- 1Department of Microbial Infection and Immunity, The Ohio State University, Columbus, OH, USA
- 2Center for Microbial Interface Biology, The Ohio State University, Columbus, OH, USA
- 3Department of Microbiology, The Ohio State University, Columbus, OH, USA
- 4Department of Medical Microbiology and Immunology, Faculty of Medicine, Mansoura University, Mansoura, Egypt
Many bacteria determine their population density using quorum sensing. The most intensively studied mechanism of quorum sensing utilizes proteins of the LuxI family to synthesize a signaling molecule of the acylhomoserine lactone (AHL) type, and a protein of the LuxR family to bind AHL and regulate transcription. Genes regulated by quorum sensing often encode functions that are most effective when a group of bacteria are working cooperatively (e.g., luminescence, biofilm formation, host interactions). Bacteria in the Escherichia, Salmonella, Klebsiella, and Enterobacter genera do not encode an AHL synthase but they do encode an AHL receptor of the LuxR family, SdiA. Instead of detecting their own AHL synthesis, these organisms use SdiA to detect the AHLs synthesized by other bacterial species. In this study, we used a genetic screen to identify AHL-responsive genes in a commensal Enterobacter cloacae strain that was isolated from a laboratory mouse. The genes include a putative type VI secretion system, copA (a copper transporter), and fepE (extends O-antigen chain length). A new transposon mutagenesis strategy and suicide vectors were used to construct an sdiA mutant of E. cloacae. The AHL-responsiveness of all fusions was entirely sdiA-dependent, although some genes were regulated by sdiA in the absence of AHL.
Introduction
Many bacteria monitor their population density (often called quorum sensing) as one of many inputs to gene regulation. The genes regulated by quorum sensing are often those that provide maximal benefit when expressed simultaneously throughout a population (Schuster et al., 2013). The classic example is the expression of luciferase by Vibrio fischeri, in which luminescence is most effective when the entire population participates (Hastings and Greenberg, 1999). Other examples include the expression of conjugation functions, biofilm formation, or various aspects of host interaction (Rutherford and Bassler, 2012).
The most intensively studied type of quorum sensing utilizes a protein of the LuxI family to synthesize a signaling molecule of the acylhomoserine lactone (AHL) type (Schaefer et al., 1996). Accumulation of the AHL results in its detection by a transcription factor of the LuxR family. In the case of V. fischeri, the LuxR-AHL complex binds upstream of the luxICDABEG operon to activate the expression of luciferase (Choi and Greenberg, 1991; Hanzelka and Greenberg, 1995). Thus, the population of bacteria cooperate to create light and illuminate their host, the squid Euprymna scolopes (Chun et al., 2008; Miyashiro and Ruby, 2012).
Homologous LuxI/LuxR regulatory systems have been identified in numerous Proteobacteria (Case et al., 2008). Some bacteria that live in mammalian intestinal tracts encode AHL synthases, although AHLs themselves have not yet been demonstrated to be present in this environment (Swearingen et al., 2012). Interestingly, a LuxR homolog, SdiA, has been identified in the Enterobacteriaceae, including the genera Escherichia, Salmonella, Enterobacter, Klebsiella, and Citrobacter. However, these organisms do not encode a cognate AHL synthase (Smith and Ahmer, 2003; Sabag-Daigle and Ahmer, 2012). Instead, it has been shown that SdiA of E. coli and Salmonella enterica detect the AHLs produced by other species of bacteria (Michael et al., 2001; Smith and Ahmer, 2003; Dyszel et al., 2010a,b; Sperandio, 2010a; Soares and Ahmer, 2011; Sheng et al., 2013). In Salmonella enterica serovar Typhimurium, SdiA positively regulates two loci, (1) the rck (resistance to complement killing) operon located on the virulence plasmid, pSLT (Ahmer et al., 1998; Michael et al., 2001; Smith and Ahmer, 2003; Abed et al., 2014); and (2) srgE (sdiA-regulated gene), a single gene horizontal acquisition that encodes an effector protein that is secreted by type III secretion system 2 (T3SS2) (Smith and Ahmer, 2003; Habyarimana et al., 2014). SdiA in EHEC functions to activate expression of genes involved in the glutamate-dependent acid resistance system (gad) and has also been found to repress the expression of flagella genes and the enterocyte effacement (LEE) locus (Van Houdt et al., 2006; Lee et al., 2008; Nikaido et al., 2008; Dyszel et al., 2010b; Hughes et al., 2010; Nguyen and Sperandio, 2012; Nguyen et al., 2013; Sheng et al., 2013). Competition assays in cattle of wild-type EHEC and an isogenic sdiA mutant indicate a defect of the sdiA mutant in colonization of rumen and the recto-anal junction (RAJ) (Hughes et al., 2010; Sheng et al., 2013). This phenotype was shown to correlate with lack of gad activation in the rumen and a failure to repress the LEE locus in the RAJ in the absence of sdiA (Hughes et al., 2010; Nguyen et al., 2013). In a plant-associated isolate of Enterobacter, an sdiA mutation derepresses the csgBAC operon leading to an overproduction of curli fimbrae (Shankar et al., 2012). The sdiA mutant has increased root colonization and biofilm formation correlating with the increased expression of curli adhesion molecules (Shankar et al., 2012).
We wanted to study the role of sdiA in a commensal member of the murine microbiota. Laboratory strains of E. coli K-12 and EHEC do not colonize mice well. Commensal strains of E. coli recovered from mice are very rare in the literature, and during microbiome studies E. coli has been found to be rare or non-existent in mice depending on strain and vendor. In this study, we performed a genetic screen to identify AHL-responsive genes of an Enterobacter cloacae strain that was isolated from laboratory mice (Ali et al., 2014). We utilized a transposon to create chromosomal luxCDABE fusions in a wild-type background, with sdiA at its natural position in the chromosome. We screened these fusions to identify those that are AHL-responsive. A new suicide vector and novel mutagenesis strategy were then used to mutate sdiA in each fusion strain. The AHL-responsiveness of all of the fusions was entirely sdiA-dependent, but a few genes were regulated by sdiA, including one gene repressed by sdiA, largely in the absence of AHLs. This ligand-independent activity of SdiA has important implications for our understanding of the role of this LuxR solo (Patankar and Gonzalez, 2009; Subramoni and Venturi, 2009).
Materials and Methods
Bacterial Strains and Media
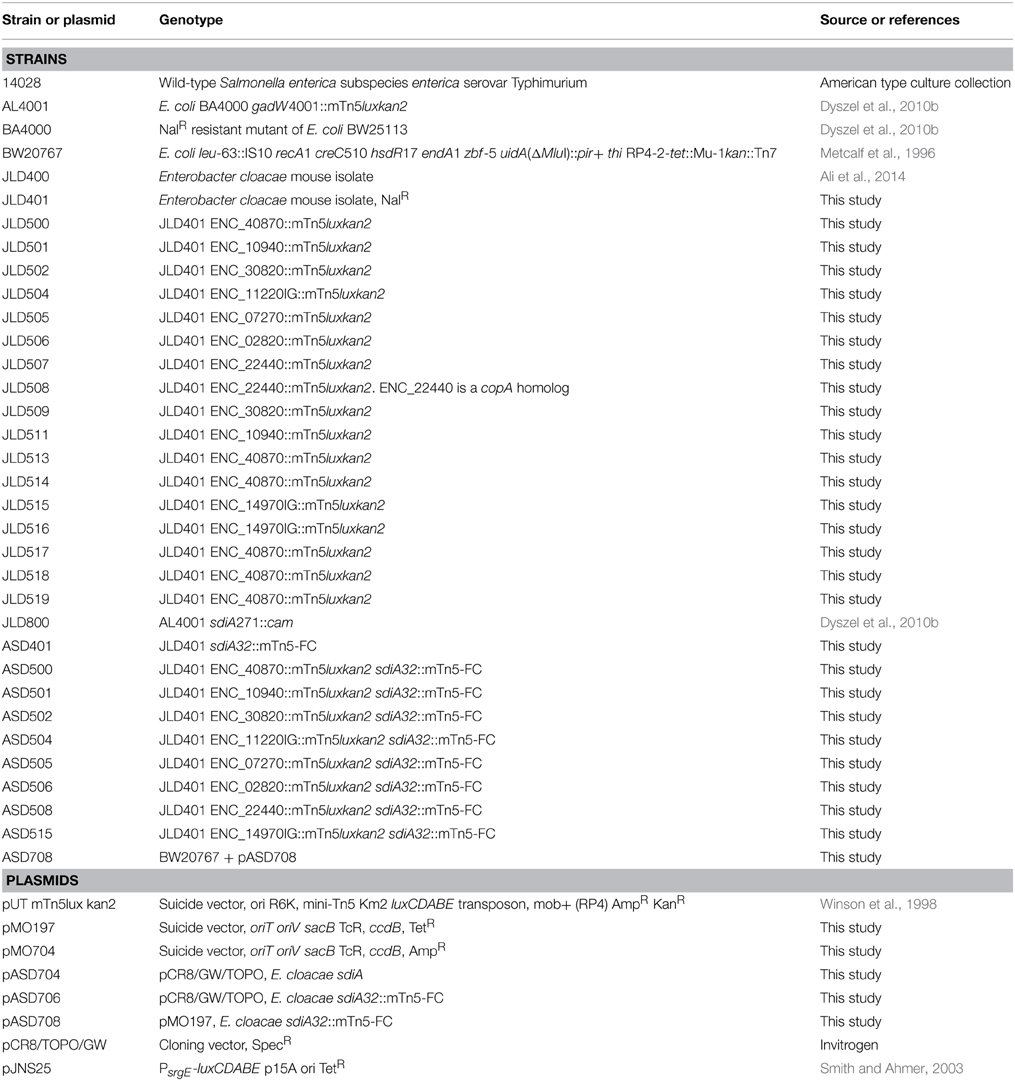
Bacterial strains are listed in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) broth or on LB agar unless otherwise stated. LB motility agar was also used (LB broth + 0.3% agar). Chloramphenicol (cam), kanamycin (kan), tetracycline (tet), ampicillin (amp), and nalidixic acid (nal) were used at 20, 50, 10, 200, and 50 μg/ml, respectively. N-(3-oxo-hexanoyl)-L-homoserine lactone (oxoC6) was obtained from Sigma-Aldrich and dissolved in ethyl acetate that had been acidified by the addition of 0.1 ml glacial acetic acid per liter (EA) (Pearson et al., 1994). The stock concentration of oxoC6 was 1 mM and it was used at a final concentration of 1 μM. Solvent controls were performed by using EA alone at 0.1%.
Constructing Transposon Based Luciferase Fusions and Screening for AHL Responsiveness in E. cloacae
Transposon mutagenesis was performed by mating BW20767+pUTmTn5luxkan2 (Winson et al., 1998) and JLD401, a spontaneous nalidixic acid resistant mutant of Enterobacter cloacae strain JLD400. The two strains were plated on LB plates at 37°C overnight. Cells were then scraped with sterile PBS and plated on LB kan nal. 10,000 single colonies were patched into 96-well plates with 0.3% motility agar in the presence of oxoC6 or the solvent control, EA, at 37°C for 9 h. Plates were read with a Wallac Victor3 (Perkin Elmer) plate reader. Those wells that had greater than 3-fold difference after 9 h were streaked for isolation on LB kan nal plates at 37°C overnight. For confirmation, one colony from each plate was inoculated into LB kan nal broth or 0.3% motility agar in 96-well format in the presence of oxoC6, or the solvent control, EA. Plates were read on the Victor plate reader every 3 h. Those fusions that demonstrated greater than 2.5-fold AHL-dependent induction after 9 h were saved for future studies.
Identification of Transposon Insertion Sites
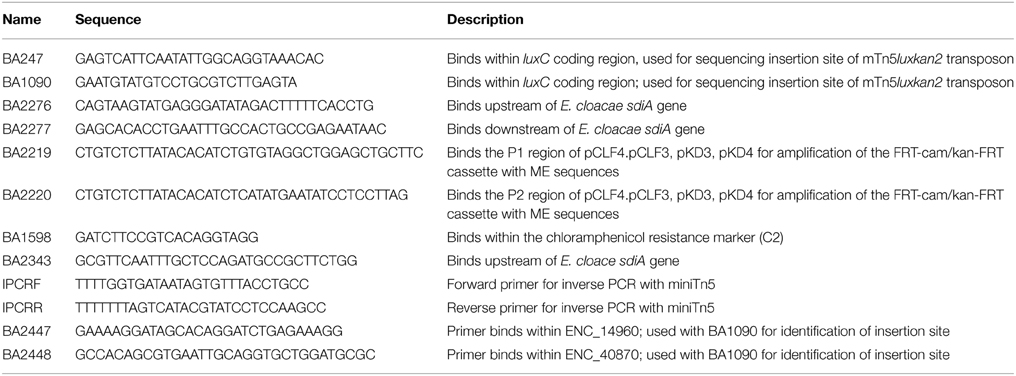
Genomic DNA was isolated from overnight cultures of the transposon insertion mutants using the GenElute™ Bacterial Genomic DNA Isolation kit (Sigma Aldrich, St. Louis, MO). The transposon insertion site in the genomic DNA was sequenced using Sanger sequencing with two different primers, BA247 and BA1090 (Table 2). Both sequencing primers bind within the luxC coding region oriented out of the transposon. Oligonucleotides were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). DNA sequencing was performed by the Plant Microbe Genomics Facility at The Ohio State University. The sequence adjacent to the transposon insertion site was used for BLASTN searches using the BLAST program at the National Center for Biotechnology Information (NCBI).
Not all insertion points were identified using Sanger genomic DNA sequencing. The rest were identified using inverse PCR. Genomic DNA was digested with NlaIII (NEB) for 3 h at 37°C. The enzyme was inactivated for 20 min at 65°C, then T4 DNA ligase (NEB) was added to the digestion reaction in a total reaction volume of 200 μL at 16°C overnight. The ligation reaction was purified using the QiaQuick PCR purification kit (Qiagen), digested with XmnI for 3 h at 37°C, then heat inactivated at 65°C for 20 min. This digest was used in a PCR reaction using the primers IPCRF and IPCRR (Table 2) using Taq DNA polymerase (NEB). The PCR product was sequenced using the IPCRF primer at the Plant Microbe Genomics Facility at The Ohio State University.
Liquid and Motility Agar Assays for Lux Fusions
Strains were grown in LB kan for sdiA+ strains or LB kan cam for sdiA mutants, and grown at 37°C shaking overnight. They were then subcultured 1:100 in triplicate into either LB broth or LB motility agar containing the appropriate antibiotics and either 1 μM oxoC6 or 0.1% EA as the solvent control and then placed in the well of a black clear bottom 96-well plate. The plate was grown with shaking at 37°C and time points were taken by placing the 96-well plate in the Wallac Victor plate reader. For broth cultures, both OD590 and luminescence were measured. For the motility agar assays, only the luminescence was measured. For the AHL concentration sensitivity assay, an sdiA-regulated fusion was assayed in LB broth or motility agar with 10-fold dilutions of either oxoC6 or oxoC8 starting at a concentration of 1 μM. AHL-dependent changes in gene expression were analyzed using a Two-Way ANOVA over the time course. P-values were marked accordingly (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 = ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p ≤ 0.00005) for all fusions in all growth conditions for at least one time point.
Construction of an sdiA Mutant of E. cloacae, Using a New Suicide Vector and Transposon Mutagenesis Strategy
The new suicide vectors are derivatives of pDMS197 (Edwards et al., 1998) and pGP704 (Miller and Mekalanos, 1988), respectively, that have been modified for the Gateway cloning system of Invitrogen, which uses phage attachment sites for in vitro recombination reactions. To modify these vectors, a blunt-ended DNA fragment (Reading Frame Cassette C) obtained from Invitrogen containing attR-camr-ccdB-attR was ligated into the SmaI site of pGP704 and pDMS197, resulting in pMO704 and pMO197, respectively (Figure 1).
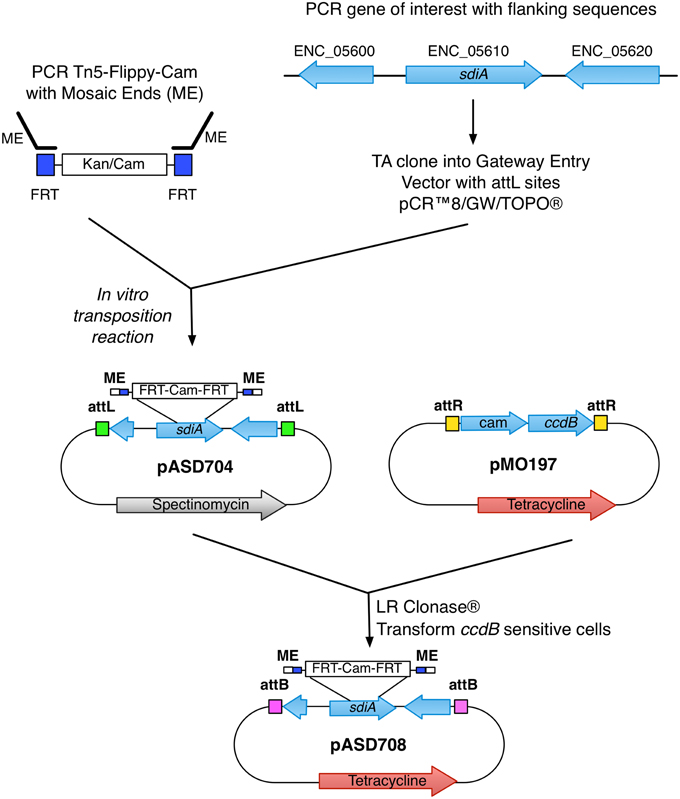
Figure 1. Generation of the sdiA mutant in E. cloacae. Schematic of the construction and generation of the sdiA mutant in E. cloacae using the mTn5-FC and novel suicide vector pMO197.
With the suicide vectors completed, the sdiA gene of E. cloacae was amplified by PCR using primers BA2276 and BA2277 (Table 2) and cloned into the Gateway entry vector pCR8/GW/TOPO creating pASD704. This vector has attL sites flanking the inserted PCR product. Mutations were created in this cloned E. cloacae sdiA gene using in vitro transposon mutagenesis. We utilized a new mTn5 derivative that we named mTn5-FC where FC stands for flippy-cam (Figure 1). Essentially, the DNA sequences of the optimized mosaic ends of Tn5 (Goryshin and Reznikoff, 1998) are appended to PCR primers (BA2219 and BA2220) that are used to amplify the FRT-camr-FRT cassette from pCLF3 (Santiviago et al., 2009). The resulting PCR product is the transposon. To mutagenize the sdiA gene on pASD704, plasmid DNA and transposon DNA were mixed in the presence of transposase enzyme in an in vitro transposition reaction. This reaction mix was then transformed into DH5αλ pir and plated on LB spec cam. The resulting colonies were screened for transposon insertions in the sdiA gene using PCR with primers BA2276 and BA1598. One insertion of interest, named sdiA32::mTn5-FC (pASD706), was then used in an “LR” cloning reaction to recombine the mutated sdiA gene into the new pMO197 suicide vector by transforming BW20767, creating pASD708. This vector was then mobilized into each of the E. cloacae strains, selecting for replacement of the wild-type sdiA allele with the mutant allele, which was verified by PCR using primers BA2343 and BA1598.
Results
Identification of AHL-Responsive Transcriptional Fusions in E. cloacae
A murine isolate of Enterobacter cloacae strain, JLD400, was found to be genetically tractable and sensitive to all antibiotics tested except ampicillin (Ali et al., 2014). It is easily electroporated and readily serves as a recipient in RP4-mediated conjugation. A mutant resistant to nalidixic acid was isolated after passage on LB with nalidixic acid (JLD401, Table 1).
In order to identify genes that are regulated in response to AHLs in E. cloacae, we constructed random transcriptional fusions to the luciferase genes of Photorhabdus luminescens (luxCDABE) using the transposon mTn5-luxCDABE (Winson et al., 1998). Previous screens for sdiA-regulated targets in Salmonella have utilized plasmid-borne sdiA, which bypasses the AHL requirement, but this approach has been shown to have pleotropic effects in E. coli (Ahmer, 2004; Dyszel et al., 2010b). Therefore, we took an alternate strategy in which we mutagenized the wild-type strain with the mTn5-luxCDABE transposon while sdiA remained in its native position in the chromosome and screened 10,000 mutants for responsiveness to synthetic AHL (oxoC6, which is detected by SdiA of E. coli and Salmonella). Seventeen insertions were identified that demonstrated an increase in luminescence greater than 2.5-fold.
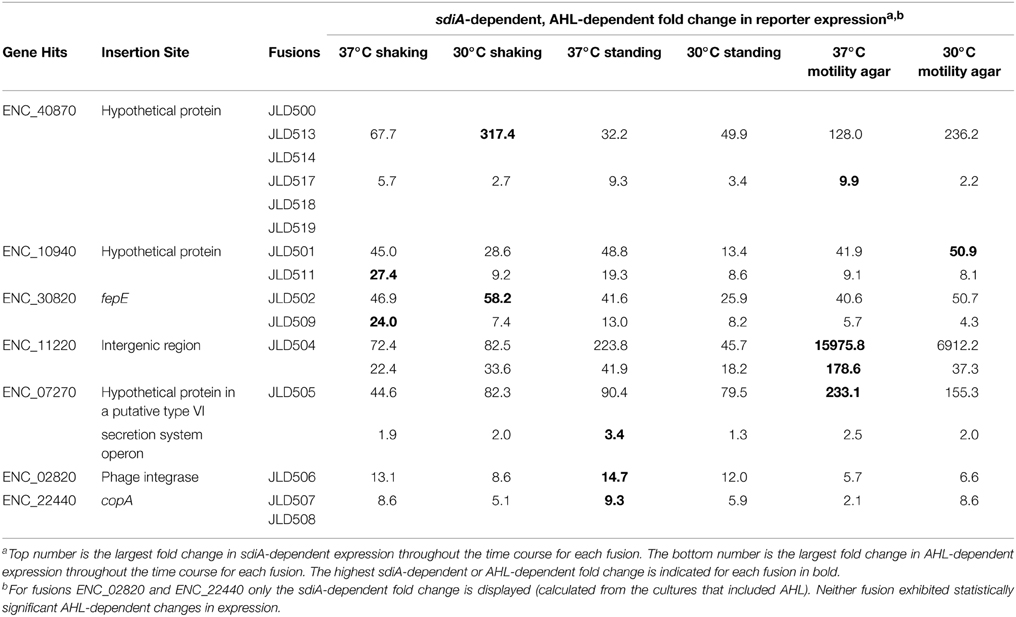
The transposon insertion point was identified for all 17 insertions using either of two methods: (1) sequencing genomic DNA using two different sequencing primers that bind within the transposon sequence and are oriented outward, or (2) inverse PCR and subsequent sequencing of the product. After identification of the transposon insertion sites, a confirmatory PCR was performed using a primer within the transposon and another within the putative AHL-responsive gene. A positive PCR reaction confirms that the transposon insertion is in the correct location, but we also sequenced the resulting PCR product to further define the transposon insertion site. All 17 AHL-responsive fusions were located within 8 unique genes (Table 3, Figure 2). The genome sequence of our Enterobacter cloacae isolate is not known, but BLAST searches revealed that the majority of these sequences were most similar to the genome sequence of Enterobacter cloacae subspecies cloacae NCTC 9394 (FP929040). The transposon insertions of six strains (JLD500, JLD513, JLD514, JLD517, JLD518, and JLD519) were within ENC_40870, which encodes a hypothetical protein only present in Enterobacter cloacae. JLD501 and JLD511 each contained an insertion in ENC_10940, which encodes a hypothetical protein with a secretion signal predicted by SignalP within the first 20 amino acids (Petersen et al., 2011). JLD502 and JLD509 each contained an insertion in ENC_30820, which encodes a homolog of FepE, a protein that increases the length of O antigen chains (Murray et al., 2003; Crawford et al., 2012, 2013). JLD504 has an insertion within the intergenic region of ENC_11220, which encodes a hypothetical protein. JLD505 contained an insertion in ENC_07270, which encodes a hypothetical protein within an operon that encodes a putative type VI secretion system (Durand et al., 2014; Li et al., 2015). JLD506 contains an insertion in ENC_02820, which encodes a prophage integrase. JLD507 and JLD508 each contained an insertion in ENC_22440, which encodes a homolog of CopA, a putative copper-translocating P-type ATPase (Rensing and Grass, 2003; Osman and Cavet, 2011). JLD515 and JLD516 have insertions within the promoter region of ENC_14970, encoding a putative signal transduction protein containing a sensor and diguanylate phosphodiesterase (EAL) domain (Römling et al., 2013).
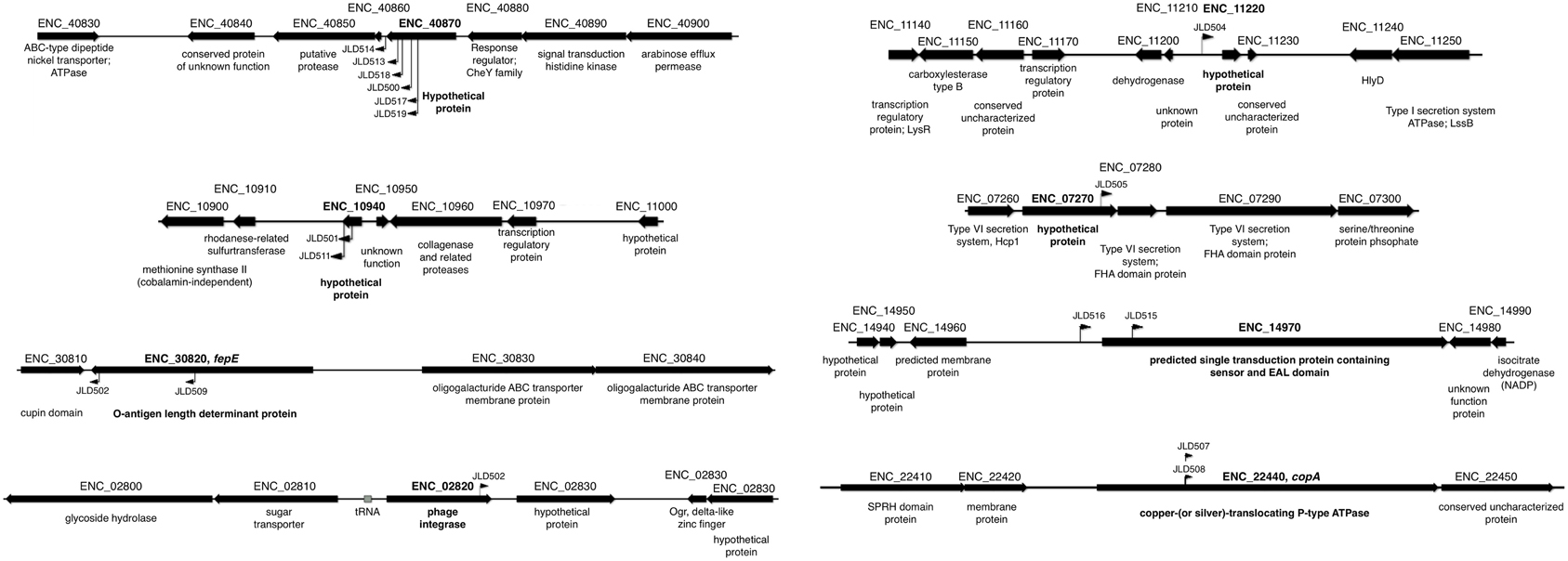
Figure 2. Insertion points of AHL-regulated genes in E. cloacae. Diagram of the location of AHL-regulated mTn5-luxCDABE insertions. The mTn5-luxCDABE insertion points and orientation are indicated by flags.
In Salmonella, we have observed differences in the behavior of sdiA-regulated fusions at 30°C compared to 37°C, and in motility agar compared to broth or agar plates (Smith and Ahmer, 2003). More ligand-independent SdiA activity is observed at lower temperatures, and more activity in general is observed in motility agar than in broth or agar plates. For E. cloacae, therefore, we chose one representative fusion-containing strain for each AHL-responsive locus identified, and tested these representatives under each of these conditions (Figures 3–8). Unlike sdiA-regulated fusions in Salmonella, the E. cloacae fusions did not become more ligand-independent at 30°C compared to 37°C. Instead, some fusions were largely ligand-independent under all conditions, while the remainder were ligand-dependent under all conditions. We also tested the response of these fusions to a series of AHL concentrations using oxoC6 and oxoC8. The AHL detection limits of E. cloacae are similar to those of E. coli and Salmonella (Figure 9).
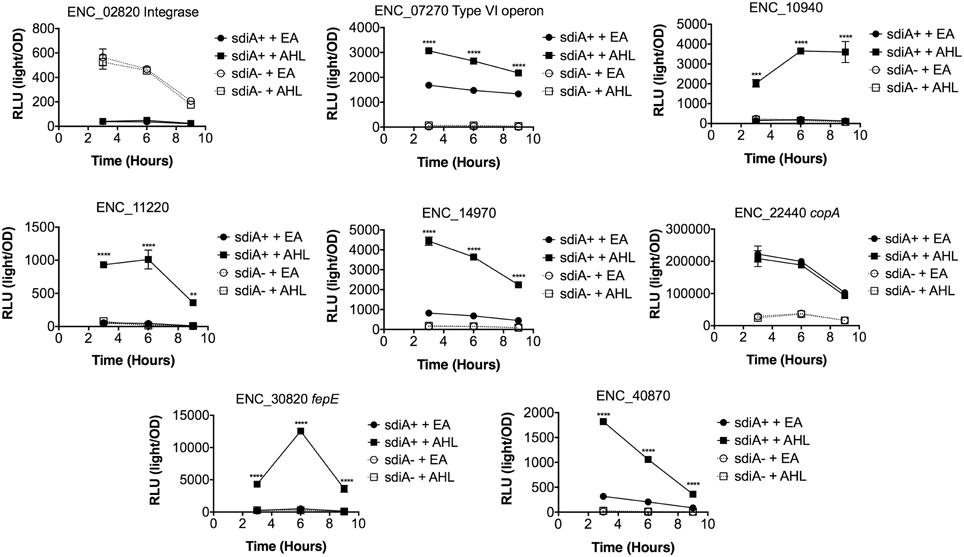
Figure 3. Regulation of AHL-regulated genes in E. cloacae in LB broth at 37°C shaking. Expression of mTn5luxCDABE fusion strains in either the wild-type (closed symbols) or sdiA mutant backgrounds (open symbols) with either 1 μM oxoC6 (squares) or 0.1% ethyl acetate (EA) solvent control (circles) in LB broth. Luminescence is reported in relative light units (light/OD590). Data was collected at 3, 6, and 9 h time points. All data points are the average of three technical replicates and error bars indicate SEM. This is a representative graph of three independent biological replicates. The statistical significance of AHL-dependent changes in gene expression are designated with p-values (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 = ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p = 0.00005) for all fusions in all growth conditions for at least one time point. The sdiA+ and sdiA mutant strains for each fusion are indicated in parentheses: ENC_02820 (JLD506/ASD506), ENC_07270 (JLD505/ASD505), ENC_10940 (JLD501/ASD501), ENC_11220 (JLD504/ASD504), ENC_14970 (JLD515/ASD515), ENC_22440 (JLD508/ASD508), ENC_30820 (JLD502/ASD502), and ENC_40870 (JLD500/ASD500).
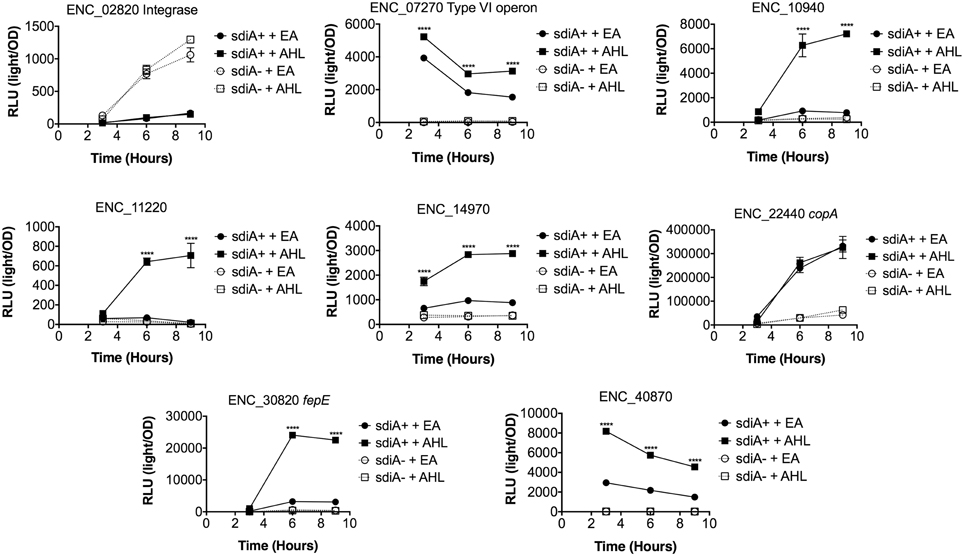
Figure 4. Regulation of AHL-regulated genes in E. cloacae in LB broth at 30°C shaking. Expression of mTn5luxCDABE fusion strains in either the wild-type (closed symbols) or sdiA mutant backgrounds (open symbols) with either 1 μM oxoC6 (squares) or 0.1% ethyl acetate (EA) solvent control (circles) in LB broth. Luminescence is reported in relative light units (light/OD590). Data was collected at 3, 6, and 9 h time points. All data points are the average of three technical replicates and error bars indicate SEM. This is a representative graph of three independent biological replicates. The statistical significance of AHL-dependent changes in gene expression are designated with p-values (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 = ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p = 0.00005) for all fusions in all growth conditions for at least one time point. The sdiA+ and sdiA mutant strains for each fusion are indicated in parentheses: ENC_02820 (JLD506/ASD506), ENC_07270 (JLD505/ASD505), ENC_10940 (JLD501/ASD501), ENC_11220 (JLD504/ASD504), ENC_14970 (JLD515/ASD515), ENC_22440 (JLD508/ASD508), ENC_30820 (JLD502/ASD502), and ENC_40870 (JLD500/ASD500).
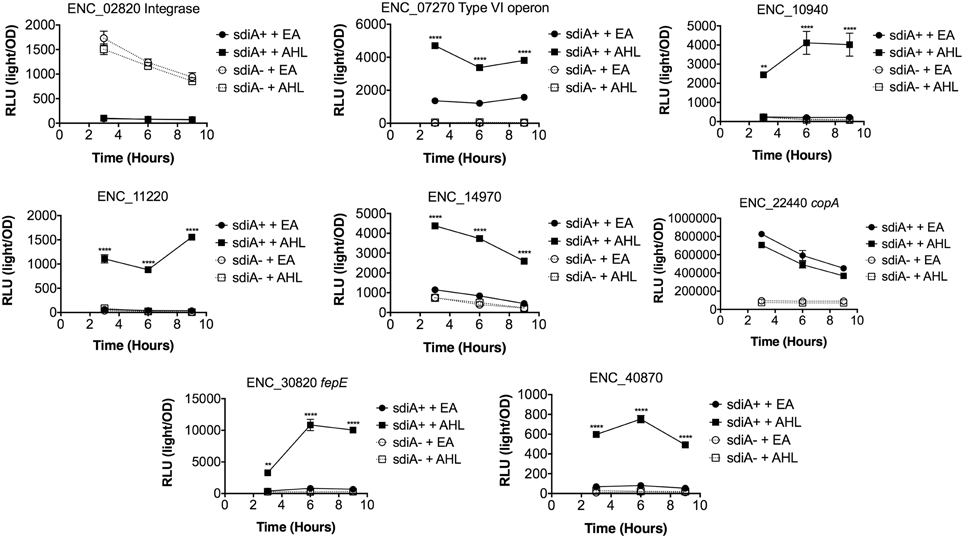
Figure 5. Regulation of AHL-regulated genes in E. cloacae in LB broth at 37°C standing. Expression of mTn5luxCDABE fusion strains in either the wild-type (closed symbols) or sdiA mutant backgrounds (open symbols) with either 1 μM oxoC6 (squares) or 0.1% ethyl acetate (EA) solvent control (circles) in LB broth. Luminescence is reported in relative light units (light/OD590). Data was collected at 3, 6, and 9 h time points. All data points are the average of three technical replicates and error bars indicate SEM. This is a representative graph of three independent biological replicates. The statistical significance of AHL-dependent changes in gene expression are designated with p-values (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 = ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p ≤ 0.00005) for all fusions in all growth conditions for at least one time point. The sdiA+ and sdiA mutant strains for each fusion are indicated in parentheses: ENC_02820 (JLD506/ASD506), ENC_07270 (JLD505/ASD505), ENC_10940 (JLD501/ASD501), ENC_11220 (JLD504/ASD504), ENC_14970 (JLD515/ASD515), ENC_22440 (JLD508/ASD508), ENC_30820 (JLD502/ASD502), and ENC_40870 (JLD500/ASD500).
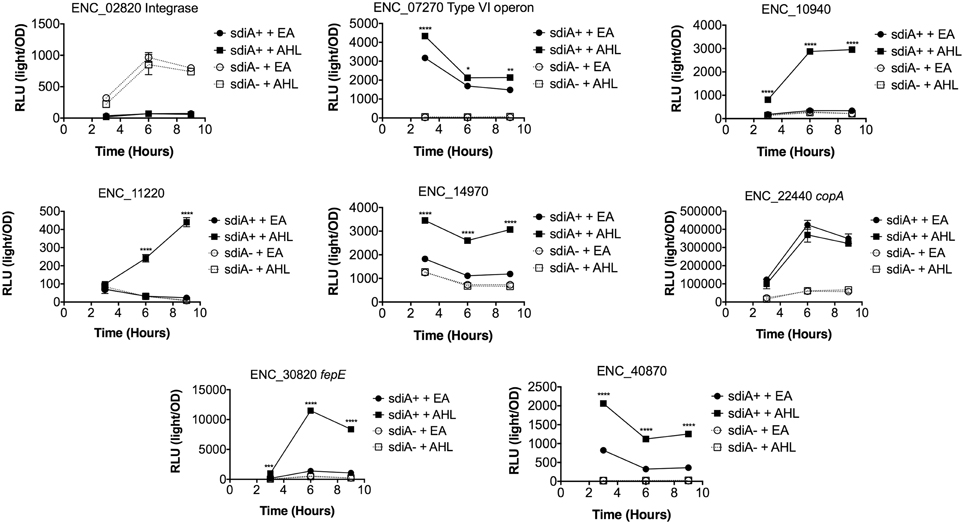
Figure 6. Regulation of AHL-regulated genes in E. cloacae in LB broth at 30°C standing. Expression of mTn5luxCDABE fusion strains in either the wild-type (closed symbols) or sdiA mutant backgrounds (open symbols) with either 1 μM oxoC6 (squares) or 0.1% ethyl acetate (EA) solvent control (circles) in LB broth. Luminescence is reported in relative light units (light/OD590). Data was collected at 3, 6, and 9 h time points. All data points are the average of three technical replicates and error bars indicate SEM. This is a representative graph of three independent biological replicates. The statistical significance of AHL-dependent changes in gene expression are designated with p-values (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 = ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p = 0.00005) for all fusions in all growth conditions for at least one time point. The sdiA+ and sdiA mutant strains for each fusion are indicated in parentheses: ENC_02820 (JLD506/ASD506), ENC_07270 (JLD505/ASD505), ENC_10940 (JLD501/ASD501), ENC_11220 (JLD504/ASD504), ENC_14970 (JLD515/ASD515), ENC_22440 (JLD508/ASD508), ENC_30820 (JLD502/ASD502), and ENC_40870 (JLD500/ASD500).
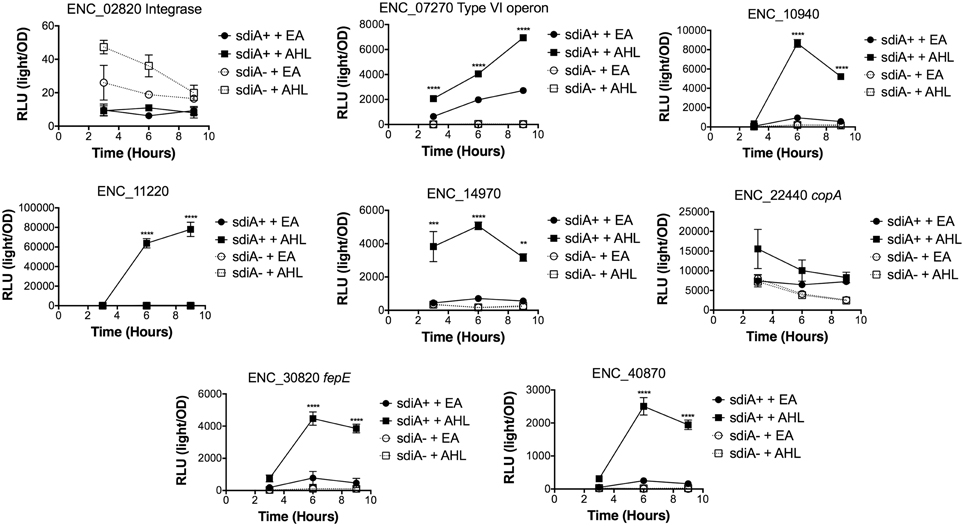
Figure 7. Regulation of AHL-regulated genes in E. cloacae in motility agar at 37°C. Expression of mTn5luxCDABE fusion strains in either the wild-type (closed symbols) or sdiA mutant backgrounds (open symbols) with either 1 μM oxoC6 (squares) or 0.1% ethyl acetate (EA) solvent control (circles) in LB broth. Luminescence is reported in relative light units (light/OD590). Data was collected at 3, 6, and 9 h time points. All data points are the average of three technical replicates and error bars indicate SEM. This is a representative graph of three independent biological replicates. The statistical significance of AHL-dependent changes in gene expression are designated with p-values (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 ≤ ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p = 0.00005) for all fusions in all growth conditions for at least one time point. The sdiA+ and sdiA mutant strains for each fusion are indicated in parentheses: ENC_02820 (JLD506/ASD506), ENC_07270 (JLD505/ASD505), ENC_10940 (JLD501/ASD501), ENC_11220 (JLD504/ASD504), ENC_14970 (JLD515/ASD515), ENC_22440 (JLD508/ASD508), ENC_30820 (JLD502/ASD502), and ENC_40870 (JLD500/ASD500).
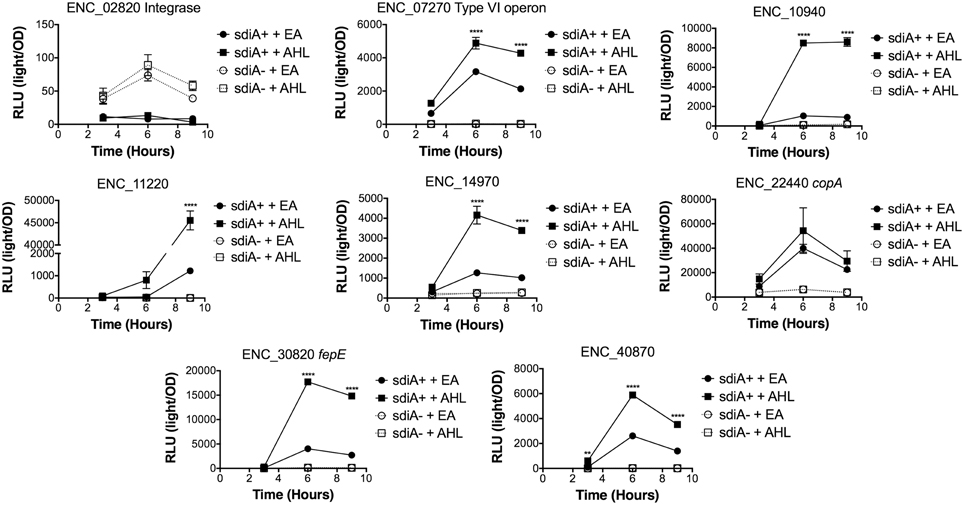
Figure 8. Regulation of AHL-regulated genes in E. cloacae in motility agar at 30°C. Expression of mTn5luxCDABE fusion strains in either the wild-type (closed symbols) or sdiA mutant backgrounds (open symbols) with either 1 μM oxoC6 (squares) or 0.1% ethyl acetate (EA) solvent control (circles) in LB broth. Luminescence is reported in relative light units (light/OD590). Data was collected at 3, 6, and 9 h time points. All data points are the average of three technical replicates and error bars indicate SEM. This is a representative graph of three independent biological replicates. The statistical significance of AHL-dependent changes in gene expression are designated with p-values (≤0.05 = *, ≤0.005 = **, ≤0.0005 = ***, ≤0.00005 = ****). The sdiA-dependent changes in gene expression were larger than the AHL-dependent changes and were statistically significant (p ≤ 0.00005) for all fusions in all growth conditions for at least one time point. The sdiA+ and sdiA mutant strains for each fusion are indicated in parentheses: ENC_02820 (JLD506/ASD506), ENC_07270 (JLD505/ASD505), ENC_10940 (JLD501/ASD501), ENC_11220 (JLD504/ASD504), ENC_14970 (JLD515/ASD515), ENC_22440 (JLD508/ASD508), ENC_30820 (JLD502/ASD502), and ENC_40870 (JLD500/ASD500).
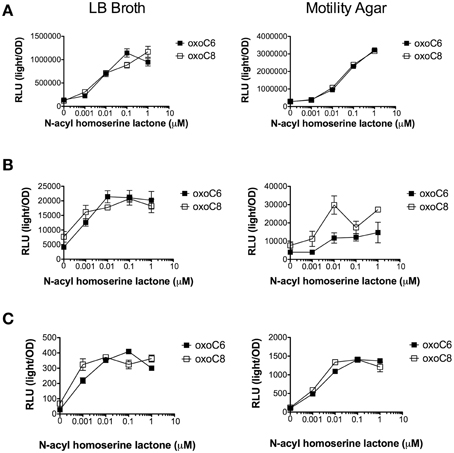
Figure 9. AHL concentration dependent response of Salmonella, E. coli, and Enterobacter. (A) Expression of srgE-luxCDABE fusion (pJNS25) in S. Typhimurium 14028. (B) Expression of the gadW::Tn5luxCDABE fusion in E. coli K-12 AL4001. (C) Expression of the ENC_40870::Tn5luxCDABE fusion in E. cloacae JLD500. Assays were done in either LB liquid broth with shaking or motility agar (0.3% agar) at 37°C. Relative light units (light/OD590) after 6 h of growth are indicated for oxoC6 (black squares) or oxoC8 (open squares). All data points are the mean of three biological replicates and error bars indicate SEM.
AHL-responses are sdiA-dependent
We hypothesized that the AHL-dependent responses of the lux fusions were dependent on the sdiA gene encoded on the chromosome of E. cloacae. To test this hypothesis, we constructed an sdiA mutation in each of the mTn5-luxCDABE fusion strains using two new suicide vectors and a new transposon mutagenesis strategy (see Materials and Methods). Indeed, mutation of sdiA in each fusion strain eliminated any responsiveness to AHL in both liquid culture and motility agar (Table 3). In one case, sdiA in E. cloacae acts as a negative regulator of expression. We observed that sdiA is required for repression of ENC_02820 since the sdiA mutant strain produced more light than the wild-type strain regardless of the presence of AHL, in all of the conditions tested (Figures 3–8).
Discussion
SdiA is a LuxR homolog that detects the AHLs produced by other bacteria (Michael et al., 2001; Smith and Ahmer, 2003). To date, SdiA regulon members in S. enterica serovar Typhimurium, E. coli K-12, EHEC and a plant-pathogenic isolate of Enterobacter cloacae have been described (Ahmer et al., 1998; Kanamaru et al., 2000; Wei et al., 2001; Suzuki et al., 2002; Smith and Ahmer, 2003; Van Houdt et al., 2006; Lee et al., 2007; Ghosh et al., 2009; Dyszel et al., 2010b; Hughes et al., 2010; Sharma et al., 2010; Shankar et al., 2012; Sharma and Bearson, 2013). Here we report the identification of AHL-responsive and sdiA-dependent genes in a mouse isolate of E. cloacae. We have previously shown that this E. cloacae isolate is not pathogenic and competes with Salmonella for colonization of mice (Ali et al., 2014). To identify AHL-responsive genes in this organism, we used a transposon-based genetic screen in which the expression of luciferase by individual mTn5-luxCBADE mutants was measured in the presence and absence of AHL. The sdiA gene was then mutated in each strain, and the response of every fusion was found to be sdiA-dependent (Figures 3–8). This suggests that SdiA is the only AHL receptor in this isolate of E. cloacae.
In Salmonella, there is very little SdiA activity in the absence of AHL at 37°C, although some is observed at 30°C (Smith and Ahmer, 2003; Sabag-Daigle et al., 2012). In E. coli there seems to be more SdiA activity in the absence of AHL (Dyszel et al., 2010b; Hughes et al., 2010; Sperandio, 2010b). Other work in E. coli has shown that SdiA binds target genes in vivo in the absence of AHL (Ishihama et al., 2014; Shimada et al., 2014). AHL-independent activity of SdiA was also noted with some, but not all, plasmids used as AHL biosensors in E. coli (Lindsay and Ahmer, 2005). However, the E. cloacae regulon identified here is very unusual in that SdiA is demonstrating high levels of AHL-independent activity for some fusions but not others. For instance, substantial AHL-independent SdiA activity is observed with ENC_22440 and ENC_07270 (Figures 3–8). Another fusion was repressed by sdiA and this was also independent of AHL (ENC_02820) (Figures 3–8). It appears that we were fortunate to identify these particular fusions using AHL-responsiveness as the first screen. The LuxR homolog TraR requires AHL for proper structural folding in order to oligomerize into a fully functional dimer structure capable of binding its target promoters (Zhu and Winans, 2001). However, the ligand-independent activity of SdiA suggests that SdiA is properly folded and able to bind target promoters even in the absence of AHL. This may be due to the folding of SdiA around endogenous 1-octanoyl-rac-glycerol (Nguyen et al., 2015). The mechanistic differences between ligand-dependent and -independent regulation of genes by SdiA is an interesting topic for further studies.
The role(s) for the SdiA regulon in this isolate of E. cloacae is unclear (Figures 2, 10). The fepE, copA, and type VI secretion genes could be envisioned to have direct interactions with the host, direct interactions with other microbes, or in general survival in the intestinal tract. Alternatively, these genes may play a role outside the host in other environments. In E. coli, the induction of lambda prophage is enhanced by AHL in an sdiA-dependent manner (Ghosh et al., 2009). The fusion that was repressed by sdiA in this study encodes a putative phage integrase, although it does not appear to be encoded within a prophage (Figure 2). It would be interesting to determine if SdiA plays a role in phage biology of E. cloacae.
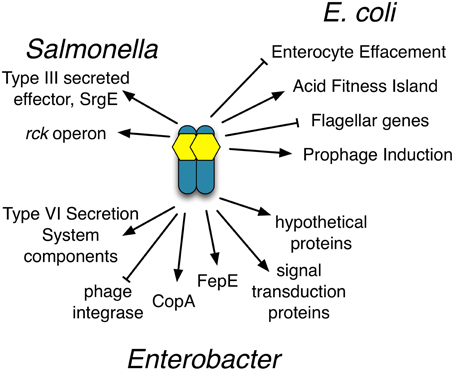
Figure 10. Compilation of the SdiA regulon members in Salmonella, EHEC and Enterobacter. Diagram of known SdiA regulon members in Salmonella, EHEC and Enterobacter. Arrows indicate whether SdiA increases the expression of the regulon member and blunt arrows indicate SdiA decreases the expression of the regulon member, either directly or indirectly.
In an isolate of E. cloacae that promotes the growth of rice roots, SdiA represses biofilm formation and rice root colonization (Shankar et al., 2012). This phenotype is at least partially due to the sdiA-dependent repression of the genes encoding curli fimbriae (Shankar et al., 2012). Repression of biofilm formation has been observed in E. coli as well (Lee et al., 2009; Sharma et al., 2010). Interestingly, we did not isolate curli genes in this study. It is not known if curli genes are not regulated by sdiA in this isolate, or if we simply missed them, either by chance or due to growth conditions. It would be interesting to determine if the genes identified in this study are regulated by sdiA in the E. cloacae plant isolate and to determine if the sdiA regulon has diverged between the plant and mouse isolates, or if the regulon has remained largely the same.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The project described was supported by Award Numbers R01AI073971, R01AI097116, and 1R01AI116119 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
Abed, N., Grépinet, O., Canepa, S., Hurtado-Escobar, G. A., Guichard, N., Wiedemann, A., et al. (2014). Direct regulation of the pefI-srgC operon encoding the Rck invasin by the quorum-sensing regulator SdiA in Salmonella Typhimurium. Mol. Microbiol. 94, 254–271. doi: 10.1111/mmi.12738
Ahmer, B. M. M. (2004). Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol. Microbiol. 52, 933–945. doi: 10.1111/j.1365-2958.2004.04054.x
Ahmer, B. M., van Reeuwijk, J., Timmers, C. D., Valentine, P. J., and Heffron, F. (1998). Salmonella typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180, 1185–1193.
Ali, M. M., Newsom, D. L., González, J. F., Sabag-Daigle, A., Stahl, C., Steidley, B., et al. (2014). Fructose-asparagine is a primary nutrient during growth of salmonella in the inflamed intestine. PLoS Pathog. 10:e1004209. doi: 10.1371/journal.ppat.1004209
Case, R. J., Labbate, M., and Kjelleberg, S. (2008). AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2, 345–349. doi: 10.1038/ismej.2008.13
Choi, S. H., and Greenberg, E. P. (1991). The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc. Natl. Acad. Sci. U.S.A. 88, 11115–11119.
Chun, C. K., Troll, J. V., Koroleva, I., Brown, B., Manzella, L., Snir, E., et al. (2008). Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc. Natl. Acad. Sci. U.S.A. 105, 11323–11328. doi: 10.1073/pnas.0802369105
Crawford, R. W., Keestra, A. M., Winter, S. E., Xavier, M. N., Tsolis, R. M., Tolstikov, V., et al. (2012). Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog. 8:e1002918. doi: 10.1371/journal.ppat.1002918
Crawford, R. W., Wangdi, T., Spees, A. M., Xavier, M. N., Tsolis, R. M., and Bäumler, A. J. (2013). Loss of very-long O-antigen chains optimizes capsule-mediated immune evasion by Salmonella enterica serovar Typhi. MBio 4:e00232–13. doi: 10.1128/mBio.00232-13
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Dyszel, J. L., Smith, J. N., Lucas, D. E., Soares, J. A., Swearingen, M. C., Vross, M. A., et al. (2010a). Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J. Bacteriol. 192, 29–37. doi: 10.1128/JB.01139-09
Dyszel, J. L., Soares, J. A., Swearingen, M. C., Lindsay, A., Smith, J. N., and Ahmer, B. M. M. (2010b). E. coli K-12 and EHEC genes regulated by SdiA. PLoS ONE 5:e8946. doi: 10.1371/journal.pone.0008946
Edwards, R. A., Keller, L. H., and Schifferli, D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157.
Ghosh, D., Roy, K., Williamson, K. E., Srinivasiah, S., Wommack, K. E., and Radosevich, M. (2009). Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75, 7142–7152. doi: 10.1128/AEM.00950-09
Goryshin, I. Y., and Reznikoff, W. S. (1998). Tn5 in vitro transposition. J. Biol. Chem. 273, 7367–7374.
Habyarimana, F., Sabag-Daigle, A., and Ahmer, B. M. M. (2014). The sdiA-regulated gene, srgE, encodes a Type 3 secreted effector. J. Bacteriol. 196, 2301–2312. doi: 10.1128/JB.01602-14
Hanzelka, B. L., and Greenberg, E. P. (1995). Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177, 815–817.
Hastings, J. W., and Greenberg, E. P. (1999). Quorum sensing: the explanation of a curious phenomenon reveals a common characteristic of bacteria. J. Bacteriol. 181, 2667–2668.
Hughes, D. T., Terekhova, D. A., Liou, L., Hovde, C. J., Sahl, J. W., Patankar, A. V., et al. (2010). Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U.S.A. 107, 9831–9836. doi: 10.1073/pnas.1002551107
Ishihama, A., Kori, A., Koshio, E., Yamada, K., Maeda, H., Shimada, T., et al. (2014). Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli. J. Bacteriol. 196, 2718–2727. doi: 10.1128/JB.01579-14
Kanamaru, K., Kanamaru, K., Tatsuno, I., Tobe, T., and Sasakawa, C. (2000). SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38, 805–816. doi: 10.1046/j.1365-2958.2000.02171.x
Lee, J., Jayaraman, A., and Wood, T. K. (2007). Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7:42. doi: 10.1186/1471-2180-7-42
Lee, J., Maeda, T., Hong, S. H., and Wood, T. K. (2009). Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75, 1703–1716. doi: 10.1128/AEM.02081-08
Lee, J., Zhang, X.-S., Hegde, M., Bentley, W. E., Jayaraman, A., and Wood, T. K. (2008). Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2, 1007–1023. doi: 10.1038/ismej.2008.54
Li, J., Yao, Y., Xu, H. H., Hao, L., Deng, Z., Rajakumar, K., et al. (2015). SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. doi: 10.1111/1462-2920.12794. [Epub ahead of print].
Lindsay, A., and Ahmer, B. M. M. (2005). Effect of sdiA on biosensors of N-acylhomoserine lactones. J. Bacteriol. 187, 5054–5058. doi: 10.1128/JB.187.14.5054-5058.2005
Metcalf, W. W., Jiang, W., Daniels, L. L., Kim, S. K., Haldimann, A., and Wanner, B. L. (1996). Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35, 1–13. doi: 10.1006/plas.1996.0001
Michael, B., Smith, J. N., Swift, S., Heffron, F., and Ahmer, B. M. (2001). SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001
Miller, V. L., and Mekalanos, J. J. (1988). A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170, 2575–2583.
Miyashiro, T., and Ruby, E. G. (2012). Shedding light on bioluminescence regulation in Vibrio fischeri. Mol. Microbiol. 84, 795–806. doi: 10.1111/j.1365-2958.2012.08065.x
Murray, G. L., Attridge, S. R., and Morona, R. (2003). Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47, 1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x
Nguyen, Y., Nguyen, N. X., Rogers, J. L., Liao, J., MacMillan, J. B., Jiang, Y., et al. (2015). Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. MBio 6:e02429–14. doi: 10.1128/mBio.02429-14
Nguyen, Y. N., Sheng, H., Dakarapu, R., Falck, J. R., Hovde, C. J., and Sperandio, V. (2013). The acyl-homoserine lactone synthase YenI from Yersinia enterocolitica modulates virulence gene expression in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 81, 4192–4199. doi: 10.1128/IAI.00889-13
Nguyen, Y., and Sperandio, V. (2012). Enterohemorrhagic E. coli (EHEC) pathogenesis. Front. Cell. Infect. Microbiol. 2:90. doi: 10.3389/fcimb.2012.00090
Nikaido, E., Yamaguchi, A., and Nishino, K. (2008). AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J. Biol. Chem. 283, 24245–24253. doi: 10.1074/jbc.M804544200
Osman, D., and Cavet, J. S. (2011). Metal sensing in Salmonella: implications for pathogenesis. Adv. Microb. Physiol. 58, 175–232. doi: 10.1016/B978-0-12-381043-4.00005-2
Patankar, A. V., and Gonzalez, J. E. (2009). Orphan LuxR regulators of quorum sensing. FEMS Microbiol. Rev. 33, 739–756. doi: 10.1111/j.1574-6976.2009.00163.x
Pearson, J. P., Gray, K. M., Passador, L., Tucker, K. D., Eberhard, A., Iglewski, B. H., et al. (1994). Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U.S.A. 91, 197–201.
Petersen, T. N., Brunak, S., Heijne von, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Rensing, C., and Grass, G. (2003). Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol. Rev. 27, 197–213. doi: 10.1016/S0168-6445(03)00049-4
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Sabag-Daigle, A., and Ahmer, B. M. M. (2012). ExpI and PhzI are descendants of the long lost cognate signal synthase for SdiA. PLoS ONE 7:e47720. doi: 10.1371/journal.pone.0047720
Sabag-Daigle, A., Soares, J. A., Smith, J. N., Elmasry, M. E., and Ahmer, B. M. M. (2012). The acyl homoserine lactone receptor, SdiA, of Escherichia coli and Salmonella enterica serovar Typhimurium does not respond to indole. Appl. Environ. Microbiol. 78, 5424–5431. doi: 10.1128/AEM.00046-12
Santiviago, C. A., Reynolds, M. M., Porwollik, S., Choi, S.-H., Long, F., Andrews-Polymenis, H. L., et al. (2009). Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477. doi: 10.1371/journal.ppat.1000477
Schaefer, A. L., Hanzelka, B. L., Eberhard, A., and Greenberg, E. P. (1996). Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 178, 2897–2901.
Schuster, M., Sexton, D. J., Diggle, S. P., and Greenberg, E. P. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
Shankar, M., Ponraj, P., Illakkiam, D., Rajendhran, J., and Gunasekaran, P. (2012). Inactivation of the transcriptional regulator-encoding gene sdiA enhances rice root colonization and biofilm formation in Enterobacter cloacae GS1. J. Bacteriol. 195, 39–45. doi: 10.1128/JB.01236-12
Sharma, V. K., and Bearson, S. M. D. (2013). Evaluation of the impact of quorum sensing transcriptional regulator SdiA on long-term persistence and fecal shedding of Escherichia coli O157:H7 in weaned calves. Microb. Pathog. 57, 21–26. doi: 10.1016/j.micpath.2013.02.002
Sharma, V. K., Bearson, S. M. D., and Bearson, B. L. (2010). Evaluation of the effects of sdiA, a luxR homologue, on adherence and motility of Escherichia coli O157: H7. Microbiology 156(Pt 5), 1303–1312. doi: 10.1099/mic.0.034330-0
Sheng, H., Nguyen, Y. N., Hovde, C. J., and Sperandio, V. (2013). SdiA aids enterohemorrhagic Escherichia coli carriage by cattle fed a forage or grain diet. Infect. Immun. 81, 3472–3478. doi: 10.1128/IAI.00702-13
Shimada, T., Shimada, K., Matsui, M., Kitai, Y., Igarashi, J., Suga, H., et al. (2014). Roles of cell division control factor SdiA: recognition of quorum sensing signals and modulation of transcription regulation targets. Genes Cells. 19, 405–418. doi: 10.1111/gtc.12139
Smith, J. N., and Ahmer, B. M. M. (2003). Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185, 1357–1366. doi: 10.1128/JB.185.4.1357-1366.2003
Soares, J. A., and Ahmer, B. M. M. (2011). Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr. Opin. Microbiol. 14, 188–193. doi: 10.1016/j.mib.2011.01.006
Sperandio, V. (2010a). SdiA bridges chemical signaling between Salmonella enterica serovar Typhimurium and Yersinia enterocolitica in mice. J. Bacteriol. 192, 21–22. doi: 10.1128/JB.01359-09
Sperandio, V. (2010b). SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen. Gut. Microbes 1, 432–435. doi: 10.4161/gmic.1.6.14177
Subramoni, S., and Venturi, V. (2009). LuxR-family “solos”: bachelor sensors/regulators of signalling molecules. Microbiology 155(Pt 5), 1377–1385. doi: 10.1099/mic.0.026849-0
Suzuki, K., Wang, X., Weilbacher, T., Pernestig, A.-K., Melefors, O., Georgellis, D., et al. (2002). Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J. Bacteriol. 184, 5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002
Swearingen, M. C., Sabag-Daigle, A., and Ahmer, B. M. M. (2012). Are there acyl-homoserine lactones within mammalian intestines? J. Bacteriol. 195, 173–179. doi: 10.1128/JB.01341-12
Van Houdt, R., Aertsen, A., Moons, P., Vanoirbeek, K., and Michiels, C. W. (2006). N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256, 83–89. doi: 10.1111/j.1574-6968.2006.00103.x
Wei, Y., Lee, J. M., Smulski, D. R., and LaRossa, R. A. (2001). Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183, 2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001
Winson, M. K., Swift, S., Hill, P. J., Sims, C. M., Griesmayr, G., Bycroft, B. W., et al. (1998). Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163, 193–202.
Keywords: SdiA, LuxR solo, Enterobacter, regulon, acylhomoserine lactone, transposon mutagenesis, suicide vector, quorum sensing
Citation: Sabag-Daigle A, Dyszel JL, Gonzalez JF, Ali MM and Ahmer BMM (2015) Identification of sdiA-regulated genes in a mouse commensal strain of Enterobacter cloacae. Front. Cell. Infect. Microbiol. 5:47. doi: 10.3389/fcimb.2015.00047
Received: 12 March 2015; Accepted: 12 May 2015;
Published: 27 May 2015.
Edited by:
Matthew S. Francis, Umeå University, SwedenReviewed by:
Jun Zhu, University of Pennsylvania, USABradley L. Bearson, USDA Agricultural Research Service, USA
Akira Ishihama, Hosei University, Japan
Copyright © 2015 Sabag-Daigle, Dyszel, Gonzalez, Ali and Ahmer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian M. M. Ahmer, Department of Microbial Infection and Immunity, The Ohio State University, 460 West 12th Ave, 710 Biological Research Tower, Columbus, OH 43210, USA,YWhtZXIuMUBvc3UuZWR1
 Anice Sabag-Daigle
Anice Sabag-Daigle Jessica L. Dyszel
Jessica L. Dyszel Juan F. Gonzalez
Juan F. Gonzalez Mohamed M. Ali
Mohamed M. Ali Brian M. M. Ahmer
Brian M. M. Ahmer