- 1Department of Microbiology and Immunology, The University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA
- 2Department of Pharmaceutical Sciences, The University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA
Streptococcus pyogenes is a significant pathogen of humans, annually causing over 700,000,000 infections and 500,000 deaths. Virulence in S. pyogenes is closely linked to mobile genetic elements like phages and chromosomal islands (CI). S. pyogenes phage-like chromosomal islands (SpyCI) confer a complex mutator phenotype on their host. SpyCI integrate into the 5′ end of DNA mismatch repair (MMR) gene mutL, which also disrupts downstream operon genes lmrP, ruvA, and tag. During early logarithmic growth, SpyCI excise from the bacterial chromosome and replicate as episomes, relieving the mutator phenotype. As growth slows and the cells enter stationary phase, SpyCI reintegrate into the chromosome, again silencing the MMR operon. This system creates a unique growth-dependent and reversible mutator phenotype. Additional CI using the identical attachment site in mutL have been identified in related species, including Streptococcus dysgalactiae subsp. equisimilis, Streptococcus anginosus, Streptococcus intermedius, Streptococcus parauberis, and Streptococcus canis. These CI have small genomes, which range from 13 to 20 kB, conserved integrase and DNA replication genes, and no identifiable genes encoding capsid proteins. SpyCI may employ a helper phage for packaging and dissemination in a fashion similar to the Staphylococcus aureus pathogenicity islands (SaPI). Outside of the core replication and integration genes, SpyCI and related CI show considerable diversity with the presence of many indels that may contribute to the host cell phenotype or fitness. SpyCI are a subset of a larger family of streptococcal CI who potentially regulate the expression of other host genes. The biological and phylogenetic analysis of streptococcal chromosomal islands provides important clues as to how these chromosomal islands help S. pyogenes and other streptococcal species persist in human populations in spite of antibiotic therapy and immune challenges.
Introduction
Streptococcus pyogenes is a significant human pathogen, annually causing over 700,000,000 infections and 500,000 deaths (Carapetis et al., 2005). Genome sequencing has revealed that prophages and other mobile genetic elements are important features of Streptococcus pyogenes (group A streptococcus) chromosomes, sometimes contributing up to 10% of the total DNA (Desiere et al., 2001; Ferretti et al., 2001; Banks et al., 2002; Canchaya et al., 2002). These genome prophages follow a typical lambdoid gene arrangement, with sequentially organized modules for integration and lysogeny, DNA replication, transcriptional regulation, DNA packaging and head assembly, tail and tail fiber assembly, and lysis (Desiere et al., 2001; Banks et al., 2002; Canchaya et al., 2002; Brussow et al., 2004). In S. pyogenes and many other pathogens, these essential phage genes are often followed by one or more virulence genes such as toxins (Brussow et al., 2004). Numerous genes on S. pyogenes chromosomes are the targets for site-specific integration by these mobile genetic elements, and for some of these genes, integration has the potential to interrupt or alter their transcription (McShan and Ferretti, 2007). Of these targeted genes, one location stands out both for its frequency of occupation by a chromosomal island (CI) as well as the potential phenotypic impact integration would have on the cell: the operon encoding the genes for DNA mismatch repair (MMR). We have characterized phage-like CI in S. pyogenes that integrate into MMR gene mutL, silencing this gene and the other downstream genes of the operon (Scott et al., 2008, 2012). The integration of the S. pyogenes Chromosomal Island M1 (SpyCIM1) into the chromosome induces a complex mutator phenotype that results from the interruption of the operon and downstream DNA repair genes (Scott et al., 2008, 2012). Largely due to the extensive and ongoing efforts to sequence the genomes of many species of bacteria, additional islands integrated into mutL also have been identified in other Streptococcus species. This review will examine the CI identified so far, their known or potential impacts on host phenotype and survival, and implications for the evolution of this group and their host bacteria. A note concerning nomenclature: when referred to collectively as a group, S. pyogenes phage-like CI are referred to as SpyCI, but a specific CI is identified so to associated it with a particular strain or isolate (e.g., SpyCIM1, SpyCIM49, etc.). The same convention is applied to CI from other streptococcal species.
SpyCIM1 and the Host Mutator Phenotype
Typically, site-specific recombination occurs between bacterial and phage genomes such that the transcription of the targeted host gene is unimpeded by the presence of the prophage (Fouts, 2006). This maintenance of gene function is accomplished by two factors: (1) duplication of the host DNA sequence at the site of crossover by a portion of the phage DNA and (2) integration at the 3′ end of the targeted gene so that the duplication can complete the original bacterial ORF (Fouts, 2006; Louie et al., 2007; McShan and Ferretti, 2007). By contrast, integration into the 5′ end or the middle of a gene could result in the disruption of normal transcription with a concomitant loss of gene function. Occasional examples of prophages altering the expression of host genes have been reported in Escherichia coli and Staphylococcus aureus (Mason and Allen, 1975; Lee and Iandolo, 1986; Thomas and Drabble, 1986; Coleman et al., 1991; Campbell et al., 1992), but these occurrences have been notable in part because of their rarity. By contrast, genome sequencing has revealed that S. pyogenes prophages frequently target attachment sites positioned at the promoter or 5′ end of genes that, following integration, potentially could alter gene expression, or create polar mutations (McShan and Ferretti, 2007). Of these mobile genetic elements with the potential to alter gene expression, the phage-like chromosomal island that frequently targets and regulates the DNA mismatch repair (MMR) operon of S. pyogenes is perhaps the most remarkable.
MMR has been extensively studied in E. coli where the system recognizes base pair mismatches in nascent, hemimethylated DNA and directs strand-specific repair. The newly synthesized unmethylated DNA strand, which contains the mismatch, is cleaved by the MMR system and repair is initiated by re-synthesis of the cleaved region. Gram-positive bacteria and eukaryotes do not rely upon methylation for strand recognition, probably instead relying upon modification of the beta clamp of DNA polymerase III for strand discrimination (Li, 2008). LeClerc and co-workers were the first to observe that the mutator phenotype was present in wild populations of E. coli and Salmonella enterica at unexpectedly high frequencies and that these phenotypes mapped to mutations in MMR genes (LeClerc et al., 1996). It was subsequently found that mutators were present in both pathogenic and non-pathogenic E. coli, suggesting that this trait conferred a selective advantage upon the cell in spite of the risk of increased frequencies of deleterious mutations (Matic et al., 1997). Subsequent studies showed that MMR mutants are frequently isolated from clinical strains of many species. For example, antibiotic treatment of Pseudomonas aeruginosa infections in cystic fibrosis patients correlates with the rapid appearance of drug-resistant MMR mutants (LeClerc and Cebula, 2000; Oliver et al., 2002). Other examples have been found in Neisseria meningitis, Helicobacter pylori, Haemophilus influenzae, and Staphylococcus aureus (Bjorkholm et al., 2001; Richardson et al., 2002; Bayliss et al., 2004; Prunier and Leclercq, 2005; Trong et al., 2005). The frequency of such mutator strains is often high: for example, 20% of P. aeruginosa strain from cystic fibrosis patients and over 50% of epidemic-associated serogroup A N. meningitis strain are mutators (Oliver et al., 2002; Richardson et al., 2002). The MMR system can act as a barrier for genetic diversity and bacteriophage transduction, thus inhibition of MMR removes this barrier and promotes diversification through homeologous recombination (Limia et al., 1998; Kataja et al., 1999; Matic et al., 2000). However, in all of these species, the MMR defects result from mutations that render mutS or mutL permanently defective. The resulting mutator phenotype is a double-edged sword, however. The advantages gained by a cell in rapidly acquiring favorable mutations like antibiotic resistance are balanced by the possibility of deleterious mutations arising that diminishes cell viability. In S. pyogenes, a remarkable solution has evolved to achieve a mutator phenotype while minimizing the potential risks: a growth-dependent molecular switch controlled by a CI, which allows the cells to be phenotypically wild type when resources are abundant but switching to a mutator phenotype when facing challenges like limited nutrient availability.
The MMR operon of S. pyogenes M1 strain SF370 (Figure 1) is composed of the genes mutS, mutL, lmrP, ruvA, and tag, which encode MMR, a multidrug efflux pump of the major facilitator family, a Holliday junction resolvase, and base excision repair glycosylase, respectively, (Ferretti et al., 2001). These genes are grouped on a polycistronic mRNA that is controlled by a single promoter upstream of mutS. Analysis of the M1 genome showed that the phage-like chromosomal island SpyCIM1 was integrated between mutS and mutL, and this integration was subsequently found to interrupt the expression of mutL and the downstream genes (Scott et al., 2008, 2012). SpyCIM1 is not a static element, permanently residing in the bacterial genome like a typical prophage, but rather a dynamic element that excises from chromosome during early logarithmic growth and replicates as a circular episome (Scott et al., 2008). As the bacterial population reaches the end of logarithmic phase and enters stationary phase, SpyCIM1 re-integrated into its unique attachment site (attB) at the beginning of mutL (Figure 1, insert). Thus, SpyCIM1 acts as a growth-dependent molecular switch to control the expression of MMR. The outcome of this switch is that the S. pyogenes cell alternates between a mutator and wild type phenotype in response to growth: during rapid cell division and DNA replication, the integrity of the genome is maintained by an active MMR system while during stationary phase or other periods of infrequent cell division, mutations may accumulate at a higher rate.
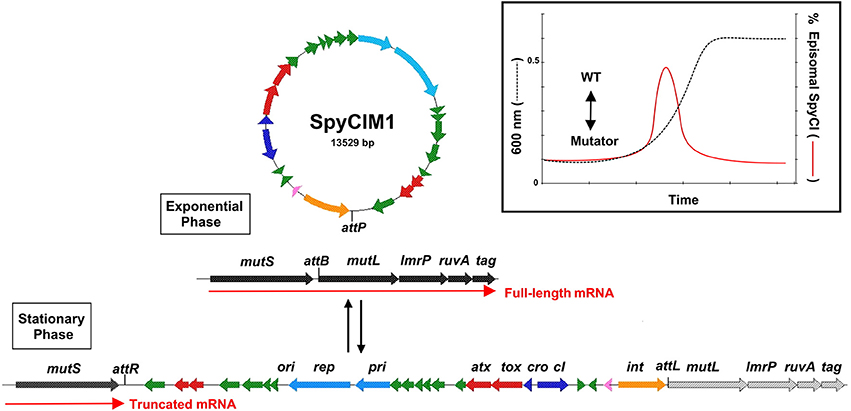
Figure 1. SpyCIM1 regulates the MMR operon through dynamic site-specific excision and integration. The molecular switch controlled by SpyCIM1 is shown. The MMR operon of S. pyogenes is comprised of genes encoding DNA mismatch repair (mutS and mutL), multidrug efflux (lmrP), a Holliday-junction resolvase (ruvA), and base excision repair (tag). During exponential phase, SpyCIM1 excises from the chromosome, circularizes, and replicates as an episome, restoring transcription of the entire MMR operon (WT). Excision and mobilization occurs early in logarithmic growth in response to yet unknown cellular signals (Insert; adapted from Scott et al., 2008). As logarithmic growth continues, SpyCIM1 re-integrates into mutL at attB, and by the time the culture reaches stationary phase, the integration process has completed, again blocking transcription of the MMR operon. WT, Wild type phenotype associated with unimpeded expression of the MMR operon. Color key of predicted gene functions: Green, genes of unknown function; red, possible toxin-antitoxin maintenance genes; light blue, DNA replication; dark blue, control of lysogeny; pink, transmembrane peptide; orange, site-specific integrase.
This molecular switch controls additional operon genes downstream of mutL. The next gene, lmrP, encodes a putative multidrug resistance efflux pump (MDR) of the major facilitator family (Bolhuis et al., 1995; Putman et al., 2001). In Lactococcus lactis, the gene was characterized as an ATP dependent pump that extrudes multiple drugs across the membrane, preventing toxic accumulations of these chemicals in the bacteria (Bolhuis et al., 1995). Presently, the natural substrate for LmrP in S. pyogenes is unknown, as is why a multidrug efflux pump is transcriptionally linked with a DNA repair operon in group A and related species of streptococci (Supplemental Table 1). However, the ability to regulate expression of this gene may have selective advantage for the streptococcus. In Listeria monocytogenes an LmrP homolog (mdrM) exists whose expression or inhibition can control the magnitude of the host cytosolic response to infection, and loss of MdrM protein function leads to a 3-fold reduction in IFN-β response to infection (Crimmins et al., 2008). It may be that inhibition of LmrP expression in S. pyogenes similarly provides a mechanism of innate immunity avoidance. Indeed, if true, SpyCI regulation of the LmrP in S. pyogenes may have a large biological impact given how a MDR can influence multiple processes in a cell by removal of toxic or inhibitory substances.
The next gene on the polycistronic message, ruvA, encodes an ATP-dependent helicase that promotes branch migration of Holliday junctions during homologous genetic recombination and recombinant repair of damaged DNA. The loss of RuvA function leads to increased sensitivity to UV damage as irradiation induced DNA lesions lead to arrested replication forks (Iwasaki et al., 1989; Tsaneva et al., 1992; Kaplan and O'Donnell, 2006). This increase in sensitivity to UV irradiation is clearly observed in S. pyogenes strains carrying SpyCI (Scott et al., 2008, 2012).
The last gene on the operon, tag, encodes a 3-methyladenine DNA glycosylase I which is involved in base excision repair (Bjelland et al., 1993). This enzyme is important in recognizing and purging aberrant and modified bases from damage induced by DNA damaging elements such as the alkylating agent ethyl methanesulfonate (Wyatt et al., 1999). Loss of the 3-methyladenine DNA glycosylase greatly increases the spontaneous mutation rate associated with single nucleotide substitution (Kaasen et al., 1986; Bjelland et al., 1993; Wyatt et al., 1999). The loss of gene expression from mutL to tag causes the cell to exhibit a complex mutator phenotype that impacts several DNA repair or maintenance systems (Scott et al., 2008, 2012). Indeed, the silencing of this operon may necessarily need to be reversed occasionally to maintain cell viability, given our observation that permanent loss of the ability to excise from the bacterial chromosome lead to the use of a new promoter to express these genes in M5 strain Manfredo (Scott et al., 2012).
SpyCI are frequent genetic elements in S. pyogenes genomes (Table 1). M serotypes associated with SpyCI carriage currently include M1, M2, M4, M5, M6, M18, M25, M28, M31, M37, M49, M53, M59, M78, and M123 (Scott et al., 2008, 2012; Suvorov et al., 2009). Currently, it is not known whether SpyCI infect only a subset of S. pyogenes serotypes, perhaps defined by surface targets for phage attachment, or whether most serotypes may serve as SpyCI hosts and the current sample size is simply too small. Other factors such as phage immunity proteins or the dependence of SpyCI on helper phages with limited host ranges may also influence the dissemination of these chromosomal islands. At least from the standpoint of integration, virtually all group A streptococci could serve as a host for SpyCI since the attB DNA sequence at the beginning of mutL is highly conserved.
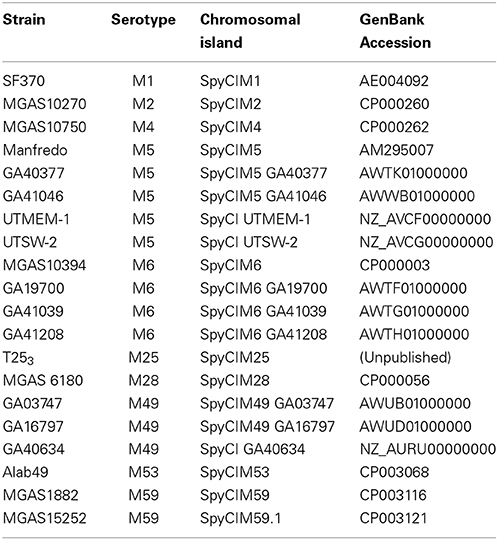
Table 1. S. pyogenes strains with SpyCI integrated into DNA MMR gene mutL that have been identified by genome sequencing.
In general, the presence of a SpyCI in a given S. pyogenes strain correlates with a higher mutation rate and UV sensitivity when compared to strains lacking this chromosomal island (Scott et al., 2012). Different SpyCI+ strains do show a range of mutation rates, however (Figure 2). When compared to SpyCI-free strain NZ131, genome strains with the chromosomal island showed mutation rates that ranged between 5 and 167 times higher. Similarly, resistance to UV irradiation also showed a range of sensitivities. This strain-to-strain variation may reflect differences in SpyCI regulation that determine whether the chromosomal island tends to be integrated into mutL or excised as an episome (i.e., how frequently the MMR operon is transcriptionally active). Variations in the operator controlling the repressor and antirepressor may play a role in this decision to remain integrated or extrachromosomal as well as variations in other DNA repair genes that affect the overall cell mutation rate (Scott et al., 2012). The one exception to this general trend was found in serotype M5 strain Manfredo, which has a 128 bp deletion in the SpyCI integrase gene that renders it inactive but a mutation rate that was a 1000-fold lower than NZ131 (Figure 2). So, in spite of the fact that SpyCIM5 was permanently integrated into the Manfredo chromosome, this strain was wild type for the MMR operon. This paradox was resolved by the discovery of a novel promoter within the SpyCIM5 integrase pseudogene that rescued the expression of mutL and the downstream genes. Interestingly, expression from this novel promoter was depressed by mitomycin C treatment, which was in contrast to the activation of the MMR in SF370 and other strains with a SpyCI capable of excision from mutL. It remains unknown whether this apparent mechanism of gene expression control is the result of natural selection or merely a circumstantial byproduct of evolution of this compensatory promoter (Scott et al., 2012).
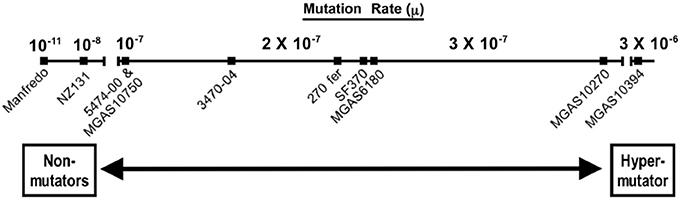
Figure 2. The mutation rates of SpyCI+ S. pyogenes genome and clinical strains. Strains harboring SpyCI integrated into mutL have phenotypes that range from non-mutator in Manfredo, where a novel CI promoter in the defective integrase gene rescues expression of mutL, to a hypermutator in M6 genome strain MGAS10394. Strain NZ131 does not carry a SpyCI and is presented as a wild type strain with regards to the MMR operon. The mutation rate is calculated as mutations per generation. The figure was drawn using the data from Scott et al. (2012).
Comparisons of mutation rates between strains possessing or lacking SpyCI is informative, but the conclusions are inferential since other genes not in this operon may influence the observed final phenotype. Therefore, to directly assess the effect of SpyCIM1 carriage alone, the chromosomal island was cured from the M1 SF370 genome to create an isogenic derivative, and comparisons of the two strains show that island integration is responsible for a 200-fold increased mutation rate, increased sensitivity to ethidium bromide, increased UV irradiation sensitivity, and higher rates of single point mutations (Euler et al., submitted). As the island excises in response to growth, this dynamic regulation of the operon allows the organism to maintain genetic fidelity in optimal conditions while selectively increasing mutation rates during stressful conditions (Scott et al., 2008).
Conservation and Diversity of SpyCI Genes
SpyCI and other phage-like chromosomal islands may have originated from defective prophages, but their biology suggests an even more complex origin. The defining gene of SpyCI is the integrase (int), whose expression and regulation controls the molecular switch for the MMR operon. Within the known SpyCI, int is highly conserved at both the gene and protein level (Scott et al., 2008, 2012). The SpyCI integrase genes form a distinct group within the S. pyogenes prophage integrases (McShan, 2005), although the integrase encoded by S. pyogenes strain NZ131 prophage NZ131.1 is 62% identical to the SpyCIM1 integrase at the amino acid level, suggesting that these genes may have had a recent common origin (McShan et al., 2008). The integration site for prophage NZ131.1 is near the promoter of hypothetical protein Spy49_0371 (McShan et al., 2008), which is unrelated to the MMR operon.
Typical for prophages and phage-like CI, two genes encoding predicted DNA-binding proteins are found upstream of int and arranged in opposite orientations flanking a probable operator site. These genes, which encode the predicted repressor and antirepressor, are the likely candidates for the control of SpyCI integration and excision. Excision of SpyCI can be induced by mitomycin C treatment (Scott et al., 2008, 2012), suggesting that the mostly uncharacterized S. pyogenes SOS DNA repair pathway can trigger SpyCI excision and may lead to packaging by a helper prophage for dissemination to new host cells (Nguyen, unpublished observations). However, the cellular signals that induce the normal cycle of SpyCI excision and re-integration during growth, which presumably is triggered by repressor cleavage, are yet unknown. Interestingly, these genes and their operators vary between the different SpyCI, and these differences may contribute to the range of mutation rates seen between the different S. pyogenes genome strain that harbor these CI (Scott et al., 2012). That is, SpyCI repressors that are more sensitive to cleavage would favor the episomal form of the SpyCI and the appearance of the wild type phenotype with respect to MMR while repressors that are more stable would favor integration and the mutator phenotype. The operator DNA sequences of SpyCIM1, SpyCIM2, SpyCIM28, and SpyCIM53 are all nearly identical while the lysogeny module of the hypermutator M6 strain MGAS10394 is essentially the same as the emerging hypervirulent strain MGAS15252 (Fittipaldi et al., 2012; Scott et al., 2012). As we previously pointed out, the SpyCI controlled mutator phenotype in MGAS15252 may contribute to the many striking phenotypic changes that are associated with this strain, which include enhanced transmission by skin contact, significantly impaired ability to grow in saliva, and a tendency not to colonize the oropharynx (Scott et al., 2012).
Other SpyCI genes are conserved, forming part of the core set of genes that identify these mobile genetic elements. The predicted primase and replicase genes (Figure 1) are closely related to homologs from Streptococcus thermophilus plasmid pSt106, which are essential for plasmid replication (Geis et al., 2003). While the molecular details of these genes in SpyCI DNA replication and extrachromosomal maintenance remain to be determined, their high degree of conservation within these CI argues their essential role (Scott et al., 2008), presumable to ensure DNA replication of episomal SpyCI. Other SpyCI genes show various degrees of conservation. One universally conserved gene, which was not originally annotated due to its small size, is positioned immediately upstream of int and encodes a small transmembrane domain peptide of unknown function (Figure 1). If this ORF really represents an expressed gene, then its product has the potential to alter the surface properties of the streptococcal cell, potentially altering its antigenicity, functioning as an environmental sensor, or providing a protection mechanism against lytic phages by interfering with their attachment. Other genes, based upon homology with known homologs, may function like a toxin-antitoxin pair to prevent SpyCI eliminations (labeled tox and atx in Figure 1). None of the SpyCI-encoded genes include an identifiable DNA polymerase or a terminase subunit, which plays an important role in the helper phage packaging process of the Staphylococcus aureus pathogenicity islands (SaPI) (Novick et al., 2010). Most of the remaining ORFs in the SpyCI encode products of unknown function, and much variation exists between the individual members (Scott et al., 2008). The driving forces behind the genetic diversity of SpyCI from different S. pyogenes isolates are poorly understood at present.
Mismatch Repair Chromosomal Islands of Other Streptococcus Species
The transcriptional linking of mutS and mutL is not universal in prokaryotes, although analysis of publicly available genomes shows that many streptococcal species do group these genes as a unit. This group includes, in addition to S. pyogenes, Streptococcus mutans, Streptococcus equi subspecies equi and subsp. zooepidemicus, Streptococcus agalactiae, Streptococcus uberis, and Streptococcus thermophilus (Table 2). Interestingly, Streptococcus pneumoniae does not follow this pattern, encoding MMR genes hexA and hexB at distant sites on its chromosome. Genes ruvA and tag are positioned near hexB on the S. pneumoniae chromosome and are probably co-transcribed with each other but not with hexB. The gene composition of the operon found in S. pyogenes and other related streptococci may represent an instance where evolution has selected for an arrangement that simplifies the control of expression for several housekeeping genes. This arrangement, however, has allowed mobile genetic elements like the SpyCI to assume a unique regulatory role.
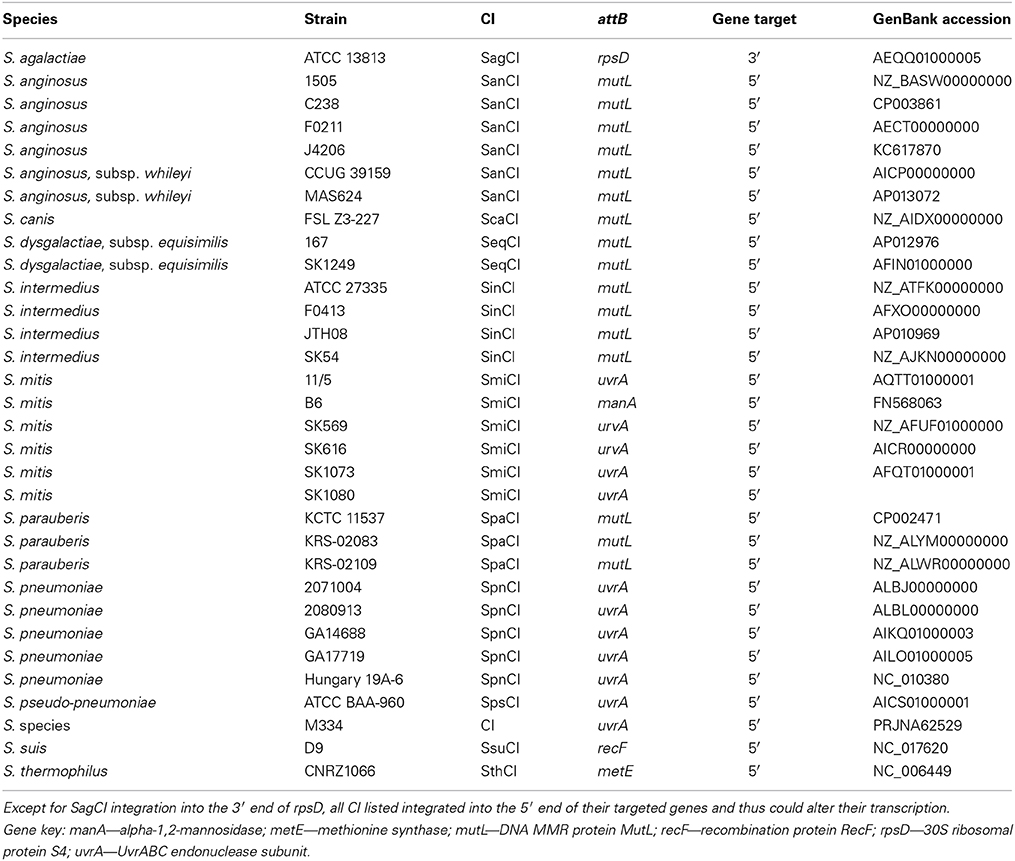
Table 2. Chromosomal islands identified in other streptococcal species that target mutL or other genes for integration.
Using the SpyCIM1 integrase gene as the query, a TBLASTN search (Altschul et al., 1997) of the available complete or partial bacterial genomes revealed many related islands in Streptococcus species that are integrated into the same attachment site in mutL, including Streptococcus anginosus, Streptococcus intermedius, Streptococcus dysgalactiae subsp. equisimilus, Streptococcus canis, and Streptococcus parauberis (Table 2). In these species, the CI may potentially regulate the MMR operon much like how SpyCIM1 does. Ignoring the defective SpyCIM5 integrase that has a 128 bp deletion in the gene (Scott et al., 2012), these integrases share at least 64.0% amino acid sequence identity (Supplemental Figure 1). Phylogenetic tree analysis of the integrases shows close similarity as well with little sequence distance (Figure 3). Perhaps this is not surprising, as these islands have conserved core sequences required for integration into streptococcal mutL, which itself provides a conserved target (Figure 3, insert). However, despite strong conservation of the integrase, other regions of these CI show considerable genetic diversity between species. Genomic alignment of the islands revealed four distinct groups within the MMR islands (Figure 4). The diversity and frequency of these islands is remarkable with chromosomal islands found in streptococcal species associated with human disease, streptococcosis in flounder and dairy bovine mastitis (Nho et al., 2011; Lefébure et al., 2012).
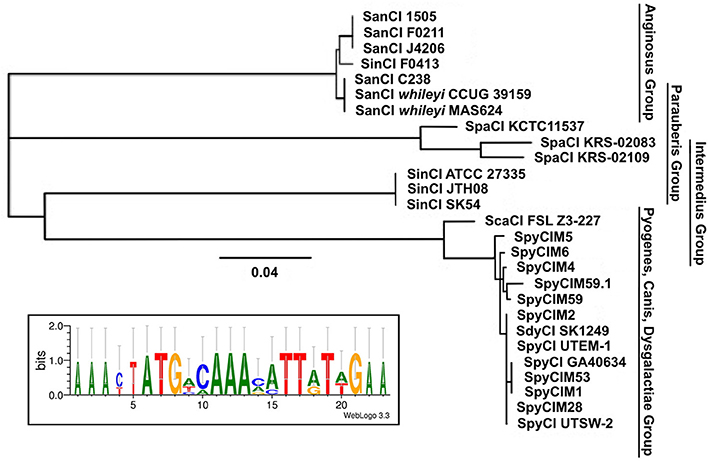
Figure 3. Phylogenetic tree of the streptococcal CI integrases that target MMR gene mutL. Analysis of the chromosomal island integrases amino acid sequences that target mutL was used to construct a phylogenetic tree showing the four known major groups. The insert shows the consensus alignment of the mutL. Strain details are given in Tables 1, 2. The proteins encoded by the CI integrase genes were aligned and the phylogenetic tree created using Geneious v. 6.1.7 (Drummond et al., 2012). The consensus of the mutL attachment site was created using WebLogo (Crooks et al., 2004).
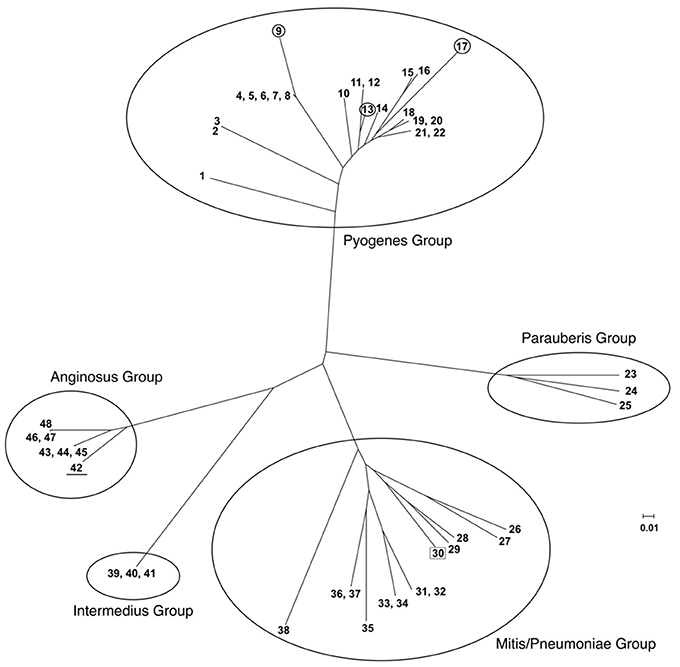
Figure 4. Phylogenetic tree of streptococcal chromosomal islands integrated into DNA repair genes mutL. Full genome trees are derived. In the Pyogenes group, all of which integrate into mutL, the circled members of the tree are CI found in the closely related species S. dysgalactiae subsp. equisimilis (SeqCI; nos. 9 and 13) and S. canis (ScaCI; no. 17). The SmiCI that integrates into manA in the Mitis/Pneumoniae group is boxed for identification; the remainder of the group integrates into uvrA. The members of the Anginosus and Intermedius groups integrate into mutL and have a conserved integrase gene; however, the remainder of their chromosomes is divergent, causing them to form separate phylogenetic groups. The one exception is that the S. intermedius strain F0413 SinCI (#42; underlined) is closely related to the SanCI and forms part of that group. The detailed legend for the individual members of the tree is presented in Supplemental Table 2.
The MMR operon, as organized in S. pyogenes, is present in many other species of the genus streptococcus. However, there is one major differentiating characteristic in this operon that creates a division between these other species: the presence or absence of the gene for MDR LmrP (Figure 5 and Supplemental Table 1). Groups A, B, C, and G streptococci have lmrP inserted between mutL and ruvA as do a number of other species including S. iniae, S. uberis, and S. parauberis. While S. mutans and most viridans streptococci do not have lmrP, a few, like S. oralis, do. The presence or absence of this MDR gene, as well as its regulation by a chromosomal island, raises some interesting biological questions. Why is a gene for a drug efflux pump transcriptionally linked to DNA repair genes? Further, what is to be gained, if anything, by the cell through inhibition of expression of this gene? Indeed, why is this gene dispensable in some species but present in others? The answers to these questions will come as the role of this MDR protein in the biology and virulence of S. pyogenes and other species is determined. SpyCI-like CI that target mutL usually are found in species that have lmrP; however, S. anginosus and S. intermedius, both members of the milleri group that lack lmrP, have acquired CI that integrate into this gene.
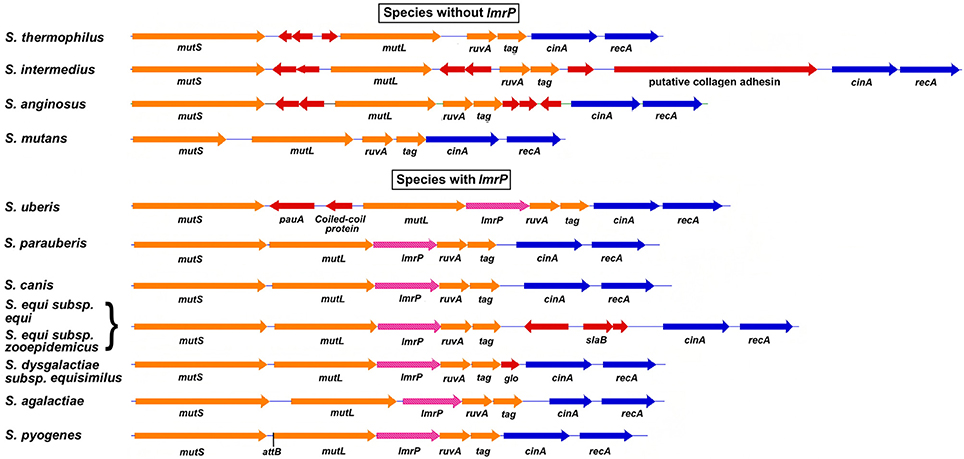
Figure 5. The MMR operon region from selected streptococcal species. The MMR operon in S. pyogenes and related streptococci is composed of a core group of genes involved in MMR, Holliday junction resolution and BER (mutS, mutL, ruvA, and tag). Indels are frequently found to supplement this basic genetic unit: MDR gene lmrP forms an additional part of this core in groups A, B, C, and G streptococci. Some indels encode proteins of unknown function, but others appear to encode potential virulence factors such as pauA (streptokinase) in S. uberis and the collagen-like adhesin in S. intermedius. For an extended list of streptococcal species that are differentiated by the presence or absence of lmrP, see Supplemental Table 1. The core genes of the streptococcal MMR operon are colored orange; MDP gene lmrP is magenta, and indels are red. The attachment site for SpyCI in S. pyogenes is indicated (attB). Legend for indel genes: glo—glyoxalase family protein; slaB—phospholipase A2 SlaB; pauA—streptokinase.
In addition to CI identified by genome sequencing, another S. anginosus CI was identified from a clinical source. A daptomycin resistant strain of S. anginosus (strain J4206) was isolated from a patient with bacteremia and septic shock (Palacio et al., 2011) and was found to have a CI integrated into mutL. This CI (SanCI J4206) has been sequenced in our laboratory and its impact on the host mutator phenotype assessed (manuscript in preparation). Daptomycin resistance results from multistep genetic changes and is thought to be rare (Tran et al., 2013). As the related SpyCI are known to confer a mutator phenotype in their host, SanCI J4206 could putatively contribute to the emergence of daptomycin resistance in S. anginosus through hypermutability. Previously, daptomycin resistance has not observed in an in vitro study of 106 S. anginosus isolates (Streit et al., 2005).
It is possible that SpyCI, as well as other Gram-positive phage-like chromosomal islands (Novick et al., 2010), have a complex evolutionary history and their genetic material may have originated from disparate sources. While each chromosomal island shows considerable diversity (Scott et al., 2008), several genes, notably the integrase, primase, and replicase genes, are highly conserved, providing clues to the minimal genome composition needed for a functional CI.
Streptococcal CI with Other Gene Integration Targets
Streptococcal chromosomal islands have been identified by genome sequencing that target genes other than mutL (Table 2 and Figure 6). Although the biological impact of integration into any of these genes is not yet known, some predictions may be made based upon whether the 5′ or 3′ end of the ORF is the point of site-specific recombination. For example, in three of the nine currently available genomes of S. agalactiae, a SagCI is found integrated into rpsD, which encodes the 30S ribosomal protein S4. Given the essential role of this protein in ribosome function, it is perhaps no surprise that this chromosomal island integrates into the 3′ end of the rpsD ORF so transcription is unimpeded. Other genes, by contrast, could be regulated in their expression patterns to the benefit of the cell, at least under certain environmental or physiological conditions. For example, the chromosomal island from the B6 strain of Streptococcus mitis (SmiCIB6) integrates into the 5′ end of the gene encoding alpha-1,2-mannosidase (manA) while the Streptococcus thermophilus CI (SthCI) integrates into metE, encoding methionine synthase. In both cases, a SpyCI-like switch could activate or silence these genes to optimize host fitness. Regulation of metE expression could alter the relative intracellular levels of homocysteine and methionine (Matthews et al., 1998), which under some conditions might be favorable to the cell. Similarly, S. mitis B6 may only need the action of alpha-1,2-mannosidase when a need arises to use glycan or glycoproteins as a carbon source as observed in other oral streptococci (Tarelli et al., 1998).
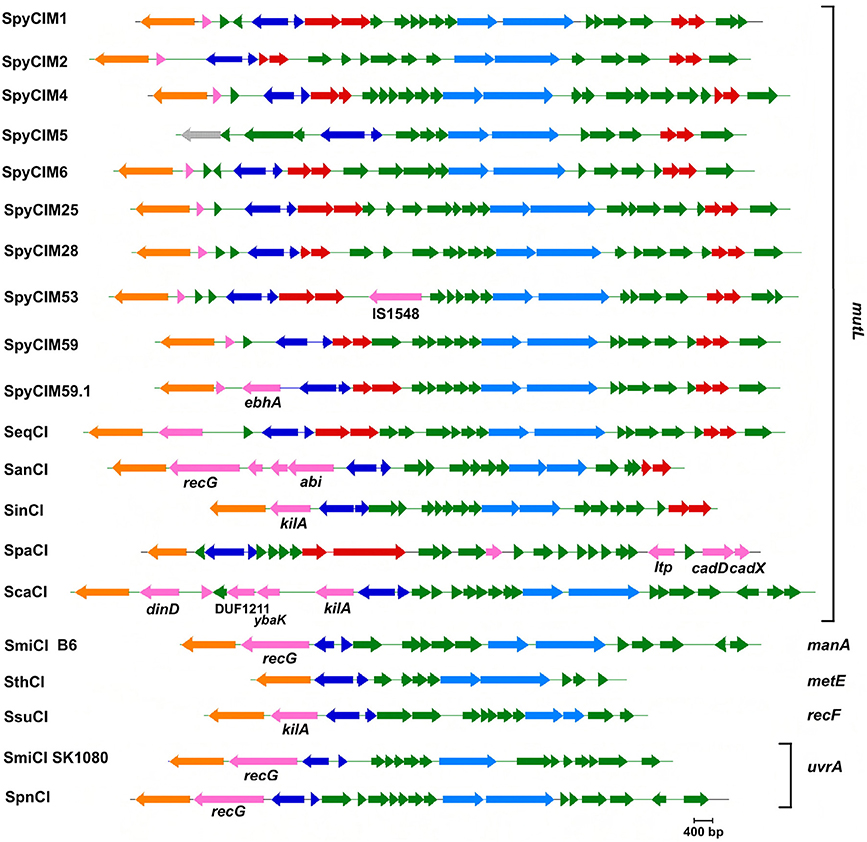
Figure 6. The CI of S. pyogenes and related species. The identification of each CI is shown to the left of its map, while the integration site (attB) is shown to the right of the map (see Tables 1, 2 for identification of CI and attB sites). INDELs with identifiable homologous genes are labeled by that homolog. Color key: orange—site-specific integrase; dark blue—control of lysogeny; light blue—DNA replication; red—maintenance; pink—INDELs; green—unknown function; gray—pseudogene. INDEL key: IS1548—insertion element 1548; ebhA—transmembrane surface adhesin; recG—recombination protein RecG superfamily; abi—abortive phage infection protein; kilA—plasmid maintenance protein; ltp—host cell surface-exposed lipoprotein; cadD and cadX—cadmium resistance proteins; dinD—DNA damage inducible protein; DUF1211—domain of unknown function; ybaK—prolyl tRNA synthetase. Maps were created using Gene Construction Kit (Textco BioSoftware, West Lebanon, NH).
The S. mitis SmiCI provide an interesting glimpse into the evolution and diversification of phage-like CI. As discussed above, SmiCI B6 integrates into manA. However, the related SmiCI from other S. mitis strains as well as the SpnCI from S. pneumoniae target the probable operator controlling the expression of the gene (uvrA) encoding the A subunit of the UvrABC excinuclease that is a key component of nucleotide excision repair (NER). Remarkably, the SmiCI B6 integrase has 98% homology at the protein level with the other SmiCI and the SpnCI integrases (Figure 7), even though they target separate DNA genes (manA and uvrA, respectively). This observation suggests that only a relatively few amino acid changes were necessary to expand the gene repertoire of this CI, creating new possibilities for altering host expression patterns. The SmiCI and SpnCI targeting of uvrA, a key gene for another essential DNA repair pathway (NER), suggests that ability to selectively adopt a mutator phenotype in a regulated fashion provides a selectable advantage for these cells. Based upon the example of SpyCI in S. pyogenes, one could predict that the SpnCI and SmiCI will have a cycle of excision and re-integration in response to the growth state of the cell or some environmental sensor.
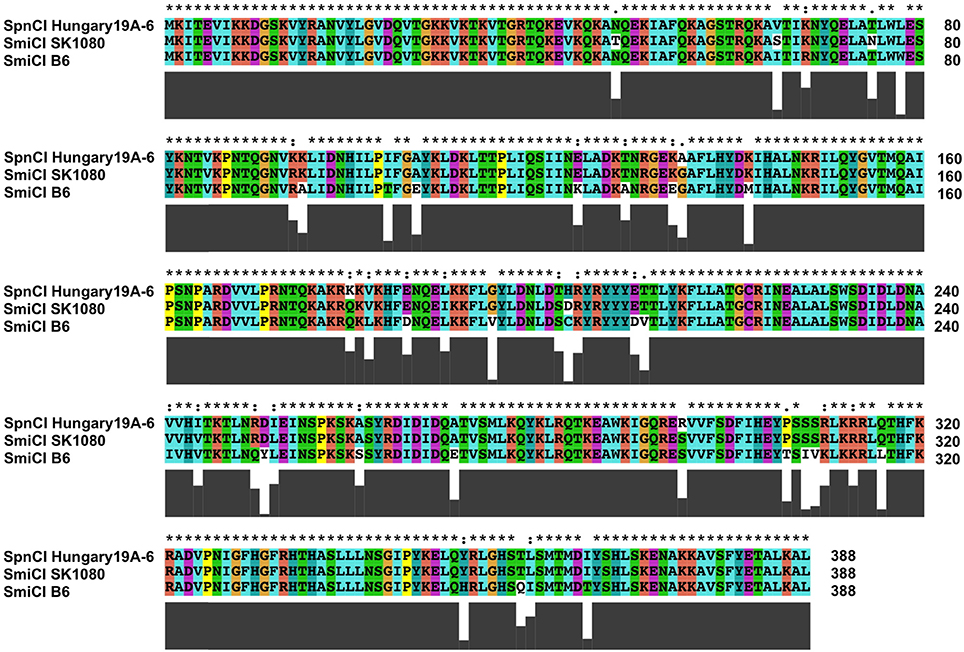
Figure 7. Alignment of the SmiCI and SpnCI integrases that target either manA or uvrA. The integrase proteins encoded by the S. mitis SmiCI B6, the S. mitis SmiCI SK1080, and S. pneumoniae Hungary19A-6 SpnCI were aligned using ClustalX (Jeanmougin et al., 1998). The integrases from SpnCI Hungary19A-6 and SmiCI SK1080 both recognize an identical attachment site in the promoter of uvrA, while the SmiCI B6 integrase recognizes manA. Above the alignment, an asterisk (*) indicates identity between the three proteins, a colon (:) indicates a strongly conservative amino acid substitution, and a dot (.) indicates a weakly conservative amino acid substitution
Indels and CI-Mediated Phenotypes
Many members of the streptococcal phage-like CI have notable indels (insertions and deletions) in their genomes that may contribute to the host phenotype (Figure 6). We previously reported on the impact of the 128 bp deletion in the SpyCIM5 integrase gene on S. pyogenes strain Manfredo and the solution the cell adopted to prevent permanent silencing of mutL and the downstream genes of the operon (Scott et al., 2012). The SpyCIM53 element from strain Alab49 (Bessen et al., 2011) is genetically quite related to SpyCiM1 from strain SF370 but has acquired the insertion element IS1548 (Figures 4, 6). The recently reported severe disease associated, epidemic strain MGAS15252 (Fittipaldi et al., 2012) contains SpyCIM59.1, a CI with an indel (ebhA) predicted to encode extracellular matrix-binding protein with a transmembrane domain, which could be a potential virulence factor.
The phage-like CI from the non-pyogenes streptococcal species often contain indels that may contribute to host fitness or prevent elimination of the CI. A gene encoding an ATPase related to the RecG superfamily family of proteins is found in the S. anginosus SanCI as well as the phylogenetically separate SmiCI from S. mitis and SpnCI from S. pneumoniae (Figures 4, 6). Protein RecG provides a means of rescuing stalled replication forks in E. coli (Briggs et al., 2004), and these CI-encoded homologs might offset some aspects of the mutator phenotype caused by the integration of the CI into mutL (SanCI) or uvrA (SmiCI and SpnCI). Another CI indel that frequently appears is a homolog of the KilA family of proteins (found in the SinCI, ScaCI, and SsuCI genomes). The kilA gene was identified in the broad-host-range E. coli plasmid RK2 and originally detected by the potential lethality of its gene product to the host cell (Goncharoff et al., 1991). Based upon previously reported criteria (Makarova et al., 2009), a number of potential toxin-antitoxin genes are found in the streptococcal phage-like CI that may function to prevent elimination of the CI from the host during replication and cell division (Figure 6), and these KilA homologs may also contribute to CI maintenance.
Other streptococcal phage-like CI indels may improve host fitness to environmental challenges. Resistance to cadmium and other heavy metals in microbes occurs frequently, which interestingly is often co-selected with antibiotic resistance and may contribute to the persistence of resistance genes in the environment (Baker-Austin et al., 2006). The S. parauberis SpaCI has acquired the cadDX cadmium resistance operon that is related to ones found in S. aureus and Streptococcus salivarius (O'Brien et al., 2002; Chen et al., 2008). Many heavy metal resistance mechanisms are able to act upon multiple substrates (Baker-Austin et al., 2006), and the principal substrate for resistance conferred by this SpaCI operon in S. parauberis is unknown. However, since cadD encodes a transmembrane protein, its gene product will alter the surface properties of the host cell, which could not only confer resistance but also alter antigenicity and charge of the cell membrane. The SpaCI also encodes a second protein with the potential for altering the surface of its host: Ltp, a member of a family of phage encoded lipoproteins that are involved in superinfection exclusion. Proteins of this family have been shown to act at the stage of DNA release from the phage head into the cell (Neve et al., 1998), thus interfering with the lytic cycle of phages that might infect S. parauberis. Other streptococcal phage-like CI also carry genes that may protect their host cell from infection by invading lytic phages or induction of endogenous prophages to the lytic cycle. The SanCI of S. anginosus encode the gene abi, which encodes a protein that is a member of a protein family that mediates bacteriophage resistance by causing abortive infection in Lactococcus species (Anba et al., 1995; Bidnenko et al., 1995). The SanCI encoded Abi protein has a predicted helix-turn-helix DNA binding motif, which in these proteins is thought to play a role in the interference of the phage life cycle through altering the transcriptional program of the virus. In addition to the kilA gene discussed above, the S. canis ScaCI contains several other indels that may promote host fitness through protection of cell metabolism. Located immediately upstream of int is a gene encoding a homolog of the DinD DNA damage induced protein. In E. coli, the DinD protein inhibits RecA-mediated DNA strand exchange, which may limit unwanted homologous recombination (Uranga et al., 2011). Another ScaCI gene (ybaK) also may encode a system to increase host cell fitness. Members of the YbaK protein family have deacylase domains are trans-acting amino acid-editing class II prolyl tRNA synthetases, whose primary function is to hydrolyze mischarged cysteinyl-tRNAPro, thus ensuring the fidelity of translation and prevent accumulation of mistranslated proteins (Kumar et al., 2012; Das et al., 2014).
CI as Regulatory Elements of the Host Phenotype
While the regulation of MMR by SpyCI in S. pyogenes and other streptococci is a remarkable evolutionary adaptation, it is one of a number of examples of how phages and phage-like mobile genetic elements have evolved to form a beneficial relationship with their host bacterium. In E. coli, several examples of integrated phage-like elements affecting gene sequence or expression have been reported. In strain K-12, a P-4-like cryptic prophage controls expression of the AlpA transcriptional regulator by site-specific recombination. Overexpression of alpA leads to suppression of capsule overproduction and UV sensitivity in cells defective for the Lon protease (Kirby et al., 1994; Trempy et al., 1994). The integration of the cryptic prophage suppresses alpA expression and restores normal capsule production and UV sensitivity. Unlike the dynamic cycle of MMR control by SpyCIM1, excision of the cryptic prophage leads to the elements loss and a permanent conversion of cell phenotype (Kirby et al., 1994). Similarly, integration of phage lambda into a secondary attachment site at the guaB promoter inhibits the expression of inosine monophosphate (IMP) dehydrogenase (Thomas and Drabble, 1986). In this case, though, the selective benefit of the resulting inhibition of de novo purine biosynthesis following integration is uncertain and may be just an unfavorable event that would be unstable over time. The integration of Lambdoid phage 21 replaces 165 bp of the 3′ end of the isocitrate dehydrogenase gene icd to provide an alternative ending (Campbell et al., 1992), which again is an event of unknown biological impact that may warrant further study.
A number of Gram-positive examples of gene control by mobile phage-like elements have also been reported. The sigma K intervening element (skin) of Bacillus subtilis is a 48 kB prophage-like element that integrates into sigK, separating the gene into regions historically known as spoIIIC and spoIVCB (Kunkel et al., 1990). Excision of the skin element from the chromosome leads to reconstitution of the sigK gene, which during sporulation encodes the mother-cell-specific sK factor (Stragier et al., 1989). Likewise, a smaller, 14.6 kb element (skinCd) was found to interrupt sigK in Clostridium difficile; unlike in B. subtilis where skin may be deleted without major impact upon sporulation, the C. difficile skinCd is required for efficient completion of this event (Haraldsen and Sonenshein, 2003). Interestingly, phylogenetic analysis shows that these skin elements arose independently in Bacillus and Clostridium, leading to the speculation that this unusual form of sigK regulation may have some specific selective advantage in the regulation of sporulation. In Listeria monocytogenes, a recent report showed that the DNA uptake competence system, considered non-functional, has a temperate prophage integrated into comK (Rabinovich et al., 2012). Surprisingly, the L. monocytogenes Com system promoted bacterial escape from macrophages, and the regulation of this system depended upon the activation of comK following prophage excision, which was specifically induced during intracellular growth, reminiscent of the activation of SpyCIM1 activation at the onset of exponential phase (Scott et al., 2008). Recently, a phage-like chromosomal island from the genome of Enterococcus faecalis V583 has been described and its requirement for a helper phage for packaging demonstrated (Matos et al., 2013). The integration target site for this element is identified as the promoter of a xanthine/uracil permeases family of genes; however, the precise function of this gene in E. faecalis or the impact of the chromosomal island integration into this gene remains unknown.
The relation between mobile genetic elements and host gene expression is a key component of the biology of S. aureus, which in many ways resembles these relationships in S. pyogenes. These elements range from typical lambdoid prophages to the SaPI phage-like chromosomal islands. S. aureus prophages have been demonstrated to mediate gene conversion by controlling the expression of lipase, β-Lysin, Staphylokinase, and Enterotoxin A (Lee and Iandolo, 1985; Coleman et al., 1989; Zabicka et al., 1993), while the SaPI are vectors for the toxic shock syndrome toxin (TSST) (Lindsay et al., 1998; Novick et al., 2010). In addition to being vectors for TSST, the SaPI carry other genes that modify the host phenotype such as a biofilm-associated protein (BAP) (Ubeda et al., 2003) or von Willebrand factor-binding protein (Lindsay et al., 1998; Viana et al., 2010).
In their pioneering paper demonstrating the unexpectedly high frequency of the mutator phenotype in wild populations of bacteria, LeClerc and co-workers observed that “the ultimate pathogen would possess an elevated mutation rate that is transient (or conditional), providing genetic variation during the first few hours when the pathogen must survive, invade, and colonize its host” (LeClerc et al., 1996). The SpyCI of S. pyogenes and the similar CI that colonize related species of the genus streptococcus may well be an example of a system that fulfills this prediction by their unique mechanism of MMR control. The frequent occurrence of MMR defects in natural bacterial populations argues that a selective benefit exists in this phenotype, whether it stems from increased mutability or the potential for horizontal gene transfer (Matic et al., 1995). In most species of bacteria, however, the mutator phenotype is fixed and poses a distinct risk to the cell in the form of acquiring unwanted mutations that might lead to decreased viability. So far, only in the streptococci has a system been discovered that allows the cell to switch between a mutator and wild type phenotype to presumably achieve a balance between costs and benefits. The presence of other phage-like CI in the various streptococcal species that potentially target other genes for regulation suggests that these elements may be an important aspect of the biology of these low G+C% bacteria. The widespread occurrence by SpyCI and related CI in the pathogenic streptococci may be a clue to their importance to the virulence and survival of these bacteria, which may prove ultimately to be as significant as the carriage of toxigenic bacteriophages.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was made possible by an Oklahoma Center for the Advancement of Science and Technology (OCAST) grant HR11-133 and by NIH Grant Number R15A1072718 to W. M. McShan.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fcimb.2014.00109/abstract
Supplemental Figure S1 | Identity Matrix of Integrases. An amino acid sequence identity matrix of the CI integrase proteins from streptococcus species that target mutL is presented. SpyCI integrases show a high degree of similarity with each other, as well as strong similarities to ones found in S. canis and S. dysgalactiae subspecies equisimilis. High amino acid similarities suggest that core sequences are conserved and required for integration into mutL.
References
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Anba, J., Bidnenko, E., Hillier, A., Ehrlich, D., and Chopin, M. C. (1995). Characterization of the lactococcal abiD1 gene coding for phage abortive infection. J. Bacteriol. 177, 3818–3823.
Baker-Austin, C., Wright, M. S., Stepanauskas, R., and McArthur, J. V. (2006). Co-selection of antibiotic and metal resistance. Trends Microbiol. 14, 176–182. doi: 10.1016/j.tim.2006.02.006
Banks, D. J., Beres, S. B., and Musser, J. M. (2002). The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10, 515–521. doi: 10.1016/S0966-842X(02)02461-7
Bayliss, C. D., Sweetman, W. A., and Moxon, E. R. (2004). Mutations in Haemophilus influenzae mismatch repair genes increase mutation rates of dinucleotide repeat tracts but not dinucleotide repeat-driven pilin phase variation rates. J. Bacteriol. 186, 2928–2935. doi: 10.1128/JB.186.10.2928-2935.2004
Bessen, D. E., Kumar, N., Hall, G. S., Riley, D. R., Luo, F., Lizano, S., et al. (2011). Whole-genome association study on tissue tropism phenotypes in group A Streptococcus. J. Bacteriol. 193, 6651–6663. doi: 10.1128/JB.05263-11
Bidnenko, E., Ehrlich, D., and Chopin, M. C. (1995). Phage operon involved in sensitivity to the Lactococcus lactis abortive infection mechanism AbiD1. J. Bacteriol. 177, 3824–3829.
Bjelland, S., Bjoras, M., and Seeberg, E. (1993). Excision of 3-methylguanine from alkylated DNA by 3-methyladenine DNA glycosylase I of Escherichia coli. Nucleic Acids Res. 21, 2045–2049. doi: 10.1093/nar/21.9.2045
Bjorkholm, B., Sjolund, M., Falk, P. G., Berg, O. G., Engstrand, L., and Andersson, D. I. (2001). Mutation frequency and biological cost of antibiotic resistance in Helicobacter pylori. Proc. Natl. Acad. Sci. U.S.A. 98, 14607–14612. doi: 10.1073/pnas.241517298
Bolhuis, H., Poelarends, G., van Veen, H. W., Poolman, B., Driessen, A. J., and Konings, W. N. (1995). The Lactococcal lmrP gene encodes a proton motive force-dependent drug transporter. J. Biol. Chem. 270, 26092–26098. doi: 10.1074/jbc.270.44.26092
Briggs, G. S., Mahdi, A. A., Weller, G. R., Wen, Q., and Lloyd, R. G. (2004). Interplay between DNA replication, recombination and repair based on the structure of RecG helicase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359, 49–59. doi: 10.1098/rstb.2003.1364
Brussow, H., Canchaya, C., and Hardt, W. D. (2004). Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68, 560–602. doi: 10.1128/MMBR.68.3.560-602.2004
Campbell, A., Schneider, S. J., and Song, B. (1992). Lambdoid phages as elements of bacterial genomes. Genetica 86, 259–267. doi: 10.1007/BF00133724
Canchaya, C., Desiere, F., McShan, W. M., Ferretti, J. J., Parkhill, J., and Brussow, H. (2002). Genome analysis of an inducible prophage and prophage remnants integrated in the Streptococcus pyogenes strain SF370. Virology 302, 245–258. doi: 10.1006/viro.2002.1570
Carapetis, J. R., Steer, A. C., Mulholland, E. K., and Weber, M. (2005). The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694. doi: 10.1016/S1473-3099(05)70267-X
Chen, Y. Y., Feng, C. W., Chiu, C. F., and Burne, R. A. (2008). cadDX operon of Streptococcus salivarius 57.I. Appl. Environ. Microbiol. 74, 1642–1645. doi: 10.1128/AEM.01878-07
Coleman, D., Knights, J., Russell, R., Shanley, D., Birkbeck, T. H., Dougan, G., et al. (1991). Insertional inactivation of the Staphylococcus aureus beta-toxin by bacteriophage f13 occurs by site- and orientation-specific integration of the f13 genome. Mol. Microbiol. 5, 933–939. doi: 10.1111/j.1365-2958.1991.tb00768.x
Coleman, D. C., Sullivan, D. J., Russell, R. J., Arbuthnott, J. P., Carey, B. F., and Pomeray, H. M. (1989). Staphylococcus aureus bacteriophages mediating the simultaneous lysogenic conversion of beta-lysin, staphylokinase and enterotoxin A: molecular mechanism of triple conversion. J. Gen. Microbiol. 135, 1679–1697.
Crimmins, G. T., Herskovits, A. A., Rehder, K., Sivick, K. E., Lauer, P., Dubensky, T. W. Jr., et al. (2008). Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc. Natl. Acad. Sci. U.S.A. 105, 10191–10196. doi: 10.1073/pnas.0804170105
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Das, M., Vargas-Rodriguez, O., Goto, Y., Suga, H., and Musier-Forsyth, K. (2014). Distinct tRNA recognition strategies used by a homologous family of editing domains prevent mistranslation. Nucleic Acids Res. 42, 3943–3953. doi: 10.1093/nar/gkt1332
Desiere, F., McShan, W. M., van Sinderen, D., Ferretti, J. J., and Brussow, H. (2001). Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic Streptococci: evolutionary implications for prophage-host interactions. Virology 288, 325–341. doi: 10.1006/viro.2001.1085
Drummond, A., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Heled, J., et al. (2012). Geneious v6.1.7. Available online at: http://www.geneious.com
Ferretti, J. J., McShan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., et al. (2001). Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663. doi: 10.1073/pnas.071559398
Fittipaldi, N., Beres, S. B., Olsen, R. J., Kapur, V., Shea, P. R., Watkins, M. E., et al. (2012). Full-Genome dissection of an epidemic of severe invasive disease caused by a hypervirulent, recently emerged clone of group A Streptococcus. Am. J. Pathol. 180, 1522–1534. doi: 10.1016/j.ajpath.2011.12.037
Fouts, D. E. (2006). Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 34, 5839–5851. doi: 10.1093/nar/gkl732
Geis, A., El Demerdash, H. A., and Heller, K. J. (2003). Sequence analysis and characterization of plasmids from Streptococcus thermophilus. Plasmid 50, 53–69. doi: 10.1016/S0147-619X(03)00029-5
Goncharoff, P., Saadi, S., Chang, C. H., Saltman, L. H., and Figurski, D. H. (1991). Structural, molecular, and genetic analysis of the kilA operon of broad-host-range plasmid RK2. J. Bacteriol. 173, 3463–3477.
Haraldsen, J. D., and Sonenshein, A. L. (2003). Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol. Microbiol. 48, 811–821. doi: 10.1046/j.1365-2958.2003.03471.x
Iwasaki, H., Shiba, T., Nakata, A., and Shinagawa, H. (1989). Involvement in DNA repair of the ruvA gene of Escherichia coli. Mol. Gen. Genet. 219, 328–331. doi: 10.1007/BF00261196
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., and Gibson, T. J. (1998). Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23, 403–405. doi: 10.1016/S0968-0004(98)01285-7
Kaasen, I., Evensen, G., and Seeberg, E. (1986). Amplified expression of the tag + and alkA + genes in Escherichia coli: identification of gene products and effects on alkylation resistance. J. Bacteriol. 168, 642–647.
Kaplan, D. L., and O'Donnell, M. (2006). RuvA is a sliding collar that protects Holliday junctions from unwinding while promoting branch migration. J. Mol. Biol. 355, 473–490. doi: 10.1016/j.jmb.2005.10.075
Kataja, J., Huovinen, P., Skurnik, M., and Seppala, H. (1999). Erythromycin resistance genes in group A streptococci in Finland. The finnish study group for antimicrobial resistance. Antimicrob. Agents Chemother. 43, 48–52.
Kirby, J. E., Trempy, J. E., and Gottesman, S. (1994). Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J. Bacteriol. 176, 2068–2081.
Kumar, S., Das, M., Hadad, C. M., and Musier-Forsyth, K. (2012). Substrate and enzyme functional groups contribute to translational quality control by bacterial prolyl-tRNA synthetase. J. Phys. Chem. B 116, 6991–6999. doi: 10.1021/jp300845h
Kunkel, B., Losick, R., and Stragier, P. (1990). The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene. Genes Dev. 4, 525–535. doi: 10.1101/gad.4.4.525
LeClerc, J. E., and Cebula, T. A. (2000). Pseudomonas survival strategies in cystic fibrosis. Science 289, 391–392. doi: 10.1126/science.289.5478.391c
LeClerc, J. E., Li, B., Payne, W. L., and Cebula, T. A. (1996). High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274, 1208–1211. doi: 10.1126/science.274.5290.1208
Lee, C. Y., and Iandolo, J. J. (1985). Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J. Bacteriol. 164, 288–293.
Lee, C. Y., and Iandolo, J. J. (1986). Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J. Bacteriol. 166, 385–391.
Lefébure, T., Richards, V. P., Lang, P., Pavinski-Bitar, P., and Stanhope, M. J. (2012). Gene repertoire evolution of Streptococcus pyogenes inferred from Phylogenomic analysis with Streptococcus canis and Streptococcus dysgalactiae. PLoS ONE 7:e37607. doi: 10.1371/journal.pone.0037607
Li, G. M. (2008). Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98. doi: 10.1038/cr.2007.115
Limia, A., Jimenez, M. L., Delgado, T., Sanchez, I., Lopez, S., and Lopez-Brea, M. (1998). [Phenotypic characterization of erythromycin resistance in strains of the genus Streptococcus isolated from clinical specimens]. Rev. Esp. Quimioter. 11, 216–220.
Lindsay, J. A., Ruzin, A., Ross, H. F., Kurepina, N., and Novick, R. P. (1998). The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 29, 527–543. doi: 10.1046/j.1365-2958.1998.00947.x
Louie, A., Brown, D. L., Liu, W., Kulawy, R. W., Deziel, M. R., and Drusano, G. L. (2007). In vitro infection model characterizing the effect of efflux pump inhibition on prevention of resistance to levofloxacin and ciprofloxacin in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51, 3988–4000. doi: 10.1128/AAC.00391-07
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4:19. doi: 10.1186/1745-6150-4-19
Mason, R. E., and Allen, W. E. (1975). Characteristics of Staphylococcus aureus associated with lysogenic conversion to loss of beta-hemolysin production. Can. J. Microbiol. 21, 1113–1116. doi: 10.1139/m75-161
Matic, I., Radman, M., Taddei, F., Picard, B., Doit, C., Bingen, E., et al. (1997). Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277, 1833–1834. doi: 10.1126/science.277.5333.1833
Matic, I., Rayssiguier, C., and Radman, M. (1995). Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80, 507–515. doi: 10.1016/0092-8674(95)90501-4
Matic, I., Taddei, F., and Radman, M. (2000). No genetic barriers between Salmonella enterica serovar typhimurium and Escherichia coli in SOS-induced mismatch repair-deficient cells. J. Bacteriol. 182, 5922–5924. doi: 10.1128/JB.182.20.5922-5924.2000
Matos, R., Lapaque, N., Rigottier-Gois, L., Debarbieux, L., Meylheuc, T., Gonzalez-Zorn, B., et al. (2013). Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 9:e1003539. doi: 10.1371/journal.pgen.1003539
Matthews, R. G., Sheppard, C., and Goulding, C. (1998). Methylenetetrahydrofolate reductase and methionine synthase: biochemistry and molecular biology. Eur. J. Pediatr. 157(Suppl. 2), S54–S59. doi: 10.1007/PL00014305
McShan, W. M. (2005). “The bacteriophages of group A Streptococci,” in Gram-Positive Pathogens, 2nd Edn., eds V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (Washington, DC: American Society for Microbiology), 123–142.
McShan, W. M., and Ferretti, J. J. (2007). “Bacteriophages and the host phenotype,” in Bacteriophages: Genetics and Molecular Biology, ed S. McGrath (Norwich: Horizon Scientific Press), 229–250.
McShan, W. M., Ferretti, J. J., Karasawa, T., Suvorov, A. N., Lin, S., Qin, B., et al. (2008). Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190, 7773–7785. doi: 10.1128/JB.00672-08
Neve, H., Zenz, K. I., Desiere, F., Koch, A., Heller, K. J., and Brussow, H. (1998). Comparison of the lysogeny modules from the temperate Streptococcus thermophilus bacteriophages TP-J34 and Sfi21: implications for the modular theory of phage evolution. Virology 241, 61–72. doi: 10.1006/viro.1997.8960
Nho, S. W., Hikima, J., Cha, I. S., Park, S. B., Jang, H. B., del Castillo, C. S., et al. (2011). Complete genome sequence and immunoproteomic analyses of the bacterial fish pathogen Streptococcus parauberis. J. Bacteriol. 193, 3356–3366. doi: 10.1128/JB.00182-11
Novick, R. P., Christie, G. E., and Penades, J. R. (2010). The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 8, 541–551. doi: 10.1038/nrmicro2393
O'Brien, F. G., Price, C., Grubb, W. B., and Gustafson, J. E. (2002). Genetic characterization of the fusidic acid and cadmium resistance determinants of Staphylococcus aureus plasmid pUB101. J. Antimicrob. Chemother. 50, 313–321. doi: 10.1093/jac/dkf153
Oliver, A., Baquero, F., and Blazquez, J. (2002). The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43, 1641–1650. doi: 10.1046/j.1365-2958.2002.02855.x
Palacio, F., Lewis, J. S. 2nd, Sadkowski, L., Echevarria, K., and Jorgensen, J. H. (2011). Breakthrough bacteremia and septic shock due to Streptococcus anginosus resistant to daptomycin in a patient receiving daptomycin therapy. Antimicrob. Agents Chemother. 55, 3639–3640. doi: 10.1128/AAC.00231-11
Prunier, A. L., and Leclercq, R. (2005). Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J. Bacteriol. 187, 3455–3464. doi: 10.1128/JB.187.10.3455-3464.2005
Putman, M., van Veen, H. W., Degener, J. E., and Konings, W. N. (2001). The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines. Microbiology 147, 2873–2880.
Rabinovich, L., Sigal, N., Borovok, I., Nir-Paz, R., and Herskovits, A. A. (2012). Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150, 792–802. doi: 10.1016/j.cell.2012.06.036
Richardson, A. R., Yu, Z., Popovic, T., and Stojiljkovic, I. (2002). Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc. Natl. Acad. Sci. U.S.A. 99, 6103–6107. doi: 10.1073/pnas.092568699
Scott, J., Nguyen, S., King, C. J., Hendrickson, C., and McShan, W. M. (2012). Mutator phenotype prophages in the genome strains of Streptococcus pyogenes: control by growth state and by a cryptic prophage-encoded promoter. Front. Microbiol. 3:317. doi: 10.3389/fmicb.2012.00317
Scott, J., Thompson-Mayberry, P., Lahmamsi, S., King, C. J., and McShan, W. M. (2008). Phage-associated mutator phenotype in group A Streptococcus. J. Bacteriol. 190, 6290–6301. doi: 10.1128/JB.01569-07
Stragier, P., Kunkel, B., Kroos, L., and Losick, R. (1989). Chromosomal rearrangement generating a composite gene for a developmental transcription factor. Science 243, 507–512. doi: 10.1126/science.2536191
Streit, J. M., Steenbergen, J. N., Thorne, G. M., Alder, J., and Jones, R. N. (2005). Daptomycin tested against 915 bloodstream isolates of viridans group streptococci (eight species) and Streptococcus bovis. J. Antimicrob. Chemother. 55, 574–578. doi: 10.1093/jac/dki032
Suvorov, A. N., Polyakova, E. M., McShan, W. M., and Ferretti, J. J. (2009). Bacteriophage content of M49 strains of Streptococcus pyogenes. FEMS Microbiol. Lett. 294, 9–15. doi: 10.1111/j.1574-6968.2009.01538.x
Tarelli, E., Byers, H. L., Homer, K. A., and Beighton, D. (1998). Evidence for mannosidase activities in Streptococcus oralis when grown on glycoproteins as carbohydrate source. Carbohydr. Res. 312, 159–164. doi: 10.1016/S0008-6215(98)00246-8
Thomas, M. S., and Drabble, W. T. (1986). Secondary attachment site for bacteriophage lambda in the guaB gene of Escherichia coli. J. Bacteriol. 168, 1048–1050.
Tran, T. T., Panesso, D., Gao, H., Roh, J. H., Munita, J. M., Reyes, J., et al. (2013). Whole-genome analysis of a daptomycin-susceptible Enterococcus faecium strain and its daptomycin-resistant variant arising during therapy. Antimicrob. Agents Chemother. 57, 261–268. doi: 10.1128/AAC.01454-12
Trempy, J. E., Kirby, J. E., and Gottesman, S. (1994). Alp suppression of Lon: dependence on the slpA gene. J. Bacteriol. 176, 2061–2067.
Trong, H. N., Prunier, A. L., and Leclercq, R. (2005). Hypermutable and fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 49, 2098–2101. doi: 10.1128/AAC.49.5.2098-2101.2005
Tsaneva, I. R., Muller, B., and West, S. C. (1992). ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69, 1171–1180. doi: 10.1016/0092-8674(92)90638-S
Ubeda, C., Tormo, M. A., Cucarella, C., Trotonda, P., Foster, T. J., Lasa, I., et al. (2003). Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol. 49, 193–210. doi: 10.1046/j.1365-2958.2003.03577.x
Uranga, L. A., Balise, V. D., Benally, C. V., Grey, A., and Lusetti, S. L. (2011). The Escherichia coli DinD protein modulates RecA activity by inhibiting postsynaptic RecA filaments. J. Biol. Chem. 286, 29480–29491. doi: 10.1074/jbc.M111.245373
Viana, D., Blanco, J., Tormo-Mas, M. A., Selva, L., Guinane, C. M., Baselga, R., et al. (2010). Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 77, 1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x
Wyatt, M. D., Allan, J. M., Lau, A. Y., Ellenberger, T. E., and Samson, L. D. (1999). 3-methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays 21, 668–676.
Keywords: Streptococcus pyogenes, group A streptococcus, phage-like chromosomal islands, SpyCI, mutator phenotype
Citation: Nguyen SV and McShan WM (2014) Chromosomal islands of Streptococcus pyogenes and related streptococci: molecular switches for survival and virulence. Front. Cell. Infect. Microbiol. 4:109. doi: 10.3389/fcimb.2014.00109
Received: 13 May 2014; Paper pending published: 08 July 2014;
Accepted: 25 July 2014; Published online: 12 August 2014.
Edited by:
Emmanuelle Charpentier, Umeå University, SwedenReviewed by:
Michael L. Vasil, University of Colorado School of Medicine, USAMichael Otto, National Institute of Allergy and Infectious Diseases, USA
Copyright © 2014 Nguyen and McShan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William M. McShan, 1110 N. Stonewall Ave., College of Pharmacy CPB 307, Oklahoma City, OK 73117, USA e-mail:d2lsbGlhbS1tY3NoYW5Ab3Voc2MuZWR1
 Scott V. Nguyen
Scott V. Nguyen William M. McShan
William M. McShan