- 1Department of Pathology and Laboratory Medicine, University of Texas Medical School at Houston, Houston, TX, USA
- 2Division of Geographic Medicine and Infectious Diseases, Tufts Medical Center, Boston, MA, USA
Transposon insertion provides a method for near-random mutation of bacterial genomes, and has been utilized extensively for the study of bacterial pathogenesis and biology. This approach is particularly useful for organisms that are relatively refractory to genetic manipulation, including Lyme disease Borrelia. In this review, progress to date in the application of transposon mutagenesis to the study of Borrelia burgdorferi is reported. An effective Himar1-based transposon vector has been developed and used to acquire a sequence-defined library of nearly 4500 mutants in the infectious, moderately transformable B. burgdorferi B31 derivative 5A18NP1. Analysis of these transposon mutants using signature-tagged mutagenesis (STM) and Tn-seq approaches has begun to yield valuable information regarding the genes important in the pathogenesis and biology of this organism.
Lyme disease is a tick-transmitted, multi-stage bacterial infection caused by several species of the genus Borrelia, including the primary agents B. burgdorferi, B. garinii, and B. afzelii (Steere et al., 2004). These spirochetes are maintained in nature through a cycle involving transmission between small mammals and ticks of the Ixodes ricinus group. Large mammals such as deer are also required for the feeding of the adult ticks and hence egg production. Humans are accidental hosts that are infected by the bite of Borrelia-infested ticks, usually of the nymphal stage. Most infected humans develop mild fever, malaise, and a localized skin lesion called erythema migrans days to weeks after the tick bite, followed by dissemination of propagating organisms through the blood and lymphatics to other tissues. Later manifestations may include neurologic effects, joint involvement, cardiac block, and skin lesions called acrodermatitis chronica atrophicans. If untreated, infection can last for months to years; in mouse studies, organisms can be reisolated from almost any tissue throughout the lifetime of the animal. Despite this pattern of pathogenesis, B. burgdorferi produces no known toxins. Lyme disease Borrelia thus appear to represent non-toxigenic, invasive, persistent pathogens that cause disease through the induction of host inflammatory reactions, similar to Treponema pallidum (syphilis) and Mycobacterium tuberculosis (Norris et al., 2010).
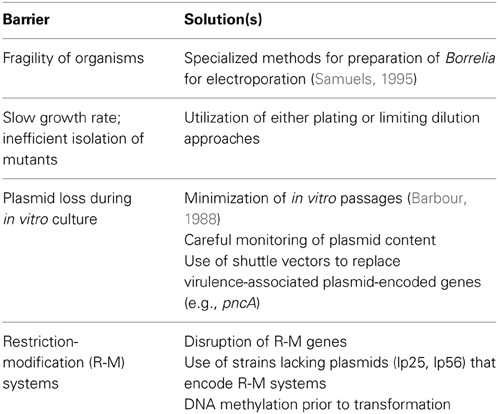
Since its initial discovery and culture in 1981, B. burgdorferi has been the subject of intensive study in an attempt to better understand the biology of this organism and thereby identify properties useful in the diagnosis, treatment, or prevention of Lyme disease. This work has resulted in tremendous progress (Rosa et al., 2005; Samuels and Radolf, 2010), particularly in terms of understanding the spirochete's molecular biology and the massive gene regulation that accompanies the transition between the disparate mammalian and tick host environments. Despite these advances, several barriers (Table 1) have hampered the ability to fulfill molecular Koch's postulates regarding the role of borrelial genes in biological processes and pathogenesis. While some of these barriers have been at least partially overcome, transformation of low-passage, infectious B. burgdorferi remains a challenge. Thus, site-directed mutagenesis of a particular gene may require 3–6 person-months for the transformation process, outgrowth of transformants, screening for mutants with appropriate insertions, and plasmid analysis. As a result, fewer than 100 of the 1739 open reading frames (ORFs) in infectious B. burgdorferi have been subjected to site-directed mutagenesis despite intensive efforts by several laboratories.
Transposon Mutagenesis of B. burgdorferi
Transposon mutagenesis is a powerful means of producing randomized gene mutations in bacterial genomes (Beaurepaire and Chaconas, 2007). In 2004, Stewart et al. (2004) reported the development of a Himar1-based transposon suicide vector called pMarGent for use in Borrelia organisms. Himar1 is a transposon of the mariner family that was originally isolated from the blowfly, Drosophila mauritiana. Lampe et al. (1999) selected point mutants of the Himar1 transposase (included so-called C9 and A7 derivatives) that exhibited increased transposition rates; the C9 variant was used in pMarGent. A modified version of pMarGent called pGKT (Figure 1) was later developed to include a second selectable marker (KanR) in the non-transposed “backbone” in addition to the gentamycin resistance gene present in the transposable element (Stewart and Rosa, 2008). This modification greatly increases the stability of the vector in E. coli and facilitates additional alterations (such as the addition of signature tags). In both pMarGent and pGKT, the transposable element consists of the B. burgdorferi constitutive promoter flgBP coupled with the gentamicin resistance cassette aacC1 and the ColE1 origin of replication flanked by two Himar1 inverted tandem repeat sequences (Figure 1). The non-transposed region of pGKT includes flgBP::C9 transposase gene and the B. burgdorferi flaB promoter with the aph kanamycin resistance cassette. mariner-based transposons insert at any 5′-TA-3′ sequence with no apparent sequence bias, and result in duplication of the TA sequence at the other end of the transposon. Stewart et al. (2004) demonstrated the apparent random insertion of the pMarGent transposable element in both the chromosome and plasmids with high transformation efficiency (>3 × 10−5) in the non-infectious B. burgdorferi clones B31-AchbC72 and A3-89. However, these strains lack both lp25 and lp56, which contain the restriction-modification genes bbe02 and bbq67 (Lawrenz et al., 2002); lp25 also contains the pncA nicotinamidase gene that is required for mammalian and tick infection (Purser et al., 2003; Deneke and Chaconas, 2008). Attempts to transform the low-passage, infectious B. burgdorferi A3 and N40 strains (which contain lp25) with pMarGent were unsuccessful (Stewart et al., 2004).
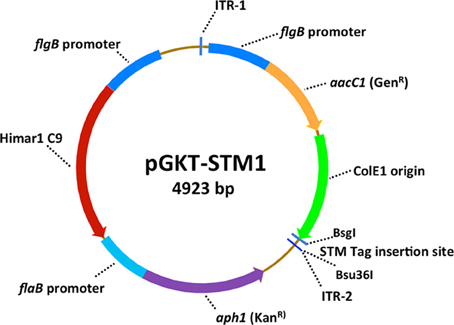
Figure 1. Diagram of the transposon vector pGKT-STM1, modified from the Borrelia-enhanced transposon deliver suicide vector pGKT of Stewart et al. (Stewart and Rosa, 2008). Features of pGKT include the use of B. burgdorferi flgB and flaA promoters to increase the efficiency of expression of the Himar1 c9 transposase and the gentamicin and kanamycin antibiotic resistance markers. The transposable element is flanked by the inverted tandem repeats ITR-1 and ITR-2, and contains the aacC1 gentamicin resistance cassette and the ColE1 origin of replication (to permit plasmid replication in E. coli and transposon insertion site rescue) (Stewart et al., 2004). The modification in pGKT-STM1 and other STM derivatives is the addition of a unique 7-bp signature tag close to ITR2.
In 2004, Kawabata et al. (2004) introduced the infectious, transformable B. burgdorferi strains 5A4NP1 and 5A18NP1. Both strains have a partial deletion and insertion of an aph KanR cassette in bbe02; 5A18NP1 also lacks the plasmid lp56 and hence bbq67. In electroporation studies with a shuttle vector, 5A4NP1 and 5A18NP1 yielded 10 colonies/μg DNA and 600 colonies/μg DNA, respectively; in contrast, the parental strains 5A4 and 5A18 (which contain functional bbe02) had <1 and 14 colonies per μg DNA. These strains thus reduce the transformation barrier while retaining full infectivity in the mouse model (Kawabata et al., 2004).
Utilization of pMarGent in combination with strain 5A18NP1 resulted in the first successful transposon mutagenesis of infectious B. burgdorferi (Botkin et al., 2006). A small library of 33 mutants was examined for transposon insertion site by E. coli rescue (Stewart et al., 2004), plasmid content, and infectivity in C3H/HeN mice. Mutations in the genes encoding IMP dehydrogenase (GuaB, involved in inosine-guanine interconversion) and the flagellar switch protein FlaG-1 were found to render B. burgdorferi non-infectious, but complementation was not attempted in these experiments. This study, although limited, indicated the feasibility of larger scale transposon mutagenesis studies.
Ordered Transposon Mutant Library
In 2007, Lin et al. (2012) began the process of accumulating a comprehensive transposon mutagenesis library utilizing signature-tagged versions of pKGT and the B. burgdorferi B31 derivative 5A18NP1. pKGT was modified to contain 12 different 7 bp “signature tags” that could be utilized to distinguish between co-infecting strains. This approach was based on earlier STM studies originated by Holden and colleagues (Hensel et al., 1995), as has been widely used for the study of bacterial-host interactions and pathogenesis. Relatively few signature tags were utilized in the construction of the B. burgdorferi library because of the inherent difficulties in producing large numbers of mutants in this organism. Each transformation typically yielded between 200 and 300 transformants, so several years was required to accumulate and characterize the number of clones estimated to yield a near saturation library. Overall, 6325 clones were isolated, and the insertion sites for 4479 clones were mapped by E. coli rescue and sequencing; the insertion sites for some transformations were not determined because of a high proportion of sibling clones. The plasmid content of the sequence-defined clones was determined using a novel, multiplex Luminex-based approach in which the samples can be analyzed in a high-throughput, 96-well format (Norris et al., 2011).
The large number of transposon mutant clones isolated precluded the use of re-cloning to assure clone purity. Although only well-isolated colonies were selected, it has been found that a fair number of the preparations contain two co-isolated clones. Colonies of B. burgdorferi are diffuse and transluscent, so this co-isolation is most likely due to the presence of overlapping colonies. It is recommended that the transposon mutants be re-cloned prior to detailed analysis and examined for the presence of other clones. The method currently in use involves PCR using primers flanking the transposon insertion site. A “wild type” PCR product the same size as obtained with 5A18NP1 parental strain in addition to a larger product (consistent with the expected transposon insertion) indicates the presence of clone(s) with insertions at other sites.
Each transposition event represents a mini-experiment, in that clones containing transposon insertions that significantly reduce fitness for in vitro growth will not survive. The distribution of transposon insertion sites thus provides a map of the genes and other elements essential for in vitro culture or replicon maintenance. The map is incomplete in the case of the existing B. burgdorferi transposon mutagenesis library, in that the library is not saturating. However, patterns can be discerned based on the lack of crippling mutations in groups of required genes. For example, the only ribosomal protein gene mutation isolated was in the last 10% of the ORF for the S21 protein, consistent with this mutation resulting a truncated but functional product. The 22 clones recovered with mutations in one of the two 23S ribosomal RNA genes had no obvious growth defect, indicating that cells are functional with only a single copy of this redundant gene. Only 34% of predicted protein-encoding genes in the chromosome had insertions, whereas 57% of predicted plasmid genes had insertions, most likely because of the higher proportion of essential genes required for in vitro culture in the chromosome. Variability occurred in the “hit rate” of both genes and replicons, with some genes and plasmids having as high as 23.06 (conserved hypothetical gene bb0017) and 18.01 (circular plasmid cp9) unique insertions per kb of DNA, respectively. The reason for this variability is not known, but conceivably may be related to improved in vitro growth characteristics in some mutants. Core gene classes that did not have debilitating mutations included those involved in DNA synthesis, transcription, translation, protein translocation, peptidoglycan synthesis, glycolysis, and pentose phosphate pathway, lipid interconversion, the V-type ATPase (presumably involved in maintaining the proton gradient across the cytoplasmic membrane), and the ubiquitous chaperones GroE and HP70 (Lin et al., 2012). Gene mutations that were permissive for in vitro growth included most of the numerous lipoprotein genes and several genes involved in chemotaxis. Whereas an intact chemotaxis apparatus is not required for culture, relatively few mutants were obtained in flagellar genes. Those mutants that were obtained in flagellar genes often produced elongated cells, indicative of defective cell division. Thus, spirochetal motility may facilitate the cell division process.
Detection of Virulence Determinants in Functional Groups
Screening of mutants for mouse infectivity by needle inoculation has been initiated using a Luminex-based STM analysis approach (Lin et al., 2012). In a typical STM experiment, 11 mutants, each with a different signature tag, were inoculated subcutaneously at 105 of each mutant into six C3H/HeN mice. Groups of three mice were analyzed at 2 and 4 weeks post-inoculation (PI), and up to five tissues (joint, heart, bladder, inoculation site, and a distal skin site) were examined from each mouse. In many experiments, the tissue specimens were each examined by both direct extraction of DNA from the tissue and by culture of the B. burgdorferi from the tissue followed by DNA extraction from the culture. A semi-quantitative, high-throughput Luminex©FlexMap™ procedure was developed for detection of B. burgdorferi STM mutants in mammalian hosts. The Luminex technology allows the assessment of up to 200 signals in a single well of a 96-well plate, thus providing a high-throughput method. Five tissues per mouse (joint, heart, bladder, inoculation site, and a distal skin site) were examined at two time points (2 and 4 weeks) with three mice each. A relatively small number of signature-tagged mutants were utilized in each STM infection experiment because of the challenges of constructing B. burgdorferi mutant libraries with multiple sequence tags, as well as the potential of a bottleneck effect. By combining the multiple mice, time points, tissues, and DNA preparation procedures, up to 60 data points per clone were obtained in a typical STM experiment (Lin et al., 2012). The data consisted of the Median Fluorescence Intensity (MFI) values obtained by Luminex analysis for each clone. Results obtained with representative infectious and non-infectious clones indicated that MFI values <100 (arbitrary units) were consistent with low infectivity. Variability in the numbers of organisms in individual tissue specimens occurs because of the paucibacillary nature of infection, and averaging of the multiple data points was found to yield more reliable results.
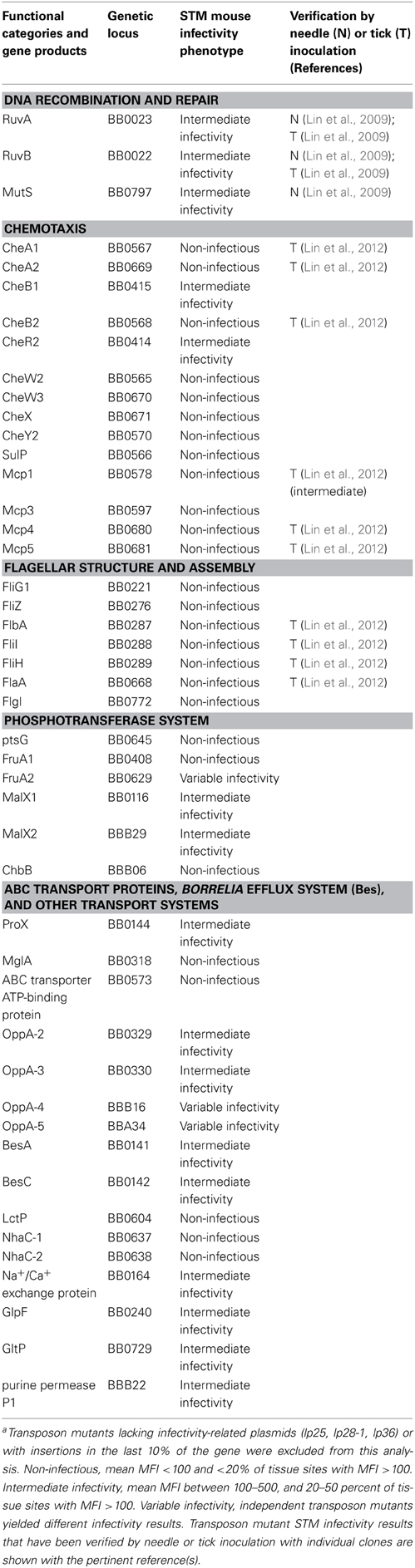
Functional genomics and pathogenomics approaches were applied to select mutants for the analysis of the roles of B. burgdorferi genes in metabolism, structure, pathogenesis, and other biological functions. The infectivity of 502 mutants in 422 genes has been tested (Lin et al., 2012), and the roles of some of these genes in pathogenesis and biological activities are being characterized more fully. Transposon mutations in genes associated with DNA recombination and repair, chemotaxis, motility, transport systems, plasmid maintenance, genes involved in gene regulation, sRNA genes, metabolic pathways, proteases, complement regulator-acquiring surface proteins (CRASPs), predicted lipoproteins, conserved hypothetical proteins (CHPs), and hypothetical proteins (HPs) of B. burgdorferi were selected for STM screening. A representative number of mutants in intergenic regions were also tested for infectivity to evaluate potential polar effects, small RNA genes, and regulatory regions (Lin et al., 2012). Some of the mutants exhibiting reduced infectivity in the STM screen are listed in Table 2. As an example, the effects of mutations in 23 genes involved in DNA recombination and repair on vlsE recombination were determined, and the results indicated that transposon mutants in ruvA, ruvB, and mutS exhibited reduced infectivity in mice and diminished vlsE recombination. Using this approach and site-directed mutagenesis, RuvA, RuvB, and MutS were the first trans-acting factors identified as necessary for vlsE recombination and antigenic variation (Dresser et al., 2009; Lin et al., 2009).
Motility and chemotaxis have long been thought to be key factors in borrelial infectivity and pathogenesis, but genetic evidence for that role has been obtained only recently (Li et al., 2010; Sze et al., 2012; Guyard et al., 2013; Sultan et al., 2013). In the STM transposon mutant analysis, 10 of 14 mutations in chemotaxis genes resulted in a severe loss of infectivity, indicating that the chemotaxis pathway is critical to infectivity (Lin et al., 2012). The genes involved in flagellar structure and assembly are also required for mouse infection. Some of these mutants, such as fliH, fliI, and flbA, had reduced motility, division defects, and structural changes in the flagellar motor. The cell division, motility, and structural defects of these mutants may all play a role in the observed low infectivity phenotype.
Transport is another key function in pathogenesis. The phosphoenolpyruvate phosphotransferase system (PEP-PTS) plays an important role in carbohydrate transportation and phosphorylation, also a potential role in gene regulation. STM results consistent with non-infectivity were observed for a transposon mutant in the gene encoding the glucose-glucoside specific IIBC component PtsG (Table 2); reduced infectivity was observed with mutants in the genes for the maltose specific PTS transporters MalX-1 and MalX-2 and the fructose-mannose transporter FruA-1. Disparate infectivity results were obtained with two mutants in the gene encoding the fructose-mannose specific IIB component FruA-2, so further analysis is needed. The Chb system is thought to play important roles in the utilization of chitobiose during infection of ticks (Tilly et al., 2001; Rhodes et al., 2010; Sze et al., 2013). Not surprisingly, inactivation of the genes encoding the chitobiose-specific components ChbA and ChbC did not affect mouse infection by needle inoculation in our system. However, the chitobiose-specific IIBC component ChbB mutant tested exhibited a no-infectivity phenotype (Lin et al., 2012); this finding will require further verification and investigation. In addition, the ABC transporter genes encoding the ATP-binding protein of the galactose transporter MglA, ProX, and an uncharacterized ATP-binding protein (bb0573) exhibited a low infectivity phenotype in the STM system. The five OppA oligopeptide ABC periplasmic binding proteins of B. burgdorferi exhibit varied infectivity phenotypes, but obviously have required specific binding properties. Genes encoding the lactose permease (LctP), Na+/H+ antiporter proteins (NhaC-1 and NhaC-2) also appear to required for mouse infection. Mutation of Borrelia efflux system genes besC and besA, Na+/Ca+ exchange protein (bb0164), glycerol uptake facilitator GlpF (bb0240), glutamate transporter (bb0729), and purine permease P1 (bbB22) resulted an intermediate infectivity phenotype (Table 2).
Screening of Virulence Genes in Infectivity-Related Plasmids
Lyme disease Borrelia species typically contain over 20 linear and circular plasmids. These replicons have been called “minichromosomes” because several are required for the complex life cycle of these organisms. Himar1-based transposon mutagenesis resulted in over 2500 sequence-defined unique insertion sites in the B. burgdorferi plasmids, disrupting 503 of 790 ORFs (68%); the proportion of ORFs disrupted in each plasmid ranged from 29 (in lp5) to 91% (cp9). A high proportion of borrelial plasmid genes do not have orthologs in other organisms, so many have no predicted function. There are also many paralogous families among the plasmid genes. Although over 30 B. burgdorferi plasmid genes have been successfully inactivated by site-directed mutagenesis (Rosa et al., 2005), the availability of the transposon mutant library will yield valuable information about these enigmatic replicons and their genes.
To date, representative mutants in genes associated with the infectivity-related plasmids cp26, lp25, lp28-1, lp36, and lp54 were analyzed using the STM process and Luminex-based technology (Lin et al., 2012). A summary of the results obtained for cp26, lp36, and lp54 are displayed in Figure 2. The percentage of positive tissue sites and mean MFI value were found to be useful parameters in summarizing the data from many animals, time points, tissue sites, and sampling methods, resulting in up to 60 data points. These two criteria yielded comparable patterns (Figure 2). The contiguous range of values sometimes does not permit a clear “±” result, resulting in an intermediate (or indeterminate) infectivity category in some instances. Nevertheless, the results obtained with mutants in these plasmids provide evidence that a high number of genes encoded in these plasmids appear to be required for full infectivity in mice. STM analysis is considered a screening procedure, and all results obtained by this method need to be confirmed by single clone inoculation and genetic complementation.
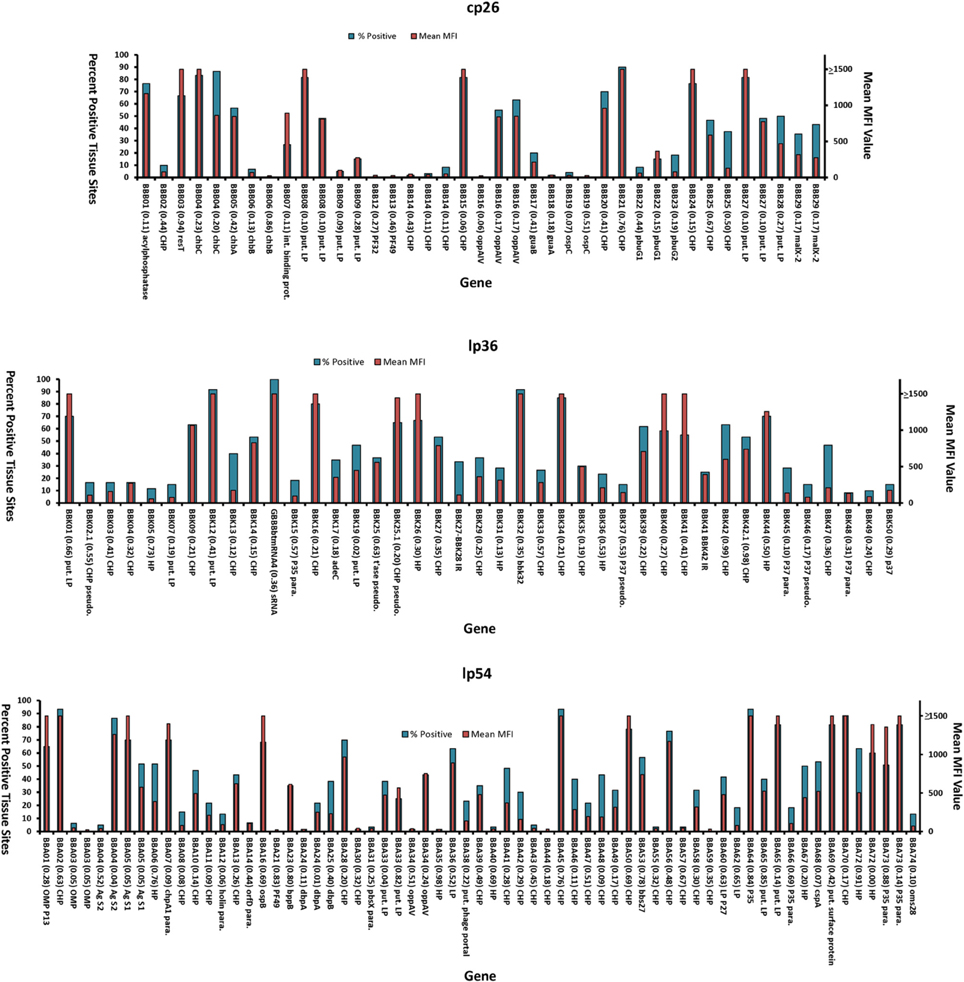
Figure 2. Mouse infectivity of transposon mutants in plasmid-encoded genes, as determined by Luminex-based STM analysis. Results obtained for genes in plasmids cp26, lp36, and lp54 are shown. For each mutant, the data represent the percent of positive tissue sites (MFI >100; blue bars) and mean MFI value (red bars) for 30–60 data points. Only clones that have all plasmids known to be required for mouse infection (cp26, lp25, lp28–1, lp36, and lp54) were analyzed. The gene designation, insertion ratio (bp from 5′ end to insertion/total bp), and description are provided. Abbreviations: HP, hypothetical protein; CHP, conserved hypothetical protein; para., paralog; pseudo., pseudogene; put. LP, putative lipoprotein; t'ase, transposase.
Transposon insertions were obtained in 26 of the 29 genes in cp26. Inactivation of 11 genes appeared to render a non-infectious phenotype based on the STM analysis, including those encoding chitobiose transporter ChbB, the PF32 and PF49 plasmid maintenance proteins, CHP BBB14, GMP synthase GuaA, and OspC (Figure 2). In addition, mutations in the genes encoding the CHP BBB02, HP gene BBB09, GuaB (inosine-5-monophosphate dehydrogenase, and purine permeases PbuG1 and PbuG2) yielded reduced STM parameter values (Figure 2) (Lin et al., 2012). Mutants in the genes encoding OspC, GuaA, GuaB, PbuG1, and PbuG2 had all been identified as virulence determinants previously (Byram et al., 2004; Grimm et al., 2004; Pal et al., 2004; Stewart et al., 2006; Tilly et al., 2006, 2009; Jewett et al., 2007a).
Of the 60 annotated genes in lp36, 38 protein-encoding genes have been disrupted. Transposon insertions were also found in one small RNA gene and two intergenic spacers. A total of 41 unique mutants have been examined for their infectivity in STM system (Figure 2). Consistent with previously published report by Jewett et al. (2007b), our result indicates that inactivation of the gene encoding adenine deaminase AdeC resulted in reduced mouse infection. Disruption of the BBK32 fibronectin-binding protein gene did not have an apparent effect on infectivity. BBK32 is involved in vascular endothelium adherence and dissemination, but is not absolutely required for infection (Li et al., 2006; Seshu et al., 2006; Norman et al., 2008; Moriarty et al., 2012); it is possible that the high infectious dose (105) used for each mutant in the STM analysis was sufficient to establish disseminated infection. Mutations in a region encompassing bbk02.1 to bbk07 (a poorly defined region in which some of the genes are no longer annotated) resulted in decreased infectivity. Mutations in several other CHP genes appear to result in reduced infectivity, including those in bbk15, and putative lipoprotein genes bbk13, bbk36, bbk37, and bbk45 through bbk50 (Figure 2) (Lin et al., 2012). In vivo expression technology (IVET) studies by Ellis et al. (2013) indicated that bbk46 is highly expressed during mouse infection, and further analysis showed that a bbk46 mutant was defective in persistent infection of mice.
Linear plasmid lp54 is one of the most highly conserved elements in the Borrelia genome. A total of 75 protein-encoding genes are encoded on lp54, and some of them are differentially expressed in tick vector and mammalian hosts. Most proteins encoded on lp54 do not have homologs in other organisms and are of unknown function (Casjens et al., 2000; Jewett et al., 2007a). Transposon mutants were obtained in 58 genes on lp54, and a high proportion of these mutants had decreased infectivity (Figure 2). Most of these are Borrelia species-specific proteins with unknown functions and several are B. burgdorferi-specific proteins. Transposon insertions in BBA03 (outer surface protein), BBA14 (OrfD paralog), BBA21 (PF49), BBA24 (DbpA), BBA30 (CHP), BBA31 (CHP), BBA35 (HP), BBA40 (HP), BBA43 (CHP), BBA44 (CHP), BBA55 (CHP), BBA57 (CHP), and BBA59 (CHP) resulted in a non-infectious phenotype by STM analysis. Additional mutants exhibited reduced STM analysis parameters, and may also have attenuated phenotypes. Disparate results were obtained for bba04, with two clones having high and low infectivity phenotypes (Figure 2) (Lin et al., 2012). Bestor et al. (2012) determined previously that a bba03 deletion mutant had a competitive disadvantage in tick-mediated infection of mice in comparison to the wild type parental strain; a deletion mutant lacking bba01-bba07 was further attenuated, indicating that other genes in this region may be required for full infectivity in mice. In other studies, site-directed disruption of the decorin binding protein DbpA was found to reduce mouse infectivity by needle inoculation but not by tick-mediated infection (Hagman et al., 2000); the transposon mutant results are consistent with the prior needle inoculation findings. Mutation in gene of ospB had little apparent effect on mouse infection, although this surface lipoprotein expressed predominantly in the tick infection and play the important role in tick midgut colonization. BBA64 is a member of the P35 paralogous protein family, and previous studies indicated that BBA64 is required for tick transmission (Gilmore et al., 2010), but mutants in this gene had only minor effects on infection by needle inoculation (Maruskova and Seshu, 2008; Maruskova et al., 2008). The STM analysis was consistent with the latter result, in that the available bba64 mutant (which had a transposon insertion in last 16% of reading frame) was fully infectious (Figure 2); in this case, however, it is possible that the mutant expresses a truncated but functional protein. Further studies with BBA64 immunization indicate that antibodies against this protein do not protect against needle or tick infection (Brandt et al., 2014). STM analysis indicated that mutation of lp54-encoded protein Oms28 (bba74) resulted in decreased infectivity (Lin et al., 2012), but these results are not consistent with prior studies with B31 A3, which apparently expresses a truncated BBA74 product (Mulay et al., 2009).
BptA, PncA, and Bbe31 encoded on lp25 had been identified as virulence determinants previously (Purser et al., 2003; Revel et al., 2005; Strother et al., 2005; Zhang et al., 2012). The transposon mutant library contains insertions in the genes encoding BptA and PncA, and these mutants exhibited reduced or non-infectivity as reported previously. In addition, mutations in genes of bbe04.1, bbe09, bbe18, bbe19, bbe24, and bbe29.1 substantially reduced infectivity, and intermediate infectivity was observed in mutants in genes of bbe07 and bbe18 (Lin et al., 2012).
In the STM studies, low infectivity was observed among lp28-1 mutants of genes encoding BBF03, BBF05, BBF10, BBF18, and BBF25. Mutation of genes of BBF04, BBF07, BBF08, BBF12, BBF19 appear to cause reduce infectivity. Interestingly, transposon insertions in intergenic spacers between bbf25-26 and bbf28-29 causes severe defect in spirochete infectivity (Lin et al., 2012). The vls locus involved in VlsE antigenic variation and immune evasion is located at one end of lp28-1 (Zhang et al., 1997). Only one transposon insertion was obtained in the vls silent cassettes, and none were obtained in the vlsE expression site (Lin et al., 2012). The silent cassette mutant was infectious, indicating that interruption of the silent cassette region is not sufficient to inhibit the vlsE recombination process.
Tn-seq
Transposon insertion site sequencing approaches such as Tn-seq is a powerful genetic screening technique in which high-throughput sequencing is used as a detection method in transposon mutagenesis screens to identify the contribution of individual genes to bacterial fitness under specific growth conditions (van Opijnen and Camilli, 2013). A library of transposon mutants (input pool) can be utilized to infect animals or subjected to in vitro selection conditions, and the resulting organisms (output pool) collected. Amplification of the transposon insertion site region and massively parallel sequencing provides a high-throughput comparison of the input pool and output pool. In comparison with STM approaches, Tn-seq has several advantages (Table 3), including the ability to examine an entire library in one set of replicate experiments without prior characterization of the insertion sites. For Borrelia studies, a disadvantage is that the plasmid content of each clone is not known, so the low infectivity of a particular mutant may be due to concomitant plasmid loss. Thus, presence of more than one clone with a mutation in a particular gene in the input pool and the underrepresentation of those clones in the output pool may be necessary to establish the gene product as a potential virulence determinant.
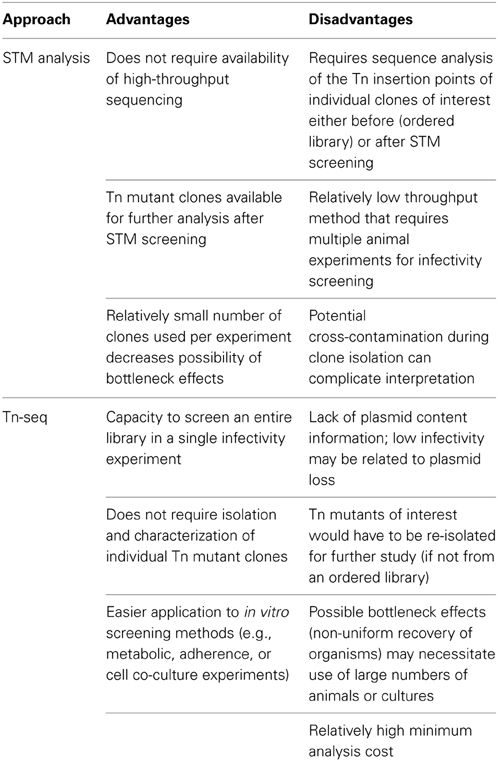
Table 3. Advantages and disadvantages of ordered library, signature-tagged mutagenesis (STM) and Tn-seq approaches.
In 2013, Troy et al. adapted Tn-seq for use in studies of B. burgdorferi using the STM transposon library (Troy et al., 2013). In a Tn-seq experiment, the transposon library is subjected to growth under a test condition. Genomic DNA is then isolated from the B. burgdorferi population before and after selection. The DNA is fragmented and cytosine tails (C-tails) are added using terminal deoxynucleotidyl transferase. The addition of a C-tail allows for the amplification of the transposon-genomic DNA junction using primers specific to the ColE1 site on the end of the transposon and the C-tail. The relative abundance of each mutant is determined by sequencing the chromosomal DNA flanking the transposon en masse using a primer that anneals to the end of the transposon. In this way, the exact location of every transposon insertion present in the library is revealed. The frequency of the insertion sequence in the library corresponds to the frequency of the transposon mutant containing that insertion in the original bacterial populations. The fitness of each insertion mutant in the test condition is determined by comparing the relative frequency of each mutant before and after growth under the selective pressure. Sequences that decrease in frequency in the library after selection have insertions in genes contributing to bacterial growth in the test condition. The quantitative nature of the high-throughput sequencing enables the discovery of unknown growth determinants including the differentiation of factors with partial phenotypes and/or those acting in redundant pathways. Furthermore, due to the non-specific nature of the genome library preparation, this Tn-seq technique can be used with any transposon as long as the sequence of the 3′ end of the transposon is known.
Tn-seq is particularly amenable to in vitro screens as unlike STM experiments that are limited to a small subset of mutants in each experiment, the entire transposon library can be tested in a single competition assay. Tn-seq has also been used to screen in vivo fitness. However, a potential complication of animal screens is a population bottleneck at the site of infection that results in the stochastic loss of a significant portion of the injected B. burgdorferi by 3 days post-infection. If all transposon mutants containing a particular insertion are lost at the inoculation site due to the bottleneck rather than decreased fitness, the disrupted gene may be misidentified as contributing to bacterial survival during infection. However, experiments by Troy et al. have demonstrated that the effects of this bottleneck can be circumvented, at least in part, by combining results from the B. burgdorferi populations recovered from multiple mice infected with the same inoculum (Troy et al., 2013). Part of the bottleneck effect is due to innate immune mechanisms (resulting in a non-specific reduction of the inoculated population), as demonstrated by the reduced magnitude of the bottleneck in MyD88−/− mice in comparison with wild type mice.
Conclusions
Results obtained to date with B. burgdorferi transposon mutants indicate that a high proportion of genes are required for mouse infection. The assessment of multiple tissue sites at two different time points using Luminex FlexMap technology provides a robust data set. Tn-seq has multiple advantages and is expected to greatly accelerate the application of transposon mutant libraries to studies of infectivity and other biological processes. Both STM and Tn-seq are considered screening methods, so the results must be confirmed by studies with isolated transposon mutants (or independently derived mutants) and genetic complemention. In addition, polar effects on downstream genes should be verified for observed reduced infectivity phenotype. All mutants should be re-cloned prior to more thorough studies due to the potential occurrence of co-isolation of two or more clones in a single colony. Finally, cross-complementation may occur between clones in the transposon library. We believe that the continued analysis of the mutant library will lead to a more comprehensive delineation of the virulence determinants of B. burgdorferi and will improve our understanding of the novel pathogenic and biologic properties of Lyme Borrelia organisms and other invasive, non-toxigenic, persistent pathogens.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Studies described in this article were supported by the United States National Institutes of Health, Institute of Allergy and Infectious Diseases through grants R01 AI059048 (Steven J. Norris and Tao Lin) and R21 AI103905 (Linden T. Hu, Tao Lin, and Steven J. Norris).
References
Barbour, A. G. (1988). Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J. Clin. Microbiol. 26, 475–478.
Beaurepaire, C., and Chaconas, G. (2007). Topology-dependent transcription in linear and circular plasmids of the segmented genome of Borrelia burgdorferi. Mol. Microbiol. 63, 443–453. doi: 10.1111/j.1365-2958.2006.05533.x
Bestor, A., Rego, R. O., Tilly, K., and Rosa, P. A. (2012). Competitive advantage of Borrelia burgdorferi with outer surface protein BBA03 during tick-mediated infection of the mammalian host. Infect. Immun. 80, 3501–3511. doi: 10.1128/IAI.00521-12
Botkin, D. J., Abbott, A. N., Stewart, P. E., Rosa, P. A., Kawabata, H., Watanabe, H., et al. (2006). Identification of potential virulence determinants by Himar1 transposition of infectious Borrelia burgdorferi B31. Infect. Immun. 74, 6690–6699. doi: 10.1128/IAI.00993-06
Brandt, K. S., Patton, T. G., Allard, A. S., Caimano, M. J., Radolf, J. D., and Gilmore, R. D. (2014). Evaluation of the Borrelia burgdorferi BBA64 protein as a protective immunogen in mice. Clin. Vaccine Immunol. 21, 526–533. doi: 10.1128/CVI.00824-13
Byram, R., Stewart, P. E., and Rosa, P. (2004). The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J. Bacteriol. 186, 3561–3569. doi: 10.1128/JB.186.11.3561-3569.2004
Casjens, S., Palmer, N., van Vugt, R., Huang, W. M., Stevenson, B., Rosa, P., et al. (2000). A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35, 490–516. doi: 10.1046/j.1365-2958.2000.01698.x
Deneke, J., and Chaconas, G. (2008). Purification and properties of the plasmid maintenance proteins from the Borrelia burgdorferi linear plasmid lp17. J. Bacteriol. 190, 3992–4000. doi: 10.1128/JB.00057-08
Dresser, A. R., Hardy, P. O., and Chaconas, G. (2009). Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog. 5:e1000680. doi: 10.1371/journal.ppat.1000680
Ellis, T. C., Jain, S., Linowski, A. K., Rike, K., Bestor, A., Rosa, P. A., et al. (2013). In vivo expression technology identifies a novel virulence factor critical for Borrelia burgdorferi persistence in mice. PLoS Pathog. 9:e1003567. doi: 10.1371/journal.ppat.1003567
Gilmore, R. D., Howison, R. R., Dietrich, G., Patton, T. G., Clifton, D. R., and Carroll, J. A. (2010). The bba64 gene of Borrelia burgdorferi, the Lyme disease agent, is critical for mammalian infection via tick bite transmission. Proc. Natl. Acad. Sci. U.S.A. 107, 7515–7520. doi: 10.1073/pnas.1000268107
Grimm, D., Tilly, K., Byram, R., Stewart, P. E., Krum, J. G., Bueschel, D. M., et al. (2004). Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U.S.A. 101, 3142–3147. doi: 10.1073/pnas.0306845101
Guyard, C., Raffel, S. J., Schrumpf, M. E., Dahlstrom, E., Sturdevant, D., Ricklefs, S. M., et al. (2013). Periplasmic flagellar export apparatus protein, FliH, is involved in post-transcriptional regulation of FlaB, motility and virulence of the relapsing fever spirochete Borrelia hermsii. PLoS ONE 8:e72550. doi: 10.1371/journal.pone.0072550
Hagman, K. E., Yang, X., Wikel, S. K., Schoeler, G. B., Caimano, M. J., Radolf, J. D., et al. (2000). Decorin-binding protein A (DbpA) of Borrelia burgdorferi is not protective when immunized mice are challenged via tick infestation and correlates with the lack of DbpA expression by B. burgdorferi in ticks. Infect. Immun. 68, 4759–4764. doi: 10.1128/IAI.68.8.4759-4764.2000
Hensel, M., Shea, J. E., Gleeson, C., Jones, M. D., Dalton, E., and Holden, D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403. doi: 10.1126/science.7618105
Jewett, M. W., Byram, R., Bestor, A., Tilly, K., Lawrence, K., Burtnick, M. N., et al. (2007a). Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66, 975–990. doi: 10.1111/j.1365-2958.2007.05969.x
Jewett, M. W., Lawrence, K., Bestor, A. C., Tilly, K., Grimm, D., Shaw, P., et al. (2007b). The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol. Microbiol. 64, 1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x
Kawabata, H., Norris, S. J., and Watanabe, H. (2004). BBE02 disruption mutants of Borrelia burgdorferi B31 have a highly transformable, infectious phenotype. Infect. Immun. 72, 7147–7154. doi: 10.1128/IAI.72.12.7147-7154.2004
Lampe, D. J., Akerley, B. J., Rubin, E. J., Mekalanos, J. J., and Robertson, H. M. (1999). Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. U.S.A. 96, 11428–11433. doi: 10.1073/pnas.96.20.11428
Lawrenz, M. B., Kawabata, H., Purser, J. E., and Norris, S. J. (2002). Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious B. burgdorferi. Infect. Immun. 70, 4798–4804. doi: 10.1128/IAI.70.9.4798-4804.2002
Li, C., Xu, H., Zhang, K., and Liang, F. T. (2010). Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol. Microbiol. 75, 1563–1576. doi: 10.1111/j.1365-2958.2010.07078.x
Li, X., Liu, X., Beck, D. S., Kantor, F. S., and Fikrig, E. (2006). Borrelia burgdorferi lacking BBK32, a fibronectin-binding protein, retains full pathogenicity. Infect. Immun. 74, 3305–3313. doi: 10.1128/IAI.02035-05
Lin, T., Gao, L., Edmondson, D. G., Jacobs, M. B., Philipp, M. T., and Norris, S. J. (2009). Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog. 5:e1000679. doi: 10.1371/journal.ppat.1000679
Lin, T., Gao, L., Zhang, C., Odeh, E., Jacobs, M. B., Coutte, L., et al. (2012). Analysis of an ordered, comprehensive STM mutant library in infectious Borrelia burgdorferi: insights into the genes required for mouse infectivity. PLoS ONE 7:e47532. doi: 10.1371/journal.pone.0047532
Maruskova, M., Esteve-Gassent, M. D., Sexton, V. L., and Seshu, J. (2008). Role of the BBA64 locus of Borrelia burgdorferi in early stages of infectivity in a murine model of Lyme disease. Infect. Immun. 76, 391–402. doi: 10.1128/IAI.01118-07
Maruskova, M., and Seshu, J. (2008). Deletion of BBA64, BBA65, and BBA66 loci does not alter the infectivity of Borrelia burgdorferi in the murine model of Lyme disease. Infect. Immun. 76, 5274–5284. doi: 10.1128/IAI.00803-08
Moriarty, T. J., Shi, M., Lin, Y. P., Ebady, R., Zhou, H., Odisho, T., et al. (2012). Vascular binding of a pathogen under shear force through mechanistically distinct sequential interactions with host macromolecules. Mol. Microbiol. 86, 1116–1131. doi: 10.1111/mmi.12045
Mulay, V. B., Caimano, M. J., Iyer, R., Dunham-Ems, S., Liveris, D., Petzke, M. M., et al. (2009). Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J. Bacteriol. 191, 2783–2794. doi: 10.1128/JB.01802-08
Norman, M. U., Moriarty, T. J., Dresser, A. R., Millen, B., Kubes, P., and Chaconas, G. (2008). Molecular mechanisms involved in vascular interactions of the Lyme disease pathogen in a living host. PLoS Pathog. 4:e1000169. doi: 10.1371/journal.ppat.1000169
Norris, S. J., Coburn, J., Leong, J. M., Hu, L. T., and Höök, M. (2010). “Pathobiology of Lyme disease Borrelia,” in Borrelia: Molecular and Cellular Biology, eds D. S. Samuels and J. D. Radolf, (Hethersett, Norwich: Caister Academic Press), 299–331.
Norris, S. J., Howell, J. K., Odeh, E. A., Lin, T., Gao, L., and Edmondson, D. G. (2011). High-throughput plasmid content analysis of Borrelia burgdorferi B31 by using Luminex multiplex technology. Appl. Environ. Microbiol. 77, 1483–1492. doi: 10.1128/AEM.01877-10
Pal, U., Yang, X., Chen, M., Bockenstedt, L. K., Anderson, J. F., Flavell, R. A., et al. (2004). OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Invest. 113, 220–230. doi: 10.1172/JCI200419894
Purser, J. E., Lawrenz, M. B., Caimano, M. J., Howell, J. K., Radolf, J. D., and Norris, S. J. (2003). A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48, 753–764. doi: 10.1046/j.1365-2958.2003.03452.x
Revel, A. T., Blevins, J. S., Almazan, C., Neil, L., Kocan, K. M., de la Fuente, J., et al. (2005). bptA (bbe16) is essential for the persistence of the Lyme disease spirochete, Borrelia burgdorferi, in its natural tick vector. Proc. Natl. Acad. Sci. U.S.A. 102, 6972–6977. doi: 10.1073/pnas.0502565102
Rhodes, R. G., Atoyan, J. A., and Nelson, D. R. (2010). The chitobiose transporter, chbC, is required for chitin utilization in Borrelia burgdorferi. BMC Microbiol. 10:21. doi: 10.1186/1471-2180-10-21
Rosa, P. A., Tilly, K., and Stewart, P. E. (2005). The burgeoning molecular genetics of the Lyme disease spirochaete. Nat. Rev. Microbiol. 3, 129–143. doi: 10.1038/nrmicro1086
Samuels, D. S. (1995). Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47, 253–259.
Samuels, D. S., and Radolf, J. D. (2010). Borrelia: Molecular Biology, Host Interaction and Pathogenesis. Hethersett, Norwich: Caister Academic Press.
Seshu, J., Esteve-Gassent, M. D., Labandeira-Rey, M., Kim, J. H., Trzeciakowski, J. P., Hook, M., et al. (2006). Inactivation of the fibronectin-binding adhesin gene bbk32 significantly attenuates the infectivity potential of Borrelia burgdorferi. Mol. Microbiol. 59, 1591–1601. doi: 10.1111/j.1365-2958.2005.05042.x
Steere, A. C., Coburn, J., and Glickstein, L. (2004). The emergence of Lyme disease. J. Clin. Invest. 113, 1093–1101. doi: 10.1172/JCI21681
Stewart, P. E., Hoff, J., Fischer, E., Krum, J. G., and Rosa, P. A. (2004). Genome-wide transposon mutagenesis of Borrelia burgdorferi for identification of phenotypic mutants. Appl. Environ. Microbiol. 70, 5973–5979. doi: 10.1128/AEM.70.10.5973-5979.2004
Stewart, P. E., and Rosa, P. A. (2008). Transposon mutagenesis of the Lyme disease agent Borrelia burgdorferi. Methods Mol. Biol. 431, 85–95. doi: 10.1016/j.idc.2007.12.013
Stewart, P. E., Wang, X., Bueschel, D. M., Clifton, D. R., Grimm, D., Tilly, K., et al. (2006). Delineating the requirement for the Borrelia burgdorferi virulence factor OspC in the mammalian host. Infect. Immun. 74, 3547–3553. doi: 10.1128/IAI.00158-06
Strother, K. O., Broadwater, A., and de Silva, A. (2005). Plasmid requirements for infection of ticks by Borrelia burgdorferi. Vector Borne Zoonotic Dis. 5, 237–245. doi: 10.1089/vbz.2005.5.237
Sultan, S. Z., Manne, A., Stewart, P. E., Bestor, A., Rosa, P. A., Charon, N. W., et al. (2013). Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infect. Immun. 81, 2012–2021. doi: 10.1128/IAI.01228-12
Sze, C. W., Smith, A., Choi, Y. H., Yang, X., Pal, U., Yu, A., et al. (2013). Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi. Infect. Immun. 81, 1775–1787. doi: 10.1128/IAI.00050-13
Sze, C. W., Zhang, K., Kariu, T., Pal, U., and Li, C. (2012). Borrelia burgdorferi needs chemotaxis to establish infection in mammals and to accomplish its enzootic cycle. Infect. Immun. 80, 2485–2492. doi: 10.1128/IAI.00145-12
Tilly, K., Bestor, A., Dulebohn, D. P., and Rosa, P. A. (2009). OspC-independent infection and dissemination by host-adapted Borrelia burgdorferi. Infect. Immun. 77, 2672–2682. doi: 10.1128/IAI.01193-08
Tilly, K., Elias, A. F., Errett, J., Fischer, E., Iyer, R., Schwartz, I., et al. (2001). Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183, 5544–5553. doi: 10.1128/JB.183.19.5544-5553.2001
Tilly, K., Krum, J. G., Bestor, A., Jewett, M. W., Grimm, D., Bueschel, D., et al. (2006). Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74, 3554–3564. doi: 10.1128/IAI.01950-05
Troy, E. B., Lin, T., Gao, L., Lazinski, D. W., Camilli, A., Norris, S. J., et al. (2013). Understanding barriers to Borrelia burgdorferi dissemination during infection using massively parallel sequencing. Infect. Immun. 81, 2347–2357. doi: 10.1128/IAI.00266-13
van Opijnen, T., and Camilli, A. (2013). Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442. doi: 10.1038/nrmicro3033
Zhang, J. R., Hardham, J. M., Barbour, A. G., and Norris, S. J. (1997). Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89, 275–285. doi: 10.1016/S0092-8674(00)80206-8
Keywords: Borrelia burgdorferi, Lyme disease, transposon mutagenesis, microbial pathogenesis, bacterial physiology, mouse models
Citation: Lin T, Troy EB, Hu LT, Gao L and Norris SJ (2014) Transposon mutagenesis as an approach to improved understanding of Borrelia pathogenesis and biology. Front. Cell. Infect. Microbiol. 4:63. doi: 10.3389/fcimb.2014.00063
Received: 19 February 2014; Accepted: 25 April 2014;
Published online: 20 May 2014.
Edited by:
Catherine Ayn Brissette, University of North Dakota School of Medicine and Health Sciences, USAReviewed by:
Janakiram Seshu, The University of Texas at San Antonio, USAPeter Kraiczy, University Hospital of Frankfurt, Germany
Copyright © 2014 Lin, Troy, Hu, Gao and Norris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven J. Norris, Department of Pathology and Laboratory Medicine, University of Texas Medical School at Houston, PO Box 20708, Houston, TX, USA e-mail:c3RldmVuLmoubm9ycmlzQHV0aC50bWMuZWR1
 Tao Lin
Tao Lin Erin B. Troy2
Erin B. Troy2 Linden T. Hu
Linden T. Hu Lihui Gao
Lihui Gao Steven J. Norris
Steven J. Norris