (2002) inactivated icaR in strain CSF41498, a clinical isolate of S. epidermidis that produced a weak biofilm when grown in BHI broth at 37°C. Insertional inactivation of icaR significantly increased icaA expression, indicating that icaR may function as a repressor of icaADBC. Transcription of the icaR gene was unaffected in icaR mutants indicating that icaR is not autoregulated, a trait that is conserved in S. aureus (Jefferson et al., 2003). In CSF41498, transcription of icaA was inducible by high NaCl, high glucose, or ethanol, whereas icaR expression was reduced by ethanol, but was unaffected by NaCl or glucose. Expression of icaA was unaffected by ethanol in CSF41498 icaR::Ermr, but was increased by NaCl or glucose in the same strain. It was concluded that regulation of ica expression by ethanol was icaR-dependent, whereas regulation by NaCl-glucose was icaR-independent. IcaR provided in trans was able to complement the icaR::Ermr mutation and repress transcription of icaA. Complementation of icaR was modulated by ethanol.
The icaR gene has also been shown to be a negative regulator of icaADBC in S. aureus. The predicted S. aureus 8325-4 IcaR protein has 65.6% identity and 90.9% similarity with IcaR from S. epidermidis RP62A (Heilmann et al., 1996b; Mack et al., 2000). Jefferson et al. (2003) demonstrated that IcaR can bind to a DNA region immediately 5′ to icaA and that a short nucleotide sequence in the icaA-icaR intergenic region could affect expression of icaADBC. A spontaneous mutant of S. aureus MN8, called MN8m, was isolated which exhibited constitutive hyperproduction of PIA/PNAG and enhanced biofilm formation (McKenney et al., 1999; Jefferson et al., 2003). The mutation in MN8m responsible for hyperproduction of PIA/PNAG was determined to be a 5 bp (TATTT) deletion within the icaA-icaR intergenic region (Figure 1B). The 5 bp deletion increased icaADBC transcription but had no effect on icaR expression. Substitution of the TATTT sequence with ATAAA resulted in the same phenotype as the original deletion.
DNase I protection experiments did show that recombinant IcaR protected a 42 bp region upstream of the icaA gene (Figure 1B) (Jefferson et al., 2003). The TATTT sequence, however, played no role in IcaR binding. The latter result seemed to indicate that another DNA binding protein utilizes the TATTT sequence to regulate icaADBC expression. It was postulated that deletion of the TATTT sequence might alter an intrinsic bend in ica DNA, but this possibility was not directly tested. Interestingly, the TATTT sequence lies between two putative binding sites for SarA protein which is a positive activator of icaADBC (Tormo et al., 2005). Thus it is possible that the 5 bp deletion affects SarA binding to the ica intergenic region. Precisely how the deletion affects icaADBC transcription has not yet been determined, however.
Subsequent work by these same authors demonstrated that icaR is a repressor of ica transcription and that the protein is functional in MN8 and MN8m (Jefferson et al., 2004). Deletion of icaR increased icaADBC expression by 100-fold and PNAG production by 10-fold. PIA/PNAG production by strain MN8 requires exogenous glucose, whereas MN8 icaR overproduces PNAG in the absence of glucose, leading the authors to conclude that in S. aureus, upregulation of PNAG by glucose is at least in part due to alleviation of IcaR-mediated repression of icaADBC. Recall that glucose reportedly did not affect icaR expression in S. epidermidis CSF41498 (Conlon et al., 2002). Deletion of the icaR gene in S. aureus 8325-4 also resulted in hyperexpression of icaADBC and increased PNAG production (Cue et al., 2009). As described below, several different regulators appear to affect icaADBC expression by repression or upregulation of icaR.
The crystal structure of IcaR from S. epidermidis was recently reported (Jeng et al., 2008). Like other proteins in the TetR family, IcaR is primarily α-helical. Three α helices form an N-terminal DNA binding domain with α helices two and three forming a helix-turn-helix motif. The native IcaR protein exists primarily as a homodimer. Dimerization is mediated by a large helix bundle formed by α helices 4–9 near the C-terminus of IcaR. Electromobility shift assays (EMSAs) performed with recombinant IcaR revealed a 28 bp ica operator centered 17/18 nucleotides 5′ to the icaA start codon. This agrees well with the localization of the IcaR binding site in the S. aureus operator (Jefferson et al., 2003). Two IcaR dimers bind cooperatively to the operator with the two dimers binding to opposite faces of the DNA. It was proposed that the binding of one icaR dimer may alter the DNA conformation thereby allowing binding of a second dimer (Jeng et al., 2008).
The ability of certain antibiotics to inhibit IcaR binding to DNA was also investigated, in part, because DNA binding by some members of the TetR family has been shown to be inhibited by certain antibiotics. Tetracycline, which can induce ica expression in S. epidermidis, did not affect DNA binding by IcaR, however, two aminoglycoside antibiotics, streptomycin and gentamicin, were shown to inhibit IcaR binding to DNA, presumably by directly altering the conformation of the protein (Rachid et al., 2000b; Jeng et al., 2008).
The Sar protein family
The Sar (staphylococcal accessory regulator) family of proteins is composed of at least 11 different proteins some of which (e.g., SarA and SarR, SarX) are found in both S. aureus and S. epidermidis. The various Sar proteins have been categorized as fitting into one of three subfamilies (Cheung et al., 2008). The first subfamily, which includes SarA and SarX, are generally small, about 15 kDa, basic proteins with a single DNA binding domain that probably bind DNA as homodimers. Proteins in the second subfamily have two homologous DNA binding domains and likely bind DNA as monomers. The final subfamily is comprised of proteins that seem closely related to MarR protein (Liu et al., 2001, 2006; Manna and Cheung, 2001; Li et al., 2003; Ballal and Manna, 2009).
SarA.
SarA is arguably the most well studied of the various Sar proteins. The SarA protein is a 124 amino acid residue protein that has a calculated PI of around 9. SarA is a dimeric protein with a central core region comprised of a winged-helix DNA binding domain where the helix-turn-helix domain recognizes the major groove and the winged region interacts with the minor groove. Dimerization appears to be mediated by a conserved α-helical region near the N-terminus of the protein. Structure function studies have suggested that multiple SarA dimers may bind a single target sequence and that the association of multiple dimers is fostered by Ca++ binding. It has been proposed that a SarA homodimer can bind a target site and recruit additional homodimers to the site (Liu et al., 2006).
The SarA protein can function as either an activator or repressor of transcription (Bayer et al., 1996; Beenken et al., 2003; Tormo et al., 2005; Oscarsson et al., 2006). SarA is a global regulatory protein affecting expression of many genes in S. aureus including many genes involved in pathogenesis thus making SarA a major virulence factor. Among the genes under positive regulation by SarA is the agr (accessory gene regulator) locus. The agr locus contains two divergent promoters that produce two transcripts. One transcript, RNAII, encodes four proteins that constitute a quorum sensing system. The second transcript, RNAIII, is a regulatory RNA and also encodes δ-toxin. The agr system in general is involved in the switch from synthesis of cell surface proteins during exponential growth to synthesis of toxins and degradative proteins in the postexponential to stationary growth phases. Expression of agr can reduce the capacity of S. aureus to form biofilms (Vuong et al., 2000; Cafiso et al., 2007; Coelho et al., 2008; Beenken et al., 2010). Due to the fact that SarA is a positive activator of agr, and because agr can repress biofilm formation, it might be anticipated that mutation of sarA would increase biofilm. It appears this is not the case, however, as sarA mutants have a reduced capacity to form biofilms (Valle et al., 2003; Handke et al., 2007; Tsang et al., 2008; Beenken et al., 2010). This is perhaps not surprising in that SarA affects biofilm formation by affecting expression of multiple targets. For example, mutation of SarA results in increased expression of proteinases and nucleases, both of which have a negative impact on biofilm (Beenken et al., 2010). SarA also appears to enhance biofilm formation more directly by increasing ica expression (Valle et al., 2003).
Valle et al. (2003) screened a library of Tn917 insertion mutants to identify biofilm-defective mutants of S. aureus. Some of the mutants had Tn917 insertions within sarA. Subsequently, they deleted or insertionally inactivated sarA in four unrelated S. aureus strains, all of the mutants failed to produce a biofilm. Deletion of agr in the wild type strains did not affect biofilm formation, indicating that the effect of the sarA mutations was independent of agr. A series of experiments was performed to determine if increased protease production accounted for the phenotype of the sarA mutants. The authors concluded that enhanced proteolysis could not account for the biofilm deficient phenotype. This is somewhat at odds with some of the studies described above. In the Valle et al. (2003) study, sarA mutations did significantly decrease ica transcription and PIA/PNAG production, but this study did not determine whether SarA can bind to the ica promoter. Subsequent studies, however, did establish that SarA could bind the ica promoter (Tormo et al., 2005).
S. epidermidis also encodes SarA which is 84% identical to SarA from S. aureus (Fluckiger et al., 2005). Tormo et al. (2005) deleted sarA in two different clinical isolates of S. epidermidis and reported that both mutants were deficient in biofilm formation. PIA/PNAG production and transcription of icaA were both significantly reduced but not abolished in the sarA deleted strains. Transcription of icaR was unaffected by deletion of sarA. As has been observed for S. aureus, proteinase production was increased in sarA mutants, which likely contributed to the mutants' inabilities to form biofilms. Recombinant SarA protein was shown to bind with comparable affinities to the icaR-icaA promoter regions of S. aureus and S. epidermidis. The icaR-icaA promoter regions of both species contain multiple SarA consensus binding sites (Figure 1B).
It is important to note that protein phosphorylation/dephosphorylation plays an important role in biofilm formation and SarA activity. Two S. aureus serine/threonine kinases, Stk1/PknB and SA0077, can both phosphorylate SarA. Phosphorylation by Stk1/PknB seems to increase the affinity of SarA for some promoters and decrease it's affinity for other promoters (Didier et al., 2010). In S. epidermidis, Stk is required for biofilm formation and plays a major role in icaADBC expression (Liu et al., 2011). The sarA, agr, and sigB genes are all regulated by Stk1/PknB in S. aureus (Tamber et al., 2010), thus phosphorylation of SarA and possibly other regulatory proteins, seems likely to significantly affect ica expression.
SarX.
SarX was first discovered in S. aureus by virtue of its homology with other Sar family proteins (Manna and Cheung, 2006). It is a 119 amino acid protein representative of the single domain class of Sar proteins. Manna and Cheung (2006) demonstrated that the SarX protein of S. aureus RN6390 binds to the agr promoter, repressing synthesis of RNAII and RNAIII, thereby indirectly repressing exoprotein synthesis. Subsequently, Rowe et al. (2010) demonstrated that SarX from S. epidermidis strain CSF41498 bound to its cognate agr promoter and repressed agr transcription.
Reportedly, sarX did not affect biofilm formation in S. aureus RN6390, but did promote biofilm formation by S. epidermidis CSF41498 in an ica-dependent manner (Rowe, 2010, C.Y.L. and J.P.O., unpublished data). Expression of S. epidermidissarX on a multicopy plasmid not only complemented a sarX mutation, but also enhanced biofilm formation by the wild type strain (Rowe et al., 2010). Expression of sarX increased icaA transcription as well as PNAG production, but expression of icaR was unaffected by sarX. A purified maltose binding-SarX fusion protein bound to ica promoter DNA generating a ladder of protein-DNA complexes. A similar pattern was previously shown for SarX binding to the agr promoter region (Manna and Cheung, 2006). To account for the observed laddering, it has been suggested that either the ica and agr promoters each contain multiple SarX binding sites or that SarX oligomers form on bound DNA. Thus sarX appears to directly affect icaADBC transcription, at least in S. epidermidis CSF41498. Modulation of agr expression by SarX is apparently inadequate to affect biofilm formation in S. aureus.
Interestingly, sarX is located immediately downstream from the rbf gene in both S. aureus and S. epidermidis (see below) and is under positive regulation by rbf in S. aureus strains 8325-4 and UAMS-1. The rbf gene has been shown to upregulate biofilm formation and icaADBC expression and may do so, at least in part, by increasing sarX transcription (Lim et al., 2004; Cue et al., 2009).
SarZ.
SarZ has also been shown to affect ica expression in S. epidermidis 1457. Wang et al. (2008) utilized a novel biofilm screening assay, involving separate and consecutive screens, to isolate biofilm-defective mutants with Tn917 insertions. The screen resulted in the isolation of two mutants both of which had Tn917 inserted in sarZ. The mutants had defects in primary attachment as well as biofilm accumulation. PIA/PNAG production and icaADBC expression were both reduced in sarZ mutants. Moreover, sarZ was shown to contribute to virulence in both rat and mouse models of biofilm-associated infection. Microarray studies revealed that the sarZ regulon is comprised of at least 80 genes thus decreases in ica expression may not completely account for the biofilm negative phenotype of sarZ mutants. As an example, three genes encoding proteinases were all upregulated in the sarZ mutant (Wang et al., 2008). Increased proteinase activity seems likely to account, in part, for the mutant phenotype.
TcaR
TcaR is a member of the MarR family of transcription factors and is encoded by both S. aureus and S. epidermidis. A role for TcaR in ica expression was first revealed by Jefferson et al. (2004) who used DNA affinity chromatography to identify S. aureus proteins capable of binding to a DNA fragment containing the icaA-icaR promoter region. Topoisomerase IV, SarA and DNA-binding protein II were also recovered in the same experiment. Purified TcaR did not produce a distinct footprint with ica DNA, however, and produced a ladder of complexes in EMSA experiments. These results suggested that either there are multiple TcaR binding sites in ica DNA or that TcaR oligomerizes once bound to DNA (Jefferson et al., 2004). Of these, the former possibility seems the most probable (Chang et al., 2010).
Northern analysis indicated that inactivation of tcaR increased transcription of icaADBC fivefold in three different strains of S. aureus, indicating that tcaR is a negative regulator of ica. Surprisingly, deletion of tcaR did not affect bacterial binding to polystyrene nor did it affect PIA/PNAG production, whereas, deletion of icaR affected both attachment and PIA/PNAG. When coupled with an icaR deletion mutation, deletion of tcaR increased ica expression fivefold over the single icaR mutant and 500-fold over the wild type. The icaR mutation alone augmented icaA transcription approximately 100-fold. Bacterial adherence and PNAG production were also increased in an icaRtcaR double mutant, relative to an icaR single mutant (Jefferson et al., 2004).
TcaR binds DNA as a dimer and displays non-cooperative binding to the ica promoter region (Chang et al., 2010). TcaR from S. epidermidis binds to at least one of three consecutive 33 bp pseudopalindromic sequences located immediately upstream of icaA. TcaR seems to have the highest affinity for the most proximal binding site which is only a few bps away from the icaR binding site. It is not known whether TcaR and IcaR can bind simultaneously to ica DNA. A number of antibiotics were shown to inhibit DNA binding by TcaR, this was believed to be due to antibiotic induced changes in the conformation of the TcaR DNA binding domain. Three aminoglycoside antibiotics, kanamycin, gentamicin, and streptomycin, were shown to inhibit DNA binding by TcaR and to promote biofilm formation by S. epidermidis RP62A. However other antibiotics, such as β-lactams, disrupted DNA binding but had no significant effect on biofilm formation. It was proposed that low concentrations of some antibiotics, by virtue of their abilities to disrupt DNA binding by TcaR and IcaR, may derepress icaADBC which, in turn, would confer increased antibiotic resistance due to biofilm formation.
σB
σB is an alternative sigma factor found in staphylococci and other Gram-positive bacteria that plays a key role in the response to environmental stress (Conlon et al., 2004). In S. aureus, σB is activated by signal transduction in response to high temperature, high osmolarity, antibiotics, or extreme pH. Transcription of the sigB operon is driven by three distinct promoters. The first is a σA-dependent promoter that produces a transcript encoding rsbUVW and sigB. The second is a σB-dependent promoter that drives synthesis of a shorter transcript lacking rsbU. The third promoter is the mazEF promoter which drives transcription of a 3.7 kb mRNA encoding the toxin-antitoxin pair, MazEF, as well as RsbUVW-σB. Transcription from pmazEF is enhanced by heat shock and exposure to tetracycline or erythromycin (Donegan and Cheung, 2009). Full expression of sigB appears to require all three promoters.
The activity of σB is controlled by a network of kinases and phosphatases. In the absence of stress, σB is inactive due to its association with an anti-σ factor, RsbW. RsbW also functions to phosphorylate and thereby inactivate the anti-anti-σ factor RsbV. Under stress conditions, RsbU, a phosphatase, dephosphorylates RsbV. RsbV can then bind RsbW, disrupting the latter's association with σB. The released sigma factor can then associate with the core RNA polymerase (Knobloch et al., 2004).
σB has been shown to regulate in excess of 200 genes, including a number of genes involved in biofilm formation (Bischoff et al., 2004; Pane-Farre et al., 2006; Nielsen et al., 2011). Rachid et al. (2000a) reported a role for σB in S. aureus biofilm formation in a clinical isolate and the laboratory strain RN4220. In these experiments, σB was found to be required for induction of ica transcription and biofilm formation in response to high NaCl. Cerca et al. (2008) looked at icaADBC and icaR expression in σB-deleted derivatives of S. aureus strains SA113 and Newman (Cerca et al., 2008). Surprisingly, σB was found to be a positive regulator of both icaR and icaADBC. This unexpected result was proposed to be possibly due to rather weak repression of icaADBC by icaR in the strains used in the study.
As described above, Valle et al. (2003) reported that sarA was critical for ica expression and biofilm formation in multiple, unrelated S. aureus clinical isolates. Additionally, they reported that mutation of σB in the same strains had no significant effect on ica expression or biofilm. Remarkably, inactivation of σB in sarA mutant strains increased PIA/PNAG production and biofilm formation relative to the single sarA mutant strains. The latter occurred even though icaA expression in sarA-sigB double mutants was significantly less than in sarA mutants. It was proposed that σB might upregulate expression of a factor involved in turnover of PIA/PNAG.
S. epidermidis also possesses a sigB operon similar in size, organization, and function as sigB in S. aureus. S. epidermidis σB has been shown to affect biofilm production both in vivo and in vitro (Knobloch et al., 2004; Handke et al., 2007; Pintens et al., 2008). RsbU was shown to function as a negative regulator of icaADBC in S. epidermidis strains 1457 and 8400 (Knobloch et al., 2004). This effect was shown to be due to a reduction of σB expression that, in turn, increased icaR expression. Increased icaR expression decreased icaADBC expression and biofilm. As was observed for S. aureus, high NaCl concentration did not induce biofilm in a sigB mutant. The biofilm defect in sigB mutants could be overcome, however, by growth in the presence of subinhibitory concentrations of ethanol. The effect of ethanol was due to σB-independent repression of icaR. The σB defect could also be overcome by multiple copies of icaADBC. The repression of icaR via ethanol has been speculated to involve an unknown intermediate factor. Notably, sarA was expressed in sigB mutants via the σB-independent sarA promoters, P1 and P2, thus decreased icaADBC expression was apparently not due to loss of SarA.
Upregulation of biofilm by anaerobiosis also involves σB. Anaerobic activation of icaADBC was σB-dependent and was concomitant with σB-dependent repression of icaR. σB appears to play a more important role in ica regulation under anaerobic conditions than it does under aerobic conditions, at least in S. epidermidis 1457 (Cotter et al., 2009). None of the aforementioned studies found any evidence for direct regulation of ica genes by σB in either S. aureus or S. epidermidis. This coupled with the lack of a σB consensus promoter sequence in the ica intergenic region, implies σB regulation is indirect.
The role of σB in biofilm expression is further complicated by the fact that σB can also activate ica-independent biofilm formation. σB has also been shown to function in ica-independent biofilm formation in S. aureus USA300 LAC, a CA-MRSA isolate (Lauderdale et al., 2009). In general, σB can promote biofilm by repressing the production of proteases and toxins, an effect that is manifest through decreasing expression of RNAIII as well as through positive activation of sarA. In USA300 LAC, loss of σB increased the level of RNAIII and mutation of agr restored biofilm formation in a sigB mutant. The addition of proteinase inhibitors to growth media also restored biofilm in a σB deficient strain. Thus another role of σB appears to be repression of agr that, in turn, represses the production of proteinases that are involved in the disassembly of biofilm.
Rbf
Rbf (regulator of biofilm) is a transcriptional regulatory protein found to play an important role in biofilm formation in both S. aureus 8325-4 and S. epidermidis CSF 41498 (Lim et al., 2004; Rowe, 2010). Rbf is a member of the AraC/XylS family of transcriptional regulators, a family that bears a highly conserved 100 amino acid region forming a dual, helix-turn-helix DNA binding motif. The dual helix-turn-helix is usually localized in the C-terminal region of a protein (Gallegos et al., 1997). The two helix-turn-helix domains within a monomer are believed to interact with consecutive major grooves of DNA, thus their binding sites are typically longer than they are for classical HTH proteins (Schleif, 2010). There are hundreds of different AraC/XylS-like proteins, many of which have been identified in sequence data bases by virtue of possessing the dual HTH motif (Gallegos et al., 1997). S. aureus is predicted to encode at least 6 AraC/XylS-like proteins at least two of which, Rbf and Rsp, regulate biofilm formation (Lim et al., 2004; Lei et al., 2011).
Typical AraC/XylS proteins are relatively small, 250–300 amino acids long, and many have effector binding sites in the N-terminal domains of the proteins. The effector binding sites and DNA binding regions are typically separated by a linker region. The effector binding domain often regulates the DNA binding activity (Gallegos et al., 1997). For example, DNA binding to the araBAD promoter by the AraC protein of E. coli, is affected by binding of arabinose and some other sugars. In the presence of arabinose, AraC binds to two half-sites, called I1 and I2, which lie just upstream of araBAD. In this conformation AraC can interact with RNA polymerase and promote transcription of araBAD. In the absence of arabinose, AraC binds to the I1 half-site and to a third half-site, O1, which lays 210 bp upstream of I1. In this conformation a DNA loop is formed and transcription of araBAD is repressed (Schleif, 2010).
Although Rbf protein has the conserved DNA binding motif, it is an atypical AraC-like protein. First, it is significantly larger than most other AraC proteins, approximately 716 amino acid residues in S. aureus 8325-4. Additionally, the DNA binding motif of Rbf is located near the N-terminus of the protein. Expression of rbf is likely to be complex as the promoter-regulatory region contains putative binding sites for σB and saeR (C.Y.L. unpublished). The rbf gene was originally identified by screening of a transposon insertion library of strain 8325-4 for biofilm-defective mutants (Lim et al., 2004). Loss of rbf led to a defect in biofilm formation in response to high NaCl and glucose, but did not affect ethanol-induced biofilm. Inactivation of rbf did not affect initial attachment of staphylococci to polystyrene but did severely inhibit multicellular aggregation. Extensive macrocellular clumping was observed when Rbf from either S. aureus or S. epidermidis was expressed from a multicopy plasmid (Lim et al., 2004; Rowe, 2010). Additionally, multicopy rbf increased biofilm formation in S. aureus via increasing intercellular aggregation. The protein was also found to play a significant role in biofilm formation in vivo (Luong et al., 2009).
In a subsequent study, microarray experiments were performed to determine the rbf-regulon in a clinical isolate of S. aureus strain UAMS-1 (Cue et al., 2009). Expression of Rbf from a multicopy plasmid was found to increase expression of six genes and reduce expression of 35 genes. A number of the rbf-regulated genes could, potentially at least, affect biofilm formation. The tagB gene, which encodes teichoic acid biosynthesis gene B, is upregulated by rbf. Four genes likely to affect cell lysis and DNA release in biofilms, lytSR, lrgAB are all repressed by rbf. The lytSR genes encode a two component system that upregulates lrgAB. The lrgA gene product function as an antiholin that can inhibit cell lysis and the release of DNA into the environment (Sadykov and Bayles, 2011). Thus by inhibiting lytSR expression, Rbf would be predicted to increase the level of extracellular DNA in biofilm. SarX, a protein observed to enhance icaADBC expression in S. epidermidis (Rowe et al., 2010) is regulated by rbf in S. aureus UAMS-1. The gene encoding KdpD, a histidine kinase affecting luxS expression, is also regulated by rbf (Cue et al., 2009).
The microarray experiments performed with UAMS-1 produced two surprising results. The first was that deletion of rbf had no effect on gene expression. This was apparently due to the fact that the rbf gene in UAMS-1 has a 2 bp insertion near the N-terminal coding region. Thus, although rbf is transcribed, no Rbf would be synthesized. The second surprise was that multi-copy rbf increased icaADBC transcription about five to sixfold. This was surprising due to the fact that Lim et al. (2004) reported that expression of an icaA-xylE fusion was unaffected by inactivation of rbf in 8325-4. Real time quantitative PCR experiments, as well as PIA/PNAG, assays confirmed that rbf does positively regulate icaADBC transcription in 8325-4 and UAMS-1. The reason Rbf failed to increase transcription of the icaA-XylE fusion is unclear at this time.
The microarray experiments also revealed that rbf can reduce icaR transcription, a finding confirmed by qRT-PCR experiments. Thus, it appears that rbf activates icaADBC expression, at least in part, via inhibiting expression of icaR. Most AraC/XylS proteins act as activators of transcription and some, such as AraC, also act as repressors (Gallegos et al., 1997). It is possible that rbf affects ica by direct activation of icaADBC, in addition to repression of icaR. Experiments to test for direct binding of Rbf to the ica promoter have yielded only negative results, suggesting that rbf regulation may be indirect (Cue et al., 2009; Rowe, 2010). Recombinant Rbf also failed to bind to the promoter regions of other genes, (i.e., sarA, sarX, sarZ, spx, and srrA) that regulate ica (Rowe, 2010).
Rbf in S. epidermidis CSF41498 is 46% homologous and 65% similar to Rbf from S. aureus with the highest similarity being in the putative DNA-binding domains (Rowe et al., 2010). Expression of the CSF414498 rbf gene in S. aureus increased macroscopic cell clumping, biofilm formation and icaA expression. Site-specific mutagenesis of the rbf DNA-binding domain, resulted in the loss of Rbf -induced cell clumping and biofilm. As was observed for 8325-4, mutation of rbf in CSF41498 reduced biofilm formation in response to high NaCl and glucose, but not in response to ethanol. Initial attachment by S. epidermidis was unaffected by rbf. Interestingly, the cloned rbf gene did not fully complement the biofilm defect of CSF41498 rbf, leading Rowe (2010) to propose that overexpression of Rbf may have negative effects on biofilm formation. It has been demonstrated that Rbf could bind specifically to the sarR promoter of S. epidermidis and that rbf had a modest effect on sarR transcription in stationary phase cultures. Furthermore, it was found that SarR protein could bind specifically to the ica promoter. While these data support a model where Rbf could regulate ica expression through SarR, mutation of sarR had no significant effect on icaADBC expression or biofilm formation, thus the significance of the DNA binding studies is unclear at present (Rowe, 2010).
LuxS
LuxS is part of a quorum sensing system found in numerous species of Gram-negative and Gram-positive bacteria, including S. aureus and S. epidermidis (Doherty et al., 2006; Li et al., 2008). LuxS is required for the synthesis of autoinducer 2 (AI-2) a family of small, diffusible compounds that can penetrate cell membranes. Unlike other quorum-sensing systems luxS is not species-specific, rather, AI-2 produced by one species can affect gene expression in multiple bacterial species.
Several groups have determined that luxS is a negative regulator of biofilm formation in S. epidermidis (Kong et al., 2006; Xu et al., 2006; Li et al., 2008). AI-2 can be found in culture supernatants of S. epidermidis and is secreted optimally during log and early stationary phases of growth. AI-2 present in culture supernatants of S. epidermidis can activate expression of AI-2 responsive genes in E. coli DH5α (Xu et al., 2006).
Deletion of lux in S. epidermidis strain 1457 was found to enhance biofilm formation. This effect seemed largely due to changes in ica expression as transcription of icaC increased over fourfold and PIA/PNAG synthesis was enhanced about threefold in the luxS strain. Expression of ica returned to the wild type level in a complemented strain and when exogenous AI-2 was added to cultures of the luxS mutant. Whether icaR expression was affected by deletion of luxS was not reported. The luxS mutant was found to more virulent than the wild type strain in a rat intravascular, central-venous-catheter-associated model, presumably due to increased PIA/PNAG production by the mutant strain (Xu et al., 2006).
While luxS appears to be an important ica regulator in S. epidermidis, the role of luxS in S. aureus biofilm formation is less clear. Doherty et al. (2006) found no role for luxS in the expression of virulence traits in strain RN6390, including biofilm formation. LuxS is reportedly inactivated by serine/threonine phosphorylation in S. aureus, but the effects on biofilm formation have not been reported (Cluzel et al., 2010). It has been reported, however, that a furanone derived from a marine algae could promote biofilm formation by S. aureus strain Newman and S. epidermidis strain 1457. In S. epidermidis, enhanced biofilm formation correlated with reduced luxS expression and increased PIA/PNAG production. It was not reported whether the furanone affected biofilm formation by the same mechanism in S. aureus (Kuehl et al., 2009).
Spx
The Spx protein is a global transcriptional regulator that is itself subject to regulation by the energy-dependent ClpXP proteinase complex in a number of Gram-positive bacteria. Spx appears to function in S. aureus in much the same manner as Spx in Bacillus subtilis. In the latter organism, Spx acts as both a transcriptional activator and a repressor. Spx binds directly to the α subunit of RNA polymerase thereby potentially blocking the interaction between RNA polymerase and other transcription factors. Spx can also directly affect promoter recognition by RNA polymerase (Nakano et al., 2010). Spx is a redox sensitive regulator that can activate genes, such as those encoding thioredoxin and thioredoxin reductase, important in the cellular response to oxidative stress. The N-terminal region of Spx contains two cysteine residues that form an intramolecular disulfide bond under thiol-oxidizing conditions. Oxidized Spx can associate with RNA polymerase and direct transcription of select B. subtilis genes (Nakano et al., 2003, 2005).
In S. aureus 8325-4, Spx plays an important role in response to stress as a spx mutant is hypersensitive to high and low temperatures, high osmolarity, and oxidative stress. Transcription of txrB, the gene encoding thioredoxin reductase, requires spx in strain 8325-4. ClpP and Spx also affect biofilm formation. In a spx mutant, initial attachment, cell aggregation and biofilm formation were all enhanced. Transcription of icaADBC was increased in spx mutants while icaR transcription was decreased. Thus the normal role of Spx with regard to ica expression is to repress icaADBC via enhancement of icaR transcription. Precisely how Spx enhances icaR expression is unclear. It should be noted that, while an S. aureusspx null mutant could be constructed, the mutant exhibited growth defects even in the absence of stress (Pamp et al., 2006).
In S. epidermidis, clpP mutants reportedly accumulate high levels of Spx and exhibit defects in initial attachment and biofilm formation (Wang et al., 2010). The spx gene may be essential in S. epidermidis as knockout of the gene was not achieved in S. epidermidis 1457. However, a knockdown plasmid construct carrying an antisense spx RNA could promote biofilm formation, icaADBC transcription and PIA production. Unlike the case with S. aureus, spx did not affect expression of icaR nor did spx affect initial attachment. S. aureus and S. epidermidis also differ in that the S. epidermidis Spx does not regulate trxB (Wang et al., 2010).
SrrAB
SrrAB is a two-component regulatory system responsive to anaerobiosis (Ulrich et al., 2007). An srrAB mutant of S. aureus strain SA113 exhibited downregulation of icaA transcription and PIA/PNAG expression under anaerobic condition. SrrAB did not affect icaR expression. Phosphorylated SrrA protein bound to a 100 bp DNA segment immediately upstream of the icaADBC promoter. SrrAB is important for anaerobic growth and protection of staphylococci from killing by neutrophils under anaerobic conditions (Ulrich et al., 2007).
Ygs
Ygs is a general stress response protein identified by transposon mutagenesis of S. epidermidis strain 1457. Strains with mutation of ygs show decreased survival upon exposure to a variety of stressful conditions including high temperature, high osmolarity, pH, and ethanol exposure. Loss of ygs disrupted biofilm formation but not primary attachment. The biofilm defect seemed to be due to decreased icaADBC expression and PIA/PNAG production in the mutant strains, but expression of icaR was unaffected by Ygs. Ygs also played a significant role in biofilm formation in vivo and pathogenesis in rats (Wang et al., 2011).
GdpS
Holland et al. (2008) reported that a novel staphylococcal protein, GdpS (GGDEF domain protein from staphylococcus), plays a role in ica expression in S. epidermidis CSF41498. These authors identified gdpS by searching data bases for proteins with homology to diguanylate cyclases that bear a conserved GGDEF domain. These enzymes are responsible for the synthesis of cyclic-dimeric-GMP in many bacterial species. c-di-GMP allosterically activates enzymes involved in exopolymer synthesis. Staphylococci have only a single GGDEF domain protein, GdpS, which reportedly lacks cyclase activity (Holland et al., 2008). Despite this, gdpS was shown to enhance biofilm formation, icaADBC expression and PIA/PNAG production in media supplemented with NaCl. Expression of icaR was unaffected by mutation of gdpS. S. aureus also encodes gdpS which is important in icaADBC expression (Tu Quoc et al., 2007), but like the S. epidermidis protein, lacks cyclase activity. It is unclear precisely how gdpS regulates ica expression.
CcpA, the TCA cycle and ica expression
Vuong et al. (2005) noted that many of the same conditions that induce PIA/PNAG production, i.e., high osmolarity, high temperature, ethanol, etc., are also known to inhibit the tricarboxylic acid, or TCA, cycle. They proposed that ica expression may respond to the metabolic state of the cell via alterations in the levels of TCA cycle intermediates. They did show that PIA/PNAG production could be upregulated by exposing cultures of S. epidermidis to fluorocitrate, an inhibitor of the TCA cycle enzyme aconitase (citrate (isocitrate) hydroxylase). Subsequently, the same group inactivated the gene coding aconitase (acnA) in S. epidermidis 1457 to study its effect on biofilm and ica expression (Sadykov et al., 2008). TCA activity was blocked and icaADBC expression was increased by the acnA mutation. Inactivation of the TCA cycle increased the intracellular concentration of the immediate biosynthetic precursor of PIA/PNAG, UDP-N-acetylglucosamine. Moreover, transcripts of genes encoding enzymes for the synthesis of UDP-N-acetylglucosamine from glucose-6-phosphate were all increased. Thus a major effect of acnA inactivation is a rerouting of carbon into N-acetylglucosamine biosynthesis.
The level of icaADBC transcript increased dramatically as a result of acnA inactivation. Surprisingly, the expression of icaR, sarA, and sigB were all increased in the acnA mutant. To determine whether any of these regulators affected PIA/PNAG production in response to TCA cycle disruption, the effects of fluorocitrate on PIA/PNAG was determined for icaR, sigB, and sarA mutants. Fluorocitrate increased PIA/PNAG in both the icaR and sigB mutant strains, indicating that neither of these regulators responds to TCA-induced metabolic changes. Fluorocitrate did not significantly affect PIA/PNAG production in a sarA mutant, however, making SarA a candidate for a TCA cycle-responsive regulator (Sadykov et al., 2008).
Sadykov et al. (2008) noted, however, that the aconitase mutant accumulated higher levels of branched chain amino acids than the wild type strain. This result suggested that CodY, a transcriptional regulatory protein that is responsive to branched chain amino acids, could be involved in icaADBC regulation. The authors also noted that the carbon catabolite repression protein, CcpA, may respond to higher intracellular levels of fructose-6-phosphate and increase icaADBC expression. CodY and CcpA are both regulators of icaADBC in S. aureus (Majerczyk et al., 2008; Seidl et al., 2008).
As mentioned above, CcpA has been shown to be an activator of icaADBC in S. aureus (Seidl et al., 2008) and, more recently, in S. epidermidis (Sadykov et al., 2011). CcpA is the primary mediator of carbon catabolite repression in staphylococci and is known to function as either a repressor or activator of transcription. Repression of TCA cycle genes is a common response to high concentrations of glucose in culture media, a response that among Gram-positive bacteria is mediated by CcpA. The activity of CcpA is regulated by intracellular levels of glucose-6-phosphate and fructose-1,6-bisphosphate, both of which affect phosphorylation of histidine-containing protein (Hpr). Phosphorylated Hpr can complex with CcpA affecting the interaction of the latter with DNA, typically causing CcpA to act as a repressor (Fujita, 2009).
Glucose can induce biofilm formation by S. aureus strain SA113 (Seidl et al., 2008). Induction of biofilm formation by glucose is dependent upon CcpA. Deletion of ccpA in both S. aureus SA113 and DSM20231 blocked biofilm formation but not initial attachment to polystyrene. CcpA was found to affect icaA transcription but was also required for expression of cidA. The latter is a putative holin protein that contributes to the release of bacterial DNA in biofilms (Ranjit et al., 2011). Biofilms formed by a SA113 ccpA mutant were more susceptible to disruption by exogenous DNase than were biofilms formed by SA113. Transcription of other regulatory genes, sarA, arlRS, mgrA, and rbf, were unaffected by deletion of ccpA. Based in part on the work with S. epidermidis, the effect of CcpA on transcription of citZ and citB was investigated. CitB is the S. aureus homolog of AcnA while citZ encodes citrate synthase. Both citB and citZ were repressed by ccpA in strain SA113 thereby linking CcpA with TCA cycle regulation. Based upon studies with Bacillus subtilis CcpA, a putative binding site for CcpA was found upstream of the citZ open reading frame. No such site was found upstream of citB. These findings suggested that CcpA may regulate citZ directly and citB indirectly (Seidl et al., 2008). Thus CcpA appears to play an important role in regulating biofilm in the presence of high glucose. The effect of CcpA on ica is likely indirect and a consequence of downregulation of the TCA cycle, in part, through repression of citB and citZ (Seidl et al., 2008).
CcpA has also been found to coordinate the TCA cycle and biofilm formation in S. epidermidis 1457 (Sadykov et al., 2008, 2011). Deletion of ccpA resulted in increases in aconitase and citrate synthase activity as well as acnA and citZ transcripts. CcpA proved critical for biofilm production in glucose-containing media. Deletion of acnA resulted in the upregulation of genes involved in PIA/PNAG synthesis including icaD, glmU (encoding glucosamine-1-phosphate N-acetyltransferase), pfkA (6-phosphofruktokinase), and glnA (glutamine synthetase). The increased expression of icaD and pfkA were ccpA-dependent and were manifest in 2 and 6 h cultures. Expression of glmU was similarly regulated except that expression was only evident in 6 h cultures. The acnA mutation increased the level of the glnA transcript after 6 h, but not after 2 h of incubation and was independent of CcpA. CcpA binding sites were located 5′ to both the acnA and glmU genes. The authors argued that CcpA both regulates TCA cycle activity and conveys signals associated with the TCA cycle to PIA/PNAG biosynthetic genes (Sadykov et al., 2011).
Summary
It is obvious from the long list of factors that affect ica expression that regulation is extremely complex and multifactorial. This seems especially true for S. aureus as many isolates do not produce PIA/PNAG in vitro even though nearly all S. aureus isolates encode icaADBC. Moreover, the relative importance of the various factors seems to differ considerably between different strains as well as between species. In some instances, researchers have identified regulatory proteins that act directly on ica DNA. However in other instances regulators appear to act indirectly, via affecting the expression or activity of hypothetical proteins that, in turn, interact with ica DNA. Even in cases where proteins have been shown to bind ica DNA, precisely how these factors regulate transcription is not completely clear. It seems highly likely that multiple regulatory factors are co-expressed during infection and we have virtually no information on how these factors may interact with DNA and/or other macromolecules to regulate gene expression. Moreover, it remains unknown how PIA/PNAG synthesis is induced during infection.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Statements
Acknowledgments
This work was supported by grants AI37027 and AI067857 from the National Institute of Allergy and Infectious Diseases.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
BallalA.MannaA. C. (2009). Expression of the sarA family of genes in different strains of Staphylococcus aureus. Microbiology155, 2342–2352. 10.1099/mic.0.027417-0
2
BayerM. G.HeinrichsJ. H.CheungA. L. (1996). The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178, 4563–4570.
3
BeenkenK. E.BlevinsJ. S.SmeltzerM. S. (2003). Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71, 4206–4211. 10.1128/IAI.71.7.4206-4211.2003
4
BeenkenK. E.MrakL. N.GriffinL. M.ZielinskaA. K.ShawL. N.RiceK. C.HorswillA. R.BaylesK. W.SmeltzerM. S. (2010). Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One5:e10790. 10.1371/journal.pone.0010790
5
BischoffM.DunmanP.KormanecJ.MacapagalD.MurphyE.MountsW.Berger-BachiB.ProjanS. (2004). Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186, 4085–4099. 10.1128/JB.186.13.4085-4099.2004
6
BolesB. R.HorswillA. R. (2011). Staphylococcal biofilm disassembly. Trends Microbiol. 19, 449–455. 10.1016/j.tim.2011.06.004
7
CafisoV.BertuccioT.SantagatiM.DemelioV.SpinaD.NicolettiG.StefaniS. (2007). agr-Genotyping and transcriptional analysis of biofilm-producing Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 51, 220–227. 10.1111/j.1574-695X.2007.00298.x
8
CaiazzaN. C.O'TooleG. A. (2003). Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 185, 3214–3217. 10.1128/JB.185.10.3214-3217.2003
9
CercaN.BrooksJ. L.JeffersonK. K. (2008). Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190, 6530–6533. 10.1128/JB.00482-08
10
ChangY. M.JengW. Y.KoT. P.YehY. J.ChenC. K.WangA. H. (2010). Structural study of TcaR and its complexes with multiple antibiotics from Staphylococcus epidermidis. Proc. Natl. Acad. Sci. U.S.A. 107, 8617–8622. 10.1073/pnas.0913302107
11
CheungA. L.NishinaK. A.TrotondaM. P.TamberS. (2008). The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40, 355–361. 10.1016/j.biocel.2007.10.032
12
CluzelM. E.Zanella-CleonI.CozzoneA. J.FuttererK.DuclosB.MolleV. (2010). The Staphylococcus aureus autoinducer-2 synthase LuxS is regulated by Ser/Thr phosphorylation. J. Bacteriol. 192, 6295–6301. 10.1128/JB.00853-10
13
CoelhoL. R.SouzaR. R.FerreiraF. A.GuimaraesM. A.Ferreira-CarvalhoB. T.FigueiredoA. M. (2008). agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiology154, 3480–3490. 10.1099/mic.0.2007/016014-0
14
ConlonK. M.HumphreysH.O'GaraJ. P. (2002). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184, 4400–4408. 10.1128/JB.184.16.4400-4408.2002
15
ConlonK. M.HumphreysH.O'GaraJ. P. (2004). Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186, 6208–6219. 10.1128/JB.186.18.6208-6219.2004
16
CostertonJ. W.StewartP. S.GreenbergE. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science284, 1318–1322.
17
CotterJ. J.O'GaraJ. P.MackD.CaseyE. (2009). Oxygen-mediated regulation of biofilm development is controlled by the alternative sigma factor sigma(B) in Staphylococcus epidermidis. Appl. Environ. Microbiol. 75, 261–264. 10.1128/AEM.00261-08
18
CramtonS. E.GerkeC.SchnellN. F.NicholsW. W.GotzF. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433.
19
CucarellaC.SolanoC.ValleJ.AmorenaB.LasaI.PenadesJ. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. 10.1128/JB.183.9.2888-2896.2001
20
CucarellaC.TormoM. A.UbedaC.TrotondaM. P.MonzonM.PerisC.AmorenaB.LasaI.PenadesJ. R. (2004). Role of biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 72, 2177–2185. 10.1128/IAI.72.4.2177-2185.2004
21
CueD.LeiM. G.LuongT. T.KuechenmeisterL.DunmanP. M.O'DonnellS.RoweS.O'GaraJ. P.LeeC. Y. (2009). Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J. Bacteriol. 191, 6363–6373. 10.1128/JB.00913-09
22
DidierJ. P.CozzoneA. J.DuclosB. (2010). Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol. Lett. 306, 30–36. 10.1111/j.1574-6968.2010.01930.x
23
DohertyN.HoldenM. T.QaziS. N.WilliamsP.WinzerK. (2006). Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J. Bacteriol. 188, 2885–2897. 10.1128/JB.188.8.2885-2897.2006
24
DoneganN. P.CheungA. L. (2009). Regulation of the mazEF toxin-antitoxin module in Staphylococcus aureus and its impact on sigB expression. J. Bacteriol. 191, 2795–2805. 10.1128/JB.01713-08
25
FeyP. D.OlsonM. E. (2010). Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5, 917–933. 10.2217/fmb.10.56
26
FitzpatrickF.HumphreysH.O'GaraJ. P. (2005). The genetics of staphylococcal biofilm formation–will a greater understanding of pathogenesis lead to better management of device-related infection?Clin. Microbiol. Infect. 11, 967–973. 10.1111/j.1469-0691.2005.01274.x
27
FluckigerU.UlrichM.SteinhuberA.DoringG.MackD.LandmannR.GoerkeC.WolzC. (2005). Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73, 1811–1819. 10.1128/IAI.73.3.1811-1819.2005
28
FujitaY. (2009). Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci. Biotechnol. Biochem. 73, 245–259. 10.1271/bbb.80479
29
GallegosM. T.SchleifR.BairochA.HofmannK.RamosJ. L. (1997). Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61, 393–410.
30
GeogheganJ. A.CorriganR. M.GruszkaD. T.SpezialeP.O'GaraJ. P.PottsJ. R.FosterT. J. (2010). Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192, 5663–5673. 10.1128/JB.00628-10
31
GillaspyA. F.WorrelV.OrvisJ.RoeB. A.DyerD. W.IandoloJ. J. (2006). “The Staphylococcus aureus NCTC 8325 genome,” in Gram-Positive Pathogens 2nd edn, ed FischettiV. A. (Washington, DC: ASM Press, American Society for Microbiology), 381–412.
32
GotzF. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. 10.1046/j.1365-2958.2002.02827.x
33
GrossM.CramtonS. E.GotzF.PeschelA. (2001). Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69, 3423–3426. 10.1128/IAI.69.5.3423-3426.2001
34
HandkeL. D.SlaterS. R.ConlonK. M.O'DonnellS. T.OlsonM. E.BryantK. A.RuppM. E.O'GaraJ. P.FeyP. D. (2007). SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 53, 82–91. 10.1139/w06-108
35
HeilmannC.GerkeC.Perdreau-RemingtonF.GotzF. (1996a). Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64, 277–282.
36
HeilmannC.SchweitzerO.GerkeC.VanittanakomN.MackD.GotzF. (1996b). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. 10.1111/j.1365-2958.1996.tb02548.x
37
HeilmannC.HussainM.PetersG.GotzF. (1997) Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024. 10.1046/j.1365-2958.1997.4101774.x
38
HennigS.Nyunt WaiS.ZiebuhrW. (2007). Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int. J. Med. Microbiol. 297, 117–122. 10.1016/j.ijmm.2006.12.001
39
HirschhausenN.SchlesierT.SchmidtM. A.GotzF.PetersG.HeilmannC. (2010). A novel staphylococcal internalization mechanism involves the major autolysin Atl and heat shock cognate protein Hsc70 as host cell receptor. Cell. Microbiol. 12, 1746–1764. 10.1111/j.1462-5822.2010.01506.x
40
HollandL. M.O'DonnellS. T.RyjenkovD. A.GomelskyL.SlaterS. R.FeyP. D.GomelskyM.O'GaraJ. P. (2008). A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J. Bacteriol. 190, 5178–5189. 10.1128/JB.00375-08
41
HoustonP.RoweS. E.PozziC.WatersE. M.O'GaraJ. P. (2011). Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79, 1153–1165. 10.1128/IAI.00364-10
42
HusebyM. J.KruseA. C.DigreJ.KohlerP. L.VockeJ. A.MannE. E.BaylesK. W.BohachG. A.SchlievertP. M.OhlendorfD. H.EarhartC. A. (2010). Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 107, 14407–14412. 10.1073/pnas.0911032107
43
HusebyM.ShiK.BrownC. K.DigreJ.MengistuF.SeoK. S.BohachG. A.SchlievertP. M.OhlendorfD. H.EarhartC. A. (2007). Structure and biological activities of beta toxin from Staphylococcus aureus. J. Bacteriol. 189, 8719–8726. 10.1128/JB.00741-07
44
IwaseT.UeharaY.ShinjiH.TajimaA.SeoH.TakadaK.AgataT.MizunoeY. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature465, 346–349. 10.1038/nature09074
45
JeffersonK. K.CramtonS. E.GotzF.PierG. B. (2003). Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48, 889–899. 10.1046/j.1365-2958.2003.03482.x
46
JeffersonK. K.PierD. B.GoldmannD. A.PierG. B. (2004). The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186, 2449–2456. 10.1128/JB.186.8.2449-2456.2004
47
JengW. Y.KoT. P.LiuC. I.GuoR. T.LiuC. L.ShrH. L.WangA. H. (2008). Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 36, 1567–1577. 10.1093/nar/gkm1176
48
KnoblochJ. K.JagerS.HorstkotteM. A.RohdeH.MackD. (2004). RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect. Immun. 72, 3838–3848. 10.1128/IAI.72.7.3838-3848.2004
49
KongK. F.VuongC.OttoM. (2006). Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296, 133–139. 10.1016/j.ijmm.2006.01.042
50
KuehlR.Al-BatainehS.GordonO.LuginbuehlR.OttoM.TextorM.LandmannR. (2009). Furanone at subinhibitory concentrations enhances staphylococcal biofilm formation by luxS repression. Antimicrob. Agents Chemother. 53, 4159–4166. 10.1128/AAC.01704-08
51
LauderdaleK. J.BolesB. R.CheungA. L.HorswillA. R. (2009). Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77, 1623–1635. 10.1128/IAI.01036-08
52
LeiM. G.CueD.RouxC. M.DunmanP. M.LeeC. Y. (2011). Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J. Bacteriol. 193, 5231–5241. 10.1128/JB.05454-11
53
LiM.VillaruzA. E.VadyvalooV.SturdevantD. E.OttoM. (2008). AI-2-dependent gene regulation in Staphylococcus epidermidis. BMC Microbiol. 8, 4. 10.1186/1471-2180-8-4
54
LiR.MannaA. C.DaiS.CheungA. L.ZhangG. (2003). Crystal structure of the SarS protein from Staphylococcus aureus. J. Bacteriol. 185, 4219–4225. 10.1128/JB.185.14.4219-4225.2003
55
LimY.JanaM.LuongT. T.LeeC. Y. (2004). Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186, 722–729. 10.1128/JB.186.3.722-729.2004
56
LiuQ.FanJ.NiuC.WangD.WangJ.WangX.VillaruzA. E.LiM.OttoM.GaoQ. (2011). The eukaryotic-type serine/threonine protein kinase Stk is required for biofilm formation and virulence in Staphylococcus epidermidis. PLoS One6:e25380. 10.1371/journal.pone.0025380
57
LiuY.MannaA.LiR.MartinW. E.MurphyR. C.CheungA. L.ZhangG. (2001). Crystal structure of the SarR protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 98, 6877–6882. 10.1073/pnas.121013398
58
LiuY.MannaA. C.PanC. H.KriksunovI. A.ThielD. J.CheungA. L.ZhangG. (2006). Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 103, 2392–2397. 10.1073/pnas.0510439103
59
LuongT. T.LeiM. G.LeeC. Y. (2009). Staphylococcus aureus Rbf activates biofilm formation in vitro and promotes virulence in a murine foreign body infection model. Infect. Immun. 77, 335–340. 10.1128/IAI.00872-08
60
MackD.FischerW.KrokotschA.LeopoldK.HartmannR.EggeH.LaufsR. (1996). The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183.
61
MackD.RohdeH.DobinskyS.RiedewaldJ.NedelmannM.KnoblochJ. K.ElsnerH. A.FeuchtH. H. (2000). Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68, 3799–3807. 10.1128/IAI.68.7.3799-3807.2000
62
MajerczykC. D.SadykovM. R.LuongT. T.LeeC.SomervilleG. A.SonensheinA. L. (2008). Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190, 2257–2265. 10.1128/JB.01545-07
63
MannaA.CheungA. L. (2001). Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69, 885–896. 10.1128/IAI.69.2.885-896.2001
64
MannaA. C.CheungA. L. (2006). Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188, 4288–4299. 10.1128/JB.00297-06
65
MarshallM. J.BohachG. A.BoehmD. F. (2000). Characterization of Staphylococcus aureus beta-toxin induced leukotoxicity. J. Nat. Toxins9, 125–138.
66
McKenneyD.HubnerJ.MullerE.WangY.GoldmannD. A.PierG. B. (1998). The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66, 4711–4720.
67
McKenneyD.PouliotK. L.WangY.MurthyV.UlrichM.DoringG.LeeJ. C.GoldmannD. A.PierG. B. (1999). Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science284, 1523–1527.
68
MerinoN.Toledo-AranaA.Vergara-IrigarayM.ValleJ.SolanoC.CalvoE.LopezJ. A.FosterT. J.PenadesJ. R.LasaI. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191, 832–843. 10.1128/JB.01222-08
69
NakanoM. M.LinA.ZuberC. S.NewberryK. J.BrennanR. G.ZuberP. (2010). Promoter recognition by a complex of Spx and the C-terminal domain of the RNA polymerase alpha subunit. PLoS One5:e8664. 10.1371/journal.pone.0008664
70
NakanoS.ErwinK. N.RalleM.ZuberP. (2005). Redox-sensitive transcriptional control by a thiol/disulphide switch in the global regulator, Spx. Mol. Microbiol. 55, 498–510. 10.1111/j.1365-2958.2004.04395.x
71
NakanoS.Kuster-SchockE.GrossmanA. D.ZuberP. (2003). Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 100, 13603–13608. 10.1073/pnas.2235180100
72
NielsenJ. S.ChristiansenM. H.BondeM.GottschalkS.FreesD.ThomsenL. E.KallipolitisB. H. (2011). Searching for small sigmaB-regulated genes in Staphylococcus aureus. Arch. Microbiol. 193, 23–34. 10.1007/s00203-010-0641-1
73
NuryastutiT.KromB. P.AmanA. T.BusscherH. J.van der MeiH. C. (2011). Ica-expression and gentamicin susceptibility of Staphylococcus epidermidis biofilm on orthopedic implant biomaterials. J. Biomed. Mater. Res. A96, 365–371. 10.1002/jbm.a.32984
74
O'GaraJ. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. 10.1111/j.1574-6968.2007.00688.x
75
O'NeillE.PozziC.HoustonP.HumphreysH.RobinsonD. A.LoughmanA.FosterT. J.O'GaraJ. P. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. 10.1128/JB.00167-08
76
OscarssonJ.KanthA.Tegmark-WisellK.ArvidsonS. (2006). SarA is a repressor of hla (alpha-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188, 8526–8533. 10.1128/JB.00866-06
77
OttoM. (2008). Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322, 207–228.
78
OttoM. (2009). Staphylococcus epidermidis–the “accidental” pathogen. Nat. Rev. Microbiol. 7, 555–567. 10.1038/nrmicro2182
79
PampS. J.FreesD.EngelmannS.HeckerM.IngmerH. (2006). Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J. Bacteriol. 188, 4861–4870. 10.1128/JB.00194-06
80
Pane-FarreJ.JonasB.ForstnerK.EngelmannS.HeckerM. (2006). The sigmaB regulon in Staphylococcus aureus and its regulation. Int. J. Med. Microbiol. 296, 237–258. 10.1016/j.ijmm.2005.11.011
81
PattiJ. M.AllenB. L.McGavinM. J.HookM. (1994). MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617. 10.1146/annurev.mi.48.100194.003101
82
PintensV.MassonetC.MerckxR.VandecasteeleS.PeetermansW. E.KnoblochJ. K.van EldereJ. (2008). The role of sigmaB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology154, 2827–2836. 10.1099/mic.0.2007/015768-0
83
QinZ.OuY.YangL.ZhuY.Tolker-NielsenT.MolinS.QuD. (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology153, 2083–2092. 10.1099/mic.0.2007/006031-0
84
RachidS.OhlsenK.WallnerU.HackerJ.HeckerM.ZiebuhrW. (2000a). Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182, 6824–6826. 10.1128/JB.182.23.6824-6826.2000
85
RachidS.OhlsenK.WitteW.HackerJ.ZiebuhrW. (2000b). Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44, 3357–3363.
86
RanjitD. K.EndresJ. L.BaylesK. W. (2011). Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J. Bacteriol. 193, 2468–2476. 10.1128/JB.01545-10
87
RogersK. L.RuppM. E.FeyP. D. (2008). The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl. Environ. Microbiol. 74, 6155–6157. 10.1128/AEM.01017-08
88
RohdeH.BurdelskiC.BartschtK.HussainM.BuckF.HorstkotteM. A.KnoblochJ. K.HeilmannC.HerrmannM.MackD. (2005). Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895. 10.1111/j.1365-2958.2005.04515.x
89
RoweS. E. (2010). Contribution of Rbf and SarX to Biofilm Regulation in Staphylococcus epidermidis. Ph.D. thesis, Dublin: University College Dublin.
90
RoweS. E.MahonV.SmithS. G.O'GaraJ. P. (2010). A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology157, 1042–1049. 10.1099/mic.0.046581-0
91
SadykovM. R.BaylesK. W. (2011). The control of death and lysis in staphylococcal biofilms: a coordination of physiological signals. Curr. Opin. Microbiol. 15, 1–5. 10.1016/j.mib.2011.12.010
92
SadykovM. R.HartmannT.MattesT. A.HiattM.JannN. J.ZhuY.LedalaN.LandmannR.HerrmannM.RohdeH.BischoffM.SomervilleG. A. (2011). CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology157, 3458–3468. 10.1099/mic.0.051243-0
93
SadykovM. R.OlsonM. E.HalouskaS.ZhuY.FeyP. D.PowersR.SomervilleG. A. (2008). Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J. Bacteriol. 190, 7621–7632. 10.1128/JB.00806-08
94
SchleifR. (2010). AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34, 779–796. 10.1111/j.1574-6976.2010.00226.x
95
SeidlK.GoerkeC.WolzC.MackD.Berger-BachiB.BischoffM. (2008). Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 76, 2044–2050. 10.1128/IAI.00035-08
96
StoodleyP.SauerK.DaviesD. G.CostertonJ. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. 10.1146/annurev.micro.56.012302.160705
97
TamberS.SchwartzmanJ.CheungA. L. (2010). Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect. Immun. 78, 3637–3646. 10.1128/IAI.00296-10
98
TormoM. A.MartiM.ValleJ.MannaA. C.CheungA. L.LasaI.PenadesJ. R. (2005). SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 187, 2348–2356. 10.1128/JB.187.7.2348-2356.2005
99
TsangL. H.CassatJ. E.ShawL. N.BeenkenK. E.SmeltzerM. S. (2008). Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One3:e3361. 10.1371/journal.pone.0003361
100
Tu QuocP. H.GenevauxP.PajunenM.SavilahtiH.GeorgopoulosC.SchrenzelJ.KelleyW. L. (2007). Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect. Immun. 75, 1079–1088. 10.1128/IAI.01143-06
101
UlrichM.BastianM.CramtonS. E.ZieglerK.PragmanA. A.BragonziA.MemmiG.WolzC.SchlievertP. M.CheungA.DoringG. (2007). The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65, 1276–1287. 10.1111/j.1365-2958.2007.05863.x
102
ValleJ.Toledo-AranaA.BerasainC.GhigoJ. M.AmorenaB.PenadesJ. R.LasaI. (2003). SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. 10.1046/j.1365-2958.2003.03493.x
103
Vergara-IrigarayM.Maira-LitranT.MerinoN.PierG. B.PenadesJ. R.LasaI. (2008). Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology154, 865–877. 10.1099/mic.0.2007/013292-0
104
VuongC.KidderJ. B.JacobsonE. R.OttoM.ProctorR. A.SomervilleG. A. (2005). Staphylococcus epidermidis polysaccharide intercellular adhesin production significantly increases during tricarboxylic acid cycle stress. J. Bacteriol. 187, 2967–2973. 10.1128/JB.187.9.2967-2973.2005
105
VuongC.SaenzH. L.GotzF.OttoM. (2000). Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182, 1688–1693. 10.1086/317606
106
WangC.FanJ.NiuC.WangC.VillaruzA. E.OttoM.GaoQ. (2010). Role of spx in biofilm formation of Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 59, 152–160. 10.1111/j.1574-695X.2010.00673.x
107
WangL.LiM.DongD.BachT. H.SturdevantD. E.VuongC.OttoM.GaoQ. (2008). SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J. Infect. Dis. 197, 1254–1262. 10.1086/586714
108
WangX.NiuC.SunG.DongD.VillaruzA. E.LiM.WangD.WangJ.OttoM.GaoQ. (2011). ygs is a novel gene that influences biofilm formation and the general stress response of Staphylococcus epidermidis. Infect. Immun. 79, 1007–1015. 10.1128/IAI.00916-10
109
XuL.LiH.VuongC.VadyvalooV.WangJ.YaoY.OttoM.GaoQ. (2006). Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74, 488–496. 10.1128/IAI.74.1.488-496.2006
110
ZiebuhrW.KrimmerV.RachidS.LossnerI.GotzF.HackerJ. (1999). A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32, 345–356. 10.1046/j.1365-2958.1999.01353.x
Summary
Keywords
Staphylococcus aureus, Staphylococcus epidermidis, biofilm, intercellular adhesion locus, ica, PIA, PNAG
Citation
Cue D, Lei MG and Lee CY (2012) Genetic regulation of the intercellular adhesion locus in staphylococci. Front. Cell. Inf. Microbio. 2:38. doi: 10.3389/fcimb.2012.00038
Received
09 January 2012
Accepted
05 March 2012
Published
26 March 2012
Volume
2 - 2012
Edited by
David Heinrichs, University of Western Ontario, Canada
Reviewed by
Motoyuki Sugai, Hiroshima University, Japan; Ambrose Cheung, Dartmouth Medical School, USA
Copyright
© 2012 Cue, Lei and Lee.
This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Chia Y. Lee, Department of Microbiology and Immunology, University of Arkansas for Medical Sciences, 4301 Markam Street, Slot 511, Little Rock, AR 72205, USA. e-mail: clee2@uams.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.