- 1Department of Microbiology, School of Natural Sciences, National University of Ireland, Galway, Ireland
- 2Department of Clinical Microbiology, Royal College of Surgeons in Ireland, Dublin, Ireland
Antibiotic resistance and biofilm-forming capacity contribute to the success of Staphylococcus aureus as a human pathogen in both healthcare and community settings. These virulence factors do not function independently of each other and the biofilm phenotype expressed by clinical isolates of S. aureus is influenced by acquisition of the methicillin resistance gene mecA. Methicillin-sensitive S. aureus (MSSA) strains commonly produce an icaADBC operon-encoded polysaccharide intercellular adhesin (PIA)-dependent biofilm. In contrast, the release of extracellular DNA (eDNA) and cell surface expression of a number of sortase-anchored proteins, and the major autolysin have been implicated in the biofilm phenotype of methicillin-resistant S. aureus (MRSA) isolates. Expression of high level methicillin resistance in a laboratory MSSA strain resulted in (i) repression of PIA-mediated biofilm production, (ii) down-regulation of the accessory gene regulator (Agr) system, and (iii) attenuation of virulence in murine sepsis and device infection models. Here we review the mechanisms of MSSA and MRSA biofilm production and the relationships between antibiotic resistance, biofilm and virulence gene regulation in S. aureus.
Biofilm Formation by Staphylococci and its Medical Significance
Implantable medical devices have revolutionized modern healthcare; however attachment to these devices by surface adhering bacteria resulting in biofilm formation and device related infections (DRIs) substantially impact patient morbidity and mortality. Biofilms formed by staphylococci have for many decades been recognized as the most frequent cause of biofilm-associated infections with Staphylococcus epidermidis and S. aureus being among the most common etiologic agents of DRIs (Davies, 2003; Otto, 2013). Infections associated with biofilms are difficult to treat because the biofilm matrix and phenotypic characteristics of the bacteria confer resistance to the host immune response and the action of antimicrobial drugs (O'Gara and Humphreys, 2001).
All implanted medical devices are susceptible to colonization by staphylococci and staphylococcal biofilm-associated infections have been associated with devices ranging from implanted catheters to prosthetic heart valves, cardiac pacemakers, contact lenses, cerebrospinal fluid shunts, joint replacements and intravascular lines (Donlan and Costerton, 2002; Hall-Stoodley et al., 2004). Damaged host tissue is also a risk factor for developing biofilm-associated infection. S. epidermidis and S. aureus, which are part of the normal skin flora are opportunistic pathogens that can cause deep-seated skin infections for example in burn or post-operative wounds (Hammond et al., 2011). They are also known to form biofilms on damaged heart valves, leading to the development of life-threatening infective endocarditis (Claret et al., 2007; Nethercott et al., 2013).
S. epidermidis, unlike the more virulent S. aureus, relies almost exclusively on its ability to colonize and form biofilms on implanted medical devices to cause clinical infections (O'Gara and Humphreys, 2001; Christner et al., 2012) and a strong correlation has been reported between the pathogenicity of S. epidermidis and its ability to colonize implanted medical devices (Vuong and Otto, 2002). The ability of S. aureus to form biofilms on implanted medical devices or damaged host tissue is also a key virulence factor for this pathogen, especially in healthcare settings where antibiotic usage is high and such biofilm formation represents a survival mechanism for the bacteria (Høiby et al., 2010). Among the most studied mechanism of biofilm formation is the production of the icaADBC operon-encoded polysaccharide intercellular adhesion (PIA) or poly-N-acetylglucosamine (PNAG) by both S. aureus and S. epidermidis (O'Gara, 2007; Joo and Otto, 2012). However, biofilm formation independent of the ica operon has also been described in both S. epidermidis and S. aureus (Hussain et al., 1997; Fitzpatrick et al., 2005; Tormo et al., 2005; Qin et al., 2007; O'Neill et al., 2008; Schroeder et al., 2009; Shahrooei et al., 2009; Geoghegan et al., 2010).
In this review, we describe different mechanisms used by methicillin-sensitive S. aureus (MSSA) and methicillin-resistant S. aureus (MRSA) isolates to form biofilm and how the acquisition or loss of methicillin resistance impacts on, not only the biofilm phenotype of S. aureus, but also on global gene regulation and virulence.
Biofilm Formation by Methicillin-Sensitive S. aureus
The first described mechanism of S. aureus biofilm formation involved PIA/PNAG production (Cramton et al., 1999, 2001) that had earlier been described in S. epidermidis (Mack et al., 1994; Heilmann et al., 1996a,b; Heilmann and Götz, 1998). PIA is a glycan of β-1,6-linked 2-acetamido-2-deoxy-D-glucopyranosyl residues with a net positive charge that promotes intercellular aggregation and attachment of cells to inert surfaces (Rohde et al., 2010). The ica operon consists of four biosynthesis genes, icaA, icaD, icaB, and icaC and a divergently transcribed repressor, icaR (Conlon et al., 2002; Götz, 2002). The majority of research on the activity of the ica locus has been conducted in S. epidermidis but there is high nucleotide sequence homology between the ica loci of S. epidermidis and S. aureus and 78% identity at the amino acid level (Cramton et al., 1999). An early study demonstrated that a transposon mutation in the ica operon of S. epidermidis impaired biofilm formation and PIA production (Heilmann et al., 1996b). Subsequent studies showed that S. aureus also commonly uses icaADBC-encoded PIA as a mechanism of biofilm formation (Cramton et al., 1999, 2001).
Carriage of the ica locus is a characteristic of most clinical S. aureus strains (Cramton et al., 1999; Fowler et al., 2001; Rohde et al., 2001). One of the first reports of PNAG-independent S. aureus biofilm production was described in bovine mastitis isolates, which formed biofilm mediated by the biofilm associated protein (Bap) (Cucarella et al., 2001, 2004). An early study with the human S. aureus isolate UAMS-1, also revealed an icaADBC-independent biofilm phenotype under in vitro and in vivo conditions (Beenken et al., 2004).
Investigations into PIA-dependent and PIA-independent mechanisms of biofilm formation identified a correlation between methicillin susceptibility and biofilm in S. aureus (Fitzpatrick et al., 2005; O'Neill et al., 2007, 2008; Houston et al., 2011). This relationship between the biofilm phenotype and β-lactam susceptibility was first investigated in S. epidermidis by Mempel et al. who reported that variable levels of PIA production were significantly associated with different levels of β-lactam susceptibility in phenotypic variants (Mempel et al., 1994, 1995). The significant association between methicillin susceptibility in S. aureus and ica-dependent biofilm formation was first reported when PIA production was found to be essential for biofilm formation by MSSA but not MRSA (O'Neill et al., 2007). MSSA biofilms are significantly induced in growth media supplemented with NaCl, which is known to activate ica operon expression (Fitzpatrick et al., 2006; O'Neill et al., 2007). Furthermore, MSSA biofilms are susceptible to sodium metaperiodate treatment (which oxidizes polysaccharide bonds) and are resistant to treatment with proteinase K (O'Neill et al., 2007). Deletion of the ica locus abolished biofilm-forming capacity among clinical MSSA isolates that are amenable to genetic manipulation, as did mutation of the staphylococcal accessory regulator sarA (Valle et al., 2003; O'Neill et al., 2007).
Building on an original finding that four clinical MRSA isolates were capable of producing icaADBC-independent biofilm (Fitzpatrick et al., 2005), our laboratory reported differences in the environmental regulation of biofilm formation among 32 S. aureus isolates from intensive care units (15 MSSA and 17 MRSA strains) and found that NaCl-induced, PIA-dependent biofilm was more likely to be associated with MSSA biofilm formation (Fitzpatrick et al., 2006). A follow-up study of a large collection of 212 S. aureus isolates from device-related infections (114 MRSA and 98 MSSA) representing five clonal complexes (CC5, CC8, CC22, CC30, and CC45) further demonstrated that MSSA strains were more likely to produce NaCl-induced, PIA-mediated biofilm whereas MRSA biofilm was induced in media supplemented with glucose and was PIA-independent (O'Neill et al., 2007).
Biofilm Formation by MRSA Strains
The ica locus was found to be redundant for MRSA biofilm formation (O'Neill et al., 2007). Unlike the NaCl-induced biofilm expressed by MSSA clinical isolates, biofilm formation by MRSA isolates was significantly more likely to be induced by the addition of glucose to the growth medium, which is associated with the acidification of the culture media (O'Neill et al., 2007). MRSA biofilms are resistant to sodium metaperiodate treatment but are susceptible to proteinase K treatment, implicating protein adhesins in this biofilm phenotype (O'Neill et al., 2007).
As observed in MSSA isolates, mutation in sarA also abolished biofilm formation by clinical MRSA isolates (Beenken et al., 2004; O'Neill et al., 2007). SarA is a known repressor of four major extracellular proteases, namely SspA, SspB, Aur, and ScpA (Karlsson et al., 2001) and it is proposed that impaired glucose-induced biofilm formation by MRSA sarA mutants may be associated with upregulation of protease activity, which can inhibit S. aureus biofilm production (Marti et al., 2010).
A deletion in the accessory gene regulator (agr) system enhanced biofilm formation by clinical MRSA strains but had no significant effect on biofilms formed by MSSA strains (O'Neill et al., 2007). In contrast to sarA, agr positively regulates expression of the four major proteases (Shaw et al., 2004). Additionally, mutations in the agr-regulated Aur metalloprotease and the SplABCDEF serine proteases increased biofilm formation and reduced detachment from established biofilms (Boles and Horswill, 2008). Similarly, the serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF) enhanced biofilm formation, further implicating increased protease activity as the mechanism of agr-mediated dispersal from biofilms (Boles and Horswill, 2008). The positive impact of an agr mutation on MRSA biofilm formation is largely consistent with other research showing that deletion of agr can enhance S. aureus biofilm formation (Vuong et al., 2000; Beenken et al., 2003), whilst a separate study demonstrated that agr deletion does not significantly alter icaADBC expression or PIA production (Vuong et al., 2003). Interestingly, the reduced pH of cultures grown in media supplemented with glucose, was associated with repression of the agr system (Regassa et al., 1992). Furthermore, activation of the agr system was shown to initiate biofilm dispersal (Boles and Horswill, 2008). The agr locus encodes two divergent transcripts, RNAII and RNAIII driven by the P2 and P3 promoters, respectively (Novick et al., 1993). RNAII comprises four genes, agrBDCA, that encode for the synthesis of secreted autoinducing peptide AIP. At threshold concentrations, AIP binds to the sensor histidine kinase AgrC, which phosphorylates AgrA and activates the P2 and P3 promoters, resulting in upregulated agr expression and increased transcription of the RNAIII effector molecule. Exogenous addition of AIP to biofilms induced dispersal from established biofilms by upregulating the agr system and subsequently increasing protease production (Boles and Horswill, 2008). The enhanced biofilm phenotype observed in agr mutants is also attributed to the loss of production of the RNAIII-encoded delta-toxin, which has surfactant properties (Vuong et al., 2000, 2004). Biofilm detachment by agr+ strains is proposed to require the formation of a film of amphipathic delta-toxin molecules expressed at the biofilm/fluid interface which can inhibit the hydrophobic interactions between bacterial cell surfaces, thus lowering surface tension and causing cell detachment from the biofilm matrix (Vuong et al., 2004).
Role of LPXTG-Anchored Surface Proteins in MRSA Biofilm Formation
S. aureus expresses 28 surface proteins, 21 of which are predicted to contain LPXTG binding motifs required for normal display on the cell surface (Ton-That et al., 2000; Mazmanian et al., 2001). Anchoring of these LPXTG motif-containing proteins to peptidoglycan is catalyzed by Sortase, an extracellular transpeptidase encoded by the srtA gene (Mazmanian et al., 2000). A srtA mutation affected biofilm accumulation by clinical MRSA strains from three clonal complexes and had no impact on ica-dependent biofilms formed by MSSA strains (O'Neill et al., 2008). As noted earlier Bap was the first LPXTG-anchored surface protein implicated in ica-independent S. aureus biofilm formation but is rarely found in human isolates (Cucarella et al., 2001; Tormo et al., 2005).
The fibronectin-binding proteins (FnBPs) were subsequently shown to play an important role in ica-independent biofilm formation by human MRSA isolates (O'Neill et al., 2008; Shanks et al., 2008; Vergara-Irigaray et al., 2009). The FnBPs are multi-functional surface proteins with an N-terminal A domain and a C-terminal wall spanning LPXTG-anchoring domain separated by tandem repeats involved in binding to fibronectin (Schwarz-Linek et al., 2003; Meenan et al., 2007). Both FnBP proteins, FnBPA and FnBPB, are involved in the accumulation phase of MRSA biofilm formation in hospital and community isolates under static and flow conditions, with each individual protein capable of complementing the biofilm defect of a double fnbpAB mutant (O'Neill et al., 2008; McCourt et al., 2014). The FnBPs do not contribute to PIA-mediated biofilm formation by clinical MSSA strains (O'Neill et al., 2008). Expression of the FnBPs was constitutive in a MRSA isolate producing an FnBP-dependent biofilm but restricted to the exponential growth phase in MSSA isolates that form PIA biofilms (Geoghegan et al., 2013). The N-terminal of the FnBPA protein, specifically residues 166–498 comprising the N2 and N3 subdomains of the A domain, has been implicated in the biofilm phenotype via a Zn(2+)-dependent mechanism (O'Neill et al., 2008; Geoghegan et al., 2013). A separate study from the Lasa laboratory, demonstrated that MRSA strain 132 was capable of producing either acid stress-induced FnBP-dependent biofilm or osmotic stress-induced exopolysaccharide-mediated biofilm (Vergara-Irigaray et al., 2009). Furthermore, the 132 fnbpAB mutant displayed significantly impaired colonization and biofilm formation compared to the isogenic 132 ica mutant in a murine model of subcutaneous catheter-related infection (Vergara-Irigaray et al., 2009). Recently the formation of bacterial biofilm-like aggregates in human synovial fluid by MRSA and MSSA strains was attributed to expression of the FnBPs as well as the fibrinogen binding proteins ClfA and ClfB, identifying biofilm and in particular LPXTG-wall anchored proteins as important virulence determinants in staphylococcal joint infections (Dastgheyb et al., 2014).
In MRSA strains where the FnBPs have not (yet) been implicated in protein dependent biofilm, a number of other LPXTG proteins may be important. The accumulation associated protein (Aap), which is 54% identical to the SasG protein of S. aureus (Corrigan et al., 2007) was first identified in S. epidermidis and is known to play a well-defined role in the biofilm phenotype (Hussain et al., 1997; Rohde et al., 2005; Conrady et al., 2008, 2013; Conlon et al., 2014; Schaeffer et al., 2015). SasG, which is found in approximately 50% of human clinical isolates, has also been implicated in the intercellular accumulation phase of ica-independent biofilm formation (Corrigan et al., 2007; Sung et al., 2008). SasG promotes intercellular aggregation via homo-oligomerization of the SasG protein (Kuroda et al., 2008) with biofilm formation promoted by the repeated B domains of the SasG protein (Geoghegan et al., 2010). The B repeat regions promote ica-independent biofilm formation in a Zn(2+)-dependent manner through the formation of extended fibrils on the cell surface (Geoghegan et al., 2010; Gruszka et al., 2012). Protein A also promotes polysaccharide-independent biofilm but does not need to be anchored to the cell wall via its LPXTG motif to do so, with exogenous Protein A capable of promoting biofilm accumulation (Merino et al., 2009). However, the role or otherwise of SasG and protein A to staphylococcal biofilm-associated infections has not yet been investigated. Readers are referred to a companion review in this series focusing on staphylococcal proteinaceous biofilms by Pietro Speziale.
Role of the Major Autolysin and Extracellular DNA in MRSA Biofilm Formation
The major autolysin, Atl, which is involved in daughter cell separation, cell wall homeostasis and peptidoglycan turnover (Yamada et al., 1996; Biswas et al., 2006) plays a role in the early stages of ica-independent biofilm formation by clinical MRSA strains (Houston et al., 2011). This bifunctional peptidoglycan hydrolase was first implicated in the primary attachment of S. epidermidis to polystyrene (Heilmann et al., 1997; Biswas et al., 2006), with atlE mutants exhibiting significantly attenuated virulence in a rat central venous catheter infection model (Rupp et al., 2001). AtlE of S. epidermidis also possesses vitronectin-binding activity implicated in AtlE-mediated biofilm formation on plasma protein-coated polymer surfaces (Heilmann et al., 1997). Clinical MRSA strains that produce ica-independent biofilms require Atl for primary attachment to polystyrene surfaces (Houston et al., 2011).
The major autolysin of S. aureus is expressed as a 137.5 kDa pro-protein which is proteolytically processed to yield a 3.1 kDa signal sequence, a 17.6 kDa propeptide and two enzymatically active regions, a 63.3 kDa N-acetylmuramyl-L-alanine amidase enzyme and a 53.6 kDa endo-β-N-acetylglucosaminidase enzyme (Oshida et al., 1995). Catalytic activity of the amidase region is required for PIA-independent proteinaceous biofilm formation by hospital acquired MRSA (HA-MRSA) strains (Houston et al., 2011). The protease inhibitor PMSF, which increases the release of extracellular autolytic enzymes (Fournier and Hooper, 2000), and polyanethole sodium sulfanate, which inhibits autolytic activity without impairing growth (Wecke et al., 1986; Yabu and Kaneda, 1995), both prevented biofilm formation by HA-MRSA strains indicating a vital role for both the unprocessed Atl protein and the active amidase and glucosaminidase enzymes in early MRSA biofilm formation (Houston et al., 2011).
Construction of enzymatically inactive point mutations within the active regions of Atl revealed Atl-mediated cell lysis and the release of extracellular DNA (eDNA) by both the amidase and glucosaminidase regions as the mechanisms of Atl-mediated biofilm formation (Bose et al., 2012). Several studies have reported a role for eDNA in the ica-independent healthcare associated and community acquired MRSA (CA-MRSA) biofilm phenotypes (Izano et al., 2008; Lauderdale et al., 2010; Houston et al., 2011). In S. aureus, addition of DNase I to the culture media inhibited PIA-independent biofilm formation by HA-MRSA but did not significantly disperse mature MRSA biofilms implicating eDNA in the attachment and/or early stage of biofilm development (Houston et al., 2011). In the CA-MRSA strain USA300, the secreted thermostable nuclease enzyme, Nuc has also been shown to negatively impact on biofilm formation (Kiedrowski et al., 2011). In a mouse model of catheter infections, however, mutations in nuc and a second nuclease gene nuc2 of the UAMS-1 MSSA strain were associated with reduced biofilm (Beenken et al., 2012) indicating that additional studies with MSSA and MRSA isolates, are needed to fully elucidate the role of these enzymes and extracellular nucleic acid in the biofilm phenotype in vivo.
A study of ica-independent mechanisms of biofilm formation by S. aureus revealed that altered levels of autolysis were associated with defective biofilm production (Boles et al., 2010). Recently cytoplasmic proteins released during the stationary phase of growth, which may be a consequence of autolysis, have been shown to be part of the biofilm matrix of S. aureus HG003 (Foulston et al., 2014). Interestingly the release of cytoplasmic proteins during stationary phase appears to be in response to decreasing pH, which can be triggered by the addition of excess glucose to the growth media (Foulston et al., 2014) and it is worth noting that these same growth conditions promote Atl/FnBP-mediated MRSA biofilm formation (O'Neill et al., 2008). A simplified model of MSSA and MRSA biofilm mechanisms together with scanning electron micrographs of SH1000 (MSSA) and BH1CC (MRSA) biofilms is presented in Figure 1.
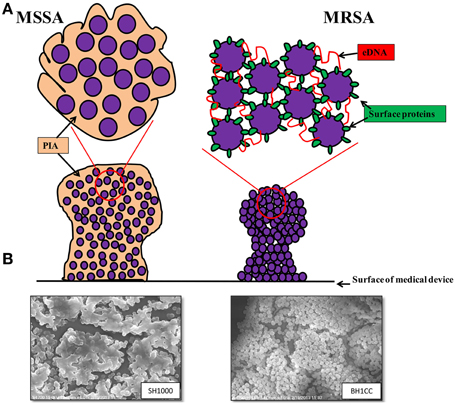
Figure 1. (A) Graphic representations of the biofilm phenotypes expressed by MSSA and MRSA. MSSA strains form ica-dependent, PIA-mediated biofilms whereas MRSA strains form biofilms independent of PIA and require surface proteins such as the fibronectin binding proteins, Atl-mediated cell lysis and eDNA for colonization of surfaces and biofilm accumulation. (B) Scanning electron micrographs (3500 × magnification) of biofilms formed by MSSA strain SH1000 grown in BHI media supplemented with 4% NaCl (left) and MRSA strain BH1CC grown in BHI supplemented with 1% glucose (right).
How does Methicillin Resistance Affect the Biofilm Phenotype and Virulence?
Methicillin Resistance and Biofilm
The mechanism(s) underpinning the association between the MRSA and MSSA biofilm phenotypes and utilization of polysaccharide and protein adhesins, respectively remains unclear. However, the level of resistance to beta-lactam antibiotics appears to be important for the biofilm phenotype. For instance, excision of the SCCmec element from the hospital MRSA strain BH1CC was associated with reduced FnBP-mediated biofilm forming capacity, presumably due to up-regulated protease activity (Pozzi et al., 2012; Rudkin et al., 2012). The relationship between methicillin resistance and biofilm is further complicated by the observation that MRSA strains can express either low level heterogeneous resistance (HeR) to methicillin or high-level, homogeneous resistance (HoR) (Keaton et al., 2013; Mwangi et al., 2013). Carriage of the mecA gene alone does not confer the HoR phenotype and additional genetic events are also needed (Pozzi et al., 2012; Mwangi et al., 2013; Dordel et al., 2014). In the laboratory isolation of homogeneously resistant strains from HeR strains is readily achieved by plating cell suspensions on media supplemented with high concentrations of oxacillin, the clinically used derivative of methicillin (100 μg/ml) (Pozzi et al., 2012; Dordel et al., 2014). In clinical isolates of MRSA the ica locus is present and expressed but PIA does not appear to be produced (O'Neill et al., 2007). In an engineered derivative of the laboratory strain 8325-4 carrying the mecA gene on a plasmid and expressing HoR, the ica operon was repressed >300-fold compared to its MSSA parent and was associated with a switch from PIA-dependent to proteinaceous biofilm (Pozzi et al., 2012). In contrast expression of HeR in 8325-4 did not significantly alter the biofilm phenotype (Pozzi et al., 2012). Extracellular protease activity was reduced in the 8325-4 HoR strain and 8235-4 HoR biofilms were dispersed with proteinase K implicating a protein adhesin in this phenotype (Pozzi et al., 2012). However, because this biofilm phenotype was independent of PIA/PNAG, the major autolysin or any of the LPXTG cell wall anchored proteins whereas clinical MRSA isolates express an FnBP/eDNA dependent biofilm, it appears that expression of homogeneous methicillin resistance is associated with at least two distinct biofilm phenotypes. It remains to be determined whether PBP2a expression exerts direct or indirect (e.g., via altered cell wall architecture) effects on biofilm production. In a murine device infection model 8325-4 and 8325-4 HoR were recovered in similar numbers from implanted catheters, suggesting that PIA/PNAG and protein adhesin-mediated biofilms may be equally effective for device colonization in vivo (Pozzi et al., 2012).
Oxacillin Resistance and Virulence
High level PBP2a expression in MRSA is associated with repression of the Agr quorum sensing operon (Pozzi et al., 2012; Rudkin et al., 2012). Agr repression in MRSA is directly linked to mecA expression and the subsequent changes in cell wall architecture are accompanied by reduced cytotoxin production and attenuated virulence. The repression of Agr blocks the co-ordinated switch-on of toxin and enzyme secretion during the latter stages of bacterial growth in vitro, leaving the cell arrested in the surface protein expression stage of the Agr regulatory cycle (Rudkin et al., 2012). As noted above, increased surface protein expression and repression of extracellular protease production are also consistent with the protein-mediated MRSA biofilm phenotype (Pozzi et al., 2012).
Murine infection model studies showed that mice infected with the 8325-4 HoR strain were more likely to survive than mice infected with 8325-4 (Pozzi et al., 2012). The 8325-4 HoR strain did not significantly disseminate beyond the surrounding peri-catheter tissue whereas high numbers of the 8235-4 MSSA strain were recovered from the kidneys, liver and spleen (Pozzi et al., 2012). In a murine bacteremia model, mice infected with ΔSCCmec and ΔmecA mutants of the HA-MRSA strain BH1CC consistently lost more weight during the course of infection and suffered >60% mortality compared to the wild type (Rudkin et al., 2012). These data have led to the hypothesis that MRSA strains have sacrificed virulence for high level antibiotic resistance. Supporting this idea, the infectivity and lethality of MRSA strains was also reduced in guinea pig and murine models of infection compared to MSSA strains (Mizobuchi et al., 1994). Interestingly, MRSA and MSSA isolates were equally virulent in immunocompromised animals (Mizobuchi et al., 1994). Furthermore, a study of 104 patients with S. aureus bacteremia found that MSSA bacteremia correlated with significantly higher rates of infective endocarditis than MRSA (Abraham et al., 2004). Similarly, MSSA infections have been associated with a significantly higher illness severity score from invasive disease than MRSA clones (Wehrhahn et al., 2012).
The phenol soluble modulin mec (psm-mec) locus located adjacent to mecA on type II SCCmec elements promotes biofilm formation in MRSA strains (Kaito et al., 2011, 2013) and has also been implicated in reduced virulence of MRSA strains in murine models of infection (Kaito et al., 2008, 2011; Queck et al., 2009). The psm-mec locus was directly implicated in inhibiting translation of the agrA transcript (Kaito et al., 2013). Interestingly carriage of psm-mec was only associated with HA-MRSA strains and was absent in the more virulent CA-MRSA isolates and mutation or deletion of the locus from HA-MRSA increased virulence capacity in murine infection models (Kaito et al., 2013). In addition to PSM-mec, phenol soluble modulins (PSMs) generally have surfactant qualities and are known to be centrally involved in biofilm structuring and detachment, including dissemination of biofilm-associated infection (Wang et al., 2011; Periasamy et al., 2012). In this context, repression of the Agr system including PSM-mec in MRSA is consistent with enhanced biofilm formation.
Oxacillin resistance has pleiotropic effects on S. aureus, changing the biofilm phenotype, altering global gene regulation, reducing toxin production and ultimately reducing virulence. However, the importance of MRSA strains as pathogens in healthcare and community setting reflects the high degree of adaptation of S. aureus to methicillin resistance and the associated cellular changes. The attenuated virulence associated with high level resistance in HA-MRSA strains, is consistent with their confinement within healthcare settings (Collins et al., 2010; Rudkin et al., 2012), whereas the resistance/virulence equilibrium in CA-MRSA strains has clearly supported their ability in infect otherwise healthy individuals.
Nucleotide Signaling and Methicillin Resistance
The mechanisms underpinning the relationship between antibiotic resistance and virulence in S. aureus remain to be elucidated but studies over recent years implicate nucleotide signaling in these phenotypes. Mutation of the S. aureus diadenylate cyclase gene dacA, which reduced c-di-AMP levels, resulted in the conversion of a HoR MRSA to a HeR strain (Dengler et al., 2013). Conversely mutations in the gdpP-encoded c-di-AMP phosphodiesterase, which resulted in increased c-di-AMP levels were accompanied by HoR to methicillin (Corrigan et al., 2011; Pozzi et al., 2012) or increased tolerance to beta-lactam antibiotics (Griffiths and O'Neill, 2012). In Bacillus subtilis, the activity of the GdpP homolog YybT is strongly repressed by the stringent response alarmone ppGpp, which is synthesized by the RelA enzyme (Rao et al., 2010) and Mwangi et al. reported that activation of the stringent response and constitutive ppGpp production was accompanied by homogeneous methicillin resistance (Mwangi et al., 2013). Proteomic analysis has revealed that aminoacyl-tRNA biosynthesis was repressed by subinhibitory concentrations of oxacillin in both MSSA and MRSA (Liu et al., 2014). Reduced levels of aminoacyl-tRNA may trigger the stringent response, which is normally activated by amino acid starvation and specifically the concomitant reduction in charged tRNA levels. A recent genomics analysis of HoR strains derived from a range of clinical HeR isolates identified mutations in 27 genes and 3 intergenic regions, most of which are potentially involved in the stringent response (Dordel et al., 2014). Collectively these data implicate the stringent response in the HoR phenotype and may suggest that ppGpp-mediated repression of GdpP activity leads to reduced c-di-AMP levels and in turn modulates resistance to beta-lactam antibiotics, biofilm and virulence. Identification of c-di-AMP targets may help elucidate the mechanism c-di-AMP-controlled beta-lactam resistance and initial studies to identify c-di-AMP target proteins identified in S. aureus and Listeria monocytogenes suggest a pleiotrophic role for this nucleotide second messenger (Corrigan et al., 2013; Sureka et al., 2014).
Concluding Thoughts
Biofilm formation is a key virulence factor of staphylococci and distinct mechanisms are employed by MSSA and MRSA for biofilm formation. Clinical MSSA strains predominantly form biofilm dependent on the icaADBC operon and PIA production whereas MRSA strains form biofilms independent of PIA (O'Neill et al., 2007). There are important roles for LPXTG-anchored surface proteins, the major autolysin and eDNA during MRSA biofilm formation in the absence of PIA (O'Neill et al., 2008; Houston et al., 2011). Acquisition of methicillin resistance appears to repress polysaccharide-type biofilm production and promote formation of proteinaceous-type biofilms (Pozzi et al., 2012). While the different mechanisms of biofilm formation employed by MSSA and MRSA do not appear to impair the ability of either to colonize implanted biomaterial in vivo, the acquisition of methicillin resistance by HA-MRSA is associated with an overall downregulation of virulence gene expression (Pozzi et al., 2012; Rudkin et al., 2012, 2014). Despite this, HA-MRSA strains remain a significant cause of morbidity and mortality for hospitalized patients perhaps reflecting their successful adaptation to a specific niche within the hospital setting where there is a large pool of immunocompromised individuals that often require implanted medical devices and intensive antibiotic treatments (Pozzi et al., 2012). Understanding how MRSA has remained a successful pathogen in the hospital environment, will inform the development of novel therapeutics. Indeed treating MRSA strains with oxacillin may still have therapeutic potential (Rudkin et al., 2014). In CA-MRSA strains, the lower levels of PBP2a expression compared to HA-MRSA is associated with maintenance of high level toxin production. Using oxacillin to increase PBP2a expression can repress Agr activity and secretion of cytolytic toxins, indicating therapeutic potential (Rudkin et al., 2014). Further investigations to assess the global impact of oxacillin treatment on MRSA gene expression are needed but these data illustrate how advances in our understanding of antibiotic resistance and biofilm might be exploited in the development of new strategies to better prevent and treat S. aureus infections.
Conflict of Interest Statement
The Associate Editor, Joan Goeghegan declares that, despite having collaborated with authors Hannah McCarthy and James P. O'Gara, the review process was handled objectively and no conflict of interest exists. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research in the author's laboratories is funded by grants from the Irish Health Research Board, the Healthcare Infection Society (UK), and the Irish Research Council.
References
Abraham, J., Mansour, C., Veledar, E., Khan, B., and Lerakis, S. (2004). Staphylococcus aureus bacteremia and endocarditis: the Grady Memorial Hospital experience with methicillin-sensitive S aureus and methicillin-resistant S aureus bacteremia. Am. Heart J. 147, 536–539. doi: 10.1016/j.ahj.2003.09.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beenken, K. E., Blevins, J. S., and Smeltzer, M. S. (2003). Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71, 4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beenken, K. E., Dunman, P. M., McAleese, F., Macapagal, D., Murphy, E., Projan, S. J., et al. (2004). Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186, 4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beenken, K. E., Spencer, H., Griffin, L. M., and Smeltzer, M. S. (2012). Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect. Immun. 80, 1634–1638. doi: 10.1128/IAI.06134-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Biswas, R., Voggu, L., Simon, U. K., Hentschel, P., Thumm, G., and Gotz, F. (2006). Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259, 260–268. doi: 10.1111/j.1574-6968.2006.00281.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boles, B. R., and Horswill, A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4:e1000052. doi: 10.1371/journal.ppat.1000052
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Boles, B. R., Thoendel, M., Roth, A. J., and Horswill, A. R. (2010). Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 5:e10146. doi: 10.1371/journal.pone.0010146
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bose, J. L., Lehman, M. K., Fey, P. D., and Bayles, K. W. (2012). Contribution of the Staphylococcus aureus Atl AM and GL murein hydrolase activities in cell division, autolysis, and biofilm formation. PLoS ONE 7:e42244. doi: 10.1371/journal.pone.0042244
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christner, M., Heinze, C., Busch, M., Franke, G., Hentschke, M., Bayard duhring, S., et al. (2012). sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol. Microbiol. 86, 394–410. doi: 10.1111/j.1365-2958.2012.08203.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Claret, L., Miquel, S., Vieille, N., Ryjenkov, D. A., Gomelsky, M., and Darfeuille-Michaud, A. (2007). The flagellar sigma factor FliA regulates adhesion and invasion of Crohn disease-associated Escherichia coli via a cyclic dimeric GMP-dependent pathway. J. Biol. Chem. 282, 33275–33283. doi: 10.1074/jbc.M702800200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collins, J., Rudkin, J., Recker, M., Pozzi, C., O'Gara, J. P., and Massey, R. C. (2010). Offsetting virulence and antibiotic resistance costs by MRSA. ISME J. 4, 577–584. doi: 10.1038/ismej.2009.151
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conlon, B. P., Geoghegan, J. A., Waters, E. M., McCarthy, H., Rowe, S. E., Davies, J. R., et al. (2014). Role for the a domain of unprocessed Accumulation-Associated Protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J. Bacteriol. 196, 4268–4275. doi: 10.1128/JB.01946-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conlon, K. M., Humphreys, H., and O'Gara, J. P. (2002). icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184, 4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conrady, D. G., Brescia, C. C., Horii, K., Weiss, A. A., Hassett, D. J., and Herr, A. B. (2008). A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 105, 19456–19461. doi: 10.1073/pnas.0807717105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conrady, D. G., Wilson, J. J., and Herr, A. B. (2013). Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 110, E202–E211. doi: 10.1073/pnas.1208134110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corrigan, R. M., Abbott, J. C., Burhenne, H., Kaever, V., and Grundling, A. (2011). c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7:e1002217. doi: 10.1371/journal.ppat.1002217
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corrigan, R. M., Campeotto, I., Jeganathan, T., Roelofs, K. G., Lee, V. T., and Gründling, A. (2013). Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. U.S.A. 110, 9084–9089. doi: 10.1073/pnas.1300595110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Corrigan, R. M., Rigby, D., Handley, P., and Foster, T. J. (2007). The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446. doi: 10.1099/mic.0.2007/006676-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cramton, S. E., Gerke, C., Schnell, N. F., Nichols, W. W., and Götz, F. (1999). The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67, 5427–5433.
Cramton, S. E., Ulrich, M., Gotz, F., and Doring, G. (2001). Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69, 4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cucarella, C., Solano, C., Valle, J., Amorena, B., Lasa, I., and Penades, J. R. (2001). Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183, 2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cucarella, C., Tormo, M. A., Ubeda, C., Trotonda, M. P., Monzón, M., Peris, C., et al. (2004). Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 72, 2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dastgheyb, S., Parvizi, J., Shapiro, I. M., Hickok, N. J., and Otto, M. (2014). Effect of biofilms on recalcitrance of staphylococcal joint infection to antibiotic treatment. J. Infect. Dis. doi: 10.1093/infdis/jiu514. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davies, D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2, 114–122. doi: 10.1038/nrd1008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dengler, V., McCallum, N., Kiefer, P., Christen, P., Patrignani, A., Vorholt, J. A., et al. (2013). Mutation in the C-Di-AMP Cyclase dacA affects fitness and resistance of methicillin resistant Staphylococcus aureus. PLoS ONE 8:e73512. doi: 10.1371/journal.pone.0073512
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dordel, J., Kim, C., Chung, M., Pardos de la Gandara, M., Holden, M. T., Parkhill, J., et al. (2014). Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio 5:e01000. doi: 10.1128/mBio.01000-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fitzpatrick, F., Humphreys, H., and O'Gara, J. P. (2006). Environmental regulation of biofilm development in methicillin-resistant and methicillin-susceptible Staphylococcus aureus clinical isolates. J. Hosp. Infect. 62, 120–122. doi: 10.1016/j.jhin.2005.06.004
Fitzpatrick, F., Humphreys, H., and O'Gara, J. P. (2005). Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 43, 1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Foulston, L., Elsholz, A. K., Defrancesco, A. S., and Losick, R. (2014). The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 5:e01667-14. doi: 10.1128/mBio.01667-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fournier, B., and Hooper, D. C. (2000). A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182, 3955–3964. doi: 10.1128/JB.182.14.3955-3964.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fowler, V. G. J., Fey, P. D., Reller, L. B., Chamis, A. L., Corey, G. R., and Rupp, M. E. (2001). The intercellular adhesin locus ica is present in clinical isolates of Staphylococcus aureus from bacteremic patients with infected and uninfected prosthetic joints. Med. Microbiol. Immunol. 189, 127–131. doi: 10.1007/s430-001-8018-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geoghegan, J. A., Corrigan, R. M., Gruszka, D. T., Speziale, P., O'Gara, J. P., Potts, J. R., et al. (2010). Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192, 5663–5673. doi: 10.1128/JB.00628-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geoghegan, J. A., Monk, I. R., O'Gara, J. P., and Foster, T. J. (2013). Subdomains N2N3 of fibronectin binding protein a mediate Staphylococcus aureus biofilm formation and adherence to fibrinogen using distinct mechanisms. J. Bacteriol. 195, 2675–2683. doi: 10.1128/JB.02128-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Götz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Griffiths, J. M., and O'Neill, A. J. (2012). Loss of function of the gdpP protein leads to joint beta-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 579–581. doi: 10.1128/AAC.05148-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gruszka, D. T., Wojdyla, J. A., Bingham, R. J., Turkenburg, J. P., Manfield, I. W., Steward, A., et al. (2012). Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc. Natl. Acad. Sci. U.S.A. 109, E1011–E1018. doi: 10.1073/pnas.1119456109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hall-Stoodley, L., Costerton, J. W., and Stoodley, P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. doi: 10.1038/nrmicro821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hammond, A. A., Miller, K. G., Kruczek, C. J., Dertien, J., Colmer-Hamood, J. A., Griswold, J. A., et al. (2011). An in vitro biofilm model to examine the effect of antibiotic ointments on biofilms produced by burn wound bacterial isolates. Burns 37, 312–321. doi: 10.1016/j.burns.2010.09.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heilmann, C., Gerke, C., Perdreau-Remington, F., and Götz, F. (1996a). Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64, 277–282.
Heilmann, C., and Götz, F. (1998). Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentralbl. Bakteriol. 287, 69–83. doi: 10.1016/S0934-8840(98)80149-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heilmann, C., Hussain, M., Peters, G., and Götz, F. (1997). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Götz, F. (1996b). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Høiby, N., Bjarnsholt, T., Givskov, M., Molin, S., and Ciofu, O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Houston, P., Rowe, S. E., Pozzi, C., Waters, E. M., and O'Gara, J. P. (2011). Essential role for the major autolysin in the fibronectin-binding protein-mediated Staphylococcus aureus biofilm phenotype. Infect. Immun. 79, 1153–1165. doi: 10.1128/IAI.00364-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hussain, M., Herrmann, M., Von eiff, C., Perdreau-Remington, F., and Peters, G. (1997). A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65, 519–524.
Izano, E. A., Amarante, M. A., Kher, W. B., and Kaplan, J. B. (2008). Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476. doi: 10.1128/AEM.02073-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Joo, H. S., and Otto, M. (2012). Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem. Biol. 19, 1503–1513. doi: 10.1016/j.chembiol.2012.10.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaito, C., Omae, Y., Matsumoto, Y., Nagata, M., Yamaguchi, H., Aoto, T., et al. (2008). A novel gene, fudoh, in the SCCmec region suppresses the colony spreading ability and virulence of Staphylococcus aureus. PLoS ONE 3:e3921. doi: 10.1371/journal.pone.0003921
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaito, C., Saito, Y., Ikuo, M., Omae, Y., Mao, H., Nagano, G., et al. (2013). Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 9:e1003269. doi: 10.1371/journal.ppat.1003269
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaito, C., Saito, Y., Nagano, G., Ikuo, M., Omae, Y., Hanada, Y., et al. (2011). Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence. PLoS Pathog. 7:e1001267. doi: 10.1371/journal.ppat.1001267
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karlsson, A., Saravia-Otten, P., Tegmark, K., Morfeldt, E., and Arvidson, S. (2001). Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69, 4742–4748. doi: 10.1128/IAI.69.8.4742-4748.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keaton, M. A., Rosato, R. R., Plata, K. B., Singh, C. R., and Rosato, A. E. (2013). Exposure of clinical MRSA heterogeneous strains to beta-lactams redirects metabolism to optimize energy production through the TCA cycle. PLoS ONE 8:e71025. doi: 10.1371/journal.pone.0071025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kiedrowski, M. R., Kavanaugh, J. S., Malone, C. L., Mootz, J. M., Voyich, J. M., Smeltzer, M. S., et al. (2011). Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE 6:e26714. doi: 10.1371/journal.pone.0026714
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuroda, M., Ito, R., Tanaka, Y., Yao, M., Matoba, K., Saito, S., et al. (2008). Staphylococcus aureus surface protein SasG contributes to intercellular autoaggregation of Staphylococcus aureus. Biochem. Biophys. Res. Commun. 377, 1102–1106. doi: 10.1016/j.bbrc.2008.10.134
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lauderdale, K. J., Malone, C. L., Boles, B. R., Morcuende, J., and Horswill, A. R. (2010). Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J. Orthop. Res. 28, 55–61. doi: 10.1002/jor.20943
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, X., Hu, Y., Pai, P. J., Chen, D., and Lam, H. (2014). Label-free quantitative proteomics analysis of antibiotic response in Staphylococcus aureus to oxacillin. J. Proteome Res. 13, 1223–1233. doi: 10.1021/pr400669d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mack, D., Nedelmann, M., Krokotsch, A., Schwarzkopf, A., Heesemann, J., and Laufs, R. (1994). Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62, 3244–3253.
Marti, M., Trotonda, M. P., Tormo-Mas, M. A., Vergara-Irigaray, M., Cheung, A. L., Lasa, I., et al. (2010). Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 12, 55–64. doi: 10.1016/j.micinf.2009.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mazmanian, S. K., Liu, G., Jensen, E. R., Lenoy, E., and Schneewind, O. (2000). Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. U.S.A. 97, 5510–5515. doi: 10.1073/pnas.080520697
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mazmanian, S. K., Ton-That, H., and Schneewind, O. (2001). Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40, 1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCourt, J., O'Halloran, D. P., McCarthy, H., O'Gara, J. P., and Geoghegan, J. A. (2014). Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 353, 157–164. doi: 10.1111/1574-6968.12424
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meenan, N. A., Visai, L., Valtulina, V., Schwarz-linek, U., Norris, N. C., Gurusiddappa, S., et al. (2007). The tandem beta-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 282, 25893–25902. doi: 10.1074/jbc.M703063200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mempel, M., Feucht, H., Ziebuhr, W., Endres, M., Laufs, R., and Gruter, L. (1994). Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 38, 1251–1255. doi: 10.1128/AAC.38.6.1251
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mempel, M., Müller, E., Hoffmann, R., Feucht, H., Laufs, R., and Grüter, L. (1995). Variable degree of slime production is linked to different levels of beta-lactam susceptibility in Staphylococcus epidermidis phase variants. Med. Microbiol. Immunol. 184, 109–113. doi: 10.1007/BF00224346
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Merino, N., Toledo-Arana, A., Vergara-Irigaray, M., Valle, J., Solano, C., Calvo, E., et al. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 191, 832–843. doi: 10.1128/JB.01222-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mizobuchi, S., Minami, J., Jin, F., Matsushita, O., and Okabe, A. (1994). Comparison of the virulence of methicillin-resistant and methicillin-sensitive Staphylococcus aureus. Microbiol. Immunol. 38, 599–605. doi: 10.1111/j.1348-0421.1994.tb01829.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mwangi, M. M., Kim, C., Chung, M., Tsai, J., Vijayadamodar, G., Benitez, M., et al. (2013). Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Resist. 19, 153–159. doi: 10.1089/mdr.2013.0053
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nethercott, C., Mabbett, A. N., Totsika, M., Peters, P., Ortiz, J. C., Nimmo, G. R., et al. (2013). Molecular characterization of endocarditis-associated Staphylococcus aureus. J. Clin. Microbiol. 51, 2131–2138. doi: 10.1128/JCM.00651-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Novick, R. P., Ross, H. F., Projan, S. J., Kornblum, J., Kreiswirth, B., and Moghazeh, S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975.
O'Gara, J. P. (2007). ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188. doi: 10.1111/j.1574-6968.2007.00688.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O'Gara, J. P., and Humphreys, H. (2001). Staphylococcus epidermidis biofilms: importance and implications. J. Med. Microbiol. 50, 582–587.
O'Neill, E., Pozzi, C., Houston, P., Humphreys, H., Robinson, D. A., Loughman, A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190, 3835–3850. doi: 10.1128/JB.00167-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
O'Neill, E., Pozzi, C., Houston, P., Smyth, D., Humphreys, H., Robinson, D. A., et al. (2007). Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45, 1379–1388. doi: 10.1128/JCM.02280-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oshida, T., Sugai, M., Komatsuzawa, H., Hong, Y. M., Suginaka, H., and Tomasz, A. (1995). A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U.S.A. 92, 285–289. doi: 10.1073/pnas.92.1.285
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Otto, M. (2013). Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35, 4–11. doi: 10.1002/bies.201200112
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Periasamy, S., Joo, H. S., Duong, A. C., Bach, T. H., Tan, V. Y., Chatterjee, S. S., et al. (2012). How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1281–1286. doi: 10.1073/pnas.1115006109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pozzi, C., Waters, E. M., Rudkin, J. K., Schaeffer, C. R., Lohan, A. J., Tong, P., et al. (2012). Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 8:e1002626. doi: 10.1371/journal.ppat.1002626
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qin, Z., Ou, Y., Yang, L., Zhu, Y., Tolker-Nielsen, T., Molin, S., et al. (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153, 2083–2092. doi: 10.1099/mic.0.2007/006031-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Queck, S. Y., Khan, B. A., Wang, R., Bach, T. H., Kretschmer, D., Chen, L., et al. (2009). Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 5:e1000533. doi: 10.1371/journal.ppat.1000533
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rao, F., See, R. Y., Zhang, D., Toh, D. C., Ji, Q., and Liang, Z. X. (2010). YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285, 473–482. doi: 10.1074/jbc.M109.040238
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Regassa, L. B., Novick, R. P., and Betley, M. J. (1992). Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect. Immun. 60, 3381–3388.
Rohde, H., Burdelski, C., Bartscht, K., Hussain, M., Buck, F., Horstkotte, M. A., et al. (2005). Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Frankenberger, S., Zahringer, U., and Mack, D. (2010). Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 89, 103–111. doi: 10.1016/j.ejcb.2009.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Knobloch, J. K., Horstkotte, M. A., and Mack, D. (2001). Correlation of Staphylococcus aureus icaADBC genotype and biofilm expression phenotype. J. Clin. Microbiol. 39, 4595–4596. doi: 10.1128/JCM.39.12.4595-4596.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rudkin, J. K., Edwards, A. M., Bowden, M. G., Brown, E. L., Pozzi, C., Waters, E. M., et al. (2012). Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J. Infect. Dis. 205, 798–806. doi: 10.1093/infdis/jir845
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rudkin, J. K., Laabei, M., Edwards, A. M., Joo, H. S., Otto, M., Lennon, K. L., et al. (2014). Oxacillin alters the toxin expression profile of community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 1100–1107. doi: 10.1128/AAC.01618-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rupp, M. E., Fey, P. D., Heilmann, C., and Götz, F. (2001). Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183, 1038–1042. doi: 10.1086/319279
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schaeffer, C. R., Woods, K. M., Longo, G. M., Kiedrowski, M. R., Paharik, A. E., Büttner, H., et al. (2015). Accumulation-associated protein (Aap) enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 83, 214–226. doi: 10.1128/IAI.02177-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schroeder, K., Jularic, M., Horsburgh, S. M., Hirschhausen, N., Neumann, C., Bertling, A., et al. (2009). Molecular characterization of a novel Staphylococcus aureus surface protein (SasC) involved in cell aggregation and biofilm accumulation. PLoS ONE 4:e7567. doi: 10.1371/journal.pone.0007567
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schwarz-Linek, U., Werner, J. M., Pickford, A. R., Gurusiddappa, S., Kim, J. H., Pilka, E. S., et al. (2003). Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423, 177–181. doi: 10.1038/nature01589
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shahrooei, M., Hira, V., Stijlemans, B., Merckx, R., Hermans, P. W., and Van eldere, J. (2009). Inhibition of Staphylococcus epidermidis biofilm formation by rabbit polyclonal antibodies against the SesC protein. Infect. Immun. 77, 3670–3678. doi: 10.1128/IAI.01464-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shanks, R. M., Meehl, M. A., Brothers, K. M., Martinez, R. M., Donegan, N. P., Graber, M. L., et al. (2008). Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect. Immun. 76, 2469–2477. doi: 10.1128/IAI.01370-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shaw, L., Golonka, E., Potempa, J., and Foster, S. J. (2004). The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150, 217–228. doi: 10.1099/mic.0.26634-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sung, J. M., Lloyd, D. H., and Lindsay, J. A. (2008). Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology 154, 1949–1959. doi: 10.1099/mic.0.2007/015289-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sureka, K., Choi, P. H., Precit, M., Delince, M., Pensinger, D. A., Huynh, T. N., et al. (2014). The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell 158, 1389–1401. doi: 10.1016/j.cell.2014.07.046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ton-That, H., Mazmanian, S. K., Faull, K. F., and Schneewind, O. (2000). Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J. Biol. Chem. 275, 9876–9881. doi: 10.1074/jbc.275.13.9876
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tormo, M. A., Knecht, E., Götz, F., Lasa, I., and Penadés, J. R. (2005). Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151, 2465–2475. doi: 10.1099/mic.0.27865-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Valle, J., Toledo-Arana, A., Berasain, C., Ghigo, J. M., Amorena, B., Penades, J. R., et al. (2003). SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48, 1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vergara-Irigaray, M., Valle, J., Merino, N., Latasa, C., Garcia, B., Ruiz de los Mozos, I., et al. (2009). Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 77, 3978–3991. doi: 10.1128/IAI.00616-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Gerke, C., Somerville, G. A., Fischer, E. R., and Otto, M. (2003). Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188, 706–718. doi: 10.1086/377239
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Kocianova, S., Yao, Y., Carmody, A. B., and Otto, M. (2004). Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505. doi: 10.1086/424487
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., and Otto, M. (2002). Staphylococcus epidermidis infections. Microbes Infect. 4, 481–489. doi: 10.1016/S1286-4579(02)01563-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Saenz, H. L., Götz, F., and Otto, M. (2000). Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182, 1688–1693. doi: 10.1086/317606
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, R., Khan, B. A., Cheung, G. Y., Bach, T. H., Jameson-Lee, M., Kong, K. F., et al. (2011). Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248. doi: 10.1172/JCI42520
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wecke, J., Lahav, M., Ginsburg, I., Kwa, E., and Giesbrecht, P. (1986). Inhibition of wall autolysis of staphylococci by sodium polyanethole sulfonate “liquoid.” Arch. Microbiol. 144, 110–115. doi: 10.1007/BF00414719
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wehrhahn, M. C., Robinson, J. O., Pascoe, E. M., Coombs, G. W., Pearson, J. C., O'Brien, F. G., et al. (2012). Illness severity in community-onset invasive Staphylococcus aureus infection and the presence of virulence genes. J. Infect. Dis. 205, 1840–1848. doi: 10.1093/infdis/jis279
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yabu, K., and Kaneda, S. (1995). Salt-induced cell lysis of Staphylococcus aureus. Curr. Microbiol. 30, 299–303. doi: 10.1007/BF00295505
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: biofilm, LPXTG proteins, Atl, PIA, mecA, methicillin resistance, c-di-AMP, Staphylococcus aureus
Citation: McCarthy H, Rudkin JK, Black NS, Gallagher L, O'Neill E and O'Gara JP (2015) Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 5:1. doi: 10.3389/fcimb.2015.00001
Received: 06 November 2014; Accepted: 05 January 2015;
Published online: 28 January 2015.
Edited by:
Joan A. Geoghegan, Trinity College Dublin, IrelandReviewed by:
Michael Otto, National Institute of Allergy and Infectious Diseases, USAIñigo Lasa, Universidad Pública de Navarra, Spain
Copyright © 2015 McCarthy, Rudkin, Black, Gallagher, O'Neill and O'Gara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James P. O'Gara, Department of Microbiology, School of Natural Sciences, National University of Ireland, Galway, Ireland e-mail:amFtZXNwLm9nYXJhQG51aWdhbHdheS5pZQ==
 Hannah McCarthy
Hannah McCarthy Justine K. Rudkin
Justine K. Rudkin Nikki S. Black
Nikki S. Black Laura Gallagher1
Laura Gallagher1 Eoghan O'Neill
Eoghan O'Neill James P. O'Gara
James P. O'Gara