- 1Department of Microbiology and Immunology, University of Maryland School of Medicine, Institute for Genome Sciences, Baltimore, MD, USA
- 2Translational Genomics Research Institute, Flagstaff, AZ, USA
- 3Department of Microbiology and Immunology, Uniformed Services University of the Health Sciences, Bethesda, MD, USA
- 4WHO Collaborating Centre for Reference and Research on Escherichia and Klebsiella, Statens Serum Institut, Copenhagen S, Denmark
Infection by Escherichia coli and Shigella species are among the leading causes of death due to diarrheal disease in the world. Shiga toxin-producing E. coli (STEC) that do not encode the locus of enterocyte effacement (LEE-negative STEC) often possess Shiga toxin gene variants and have been isolated from humans and a variety of animal sources. In this study, we compare the genomes of nine LEE-negative STEC harboring various stx alleles with four complete reference LEE-positive STEC isolates. Compared to a representative collection of prototype E. coli and Shigella isolates representing each of the pathotypes, the whole genome phylogeny demonstrated that these isolates are diverse. Whole genome comparative analysis of the 13 genomes revealed that in addition to the absence of the LEE pathogenicity island, phage-encoded genes including non-LEE encoded effectors, were absent from all nine LEE-negative STEC genomes. Several plasmid-encoded virulence factors reportedly identified in LEE-negative STEC isolates were identified in only a subset of the nine LEE-negative isolates further confirming the diversity of this group. In combination with whole genome analysis, we characterized the lambdoid phages harboring the various stx alleles and determined their genomic insertion sites. Although the integrase gene sequence corresponded with genomic location, it was not correlated with stx variant, further highlighting the mosaic nature of these phages. The transcription of these phages in different genomic backgrounds was examined. Expression of the Shiga toxin genes, stx1 and/or stx2, as well as the Q genes, were examined with quantitative reverse transcriptase polymerase chain reaction assays. A wide range of basal and induced toxin induction was observed. Overall, this is a first significant foray into the genome space of this unexplored group of emerging and divergent pathogens.
Introduction
Shiga toxin-producing Escherichia coli (STEC) isolates can colonize the intestinal tract in animals and humans, and in humans are associated with diarrheal symptoms ranging from mild diarrhea to severe hemorrhagic colitis (Kaper et al., 2004; Manning et al., 2008). Hemolytic uremic syndrome (HUS), although arising in only a minority of colonized individuals, is a serious and sometimes fatal complication resulting from elaboration of the Shiga toxins (Stx; Karch et al., 1999; Kaper et al., 2004). Many STEC disease outbreaks have been caused by a subset of STEC isolates, Locus of Enterocyte Effacement (LEE)-positive STEC, that harbor the LEE pathogenicity island and one or more stx genes (Yoon and Hovde, 2008). These isolates have often been designated enterohemorrhagic E. coli (EHEC), but the current study will use a genomic designation of LEE-positive STEC. The genes carried in the LEE pathogenicity island encode a type III secretion system that transports effector molecules into the host cells (Kaper et al., 2004). LEE-positive O157:H7 has been responsible for the majority of STEC disease outbreaks in the United States; however, non-O157 LEE-positive STEC serogroups are prevalent in other countries and are increasingly found associated with outbreaks in the United States (Brooks et al., 2005; Johnson et al., 2006; Gould et al., 2009). Although the LEE pathogenicity island is known to be an important virulence factor, LEE-negative STEC isolates from diverse serogroups have been found to cause the same severe diarrheal symptoms and HUS (Johnson et al., 2006; Mellmann et al., 2008; Newton et al., 2009; Kappeli et al., 2011). With the exception of the recent O104:H4 outbreak that occurred in Germany (Rasko et al., 2011), non-O157 STEC isolates have received much less scrutiny at the whole genome level than their LEE-positive counterparts.
Shiga toxin, the crucial virulence factor attributed to the progression of HUS, can be identified in two major antigenic forms, Stx1 and Stx2, with Stx2 identified as the more potent form (Boerlin et al., 1999; Friedrich et al., 2002). However, stx1 and stx2 allele variants have been identified; LEE-negative STEC, in particular, have been determined to often carry these diverse toxin subtypes (Zhang et al., 2002; Burk et al., 2003; Orth et al., 2007; De Sablet et al., 2008; Slanec et al., 2009). Scant information exists on the potency of the different allelic forms, but one report concluded that both in vitro and in vivo potencies of Stx2a and Stx2d were greater than Stx2b and Stx2c (Fuller et al., 2011). In addition to the potency of the particular encoded Stx, the amount of Stx produced is thought to play a role in virulence (De Sablet et al., 2008; Neupane et al., 2011). Stx genes are encoded by lambdoid bacteriophages and enhanced levels of stx expression has been observed for some isolates in prophage inducing conditions (Zhang et al., 2000; Ritchie et al., 2003). Considerable heterogeneity in both basal and induced levels of stx2 expression has been reported among LEE-positive O157:H7 isolates (Ritchie et al., 2003; De Sablet et al., 2008; Zhang et al., 2010; Neupane et al., 2011). In comparison, less information is available regarding levels of stx expression for LEE-negative STEC isolates.
Qualitatively, lambdoid bacteriophages are composed of non-homologous DNA segments, or modules, that have been exchanged between various prophages, leading to broad genetic diversity even within single isolates (Johansen et al., 2001; Brussow et al., 2004; Casjens, 2005). For example, substantial phage sequence diversity has been noted among the 11 lambdoid prophages within the genome of the LEE-positive O157:H7 Sakai isolate (Brussow et al., 2004), and other LEE-positive O157:H7 isolates (Johansen et al., 2001; Ogura et al., 2006). Although sequence divergence of stx-encoding phages has been identified, the gene structure of the stx cassettes is less well known, and has been determined for only a few LEE-negative STEC isolates. Along with the assortment of mosaic structures, a variety of chromosomal insertion locations have been identified for stx-encoding phages in LEE-positive STEC isolates. These insertion sites include wrbA, yecE, torS/T, sbcB, yehV, argW, ssrA, and prfC (Ogura et al., 2007). Interestingly, the insertion sites of the stx phages in the genomes of the majority of LEE-negative STEC isolates are often different than those determined for LEE-positive STEC isolates, and remain largely unidentified (Garcia-Aljaro et al., 2006, 2009; Prager et al., 2011).
Although production of Shiga toxin is essential for the progression of infection to HUS, STEC utilize many other virulence mechanisms during colonization of the human intestine (Yoon and Hovde, 2008). The tight adherence of the bacterial cell to the colonic epithelium resulting from expression of the eae encoded Intimin and Tir proteins encoded by the LEE pathogenicity island is considered an important step in infection. The LEE-positive STEC also utilize other chromosomally encoded adhesins and typically express multiple fimbriae (Toma et al., 2004; Farfan and Torres, 2011). LEE-positive STEC genomes also carry genes encoding autotransporter (AT) proteins that have been associated with virulence (Wells et al., 2010). Many AT proteins expressed by pathogenic E. coli have been characterized and determined to either to function as proteases, adhesins, hemagglutinins, or to promote autoaggregation or biofilm formation (Wells et al., 2010). LEE-negative STEC isolates must utilize factors other than the Intimin/Tir complex to adhere, thus the question arises as to whether they only make use of factors already identified in LEE-positive STEC genomes or also use as yet undiscovered chromosomally encoded adherence factors. The long polar fimbrial gene cluster, designated lpfO113, was identified in the LEE-negative STEC O113:H21 isolate EH41 (Doughty et al., 2002), and subsequently identified in other LEE-negative STEC isolates, as well as some non-O157 LEE-positive STEC isolates (Doughty et al., 2002; Toma et al., 2004). Along with chromosomally encoded virulence factors, pathogenic E. coli often harbor a large virulence plasmid encoding a variety of additional virulence factors. Although there is heterogeneity between virulence plasmids carried by a particular E. coli pathotype, the plasmids display a greater level of similarity within the pathotype than between pathotypes (Johnson and Nolan, 2009). A single LEE-negative STEC O113:H21 isolate, designated EH41, harbors a virulence plasmid of ∼166 kb, designated pO113 (Newton et al., 2009). Both pO157, commonly carried by O157:H7 isolates, and pO113 carry the ehxA gene encoding enterohemolysin and an espP gene encoding a serine protease autotransporter of Enterobacteriaceae (SPATE; Newton et al., 2009; Ogura et al., 2009). The STEC autoagglutinating adhesion, encoded by saa, has been suggested to be unique to LEE-negative STEC isolates (Paton et al., 2001; Toma et al., 2004; Cergole-Novella et al., 2007; Wu et al., 2010) and is encoded on pO113. Additional genes carried on pO113, reported to be unique to LEE-negative STEC, are epeA, sab, and subAB (Paton and Paton, 2005; Cergole-Novella et al., 2007; Herold et al., 2009; Newton et al., 2009; Bugarel et al., 2010; Irino et al., 2010; Wu et al., 2010) encoding, respectively, a SPATE exhibiting protease and mucinase activity (Leyton et al., 2003), an AT family protein contributing to adherence and biofilm formation (Herold et al., 2009) and the subtilase cytotoxin; this virulence factor is an AB5 family toxin that displays cytotoxicity in Vero cell assays and is lethal to mice (Paton et al., 2004).
Multilocus sequence typing (MLST) based on housekeeping genes has demonstrated that LEE-negative STEC isolates are evolutionarily divergent (Tarr et al., 2008; Newton et al., 2009; Steyert et al., 2011). Whereas whole genome comparative analysis has been predominately focused on LEE-positive STEC (Ogura et al., 2007, 2009; Eppinger et al., 2011b). The current study focuses on a diverse set of nine LEE-negative STEC carrying various stx alleles, and includes a comparison with four complete reference LEE-positive STEC isolates. The genome-wide comparison allowed for identification of genes located outside the LEE pathogenicity island that are shared in the four LEE-positive STEC genomes, but not in the nine LEE-negative STEC, as well as virulence profile comparisons and identification of sequence regions unique to each isolate. Additionally, we characterized the stx phages in the LEE-negative STEC isolates in terms of chromosomal insertion site, genetic sequence, and structure, and levels of basal and induced stx expression. Insertion sites not previously reported for stx-encoding phages were identified. We were also able to demonstrate that in the more highly virulent of the nine isolates examined, despite carrying different stx alleles, the phages share similar Q protein sequences and genetic structure directly upstream of the stxAB genes.
Materials and Methods
Bacterial Isolates and Growth Conditions
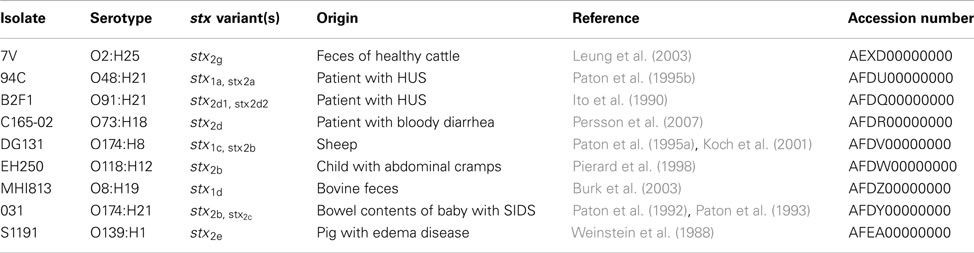
Nine LEE-negative STEC isolates were examined in this study; the isolate names, serotypes, and origins are listed in Table 1. These particular isolates were chosen to represent LEE-negative STEC with diverse serotypes and stx allele variants as part of a Genomic Sequencing Center for Infectious Diseases (GSCID) project1. Bacteria were cultured in Luria–Bertani (LB) broth at 37°C.
Genomic DNA Extraction, Sequencing, and Assembly
Genomic DNA was isolated from an overnight culture using the Sigma GenElute kit (Sigma-Aldrich) and was sequenced at the University of Maryland School of Medicine, Institute for Genome Sciences, Genome Resource Center2. The genome sequence was generated using 3 kb insert paired-end libraries on the 454 Titanium FLX (Roche) and the raw paired-end sequence reads were assembled with Celera v. 6.0 (wgs-assembler.sourceforge.net). The raw sequence reads are available for each genome sequenced in this study3.
Phylogenetic Analysis Based on Whole Genome Alignment
The sequence data for E. coli/Shigella genomes (Table A1 in Appendix) were downloaded from GenBank and combined with sequence data from the nine LEE-negative STEC isolates in this study for a total of 39 genomes. The genome sequences were aligned with Mugsy (Angiuoli and Salzberg, 2011), and the genomic core alignment, which consisted of ∼2.5 Mb, was parsed from the Mugsy output using methods described previously (Sahl et al., 2011). A phylogenetic tree was inferred using FastTree2 (Price et al., 2010) with E. fergusonii isolate 35469 as the outgroup.
Whole Genome Sequence Comparison
The sequences of the nine LEE-negative STEC genomes were compared in detail to four complete reference LEE-positive STEC genomes (Table A1 in Appendix). These reference isolates were LEE-positive O157:H7 EDL933 (Perna et al., 2001), O111:H- str. 11128 (Ogura et al., 2009), O26:H11 str. 11368 (Ogura et al., 2009), and O103:H2 str. 12009 (Ogura et al., 2009). The shared genomic sequence regions between the 13 isolates were identified using Mugsy (Angiuoli and Salzberg, 2011) as defined above. Sequence regions uniquely shared by subsets of the 13 genomes, or by a single genome, were identified from the Mugsy output using scripts from bx-python4 combined with custom python scripts. Putative unique regions were then further characterized using BLAST (Altschul et al., 1997) against the entire sequence set to verify uniqueness of the alignments.
BLAST Score Ratio Analysis
BLAST score ratio analysis of selected virulence factors was performed as previously described (Rasko et al., 2005). BLAST score ratio (BSR) analysis identifies the level of relatedness between peptide sequences by dividing the protein query BLAST score by the reference BLAST score. The normalized BSR values were visualized using the MultiExperiment Viewer (Saeed et al., 2003).
PCR Screens for Genes of Interest
Genomic DNA from two collections of E. coli isolates was screened by PCR for the presence of genes of interest. These collections consisted of 73 isolates from the environmental E. coli ECOR set (all stx-negative, Ochman and Selander, 1984) and the diarrheagenic DECA set containing 79 isolates5. The gDNA was interrogated for the genes saa, perC1, and a gene coding for a hypothetical protein (ECO103_2361 from O103:H2 isolate 12009) using primer pairs saa1, perC1, and hyp, respectively (Table 2). These primers were designed to anneal to conserved regions of the genes after examining MUSCLE alignments for regions with no polymorphism. In addition, the LEE-negative STEC isolate 87-1714 was included in the PCR screen as a control (Tarr et al., 2008; Newton et al., 2009; Steyert et al., 2011). Each 20 μL reaction included 30 cycles consisting of 95°C for 30 s, 53°C for 30 s, and 72°C for 40 s. The E. coli K12 isolate MG1655 was employed as a negative control, and STEC O48:H21 94C and LEE-positive O157:H7 EDL933 were used as positive controls for saa and the other two genes, respectively.
Shiga Toxin Containing Phage Sequences and Insertion Sites
The insertion sites of the phages carrying the Shiga toxin genes were bioinformatically determined for each isolate. The stx genes were located in the assembled contigs and the adjacent sequence surrounding the stx genes was extracted and subjected to coding sequence (CDS) analysis6. The phage integrase gene and genes adjacent to the integrase gene were identified using BLASTp where possible. The gene adjacent to the integrase was designated as the phage insertion site. The stx phage sequences were compared using Mauve (Darling et al., 2010). In some cases contigs were bioinformatically linked where appropriate to obtain complete phage sequences. Although the integration site was determined for all stx phages, the 3′ end of the phage could not be conclusively identified in three cases.
Integrase, Q, and Shiga Toxin Gene Phylogeny
Phylogenetic analysis was performed on stx gene sequences extracted from the LEE-negative STEC genomes and the four reference LEE-positive genomes. Q genes carried by the stx phages were identified by BLASTp and were aligned with MUSCLE (Edgar, 2004), to the Q gene sequence identified in the STEC EDL933 isolate stx1- and stx2-encoding phages. Integrase gene sequences were identified from reference genomes and the LEE-negative STEC genomes in this study. Sequence surrounding BLAST alignments was extracted and integration sites of insertion elements were determined as described above in an iterative process to provide the most complete dataset. For analysis of each of the stx, Q, and integrase gene phylogenies, the sequences were aligned using MUSCLE (Edgar, 2004) and a phylogeny was inferred with FastTree (Price et al., 2009).
Mitomycin C Phage Induction
Overnight cultures of each STEC isolate were diluted 1:500 into fresh LB broth and grown to an OD600 of ∼0.35, then divided into separate cultures of equal volume. Mitomycin C at a final concentration of 0.5 μg/mL was added to one of the cultures. The induced and control cultures for each isolate were incubated at 37°C with shaking for 2 h, followed immediately by RNA extraction. The experiment was performed in triplicate for each isolate.
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from 8 mL cultures using the RiboPure Bacteria Kit (Ambion) and treated with DNaseI (Ambion). The RNA concentration was measured using a ND-1000 Spectrophotometer (NanoDrop). SuperScript III Reverse Transcriptase (Invitrogen) with random hexamers was used to prepare cDNA from 1 μg total RNA for each sample. The resulting cDNA, diluted 1:50, was used in quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) reactions performed using Power SYBR Green PCR Master Mix (Applied Biosystems) in a 7900HT Fast Real-Time PCR System (Applied Biosystems). Each 10 μL qRT-PCR reaction contained 2.5 μL cDNA template, 2X SYBR Green mix, and gene specific primers at a concentration of 0.2 μM each. All qRT-PCR reactions were carried out in triplicate for each of the three biological replicates for each condition, and included 40 cycles consisting of 95°C for 15 s followed by 60°C for 1 min. Fluorescence was monitored in a dissociation stage as products were heated from 60 to 95°C to verify primer specificity by melting curve analysis. Transcripts encoding the target genes stxA1, stxA2, and Q, along with the reference gene, rpoA, were detected using primer pairs listed in Table 2. Efficiencies for qPCR reactions were determined using LinRegPCR (Ramakers et al., 2003), and relative expression levels of the target genes in induced versus control cultures for each isolate were calculated from Ct results and efficiencies using the Pfaffl method (Pfaffl, 2001). Basal level target gene expression for each isolate relative to EDL933 were also calculated from results obtained from control cultures.
Notably, the primers annealing to the A subunit of the Shiga toxin genes were designed to be specific for either stx1 or stx2; this was verified by examining isolates carrying stx1 or stx2 or in combination. The Q gene primers were designed to be specific for a particular cluster of Q gene sequences as described below; however, there are cases where two Q genes with similar sequence are present in a single genome. For example, there is a similar Q gene associated with the stx1 and stx2 genes in EDL933, thus measured transcript abundance cannot distinguish between Q mRNA from the two phages. This is also true for isolates EH250 and 7V.
Results
Isolate Diversity
The nine LEE-negative STEC isolates examined in this study display both whole genome phylogenetic diversity and variation in the Shiga toxin alleles they harbor. A phylogeny was inferred from the conserved genomic core (∼2.5 Mbp) of a diverse set of 39 E. coli/Shigella genome sequences including representatives of all the major pathotypes (Figure 1). The phylogeny demonstrates that the LEE-negative STEC do not form a tight phylogenetic grouping suggesting that they have evolved multiple times and acquired the stx phage multiple times. Additionally, the phylogenetic analysis identified the early evolutionary divergence of the 7V isolate, which had been noted previously by MLST (Tarr et al., 2008; Newton et al., 2009; Walk et al., 2009; Steyert et al., 2011). The MHI813 isolate is more closely related to the EHEC 1 clonal group containing the O157:H7 isolates, while DG131 is the isolate most closely related to the EHEC 2 clonal group. The remaining isolates were distributed throughout the phylogeny. In general, the stx gene phylogeny (Figure A1 in Appendix) does not parallel the result found for whole genome phylogenetic analysis. This is not unexpected since stx genes are carried on mobile genetic elements.
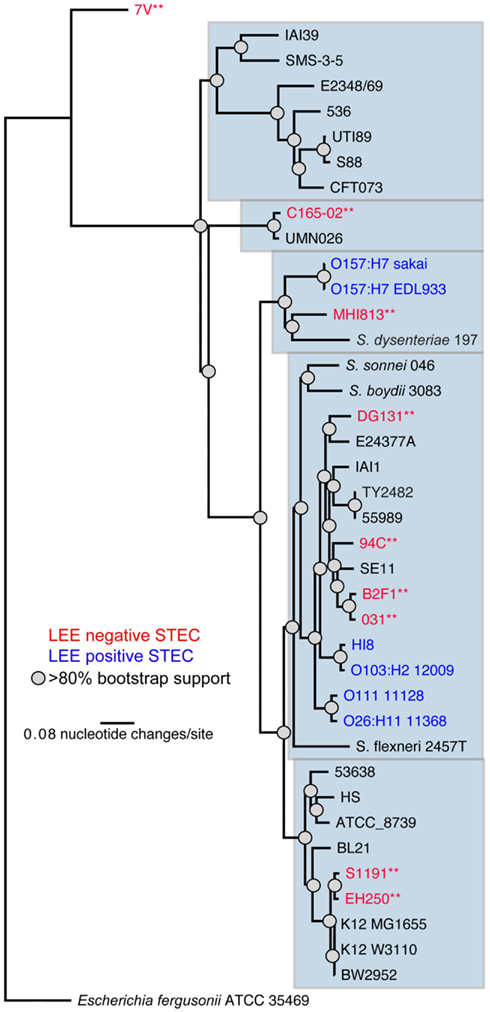
Figure 1. A whole genome phylogeny of nine LEE-negative (red) and four LEE-positive (blue) STEC compared in this study. Whole genome sequences for the LEE-negative STEC sequenced in this study (indicated by asterisks) was combined with sequence data obtained from GenBank for E. coli/Shigella genomes representing the major pathotypes (Table A1 in Appendix), and aligned based on concatenated regions of shared sequence as determined from analysis using Mugsy (Angiuoli and Salzberg, 2011). The phylogenetic tree was inferred with E. fergusonii isolate 35469 as the outgroup.
Whole Genome Sequence Comparison
Comparative genomics was utilized to determine whether there were any genes shared by all the LEE-negative STEC isolates that were not in the reference LEE-positive STEC genomes, and conversely, whether the LEE pathogenicity island was the only feature that distinguished LEE-positive from LEE-negative STEC. In addition to the nine LEE-negative STEC isolates, four representative LEE-positive STEC genomes were included in the comparative analysis including one from the EHEC 1 clonal group, O157:H7 str. EDL933, two from the EHEC 2 clonal group, O111:H-str. 11128, and O26:H11 str. 11368, and one that is a member of neither group, O103:H2 str. 12009. Whole genome comparative analysis was performed on this set of 13 genomes and identified a shared core alignment length of ∼3.66 Mb. This core sequence size is greater than the ∼2.5 Mb identified when including the 39 isolates used to construct the E. coli phylogeny in Figure 1. The whole genome comparison revealed no genomic regions (>500 bp) that are common to all nine LEE-negative STEC and absent in the four LEE-positive STEC genomes. Conversely, in addition to the LEE pathogenicity island, there were six genomic regions identified in all four LEE-positive STEC genomes that were not present in any of the LEE-negative STEC genomes. These include the five non-LEE encoded effectors espK, espN, espX7, nleA, and nleG, along with two other phage-encoded genes; one gene encodes the transcriptional regulator PerC1 (also termed PchABC in STEC), a homolog of PerC in EPEC, while the other encodes a hypothetical protein (locus tag ECO103_2361 in isolate 12009 and further referred to as hyp).
To determine whether the 7V isolate, having diverged earlier from other E. coli genomes, was lacking genes that were present in the other 12 genomes. The whole genome comparison revealed that the 7V isolate lacked an 8.9 kb cluster containing seven genes; these genes were identified as Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated proteins (Barrangou et al., 2007). The reverse analysis (i.e., unique in 7V when compared to the other genomes) identified that 7V contains 120 blocks of sequence >300 bases each, totaling ∼298 kb, that are unique. This quantity of unique sequence was greater than any of the other LEE-negative STEC isolates included in this analysis (Table 3). The number of unique sequence regions >300 bp and total length of unique sequence, along with selected possible virulence factors identified by BLASTX contained in the sequence blocks, are listed in Table 3. Although some putative virulence factors were identified, the majority of the sequence regions contain hypothetical proteins. Other unique regions with predicted functions include phage structural genes and some metabolic-related genes. For example a gene cluster coding for proteins involved in propanediol utilization was discovered in the C165-02 isolate. Overall, the LEE-negative STEC isolates are phylogenetically diverse and each isolate contains features that may contribute to virulence; however further functional analysis will be required to determine the role in virulence, if any.
Virulence Profiles of the LEE-Negative STEC Isolates
Since some of the LEE-negative isolates included in this study cluster more closely with pathotypes other than LEE-positive STEC (Figure 1), we queried the nine LEE-negative STEC genomes using the BSR for virulence factors that are typically associated with pathotypes other than LEE-positive STEC [BfpA (EPEC), AggR (EAEC), PapA (UPEC), STa, STb, LT-A, and LT-B(ETEC); Kaper et al., 2004]. The results revealed the presence of enterotoxin genes typically associated with enterotoxigenic E. coli in four of the LEE-negative STEC genomes; the 7V genome encodes a homolog of the heat stable enterotoxin STa, while the S1191 and C165-02 genomes encode STb. The genome of the C165-02 isolate also contains a gene similar to that encoding the B subunit of the heat-labile enterotoxin LT-IIa, while the MHI813 isolate carries a homolog of the gene encoding the A subunit of LT-IIb. These observed intersections of pathotype virulence factors highlight the diversity of E. coli as a species as well as the LEE-negative STEC.
There are few putative virulence factors have been definitively associated exclusively with LEE-negative STEC. Using a BSR analysis we examined the presence and level of sequence similarity of LEE-positive and LEE-negative STEC virulence factors in the 13 genomes (Figure 2). The analysis can be broadly divided into groups of virulence genes: toxins, adhesins, fimbriae, ATs, and plasmid associated genes from pO157 (marker of O157:H7) and pO113 (marker of some LEE-negative STEC; Figure 2). As predicted, all isolates encode one or more of the Shiga toxins (Table 1, Figure 2). The LEE encoded adhesion, intimin, is restricted to the LEE-positive STEC isolates and lacking in the LEE-negative STEC isolates, whereas common fimbriae and ATs are distributed in all types of STEC. The plasmid features appear to be more restricted, but not exclusive, with the LEE-positive STEC isolates containing more pO157 features and the pO113 features being more common among the LEE-negative STEC (Figure 2). Features previously predicted to be restricted to LEE-negative STEC include the adhesin protein Saa, encoded on pO113 (Paton et al., 2001). We confirm that this feature is restricted to the LEE-negative STEC, but is not found widely among LEE-negative STEC isolates. In addition to saa, the genes sab, epeA, and subAB, have been reportedly observed only in LEE-negative STEC isolates (Paton and Paton, 2005; Cergole-Novella et al., 2007; Herold et al., 2009; Newton et al., 2009; Bugarel et al., 2010; Irino et al., 2010; Wu et al., 2010). As with saa, these genes are present in several of the LEE-negative STEC, but not all. As above, these findings further support the diversity of the LEE-negative STEC isolates within E. coli.
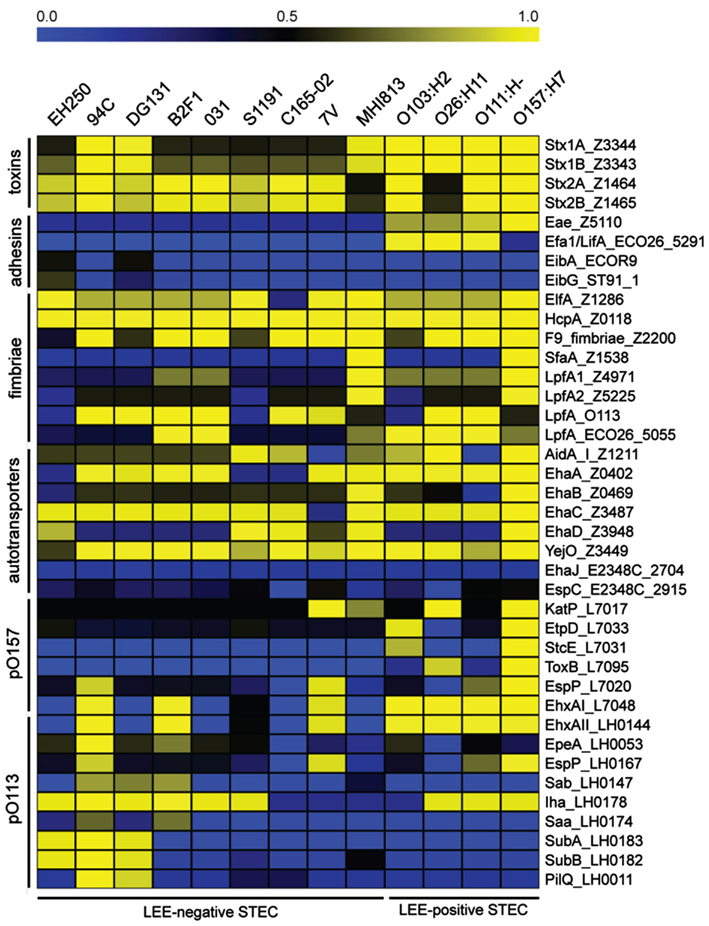
Figure 2. A virulence gene profile based on BLAST score ratio (BSR) analysis. BSR analysis was performed on the genomes to determine the presence and level of protein sequence identity of selected virulence factors. Unless an E. coli isolate is otherwise indicated in the gene label, reference protein sequences were taken from the LEE-positive O157:H7 EDL933 isolate with the exception of the proteins encoded on pO113, which were taken from STEC O113:H21 isolate EH41. Yellow indicates a higher level of similarity, blue indicates a lower level of similarity, and black indicates ∼50% identity over the length of the sequence queried.
Distribution of Genes of Interest in E. coli Collections
Since a limited number of genomes were used in the whole genome analysis, we determined the frequency of the saa, perC1, and hyp genes in a larger collection of E. coli genomes. Polymerase chain reaction assays were developed for each of these features, and the prevalence was determined in both the E. coli ECOR (environmental) and DECA (diarrheagenic) collections7. Among the environmental isolates, only the ECOR37 isolate encodes the LEE pathogenicity island, perC1 and hyp. Seven other ECOR isolates (7/72, 9.7%) also carry hyp, but no other isolate contained perC1. In the DECA collection, 100% of the EHEC (LEE+/stx+) genomes (18 of 78 total isolates) harbor both perC1 and hyp, whereas none of the EPEC1 clonal group carries either of the genes. However, the perC1 gene was found in 100% (24/24), and hyp in 58% (14/24) of the remaining LEE-positive stx-negative isolates whereas these two genes were present in only 1 of 25 LEE-negative stx-negative isolates. The reported absence of the saa gene in LEE-positive STEC genomes prompted us to include saa in our PCR analysis, which demonstrated the absence of saa in all isolates in both the ECOR and DECA collections. These analyses support the previous assertion that Saa is LEE-negative STEC restricted, and that LEE-positive STEC genomes contain perC1 and hyp, but that these genes are not highly conserved among E. coli in general.
Insertion Sequence Sites and Integrase Gene Phylogeny
Several common stx phage insertion sites such as wrbA, yecE, yehV, argW, ssrA, and prfC have been reported in LEE-positive STEC genomes (Ogura et al., 2007). Those sites, however, were determined to be unoccupied in many LEE-negative STEC isolates and thus the insertion sites of the stx phages in these isolates were essentially unknown (Garcia-Aljaro et al., 2006, 2009; Prager et al., 2011). The stx phage insertion sites, as well as the genomic locations of other identifiable phages, were determined in the LEE-negative isolates by examining the integrase genes. Unless a particular insertion site is already occupied, insertion sequences can integrate at preferred locations having a DNA sequence specificity associated with the encoded integrase (Groth and Calos, 2004; Serra-Moreno et al., 2007). The results demonstrate that the stx phages are located at a variety of sites in the LEE-negative genomes in this study, many of which appear to be novel insertion sites for stx phages (Figure 3). However, this was not because the more widely known insertion sites were already occupied, but rather because of the variety of integrase proteins carried on the phages. The phage integrase sequences were examined, and as expected, integrase phylogeny reveals clusters of genes that utilize the same insertion site (Figure A2 in Appendix). As displayed in Figure A2 in Appendix, there are integrases that are more commonly associated with stx-encoding phages; however the integrase sequences are phylogenetically diverse, and no association between a particular stx variant and integrase was observed. There are 59 phage insertion sites that have been identified in the 13 genomes examined, but some appear to be more frequently occupied than others (Figure 3). There also does not appear to be an association between phage occupation and phylogeny, as no correlation is seen when the phylogenetic analysis in Figure 1 is combined with the phage insertions sites in Figure 3. This confirms that the phage insertions are governed by the phage integrases and not the core genome, other than containing the insertion site.
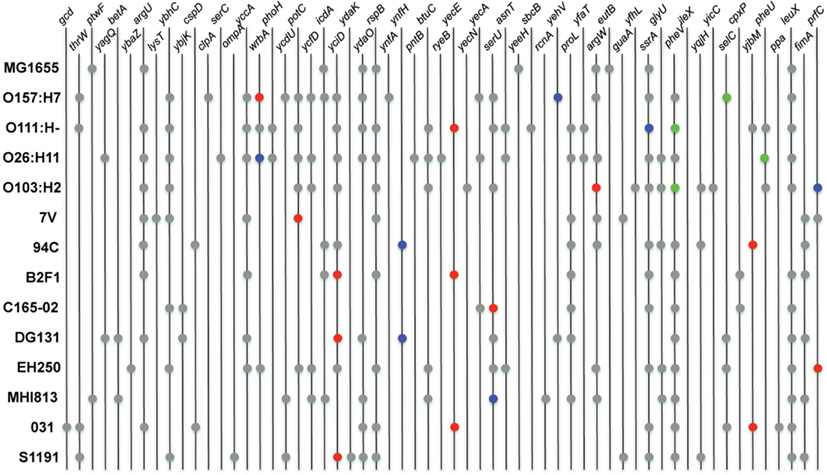
Figure 3. Chromosomal location of phage integration. Locations of phage were determined by identifying integrase genes in the genomes of the LEE-negative STEC isolates. Insertion sites were obtained from GenBank for the four reference LEE-positive STEC isolates and E. coli MG1655 K12. Prophages encoding stx1 and stx2 are represented in blue and red, respectively. The LEE pathogenicity island is indicated by green, and locations of all other insertion elements are represented in gray.
Stx-Containing Phage Sequence Diversity
Lambda phages are known to often undergo a significant amount of genetic exchange (Johansen et al., 2001; Brussow et al., 2004; Casjens, 2005). Comparison of the 20 stx phage sequences contained in the 13 genomes allowed examination of the potential diversity of the stx phages. Complete phage sequences were obtained for the majority of the phages; however, in some draft genomes phage sequences were not contiguous and phages were reconstructed from multiple contigs (Figure A3 in Appendix). In Figure A3 in Appendix, the colored blocks indicate regions of homology and the stx genes are indicated by the asterisk. The analysis clearly demonstrates the mosaic nature of the stx phages. Furthermore, phages sharing either insertion site or stx gene variant often contain extensive non-homologous regions. For example, the B2F1 stx2d2, DG131 stx2b, and S1191 stx2e-encoding phages share the yciD insertion site, but display very little sequence homology within the phage. These comparisons suggest a significant degree of diversity among stx-containing phages.
Shiga Toxin Transcription
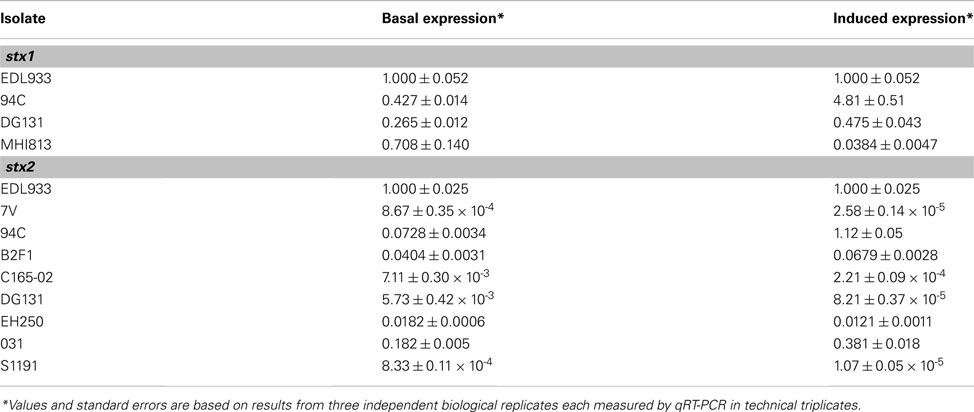
Potential Shiga toxin induction and production are important as severe complications such as HUS result from the Shiga toxin produced by the bacteria during infection (Karch et al., 1999; Kaper et al., 2004). To determine if the phages in the LEE-negative STEC could be induced to express greater levels of stx transcript, mid-log phase cultures were incubated for 2 h either in the presence or absence of mitomycin C, and stx gene expression was determined by qRT-PCR. Primers were designed to be specific to either stx1 or stx2 alleles; the expression of stx1 and stx2 were measured separately in isolates carrying both Shiga toxin types. Two isolates, B2F1 and 031, each harbor 2 distinct stx2 alleles; however, due to sequence similarity the signal from each stx2 gene allele could not be determined for these isolates. Levels of stx transcripts in induced cultures were normalized to stx mRNA levels from untreated cultures for each isolate (Figure 4A). The most highly induced stx gene was 94C stx2a, where the level of induction was over 10 times greater than that observed for EDL933 stx2. Not only is stx2 more highly induced in the 94C isolate compared to EDL933, but stx1 is as well. The results demonstrate that the induction level of the stx genes in isolates B2F1 and 031 is also greater than for EDL933 stx2, but it is not clear if this is due to one of the stx genes or both. Elevated levels of stx mRNA were not observed under inducing conditions for five isolates. Overall, there does not appear to be a consistent stx induction pattern based on STEC genome phylogeny or phage insertion site. Our results also reveal a wide variation in basal level expression of the stx genes in the isolates studied. Calculations of the basal and induced expression levels of the stx1 and stx2 alleles carried in the LEE-negative STEC isolates relative to those carried by EDL933 are reported in Table A2 in Appendix. From these results it becomes evident that the stx2 genes are expressed at similar levels in the 94C and EDL933 isolates when induced.
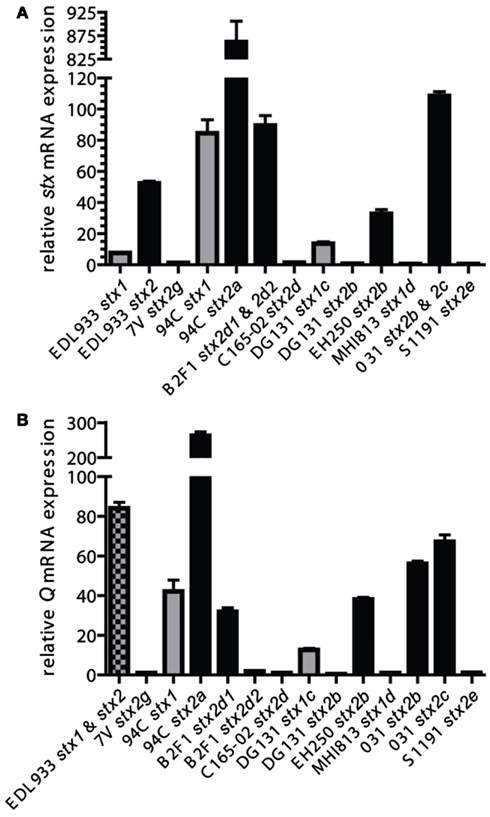
Figure 4. A comparison of induced stx and Q gene expression. Mid-log phase cultures were incubated for 2 h either in the presence or absence of mitomycin C and relative mRNA levels were determined with qRT-PCR. stx(A) and Q (B) mRNA expression comparisons were made of mitomycin C-treated cultures relative to un-induced cultures (value of 1 signifies no induction for that particular stx in the isolate). Values and standard errors are presented and are based on results from three independent biological replicates each measured with technical triplicates. Results are displayed in gray for stx1-encoding phages, black for stx2-encoding phages, and checkered where the expression from the stx1 and stx2 phages could not be distinguished. The Q genes associated with the stx2b and stx2g phages in isolates EH250 and 7V, respectively, were each found to be associated with another phage in the isolate, thus the measured Q expression might have a contribution from that Q gene as well.
Q Antiterminator Phylogeny and Transcription
Expression of stx genes within lambdoid phages is believed to be largely under the control of the Q antiterminator protein (Brussow et al., 2004). In lambdoid phages the Q gene transcription is increased under inducing conditions allowing for increased transcription of the stx genes that are downstream of the Q binding site (Brussow et al., 2004). The variety of genetic structures within the Shiga toxin cassettes in the phages can be observed when examining the genes upstream of the Q gene through the endolysin gene for each stx-encoding phage (Figures 5A,B for stx1 and stx2-encoding phages, respectively). Interestingly, the phage gene organization in the vicinity of the Shiga toxin genes 94C stx2a, B2F1 stx2d1, and 031 stx2c is quite similar and these three phages display the greatest stx expression induction. However the genetic architecture does not appear to be the only factor affecting stx expression. To further examine the involvement of the Q protein in the regulation of stx, the Q gene sequences associated with each stx-encoding phage were aligned and an inferred phylogeny based on the alignment confirms the broad phylogenetic diversity observed with the whole genome phylogeny (Figure 5C). Interestingly, the three isolates exhibiting the highest level of stx induction share similar Q proteins (94C stx2a, 031 stx2c, and B2F1 stx2d1). This suggests that the primary sequence of Q may play a role in the regulation of Shiga toxin, however further experimental evidence is required.
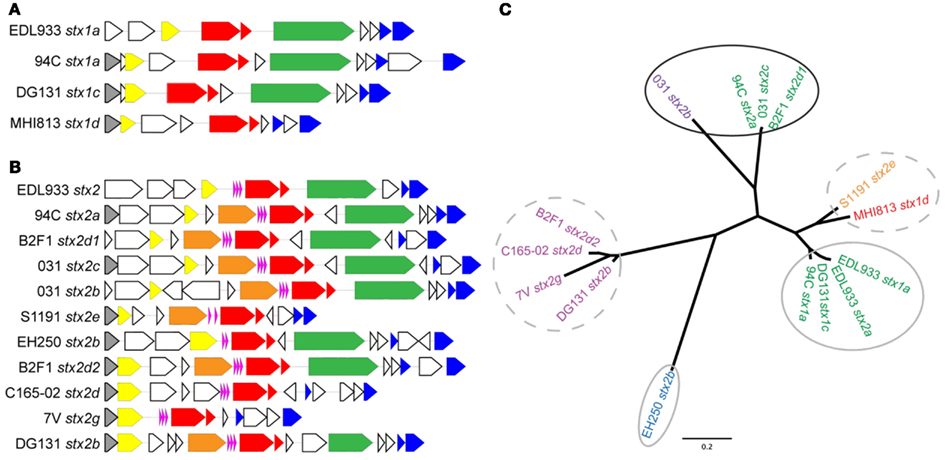
Figure 5. Gene organization flanking the stx genes and Q gene phylogeny for the stx phages in the LEE-negative STEC isolates and LEE-positive O157:H7 EDL933. Gene organization comparisons are shown for (A) stx1-encoding phages and (B) stx2-encoding phages. The colors correspond to the following gene designations: gray, rusA; yellow, Q; orange, DNA methylase; pink, tRNA genes; red, stxAB; green, yjhS; blue, lysis S, and endolysin genes; white, all other genes, predominantly encoding hypothetical proteins. A cluster diagram based on the Q gene sequences was determined (C) and primers (Table 2) were designed to be specific for each cluster according to the colors: Q1 green, Q2 purple, Q3 turquoise, Q4 blue, Q5 magenta, Q6a orange, and Q6b red. Clusters circled by a solid black line denote a high level of stx induction, gray circles denote intermediate level induction, and broken lines denote lack of induction.
To determine if the induction of the Q genes with mitomycin C correlates with the stx gene expression, specific primers were designed for each cluster of Q gene sequences in an attempt to maximize qRT-PCR efficiency and minimize potential signal from Q genes associated with phages in the genome other than the specific stx-encoding phage. In the isolates EDL933, EH250, and 7V, the contribution to the Q gene qRT-PCR signal from two phages (both stx-encoding in EDL933) cannot be distinguished, but independent determination of Q expression in the stx-encoding phages was possible for all other isolates. The induction pattern for Q gene expression parallels the stx gene expression, but there is not a perfect quantitative correlation (Figure 4B), suggesting other factors may be involved. These studies confirm that the stx2d1 gene expression is inducible in isolate B2F1, but not stx2d2 gene expression (Teel et al., 2002). Our results also indicate that basal level stx and Q gene expression are not correlated (data not shown), thus expression of stx is at least partially dependent on some factor other than levels of Q transcripts produced under non-inducing conditions.
Discussion
Recently, there has been an increased interest in characterizing LEE-negative STEC isolates because certain isolates have been associated with diarrheal symptoms and HUS, as results from infection with certain LEE-positive STEC isolates (Johnson et al., 2006; Mellmann et al., 2008; Newton et al., 2009; Kappeli et al., 2011). Detailed characterization of LEE-negative STEC has indicated that the association to HUS is especially significant for the activatable stx2d subtype (Bielaszewska et al., 2006) and that other toxin subtypes are primarily associated with a milder course of disease (Friedrich et al., 2002; Persson et al., 2007). A limited number of reports have partially characterized these stx-encoding phages and detailed PCR screens for virulence factors associated with LEE-negative STEC isolates (Muniesa et al., 2000; Recktenwald and Schmidt, 2002; Teel et al., 2002; Cergole-Novella et al., 2007; Beutin et al., 2008; Newton et al., 2009; Wu et al., 2010; Prager et al., 2011), but there remains a paucity of whole genome studies. To fill this knowledge gap a comparative genomics study of nine phylogenetically diverse LEE-negative STEC isolates and four reference LEE-positive STEC isolates was undertaken. Utilizing a gene-independent whole genome alignment method we determined that as a subset of STEC, the LEE-negative STEC isolates, do not share any genes in common that are lacking in all the LEE-positive STEC genomes examined. The phylogenetic diversity of the LEE-negative STEC may preclude the identification of a molecular marker that can differentiate the LEE-negative STEC isolates as a group from all other E. coli (Figure 1). Traditionally, LEE-positive STEC isolates are defined as STEC that carry the LEE pathogenicity island in their genome. Our results suggest that genes encoded outside the LEE such as the non-LEE encoded effectors espK, espN, espX7, nleA, and nleG, as well as the perC1 gene (also termed pchABC) and a hypothetical gene marker, hyp, may be suitable biomarkers for LEE-positive STEC. Indeed, the presence of perC1 and hyp in an additional 18 LEE-positive STEC genomes examined, and the lack of these genes in a selection of LEE-negative genomes, suggest that these may be reliable LEE-positive STEC biomarkers. Nonetheless, the set of LEE-negative isolates queried will need to be expanded for a more conclusive result.
The definition of a pathotype of E. coli based on a single feature, especially one encoded on a mobile element such as the phage-borne Shiga toxin genes, is likely to reveal highly diverse host isolate backgrounds when examined on a genomic scale. The whole genome phylogeny based on conserved core sequence, utilizing approximately half the genome, determined that the majority of the LEE-negative isolates are more similar to other E. coli pathotypes than to LEE-positive STEC (Figure 1). The 7V isolate also appears to be on a deep rooting branch of this phylogeny, previously described as a “cryptic lineage” (Walk et al., 2009). Although the 7V isolate is not phylogenetically related to any prototype ETEC isolates, we determined that it does harbor the heat stable enterotoxin gene STa (ST-IA). These results confirm a recent report that identified the genes encoding STa and KatP carried on the 7V plasmid (Prager et al., 2011). The S1191 and C165-02 isolates also appear to have a STEC/ETEC intermediate pathotype based on virulence factors, as their genomes encode both Stx and heat stable enterotoxin b, STb. Additionally, the C165-02 genome encodes the gene for the B subunit of LT-IIa, whereas the gene coding for the A subunit of LT-IIb was found in the MHI813 genome. As these features are usually plasmid-borne, it is possible that these isolates contain a novel virulence plasmid that is different than pO157, pO113, or the 7V plasmid, but since these are draft genomes it also does not preclude chromosomal insertion of these virulence factors. Without more detailed information from sequencing the isolated plasmids, the comparative genomic analyses suggest that there is a variety of substantially different virulence plasmids harbored by LEE-negative STEC isolates that, in some cases, encode enterotoxin genes.
Without the LEE pathogenicity island, LEE-negative STEC must adhere to the intestinal epithelium by means other than the tight binding brought about by the Intimin/Tir complex (Mellies et al., 2007). The focused analysis on the presence/absence of multiple fimbriae and ATs, some of which may function as adhesions, in the 13 genomes examined, identified further variability (Figure 2). While some of the traditional adhesins were identified in the core of the LEE-negative STEC, additional isolate – specific adhesins and fimbrial genes were identified (Table 3). In fact, additional adherence factors were identified in each of the LEE-negative STEC genomes (Table 3). The combined results of the whole genome sequence comparison, virulence factor profiling analysis and the identification of factors encoded in the isolate-specific sequence regions indicate that there is no common adherence factor in all LEE-negative STEC isolates, but rather that each isolate encodes a particular assortment of adherence factors that allows pathogenic success.
In general, analysis of LEE-positive STEC genomes has revealed the presence of a great number of prophages in each genome, some of which contain virulence-associated genes (Schmidt and Hensel, 2004; Asadulghani et al., 2009). The genomic location of insertion elements and phages in the LEE-negative STEC genomes were cataloged (Figure 3). By inspecting the various insertion site occupancies in the genomes, it is clear that while some genomic sites are occupied by phage more frequently, there appears to be no discernable pattern of phage insertion that correlates to the phylogenetic relationship. Most sites occupied by prophages in the nine LEE-negative STEC genomes are also utilized in at least one of the four LEE-positive STEC genomes, but a few novel insertion sites are identified. We determined that insertion elements are predominately inserted at specific genomic locations that can be correlated to the integrase gene carried on the mobile genetic element (Figure A2 in Appendix). Of note the absence of the LEE pathogenicity island in the LEE-negative STEC genomes is not due to lack of availability of the usual insertion sites adjacent to selC, pheV, or pheU (Figure 3). Interestingly, the pheV site is occupied in all LEE-negative STEC isolates, except 7V. The pheU site is unoccupied in all nine LEE-negative genomes and the selC site is occupied in only the DG131, EH250, and 031 genomes (Figure 3). Thus the LEE pathogenicity island could potentially insert in any of these genomes, but has not.
A comparison of stx phage sequences demonstrates the modular structure and sequence heterogeneity present even between phages encoding the same stx allele variant (Figures 5 and A3 in Appendix). This heterogeneity, especially in the integrase genes has led to the insertion of stx-encoding phages at a variety of genomic locations in the LEE-negative STEC isolates, such that an stx allele variant cannot necessarily be correlated with a particular genomic location. As a further example of this fact, we determined the integration site of the stx2e-encoding phage carried in the S1191 isolate to be yciD (Figure 3), whereas, yecE is the integration site of the stx2e-encoding phage in the 2771/97 isolate (Recktenwald and Schmidt, 2002). We also determined that the Q protein sequences are divergent in these two stx2e-encoding phages, and that the phage gene organization is not shared (Recktenwald and Schmidt, 2002; Beutin et al., 2008; Figure 5B). Q proteins with low sequence identity have been noted previously between LEE-positive O157:H7 stx2c-encoding phages (Eppinger et al., 2011a) and this work demonstrates the same phenomenon in the LEE-negative STEC isolates (Figure 5C). The extent to which dissimilar Q proteins and/or genetic organization upstream of the stx genes affects stx expression is not known (Brussow et al., 2004). It is of significance that in a detailed analysis of the Q gene sequences, the four Q proteins associated with phages that were not induced by mitomycin C, namely, B2F1 stx2d2, C165-02 stx2d, 7V stx2g, and DG131 stx2b, are more similar (Figure 5C). Likewise, the Q sequences corresponding to the most highly induced stx transcript cluster together. In fact, there is a general trend between the Q gene induction and the associated stx gene induction (Figures 5A,B); however further work would be required to elucidate the reason for the lack of increase in Q expression under inducing conditions noted for some of the phages included in this work.
Conflicting reports exist as to whether the Shiga toxin genotype or the level of Shiga toxin production can be used as an indicator for severity of clinical symptoms and progression to HUS associated with STEC infection (Friedrich et al., 2002; Bielaszewska et al., 2006; Orth et al., 2007; De Sablet et al., 2008; Neupane et al., 2011). Not all of the LEE-negative STEC isolates included in this work were isolated from humans, thus a complete comparison aimed at associating Shiga toxin characteristics with virulence in humans cannot be made. All of the Shiga toxins carried in the LEE-negative STEC isolates in this work are prophage-encoded and possibly inducible. Cultures of the LEE-negative STEC and LEE-positive EDL933 were incubated either with or without mitomycin C followed by qRT-PCR utilizing primers having either stxA1 or stxA2 as a target (Table 2). Heterogeneity in stx expression between isolates has been previously reported (Ritchie et al., 2003; Beutin et al., 2008; De Sablet et al., 2008; Zhang et al., 2010). Variation in basal stx expression and level of stx induction was observed among the LEE-negative isolates in this work, and a number of the stx genes did not appear to be inducible under conditions tested (Table A2 in Appendix and Figure 4A). The isolates demonstrating the greatest induction of stx2 are EDL933, 94C, B2F1, and 031. This induction may be related to the severe clinical outcome associated with each isolate (Table 1) and the potential to exacerbate the disease with the administration of antibiotics. Overall the Q gene induction matched the trend of the associated stx gene, suggesting that there was a phage-based regulation of the toxin.
In conclusion, this study highlights the broad phylogenetic diversity of LEE-negative STEC isolates as well as the stx-encoding prophages harbored in their genomes. Our genome-wide comparative results indicate that LEE-negative STEC isolates as a group vary significantly in the assortment of adhesins and other virulence factors they encode. Sequence comparisons of the stx-encoding prophages demonstrate the extensive variation in terms of overall mosaic structures, stx allele variants, integrase sequence, Q antiterminator homologs and even the gene organization flanking the stxAB genes. These results suggests that extensive genetic exchange has taken place between phages and the possibility may arise from continued genetic exchange. Various genomic insertion sites of the stx-encoding phages in the LEE-negative STEC isolates were identified, revealing five sites not previously reported to be utilized by stx-encoding phages. The qRT-PCR results of the stx and Q genes determined that stx expression levels are increased in isolates in which Q expression levels are also increased under inducing conditions. Finally, this study demonstrates that the overall genome content, phage location and combination of potential virulence factors are variable in the LEE-negative STEC, requiring a larger set of isolates and further functional analyses before conclusions about this group can be made.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number HHSN272200900009C and NIH grant numbers AI089894, AI20148, and 1RC4AI092828.
Footnotes
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Angiuoli, S. V., and Salzberg, S. L. (2011). Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342.
Asadulghani, M., Ogura, Y., Ooka, T., Itoh, T., Sawaguchi, A., Iguchi, A., et al. (2009). The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5, e1000408. doi:10.1371/journal.ppat.1000408
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712.
Beutin, L., Kruger, U., Krause, G., Miko, A., Martin, A., and Strauch, E. (2008). Evaluation of major types of Shiga toxin 2E-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl. Environ. Microbiol. 74, 4806–4816.
Bielaszewska, M., Friedrich, A. W., Aldick, T., Schurk-Bulgrin, R., and Karch, H. (2006). Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43, 1160–1167.
Boerlin, P., Mcewen, S. A., Boerlin-Petzold, F., Wilson, J. B., Johnson, R. P., and Gyles, C. L. (1999). Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37, 497–503.
Brooks, J. T., Sowers, E. G., Wells, J. G., Greene, K. D., Griffin, P. M., Hoekstra, R. M., et al. (2005). Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192, 1422–1429.
Brussow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602.
Bugarel, M., Beutin, L., Martin, A., Gill, A., and Fach, P. (2010). Micro-array for the identification of Shiga toxin-producing Escherichia coli (STEC) seropathotypes associated with hemorrhagic colitis and hemolytic uremic syndrome in humans. Int. J. Food Microbiol. 142, 318–329.
Burk, C., Dietrich, R., Acar, G., Moravek, M., Bulte, M., and Martlbauer, E. (2003). Identification and characterization of a new variant of Shiga toxin 1 in Escherichia coli ONT:H19 of bovine origin. J. Clin. Microbiol. 41, 2106–2112.
Casjens, S. R. (2005). Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8, 451–458.
Cergole-Novella, M. C., Nishimura, L. S., Dos Santos, L. F., Irino, K., Vaz, T. M., Bergamini, A. M., et al. (2007). Distribution of virulence profiles related to new toxins and putative adhesins in Shiga toxin-producing Escherichia coli isolated from diverse sources in Brazil. FEMS Microbiol. Lett. 274, 329–334.
Darling, A. E., Mau, B., and Perna, N. T. (2010). Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147. doi:10.1371/journal.pone.0011147
De Sablet, T., Bertin, Y., Vareille, M., Girardeau, J. P., Garrivier, A., Gobert, A. P., et al. (2008). Differential expression of stx2 variants in Shiga toxin-producing Escherichia coli belonging to seropathotypes A and C. Microbiology 154, 176–186.
Doughty, S., Sloan, J., Bennett-Wood, V., Robertson, M., Robins-Browne, R. M., and Hartland, E. L. (2002). Identification of a novel fimbrial gene cluster related to long polar fimbriae in locus of enterocyte effacement-negative strains of enterohemorrhagic Escherichia coli. Infect. Immun. 70, 6761–6769.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Eppinger, M., Mammel, M. K., Leclerc, J. E., Ravel, J., and Cebula, T. A. (2011a). Genome signatures of Escherichia coli O157:H7 isolates from the bovine host reservoir. Appl. Environ. Microbiol. 77, 2916–2925.
Eppinger, M., Mammel, M. K., Leclerc, J. E., Ravel, J., and Cebula, T. A. (2011b). Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U.S.A. 108, 20142–20147.
Farfan, M. J., and Torres, A. G. (2011). Molecular mechanisms mediating colonization of Shiga toxin-producing Escherichia coli strains. Infect. Immun. 80, 903–913.
Friedrich, A. W., Bielaszewska, M., Zhang, W. L., Pulz, M., Kuczius, T., Ammon, A., et al. (2002). Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185, 74–84.
Fuller, C. A., Pellino, C. A., Flagler, M. J., Strasser, J. E., and Weiss, A. A. (2011). Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79, 1329–1337.
Garcia-Aljaro, C., Muniesa, M., Jofre, J., and Blanch, A. R. (2006). Newly identified bacteriophages carrying the stx2g Shiga toxin gene isolated from Escherichia coli strains in polluted waters. FEMS Microbiol. Lett. 258, 127–135.
Garcia-Aljaro, C., Muniesa, M., Jofre, J., and Blanch, A. R. (2009). Genotypic and phenotypic diversity among induced, stx2-carrying bacteriophages from environmental Escherichia coli strains. Appl. Environ. Microbiol. 75, 329–336.
Gould, L. H., Bopp, C., Strockbine, N., Atkinson, R., Baselski, V., Body, B., et al. (2009). Recommendations for diagnosis of shiga toxin – producing Escherichia coli infections by clinical laboratories. MMWR Recomm. Rep. 58, 1–14.
Groth, A. C., and Calos, M. P. (2004). Phage integrases: biology and applications. J. Mol. Biol. 335, 667–678.
Herold, S., Paton, J. C., and Paton, A. W. (2009). Sab, a novel autotransporter of locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli O113:H21, contributes to adherence and biofilm formation. Infect. Immun. 77, 3234–3243.
Irino, K., Vieira, M. A., Gomes, T. A., Guth, B. E., Naves, Z. V., Oliveira, M. G., et al. (2010). Subtilase cytotoxin-encoding subAB operon found exclusively among Shiga toxin-producing Escherichia coli strains. J. Clin. Microbiol. 48, 988–990.
Ito, H., Terai, A., Kurazono, H., Takeda, Y., and Nishibuchi, M. (1990). Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8, 47–60.
Johansen, B. K., Wasteson, Y., Granum, P. E., and Brynestad, S. (2001). Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147, 1929–1936.
Johnson, K. E., Thorpe, C. M., and Sears, C. L. (2006). The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43, 1587–1595.
Johnson, T. J., and Nolan, L. K. (2009). Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73, 750–774.
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140.
Kappeli, U., Hachler, H., Giezendanner, N., Beutin, L., and Stephan, R. (2011). Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000-2009. Emerging Infect. Dis. 17, 180–185.
Karch, H., Bielaszewska, M., Bitzan, M., and Schmidt, H. (1999). Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn. Microbiol. Infect. Dis. 34, 229–243.
Koch, C., Hertwig, S., Lurz, R., Appel, B., and Beutin, L. (2001). Isolation of a lysogenic bacteriophage carrying the stx(1(OX3)) gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39, 3992–3998.
Leung, P. H., Peiris, J. S., Ng, W. W., Robins-Browne, R. M., Bettelheim, K. A., and Yam, W. C. (2003). A newly discovered verotoxin variant, VT2g, produced by bovine verocytotoxigenic Escherichia coli. Appl. Environ. Microbiol. 69, 7549–7553.
Leyton, D. L., Sloan, J., Hill, R. E., Doughty, S., and Hartland, E. L. (2003). Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect. Immun. 71, 6307–6319.
Manning, S. D., Motiwala, A. S., Springman, A. C., Qi, W., Lacher, D. W., Ouellette, L. M., et al. (2008). Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U.S.A. 105, 4868–4873.
Mellies, J. L., Barron, A. M., and Carmona, A. M. (2007). Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect. Immun. 75, 4199–4210.
Mellmann, A., Bielaszewska, M., Kock, R., Friedrich, A. W., Fruth, A., Middendorf, B., et al. (2008). Analysis of collection of hemolytic uremic syndrome-associated enterohemorrhagic Escherichia coli. Emerging Infect. Dis. 14, 1287–1290.
Muniesa, M., Recktenwald, J., Bielaszewska, M., Karch, H., and Schmidt, H. (2000). Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68, 4850–4855.
Neupane, M., Abu-Ali, G. S., Mitra, A., Lacher, D. W., Manning, S. D., and Riordan, J. T. (2011). Shiga toxin 2 overexpression in Escherichia coli O157:H7 strains associated with severe human disease. Microb. Pathog. 51, 466–470.
Newton, H. J., Sloan, J., Bulach, D. M., Seemann, T., Allison, C. C., Tauschek, M., et al. (2009). Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerging Infect. Dis. 15, 372–380.
Ochman, H., and Selander, R. K. (1984). Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157, 690–693.
Ogura, Y., Kurokawa, K., Ooka, T., Tashiro, K., Tobe, T., Ohnishi, M., et al. (2006). Complexity of the genomic diversity in enterohemorrhagic Escherichia coli O157 revealed by the combinational use of the O157 Sakai OligoDNA microarray and the Whole Genome PCR scanning. DNA Res. 13, 3–14.
Ogura, Y., Ooka, T., Asadulghani Terajima, J., Nougayrede, J. P., Kurokawa, K., Tashiro, K., et al. (2007). Extensive genomic diversity and selective conservation of virulence-determinants in enterohemorrhagic Escherichia coli strains of O157 and non-O157 serotypes. Genome Biol. 8, R138.
Ogura, Y., Ooka, T., Iguchi, A., Toh, H., Asadulghani, M., Oshima, K., et al. (2009). Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106, 17939–17944.
Orth, D., Grif, K., Khan, A. B., Naim, A., Dierich, M. P., and Wurzner, R. (2007). The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn. Microbiol. Infect. Dis. 59, 235–242.
Paton, A. W., Beutin, L., and Paton, J. C. (1995a). Heterogeneity of the amino-acid sequences of Escherichia coli Shiga-like toxin type-I operons. Gene 153, 71–74.
Paton, A. W., Bourne, A. J., Manning, P. A., and Paton, J. C. (1995b). Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect. Immun. 63, 2450–2458.
Paton, A. W., and Paton, J. C. (2005). Multiplex PCR for direct detection of Shiga toxigenic Escherichia coli strains producing the novel subtilase cytotoxin. J. Clin. Microbiol. 43, 2944–2947.
Paton, A. W., Paton, J. C., Heuzenroeder, M. W., Goldwater, P. N., and Manning, P. A. (1992). Cloning and nucleotide sequence of a variant Shiga-like toxin II gene from Escherichia coli OX3:H21 isolated from a case of sudden infant death syndrome. Microb. Pathog. 13, 225–236.
Paton, A. W., Paton, J. C., and Manning, P. A. (1993). Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb. Pathog. 15, 77–82.
Paton, A. W., Srimanote, P., Talbot, U. M., Wang, H., and Paton, J. C. (2004). A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200, 35–46.
Paton, A. W., Srimanote, P., Woodrow, M. C., and Paton, J. C. (2001). Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69, 6999–7009.
Perna, N. T., Plunkett, G. III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., et al. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–533.
Persson, S., Olsen, K. E., Ethelberg, S., and Scheutz, F. (2007). Subtyping method for Escherichia coli shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45, 2020–2024.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45.
Pierard, D., Muyldermans, G., Moriau, L., Stevens, D., and Lauwers, S. (1998). Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36, 3317–3322.
Prager, R., Fruth, A., Busch, U., and Tietze, E. (2011). Comparative analysis of virulence genes, genetic diversity, and phylogeny of Shiga toxin 2g and heat-stable enterotoxin STIa encoding Escherichia coli isolates from humans, animals, and environmental sources. Int. J. Med. Microbiol. 301, 181–191.
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650.
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490. doi:10.1371/journal.pone.0009490
Ramakers, C., Ruijter, J. M., Deprez, R. H., and Moorman, A. F. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339, 62–66.
Rasko, D. A., Myers, G. S., and Ravel, J. (2005). Visualization of comparative genomic analyses by BLAST score ratio. BMC Bioinform. 6, 2. doi:10.1186/1471-2105-6-2
Rasko, D. A., Webster, D. R., Sahl, J. W., Bashir, A., Boisen, N., Scheutz, F., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717.
Recktenwald, J., and Schmidt, H. (2002). The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage phiP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70, 1896–1908.
Ritchie, J. M., Wagner, P. L., Acheson, D. W., and Waldor, M. K. (2003). Comparison of Shiga toxin production by hemolytic-uremic syndrome-associated and bovine-associated Shiga toxin-producing Escherichia coli isolates. Appl. Environ. Microbiol. 69, 1059–1066.
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378.
Sahl, J. W., Steinsland, H., Redman, J. C., Angiuoli, S. V., Nataro, J. P., Sommerfelt, H., et al. (2011). A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79, 950–960.
Schmidt, H., and Hensel, M. (2004). Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17, 14–56.
Serra-Moreno, R., Jofre, J., and Muniesa, M. (2007). Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J. Bacteriol. 189, 6645–6654.
Slanec, T., Fruth, A., Creuzburg, K., and Schmidt, H. (2009). Molecular analysis of virulence profiles and Shiga toxin genes in food-borne Shiga toxin-producing Escherichia coli. Appl. Environ. Microbiol. 75, 6187–6197.
Steyert, S. R., Rasko, D. A., and Kaper, J. B. (2011). Functional and phylogenetic analysis of ureD in Shiga toxin-producing Escherichia coli. J. Bacteriol. 193, 875–886.
Tarr, C. L., Nelson, A. M., Beutin, L., Olsen, K. E., and Whittam, T. S. (2008). Molecular characterization reveals similar virulence gene content in unrelated clonal groups of Escherichia coli of serogroup O174 (OX3). J. Bacteriol. 190, 1344–1349.
Teel, L. D., Melton-Celsa, A. R., Schmitt, C. K., and O’Brien, A. D. (2002). One of two copies of the gene for the activatable shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70, 4282–4291.
Toma, C., Martinez Espinosa, E., Song, T., Miliwebsky, E., Chinen, I., Iyoda, S., et al. (2004). Distribution of putative adhesins in different seropathotypes of Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 42, 4937–4946.
Walk, S. T., Alm, E. W., Gordon, D. M., Ram, J. L., Toranzos, G. A., Tiedje, J. M., et al. (2009). Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75, 6534–6544.
Weinstein, D. L., Jackson, M. P., Samuel, J. E., Holmes, R. K., and O’Brien, A. D. (1988). Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170, 4223–4230.
Wells, T. J., Totsika, M., and Schembri, M. A. (2010). Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology 156, 2459–2469.
Wu, Y., Hinenoya, A., Taguchi, T., Nagita, A., Shima, K., Tsukamoto, T., et al. (2010). Distribution of virulence genes related to adhesins and toxins in shiga toxin-producing Escherichia coli strains isolated from healthy cattle and diarrheal patients in Japan. J. Vet. Med. Sci. 72, 589–597.
Yoon, J. W., and Hovde, C. J. (2008). All blood, no stool: enterohemorrhagic Escherichia coli O157:H7 infection. J. Vet. Sci. 9, 219–231.
Zhang, W., Bielaszewska, M., Kuczius, T., and Karch, H. (2002). Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx(1c)) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40, 1441–1446.
Zhang, X., Mcdaniel, A. D., Wolf, L. E., Keusch, G. T., Waldor, M. K., and Acheson, D. W. (2000). Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181, 664–670.
Zhang, Y., Laing, C., Zhang, Z., Hallewell, J., You, C., Ziebell, K., et al. (2010). Lineage and host source are both correlated with levels of Shiga toxin 2 production by Escherichia coli O157:H7 strains. Appl. Environ. Microbiol. 76, 474–482.
Appendix
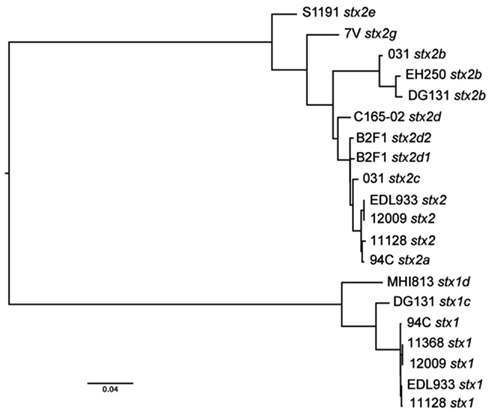
Figure A1. Shiga toxin gene phylogeny. A phylogenetic tree was constructed from an alignment of concatenated stxA and stxB gene subunits for each of the Shiga toxins encoded in the 13 isolates compared in this study.
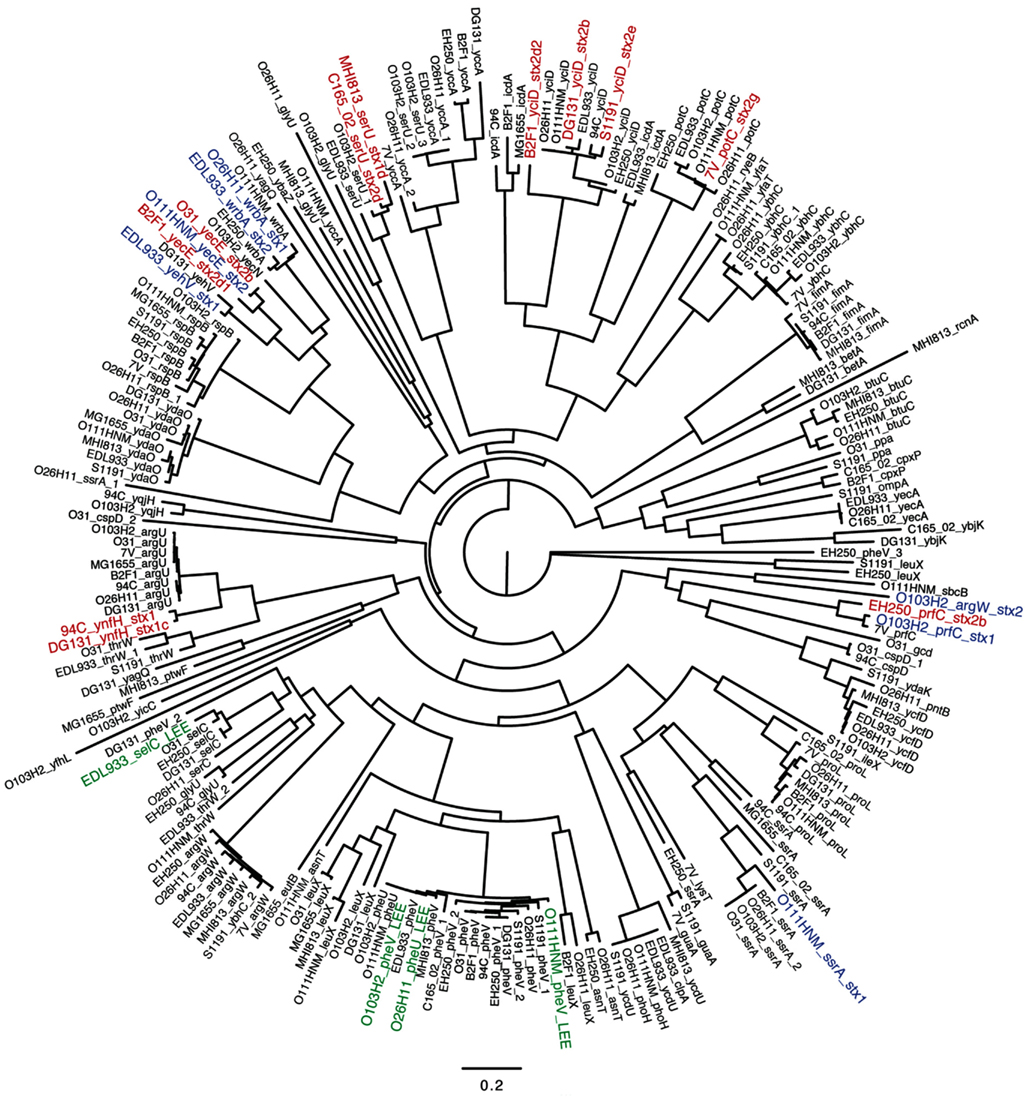
Figure A2. Relationship between integrase gene phylogeny and chromosomal location of insertion elements. Integrase gene sequences were extracted from the LEE-negative STEC genomes and the gene adjacent to the integrase gene was designated as the insertion site. Integrase gene sequences were obtained from GenBank for the E. coli K12 MG1655 genome along with the four reference LEE-positive STEC genomes. A phylogenetic tree was inferred from an alignment of the integrase genes, and displays the predominant correlation between integrase gene sequence and chromosomal location of the insertion element. Integrase genes extracted from stx-encoding phages in the LEE-negative STEC genomes are depicted in red, while those from the reference LEE-positive STEC genomes are depicted in blue and the integrase genes associated with the LEE pathogenicity island are denoted in green. An integrase gene could not be identified in the STEC 94C stx2a and STEC O31 stx2c prophages, thus those phages are not included in this analysis.
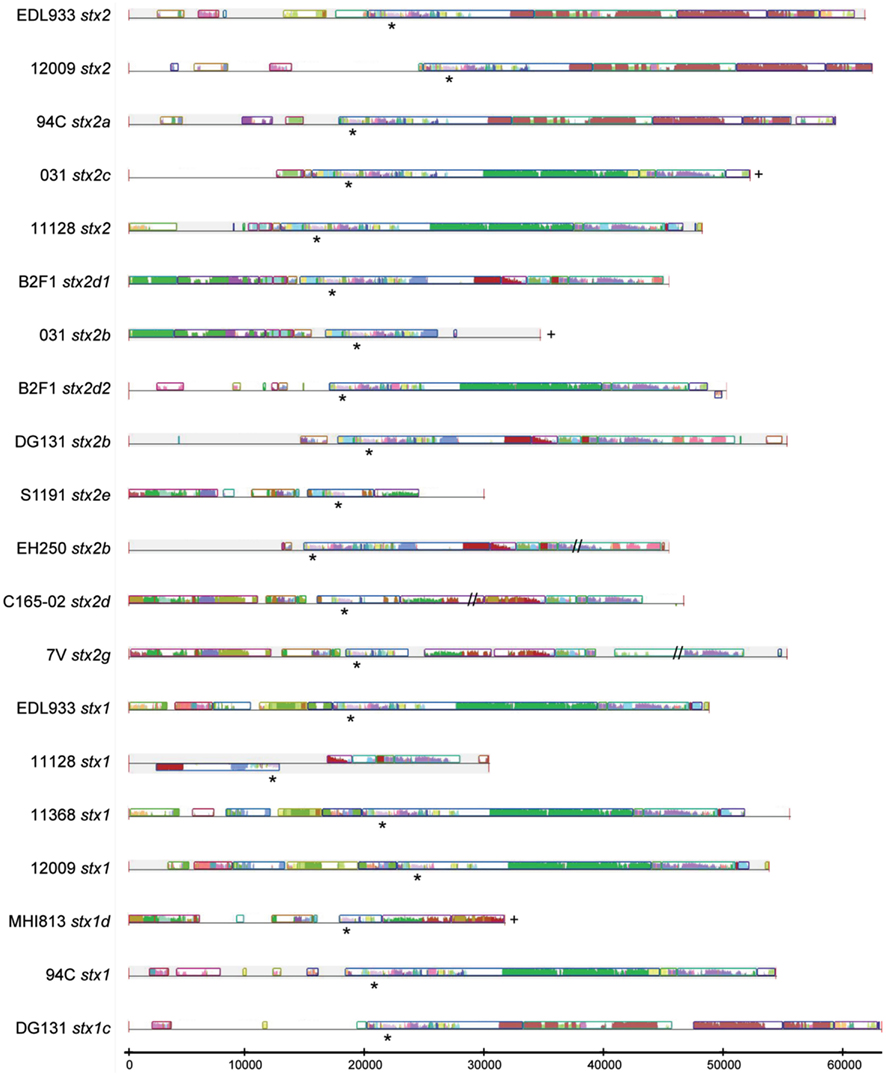
Figure A3. Sequence comparison of the stx-encoding prophages. Phage sequences extracted from the genomes of the nine LEE-negative STEC isolates and obtained from GenBank for the four reference LEE-positive STEC genomes were subjected to sequence analysis using Mauve (Darling et al., 2010). Similar color denotes regions of shared sequence and the height of the bars denotes level of similarity of the shared sequence regions. Regions where there is a line, but no colored bars, indicate a lack of homology with any of the other phages in the comparison. The location of the stx genes is identified with an asterisk (*), the plus (+) signifies that the 3′ end of phage could not be determined unambiguously from the sequence data, and the double hash (//) denotes a gap in known sequence data.
Keywords: Escherichia coli, microbial genomics, Shiga toxin, evolution, phage
Citation: Steyert SR, Sahl JW, Fraser CM, Teel LD, Scheutz F and Rasko DA (2012) Comparative genomics and stx phage characterization of LEE-negative Shiga toxin-producing Escherichia coli. Front. Cell. Inf. Microbio. 2:133. doi: 10.3389/fcimb.2012.00133
Received: 29 April 2012; Paper pending published: 21 May 2012;
Accepted: 11 October 2012; Published online: 07 November 2012.
Edited by:
Nora Lía Padola, Faculty of Veterinary Sciences-Universidad Nacional del Centro de la Provincia de Buenos Aires, ArgentinaReviewed by:
Alfredo G. Torres, University of Texas Medical Branch, USAShu-Lin Liu, Harbin Medical University, China
Copyright: © 2012 Steyert, Sahl, Fraser, Teel, Scheutz and Rasko. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: David A. Rasko, Department of Microbiology and Immunology, University of Maryland School of Medicine, Institute for Genome Sciences, 801 West Baltimore Street, Suite 619, Baltimore, MD 21201, USA. e-mail:ZHJhc2tvQHNvbS51bWFyeWxhbmQuZWR1
†Present address: Jason W. Sahl, Translational Genomics Research Institute, 3051 West Shamrell Blvd., Suite 106, Flagstaff, AZ 86001, USA.


 Claire M. Fraser1
Claire M. Fraser1